SUMMARY
Escherichia albertii is a newly emerging enteric pathogen that has been associated with gastroenteritis in humans. Recently, E. albertii has also been detected in healthy and sick birds, animals, chicken meat and water. In the present study, the prevalence and characteristics of the eae-positive, lactose non-fermenting E. albertii strains in retail raw meat in China were evaluated. Thirty isolates of such strains of E. albertii were identified from 446 (6·73%) samples, including duck intestines (21·43%, 6/28), duck meat (9·52%, 2/21), chicken intestines (8·99%, 17/189), chicken meat (5·66%, 3/53), mutton meat (4·55%, 1/22) and pork meat (2·44%, 1/41). None was isolated from 92 samples of raw beef meat. Strains were identified as E. albertii by phenotypic properties, diagnostic PCR, sequence analysis of the 16S rRNA gene, and housekeeping genes. Five intimin subtypes were harboured by these strains. All strains possessed the II/III/V subtype group of the cdtB gene, with two strains carrying another copy of the I/IV subtype group. Pulsed-field gel electrophoresis showed high genetic diversity of E. albertii in raw meats. Our findings indicate that E. albertii can contaminate various raw meats, posing a potential threat to public health.
Key words: Escherichia albertii, cytolethal distending toxin (CDT), intimin, raw meat
INTRODUCTION
Escherichia albertii is a newly described and emerging diarrhoeagenic pathogen, which is associated with sporadic infections and outbreaks in humans and birds [1–7]. E. albertii was originally recovered from the faeces of Bangladeshi children with accompanying symptoms of diarrhoea, vomiting, fever, mild dehydration and abdominal distention, and was initially identified as Hafnia alvei [1]. Nevertheless, subsequent analysis based on phenotypic characterization, 16S rDNA sequencing and DNA–DNA hybridization, indicated that these H. alvei-like strains belong to a new Escherichia species, named E. albertii [8].
E. albertii belongs to the attaching and effacing (A/E) group of pathogens, which form A/E lesions on intestinal epithelial cell surfaces by the combined action of an outer membrane protein, intimin, encoded by eae gene, and other type III secretion system effectors of the locus of enterocyte effacement pathogenicity island [9]. Currently, more than 30 distinct intimin subtypes have been reported and E. albertii possess intimin subtypes that are rarely described or have not been described in E. coli [3]. Donato et al. found that E. albertii can express enterohaemorrhagic E. coli O157:H7 type III secretion system effectors EspE and EspF and cause a redistribution of the tight junction protein zona occludens-1 of the polarized epithelial MDCK-I and T84 cells [10]. In addition to intimin, cytolethal distending toxin (CDT) has also been reported as a putative virulence factor in E. albertii [11], which is a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages [12]. CDT consists of a heterotrimeric complex of three subunits designated CdtA, CdtB and CdtC [12]. CdtB has DNase I activity, and is responsible for CDT-induced cell cycle arrest [13, 14]. Subunits CdtA and CdtC associate with CdtB to translocate it into the host cell [15]. CdtB has distinct subtypes, and most E. albertii strains possess a cdtB belonging to the II/III/V subtype group [3]. Shiga toxins (Stx) are the most significant virulence factors of Stx-producing E. coli in human infections and Stx2 are classified into seven subtypes (Stx2a to Stx2g) [16]. Stx2f, originally identified in E. coli strains isolated from the faeces of feral pigeons [17], has also been detected in some E. albertii strains [3, 5]; however, the clinical significance of Stx2f-producing E. albertii is unknown. Recently, a stx2a-positive E. albertii strain was identified from a patient with bloody diarrhoea [18].
The specific biochemical characteristics of E. albertii are poorly defined; therefore, no commercial system currently includes this species in its database. Thus, it remains difficult to discriminate E. albertii from E. coli and other members of the Enterobacteriaceae using routine bacterial identification systems, which often misidentify E. albertii strains as Hafnia, Salmonella, Yersinia ruckeri, and, in particular, E. coli, with high probability [19]. The lack of motility and the inability to ferment d-xylose and lactose have been reported to be the common biochemical properties of confirmed E. albertii strains, which can help to distinguish E. albertii from other Escherichia species [3, 6, 8, 19–21]. Hyma et al. devised a diagnostic multiplex polymerase chain reaction (PCR) to detect conserved sequences that distinguish members of the E. albertii lineage from E. coli and Shigella spp. strains. This method was based on nucleotide polymorphisms of the housekeeping genes lysP and mdh in the E. albertii lineage [11]. Multiplex PCR is independent of biochemical and antigenic phenotypes and, therefore, provides a useful method to screen E. albertii strains [2, 4–6, 21, 22].
Although E. albertii is found in humans, animals, birds and water, the prevalence, epidemiology and clinical relevance of the species are still poorly defined, in part because it is likely to either remain unidentified or be misidentified. E. albertii may be one of the unknown aetiological agents that contribute to the estimated 62 million cases of foodborne illnesses and 3200 deaths in the United States [23]. An eae-positive isolate of H. alvei was isolated from minced meat in Sweden, but it was doubtful whether this H. alvei strain was actually E. albertii [24]. The lack of identification of this pathogen has resulted in limited investigations into its incidence in foodstuffs. Current E. albertii studies are mainly based on the type strains and reference strains, which show the presence of the eae gene, lack of motility, and an inability to ferment d-xylose and lactose. Yet, the phenotypic and genetic descriptions tend to vary as a greater number of E. albertii isolates are identified from different sources. In this study, we investigated the prevalence and characteristics of eae-positive, lactose non-fermenting phenotype E. albertii strains which represent a substantial proportion of E. albertii in retail raw meats collected from Zigong city, Sichuan province, China.
MATERIALS AND METHODS
Sample collection
A total of 446 raw meat and intestine samples were collected from supermarkets and farmers' markets in Zigong city, Sichuan province, China, from March 2013 to July 2014. The samples included beef meat (n = 92), pork meat (n = 41), mutton meat (n = 22), chicken meat (n = 53), chicken intestines (n = 189), duck meat (n = 21), and duck intestines (n = 28). The samples were transported in ice-bags to the laboratory of Zigong Centre for Disease Control and Prevention within 4 h and tested immediately.
Screening eae-positive samples
Twenty-five grams of each minced raw meat or sliced intestine sample was enriched with 225 ml EC broth (Oxoid, UK) and incubated at 20 °C for 24–36 h on a shaking platform (220 rpm). One microlitre of each enrichment sample was centrifuged at 1500 g for 1 min to pellet large meat debris. The supernatant was centrifuged at 13000 g for 2 min and the pellet was suspended in 100 μl lysis buffer [100 mm NaCl, 10 mm Tris-HCl (pH 8·3), 1 mm EDTA (pH 9·0), and 1% Triton X-100], boiled for 10 min, and centrifuged again. The resulting supernatant was used as a template to test the presence of eae by PCR, using previously published primers (Supplementary Table S1) [2].
Isolation of lactose non-fermenting strains
One loopful of each eae-positive enrichment culture was directly streaked on MacConkey agar (Oxoid, UK) and CHROMagar ECC agar (CHROMagar, France). After overnight incubation at 36 °C, colourless, round and moist, presumptive E. albertii colonies on both agars were chosen to test for the presence of eae. The eae-positive colonies were plated on Luria–Bertani (LB) agar (Oxoid) and incubated overnight for further identification. If more than one eae-positive lactose non-fermenting isolate was identified, only one from each sample was retained for further investigation.
Phenotypic tests
Carbohydrate-fermenting abilities were determined after 3 days of incubation at 36 °C in Andrade peptone water containing lactose or d-xylose (Land Bridge, China). In vitro motility was determined on motility agar plates (0·3% LB agar) incubated at 36 °C for up to 48 h. Additional biochemical characterization was performed using the API 20E system (bioMérieux, France).
Diagnostic multiplex PCR and 16S rDNA sequencing
All eae-positive, lactose non-fermenting strains were screened by diagnostic multiplex PCR for the E. albertii lineage-specific housekeeping genes lysP and mdh. The clpX gene, which is conserved in all E. coli, Shigella spp. and the E. albertii lineages, was used as a positive control. The PCR primers and conditions used were as described previously (Supplementary Table S1) [6, 11].
16S rRNA genes were amplified and sequenced according to published protocols [25] and nucleotide sequences were aligned using ClustalW. Phylogenetic trees were constructed using the neighbour-joining algorithm with 1000 bootstrap resamplings to assess the relationships between individual pathogens using MEGA 6 (www.megasoftware.net) [26].
Multi-locus sequence typing (MLST)
All eae-positive, lactose non-fermenting strains were analysed by MLST to determine their phylogenetic relationships according to the E. coli MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). A neighbour-joining dendrogram was constructed, based on the concatenated nucleotide sequences of the seven genes and the maximum composite likelihood model using MEGA 6. The genome-sequenced E. coli, E. fergusonii, E. albertii, Shigella spp., and Salmonella enterica serovar Typhi strains were included in the phylogenetic representation, as described by Ooka et al. [3].
Detection of virulence genes and sequence analysis
In addition to eae, eight diarrhoeagenic E. coli marker genes, i.e. stx1, stx2, ipaH, aggR, elt, estIa, estIb and daaD, were screened with published primers [27] and the stx2f subtype was determined as described by Schmidt et al. [17]. The entire eae gene was amplified by PCR using the cesT-F9/eae-R3 primer pair for the 5′ half of the gene, and the eae-F1/escD-R1 primer pair for the 3′ half of the gene [28]. PCR amplicons were sequenced using the above-mentioned primers and an additional sequence primer (1669–1688) was used to fully sequence the 3′ half of the eae gene, as described by Ooka et al. [3].
All sequencing results were checked and assembled using SeqMan II software (DNASTAR Inc., USA). Using the ClustalW program in MEGA 6, the predicted amino acid sequences obtained from this study were aligned with those of the reference intimin subtypes downloaded from GenBank. A phylogenetic tree was constructed as detailed above.
cdtB type I/IV was detected by the PCR primer pair CDT-s2 and CDT-as2 and type II/ III/ V by PCR primer pair CDT-s1 and CDT-as1, as described previously [29]. The partial amino acid sequences of the CdtB subunit were used to construct a neighbour-joining tree.
Pulsed-field gel electrophoresis (PFGE)
DNA extracts of E. albertii strains were digested with XbaI and fragments separated on a 1% agarose gel using a CHEF-DR III PFGE apparatus (Bio-Rad, USA). The pulse times were ramped from 2·2 to 54·2 s over 18 h, according to the protocol for E. coli O157:H7 from PulseNet, USA (http://www.cdc.gov/pulsenet/pathogens/index.html). Gel images were captured with a Gel Doc™ XR+ system (Bio-Rad, USA), converted to TIFF files, and analysed by BioNumerics v. 4·0 (Applied Maths, Belgium). An unweighted pair-group method with arithmetic mean dendrogram was constructed using BioNumerics software.
The nucleotide sequences determined in this study have been submitted to GenBank. Their accession numbers are KP197062–KP197093 for 16S rRNA genes; KP015856–KP016011 and KP064411–KP064472 for the seven housekeeping genes; KP197094–KP197126 for the eae genes; and KP197127-KP197158 for the cdtB genes.
RESULTS
Prevalence of eae-positive, lactose non-fermenting strains in raw meat
Thirty isolates from 446 (6·73%) samples were identified as eae-positive, lactose non-fermenting strains. Seventeen (56·7%) were recovered from chicken intestines and six (20·0%) from duck intestines. The sources of the remaining isolates are given in Table 1; none was isolated from the 92 raw beef meat samples.
Table 1.
Prevalence of Escherichia albertii in raw meat and intestinal samples
| Samples | No. of samples | No. of E. albertii strains (%) |
|---|---|---|
| Beef meat | 92 | 0 (0) |
| Pork meat | 41 | 1 (2·44) |
| Mutton meat | 22 | 1 (4·55) |
| Chicken meat | 53 | 3 (5·66) |
| Duck meat | 21 | 2 (9·52) |
| Chicken intestines | 189 | 17 (8·99) |
| Duck intestines | 28 | 6 (21·43) |
| Total | 446 | 30 (6·73) |
All isolates were oxidase-negative, fermented glucose but not lactose or d-xylose, and were non-motile. Variation was observed in utilization of β-galactosidase, production of indole or acetoin, and fermentation of d-manitol, d-sorbitol or d-melibiose. All strains were identified phenotypically as E. coli with different probabilities.
None of the strains tested positive for stx1, stx2, ipaH, aggR, elt, estIa, estIb or daaD genes and all were positive for the E. albertii-specific alleles of lysP and mdh, and for the clpX gene conserved in E. coli, Shigella spp., and E. albertii lineages, which tentatively identified these strains as E. albertii.
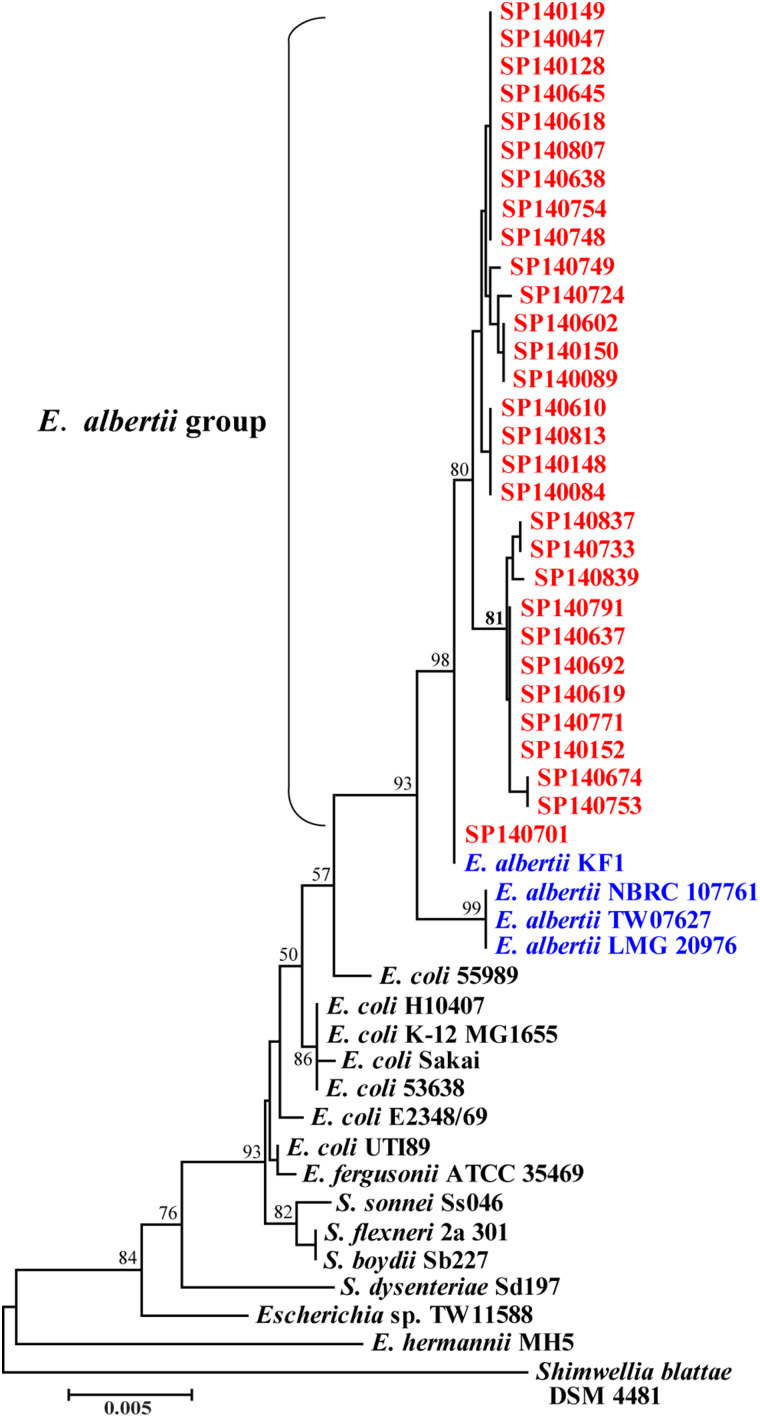
Sequencing of a 1506 bp amplicon of the 16S rRNA genes revealed ten distinct sequence types in the 30 E. albertii strains with a percentage similarity ranging from 99·7% to 100%, with 99·6% identity to the E. albertii type strain LMG 20976 (Fig. 1). One strain (SP140701) from chicken meat was identical to E. albertii KF1, a clinical strain. These results support the identification of the strains as E. albertii.
Fig. 1.
Phylogenetic tree based on a 1404 bp portion of the 16S rRNA gene of Escherichia albertii strains isolated in this study, the E. albertii type strain LMG 20976 and other related genome-sequenced bacterial species.
MLST analysis and virulence gene sequence
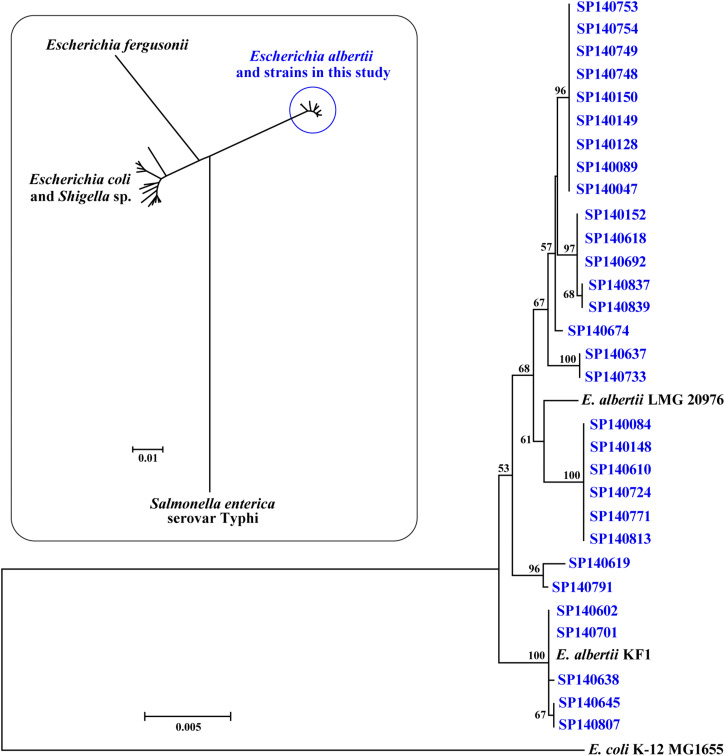
By MLST all strains from meat and intestine samples were highly divergent from E. coli/Shigella spp., E. fergusonii and Salmonella enterica serotype Typhi stains, and formed a separate lineage with E. albertii reference strains (Fig. 2, inset). Eleven different sequence types, with pairwise differences of 0·0003–0·0079, were identified in the test E. albertii strains, each type comprising 1–9 strains. All sequence types were closely related to the E. albertii type strain LMG 20976, but were less related to E. coli K-12 MG1655 (Fig. 2). Two strains (SP140602 and SP140701) were of identical sequence type to E. albertii KF1.
Fig. 2.
Neighbour-joining dendrogram of Escherichia albertii strains and reference strains based on the 3423 bp concatenated partial nucleotide sequences of seven housekeeping genes. Inset is an image of the relationship of the strains isolated in this study and reference strains of E. albertii, E. coli/Shigella spp., E. fergusonii and Salmonella enterica serovar Typhi.
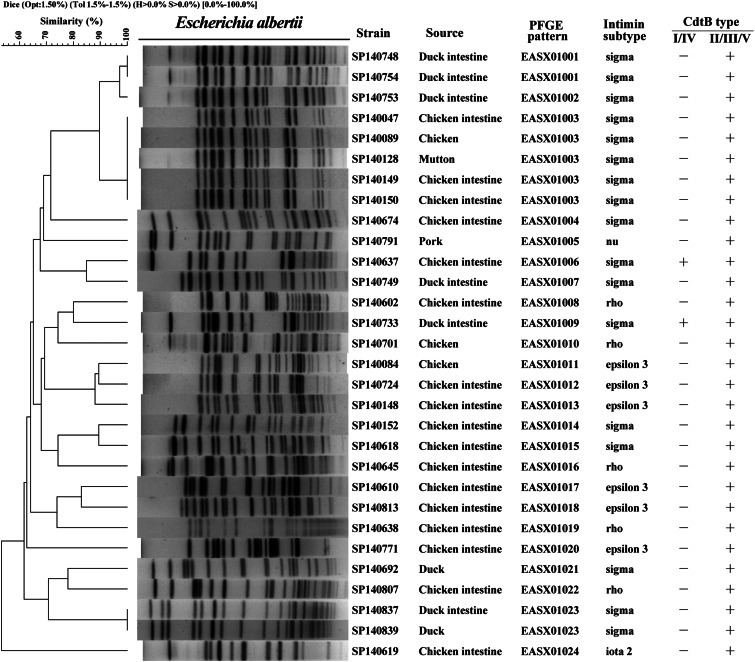
The phylogenetic tree of the amino acid sequences of the intimin protein encoded by the eae gene in all meat and intestine sample strains showed identical or close relation to five known intimin subtypes, σ, ρ, ι2, ε3, ν (Supplementary Fig. S1A). Seventeen strains contained the σ subtype and were isolated from all meat samples excepting those of beef meat and pork meat. Five and six strains from chicken meat or chicken intestines samples were subtyped as ρ and ε3, respectively; one strain from chicken intestines was of the ι2 subtype and the single pork meat isolate was of the ν subtype (Fig. 3).
Fig. 3.
Dendrogram of pulsed-field gel electrophoresis (PFGE) profiles of 30 Escherichia albertii strains from meat and intestinal samples; strain designations, animal source, PFGE patterns, intimin subtypes and CdtB types.
All test strains were PCR-positive for cdtB using the CDT-s1/CDT-as1 primer pair and two strains, SP140637 from chicken intestines and SP140733 from duck intestines were also positive for cdtB using the CDT-s2/CDT-as2 primer pair. Sequencing and alignment of the CDT-s1/CDT-as1 PCR products showed 94–100% nucleotide and 95–100% amino acid identities and they were closely related to types II, III and V cdtB reference alleles. Sequencing and alignment of the CDT-s2/CDT-as2 PCR product showed that the sequences from the two strains were identical to each other and the type I cdtB reference allele (Fig. S1B).
Twenty-four different PFGE patterns were distinguishable (<97% similarity) in the strains indicating a high degree of genetic diversity in this collection of E. albertii (Fig. 3).
Strains did not appear to cluster on the basis of their source, intimin subtype or CdtB type. Three PFGE patterns (EASX01001, EASX01003, EASX01023) contained two, five and two strains, respectively; the remaining patterns were each represented by single strains. The five strains of pattern EASX01003 from different sources (chicken intestines, chicken meat, mutton meat) had the same intimin subtype and CdtB type. Strains SP140748 and SP140754 from duck intestines were identical by PFGE,16S rRNA gene sequence, MLST, eae gene and cdtB gene sequences, indicating that the strains may belong to the same E. albertii clone (EASX01001, Fig. 3). Strain SP140753 from duck intestine had a similar pattern to EASX01001, identical MLST, eae gene and cdtB gene sequences, but different 16S rRNA gene sequence.
DISCUSSION
E. albertii has been recently recognized as a potential enteric human pathogen. These organisms isolated from diarrhoeal patients were originally identified as enteropathogenic E. coli or enterohaemorrhagic E. coli by routine diagnostic protocols [3, 5]. E. albertii strains were isolated from outbreaks of gastroenteritis in Japan [2, 4, 7] and were implicated as the probable cause of death of wild birds in Alaska, USA [6], although also found in healthy wild birds from Korea [22]. E. albertii has been detected in other bacterial opportunistic pathogens in the drinking water distribution system of a hospital in Budapest, Hungary [30], and in different environmental waters from Québec, Canada [31], suggesting that water could be a potential source of this pathogen.
To date, relatively few studies have investigated the prevalence of E. albertii in foodstuffs of animal origin. We isolated the eae-positive, lactose non-fermenting strains from a variety of raw meats and intestines, but none was recovered from beef meat samples. Phenotypic and biochemical analyses, diagnostic PCR, 16S rDNA sequencing, and MLST analyses classified the isolates as E. albertii, indicating that a wide range of raw meats could be a source of the species. Lindberg et al. reported an eae-positive strain of H. alvei isolated from minced meat in Sweden, which might have been E. albertii [24], and recently, E. albertii isolates were also detected in chicken rinses from three out of five regions of the United States, and in chicken giblets (liver) collected in Japan [32, 33]. The contaminant sources of raw meats are unclear; however, our limited investigation showed a higher prevalence of this pathogen in chicken intestines and duck intestines, suggesting that E. albertii could colonize the intestines of these animals. Ooka et al. analysed 278 eae-positive strains isolated from humans, animals, and the environment in Japan, and found a strain of E. albertii from a domestic cat [3]. The shedding of E. albertii from animal faeces requires further investigation.
We found that more colourless (lactose non-fermenting) colonies appeared on MacConkey agar after the sample was enriched at 20 °C for 24–36 h compared to 37 °C (data not shown) and this may reflect the low tolerance of E. albertii to heat, acid, and pressure which is reported to be significantly lower than for wild-type E. coli O157:H7 [34]. The growth of E. albertii is uncertain when it competes with other bacteria, especialy E. coli. In the present study, diagnostic PCR proved useful for rapid identification of the E. albertii strains. However, the specificity of the primers has recently been questioned and may not be optimal for the detection of E. albertii in food samples [35].
The protein intimin is a critical virulence factor of E. albertii, which is characterized by intimin subtypes that are rare or have not been previously described in E. coli [3]. Ooka et al. found that 26 E. albertii strains from humans, birds and a cat were grouped into nine known intimin subtypes and five new subtypes [3] and other investigations have shown that E. albertii strains from birds possess a variety of intimin subtypes, some novel and some similar to those previously reported [6, 22]. Five intimin subtypes, σ, ρ, ι2, ε3, and ν, were identified in this study and with the exception of ι2 were reported previously in E. albertii strains in several other studies [3, 6, 22]. The ι2 subtype was identified in Shigella boydii serotype 13, which is regarded as within the E. albertii lineage [11]. Additionally, all 30 E. albertii strains possessed the II/III/V subtype group cdtB gene, and two strains possessed another copy of the I/IV subtype group cdtB gene, which is in agreement with previous findings [3, 6].
In conclusion, heterogeneous eae-positive, lactose non-fermenting E. albertii strains were recovered from a variety of retail animal meat samples and intestines collected in Zigong city, Sichuan province, China. To the best of our knowledge, this is the first investigation of this newly recognized pathogen in China, and also the largest-scale survey into the prevalence of E. albertii in raw meats worldwide. The transmission of E. albertii from foodstuffs of animal origin to humans is possible, which may pose a potential threat to public health. Our study mainly focused on the type strains; other various phenotypic/genotypic strains might be missed, which may lead to an underestimation of its prevalence in retail raw meats. Further investigations are needed to assess the occurrence, pathogenicity and clonal diversity of all phenotypic/genotypic variants, and their public health significance in causing human disease in China.
ACKNOWLEDGEMENTS
We thank Dr Tadasuke Ooka and Dr Tetsuya Hayashi (University of Miyazaki, Japan) for their help in MLST analysis. This work was supported by grants from the National Natural Science Foundation of China (81371762, 81290345), the China Mega-Project for Infectious Disease (2013ZX10004-001, 2012ZX10004-215), and the Key Science and Technology Project of Zigong, China (2013S06).
SUPPLEMENTARY MATERIAL
For supplementary material accompanying this paper visit http://doi.org/10.1017/S0950268815001120.
click here to view supplementary material
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Albert MJ, et al. Hafnia alvei, a probable cause of diarrhea in humans. Infection and Immunity 1991; 59: 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konno T, et al. Isolation and identification of Escherichia albertii from a patient in an outbreak of gastroenteritis. Japanese Journal of Infectious Diseases 2012; 65: 203–207. [DOI] [PubMed] [Google Scholar]
- 3.Ooka T, et al. Clinical significance of Escherichia albertii. Emerging Infectious Diseases 2012; 18: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asoshima N, et al. Identification of Escherichia albertii as a causative agent of a food-borne outbreak occurred in 2003. Japanese Journal of Infectious Diseases 2014; 67: 139–140. [DOI] [PubMed] [Google Scholar]
- 5.Murakami K, et al. Shiga toxin 2f-producing Escherichia albertii from a symptomatic human. Japanese Journal of Infectious Diseases 2014; 67: 204–208. [DOI] [PubMed] [Google Scholar]
- 6.Oaks JL, et al. Escherichia albertii in wild and domestic birds. Emerging Infectious Diseases 2010; 16: 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ooka T, et al. Human gastroenteritis outbreak associated with Escherichia albertii, Japan. Emerging Infectious Diseases 2013; 19: 144–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huys G, et al. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. International Journal of Systematic and Evolutionary Microbiology 2003; 53: 807–810. [DOI] [PubMed] [Google Scholar]
- 9.Kenny B. Mechanism of action of EPEC type III effector molecules. International Journal of Medical Microbiology 2002; 291: 469–477. [DOI] [PubMed] [Google Scholar]
- 10.Donato KA, et al. Escherichia albertii and Hafnia alvei are candidate enteric pathogens with divergent effects on intercellular tight junctions. Microbial Pathogenesis 2008; 45: 377–385. [DOI] [PubMed] [Google Scholar]
- 11.Hyma KE, et al. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. Journal of Bacteriology 2005; 187: 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinadasa RN, et al. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology 2011; 157: 1851–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elwell CA, Dreyfus LA. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Molecular Microbiology 2000; 37: 952–963. [DOI] [PubMed] [Google Scholar]
- 14.Lara-Tejero M, Galan JE. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 2000; 290: 354–357. [DOI] [PubMed] [Google Scholar]
- 15.Nesic D, Hsu Y, Stebbins CE. Assembly and function of a bacterial genotoxin. Nature 2004; 429: 429–433. [DOI] [PubMed] [Google Scholar]
- 16.Scheutz F, et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. Journal of Clinical Microbiology 2012; 50: 2951–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt H, et al. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Applied and Environmental Microbiology 2000; 66: 1205–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandal LT, et al. Shiga toxin 2a in Escherichia albertii. Journal of Clinical Microbiology 2015; 53: 1454–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbott SL, et al. Biochemical properties of a newly described Escherichia species, Escherichia albertii. Journal of Clinical Microbiology 2003; 41: 4852–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janda JM, Abbott SL, Albert MJ. Prototypal diarrheagenic strains of Hafnia alvei are actually members of the genus Escherichia. Journal of Clinical Microbiology 1999; 37: 2399–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nimri LF. Escherichia albertii, a newly emerging enteric pathogen with poorly defined properties. Diagnostic Microbiology and Infectious Disease 2013; 77: 91–95. [DOI] [PubMed] [Google Scholar]
- 22.Oh JY, et al. Epidemiological investigation of eaeA-positive Escherichia coli and Escherichia albertii strains isolated from healthy wild birds. Journal of Microbiology 2011; 49: 747–752. [DOI] [PubMed] [Google Scholar]
- 23.Mead PS, et al. Food-related illness and death in the United States. Emerging Infectious Diseases 1999; 5: 607–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindberg AM, et al. Enterobacteriaceae found in high numbers in fish, minced meat and pasteurised milk or cream and the presence of toxin encoding genes. International Journal of Food Microbiology 1998; 39: 11–17. [DOI] [PubMed] [Google Scholar]
- 25.Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. Journal of Microbiological Methods 2003; 55: 541–555. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, et al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 2013; 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guion CE, et al. Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR. Journal of Clinical Microbiology 2008; 46: 1752–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacher DW, Steinsland H, Whittam TS. Allelic subtyping of the intimin locus (eae) of pathogenic Escherichia coli by fluorescent RFLP. FEMS Microbiology Letters 2006; 261: 80–87. [DOI] [PubMed] [Google Scholar]
- 29.Toth I, et al. Production of cytolethal distending toxins by pathogenic Escherichia coli strains isolated from human and animal sources: establishment of the existence of a new cdt variant (type IV). Journal of Clinical Microbiology 2003; 41: 4285–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felfoldi T, et al. Detection of potentially pathogenic bacteria in the drinking water distribution system of a hospital in Hungary. Clinical Microbiology and Infection 2010; 16: 89–92. [DOI] [PubMed] [Google Scholar]
- 31.Maheux AF, et al. Characterization of Escherichia fergusonii and Escherichia albertii isolated from water. Journal of Applied Microbiology 2014; 117: 597–609. [DOI] [PubMed] [Google Scholar]
- 32.Lindsey RL, et al. Evaluating the occurrence of Escherichia albertii in chicken carcass rinses by PCR, Vitek analysis, and sequencing of the rpoB gene. Applied and Environmental Microbiology 2015; 81: 1727–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maeda E, et al. Detection of Escherichia albertii from chicken meat and giblets. Journal of Veterinary Medical Science. Published online: 8 March 2015. doi: 10.1292/jvms.14-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma M, et al. Sensitivity of Escherichia albertii, a potential food-borne pathogen, to food preservation treatments. Applied and Environmental Microbiology 2007; 73: 4351–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda E, et al. Nonspecificity of primers for Escherichia albertii detection. Japanese Journal of Infectious Diseases 2014; 67: 503–505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://doi.org/10.1017/S0950268815001120.
click here to view supplementary material