SUMMARY
Fosfomycin has become a therapeutic option in urinary tract infections. We identified 57 fosfomycin-resistant Escherichia coli from 465 urine-derived extended-spectrum β-lactamase (ESBL)-producing isolates from a Chinese hospital during 2010–2014. Of the 57 fosfomycin-resistant isolates, 51 (89·5%) carried fosA3, and one carried fosA1. Divergent pulsed-field gel electrophoresis profiles and multi-locus sequence typing results revealed high clonal diversity in the fosA3-positive isolates. Conjugation experiments showed that the fosA3 genes from 50 isolates were transferable, with IncFII or IncI1 being the most prevalent types of plasmids. The high prevalence of fosA3 was closely associated with that of blaCTX-M. Horizontal transfer, rather than clonal expansion, might play a central role in dissemination. Such strains may constitute an important reservoir of fosA3 and blaCTX-M, which may well be readily disseminated to other potential human pathogens. Since most ESBL-producing E. coli have acquired resistance to fluoroquinolones worldwide, further spread of fosA3 in such E. coli isolates should be monitored closely.
Key words: Escherichia coli, ESBL, fosfomycin, fosA3, urinary tract infection
INTRODUCTION
In China, extended-spectrum β-lactamase (ESBL) production has been increasingly prevalent in strains of Escherichia coli, the major aetiological agent of urinary tract infections (UTIs) [1]. Options for effective antibiotic treatment of infections, including UTIs, are limited owing to the frequent occurrence of expanded-spectrum cephalosporin-resistant and carbapenem-resistant, Gram-negative bacteria of the family Enterobacteriaceae [2]. Use of older antibiotics such as fosfomycin has therefore been proposed as an alternative treatment of such infections [3].
Fosfomycin is an organic phosphonate agent that inhibits cell wall synthesis by irreversibly inhibiting MurA, which is responsible for the initial step of peptidoglycan biosynthesis [4]. Fosfomycin exhibits a broad spectrum of antimicrobial activity, including rapid bactericidal effects against several Gram-negative rods, particularly E. coli, and also has good activity against Staphylococcus aureus [4]. Successful treatment of infections, especially UTIs, with fosfomycin has been documented in Japan [5], and thus it is receiving renewed attention as an alternative agent for the treatment of UTIs caused by ESBL-producing E. coli [6].
To date, fosfomycin resistance in E. coli has primarily involved either reduced uptake of the drug due to mutations in chromosomally encoded transporters [7], or enzymatic inactivation by plasmid-mediated glutathione S-transferases (PMGST) such as FosA3, FosA4, and FosC2 [5, 8–11]. It has also been reported that fosfomycin-resistant isolates are more likely to be ESBL producers than fosfomycin-susceptible isolates [5]. However, information on the prevalence of plasmid-mediated fosfomycin resistance genes in ESBL-producing urinary E. coli strains is lacking, with only limited knowledge of the molecular characteristics and prevalence of fosA3 and the ESBL genes blaCTX-M in strains in China.
The purpose of this study was to examine the occurrence of fosfomycin-resistant E. coli in ESBL-producing, urinary E. coli isolates, and to identify the distribution of PMGST and ESBL determinants. Furthermore, the genetic relatedness in fosA3-positive strains, transferability of fosA3, and replicon types of fosA3-carrying plasmids were analysed.
MATERIALS AND METHODS
Bacterial isolates
A total of 821 non-repetitive urinary E. coli isolates were streaked from the Strain Library of the Department of Laboratory Medicine, Nanjing Drum Tower Hospital. ESBL production was confirmed phenotypically, using both cefotaxime and ceftazidime alone or in combination with clavulanic acid. The susceptibility of strains to fosfomycin was tested by the disk diffusion method described previously [12], using Mueller–Hinton agar plates (Oxoid, UK) containing 25 mg/l glucose-6-phosphate (G6P). E. coli ATCC25922 was used as the quality control strain in antimicrobial susceptibility testing.
Detection of genes for fosfomycin resistance and ESBL production
Genes reported to be involved in fosfomycin resistance in Enterobacteriaceae, including fosA, fosB, fosC, and fosX, as well as the subtypes fosA1, fosA2, fosA3, fosA4, and fosC2, were detected by PCR and DNA sequencing analyses according to previously described protocols [5, 8–11]. The presence of bla genes for ESBL production (blaCTX-M, blaTEM, blaSHV) was assessed in each of the 57 ESBL-positive strains, following a previously described protocol [13].
Phylogenetic grouping
Phylogenetic grouping of fosfomycin-resistant E. coli isolates was conducted via triplex PCR, using six primers in a single reaction [14]. The amplification of three DNA markers (chuA, yjaA, TSPE4.C2) generated fragments of 279, 211, and 152 bp, respectively. This allowed E. coli isolates to be classified into the phylogenetic groups A, B1, B2, or D. E. coli strains ECOR 20 (yjaA positive), ECOR 48 (chuA positive), ECOR 58 (TSPE4.C2 positive), and ECOR 62 (chuA, yjaA, and TSPE4.C2 positive) were used as the positive controls, and E. coli strain ECOR 4 was used as the negative control. All controls were kindly provided by Statens Serum Institute, Denmark.
Genetic relatedness by pulsed-field gel electrophoresis (PFGE)
The fosA3-positive E. coli isolates were characterized by PFGE using the CHEF Mapper System (Bio-Rad Laboratories, USA) as described previously [15]. Briefly, the chromosomal DNA of E. coli isolates was subjected to digestion with XbaI for 2 h at 37 °C. Electrophoresis was conducted at 6·0 V/cm and 14 °C for 19 h with an angle of 120°. The switch time was increased from 2·2 s to 54·2 s at a gradient of 6 V/cm. Salmonella enterica serovar Braenderup HP812 (kindly provided by the Centers for Disease Control and Prevention, USA) was used in parallel as a molecular weight standard. The results were analysed and interpreted using Bionumerics software v. 6.5 (Applied Maths, Belgium). The Dice similarity coefficient on the basis of the unweighted-pair group method using average linkages (UPGMA) with a 1·5% band tolerance was used. Furthermore, cut-off lines at 80% were used to analyse genetic relatedness.
Multi-locus sequence typing (MLST)
The fosA3-positive E. coli isolates were assessed for sequence types (STs) according to the MLST scheme developed for E. coli by the University College Cork (http://mlst.ucc.ie/mlst/dbs/Ecoli). Briefly, the housekeeping genes adk, fumC, gyrB, icd, mdh, purA, and recA were analysed using the primer sequences and amplification conditions available at http://mlst.warwick.ac.uk/mlst/.
Conjugation experiments
Conjugation experiments were performed using azide-resistant E. coli J53 as a recipient strain by the broth mating method. Trans-conjugants were selected on trypticase soy agar plates supplemented with 150 mg/l sodium azide, 40 mg/l fosfomycin, and 25 mg/l G6P. The presence of fos genes in phenotypically selected ESBL producers harbouring blaTEM, blaSHV, or blaCTX-M was assessed by PCR as described previously [16].
PCR-based replicon typing
DNA was extracted from 51 trans-conjugants, and main plasmid incompatibility groups, including F, FIA, FIB, FIC, HI1, HI2, I1-Ic, L/M, N, P, W, T, A/C, K, B/O, X, Y, and FII, were determined using the PCR-based replicon typing scheme, as described by Carattoli et al. [17].
Ethical statement
All procedures were performed in compliance with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
RESULTS
Susceptibility of ESBL-producing strains to fosfomycin
In total, 465 out of 821 E. coli isolates were found to be ESBL producers. Antimicrobial susceptibility testing revealed that fosfomycin exhibited good antibacterial activity towards ESBL-producing urinary E. coli strains, demonstrating effectiveness against 87·7% (408/465) of strains. The average fosfomycin resistance rate of ESBL-producing E. coli associated with UTIs was about 10% over the 5 years (2010–2014).
Prevalence of plasmid-mediated fosfomycin resistance genes and ESBL genes in fosfomycin-resistant E. coli
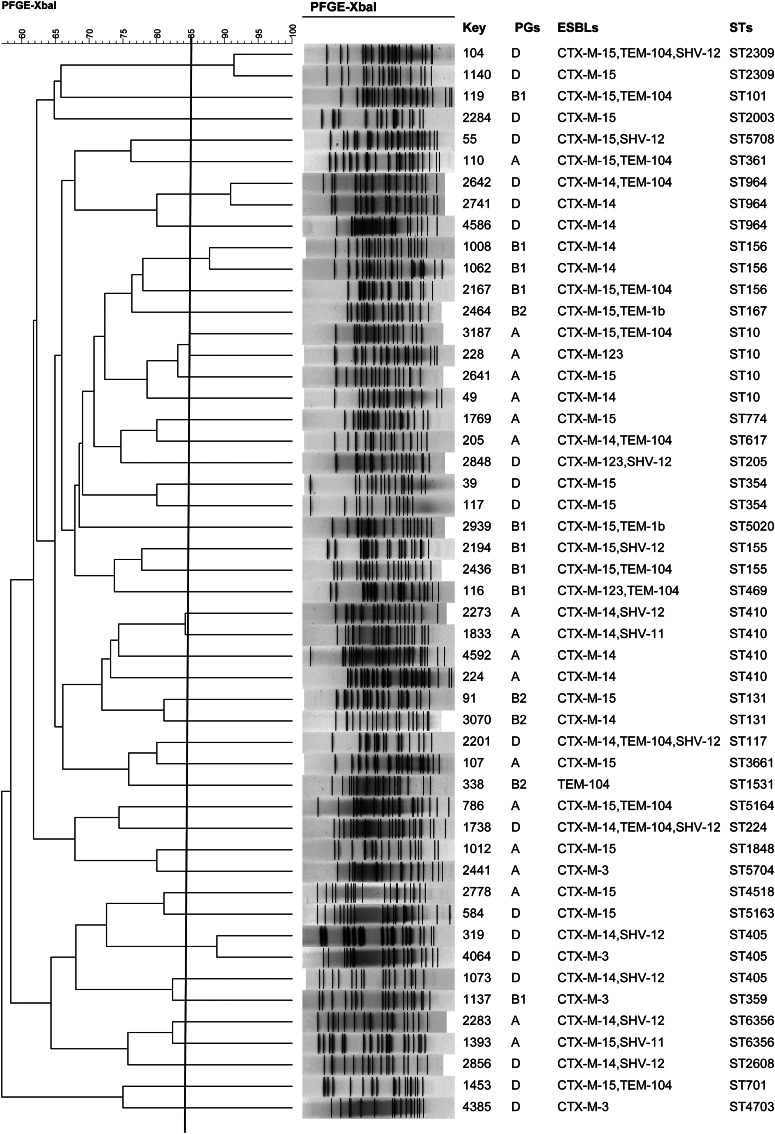
Molecular analysis showed that 89·5% (51/57) of the fosfomycin-resistant isolates were positive for fosA3, whereas only one was fosA1-positive; other fosfomycin resistance determinants were not identified. Fifty-five isolates were also blaCTX-M-positive, 26 harboured blaCTX-M-15, 22 blaCTX-M-14, 4 blaCTX-M-3, and three harboured blaCTX-M-123 (Fig. 1). In addition, 17 isolates carried blaTEM variants (14 blaTEM-104 and three blaTEM-1b) and 13 carried blaSHV variants (nine blaSHV-12 and four blaSHV-11) (Fig. 1).
Fig. 1.
Dendrogram of PFGE profiles of 50 fosA3-carrying E. coli isolates, phylogentic groups (PGs), extended-spectrum β-lactamases (ESBLs), and sequence types (STs).
Phylogenetic groups
Of the 57 fosfomycin-resistant isolates in phylogenetic groups, 19 were classified as group D, 18 group A, 12 group B1, and eight group B2.
Genetic relatedness of fosA3-positive isolates determined by PFGE and MLST
The 50 strains harbouring fosA3 exhibited 44 different PFGE profiles and one strain was not typable. MLST revealed 37 STs and major STs were ST410 (n = 4 strains), ST10 (n = 4), ST405 (n = 3), ST156 (n = 3), and ST964 (n = 3), which together comprised 33·3% of the strains analysed. Similar or identical PFGE profiles were observed within ST10 clones, ST156, ST354, ST405, ST964, and ST2309. This level of genetic diversity indicates that most of the fosA3-carrying isolates were clonally unrelated (Fig. 1).
Transferability and replicon typing of fosA3 plasmids
Conjugation assays revealed that the fosA3 genes were transmissible. Moreover, blaCTX-M and blaTEM genes were able to be transferred simultaneously, indicating genetic linkage between fosA3 and blaCTX-M. Plasmids carrying fosA3 from 50 isolates were successfully transferred by conjugation. These 50 plasmids consisted of 39 that were replicon type IncFII, nine that were IncI1, four that were IncN, two that were IncA/C, and one that was IncP. In addition, plasmids from four isolates were fused, containing both the IncN and IncFII replication origins.
DISCUSSION
Fosfomycin has been extensively used in several European countries since 1988 for the treatment of uncomplicated UTIs [4], but it was not approved for clinical use in China until recently. This is the first investigation of the prevalence of fosfomycin resistance (fos) genes in ESBL-producing urinary E. coli isolates in mainland China.
The strains in our study displayed a rate of resistance to fosfomycin of about 10%, which is higher than that previously reported [18]. However, our data indicate that fosfomycin should still be considered for the treatment of patients with infections due to ESBL-producing E. coli in China if they exhibit high resistance rates to other commonly used antimicrobial agents, including cephalosporins and fluoroquinolones [19]. This is because a previous study reported that fosfomycin retains its activity against both Gram-positive and Gram-negative multiple-drug-resistant (MDR) and extremely-drug-resistant (XDR) bacteria [4]. To date, fosfomycin has not been used for clinical treatment in our hospital, and so we speculate that the observed resistance may be co-selected by antimicrobials other than fosfomycin.
Our study found that blaCTX-M-15 and blaCTX-M-14 were the main ESBL-encoding genes detected in fosfomycin-resistant urinary E. coli strains. This is in line with the results of previous studies investigating the global prevalence of bla genes in ESBL producers [20]. It should be noted that we also detected blaCTX-M-123, which has been identified as a novel hybrid of the blaCTX-M-1 and blaCTX-M-9 β-lactamases recovered from E. coli isolates in China [21]. In parallel, there was a high prevalence of the blaTEM-104 variant in our study which to the best of our knowledge, has been identified in a MDR avian pathogenic E. coli strain isolated from septicaemic broilers in Egypt [22]. This is therefore the first report of TEM-104 variants in clinical urinary E. coli isolates in China.
It has been previously reported that fosA3 is the most prevalent PMGST in E. coli isolates of both clinical and non-clinical (healthy persons, companion and food animals) origins in several Asian countries (China, South Korea, Japan) [5, 23–27]. Thus, the high prevalence of fosA3 found here is consistent with these reports, and confirms that fosA3 is the primary mechanism of fosfomycin resistance in mainland China. Moreover, all but one of the 51 fosA3-positive isolates in our study were CTX-M producers, suggesting a high degree of association between the two resistance determinants. Indeed, the high transferability of these two genes via plasmids with identical replicon types further indicates that the two genes may be simultaneously disseminated by plasmids [23, 26]. The implication of this is that there is a high risk for their widespread dissemination and suggests a critical need for close monitoring of such strains.
UTI-causing E. coli isolates have been closely associated with phylogroups D and B2 in China [27]. D was the main phylogroup in the ESBL-producing urinary E. coli isolates in this study, consistent with previous reports [27, 28]. This indicates that group D may contribute more to MDR and UTI infections in China than other phylogroups. Phylogroups A and B1, however, were more common in our study than B2. Since phylogroups A and B1 have been reported in animal or human commensal E. coli strains [29, 30], this provides evidence that animals may be the source of some UTI-causing E. coli isolates [31].
Clonal diversity in fosA3- and blaCTX-M-harbouring E. coli from humans, as revealed by both PFGE and MLST, indicates that the spread of fosA3 in ESBL-producing E. coli is not attributable to clonal transfer of FosA3 producers in patients. In addition, MLST results suggest that several clonal strains involved in the dissemination of blaCTX-M-positive E. coli, such as ST450, also carry fosA3 [23]. IncFII, IncI1, and IncN plasmids carrying fosA3 as well as blaCTX-M β-lactamase genes have previously been reported in E. coli from chickens, pets, livestock, and other animals in China [26, 32, 33]. Furthermore, fosA3 and blaKPC-2 genes were found to be able to spread together worldwide through IncP plasmid transfer [34, 35]. The high transferability of plasmids carrying fosA3 and multiple replicons found here provide further evidence of the high potential for transfer of fosfomycin resistance gene fosA3. Recently, fosA3 has been found on an epidemic plasmid carrying blaCTX-M-65 and rmtB [36], Of particular concern, the gene has also been identified on a novel IncR-F33:A-:B- plasmid harbouring blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB that was isolated from an epidemic Klebsiella pneumoniae ST11 strain in China [37]. Therefore, close monitoring and continued surveillance of patterns of fosfomycin resistance are necessary in order to prevent further dissemination of fosA3 genes.
One limitation of this study stems from the fact that only ESBL producers in the 821 E. coli urinary isolates were screened for fosfomycin-resistance genes based on the strong association between the presence of fosA3 and that of blaCTX [23, 26]. However, the high prevalence of fosA3 demonstrates the rapid spread of fosfomycin resistance in this region.
In summary, the high prevalence of fosfomycin resistance observed in ESBL-producing urinary E. coli isolates recovered during 2010–2014 is mainly attributed to the widespread occurrence of plasmid-mediated fosA3 genes. The dissemination of the fosA3 gene is closely associated with that of blaCTX-M. Rather than clonal expansion of fosA3-harbouring E. coli lineages, horizontal transfer of plasmid-mediated mobile elements carrying fosA3 played a central role in the spread of E. coli harbouring both fosA3 and blaCTX-M in our hospital.
ACKNOWLEDGEMENTS
The authors thank the EU Reference Laboratory for antimicrobial resistance and the National Food Institute of Technical University of Denmark for providing positive controls for our experiments. This study was supported by the Youth Fund of Jiangsu Province (grant no. BK20140099) and The Fundamental Research Funds for the Central Universities (grant no. 021414340283).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Jean SS, et al. Epidemiology and antimicrobial susceptibility profiles of pathogens causing urinary tract infections in the Asia-Pacific region: results from the Study for Monitoring Antimicrobial Resistance Trends (SMART), 2010–2013. International Journal of Antimicrobial Agents 2016; 47: 328–334. [DOI] [PubMed] [Google Scholar]
- 2.Martirosov DM, Lodise TP. Emerging trends in epidemiology and management of infections caused by carbapenem-resistant Enterobacteriaceae. Diagnostic Microbiology and Infectious Disease 2016; 85: 266–275. [DOI] [PubMed] [Google Scholar]
- 3.Bergen PJ, et al. ‘Old’ antibiotics for emerging multidrug-resistant bacteria. Current Opinion in Infectious Diseases 2012; 25: 626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falagas ME, et al. Fosfomycin. Clinical Microbiology Reviews 2016; 29: 321–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wachino J, et al. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrobial Agents and Chemotherapy 2010; 54: 3061–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neuner EA, et al. Experience with fosfomycin for treatment of urinary tract infections due to multidrug-resistant organisms. Antimicrobial Agents and Chemotherapy 2012; 56:5744–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahata S, et al. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. International Journal of Antimicrobial Agents 2010; 35: 333–337. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, et al. Prevalence of the fosfomycin-resistance determinant, fosB3, in Enterococcus faecium clinical isolates from China. Journal of Medical Microbiology 2014; 63: 1484–1489. [DOI] [PubMed] [Google Scholar]
- 9.Kitanaka H, et al. Novel integron mediated fosfomycin resistance gene fosK. Antimicrobial Agents and Chemotherapy 2014; 58: 4978–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Identification of a novel osXCC gene conferring fosfomycin resistance in Campylobacter. Journal of Antimicrobial Chemotherapy 2015; 70: 1261–1263. [DOI] [PubMed] [Google Scholar]
- 11.Ma Y, et al. Characterization of fosA5, a new plasmid-mediated fosfomycin resistance gene in Escherichia coli. Letters in Applied Microbiology 2015; 60: 259–264. [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute (CLSI). 24th Informational Supplement, M100-S24. Performance Standards for Antimicrobial Susceptibility Testing. CLSI; Wayne, PA: 2014.
- 13.Dallenne C, et al. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. Journal of Antimicrobial Chemotherapy 2010; 65: 4905. [DOI] [PubMed] [Google Scholar]
- 14.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Applied and Environmental Microbiology 2000; 66: 4555–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribot EM, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathogend and Disease 2006; 3: 59–67. [DOI] [PubMed] [Google Scholar]
- 16.Wang M, et al. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrobial Agents and Chemotherapy 2003; 47: 2242–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carattoli A, et al. Identification of plasmids by PCR-based replicon typing. Journal of Microbiological Methods 2005; 63: 219–228. [DOI] [PubMed] [Google Scholar]
- 18.Lai B, et al. In vitro susceptibility of Escherichia coli strains isolated from urine samples obtained in mainland China to fosfomycin trometamol and other antibiotics: a 9-year surveillance study (2004–2012). BMC Infectious Diseases 2014; 14: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, et al. Antimicrobial resistance monitoring of gram-negative bacilli isolated from 15 teaching hospitals in 2014 in China. Zhonghua Nei Ke Za Zhi 2015; 54: 837–845. [PubMed] [Google Scholar]
- 20.Ghafourian S, et al. Extended spectrum beta-lactamases: definition, classification and epidemiology. Current Issues in Molecular Biology 2015; 17: 11–21. [PubMed] [Google Scholar]
- 21.He D, et al. CTX-M-123, a novel hybrid of the CTX-M-1 and CTX-M-9 group β-lactamases recovered from Escherichia coli isolates in China. Antimicrobial Agents and Chemotherapy 2013; 57: 4068–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed AM, Shimamoto T, Shimamoto T. Molecular characterization of multidrug-resistant avian pathogenic Escherichia coli isolated from septicemic broilers. International Journal of Medical Microbiology 2013; 303: 475–483. [DOI] [PubMed] [Google Scholar]
- 23.Sato N, et al. First detection of fosfomycin resistance gene fosA3 in CTX-M-producing Escherichia coli isolates from healthy individuals in Japan. Microbial Drug Resistance 2013; 19: 477–482. [DOI] [PubMed] [Google Scholar]
- 24.Lee SY, et al. Prevalence of acquired fosfomycin resistance among extended-spectrum beta-lactamase–producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. Journal of Antimicrobial Chemotherapy 2012; 67: 2843–2847. [DOI] [PubMed] [Google Scholar]
- 25.Ho PL, et al. Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. Journal of Medical Microbiology 2013; 62: 1707–1713. [DOI] [PubMed] [Google Scholar]
- 26.Hou J, et al. Dissemination of the fosfomycin resistance gene fosA3with CTX-M beta-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrobial Agents and Chemotherapy 2012; 56: 2135–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao X, et al. Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. Journal of Clinical Microbiology 2011; 49: 2496–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navidinia M, et al. Phylogenetic grouping and pathotypic comparison of urine and fecal Escherichia coli isolates from children with urinary tract infection. Brazilan Journal of Microbiology 2014; 45: 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakobsen L, Hammerum AM, Frimodt-Møller N. Detection of clonal group A Escherichia coli isolates from broiler chickens, broiler chicken meat, community-dwelling humans, and urinary tract infection (UTI) patients and their virulence in a mouse UTI model. Applied and Environmental Microbiology 2010; 76: 8281–8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-Cerero L, et al. Characterization of clinical and food animal Escherichia coli isolates producing CTX-M-15 extended-spectrum β-lactamase belonging to ST410 phylogroup A. International Journal of Antimicrobial Agents 2011; 37: 365–367. [DOI] [PubMed] [Google Scholar]
- 31.Osugui L, et al. Virulence genotypes, antibiotic resistance and the phylogenetic background of extraintestinal pathogenic Escherichia coli isolated from urinary tract infections of dogs and cats in Brazil. Veterinary Microbiology 2014; 171: 242–247. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, et al. F33: A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and bla CTX-M-55/-14/-65 in Escherichia coli from chickens in China. Frontiers in Microbiology 2014; 5: 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho PL, et al. Dissemination of plasmid-mediated fosfomycin resistance fosA3 among multidrug-resistant Escherichia coli from livestock and other animals. Journal of Applied Microbiology 2013; 114: 695–702. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Y, et al. Dissemination of a clone carrying a fosA3-harbouring plasmid mediates high fosfomycin resistance rate of KPC-producing Klebsiella pneumoniae in China. International Journal of Antimicrobial Agents 2015; 45: 66–70. [DOI] [PubMed] [Google Scholar]
- 35.Li G, et al. First report of a clinical, multidrug-resistant Enterobacteriaceae isolate coharboring fosfomycin resistance gene fosA3 and carbapenemase gene blaKPC-2 on the same transposon, Tn1721. Antimicrobial Agents and Chemotherapy 2015; 59: 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He L, et al. Complete nucleotide sequence of pHN7A8, an F33:A-:B-type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from China. Journal of Antimicrobial Chemotherapy 2013; 68: 46–50. [DOI] [PubMed] [Google Scholar]
- 37.Xiang DR, et al. Complete sequence of a novel IncR-F33:A-:B-plasmid, pKP1034, harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an epidemic Klebsiella pneumoniae sequence Type 11 strain in China. Antimicrobial Agents and Chemotherapy 2015; 60: 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]