Abstract
Eukaryotic chromosomes contain regions of varying accessibility, yet DNA replication factors must access all regions. The first replication step is loading MCM complexes to license replication origins during the G1 cell cycle phase. It is not yet known how mammalian MCM complexes are adequately distributed to both accessible euchromatin regions and less accessible heterochromatin regions. To address this question, we combined time-lapse live-cell imaging with immunofluorescence imaging of single human cells to quantify the relative rates of MCM loading in euchromatin and heterochromatin throughout G1. We report here that MCM loading in euchromatin is faster than that in heterochromatin in early G1, but surprisingly, heterochromatin loading accelerates relative to euchromatin loading in middle and late G1. This differential acceleration allows both chromatin types to begin S phase with similar concentrations of loaded MCM. The different loading dynamics require ORCA-dependent differences in origin recognition complex distribution. A consequence of heterochromatin licensing dynamics is that cells experiencing a truncated G1 phase from premature cyclin E expression enter S phase with underlicensed heterochromatin, and DNA damage accumulates preferentially in heterochromatin in the subsequent S/G2 phase. Thus, G1 length is critical for sufficient MCM loading, particularly in heterochromatin, to ensure complete genome duplication and to maintain genome stability.
INTRODUCTION
Eukaryotic cell cycle progression is a highly orchestrated and strictly regulated process. One key event during the cell cycle is DNA replication, and it must be tightly controlled to ensure complete and precise genome duplication [for reviews, see (1,2)]. DNA replication in mammalian cells initiates at discrete sites called replication origins that are not strictly defined by DNA sequence, but rather by other aspects of chromatin (3–5). In G1 phase, origin DNA is ‘licensed’ by the loading of MCM complexes that will later be activated in S phase to form the core of the replicative helicase (6–8). Successful genome duplication requires many active DNA replication origins per chromosome; in mammalian cells, thousands of origins are licensed in each G1, and then a subset is activated or ‘fired’ in S phase (9). Origins that are licensed but not fired are dormant origins, and they are induced to fire near stalled forks to ensure complete replication (10,11). When too few licensed origins are available in a local genomic region, the resulting incomplete replication promotes chromosome breaks and genome instability (12,13). Thus, successful replication requires sufficient origin licensing in all genomic regions.
Chromosomes are not uniform substrates for replication, however, and chromatin structure and DNA accessibility vary widely among different genomic regions. If MCM loading is too unevenly distributed, then some regions become vulnerable to under-replication. For example, common fragile sites are more likely to be under-replicated in part because of large inter-origin distances that flank these sites and few available licensed dormant origins (14–16). Transcription of large genes at fragile sites removes potential replication complexes from gene bodies and contributes to the paucity of active origins (17). The reasons for large distances between origins near common fragile sites include sparse MCM loading during the preceding G1 (16), but the causes for regions of low MCM loading are not yet fully understood. Another possibility is that some regions are licensed slowly or later during G1, and these differential licensing dynamics create a risk for local underlicensing when S phase begins.
To date, little is known about intranuclear MCM loading dynamics during G1 in any system. Kuipers et al. investigated human MCM dynamics by FRAP (fluorescence recovery after photobleaching) in live human cells, and they found that MCM proteins are stably and cumulatively loaded onto chromatin throughout G1 phase and are only unloaded by replication during S phase (18). Another study by Symeonidou et al. used a similar method to reveal distinct modes of human MCM chromatin association in different cell cycle phases. They revealed transient interaction of MCM with chromatin in mitosis, stable MCM binding during G1 phase and significantly increased loading in late G1 (19). A later study by Powell et al. found that cyclin E/CDK2 activity is required for maximal loading of fruit fly MCM in late G1 phase, suggesting different dynamics before and after activation of cyclin E/CDK2 (20). These studies are a strong basis of our understanding that MCM loading is essentially unidirectional and requires replication to unload, and each study showed MCM loading increasing throughout G1 (18–20). However, the relative dynamics of loading within different nuclear regions was not addressed, nor could it easily have been with the techniques employed. Are all genomic regions licensed at the same speed, or is there a temporal hierarchy among regions? If licensing proceeds by a preferred temporal order, then are regions that are licensed last more likely to be underlicensed and then under-replicated? We hypothesized that different chromatin environments influence licensing dynamics, and that those dynamics, in turn, impact DNA replication.
Chromatin can be divided into two distinct environments, heterochromatin and euchromatin. Euchromatin is loosely packed and associated with transcriptional activity, whereas heterochromatin is more condensed and generally transcriptionally repressed (21–23). Thus far, the link between replication and chromatin states has primarily been explored in the context of replication timing studies. Origins fire at different times in S phase, and a general correlation between less accessible heterochromatin and replication later in S phase has been reported in many species and cell types (24–26). However, quantifying origin licensing dynamics, as opposed to replication dynamics, in different chromatin environments in G1 phase is still a knowledge gap. Replication timing programs have been suggested as mechanisms that avoid replication stress and maintain genome and chromatin organization (27–29), but the relationship between G1 phase MCM loading dynamics and S phase replication dynamics is still unknown. The relative local amounts of loaded MCM complexes and MCM loading factors, such as the origin recognition complex (ORC), have been implicated in establishing replication timing programs within S phase (30). Studies in yeast and human cells reported that ORC or MCM density is (imperfectly) predictive of replication timing, but they did not address the process leading to MCM loading density differences (31–33). These studies examined overall ORC and MCM binding rather than MCM loading throughout G1 progression. Although replication timing correlates with MCM loading density, neither is timing within S phase a direct measure of origin licensing because origin firing time can be influenced by other factors in S phase (24,34,35), nor does timing analysis itself explain how differences in MCM loading arise during G1 phase in the first place.
It is also currently unknown how MCM loading is adequately distributed during G1 phase to ensure full genome duplication. Since heterochromatin is considered a repressive and less accessible environment, MCM loading may be less efficient in heterochromatin compared to MCM loading in euchromatin. MCM and its loading factor ORC are indeed more concentrated in genomic regions associated with active chromatin marks, suggesting that euchromatin is particularly permissive for origin licensing, although the precise mechanisms driving licensing enrichment in euchromatin are not yet clear (32). Nonetheless, some mechanistic links between MCM loading efficiency and different chromatin features or chromatin-modifying enzymes are known (16,33,36,37). MCM loading is generally enriched at sites with low nucleosome density (16,38,39). Several chromatin-modifying factors are reported to promote MCM loading, such as the chromatin remodeler SNF2H (40), the HBO1 histone acetylase (41,42), the histone variant H2A.Z (43), the histone H4 lysine 20 methyltransferase PR-Set7 (44) and the heterochromatin binding protein ORCA (45,46). In contrast, the Sir2 histone deacetylase suppresses MCM loading at some budding yeast origins (47). However, these studies did not address the dynamics or distribution of mammalian MCM loading during G1.
Although euchromatin is efficiently licensed, a substantial fraction of mammalian DNA resides in heterochromatin that must also be replicated during each cell cycle. In mammalian cells, the longest inter-origin lengths typically observed were ∼400–600 kb (48,49), yet heterochromatin regions can be as long as multiple megabases, including the extreme example of the inactive X chromosome that is largely heterochromatic, but is still replicated every S phase (50,51). Thus, at least some origins in heterochromatin must be licensed during G1 to ensure complete replication in S phase. Importantly, once S phase begins, the MCM loading factors (ORC, CDC6 and CDT1) are degraded or inactivated to prevent any more MCM loading after the end of G1 (52–54). This strict separation of origin licensing and origin firing avoids re-replication, a source of endogenous DNA damage and genome instability (55–57). An important consequence of blocking MCM loading factors in S phase is that all of the MCM loading needed for a complete S phase must occur before the G1/S transition. Thus, the amount and distribution of MCM loading at the end of G1 phase determine the likelihood of a successful S phase (58). However, it is still unknown how the entire genome—both euchromatin and heterochromatin—receives sufficient MCM before S phase starts.
Here, we combined live-cell imaging with fixed-cell imaging to quantify human MCM loading in both heterochromatin and euchromatin as G1 progresses. We discovered that the MCM loading rate is higher in euchromatin than in heterochromatin during early G1. However, the loading rate in heterochromatin accelerates in middle and late G1 so that both euchromatin and heterochromatin achieve similar final loading concentrations throughout the nucleus by the end of G1. Because MCM loading in heterochromatin is later during G1, and heterochromatin typically replicates later in S phase, cells that start S phase prematurely experience under-replication and DNA damage preferentially in heterochromatin. These findings quantify MCM loading dynamics with high temporal resolution to reveal a source of unique vulnerability to genome instability specifically in heterochromatin.
MATERIALS AND METHODS
Cell culture
HEK293T and RPE1-hTERT cells were originally obtained from the ATCC and confirmed to be mycoplasma negative. HEK293T and RPE1-hTERT were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 2 mM l-glutamine and 10% fetal bovine serum (FBS) and incubated in 5% CO2 at 37°C. All cell lines were authenticated by STR profiling (Genetica Cell Line Service, Burlington, NC), passaged with trypsin and not allowed to reach confluence. HEK293T cells were only used for lentivirus packaging. RPE1-hTRET cells were used for the imaging and other experiments.
Cloning
All constructs were generated using either the Gateway cloning method or Gibson assembly following standard protocols as described before (59). PCR fragments were amplified using Q5 polymerase (New England Biolabs). DNA fragments were isolated using the QIAprep Spin Miniprep Kit (Qiagen). Plasmids were transformed into either DH5α or Stbl2 Escherichia coli strains for propagation. pENTR constructs were combined with the expression constructs: pInducer20 (Addgene, plasmid #44012). Plasmids were validated via sequencing (Eton Biosciences) for the desired insert using appropriate primers. YFP-tagged ORC1 was a gift from Supriya Prasanth (60), and was cloned to pInducer20-neo using Gateway cloning. The CDK activity reporter plasmid CSII-EF zeo DHB-mCherry was a gift from S. Spencer (University of Colorado–Boulder, Boulder, CO).
Cell line construction and inducible protein production
To generate self-inactivating lentivirus stocks, PCNA-mTurq2, DHB-mCherry, pInducer20-mVenus-MCM3, pInducer20-YFP-ORC1 or pInducer20-cyclin E1 (59) were individually cotransfected with appropriate envelope and packaging plasmids (61) into HEK293T using 50 μg/ml Polyethylenimine-Max (Aldrich Chemistry).
To generate RPE1-hTERT cells expressing PCNA-mTurq2 and DHB-mCherry, RPE1-hTert cells were transduced with lentivirus packaged from PCNA-mTurq2 and DHB-mCherry in the presence of 8 μg/ml polybrene (Millipore, Burlington, MA) for 24 h. Five hundred cells were seeded in a 15-cm dish, and individual double positive clones were hand-picked and screened by immunoblotting and flow cytometry.
For MCM loading rate experiments in Figure 3, RPE cells expressing PCNA-mTurq2 and DHB-mCherry were transduced with lentivirus packaged from pInducer20-mVenus-MCM3 in the presence of 8 μg/ml polybrene (Millipore, Burlington, MA) into RPE cells expressing PCNA-mTurq2 and DHB-mCherry for 24 h. Transduced cells were selected with 500 μg/ml neomycin (Gibco) for 1 week. Five hundred cells were seeded in a 15-cm dish, and individual positive clones were hand-picked and screened by immunoblotting and evaluated by flow cytometry. To overproduce mVenus-MCM3, cells were treated with 500 ng/ml doxycycline (dox) for 2 h in 10% FBS, DMEM and l-glutamine.
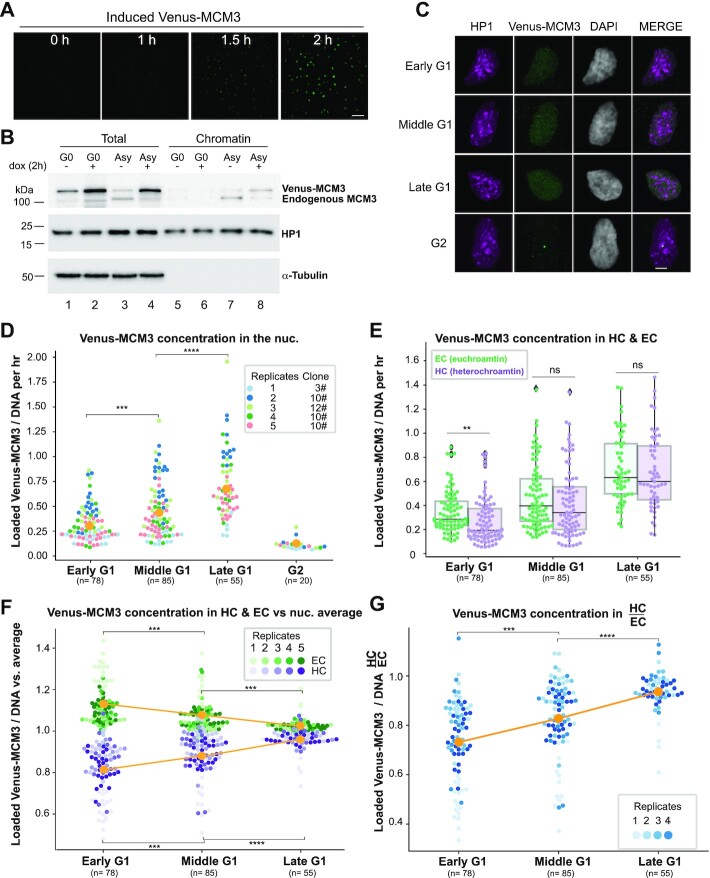
Figure 3.
The rate of MCM loading in early G1 is faster in euchromatin than in heterochromatin. (A) Selected images are shown from time-lapse imaging of RPE cells expressing PCNA-mTurquoise and DHB-mCherry reporters and dox-inducible mVenus-MCM3. mVenus-MCM3 expression was recorded every 10 min after the addition of 500 ng/ml dox; hours since induction are indicated; scale bar: 50 μm. (B) Cells were made quiescent (G0) by contact inhibition or left to proliferate asynchronously (‘Asy’) and then treated with 500 ng/ml dox for 2 h before harvesting. Whole cell lysates (‘total’) or chromatin fractions were analyzed by immunoblotting. (C) Projections of 3D immunofluorescence images of representative cells after live-cell imaging as in Figure 1: HP1 (magenta), Venus-MCM3 detected with anti-GFP antibody (green) and DNA stained with DAPI (gray); scale bar: 5 μm. Cells were treated with 500 ng/ml dox 2 h before the end of live-cell imaging. Quantification of loaded mVenus-MCM3 concentration (i.e. normalized to DNA) in whole nuclei (D) and in heterochromatin and euchromatin (E) after induction for 2 h. Boxplots in (E) show median and interquartile ranges, n (number of cells) is indicated in the figure and five biological replicates are shown. P-values are 0.008 (early G1), 0.219 (middle G1) and 0.595 (late G1). (F) Loaded Venus-MCM3 concentration in heterochromatin (lower purple dots) or euchromatin (higher green dots) relative to the average loaded Venus-MCM3 concentration in whole nuclei; mean is plotted in orange. (G) Ratio of Venus-MCM3 concentration in heterochromatin to euchromatin; mean is plotted in orange. For all comparisons, one-way ANOVA, Tukey post-hoc test and n (number of cells) is indicated on the figure panels. P-value ranges are indicated as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001.
For ORC1 loading experiments in Supplementary Figure S6, RPE cells expressing PCNA-mTurq2 and DHB-mCherry were transduced with lentivirus packaged from pInducer20-YFP-ORC1 in the presence of 8 μg/ml polybrene (Millipore, Burlington, MA) into RPE cells expressing PCNA-mTurq2 and DHB-mCherry for 24 h. Transduced cells were selected with 500 μg/ml neomycin (Gibco) for 1 week. Five hundred cells were seeded in a 15-cm dish, and individual positive clones were hand-picked and screened by immunoblotting and evaluated by flow cytometry. To overproduce YFP-ORC1, cells were treated with 100 ng/ml dox for 24 h in complete medium.
For G1 shortening experiments in Figures 6 and 7, RPE cells expressing PCNA-mTurq2 and DHB-mCherry were transduced with lentivirus packaged from pInducer20-cyclin E1 in the presence of 8 μg/ml polybrene (Millipore, Burlington, MA) for 24 h. Transduced cells were selected with 500 μg/ml neomycin (Gibco) for 1 week. Five hundred cells were seeded in a 15-cm dish, and individual positive clones were hand-picked and screened by immunoblotting and flow cytometry. To overproduce cyclin E1, cells were treated with 15 ng/ml dox (Calbiochem, San Diego, CA) for 7 or 18 h in complete medium as indicated.
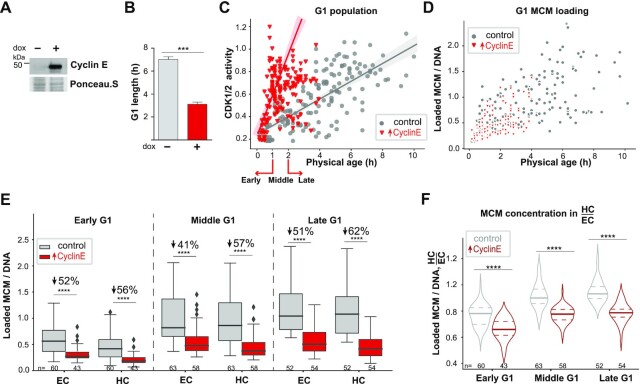
Figure 6.
Preferential heterochromatin underlicensing in shortened G1 cells. (A) Ectopic cyclin E expression was induced by treatment with 15 ng/ml dox for 7 h prior to analysis by immunoblotting. (B) G1 length of cells expressing PCNA-mTurquoise, DHB-mCherry and dox-inducible cyclin E in (A) measured by live-cell imaging of the PCNA S phase reporter; t-test, n (number of cells) = 40 for each group. (C) CDK1/2 activity relative to physical age in control (gray dots) or cyclin E-expressing (red triangles) G1 phase cells treated as in (B). (D) Loaded MCM3 concentration relative to physical age in control (gray dots) or cyclin E-expressing (red triangles) G1 cells as in (B). (E) Loaded MCM concentration in heterochromatin and euchromatin in G1 subphases in control or cyclin E-expressing cells. G1 subphases for (E) and (F) were defined for control cells as in Figure 1 and for cyclin E-overproducing cells by physical age as indicated in (C). Boxplots show median and interquartile ranges, one-way ANOVA, Tukey post-hoc test and n (number of cells) is indicated in the figure. (F) Quantification of the ratio of loaded MCM concentration in heterochromatin to euchromatin in control or cyclin E-expressing G1 cells. Violin plots indicate the median and interquartile ranges, one-way ANOVA, Tukey post-hoc test and n (number of cells) is indicated in the figure. In all panels, P-value ranges are indicated as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001.
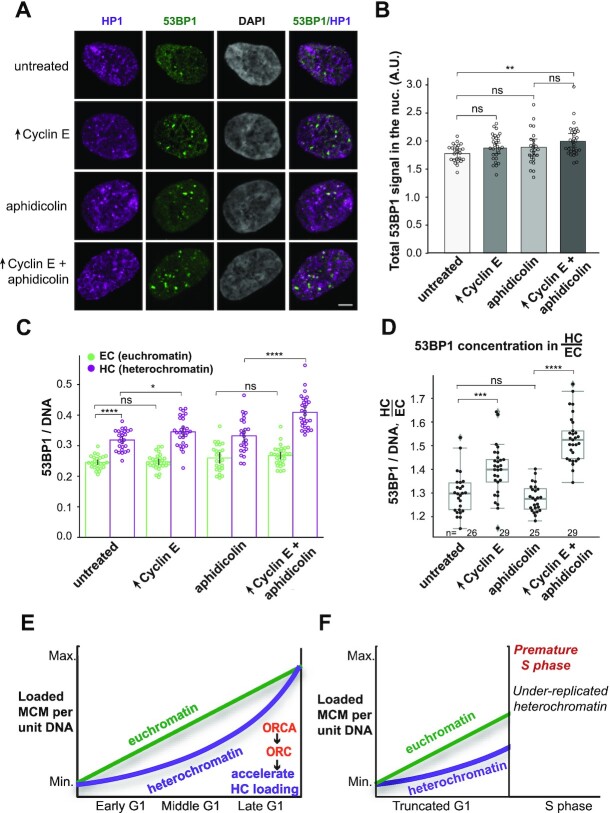
Figure 7.
Heterochromatin is more vulnerable than euchromatin to under-replication and DNA damage. (A) Projections of 3D confocal immunofluorescence images of representative late S phase cells expressing PCNA-mTurquoise, DHB-mCherry and dox-inducible cyclin E after live-cell imaging as in Figure 1. Cells were treated with 500 ng/ml dox for 18 h before imaging and with vehicle or 25 ng/ml aphidicolin for the final 4 h of imaging as indicated. Soluble proteins were extracted prior to fixation and immunostaining for endogenous 53BP1 (green), HP1 (magenta) and DNA (DAPI, gray); scale bar: 5 μm. Cells with a G1 phase shorter than 4 h were selected for analysis of the immediate next late S phase using PCNA variance as an indicator of S phase progression. (B) Quantification of total bound 53BP1 signal in late S phase nuclei and (C) the concentration of 53BP1 signal in heterochromatin (purple, right in each pair) or euchromatin (green, left in each pair). (D) Ratio of 53BP1 concentration in heterochromatin to euchromatin; n (number of cells) is indicated in the figure. Two replicates are shown. In all panels, P-value ranges are indicated as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001. (E) Illustration of normal MCM loading dynamics in euchromatin versus heterochromatin. In early G1 phase, heterochromatin loads MCM more slowly than euchromatin, but differences in loading rate acceleration allow heterochromatin to catch up in middle to late G1 phase. ORCA promotes ORC recruitment to heterochromatin and aids in accelerated MCM loading in heterochromatin. (F) Illustration of heterochromatin underlicensing from premature G1/S transition. When cells prematurely enter S phase, heterochromatin is more underlicensed than euchromatin and subject to under-replication and DNA damage.
Live-cell imaging
Cells were plated on fibronectin-coated (1 μg/cm2, Sigma) F1141 glass‐bottom plates (Cellvis) with FluoroBrite™ DMEM (Invitrogen) supplemented with 10% FBS, 4 mM l‐glutamine and penicillin/streptomycin. Fluorescence images were acquired using a Nikon Ti Eclipse inverted microscope with Plan Apochromat dry objective lenses 20× [numerical aperture (NA) 0.75] or 40× (NA 0.95). Images were captured using an Andor Zyla 4.2 sCMOS detector with 12-bit resolution. Autofocus was provided by the Nikon Perfect Focus System, and a custom enclosure (Okolabs) was used to maintain constant temperature (37°C) and atmosphere (5% CO2) in a humidified chamber. All filter sets were from Chroma: CFP—436/20, 455 and 480/40 nm (excitation, beam splitter and emission filter); YFP—500/20, 515 and 535/30 nm; and mCherry—560/40, 585 and 630/75 nm. Images were collected every 10 min using NIS-Elements AR software. No photobleaching or phototoxicity was observed in cells imaged by this protocol. The relative microscope coordinates from the live-cell imaging microscope were saved for matching to fixed-cell images as needed.
Tracking and segmentation
Individual cells were segmented and tracked in time-lapse videos by a user-assisted approach as previous described (62). In brief, all videos were pre-processed using rolling ball background subtraction. Individual cells in the video were manually tracked using a set of in-house developed ImageJ scripts. Using user-defined tracks, nuclear regions of interest (ROIs) were segmented automatically based on intensity of PCNA followed by separation of touching nuclei by a watershed algorithm. In cases of failed segmentation, the user could manually define polygons as replacement ROIs. The same set of ROIs was used to analyze all fluorescence channels.
PCNA variance and CDK activity
PCNA variance was quantified as described previously (62). The PCNA pattern was analyzed within nuclear ROIs, and then images were processed in a series of steps implemented in Fiji (ImageJ): (i) image procession: image smoothing, edge enhancement and nuclear region reduction; and (ii) quantification of processed PCNA signal: a sum of mean and standard deviation of variance image showed the highest contrast at the beginning and the end of the S phase and was therefore used for cell cycle phase delineation. CDK1/2 activity was quantified as the ratio of cytoplasmic to nuclear mean intensity of the DHB-mCherry sensor (cytoplasm quantified in a 15-pixel ring outside the nuclear segmentation).
siRNA transfections
For siRNA treatment of RPE cells, Dharmafect 4 (Dharmacon) was mixed in Optimem (Gibco) with the appropriate siRNA according to the manufacturer’s instructions, then diluted with DMEM, 10% FBS and l-glutamine, and added to cells after aspirating the original medium. The next day, the siRNA mix was aspirated and replaced with fresh DMEM, 10% FBS and l-glutamine, collecting samples 48 h after the start of siRNA treatment. Generally, the siRNA was siControl (luciferase) at 100 nM or a mixture of two MCM3 siRNAs (2859 and 2936 at 100 nM each) or siORCA (100 nM). The luciferase siRNA and MCM siRNA were synthesized by Sigma (59). The ORCA siRNA was synthesized by Dharmacon (46).
siControl (luciferase): 5′-cuuacgcugaguacuucga
siMCM3-2859: 5′-augacuauugcaucuucauug
siMCM3-2936: 5′-aacauaugacuucugaguacu
siORCA: 5′-ccaaccaggacuacgaauu
Immunofluorescence for chromatin-associated proteins
Cells grown in multiwell glass-bottomed dishes were washed with phosphate-buffered saline (PBS) immediately after live-cell imaging; CSK buffer (300 mM sucrose, 300 mM NaCl, 3 mM MgCl2, 10 mM PIPES, pH 7.0) with 0.5% Triton X-100 and protease and phosphatase inhibitors (0.1 mM AEBSF, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 10 μg/ml phosvitin, 1 mM β-glycerol phosphate, 1 mM sodium orthovanadate) was added to each well for 5 min on ice. Then, CSK buffer with soluble proteins was discarded, and cells were fixed with 1 ml 4% paraformaldehyde for 15 min and washed with PBS twice. Cells were blocked in 1% bovine serum albumin (BSA) in PBS for 1 h, and incubated with the primary antibody overnight at 4°C. Primary antibodies used were as follows: Mcm2 (1:1000, BD Biosciences, Cat#610700), Mcm3 (1:1000, Bethyl Laboratories, Cat#A300-192A), histone H3K9me3 (1:5000, Active Motif, Cat#39062), histone H3K9me3 (1:3000, Active Motif, Cat#61014), HP1 (1:500, Santa Cruz Biotechnology, Cat#sc-515341), histone H4ac (1:3000, Active Motif, Cat#39244), ORC4 (1:100, Santa Cruz Biotechnology, Cat#136331), GFP (1:1000, Invitrogen, Cat#A11122), 53BP1 (1:1000, Novus, Cat#NB100304) and RPA (1:200, Cell Signaling Technology, Cat#2208). Cells were incubated with secondary Alexa Fluor series antibodies (all 1:500, Invitrogen) for 1 h at room temperature and then with 1 μg/ml DAPI for 5 min. Secondary antibodies were all obtained from Jackson ImmunoResearch: donkey anti-mouse-Alexa 488 (1:500, Cat#715-545-150), donkey anti-rabbit-Alexa 594 (1:500, Cat#711-585-158), donkey anti-rabbit-Alexa 488 (1:500, Cat#711-545-152), goat anti-mouse-Alexa 594 (1:500, Cat#715-585-150) and donkey anti-rat-Alexa 647(1:500, Cat#712-605-150).
For confocal microscopy after live-cell imaging, the x–y coordinates from live imaging were recorded and used to locate the same fields on the confocal microscope. Individual G1 cells in the final frame of live imaging were identified as post-mitosis and pre-S phase based on PCNA localization [as in (62)]. The entire area was identified from the x–y coordinates on the confocal microscope and scanned for DAPI (nuclei) to generate a map of nuclear positions. Z stack images (16-bit) were collected using a Zeiss 880 upright confocal microscope with a 63 oil immersion objective lens (Pln Apo 63×/1.4 NA). The pixel size was 0.07 μm and the x–y resolution was 380 × 380 for each slice. The distance between two slices was 0.2 μm. Images were acquired in an automated fashion with the ZEN acquisition software (Zeiss). No photobleaching was observed during acquisition of the stacks.
Analysis of 3D confocal images
Otsu thresholding was performed on each full nucleus using the DAPI channel. Image segmentation was performed using an ImageJ script. The top 20% brightest pixels in the HP1 or H3K9me3 (histone H3 lysine 9 trimethylation) images were used to generate a mask for heterochromatic regions in each Z stack image, unless otherwise stated. Euchromatin was defined as the 80% of pixels outside the HP1 mask, unless otherwise stated. All fluorescence intensity signals in heterochromatin pixels or in euchromatin pixels for each channel were calculated using custom Python scripts (v3.7.1) in Jupyter Notebooks (v6.1.4). Signal intensities were gathered from individual Z stack images in grayscale. Unprocessed intensities (without background subtraction) are reported in figures as arbitrary units and, where indicated, normalized to heterochromatin and euchromatin DAPI intensities as indicated. RPA foci were counted within nuclear ROIs in the channel to be quantified using the 3D Objects Counter plugin in ImageJ. Data were visualized using Jupyter Notebooks [Python graphical libraries Matplotlib (63) and Seaborn (64) and GraphPad Prism (v8)]. All custom scripts are available via GitHub (https://github.com/purvislab/MCM_project).
Total lysate and chromatin fractionation
Cells were collected by trypsinization. Total protein lysates for immunoblotting were generated as described previously (59): cells were lysed on ice for 20 min in CSK buffer with 0.5% Triton X-100 and protease and phosphatase inhibitors. Cell lysates were centrifuged, and the supernatants kept for a total protein Bradford assay (Bio-Rad, Hercules, CA) using a BSA standard curve. Chromatin fractionation for immunoblotting was performed as described previously (59) using CSK buffer with 1 mM ATP, 5 mM CaCl2, 0.5% Triton X-100 and protease and phosphatase inhibitors to isolate insoluble proteins and S7 nuclease (Roche) to release DNA-bound proteins. A protein assay was performed for chromatin fraction quantification.
Immunoblotting
Samples were diluted with SDS loading buffer and boiled. Proteins were separated on SDS-polyacrylamide gels, and then transferred onto polyvinylidene difluoride membranes (Thermo Fisher, Waltham, MA). Membranes were blocked in 5% milk in Tris-buffered saline–0.1% Tween 20 (TBST) at room temperature for 1 h. Then, membranes were incubated in primary antibody overnight at 4°C in 2.5% milk in TBST. The next day, blots were washed with TBST, then incubated in secondary antibody conjugated to horseradish peroxidase in 2.5% milk in TBST for 1 h and washed with TBST for three times. For detection, membranes were incubated with ECL Prime (Amersham, UK) and exposed to autoradiography film (Denville, Holliston, MA) or detected by a ChemiDoc imaging system (Bio-Rad). Ponceau S staining for total protein (Sigma-Aldrich) was typically used as a loading control. The following primary antibodies were used for immunoblotting: Mcm2 (BD Biosciences, San Jose, CA, Cat#610700), Mcm3 (Bethyl Laboratories, Montgomery, TX, Cat#A300-192A), Cdt1 (Santa Cruz Biotechnology, Cat#sc-365305), histone H3 (GeneScript, Piscataway, NJ, Cat#A01502), histone H3ac (Millipore Sigma, Carlsbad, CA, Cat#06-599), histone H3K9me3 (Active Motif, Carlsbad, CA, Cat#39062), α-tubulin (Sigma-Aldrich, Cat#9026). PCNA (Santa Cruz Biotechnology, Cat#sc-25280) and HP1 (Santa Cruz Biotechnology, Cat#sc-515341). The secondary antibodies were used for immunoblotting: anti-rabbit IgG HRP-conjugated (1:10 000, Jackson ImmunoResearch) and goat anti-mouse IgG HRP-conjugated (1:10 000, Jackson ImmunoResearch).
Flow cytometry
Analysis of chromatin-bound MCM was performed as described in (59). Briefly, cells were treated with 10 μM EdU (Santa Cruz Biotechnology) for 30 min before collection. Cells were collected and then lysed on ice for 8 min in CSK buffer (10 mM PIPES, pH 7.0, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2) with 0.5% Triton X-100 with protease and phosphatase inhibitors. Cells were washed with 1% BSA–PBS and then fixed in 4% paraformaldehyde in PBS for 15 min at room temperature. For EdU detection, samples were incubated in PBS with 1 mM CuSO4, 1 μM Alexa 647-azide (Life Technologies) and 100 mM ascorbic acid (fresh) for 30 min at room temperature in the dark. Then, cells were labeled in primary antibody and incubated at 37°C for 1 h in the dark. Next, cells were resuspended in secondary antibody and incubated at 37°C for 1 h in the dark. Finally, cells were resuspended in 1% BSA–PBS + 0.1% NP-40 with 1 μg/ml DAPI (Life Technologies) and 100 μg/ml RNase A (Sigma-Aldrich) and incubated overnight at 4°C in the dark. Data were collected on an Attune NxT flow cytometer (Thermo Fisher Scientific) and analyzed using FCS Express 7 software (De Novo Software). Control samples were prepared omitting primary antibody or EdU detection to define thresholds of detection as in (59). The following antibodies were used: primary—Mcm2 (1:100, BD Biosciences, Cat#610700) or ORC4 (1:50, Santa Cruz Biotechnology, Cat#136331); and secondary—donkey anti-mouse-Alexa 488 (Jackson ImmunoResearch). A representative gating scheme is provided in Supplementary Figure S8A–D.
Cell synchronization and treatments
To synchronize cells in quiescence (G0), RPE1-hTert cells were grown to 100% confluency and incubated for another 48 h in 10% FBS, DMEM and l-glutamine. G0 cells were then released by passaging 1:10 with trypsin to new dishes in 10% FBS, DMEM and l-glutamine; cells were collected at different time points (16, 18, 20, 23, 26, 28 and 30 h) to enrich for different phases of the cell cycle. For aphidicolin (Sigma) treatment, asynchronous RPE cells were treated with 25 ng/ml or vehicle control for 4 h.
Statistical analysis
Bar graphs represent means, and error bars indicate standard error of the mean, unless otherwise noted. The number and type of replicates are indicated in the figure legends. Significance tests were performed using a one-way ANOVA test, as indicated in the figure legends, unless otherwise specified. Statistical significance is indicated as asterisks in figures: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001. GraphPad Prism v.8.0 and Python were used for statistical analysis.
RESULTS
An experimental system for analyzing subnuclear MCM loading dynamics within G1 phase
To analyze MCM loading dynamics, we sought to quantify MCM loading at different points in G1 phase. MCM complexes can begin to load during telophase of mitosis, and loading continues through G1 phase, peaking just before the G1/S transition; MCM complexes are unloaded during S phase (65,66). The total levels of MCM subunits are constant throughout the cell cycle, but the levels of DNA-loaded MCM change as cells progress through the cell cycle (67). To quantify and analyze the dynamics of subnuclear MCM loading in different stages of G1 phase, we developed a method to correlate intranuclear MCM loading positions in individual human cells with cell cycle progression (Figure 1A and B). We first defined subphases of G1 progression, and we chose an untransformed human epithelial cell line, retinal pigmented epithelial cells (RPE1-hTert), as a model. We created a derivative of these cells that stably expresses two previously described reporters: one to detect S phase entry and exit based on PCNA localization [PCNA-mTurq2 (68)] and one that reports CDK1/CDK2 activity from the phosphorylation-dependent cytoplasmic localization of a fragment of DNA helicase B fused to mCherry (69,70). We performed time-lapse imaging on these asynchronously proliferating cells to define the cell cycle age of individual cells (time elapsed since the previous mitosis), cell cycle phase (G1, S, G2, M) and CDK1/2 activity for each cell.
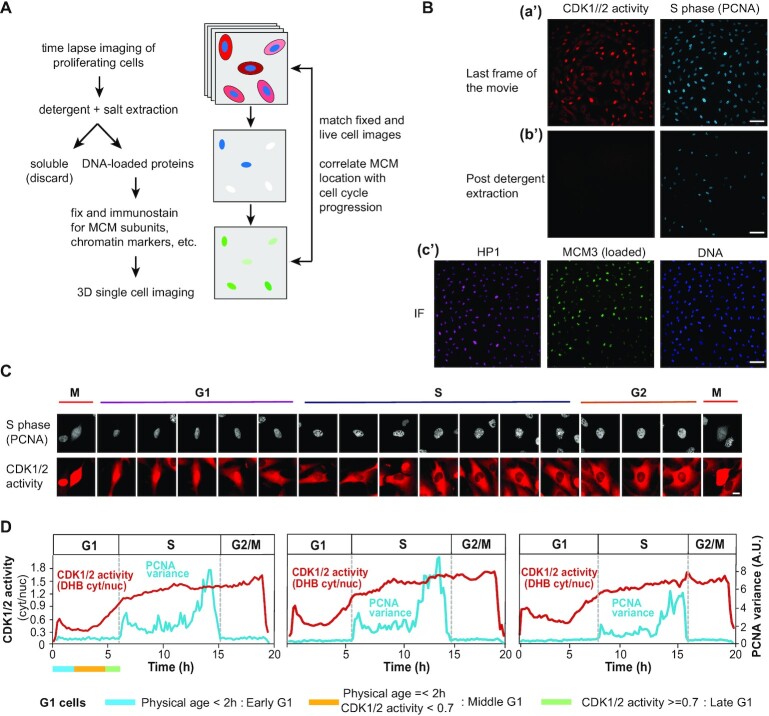
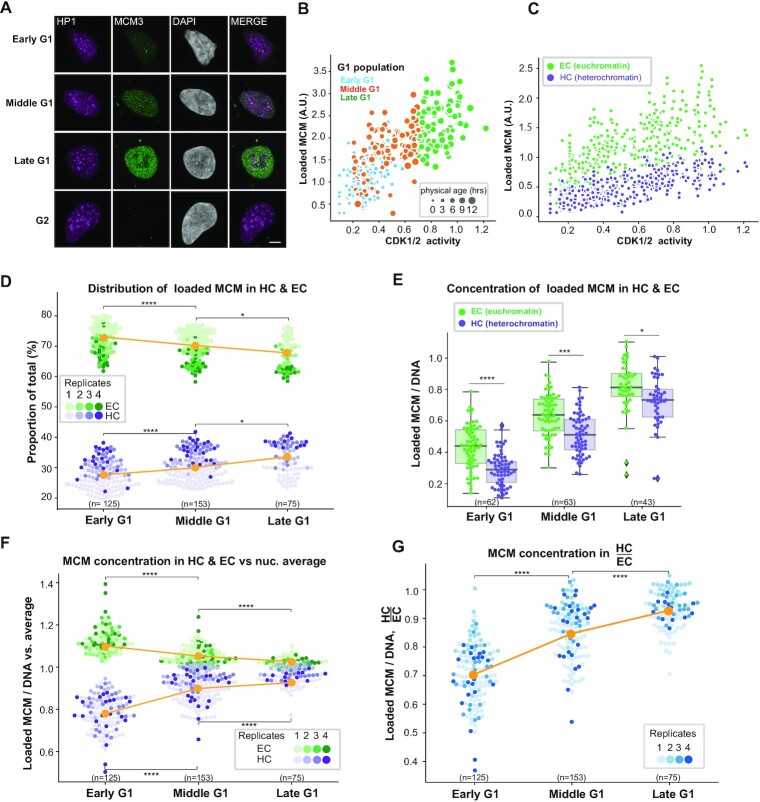
Figure 1.
An experimental system for analyzing subnuclear MCM loading dynamics within G1 phase. (A) Workflow. RPE1-hTert cells expressing PCNA-mTurquoise and the DHB-mCherry CDK1/2 activity reporter were subjected to time-lapse live-cell imaging, then soluble proteins were extracted with nonionic detergent and salt, and cells were fixed immediately after imaging for confocal immunofluorescence staining. (B) Representative example from combining live-cell imaging with fixed-cell imaging. (a′) Last frames from wide-field time-lapse imaging of cells expressing CDK1/2 activity and S phase reporters. (b′) Images collected with the same microscope settings after detergent extraction and fixation; scale bar represents 100 μm. (The CDK activity reporter is soluble.) (c′) Immunofluorescence of extracted and fixed cells after live-cell imaging. Cells were stained for bound HP1 (heterochromatin marker) and loaded MCM3 (MCM2–7 complex marker) and imaged by confocal microscopy; scale bar represents 100 μm. (C) Selected images from wide-field time-lapse imaging of one cell. Images were captured every 10 min for one cell cycle, and selected frames from one of the 50 cells are shown. The scale bar is 10 μm and applies to all images. Images were brightness/contrast adjusted. (D) Top: Traces of PCNA variance and CDK1/2 activity for three cells. CDK1/2 activity is the ratio of mean cytoplasmic DHB-mCherry reporter fluorescence divided by mean nuclear DHB-mCherry fluorescence. Hours are time since mitosis. Bottom: Defining G1 subphases by both physical age and CDK1/2 activity. G1 cells younger than 2 h after mitosis are early G1 cells; G1 cells older than 2 h with CDK activity <0.7 are middle G1 phase cells; and G1 phase cells with CDK activity ≥0.7 but not yet in S phase by PCNA variance are late G1 cells.
We defined G1 cells by identifying cells post-mitosis and pre-S phase using the variance of PCNA-mTurq2 localization, as previously described (62). PCNA-mTurq2 was present throughout the cell cycle, diffusely distributed in G1 nuclei, punctate during S phase and diffuse again in G2. To automatically detect S phase boundaries, we calculated the variance of PCNA intensity across the nucleus. The rapid increase in PCNA variance indicates the onset of S phase, whereas the steep drop in PCNA variance indicates the end of S phase (Figure 1C and D). In normal RPE1-hTert cells, G1 length varies from 5 to 12 h, with most cells spending 7–8 h in G1 (71). We defined cells younger than 12 h with no significant increase in PCNA variance as G1 cells. As a measure of CDK2 activity, we monitored the relative localization the DHB-mCherry reporter. CDK2 is activated by cyclin E during G1, and CDK-dependent DHB-mCherry reporter phosphorylation induces translocation from the nucleus to the cytoplasm (69). Based on the normal timing of CDK2 and CDK1 activation and previous reports, we attribute reporter translocation in late G1 and early S phases to CDK2 activity and G2 phase reporter localization to a combination of CDK2 and CDK1 activities (70); the reporter is not responsive to CDK4 or CDK6 activity (69,72). We quantified the ratio of cytoplasmic to nuclear reporter localization throughout the cell cycle. Of note, the CDK reporter activity increases nonlinearly over time in G1 (Figure 1D). We tracked complete cell cycles of 50 cells and quantified their CDK1/2 activity when they entered S phase; the cytoplasmic to nuclear localization range was from 0.9 to 1.3, and the majority of cells enter S phase with the CDK1/2 reporter activity around 1.1 (Figure 1D and Supplementary Figure S1A).
We categorized G1 cells into three groups according to both their physical age in hours and their CDK1/2 activity. Because the nuclear membrane is undergoing maturation in early G1, and many nuclear components (including the CDK reporter) are still partly cytoplasmic, we categorized early G1 cells by their physical age; cells younger than 2 h are early G1 cells. However, the large intercellular variation in G1 length meant that CDK activity correlated with G1 progression better than physical age in middle and late G1 phases. For example, sisters with different G1 lengths can have very different CDK activities and MCM loading amounts at the same physical age (Supplementary Figure S1B). We therefore used CDK activity to define the ‘molecular age’ of middle and late G1 cells instead of simply subdividing G1 phase according to time since mitosis. Previous studies have measured CDK1/2 reporter ∼0.7 near the point in G1 when cells are committed to S phase, so we chose 0.7 as a mark between middle and late G1 (69,70). G1 cells older than 2 h but with CDK1/2 activity still below 0.7 are middle G1 phase cells, and G1 phase cells with CDK1/2 activity above 0.7 are late G1 cells (Figure 1D). The physical age distribution for the different categories of G1 cells is shown in Supplementary Figure S1C.
To quantify MCM loading, we extracted the cells with nonionic detergent in the presence of 300 mM salt to remove soluble MCM immediately after the live-cell imaging was stopped. Nonextractable, salt-resistant MCM complexes are strongly correlated with DNA replication origins and replication competence in vitro (73,74) and in vivo (18,59). We then fixed the remaining chromatin-bound proteins for immunofluorescence using anti-MCM3 antibody as a marker of the MCM2–7 complex, anti-HP1 antibody as a marker of heterochromatin (75,76) and DAPI for DNA content (Figure 1B). We selected HP1 as a marker of heterochromatin based on its binding to the heterochromatin feature, H3K9me3, and its general association with compacted and less accessible DNA (75,77,78), and we used an antibody that detects three of the four HP1 isoforms. We performed confocal imaging and quantified the colocalization of loaded MCM and HP1 in Z stack images of G1 cells (Supplementary Video S1). We then matched each G1 cell in the confocal images to its history in the time-lapse video to define both physical and molecular ages. This approach is suitable for analyzing unperturbed proliferating populations to identify cell cycle-related changes that would be difficult to detect by immunoblotting or flow cytometry.
As controls, we validated the MCM3 antibody specificity by the loss of immunostaining of both total and loaded MCM3 in MCM3-depleted cells (Supplementary Figure S1D and E). Anti-MCM3 staining showed strong colocalization with MCM2, which is expected from two subunits of the MCM2–7 complex (Supplementary Figure S1F). MCM3 also colocalized with an ectopically expressed YFP–ORC1 fusion in G1 cells indicating that MCM detected this way is at sites of origin licensing (Supplementary Figure S1G). Furthermore, our HP1 staining was largely mutually exclusive with a marker of euchromatin, histone H4 acetylation (H4ac), and generally colocalized with H3K9me3, another established heterochromatin marker (Supplementary Figure S2A–D).
Differential dynamics of MCM loading in euchromatin and heterochromatin
By the analysis outlined earlier, MCM signal was very low on chromatin in G2 cells, but total nuclear MCM loading signal increased throughout G1 (Figure 2A and B, and Supplementary Figure S2E and F). We hypothesized that the dynamics of intranuclear MCM loading during G1 phase are affected by chromatin environments. To test this idea, we used the HP1 immunostaining to distinguish heterochromatin from euchromatin and quantified loaded MCM in heterochromatin and euchromatin as a function of CDK activity. As expected, the HP1 signal was unevenly distributed in nuclei and was concentrated in discrete foci (79,80) (Figure 2A). It has been reported that for many eukaryotes, heterochromatin constitutes 10–20% of the genome (81–83). These reports prompted us to set an initial threshold for identifying heterochromatin in the individual confocal Z stack images as the pixels with the 20% brightest HP1 signals, whereas euchromatin is the remaining 80% of nuclear pixels. As expected, the HP1 locations were more intensely stained with DAPI, which is an indicator of condensed chromatin (Supplementary Figure S2G). Neither total HP1 nor the distribution of HP1 on heterochromatin and euchromatin changed during G1 phase (Supplementary Figure S2H and I).
Figure 2.
Differential dynamics of MCM loading in euchromatin and heterochromatin. (A) Projections of 3D confocal immunofluorescence images of representative cells after live-cell imaging as in Figure 1: endogenous HP1 (magenta), endogenous MCM3 (green) and DNA stained with DAPI (gray); scale bar: 5 μm. All analyses used RPE1-hTert cells expressing PCNA-mTurquoise and the DHB-mCherry CDK1/2 activity reporter. (B) Total loaded MCM3 immunofluorescence signal (y-axis) relative to CDK1/2 activity defined by the cytoplasmic versus nuclear localization of the reporter (x-axis). Cells are color coded for early (blue), middle (orange) and late (green) G1 cells defined in Figure 1. The size of data points for single cells corresponds to time since mitosis. (C) The total amount of loaded MCM3 colocalized with heterochromatin (high HP1 regions, lower purple dots) and euchromatin (low HP1 regions, higher green dots) within each cell relative to CDK1/2 activity. All five replicates are shown, n = 446. (D) Distribution of loaded MCM3 in heterochromatin (purples, lower) and euchromatin (greens, higher) in early, middle and late G1 cells as proportions of the total MCM signal per cell. Four replicates are shown in different shades; means are plotted in orange. (E) MCM3 concentration normalized to DNA/DAPI in heterochromatin (purple) and euchromatin (green) in G1 subphases. Boxplots show median (solid line) and interquartile ranges (box ends); whiskers mark the minimum or maximum. (F) MCM concentration in heterochromatin (lower purple dots) or euchromatin (upper green dots) relative to the average loaded MCM concentration in whole nuclei; mean is plotted in orange. Four replicates are shown with different shades. (G) Ratio of MCM3 concentration in heterochromatin to euchromatin; mean is plotted in orange. Four replicates are shown with different shades. For all comparisons, one-way ANOVA, Tukey post-hoc test and n (number of cells) is indicated on the figure panels or in the legend. In all panels, P-value ranges are indicated as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001.
Our aggregate analysis of cells from multiple independent experiments revealed intercellular variability in total MCM loading signal (the sum of euchromatin and heterochromatin) among cells with similar CDK activity (Figure 2B). Within this heterogeneity, we nonetheless measured distinct and reproducible trends in MCM loading when we analyzed heterochromatin and euchromatin separately (Figure 2C). For both euchromatin and heterochromatin, MCM loading increases throughout G1 indicating a positive loading rate for the entire G1 phase. The total MCM signal is higher in euchromatin than in heterochromatin in keeping with their respective 80%/20% shares of nuclear volume.
Although the MCM loading signal increased for both euchromatin and heterochromatin, the relative proportion of MCM loading in heterochromatin versus euchromatin changed during G1 (Figure 2D); the relative amount of heterochromatin itself did not change during G1 (Supplementary Figure S2H and I). We classified G1 subphases by the same criteria as defined in Figure 1D, and found that in early G1, euchromatin accounted for a larger proportion of the total MCM that had been loaded than it did by middle and late G1. As a result, the relative proportions of loading on the two chromatin types converged between early and late G1 so that the difference between them was smaller (Figure 2D). This change in proportion plus the general MCM loading increase in both chromatin types throughout G1 suggested that heterochromatin loading increases faster in middle and late G1 than the increase in euchromatin loading.
A feature of heterochromatin is its higher chromatin density, i.e. more DNA and associated protein content per unit volume (84,85). The intensity of DAPI-stained DNA was indeed higher in the regions with the 20% brightest HP1 signals than in the rest of the nucleus (Supplementary Figure S2G). This observation supports our classification of HP1 localization as generally marking heterochromatin, although we acknowledge that HP1 can also be found in euchromatic locations (76). To fairly compare MCM loading in heterochromatin to euchromatin, we normalized MCM loading to DNA content at each location to derive the concentration of loaded MCM per unit DNA (loaded MCM/DNA). Similar to measurements of total loaded MCM, the overall concentration of loaded MCM increased during G1 progression (Figure 2E and Supplementary Figure S2J). Interestingly, the concentration of loaded MCM on heterochromatin was lower than that on euchromatin in early G1, but the difference between them had narrowed by the end of G1 phase (Figure 2E). As another means to visualize these differences, we compared the concentration of loaded MCM in heterochromatin and euchromatin to the average concentration of loaded MCM across each whole nucleus. Euchromatin loading starts out higher than the nuclear average early in G1, but the euchromatin and heterochromatin concentrations at their respective locations approach each other by the end of G1 (Figure 2F). We observed similar dynamics using another MCM subunit, MCM2, and H3K9me3 as a heterochromatin marker (Supplementary Figure S3A–F). Finally, we calculated the ratio of the loaded MCM concentrations in heterochromatin versus euchromatin and found that this ratio increases from a low of ∼0.7 in early G1 to ∼0.9 in late G1, indicating near-equivalent origin licensing for both chromatin types by the end of G1 (Figure 2G).
As a test of our heterochromatin definition threshold, we randomly selected a set of 20% pixels for similar analysis. Although total MCM signal increased at all locations during G1, neither the proportion of total MCM nor the relative concentration of loaded MCM in these randomly selected pixels changed over the course of G1 progression (Supplementary Figure S4A and B). We also tested whether choosing different thresholds for defining heterochromatin affected the overall results. When we analyzed MCM loading using a threshold for defining heterochromatin as the 10% or 50% brightest HP1 signals instead of 20%, we found the same relative dynamics, but at different absolute values (Supplementary Figure S4C–F). Defining heterochromatin as the 50% brightest HP1 signals without normalizing to DNA reversed the position of heterochromatin and euchromatin on the y-axis, but their increase/decrease over G1 progression remained the same, and they converged when the signals were normalized to DNA concentrations (Supplementary Figure S4E and F). For all other analyses, we continued to use a 20% brightest HP1 pixel threshold.
The rate of MCM loading in early G1 is faster in euchromatin than in heterochromatin
Loaded MCM complexes are very stably associated with DNA and are essentially only unloaded in S phase (18) or very locally displaced by active transcription (20,32,86). The remarkable stability of loaded MCM means that licensing in G1 is largely unidirectional, and the loaded MCM we detect in late G1 is the sum of all the loading that has occurred since early G1. We were therefore limited to inferring endogenous MCM loading rates in euchromatin versus heterochromatin rather than directly measuring the rates. To compare actual MCM loading rates in heterochromatin and euchromatin, we integrated a dox-inducible mVenus-MCM3 construct into the RPE1-hTert cells with the cell cycle phase and CDK activity reporters. We induced expression for brief periods and detected a strong signal increase by both fluorescence and immunoblotting after 2 h of induction (Figure 3A and Supplementary Figure S5A). In the absence of dox, Venus-MCM3 was produced at only ∼10% of the amount produced after 2 h of induction by immunoblotting and was undetectable by microscopy (Figure 3A and B, compare lanes 3 and 4; Supplementary Figure S5A, compare lanes 1 and 3). To test whether the Venus-MCM3 fusion is loaded normally, we compared the dynamics of induced Venus-MCM3 with endogenous MCM3 during the cell cycle. We probed immunoblots of chromatin-enriched fractions and found that the ectopic MCM3 fusion has a similar G1 loading and S phase unloading pattern as endogenous MCM3 (Supplementary Figure S5B). The MCM3 fusion may compete with endogenous MCM3 for other endogenous MCM subunits required for loading on chromatin, a phenomenon previously reported by Kuipers et al. (18). We detected very little loaded Venus-MCM3 or endogenous MCM3 in quiescent (G0) cells as expected (Figure 3B, lanes 5 and 6). Thus, Venus-MCM3 is a bona fide reporter for MCM loading.
We then imaged asynchronously proliferating live cells and induced Venus-MCM3 expression for the final 2 h of imaging prior to extraction and immunostaining. In this way, we measure MCM loading over a defined period of time because we restrict our analysis to the newly synthesized Venus-MCM3. We detected only faint Venus fluorescence after 1 h of induction, and almost all of the detectable Venus-MCM3 accumulated during the second hour (Figure 3A). We stained for loaded Venus-MCM3 using an anti-GFP antibody (anti-GFP antibodies detect Venus, which is a modified form of GFP) and HP1 as in Figure 2 and derived relative rates of Venus-MCM3 loading per hour (Figure 3C and D). By this analysis, the overall MCM loading rate increases during G1 progression and is highest in late G1 (Figure 3D). We then compared relative Venus-MCM3 loading rates in heterochromatin and euchromatin. In early G1, the MCM loading rate was significantly higher in euchromatin than in heterochromatin, and this difference disappeared gradually in middle G1 and late G1 phases (Figure 3E). Similar to endogenous MCM (Figure 2F), in early G1 Venus-MCM3 euchromatin loading was more concentrated and heterochromatin loading less concentrated than the nuclear average, but the concentrations of loaded MCM in both chromatin types were similar to each other by late G1 (Figure 3F). The ratio of Venus-MCM3 loading in heterochromatin to euchromatin also increased as G1 progressed (Figure 3G). Taken together, we conclude that the rate of MCM loading per hour increases during G1, and early G1 favors fast euchromatin loading. Strikingly, although the loading rate increases for both heterochromatin and euchromatin in middle and late G1, heterochromatin loading accelerates more than euchromatin does to achieve similar rates and concentrations by the end of G1.
Global histone hyperacetylation improves early G1 heterochromatin MCM loading
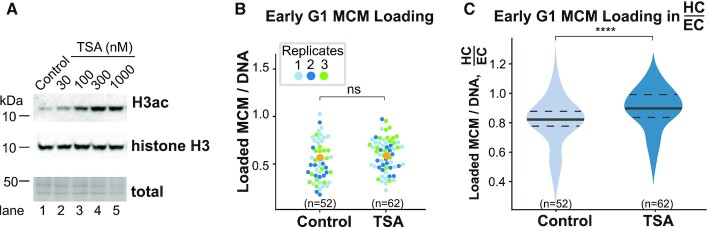
We then sought to determine whether manipulating chromatin in cells would cause a corresponding change in MCM loading dynamics. Since many of the largest differences between heterochromatin and euchromatin were apparent in early G1, we hypothesized that narrowing the difference between euchromatin and heterochromatin properties would promote more equal loading in early G1. To test this idea, we treated asynchronously proliferating cells for just 3 h with increasing concentrations of the histone deacetylase inhibitor, trichostatin A (TSA), to induce histone hyperacetylation and presumably a higher proportion of open chromatin (87). We prioritized short treatment to reduce the confounding effects of perturbed gene expression. We then analyzed early G1 cells as defined in Figure 1D. We found that 300 nM TSA induced a strong increase in global histone H3 acetylation after 3 h (Figure 4A, compare lanes 1 and 4). When we analyzed overall MCM loading (all types of chromatin, throughout G1) after TSA treatment, we detected no difference between control and treated cells (Figure 4B); brief TSA treatment did not affect HP1 levels or general localization (unpublished observation). On the other hand, when we analyzed the distribution of MCM loading in early G1, we found that, as before, MCM loading favored euchromatin in control cells (ratio of HC and EC well below 1), but brief TSA treatment significantly reduced the disadvantage for heterochromatin (Figure 4C). These results indicate that the differences in MCM loading dynamics rely on global chromatin acetylation status.
Figure 4.
Global histone hyperacetylation improves early G1 heterochromatin MCM loading. (A) Representative immunoblot of histone H3 acetylation in untreated and TSA-treated asynchronously proliferating RPE1-hTert cells (expressing PCNA-mTurquoise and DHB-mCherry). Cells were treated with the indicated concentrations of TSA for 3 h. Quantification of total loaded MCM3 concentration (B), and the ratio of loaded MCM3 concentration in heterochromatin to euchromatin (C) in early G1 cells that had been treated with 300 nM TSA for 3 h. Three replicates are shown. One-way ANOVA test and n (number of cells) is indicated in the figure. ****P ≤ 0.0001.
ORCA-dependent ORC loading dynamics support faster MCM heterochromatin loading
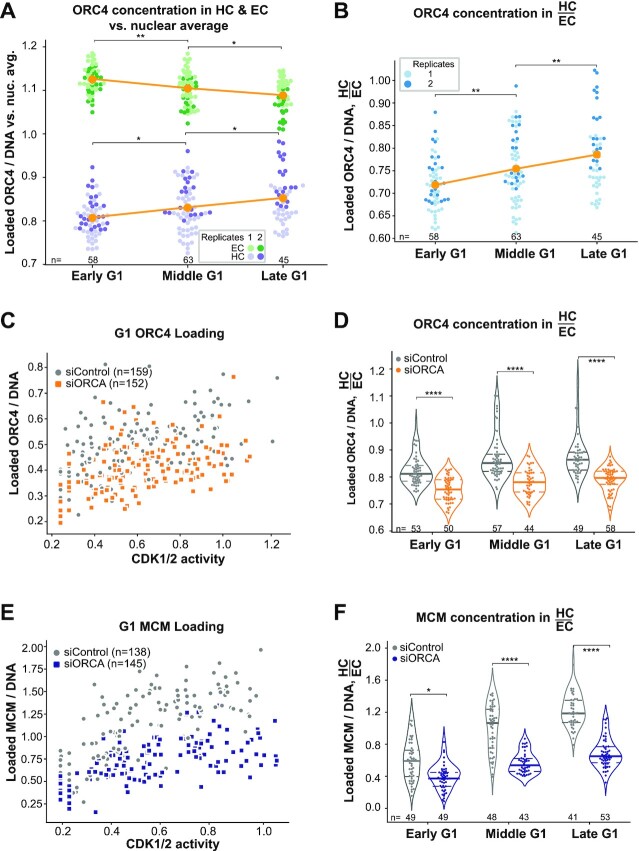
In eukaryotic cells, MCM loading to license origins starts with the DNA binding complex, ORC, which consists of six subunits, ORC1–ORC6. Mammalian ORC selects the sites for MCM loading by mechanisms that are still incompletely understood but include interactions with chromatin features and chromatin binding proteins (88). ORC then cooperates with the CDC6 and CDT1 proteins to directly load MCM complexes onto DNA (1,2,89). We hypothesized that the specific MCM loading dynamics in different G1 subphases could be dictated by the distribution of loaded ORC. To test that idea, we analyzed the distribution of endogenous loaded ORC4 as a marker of the ORC complex. The overall ORC4 concentration per unit DNA moderately increased from early G1 to late G1 phase indicating progressive accumulation of ORC on chromatin during G1 (Supplementary Figure S6A and B). Interestingly and like MCM, the concentration of ORC4 per unit DNA in heterochromatin and euchromatin relative to the nuclear average was widely different in early G1 again strongly favoring euchromatin over heterochromatin, but these values converged in late G1 (Figure 5A). Like MCM loading, the ratio of loaded ORC4 in heterochromatin to euchromatin increased from early to late G1 (Figure 5B). We observed similar trends in a stably expressed YFP–ORC1 fusion, although the effects were more modest than for endogenous ORC4 (Supplementary Figure S6C–F). The ORC1 subunit can independently bind chromatin through its BAH domain (88), raising the possibility that the ORC1 we detect may localize to some chromatin sites independently of the full ORC. Taken together, these results are consistent with a relative increase in loaded ORC concentration on heterochromatin in late G1 that could support accelerated MCM loading on heterochromatin in late G1.
Figure 5.
ORCA-dependent ORC loading dynamics support faster MCM heterochromatin loading. (A) Loaded ORC4 concentration (ORC4 signal normalized to DNA/DAPI) in heterochromatin (purple, lower dots) and euchromatin (green, higher dots) relative to the overall loaded ORC4 concentration in each nucleus in RPE1-hTert cells (expressing PCNA-mTurquoise and DHB-mCherry); mean is plotted in orange. Two replicates are shown. (B) Ratio of loaded ORC4 concentration in heterochromatin to euchromatin; mean is plotted in orange. Two replicates are shown. ORC4 concentration (C) and the ratio of ORC4 concentration in heterochromatin to euchromatin (D) in G1 cells treated with 100 nM control siRNA or ORCA siRNA for 48 h. Two biological replicates are shown. MCM3 concentration (E) and the ratio of endogenous MCM3 concentration in heterochromatin to euchromatin (F) in G1 cells treated with 100 nM control siRNA or ORCA siRNA for 48 h. Two biological replicates are shown. For all comparisons, one-way ANOVA, Tukey post-hoc test and n (number of cells) is indicated on the figure panels. In all panels, P-value ranges are indicated as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001.
To understand how ORC loading dynamics change during G1, we turned our attention to the role of the ORCA/LRWD1 protein. ORCA interacts with both ORC and heterochromatin markers, and ORCA plays a role in both ORC recruitment to heterochromatin loci and heterochromatin organization (45,90). We considered that ORCA may be required for the dynamics of both ORC and MCM loading as G1 progresses. We used RNAi to reduce ORCA and observed the previously reported reduction in ORC chromatin association and the heterochromatin marker H3K9me3 [Supplementary Figure S7A, compare lanes 3 and 4 (46,91)]. ORCA depletion for 72 h caused a moderate increase in S phase cells (Supplementary Figure S7B). As measured by analytical flow cytometry of cells extracted to remove soluble proteins, ORCA depletion also specifically decreased the level of chromatin-loaded ORC4 in G1 phase cells (Supplementary Figure S7C and D). By quantitative immunofluorescence, ORCA depletion reduced the concentrations of both ORC4 (Figure 5C) and MCM (Figure 5E) on DNA in all G1 subphases (Supplementary Figure S7E and F). Importantly, ORCA depletion disrupted not only the total increase of ORC4 loading, but also the normal relative increase in the ratio of heterochromatin to euchromatin ORC4 loading (Figure 5D). Correspondingly, ORCA depletion disrupted total MCM loading (Figure 5E) and the relative increase in MCM loading in heterochromatin versus euchromatin (Figure 5F). These results indicate that ORCA is required for the relative increase in ORC-dependent MCM loading on heterochromatin versus euchromatin during G1 progression.
Preferential heterochromatin underlicensing in shortened G1 cells
Because we established that MCM loading in euchromatin is advanced relative to heterochromatin in early G1 (Figure 2), we hypothesized that a premature G1/S transition will have a preferential negative impact on heterochromatin replication. Our reasoning is that an early G1/S transition could occur when euchromatin is more fully licensed but heterochromatin licensing is less complete. To test this hypothesis, we expressed human cyclin E from a dox-inducible promoter. Cyclin E normally accumulates in late G1, where it promotes S phase entry and progression by activating CDK2 (92,93). We and others had previously shown that cyclin E overproduction shortens G1 and can cause cells to enter S phase with less total loaded MCM [Supplementary Figure S8E and F (59,94,95)]. Treating cells with dox for 7 h strongly induced cyclin E and significantly shortened G1 length from a mean of 7 to 3 h (Figure 6A and B). Under these conditions, cyclin E overexpression also induced high CDK1/2 reporter activity at much younger physical ages (Figure 6C). In these cells, the overall concentration of loaded MCM achieved within these shorter G1 phases was also clearly less than that in control cells, but cyclin E overproduction did not greatly affect the overall rate of MCM loading (Figure 6D and E). This underlicensing was more profound in heterochromatin than in euchromatin especially in middle and late G1 cells (Figure 6E). Since we had manipulated CDK1/2 activity by cyclin E overexpression, we categorized G1 subphases by their physical age rather than molecular age for cyclin E-overproducing cells as indicated on the x-axis of Figure 6C, but using only physical age produced similar results (data not shown). Interestingly, when we compared euchromatin and heterochromatin, we found that heterochromatin MCM loading did not reach the same concentration as euchromatin loading even in late G1 cells, whereas in the control cells both types of chromatin had similar MCM loading in late G1 as before (Figure 6F). Compared to control G1 cells, loaded MCM distribution in cyclin E-overproducing cells favored euchromatin throughout all G1 subphases, and the two chromatin types never reached parity. These results support the conclusion that a premature G1/S transition results in underlicensed heterochromatin once S phase has begun.
Heterochromatin is more vulnerable than euchromatin to under-replication and DNA damage
Reduced origin licensing in G1 phase could leave segments of DNA under-replicated in S phase, thus threatening genome stability. Based on our findings that heterochromatin MCM loading is relatively slower in G1 phase, we postulated that the consequences of a premature G1/S transition include preferential heterochromatin under-replication during the following S phase resulting in more replication stress and DNA damage in heterochromatin. We therefore examined the recruitment of p53 binding protein 1 (53BP1), a biomarker for local replication stress and DNA double-strand breaks (96,97), to different chromatin types in very late S phase immediately after cyclin E induction. We first analyzed overall 53BP1 loading per nucleus in control and cyclin E-overproducing cells (similar to our analysis of MCM and ORC loading) and detected no differences between the two groups (Figure 7A and B, untreated and ‘↑Cyclin E’). We also treated control and cyclin E-overproducing cells briefly (4 h) with a low dose (25 ng/ml) of aphidicolin, a DNA polymerase inhibitor that induces replication stress and DNA damage. Under these conditions, aphidicolin treatment did not induce significant global DNA damage in control cells, but it did modestly increase overall DNA damage in cyclin E-overproducing cells as measured by 53BP1 recruitment (Figure 7A and B, ‘Control’ compared to ‘↑Cyclin E + aphidicolin’).
To explore the intranuclear DNA damage distribution between the different chromatin types, we analyzed the location of 53BP1 signals in heterochromatin and euchromatin. We noted that even in untreated cells, the concentration of 53BP1 in heterochromatin was higher than that in euchromatin (Figure 7C). Cyclin E overproduction for 18 h (<1 full RPE1-hTert cell cycle) caused even more 53BP1 recruitment to heterochromatin, but strikingly had no effect on euchromatin (Figure 7C). This effect was exacerbated by low-dose aphidicolin treatment for 4 h (Figure 7C). As we did for MCM distribution, we calculated the distribution of 53BP1 concentrations. 53BP1 recruitment induced after cyclin E overproduction occurred preferentially in heterochromatin, and this difference was exacerbated by aphidicolin treatment (Figure 7D). Furthermore, we analyzed the chromatin association and focal localization of the single-stranded DNA binding protein, replication protein A (RPA), specifically in the first G2 phase following cyclin E induction and/or aphidicolin treatment. Notably, cyclin E-overproducing cells accumulated many RPA foci in G2 phase, suggesting the persistence or exposure of ssDNA from incomplete replication (Supplementary Figure S9A–C). Aphidicolin alone did not induce G2 phase RPA foci but did enhance the effects of cyclin E overproduction. Overall, RPA signal was also preferentially distributed to heterochromatin over euchromatin (Supplementary Figure S9D and E). Altogether, these data strongly suggest that precocious S phase entry is particularly detrimental for heterochromatin replication, and that this disadvantage for heterochromatin is at least partly because these regions are more underlicensed than euchromatic regions (Figure 7E and F). Preferential underlicensing in heterochromatin is itself attributable to the unique inherent dynamics of MCM loading in heterochromatin compared to euchromatin within G1 phase.
DISCUSSION
In this study, we combined time-lapse imaging with fixed-cell immunostaining to define the dynamics of human MCM loading within G1 phase. To our knowledge, this is the first quantitative analysis of MCM loading in single cells at such high temporal resolution with respect to both physical age (in hours since cell division) and ‘molecular age’ defined by CDK2 activity. Several previous studies analyzed MCM loading dynamics during G1, either by using FRAP through live-cell imaging in mammalian cells or by using immunoblot through synchronization and CDK2 manipulation to infer G1 subphases in fruit fly cells. Those studies did not aim to measure the MCM loading dynamics in different chromatin compartments, however. Using our strategy, the dynamics of MCM loading can be analyzed relative to any marker for which there are suitable detection reagents. Identifying where and how MCM is loaded within distinct genomic regions in G1 typically poses technical challenges because MCM is at all replication forks during S phase. Moreover, as in most eukaryotes, human origins are highly flexible and variable between individual cells, which reduces the information obtained from analyzing cell populations. We instead developed methods to quantify relative rates of MCM loading in single G1 cells by comparing two primary genomic environments, heterochromatin and euchromatin. This strategy also avoids the potential pitfalls of artificial cell cycle synchronization or the averaging effect of analyzing bulk cell populations. Here, we report the first comprehensive analysis of G1 MCM loading dynamics in different intranuclear environments, the mechanism driving those dynamics and the downstream consequences those dynamics can have.
We uncovered a preference for MCM loading in euchromatin, but surprisingly, this preference is largely confined to cells in early G1 phase. Based on the long history of the transcriptional regulation field, it is intuitive that the more accessible euchromatin regions have an advantage over less accessible heterochromatin. MCM loading may be easier or simpler in a more accessible chromatin environment, and thus faster. What was not anticipated at the outset of this study is that the rate of heterochromatin loading increases relative to euchromatin loading during middle and late G1 subphases. As a result, heterochromatin loading lags behind euchromatin in early G1, but differences in acceleration allow heterochromatin to ‘catch up’ with euchromatin by the end of G1. Other studies of MCM binding sites in mammalian cells also report similar overall loading in both transcriptionally active and repressed genomic loci that generally correlate with euchromatin and heterochromatin, respectively (16,32). Our findings help explain how those similar levels are achieved.
Thus far, attention to the differences between the two chromatin types in DNA replication has been almost exclusively focused on the timing of origin firing within S phase itself. Origin firing time is a combined effect of origin licensing in G1 to define all potential origins and the recruitment of origin initiation factors in a process that is also influenced by chromatin (45,47,98). In many species, replication timing in S phase shows a general (though not exclusive) pattern of early euchromatin replication and late heterochromatin replication (99,100). Differences in the density of G1 MCM loading at different individual loci have been implicated in these S phase replication timing differences; more ORC or MCM loading in G1 correlates with early firing origins (31–33). Early firing in S and early G1 origin licensing may both be consequences of general DNA accessibility in euchromatin. We also speculate that the higher density of MCM loading in early replicating regions is partly from loading very quickly early in G1 and then having the rest of G1 phase to add additional MCM complexes at the same sites.
The molecular mechanism driving differential loading rates correlates with differences in ORC chromatin binding, and the ORC and MCM chromatin binding differences themselves require the ORCA/LRWD1 protein (Figure 7E). ORCA plays multiple roles in chromosome biology, including ORC recruitment to heterochromatin through a direct interaction with H3K9me3 (91). Thus, only some genomic regions (heterochromatin) rely on help from ORCA-mediated ORC recruitment. And more importantly, the percentage of ORCA binding sites that colocalized with H3K9me3 increases throughout G1 phase, indicating that ORCA helps heterochromatin more in late G1 cells (45). We envision that accessible euchromatic regions recruit ORC for MCM loading without much assistance, but less accessible regions require helper factors. These additional factors may be specialized for different types of chromatin or may cooperate with one another to ensure MCM loading in even the most inaccessible regions. For example, trimethylation of histone H4 lysine 20 has also been implicated in heterochromatin licensing at a subset of ORCA-bound sites (101). These mechanisms enhance MCM loading in heterochromatin regions. More equitable licensing distribution can also be promoted by factors that suppress MCM loading in euchromatin. Two recent studies of yeast origins described mechanisms to reduce disparities in MCM loading levels among different origins (47,102); analogous mechanisms may also operate in mammalian cells. We are also intrigued by the apparent weaker euchromatin loading acceleration between early G1 and later G1 times relative to the acceleration in heterochromatin. It is possible that a passive mechanism also improves MCM loading in heterochromatin later in G1 phase because the more accessible euchromatin sites are already nearly occupied (103–105).
Because heterochromatin reaches full licensing closer to the end of G1 phase than euchromatin does, heterochromatin is more vulnerable to any change that causes premature S phase entry (Figure 7E). Early S phase entry may happen stochastically from random fluctuations in gene expression leading to early cyclin E/CDK2 activation, or it may happen chronically if cells acquire genetic or epigenetic alterations that shorten G1 phase, such as oncogene activation or tumor suppressor loss. Importantly, it is the rate of licensing combined with the length of G1 phase that determines how much overall underlicensing cells experience, and the relative rates in different regions determine where underlicensing will be most severe. We note that cyclin E overproduction truncated G1 without affecting the apparent MCM loading rate (Figure 6D). We demonstrate that euchromatin is also somewhat underlicensed in cells with artificially short G1 phases, but not to the extent that it increased genome damage in euchromatin in the subsequent S phase. On the other hand, heterochromatin is much more underlicensed and the increase in under-replication and DNA damage was confined to heterochromatin (Figure 7F). This concentration of under-replication in heterochromatin is presumably from both underlicensing in G1 and late origin firing in S phase, which leaves even less time for replication to finish (99,100). Highly compacted heterochromatin could also be a barrier for timely DNA repair factor recruitment (106–108).
Interestingly, a period of cyclin E overproduction in a prior report caused some large genomic deletions (109). In that study, those deleted sequences were primarily late replicating (13 of the 16 deletions analyzed) and, based on replication timing, we presume are associated with heterochromatin and delayed origin licensing. A study by Macheret and Halazonetis of the effects of cyclin E overproduction mapped induced DNA damage near highly transcribed genes by sequencing methods, and those locations are presumably euchromatic by virtue of active transcription (86). That study analyzed cyclin E-induced origin firing within active genes in two tumor-derived cell lines, but the authors also specifically tested untransformed cells and detected no intragenic oncogene-induced origin firing in RPE1 cells, the same cells used here. In addition, we analyzed the effects of shortening G1 on the very next S phase, whereas Macheret and Halazonetis overproduced cyclin E for multiple days before analysis of chromosome translocations. It is possible that we induced very low levels of euchromatin damage in the first S phase after cyclin E overproduction, but that damage is much less than that in the heterochromatin and not detected by our microscopy method. Our results suggest that the final stage of G1 is crucial for heterochromatin to become fully licensed and therefore fully replicated.
Finally, the distribution of euchromatin and heterochromatin varies by cell type. We predict that differences in facultative heterochromatin that distinguish one cell type from another are also among the regions most vulnerable to late origin licensing and under-replication in those cell types. We also presume that constitutive heterochromatin is hypersensitive to underlicensing in most cell types, but that notion remains to be explored. Licensing dynamics may be unique in centromeres, telomeres or other distinct chromatin subregions, and additional investigations using other localization markers can reveal those dynamics. Both chromatin structure and G1 length are altered in many cancers (110–112). The insights gained from this study can contribute to understanding both the source and location of genome instability in cells with such perturbations.
DATA AVAILABILITY
The analysis of loaded MCM/ORC distribution is available in GitHub repository (https://github.com/purvislab/MCM_project).
Flow cytometry data for cell cycle distribution, MCM loading and ORC loading have been deposited in the FlowRepository with the repository ID FR-FCM-Z3TN.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Supriya Prasanth and Sabrina Spencer for the generous gifts of antibodies and reagents, and Abid Khan, Robert Duronio and Cook lab members for discussion and comments on the manuscript. We thank Jeffrey Jones for research support assistance. The UNC Hooker Imaging Core and the UNC Flow Cytometry Core Facility are supported in part by a National Institutes of Health Cancer Core Support Grant to the UNC Lineberger Comprehensive Cancer Center (CA016086).
Contributor Information
Liu Mei, Department of Biochemistry & Biophysics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Katarzyna M Kedziora, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Bioinformatics and Analytics Research Collaborative (BARC), University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Eun-Ah Song, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Jeremy E Purvis, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Jeanette Gowen Cook, Department of Biochemistry & Biophysics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01GM102413, R01GM083024 and R35GM141833 to J.G.C. in part]; National Science Foundation [to J.E.P.]; National Institute of General Medical Sciences [R01-GM138834 to J.E.P.]; North Carolina Biotechnology Center [2017-IDG-1025, in part]; National Institute of Health [1-UM2AI30836-01, in part]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Parker M.W., Botchan M.R., Berger J.M.. Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2017; 52:107–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siddiqui K., On K.F., Diffley J.F.. Regulating DNA replication in eukarya. Cold Spring Harb. Perspect. Biol. 2013; 5:a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith O.K., Aladjem M.I.. Chromatin structure and replication origins: determinants of chromosome replication and nuclear organization. J. Mol. Biol. 2014; 426:3330–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Prioleau M.N., MacAlpine D.M.. DNA replication origins—where do we begin?. Genes Dev. 2016; 30:1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ekundayo B., Bleichert F.. Origins of DNA replication. PLoS Genet. 2019; 15:e1008320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Remus D., Diffley J.F.. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 2009; 21:771–777. [DOI] [PubMed] [Google Scholar]
- 7. Evrin C., Clarke P., Zech J., Lurz R., Sun J., Uhle S., Li H., Stillman B., Speck C.. A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl Acad. Sci. U.S.A. 2009; 106:20240–20245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yeeles J.T., Deegan T.D., Janska A., Early A., Diffley J.F.. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015; 519:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petryk N., Kahli M., d’Aubenton-Carafa Y., Jaszczyszyn Y., Shen Y., Silvain M., Thermes C., Chen C.L., Hyrien O.. Replication landscape of the human genome. Nat. Commun. 2016; 7:10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ge X.Q., Jackson D.A., Blow J.J.. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 2007; 21:3331–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawabata T., Luebben S.W., Yamaguchi S., Ilves I., Matise I., Buske T., Botchan M.R., Shima N.. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell. 2011; 41:543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ibarra A., Schwob E., Mendez J.. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl Acad. Sci. U.S.A. 2008; 105:8956–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pruitt S.C., Qin M., Wang J., Kunnev D., Freeland A.. A signature of genomic instability resulting from deficient replication licensing. PLoS Genet. 2017; 13:e1006547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Letessier A., Millot G.A., Koundrioukoff S., Lachages A.M., Vogt N., Hansen R.S., Malfoy B., Brison O., Debatisse M.. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011; 470:120–123. [DOI] [PubMed] [Google Scholar]
- 15. Miotto B., Ji Z., Struhl K.. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc. Natl Acad. Sci. U.S.A. 2016; 113:E4810–E4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sugimoto N., Maehara K., Yoshida K., Ohkawa Y., Fujita M.. Genome-wide analysis of the spatiotemporal regulation of firing and dormant replication origins in human cells. Nucleic Acids Res. 2018; 46:6683–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brison O., El-Hilali S., Azar D., Koundrioukoff S., Schmidt M., Nahse V., Jaszczyszyn Y., Lachages A.M., Dutrillaux B., Thermes C.et al.. Transcription-mediated organization of the replication initiation program across large genes sets common fragile sites genome-wide. Nat. Commun. 2019; 10:5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuipers M.A., Stasevich T.J., Sasaki T., Wilson K.A., Hazelwood K.L., McNally J.G., Davidson M.W., Gilbert D.M.. Highly stable loading of Mcm proteins onto chromatin in living cells requires replication to unload. J. Cell Biol. 2011; 192:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Symeonidou I.E., Kotsantis P., Roukos V., Rapsomaniki M.A., Grecco H.E., Bastiaens P., Taraviras S., Lygerou Z.. Multi-step loading of human minichromosome maintenance proteins in live human cells. J. Biol. Chem. 2013; 288:35852–35867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Powell S.K., MacAlpine H.K., Prinz J.A., Li Y., Belsky J.A., MacAlpine D.M.. Dynamic loading and redistribution of the Mcm2–7 helicase complex through the cell cycle. EMBO J. 2015; 34:531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grewal S.I., Moazed D.. Heterochromatin and epigenetic control of gene expression. Science. 2003; 301:798–802. [DOI] [PubMed] [Google Scholar]
- 22. Schubeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J.et al.. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004; 18:1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huisinga K.L., Brower-Toland B., Elgin S.C.. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 2006; 115:110–122. [DOI] [PubMed] [Google Scholar]
- 24. Rivera-Mulia J.C., Gilbert D.M.. Replication timing and transcriptional control: beyond cause and effect—part III. Curr. Opin. Cell Biol. 2016; 40:168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fragkos M., Ganier O., Coulombe P., Mechali M.. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015; 16:360–374. [DOI] [PubMed] [Google Scholar]
- 26. Goren A., Cedar H.. Replicating by the clock. Nat. Rev. Mol. Cell Biol. 2003; 4:25–32. [DOI] [PubMed] [Google Scholar]
- 27. Klein K.N., Zhao P.A., Lyu X., Sasaki T., Bartlett D.A., Singh A.M., Tasan I., Zhang M., Watts L.P., Hiraga S.I.et al.. Replication timing maintains the global epigenetic state in human cells. Science. 2021; 372:371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Foti R., Gnan S., Cornacchia D., Dileep V., Bulut-Karslioglu A., Diehl S., Buness A., Klein F.A., Huber W., Johnstone E.et al.. Nuclear architecture organized by Rif1 underpins the replication-timing program. Mol. Cell. 2016; 61:260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mantiero D., Mackenzie A., Donaldson A., Zegerman P.. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 2011; 30:4805–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Foss E.J., Sripathy S., Gatbonton-Schwager T., Kwak H., Thiesen A.H., Lao U., Bedalov A.. Chromosomal Mcm2–7 distribution and the genome replication program in species from yeast to humans. PLoS Genet. 2021; 17:e1009714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Das S.P., Borrman T., Liu V.W., Yang S.C., Bechhoefer J., Rhind N.. Replication timing is regulated by the number of MCMs loaded at origins. Genome Res. 2015; 25:1886–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kirstein N., Buschle A., Wu X., Krebs S., Blum H., Kremmer E., Vorberg I.M., Hammerschmidt W., Lacroix L., Hyrien O.et al.. Human ORC/MCM density is low in active genes and correlates with replication time but does not delimit initiation zones. eLife. 2021; 10:e62161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dukaj L., Rhind N.. The capacity of origins to load MCM establishes replication timing patterns. PLoS Genet. 2021; 17:e1009467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soudet J., Gill J.K., Stutz F.. Noncoding transcription influences the replication initiation program through chromatin regulation. Genome Res. 2018; 28:1882–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Du Q., Bert S.A., Armstrong N.J., Caldon C.E., Song J.Z., Nair S.S., Gould C.M., Luu P.L., Peters T., Khoury A.et al.. Replication timing and epigenome remodelling are associated with the nature of chromosomal rearrangements in cancer. Nat. Commun. 2019; 10:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marks A.B., Smith O.K., Aladjem M.I.. Replication origins: determinants or consequences of nuclear organization. Curr. Opin. Genet. Dev. 2016; 37:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aladjem M.I., Redon C.E.. Order from clutter: selective interactions at mammalian replication origins. Nat. Rev. Genet. 2017; 18:101–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eaton M.L., Galani K., Kang S., Bell S.P., MacAlpine D.M.. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010; 24:748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cayrou C., Ballester B., Peiffer I., Fenouil R., Coulombe P., Andrau J.C., van Helden J., Mechali M.. The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res. 2015; 25:1873–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sugimoto N., Yugawa T., Iizuka M., Kiyono T., Fujita M.. Chromatin remodeler sucrose nonfermenting 2 homolog (SNF2H) is recruited onto DNA replication origins through interaction with Cdc10 protein-dependent transcript 1 (Cdt1) and promotes pre-replication complex formation. J. Biol. Chem. 2011; 286:39200–39210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miotto B., Struhl K.. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell. 2010; 37:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miotto B., Struhl K.. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 2008; 22:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Long H., Zhang L., Lv M., Wen Z., Zhang W., Chen X., Zhang P., Li T., Chang L., Jin C.et al.. H2A.Z facilitates licensing and activation of early replication origins. Nature. 2020; 577:576–581. [DOI] [PubMed] [Google Scholar]
- 44. Tardat M., Brustel J., Kirsh O., Lefevbre C., Callanan M., Sardet C., Julien E.. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 2010; 12:1086–1093. [DOI] [PubMed] [Google Scholar]
- 45. Wang Y., Khan A., Marks A.B., Smith O.K., Giri S., Lin Y.C., Creager R., MacAlpine D.M., Prasanth K.V., Aladjem M.I.et al.. Temporal association of ORCA/LRWD1 to late-firing origins during G1 dictates heterochromatin replication and organization. Nucleic Acids Res. 2017; 45:2490–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shen Z., Sathyan K.M., Geng Y., Zheng R., Chakraborty A., Freeman B., Wang F., Prasanth K.V., Prasanth S.G.. A WD-repeat protein stabilizes ORC binding to chromatin. Mol. Cell. 2010; 40:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hoggard T., Muller C.A., Nieduszynski C.A., Weinreich M., Fox C.A.. Sir2 mitigates an intrinsic imbalance in origin licensing efficiency between early- and late-replicating euchromatin. Proc. Natl Acad. Sci. U.S.A. 2020; 117:14314–14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Conti C., Sacca B., Herrick J., Lalou C., Pommier Y., Bensimon A.. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol. Biol. Cell. 2007; 18:3059–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cayrou C., Coulombe P., Vigneron A., Stanojcic S., Ganier O., Peiffer I., Rivals E., Puy A., Laurent-Chabalier S., Desprat R.et al.. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011; 21:1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gomez M., Brockdorff N.. Heterochromatin on the inactive X chromosome delays replication timing without affecting origin usage. Proc. Natl Acad. Sci. U.S.A. 2004; 101:6923–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zylicz J.J., Heard E.. Molecular mechanisms of facultative heterochromatin formation: an X-chromosome perspective. Annu. Rev. Biochem. 2020; 89:255–282. [DOI] [PubMed] [Google Scholar]
- 52. DePamphilis M.L., Blow J.J., Ghosh S., Saha T., Noguchi K., Vassilev A.. Regulating the licensing of DNA replication origins in metazoa. Curr. Opin. Cell Biol. 2006; 18:231–239. [DOI] [PubMed] [Google Scholar]
- 53. Truong L.N., Wu X.. Prevention of DNA re-replication in eukaryotic cells. J. Mol. Cell Biol. 2011; 3:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Blow J.J., Dutta A.. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005; 6:476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Munoz S., Bua S., Rodriguez-Acebes S., Megias D., Ortega S., de Martino A., Mendez J.. In vivo DNA re-replication elicits lethal tissue dysplasias. Cell Rep. 2017; 19:928–938. [DOI] [PubMed] [Google Scholar]
- 56. Neelsen K.J., Zanini I.M., Mijic S., Herrador R., Zellweger R., Ray Chaudhuri A., Creavin K.D., Blow J.J., Lopes M.. Deregulated origin licensing leads to chromosomal breaks by rereplication of a gapped DNA template. Genes Dev. 2013; 27:2537–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Green B.M., Finn K.J., Li J.J.. Loss of DNA replication control is a potent inducer of gene amplification. Science. 2010; 329:943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Limas J.C., Cook J.G.. Preparation for DNA replication: the key to a successful S phase. FEBS Lett. 2019; 593:2853–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Matson J.P., Dumitru R., Coryell P., Baxley R.M., Chen W., Twaroski K., Webber B.R., Tolar J., Bielinsky A.K., Purvis J.E.et al.. Rapid DNA replication origin licensing protects stem cell pluripotency. eLife. 2017; 6:e30473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hemerly A.S., Prasanth S.G., Siddiqui K., Stillman B.. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009; 323:789–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu K., Ma H., McCown T.J., Verma I.M., Kafri T.. Generation of a stable cell line producing high-titer self-inactivating lentiviral vectors. Mol. Ther. 2001; 3:97–104. [DOI] [PubMed] [Google Scholar]
- 62. Grant G.D., Kedziora K.M., Limas J.C., Cook J.G., Purvis J.E.. Accurate delineation of cell cycle phase transitions in living cells with PIP-FUCCI. Cell Cycle. 2018; 17:2496–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007; 9:90–95. [Google Scholar]
- 64. Waskom M.L. Seaborn: statistical data visualization. J. Open Source Softw. 2021; 6:3021. [Google Scholar]
- 65. Mendez J., Stillman B.. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell Biol. 2000; 20:8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dimitrova D.S., Prokhorova T.A., Blow J.J., Todorov I.T., Gilbert D.M.. Mammalian nuclei become licensed for DNA replication during late telophase. J. Cell Sci. 2002; 115:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Haland T.W., Boye E., Stokke T., Grallert B., Syljuasen R.G.. Simultaneous measurement of passage through the restriction point and MCM loading in single cells. Nucleic Acids Res. 2015; 43:e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Leonhardt H., Rahn H.P., Weinzierl P., Sporbert A., Cremer T., Zink D., Cardoso M.C.. Dynamics of DNA replication factories in living cells. J. Cell Biol. 2000; 149:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Spencer S.L., Cappell S.D., Tsai F.C., Overton K.W., Wang C.L., Meyer T.. The proliferation–quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013; 155:369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schwarz C., Johnson A., Koivomagi M., Zatulovskiy E., Kravitz C.J., Doncic A., Skotheim J.M.. A precise Cdk activity threshold determines passage through the restriction point. Mol. Cell. 2018; 69:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chao H.X., Fakhreddin R.I., Shimerov H.K., Kedziora K.M., Kumar R.J., Perez J., Limas J.C., Grant G.D., Cook J.G., Gupta G.P.et al.. Evidence that the human cell cycle is a series of uncoupled, memoryless phases. Mol. Syst. Biol. 2019; 15:e8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu C., Konagaya Y., Chung M., Daigh L.H., Fan Y., Yang H.W., Terai K., Matsuda M., Meyer T.. Altered G1 signaling order and commitment point in cells proliferating without CDK4/6 activity. Nat. Commun. 2020; 11:5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Donovan S., Harwood J., Drury L.S., Diffley J.F.. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. U.S.A. 1997; 94:5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Edwards M.C., Tutter A.V., Cvetic C., Gilbert C.H., Prokhorova T.A., Walter J.C.. MCM2–7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 2002; 277:33049–33057. [DOI] [PubMed] [Google Scholar]
- 75. Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T.. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001; 410:116–120. [DOI] [PubMed] [Google Scholar]
- 76. Kwon S.H., Workman J.L.. The heterochromatin protein 1 (HP1) family: put away a bias toward HP1. Mol. Cells. 2008; 26:217–227. [PubMed] [Google Scholar]
- 77. Nestorov P., Tardat M., Peters A.H.. H3K9/HP1 and Polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr. Top. Dev. Biol. 2013; 104:243–291. [DOI] [PubMed] [Google Scholar]
- 78. Sanulli S., Gross J.D., Narlikar G.J.. Biophysical properties of HP1-mediated heterochromatin. Cold Spring Harb. Symp. Quant. Biol. 2019; 84:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kourmouli N., Theodoropoulos P.A., Dialynas G., Bakou A., Politou A.S., Cowell I.G., Singh P.B., Georgatos S.D.. Dynamic associations of heterochromatin protein 1 with the nuclear envelope. EMBO J. 2000; 19:6558–6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Taddei A., Roche D., Sibarita J.B., Turner B.M., Almouzni G.. Duplication and maintenance of heterochromatin domains. J. Cell Biol. 1999; 147:1153–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W.et al.. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 82. Sandritter W., Kiefer G., Kiefer R., Salm R., Moore G.W., Grimm H.. DNA in heterochromatin cytophotometric pattern recognition image analysis among cell nuclei in duct epithelium and in carcinoma of the human breast. Beitr. Pathol. 1974; 151:87–96. [DOI] [PubMed] [Google Scholar]
- 83. Miklos G.L., John B.. Heterochromatin and satellite DNA in man: properties and prospects. Am. J. Hum. Genet. 1979; 31:264–280. [PMC free article] [PubMed] [Google Scholar]
- 84. Dillon N. Heterochromatin structure and function. Biol. Cell. 2004; 96:631–637. [DOI] [PubMed] [Google Scholar]
- 85. Grewal S.I., Jia S.. Heterochromatin revisited. Nat. Rev. Genet. 2007; 8:35–46. [DOI] [PubMed] [Google Scholar]
- 86. Macheret M., Halazonetis T.D.. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature. 2018; 555:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Toth K.F., Knoch T.A., Wachsmuth M., Frank-Stohr M., Stohr M., Bacher C.P., Muller G., Rippe K.. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J. Cell Sci. 2004; 117:4277–4287. [DOI] [PubMed] [Google Scholar]
- 88. Kuo A.J., Song J., Cheung P., Ishibe-Murakami S., Yamazoe S., Chen J.K., Patel D.J., Gozani O.. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier–Gorlin syndrome. Nature. 2012; 484:115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Petropoulos M., Champeris Tsaniras S., Taraviras S., Lygerou Z.. Replication licensing aberrations, replication stress, and genomic instability. Trends Biochem. Sci. 2019; 44:752–764. [DOI] [PubMed] [Google Scholar]
- 90. Shen Z., Chakraborty A., Jain A., Giri S., Ha T., Prasanth K.V., Prasanth S.G.. Dynamic association of ORCA with prereplicative complex components regulates DNA replication initiation. Mol. Cell Biol. 2012; 32:3107–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Giri S., Aggarwal V., Pontis J., Shen Z., Chakraborty A., Khan A., Mizzen C., Prasanth K.V., Ait-Si-Ali S., Ha T.et al.. The preRC protein ORCA organizes heterochromatin by assembling histone H3 lysine 9 methyltransferases on chromatin. eLife. 2015; 4:e06496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Koff A., Giordano A., Desai D., Yamashita K., Harper J.W., Elledge S., Nishimoto T., Morgan D.O., Franza B.R., Roberts J.M.. Formation and activation of a cyclin E–cdk2 complex during the G1 phase of the human cell cycle. Science. 1992; 257:1689–1694. [DOI] [PubMed] [Google Scholar]
- 93. Lew D.J., Dulic V., Reed S.I.. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991; 66:1197–1206. [DOI] [PubMed] [Google Scholar]
- 94. Ekholm-Reed S., Mendez J., Tedesco D., Zetterberg A., Stillman B., Reed S.I.. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J. Cell Biol. 2004; 165:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Resnitzky D., Gossen M., Bujard H., Reed S.I.. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell Biol. 1994; 14:1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mochan T.A., Venere M., DiTullio R.A. Jr, Halazonetis T.D.. 53BP1, an activator of ATM in response to DNA damage. DNA Repair. 2004; 3:945–952. [DOI] [PubMed] [Google Scholar]
- 97. Wang B., Matsuoka S., Carpenter P.B., Elledge S.J.. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002; 298:1435–1438. [DOI] [PubMed] [Google Scholar]
- 98. Kurat C.F., Yeeles J.T.P., Patel H., Early A., Diffley J.F.X.. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell. 2017; 65:117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Nathanailidou P., Taraviras S., Lygerou Z.. Chromatin and nuclear architecture: shaping DNA replication in 3D. Trends Genet. 2020; 36:967–980. [DOI] [PubMed] [Google Scholar]
- 100. Rhind N., Gilbert D.M.. DNA replication timing. Cold Spring Harb. Perspect. Biol. 2013; 5:a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Brustel J., Kirstein N., Izard F., Grimaud C., Prorok P., Cayrou C., Schotta G., Abdelsamie A.F., Dejardin J., Mechali M.et al.. Histone H4K20 tri-methylation at late-firing origins ensures timely heterochromatin replication. EMBO J. 2017; 36:2726–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hoggard T., Hollatz A.J., Cherney R.E., Seman M.R., Fox C.A.. The Fkh1 forkhead associated domain promotes ORC binding to a subset of DNA replication origins in budding yeast. Nucleic Acids Res. 2021; 49:10207–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Prasanth S.G., Shen Z., Prasanth K.V., Stillman B.. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc. Natl Acad. Sci. U.S.A. 2010; 107:15093–15098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Giri S., Chakraborty A., Sathyan K.M., Prasanth K.V., Prasanth S.G.. Orc5 induces large-scale chromatin decondensation in a GCN5-dependent manner. J. Cell Sci. 2016; 129:417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Chou H.C., Bhalla K., Demerdesh O.E., Klingbeil O., Hanington K., Aganezov S., Andrews P., Alsudani H., Chang K., Vakoc C.R.et al.. The human origin recognition complex is essential for pre-RC assembly, mitosis, and maintenance of nuclear structure. eLife. 2021; 10:e61797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Goodarzi A.A., Jeggo P., Lobrich M.. The influence of heterochromatin on DNA double strand break repair: getting the strong, silent type to relax. DNA Repair. 2010; 9:1273–1282. [DOI] [PubMed] [Google Scholar]
- 107. Goodarzi A.A., Jeggo P.A.. The heterochromatic barrier to DNA double strand break repair: how to get the entry visa. Int. J. Mol. Sci. 2012; 13:11844–11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lorkovic Z.J., Berger F.. Heterochromatin and DNA damage repair: use different histone variants and relax. Nucleus. 2017; 8:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Teixeira L.K., Wang X., Li Y., Ekholm-Reed S., Wu X., Wang P., Reed S.I.. Cyclin E deregulation promotes loss of specific genomic regions. Curr. Biol. 2015; 25:1327–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Nguyen C.T., Gonzales F.A., Jones P.A.. Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Res. 2001; 29:4598–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Brock M.V., Herman J.G., Baylin S.B.. Cancer as a manifestation of aberrant chromatin structure. Cancer J. 2007; 13:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Michalak E.M., Burr M.L., Bannister A.J., Dawson M.A.. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 2019; 20:573–589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analysis of loaded MCM/ORC distribution is available in GitHub repository (https://github.com/purvislab/MCM_project).
Flow cytometry data for cell cycle distribution, MCM loading and ORC loading have been deposited in the FlowRepository with the repository ID FR-FCM-Z3TN.