Abstract
Preterm birth contributes significantly to neonatal mortality and morbidity. Despite its global significance, there has only been limited progress in preventing preterm birth. Spontaneous preterm birth (sPTB) results from a wide variety of pathological processes. Although many non-genetic risk factors influence the timing of gestation and labor, compelling evidence supports the role of substantial genetic and epigenetic influences and their interactions with the environment contributing to sPTB. To investigate a common and complex disease such as sPTB, various approaches such as genome-wide association studies, whole-exome sequencing, transcriptomics, and integrative approaches combining these with other ‘omics studies have been used. However, many of these studies were typically small or focused on a single ethnicity or geographic region with limited data, particularly in populations at high risk for sPTB, or lacked a robust replication. These studies found many genes involved in the inflammation and immunity-related pathways that may affect sPTB. Recent studies also suggest the role of epigenetic modifications of gene expression by the environmental signals as a potential contributor to the risk of sPTB. Future genetic studies of sPTB should continue to consider the contributions of both maternal and fetal genomes as well as their interaction with the environment.
Keywords: genes, inflammation, environment, GWAS, genome, transcriptome, epigenome, WES, gestation, spontaneous, RNA-seq
Introduction:
The consequences of being born prematurely remain one of the most significant health burdens to society despite increased clinical and research attention. Preterm birth is defined as being born <37 weeks of completed gestation based on the first day of a woman’s last menstrual period. In 2002, 34.3 % of all infant deaths in the US were attributed to preterm birth, and 95% of those deaths occurred among those born at <32 weeks of gestation or with birth weight < 1500g.1 Though infant mortality attributed to preterm birth progressively reduced to <17% in the US,2 still an estimated 15 million neonates are born prematurely worldwide every year, and approximately 1 million of those children die due to complications of preterm birth.3 This results in preterm birth being the leading cause of under-five mortality worldwide.4
Preterm birth is relatively common, ranging from ~5% in European countries to ~18 % in low and middle-income African and South Asian countries, and preterm birth rates continue to rise globally.5 In the US, the rate of preterm birth increased steadily from 9.63% in 2015 to 10.1% in 2020.6 Preterm birth is associated with a significant burden on the healthcare system with an average lifetime incremental cost of $65,000 per preterm birth in the US.7,8 In addition, families of preterm infants often experience considerable psychological and financial hardship.9 Though survival of preterm infants has continued to improve, preterm neonates who survive have many short and long-term morbidities which can be life-long.10,11 They also suffer from various disabilities which become apparent later in childhood and young adulthood, such as school difficulties and behavioral problems.12 In addition, these infants are at increased risk of various adult-onset metabolic diseases such as obesity, diabetes, and hypertension.13–15 The ideal way to improve the overall health of these preterm infants would be to prevent preterm birth. Despite its global significance, there has been limited progress in preventing prematurity, likely due to failure in understanding the normal control mechanisms for pregnancy, initiation of labor, and the pathways through which these mechanisms are disrupted, leading to preterm birth.
Delivery of a healthy newborn at term gestation depends on numerous mechanisms, many of which involve inflammatory pathways.16 It has been suggested that although term labor is a physiological activation of these pathways, preterm labor results from pathological activation of the same pathways at an earlier time.17 To support this concept, a significant degree of overlapping transcriptomic regulation in the immunological pathways was noted in maternal blood from women with labor at term and before delivery in women that ultimately delivered preterm.18 Preterm births can be classified as spontaneous (due to preterm labor with intact membrane or preterm premature rupture of membranes) or iatrogenic/medically-induced (e.g., cesarean section or labor induction due to maternal or fetal conditions that compromise the health of the mother or infant).19 Spontaneous preterm birth (sPTB) accounts for ~65–70% of all cases of preterm birth, and about 50% of these occur in apparently low-risk pregnancies.19
Although the pathogenesis of preterm birth is not well understood, multiple risk factors have been associated with an increased incidence of preterm birth (Fig 1).19 In the US, disparities in preterm birth are evidenced by the higher rate of preterm birth in non-Hispanic Black women at 14.39% vs. 9.26% in non-Hispanic white women, even after adjusting for maternal socioeconomic status and education.6 Increased preterm birth rates are also associated with non-Hispanic Black paternal race.20 Increasingly recognized as contributing factors in the minority, particularly Black, pregnancies, are the pervasive consequences of social determinants including racism.21 Ideally, identifying variously modifiable and non-modifiable risk factors associated with preterm birth prior to conception or early in pregnancy provides an opportunity to initiate interventions that can prevent complications related to preterm birth. Various interventions such as nutritional supplementation, adequate pregnancy weight gain, tocolytics, bed rest to delay labor, home uterine monitoring for fetal distress, cervical cerclage for short cervix, treatment of bacterial vaginosis, and antibiotic treatment for chorioamnionitis have been implemented to prevent or treat preterm labor. However, they have proven to be of little or no benefit.22 Progesterone supplementation in high-risk pregnant women with a history of preterm delivery or a short cervix at mid-gestation has been found to reduce preterm birth risk, but the mechanism by which this occurs remains unclear and has not been replicated in many populations.23–25 The older trials, which showed benefits had unusually high preterm birth rates in the control group, and recent trials which did not show a benefit had a much lower rate of preterm birth rates in controls. In a meta-analysis of individual participant data from randomized trials evaluating progesterone for preventing preterm birth in singleton pregnancies with either a previous spontaneous preterm birth or cervical shortening in the current pregnancy (31 trials, n = 11,644), vaginal progesterone [RR 0·78, 95% CI (0·68–0·90)] and oral progesterone [RR: 0·60, (0·40–0·90)] significantly reduced preterm birth (<34weeks), but results were not significant for hydroxyprogesterone caproate [RR 0·83, (0·68–1·01)]. No benefit was found for multi-fetal pregnancies.26
Figure 1: Causes and interaction among various factors leading to preterm birth.
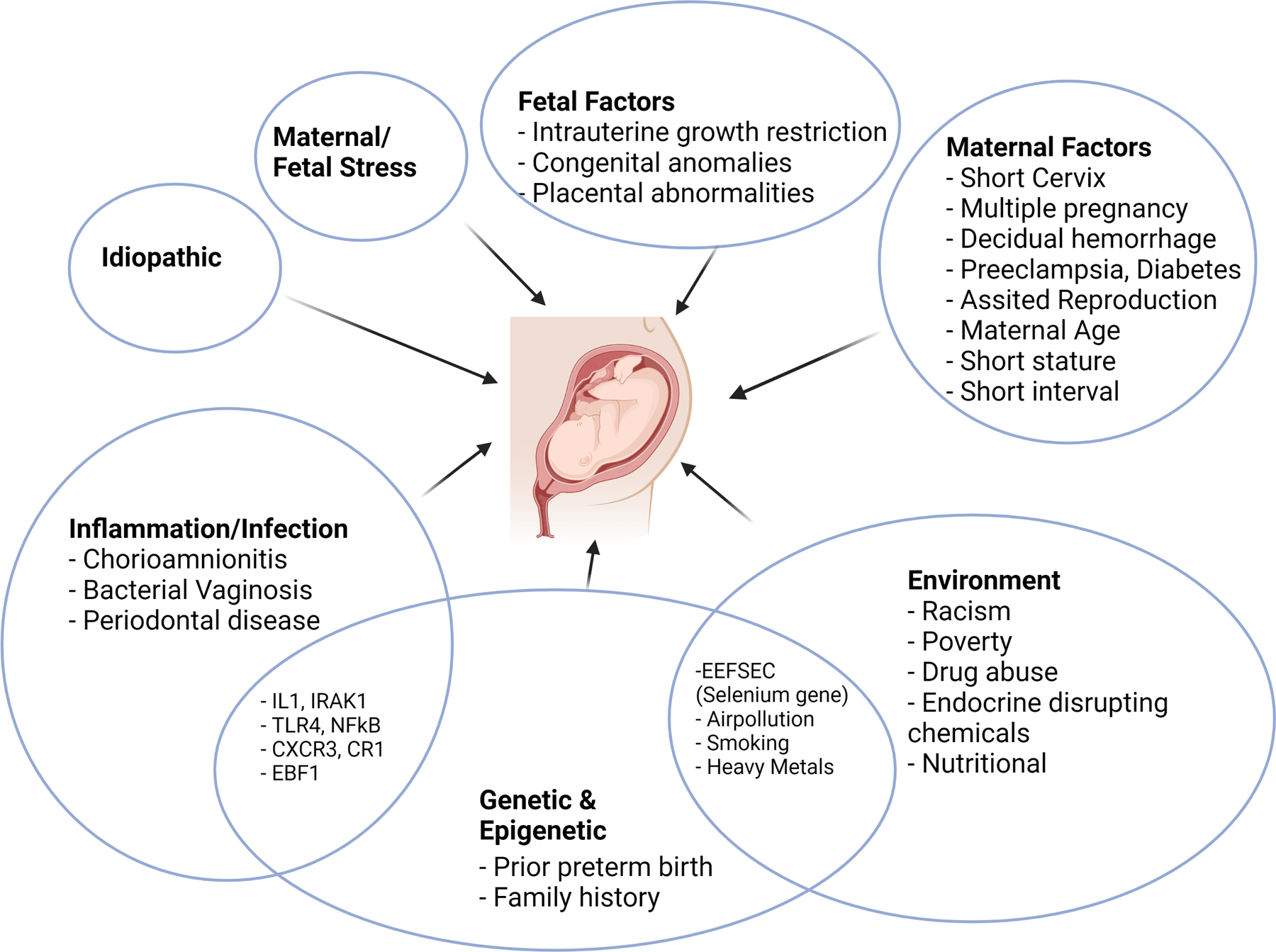
Preterm birth is a result of various factors, many of which overlap, resulting in the common outcome of preterm delivery. Several genes associated with inflammation and infection have been implicated with preterm birth. Many of the environmental factors likely lead to genetic and epigenetic changes resulting in preterm birth.
Many sociodemographic, nutritional, biologic, genetic, and environmental factors are associated with an increased risk of sPTB.19,27 The complex interactions of these contributors with both the mother and fetus have made disentangling causation challenging.28 Although the timing of labor is influenced by many non-genetic risk factors; there is strong evidence for a substantial genetic and epigenetic component. This review will focus on various genetic and epigenetic determinants of preterm birth to gain new insights into pathways that mediate not only primary genetic etiologies but also those that are dysregulated by environmental exposures.
Family-based and Epidemiological Evidence for Preterm Birth
Twin and Family Studies
Substantial evidence, although indirect, suggested that genetics plays an important role in determining gestational duration and risk of preterm birth.29 A history of sPTB in a mother is a significant risk factor for subsequent preterm birth, and recurrences often occur at the same or earlier gestational age.30,31 There is a 5-fold increase in delivering preterm in subsequent pregnancies if one of the mother’s previous infants was born preterm, which increases up to 18-fold if the previous delivery was at less than 29 weeks of gestation.32 Epidemiological studies of large population-based cohorts reveal that mothers who are born preterm, have sisters who were also born preterm or delivered preterm and have an increased risk of themselves delivering preterm.30,32,33 Twin studies and segregation analysis of traits of families demonstrate significant genetic contribution to preterm birth with the heritability estimates of maternal genetic contribution ranging from 15 to 40%.34–37 (Table 1). These estimates may be affected by confounding effects of the fetal genome or similar lifestyle factors of mother and daughter, though attempts have been made to control for these lifestyle confounders by utilizing sisters-in-law as controls. Some studies have suggested that the fetal genome contributes ~5 to 11% genetic variation to gestational age at delivery. 38,39 However, the fetal contribution was negligible in sPTB compared to 14% in medically-induced deliveries.38 There is also inconsistency in how preterm birth is defined in these studies (Table 1). Some studies used preterm birth (<37 weeks) as a categorical variable,38 or did not differentiate between medically-induced or spontaneous preterm birth.40–43 Since genetics and environment homogenously affect across the range of gestational age during pregnancy, considering gestational duration as a continuous variable could be more useful than using a dichotomized outcome in genetic studies.39 Similarly, there is no convincing evidence that parental imprinting influences sPTB or gestational duration. There is also negligible to relatively small (~6%) contribution from the paternal genome.40,44,45 Overall, these studies along with single nucleotide polymorphisms (SNP) based heritability estimation in mother/child pairs overwhelmingly suggest a well-substantiated and important contribution by the maternal genome and a much smaller contribution from the fetal genome for the gestational duration or preterm birth.46,47
Table 1:
Twin studies and segregation analysis of traits of families demonstrating the maternal and fetal genetic contributions
| Study | Type of Study | Fetal genes | Maternal genes | Limitations |
|---|---|---|---|---|
| Boyd et al.33 | Population epidemiology | − | + | No distinction between sPTB and medically-induced |
| Clausson et al.42 | Twin mothers study | NA | + | No distinction between sPTB and medically-induced |
| Kistka et al.41 | Twin mothers study | − | ++ | No distinction between sPTB and medically-induced |
| Lunde et al.36 | Population-based (parent-infant pair) | + | + | Excluded births <35 weeks, used gestational age as quantitative trait |
| Plunkett et al.37 | Segregation analysis (mother-infant pair) | + | ++ | Potential confounding between maternal & fetal estimates |
| Svensson et al.39 | Population epidemiology (Children of siblings) | − | ++ | Categorically defined preterm birth (<37 weeks) |
| Treloar et al44 | Twin mothers study | NA | ++ | No distinction between sPTB and medically-induced |
| Wilcox et al.46 | Population epidemiology (mother-infant pair) | − | ++ | Categorically defined preterm birth (<37 weeks) |
| Wu et al.38 | Population epidemiology (mother-infant pair) | − | ++ | Not able to control for environmental risk factors |
| York et al.40 | Twin mothers study | + | + | Excluded births <30 weeks |
No evidence of genetic contribution
Moderate genetic contribution
Strong genetic contribution
NA, Not available
Genome-wide approaches to preterm birth
Genome-wide linkage studies
Family-based linkage studies allowed the identification of a locus of interest based upon the linkage with a trait. Genome-wide linkage studies (GWL) involve either single large pedigrees or a large number of nuclear families to identify location of disease genes with large effects. In a study of Finnish families with multiple sPTB, linkage of sPTB with gene encoding insulin-like growth factor 1 (IGF1) receptor and androgen receptor (AR) in the fetal genome was found. These results were replicated in case-control studies of nuclear families from Finland.48,49 IGF1 expression in placental and fetal tissues has been reported. IGF1 plays an essential role in fetal growth and regulates multiple downstream signaling pathways involved in inflammation and critical cellular processes such as mitochondrial biogenesis.50 Low IGF1 levels have been associated with various preterm neonatal morbidities as well as dysregulated lipid metabolism, cardiovascular disease, and diabetes, common in preterm infants in adulthood.51,52 IGF1 has anti-inflammatory and antioxidant effects, and downregulation of IGF1 receptor expression increases cellular stress and cytokine (IL-6 and CCL2) production.53 All these taken together support a causal role of IGF1 in preterm birth pathogenesis. Decreased AR signaling leads to apoptosis via activation of Caspase-3.54 This decreased AR signaling might lead to sPTB via apoptosis of the fetal membrane in the placenta.55 In addition, interactions of IGF1 and AR genes may affect the onset of spontaneous preterm labor.48 Furthermore, a fetal chemokine receptor CXCR3 variant was associated with sPTB.56 CXCR3 receptor plays a critical role in cell-mediated immunity and is expressed in the placenta and fetal membranes. In CXCR3-deficient preterm birth mice, sPTB-associating cytokines were not increased in amniotic fluid,57, and prevented fetal wastage after Listeria infection and depletion of maternal Tregs.58 In another GWL study involving Mexican Americans, PAI-2, a member of the plasminogen activator system was linked to sPTB. This gene was previously found to be significantly associated with sPTB in a case-control study of the Australian population.59 This plasminogen activator system is associated with various reproductive processes such as placental development and functioning, hemostasis, and labor-associated rupture of fetal membranes.60 It should also be noted that for common and complex disorders, such as sPTB, the results of GWL studies are hard to reproduce.61
Genome-wide association studies
The genome-wide association study (GWAS) has been the most common genomic approach to investigating complex diseases. GWAS focuses on associations between single-nucleotide polymorphisms (SNPs) and human traits and diseases. Genomic studies have recently expanded to include whole-exome sequence (WES) and whole-genome sequence (WGS) analyses. The GWAS approach is “hypothesis-free”, and it systematically screens the whole genome without prior preference for specific regions or genes. These approaches offer the advantage to overcome difficulties imposed by the incomplete understanding of the disease pathophysiology such as in sPTB. GWAS is an alternative to family-based linkage studies and is better at detecting weak genetic effects. Since the effect sizes of most risk variants associated with a complex disease are small and the statistical significance thresholds are stringent in GWAS, large samples of cases and controls, or cohort studies, are needed for a robust analysis. Independent replications are also required to confirm the results and avoid false-positive associations.62 Thus, though candidate gene studies have associated approximately 119 candidate genes with sPTB, most of these studies suffered from a lack of replication, even within the same population.63
Maternal GWAS
The first GWAS study used maternal genome derived from the Danish National Birth Cohort (n =2000) and found no evidence of genetic association with sPTB in this European population.64 The first GWAS study from the US (n=2040) of the maternal genome and early sPTB (<34 weeks) comprising mixed racial distribution identified multiple SNPs associated with sPTB. However, these results could not be replicated in a validation cohort.65,66 In a separate Norwegian cohort of mothers with sPTB (n=1921), no genome-wide significant associations with gestational age were found. However, genes involved in the inflammation/infection pathway (TLR4, NFKB1, ABCA1, MMP9) that contribute to gestational age were found using a gene-set enrichment analysis of GWAS results.67 The negative results of these studies were not surprising given the small sample size.
In a large GWAS of 43,568 women of European descent with self-reported sPTB, variants at six loci in the maternal genome (EBF1, EEFSEC, AGTR2, WNT4, ADCY5, and RAP2C) were found to be associated with gestational duration, and three loci (EBF1, EEFSEC, and AGTR2) with preterm birth as a dichotomous trait (<37 weeks).68 These findings were replicated in a separate Northern European cohort with spontaneous preterm birth (n=8643). Analysis of mother-infant dyads showed that these findings likely resulted from the action of the maternal genome. EBF1 is essential for B cell development and control of blood pressure.69,70 Recent studies have identified the critical role of B cells in birth timing and preterm birth in animals and humans.71–73 In mice, the expression level of EBF1 at the mRNA level is downregulated in splenic B cells during normal pregnancies.74 Low EBF1 mRNA has been associated with an increased risk of sPTB in humans due to altered maternal-fetal immune and cell cycle/apoptosis pathways.75 The EEFSEC gene, which encodes the selenocysteine tRNA (tRNASeleno)-specific eukaryotic elongation factor plays a critical role in incorporating selenium in the form of selenocysteine into selenoproteins. Selenoproteins serve critical cellular homeostatic functions in maintaining redox status and antioxidant defenses and modulating inflammatory responses and have been implicated in various reproductive and obstetric health disorders.76 AGTR2 plays a role in modulating uteroplacental circulation, and it has been suggested it may harbor variants that contribute to the risk of preeclampsia.77 Women with preeclampsia as an indication for their delivery were excluded from this GWAS dataset, suggesting that this association indicates the risk of sPTB rather than preeclampsia.68 Functional analysis showed that an implicated variant in WNT4 alters the binding of the estrogen receptor. Estrogen is known to play a key role in vascularization during the formation of the decidua. Higher expression of negative regulators of WNT signaling SFRP1/SFRP3 is found in preterm human placenta compared to term controls.78 Thus, WNT4 is likely involved in uterine & placental development and vascular control. However, further studies are needed to implicate these genes in the causal pathogenesis of preterm birth.
In another GWAS, Tiensuu et al. analyzed 247 mothers with sPTB and compared it with 419 term controls. They found that the fetal SLIT2 variant and both SLIT2 and ROBO1 expression in placenta and trophoblast cells are associated with sPTB. The minor allele of SLIT2, SNP rs116461311, was overrepresented in very preterm infants (< 32 weeks). SLIT2-ROBO1 signaling is linked with the regulation of genes involved in inflammation, decidualization, and fetal growth.79
Offspring GWAS
One of the first studies of the fetal genome using a Scandinavian cohort (n=3022) did not find any significant SNPs associated with sPTB.80 In another preliminary GWAS study using fetal genome in those with sPTB <34 weeks (n=1851), Zhang et al. identified two significant variants.65,66 However, the study included highly heterogeneous groups and failed to replicate them in independent samples. A larger GWAS (n=1,349 cases and 12,595 ancestry-matched controls) with five ancestral groups investigating the fetal genome in sPTB between 25–30 weeks of gestation found two significant intergenic loci associated with sPTB.81 However, each association was only observed in one of the five ancesteral groups and could not be replicated in any external samples.
A well-powered GWAS of infants of European descent (n = 84,689) with 4775 of whom were born by sPTB (<37 weeks) identified a fetal locus on chromosome 2q13 associated with gestational duration.82 Genes at this locus include several Interleukin-1 (IL-1) family members. Recently, IL-1 and IL-1 receptor-associated kinase 1 (IRAK1) have been identified as critical mediators of preterm birth.16,83 This association was replicated in 9,291 additional infants. No association was seen at the 2q13 locus in analyses of 1139 early preterm birth infants (<34 weeks). Further analysis showed that genetic variation at the locus was most strongly associated with the timing of labor in the later stages of pregnancy. This finding suggests that the 2q13 locus may be downstream of a primary activating signal and serves to accelerate labor once initiated.
The association of preterm birth with neonatal morbidity and mortality makes sPTB likely subjected to negative selection. Thus, the effect size is expected to be weak for common alleles that influence liability to a phenotype.84 In addition, GWAS studies on preterm birth have so far been underpowered. However, the finding of preterm birth-associated alleles in these GWAS studies suggests that further increasing the sample size in GWAS will reveal new loci and define the genetic pathways for birth timing and sPTB.
Whole exome and whole genome analyses
As demonstrated in most GWAS findings, in common complex phenotypes, like sPTB, disease associating genetic mutations are often outside the coding regions of genes. Often, the responsible gene is not clear and the mechanism for altered regulation is difficult to determine. Thus, Whole Exome Sequencing (WES) or Whole Genome Sequencing (WGS) analyses evaluating families with a high prevalence of sPTB have the potential to identify causal, highly penetrant variants more clearly.
In the first study of WES and sPTB of 10 Finnish mothers with multiple sPTB of which two were mother-daughter pairs, novel variants in complement and coagulation cascade pathways were identified. These findings were further tested in a large sample (n = 565) and found significant associations in three complement receptor 1 (CR1) SNPs.85 CR1 encodes complement C3b/C4b Receptor 1 which is located on the surface of the erythrocytes. This CR1 SNP is associated with decreased erythrocyte sedimentation rate, which indicates a higher risk of systemic inflammation due to non-clearance of immune complexes.85 Another WES study compared variants identified by targeted sequencing of 32 Finnish women with 2–3 generations of preterm birth (< 34 weeks) with 16 term controls. IGF1, ATM, and IQGAP2 were most frequently identified. These genes are involved in growth, metabolic, and inflammation pathways.86
A recent WES analysis of 17 Finnish mothers with sPTB found damaging variants in genes involving the steroid receptor-signaling pathways. The results were confirmed in a replication cohort of 93 Danish sister pairs with a history of sPTB. A gene in this pathway, heat shock protein family A (Hsp70) member 1 like (HSPA1L) which contained two likely damaging missense alleles was identified in four different Finnish families. Heat shock proteins are involved in stress response, including activation of the immune response. HSPA1L variants were further validated using imputed GWAS of European ancestry (n=40,000).87 A meta-analysis using pathway analysis indicated an association of HSPA1L with sPTB.88 In vitro functional experiments showed a link between HSPA1L activity and decidualization of the endometrium.87
In the US, WES analysis on the fetal genome of 49 African American mothers with preterm premature rupture of membranes (pPROM) and 20 controls identified damaging/potentially damaging rare variants in fibrillar collagen genes, which are known to contribute to fetal membrane strength and integrity.89 The following WES analysis in 76 African American mothers with pPROM and 43 term controls identified damaging mutations in innate immunity and host defense genes,90 and in genes that encode anti-microbial proteins.91
In a study using an automated pipeline developed for detecting mutations in the mitochondrial genome (mtDNA) and using low-coverage whole-genome sequencing data from an sPTB cohort (n=929) from diverse ethnic backgrounds (average gestational age = 27 weeks), variants that may contribute to sPTB were identified. These included haplogroups and a large number of mtDNA variants, including 8 samples carrying known pathogenic variants and 47 samples carrying rare mtDNA variants.92
Transcriptomic analysis of preterm birth
The transcriptome is an array of all RNA (particularly mRNA) transcripts derived genes and produced in a particular cell or tissue. Studies of the transcriptomes in sPTB have proven to be challenging. Potent technology for transcriptome analysis such as RNA sequencing (RNA-Seq) can help identify the molecular landscape of preterm birth and improve understanding of the physiology and pathology of term and preterm labor. In an RNA seq study (n=24) of placental membranes from severe sPTB (<33 weeks), multiple inflammatory and immunological pathways were noted to be upregulated.93 Transcriptomic analyses of preterm infants (n =32) born due to infection or sPTB revealed a unique expression signature which included the upregulation of genes in IGF signaling and inflammation pathways. A recent RNA-seq study in male and female placentas from women with sPTB (<36 weeks) showed alterations with fetal sex disparities in the genes and canonical pathways critical for regulating inflammation, oxidative stress, detoxification, mitochondrial function, energy metabolism, and extracellular matrix.94 In a network analysis of the placenta transcriptome, the SOD1 gene was shown to be down-regulated in the preterm birth placenta.95 Antenatal steroids given to mothers with impending preterm delivery transiently up-regulates SOD1 gene expression which helps to counteract increased production of reactive oxygen species, emphasizing its importance in improving preterm neonatal outcomes.96 Further studies are needed to understand the transcriptomic changes and molecular etiology of sPTB.
Integrative Genomic approach
Common maternal SNPs explain approximately 23% of the phenotypic variance in preterm birth.68 Thus, other sources that could explain preterm birth phenotypic variation need to be explored. Given the complexity of various pathways involved in human pregnancy, integrative approaches that utilizes diverse data types and analyses can help identify the genetic and environmental interactions influencing sPTB.97
Combining genetic and proteomic analysis, Haapalainen et al. analyzed SNPs in 10 fetal genes encoding for placental proteins associated with the duration of pregnancy (n = 77). Of these, only one SNP within CPPED1 was associated with induction of term labor. CPPED1 affects gene expression related to inflammation and blood vessel development.98 To identify preterm birth-associated genes and pathways, another study integrated WGS, RNA-seq, and DNA methylation data from 270 cases with preterm birth and 521 controls of family trios (mother, father, and neonate). They identified 72 candidate biomarker genes for very early preterm birth (<28 weeks, n = 44). All three data types (WGS, RNA-seq, and DNA methylation) identified preterm birth-associated genes RAB31 and RBPJ. These genes are involved in EGFR (epidermal growth factor receptor) and prolactin signaling pathways, inflammation- and immunity-related pathways, chemokine signaling, IFN-γ signaling, and Notch1 signaling, all of which are linked to preterm birth.99 This study replicated and identified four of the six genes described by Zhang et al. mentioned above,68 albeit in different SNPs (loci) associated with these genes and at a less stringent statistical threshold (FDR < 10%) given their lower and diverse sample size. Associations of heat shock protein (HSPA1L, SEC63, SACS) and nuclear receptor genes (AR) with sPTB have been found using multiple sPTB datasets based on GWASs, WES, and placental transcriptomics of maternal, fetal, and placental samples (Table 2).100
Table 2:
List of genes identified through various omics approaches and implicated with preterm birth
| Genomic | Transcriptomic | Epigenomic |
|---|---|---|
| RAB31 | RAB31 | RAB31 |
| RBPJ | RBPJ | RBPJ |
| Heat shock protein family | Heat shock protein family | TTN |
| Nuclear receptor genes (AR) | Nuclear receptor genes (AR) | |
| Immune signaling (IL1, TLR4, NFKB1) | Immune signaling (IL1) | |
| IGF signaling | IGF signaling | |
| EBF1, EEFSEC, AGTR2 | SOD1 | |
| CR1 | ||
| PAI-2 | ||
| SLIT2-ROBO1 | ||
| COL24A1 (gene × environment) | ||
Dissecting maternal and fetal genetic effects on pregnancy outcomes
Multiple epidemiological studies have shown that various maternal physical and physiological traits are associated with birth outcomes. These studies have shown maternal height to be positively associated with gestational duration, birth weight, and birth length,101,102 elevated maternal blood pressure with reduced birth weight,103 and higher maternal blood glucose with higher birth weight,104 To explain these associations various mechanisms have been proposed (see Zhang et al.28 for detailed review).
To further understand these mechanisms and distinguish the effect of the maternal intrauterine environment from direct fetal genetic effects, investigators examined the relationship between maternal height with fetal growth measures and gestational age using a haplotype-based Mendelian randomization analysis of mother-infant pairs.105 They found that higher maternal height causally increases with gestational duration. In a recent study, they further expanded the analysis and examined the causal effects of additional maternal phenotypes on birth outcomes. 106 They continue to find maternal height to be positively associated with longer gestational duration as well as larger birth size. Through maternal effect, alleles that caused higher blood pressure were associated with shorter gestational duration, and higher maternal BMI and glucose levels were positively associated with birth weight. Elevated blood pressure alleles were associated with reduced fetal growth through fetal effect. In the fetus, alleles associated with higher metabolic risks (type 2 diabetes) were associated with decreased birth weight. They also found rapid fetal growth was associated with shorter gestational duration and elevated maternal blood pressure.106. These maternal and fetal genetic effects explain the observed associations between the maternal phenotypes and birth outcomes and the life-long associations between these birth outcomes and adult phenotypes.
Environmental exposure and preterm birth:
It has been suggested that temporal changes in the environment may explain the intergenerational variation and correlation in gestational age between relatives.37,107 While the contribution of genetic heritability of sPTB is significant, multiple studies have shown that environmental factors contribute to the largest difference in timing of birth.39,108 Studies have linked maternal smoking during pregnancy to preterm birth and low birth weight.109–111 Ambient air pollution including particulate matter has shown to be associated with preterm birth suggesting that climate change could lead to increased preterm birth.112,113 Exposure to heavy metals (cadmium, chromium, arsenic, lead, and nickel), 114 and endocrine-disrupting chemicals such as phthalates have been linked to preterm birth.115
The potential epigenetic modifications of genes could explain the strong familial aggregation and cross-generational risk of preterm birth. However, most genetic studies on sPTB have failed to consider the genetic and environmental interactions which could be one of the reasons for the lack of replication in genetic studies. Examining only the direct associations of traits without accounting for environmental exposures may result in missing relevant genes which influence sPTB.116 To overcome this, a genome-wide gene × environment interaction analysis to explore the “missing heritability” of preterm birth in 1,733 African-American women (n = 698 preterm birth & 1,035 of term birth) showed that maternal COL24A1 variants have significant genome-wide interaction with maternal pre-pregnancy overweight/obesity on preterm birth risk. The interaction effect size and direction were comparable across all subtypes of preterm birth, including spontaneous, medically indicated, early preterm birth (<32 weeks), late preterm birth (32–37 weeks), and preterm birth with chorioamnionitis. This interaction was further replicated in African-American mothers from an independent cohort and in a meta-analysis but failed to be replicated in Caucasians, suggesting a population-specific role of this variant.117 Altered COL24A1 expression is required for the proper functioning of the extracellular matrix and its alteration may lead to various pathological disorders leading to sPTB.118 Though further studies should account for gene-environment interaction, designing a robust gene × environment interaction analysis remains a challenge and multiple biases and confounders have to be accounted for.119
Epigenetics
Epigenetics is defined as reversible alterations of the gene function that are not due to changes in the DNA sequence but are heritable through cell division. Differences in epigenome may account for important phenotypic differences even in the setting of identical genetics. The two primary sources of epigenetic modification are DNA methylation and histone acetylation/deacetylation.120 Epigenetic modifications occur not only in DNA but also in RNA. Since epigenetic changes occur during embryogenesis, any disturbance of the normal environment during the critical in-utero period can cause epigenetic alterations that last into the offspring’s lifetime. As such, many studies have associated epigenetic and methylation differences in various tissue types (cord blood, maternal blood, placenta) with gestational age and sPTB, and provided insight that both genetic and epigenetic factors contribute to sPTB.121–124
Maternal toxic exposure to heavy metals, air pollution, and pesticides have been correlated to a reduction in placental methylation which may lead to genomic instability and an increased number of mutational events.125 Epigenome-wide association meta-analysis studies (EWAS) have shown that many prenatal exposures associated with sPTB also change DNA methylation in cord blood. EWAS has shown reproducible associations between blood DNA methylation in newborns and maternal folate levels,126 exposure to smoking during pregnancy, 127 air pollutants,128 and exposure to heavy metals.129 Although these studies have investigated the mechanisms of one environmental toxin at a time, many studies have failed to account for an individual’s day-to-day complex toxin exposures.
Other studies have found an association of premature uterine contraction with pathogenic variants of the sarcomere gene TTN and with transcriptomic variations of sarcomeric premature uterine contraction genes. This association was regulated by epigenetic factors, including methylation and long non-coding RNAs.130 Maternal age is an independent risk factor for preterm birth. The use of chronological age assumes that individuals age at a similar rate and it does not capture inter-individual differences that may exist due to genetic background and environmental exposures. Studies have estimated biological age using genome-wide DNA methylation and found a significant relationship between a mother’s biological age and gestational age at delivery.131
Thus, studying epigenetic changes related to preterm birth would improve the understanding of various mechanisms leading to sPTB and the long-term health consequences for the offspring. Epigenomic markers could also serve as an important diagnostic tool as epigenetic reprogramming in the tissue of interest (placental) might be captured by more accessible surrogate tissue (maternal blood).124 The exposure-driven methylation differences might mediate the effects of exposures on preterm birth, but the causal epigenomic mechanism remains unclear.
Genetic studies to guide intervention
One criticism of genetic studies is that few actionable findings across complex phenotypes have emerged. One interesting, though unproven implication from Zhang et al. arises from the association of variants near eukaryotic elongation factor selenocysteine-tRNA-specific (EEFSEC) associated with sPTB and length of pregnancy.68 The identification of the selenium/selenoprotein pathway suggests the potential benefit for further evaluating the role of maternal selenium micronutrient status on sPTB risk. Selenium protects against acute pro-oxidant injury and low maternal Selenium levels have been linked with preterm birth and increased risk of neonatal morbidity and mortality.132,133 Though low plasma Se has been associated with sPTB risk, it was not found to be sufficiently predictive at individual patient level.134 In a worldwide study of 9946 singleton live births from 17 geographically diverse locations, statistically significant associations between maternal Se concentration and sPTB at some sites were observed. 135 However, this finding was not generalizable across the whole cohort and might lower the enthusiasm for the broad use of Se supplements as a general strategy to prevent sPTB. However, these results suggest there could be a potential benefit for certain high-risk, low-income geographic regions.
Future Directions
Early detection of the risk of preterm birth would be helpful to reduce the global health burden of adverse neonatal outcomes. Many issues in conducting genetic studies of preterm birth remain to be resolved as genetic variation alone is likely not sufficient to explain the risks of sPTB due to the interaction with various environmental factors. Epigenomic signatures are also to be included in preterm birth studies as they dynamically change in response to the environment. As a result, genomic, transcriptomic, and epigenomic markers can identify high-risk women even prior to or early in pregnancy compared to biochemical markers. Investigators are developing a predictive model for sPTB by using multi-omics on maternal blood collected in early pregnancy.136 Recently, there has been an interest in polygenic risk scores to improve the prediction of a person’s genetic susceptibility to common chronic diseases which can also be applied to sPTB. Polygenic risk scores can explain phenotypic variation among geographic populations based solely on risk allele frequencies.137 This can be calculated using genome-wide data, family history, and clinical variables to predict the onset of disease and the prognosis.138
Conclusion:
sPTB is a multifactorial disease involving multiple environmental and genetic risk factors involving the mother and the fetus with many genes involved in the inflammation and immunity-related pathways while others, particularly in the maternal genome, are not. Thus, genetic studies of sPTB should consider the contributions of both maternal and fetal genomes as well as the genetic and environmental interactions to modify the risk. These studies should use gestational duration as a continuous variable in addition to a binary <37-week outcome as genetics and environment affect across the range of gestational ages during pregnancy. The function of various genes identified from genomic studies of sPTB are from animal studies or from bioinformatics evidence generated from a curated knowledge base that is primarily biased based on cancer literature. Thus, the functional role of many genes implicated in sPTB remains elusive. Many reported studies also failed replication. This could be due to small sample size, heterogeneity in classifying sPTB, or unaccounted environmental influences within the same population.
Most genetic studies have been conducted on samples from European ancestries. Since preterm birth affects the worldwide population, further genetic studies should focus on other ancestries and ethnic groups, especially those representing the high-risk populations with attention to environmental influences including climate, toxins, nutrition, and infectious exposures. Large population-based genomic studies and meta-analyses of the aggregated datasets from various studies can help address the issues of low sample size, increase the power to detect true associations, and can reduce false-positive associations. More comprehensively identifying the genomic loci associated with sPTB will provide further insight into regulatory pathways, that can be dissected through analysis of human tissues, humanized animal models, organoids, and tissue-on-chip approaches that are currently emerging related to reproductive tissues. These new opportunities to take associations into understanding causal mechanisms will foster new biological insight and offer new opportunities for early prediction and intervention of preterm birth.
Funding:
Supported by the National Institute of Health (1R01HD101669-01A1 to GZ and KL2 to VGJ), The Lung Health Center Pilot Grant, the University of Alabama at Birmingham and The Kaul Pediatric Research Award, Children’s of Alabama (to VGJ), and the Bill and Melinda Gates Foundation (to LJM, GZ, NM) March of Dimes Prematurity Research Center Ohio Collaborative (to LJM, GZ, NM) and the Burroughs Wellcome Fund (to GZ).
Abbreviations:
- sPTB
spontaneous preterm birth
- GWAS
genome-wide association study
- WES
whole-exome sequence
- WGS
whole-genome sequence
- GWL
Genome-wide linkage studies
- SNP
Single nucleotide polymorphisms
Footnotes
Conflict of interest: LJM consults for Mirvie, Inc., a biotech company involved in preterm birth diagnostics, and Cognitive Care, Inc., an AI/software diagnostics company
Ethics: The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to. No ethical approval was required as this is a review article with no original research data.
References
- 1.Callaghan WM, MacDorman MF, Rasmussen SA, Qin C, Lackritz EM. The contribution of preterm birth to infant mortality rates in the United States. Pediatrics. 2006;118(4):1566–1573. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Murphy S, Kochanek K, Arias E. Mortality in the United States, 2018. NCHS Data Brief, no. 355. 2020. [PubMed]
- 3.Blencowe H, Cousens S, Chou D, et al. Born too soon: the global epidemiology of 15 million preterm births. Reprod Health. 2013;10 Suppl 1:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet. 2015;385(9966):430–440. [DOI] [PubMed] [Google Scholar]
- 5.Blencowe H, Cousens S, Oestergaard MZ, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. The lancet. 2012;379(9832):2162–2172. [DOI] [PubMed] [Google Scholar]
- 6.Martin JA, Hamilton BE, Osterman MJK. Births in the United States, 2019. NCHS Data Brief. 2020(387):1–8. [PubMed] [Google Scholar]
- 7.2020 MARCH OF DIMES REPORT CARD. 2020; https://www.marchofdimes.org/materials/MOD2020_REPORT_CARD_and_POLICY_ACTIONS_BOOKLET_FIN.pdf. Accessed 12/10/20, 2020.
- 8.Korvenranta E, Lehtonen L, Rautava L, et al. Impact of very preterm birth on health care costs at five years of age. Pediatrics. 2010;125(5):e1109–e1114. [DOI] [PubMed] [Google Scholar]
- 9.Singer LT, Salvator A, Guo S, Collin M, Lilien L, Baley J. Maternal psychological distress and parenting stress after the birth of a very low-birth-weight infant. JAMA : the journal of the American Medical Association. 1999;281(9):799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mwaniki MK, Atieno M, Lawn JE, Newton CR. Long-term neurodevelopmental outcomes after intrauterine and neonatal insults: a systematic review. The Lancet. 2012;379(9814):445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Dommelen P, Verkerk PH, van Straaten HL, et al. Hearing loss by week of gestation and birth weight in very preterm neonates. The Journal of pediatrics. 2015;166(4):840–843. e841. [DOI] [PubMed] [Google Scholar]
- 12.Saigal S, Doyle LW. An overview of mortality and sequelae of preterm birth from infancy to adulthood. The Lancet. 2008;371(9608):261–269. [DOI] [PubMed] [Google Scholar]
- 13.Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261(5):412–417. [DOI] [PubMed] [Google Scholar]
- 14.Järvelin MR, Sovio U, King V, et al. Early life factors and blood pressure at age 31 years in the 1966 northern Finland birth cohort. Hypertension. 2004;44(6):838–846. [DOI] [PubMed] [Google Scholar]
- 15.Sipola-Leppänen M, Vääräsmäki M, Tikanmäki M, et al. Cardiometabolic risk factors in young adults who were born preterm. Am J Epidemiol. 2015;181(11):861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain VG, Kong F, Kallapur SG, et al. IRAK1 Is a Critical Mediator of Inflammation-Induced Preterm Birth. J Immunol. 2020;204(10):2651–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero R, Espinoza J, Kusanovic JP, et al. The preterm parturition syndrome. BJOG : an international journal of obstetrics and gynaecology. 2006;113 Suppl 3:17–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez-Lopez N, Romero R, Galaz J, et al. Transcriptome changes in maternal peripheral blood during term parturition mimic perturbations preceding spontaneous preterm birthdagger. Biology of reproduction. 2022;106(1):185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet (London, England). 2008;371(9606):75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green CA, Johnson JD, Vladutiu CJ, Manuck TA. The association between maternal and paternal race and preterm birth. Am J Obstet Gynecol MFM. 2021;3(4):100353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krieger N, Van Wye G, Huynh M, et al. Structural Racism, Historical Redlining, and Risk of Preterm Birth in New York City, 2013–2017. American journal of public health. 2020;110(7):1046–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith V, Devane D, Begley CM, Clarke M, Higgins S. A systematic review and quality assessment of systematic reviews of randomised trials of interventions for preventing and treating preterm birth. Eur J Obstet Gynecol Reprod Biol. 2009;142(1):3–11. [DOI] [PubMed] [Google Scholar]
- 23.Schmouder VM, Prescott GM, Franco A, Fan-Havard P. The rebirth of progesterone in the prevention of preterm labor. Ann Pharmacother. 2013;47(4):527–536. [DOI] [PubMed] [Google Scholar]
- 24.Society for Maternal-Fetal Medicine Publications Committee. Electronic address pso. SMFM Statement: Use of 17-alpha hydroxyprogesterone caproate for prevention of recurrent preterm birth. American journal of obstetrics and gynecology. 2020;223(1):B16–B18. [DOI] [PubMed] [Google Scholar]
- 25.Norman JE, Marlow N, Messow CM, et al. Vaginal progesterone prophylaxis for preterm birth (the OPPTIMUM study): a multicentre, randomised, double-blind trial. Lancet. 2016;387(10033):2106–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Group E. Evaluating Progestogens for Preventing Preterm birth International Collaborative (EPPPIC): meta-analysis of individual participant data from randomised controlled trials. Lancet. 2021;397(10280):1183–1194. [DOI] [PubMed] [Google Scholar]
- 27.Muglia LJ, Katz M. The enigma of spontaneous preterm birth. N Engl J Med. 2010;362. [DOI] [PubMed] [Google Scholar]
- 28.Zhang G, Srivastava A, Bacelis J, Juodakis J, Jacobsson B, Muglia LJ. Genetic studies of gestational duration and preterm birth. Best practice & research Clinical obstetrics & gynaecology. 2018;52:33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bezold KY, Karjalainen MK, Hallman M, Teramo K, Muglia LJ. The genomics of preterm birth: from animal models to human studies. Genome Med. 2013;5(4):34–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharya S, Raja EA, Mirazo ER, et al. Inherited predisposition to spontaneous preterm delivery. Obstet Gynecol. 2010;115(6):1125–1133. [DOI] [PubMed] [Google Scholar]
- 31.Koire A, Chu DM, Aagaard K. Family history is a predictor of current preterm birth. Am J Obstet Gynecol MFM. 2021;3(1):100277. [DOI] [PubMed] [Google Scholar]
- 32.Boyd HA, Poulsen G, Wohlfahrt J, Murray JC, Feenstra B, Melbye M. Maternal contributions to preterm delivery. American journal of epidemiology. 2009;170(11):1358–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherf Y, Sheiner E, Vardi IS, Sergienko R, Klein J, Bilenko N. Recurrence of Preterm Delivery in Women with a Family History of Preterm Delivery. Am J Perinatol. 2017;34(4):397–402. [DOI] [PubMed] [Google Scholar]
- 34.Bezold KY, Karjalainen MK, Hallman M, Teramo K, Muglia LJ. The genomics of preterm birth: from animal models to human studies. Genome Med. 2013;5(4):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lunde A, Melve KK, Gjessing HK, Skjaerven R, Irgens LM. Genetic and environmental influences on birth weight, birth length, head circumference, and gestational age by use of population-based parent-offspring data. Am J Epidemiol. 2007;165. [DOI] [PubMed] [Google Scholar]
- 36.Plunkett J, Feitosa MF, Trusgnich M, et al. Mother’s genome or maternally-inherited genes acting in the fetus influence gestational age in familial preterm birth. Hum Hered. 2009;68(3):209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu W, Witherspoon DJ, Fraser A, et al. The heritability of gestational age in a two-million member cohort: implications for spontaneous preterm birth. Human genetics. 2015;134(7):803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svensson AC, Sandin S, Cnattingius S, et al. Maternal effects for preterm birth: a genetic epidemiologic study of 630,000 families. American journal of epidemiology. 2009;170(11):1365–1372. [DOI] [PubMed] [Google Scholar]
- 39.York TP, Eaves LJ, Lichtenstein P, et al. Fetal and maternal genes’ influence on gestational age in a quantitative genetic analysis of 244,000 Swedish births. American journal of epidemiology. 2013;178(4):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kistka ZAF, DeFranco EA, Ligthart L, et al. Heritability of parturition timing: an extended twin design analysis. American journal of obstetrics and gynecology. 2008;199(1):43.e41–43.e45. [DOI] [PubMed] [Google Scholar]
- 41.Clausson B, Lichtenstein P, Cnattingius S. Genetic influence on birthweight and gestational length determined by studies in offspring of twins. BJOG: An International Journal of Obstetrics and Gynaecology. 2000;107(3):375–381. [DOI] [PubMed] [Google Scholar]
- 42.Boyd HA, Poulsen G, Wohlfahrt J, Murray JC, Feenstra B, Melbye M. Maternal contributions to preterm delivery. American journal of epidemiology. 2009;170(11):1358–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treloar SA, Macones GA, Mitchell LE, Martin NG. Genetic influences on premature parturition in an Australian twin sample. Twin Res. 2000;3(2):80–82. [DOI] [PubMed] [Google Scholar]
- 44.Wu W, Witherspoon DJ, Fraser A, et al. The heritability of gestational age in a two-million member cohort: implications for spontaneous preterm birth. Human genetics. 2015;134(7):803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilcox AJ, Skjaerven R, Lie RT. Familial Patterns of Preterm Delivery: Maternal and Fetal Contributions. American journal of epidemiology. 2008;167(4):474–479. [DOI] [PubMed] [Google Scholar]
- 46.Srivastava AK, Juodakis J, Sole-Navais P, et al. Haplotype-based heritability estimations reveal gestational duration as a maternal trait and fetal size measurements at birth as fetal traits in human pregnancy. bioRxiv. 2020:2020.2005.2012.079863. [Google Scholar]
- 47.Eaves LJ, Pourcain BS, Smith GD, York TP, Evans DM. Resolving the effects of maternal and offspring genotype on dyadic outcomes in genome wide complex trait analysis (“M-GCTA”). Behav Genet. 2014;44(5):445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karjalainen MK, Huusko JM, Ulvila J, et al. A potential novel spontaneous preterm birth gene, AR, identified by linkage and association analysis of X chromosomal markers. PloS one. 2012;7(12):e51378–e51378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haataja R, Karjalainen MK, Luukkonen A, et al. Mapping a new spontaneous preterm birth susceptibility gene, IGF1R, using linkage, haplotype sharing, and association analysis. PLoS genetics. 2011;7(2):e1001293–e1001293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keleher MR, Erickson K, Smith HA, et al. Placental Insulin/IGF-1 Signaling, PGC-1alpha, and Inflammatory Pathways Are Associated With Metabolic Outcomes at 4–6 Years of Age: The ECHO Healthy Start Cohort. Diabetes. 2021;70(3):745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aguirre GA, De Ita JR, de la Garza RG, Castilla-Cortazar I. Insulin-like growth factor-1 deficiency and metabolic syndrome. J Transl Med. 2016;14:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hellstrom A, Engstrom E, Hard AL, et al. Postnatal serum insulin-like growth factor I deficiency is associated with retinopathy of prematurity and other complications of premature birth. Pediatrics. 2003;112(5):1016–1020. [DOI] [PubMed] [Google Scholar]
- 53.He JR, Lai YM, Liu HH, et al. Maternal IGF1 and IGF1R polymorphisms and the risk of spontaneous preterm birth. J Clin Lab Anal. 2017;31(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee MS, Lee SO, Kim SH, Lee EO, Lee HJ. Anti-Cancer Effect of Lambertianic Acid by Inhibiting the AR in LNCaP Cells. Int J Mol Sci. 2016;17(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarquini F, Picchiassi E, Coata G, et al. Induction of the apoptotic pathway by oxidative stress in spontaneous preterm birth: Single nucleotide polymorphisms, maternal lifestyle factors and health status. Biomed Rep. 2018;9(1):81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karjalainen MK, Ojaniemi M, Haapalainen AM, et al. CXCR3 Polymorphism and Expression Associate with Spontaneous Preterm Birth. J Immunol. 2015;195(5):2187–2198. [DOI] [PubMed] [Google Scholar]
- 57.Karjalainen MK. CXCR3 polymorphism and expression associate with spontaneous preterm birth. J Immunol. 2015;195. [DOI] [PubMed] [Google Scholar]
- 58.Chaturvedi V, Ertelt JM, Jiang TT, et al. CXCR3 blockade protects against Listeria monocytogenes infection-induced fetal wastage. The Journal of clinical investigation. 2015;125(4):1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibson CS, MacLennan AH, Dekker GA, et al. Genetic polymorphisms and spontaneous preterm birth. Obstet Gynecol. 2007;109(2 Pt 1):384–391. [DOI] [PubMed] [Google Scholar]
- 60.Bogic LV, Ohira RH, Yamamoto SY, Okazaki KJ, Millar K, Bryant-Greenwood GD. Tissue plasminogen activator and its receptor in the human amnion, chorion, and decidua at preterm and term. Biology of reproduction. 1999;60(4):1006–1012. [DOI] [PubMed] [Google Scholar]
- 61.Altmuller J, Palmer LJ, Fischer G, Scherb H, Wjst M. Genomewide scans of complex human diseases: true linkage is hard to find. American journal of human genetics. 2001;69(5):936–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manolio TA. Genomewide association studies and assessment of the risk of disease. The New England journal of medicine. 2010;363(2):166–176. [DOI] [PubMed] [Google Scholar]
- 63.Sheikh IA, Ahmad E, Jamal MS, et al. Spontaneous preterm birth and single nucleotide gene polymorphisms: a recent update. BMC Genomics. 2016;17(Suppl 9):759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu W, Clark E, Manuck T, Esplin M, Varner M, Jorde L. A Genome-Wide Association Study of spontaneous preterm birth in a European population [version 1; peer review: 2 approved with reservations]. F1000Research. 2013;2(255). [Google Scholar]
- 65.Zhang H, Baldwin DA, Bukowski RK, et al. A genome-wide association study of early spontaneous preterm delivery. Genet Epidemiol. 2015;39(3):217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H A genome-wide association study of early spontaneous preterm delivery. Genet Epidemiol. 2015;39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bacelis J, Juodakis J, Sengpiel V, et al. Literature-Informed Analysis of a Genome-Wide Association Study of Gestational Age in Norwegian Women and Children Suggests Involvement of Inflammatory Pathways. PloS one. 2016;11(8):e0160335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang G, Feenstra B, Bacelis J, et al. Genetic Associations with Gestational Duration and Spontaneous Preterm Birth. The New England journal of medicine. 2017;377(12):1156–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.International Consortium for Blood Pressure Genome-Wide Association S, Ehret GB, Munroe PB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gyory I, Boller S, Nechanitzky R, et al. Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev. 2012;26(7):668–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang B, Faucette AN, Pawlitz MD, et al. Interleukin-33-induced expression of PIBF1 by decidual B cells protects against preterm labor. Nature medicine. 2017;23(1):128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leng Y, Romero R, Xu Y, et al. Are B cells altered in the decidua of women with preterm or term labor? American journal of reproductive immunology : AJRI : official journal of the American Society for the Immunology of Reproduction and the International Coordination Committee for Immunology of Reproduction. 2019;81(5):e13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bommer I, Juriol L, Muzzio D, et al. Characterization of murine amniotic fluid B cells in normal pregnancy and in preterm birth. Reproduction. 2019;158(4):369–376. [DOI] [PubMed] [Google Scholar]
- 74.Valeff N, Muzzio DO, Matzner F, et al. B cells acquire a unique and differential transcriptomic profile during pregnancy. Genomics. 2021;113(4):2614–2622. [DOI] [PubMed] [Google Scholar]
- 75.Zhou G, Holzman C, Heng YJ, Kibschull M, Lye SJ, Vazquez A. EBF1 Gene mRNA Levels in Maternal Blood and Spontaneous Preterm Birth. Reprod Sci. 2020;27(1):316–324. [DOI] [PubMed] [Google Scholar]
- 76.Mistry HD, Broughton Pipkin F, Redman CW, Poston L. Selenium in reproductive health. American journal of obstetrics and gynecology. 2012;206(1):21–30. [DOI] [PubMed] [Google Scholar]
- 77.Zhou A, Dekker GA, Lumbers ER, et al. The association of AGTR2 polymorphisms with preeclampsia and uterine artery bilateral notching is modulated by maternal BMI. Placenta. 2013;34(1):75–81. [DOI] [PubMed] [Google Scholar]
- 78.Zmijanac Partl J, Karin V, Skrtic A, et al. Negative regulators of Wnt signaling pathway SFRP1 and SFRP3 expression in preterm and term pathologic placentas. J Matern Fetal Neonatal Med. 2018;31(22):2971–2979. [DOI] [PubMed] [Google Scholar]
- 79.Tiensuu H, Haapalainen AM, Karjalainen MK, et al. Risk of spontaneous preterm birth and fetal growth associates with fetal SLIT2. PLoS genetics. 2019;15(6):e1008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Myking S, Boyd HA, Myhre R, et al. X-chromosomal maternal and fetal SNPs and the risk of spontaneous preterm delivery in a Danish/Norwegian genome-wide association study. PloS one. 2013;8(4):e61781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rappoport N, Toung J, Hadley D, et al. A genome-wide association study identifies only two ancestry specific variants associated with spontaneous preterm birth. Sci Rep. 2018;8(1):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu X, Helenius D, Skotte L, et al. Variants in the fetal genome near pro-inflammatory cytokine genes on 2q13 associate with gestational duration. Nat Commun. 2019;10(1):3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Presicce P, Park CW, Senthamaraikannan P, et al. IL-1 signaling mediates intrauterine inflammation and chorio-decidua neutrophil recruitment and activation. JCI Insight. 2018;3(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wadon M, Modi N, Wong HS, Thapar A, O’Donovan MC. Recent advances in the genetics of preterm birth. Ann Hum Genet. 2020;84(3):205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McElroy JJ. Maternal coding variants in complement receptor 1 and spontaneous idiopathic preterm birth. Hum Genet. 2013;132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uzun A, Schuster J, McGonnigal B, Schorl C, Dewan A, Padbury J. Targeted Sequencing and Meta-Analysis of Preterm Birth. PloS one. 2016;11(5):e0155021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huusko JM, Karjalainen MK, Graham BE, et al. Whole exome sequencing reveals HSPA1L as a genetic risk factor for spontaneous preterm birth. PLoS genetics. 2018;14(7):e1007394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Capece A, Vasieva O, Meher S, Alfirevic Z, Alfirevic A. Pathway analysis of genetic factors associated with spontaneous preterm birth and pre-labor preterm rupture of membranes. PloS one. 2014;9(9):e108578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Modi BP, Teves ME, Pearson LN, et al. Rare mutations and potentially damaging missense variants in genes encoding fibrillar collagens and proteins involved in their production are candidates for risk for preterm premature rupture of membranes. PloS one. 2017;12(3):e0174356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Modi BP, Teves ME, Pearson LN, et al. Mutations in fetal genes involved in innate immunity and host defense against microbes increase risk of preterm premature rupture of membranes (PPROM). Mol Genet Genomic Med. 2017;5(6):720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Modi BP, Parikh HI, Teves ME, et al. Discovery of rare ancestry-specific variants in the fetal genome that confer risk of preterm premature rupture of membranes (PPROM) and preterm birth. BMC Med Genet. 2018;19(1):181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang Z, Slone J, Wang X, et al. Validation of low-coverage whole-genome sequencing for mitochondrial DNA variants suggests mitochondrial DNA as a genetic cause of preterm birth. Human mutation. 2021;42(12):1602–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pereyra S, Sosa C, Bertoni B, Sapiro R. Transcriptomic analysis of fetal membranes reveals pathways involved in preterm birth. BMC Med Genomics. 2019;12(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lien YC, Zhang Z, Cheng Y, et al. Human Placental Transcriptome Reveals Critical Alterations in Inflammation and Energy Metabolism with Fetal Sex Differences in Spontaneous Preterm Birth. Int J Mol Sci. 2021;22(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin P, Lai X, Wu L, Liu W, Lin S, Ye J. Network analysis reveals important genes in human placenta. J Obstet Gynaecol Res. 2021;47(8):2607–2615. [DOI] [PubMed] [Google Scholar]
- 96.Mahzabin T, Pillow JJ, Pinniger GJ, et al. Influence of antenatal glucocorticoid on preterm lamb diaphragm. Pediatric research. 2017;82(3):509–517. [DOI] [PubMed] [Google Scholar]
- 97.Eidem HR, McGary KL, Capra JA, Abbot P, Rokas A. The transformative potential of an integrative approach to pregnancy. Placenta. 2017;57:204–215. [DOI] [PubMed] [Google Scholar]
- 98.Haapalainen AM, Karjalainen MK, Daddali R, et al. Expression of CPPED1 in human trophoblasts is associated with timing of term birth. Journal of cellular and molecular medicine. 2018;22(2):968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Knijnenburg TA, Vockley JG, Chambwe N, et al. Genomic and molecular characterization of preterm birth. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(12):5819–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huusko JM, Tiensuu H, Haapalainen AM, et al. Integrative genetic, genomic and transcriptomic analysis of heat shock protein and nuclear hormone receptor gene associations with spontaneous preterm birth. Sci Rep. 2021;11(1):17115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Addo OY, Stein AD, Fall CH, et al. Maternal height and child growth patterns. The Journal of pediatrics. 2013;163(2):549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Myklestad K, Vatten LJ, Magnussen EB, Salvesen KA, Romundstad PR. Do parental heights influence pregnancy length?: A population-based prospective study, HUNT 2. BMC pregnancy and childbirth. 2013;13:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Steer PJ, Little MP, Kold-Jensen T, Chapple J, Elliott P. Maternal blood pressure in pregnancy, birth weight, and perinatal mortality in first births: prospective study. BMJ. 2004;329(7478):1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Group HSCR Metzger BE, Lowe LP, et al. Hyperglycemia and adverse pregnancy outcomes. The New England journal of medicine. 2008;358(19):1991–2002. [DOI] [PubMed] [Google Scholar]
- 105.Zhang G, Bacelis J, Lengyel C, et al. Assessing the Causal Relationship of Maternal Height on Birth Size and Gestational Age at Birth: A Mendelian Randomization Analysis. PLoS Med. 2015;12(8):e1001865–e1001865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen J, Bacelis J, Sole-Navais P, et al. Dissecting maternal and fetal genetic effects underlying the associations between maternal phenotypes, birth outcomes, and adult phenotypes: A mendelian-randomization and haplotype-based genetic score analysis in 10,734 mother-infant pairs. PLoS Med. 2020;17(8):e1003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Modzelewska D, Sole-Navais P, Zhang G, Muglia LJ, Nilsson S, Jacobsson B. Importance of the environment for gestational duration variability and correlation between relatives - results from the Medical Swedish Birth Registry, 1973–2012. PloS one. 2020;15(7):e0236494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dolan SM, Hollegaard MV, Merialdi M, et al. Synopsis of preterm birth genetic association studies: the preterm birth genetics knowledge base (PTBGene). Public Health Genomics. 2010;13(7–8):514–523. [DOI] [PubMed] [Google Scholar]
- 109.Shah NR, Bracken MB. A systematic review and meta-analysis of prospective studies on the association between maternal cigarette smoking and preterm delivery. American journal of obstetrics and gynecology. 2000;182(2):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu B, Xu G, Sun Y, et al. Maternal cigarette smoking before and during pregnancy and the risk of preterm birth: A dose-response analysis of 25 million mother-infant pairs. PLoS Med. 2020;17(8):e1003158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Philips EM, Santos S, Trasande L, et al. Changes in parental smoking during pregnancy and risks of adverse birth outcomes and childhood overweight in Europe and North America: An individual participant data meta-analysis of 229,000 singleton births. PLoS Med. 2020;17(8):e1003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bekkar B, Pacheco S, Basu R, DeNicola N. Association of Air Pollution and Heat Exposure With Preterm Birth, Low Birth Weight, and Stillbirth in the US: A Systematic Review. JAMA Netw Open. 2020;3(6):e208243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brumberg HL, Karr CJ, Council On Environmental H. Ambient Air Pollution: Health Hazards to Children. Pediatrics. 2021;147(6). [DOI] [PubMed] [Google Scholar]
- 114.Porpora MG, Piacenti I, Scaramuzzino S, Masciullo L, Rech F, Benedetti Panici P. Environmental Contaminants Exposure and Preterm Birth: A Systematic Review. Toxics. 2019;7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ferguson KK, McElrath TF, Meeker JD. Environmental phthalate exposure and preterm birth. JAMA pediatrics. 2014;168(1):61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bhattacharjee E, Maitra A. Spontaneous preterm birth: the underpinnings in the maternal and fetal genomes. NPJ Genom Med. 2021;6(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hong X, Hao K, Ji H, et al. Genome-wide approach identifies a novel gene-maternal pre-pregnancy BMI interaction on preterm birth. Nat Commun. 2017;8:15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Strauss JF 3rd, . Extracellular matrix dynamics and fetal membrane rupture. Reprod Sci. 2013;20(2):140–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Boffetta P, Winn DM, Ioannidis JP, et al. Recommendations and proposed guidelines for assessing the cumulative evidence on joint effects of genes and environments on cancer occurrence in humans. International journal of epidemiology. 2012;41(3):686–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dean SV, Mason E, Howson CP, Lassi ZS, Imam AM, Bhutta ZA. Born too soon: care before and between pregnancy to prevent preterm births: from evidence to action. Reproductive health. 2013;10 Suppl 1(Suppl 1):S3–S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Parets SE, Bedient CE, Menon R, Smith AK. Preterm birth and its long-term effects: methylation to mechanisms. Biology (Basel). 2014;3(3):498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schroeder JW, Conneely KN, Cubells JC, et al. Neonatal DNA methylation patterns associate with gestational age. Epigenetics. 2011;6(12):1498–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Parets SE, Conneely KN, Kilaru V, et al. Fetal DNA Methylation Associates with Early Spontaneous Preterm Birth and Gestational Age. PloS one. 2013;8(6):e67489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Park B, Khanam R, Vinayachandran V, Baqui AH, London SJ, Biswal S. Epigenetic biomarkers and preterm birth. Environ Epigenet. 2020;6(1):dvaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Arai Y, Ohgane J, Yagi S, et al. Epigenetic assessment of environmental chemicals detected in maternal peripheral and cord blood samples. J Reprod Dev. 2011;57(4):507–517. [DOI] [PubMed] [Google Scholar]
- 126.Joubert BR, den Dekker HT, Felix JF, et al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun. 2016;7:10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Joubert BR, Felix JF, Yousefi P, et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. American journal of human genetics. 2016;98(4):680–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gruzieva O, Xu CJ, Breton CV, et al. Epigenome-Wide Meta-Analysis of Methylation in Children Related to Prenatal NO2 Air Pollution Exposure. Environmental health perspectives. 2017;125(1):104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cardenas A, Rifas-Shiman SL, Godderis L, et al. Prenatal Exposure to Mercury: Associations with Global DNA Methylation and Hydroxymethylation in Cord Blood and in Childhood. Environmental health perspectives. 2017;125(8):087022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang J, Luo X, Pan J, et al. (Epi)genetic variants of the sarcomere-desmosome are associated with premature utero-contraction in spontaneous preterm labor. Environment international. 2021;148:106382. [DOI] [PubMed] [Google Scholar]
- 131.Lancaster EE, Lapato DM, Jackson-Cook C, Strauss JF 3rd, Roberson-Nay R, York TP. Maternal biological age assessed in early pregnancy is associated with gestational age at birth. Sci Rep. 2021;11(1):15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rayman MP, Wijnen H, Vader H, Kooistra L, Pop V. Maternal selenium status during early gestation and risk for preterm birth. CMAJ. 2011;183(5):549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Modzelewska D, Sole-Navais P, Brantsaeter AL, et al. Maternal Dietary Selenium Intake during Pregnancy and Neonatal Outcomes in the Norwegian Mother, Father, and Child Cohort Study. Nutrients. 2021;13(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Care AG, Gupta JK, Goodfellow L, et al. Maternal selenium levels and whole genome screen in recurrent spontaneous preterm birth population: A nested case control study. European journal of obstetrics, gynecology, and reproductive biology. 2021;265:203–211. [DOI] [PubMed] [Google Scholar]
- 135.Monangi N, Xu H, Khanam R, et al. Association of maternal prenatal selenium concentration and preterm birth: a multicountry meta-analysis. BMJ Glob Health. 2021;6(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tarca AL, Pataki BA, Romero R, et al. Crowdsourcing assessment of maternal blood multi-omics for predicting gestational age and preterm birth. Cell Rep Med. 2021;2(6):100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Graham BE, Plotkin B, Muglia L, Moore JH, Williams SM. Estimating prevalence of human traits among populations from polygenic risk scores. Hum Genomics. 2021;15(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19(9):581–590. [DOI] [PubMed] [Google Scholar]


