Abstract
Two-component signaling proteins are involved in transducing environmental stimuli into intracellular signals. Information is transmitted through a phosphorylation cascade that consists of a histidine protein kinase and a response regulator protein. Generally, response regulators are made up of a receiver domain and an output domain. Phosphorylation of the receiver domain modulates the activity of the output domain. The mechanisms by which receiver domains control the activities of their respective output domains are unknown. To address this question for the PhoB protein from Escherichia coli, we have employed two separate genetic approaches, deletion analysis and domain swapping. In-frame deletions were generated within the phoB gene, and the phenotypes of the mutants were analyzed. The output domain, by itself, retained significant ability to activate transcription of the phoA gene. However, another deletion mutant that contained the C-terminal α-helix of the receiver domain (α5) in addition to the entire output domain was unable to activate transcription of phoA. This result suggests that the α5 helix of the receiver domain interacts with and inhibits the output domain. We also constructed two chimeric proteins that join various parts of the chemotaxis response regulator, CheY, to PhoB. A chimera that joins the N-terminal ∼85% of CheY's receiver domain to the β5-α5 loop of PhoB's receiver domain displayed phosphorylation-dependent activity. The results from both sets of experiments suggest that the regulation of PhoB involves the phosphorylation-mediated modulation of inhibitory contacts between the α5 helix of its unphosphorylated receiver domain and its output domain.
Two-component signal transduction proteins are commonly employed by bacteria to respond to changes in environmental conditions (11, 32, 38). In their simplest forms, two-component systems consist of histidine kinases and response regulators. Histidine kinases transduce environmental cues into intracellular signals by interacting with and modifying response regulator proteins. Signal processing involves the transfer of phosphate between a histidine residue within the kinase and an aspartate residue located within the response regulator.
A large family of response regulator proteins has been identified through genetic and genomic analyses of many bacteria (32, 42). These proteins generally consist of multiple domains and are characterized by a conserved receiver domain, which contains the site of aspartyl phosphorylation, and an output domain, which regulates transcription. Response regulators have been subdivided into families based on their output domains (31, 42). The pattern of conserved residues within the receiver domain defines this superfamily and strongly supports the idea that these domains have a common structure and potentially employ a common mechanism of activation. The three-dimensional structures of several receiver domains have been determined (CheY, NtrC, FixJ, SpoOF, NarL, CheB, and PhoB) (2, 3, 5, 7, 27, 35, 36, 41, 43). In each of these proteins, the receiver domain has a doubly wound α/β topology consisting of a central five-stranded parallel β-sheet (β1 to β5) surrounded by five α-helices (α1 to α5). A prominent feature of the receiver domain is an acidic pocket, which is found at the C-terminal edge of the β-sheet. This pocket contains the phosphoaccepting aspartate residue. The structures of intact multidomain response regulators NarL and CheB have recently been determined (2, 5). Although the structures of all receiver domains are similar, these proteins do not have the same domain-packing arrangements.
The mechanism(s) by which the phosphorylation signal originating within the receiver domain is propagated to the output domain is not known. However, several recent studies of activated receiver domains have demonstrated a common structural change involving the repositioning of a conserved tyrosine or phenylalanine residue in β5 from a solvent-exposed position into a hydrophobic pocket (3, 4, 9, 15). This conserved change leads to slightly different structural alterations in each of the receiver domains studies.
A well-characterized adaptive response in Escherichia coli that employs a two-component signaling pathway is triggered by inorganic phosphate (Pi) limitation (44). The phosphate response permits cells to acquire Pi with high affinity and to utilize alternate phosphorus sources. The genes under phosphate control are positively regulated and are called the Pho regulon. When Pi becomes limiting, transcription is initiated from the promoters of the regulon; for example, the expression of alkaline phosphatase, the product of the phoA gene, is stimulated more than 150-fold (45).
The signaling proteins that operate on the cytoplasmic side of the inner membrane are two-component regulators PhoR and PhoB. PhoR is a histidine kinase that receives environmental input from the high-affinity phosphate transporter (20, 21). When phosphate levels are low, PhoR donates a phosphoryl group to a conserved aspartate residue within response regulator PhoB (18). PhoB is a soluble 229-amino-acid protein that consists of two domains: an ∼125-amino-acid N-terminal receiver domain and an ∼100-amino-acid C-terminal output domain that binds DNA and interacts with the ς70 subunit of RNA polymerase (16, 34). The output domain is a member of the winged-helix-turn-helix family of transcription factors, which is represented by OmpR from E. coli (22, 23). The three-dimensional structure of the output domain of PhoB was recently solved (30). Upon phosphorylation, PhoB forms a dimer and its affinity for target DNA sequences, called pho boxes, is increased, which leads to enhanced levels of transcription (8, 18, 24).
We have recently demonstrated that the receiver domain of PhoB negatively regulates its output domain (6). We have shown that the liberated output domain of PhoB binds to pho box DNA more tightly and activates transcription better than the intact unphosphorylated protein. In this paper, we extend those studies to show that the α5 helix of the receiver domain is involved in the interdomain interactions that negatively control the output domain of PhoB. We also provide data that suggest that the phosphorylation-generated activation signal requires the β5-α5 loop and the α5 helix to be propagated to the output domain.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The E. coli strains and plasmids used in this study are listed in Table 1. Cells were grown in either Luria-Bertani (LB) medium which was supplemented with ampicillin (100 μg/ml) or in modified glucose-morpholinepropanesulfonic acid (MOPS) minimal medium containing 5.0 mM KH2PO4 and ampicillin (100 μg/ml) (25, 28).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F−recA1 endA1 supE44 hsdR17 deoR thi-1 relA1 gyr96 φ80dlacZΔM15 Δ(lacZYA-argF)U169 | 33 |
| DWE1 | phoB23 Kmrleu lacY trp his argG strA ilv metA (or metB) thi recA1 Tetr | 48 |
| PS2001 | ΔcheBcheYcheZ; wild type for the Pho regulon | 1 |
| PS2002 | ΔcheA-cheZ; wild type for the Pho regulon | 1 |
| BL21(DE3) pLysS | dcm ompT hsdS gal λ(DE3) Camr | 39 |
| Plasmids | ||
| pDE1 | Apr; pUC19 carrying a 1.4-kb DNA fragment containing the phoB gene | This work |
| pMP40 | Apr; pDE1 carrying phoB Δ4-122 (BR289, BF646)a | This work |
| pMP7 | Apr; pDE1 carrying phoB Δ130-227 (BR666, BF960) | This work |
| pMP8 | Apr; pDE1 carrying phoB Δ4-110 (BR289, BF610) | This work |
| pMP17 | Apr; pDE1 carrying phoB Δ125-131 (BR655, BF673) | This work |
| pMP48 | Apr; pDE1 carrying phoB Δ4-113 (BR289, BF619) | This work |
| pMP49 | Apr; pDE1 carrying phoB Δ4-116 (BR289, BF628) | This work |
| pMP46 | Apr; pDE1 carrying phoB Δ4-104 (BR289, BF592) | This work |
| pMP44 | Apr; pDE1 carrying phoB Δ4-98 (BR289, BF574) | This work |
| pMP42 | Apr; pDE1 carrying phoB Δ4-92 (BR289, BF556) | This work |
| pMP41 | Apr; pDE1 carrying phoB Δ4-89 (BR289, BF547) | This work |
| pT7-Ch1 | Apr; Ch1 expression under the control of the T7 promoter | This work |
| pT7-Ch3 | Apr; Ch3 expression under the control of the T7 promoter | This work |
| pMLB1120-Ch1 | Apr; Ch1 expression under the control of the lac promoter | This work |
| pMLB1120-Ch3 | Apr; Ch3 expression under the control of the lac promoter | This work |
The primers that were used to generate the deletions are shown in parentheses (see Table 2).
Plasmid pDE1 was constructed by inserting a 1.37-kbp PCR fragment containing the phoB locus into the multiple cloning site of pUC19 (47). The PCR product was generated by amplifying chromosomal DNA using primers BF1 and RR1372 (Table 2).
TABLE 2.
List of oligonucleotides
| Oligonucleotide | Sequencea (5′–3′) |
|---|---|
| For construction of pDE1 | |
| BF1 | CCAGTCAAGAAAAGCCTGAT |
| RR1372 | CCGTGGTCAGCACCACCGCG |
| For deletion mutagenesis | |
| BR289 | GCGGGATCCTCTCGCCATGATTTGCCCTGTTG |
| BR666 | GCGGGATCCTTCCACCGCCATTGGCGAAATAC |
| BR655 | GCGGGATCCCGAAATACGGCGCATTACCGCTTT |
| BF547 | CGCGGATCCGATCGCGTGCGCGGCCTTGAAA |
| BF556 | CGCGGATCCCGCGGCCTTGAAACCGGCGCGG |
| BF574 | CGCGGATCCGCGGATGACTATATCACCAAGC |
| BF592 | CGCGGATCCAAGCCGTTTTCGCCGAAGGAGC |
| BF610 | GCGGGATCCGAGCTGGTGGCGCGAATCAAAG |
| BF619 | CGCGGATCCGCGCGAATCAAAGCGGTAATGC |
| BF628 | CGCGGATCCAAAGCGGTAATGCGCCGTATTT |
| BF646 | GCGGGATCCATTTCGCCAATGGCGGTGGAAG |
| BF673 | GCGGGATCCATTGAGATGCAGGGATTAAGTC |
| BF960 | GCGGGATCCCGCTTTTAACGCCTTGCTCATC |
| For chimera construction | |
| A | GCCGCTAGCCATATGGCGGATAAAGAGCTT |
| B | GCCGCCTAGGATCCAAGGCGTTAAAAGCGGG |
| C for Ch1 | GAACGGTTTTACGACATAACC |
| C for Ch3 | CAGTTTCTCAAAGATTTTGTT |
| D for Ch1 | GGTTATGTCGTAAAACCGTTCTCGCCGAAGGCGCTGGTGGCG |
| D for Ch3 | AACAAAATCTTTGAGAAACTGTCGCCAATGGCGGTGGAAGAG |
The NdeI restriction site is underlined, and the BamHI restriction sites are in boldface.
Deletion mutagenesis.
Inverse PCR was performed on plasmid pDE1 to generate all deletions used in this study. All synthetic oligonucleotides were purchased from Life Technologies (Rockville, Md.) and are listed in Table 2. The primers were designed to flank the region to be deleted, and each contained a BamHI site so that the linear PCR product could be ligated after restriction digestion with BamHI. The digested fragments were joined using T4 ligase to create each of the pMP plasmids. The design of each of the deletions was based on the predicted secondary structure of PhoB from the crystal structures of the homologous proteins CheY and OmpR (22, 37). Mutagenized plasmid DNA was transformed into E. coli DH5α for plasmid maintenance and into E. coli DWE1 for phenotypic evaluation. Deletion mutagenesis was verified by DNA sequencing using a LI-COR (Lincoln, Nebr.) 4000L automated sequencer. The sequence information was compared to the phoB sequence previously published (19).
Alkaline phosphatase assays.
Alkaline phosphatase assays were performed as described previously (48).
Construction of chimeric genes.
The cheY/phoB chimeric genes were constructed using a “gene SOEing” process previously described (12). Gene fragments were generated from cheY and phoB, which contained an overlapping 15-bp sequence. For each construct, four primers were used (Table 2), an A::C primer pair for CheY and a D::B primer pair for PhoB. The 15-bp complementary region was created by using a 30-mer for the D primer that contained at its 5′ end 15 bases complementary to the C primer for CheY and 15 residues complementary to the PhoB coding sequence at its 3′ end. The C and D primers specify the location of the splice site between CheY and PhoB. The A primer contains an internal NdeI site that provides the start codon for cheY, and the B primer contains an internal BamHI site downstream of the phoB gene termination codon. The A::C and D::B amplification products were separated from primers by agarose gel electrophoresis and were purified using the Qiaex II resin from Qiagen Inc. (Valencia, Calif.). They were then combined, denatured, and reannealed under PCR conditions. The overlaps were extended with Taq polymerase, and the new chimeric gene was further amplified using the A::B primer pair. This amplification product was purified following agarose gel electrophoresis using Qiaex II resin, was digested with NdeI and BamHI, and was cloned into expression vector pJES307 (26) to give plasmids pT7-Ch1 and pT7-Ch3.
To make pMLB1120-Ch1 and pMLB1120-Ch3, the chimeric genes were excised from the pT7 constructs with XbaI and cloned into the single XbaI site of pUC18 to give pUC-Ch1 and pUC-Ch3, respectively. The chimeric genes were then excised from these plasmids with EcoRI and HindIII and cloned into the respective sites of pMLB1120.215 (37).
Overexpression, purification, and analysis of chimeric proteins.
The expression and purification of insoluble proteins were performed as previously described (26). The phosphotransfer assays were conducted by incubating [32P]phospho-CheA in the presence of various phosphoacceptors. CheA was phosphorylated at room temperature for 15 min in a 20-μl reaction mixture containing 10 μM CheA, 10 mM MgCl2, 25 mM Tris-HCl, pH 7.2, and 0.2 μM [γ-32P]ATP. Phosphotransfer reactions were initiated by adding 2.5 μl of the CheA phosphorylation reaction mixture to a tube containing 6 μl of a phosphoacceptor. These reaction mixtures were incubated for 2 min at room temperature, and the reactions were stopped by the addition of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer containing 25 mM EDTA. The final concentrations of the phosphoacceptors in the phosphotransfer reaction mixtures were as follows: CheY, 12 μM; PhoB, 15 μM; Ch1, 2 μM; Ch3, 12 μM. Samples were separated on an SDS–15% polyacrylamide gel, after which the gel was dried and exposed to X-ray film for autoradiography.
Western immunoblotting.
E. coli DWE1 cells containing various plasmids were grown overnight in LB media supplemented with ampicillin (100 μg/ml). Equivalent amounts of cellular protein, adjusted according to the optical densities of the overnight cultures, were separated on an SDS–15% PAGE gel and transferred onto a nitrocellulose membrane using the Mini Trans-Blot transfer cell (Bio-Rad) according to the manufacturer's instructions. The membrane was blocked overnight at 25°C in TBS (20 mM Tris-HCl [pH 7.5], 500 mM NaCl)–3% gelatin–5% (wt/vol) nonfat dried milk and then incubated with anti-PhoB rabbit polyclonal antiserum for 2 h. Proteins were detected using the Bio-Rad Immun-Star chemiluminescent protein detection system as indicated by the manufacturer. The membranes were then wrapped in plastic wrap and exposed to X-ray film and then developed in an automated film processor.
RESULTS
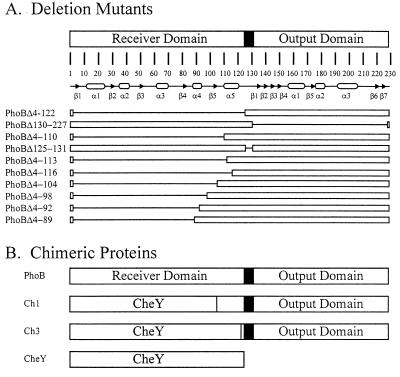
To better understand the mechanism of activation of the PhoB protein, several in-frame deletion mutations were introduced into the phoB gene using inverse PCR and the phenotypes of these mutants were examined. Each mutant was tested for the ability to activate transcription of the phoA gene. Initially, four deletion mutations were created (Fig. 1A). The corresponding proteins are designated PhoBΔ4-122, PhoBΔ130-227, PhoBΔ4-110, and PhoBΔ125-131 and are encoded on plasmids pMP40, pMP7, pMP8, and pMP17, respectively. The nomenclature for each of the mutant proteins indicates which residues of PhoB have been deleted. For example, PhoBΔ4-122 consists of the first three amino acids of PhoB, followed by Gly-Ser (from the introduction of a BamHI site at the point in the plasmid corresponding to the site of the deletion; see Materials and Methods), followed by residues 123 to 229. PhoBΔ4-122 and PhoBΔ130-227 lack the receiver and output domains, respectively. PhoBΔ4-110 lacks 80% of the receiver domain but retains the α5 helix of the receiver domain plus the entire output domain. PhoBΔ125-131 retains both domains but is missing the predicted interdomain linker.
FIG. 1.
Structures of the deletion and chimeric proteins used in this study. (A) The domain structure of PhoB is represented as two white rectangles separated by a black linker region. The amino acid numbers are shown above the map of the secondary structures of PhoB (arrows, β-strands; ovals, α-helices) (30, 35). For the 10 deletion proteins the white bars represent the protein segments that remain whereas the lines correspond to the deleted segments. The name of each protein designates which amino acid residues have been deleted from PhoB. For example, PhoBΔ4-122 contains residues 1 to 3 of PhoB, followed by Gly-Ser (from an inserted BamHI site in the coding sequence), followed by residues 123 to 229. (B) Schematic representation of the chimeric proteins used in this study. Ch1 joins the N-terminal 108 residues of CheY to the C-terminal 125 residues of PhoB. Ch3 joins the N-terminal 127 amino acid residues of CheY to the C-terminal 106 residues of PhoB.
The induction of alkaline phosphatase by mutant proteins.
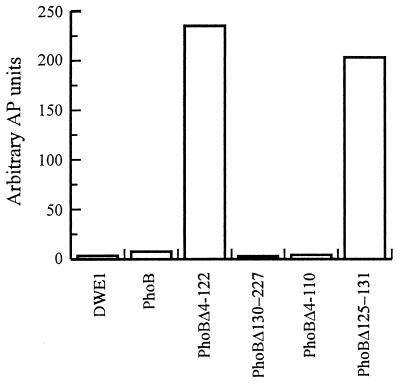
Plasmids expressing PhoB or the four deletion derivatives were introduced into the phoB mutant strain, DWE1, and the levels of alkaline phosphatase produced by these strains were determined following growth in phosphate-sufficient media (Fig. 2). DWE1 contains the phoB23 allele, which has a transition mutation in the ninth codon of the phoB gene that results in the conversion of a glutamate residue to a lysine (46). Under these growth conditions the Pho regulon remains uninduced in wild-type cells and, as expected, the strain harboring pDE1 (which contains the full-length phoB gene) produced very low levels of alkaline phosphatase. In contrast, the strain producing the PhoBΔ4-122 protein, consisting of only the output domain, induced alkaline phosphatase to high levels. These results confirm our previous findings that the unphosphorylated receiver domain of PhoB inhibits the activity of the output domain (6). As anticipated, PhoBΔ130-227, consisting of only the receiver domain, was unable to activate transcription. Expression of the PhoBΔ125-131 protein also induced the synthesis of alkaline phosphatase, but to slightly lower levels than did expression of PhoBΔ4-122. This observation suggests that in PhoB a functional interdomain linker is required for the receiver domain to inhibit the output domain. This interdomain linker is probably required to correctly position the two domains relative to each other.
FIG. 2.
The transcriptional activation activities of PhoB deletion proteins were determined by measuring the amounts of alkaline phosphatase (AP) synthesized in phosphate-sufficient medium. E. coli DWE1 cells were transformed with plasmids encoding PhoB deletion proteins. The genes for PhoB, PhoBΔ4-122, PhoBΔ130-227, PhoBΔ4-110, and PhoBΔ125-131 were contained on plasmids pDE1, pMP40, pMP7, pMP8, and pMP40, respectively. The cells were grown overnight in LB medium containing ampicillin, and alkaline phosphatase assays were performed.
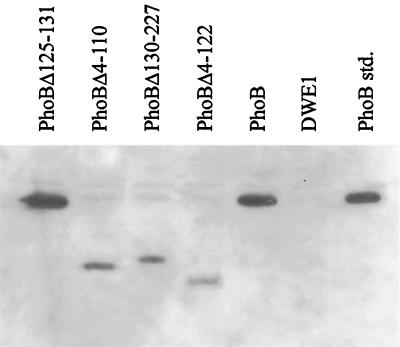
Surprisingly, the expression of the PhoBΔ4-110 protein does not induce alkaline phosphatase. This deletion protein has a sequence identical to that of PhoBΔ4-122 except that it also contains an additional 12 amino acid residues constituting the α5 helix of the receiver domain. Three potential explanations for the lack of activity in the strain expressing the PhoBΔ4-110 protein are that the protein was not produced (or was rapidly degraded), that intragenic complementation occurred, or that the amino acid residues encoding the α5 helix interacted with the output domain to inhibit its ability to stimulate transcription. To investigate the first possibility, Western immunoblotting was performed. Strains were grown overnight in phosphate-sufficient media and prepared for SDS-PAGE and subsequent transfer onto nitrocellulose. As can be seen in Fig. 3, all of the proteins were expressed, although not to equivalent levels. Since the important comparison of activities is between PhoBΔ4-110 and PhoBΔ4-122, it should be noted that these two proteins were produced in similar amounts. The likelihood of intragenic complementation is small because of the low level of expression of the phoB23 gene compared to that of the allele carried by the plasmid. Taken together, these results show that in PhoBΔ4-110 the presence of the amino acids forming the α5 helix of the receiver domain inhibits the activity of the output domain.
FIG. 3.
Western immunoblot analysis of PhoB, PhoBΔ4-122, PhoBΔ130-227, PhoBΔ4-110, and PhoBΔ125-131. E. coli DWE1 cells were transformed with plasmids encoding PhoB and the four deletion mutants. The cells were grown overnight in LB medium containing ampicillin, were collected by centrifugation, and were lysed in SDS-PAGE sample buffer. Equal amounts of cellular extracts were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and detected with a chemiluminescence detection system using rabbit anti-PhoB polyclonal sera. Purified PhoB was run as a standard (48).
Deletion analysis of the α5 helix.
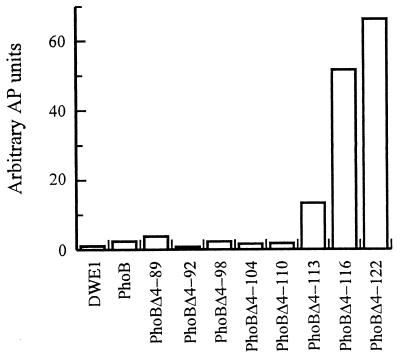
An additional series of deletions was created to better define the amount of the receiver domain that was required to silence the output domain. Several of these deletions removed residues from the α5 helix, whereas other deletions extended the amount of PhoB from the α5 helix. PhoBΔ4-113 and -Δ4-116 lacked 3 and 6 amino acid residues, respectively, whereas PhoBΔ4-104, -Δ4-98, -Δ4-92, and -Δ4-89 contained an additional 6, 12, 18, and 21 residues, respectively (Fig. 1A). Plasmids encoding these deletion mutations were introduced into DWE1 cells, and their phenotypes were determined. There was no alkaline phosphatase induction in DWE1 cells expressing mutant proteins that extended the α5 helix (Fig. 4). In contrast, as the α5 helix was deleted, inhibition of the output domain was decreased and the proteins behaved similarly to PhoBΔ4-122. These results demonstrate that the minimum amount of the receiver domain that is required to silence the output domain is the entire α5 helix. These observations raise the possibility that in full-length PhoB the α5 helix of the receiver domain interacts with the output domain in a specific manner. It is the modulation of this interaction through conformational changes that is triggered by phosphorylation of Asp53, which controls the activity of the output domain.
FIG. 4.
The transcriptional activation activities of a series of PhoB deletion proteins localize the inhibitory region of the receiver domain to the α5 helix. E. coli DWE1 cells were transformed with plasmids encoding PhoB deletion proteins. The genes for PhoB, PhoBΔ4-89, PhoBΔ4-92, PhoBΔ4-98, PhoBΔ4-104, PhoBΔ4-110, PhoBΔ4-113, PhoBΔ4-116, and PhoBΔ4-122 were contained on plasmids pDE1, pMP41, pMP42, pMP44, pMP46, pMP8, pMP48, pMP49, and pMP40, respectively. The cells were grown overnight in glucose-MOPS minimal medium containing 5.0 mM KH2PO4, and alkaline phosphatase (AP) assays were performed.
Design and construction of chimeric proteins.
To more fully understand the role of the α5 helix in controlling the activity of the output domain of PhoB, two chimeric proteins in which portions of CheY were swapped for homologous regions of PhoB were constructed. The first chimera, Ch1, has a splice site at the end of the β5 β-strand at the conserved Lys-Pro-Phe triplet (residues 105 to 107 of PhoB) and maintains the β5-α5 loop and the entire α5 helix from PhoB (Fig. 1B). The design of this construct is based on the idea that regions of amino acid identity between CheY and PhoB may result from structural or functional constraints and that it may be necessary to maintain these identities to generate a functional protein. The second chimera, Ch3, has a splice site downstream of the α5 helix and substitutes the entire response regulator domain of CheY for that of PhoB. The chimeric genes were created by extending engineered overlaps in PCR products that contain the cheY and phoB gene fragments (12). The analysis of these proteins was designed to focus on the role of the α5 helix in propagating an input signal into an appropriate output response. Phosphorylation of the receiver domain by the CheA protein provided the input, whereas the regulated production of alkaline phosphatase was the output.
Phosphorylation with phospho-CheA.
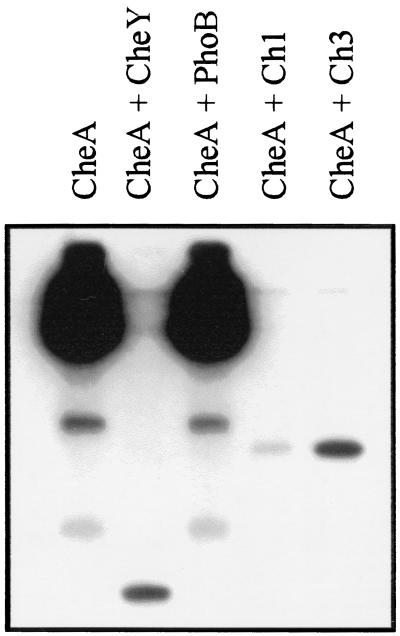
To determine whether the addition of an output domain to CheY would prevent proper interactions with CheA, we conducted phosphotransfer assays. The two chimeric genes were cloned into a T7 expression vector from which high levels of protein expression were obtained. Since most of the overexpressed protein was insoluble, the chimeric proteins were purified from inclusion bodies. Phosphotransfer reactions were initiated by adding an aliquot of each protein to a sample of [32P]phospho-CheA. The reactions were analyzed by SDS-PAGE and autoradiography (Fig. 5). The reaction mixture containing only CheA showed, in addition to the full-length phosphoprotein, two low-abundance, faster-migrating bands. These bands were most likely proteolytic fragments of CheA that were either capable of autophosphorylation or substrates for transphosphorylation from CheA and could also serve as phosphodonors since they and the full-length band disappeared upon incubation with CheY. Dephosphorylation of phospho-CheA and the subsequent phosphotransfer to the CheY moiety were observed with both of the chimeric proteins. However, no phosphotransfer or phospho-CheA dephosphorylation was observed when phospho-CheA was incubated with PhoB. These results show that both chimeric proteins are functionally active in receiving input from CheA. Note that the relative intensities of the phospho-Ch1 and phospho-Ch3 bands in Fig. 5 reflect the amounts of each protein in the phosphotransfer reactions and not the stability of the phosphorylated proteins.
FIG. 5.
Phosphotransfer reactions between CheA and chimeric proteins Ch1 and Ch3. CheA was phosphorylated with [γ-32P]ATP. Aliquots of [γ-32P]phospho-CheA were mixed with the indicated phosphoacceptors, incubated for 2 min at room temperature, and analyzed by SDS-PAGE and autoradiography. The final concentrations of the phosphoacceptors in the phosphotransfer reactions were as follows: CheY, 12 μM; PhoB, 15 μM; Ch1, 2 μM; Ch3, 12 μM.
Activation by the chemotaxis signaling pathway.
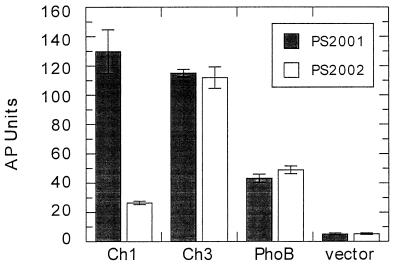
To test whether the phosphorylation signal within the receiver domains of Ch1 and Ch3 could be transmitted to their output domains, we examined the ability of Ch1 and Ch3 to induce the expression of the chromosome-located alkaline phosphatase gene when presented with a chemotactic stimulus. The two chimeric constructs were subcloned into a regulatable expression vector and transformed into the appropriate tester strains. These strains were selected to assay the activation of CheY by the methylated chemotaxis protein-CheA pathway (1). E. coli PS2001 constitutively activates CheA and produces high levels of phospho-CheY when CheY is encoded on a plasmid, whereas E. coli PS2002 has a deletion of most chemotaxis genes and cannot activate CheY when it is encoded on a plasmid. By comparing the levels of alkaline phosphatase produced in the strains expressing either Ch1 or Ch3 it is possible to determine their levels of phosphorylation-dependent activation.
The PS2001 strain expressing Ch1 showed approximately a fourfold increase in the level of alkaline phosphatase compared to the PS2002 strain (Fig. 6). The two tester strains expressing Ch3 produced equivalent levels of alkaline phosphatase, indicating that there was no phosphorylation-dependent regulation of output function. It is important to note that the levels of alkaline phosphatase produced in the strains expressing Ch3 were elevated compared to that produced in the PS2002 strain expressing Ch1. These results are consistent with those obtained above for the strain expressing PhoBΔ125-131 and suggest that regulation of the output domain by the receiver domain involves inhibition of the output domain by a correctly matched and correctly positioned receiver domain. The α5 helix in the receiver domain of Ch3 originates from CheY and is unable to silence the activity of the output domain from PhoB. PS2001 and PS2002 strains expressing native PhoB also did not display differential expression of alkaline phosphatase, thereby showing the specificity of the phosphorylation pathway from CheA to the CheY receiver structure. Taken together, these results show that the signal that was generated through phosphorylation of the receiver domain in Ch1 was propagated to the output domain whereas, in the Ch3 protein, it was not. From our results, signal propagation to the output domain of PhoB requires the β5-α5 loop and the α5 helix of the receiver domain.
FIG. 6.
Alkaline phosphatase (AP) assay to measure the output activities of chimeric proteins Ch1 and Ch3. The genes for Ch1, Ch3, and PhoB were cloned into vector pMLB1120.215 and transformed into either PS2001 or PS2002. The data show means and standard deviations of assays performed in triplicate.
DISCUSSION
This study reports experiments on the mechanism by which the receiver domain of response regulator PhoB controls the activity of its output domain. We propose that the α5 helix of the receiver domain participates in interdomain interactions that control the activity of the output domain and that these interactions are modulated through phosphorylation of the receiver domain. This proposal is based on two different lines of experimental evidence. The first line is based on results from a deletion analysis of the PhoB protein. A deletion of the entire receiver domain generated a constitutively active protein (PhoBΔ4-122). The addition of the α5 helix from the receiver domain to the output domain (found in PhoBΔ4-110) silenced the activity of the output domain. Our experiments cannot distinguish whether this silencing results from blocked DNA-binding and/or RNA polymerase interactions or by locking the output domain in an inactive conformation. If PhoB is like the NarL and CheB response regulators, then inhibition is achieved by blocking the active site of its output domain (2, 5).
The second line of investigation involved domain swapping experiments using the CheY protein from Salmonella enterica serovar Typhimurium and the PhoB protein from E. coli. Two chimeric CheY/PhoB proteins in which either 85% (Ch1) or 98% (Ch3) of the receiver domain of PhoB was replaced by the corresponding regions of CheY were constructed. Ch1 maintains the β5-α5 loop and the α5 helix from PhoB's receiver domain, whereas Ch3 does not. These proteins were used to test whether an input signal could be transduced into an appropriate output response. Of the two chimeras examined, only the Ch1 protein transduced the input signal into the appropriate response. This result supports the proposal that the β5-α5 loop and the α5 helix from the receiver domain of PhoB are required to propagate the phosphorylation-triggered signal from the receiver domain to the output domain. The Ch3 protein was constitutively active, consistent with the idea that, in the unphosphorylated receiver, the α5 helix directly participates in interdomain interactions that silence the output domain. By the incorporation of the α5 helix from CheY into Ch3, the interdomain interactions are abrogated and the inhibition imposed on the output domain by this helix does not occur, which leaves the output domain active.
Expression of the PhoBΔ125-131 protein in cells grown in high-phosphate media resulted in production of alkaline phosphatase. PhoBΔ125-131 consists of the receiver and output domains but is missing the interdomain linker region. We propose that the interdomain linker of PhoB is important for the correct positioning of the α5 helix relative to the output domain. It has previously been shown that the linker region of OmpR is essential in relaying conformational changes between its two domains (14, 40).
The levels of alkaline phosphatase that were induced by the chimeric constructs were only one-fifth of those routinely observed in our laboratory when wild-type cells are grown in phosphate-limiting media (data not shown). Part of this difference in expression levels may be due to the lack of a positive regulatory circuit in the tester cells in which phospho-PhoB induces its own expression (19). In the experiments reported in this study, the expression of the chimeric constructs was under the control of a lac promoter and the levels of protein should remain constant upon induction. In addition, the increase of expression between uninduced cells, PS2002(pCh1), and induced cells, PS2001(pCh1), was only four- or fivefold and was much lower than that observed for wild-type cells grown in phosphate-sufficient and phosphate-limiting media (45). A potential explanation for these results is that the α5 helix of the receiver domain is not the only component involved in interdomain interactions and that other parts of PhoB's receiver domain (perhaps other helices and/or loops) are necessary for complete induction. Another difference could be in the half-lives of the phosphorylated proteins; phospho-PhoB has a half-life of approximately 15 min, whereas phospho-CheY has a half-life of ∼15 s (10, 24). This difference may alter the relative amounts of activated proteins within the cells and influence the amount of induction.
Recent studies on the activation of the NtrC protein have suggested that the α4 helix of its receiver domain is involved in an interdomain interaction that propagates the phosphorylation-induced signal (13, 15, 17, 29). In FixJ, the propagation signal is transmitted through the α4-β5 surface (3). The interdomain interface for the NarL protein, which must be modulated for activation to occur, involves the α2-β3, α3-β4, and α4-β5 loops as well as the end of α5 (2); In CheB, it is the α4-β5-α5 surface that constitutes the interdomain interface (5). Taken together with the work presented in this study, these results show that different response regulators employ different molecular surfaces for their interdomain interactions and imply that slightly different signal propagation strategies may be used to control the activities of different output domains.
We propose that response regulator proteins are composed of three functional units: a phosphorylation-triggered switch, a relay, and an output domain. The switch receives input either from a cognate kinase or from a small-molecule phosphodonor (25). This information is transmitted to the relay structure through a conserved conformational change that involves the repositioning of the conserved tyrosine or phenylalanine residue in β5 from a solvent-exposed position into a hydrophobic pocket (3, 4, 9). We propose that this conformational change is at least somewhat conserved because the CheY moiety of Ch1 functioned with the relay unit from PhoB. The relay interprets the conformational change and propagates this information to the output domain.
ACKNOWLEDGMENTS
We thank Mike Surette for the kind gift of PS2001 and PS2002. We also thank members of the McCleary laboratory for helpful comments in the preparation of this report.
This work was supported by Public Health Service grant GM53981 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Alon U, Camarena L, Surette M G, Arcas B A, Liu Y, Leibler S, Stock J B. Response regulator output in bacterial chemotaxis. EMBO J. 1998;17:4238–4248. doi: 10.1093/emboj/17.15.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 3.Birck C, Mourey L, Gouet P, Fabry B, Schumacker J, Rousseau P, Kahn D, Samama J-P. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure. 1999;7:1505–1515. doi: 10.1016/s0969-2126(00)88341-0. [DOI] [PubMed] [Google Scholar]
- 4.Cho H S, Lee S Y, Yan D, Pan X, Parkinson J S, Kustu S, Wemmer D E, Pelton J G. NMR structure of activated CheY. J Mol Biol. 2000;297:543–551. doi: 10.1006/jmbi.2000.3595. [DOI] [PubMed] [Google Scholar]
- 5.Djordjevic S, Goudreau P N, Xu Q, Stock A M, West A H. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc Natl Acad Sci USA. 1998;95:1381–1386. doi: 10.1073/pnas.95.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellison D W, McCleary W R. The unphosphorylated receiver domain of PhoB silences the activity of its output domain. J Bacteriol. 2000;182:6592–6597. doi: 10.1128/jb.182.23.6592-6597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feher V A, Zapf J W, Hoch J A, Dahlquist F W, Whiteley J M, Cavanagh J. 1H, 15N, and 13C backbone chemical shift assignments, secondary structure, and magnesium-binding characteristics of the Bacillus subtilis response regulator, SpoOF, determined by heteronuclear high-resolution NMR. Protein Sci. 1995;4:1801–1814. doi: 10.1002/pro.5560040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiedler U, Weiss V. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induces dimerization of the receiver modules. EMBO J. 1995;14:3696–3705. doi: 10.1002/j.1460-2075.1995.tb00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halkides C J, McEvoy M M, Casper E, Matsumura P, Volz K, Dahlquist F W. The 1.9 A resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY. Biochemistry. 2000;39:5280–5286. doi: 10.1021/bi9925524. [DOI] [PubMed] [Google Scholar]
- 10.Hess J F, Oosawa K, Kaplan N, Simon M I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 11.Hoch J A, Silhavy T J. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 12.Horton R M, Cai Z, Ho S N, Pease L R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 13.Hwang I, Thorgeirsson T, Lee J, Kustu S, Shin Y K. Physical evidence for a phosphorylation-dependent conformational change in the enhancer-binding protein NtrC. Proc Natl Acad Sci USA. 1999;96:4880–4885. doi: 10.1073/pnas.96.9.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenney L J, Bauer M D, Silhavy T J. Phosphorylation-dependent conformational changes in OmpR, an osmoregulatory DNA-binding protein of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:8866–8870. doi: 10.1073/pnas.92.19.8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kern D, Volkman B F, Luginbuhl P, Nohaile M J, Kustu S, Wemmer D E. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature (London) 1999;402:894–898. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- 16.Kumar A, Grimes B, Fujita N, Makino K, Malloch R A, Hayward R S, Ishihama A. Role of the ς70 subunit of Escherichia coli RNA polymerase in transcription activation. J Mol Biol. 1994;235:405–413. doi: 10.1006/jmbi.1994.1001. [DOI] [PubMed] [Google Scholar]
- 17.Lee J, Owens J T, Hwang I, Meares C, Kustu S. Phosphorylation-induced signal propagation in the response regulator NtrC. J Bacteriol. 2000;182:5188–5195. doi: 10.1128/jb.182.18.5188-5195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol. 1989;210:551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- 19.Makino K, Shinagawa H, Amemura M, Nakata A. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J Mol Biol. 1986;190:37–44. doi: 10.1016/0022-2836(86)90073-2. [DOI] [PubMed] [Google Scholar]
- 20.Makino K, Shinagawa H, Amemura M, Nakata A. Nucleotide sequence of the phoR gene, a regulatory gene for the phosphate regulon of Escherichia coli. J Mol Biol. 1986;192:549–556. doi: 10.1016/0022-2836(86)90275-5. [DOI] [PubMed] [Google Scholar]
- 21.Makino K, Shinagawa H, Nakata A. Regulation of the phosphate regulon of Escherichia coli K-12: regulation and role of the regulatory gene phoR. J Mol Biol. 1985;184:231–240. doi: 10.1016/0022-2836(85)90376-6. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Hackert E, Stock A M. The DNA-binding domain of OmpR: crystal structure of a winged helix transcription factor. Structure. 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Hackert E, Stock A M. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol. 1997;269:301–312. doi: 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- 24.McCleary W R. The activation of PhoB by acetylphosphate. Mol Microbiol. 1996;20:1155–1163. doi: 10.1111/j.1365-2958.1996.tb02636.x. [DOI] [PubMed] [Google Scholar]
- 25.McCleary W R, Stock J B. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 26.McCleary W R, Zusman D R. Purification and characterization of the Myxococcus xanthus FrzE protein shows that it has autophosphorylation activity. J Bacteriol. 1990;172:6661–6668. doi: 10.1128/jb.172.12.6661-6668.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moy F J, Lowry D F, Matsumura P, Dahlquist F W, Krywko J E, Domaille P J. Assignments, secondary structure, global fold, and dynamics of chemotaxis Y protein using three- and four-dimensional heteronuclear (13C, 15N) NMR spectroscopy. Biochemistry. 1994;33:10731–10742. doi: 10.1021/bi00201a022. [DOI] [PubMed] [Google Scholar]
- 28.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nohaile M, Kern D, Wemmer D, Stedman K, Kustu S. Structural and functional analyses of activating amino acid substitutions in the receiver domain of NtrC: evidence for an activating surface. J Mol Biol. 1997;273:299–316. doi: 10.1006/jmbi.1997.1296. [DOI] [PubMed] [Google Scholar]
- 30.Okamura H, Hanaoka S, Nagadoi A, Makino K, Nishimura Y. Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box. J Mol Biol. 2000;295:1225–1236. doi: 10.1006/jmbi.1999.3379. [DOI] [PubMed] [Google Scholar]
- 31.Pao G M, Saier M H., Jr Response regulators of bacterial signal transduction systems: selective domain shuffling during evolution. J Mol Evol. 1995;40:136–154. doi: 10.1007/BF00167109. [DOI] [PubMed] [Google Scholar]
- 32.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Shinagawa H, Makino K, Amemura M, Nakata A. Structure and function of the regulatory genes for the phosphate regulon in Escherichia coli. In: Torriani-Gorini A, Rothman F G, Silver S, Wright A, Yagil E, editors. Phosphate metabolism and cellular regulation in microorganisms. Washington, D.C.: American Society for Microbiology; 1987. pp. 20–25. [Google Scholar]
- 35.Sola M, Gomis-Ruth F X, Serrano L, Gonzalez A, Coll M. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J Mol Biol. 1999;285:675–687. doi: 10.1006/jmbi.1998.2326. [DOI] [PubMed] [Google Scholar]
- 36.Stock A M, Martinez-Hackert E, Rasmussen B F, West A H, Stock J B, Ringe D, Petsko G A. Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 37.Stock A M, Mottonen J M, Stock J B, Schutt C E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature (London) 1989;337:745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- 38.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and the regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 40.Tran V K, Oropeza R, Kenney L J. A single amino acid substitution in the C terminus of OmpR alters DNA recognition and phosphorylation. J Mol Biol. 2000;299:1257–1270. doi: 10.1006/jmbi.2000.3809. [DOI] [PubMed] [Google Scholar]
- 41.Volkman B F, Nohaile M J, Amy N K, Kustu S, Wemmer D E. Three-dimensional structure of the N-terminal receiver domain of NTRC. Biochemistry. 1995;34:1413–1424. doi: 10.1021/bi00004a036. [DOI] [PubMed] [Google Scholar]
- 42.Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 43.Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 44.Wanner B L. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1357–1381. [Google Scholar]
- 45.Wanner B L, Chang B D. The phoBR operon in Escherichia coli K-12. J Bacteriol. 1987;169:5569–5574. doi: 10.1128/jb.169.12.5569-5574.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamada M, Makino K, Amemura M, Shinagawa H, Nakata A. Regulation of the phosphate regulon of Escherichia coli: analysis of mutant phoB and phoR genes causing different phenotypes. J Bacteriol. 1989;171:5601–5606. doi: 10.1128/jb.171.10.5601-5606.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 48.Zundel C J, Capener D C, McCleary W R. Analysis of the conserved acidic residues in the regulatory domain of PhoB. FEBS Lett. 1998;441:242–246. doi: 10.1016/s0014-5793(98)01556-7. [DOI] [PubMed] [Google Scholar]