Abstract
The CepR-CepI quorum-sensing system has been shown to regulate production of the siderophore ornibactin, extracellular proteases, and N-octanoyl-homoserine-l-lactone (OHL) in Burkholderia cepacia strain K56-2. To examine the effect of cepIR on production of other siderophores, cepR mutants were constructed in strains that produce pyochelin in addition to salicylic acid and ornibactins. Pc715j-R1 (cepR::tp) hyperproduced ornibactin but produced parental levels of pyochelin and salicylic acid, suggesting that CepR is a negative regulator of ornibactin synthesis but not pyochelin or salicylic acid. Pc715j-R1 was also protease deficient and OHL negative. The effects of cepR on ornibactin biosynthetic genes were examined by constructing cepR pvdA-lacZ and cepR pvdD-lacZ mutants and monitoring β-galactosidase activity. There was an increase in expression of pvdA in the cepR mutant compared to the level in its parent strain in both low- and high-iron media during stationary phase. When the outer membrane protein profiles of a cepR mutant and the wild-type strain were compared on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, there did not appear to be any difference in levels of expression of the ornibactin receptor. Experiments with cepI-lacZ and cepR-lacZ transcriptional fusions indicated that cepI was not expressed in the cepR mutant and that cepR acts as a negative regulator of its own expression. By a thin-layer chromatography assay for N-acyl homoserine lactones, OHL and N-hexanoyl-l-homoserine lactone (HHL) were detectable in K56-2 and Pc715j, both wild-type strains. OHL was not detectable and HHL was only weakly detectable in the cepI and cepR mutants. These results suggest that CepR is both a positive and negative transcriptional regulator and that CepR may influence the expression of ornibactin biosynthetic genes in addition to the expression of the cepIR quorum-sensing system.
Burkholderia cepacia is an opportunistic pathogen that primarily infects immunocompromised patients with cystic fibrosis (CF), chronic granulomatous disease, cancer, or burns or patients with indwelling devices (12). B. cepacia infections are a particular cause for concern in CF patients. First, although B. cepacia infections have varied outcomes, a significant number of B. cepacia-infected patients (20 to 35%) experience cepacia syndrome, which often culminates in death (14, 42). Second, there is increasing epidemiological evidence for patient-to-patient transmission (18). Since several epidemics have been shown to be caused by a single lineage, there is a possibility that some strains have an increased capacity for virulence and transmission (41). Last, the inherent multidrug resistance of B. cepacia (27) makes treatment very difficult and eradication almost impossible.
Strains of B. cepacia have recently been divided into a group of five distinct genomovars, referred to as the B. cepacia complex (43). The term genomovar denotes a group of strains with phenotypic similarity but genotypic uniqueness. Of the genomovars (numbered I through V), genomovars II, IV, and V have been renamed Burkholderia multivorans, Burkholderia stabilis, and Burkholderia vietnamiensis, respectively (43, 44). The majority of CF-transmissible or epidemic strains belong to B. cepacia genomovar III (43, 44).
Quorum sensing is a mechanism for regulating virulence factor production that involves the production and detection of signaling molecules (N-acyl homoserine lactones [AHLs]) (for reviews, see references 10, 15, and 28). The luxI gene family encodes an autoinducer synthase responsible for generation of the signaling molecule, which accumulates until a threshold concentration is attained. At this point, luxR-type transcriptional regulators bind the autoinducer and subsequently activate or repress the expression of target virulence genes. We have recently described a quorum-sensing system consisting of the cepI and cepR genes in a B. cepacia genomovar III strain (17). An N-acyl homoserine lactone (HSL) was purified from wild-type but not cepI mutant culture supernatants and identified as N-octanoyl-l-HSL (OHL). The initial characterization of cepI and cepR mutants revealed a lack of protease and OHL production, suggesting that protease and OHL production are positively regulated, and increased production of the siderophore ornibactin, suggesting that ornibactin is negatively regulated (17). A 20-bp lux box-like sequence that partially overlaps the −35 region of the putative cepI promoter was identified, suggesting that CepR binds to the cepI promoter to activate cepI expression (17). The majority of quorum-sensing systems are known to act as positive activators of target genes, although it is now recognized that LuxR-type proteins can act as transcriptional repressors (6, 45, 46). CepR appears to function as both a positive and a negative regulator of B. cepacia genes.
B. cepacia produces at least four types of siderophores: salicylic acid, pyochelin, cepabactin, and ornibactins (21, 22, 35, 38, 40), with salicylic acid and ornibactins being predominant in clinical isolates (3). Ornibactin-mediated iron acquisition has been shown to be important in both chronic and acute models of respiratory infection (36, 37). Pyochelin production was associated with 62% of clinical isolates, and its production correlated with severe pulmonary disease in CF patients (3, 35). Cepabactin is produced by few clinical isolates (11%) (3). Strain K56-2 produces ornibactins, salicylic acid, and negligible amounts of pyochelin (3). K56-2 cepR or cepI mutations resulted in hyperproduction of ornibactin but not salicylic acid.
The objectives of the present study were (i) to determine the role of cepR in the regulation of other B. cepacia siderophores and (ii) to further study the mechanisms of positive and negative regulation of cepI and cepR target genes in B. cepacia. Since the protease gene(s) has not been cloned and characterized, we have examined the role of cepR in the regulation of ornibactin biosynthesis and uptake genes and in the regulation of the cepI and cepR quorum-sensing genes.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. For genetic manipulations, Escherichia coli DH5α and B. cepacia K56-2 were grown at 37°C in Luria-Bertani (LB) broth (Life Technologies) or Bacto-Terrific broth (1.2% tryptone, 2.4% yeast extract, 0.4% glycerol, 17 mM KH2PO4, 72 mM K2HPO4) or on 1.5% LB agar plates. The following concentrations of antibiotics were used when necessary: 100 μg of ampicillin, 15 μg of tetracycline, 20 μg of gentamicin, and 1.5 mg of trimethoprim per ml for E. coli and 300 μg of tetracycline and 100 μg of trimethoprim per ml for B. cepacia. For ornibactin production and protease and Chrome Azurol S (CAS) assays, cultures were grown in succinate medium supplemented with ornithine (10 mM) (23) at 37°C. For pyochelin and salicylic acid assays, cultures were grown in deferrated CAA medium (35) at 37°C. For β-galactosidase assays of cepI-lacZ or cepR-lacZ fusions, cultures were grown in tryptic soy broth medium at 37°C. For β-galactosidase assays of pvdA-lacZ or pvdD-lacZ fusions, cultures were grown in TSBD-C medium (low iron) or TSBD-C supplemented with 5 μM FeCl3 (37). All medium components were products of Difco (Detroit, Mich.) unless otherwise stated.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | φ80dlacZΔM15 (lacZYA-argF) recA1 endA gyrA96 thi-1 hsdR17 supE44 relA1 deoR U169 | Life Technologies |
| SM10 | Mobilizing strain, RP4 integrated in chromosome, Kmr | 34 |
| HB101 | supE44 hsdS20 (rB mB) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 29 |
| B. cepacia | ||
| K56-2 | CF respiratory isolate, genomovar III, BCESM+cblA+ | 17, 19 |
| K56-R2 | cepR::Tn5-OT182 derivative of K56-2, Tcr | 17 |
| K56-I2 | cepI::tp derivative of K56-2, Tpr | 17 |
| Pc224c | CF respiratory isolate (Cleveland, Ohio) | 3 |
| Pc224c-R1 | cepR::tp derivative of Pc224c, Tpr | This study |
| Pc715j | CF respiratory isolate, genomovar III | 3, 20 |
| Pc715j-R1 | cepR::tp derivative of Pc715j, Tpr | This study |
| ATCC 17759 | Soil isolate, genomovar I | 19 |
| ATCC 25416 | Onion isolate, genomovar I | 19 |
| P109 | CF respiratory isolate (Seattle, Wash.) | 3 |
| Pc22-20 | Plant isolate | 3 |
| 34192 | CF respiratory isolate (Calgary, Alberta, Canada) | 3 |
| 34930 | CF respiratory isolate (Calgary) | 3 |
| T10 | pvdD::Tn5OT182 derivative of K56-2, Tcr | 37 |
| T10-R10 | pvdD::Tn5OT182, cepR::tp Tcr Tpr | This study |
| I117 | pvdA::Tn5OT182 derivative of K56-2, Tcr | 37 |
| I117-R21 | pvdA::Tn5OT182, cepR::tp Tcr Tpr | This study |
| A. tumefaciens A136 | Ti plasmidless host | C. Fuqua |
| Plasmids | ||
| pNOT19 | Modified pUC19 cloning vector, Apr | 30 |
| pUCP28T | Broad-host-range vector, IncP OriT, pRO1600 ori, Tpr | 31 |
| p34E-Tp | Source of trimethoprim cassette, Tpr | 5 |
| pRK2013 | ColE1 Tra (RK2)+ Kmr | 7 |
| pEX18Tc | Suicide vector, sacB Tcr | 13 |
| pUCP26 | Broad-host-range vector, IncP pRO1600 ori, Tcr | 47 |
| pZ1918G | Source of lacZ reporter, Apr Gmr | H. P. Schweizer |
| pCRR2.1TOPO | Cloning vector for PCR products | Invitrogen |
| pSLA3.2 | pUCP28T with 3.2-kb SphI fragment containing cepIR, Tpr | 17 |
| pSLR101 | pNOT19 with 1.6-kb KpnI-SphI fragment from pSLA3.2 containing cepR, Apr | This study |
| pSLR101-T | pSLR101 with tp cassette cloned in PstI site, Apr Tpr | This study |
| pSLS200 | pUCP28T with 2.3-kb SphI-PstI fragment from pSLA3.2 containing cepI and a part of cepR, Tpr | This study |
| pSLR111 | cepR-lacZ transcriptional fusion with lacZ-Gmr cassette cloned in PstI site of pSLS200, Tpr | This study |
| pSLS210 | pUCP28T with 0.8-kb PstI-BamHI fragment containing cepI promoter, Tpr | This study |
| pSLS222 | cepI-lacZ transcriptional fusion with lacZ-Gmr cassette cloned in BamHI site of pSLS210, Tpr | This study |
| pEXCEPR | pEX18Tc with tp-inactivated cepR fragment from pSLR101-T, Tpr Apr | This study |
| pCF218 | IncP traR | 49 |
| pCF372 | traI-lacZ | 9 |
BCESM, B. cepacia epidemic strain marker.
Construction of cepR mutants by allelic exchange.
The cepR gene in pSLR101 was insertionally inactivated by the introduction of the trimethoprim cassette from p34E-Tp into an internal PstI site (pSLR101-T). The inactivated cepR fragment was amplified by PCR using the primers pNOT19-1 (5′-GGCATGCGCAAGGCGATTAAGTTGG-3′) and pNOT19-2 (5′-GGCATGCCTTTATGCTTCCGGCTCG-3′), which incorporate SphI linkers, and cloned into the SphI site within the suicide vector pEX18Tc (13). This construct, designated pEXCEPR, was transferred to B. cepacia Pc224c or Pc715j by conjugation using either triparental matings with pRK2013 (7) as the mobilizing plasmid or biparental matings with E. coli SM10 as the donor strain (34). Transconjugants were plated onto Pseudomonas isolation agar plates containing 100 μg of trimethoprim per ml to select for single-crossover events in B. cepacia. Tpr transconjugants were streaked for isolated colonies on LB agar plates containing 100 μg of trimethoprim per ml and 5% sucrose to select for the loss of the vector sequence and screened to confirm tetracycline sensitivity. The insertional inactivation of cepR was confirmed with Southern hybridization analysis.
DNA manipulations.
Molecular biology techniques were performed as generally described by Sambrook et al. (29). Restriction enzymes and oligonucleotide primers were purchased from Life Technologies. T4 DNA ligase was purchased from Promega Corp. (Madison, Wis.). Genomic DNA was isolated as described by Ausubel et al. (1). PCR products were cloned using the pCRR2.1 TOPO cloning system as recommended by the manufacturer (Invitrogen, Carlsbad, Calif.). Plasmids were introduced by electroporation using a Gene Pulser (Bio-Rad, Richmond, Calif.) according to the manufacturer's recommendations into E. coli and into B. cepacia as previously described (4).
Siderophore, protease, and autoinducer bioassays.
Siderophore activity was measured using CAS assays (32). On CAS agar, siderophores remove iron from the CAS dye complex, resulting in a blue-to-orange color change in zones surrounding the colonies. The same dye complex was used to quantitate siderophore activity in culture supernatant fluid by measuring the increase in orange color at an optical density at 630 nm (OD630) as previously described (17). Ornibactins were purified by Sephadex LH-20 chromatography and quantitated using the CAS assay as previously described (3, 17). Pyochelin and salicylic acid were isolated and quantitated by thin-layer chromatography (TLC) as previously described (35, 38).
For protease assays, cultures were grown overnight, normalized to a turbidity of 0.3 at 600 nm, and spotted (3 μl) onto dialyzed brain heart infusion (D-BHI) agar containing 1.5% skim milk (39). The plates were incubated for 2 days at 37°C and examined for clear zones surrounding the colonies.
The cepI reporter strain K56-I2 does not produce autoinducer (OHL) or protease but responds to OHL produced by test strains inoculated at right angles to the reporter. The ability of test strains to produce autoinducers was detected by the restoration of protease production at the junction of the test and reporter strains on D-BHI–skim milk agar.
Agrobacterium tumefaciens A136 (pCF218)(pCF372) was used as a reporter to detect AHLs with 3-oxo-, 3-hydroxy-, and 3-unsubstituted side chains as previously described (33). Plasmid pCF218 contains traR, and pCF372 contains a traI-lacZ fusion (9, 49). AHLs were extracted from 20 ml of culture twice with equal volumes of acidified ethyl acetate (0.1 ml of glacial acetic acid per liter). Ethyl acetate was removed by rotary evaporation. The residue was resuspended in 2 ml of ethyl acetate, dried over N2 gas, and resuspended in 100 μl of acidified ethyl acetate. TLC bioassays were performed as described previously (33) with minor modifications. Samples were spotted onto a 20- by 20-cm C18 reversed-phase TLC plate (Whatman) and chromatographed using methanol-water (60:40, vol/vol) as a solvent. The plates were overlaid with a 20-ml A. tumefaciens A136 culture grown as follows. A 3-ml overnight culture was subcultured at a 1/100 dilution into 30 ml of LB broth and grown to log phase. Cells were pelleted by centrifugation, resuspended in 20 ml of AT medium (10.7g of KH2PO4, 160 mg of MgSO4 · 7H2O, 10 mg of CaCl2 · 2H2O, 5 mg of FeSO4 · 7H2O, 2 mg of MnSO4 · 7H2O, 2 g of (NH4)2SO4, 0.5% glucose, 1 liter of H20), and incubated for 30 min. This culture was then added to 150 ml of 0.7% AT agar and 60 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside per ml. TLC plates were incubated for 24 to 48 h at 30°C. Synthetic N-hexanoyl-HSL (HHL) and OHL (Sigma-Aldrich, Oakville, Ontario, Canada) were used as reference standards.
Construction of cepR-lacZ and cepI-lacZ transcriptional fusions.
The lacZ-Gmr fragment from pZ1918G was ligated into the PstI site of pSLS200, which contains a 2.3-kb SphI-PstI fragment that encodes the entire cepI gene and the 5′ end of cepR, resulting in a transcriptional cepR-lacZ fusion (pSLR111). The cepI and lac vector promoters are in the orientation opposite to that of the cepR promoter, which is solely responsible for lacZ expression. A 769-bp fragment containing the cepI promoter was PCR amplified using the primers IN-CEPI (5′-GCGGATCCACCAGACGCCCATCTACCTGCTTCG-3′) and EX-CEPI (5′-GCCTGCAGGGCACAACGACGCCTATCATGC-3′), which incorporate BamHI and PstI linkers, respectively. The PCR product was cloned as a PstI-BamHI fragment into pUCP28T (pSLS210). The lacZ-Gmr fragment was ligated into the BamHI site of pSLS210, resulting in the cepI-lacZ transcriptional fusion (pSLS222). The lac vector promoter is in the opposite direction of the cepI promoter, which is solely responsible for lacZ expression.
β-Galactosidase activity was measured as previously described (26). Overnight cultures were subcultured in 50 ml of medium containing 100 μg of trimethoprim per ml to an OD600 of 0.01. One-milliliter aliquots were removed throughout growth and assayed for enzyme activity as previously described (26). β-Galactosidase activity is expressed in Miller units.
Isolation of outer membrane proteins.
Outer membrane proteins were isolated as previously described (33) from cultures grown for 16 to 24 h (stationary phase) after overnight cultures were subcultured in fresh medium to an OD600 of 0.01. Protein samples (10 μg) were electrophoresed by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (16).
RESULTS
CepR regulates ornibactin but not pyochelin or salicylic acid production in B. cepacia.
Previously, the effects of a cepR::Tn5-OT182 insertion were determined in strain K56-2, which produces ornibactin and salicylic acid, negligible amounts of pyochelin, and no cepabactin (17). We attempted to construct cepR mutants by allelic exchange in other strains that produce pyochelin and cepabactin as well as ornibactin and salicylic acid in order to determine if pyochelin or cepabactin is regulated by CepR. CepR::tp mutants were successfully constructed with strains Pc715j and Pc224c, which produce ornibactin, salicylic acid, and pyochelin (3), and designated Pc715j-R1 and Pc224c-R1. It was not possible to obtain double-crossover cepR::tp recombinants using the following strains: ATCC 25416, ATCC 17759, P109, Pc22-20, 34192, and 34930, which also produce cepabactin (3).
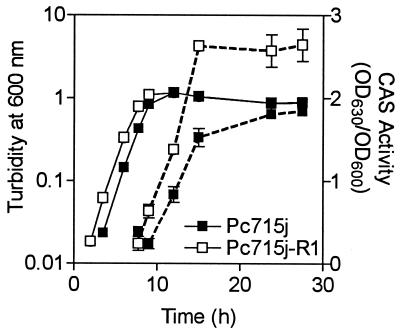
The siderophore, protease, and autoinducer phenotypes of Pc715j-R1 were characterized (i) to confirm the role of CepR in autoinducer and protease production and (ii) to determine if pyochelin synthesis is regulated by CepR. CAS activity was measured in culture fluids throughout the growth of Pc715j and Pc715j-R1 (Fig. 1). Siderophore production is growth phase dependent in that siderophore activity is not observed until late log phase and is maximal in stationary phase. The cepR mutant had significantly greater CAS activity than the respective parent strain during stationary phase, indicating that one or more siderophores were being hyperproduced (Fig. 1). Consistent with this observation, Pc715j-R1 produced 50%-larger zones on CAS agar than those of Pc715j (Table 2). Since CAS activity is a measure of total siderophore activity, the yields of individual siderophores in culture supernatants were also quantitated. Pc715j-R1 produced parental levels of pyochelin and salicylic acid but nearly 100% more ornibactin than Pc715j (Table 2). These data suggest that CepR is a negative regulator of ornibactin synthesis but not of pyochelin or salicylic acid synthesis.
FIG. 1.
Effect of cepR on CAS activity in B. cepacia. CAS activity was monitored throughout growth in succinate medium. ■, Pc715j (wild type); □, Pc715j-R1 (cepR). Growth (turbidity at 600 nm) is shown as solid lines, and CAS activity (OD630/OD600) is shown as dashed lines. All values shown are the means ± standard deviations of values from triplicate experiments. The difference between Pc715j and Pc715jR1 is significant at all time points between 9 and 30 h (P < 0.05; t test for unpaired observations).
TABLE 2.
Effect of a cepR mutation on siderophore, protease, and autoinducer productiona
| Strain | Genotype | Length of CAS agar zonesb (mm) | Ornibactinc (μg/ml/OD600 unit) | Pyochelind (μg/ml/OD600 unit) | Salicylic acidd (μg/ml/OD600 unit) | Radius of protease zonee (mm) | cepI bioassay resultf |
|---|---|---|---|---|---|---|---|
| Pc715j | Wild type | 7.3 ± 0.3 | 53.9 ± 20.5 | 5.4 ± 0.8 | 0.5 ± 0.2 | 8.3 ± 0.3 | + |
| Pc715j-R1 | cepR::tp | 10.8 ± 0.3* | 99.1 ± 19.7 | 4.1 ± 2.0 | 0.3 ± 0.2 | 6.8 ± 0.3* | − |
All values are the means of results of triplicate experiments ± standard deviations, unless indicated otherwise. *, value is significantly different from that for the parent Pc715j in an unpaired t test (P < 0.05).
The CAS zone is the distance from the edge of the orange zone to the edge of the colony after 48 h.
Values are means ± standard deviations of results of duplicate experiments.
Amount of siderophore in CAA cultures grown for 24 h.
The protease zone radius is the distance from the edge of the colony to the edge of the zone of clearance on D-BHI plus 1.5% skim milk agar at 48 h.
Cross-feeds the cepI reporter K56-I2 and restores protease production.
Interestingly, Pc715j-R1 produced significantly less protease activity than that of the parent strain Pc715j (Table 2), whereas K56-R2 had no detectable protease in this assay. It is possible that Pc715j produces more than one protease, only one of which is regulated by CepR. Pc715j did not produce detectable autoinducer activity in the bioassay with the cepI reporter K56-12 (Table 2), indicating that cepR expression is required for OHL production. The phenotypes observed in the cepR mutant Pc224c-R1 were similar to those of Pc715j-R1 (data not shown).
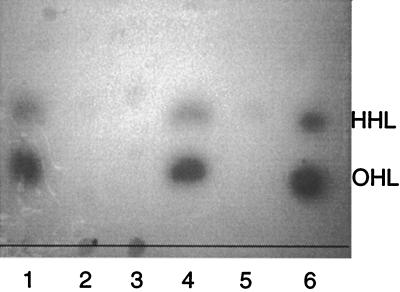
Using an A. tumefaciens traI::lacZ reporter assay, K56-2 and Pc715j produced detectable AHLs that comigrated on TLC plates with synthetic OHL and HHL (Fig. 2). The cepR mutants, K56-R2 and Pc715j-R1, and the cepI mutant, K56-I2, produced very small amounts of HHL, and OHL was only weakly detectable if the plates were incubated for several days. These data indicate that both the cepI and cepR genes are required for OHL synthesis.
FIG. 2.
TLC of acyl-HSLs. Ethyl acetate extracts were chromatographed on C18 reversed-phase TLC plates developed with methanol-water (60:40, vol/vol). The spots were visualized using the A. tumefaciens reporter strain. Lane 1, K56-2; lane 2, K56-I2; lane 3, K56-R2; lane 4, Pc715j; lane 5, Pc715j-R1; lane 6, synthetic OHL and HHL standards.
The effect of a cepR mutation on genes involved in ornibactin synthesis and uptake.
Since ornibactin yields are increased in cepR mutants, we hypothesized that ornibactin biosynthetic genes are negatively regulated by CepR at the level of transcription. If our hypothesis is true, the hyperproduction of ornibactin would be a result of the increased expression of ornibactin biosynthetic genes in cepR mutants. Previously we described two Tn5-OT182 mutants, I117 and T10, which contain transposon insertions in ornibactin biosynthetic genes (37). Strain I117 has a Tn5-OT182 insertion in the pvdA gene that encodes l-ornithine N5-oxygenase, which catalyzes the hydroxylation of l-ornithine, forming the hydroxamate ligand in the peptide moiety of ornibactin. T10 contains an insertion in the pvdD gene that shares homology with nonribosomal peptide synthetases (37). The Tn5-OT182 mutants I117 and T10 contain chromosomally borne transcriptional lacZ fusions due to the presence of a promoterless lacZ gene within the transposon (37).
We constructed cepR mutants by allelic exchange in I117 and T10, which resulted in cepR pvdA and cepR pvdD mutants, respectively. β-Galactosidase activity was monitored throughout growth in low-iron medium and iron-supplemented medium (5 μM FeCl3) to examine the effects of cepR on pvdA and pvdD expression. During stationary phase there was a 20% increase in pvdA expression in I117-R21 in low-iron medium and a 62% increase in pvdA expression in iron-supplemented medium compared to levels in I117 (P < 0.02, t test for unpaired observations) (Table 3). There was a 35% increase in pvdD expression in T10-R10 compared to that in T10 in iron-supplemented medium during stationary phase (P < 0.02, t test for unpaired observations), but there was no significant difference between these strains in low-iron medium. The increase in pvdA expression correlates with the increased levels of CAS activity in the cepR mutants, K56-R2 and Pc715j-R1 (Table 2). (17).
TABLE 3.
Effect of cepR on pvdA and pvdD expression in B. cepacia
| Strain | β-Galactosidase activity (Miller units)a in:
|
|
|---|---|---|
| TSBD-C medium | TSBD-C + FeCl3 medium | |
| I117 | 11,324 ± 548 | 270 ± 37 |
| I117-R21 | 13,526 ± 330b | 439 ± 69b |
| T10 | 8,290 ± 113 | 1,469 ± 200 |
| T10-R10 | 9,678 ± 357 | 1,986 ± 112c |
β-Galactosidase activity was determined from stationary-phase cultures (22 h) normalized for absorbance at A600. Values are the means ± standard deviations of results of triplicate assays.
Significantly different than the value for I17 (P < 0.02; t test for unpaired observations).
Significantly different than the value for T10 (P < 0.02; t test for unpaired observations).
The orbA gene encoding the 78.5-kDa ornibactin outer membrane receptor protein is located downstream of the pvdA gene (36). To determine if other genes involved in ornibactin-mediated iron acquisition were regulated by cepIR, outer membrane proteins were isolated from K56-I2 (cepI::tp) and K56-R2 and compared by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. There did not appear to be any difference in levels of expression of the ornibactin receptor in the cepR and cepI mutants (data not shown).
The effect of a cepR mutation on cepI and cepR expression.
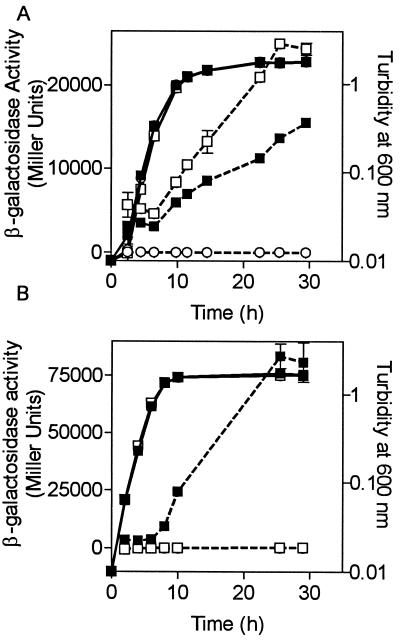
To determine the effects of CepR on cepI and cepR expression, transcriptional cepR-lacZ and cepI-lacZ fusions were constructed in pUCP28T and introduced into K56-2 and K56-R2. β-Galactosidase activity was monitored throughout growth (Fig. 3). In K56-2, the levels of expression of both cepR and cepI were low in early log phase but increased rapidly during late log and stationary phase (Fig. 3). This expression pattern is typical of quorum-sensing-regulated genes. Two important observations were made regarding cepIR expression in a cepR mutant background. In K56-R2, cepR expression increased twofold in the absence of functional cepR, indicating that cepR is autoregulated. cepI expression was abolished in the absence of functional cepR, indicating that cepI expression is positively regulated by CepR at the level of transcription (Fig. 3). Therefore, cepR is involved in regulation of both its own and cepI expression, in addition to that of genes involved in ornibactin biosynthesis.
FIG. 3.
Effect of cepR on cepR and cepI expression. β-Galactosidase activity was monitored throughout growth in tryptic soy broth plus 100 μg of trimethoprim per ml. (A) ■, K56-2 (pSLR111, cepR-lacZ); □, K56-R2 (pSLR111, cepR-lacZ); ○, K56-2 (pUCP28T). The difference between K56-2 and K56-R2 is significant at all time points between 4.5 and 29.5 h (P < 0.02; t test for unpaired observations). (B) ■, K56-2 (pSLS222, cepI-lacZ); □, K56-R2 (pSLS222, cepI-lacZ). The difference between K56-2 and K56-R2 is significantly different at all time points between 4.5 and 29.5 h (P < 0.02; t test for unpaired observations). Growth (turbidity at 600 nm) is indicated by solid lines, and β-galactosidase activity (Miller units) is indicated by dashed lines. All values are the means ± standard deviations of values from triplicate experiments.
The expression of the cepR-lacZ fusion in K56-2 was examined in cultures grown for 19 h in medium with and without iron supplementation to determine the effect of iron on cepR expression. There was 6,956 ± 1,276, 9,644 ± 472, and 10,178 ± 557 Miller units of β-galactosidase activity (values are means ± standard deviations) in cultures of K56-2 (pSLR211) grown in medium with 0, 5, and 10 μM FeCl3, respectively. The level of cepR expression was significantly higher in cultures supplemented with iron (P < 0.05). This result suggests that iron increases cepR expression, which may lead to higher levels of pvdA and pvdD repression in the parent strain. This result agrees with the observation that there is a greater effect of a cepR mutation on pvdA and pvdD expression in the presence of iron in the culture medium than in the absence of iron.
DISCUSSION
Analysis of multiple strains and their respective cepR mutants has shown that cepI and cepR comprise a functional quorum-sensing system that consistently regulates protease, ornibactin, OHL, and HHL production. Construction of gene replacement mutants in pyochelin-producing strains made it possible to determine that pyochelin biosynthesis is not regulated by cepIR. The effect of cepIR on siderophore production appears to be specific for ornibactin biosynthesis. Attempts to construct cepR::tp mutants with several strains previously reported to produce cepabactin were unsuccessful. Therefore, it was not possible to determine if cepabactin is regulated by cepIR. Many strains of the B. cepacia complex are not very amenable to genetic manipulations (19). K56-2 was the only previously reported strain to be successfully used for gene replacement studies (17, 19, 37). In this study, we have identified two additional B. cepacia complex strains that may be used for gene replacement studies.
We have determined that the hyperproduction of ornibactins in a cepR mutant is due to the increased expression of pvdA and possibly pvdD. These experiments suggest that cepR directly regulates ornibactin production at the level of transcription of ornibactin biosynthetic genes, although it is possible that this regulation occurs through an intermediate regulator. Since there are so few examples of negative regulation by quorum-sensing transcriptional regulators, it is difficult to understand the significance of a modest level of repression of ornibactin production by CepR. The negative regulation of ornibactin biosynthesis may serve as a fine-tuning mechanism to reduce energy expended synthesizing ornibactin at high cell densities in an environment in which the concentration of ornibactin is probably sufficient for iron acquisition.
Alternatively, cell density may serve as an additional environmental signal to limit the uptake of iron. The accumulation of intracellular iron can be deleterious due to the formation of hydroxyl radicals in the Haber-Weiss and Fenton reactions (2). When present in high concentrations, iron is the predominant environmental signal leading to the Fur-mediated repression of iron acquisition pathways. It is known that other conditions lead to the downregulation of iron acquisition pathways such as oxidative stress mediated by OxyR and SoxRS (48). The negative regulation of ornibactin production by cepR, when cells are growing at high cell densities, regardless of the iron concentration in the medium, may provide an additional level of control of siderophore-mediated uptake.
The roles of cepR in the control of the autoinducer synthase gene cepI and in its own expression were examined. Previously, we had been unable to detect OHL activity in stationary-phase culture supernatants of the cepR mutant K56-R2 (17). In this study using a different reporter assay, we determined that K56-2 and Pc715j produce both OHL and HHL. Production of these two AHLs has also been detected in another strain of B. cepacia using a Chromobacterium violaceum biosensor TLC assay (11). K56-2 and Pc715j cepI and cepR mutants produced little if any OHL and HHL, suggesting that cepI directs the synthesis of both AHLs and that cepI is positively regulated by CepR. This possibility was confirmed by demonstrating that there was no expression of a plasmid-borne cepI-lacZ fusion in a cepR mutant. Transcriptional fusions on high-copy-number plasmids were used for this study; however, Pesci et al. (24) have shown that Pseudomonas aeruginosa lasR expression patterns did not vary when lasR-lacZ fusions were expressed on high- or low-copy-number plasmids. This type of positive feedback control of autoinducer production is thought to provide a mechanism of signal amplification in which AHL signals are upregulated, allowing for more activation of the cognate R protein and subsequent activation of virulence genes. Although commonly employed, the positive control of autoinducer production is not an absolute requirement since several luxI homologs are not controlled by their respective R proteins (8).
Interestingly, the cepR gene was shown to negatively regulate its own expression, but the mechanism of repression is unknown. The repression effect of CepR on cepR expression would possibly be greater if expression was monitored from a single-copy chromosomal cepR-lacZ reporter since the effects of CepR would not be diluted by additional copies of the cepR promoter. Pantoea stewartii subsp. stewartii (formerly Erwinia stewartii) esaR is autoregulated in a similar pattern (45). von Bodman and Farrand proposed that the lux box overlapping the −10 region in the esaR promoter serves as an EsaR binding site that might occlude the RNA polymerase binding site and prevent its own expression (45). In fact, extracts of E. coli expressing esaR retard the mobility of a 61-bp oligonucleotide containing this lux box element (25). Although the significance of autoregulation of LuxR proteins is unknown, it is a possibility that the negative autoregulation of luxR genes may function to turn off the quorum-sensing response.
Using transcriptional lacZ fusions, we have provided evidence that cepR regulates pvdA and cepIR at the transcriptional level. To prove conclusively that CepR directly regulates these genes, further studies are required to purify CepR and examine its ability to bind to promoter regions of CepR-regulated genes.
ACKNOWLEDGMENTS
This study was supported by a grant from the Canadian Cystic Fibrosis Foundation. S.L. is the recipient of a studentship award from the Alberta Heritage Foundation for Medical Research.
We thank M. B. Visser for excellent technical assistance and D. Storey and D. Erickson for advice on the TLC autoinducer bioassays.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 2.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darling P, Chan M, Cox A D, Sokol P A. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect Immun. 1998;66:874–877. doi: 10.1128/iai.66.2.874-877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dennis J J, Sokol P A. Electrotransformation of Pseudomonas. In: Nickoloff J A, editor. Electroporation and electrofusion of microorganisms. Clifton, N.J: Humana Press; 1995. pp. 125–133. [DOI] [PubMed] [Google Scholar]
- 5.DeShazer D, Woods D E. Broad-host-range cloning and cassette vectors based on the R388 trimethoprim resistance gene. BioTechniques. 1996;20:762–764. doi: 10.2144/96205bm05. [DOI] [PubMed] [Google Scholar]
- 6.Egland K A, Greenberg E P. Conversion of the Vibrio fischeri transcriptional activator, LuxR, to a repressor. J Bacteriol. 2000;182:805–811. doi: 10.1128/jb.182.3.805-811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuqua C, Eberhard A. Signal generation in autoinduction systems: synthesis of acylated homoserine lactones by LuxI-type proteins. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. pp. 211–230. [Google Scholar]
- 9.Fuqua C, Winans S C. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J Bacteriol. 1996;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 11.Geisenberger O, Givskov M, Riedel K, Hoiby N, Tummler B, Eberl L. Production of N-acyl-l-homoserine lactones by P. aeruginosa isolates from chronic lung infections associated with cystic fibrosis. FEMS Microbiol Lett. 2000;184:273–278. doi: 10.1111/j.1574-6968.2000.tb09026.x. [DOI] [PubMed] [Google Scholar]
- 12.Govan J R, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 14.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 15.Kleerebezem M, Quadri L E, Kuipers O P, de Vos W M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lewenza S, Conway B, Greenberg E P, Sokol P A. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J Bacteriol. 1999;181:748–756. doi: 10.1128/jb.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LiPuma J J. Burkholderia cepacia epidemiology and pathogenesis: implications for infection control. Curr Opin Pulm Med. 1998;4:337–341. doi: 10.1097/00063198-199811000-00005. [DOI] [PubMed] [Google Scholar]
- 19.Mahenthiralingam E, Coenye T, Chung J W, Speert D P, Govan J R, Taylor P, Vandamme P. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol. 2000;38:910–913. doi: 10.1128/jcm.38.2.910-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKevitt A I, Bajaksouzian S, Klinger J D, Woods D E. Purification and characterization of an extracellular protease from Pseudomonas cepacia. Infect Immun. 1989;57:771–778. doi: 10.1128/iai.57.3.771-778.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer J M, Hohnadel D, Halle F. Cepabactin from Pseudomonas cepacia, a new type of siderophore. J Gen Microbiol. 1989;135:1479–1487. doi: 10.1099/00221287-135-6-1479. [DOI] [PubMed] [Google Scholar]
- 22.Meyer J M, Van V T, Stintzi A, Berge O, Winkelmann G. Ornibactin production and transport properties in strains of Burkholderia vietnamiensis and Burkholderia cepacia (formerly Pseudomonas cepacia) Biometals. 1995;8:309–317. doi: 10.1007/BF00141604. [DOI] [PubMed] [Google Scholar]
- 23.Meyer J-M, Abdallah M A. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physico-chemical properties. J Gen Microbiol. 1978;107:319–328. [Google Scholar]
- 24.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierson L S, Wood D W, von Bodman S B. Quorum sensing in plant-associated bacteria. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. pp. 101–115. [Google Scholar]
- 26.Platt T, Muller-Hill B, Miller J H. Analysis of the lac operon enzymes. In: Miller J H, editor. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 27.Prince A. Antibiotic resistance of Pseudomonas species. J Pediatr. 1986;108:830–834. doi: 10.1016/s0022-3476(86)80753-3. [DOI] [PubMed] [Google Scholar]
- 28.Salmond G P, Bycroft B W, Stewart G S, Williams P. The bacterial ‘enigma’: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 30.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 31.Schweizer H P, Klassen T, Hoang T. Improved methods for gene analysis and expression in Pseudomonas spp. In: Nakazawa T, Furukawa K, Hass D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C.: ASM Press; 1996. pp. 229–237. [Google Scholar]
- 32.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 33.Shaw P D, Ping G, Daly S L, Cha C, Cronan J E, Jr, Rinehart K L, Farrand S K. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 35.Sokol P A. Production and utilization of pyochelin by clinical isolates of Pseudomonas cepacia. J Clin Microbiol. 1986;23:560–562. doi: 10.1128/jcm.23.3.560-562.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokol P A, Darling P, Lewenza S, Corbett C R, Kooi C D. Identification of a siderophore receptor required for ferric-ornibactin uptake in Burkholderia cepacia. Infect Immun. 2000;68:6554–6660. doi: 10.1128/iai.68.12.6554-6560.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sokol P A, Darling P, Woods D E, Mahenthiralingam E, Kooi C. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N5-oxygenase. Infect Immun. 1999;67:4443–4455. doi: 10.1128/iai.67.9.4443-4455.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokol P A, Lewis C J, Dennis J J. Isolation of a novel siderophore from Pseudomonas cepacia. J Med Microbiol. 1992;36:184–189. doi: 10.1099/00222615-36-3-184. [DOI] [PubMed] [Google Scholar]
- 39.Sokol P A, Ohman D E, Iglewski B H. A more sensitive plate assay for detection of protease production by Pseudomanas aeruginosa. J Clin Microbiol. 1979;9:538–540. doi: 10.1128/jcm.9.4.538-540.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephan H, Freund S, Beck W, Jung G, Meyer J M, Winkelmann G. Ornibactins—a new family of siderophores from Pseudomonas. Biometals. 1993;6:93–100. doi: 10.1007/BF00140109. [DOI] [PubMed] [Google Scholar]
- 41.Sun L, Jiang R Z, Steinbach S, Holmes A, Campanelli C, Forstner J, Sajjan U, Tan Y, Riley M, Goldstein R. The emergence of a highly transmissible lineage of cb1+Pseudomonas (Burkholderia) cepacia causing CF centre epidemics in North America and Britain. Nat Med. 1995;1:661–666. doi: 10.1038/nm0795-661. [DOI] [PubMed] [Google Scholar]
- 42.Tablan O C, Martone W J, Doershuk C F, Stern R C, Thomassen M J, Klinger J D, White J W, Carson L A, Jarvis W R. Colonization of the respiratory tract with Pseudomonas cepacia in cystic fibrosis: risk factors and outcomes. Chest. 1987;91:527–532. doi: 10.1378/chest.91.4.527. [DOI] [PubMed] [Google Scholar]
- 43.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 44.Vandamme P, Mahenthiralingam E, Holmes B, Coenye T, Hoste B, De Vos P, Henry D, Speert D P. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV) J Clin Microbiol. 2000;38:1042–1047. doi: 10.1128/jcm.38.3.1042-1047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Bodman S B, Farrand S K. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J Bacteriol. 1995;177:5000–5008. doi: 10.1128/jb.177.17.5000-5008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Bodman S B, Majerczak D R, Coplin D L. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc Natl Acad Sci USA. 1998;95:7687–7692. doi: 10.1073/pnas.95.13.7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.West S E, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 48.Zheng M, Doan B, Schneider T D, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu J, Beaber J W, More M I, Fuqua C, Eberhard A, Winans S C. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J Bacteriol. 1998;180:5398–5405. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]