Summary
Milk fortifiers help meet the nutritional needs of preterm infants receiving their mother’s own milk (MOM) or donor human milk. We conducted a randomized clinical trial (NCT03214822) in 30 very low birth weight premature neonates comparing bovine-derived human milk fortifier (BHMF) versus human-derived fortifier (H2MF). We found that fortifier type does not affect the overall microbiome, although H2MF infants were less often colonized by an unclassified member of Clostridiales Family XI. Secondary analyses show that MOM intake is strongly associated with weight gain and microbiota composition, including Bifidobacterium, Veillonella, and Propionibacterium enrichment. Finally, we show that while oxidative stress (urinary F2-isoprostanes) is not affected by fortifier type or MOM intake, fecal calprotectin is higher in H2MF infants and lower in those consuming more MOM. Overall, the source of human milk (mother versus donor) appears more important than the type of milk fortifier (human versus bovine) in shaping preterm infant gut microbiota.
Keywords: gut microbiome, very low birth weight infants, human milk fortifiers, oxidative stress, mother’s own milk, human donor milk, gut inflammation, calprotectin
Graphical abstract
Highlights
-
•
Milk fortifier type (human versus bovine) has little impact on the preterm microbiome
-
•
Milk source (mother versus donor) is strongly associated with microbiome composition
-
•
Feeding mother’s own milk is linked to better weight gain and less gut inflammation
Kumbhare et al. demonstrate that the type of milk fortifier (bovine versus human) has no major impact on the gut microbiome of very low birth weight (VLBW) infants, whereas the source of human milk (mother versus donor) is strongly associated with the gut microbiome, healthier weight gain, and reduced gut inflammation.
Introduction
While human milk provides optimal nutrition for full-term infants, its nutrient density is inadequate for those born preterm.1,2 Formula made from bovine milk can provide higher levels of energy and protein3 but lacks many of the bioactive components found in human milk,4 including the “personalized” components found only in the mother’s own milk (MOM).5,6 Formula may also trigger inflammation and increase the risk of necrotizing enterocolitis.7 Furthermore, considering the fluid restrictions due to relatively low feeding tolerance in preterm infants, it is essential to utilize feeding regimes that provide an optimal volume with high “nutrient-to-calorie ratio.” Thus, human milk fortifiers (HMFs) are commonly used to increase the caloric density of human milk1 fed to fragile neonates. However, standard HMFs are derived from bovine milk and may still trigger inflammation and oxidative stress.8,9 Recently, human-derived HMF (H2MF) has been developed as an alternative to standard bovine-derived HMF (BHMF),1 but its impact on oxidative stress and inflammation is not known.
Apart from influencing gut inflammation and oxidative stress, early nutrition drives gut microbiome development. Preterm infants have an immature gastrointestinal tract and delayed maturation of the gut microbiota,10 which has been implicated in the pathogenesis of necrotizing enterocolitis11 and numerous chronic conditions.12 Exclusive human-milk-based nutrition regimes using H2MF may prevent some of these conditions and improve overall feeding in preterm infants,13, 14, 15, 16 although not all studies have found these clinical benefits.17 Due to the lack of microbiome analyses in prior studies, it is unclear whether any clinical benefits of H2MF are accompanied and perhaps mediated by favorable changes to the gut microbiota.
In addition to the type of fortifier, the source of human milk may also influence infant physiology and the developing microbiome. Donor human milk (DHM) is increasingly used for preterm infants when MOM is not available, offering several clinical benefits over infant formula.2,13 However, DHM is pasteurized and frozen,18,19 resulting in lower “total antioxidant capacity”20 than MOM, especially in early lactation.21 These differences may have an impact on infant microbiota and metabolism,22, 23, 24 but this has not been widely studied in the context of very low birth weight (VLBW) preterm infants receiving different types of fortifiers.
To address these knowledge gaps, we performed a randomized controlled trial to compare the effects of BHMF versus H2MF on gut microbiome development, oxidative stress, and gut inflammation in human-milk-fed VLBW preterm neonates. We also assessed the association of these outcomes with the proportion of MOM versus DHM intake.
Results
Demographics and clinical characteristics
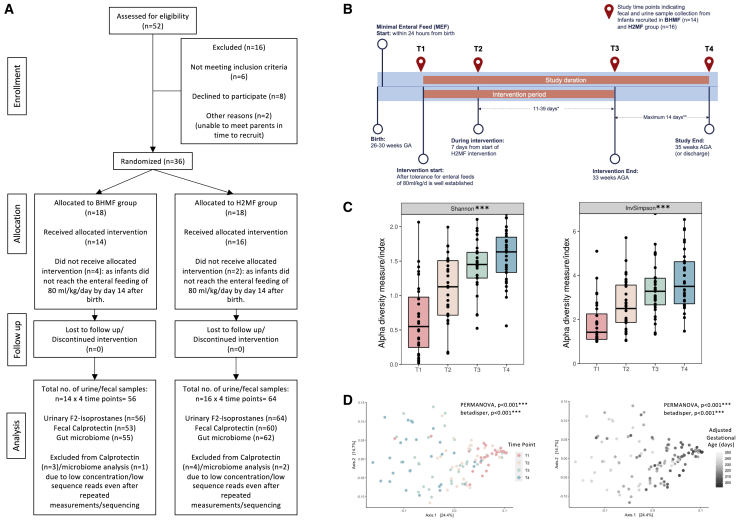
Of 36 infants randomized, 30 received their allocated intervention and completed the study (14 BHMF and 16 H2MF) (Figures 1A and 1B). Six infants were excluded before the intervention was initiated because they did not achieve the enteral feeding requirement by 14 days. Except for parity (marginally higher in the H2MF group), there were no differences in maternal demographics between groups (Table 1). Infant demographics and dietary factors were also balanced between groups, and there were no differences in terms of Apgar scores, weight gain, or the number of days on antibiotics, ventilation, or oxygen support (Table 1).
Figure 1.
Flowchart and illustration of changes in gut bacterial diversity and community structure in VLBW infants over time
(A) Participant flow chart: consolidated standards of reporting trials diagram.
(B) Study design and sample collection time points (T1–T4). Infants born from 26 to 30 weeks were recruited into the study, received their assigned fortifier (BHMF or H2MF) until 33 weeks AGA, and were followed until 35 weeks AGA or hospital discharge. Therefore, the intervention (T1 to T3) and follow-up (T3 to T4) periods ranged in duration.
(C) Alpha diversity indicating species diversity within samples (n = 30) across time.
(D) Principal coordinate analysis based on Bray-Curtis dissimilarity (Jaccard distances in Figure S1 and phylum/genus level changes over time in Figures S2A and S2B) showing diversity between samples across four study time points (left) and with adjusted gestational age in days (right). T1, study day 0 (before fortification); T2, study day 7 (during fortification); T3, week 33 AGA (end of fortification); and T4, week 35 AGA (follow-up after fortification). Statistical significance: ∗∗∗p < 0.001 across all study time points (Kruskal-Wallis test) for alpha diversity or by permutational multivariate analysis of variance (PERMANOVA) for beta diversity and betadisper test (permutation test for homogeneity of multivariate dispersions; 99,999 permutations with strata by infant ID to account for repeated measures). The p values were adjusted for multiple comparisons using false discovery rate (FDR) correction. AGA, adjusted gestational age; BHMF, bovine-derived human milk fortifier; H2MF, human-derived human milk fortifier; VLBW, very low birth weight.
Table 1.
Demographics and characteristics of participants
| Characteristic | BHMF (n = 14) | H2MF (n = 16) | p |
|---|---|---|---|
| Mother | |||
| Maternal age in years | 31.7 ± 5.2 | 30.6 ± 5.6 | 0.57 |
| Maternal parity | 1 (1, 1) | 1 (1, 3.2) | 0.04a |
| Maternal gravidity | 2 (1.2, 3) | 3 (2, 6) | 0.13 |
| Infant | |||
| Sex assigned at birth, n (%) | |||
| Male | 6 (43) | 9 (56) | 0.71 |
| Female | 8 (57) | 7 (44) | |
| Mode of delivery, n (%) | |||
| Vaginal | 2 (14) | 4 (25) | 0.17 |
| C-section | 12 (86) | 12 (75) | |
| Age (days) | |||
| Gestational age at birth | 197 ± 7 | 196 ± 8 | 0.80 |
| AGA at study time points | |||
| T1 | 204 ± 7 | 203 ± 8 | 0.70 |
| T2 | 214 ± 10 | 212 ± 8 | 0.61 |
| T3 | 231 ± 1 | 231 ± 1 | 0.51 |
| T4 | 244 ± 8 | 245 ± 1 | 0.41 |
| Weight (g) | |||
| Birth | 1,039 ± 131 | 1,016 ± 180 | 0.68 |
| T1 | 1,010 ± 142 | 999 ± 165 | 0.84 |
| T2 | 1,106 ± 130 | 1,114 ± 171 | 0.89 |
| T3 | 1,581 ± 197 | 1,555 ± 304 | 0.78 |
| T4 | 1,957 ± 216 | 1,920 ± 414 | 0.75 |
| Weight gain (since birth) | |||
| T1 | −29 ± 53 | −16 ± 65 | 0.55 |
| T2 | 67 ± 62 | 98 ± 71 | 0.21 |
| T3 | 542 ± 180 | 539 ± 207 | 0.97 |
| T4 | 918 ± 221 | 904 ± 330 | 0.89 |
| Weight for age Z score | |||
| Birth | −0.0 (−0.3, 0.5) | −0.0 (−0.6, 0.6) | 0.89 |
| T1 | −0.7 (−0.9, −0.3) | −0.6 (−1.1, −0.2) | 0.98 |
| T2 | −0.9 (−1.4, −0.5) | −0.9 (−1.3, −0.5) | 0.87 |
| T3 | −1.0 (−1.4, −0.5) | −0.9 (−1.8, −0.5) | 0.76 |
| T4 | −1.4 (−1.5, −0.9) | −1.4 (−2.2, −0.4) | 0.71 |
| Change in Z score (since birth) | |||
| T1 | −0.6 (−0.8, −0.4) | −0.6 (−0.7, −0.3) | 0.47 |
| T2 | −1.0 (−1.2, −0.7) | −0.8 (−1.1, −0.5) | 0.28 |
| T3 | −1.0 (−1.2, −0.6) | −1.0 (−1.3, −0.5) | 0.85 |
| T4 | −1.1 (−1.6, −0.7) | −1.3 (−1.7, −0.7) | 0.80 |
| Diet | |||
| Total enteral volume (since birth) | |||
| T1 | 67 ± 33 | 54 ± 33 | 0.27 |
| T2 | 128 ± 25 | 109 ± 40 | 0.12 |
| T3 | 174 ± 23 | 160 ± 41 | 0.25 |
| T4 | 206 ± 21 | 194 ± 45 | 0.34 |
| MOM proportion (% since birth) | |||
| % MOM (T1) | 79.3 (32.5, 100) | 100 (37.2, 100) | 0.54 |
| % MOM (T2) | 69.9 (26.9, 99.5) | 80.8 (46.2, 100) | 0.72 |
| % MOM (T3) | 57.2 (27.3, 94.2) | 63.9 (41.6, 95.4) | 0.69 |
| % MOM (T4) | 57.3 (17.9, 90.4) | 51.9 (36.4, 91.5) | 0.69 |
| Clinical | |||
| Received antibiotics | |||
| % days on antibiotics (total study duration) | 0 (0, 6.9) | 0 (0, 11.4) | 0.71 |
| T1–T2 | 0 (0, 0) | 0 (0, 15.7) | 0.12 |
| T1–T3 | 0 (0, 7.5) | 0 (0, 21.0) | 0.65 |
| T1–T4 | 0 (0, 6.9) | 0 (0, 11.4) | 0.71 |
| Prenatal (antenatal) steroids, n (%) | 12 (86) | 12 (75) | 0.66 |
| Received (postnatal) steroids, n (%) | 1 (8) | 2 (12) | 1.00 |
| Apgars 5-min score, median (IQR) | 7.5 (6.2, 8.0) | 8.0 (5.5, 9.0) | 0.46 |
| Total no. of days on ventilation, mean ± SD | 32.6 ± 23.3 | 29.6 ± 18.2 | 0.70 |
| Total no. of surfactant doses, median (IQR) | 1 (0.2, 2.0) | 1 (0, 1.2) | 0.42 |
| Need for oxygen support at 36 weeks, n (%) | 5 (36) | 4 (27) | 0.69 |
| Retinopathy, n (%) | 2 (14) | 5 (31) | 0.39 |
| Late-onset sepsis, n (%) | 2 (14) | 6 (37) | 0.25 |
Very low birth weight infants recruited into the study were randomized to receive standard bovine-derived human milk fortifiers (BHMF) or human-derived human milk fortifiers (H2MF) during the intervention period. Values are means ± SD or median (IQR) or n (%). Percentages reflect the proportion of non-missing data. Weight gain refers to weight at the study time point—birth weight. Weight-for-age Z scores were calculated using 2013 Fenton growth charts for preterm infants.25 Total enteral volume (TEV) is the cumulative average of TEV received by infants until a particular study time point from birth. % MOM volume is the average percentage of mother’s own milk (MOM) (proportion of MOM in the TEV used to prepare the feed prior to fortification) received by the infant until a particular study time point from birth. % days on antibiotics is the percentage of the number of days on antibiotics during total study duration or until a particular study time point. Nutritional support values indicate the proportion of infants from the particular group who received one of the other nutritional supports along with the human milk fortifiers during the study: liquid protein fortifier (LPF) 1–4 mL/100 mL; total parenteral nutrition (TPN), D10W, SMOF_Lipid 20, ECP (EnfaCare premature formula) 1 mg/100 mL; Nutramigen powder 2.5 mg. The p values are from Pearson’s chi-square test or Fisher’s exact test (for categorical variables) or t test or Wilcoxon sum rank test (for ordinal/numerical variables).
AGA, adjusted gestational age; MOM, mother’s own milk; T1, study day 0 (before fortification); T2, study day 7 (during fortification); T3, week 33 AGA (end of fortification); T4, week 35 AGA (follow-up after fortification).
p < 0.05
Changes in gut bacterial diversity and composition in VLBW infants over time
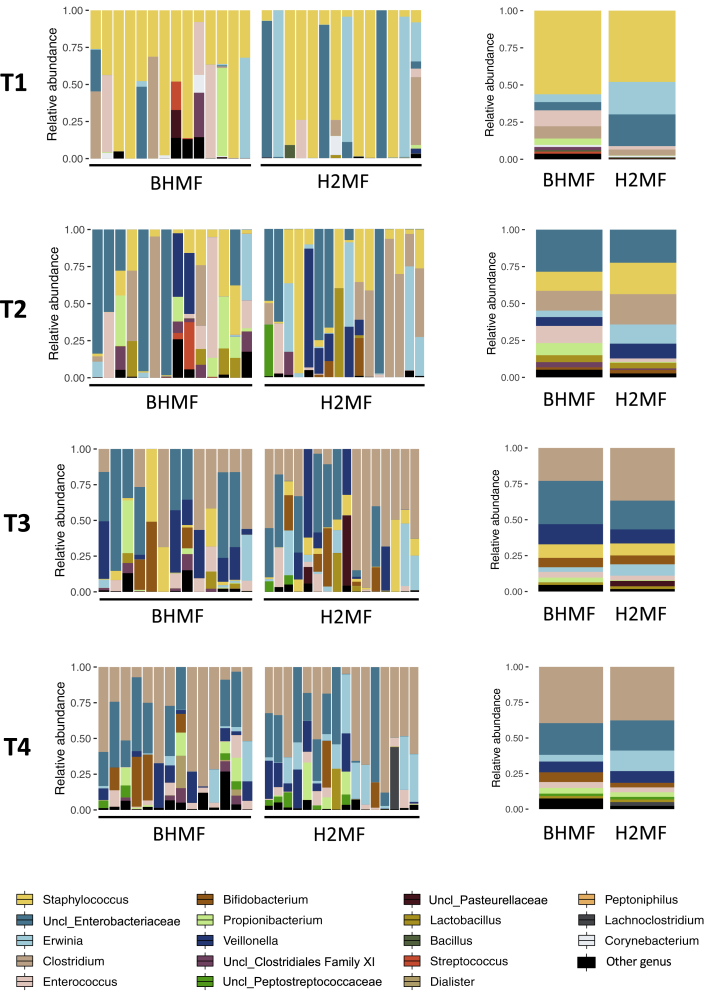
Generally, in all infants, microbiome diversity (p < 0.01) (Figure 1C) and variability increased (PERMANOVA, p < 0.001; betadisper, p < 0.01) (Figures 1D, S1A, and S1B) with age. The early phylum-level gut microbial composition (before the intervention, T1) was primarily composed of Firmicutes, Proteobacteria, and Actinobacteria. At the genus level, Staphylococcus, an unclassified genus of Enterobacteriaceae, and Erwinia were the most abundant taxa (Figure S2B). Many Firmicutes (Staphylococcus, Bacillus, unclassified Peptostreptococcaceae, and Lactobacillales) and Proteobacteria (Acinetobacter, unclassified Pasteurellaceae, and Comamonas) declined over time (Figures S2A and S2B). In contrast, Bifidobacterium, Propionibacterium, and Streptococcus marginally increased over time, while Veillonella and Clostridium increased substantially (Figures S2B).
Impact of milk fortifier type on gut microbiome, oxidative stress, and gut inflammation
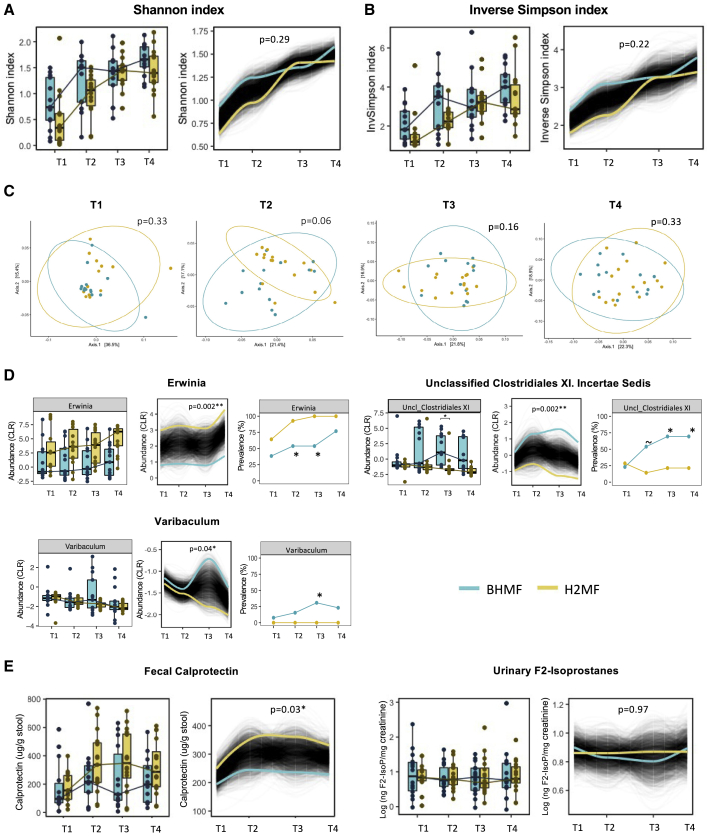
There was a high degree of interindividual variation in microbiota composition within both groups across all time points (Figures 2, S2, and S3). Fortifier type did not have an impact on alpha diversity, which increased similarly in both groups over time (Figures 3A and 3B), or overall microbial community structure (except marginally at T2, r2 = 0.08, p = 0.06; which was attributed to the differences in beta dispersion, p = 0.04; Figures 3C and S4A and Table S1A). An unclassified genus of Clostridiales Family XI incertae sedis was significantly enriched in the BHMF group by the end of fortification (T3 prevalence 69% in BHMF versus 21% in H2MF, p = 0.02), and this difference was sustained at least 2 weeks after fortification ended (splinectomeR trajectory p < 0.01; mixed-effects regression p = 0.04) (Figures 3D, S5, S6, and S7). To a lesser extent, Varibaculum was also enriched (not detected in any H2MF infants; splinectomeR p = 0.04), but only during the period of fortification (Figure 3D, Tables S1B and S1C). Erwinia was more prevalent and abundant in the H2MF group (splinectomeR p < 0.01); however, these differences were already evident at baseline, indicating they originated by chance prior to the randomized fortification intervention (Figure 3D and Table S1D).
Figure 2.
Impact of human milk fortifier type, bovine (BHMF, n = 14) or human (H2MF, n = 16), on gut bacterial composition in VLBW infants
Bar plots depict the relative abundance of gut bacterial genera (left, individual infants; right, group means). T1, study day 0 (before fortification); T2, study day 7 (during fortification); T3, week 33 AGA (end of fortification); and T4, week 35 AGA (follow-up after fortification). AGA, adjusted gestational age; BHMF, bovine-derived human milk fortifier; H2MF, human-derived human milk fortifier; VLBW, very low birth weight. Figure S3 shows results at the phylum level. The prefix “Uncl_” indicates an unclassified genus of the particular bacterial family or order.
Figure 3.
Impact of human milk fortifier type on the gut microbiome, gut inflammation, and oxidative stress in VLBW infants over time
(A and B) Longitudinal trajectories of (A) Shannon index and (B) inverse Simpson index, indicating alpha (within-sample) diversity over time in infants receiving BHMF (n = 14) or H2MF (n = 16); p values are from trend comparison using splinectomeR (999 permutations).
(C) PCoA depicts beta (between-sample) diversity based on Bray-Curtis dissimilarity (Jaccard distances in Figure S4A) at each time point; p values are from PERMANOVA (99,999 permutations). See Table S1A for betadisper test results (permutation test for homogeneity of multivariate dispersions).
(D) Genera with >10% prevalence across all samples that differed in prevalence or relative abundance (centered log-ratio [CLR] transformed) between groups over time are shown here (complete data are shown in Tables S1B–S1D, and regression analysis is shown in Figure S5). For Erwinia, the distance between groups at T1 indicates a random difference at baseline.
(E) Gut inflammation (fecal calprotectin) and oxidative stress (urinary F2-isoprostane). The p values are from Wilcoxon sum rank test (for boxplots) or using splinectomeR (for longitudinal trajectories, 999 permutations) or Fisher’s exact test (for prevalences) (∼p < 0.10, ∗p < 0.05, ∗∗p < 0.01). T1, study day 0 (before fortification); T2, study day 7 (during fortification); T3, week 33 AGA (end of fortification); and T4, week 35 AGA (follow-up after fortification). AGA, adjusted gestational age; BHMF, bovine-derived human milk fortifier; H2MF, human-derived human milk fortifier; PCoA, principal coordinate analysis; VLBW, very low birth weight.
In contrast to infants receiving standard BHMF, those receiving H2MF experienced a rise in fecal calprotectin levels across the study period (splinectomeR p = 0.03, Figure 3E). Urinary F2-isoprostanes differed significantly between groups, indicating no impact of fortifier type on oxidative stress (Figure 3E).
Association of MOM intake with gut microbiome, oxidative stress, gut inflammation, and weight gain
On average, infants in both fortifier groups received similar proportions of MOM (versus DHM) across all study time points (Table 1 and Figure S8A), and baseline demographic and clinical characteristics were comparable among infants receiving low and high proportions of MOM (Table S2). By the end of the study, infants with high MOM intake had experienced significantly higher weight gain (1,017 ± 249 g versus 789 ± 270 g, p = 0.02), translating to significantly lower declines in weight-for-age Z scores (median −0.8 [IQR −1.1, −0.5] versus −1.7 [IQR −1.9, −1.4], p < 0.001) (Table S2).
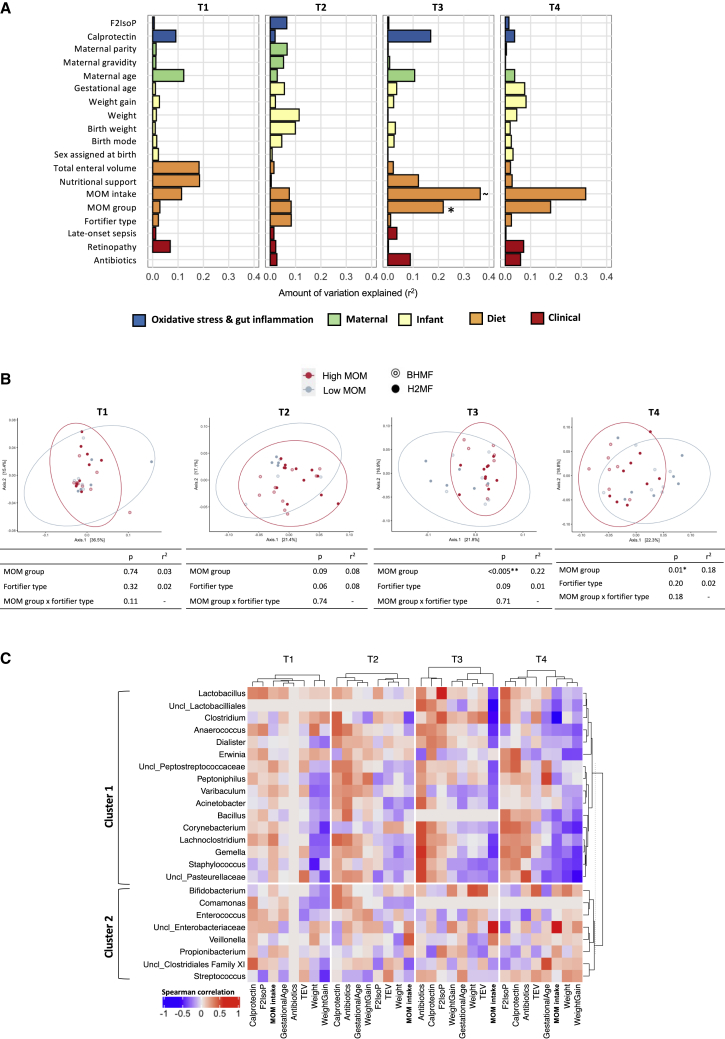
MOM intake was strongly associated with microbiome development. Among all variables assessed, MOM intake was the strongest predictor of microbiome composition at T3 and T4 (Figure 4A). For example, in univariate analyses at T3, MOM explained 22% of the microbiome variation (r2 = 0.22, p = 0.01), while fortifier type had no effect (r2 = 0.01, p = 0.99). Multivariable models accounting for both MOM intake and fortifier type further confirmed the strong association of MOM intake, but not fortifier type, with the gut microbiome composition (Figures 4B and S8C). There was no evidence of interaction between MOM intake and fortifier type (Figure 4B).
Figure 4.
Association of mother’s own milk (MOM) intake, fortifier type, and other factors with gut microbiome composition in VLBW infants
(A) Microbiome variance is explained by various factors, modeled individually by EnvFit using Bray-Curtis dissimilarity. Fortifier type, BHMF versus H2MF; MOM group, high versus low intake; F2IsoP, F2-isoprostane levels; antibiotics, percentage of days on antibiotics until a particular study time point. FDR-adjusted p values are denoted by ∼p < 0.1, ∗p < 0.05, ∗∗p < 0.01.
(B) PCoA depicting beta diversity between MOM groups (low-MOM versus high-MOM infants, see Figure S8 for details) over time, modeled from PERMANOVA (99,999 permutations) using the fortifier type and MOM group in a multivariable analysis. See Table S1A for betadisper test results (permutation test for homogeneity of multivariate dispersions).
(C) Heatmap illustrating correlations between bacterial genera relative abundances (CLR transformed) and other continuous covariates at each time point (T1–T4). Dendrogram clustering was based on pairwise distances obtained from Spearman correlations, independently for each time point (horizontal axis). Correlations represented were not significant (p > 0.05, FDR corrected). Clusters 1 and 2 on the vertical axis depict the clustering of genera based on their correlations with covariates. MOM groups were determined based on % MOM volumes that represent the proportion of MOM used to prepare the feeds prior to fortification. T1, study day 0 (before fortification: BHMF, n = 14; H2MF, n = 16; low MOM, n = 10; high MOM, n = 20); T2, study day 7 (during fortification: BHMF, n = 14; H2MF, n = 16; low MOM, n = 8; high MOM, n = 22); T3, week 33 AGA (end of fortification: BHMF, n = 14; H2MF, n = 16; low MOM, n = 13; high MOM, n = 17); and T4, week 35 AGA (follow-up after fortification: BHMF, n = 14; H2MF, n = 16; low MOM, n = 14; high MOM, n = 16). BHMF, bovine-derived human milk fortifier; H2MF, human-derived human milk fortifier; PCoA, principal coordinate analysis; VLBW, very low birth weight; F2IsoP, F2-isoprostanes; AGA, adjusted gestational age; antibiotics, percentage of days on antibiotics until a specific study time point; TEV, total enteral volume. The prefix “Uncl_” indicates an unclassified genus of the named bacterial family or order.
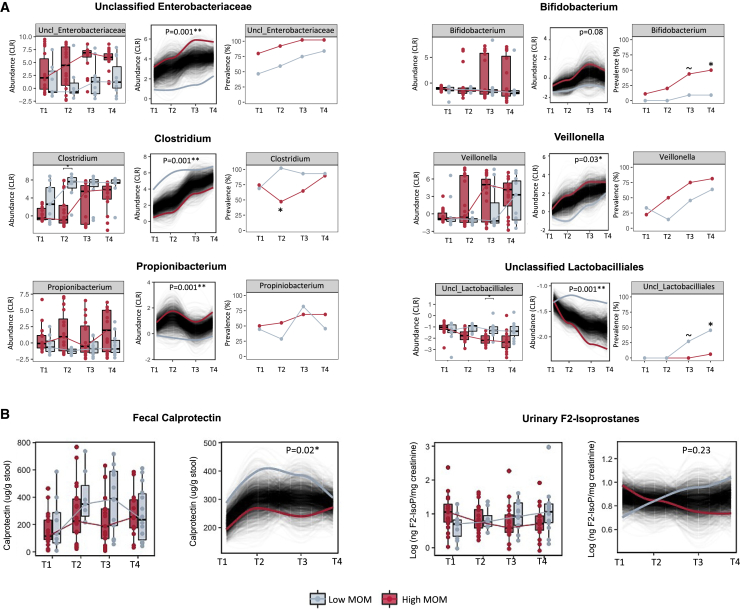
MOM-associated differences in microbiota composition included enrichment of Propionibacterium, Veillonella, and one unclassified genus in the Enterobacteriaceae family among infants with high MOM intake (Figures 5A, S9, S10, S11, and S12 and Tables S1E–S1G). Infants with low MOM intake had enrichment of Clostridium and one unclassified genus of Lactobacillales prevalent at T3 and T4. In addition, infants with low MOM intake were less likely to be colonized by Bifidobacterium (9% versus 50% prevalence for low versus high MOM, p = 0.04, Figure 5A). Low MOM intake was also associated with higher fecal calprotectin over time (p = 0.02, Figure 5B), particularly during fortification. MOM intake was not associated with F2-isoprostane levels (Figure 5B).
Figure 5.
Association of mother’s own milk (MOM) intake with microbiome trajectories, gut inflammation, and oxidative stress in VLBW infants over time
(A) Trajectories of bacterial relative abundance (CLR transformed) and prevalence over time in infants with high versus low MOM intake (see Figure S8 for details). Genera with >10% prevalence across all samples that differed in prevalence or relative abundance between groups over time are shown here (complete data are shown in Tables S1E–S1G, and regression analyses are shown in Figure S10).
(B) Gut inflammation (fecal calprotectin) and oxidative stress (urinary F2-isoprostanes) over time. The p values are from Wilcoxon sum rank test (for boxplots) or using splinectomeR (for longitudinal trajectories, 999 permutations) or Fisher’s exact test (for prevalences) (∼p < 0.10, ∗p < 0.05, ∗∗p < 0.01). MOM groups were determined based on % MOM volumes that represent the proportion of MOM used to prepare the feeds prior to fortification. T1, study day 0 (before fortification: low MOM, n = 10; high MOM, n = 20); T2, study day 7 (during fortification: low MOM, n = 8; high MOM, n = 22); T3, week 33 AGA (end of fortification: low MOM, n = 13; high MOM, n = 17); and T4, week 35 AGA (follow-up after fortification: low MOM, n = 14; high MOM, n = 16).
Correlations of MOM intake, microbiota, biomarkers, and clinical characteristics
To further investigate the importance of MOM intake, we explored associations of different bacterial taxa with biomarkers, clinical characteristics, and MOM intake as a continuous variable (cumulative proportion of feeds) while applying hierarchical clustering (Figure 4C). We observed two bacterial community clusters, partially distinguished by their correlation with MOM intake at T3 and T4. Cluster 1 comprised genera enriched with lower MOM intake, such as Clostridium, Lachnoclostrium, and Gemella. Most genera in this cluster showed negative correlations with weight gain and total enteral feeding volume and positive correlations with calprotectin, F2-isoprostanes (oxidative stress), and antibiotic usage, particularly at T3 and T4. Cluster 2 comprised several genera enriched with higher MOM intake, such as Bifidobacterium, Veillonella, and one unclassified Enterobacteriaceae genus. This cluster did not show any overall patterns of association with infant biomarkers or clinical characteristics. Notably, at T3 and T4, we also observed clustering between MOM intake, total enteral volume, and weight gain. Antibiotic usage, calprotectin, and F2-isoprostanes formed another distinct cluster. Taken together, these findings support the hypothesis that higher MOM intake could ameliorate the harmful effects of early antibiotic exposure by modulating the gut microbiome development to reduce oxidative stress and inflammation.
Discussion
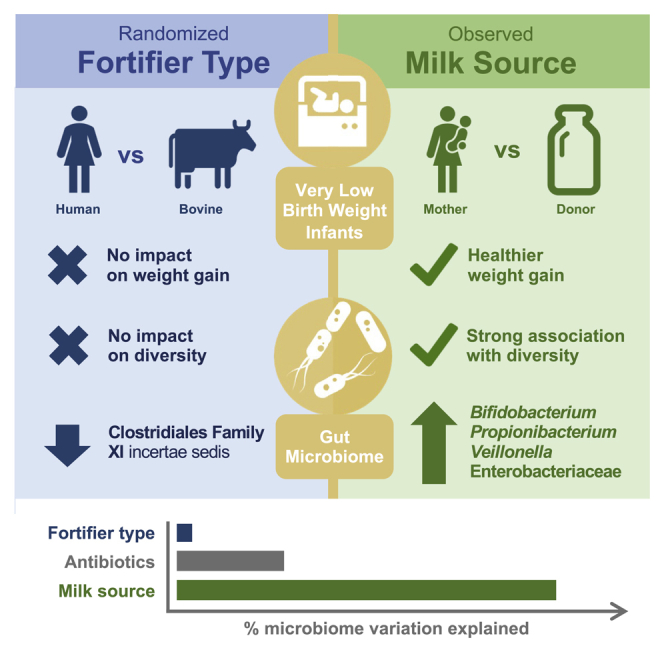
Our findings suggest that the source of human milk (own mother versus donor milk) was a major determinant of gut microbiome development in VLBW infants, whereas the type of fortifier (human versus bovine) had a minimal impact. Although a few individual taxa were affected, the type of fortifier did not substantially influence gut microbiota diversity or composition and did not affect urinary F2-isoprostanes (oxidative stress). Notably, fecal calprotectin levels were higher in those receiving H2MF. The clinical significance of this unexpected result is unclear, and warrants further investigation. In a secondary analysis, we observed strong associations between MOM intake and gut microbiome composition and development, as well as lower fecal calprotectin and greater weight gain (with significantly lower declines in weight-for-age Z scores) among infants consuming higher proportions of MOM.
Little impact of fortifier type (human versus bovine) on gut microbiome
Given the large and well-established impact of human milk versus bovine-derived infant formula on the microbiome of full-term infants,26,27 we were surprised to find relatively few differences between the microbiota of VLBW infants randomized to receive human versus bovine-derived milk fortifiers. This unexpected finding might reflect the fact that formula feeding often replaces human milk entirely, whereas fortification displaces only a small volume of human milk. Notably, liquid H2MF displaces a relatively larger volume of milk (30%–40% displacement) compared with powdered BHMF (<5% displacement), which could potentially offset some of the expected benefits of H2MF in human-milk-fed infants. It is possible that provision of human milk components (through MOM, DHM, and/or H2MF) affects the microbiome more strongly than avoidance of bovine products in BHMF. More research is needed to evaluate these hypotheses and quantify the relative impacts of MOM, DHM, formula, BHMF, and H2MF on the infant microbiome.
In our study, Varibaculum and an unclassified genus belonging to Clostridiales Family XI were the only two taxa differentially affected by fortification type, both showing depletion in the H2MF group. Interestingly, Varibaculum in the infant gut has been associated with degradation of 6′ sialyllactose, an oligosaccharide that is more abundant in bovine versus human milk;28, 29, 30 however, its functional role and clinical relevance in VLBW infants have not been studied. Depletion of Clostridiales Family XI incertae sedis often precedes nosocomial C. difficile infection31,32 in adults; however, this has not been shown in VLBW infants, where the risk factors and consequences of C. difficile infection are unclear,33 although C. difficile colonization has been linked to necrotizing enterocolitis (NEC) in this population.34,35 Overall, we observed no significant impact of the type of fortifier on the gut microbiome and growth, which aligns with the findings from a previous study that noted no impact on feeding tolerance and clinical outcomes.17
Strong association of milk source (mother versus donor) with microbiome and fecal calprotectin
While fortifier type had only a minimal impact on the developing microbiome, MOM intake was strongly associated with multiple measures of microbiome composition. MOM intake was the single most important predictor of bacterial community structure at T3 and T4 (33–35 weeks adjusted gestational age), surpassing antibiotics and birth mode, which are well-known determinants of the infant microbiome. The major role of MOM was also evident from longitudinal analyses showing that infants with higher MOM intake experienced lower levels of Clostridium over time, along with greater increases in Bifidobacterium, Veillonella, and Propionibacterium, consistent with the previous studies.23,36,37
Some of these differences were evident from the initial sampling time point, indicating the importance of MOM in the first hours and days of life for the colonization (Propionibacterium) or constraint (Clostridium) of these organisms. Higher abundance of Clostridium spp., particularly C. difficile and C. perfringens, in early stages are associated with the development of NEC in preterm infants.38,39 For other taxa (e.g., Bifidobacterium), differences emerged and increased over time, indicating a continuing role for MOM in supporting or constraining their growth as the immature gut develops in the weeks after birth. For some taxa (e.g., Veillonella), low MOM intake appeared to delay the maturation trajectory by several weeks, whereas for others (e.g., Bifidobacterium and Propionibacterium) maturation remained suppressed for the entire study period. Further research is warranted to determine whether and when these bacteria were eventually acquired by infants with low MOM intake and to understand the clinical consequences of this disruption in their microbiome development.
MOM intake tended to be negatively correlated with fecal calprotectin (a biomarker of gut inflammation), consistent with existing evidence that MOM-enriched bacteria such as Bifidobacterium and Propionibacterium can improve gut health by producing anti-inflammatory short-chain fatty acids.40 Furthermore, as others have shown,41,42 we observed greater weight gain in infants fed with high MOM proportions, suggesting a growth-promoting effect of MOM compared with DHM, perhaps mediated through the microbiome. Further research is needed to explore this hypothesis, but it is consistent with the known impact of DHM processing (pasteurization and freezing) on the maternal cells, microbes, and bioactive proteins (including lipase and adipokines) found in fresh MOM,19, 20, 21,43 and the unique composition of MOM in early lactation compared with mature milk typically provided to donor milk banks.18,43,44 Somewhat unexpectedly, we observed that infants with high MOM intake had enrichment of one unclassified member of Enterobacteriaceae, a family comprising many (but not exclusively) inflammatory species. Further analysis with strain-level resolution would be required to identify this taxon and its biological properties.
Impact of H2MF on fecal calprotectin, but not oxidative stress
We did not find any impact of the feeding regime on oxidative stress (urinary F2-isoprostanes), although differences have been reported in previous studies.8,45 Interestingly, and seemingly in contrast to our hypothesis that BHMF triggers gut inflammation, we observed 2-fold higher fecal calprotectin (a potential biomarker of gut inflammation) over time among infants fed H2MF. Previous reports found no difference in calprotectin between BHMF- and H2MF-fed infants.17 The clinical relevance of a 2-fold difference in fecal calprotectin is unclear because pathological cut-offs have not been established in VLBW infants, and typically much larger differences (5- to 20-fold) have been associated with gut pathology.46 Notably, although fecal calprotectin is an established biomarker of gut inflammation in adults, its relevance during infancy is unclear. In fact, others have observed higher fecal calprotectin in exclusively breast-fed versus formula-fed infants,47 and suggest that breast milk is a key source of calprotectin that significantly influences fecal calprotectin content.48 Moreover, it has been hypothesized that rising fecal calprotectin during the early postnatal period reflects normal gut development rather than always implicating pathological inflammation.49 Together with these previous studies, our findings indicate a need for further research to understand the role of calprotectin during early gut development and its utility as a clinical biomarker in this unique population.
Limitations of the study
Our study is limited by a relatively small sample size of 30 infants and relatively short follow-up period to 35 weeks adjusted age. We powered the trial to detect a moderate-to-large effect on the microbiome, and indeed, this sample was sufficient to detect clear associations between MOM intake and microbiota composition. The impact of H2MF was comparably much smaller, indicating that either fortifiers do not affect the overall microbiome in this population of human-milk-fed VLBW infants, or they have a relatively small impact that was not detectable in our study. It is also important to note that H2MF is a concentrated form of donor milk that displaces MOM in the feed, which needs to be accounted for while designing and interpreting future studies. Another limitation of our study is the amplicon sequencing approach. It is possible that fortifier type is associated with strain-level differences or functional changes that were not captured by 16S rRNA gene profiling. It is also important to note that our findings related to MOM intake are observational. Finally, since no infants in our study were fed infant formula, we could not compare MOM or DHM with infant formula, nor evaluate the impact of fortifier type among infants receiving formula.
Conclusions
In this randomized controlled trial of human-milk-fed VLBW infants, the type of milk fortifier (bovine versus human) had minimal impact on the gut microbiome, whereas the source of human milk (mother versus donor) was strongly associated with microbiome composition. Our findings do not provide a clear biological basis for the clinical impact of H2MF, but emphasize the importance of feeding MOM in this population of fragile infants.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Fecal samples | This paper | N/A |
| Urine samples | This paper | N/A |
| Chemicals and reagents | ||
| ZR BashingBead Lysis Tubes | Zymo Research, USA | Catalog. No. S6003-50 |
| Zymo-Spin IIC Columns | Zymo Research, USA | Catalog. No.C1011-50 |
| BioVendor Extraction buffer | BioVendor, Czech Republic | Catalog No.: C005821 |
| Critical commercial assays | ||
| Zymo Quick DNA Fecal Microbe Miniprep kit | Zymo Research, USA | Catalog. No. D6010 |
| ZymoBIOMICS Microbial Community DNA Standard | Zymo Research, USA | Catalog. No. D6305 |
| Genomic DNA from Microbial Mock Community B (Staggered, Low Concentration) | BEI Resources, USA | Catalog. No.: HM-783D |
| S100A8/A9 (Calprotectin) Human ELISA kit |
BioVendor, Czech Republic | Catalog No.: RD191217100R; RRID:AB_10953195; RRID: AB_1227456 |
| F2-Isoprostane ELISA kit | Oxford biomedical research, USA | Catalog No.: EA-85; RRID: AB_10832110 |
| Creatinine microplate assay kit | Oxford biomedical research, USA | Catalog No.: CR-01 |
| Deposited data | ||
| Fastq files from 16S rRNA gene sequencing | This paper | Accession number: NCBI SRA under BioProject ID: PRJNA690658 |
| Software and algorithms | ||
| PediTools version 1.1.1 | Chou et al.25 | https://www.peditools.org/fenton2013/ |
| R version 3.6.1 | The R Project for Statistical Computing | https://www.r-project.org/ |
| Vegan R package version 2.5-7 | CRAN R project | https://cran.r-project.org/web/packages/vegan/index.html |
| Phyloseq R package version 1.30.0 | McMurdie and Holmes50 | https://joey711.github.io/phyloseq/ |
| Microbiome R package version 1.8.0 | Lahti and Shetty51 | https://microbiome.github.io/tutorials/ |
| Splinectome R package version 0.1.0 | Shields-Cutler et al.52 | https://rrshieldscutler.github.io/splinectomeR/ |
| ComplexHeatMap R package version 2.2.0 | Gu et al.53 | https://jokergoo.github.io/ComplexHeatmap-reference/book/ |
| MaAsLin2 R package version 1.5.2 | Mallick et al.54 | https://github.com/biobakery/Maaslin2 |
| HIT DB version 1.0 | Ritari et al.55 | https://github.com/openresearchlabs/HITdb |
| DADA2 R package version 1.14.1 | Callahan et al.56 | https://benjjneb.github.io/dada2/tutorial.html |
| Decontam R package version 1.6.0 | Davis et al.57 | https://benjjneb.github.io/decontam/vignettes/decontam_intro.html |
| ggplot2 R package version 3.3.3 | Tidyverse project R | https://ggplot2.tidyverse.org/ |
Resource availability
Lead contact
Further information and requests for resources should be made directly to and will be fulfilled by, the lead contact, Dr. Meghan Azad (meghan.azad@umanitoba.ca).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Microbiome sequence data is available at the NCBI SRA under BioProject ID: PRJNA690658.
-
•
All original code has been deposited (https://github.com/shreyaskumbhare/Preterm-and-fortifiers-article-analysis.git) and is publicly available as of date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Sample size
We based our sample size on the primary outcome of microbiota composition. Based on a reported 49% decrease in microbiota diversity among infants with necrotizing enterocolitis versus healthy controls,58 we calculated that 26 infants (13 per group) would be required to detect a similar effect size, with 80% power and 5% significance. Allowing for 15% loss to follow up, we enrolled 30 infants.
Eligibility, enrollment, blinding, and allocation
All neonates born between 26 and 30 weeks gestation at the Winnipeg Health Sciences Centre between September 2017 and April 2019 were screened for eligibility on the day of birth (Figure 1A). Parents willing to participate provided written informed consent. Inclusion criteria were: birth weight <1250g, gestational age between 26 + 0 to 30+0 weeks, parenteral nutrition started by day two after birth, consent to receive DHM, and enteral feeding >80 mL/kg/day by day 14 after birth. Exclusion criteria were: major congenital malformation, intestinal perforation or stage-2 necrotizing enterocolitis, unlikely to survive the study period, probiotics at any time, or receiving antibiotics on study day 0. It was not feasible to blind clinical staff because of the different preparation protocols for BHMF and H2MF. The randomization sequence was created by a non-clinical staff member using a computer-based algorithm. Study group allocations were kept in sealed envelopes until interventions were assigned. This study used the identifier ‘mother’ in the registered protocol and recruitment materials and hence the terminology ‘Mother’s Own Milk’. We acknowledge that not all breastfeeding parents identify as ‘mothers’ and are committed to using more inclusive language in our future research.
Study design
We conducted a randomized, controlled open-label trial in VLBW premature neonates (born <1250g, 26–30 weeks gestational age) fed standard BHMF, or H2MF (ClinicalTrials.gov NCT03214822) (Figure 1A). Feces and urine were collected on study day 0 (T1: just before fortification), day 7 (T2: after 1 week of fortification), 33+0 weeks AGA (T3: end of H2MF fortification), and 35+0 weeks AGA (T4: 2 weeks after the end of H2MF fortification) (Figure 1B). Parents provided written informed consent. The study was approved by the University of Manitoba and Public Health Agency of Canada/Health Canada research ethics boards.
Intervention and control procedures
Infants randomized to BHMF received the standard feeding protocol of human milk (DHM if MOM was not available) fortified with BHMF (Enfamil™ Human Milk Fortifier, Mead Johnson, USA). Infants randomized to H2MF received human milk fortified with H2MF (Prolact+ H2MF®, Prolacta Biosciences, USA) until an adjusted gestational age (AGA) of 33+0 weeks was reached. After this period, the H2MF infants were weaned onto the standard feeding protocol of human milk with BHMF until the end of the study duration (AGA of 35 weeks or discharge).
Mother’s own milk (MOM) intake
Fortified milk fed to infants in both study groups consisted of varying amounts of MOM and DHM, depending on the availability of MOM. We calculated the cumulative proportion of MOM used in the preparation across all feedings (volume MOM/total enteral volume) and classified each infant as High (≥50%) or Low (<50%) MOM at each time point. The %MOM volumes reflect the proportion of MOM used to prepare the feeds prior to fortification, i.e. they do not account for displacement by fortifiers. We also used MOM proportion as a continuous variable.
Feeding protocols
Minimal enteral feeding (MEF; 1 mL/kg human milk [MOM or, if unavailable, DHM] every 2 hours) was started as soon as possible after birth (within 24h). Once MEF tolerance was established (typically between 72-120 hours after birth), enteral feeding was increased by 25 mL/kg/day up to 80 mL/kg/day, then fortification was initiated. Infants randomized to BHMF received the standard feeding protocol of human milk fortified with BHMF (Enfamil™ Human Milk Fortifier, Mead Johnson, USA). 100 mL of breast milk (MOM or DHM) was mixed with 4 packets of BHMF (2.84 g powder). Infants randomized to H2MF received human milk fortified with H2MF (Prolact+ H2MF®, Prolacta Biosciences, USA) until an adjusted gestational age (AGA) of 33+0 weeks was reached. H2MF fortified milk was prepared by using MOM or DHM (based on availability) with H2MF of strengths: +4, +6 and +8, each displacing 20mL, 30mL and 40mL respectively, out of the 100mL recipe. The dose increment was based on feed tolerance, nutrient needs and growth rate. After this period, the H2MF infants were weaned onto the standard feeding protocol of human milk with BHMF until the end of study duration (AGA of 35 weeks or discharge). Nutritional support was provided per standard protocols. Per the standard feeding protocol, infants received Total Parenteral Nutrition with SMOF lipid emulsion (Smoflipid® SMOF 20%, Fresenius Kabi, Sweden) and additional nutritional support as needed to meet nutrient goals, including liquid protein fortifier (LiquiProtein™; Abbott Nutrition, USA) and/or additional fortifier (Enfamil A+® Enfacare® 24 cal/oz or Nutramigen® A+® powder 2.5 g, Mead Johnson, USA). Pasteurized DHM (using holder pasteurization: heated at 62.5°C for 30 minutes) was procured from NothernStar mothers milk bank, Canada.
Sample collection and storage
Fecal and urine samples were collected on study day 0 (T1: just before fortification began), day 7 (T2: after 1 week of fortification), at 33+0 weeks AGA (T3: end of H2MF fortification), and at 35+0 weeks AGA (T4: 2 weeks after the end of H2MF fortification). The week 33 (T3) samples for the H2MF group were collected after the H2MF fortification was stopped, but prior to when they were switched to BHMF fortification. Urine samples were collected from absorbent cotton balls placed in infants’ diapers and refrigerated for no more than 24 h until transferred to the laboratory at Children’s Hospital Research Institute of Manitoba (CHRIM). The cotton balls were then centrifuged to extract liquid urine, and the urine samples aliquoted to 1.5 mL plastic tubes were then stored at −80°C until analysis. Fecal samples were also collected at the same time points from soiled diapers as soon as possible after the feces were produced. The diapers were refrigerated in sterile containers for no more than 24 h until aliquoted and transferred in −80°C freezers at CHRIM until further analysis.
Method details
Fecal DNA extraction
Two punches (using the sterile disposable biopsy punches) of frozen (−80°C) feces were transferred to a Zymo Research Bashing Bead™ Lysis Tube (Zymo Research, USA) with the genomic lysis solution and secured in a bead-beater fitted with a 2-mL tube holder assembly and processed for 10 minutes at 1200 rpm. 467 μL of genomic lysis buffer and 333.5 μL isopropanol was added to this lysate. After mixing, the lysate was transferred to the Zymo-Spin™ IIC (Zymo Research, USA) column in a clean 1.5 mL microcentrifuge tube and a 50 μL pre-heated DNA elution buffer was added to the column matrix. After 3 minutes, the assembly was subjected to centrifugation at 10,000 x g for 30 seconds and this step was repeated again after transferring the eluent onto the same column. DNA was eluted and stored in sterile 1.5 mL tubes.
16S rRNA gene sequencing-based microbiome profiling
Genomic DNA was extracted from fecal samples using Zymo Quick DNA Fecal Microbe Miniprep kit (Cat. no. D6010, Zymo Research, USA) and sequenced following amplification of the 16S rRNA V4 hypervariable region.59 Sequencing was performed in two batches on the Illumina MiSeq using V3 chemistry (600 cycles: 2 X 300 bp). Sterile DNA-free water was used as negative control and a mock community (ZymoBiomics mock community, HM-783D BEI resources mock community B: staggered, low concentrations) as a positive control during library preparation for each sequencing run and was subjected for amplicon sequencing along with the other samples.
ELISA based F2-Isoprostanes and calprotectin quantification
50–100 mg of stool samples was used for extraction by diluting (50x as per the initial weight) with the BioVendor extraction buffer (Cat. No. C005821; BioVendor, Czech Republic), homogenized by vortexing, and centrifuged at 3000 x g for 30 min. The supernatant thus obtained was diluted 200x (as per manufacturer’s instructions) before the assay. Urine samples were pretreated with glucuronidase (as per manufacturer’s instructions) to obtain free F2-Isoprostanes and then were diluted (1:4) before the assay. Biomarkers of gut inflammation (fecal Calprotectin) and oxidative stress (urinary F2-Isoprostanes) were quantified using ELISA. Calprotectin was quantified using the Human S100A8/A9 ELISA kit (BioVendor, Czech Republic). Oxidative stress was quantified using urinary F2-Isoprostane ELISA kits (EA-85, Oxford biomedical research, USA) normalized to creatinine concentrations.60 Urinary creatinine was quantified using a creatinine microplate assay kit (CR01, Oxford biomedical research, USA).
Quantification and statistical analysis
Sequence data preprocessing
Sequence reads from both sequencing batches were processed separately and merged before downstream analysis in R. Overlapping paired-end reads were preprocessed using the dada2 pipeline56 for all the samples (including controls) in R (R Core Team, 2017) for each batch. The Amplicon Sequence Variants (ASVs) were then subjected to Chimera removal.61 Unique ASVs were assigned taxonomy using the Human Intestinal 16S rRNA gene reference taxonomy Database (HITdb, V1.0).55 Potential contaminant sequences were filtered using the frequency of ASVs in negative controls.57 The merged sequence tables (both sequencing runs) were then screened for ASVs classified as Euryarchaeota, Chloroplast, Mitochondria, and those ASVs unclassified at Phylum level (n = 618). Furthermore, three low read samples (<500 reads) were removed from further analysis, resulting in sequence reads for n = 117 samples and 595 unique ASVs in total. The sparsity of the dataset was tested using microbiome package.51 ASVs that were not seen more than once in at least 10% of the samples were filtered from the total dataset. This rigorous approach reduced the number of unique ASVs from 595 to 117, removed 7% of reads from the entire dataset, and reduced the sparsity of the data, overcoming the consequences of ASVs with small mean and trivially large CV. The sequencing reads were then rarified at 7578 reads per sample (the lowest number of reads per sample in the dataset) unless otherwise specified. Data was agglomerated at genus level for downstream analysis. Microbiome longitudinal analysis (using splinectomeR52 and mixed regression model in MaAsLin254) was performed including infants with data for all time points (n = 108 samples).
Data analysis and statistical methods
Raw reads were processed to filter low-quality, chimeric and contaminant sequences; then analyzed using R. A compositional analysis approach involving Centred-Log-Ratio (CLR) transformation was employed for downstream analysis unless otherwise specified. Alpha and beta diversity analyses were performed using phyloseq.50 The effect size and significance of clinical, dietary, maternal and infant covariates was determined using the ‘EnvFit’ function in vegan.62 Multivariate analysis was performed using PERMANOVA.
Data were analyzed in R. Microbiome metrics and biomarker values across study time points were compared between groups by Kruskal–Wallis, Fischer’s exact or Wilcoxon sum rank tests using false discovery rate (FDR) correction for multiple comparisons. Beta-diversity was compared between groups using PERMANOVA, followed by a permutation test for homogeneity of multivariate dispersions (betadisper) with 99,999 permutations to test whether two groups are homogeneously dispersed (by comparing the compositional variance of samples) in relation to their taxonomic groups. For repeated measures analysis (analyzing all time points together; Figures 1D and S1) we used a blocking method by setting strata to Infant IDs in the PERMANOVA test. Spearman correlations with Benjamini-Hochberg correction were determined using the microbiome package and heat maps were plotted using ComplexHeatMap package.51,53 Longitudinal analysis was performed using SplinectomeR.52 Multivariable regression was performed using the MaAsLin2 package in R.54 MOM proportion was used as a time co-varying variable (both in categorical and continuous form). Infants who received more vs. less than 50% MOM were classified as High or Low MOM respectively (See Figure S8A). To account for the cumulative MOM consumption over time, MOM proportion was calculated at each time point by taking an average of the proportion of MOM in TEV received by the infant since birth. Z-scores (weight-for-age) were calculated using PediTools25 which uses the 2013 Fenton growth charts for preterm infants63 to report Z-scores.
Additional resources
The study protocol is registered under ClinicalTrials.gov. National Library of Medicine (U.S.) (2017, July 12) (https://clinicaltrials.gov/ct2/show/NCT03214822, Identifier: NCT03214822).
Acknowledgments
We thank Dr. Shirin Moossavi, University of Calgary, and Dattatray Mongod, the University of Pune, for their inputs in microbiome analysis. We also thank the study participants, the clinical staff, NICU dietitians and diet technicians at the Health Sciences Centre, and the research support unit staff at Children’s Hospital Research Institute of Manitoba. This research was supported by Prolacta Biosciences and a Children's Hospital Research Institute of Manitoba (CHRIM) operating grant. Prolacta Biosciences provided a discount on the human-derived human milk fortifier tested in this study, guidance on study design, and funds to support clinical research operations. A CHRIM operating grant funded biological sample analyses. Neither entity was involved in data collection, data analysis, interpretation of results, writing of the manuscript, or decision to publish. Dr. Kumbhare was partially supported by a Molly Towell Perinatal Research Foundation postdoctoral fellowship. Dr. Azad is supported by a Tier 2 Canada Research Chair in the Developmental Origins of Chronic Disease and is a Fellow of the CIFAR Humans and the Microbiome program.
Author contributions
Conceptualization, M.B.A. and M.N.; methodology, S.V.K., W.D.J., G.J., M.G., M.N., and M.B.A.; investigation, S.V.K., W.D.J., C.B., S.F., and G.V.D.; formal analysis, S.V.K.; writing – original draft, S.V.K. and M.B.A.; writing – review & editing, S.V.K., W.D.J., S.F., C.B., G.J., G.V.D., M.G., M.N., and M.B.A.; funding acquisition, M.N. and M.B.A.; resources, M.G., M.N., and M.B.A.; supervision, M.B.A.
Declaration of interests
M.B.A. has consulted for DSM Nutritional Products and serves on the Malaika Vx and Tiny Health scientific advisory boards. She has received honoraria for speaking at symposia sponsored by Medela, Prolacta Biosciences, and the Institute for Advancement of Breastfeeding and Lactation Education and has contributed without remuneration to online courses on breast milk and the infant microbiome produced by Microbiome Courses. S.V.K. is currently employed by Digbi Health (3T and AI Pvt. Ltd., India), a position taken up after concluding the research presented in this study. These entities had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the article; or decision to submit the article for publication.
Published: September 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100712.
Supplemental information
References
- 1.Arslanoglu S., Boquien C.-Y., King C., Lamireau D., Tonetto P., Barnett D., Bertino E., Gaya A., Gebauer C., Grovslien A., et al. Fortification of human milk for preterm infants: update and recommendations of the European milk bank association (EMBA) working group on human milk fortification. Front. Pediatr. 2019;7:76. doi: 10.3389/fped.2019.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dutta S., Singh B., Chessell L., Wilson J., Janes M., McDonald K., Shahid S., Gardner V.A., Hjartarson A., Purcha M., et al. Guidelines for feeding very low birth weight infants. Nutrients. 2015;7:423–442. doi: 10.3390/nu7010423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregory K. Update on nutrition for preterm and full-term infants. J. Obstet. Gynecol. Neonatal Nurs. 2005;34:98–108. doi: 10.1177/0884217504272805. [DOI] [PubMed] [Google Scholar]
- 4.Martin C.R., Ling P.-R., Blackburn G.L. Review of infant feeding: key features of breast milk and infant formula. Nutrients. 2016;8:279. doi: 10.3390/nu8050279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neu J. Mother’s own milk: how does it differ from donor milk for the baby. Breastfeed. Med. 2019;14:S3–S4. doi: 10.1089/bfm.2019.0036. [DOI] [PubMed] [Google Scholar]
- 6.Asbury M.R., Butcher J., Copeland J.K., Unger S., Bando N., Comelli E.M., Forte V., Kiss A., LeMay-Nedjelski L., Sherman P.M., et al. Mothers of preterm infants have individualized breast milk microbiota that changes temporally based on maternal characteristics. Cell Host Microbe. 2020;28:669–682.e4. doi: 10.1016/j.chom.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Shulhan J., Dicken B., Hartling L., Larsen B.M. Current knowledge of necrotizing enterocolitis in preterm infants and the impact of different types of enteral nutrition products. Adv. Nutr. 2017;8:80–91. doi: 10.3945/an.116.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friel J.K., Diehl-Jones B., Cockell K.A., Chiu A., Rabanni R., Davies S.S., Roberts L.J., 2nd Evidence of oxidative stress in relation to feeding type during early life in premature infants. Pediatr. Res. 2011;69:160–164. doi: 10.1203/PDR.0b013e3182042a07. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan S., Schanler R.J., Kim J.H., Patel A.L., Trawöger R., Kiechl-Kohlendorfer U., Chan G.M., Blanco C.L., Abrams S., Cotten C.M., et al. An exclusively human milk-based diet is associated with a lower rate of necrotizing enterocolitis than a diet of human milk and bovine milk-based products. J. Pediatr. 2010;156:562–567.e1. doi: 10.1016/j.jpeds.2009.10.040. [DOI] [PubMed] [Google Scholar]
- 10.Collado M.C., Cernada M., Neu J., Pérez-Martínez G., Gormaz M., Vento M. Factors influencing gastrointestinal tract and microbiota immune interaction in preterm infants. Pediatr. Res. 2015;77:726–731. doi: 10.1038/pr.2015.54. [DOI] [PubMed] [Google Scholar]
- 11.Torrazza R.M., Ukhanova M., Wang X., Sharma R., Hudak M.L., Neu J., Mai V. Intestinal Microbial Ecology and Environmental Factors Affecting Necrotizing Enterocolitis. PLoS One. 2013;8:e83304. doi: 10.1371/journal.pone.0083304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulet O. Potential role of the intestinal microbiota in programming health and disease. Nutr. Rev. 2015;73:32–40. doi: 10.1093/nutrit/nuv039. [DOI] [PubMed] [Google Scholar]
- 13.Cristofalo E.A., Schanler R.J., Blanco C.L., Sullivan S., Trawoeger R., Kiechl-Kohlendorfer U., Dudell G., Rechtman D.J., Lee M.L., Lucas A., Abrams S. Randomized trial of exclusive human milk versus preterm formula diets in extremely premature infants. J. Pediatr. 2013;163:1592–1595.e1. doi: 10.1016/j.jpeds.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Abrams S.A., Schanler R.J., Lee M.L., Rechtman D.J., the Prolacta Study Group Greater mortality and morbidity in extremely preterm infants fed a diet containing cow milk protein products. Breastfeed. Med. 2014;9:281–285. doi: 10.1089/bfm.2014.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen A.-C., Chung M.-Y., Chang J.H., Lin H.-C. Pathogenesis implication for necrotizing enterocolitis prevention in preterm very-low-birth-weight infants. J. Pediatr. Gastroenterol. Nutr. 2014;58:7–11. doi: 10.1097/MPG.0b013e3182a7dc74. [DOI] [PubMed] [Google Scholar]
- 16.Ghandehari H., Lee M.L., Rechtman D.J., H2MF Study Group An exclusive human milk-based diet in extremely premature infants reduces the probability of remaining on total parenteral nutrition: a reanalysis of the data. BMC Res. Notes. 2012;5:188. doi: 10.1186/1756-0500-5-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connor D.L., Kiss A., Tomlinson C., Bando N., Bayliss A., Campbell D.M., Daneman A., Francis J., Kotsopoulos K., Shah P.S., et al. Nutrient enrichment of human milk with human and bovine milk-based fortifiers for infants born weighing <1250 g: a randomized clinical trial. Am. J. Clin. Nutr. 2018;108:108–116. doi: 10.1093/ajcn/nqy067. [DOI] [PubMed] [Google Scholar]
- 18.Meier P.P., Patel A.L., Bigger H.R., Chen Y., Johnson T.J., Rossman B., Engstrom J.L. In: Diet and Nutrition in Critical Care. Rajendram R., Preedy V.R., Patel V.B., editors. Springer New York; 2015. Human milk feedings in the neonatal intensive care unit; pp. 807–822. [DOI] [Google Scholar]
- 19.Meier P., Patel A., Esquerra-Zwiers A. Donor human milk update: evidence, mechanisms, and priorities for research and practice. J. Pediatr. 2017;180:15–21. doi: 10.1016/j.jpeds.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juncker H.G., Ruhé E.J.M., Burchell G.L., van den Akker C.H.P., Korosi A., van Goudoever J.B., van Keulen B.J. The effect of pasteurization on the antioxidant properties of human milk: a literature review. Antioxidants. 2021;10:1737. doi: 10.3390/antiox10111737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collado M.C., Santaella M., Mira-Pascual L., Martínez-Arias E., Khodayar-Pardo P., Ros G., Martínez-Costa C. Longitudinal study of cytokine expression, lipid profile and neuronal growth factors in human breast milk from term and preterm deliveries. Nutrients. 2015;7:8577–8591. doi: 10.3390/nu7105415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zanella A., Silveira R.C., Roesch L.F.W., Corso A.L., Dobbler P.T., Mai V., Procianoy R.S. Influence of own mother’s milk and different proportions of formula on intestinal microbiota of very preterm newbornsPLoS One. 2019;14:e0217296. doi: 10.1371/journal.pone.0217296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ford S.L., Lohmann P., Preidis G.A., Gordon P.S., O'Donnell A., Hagan J., Venkatachalam A., Balderas M., Luna R.A., Hair A.B. Improved feeding tolerance and growth are linked to increased gut microbial community diversity in very-low-birth-weight infants fed mother’s own milk compared with donor breast milk. Am. J. Clin. Nutr. 2019;109:1088–1097. doi: 10.1093/ajcn/nqz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piñeiro-Ramos J.D., Parra-Llorca A., Ten-Doménech I., Gormaz M., Ramón-Beltrán A., Cernada M., Quintás G., Collado M.C., Kuligowski J., Vento M. Effect of donor human milk on host-gut microbiota and metabolic interactions in preterm infants. Clin. Nutr. 2021;40:1296–1309. doi: 10.1016/j.clnu.2020.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Chou J.H., Roumiantsev S., Singh R. PediTools electronic growth chart calculators: applications in clinical care, research, and quality improvement. J. Med. Internet Res. 2020;22:e16204. doi: 10.2196/16204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore R.E., Townsend S.D. Temporal development of the infant gut microbiome. Open Biol. 2019;9:190128. doi: 10.1098/rsob.190128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forbes J.D., Azad M.B., Vehling L., Tun H.M., Konya T.B., Guttman D.S., Field C.J., Lefebvre D., Sears M.R., Becker A.B., et al. Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. 2018;172:e181161. doi: 10.1001/jamapediatrics.2018.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borewicz K., Gu F., Saccenti E., Hechler C., Beijers R., de Weerth C., van Leeuwen S.S., Schols H.A., Smidt H. The association between breast milk oligosaccharides and faecal microbiota in healthy breastfed infants at two, six, and twelve weeks of age. Sci. Rep. 2020;10:4270. doi: 10.1038/s41598-020-61024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martín-Sosa S., Martín M.J., García-Pardo L.A., Hueso P. Sialyloligosaccharides in human and bovine milk and in infant formulas: variations with the progression of lactation. J. Dairy Sci. 2003;86:52–59. doi: 10.3168/jds.S0022-0302(03)73583-8. [DOI] [PubMed] [Google Scholar]
- 30.Robinson R.C. Structures and metabolic properties of bovine milk oligosaccharides and their potential in the development of novel therapeutics. Front. Nutr. 2019;6:50. doi: 10.3389/fnut.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vincent C., Stephens D.A., Loo V.G., Edens T.J., Behr M.A., Dewar K., Manges A.R. Reductions in intestinal Clostridiales precede the development of nosocomial Clostridium difficile infection. Microbiome. 2013;28:18. doi: 10.1186/2049-2618-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent C., Miller M.A., Edens T.J., Mehrotra S., Dewar K., Manges A.R. Bloom and bust: intestinal microbiota dynamics in response to hospital exposures and Clostridium difficile colonization or infection. Microbiome. 2016;14:12. doi: 10.1186/s40168-016-0156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Centers for Disease Control and Prevention . National Center for Emerging and Zoonotic Infectious Diseases, Division of Healthcare Quality and Promotion; Atlanta, GA: 2018. Clostridioides Difficile in Neonatal Intensive Care Unit Patients: A Systematic Review Centers for Disease Control and Prevention.https://www.cdc.gov/hicpac/reviews/cdiff-nicu/index.html [Google Scholar]
- 34.Mathew O.P., Bhatia J.S., Richardson C.J. An outbreak of Clostridium difficile necrotizing enterocolitis. Pediatrics. 1984;73:265–266. [PubMed] [Google Scholar]
- 35.Han V.K., Sayed H., Chance G.W., Brabyn D.G., Shaheed W.A. An outbreak of Clostridium difficile necrotizing enterocolitis: a case for oral vancomycin therapy? Pediatrics. 1983;71:935–941. [PubMed] [Google Scholar]
- 36.Parra-Llorca A., Gormaz M., Alcántara C., Cernada M., Nuñez-Ramiro A., Vento M., Collado M.C. Preterm gut microbiome depending on feeding type: significance of donor human milk. Front. Microbiol. 2018;9:1376. doi: 10.3389/fmicb.2018.01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z., Neupane A., Vo R., White J., Wang X., Marzano S.-Y.L. Comparing gut microbiome in mothers’ own breast milk- and formula-fed moderate-late preterm infants. Front. Microbiol. 2020;11:891. doi: 10.3389/fmicb.2020.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De la Cochetiere M.F., Piloquet H., des Robert C., Darmaun D., Galmiche J.P., Roze J.C. Early intestinal bacterial colonization and necrotizing enterocolitis in premature infants: the putative role of Clostridium. Pediatr. Res. 2004;56:366–370. doi: 10.1203/01.PDR.0000134251.45878.D5. [DOI] [PubMed] [Google Scholar]
- 39.Smith B., Bodé S., Skov T.H., Mirsepasi H., Greisen G., Krogfelt K.A. Investigation of the early intestinal microflora in premature infants with/without necrotizing enterocolitis using two different methods. Pediatr. Res. 2012;71:115–120. doi: 10.1038/pr.2011.1. [DOI] [PubMed] [Google Scholar]
- 40.Ríos-Covián D., Ruas-Madiedo P., Margolles A., Gueimonde M., de Los Reyes-Gavilán C.G., Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montjaux-Régis N., Cristini C., Arnaud C., Glorieux I., Vanpee M., Casper C. Improved growth of preterm infants receiving mother's own raw milk compared with pasteurized donor milk. Acta Paediatr. 2011;100:1548–1554. doi: 10.1111/j.1651-2227.2011.02389.x. [DOI] [PubMed] [Google Scholar]
- 42.Hård A.L., Nilsson A.K., Lund A.M., Hansen-Pupp I., Smith L.E.H., Hellström A. Review shows that donor milk does not promote the growth and development of preterm infants as well as maternal milk. Acta Paediatr. 2019;108:998–1007. doi: 10.1111/apa.14702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rai D., Adelman A.S., Zhuang W., Rai G.P., Boettcher J., Lönnerdal B. Longitudinal changes in lactoferrin concentrations in human milk: a global systematic review. Crit. Rev. Food Sci. Nutr. 2014;54:1539–1547. doi: 10.1080/10408398.2011.642422. [DOI] [PubMed] [Google Scholar]
- 44.Twigger A.-J., Hepworth A.R., Lai C.T., Chetwynd E., Stuebe A.M., Blancafort P., Hartmann P.E., Geddes D.T., Kakulas F. Gene expression in breastmilk cells is associated with maternal and infant characteristics. Sci. Rep. 2015;5:12933. doi: 10.1038/srep12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai C., Zhang Z., Morales M., Wang Y., Khafipour E., Friel J. Feeding practice influences gut microbiome composition in very low birth weight preterm infants and the association with oxidative stress: a prospective cohort study. Free Radic. Biol. Med. 2019;142:146–154. doi: 10.1016/j.freeradbiomed.2019.02.032. [DOI] [PubMed] [Google Scholar]
- 46.Pergialiotis V., Konstantopoulos P., Karampetsou N., Koutaki D., Gkioka E., Perrea D.N., Papantoniou N. Calprotectin levels in necrotizing enterocolitis: a systematic review of the literature. Inflamm. Res. 2016;65:847–852. doi: 10.1007/s00011-016-0963-9. [DOI] [PubMed] [Google Scholar]
- 47.Savino F., Castagno E., Calabrese R., Viola S., Oggero R., Miniero R. High faecal calprotectin levels in healthy, exclusively breast-fed infants. Neonatology. 2010;97:299–304. doi: 10.1159/000255161. [DOI] [PubMed] [Google Scholar]
- 48.Kaczmarczyk M., Löber U., Adamek K., Węgrzyn D., Skonieczna-Żydecka K., Malinowski D., Łoniewski I., Markó L., Ulas T., Forslund S.K., Łoniewska B. The gut microbiota is associated with the small intestinal paracellular permeability and the development of the immune system in healthy children during the first two years of life. J. Transl. Med. 2021;19:177. doi: 10.1186/s12967-021-02839-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Groer M., Ashmeade T., Louis-Jacques A., Beckstead J., Ji M. Relationships of feeding and mother's own milk with fecal calprotectin levels in preterm infants. Breastfeed. Med. 2016;11:207–212. doi: 10.1089/bfm.2015.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McMurdie P.J., Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leo Lahti, Sudarshan Shetty . 2017. Tools for Microbiome Analysis in R.http://microbiome.github.com/microbiome [Google Scholar]
- 52.Shields-Cutler R.R., Al-Ghalith G.A., Yassour M., Knights D. SplinectomeR enables group comparisons in longitudinal microbiome studies. Front. Microbiol. 2018;9:785. doi: 10.3389/fmicb.2018.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 54.Mallick H, McIver LJ, Rahnavard A, Ma S, Zhang Y, Nguyen LH, Tickle TL, Weingart G, Ren B, Schwager E, et al. Multivariable association discovery in population-scale meta-omics studies.Preprint at bioRxiv. 10.1101/2021.01.20.427420. [DOI] [PMC free article] [PubMed]
- 55.Ritari J., Salojärvi J., Lahti L., de Vos W.M. Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database. BMC Genom. 2015;16:1056. doi: 10.1186/s12864-015-2265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McMurtry V.E., Gupta R.W., Tran L., Blanchard E.E., 4th, Penn D., Taylor C.M., Ferris M.J. Bacterial diversity and Clostridia abundance decrease with increasing severity of necrotizing enterocolitis. Microbiome. 2015;3:11. doi: 10.1186/s40168-015-0075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma E., Ingram K.H., Milne G.L., Garvey W.T. F2-Isoprostanes reflect oxidative stress correlated with lean mass and bone density but not insulin resistance. J. Endocr. Soc. 2017;1:436–448. doi: 10.1210/js.2017-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Callahan B.J., Sankaran K., Fukuyama J.A., McMurdie P.J., Holmes S.P. Bioconductor Workflow for Microbiome Data Analysis: from raw reads to community analyses [version 2; peer review: 3 approved] F1000Res. 2016;5:1492. doi: 10.12688/f1000research.8986.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oksanen J., Blanchet F.G., Kindt R., Legendre P., O'Hara R.B. 2013. Vegan: Community Ecology Package. [Google Scholar]
- 63.Fenton T.R., Kim J.H. A systematic review and meta-analysis to revise the Fenton growth chart for preterm infants. BMC Pediatr. 2013;13:59. doi: 10.1186/1471-2431-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Microbiome sequence data is available at the NCBI SRA under BioProject ID: PRJNA690658.
-
•
All original code has been deposited (https://github.com/shreyaskumbhare/Preterm-and-fortifiers-article-analysis.git) and is publicly available as of date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.