Abstract
The sirA gene of Salmonella enterica serovar Typhimurium encodes a two-component response regulator of the FixJ family that has a positive regulatory influence on the expression of type III secretion genes involved with epithelial cell invasion and the elicitation of bovine gastroenteritis. SirA orthologs in Pseudomonas, Vibrio, and Erwinia control the expression of distinct virulence genes in these genera, but an evolutionarily conserved target of SirA regulation has never been identified. In this study we tested the hypothesis that sirA may be an ancient member of the flagellar regulon. We examined the effect of a sirA mutation on transcriptional fusions to flagellar promoters (flhD, fliE, fliF, flgA, flgB, fliC, fliD, motA, and fliA) while using fusions to the virulence gene sopB as a positive control. SirA had only small regulatory effects on all fusions in liquid medium (less than fivefold). However, in various types of motility agar plates, sirA was able to activate a sopB fusion by up to 63-fold while repressing flagellar fusions by values exceeding 100-fold. Mutations in the sirA orthologs of Escherichia coli, Vibrio cholerae, Pseudomonas fluorescens, and Pseudomonas aeruginosa result in defects in either motility or motility gene regulation, suggesting that control of flagellar regulons may be an evolutionarily conserved function of sirA orthologs. The implications for our understanding of virulence gene regulation in the gamma Proteobacteria are discussed.
The animal and plant pathogens of the gamma subdivision of Proteobacteria cause vast amounts of agricultural damage and human disease. These pathogens include members of the genera Pseudomonas, Erwinia, Escherichia, Vibrio, and Salmonella. The individual species among these genera are very diverse in some respects. They include free-living species, symbionts, commensals, plant pathogens, and animal pathogens. However, they also have striking similarities. For instance, a single locus has been identified as a transcriptional regulator of genes involved with secondary metabolism and/or virulence in all five genera. The locus is known as gacA in Pseudomonas species, varA in Vibrio cholerae, expA in Erwinia carotovora, uvrY in Escherichia coli, and sirA in Salmonella enterica serovar Typhimurium. These five genes (sirA, varA, gacA, expA, and uvrY) are orthologs based on the following criteria. They are highly conserved (each pair wise comparison shows at least 54% amino acid identity). The genomic context of each gene is conserved (each is located directly upstream of uvrC). Finally, in the four genera studied, the genes have similar functions (regulation of secondary metabolism and/or virulence; see below). For simplicity, the sirA orthologs of all species (uvrY, varA, gacA, and expA) will be referred to as sirA throughout this report.
By sequence homology, sirA orthologs encode a two-component response regulator of the FixJ family. The E. coli SirA ortholog is phosphorylated by a sensor kinase named BarA (53). Genetic evidence suggests that the SirA orthologs of Salmonella, Erwinia, and Pseudomonas are also phosphorylated by proteins orthologous to BarA of E. coli. The BarA ortholog is known as BarA in Salmonella, ExpA in Erwinia, and GacS, LemA, or PheN in Pseudomonas (7, 12, 22, 29, 31, 42, 50, 56, 57). For simplicity, the barA orthologs of all species (expA and lemA/gacS/pheN) will be referred to as barA throughout this report.
The phenotypes of sirA mutants suggest that SirA is near the top of a virulence gene regulatory cascade in each of the pathogens listed above. In V. cholerae, the sirA ortholog is required for expression of the ToxR regulon and colonization of the murine intestine (74). The sirA ortholog is required for extracellular enzyme production and plant virulence in Erwinia carotovora, Pseudomonas syringae, Pseudomonas aereofaciens, Pseudomonas marginalis, and Pseudomonas viridiflava (10, 12, 20, 42, 43). Pseudomonas aeruginosa requires the sirA ortholog for proper expression of the LasR and RhlR quorum-sensing cascade (55), which influences rpoS expression (38), extracellular virulence factor production (24, 51, 71), biofilm formation (16), twitching motility (28), and virulence in plant, animal, and nematode models (54, 61). Switching between the pathogenic wild-type and nonpathogenic phenotypic variant forms of Pseudomonas tolaasii involves a reversible DNA rearrangement within the barA (pheN) locus (30). A sirA ortholog is also required for swarming motility in Pseudomonas syringae (35) and expression of antifungal compounds and extracellular enzymes by Pseudomonas fluorescens (23, 39). In both E. coli and P. fluorescens, the sirA ortholog affects the expression of rpoS, which regulates oxidative stress resistance (49, 53, 70). In Azotobacter vinelandii, the sirA and barA orthologs regulate polymer synthesis (9).
In Salmonella serovar Typhimurium, sirA is required for optimal invasion of epithelial cells (32) and elicitation of fluid secretion and neutrophil migration into bovine ligated ileal loops (bovine gastroenteritis) (2). To do this, SirA positively regulates a pathogenicity island that encodes a type III secretion system (SPI1 for salmonella pathogenicity island 1). This secretion system directly injects effector proteins into the cytosol of host cells (11). Alteration of host cell signaling ensues, which can lead to uptake of bacteria into the host cell via macropinocytosis (6, 21). This invasion event is also associated with the elicitation of inflammation and fluid secretion into ligated bovine ileal loops (41; reviewed in references 62 and 68).
Although the biochemical details of SPI1 gene regulation are not known, genetically it appears that there is a regulatory heirarchy. SirA, which is encoded outside of SPI1, positively regulates another regulatory gene, hilA, that is encoded within SPI1 (2, 32). HilA then activates the genes that make up the structural components of the SPI1 type III secretion system and yet another regulator, invF (8). The entire sirA/hilA/invF cascade is required for the efficient expression of secreted substrates that are encoded both inside and outside of SPI1, with InvF potentially being the direct regulator (2, 8, 15, 18).
Despite the realization that sirA is required for virulence in several bacterial species, two observations led us to hypothesize that the primary function of sirA had not yet been discovered. First, sirA is found in both pathogens and non pathogens. Second, sirA is encoded within an evolutionarily conserved region of the genome, yet in every case, the virulence genes that sirA regulates are specific to each pathogen and probably acquired by horizontal transfer. This strongly suggested that sirA was present in these genomes before the acquisition of the virulence genes that it now controls. Given that flagellar regulons can influence virulence gene expression in a variety of species (13, 19, 26, 27, 33, 44, 59, 75) and that sirA is physically located between flagellar regions II and IIIa of the E. coli and S. enterica serovar Typhimurium genomes (32, 45), we hypothesized that SirA may be an ancient member of flagellar regulons. In this report, we have determined that SirA does indeed regulate flagellar promoters of serovar Typhimurium and E. coli. In addition, mutations in the sirA orthologs of V. cholerae, P. aeruginosa, and P. fluorescens result in motility defects, suggesting that SirA is a member of the flagellar regulons in these species as well.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) medium or on LB supplemented with 1.5% agar (EM Science) unless otherwise indicated. Motility assays were performed with plates containing agar concentrations varying between 0.25 and 0.35% (EM Science) in either LB medium, T medium (1% tryptone; Difco), TS medium (T plus 1% NaCl), or TSG medium (TS plus 0.2% glucose), as indicated. M9 minimal glucose medium was made as described (47). Ampicillin, tetracycline, chloramphenicol, kanamycin, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were added at 100, 20, 30, 60, and 80 μg/ml, respectively, when appropriate.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype or description | Source, construction, or reference |
|---|---|---|
| MG1655 | Wild-type Escherichia coli | E. coli Genetic Stock Center |
| RG133 | MG1655 uvrY33::Tn5 | This study |
| 14028 | Wild-type S. enterica serovar Typhimurium | American Type Culture Collection |
| BA746 | 14028 sirA3::cam | 2 |
| BA1526 | 14028 sopB1526::MudJ | 2 |
| BA1726 | 14028 sirA3::cam sopB1526::MudJ | 2 |
| 0395 | Vibrio cholerae | Stephen Calderwood (74) |
| SW33S | 0395 varA::aphA-3 | Stephen Calderwood (74) |
| PAO1 | Pseudomonas aeruginosa | Dieter Haas (55) |
| PAO6281 | PAO1 gacA::Sm/Sp | Dieter Haas (55) |
| CHAO | Pseudomonas fluorescens | Dieter Haas (39) |
| CHA89 | CHAO gacA::kan | Dieter Haas (39) |
| SM10λpir | E. coli thi-1 thr leu tonA lacY supE recA::RP4-2-tet::Mu-1kan::Tn7 integrant λpir | 48 |
| S17-1λpir | E. coli recA pro hsdR <RP4-2-tet::Mu-1kan::Tn7> λpir | 60 |
| BW20767 | E. coli RP4-2-tet::Mu-1kan::Tn7 integrant leu-63::IS10 recA1 creC510 hsdR17 endA1 zbf-5 uidA (ΔMluI):pir+thi | 46 |
| RG200 | 14028 flhD+/flhD::lacZY integrant | This study |
| RG201 | BA746 flhD+/flhD::lacZY integrant | This study |
| RG202 | 14028 fliA+/fliA::lacZY integrant | This study |
| RG203 | BA746 fliA+/fliA::lacZY integrant | This study |
| RG207 | 14028 fliC+/fliC::lacZY integrant | This study |
| RG208 | BA746 fliC+/fliC::lacZY integrant | This study |
| RG211 | 14028 fliE+/flE::lacZY integrant | This study |
| RG212 | BA746 fliE+/fliE::lacZY integrant | This study |
| RG213 | 14028 flgA+/flgA::lacZY integrant | This study |
| RG214 | BA746 flgA+/flgA::lacZY integrant | This study |
| pRE112 | Suicide vector, Camr, sacB R6K ori | 17 |
| pRE112uvrY | pRE112 uvrY+ | This study |
| pRE112uvrY33::Tn5 | pRE112 uvrY33::Tn5 | This study |
| pWSK29 | Low-copy-number cloning vector, Ampr, pSC101 | 69 |
| pBA305 | pWSK29 sirA+ | 2 |
| pSB401 | luxCDABE transcriptional fusion vector, Tetr, p15A | 72 |
| pRG19 | S. enterica serovar Typhimurium motA::luxCDABE | 1885 to 2689 of accession no. D3640 into ΔEcoRI site of pSB401 |
| pRG34 | S. enterica serovar Typhimurium fliA::luxCDABE | 250 to 899 of accession no. AB010947 into the filled-in ΔEcoRI site of pSB401 |
| pRG38 | Serovar Typhimurium flhD::luxCDABE | 499 to 1501 of accession no. D43640 into ΔEcoRI site of pSB401 |
| pRG39 | Serovar Typhimurium fliC::luxCDABE | 440 of accession no. X51740 to 619 of accession no. D13689 into ΔEcoRI site of pSB401 (≈800 bp) |
| pRG46 | Serovar Typhimurium fliD::luxCDABE | Same as pRG39 (fliC) fragment but in reverse orientation in pSB401 |
| pRG51 | Serovar Typhimurium flgA::luxCDABE | 445 of accession no. D13703 to 4200 of accession no. D25292 into ΔEcoRI site of pSB401 (≈850 bp) |
| pRG52 | Serovar Typhimurium flgB::luxCDABE | Same as pRG51 (flgA) fragment but in reverse orientation in pSB401 |
| pRG53 | Serovar Typhimurium fliE::luxCDABE | 600 of accession no. M24462 to 481 of accession no. M84993 into filled-in ΔEcoRI site of pSB401 (962 bp) |
| pRG54 | Serovar Typhimurium fliF::luxCDABE | Same as pRG53 (fliE) fragment but in reverse orientation in pSB401 |
| pBA409 | Serovar Typhimurium sopB::luxCDABE | 4 to 657 of accession no. AF021817 into ΔEcoRI site of pSB401 |
| pRG26 | E. coli flhD::luxCDABE | 1310 to 392 of accession no. 1788200 into ΔEcoRI site of pSB401 |
| pRG25 | E. coli fliA::luxCDABE | 1825 to 994 of accession no. 1788229 into ΔEcoRI site of pSB401 |
| pRG14 | E. coli motA::luxCDABE | 360 of accession no. 1788200 to 10448 of accession no. 1788189 into ΔEcoRI site of pSB401 |
| pVIK112 | Promoterless lacZY suicide vector, Kanr | 34 |
Construction of an E. coli uvrY::Tn5 mutant (RG133).
The E. coli ortholog of sirA, uvrY, was disrupted with a Tn5 insertion. To do this, the region of DNA surrounding uvrY (ca. 860 bp upstream and 200 bp downstream of uvrY) was amplified using Pfu Turbo DNA polymerase (Stratagene) with MG1655 as the template DNA. The forward primer was BA402 (ATCTCTGAGAATACGGTCAATTTCCAC), and the reverse primer was BA256 (AACCGTTACATCAATTTGCTGGATC). The resulting PCR product was cloned using pCR-Blunt II-Topo (Invitrogen). The uvrY fragment was subsequently removed by XbaI and SacI restriction and ligated into the mobilizable sacB suicide vector pRE112 (Camr) digested with XbaI and SacI to give plasmid pRE112uvrY. This plasmid was mutagenized in vitro using the EZ::TN insertion kit (Epicentre Technologies). The mutagenized plasmids were transformed into S17λpir, selecting for kanamycin and chloramphenicol resistance. Transformants with uvrY::Tn5 insertions were identified by PCR screening with the reverse primer of EZ::TN (Epicentre) and primer BA256. This screening strategy identifies only Tn5 insertions in which the Kanr gene of Tn5 is oriented opposite to uvrY. Insertion points were confirmed using DNA sequencing with the forward and reverse primers of EZ::TN (Epicentre). A single insertion in the 56th codon of uvrY was chosen for further study and designated uvrY33::Tn5. To recombine this allele into the MG1655 chromosome, SM10λpir carrying pRE112uvrY33::Tn5 was mated with MG1655, selecting for kanamycin resistance on M9 minimal medium. To select against plasmid integrants, the transformants were pooled, incubated at 37°C in shaking LB broth for 8 h, and plated on LB-kanamycin lacking NaCl but containing 5% sucrose (17). PCR screening with primers BA256 and BA402 confirmed the absence of the uvrY+ allele and the presence of the uvrY33::Tn5 allele. One isolate was designated RG133 and kept for further study.
Reporter constructions.
To examine the regulation of flagellar genes, both episomal luxCDABE and chromosomal merodiploid lacZY transcriptional fusions were constructed. pSB401 is a reporter vector containing a p15A origin of replication, a tetracycline resistance marker, and a promoterless luxCDABE operon from Photorhabdus luminescens (72). Upstream of the luciferase operon is an EcoRI fragment containing a luxI promoter from Vibrio fischeri. This fragment was removed and replaced with regulatory regions of interest. The regulatory regions were amplified using Pfu Turbo DNA polymerase (Stratagene) with 14028 as the template (Table 1). The resulting PCR products were gel purified using Qiagen gel extraction columns and cloned using pCR-Blunt II-Topo (Invitrogen). The cloning site of pCR-Blunt II-Topo is flanked by EcoRI sites, so the EcoRI fragment of each clone was gel purified and ligated into the ΔEcoRI site of pSB401. The fliA and fliE promoters contain an internal EcoRI site, so the blunt-ended PCR product was ligated directly into pSB401 that had been digested with EcoRI and filled in using the Klenow fragment of DNA polymerase. The fliF promoter DNA fragment is identical to that of fliE except that they are in opposite orientations with respect to luxCDABE. The flgA and flgB fusions, as well as the fliC and fliD fusions, are also identical DNA fragments cloned in the opposite orientation with respect to luxCDABE. The reporter plasmids were placed into the appropriate strains using electroporation with a Bio-Rad Gene Pulser II.
Chromosomal merodiploid lacZY transcriptional fusions were constructed to the promoters of flhD, fliA, fliC, fliE, and flgA. For flhD, fliC, and flgA, this was done by removing the promoter region from the appropriate pSB401-based luxCDABE fusion plasmid (pRG38, pRG39, and pRG51 respectively) by EcoRI digestion and inserting it into the EcoRI site of the suicide vector pVIK112 (34). In the case of fliA and fliE, a blunt-ended PCR product (identical to that described above for construction of pRG34 and pRG53, respectively) was ligated directly into the SmaI site of pVIK112. BW20767 carrying the resulting plasmids was mated with wild-type and sirA mutant serovar Typhimurium, selecting for plasmid integrants by kanamycin resistance on M9 minimal medium. Transconjugants were designated RG200 to RG214 (Table 1) and examined for sirA-dependent gene regulation in TS motility agar containing kanamycin and X-Gal (80 μg/ml, final concentration).
Assay of luciferase activity.
Luciferase activity was measured after growth of the bacteria under three types of conditions: shaking liquid culture, standing liquid culture, and motility agar. Shaking cultures were 5-ml cultures in tubes (18 by 150 mm) rotating at 50 rpm at a nearly horizontal angle. At various time points, the optical density of these cultures at 550 nm was measured using a Spectronic 20D+ or a Beckman DU-64 spectrophotometer. Samples (10 μl) were then taken for measurement of luciferase activity in a Turner Designs TD-20/20 luminometer. Results are expressed as relative light units per second. The standing liquid culture is a 1:50 subculture of an overnight culture which is left standing without agitation at 37°C for 6 h (40). At the 6-h point, luciferase activity was measured in a 10-μl sample with the Turner Designs TD-20/20 luminometer. All luminometer samples were oxygenated by “ratcheting” the sample tube across a tube rack prior to insertion into the luminometer.
Expression of luciferase activity in motility agar plates was imaged and quantitated using a Hamamatsu C2400-32 intensified charge-coupled device camera with an Argus 20 image processor. Images were captured with a Macintosh G4 computer and Adobe Photoshop 5.0 software. Comparison of light intensity between two strains within the same image is very accurate within a 2-log linear range. However, the optimal intensifier setting required to get each sample into the linear range varies from plate to plate. Therefore, all results are expressed as fold differences in luminescence between two strains on the same plate. Comparisons of light intensity between different images are not valid. Comparing gene expression of strains growing on the same plate also prevents plate-to-plate variations in thickness, moisture content, etc.
RESULTS
SirA dramatically affects the flagellar regulon during growth in motility agar.
There are three levels to flagellar biosynthesis. The level 1 proteins, FlhD and FlhC, form a heterotetramer that is required for transcriptional activation of the level 2 genes, which encode the hook-basal body complexes and the alternative sigma factor FliA. The FliA sigma factor allows expression of the level 3 genes, which encode the filament protein, hook-associated proteins, motor proteins, and chemotaxis proteins (36, 37). The level 3 genes are further subdivided into level 3a and level 3b to distinguish those that have some fliA-independent expression (level 3a) from those that do not (level 3b) (45). To examine the effect of a sirA mutation on the expression of these genes, we constructed plasmid-based transcriptional fusions to genes representing each level of the regulon (Table 1).
Cloning of regulatory regions into pSB401 results in fusions to a promoterless luxCDABE operon of Photorhabdus luminescens (72, 73). This operon encodes both luciferase (LuxAB) and the enzymes that synthesize the substrate (LuxCDE), so that light is produced in response to gene expression. The level 1 fusion is to the flhDC operon. Level 2 is represented by fusions to the fliA, fliE, fliF, flgA, and flgB promoters. The fliD promoter represents level 3a, and the fliC and motA promoters represent level 3b. Each plasmid-based fusion was electroporated into both wild-type and sirA mutant serovar Typhimurium strains (14028 and BA746).
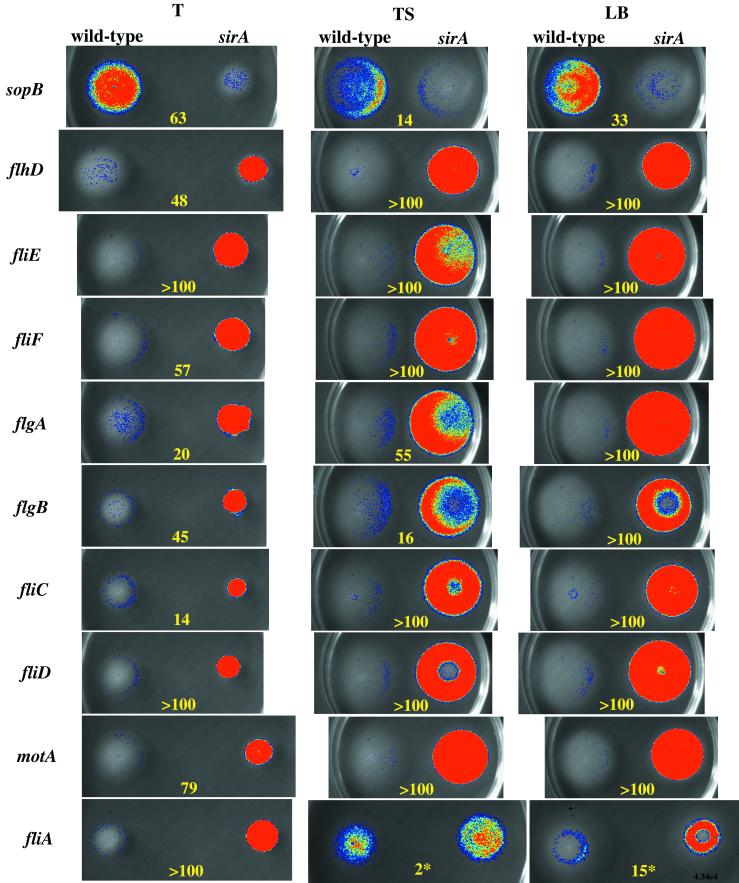
By monitoring luciferase activity in these strains, SirA was found to have repressing effects on all levels of the flagellar regulon (Fig. 1). The repressing effect was maximal while the bacteria were actively chemotaxing through motility agar (Fig. 1). Under these conditions, the sirA mutant expressed at least 100-fold more luciferase activity than the wild type from all of the level 1, 2, and 3 flagellar fusions (Fig. 1). Interestingly, despite the high levels of sirA-dependent flagellar gene regulation, the sirA mutant is nearly identical to the wild type with regard to swarm size.
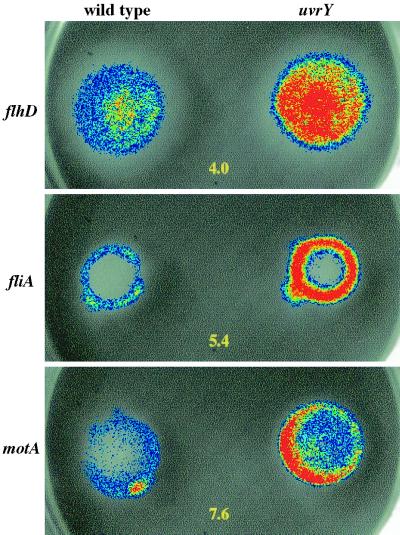
FIG. 1.
SirA-dependent regulation of serovar Typhimurium sopB and flagellar regulon components during chemotaxing through three different types of 0.3% motility agar plates (T, TS, and LB) at 37°C. Each plate compares the expression of a particular promoter fusion in wild-type serovar Typhimurium (14028) compared to the isogenic sirA mutant (BA746). The plate type is indicated at the top of each column and the promoter being tested is indicated at the left of each row. Luminescence is pseudocolored, with blue indicating low intensity and red indicating high intensity. The fold difference between each pair of strains is indicated numerically. Each plate contains tetracycline for plasmid maintenance. These results are representative of at least five independent experiments. ∗, The fliA fusion gives variable results on TS and LB motility agar. See text for details.
The fliA fusion was unique in that it did not show a simple regulatory pattern. The fliA fusion demonstrated a standard sirA-dependent repression when grown in T motility agar but was variable in both TS and LB motility agar. In TS motility agar, the fliA fusion was largely unaffected by sirA, with assay variability ranging between threefold repression and fourfold activation. In LB motility agar, the results varied widely from experiment to experiment, with values ranging between 15-fold repression and 61-fold activation by sirA (Fig. 1). No other flagellar gene fusion behaved this way, and the basis for the variability is unknown. A merodiploid chromosomal lacZY fusion to fliA is consistently repressed by sirA (see below).
SirA activates the virulence gene sopB in motility agar.
To date, SirA has never been found to have a repressing effect on any gene in any species. We wanted to determine if the repressing behavior of SirA on the flagellar fusions was due to the growth of Salmonella in motility agar or was unique to the flagellar genes. Therefore, a luciferase transcriptional fusion was constructed to the Salmonella virulence gene sopB. This fusion was placed into both wild-type and sirA mutant serovar Typhimurium, and expression was examined during growth in T, TS, and LB motility agar. In TS agar, the sopB fusion was expressed at 14-fold higher levels in the wild type than in the sirA mutant (Fig. 1). In LB, the effect was 33-fold, and in T agar, the effect was 63-fold (Fig. 1). This demonstrates that SirA positively regulates sopB regardless of growth medium and that the repressing effect of SirA is restricted to the flagellar fusions.
Regulatory effects of a sirA mutation can be complemented by plasmid-encoded sirA.
The sirA gene is directly upstream of uvrC, which raised the possibility that the effects of the sirA3::cam mutation are due to polarity on downstream genes. To confirm that the regulatory effects of the sirA3::cam mutation are not due to secondary mutations or polarity effects on downstream genes, a complementation experiment was performed. A low-copy-number plasmid encoding the sirA gene of serovar Typhimurium (or the vector control) was electroporated into a serovar Typhimurium sirA3::cam mutant (BA746) carrying the motA::luxCDABE fusion plasmid (pRG19). The presence of the sirA plasmid but not the vector control fully repressed the motA transcriptional fusion (Fig. 2). This demonstrates that sirA is responsible for the regulatory effect on motA. Complementation was used previously to confirm the regulatory role of sirA on sopB (2).
FIG. 2.
Complementation of the serovar Typhimurium sirA mutation with regard to repression of the chemotaxis gene motA. A low-copy-number plasmid encoding the Salmonella sirA gene pBA305, or the vector control, pWSK29, was electroporated into a Serovar Typhimurium sirA mutant carrying the motA::luxCDABE fusion (BA746/pRG19). Each strain was inoculated in duplicate on a 0.3% TS motility agar plate containing ampicillin and tetracycline and grown at 37°C overnight. Luminescence is pseudocolored as in Fig. 1. The presence of the sirA plasmid but not the vector control repressed the motA transcriptional fusion by greater than 100-fold.
SirA is less active in liquid media.
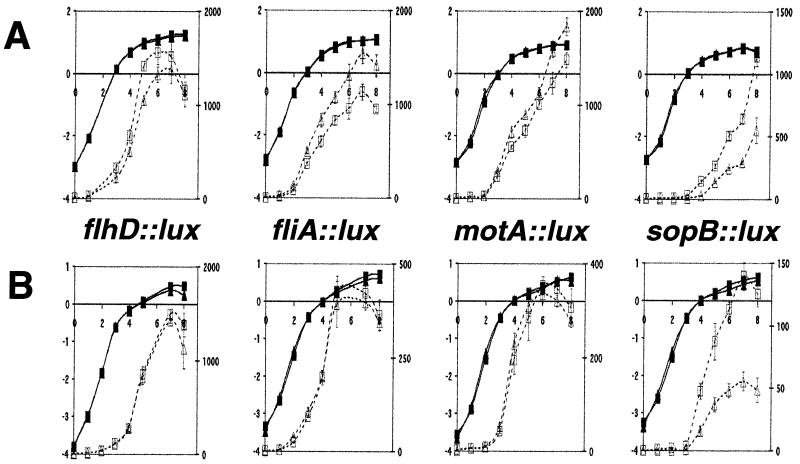
The activity of each transcriptional fusion was examined throughout the growth curve in either agitated LB broth or agitated TS medium at 37°C (Fig. 3). In both media, sirA had only small effects on virulence and flagellar gene expression. The level 2 and 3 flagellar fusions were slightly repressed by sirA, but never by more than twofold. SirA had no detectable effect on the flhD fusion under these conditions. The sopB virulence gene fusion was activated threefold by sirA. Therefore, under these conditions, the activity of SirA appears to be minimal, although the magnitude of repression of the flagellar genes mirrors the magnitude of activation of sopB.
FIG. 3.
SirA-dependent regulation of serovar Typhimurium sopB and flagellar regulon promoter fusions in shaking liquid medium. The expression of the flhD, fliA, motA, and sopB promoters was measured using plasmid-based transcriptional fusions to luciferase. Each fusion was placed into wild-type (14028) or sirA mutant (BA746) Salmonella strains. The fusion being measured is indicated. Two types of growth media were used: (A) LB and (B) TS. Solid symbols, natural log of the optical density of the culture at 550 nm, left side y axis; open symbols, luciferase activity, in RLU per second, right side y axis. Time (hours) is indicated on the x axis. Squares indicate the wild-type background, and triangles represent the sirA mutant background. Experiments were performed on three occasions. Values shown are the mean ± standard deviation of triplicate cultures from one representative experiment.
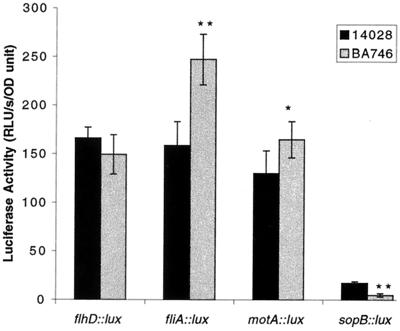
Serovar Typhimurium intestinal virulence genes require low-oxygen conditions for maximal expression (40). A common in vitro technique to obtain maximal serovar Typhimurium virulence gene expression is to allow standing subcultures to reach the late exponential or early stationary phase of growth (40). Without agitation, these cultures rapidly become microaerophilic and express the invasion genes of SPI1. We hypothesized that the effect of sirA on virulence and flagellar genes would be higher under these conditions than in the agitated LB cultures. Therefore, the reporter fusions were subcultured into LB broth and allowed to stand at 37°C without agitation for 6 h before measurement of luciferase activity. Under these conditions, sirA had very little repressing effect on the flagellar genes (less than twofold) and had a fivefold positive effect on the virulence gene sopB (Fig. 4). This fivefold activation of sopB is similar in magnitude to previous reports on the activation of secreted effector genes by SirA (2, 32, 44). Clearly the sirA gene has much larger regulatory phenotypes in motility agar than in either agitated or standing liquid medium.
FIG. 4.
Effect of sirA on the expression of sopB and the flagellar regulon under oxygen-limiting conditions. The activity of each promoter fusion was measured in serovar Typhimurium wild-type (14028) and sirA mutant (BA746) backgrounds during growth in standing LB cultures and indicated as RLU per second per optical density unit. Data are means ± standard error of three independent experiments of triplicate cultures. Statistically significant differences (∗, P < 0.05; ∗∗, P < 0.01) between wild-type and sirA mutant activity are indicated.
Chromosomal lacZY fusions are also regulated by sirA.
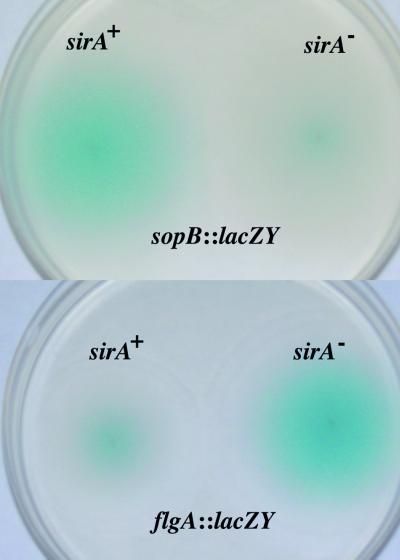
To examine the effects of sirA on the flagellar regulon with a second methodology, we constructed and tested chromosomal lacZY fusions to all levels of the serovar Typhimurium flagellar regulon: flhD (level 1), fliA, fliE, and flgA (level 2), and fliC (level 3). Examining the regulation of these genes in motility agar requires that they be able to swim, and therefore functional merodiploids were created. This was done by placing the promoter regions of these genes into the EcoRI site of the suicide vector pVIK112, which creates lacZY transcriptional fusions (34). BW20767 carrying the resulting plasmids was mated with wild-type and sirA mutant Typhimurium strains, selecting for kanamycin resistance and counterselecting for prototrophy. Transconjugants were then examined for sirA-dependent gene regulation in motility agar containing the colorimetric β-galactosidase substrate X-Gal (Fig. 5). It is difficult to quantitate the degree of blue color in the motility agar, but a qualitative assessment indicated that sirA has a repressing effect on all levels of the flagellar regulon (Fig. 5). A previously described chromosomal sopB::MudJ insertion (which creates a lacZY transcriptional fusion) was also examined in this manner (BA1526 compared to BA1726 [2]). As expected, the sopB::MudJ insertion was positively regulated by sirA in motility agar (Fig. 5). However, sirA has more dramatic effects on the plasmid-based luciferase fusions than it has on the chromosomal lacZY fusions. This could be due either to copy number effects of the plasmid-based fusions or to the accumulation of blue precipitate when using X-Gal, which would mask repressing effects. In either case, both the chromosomal lacZY fusions and the plasmid-encoded luxCDABE fusions indicate that sirA positively regulates the virulence gene sopB and negatively regulates the flagellar regulon of serovar Typhimurium.
FIG. 5.
SirA-dependent regulation of sopB and flgA chromosomal lacZY fusions during chemotaxing through 0.3% TS agar containing kanamycin and X-Gal at 37°C. Each plate compares the β-galactosidase activity of a sopB::MudJ fusion or a chromosomal merodiploid flgA+/flgA::lacZY promoter fusion in wild-type (sirA+) and sirA mutant (sirA−) Salmonella backgrounds. Fusions to flhD, fliA, fliC, and fliE demonstrated a repression similar to that seen with flgA (not shown).
SirA-dependent regulation of the flagellar regulon is evolutionarily conserved.
We hypothesized that regulation of the flagellar regulon may be an evolutionarily conserved function of SirA orthologs in the gamma proteobacteria. Therefore, we first examined the regulation of the E. coli flagellar regulon by the E. coli ortholog of sirA, which is named uvrY. Transcriptional fusions to E. coli flagellar genes and a uvrY mutant of E. coli were constructed (see Materials and Methods). The E. coli fusions were electroporated into both wild-type E. coli (MG1655) and the isogenic uvrY mutant (RG133). As was the case in serovar Typhimurium, the flagellar gene fusions of E. coli are repressed by uvrY, although the magnitude of the repression is not as great (Fig. 6). Also, like the sirA mutant of serovar Typhimurium, the uvrY mutant of E. coli does not have a motility defect.
FIG. 6.
SirA-dependent regulation of E. coli flagellar promoters during chemotaxing through 0.25% TS agar at 37°C. Each plate compares the expression of a particular promoter fusion to luxCDABE in wild-type E. coli (MG1655) compared to the isogenic uvrY (sirA) mutant (RG133). The promoter being tested is indicated at the left of each row. Luminescence is pseudocolored as in Fig. 1. The fold difference between each pair of strains is indicated numerically. Each plate contained tetracycline for plasmid maintenance. These results are representative of at least three independent experiments.
To further examine the issue of SirA orthologs being evolutionarily conserved members of flagellar regulons, we examined the motility phenotypes of sirA mutants in three other species: Pseudomonas aeruginosa and Pseudomonas fluorescens (in which the sirA ortholog is named gacA) and Vibrio cholerae (in which the sirA ortholog is named varA). All three species were found to have motility defects compared to the wild type (Fig. 7). These results are unlike those obtained with E. coli and serovar Typhimurium, in which sirA does not confer a motility defect, but suggest that sirA (gacA/varA) regulates motility genes, either positively or negatively, in these species as well. Further studies in Pseudomonas and Vibrio will be required to determine the precise role that GacA and VarA have in flagellar gene expression.
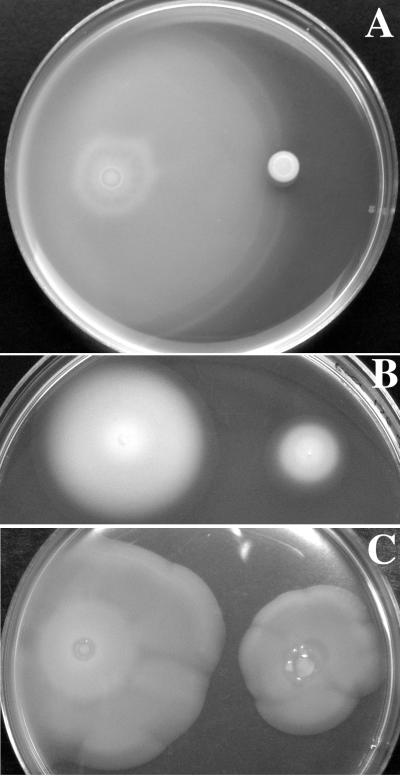
FIG. 7.
Motility phenotypes of sirA ortholog mutants of various species. (A) Pseudomonas fluorescens in 0.28% T agar at 22°C. The wild-type CHAO is on the left and the gacA (sirA) mutant CHA89 is on the right. (B) Pseudomonas aeruginosa in 0.28% TSG agar at 37°C. The wild-type PAO1 is on the left and the gacA (sirA) mutant PAO6281 is on the right. (C) Vibrio cholerae in 0.35% TS agar at 37°C. The wild-type O395 is on the left and the varA (sirA) mutant SW33S is on the right.
DISCUSSION
Numerous members of the gamma proteobacteria require sirA orthologs to cause disease. In each species, the sirA ortholog controls the expression of unique virulence genes. Because the virulence genes are unique to each species and often appear to be recent horizontal acquisitions, we hypothesized that regulation of these genes must be a relatively new function for the sirA orthologs (1). If true, sirA orthologs must have a more ancient and evolutionarily conserved function(s) that remains to be discovered (assuming that those functions have not been lost). Identification of the conserved functions of sirA orthologs is very important because it may provide clues to the environmental and/or physiological signals that lead to SirA activation. This was recently demonstrated with the phoPQ regulatory locus of S. enterica serovar Typhimurium, in which the identification of Mg2+ transporters as part of the PhoPQ regulon led to the discovery that PhoQ is a sensor of extracellular cation concentrations (25, 67). The unidentified signal(s) leading to activation of SirA orthologs is rather paradoxical. The gamma proteobacterial pathogens cause disease in organs as different as lungs and intestines and organisms as diverse as plants and animals. What signal could be common to a plant, a lung, and an intestinal tract? And why would any signal that is so common be so important?
Recently, it was discovered that the sirA orthologs of P. fluorescens and E. coli regulate the evolutionarily conserved gene rpoS (49, 53, 70). Therefore, regulation of rpoS appears to be the first example of an evolutionarily conserved function for sirA orthologs. In this study we have determined that sirA orthologs from E. coli and serovar Typhimurium have repressive effects on the flagellar genes of these species. Motility defects in sirA mutants of P. fluorescens, P. aeruginosa, and V. cholerae confirmed that control of flagellar regulons is an evolutionarily conserved function of sirA orthologs.
In S. enterica serovar Typhimurium, SirA was found to repress all levels of the flagellar regulon while activating virulence gene expression. SirA had much larger effects on virulence and flagellar fusions when the bacteria were grown in motility agar rather than in liquid medium. It is unclear whether growth in motility agar is directly stimulating SirA activity or whether the effect is indirect, potentially by removing the competitive effects of other regulators. However, the presence of high levels of SirA activity in motility agar suggests a physiological activation signal rather than a host-derived signal.
At this time, SirA has not been biochemically demonstrated to bind directly to any promoter in any species. Therefore, we do not know at what level SirA exerts its influence on the flagellar regulon. The simplest hypothesis is that SirA represses the master regulator of the flagellar regulon flhDC, which leads to decreased expression of all the flagellar gene fusions examined in this study. It is also possible that SirA only indirectly affects flhDC by controlling the expression of another regulator that directly modulates flhDC expression. Further genetic and biochemical studies are required to determine precisely how SirA affects the flagellar regulon.
There are also multiple scenarios by which SirA could simultaneously affect both motility and virulence genes. The simplest hypothesis is that SirA affects flagellar and SPI1 promoters independently. A second formal possibility is that SirA activates expression of a regulatory gene within SPI1, such as hilA, the product of which represses the flagellar regulon. While possible, this scenario seems unlikely, since both sirA and the flagellar apparatus appear to have been present in the Salmonella genome much longer than the proposed regulatory intermediate within SPI1. The third possibility is that SirA directly regulates only the flagellar regulon, and the flagellar regulon somehow affects the expression of SPI1. Although this latter hypothesis does not correlate with the positive role for fliZ in SPII expression without postulating yet another regulatory intermediate, it remains very intriguing because of recent studies in which the expression of virulence genes can be affected by mutations in flagellar genes. For instance, mutations in motility genes have been identified in numerous screens for avirulent mutants in a wide variety of species (reviewed in reference 52). However, it has largely been assumed that these mutants are avirulent simply because they cannot swim or properly chemotax to appropriate locations within their host. Only in the last few years has it become increasingly apparent that these mutants may be avirulent for reasons other than a lack of motility per se. Instead, these mutants may be avirulent because the flagellar regulon is required for the expression of virulence genes that were not previously recognized as part of the flagellar regulon. This was demonstrated in serovar Typhimurium, in which the fliZ gene and the direction of flagellar rotation were found to play a role in regulating the expression of invasion genes encoded within SPI1 (19, 33, 44). In V. cholerae it has been noted that motility phenotypes correlate with virulence gene expression (13, 26). In Xenorhabdus nematophilus, flhDC was found to be required for more than just motility and virulence in a nematode model. Instead, flhDC was also required for lipolysis and hemolysis in plate assays, suggesting that the flagellar regulon of this species regulates virulence genes in addition to motility genes (27). The most dramatic example of virulence gene expression being influenced by the flagellar regulatory cascade is found in Yersinia enterocolitica, in which the yplA gene encodes a phospholipase involved with virulence (58). This gene requires flhDC for expression, and the YplA gene product is actually secreted through the flagellar basal body (59, 75). All of these observations suggest that some component(s) of the flagellar regulon may play an active role in regulating virulence genes and/or secondary metabolism in a variety of gram-negative bacteria. Clearly there is a regulatory triad between sirA, the flagellar genes, and the virulence genes of several gamma proteobacterial species that needs to be further studied.
Interestingly, this triad appears to be similar to that of Bordetella species, which are members of the beta proteobacteria. In Bordetella, the bvgAS operon encodes the BvgA response regulator, which is phosphorylated by the sensor kinase BvgS (63–65). In the active state (the Bvg+ phase), numerous virulence genes are activated and motility genes are repressed (3–5, 14). In the Bvg− phase, the organism is motile but not virulent. While it might seem that SirA and BarA are simply distantly related orthologs of BvgA and BvgS, respectively, the evolutionary history is not so clear. In fact, E. coli encodes another locus, named evgAS, that is more likely to be orthologous to bvgAS (66). The function of evgAS is unknown. What can be concluded is that the regulatory phenotypes discovered for the barA/sirA system of Salmonella are similar to those of the bvg system of Bordetella.
ACKNOWLEDGMENTS
We are grateful to numerous labs for generously providing strains. Simon Swift provided pSB401, Bill Metcalf provided BW20767, and Virginia Kalogeraki provided pVIK112. Dieter Haas, Joyce Loper, and Stephen Calderwood provided gacA mutants of Pseudomonas and Vibrio species. We thank Glenn Young for critical reading of the manuscript and many helpful discussions.
This work was supported by a seed grant and start-up funds from the Ohio State University.
REFERENCES
- 1.Ahmer B M M, Heffron F. Salmonella typhimurium recognition of intestinal environments: response. Trends Microbiol. 1999;7:222–223. doi: 10.1016/s0966-842x(99)01524-3. [DOI] [PubMed] [Google Scholar]
- 2.Ahmer B M M, van Reeuwijk J, Watson P R, Wallis T S, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 3.Akerley B J, Cotter P A, Miller J F. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- 4.Akerley B J, Miller J F. Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J Bacteriol. 1993;175:3468–3479. doi: 10.1128/jb.175.11.3468-3479.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akerley B J, Monack D M, Falkow S, Miller J F. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J Bacteriol. 1992;174:980–990. doi: 10.1128/jb.174.3.980-990.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alpuche A C, Racoosin E L, Swanson J A, Miller S I. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. J Exp Med. 1994;179:601–608. doi: 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altier C, Suyemoto M, Ruiz A I, Burnham K D, Maurer R. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol. 2000;35:635–646. doi: 10.1046/j.1365-2958.2000.01734.x. [DOI] [PubMed] [Google Scholar]
- 8.Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 9.Castaneda M, Guzman J, Moreno S, Espin G. The GacS sensor kinase regulates alginate and poly-beta-hydroxybutyrate production in Azotobacter vinelandii. J Bacteriol. 2000;182:2624–2628. doi: 10.1128/jb.182.9.2624-2628.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chancey S T, Wood D W, Pierson L S., 3rd Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl Environ Microbiol. 1999;65:2294–2299. doi: 10.1128/aem.65.6.2294-2299.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collazo C M, Galan J E. The invasion-associated type-III protein secretion system in Salmonella—a review. Gene. 1997;192:51–59. doi: 10.1016/s0378-1119(96)00825-6. [DOI] [PubMed] [Google Scholar]
- 12.Corbell N, Loper J E. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J Bacteriol. 1995;177:6230–6236. doi: 10.1128/jb.177.21.6230-6236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Correa N E, Lauriano C M, McGee R, Klose K E. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol Microbiol. 2000;35:743–755. doi: 10.1046/j.1365-2958.2000.01745.x. [DOI] [PubMed] [Google Scholar]
- 14.Cotter P A, Miller J F. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect Immun. 1994;62:3381–3390. doi: 10.1128/iai.62.8.3381-3390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darwin K H, Miller V L. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J Bacteriol. 1999;181:4949–4954. doi: 10.1128/jb.181.16.4949-4954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 17.Edwards R A, Keller L H, Schifferli D M. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207:149–157. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 18.Eichelberg K, Galan J E. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect Immun. 1999;67:4099–4105. doi: 10.1128/iai.67.8.4099-4105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichelberg K, Galan J E. The flagellar sigma factor FliA (sigma28) regulates the expression of Salmonella genes associated with the centisome 63 type III secretion system. Infect Immun. 2000;68:2735–2743. doi: 10.1128/iai.68.5.2735-2743.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eriksson A R, Andersson R A, Pirhonen M, Palva E T. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol Plant-Microbe Interact. 1998;11:743–752. doi: 10.1094/MPMI.1998.11.8.743. [DOI] [PubMed] [Google Scholar]
- 21.Francis C L, Ryan T A, Jones B D, Smith S J, Falkow S. Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature. 1993;364:639–642. doi: 10.1038/364639a0. [DOI] [PubMed] [Google Scholar]
- 22.Frederick R D, Chiu J, Bennetzen J L, Handa A K. Identification of a pathogenicity locus, rpfA, in Erwinia carotovora subsp. carotovora subsp. carotovora that encodes a two-component sensor-regulator protein. Mol Plant-Microbe Interact. 1997;10:407–415. doi: 10.1094/MPMI.1997.10.3.407. [DOI] [PubMed] [Google Scholar]
- 23.Gaffney T D, Lam S T, Ligon J, Gates K, Frazelle A, Di M J, Hill S, Goodwin S, Torkewitz N, Allshouse A M, et al. Global regulation of expression of antifungal factors by a Pseudomonas fluorescens biological control strain. Mol Plant-Microbe Interact. 1994;7:455–463. doi: 10.1094/mpmi-7-0455. [DOI] [PubMed] [Google Scholar]
- 24.Gambello M J, Kaye S, Iglewski B H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia V E, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 26.Gardel C L, Mekalanos J J. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Givaudan A, Lanois A. flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J Bacteriol. 2000;182:107–115. doi: 10.1128/jb.182.1.107-115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glessner A, Smith R S, Iglewski B H, Robinson J B. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J Bacteriol. 1999;181:1623–1629. doi: 10.1128/jb.181.5.1623-1629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grewal S I S, Han B, Johnstone K. Identification and characterization of a locus which regulates multiple functions in Pseudomonas tolaasii, the cause of brown blotch disease of Agaricus bisporus. J Bacteriol. 1995;177:4658–4668. doi: 10.1128/jb.177.16.4658-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han B, Pain A, Johnstone K. Spontaneous duplication of a 661 bp element within a two-component sensor regulator gene causes phenotypic switching in colonies of Pseudomonas tolaasii, cause of brown blotch disease of mushrooms. Mol Microbiol. 1997;25:211–218. doi: 10.1046/j.1365-2958.1997.4411811.x. [DOI] [PubMed] [Google Scholar]
- 31.Hrabak E M, Willis D K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnston C, Pegues D A, Hueck C J, Lee A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 33.Jones B D, Lee C A, Falkow S. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect Immun. 1992;60:2475–2480. doi: 10.1128/iai.60.6.2475-2480.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalogeraki V S, Winans S C. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene. 1997;188:69–75. doi: 10.1016/s0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- 35.Kinscherf T G, Willis D K. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J Bacteriol. 1999;181:4133–4136. doi: 10.1128/jb.181.13.4133-4136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komeda Y. Fusions of flagellar operons to lactose genes on a Mu lac bacteriophage. J Bacteriol. 1982;150:16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutsukake K, Ohya Y, IIno T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 39.Laville J, Voisard C, Keel C, Maurhofer M, Defago G, Haas D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc Natl Acad Sci USA. 1992;89:1562–1566. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee C A, Jones B D, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee C A, Silva M, Siber A M, Kelly A J, Galyov E, McCormick B A. A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc Natl Acad Sci USA. 2000;97:12283–12288. doi: 10.1073/pnas.97.22.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao C H, McCallus D E, Fett W F. Molecular characterization of two gene loci required for production of the key pathogenicity factor pectate lyase in Pseudomonas viridiflava. Mol Plant-Microbe Interact. 1994;7:391–400. doi: 10.1094/mpmi-7-0391. [DOI] [PubMed] [Google Scholar]
- 43.Liao C H, McCallus D E, Fett W F, Kang Y. Identification of gene loci controlling pectate lyase production and soft-rot pathogenicity in Pseudomonas marginalis. Can J Microbiol. 1997;43:425–431. doi: 10.1139/m97-060. [DOI] [PubMed] [Google Scholar]
- 44.Lucas R L, Lostroh C P, DiRusso C C, Spector M P, Wanner B L, Lee C A. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:1872–1882. doi: 10.1128/jb.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd. ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 46.Metcalf W W, Jiang W, Daniels L L, Kim S K, Haldimann A, Wanner B L. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 47.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 48.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukhopadhyay S, Audia J P, Roy R N, Schellhorn H E. Transcriptional induction of the conserved alternative sigma factor RpoS in Escherichia coli is dependent on BarA, a probable two-component regulator. Mol Microbiol. 2000;37:371–381. doi: 10.1046/j.1365-2958.2000.01999.x. [DOI] [PubMed] [Google Scholar]
- 50.Nagasawa S, Tokishita S, Aiba H, Mizuno T. A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol Microbiol. 1992;6:799–807. doi: 10.1111/j.1365-2958.1992.tb01530.x. [DOI] [PubMed] [Google Scholar]
- 51.Ochsner U A, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ottemann K M, Miller J F. Roles for motility in bacterial-host interactions. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 53.Pernestig, A. K., O. Melefors, and D. Georgellis. 2000. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem., in press. [DOI] [PubMed]
- 54.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 55.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 56.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schechter L M, Damrauer S M, Lee C A. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- 58.Schmiel D H, Wagar E, Karamanou L, Weeks D, Miller V L. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect Immun. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmiel D H, Young G M, Miller V L. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J Bacteriol. 2000;182:2314–2320. doi: 10.1128/jb.182.8.2314-2320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simon R, Priefer U, Puhler A. A broad-host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 61.Tan M W, Rahme L G, Sternberg J A, Tompkins R G, Ausubel F M. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci USA. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsolis R M, Kingsley R A, Townsend S M, Ficht T A, Adams L G, Baumler A J. Of mice, calves, and men: comparison of the mouse typhoid model with other Salmonella infections. Adv Exp Med Biol. 1999;473:261–274. [PubMed] [Google Scholar]
- 63.Uhl M A, Miller J F. Autophosphorylation and phosphotransfer in the Bordetella pertussis BvgAS signal transduction cascade. Proc Natl Acad Sci USA. 1994;91:1163–1167. doi: 10.1073/pnas.91.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uhl M A, Miller J F. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J Biol Chem. 1996;271:33176–33180. doi: 10.1074/jbc.271.52.33176. [DOI] [PubMed] [Google Scholar]
- 65.Uhl M A, Miller J F. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 1996;15:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- 66.Utsumi R, Katayama S, Ikeda M, Igaki S, Nakagawa H, Miwa A, Taniguchi M, Noda M. Cloning and sequence analysis of the evgAS genes involved in signal transduction of Escherichia coli K-12. Nucleic Acids Symp Ser. 1992;27:149–150. [PubMed] [Google Scholar]
- 67.Vescovi E G, Ayala Y M, Di C E, Groisman E A. Characterization of the bacterial sensor protein PhoQ: evidence for distinct binding sites for Mg2+ and Ca2+ J Biol Chem. 1997;272:1440–1443. doi: 10.1074/jbc.272.3.1440. [DOI] [PubMed] [Google Scholar]
- 68.Wallis T S, Galyov E E. Molecular basis of Salmonella-induced enteritis. Mol Microbiol. 2000;36:997–1005. doi: 10.1046/j.1365-2958.2000.01892.x. [DOI] [PubMed] [Google Scholar]
- 69.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 70.Whistler C A, Corbell N A, Sarniguet A, Ream W, Loper J E. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor sigmaS and the stress response in Pseudomonas fluorescens Pf-5. J Bacteriol. 1998;180:6635–6641. doi: 10.1128/jb.180.24.6635-6641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P, Bycroft B W, et al. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winson M K, Swift S, Fish L, Throup J P, Jorgensen F, Chhabra S R, Bycroft B W, Williams P, Stewart G S. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 73.Winson M K, Swift S, Hill P J, Sims C M, Griesmayr G, Bycroft B W, Williams P, Stewart G S. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol Lett. 1998;163:193–202. doi: 10.1111/j.1574-6968.1998.tb13045.x. [DOI] [PubMed] [Google Scholar]
- 74.Wong S M, Carroll P A, Rahme L G, Ausubel F M, Calderwood S B. Modulation of expression of the ToxR regulon in Vibrio cholerae by a member of the two-component family of response regulators. Infect Immun. 1998;66:5854–5861. doi: 10.1128/iai.66.12.5854-5861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Young G M, Schmiel D H, Miller V L. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]