Abstract
Digestive system diseases arise primarily through the interplay of genetic and environmental influences; there is an urgent need in elucidating the pathogenic mechanisms of these diseases and deploy personalized treatments. Traditional and long-established model systems rarely reproduce either tissue complexity or human physiology faithfully; these shortcomings underscore the need for better models. Organoids represent a promising research model, helping us gain a more profound understanding of the digestive organs; this model can also be used to provide patients with precise and individualized treatment and to build rapid in vitro test models for drug screening or gene/cell therapy, linking basic research with clinical treatment. Over the past few decades, the use of organoids has led to an advanced understanding of the composition of each digestive organ and has facilitated disease modeling, chemotherapy dose prediction, CRISPR-Cas9 genetic intervention, high-throughput drug screening, and identification of SARS-CoV-2 targets, pathogenic infection. However, the existing organoids of the digestive system mainly include the epithelial system. In order to reveal the pathogenic mechanism of digestive diseases, it is necessary to establish a completer and more physiological organoid model. Combining organoids and advanced techniques to test individualized treatments of different formulations is a promising approach that requires further exploration. This review highlights the advancements in the field of organoid technology from the perspectives of disease modeling and personalized therapy.
Subject terms: Gastrointestinal diseases, Molecular biology
Introduction
The digestive system, a continuous anatomical structure composed of multiple organs, is responsible for swallowing and digesting food, absorbing nutrients, and discharging residual wastes.1,2 The accumulation of many external stimuli and genetic mutations contributes to the emergence and progression of digestive diseases, including infectious, inflammatory, and malignant diseases.3–8 The morbidity and mortality of certain diseases are increasing annually, despite the constant updating of treatments.9–16 Therefore, it is of great significance and urgency to clarify the etiology of digestive diseases and find new and more effective treatments. Achieving the above goals will require two foundations: on the one hand, omics technologies and bioinformatics analysis are necessary to find correlations, and many achievements have been made in this field,4,17–27 on the other hand, reliable models can be used to reveal causal relationships, find molecular targets, and test therapeutic strategies. Ultimately, and most importantly, basic research can be translated into clinical research to benefit patients.
The human digestive system is not directly accessible and translating the rapidly evolving preclinical knowledge associated with diagnostics and therapeutic interventions is not an easy task. Differences in the anatomy, biological processes and cell-type-specific expression patterns of the human digestive system make animal models a suboptimal choice to mimic the occurrence and treatment of human digestive diseases.28–31 The limitations of cell line models established from the human digestive system are as follows: primary cells established from the original tissue have a short lifespan and require strict culture conditions; if immortal or cancerous cells are used, they will lose the complex characteristics of primary tissue after a long period of artificial culture on plastic plates with non-physiological media and may not respond to interventions in the same way as host tissue would.32–34 These defects mean that, while mouse models and cell lines have provided some understanding and insight into digestive system evolution and disease over the years, the failure rate of clinical translation has been quite high.35–38 Over the past decades, many approaches have been used in the attempt to generate models of the human digestive system in vitro.34 The development of organoid culture was a major breakthrough.39,40 Organoids recapitulate many biological features of human organs, including tissue heterogeneity, spatial distribution characteristics of different cells, cell-cell and cell-matrix interactions, and some functions arise from tissue-specific cells.41–43 Organoids are more representative of in vivo physiology and bridge the gap in existing model systems with a stable system adapted to a wide range of cultures and manipulations.44–47 Different cell types of stem cell-derived organoids contain complex cues that regulate tissue formation, giving them a unique potential in the study of biological phenomena related to tissue development and stem cell differentiation.48–50 Importantly, adult stem cell (AdSC)-derived organoids from individual patients can be expanded and preserved for therapeutic testing, be used to preserve tissue properties to achieve optimal outcomes for each patient, even be served as preclinical tools for drug screening.41,50–52
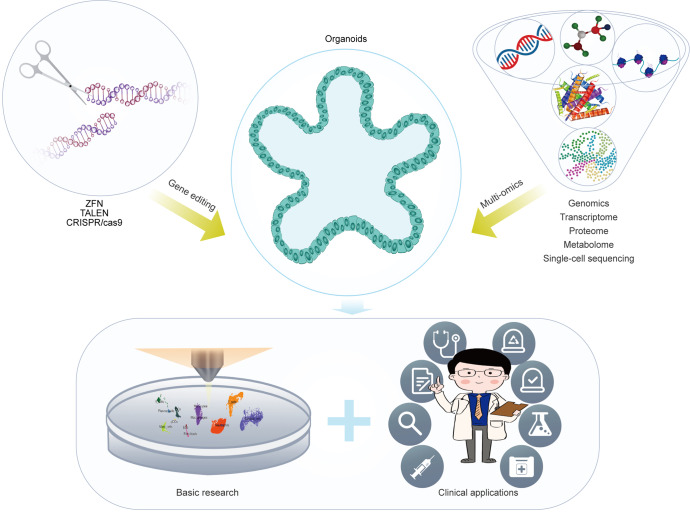
In this review, we summarize the various in vitro digestive organoid models that have been created and the compositional differences between digestive organoids generated from AdSCs and induced pluripotent stem cells (iPSCs) (Fig. 1). In addition, we summarize how they reshape or deepen our understanding of the occurrence and characteristics of digestive disease, stem cell function and regeneration, host-pathogen interactions, and the exploration and application of personalized therapy. We also discuss the current shortcomings of organoid models and directions for future improvement.
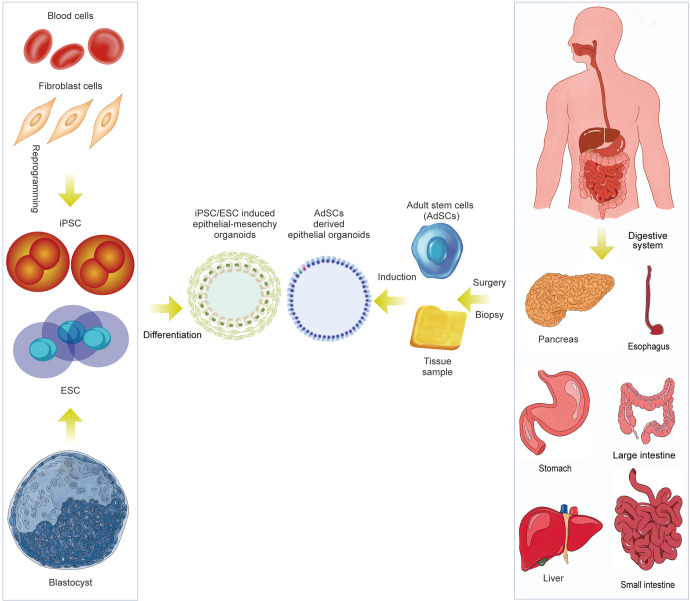
Fig. 1.
Schematic diagram summarizing the generation of digestive organoids and the manner of cell differentiation. Organoids derived from isolated epithelial stem cells and tissue samples, reprogramming of skin fibroblasts and blood cells into pluripotent stem cells (PSCs). (Induction involves germ-layer specification (endoderm) and subsequent induction and maturation by specific growth factor combinations.)
Digestive organoids: self-organizing systems of organs ex vivo
Organoids are generated from various types of stem cells, which are induced to form microscopic cell clusters and cultured in vitro as 3D structures; these clusters are also known as “mini-organs” because they mimic the in vivo structures and functions of the corresponding organs.53 Organoid formation relies on decades of research into stem cell fate determination, which is tightly coordinated by the Wnt/Notch/EGF/BMP pathway.54,55 Inspired by these studies, Cleves’ team established the first mouse gut organoid in 2009 by isolating Lgr5+ stem cells ex vivo with a combination of EGF, Wnt3a, and the BMP inhibitors Noggin or R-spondin-1 in a basal medium under a dome of matrigel.56 Their team subsequently modified the culture medium in 2011 to obtain human colonic organoids.57 Intestinal organoids are commonly termed “enteroids” when they originate from the small intestine and “colonoids” when they originate from the colon.58 The establishment of mouse- or human-derived epithelial enteroids and colonoids relies on the utilization of different culture media. For example, media containing R-spondin1, EGF, and Noggin is required for mouse intestinal organoids and does not vary with diverse proliferation and differentiation conditions. Additional Wnt3a results in indefinite growth of mouse intestinal organoids devoid of a functionally intact Wnt-secreting niche, necessitating the addition of exogenous Wnt3a to the expansion medium and the exclusion of Wnt3a from the differentiation medium. The long-term maintenance of human intestinal organoids requires more pathways, such as inhibition of the TGFβ and P38 MAPK pathways by the chemical inhibitors A83-01 and SB202190 or the addition of insulin-like growth factor 1 (IGF1) and fibroblast growth factor 2 (FGF2). Over decades of research, with continuous testing of combinations of nutritional factors, organoid models of the human digestive system have been developed into the more mature in vitro models, replicating the human esophagus, stomach, liver, pancreas, etc.59–62 These organoids are obtained from ex vivo expanded resident tissue stem cells by recreating the stem cell niche. Although each organ is unique in structure and cellular composition, organoids derived from those stem cells are very similar in terms of mitogenic factors, culture conditions, and final structure. The Wnt pathway activator Wnt-3A or R-Spondin, the BMP pathway antagonist Noggin, EGF or FGF, and TGF-β inhibitors are often needed, as well as other factors depending on the tissue origin. These multicellular organoids consist exclusively of simple hyperpolarized epithelial cells tightly surrounding the central lumen and outwardly projecting crypt-like structures.63
In addition to adult stem cell-derived organoids, digestive organoids can also be obtained from pluripotent stem cells (PSCs), including iPSCs and ESCs, in a step-by-step manner by mimicking embryonic development after implantation under a complex, coordinated set of specifications to determine the formation of morphological features.64,65 Likewise, PSC-derived organoids were first induced to develop into intestinal organoids through a gradual series of steps, in which activin-A first induced and defined endoderm formation and FGF/Wnt induced posterior endoderm patterning, subsequent hindgut specification and morphogenesis. After 4 weeks of culture, induced human intestinal organoids (iHIOs) were induced to express an intestinal stem cell marker, and they formed a polarized columnar epithelium with villus-like structures and an overall morphology similar to that of the human intestinal epithelium. iHIOs mimic the development of intestinal morphology and molecules. Later, PSC-derived organoids were also described for the large intestine.65,66 In the same manner as the endoderm, differentiation into the foregut, midgut and hindgut is finely regulated by a combination of factors. Furthermore, according to the fine-tuning regulation of the combination of factors obtained by accumulating embryonic development information on each organ, the foregut forms the esophagus, stomach, liver and pancreas, and the hindgut forms the large intestine. Thus, PSCs can form various digestive organs, such as the esophagus, stomach, intestine, liver, and pancreas, under conditions that closely mimic early patterning and morphogenesis with various specific medium components.67–73 In contrast to adult stem cells, PSCs are pluripotent, and various cellular components of the three germ layers can be obtained through different induction programs. In addition to the epithelial system, PSC-derived digestive organoids also contain a layer of mesenchymal cells.74,75 Hence, digestive organoids have extensive applications in various areas. Comprehensive protocols for establishing digestive organoids have been published, which enable the long-term expansion and passaging of organoids that can be cultured as desired for drug screening or microbial infection and mechanistic studies.76–86
The application of organoids
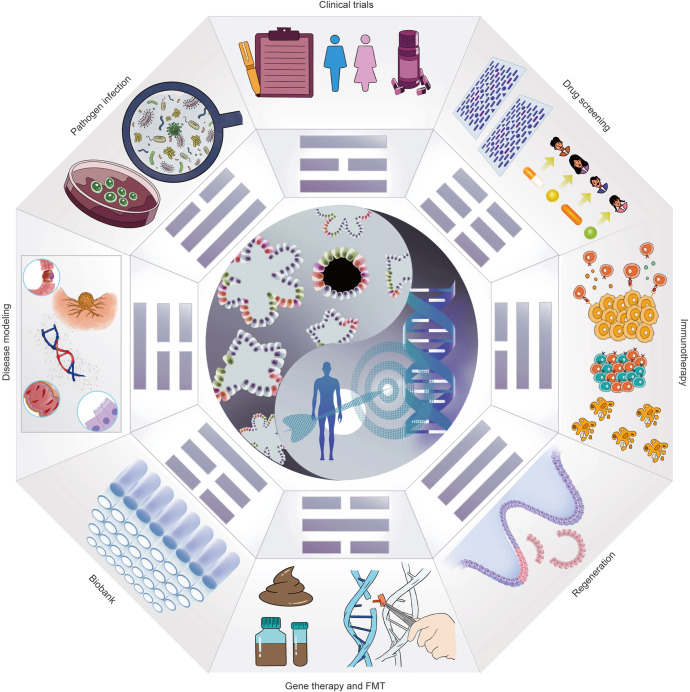
As organoid technology opens new areas of biomedical research, the establishment of long-term in vitro models of various digestive organs has increased our knowledge of digestive diseases, biobanking, clinical trials, pathogen infections, drug screening, stem cell function and regeneration, immunotherapy, gene therapy, testing for fecal microbiota transplantation (FMT), etc., all of which contribute to the development of personalized medicine (Fig. 2).
Fig. 2.
Potential applications of organoid models in precision medicine. As a versatile in vitro model, organoids can cover many applications from disease modeling to clinical trials and achieve the purpose of precise and personalized treatment. The scope of organoid research includes (1) Disease modeling. (2) Pathogenic infection. (3) Biobanking of organoids. (4) Clinical trials. (5) Stem cell function and regeneration (delivery of organoids to repair intestinal epithelium or activate the repair program of endogenous stem cells). (6) Drug screening (patient-specific high-throughput screening and discovery). (7) Immunotherapy. (8) Gene therapy and FMT
Digestive system disease modeling
The pathogenesis of digestive system diseases is complex, occurring as a series of consequences of the interweaving and interaction of genes and the environment, and this process is the sum of infectious diseases, inflammatory diseases, and malignant tumors.87–89 Abnormal expression or mutation of genes such as TP53, SMAD4 and PIK3CA mediate the occurrence and progression of diseases. In terms of environmental factors, these digestive organs tend to be prone to pathogen infections or external liquid and gaseous irritation. Notably, important aspects of anatomy and compartmentalization vary widely between species.36,37,90 Therefore, human digestive diseases can be studied most effectively by using human model systems. Organoid models derived from the digestive system can closely mimic these diseases due to their good representation of the cell composition and molecules characteristics of human organs.41 The simulation of digestive diseases by organoids can be realized mainly through two channels. First, the organoids of patients can be generated from the tissues obtained from surgery or biopsy to construct biobanks of patient-derived organoids (PDOs). PDOs have been shown to preserve their clinical markers, histomorphology, driver mutations, and even molecular and metabolite heterogeneity, which assures that organoids can respond realistically to in vivo treatments.91 Another approach is to mimic disease onset and progression by manipulating the expression of disease-causing genes or to generate mutation points by combining gene editing techniques. This method can be divided into two types: gene editing in organoid models derived from healthy/paracancerous tissue, and gene editing in organoid models derived from iPSCs. Each type has its own advantages: organoids derived from adult tissues retain the epigenetic and genetic information of adult tissues, making them more similar to the physiological state of human organs; PSC cells avoid the complex phenotypic variation of organoids from different backgrounds, allowing a variety of complex genetic manipulations and multivariate observations. Furthermore, the organoid models are also used to simulate environmental disturbances, in order to observe how those pathogens interact with the digestive organs and mediate barrier disruption, inflammation and immune responses, the genetic mutations, that ultimately cause a range of digestive diseases. At present, organoids are commonly researched in the medical field by extracting diseased tissues from patients and generating organoids to study the proportion of disease mutations that the organoids can retain and to what extent they can simulate patients’ responses to various clinical treatment strategies.51,92 Intervening in expression or activity of one or several genes or protein by a series of molecular means to study the association between corresponding genes and the occurrence or progression of diseases has been carried out mostly in the field of basic research.47,93,94 Together, these two advances form the basis of our current understanding of how organoids mimic digestive system disease and reveal its occurrence and progression.
Intestinal disease
The intestine is one of the largest vital organs of the human body, integrating many complex epithelial, immune, lymphatic, vascular, and nervous components to perform digestive and endocrine functions and regulate digestion, metabolism, and feeding behaviors.41,95,96 It can be divided into two segments, the small intestine and the large intestine. In the past decade, the generation of intestinal organoids has provided a rich reference for understanding intestinal pathology.97–99 Organoid models can effectively mimic the state of intestinal diseases, including motility disorders, malabsorptive diarrhea, inflammation, infection, and cancer.40,75,95,100 Microvillus inclusion disease (MVID) is a congenital disease of the intestinal epithelium that causes malnutrition and is characterized by diffuse villus atrophy or absence of apical microvilli.101,102 MVID cases are caused by mutations in either myosin 5B (MYO5B) or syntaxin-3.103 Loss of the microvillus phenotype was reported in a patient-tissue-derived organoid model.104,105 Deletion of MYO5B impairs precursor differentiation into organoids and partially mediates Wnt/Notch imbalance, and Notch inhibition and/or LPA treatment could be an effective treatment for MVID.106 Diacylglycerol-acyltransferase 1 (DGAT1) dysfunction often leads to another rare, intractable form of malabsorption and diarrheal disease that appears early in life.107 The main role of DGAT1 is to catalyze the conversion of diglycerides and fatty acyl-CoA to triglycerides, and DGAT1 deficiency can lead to fat intolerance and protein-losing enteropathy.108 Intestinal organoids produced from tissues of DGAT1 mutant patients showed decreased lipid catabolism and increased cell death after oleic acid treatment. Knockdown of DGAT1 in normal organoids can also replicate this phenotype, which essentially recapitulates and reflects the fat intolerance and intestinal damage caused by DGAT1 deficiency in patients.109 Multiple intestinal atresia (MIA) is a rare disease with bowel obstruction and uniform calcification in the abdominal cavity, caused by a deficiency of the tetratricopeptide repeat domain 7 A gene (TTC7A).110,111 Organoids formed from MIA patient tissue have inside-out apicobasal polarity, which is rarely observed in long-term culture; however, this reversal can be normalized by pharmacological inhibition of Rho kinase.112 Cystic fibrosis (CF) is a common, fatal, multisystem genetic disorder that presents as a chronic disease with an altered gut environment.113 It is a disease caused by impaired epithelial anion transport owing to mutations in the gene encoding cystic fibrosis transmembrane regulator protein (CFTR), resulting in impaired fluid regulation and pH imbalance in multiple organs.114,115 CFTR-dependent secretory responses can be replicated by intestinal organoids from CF patients, and it was found that increased ATP treatment of these organoids with forskolin causes CFTR-mediated chloride channel opening and induces fluid secretion from epithelial cells, resulting in swelling of intestinal organoids.116 Inflammatory bowel diseases (IBDs) come in two forms, known as ulcerative colitis (UC) and Crohn’s disease (CD); these debilitating chronic inflammatory disorders are consequences of genetic predisposition combined with accumulated dysbiosis of the gut microbiota, which mediates uncontrolled immune responses and impaired structural function of the gut epithelium.117–119 Organoid models can also be used to interrogate complex somatic cells in UC tissue as they evolve to adapt to the chronic inflammatory microenvironment.117,120–123 Intestinal organoids harvested from CD patients can recapitulate multiple patterns of disease evolution, including higher organoid reorganization capacity, DNA methylation, and changes in transcriptome and stem cell marker gene profiles.123–125 Despite a comparable mutational signature, the number of single nucleotide variants (SNVs) in UC organoids from the same patient was slightly higher than that in paired control organoids, which essentially mimics the finding that epithelial cells in UC have an increased number of mutations under inflammatory conditions in vivo.120,126 Human iPSCs from colonic fibroblasts were isolated from UC patients, followed by targeted differentiation into organoids; these organoids fully reproduced the histological and functional features of primary colonic tissue, including insufficient secretion of acidic mucus, abnormal adherent junctions in the epithelial barrier, and overexpression of the CXCL8/CXCR1 axis.121 In addition, organoids can provide a good model for the simulation of colorectal cancer progression.127–130The occurrence and development of Colorectal Cancer (CRC) is a malignant tumor and a heterogeneous disease involving a series of genomic alterations.131–134 The occurrence and development of CRC is a multifactorial, multi-pathway process that follows a progression from normal to adenoma to adenocarcinoma to liver metastasis of adenocarcinoma. Organoids derived from colonic tissue have been used to model CRC, and these PDOs recapitulate the somatic copy number alteration (SCNA) and mutational spectrum found in CRC, with ~80% of the mutations in primary tumor tissue being identified in the corresponding organoids.135 Metastasis is the main cause of death in CRC; patient-derived paired primary and metastatic tumor organoids have been selected as CRC metastasis models, which can be used for the discovery and detection of potential prognostic biomarkers and therapeutic targets of CRC. Despite shared driver mutations between primary and metastatic tumors, metastatically derived organoids after xenotransplantation exhibit higher metastatic capacity.136 When the multimutated CRC genes APC, KRAS, SMAD4, TP53 and PIK3CA were introduced into normal human intestinal organoids using CRISPR-Cas9 genome editing technology, the organoids could simulate the progression of CRC and gradually transform into invasive glandular tissues in vivo.137 Gene-edited Apc/KrasG12D/Trp53 organoids were transplanted into the distal colon and subsequently found to metastasize to the liver.138 Deletion of MLH1 from colonic epithelium-derived normal organoids using CRISPR-Cas9 technology mimics features of MSI-driven CRC.139 Several newly discovered causative genes for colorectal cancer, such as the protein tyrosine kinases (PTKs) BMX and HCK observed in adenoma precursor cells, have been suggested as potential drivers of adenomas. When these two genes were exogenously expressed in human intestinal organoids, an increase in BMX or HCK was observed to significantly improve the number and size of organoid hyperplasia within the organoid lumen, as well as multiple polyp bud protrusions and wall hyperplasia.140 In addition to the role of gene mutation, chromosomal instability is also one of the pathological causes of CRC.141 When combined with other interventions, xenografted mutant organoids with certain chromosomal rearrangements, including reversals and deletions affecting R-spondin2, form flat serrated lesions similar to sessile serrated adenoma in mice.142
Gastric disease
The stomach is composed of ordered epithelium, and the invaginated gastric unit contains acid-secreting parietal cells, mucous pit cells, enzyme-secreting primary cells, proliferating cells, and intermediate cell populations.143 Gastric cancer (GC) is a heterogeneous malignancy of the digestive system with complex and diverse mutated genes and pathogenic mechanisms.144 GC can be divided into four subtypes according to molecular typing: the chromosomally unstable (CIN) subtype, frequently featuring RTK-RAS pathway amplification and mutated in TP53; the microsatellite instability (MSI) subtype, with a hypermutated phenotype; the genomically stable (GS) subtype, with alterations in RHOA/CDH1 and manifesting as diffuse tumor morphology; and the Epstein-Barr virus (EBV)-positive subtype, which displays frequent CDKN2A dysfunction and PIK3CA mutations.145–147 The established GC organoid biobank essentially covers all four GC subtypes and captures unique molecular profiles of dysregulated genes that can be used to guide unique therapeutic targets for GC subtypes.148–150 GC organoids retain cellular markers and tumor histopathological classification, and GC-associated gene mutations and copy number variations (CNVs) are also very similar in PDOs and corresponding primary tumors.151 Some heterogeneity was noted in lymph node metastatic GC organoids; the organoids from primary tumor histology showed diffuse growth, and organoids from lymph node metastatic tumors showed glandular shapes.152 The genotype-phenotype association can be validated by the phenotypic features of gene-edited gastric-like organoids carrying multiple gastric cancer mutations, such as the migration and diffuse appearance of gastric organoids after CDH1 knockout.148 GC metastasis can also be replicated by establishing an orthotopic carcinoma organoid transplantation model and identifying the essential characteristics of stem-like TR-LGR5+ cells in the persistence and metastasis of GC.153
Esophagus diesease
The esophagus consists of four layers: inner mucosa, the underlying supporting submucosa, the muscularis propria, and the epithelium and adventitia; it acts as an anatomical conduit for the transport of food from the pharynx to the stomach.38,154 Prolonged exposure to gastroesophageal reflux can lead to Barrett’s esophagus (BE); the transition from normal squamous epithelium to abnormal specialized columnar epithelium is also known as intestinal metaplasia in the esophagus; BE is increasing in incidence and is a potential risk for esophageal adenocarcinoma (EAC).155–157 BE organoids contain periodic acid-Schiff-positive goblet cells, a hallmark of BE.158 Esophageal cancer is a malignancy with high morbidity and mortality. There are two major histological subtypes, EAC and esophageal squamous cell carcinoma (ESCC).159,160 EAC cells are morphologically similar to the secretion-producing glandular cells of the intestinal mucosa, whereas ESCC cells exhibit varying degrees of squamous cell differentiation. Organoids obtained from EAC recapitulated patient tumor histology, including p53 status and apical/basal polarity.161 Furthermore, organoids obtained from EAC also showed the same consistency between driver somatic mutational events and genome-wide mutational signatures as in the original tumor tissue, including mutations of CDKN2A and PIK3CA.161,162 The characteristic clonal heterogeneity of EACs, which contributes to chemotherapy resistance and poor patient survival, can be retained in the resulting organoids.163 Established ESCC organoids from biopsy samples show consistency with the original tumors.164
Pancreatic disease
The main function of the pancreas is to facilitate digestion and metabolism. The pancreas consists of three main cell types, namely, acinar cells, ductal cells, and endocrine cells that are responsible for stabilizing blood sugar; dysfunction of this organ can lead to diabetes and pancreatic cancer.165–167 Pancreatic ductal adenocarcinoma (PDAC) is the most lethal common malignancy, with poor prognosis and high mortality, and accounts for 90% of pancreatic cancers; PDAC is also one of the most drug-resistant cancers due to the extensive heterogeneity of mutations and the dense stromal environment.168–170 Pancreatic cancer organoids are derived from primary tumors and metastatic tumors, retaining characteristics of primary malignancies; furthermore, organoids from peritoneally disseminated nodules not only retain proliferative capacity but also form peritoneal tumors with features of metastatic pancreatic cancer.171 Seventy percent of PDAC organoids are characterized by the classical or progenitor subtype, and approximately 30% are characterized by the basal or quasi-mesenchymal subtype; commonly mutated genes, such as TP53, KRAS, SMAD4 and CDKN2A, and uncommonly mutated genes, such as PIK3CA, MAP2K1 and ERBB2, were found in PDAC-generated organoids.172,173 When CRISPR-Cas9 genome editing technology is used to modify normal organoids to mimic the pancreatic disease, such as editing PDAC driver genes in PDAC organoids; mediating KRASG12V and ERBB2 mutations; and inactivating TP53, CDKN2A and SMAD4, and these mutant organoids develop into tumors that resemble pancreatic intraepithelial neoplasia after orthotopic xenografting in immunodeficient mice.174,175 Pancreatic duct and acinar organoids derived from human iPSCs can reproduce the characteristics of the neonatal exocrine pancreas, and the PDAC-related oncogene GNASR201C is more efficient at inducing cystic growth when expressed in ducts, whereas KRASG12D is more efficient at modeling cancer in vivo when expressed in acinar cells.176
Liver diesease
The liver consists of parenchymal and nonparenchymal hepatocytes, organized into functional units called hepatic lobules; this organ carries out a variety of functions, such as digestion, detoxification, and metabolism.177 Mutations in the SERPINA1 gene mediate alpha-1-antitrypsin (A1AT) dysfunction, and patients with this mutation may develop chronic liver damage.178,179 Liver organoids from A1AT-deficient patients result in decreased A1AT secretion and increased endoplasmic reticulum stress due to misfolding of the mutated A1AT protein.180 Mutations in the Notch signaling pathway can cause Alagille syndrome, which is characterized by partial or complete atresia of the bile ducts and the bile duct function that is prevented from being established.181 Established liver organoids from Alagille syndrome patients treated with cholangiocyte differentiation mediators fail to upregulate bile markers.182 Fatty liver disease is attributed to a variety of causes, including an obese diet, a sedentary lifestyle, and the prevalence of metabolic disorders.183,184 Human hepatocyte-differentiated organoids and liver-derived intrahepatic cholangiocyte organoids can recapitulate some pathologies of fatty liver disease, such as lipid accumulation and mitochondrial damage.185 Insufficient lysosomal acid lipase activity leads to Wolman disease, a massive accumulation of lipids in liver cells with fatal steatohepatitis and fibrosis.186,187 Wolman disease-specific induced pluripotent stem cells generate liver organoids that recapitulate severe steatohepatitis and fibrosis.187 Nonalcoholic fatty liver disease can lead to steatohepatitis, which eventually leads to liver fibrosis, cirrhosis, and hepatocellular carcinoma (HCC).188 Human iPSC-derived liver organoids (HLOs) contain multiple cell types, such as stellate cells, hepatocytes, and kupffer-like cells, and have succeeded in recapitulating some features of steatohepatitis, including inflammation, lipid accumulation and fibrotic phenotypes, after oleic acid treatment.187 In a drug-induced organoid model of liver fibrosis, these organoids exhibited fibrotic features, such as human stellate cell activation, collagen secretion and deposition, when exposed to profibrotic compounds.189 Organoids generated from patients with primary sclerosing cholangitis showed changes in the expression of genes responsible for primary sclerosis.190,191 Human hepatocyte organoids were cocultured with mesenchymal cells from fetal liver tissue to recapitulate the pathophysiology of alcohol-related liver disease; hepatocyte-like cells in these organoids underwent oxidative stress, increased reactive oxygen species, and accumulated intracellular lipid droplets to induce steatosis and release inflammatory mediators after exposure to alcohol.192 Most primary liver cancers have low curative effects and high recurrence rates and can be divided into the hepatocellular carcinoma, cholangiocarcinoma and combined HC-CCA types.193 The generation of liver cancer (LC) organoids summarizes the molecular profiles of the corresponding tumor sources, including the PDO lineage of HCC, cholangiocarcinoma (CCC), and HCC/CCC combination; furthermore, lung metastases were found after transplantation into immunodeficient mice.194,195 Biopsy-derived HCC/CCC organoids also matched the mutational spectrum of primary tumor tissue and retained common driver mutations typical of HCC/CCC in ARID1A, TP53 and TSC1.195 In the study of the molecular mechanisms and related risk factors for primary liver cancer, there are also cases of organoids. For example, knockdown of arginine methylation transcription factor (PRMT6) in patient-derived nontumor liver organoids with CRISPR-Cas9 was found to increase cellular resistance to molecularly targeted drugs and chemotherapy; PRMT6 overexpression in HCC organoids attenuates tumor cell migration and invasion.196 Depletion of the tumor suppressor BAP1 in normal human cholangiocyte organoids results in impaired chromatin accessibility, loss of multiple epithelial features, and increased motility.197 Axin2 is highly expressed in HCC organoids, while RNF43/ZNRFR3 is deficient; in vitro genetic intervention produced lipid droplet aggregation in RNF43-mutated human hepatocellular carcinoma tumor organoids, whereas RNF43/ZNRF3-deficient hepatic epithelial organoids displayed not only lipid droplet aggregation but also reduced differentiation capacity.198
Microecosystem research using digestive organoids
As previously mentioned, in addition to primary tissue extraction and gene editing to mimic digestive system disease, organoids can serve as a basis for reflecting a variety of complex environmental factors that regulate cell fate changes and disease development.199,200 Here, we discuss only the simulation of microbiota and viral infection in well-studied organoid systems.
Microbiota and digestive disease
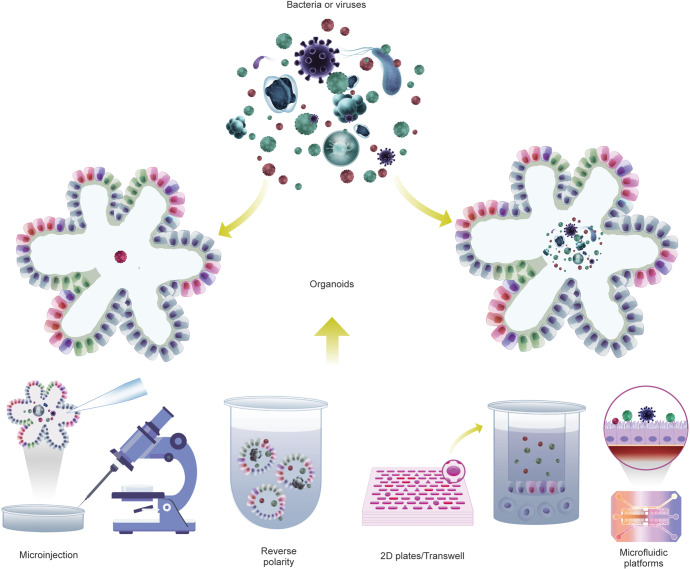
The microbiota shapes the chemical environment of the gut, the bioavailability of ingested substances, and the biological processes of tissue homeostasis; dysbiosis shift of the microbiota is expected to lead to diseases.201,202 Over the past decade, numerous studies have found that dysbiosis affects the occurrence and development of digestive system diseases.203–209 Nevertheless, numerous associations of microbes with those diseases remain correlative due to the difficulties of modeling the host-microbe relationships in a reductionist yet meaningful way that allows detailed mechanism analysis. Currently, there are two main approaches to exploring the impact of the microbiota on host physiopathology in mouse models, namely, germ-free animals and antibiotic regimens.210,211 Antibiotic treatment depletes the gut microbiota of mice with broad-spectrum antibiotic treatment. Antibiotics are usually dissolved in drinking water, provided ad libitum throughout treatment; therefore, actual doses administered may be unstable, so that the bacteria are not completely cleared from the treated mice, and levels of clearance vary between individuals. Thus, while antibiotic treatment provides an inexpensive and easy-to-use alternative to sterile models, the results obtained by these protocols have the potential for off-target drug effects, as well as incomplete or inconsistent ablation of microbes.212–214 Germ-free (GF) mice represent a system that allows the study of animals completely devoid of microbes, in which researchers introduce microbes into GF mice individually or sequentially and evaluate the effects of individual bacteria or known bacterial communities on host function impact.204 GF animal models are powerful tools for studying microbe-host interactions in health and disease.204,215 However, the high cost of GF mice and the relatively complex experimental manipulation techniques needed for these animals are barriers to the widespread adoption of this model for basic research. In addition, the species differences between animals and humans have always been an insurmountable gap. In particular, some microbes are unique to humans; therefore, conclusions from mouse models may not carry over. These dilemmas create the need for another set of model systems to help mimic the human traits that control the off-target effects of these bacteria and the perturbations of the aforementioned complexities. Organoid models derived from human tissue or cells are amenable to sterile culture methods and are a good alternative to GF mice. The 3D organoid model shows a closed, irregular luminal structure with most microbial-epithelial interactions taking place at the apical side of the epithelium. However, introducing the microbiome into the lumen of an organoid is not an easy task. There are several ways to do this (Fig. 3), including microinjection (A),61,67,85,216,217 3D organoids grown with reversed polarity (B),84,218,219 organoid-derived fragments or epithelial monolayers (C),220–224 and microfluidic platforms (D).225–229 Each approach has its advantages and disadvantages (Table 1). Directly injecting microorganisms into the lumen of differentiated or undifferentiated 3D organoids, allowing microorganisms to contact the top of epithelial cells in 3D conditions, is a popular research method. However, microinjection requires a special setup and specialized techniques, and it can affect the readout when injected bacteria leak into the basolateral region, also coculturing obligate anaerobic bacteria for a long period of time is difficult due to the lack of sufficient oxygen in the lumen of organoids. 3D organoids growing with reversed polarity (the ‘apical-out’ orientation) expose the apical side of epithelial cells directly to the medium without matrix embedding, making it accessible to microorganisms. Continuous organoid cultures in prolonged suspension were observed to assess microbial-epithelial interactions and their consequences by adding microbes directly to the culture medium. Moreover, this approach enables easy testing of interactions with bacteria or their metabolites in a high-throughput setup under multiple conditions. However, since this method does not guarantee complete polarity reversal, distinguishing between apical and basolateral interactions is difficult. Another limitation of this approach is that the mucus can be easily washed out. Fragmented organoids and organoid-derived epithelial monolayers are another set of advancements enabled by linearization of 3D organoids into 2D systems to enhance the accessibility of the apical side. Organoids of various origins are divided into small fragments or single cells and then distributed in extracellular matrix-coated dishes or Transwell plates to form organoid-derived monolayers that differentiate into different epithelial cell lineages capable of producing mucus; microbes were added directly to the culture medium, and their interactions with the epithelial cells were observed. This relatively simple experimental setup can be used to compare a variety of conditions, both for culturing under aerobic conditions and for creating an anaerobic environment in the upper chamber of a Transwell plate. For example, the air-liquid interface (ALI) culture system provides a feasible mode for culturing obligate anaerobic bacteria. The apical cavity creates anoxic conditions for anaerobic bacteria, while the basal and lateral surfaces of the organoids are supplied with oxygenated medium. This system can also support longer-term maintenance of epithelial cell monolayers and expand the study of epithelial-mesenchymal interactions due to the mitigation of oxidative stress and the addition of potential stromal elements in ALI. However, these approaches have the disadvantage of losing the three-dimensional structure of the organoids. Culturing organoid monolayers on micro-structured collagen scaffold assemblies with crypt-like invaginations is an improvement that mimics the three-dimensional spatial features defining crypt-like and villus-like structures. However, maintaining the entire system in an anaerobic chamber is expensive, and replicating models can be time consuming. Organoids on microfluidic platforms are micro-engineered systems generated based on industrial computer microchips by microfabrication methods. Current microfluidic platforms include organoids-on-a-chip, HuMiX and GuMI. Microfluidic culture systems can enhance cellular microenvironments, replicate distinct mucus layers, mimic key features of the human gut, and monitor organoid metabolism. These systems have obvious advantages in studying host-microbe interactions and some complex biological functions in vitro. However, these models are expensive, requiring special training to operate and are difficult to use in high-throughput experiments. Each of these methods of delivering bacteria to organoids has both pros and cons. These methods are also constantly being optimized and improved, and efforts are being made to develop newer methods. Regardless of the specific method, however, these approaches have all facilitated the study of organoid-bacterial interactions and provided observational insights that advance our understanding of molecular mechanisms.
Fig. 3.
Using gut organoids to study the impact of microbiota. Methods of infecting organoids with bacteria: (1) Microinjection. (2) Transwell/Plate-based 2D monolayer cultures of organoids. (3) Reversal of cell polarity of organoids. (4) Organoids-on-a-chip
Table 1.
Comparison of methods for introducing microorganisms into organoids
| Method | Description | Advantages | Disadvantages |
|---|---|---|---|
| Microinjection61,67,85,216,217 | Injecting microorganisms into the lumen of organoids using a microinjection device. |
1. The method preserves the structural integrity of the organoids, and microorganisms only contact the apical side of organoids, providing a more realistic gastrointestinal simulation environment, especially for the anaerobic bacteria. 2. The entire process of microbe-organoid interactions can be observed, including initial interactions and early host responses. 3. Quantitative experiments can be performed by controlling the MOI. 4. This method has no special requirements for organoid culture conditions and can be applied for most 3D organoids. |
1. This method requires a very specialized setup and is lacking of a standard procedure for different organoids. 2. The manual nature of the microinjection process makes it difficult to apply to high-throughput experiments, and the sequential injections resulted in asynchrony of experimental exposures. 3. The leakage of injected microorganisms toward the basolateral side can influence the readout, and the closed lumen may cause nutrient and oxygen consumption and metabolites accumulation. 4. Injections of small volumes of material are often imprecise, and differences in organoid size as well as luminal contents may cause uneven distribution of the injected material. |
| Organoid-derived fragment or epithelial monolayers220–224 | Linearizing 3D Organoids into 2D Systems such as extracellular matrix-coated dish. The organoid-derived monolayer contains various epithelial cell lineages and enables the introduction of microorganisms via direct addition to the culture media. |
1. The accessibility of the apical side of organoids is enhanced and the introduction of microorganisms can be achieved with an easily applicable setup. 2. This method can be applied to high-throughput experiments and can effectively reduce group differences caused by irrelevant variables in comparison or screening experiments. 3. Long-term co-culture with anaerobic bacteria can be achieved by incubating organoids in an aerobic environment while maintaining the apical chamber of a Transwell insert in an anaerobic environment. 4. Combined with the air-liquid interface or the microfabricated collagen scaffold array of crypt-like invaginations, this method can partially reconstruct epithelial-mesenchymal interactions and the crypt-like and villus-like structures. |
1. The inoculation process caused mechanical damage to the organoids, and 2D organoids cannot reflect the structural features of lumen. 2. Certain bacterial media such as tryptone-yeast extract-glucose (TYG) and brain heart infusion may be toxic to the monolayers during introduction. 3. The success rate of establishing functional epithelial monolayers varies between different donors, which may limit its applicability. 4. Optimization measures such as providing continuous nutrient replenishment, creating an anaerobic chamber for obligate anaerobic bacteria, combining the air-liquid interface and collagen scaffold technology are costly and time-consuming. |
| 3D organoids with growing reversed polarity84,218,219 | Making the apical surface evert to face the media and introducing microorganisms by adding them directly to the culture medium. |
1. This method simplifies the introduction of microorganisms while maintaining the 3D structure of organoids. 2. Without extracellular matrix affecting distribution, suspended apical-out organoids can be synchronously exposed to experimental agents and microorganisms. 3. Suspended organoid culture can be divided into multiple wells for different experimental conditions, which is more suitable for high-throughput experiments. |
1. This method does not guarantee complete polarity reversion, so it is difficult to distinguish between the apical and basolateral interactions. 2. The mucus can be easily washed out, making it easier for foreign substances to enter the organoids. 3. Transferring apical-out organoids to new media is a time-consuming and iterative process. 4. Apical-out organoids exhibit slower proliferation and accelerated differentiation, suggesting that some of the pathways have been altered and may interfere with host-microbe interactions. |
| Microfluidic platforms225–229 | Microfluidic platforms, including organ-on-a-chip, HuMiX and GuMI, are micro-engineered systems generated based on industrial computer microchips by microfabrication methods. Microfluidic platforms can provide organoids with precisely programmed biomimetic microenvironments and introduce microorganisms into organoids with ease and precision. |
1. This method enables microbial diversity in organoids by tuning chemical gradients, oxygen gradients, dynamic mechanical stress and even incorporating multiple cell types and connecting multiple tissue platforms. 2. Simulation of key characteristics of the human gut and reconstruction of the mucus layer provides a better model for studying microbe-host interactions. 3. Standardized and automated organoid-on-a-chip enables high-throughput experiments. 4. Long-term coculture can be achieved through the continuous supply of nutrients and scavenging of metabolites via the microfluidic platform. |
1. The complexity of organ structures and the heterogeneity of individuals determine that this method cannot fully simulate the real situation, so the applicability of this method needs to be discussed. 2. This method integrates programming, biochemistry, biomechanics, materials science and other disciplines and requires the cooperation of multiple teams and platforms, thus greatly increasing the experimental threshold and cost. |
Organoids simulating microbial interactions with the digestive organs
Several key questions to answer when studying host-microbe interactions include how microbial communities are established and maintained, patterns of microbe-host interactions and parameters that can be used to detect them, and the mechanism and targets of the microbiome contributing to disease development. Organoids cultured in growth-factor-enriched media and differentiated into various epithelial cells are emerging as a key model for deciphering pathogen invasion mechanisms due to their accurate representation of cellular heterogeneity and entry receptor expression patterns.230,231 Several pathogens associated with intestinal diseases have been studied in organoids. For example, enterohemorrhagic Escherichia coli (EHEC) can colonize a monolayer of human colonic organoids, and EHEC has been observed to reduce intestinal mucus, disrupt microvilli structure, and facilitate bacterial entry and infection of epithelial cells.232 Matrix-embedded, monolayer, or transplanted mouse organoids exposed to Shiga toxin (Stx)-producing Escherichia coli show increased transepithelial permeability; microinjection of the highly potent bacterial toxin Stx2a into human intestinal organoids can trigger abnormal upregulation of a variety of key structural proteins and tight junction proteins, epithelial injury, and apoptosis.232–234 E. coli pks+ strains are thought to cause CRC through DNA damage and chromosomal instability; after repeated microinjection of pks+ strains, the unique mutational signature in a subset of the human CRC genome can be detected by whole-genome sequencing in E. coli intervened healthy human intestinal organoids, after up to 5 months of intervention.217 Interestingly, following a similar intervention, another strain, Enterotoxigenic Bacteroides fragilis (ETBF), was found to promote CRC through a nongenomic mechanism. ETBF did not generate a distinct set of mutational signatures when incubated with human colonic organoids; instead, ETBF-induced tumors were driven by homologous errors in mismatch repair and recombinant DNA damage repair, suggesting that ETBF colonization is a potential threat for sporadic CRC or hereditary neoplastic diseases.235 Clostridium difficile (C. difficile) is an anaerobic, gram-positive, toxigenic bacterium that is a major infectious cause of nosocomial diarrhea.236 Microinjection of toxigenic C. difficile or its virulence factor TcdA into organoids resulted in loss of epithelial barrier function, marked redistribution of the Tight Junction (TJ) proteins, and downregulation of mucin 2 expression.237,238 TcdB of C. difficile infection of HIOs inhibits stem cell repair capacity and delays epithelial cell renewal.249 Infection with the major foodborne enteric pathogen Listeria affects goblet and paneth cell numbers, upregulates muc2 and lyz expression, and reduces the expression of genes related to the Notch pathway. Increased secretion of proinflammatory cytokines disrupts organoid morphology.239,240 Enterococci are the leading cause of multidrug-resistant infections.241,242 Cocultivation with Enterococcus faecalis family pore-forming toxins (Epxs) impairs HIO.243 F. nucleatum is a known pathogenic bacterium that mediates the pathogenesis of CRC by affecting cancer cell proliferation, exacerbating intestinal inflammation, or interfering with the immune microenvironment.244–247 Intervention with F. nucleatum stimulates the secretion of proinflammatory cytokines and the activation of multiple inflammatory signaling pathways, such as the NF-kB pathway, promoting intestinal inflammation in colonic epithelial cells of human colonoid monolayers.248 Clostridioides difficile Toxin B (TcdB) infection of HIOs inhibits stem cell repair capacity and delays epithelial cell renewal.249 Colonization of Helicobacter pylori is associated with GC, which causes most gastric ulcers by inducing hypersecretion of gastric acid.250–252 Gastric organoids from different sources have been used to replicate pathological conditions in vivo and to robustly mimic the gastric epithelial response to H. pylori infection in an in vitro system.253–255 H. pylori infection activates nuclear factor kappa B (NF-kB) to bind to the RAS protein activator-like 2 (RASAL2) promoters, inducing its expression and regulating tumorigenesis; RASAL2 silencing reduces nuclear β-catenin levels and disrupts organoid formation in gastric tumors.256 In a simulated acute H. pylori infection, an increase in inflammatory genes was rapidly observed by microinjection of H. pylori into the organoid lumen.221 It has also been implicated in immune regulation, with studies showing that H. pylori treatment increases PD-L1 expression in organoids; anti-PD-L1 antibodies result in H. pylori infection-induced organoid death when cocultured with autologous cytotoxic T lymphocytes and dendritic cells from patients.257 Although the most important organ affected by the intestinal microbiota is still the gastrointestinal tract, much evidence has proven that the pathological mechanism of the microbiota or its metabolites by destroying the mucosal and epithelial barrier or releasing proinflammatory factors to mediate inflammation or the immune response of other digestive organs, some of which have also been tested in organoid models.51,258–268
Viruses
Organoids have also become powerful models to simulate the occurrence of digestive system diseases mediated by viral infections. Human noroviruses (HuNoVs) cause nonbacterial acute gastroenteritis in patients of all ages and are highly contagious.269,270 Intestinal organoids were cultured in monolayers infected with norovirus strains to monitor their replication and assess the virucidal efficacy of novel disinfectants.269,271,272 In addition, human digestive organoids can also be used to identify norovirus inactivating factors.273 Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the agent that causes coronavirus disease 2019 (COVID-19), has caused a global acute respiratory disease pandemic; as a highly contagious and pathogenic coronavirus, its emergence seriously threatens human health and public safety.274–276 The SARS-CoV-2 S-glycoprotein binds to angiotensin-converting enzyme (ACE)-II, enters the lungs via trypsin, and causes a large viral load through a rapid replication mechanism, resulting in symptoms of viral pneumonia in patients.277,278 Apart from lung damage caused by SARS-CoV-2, symptoms have also occurred in multiple other organs, and many research groups have used organoid models to understand cellular responses to SARS-CoV-2 and the resulting damage.277,279,280 Intestinal organoids infected with SARS-CoV-2 from different donors showed that the levels of viral replication in small intestine and colon-derived organoids were orders of magnitude different; virus-infected organoids showed the highest infectivity to omicron variant spike protein (S), and the susceptibility to infection was correlated with the level of ACE2.281 SARS-CoV-2 altered ACE2 expression in goblet cells, and efficiently tested drugs, such as remdesivir, inhibited SARS-CoV-2 infection at low molar concentrations and rescued the morphology of the organoids.282 Interferon-induced transmembrane (IFITM)-derived polypeptide or a target antibody inhibited SARS-CoV-2 entry and replication after SARS-CoV-2 entered gut organoids.283 SARS-CoV-2 can effectively infect the human liver and biliary organoids.284–286 The SARS-CoV-2 virus replicates rapidly in infected organoids to induce proinflammatory cytokine/chemokine production, local hepatocyte and bile duct cell damage, and consequent bile acid accumulation.287 SARS-CoV-2 efficiently influences the gastric epithelium, and when gastric organoids were generated from biopsies from patients of different ages, the results showed that pediatric and late-stage fetal gastric organoids were susceptible to SARS-CoV-2 infection, and viral replication was markedly reduced in undifferentiated organoids from early fetuses and adults.219 Hepatitis B virus (HBV) is a partially double-stranded DNA virus belonging to the family Hepatoviridae, and chronic HBV infection can progress to end-stage liver diseases such as cirrhosis and HCC.288,289 Human liver organoids are a useful platform for mimicking HBV infection and related tumorigenesis because they retain the structure and gene expression patterns of human hepatocytes.290–294 HBV infection of organoids replicates the viral life cycle and virus-induced liver dysfunction.295 Organoids infected with HBV in vitro can also produce covalently closed circular DNA (cccDNA) and HBV early antigen (HBeAg), express HBV RNA and protein intracellularly, and generate infectious HBV.290 iPSC-derived liver organoids (IPSC-LOs) are more susceptible, and HBV infection not only supports HBV transmission but also downregulates hepatic gene expression, induces the release of early markers of acute liver failure (ALT/LDH), alters liver ultrastructure, and leads to hepatic dysfunction in organoids.296 EBV is etiologically associated with at least 8% of GC cases (designated EBVAGC), including distinct genetic and epigenetic GC subsets.297,298 In paired normal and cancer-derived organoids from the same patient, EBV infects only the cancer organoids.299 Organoid models have demonstrated their value in validating the safety and efficacy of antiviral treatment.272,300–302 These in vitro models are able to leverage prior knowledge of viral biology.293 Finally, the integration of genetically engineered organoids or biopsy-derived organoids to mimic specific and rare subtypes of digestive diseases could help identify effective treatments for a small subset of patients with rare diseases from whom organoids are established.303–308
Digestive system organoid models and precision and personalized therapy
In the era of precision therapy, organoid models can be used to accurately predict individual responses to treatments.309–312 The global application of organoid technology has led to unprecedented progress in many diseases; given the heterogeneous composition of tumors, no single treatment approach is suitable for all patients, and organoids derived from tumors are rapidly becoming a vital tool for the individual selection of treatments.313–316 Organoids can be used to study mutations and treatment options at different disease stages.317,318 In addition, organoids can be generated by performing multiple rounds of biopsies of patients at different time points to continuously assess their response to treatment, detect emerging resistance, and subsequently screen new treatments.51 These patient-derived organoids, or organoids that mimic disease, can be tested for medicine selection, cell therapy, immunotherapy, gene correction, or combinations of several treatments for individuals.312,313
Personalized medicine therapy
Personalized therapy is treatments performed for each patient and using a personalized disease model with similar characteristics to the primary tumor will undoubtedly give a more accurate prediction of the patient’s response to a given treatment.161,319,320 Patient-derived organoids retain donor-specific properties and disease-associated differences.172,321–323 The selection of drug therapy using organoid models can be divided into two aspects: selection of existing drugs (including the reuse and repositioning of old drugs) and screening of new drugs/chemicals. The latter can be classified as drug discovery.
Drug screening to delineate treatment strategies
AdSC-based organoid technology enables drug responsiveness testing in a fraction of the time needed for previous methods.324 Additionally, no single treatment is effective for all patients, suggesting that PDOs need to be tested to determine the best treatment regimen for the individual patient.320 Whether the levels and responses of organoids in many drugs and chemoradiotherapy are comparable to in vivo therapy has been extensively examined from multiple perspectives. Some studies showed that the chemoradiotherapy response of rectal cancer organoids was very well correlated with patients, with a specificity of 91.97%, an accuracy of 84.43%, and a sensitivity of 78.01%.129 Endoscopic biopsy of esophageal cancer generates PDO and compares clinical outcomes after neoadjuvant therapy with in vitro PDO responses.325 Organoids from metastatic CRC and GC/EAC PDOs have been used for mid-range drug screening, and parallel drug studies in organoid xenografts confirm in vitro and in vivo therapeutic response correlations.324 The response of GC PDOs to two clinically used paclitaxel (PTX) nanoformulations, albumin-bound PTX (Albu-PTX) and liposomal PTX (Lipo-PTX), was comparatively assessed using a GC PDO model that reproduced the therapeutic superiority of Lipo-PTX over Albu-PTX.151 The clinical response to drugs can be predicted from patient-derived data, which can be used to create personalized treatment strategies based on the specific response of organoids to existing treatments.326 Organoids from patients with metastatic gastrointestinal cancer receiving commonly used treatments are used to predict treatment response and tumor differences between patients and to model intra-patient tumor response heterogeneity between different chemotherapeutics.324 By obtaining PDO during a diagnostic biopsy, multiple regimens can also be tested simultaneously and applied at clinically relevant intervals.327 For individualized treatment, the most valuable direction for research is how to obtain tissue through biopsy and other means before surgery and to test radiotherapy and chemotherapy treatment as quickly as possible to guide the selection of an appropriate radiotherapy dose, chemotherapy drugs, and adjuvant therapy. Researchers have optimized intraoperative drug screening with 96-well plate-cultured organoids and made the provision of molecular typing, drug screening, and individualized adjuvant therapy guidance for each patient within one week become feasible.328 Newer techniques can be used to generate GC organoids in endoscopic biopsies of patients with advanced metastatic GC who are not suitable for surgery and then test the efficacy of multiple standard drug regimens and combinations in a relatively short period of time, providing valuable prognostic and therapeutic options for this type of patient.329 Additionally, if the dose-response curves of PDOs can be established to reproduce in vivo responses, biopsies can be processed immediately to produce organoids for high-throughput drug screening assays, simulating therapeutic responses to various conventional and investigational treatments for PDAC changes, as well as differences in individual patient responses to overall treatment.330 Organoid biobanks can be used to detect resistance to existing drugs and select alternative treatment options. Hedgehog pathway inhibitors combined with 5-fluorouracil (5-FU), or irinotecan have potent antitumor effects on drug-resistant colorectal cancer in organoids.331 A drug screening test was conducted for the treatment of oxaliplatin-resistant CRC by PDOs, and the results showed that inhibition of KLF5 by ML264 was mainly mediated by antiapoptotic effects and restored oxaliplatin sensitivity in CRC.332 Response analysis of PDAC organoids to five commonly used chemotherapeutics (5-FU, irinotecan, oxaliplatin, gemcitabine and paclitaxel) showed that one-third of PDOs were resistant to all five drugs, and half of the patients who provided these PDOs were highly sensitive to targeted drugs.172 The multicellular HCC organoid (MCHO) contains a large number of stromal cells that have some influence on drug penetration, and interference with YAP/TAZ transcriptional activity in hepatoma cells significantly increases the penetration of verteporfin into MCHO.333 Organoid technology has been shown to play a key role in gene-drug association theory in individualized and targeted therapy. For example, TP53-mutated PDOs are resistant to Nutlin-3a, an MDM2 inhibitor, KRAS-mutated organoids are extremely resistant to inhibitors of ERBB, and RNF43-mutated colon tumor organoids are dramatically susceptible to Wnt secretion inhibitors.334 ERBB2-amplified PDOs respond well to lapatinib, a dual ERBB2/EGFR inhibitor.335 Trametinib combined with celecoxib is the most effective strategy for KRAS- and TP53-mutated advanced colorectal cancer.319 Colorectal cancer organoids with RAS mutations may respond to combination therapy with MEK and pan-HER inhibitors.336 As in vitro models for drug safety evaluation, small and large intestine organoids have been used in toxicology studies to predict drug-induced gastrointestinal toxicity, such as assessing transcriptomic responses associated with viability and apoptosis following exposure to doxorubicin and gefitinib as a physiological endpoint.337,338 Human intrahepatic cholangiocyte organoids (ICOs) can recapitulate necroptosis associated with bile duct disease, providing a useful in vitro platform for biliary cytotoxicity studies and preclinical drug evaluation.191 The applicability of hepatic organoids derived from human iPSCs for drug toxicity assessment was demonstrated by comprehensive functional analysis of CYP450-mediated drug metabolism.339 These cases demonstrate the versatility of organoids and their consistency with in vivo therapy, and these validation results promote the ubiquitous role of organoids in drug selection and prediction for clinical treatment.
Drug discovery
Organoids that replicate the characteristics of in situ tissue can also be used for more accurate drug discovery models. In this approach, drugs or compounds are tested on organoids of different disease origins. These drugs or compounds include agents in preclinical testing, agents in clinical development, and agents clinically used for the other diseases at hand. Currently, there are many examples of drug discovery using organoids in preclinical or clinical practice.91,340 For drug discovery, the size of the organoid library and the size of the drug or compound library should be selected appropriately. While some drug-response models are available for processing large compound libraries, others are suitable for screening some disease types that are sensitive to single chemotherapeutic drugs or combinations of targeted drugs. In other words, some effective targeted therapeutic drugs or compounds can be identified by drug screening with different throughputs in PDOs. Clevers’ laboratory described the first mid-scale drug screen using its CRC organoid biobank; the organoid is plated and processed with an extensive library of cancer-targeting compounds.323 Huch’s laboratory initially conducted a mid-scale drug screen of 29 anticancer compounds, including those in clinical use or in development, and LC PDOs showed variable sensitivity to some of the compounds.194 Another team assessed the feasibility of large-scale drug screening using PDAC organoids by exposing PDAC PDOs to 76 targeted therapy drugs and classical chemotherapeutics.173 Some drugs in clinical development or preclinical small molecules have been identified as target drugs for a certain disease and may also be found to be suitable for the treatment of other diseases in the experiment. Organoids are used to test drugs or compounds that previously targeted other diseases, meaning that old drugs are used for new diseases; this approach eliminates many pharmacokinetic and organic toxicity tests and allows for faster clinical applications.341–343 Of course, it should be noted that it is not limited to the drugs in past and present clinical use for the purpose. The FDA-approved chronic myeloid leukemia (CML) drug omacetaxine has been shown to be the most potent small molecule in HCC PDOs.344 Through PDAC PDO screening, CHK1 inhibitors for breast and ovarian cancer (ongoing trials: Phase I or Phase II) are also effective in pancreatic cancer.171 PCSK9 is a therapeutic target for hypercholesterolemia and dyslipidemia but is also emerging as a potential target for colorectal cancer, as PCSK9 inhibitors inhibit the growth of APC/KRAS mutant CRC organoids.199 Organoid models are also a promising tool for small-molecule screening in some intractable diseases and tumor metastases to discover potential novel therapeutic agents. Fangchinoline has been shown to be a potent small molecule that can dose-dependently inhibit the growth of patient-derived organoids by directly targeting NOX4, thereby reducing non-small cell lung cancer (NSCLC) metastasis.345 The VprBP inhibitor B32B3 is able to reduce colonic organoid growth by blocking H2AAT120p and activating the normal transcriptional program.346 Human pancreatic islet organoids respond to the HIF-1α inhibitor PX-478, and long-term exposure to high glucose increases the glucose-induced insulin secretion stimulation index, suggesting that the HIF-1α inhibitor PX-478 has the potential to act as an antidiabetic agent.347 AT-rich interacting domain 1 A (ARID1A), an important subunit of the chromatin remodeling complex, carries a heterozygous mutation in most human GC cases, and tumor organoids with ARID1A heterozygosity show growth inhibition by combination therapy with TP06 and Nutlin-3, an epigenetic inhibitor and a p53 agonist, respectively, offering a new option for GC therapy.348 Investigating combination drug therapies in organoids is a good strategy to combat drug resistance and emerging diseases in the digestive system.
Stem cells and regeneration
Adult stem cells are the cornerstone of the renewal of multiple cell types in multiple tissues.48,349 To unravel the regulatory mechanisms by which stem cells act as key determinants of regeneration following tissue trauma or inflammatory injury, extensive research efforts have been conducted to elucidate the plasticity of tissue stem cells.350–355 Stem cell-derived organoids have unparalleled advantages in the field of regeneration because they contain not only stem cells but also their differentiated progeny and can be continuously cultured and stored; they can recapitulate the regenerative capacity of the epithelium and restore homeostasis after damage.98,356,357 Numerous studies have shown that organoid technology has the potential to provide alternative organ replacement strategies for various types of digestive diseases.41,358 Transplanting gut organoids into mice is also the best model to study stem cell fate determination and microenvironmental interactions, and the transplantation of normal human gut organoids onto the mouse gut surface enabled the prioritized maintenance of stem cells to reproduce human intestinal epithelial tissue in a heterograft system.65 After organoid transplantation in the Rag2/DSS colitis model, donor cells achieved “mucosal healing” and rescued the pathology of DSS colitis.351,359 Extensive resection of the small intestine can lead to malabsorption and weight loss, a condition called short bowel syndrome (SBS). In a rat model of SBS, ileal organoid transplantation yields a well-functioning small intestine and significantly mitigates intestinal failure.360 Liver transplantation is a treatment method to restore liver function in patients with irreversible liver failure.361,362 Liver organoids, similar to liver lobules, are expected to be used in regenerative medicine to generate a source of cells to overcome the current shortage of transplant organs.363–365 Liver organoids generated from expanded hepatic progenitor cells (HPCs) reconstituted hepatic interstitial structures after implantation in allogeneic mice.366 Cholangiocellular organoids can be used for the repair of human bile ducts.367 AdSCs have a remarkable ability to generate organoids, which also help identify signals that control the lineage fate of stem cell progeny and guide models of tissue plasticity during injury and regeneration.350,368 Thus, in addition to direct organoid transplantation, understanding the mechanism of stem cell plasticity for endogenous regeneration regulation is also a promising therapeutic direction.167,369–371 It is generally believed that dedifferentiation of epithelial cells to stem cells and the existence of a reserve stem cell pool are two sources of stem cell-driven regeneration.372,373 The reserve stem cell theory posits that repair and regeneration depend on the re-initiation of early transcriptional developmental programs in quiescent, viable reserve stem cell populations under homeostatic conditions. For example, p57+ cells exhibit quiescent stem cell activity, undergoing a dynamic reprogramming process of differentiation as part of constitutively activated spatiotemporal reprogramming and as facultative stem cells, supporting regeneration after injury.374 Single-cell sequencing of highly proliferating intestinal organoids obtained using a combination of chemicals and factors revealed variable expression of regenerative stem cells, such as in vivo damage-responsive Clu+ revival stem cells or Lgr5+ stem cells for tissue repair through epigenetic reprogramming.350 The theory of dedifferentiation suggests that epithelial cells are uniquely plastic, enabling them to dedifferentiate and replenish the pool of cycling cells lost upon damage; thus, by reverting to a more primitive state, the organ allows itself to remodel tissue patterns into homeostatic tissue compartments and induce regeneration.375–377 For example, in the absence of Lgr5+ stem cells, intestinal cell lines expressing intestinal alkaline phosphatase (Alpi+) dedifferentiate and further distribute and localize to the bottom of the crypt, becoming Lgr5+ stem cells.378 In vitro dedifferentiation of Alpi+ intestinal epithelial cells into lgr5+ stem cells could be reproduced by intestinal organoid models.378 Any theoretical knowledge of the emergence of stem cells that regulate injury repair in the context of injury and the molecular or signaling pathways that can be used to regulate it is beneficial for regeneration. Some factors or signaling pathways have been identified to drive the process of cell type differentiation as well as tissue regeneration.48,379,380 The Yes-associated protein 1 (YAP) and Wnt signaling pathways are involved in guiding regeneration and homeostatic tissue renewal.381 YAP is normally localized only in the nucleus in cryptic basal stem cells but becomes a nucleus in most intestinal epithelial cells in a kinase dependent Src family manner during intestinal regeneration or organoid growth after irradiation.382 CREPT is expressed in intestinal crypt Lgr5+ ISCs and maintains steady state regulatory factors; in the process of intestinal regeneration, it activates the Wnt signaling pathway.383 The use of interstitial stem cell-conditioned culture medium, including paracrine factors and the Wnt/Notch signaling pathway, was able to partially restore radiant organoids.384 These new insights into the regulation and fate determination of stem cells in organoid facilitate the development of therapies that harness regenerative capabilities to treat digestive disorders.385 Activation of endogenous stem cells is a complex process that requires the identification of various factors that affect the balance of stem cell proliferation and differentiation and the formation of finely regulated networks to control endogenous stem cell activation and promote regeneration.386 These organoids also provide valuable mechanistic insight into the development of stem cells and their niches while monitoring the development of these cells into mature functional lineages by regulating various signaling pathways, including Wnt, BMP, EGF, Notch, and FGF. Small molecule or nutritional factor manipulation of specific lineages can use organoid platforms to provide proof of concept and a deeper understanding of organ lineage specification; it can even identify potential intervention drugs.387–389 Epidermal growth factor is an important fate determinant that differentiates the surface and interior of human gastric glands and binds to BMP signaling to control pit cell differentiation from parietal or primary cells.390 The small molecule isoxazole 9 (ISX-9), a neurogenic modulator, enhances secretory progenitor cells in organoids by upregulating the transcription factor Pax4, a component of early human endocrine-specific progenitors.391 The serotonin receptor agonist Bimu8 could increase the density of L-cells in primary human colonic organoids.392 Pretreatment with L-arginine induces the stemness of the stem cell pool and preserves the gut response to TNF-α and 5-fluorouracil.393 Nuclear exportin 1 inhibitor regulates intestinal stem cell fate independently of known differentiation cues and significantly increases paneth cell abundance in organoids.394 Bile acids (BAs) and G protein-coupled bile acid receptor 1 (GPBAR1, also called TGR5) expression on gut stem cells regulates epithelial self-renewal and fate determination, cocultures of gut stem cells with BAs and TGR5 agonists improved gut organoid growth, and YAP1 and SRC inhibitors blocked TRC5-activated organoid growth.395 Utilizing multiple layers of genetic intervention on organoids platform is another approach to uncover the origin of various cell fates and determine the underlying signaling factors in the digestive system.396 Organoids with genetic knockout of protein phosphatase Mg2+/Mn2+‑dependent 1A (PPM1A) enhanced YAP/TAZ retention in the cytoplasm, leading to reduced cell proliferation and downregulation of RALY inhibition of development in an organoid model.397 CRISPR-Cas9-engineered human iPSCs with truncated HNF1αp291FsinSCs grown as 3D pancreatic organoids were shown to abolish HNF1β function and reduce pancreatic progenitor and β-cell differentiation.398 Angiotensin converting enzyme 2 (ACE2)-specific knockout gut organoids show decreased Lgr5 and Ki67 levels.399 Endoplasmic reticulum membrane protein complex subunit 3 (Emc3) is a determinant of the maintenance of gut mucous homeostasis, and the knockout of Emc3 led to enhanced endoplasmic reticulum (ER) pressure and destroyed Paneth cell function in the stem cell niche, leading to the failure of the culture of gut organoids.400 In addition to detecting the regenerative process in response to two kinds of regenerative stem cell patterns, organoid models can also detect how other cell types affect stem cell-mediated regeneration. As an integral part of the intestinal stem cell tissue microenvironment, Paneth cells not only form the boundary between the stem cell niche and epithelial organoid precursors but also provide essential Wnt3 for Lgr5+ stem cells.401,402 Immune cells, such as intraepithelial lymphocytes (IELs), also play an important role in maintaining homeostasis by secreting factors that affect the balance of stem cell self-renewal and differentiation.403 High-purity stem cells can be obtained from organoid culture; thus, new stem cell marker genes may be discovered, or the roles of cycling and quiescent stem cells may be investigated in lineage selection and differentiation; for example, CD24, EphB2, Krt17 and CD166+/GRP have been identified as novel stem cell markers using organoid cultures.355,404–406 Organoid models largely reproduce these processes and mechanisms, and further access to organoid systems can be used to identify novel physiologically relevant targets, in this case using multivariate phenotypic screens to elucidate the process of gut regeneration.
Immunotherapy
Tumor-induced immunosuppression is an important reason for evading immune surveillance attack.407 Immune checkpoint therapy (ICT) strategies targeting PD-1 and CTLA-4 can reverse tumor immunosuppression to a certain extent and achieve better therapeutic effects, but the clinical response is still low. It is necessary to further elucidate the mechanism of tumor immunosuppression and find new immunotherapy targets and strategies.408 The role of organoids in immunotherapy can be summarized in four directions: (1) reconstruction of the tumor immune microenvironment by coculture of organoids and immune cells as a platform for exploring and evaluating immunotherapy strategies;223 (2) exploration of the response of different phenotypes of cancer to immunotherapy and factors affecting the efficiency of immunotherapy by manipulating the gene expression of tumor organoids or changing the culture environment of tumor organoids in combination with gene editing technology;409–412 (3) comparison of different immune (adjuvant) therapies using tumor organoid tests to provide optimization ideas;411,413,414 and (4) the use of organoids (or cultures) to induce the generation of functional T cells as biological factories for the production of immunotherapeutic drugs.415–418 Digestive tumor organoids derived from tumor tissue or PSCs have potential applications in personalized immunotherapy.312,419,420 These organoids are cocultured with immune cells to construct or maintain complex tumor tissue microenvironment structures, which can be used for immunotherapy research for digestive system diseases and potential therapeutic development for personalized therapy.419,421,422 Organoid models of immunotherapy are slowly being built to support immune cell survival and study interactions between T cells and tumor cells.415,418 The type and source of immune cells included in the coculture can be determined according to the research scenario, for example, autologous/exogenous expanded peripheral blood mononuclear cells or specific activated immune cells derived from tumor biopsies or surgically resected human colon cancer samples.415 In a coculture of pancreatic tumor organoids with peripheral blood lymphocytes, tumor-dependent lymphocyte infiltration was observed in these models.423 Likewise, CRC and NSCLC organoids were cocultured with autologous peripheral blood mononuclear cells (PBMCs), cytotoxic T lymphocytes (CTLs) were successfully expanded, and tumoricidal effects were observed.418 Another study turned to the use of exogenous immune cells cocultured with colon cancer spheroids/organoids, and allogeneic T cells and NK cells were also able to rapidly infiltrate tumor spheroids, leading to immune-mediated tumor cell apoptosis.424 Through the successful activation of immune cells by coculture with tumor-tissue-derived organoids, one can not only observe the killing effect of immune cytotoxicity on malignant cells in vitro but also functionally test the efficacy of immunotherapy in specific patients, which is helpful to understanding potential biological principles. In an organoid model of human steatohepatitis, CD47 blockade prevented hepatic stellate cell activation and progression of steatohepatitis.425 T-cell depletion plays an important role in tumor immune escape. Identifying potential compounds that target T-cell depletion and enhance immune checkpoint inhibitor (ICI) responses to suppress immune evasion is a viable strategy. For example, dexamethasone inhibits immune evasion by decreasing PD-L1 expression and Indoleamine 2,3-dioxygenase 1 (IDO1) signaling.426 PDO models combined with gene editing techniques are used to determine which factors mediate disruption of cell-autonomous mechanisms that protect tumors from immune clearance, thereby improving the efficacy of immunotherapy. Human pancreatic cancer organoids with mutations in RNF3 are sensitive to the killing effect of CTL.427 In addition to testing the interaction and killing function between immune cells and tumor organoids, another dimension of research can be added, such as observing the impact of external stimuli on the immune environment and the effects of immunotherapy. Furthermore, these systems also showed excellent performance in examining the efficacy of combination therapies. Immune checkpoint blockade (ICB) therapies, such as PDL-1 antibodies, face the dilemma of low efficacy, and clinical and preclinical experiments have begun to test the efficacy of combination therapies. As a combination therapy, PD-1 blockers in combination with TBK1/IKK inhibitors can counteract resistance and improve the response to PD-1 blockers by helping to overcome the immunosuppressive microenvironment of immune systems.428 CDK4/6 inhibitor combined with anti-PD-1 blocking antibody treatment of tumor spheres increases the number of infiltrating T cells and enhances antitumor activity.429 Chimeric antigen receptor T-cell therapy (CAR-T), a new type of cellular immunotherapy, has shown high efficiency and wide clinical application value in the treatment of various hematological malignancies in recent years.430 This therapy generates and expands a patient’s autoimmune T cells in vitro, allowing them to more efficiently recognize specific antigens expressed by tumor cells. When reinjected into patients, these CAR-T cells can efficiently and specifically recognize and attack tumors.431 Organoids have also been shown to be effective platforms for evaluating CAR-T-cell potency and tumor specificity.417,432,433 In HER2-expressing colorectal cancer organoids obtained from patient biopsies, the addition of anti-HER2 CAR-T cells alone resulted in minimal killing, while the combination of CAR-T cells with the apoptosis antagonist birinapant induced powerful lethality of organoids.434 Colonic organoids were also used to track CAR-mediated NK-cell cytotoxicity.127 Recent studies have shown that it is possible to perform iPSC-derived T cells. These stem cell-derived T cells display full immunity and high tumor killing similar to human peripheral blood TCRαβ T cells. This novel CAR-derived T-cell circumvention of donor-derived defects will greatly promote the promotion of CAR-T therapy.435 With the continuous maturation and application of the coculture system of immune cells and organoids, it is believed that the organoid model will be a major asset to the field of immunotherapy.
Other treatments
Organoids can serve as a good platform for evaluating gene therapy, in which gene editing technologies are used to repair genetic deficiencies in patients.311,436,437 For example, the use of CRISPR gene editing technology to correct CFTR mutations in intestinal organoids of CF patients has shown that gene repair can correct the CFTR locus in patient-derived CF organoids and restore the normal response of organoids to forskolin treatment.438 Organoids can also be used as an alternative test model for FMT therapeutic strategies. Given the compelling evidence that many human diseases are associated with microbiota dysbiosis, restoring physiological microbiota balance and composition may improve disease symptoms.439–443 FMT has been tested in a variety of diseases, such as malignancies, metabolic syndrome, neurological diseases, and autoimmune diseases, and has shown surprising therapeutic effects.444–449 An important concept in this framework is the need to identify well-defined beneficial traits in the microbiome and validate the impact of beneficial bacterial combinations on individuals.450,451 Organoids would be good models to test how to determine the role of target microbiota and the effect of target microbiota.
Digestive organoid biobanks and clinical studies
Because various organoids can be easily obtained from postoperative or biopsy tissue and cultured indefinitely, many patient-derived organoids form the basis for the creation of living biobanks.323,452,453 These biobanks contain many organoids from different clinical stages and histological subtypes, determined by gene expression signatures, and culture methods are regulated and optimized according to the needs of niche factors.152,454 Human organoids can be obtained from postoperative or preoperative biopsies and contribute to understanding disease mechanisms, drug screening to predict treatment outcomes, regenerative medicine, and developing treatment options for personalized treatment regimens.455 To continuously optimize clinical treatment strategies and explore the pathogenesis of diseases, organoid biobanks for various diseases have been established in recent years, among which gene mutation-related diseases and malignant tumors are the most common.303,323,324,456 For example, organoid biobanks derived from patients with digestive system diseases have been established; the resources in these biobanks include intestine,57,319,324,415,457 stomach,61,151 pancreas171,174,458 and liver organoids.194,459 Other more extensive organoid biobanks have been established by academic institutions (HUB, https://huborganoids.nl/) and commercial entities (American Type Culture Collection (ATCC, www.attc.org), Sigma-Aldrich, DefiniGEN (https://www.definigen.com/products/intestinal/organoid/) and Cellesce (https://cellesce.com)). These human organoids bring together a large number of adult stem cell organoids derived from primitive human tissue or organoids derived from iPSCs, providing researchers with opportunities for biomedical research and drug testing.453 Another benefit of building an organoid biobank is that, by combining large samples and clinical data, it is possible to retrospectively study the response of patient samples to existing drugs. In the case of ineffectiveness, mechanistic studies and drug screening can be performed to address these causes. Given these advantages, PDO can serve as an important tool for drug screening and prognostic assessment in the clinic, demonstrating their great potential in personalized and precise treatment.
Organoid-based clinical research is another emerging hotspot, including the construction of organoid biobanks, drug screening, prognostic assessment, and disease mechanism research. Organoids can be stored for a longer period and are easier to transport than primary tissues; therefore, they are gradually becoming a new favorite for large-scale cohort studies.460 Registered clinical trials are collecting and constructing organoid biobanks for diseases such as colorectal cancer, hepatocellular carcinoma, cholangiocarcinoma, pancreatic cancer, inflammatory bowel disease and primary sclerosing cholangitis, promoting the widespread adoption of organoid construction in the clinical diagnosis and treatment process and the standardization of the construction process (Table 2). Organoids as a platform for drug screening can improve the screening efficiency and save experimental costs. The completed and ongoing clinical trials using organoids for drug screening are mainly focused on antitumor drugs. Target diseases include pancreatic, colorectal, gastric, and biliary tract cancer, and the experimental design includes exploring the response of tumor organoids to single drugs or different drug combinations, the response of specific mutation types to drugs, and exploring the consistency between treatment responses in organoids and clinical outcomes of chemotherapy. The evaluation indicators include drug effectiveness, drug tolerance, and drug side effects. Additionally, organoids derived from genetic diseases such as cystic fibrosis can also be screened for drugs.461 In addition to drug screening, organoids can be used for a wider range of preclinical assessments, including response to radiation therapy, comparison of primary and metastatic cancers, biomarker screening, and assessment of prognosis and risk of recurrence. Through prior experiments on organoids, clinicians can make evidence-based decisions to reduce unnecessary harm to patients caused by decision errors and outcome uncertainty and improve the efficiency and safety of clinical care. At present, many diseases, such as allergies and irritable bowel syndrome, lack suitable disease models462 and the emergence of organoids provides new research ideas. By constructing organoids to simulate the intestinal environment, researchers can explore the interaction between the intestinal system and the outside factors, explore the pathophysiological mechanisms and formulate treatment strategies. Relevant clinical trials include studies of microbe-host interactions, mechanisms of allergy and intestinal disorders, the characterization of intestinal stem cells in patients, the biology of innervated sensory epithelial cells, the physiopathology of necrotizing enterocolitis, and gut inflammation in the pathogenesis of hypertension. Overall, organoid-based clinical trials continue to increase in variety and number. In addition to the abovementioned four applications, organoid-based immunotherapy463 and tissue regeneration research367 have also made breakthroughs, and corresponding clinical trials and translational results are also expected to appear in the future.
Table 2.
Summary of organoid-based clinical trails
| Application | Condition or disease | Research title | ClinicalTrials.gov Identifier |
|---|---|---|---|
| Drug Screening | Pancreatic Cancer | A Prospective, Randomized, Controlled Trial of Chemotherapy for Advanced Pancreatic Cancer Based on Organoid Drug Sensitivity Test | NCT04931381 |
| Guidance of Standard of Care Treatment for Metastatic Pancreatic Cancer by Drug Screening in Patient-derived Organoids: A Single Centre, Open-label, Single Arm, Phase II Trial With Feasibility Endpoint | NCT05351983 | ||
| A Prospective, Randomized, Controlled Trial of Adjuvant Chemotherapy for Pancreatic Cancer Based on Organoid Drug Sensitivity Test | NCT04931394 | ||
| Drug Screening of Pancreatic Cancer Organoids Developed From EUS-FNA Guided Biopsy Tissues | NCT03544255 | ||
| Pharmacotyping of Patient-derived Pancreatic Cancer Organoids from Endoscopic Ultrasound-guided Biopsy as a Tool for Predicting Oncological Response | NCT05196334 | ||
| Colorectal Cancer | Prospective Observation on the Accuracy of in Vitro Screening of Colorectal Cancer Chemotherapy Drugs Based on Organoids-on-a-chip | NCT04996355 | |
| Patient-derived Organoids of RAS/RAF Wild-type Metastatic Right Colon Cancer to Test the Sensitivity and Clinical Consistency of Combined Treatment of Cetuximab. | NCT04906733 | ||
| A Prospective Multicenter Randomized Controlled Trial of the Clinical Efficacy of Neoadjuvant Therapy Based on Organoids Drug Sensitivity Versus Empirical Neoadjuvant Therapy in the Treatment of Advanced Rectal Cancer | NCT05352165 | ||
| The Culture of Advanced/Recurrent/Metastatic Colorectal Cancer Organoids and Drug Screening | NCT05304741 | ||
| Pilot Study for Ex Vivo Tailoring of Treatment in Colorectal Cancer | NCT05401318 | ||
| Gastric Cancer | The Clinical Efficacy of Patient-derived Organoid-based Drug Sensitive Neoadjuvant Chemotherapy Versus Traditional Neoadjuvant Chemotherapy in Advanced Gastric Cancer: A Prospective Multi-center Randomized Controlled Study | NCT05351398 | |
| Consistency Between Treatment Responses in Patient-Derived Organoid (PDO) Models and Clinical Outcomes of Neoadjuvant Therapy, Conversion Therapy and Palliative Therapy in Gastric Cancer | NCT05203549 | ||
| A Prospective Observational Study on the Potential Benefit of Neoadjuvant Therapy for Advanced Gastric Cancer Based on Organoid Drug Susceptibility Screening | NCT05442138 | ||
| Q-GAIN (Using Qpop to Predict Treatment for GAstroIntestinal caNcer) | NCT04611035 | ||
| Cystic Fibrosis | Response to CFTR-modulators in Intestinal Organoids of Patients with CF Having at Least One R334W Mutation | NCT04254705 | |
| Biliary Tract Cancer | A Prospective Feasibility Study of Multi-Platform Profiling Using Biospecimens From Patients With Resected Biliary Tract Cancer | NCT04561453 | |
| Preclinical Evaluation | Pancreatic Cancer | Establishing Organoids from Metastatic Pancreatic Cancer Patients, the OPT-I Study | NCT03500068 |
| Development of a Prediction Platform for Neoadjuvant Treatment and Prognosis in Pancreatic Cancer Using Ex Vivo Analysis of Organoid Culture | NCT04777604 | ||
| Development of a Prediction Platform for Adjuvant Treatment and Prognosis in Pancreatic Cancer Using Ex Vivo Analysis of Organoid Culture | NCT04736043 | ||
| Colorectal Cancer | Validation of Organoids Potential Use as a Companion Diagnostic in Predicting Neoadjuvant Chemoradiation Sensitivity in Locally Advanced Rectal Cancer | NCT03577808 | |
| Systemic Neoadjuvant and Adjuvant Control by Precision Medicine in Rectal Cancer (SYNCOPE) - Approach on High-risk Group to Reduce Metastases | NCT04842006 | ||
| Radiation Enteritis, Inflammatory Bowel Diseases | Preclinical Evaluation of Multimodal Therapeutic Strategies in Intestinal Irradiation and Inflammatory Bowel Disease from Organoids | NCT05425901 | |
| Esophageal Cancer | Chemoradioresistance in Prospectively Isolated Cancer Stem Cells in Esophageal Cancer-Organoid: RARE STEM-Organoid | NCT03283527 | |
| Esophagogastric Carcinoma | Molecular Outcome Prediction of Neoadjuvant Systemic Treatment in Esophagogastric Carcinoma | NCT03429816 | |
| Colorectal Cancer | The Exploratory Study of Patient-derived Organoids for the Prediction and Evaluation of Clinical Efficiency Effect of Colorectal Cancer Liver Metastasis | NCT05183425 | |
| Validation of the Three-dimensional Bioprinted Tumor Models as a Predictive Method of the Response to Chemotherapy for Colorectal Cancer With or Without Liver Metastases | NCT04755907 | ||
| Living biobanks | Inflammatory Bowel Disease | Prospective, monocentric cohort aiming at generating 3D organoids from human digestive samples | NCT05294107 |
| Colorectal Cancer | Feasibility of Establishing Patient-Derived Organoids for Rectal Cancer: A Biospecimen Collection Protocol | NCT04371198 | |
| Tumor Immune Microenvironment Involvement in Colorectal Cancer Chemoresistance Mechanisms: a Patient-derived Tumoroids Prospective Collection From Systemic Treatment Naive Tumors | NCT05038358 | ||
| Colorectal Cancer Metastases and Hepatocellular Carcinomas | Next Generation “ of Liver Derived-organoïd Biobank: Case of Colorectal Cancer Metastases and Hepatocellular Carcinomas | NCT05384184 | |
| Liver, Biliary and Pancreatic Cancer | A Study Designed to Develop in Vitro Models of Liver, Biliary and Pancreatic Cancer for the Investigation of Tumour Biology and Potential Therapies | NCT02436564 | |
| Primary Sclerosing Cholangitis | Characterization of Biliary Cell-derived Organoids From Bile of PSC and Non-PSC Patients | NCT04753996 | |
| Pancreatic Cancer | EUS-guided Biopsy of Pancreatic Mass Lesions for Developing Patient -Derived Cancer Models | NCT03140592 | |
| Mechanism exploration | Host-microbiota Interactions | Establishment of Human Organoid Lines as a Tool to Dissect Molecular Pathways of Host-microbiota Interactions | NCT05323357 |
| Allergy and Gastrointestinal Disorders | Establishment of Small Intestinal Human Organoids to Check the Influence of Nutrient Antigens or Therapeutic Agents | NCT03256266 | |
| Improving the Diagnosis of Food Allergy and Food Intolerance by Determining Mucosal IgE and Inflammatory Markers and Validating With Intestinal in Vitro Organoids | NCT05259826 | ||
| Bestimmung Des Einflusses Von Nahrungsmitteln Auf Die Darmschleimhaut Mit Der Konfokalen Laserendomikroskopie Und Humanen in Vitro Organoiden | NCT05056610 | ||
| Intestinal Stem Cells Characterization | Intestinal Stem Cells Characterization in Intestinal Organoid Culture from Inflammatory Bowel Disease and Intestinal Polyposis Patients | NCT02874365 | |
| Specimen acquisition | Evaluation and Comparison of the Growth Rate of Pancreatic Cancer Patient-derived Organoids Generated from Matched Fine Needle Aspirations (FNA) and Fine Needle Biopsies (FNB) | NCT03990675 | |
| The biology of innervated sensory epithelial cells | The Innervation of Human Gut Sensory Epithelial Cells | NCT02888587 | |
| The physiopathology of Necrotizing Enterocolitis | Development and Use of a Tissue and Human Enteroid Biorepository to Study the Pathophysiology of Neonatal Necrotizing Enterocolitis | NCT04549727 | |
| The gut epithelium properties in hypertension | Gut Inflammation and Gut-Gut Microbiome Interactions in the Pathogenesis of Hypertension | NCT04497727 |
Current limitations and advantages of organoids
Although organoids have been widely used in various fields, the organoids currently being produced are mainly epithelial systems. These simple components leave many studies unable to truly simulate the in vivo response, and the response of the organ should be the result of the mutual influence of various cells. Many drug effects on human tumors are often related to their microenvironment; for example, coculture of human PDAC PDOs and cancer-associated fibroblasts (CAFs) increases resistance to gemcitabine.423 In addition, the optimal composition of matrigel used for organoids is still unclear, severely limiting clinical application. In addition, the current culture systems are very different from the in vivo environment, and it is difficult to provide sufficient oxygen and nutrient supply for organoids growth. Further integration of microenvironmental components would allow digestive organoids to represent in vivo physiology more faithfully. Therefore, the major advances in organoids are more cellular components, more specific matrix components, and more suitable culture methods (Fig. 4).
Fig. 4.
Advancements in organoid cellular and functional integrity. Advancement of organoid models: improving organoid complexity by adding mesenchymal cells, immune cells, endothelial cells, nerve cells, etc.; combining organoids with 3D bioprinting, biological materials and chip technology to develop organoid models that closely resemble the physiology of the human digestive system
Construction of microenvironment and exploration of interactions between organs
The digestive organ’s microenvironment is a complex, dynamic, and multicellular system that features ongoing interactions among its components. However, existing organoid systems contain predominantly epithelial cells, low levels of maturation and cellular complexity.464 Importantly, building fully complex, well-functioning organoids enables more realistic studies of complex diseases such as cancer, syndromic diseases, and pathogen (bacterial and viral) infections. There are two ways through which organoid culture systems can achieve sufficient cellular complexity to recapitulate the function of the corresponding organs. In iPSC-derived organoids, the emergence of target cells can be induced by introducing transcription factors or exogenous mediators to promote organoid maturation. Induction of enteroendocrine cell (EEC) development can be realized by transient expression of NEUROG3 in iPSC-derived organoids.465 Intestinal microfold cells (M cells) are a dynamic epithelial cell lineage that initiates intestinal mucosal immunity; they have been successfully differentiated by adding activator of NF-κB ligand receptor (RANKL) to the culture medium, which induced the formation of M cells in organoid cultures by upregulating the transcription factor SpiB.466 TGF-β1 and Sonic Hedgehog (SHH) signaling regulate the appearance of muscularis mucosa (MM) of human gastric organoids derived from iPSCs.467 BMP activation drives the expression of villus tip genes, and loss of the Bmpr1a receptor in human organoids reduces the expression of such genes.468 When cell atlas and organoid technologies were combined to discover the effects of stem cell niche characteristics on mesenchymal-derived cells, the results suggested that the homeobox transcription factor CDX2 was required for epithelial and mesenchymal compartmentalization, while NRG1 promoted intestinal stem cell maturation.355 The enteric nervous system (ENS), part of the autonomic nervous system, has remarkable neurotransmitter diversity and complex cytoarchitecture. Human PSC-derived neural crest cells (NCCs) or blood vessels can be individually introduced to HIOs and assembled with them at appropriate times to form a functional enteric nervous system (ENS) and blood vessels in the organoids.469 Refining existing intestinal coculture systems is another way to achieve the desired grade of organoid maturity and cellular complexity. These coculture systems can be formed with organoids through two types of strategies. In the reconstituted model, organoids are cultured in extracellular matrix domes, and other cells are immersed in culture medium or the bottom layer of Transwell plates. In holistic native tumor microenvironment (TME) models without reconstitution, organoids from digested tissue can be mixed with collagen and injected into microfluidic systems or other culture devices to preserve the epithelium and its microenvironment in vitro. Complex cocultures of immune cells with organoids provide a versatile tool to study bidirectional interactions that support the delicate balance of digestive homeostasis. Coculture with innate lymphocytes (ILCs) will show how different subsets of ILCs affect intestinal epithelial barrier integrity and regeneration.470 When cocultured with interleukin 2 (IL-2)-secreting immune cells, human PSC-derived intestinal organoids rapidly mature into adult intestinal epithelial cells and increase gut-specific functional activity.471 Coculture of human intestinal stem cell-derived enterocytes and human monocyte-derived macrophages improves intestinal cell barrier function and maturation.472 Some subtypes, such as PTGER4+ intestinal macrophages, support the intestinal stem cell niche in organoids.473 A recent study went a step further, describing a platform for integrating patient-specific mature lymph node antigen-presenting cells into organoids for adaptive immunity.474 In addition to coculture of immune cells, other cell types have been extensively explored in recent decades, and a series of fruitful analyses have been performed on the degree of phenotypic mimicry and molecular mechanistic replication of in vivo function after coculture. The mesenchymal system is generally thought to be involved in stem cell maintenance. Coculture of AdSC-derived small intestine with intestinal subepithelial myofibroblasts (ISEMF) allows long-term small intestine culture in the absence of certain growth factors required for organoid culture.475 The pericryptal CD34+Gp38+αSMA- mesenchymal cells are the major intestinal producers of niche factors that promote the maintenance of stem cell populations in gut organoids.476,477 The WNT-dependent PDAC organoids grow independently of exogenous WNTs when cocultured with patient-derived CAFs.174 In Rspo-free medium, the growth of gastric organoids is supported as long as they are cocultured with stromal cells.478 Organoid cocultures were used to recapitulate portal mesenchymal cell architecture, including a periportal mesenchymal cell subset that acts as a rheostat to regulate ductal cell proliferation.479 CD90+ mesenchymal cells may enhance stem cell function due to the presence of L-arginine in the organoid and mesenchyme coculture systems, acting through mTORC1 to stimulate Wnt2b secretion.480 Pancreatic tumor spheroids were cocultured with pancreatic stellate cells in microchannels so that fibroblast activation could be observed.481 Human PDAC organoids and mouse CAFs were cocultured to identify heterogeneity within CAFs in order to guide therapy.482 The enteric nervous system is composed of many enteric glial cells (EGCs) and neurons, which are key components of the intestinal stem cell niche in regeneration and disease; these fibrillary acidic protein (GFAP)+ EGCs express multiple Wnt ligands and promote self-renewal of LGR5+ ISCs in organoid and glia coculture systems.483 Coculture with neurons similar to myofibroblasts enhances the growth of the organoids.484,485 Coculture of intestinal organoid monolayers with differentiated adipocytes induces reciprocal amplification of inflammatory responses.486 Organoids are cocultured with vascular endothelial cells and pericytes to form vascular organoids, providing hope for the further development of organoid model systems. Endothelial cells were added to a miniature device in which isolated intestinal organoid cells had been found to communicate with intestinal epithelial cells and form a blood vessel-like network.224 Vascularization can be implemented by transplantation into mice or by using microfluidic devices to connect vascular networks to intestinal organoids from a highly vascularized tissue, for example, in the case of human PSC-derived intestinal organoids implanted in mouse kidneys.487 Holistic native TME models without reconstitution favor immunotherapeutic effects or organoid transplantation for regenerative treatment of bowel disease. Tumor tissue and a mixture of stromal fibroblasts and immune components were cocultured using an air-liquid interface organoid culture system, and heterogeneous T cells from the original tumor were also preserved in these cultures.223 An in vitro model in which organoids from clinical CRC tissue were mixed with TILs from patients and F. nucleatum showed that exposure to F. nucleatum increased sensitivity to PD-L1 blockade.488 Organoids with diverse cellular components will advance organoid technology for studying the physiological and pathological functions of the digestive system. Another notable direction of progress is the establishment of a multi-organoids model, which is interesting and necessary because of the interaction of multiple organs in the process of disease development. Methods have recently been developed for the fusion of organoids and coculture of multiple organoids. For example, heart-lung-liver organoid models have been assembled in a modular manner through microengineering.489 A new protocol for generating integral multiorgan structures of the digestive system has also been reported, i.e., the fusion of two PSC-derived spheroids that correspond to the anterior and posterior gut to the creation of hepato-biliary-pancreatic organoids that establish boundary interactions between two spheroids without any extrinsic factors.490 If the timing and composition of the medium are tightly controlled, different types of organoids can be made to grow independently on the microfluidic chip but still communicate with each other. For example, fully functional tissues have been generated in microfluidic chip culture systems via the assembly of multiple organoids.491 Multilineage organoid models integrate cardiac and gut features arising from the aggregation of single mesodermal progenitors.492 Paracrine signals between adjacent tissues influence fate decisions of target organs during embryogenesis. Multi-organoids models can be used as in vitro models to simulate the effects of paracrine inputs from surrounding tissues on developmental and cellular heterogeneity during organ development. Multi-organoids model developed with microfluidic arrays were used to coculture stem cell-derived liver, intestine and stomach organoids; paracrine factors produced by intestinal organoids were detected to affect the expression of bile acid synthetase (CYP7A1) in liver organoids, which verified the interorgan interaction.226 Multiorgan metabolic diseases characterized by dynamic interactions between different organs, such as type 2 diabetes mellitus (T2DM), can be replicated through coculture interactions between different organoids. With the help of a microfluidic system consisting of two separated regions connected by a microchannel network, coculture systems can be established between liver and pancreas organoids derived from human iPSCs, recapitulating the physiological and pathological conditions of the human liver-islet axis and facilitating analytical work regarding this axis in health and disease.493 By integrating technologies such as microarray technology and multiple digestive system organoids, such as esophageal/gastric/liver/pancreatic/gut organoids, into in vitro models to bring them closer to human tissue structure and function, these advanced models can further bridge the gap between in vitro systems and animal experiments (Fig. 5).
Fig. 5.
Future trends in the development of multi-organoid models. Organoids of the esophagus, pancreas, stomach, and intestines can be generated in vitro and combined to form a multi-organoid model on a microarray to test multiorgan diseases and cancer metastasis as well as drug toxicity and efficacy
Optimization of culture conditions
Improvements in cultivation are another aspect in which progress has been made recently. These improvements have mainly been in two directions: replacing the extracellular matrix and improving organoid culture systems. On the one hand, a large number of current organoid systems rely on animal-derived extracellular matrix (mainly stroma and basement membrane extract (BME)), a protein mixture extracted from Engelbreth-Holm-Swarm mouse sarcoma.494 Matrigel/BME has no unified standard formula and shows batch-to-batch variability, which is a major roadblock for the translation of this methodology into the clinic, limiting its applicability to personalized medicine.495 Emerging matrigel-free methods for the culture of organoids can overcome these limitations; examples of matrigel-free approaches include decellularized ECM collagen, engineered ECM proteins and synthetic polymer hydrogels.496 Decellularized ECM from human or animal donors can accurately recapitulate native structure, ECM composition and vascularization during organ development, and decellularized ECM has been used to culture enteroids and HIOs derived from PSCs.63,497 Collagen I, a natural protein commonly obtained from pigs, has been used to culture human intestinal organoids.498 Synthetic hydrogels such as PDMS are attractive due to their controllable mechanical properties, functions, and erosion rates and are currently used to create organ-on-a-chip microfluidic chambers.499 Other alternatives, such as a recently developed synthetic hydrogel scaffold for use with polyethylene glycol (PEG), have shown similar results to matrigel in supporting organoid growth.500 However, these synthesized hydrogels have difficulty accurately reproducing the physicochemical properties of ECM in vivo, and the material can adsorb small hydrophobic molecules, which is disadvantageous in that it hinders drug discovery.501 There is a growing need to design and manufacture biosynthetic materials that mimic ECM function and preserve specific cellular properties while maintaining stem cell differentiation.502 Matrigel formulas made of peptides and recombinant proteins produced by genetically engineered organisms are attractive for organoid culture because their mechanical and chemical properties can be independently altered.503 Overall, the prospect of programmable recombinant protein synthesis materials has certain advantages for functionalizing the binding of biologically relevant cellular proteins or peptides, enabling the artificial ECM to reproduce the dynamic properties (erosion rate, viscoelasticity, and degradation sensitivity) as well as the correct chemical composition of the natural ECM. This artificial system with independently adjustable chemical and mechanical properties has been used to provide appropriate development and growth for various kinds of organoids. Another area that is being refined is organoid culture methods; using technologies such as 3D printing and microfluidics, problems such as structure and oxygen concentration can be solved to improve culture devices. 3D bioprinting technology includes loading tissue-specific cells into bio-inks to reconstruct various organ structures of the human body through layered printing technology.504 Another innovative technological combination involving organoids consists of “organoids-on-a-chip”. This system is a microfluidic device that grows cells and tissues in a continuous perfusion chamber. Organoids-on-a-chip can form a dynamic and controllable digestive system microenvironment. It is expected to recapitulate the physiology and pathology of living human organs in vitro and model organ-organ interactions, even serving as an alternative to animal models in the future.505 Bottom-up tissue engineering strategy building blocks can be used to generate organic cultures of multicellular superstructures, and those models may also permit the epithelium to be cultured with other cells based on a 3D structure with a type I collagen coating on a thin Matrigel base that mimics the morphology of the gut.506 Using the surfaces of tissue-engineered scaffolds to create crypt-like structures, intestinal stem cells have been induced to form tubular epithelial cells with accessible lumens and a spatial arrangement of crypt- and villus-like domains similar to those observed in vivo. By connecting an external pump system, the micro-scale intestinal tube can be perfused and the dead cells can be continuously cleared of, extending tissue life by weeks.224 In the absence of externally applied biochemical gradients, several recent approaches have created macroscopic organoids resembling periodic crypt-villus structures of native intestinal epithelium, using physiologically precise patterns and localizations conferred only by tissue geometry.99 In addition, robotic pipelines using traditional machines require fewer cells to automate work, illustrating that automation helps simplify and harmonize steps in the workflow.507 To date, fully automated 3D culture protocols for kidney organoids using liquid-handling robots have been established.508 In the future, more integrated technologies are required to combine all test conditions in one organoids culture and ultimately generate organoids with complete structure and function, reproduce the physiological functions of target organs, and further advance the state of clinical treatments.
Conclusion and perspective
Digestive organoids are 3D in vitro cultured organ models with similar structures that contain multiple cell types, including stem cells and differentiated cells.509 These organoid models can overcome the genetic variability of traditional cell culture systems and the pitfalls of interspecies dissimilarity between humans and mice, and they have the potential to provide personalized therapy for a wide range of patients, even at various preclinical stages. Thus, human organoids represent an in vitro testing model for drug screening or gene/cell therapy and serve as a bridge between in vitro and in vivo systems.510 In particular, the large-scale application of existing 3D PDO cultures has transformed in vitro cancer biology and enabled the construction of large tumor biobanks to capture the mutational diversity and histology of human cancers.40 To date, studies of digestive organoid research have covered the modeling of digestive system disease, establishment of organoid biobanks, transplantation of organoids into the injured epithelium for regeneration, repair of disease-related defects via gene editing, high-throughput drug screening, and characterizing the responses to bacterial and viral infection.511 However, although organoids have yielded many useful results in various fields of digestive system research, certain limitations of current organoids have also created bottlenecks that are currently insurmountable, requiring more breakthroughs. Emerging technologies, such as the application of CRISPR-Cas9 technology to silence or activate specific genes in organoids, will open new doors for understanding the role of specific genes in human digestive system disease.139,512–514 The distribution and function of target genes during digestive development or progression of disease can be detected in real time by gene knockin fluorescent labeling in iPSCs.514,515 Single-cell RNA sequencing (scRNA-seq) methods have changed the paradigm of biomedical science by analyzing cellular heterogeneity from multivariate data.516 scRNA-seq of organoids has allowed certain rare or abnormal cell types to be identified, and these findings can be detected in individual organoids.98,517 By deepening the combination with these new technologies, gene editing and omics (epigenetics, transcriptomics, proteomics, metabolomics, and single-cell sequencing), one can systematically and comprehensively analyze changes in molecular, cellular, and organizational structures mediated by gene mutations or environmental interventions, facilitating future basic research and clinical applications (Fig. 6). Additional microfluidic devices are moving from proof of concept to commercial tools, and easy-to-use devices have facilitated the development of high-performance sensors that can record oxygen levels, transepithelial resistance and various experimental data in real time.314 We anticipate that the implementation of organoids, standard protocols and sophisticated devices will help the scientific community discover new mechanisms of action of pathology in the digestive system and identify biomarkers and follow-up targets for precision therapy for subsequent clinical trials.
Fig. 6.
The prospect of combining high-technology approaches and digestive organoids. Gene editing and omics (epigenetics, transcriptomics, proteomics, metabolomics, and single-cell sequencing) technologies are used in combination with organoid models to systematically and comprehensively analyze molecular, cellular, and histological changes mediated by gene mutations or environmental interventions. Starting from basic research, the in vivo phenotypes and molecular targets corresponding to organoids under different conditions are continuously explored to guide the selection and testing of clinical treatment strategies
In conclusion, given the incomplete cellular composition and function of various digestive organoids, further optimization is required to enhance their application in disease modeling and personalized medicine. When organoids are derived from tissue or cells that have been directly extracted from diseased patient samples, they preserve the genetic and pathological information of the original tissue, and their 3D physiological structure and cellular composition remain relatively close to the corresponding human organ, opening a vital avenue for innovation in precision and personalized medicine. Combined with other bioengineering methods and omics techniques, organoid models will undoubtedly play an invaluable role in preclinical and clinical research.
Competing interests
The authors declare no competing interests.
References
- 1.Thompson CA, DeLaForest A, Battle MA. Patterning the gastrointestinal epithelium to confer regional-specific functions. Dev. Biol. 2018;435:97–108. doi: 10.1016/j.ydbio.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartenstein V, Martinez P. Structure, development and evolution of the digestive system. Cell Tissue Res. 2019;377:289–292. doi: 10.1007/s00441-019-03102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ananthakrishnan AN, et al. Environmental triggers in IBD: a review of progress and evidence. Nat. Rev. Gastroenterol. Hepatol. 2018;15:39–49. doi: 10.1038/nrgastro.2017.136. [DOI] [PubMed] [Google Scholar]
- 4.Aron-Wisnewsky J, et al. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2020;17:279–297. doi: 10.1038/s41575-020-0269-9. [DOI] [PubMed] [Google Scholar]
- 5.Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019;16:690–704. doi: 10.1038/s41575-019-0209-8. [DOI] [PubMed] [Google Scholar]
- 6.Gines P, et al. Liver cirrhosis. Lancet. 2021;398:1359–1376. doi: 10.1016/S0140-6736(21)01374-X. [DOI] [PubMed] [Google Scholar]
- 7.Israelsen M, et al. Metabolic and genetic risk factors are the strongest predictors of severity of alcohol-related liver fibrosis. Clin Gastroenterol Hepatol. 2022;20:1784–1794. doi: 10.1016/j.cgh.2020.11.038. [DOI] [PubMed] [Google Scholar]
- 8.Emdin CA, et al. Association of genetic variation with cirrhosis: a multi-trait genome-wide association and gene-environment interaction study. Gastroenterology. 2021;160:1620–1633 e1613. doi: 10.1053/j.gastro.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 10.Collaborators, G. B. D. O. C. The global, regional, and national burden of oesophageal cancer and its attributable risk factors in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020;5:582–597. doi: 10.1016/S2468-1253(20)30007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collaborators, G. B. D. I. B. D. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020;5:17–30. doi: 10.1016/S2468-1253(19)30333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collaborators, G. B. D. P. C. The global, regional, and national burden of pancreatic cancer and its attributable risk factors in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2019;4:934–947. doi: 10.1016/S2468-1253(19)30347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao L, et al. The emerging role of ferroptosis in non-cancer liver diseases: hype or increasing hope? Cell Death Dis. 2020;11:518. doi: 10.1038/s41419-020-2732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peery AF, et al. Burden and cost of gastrointestinal, liver, and pancreatic diseases in the United States: Update 2018. Gastroenterology. 2019;156:254–272 e211. doi: 10.1053/j.gastro.2018.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diseases GBD, Injuries C. Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396:1204–1222. doi: 10.1016/S0140-6736(20)30925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collaborators, G. B. D. C. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020;5:245–266. doi: 10.1016/S2468-1253(19)30349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd-Price J, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, et al. Integrated omics of metastatic colorectal cancer. Cancer Cell. 2020;38:734–747 e739. doi: 10.1016/j.ccell.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Perakakis N, Stefanakis K, Mantzoros CS. The role of omics in the pathophysiology, diagnosis and treatment of non-alcoholic fatty liver disease. Metabolism. 2020;111S:154320. doi: 10.1016/j.metabol.2020.154320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mars RAT, et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell. 2020;182:1460–1473 e1417. doi: 10.1016/j.cell.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yachida S, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019;25:968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 22.Wong VW, et al. Noninvasive biomarkers in NAFLD and NASH - current progress and future promise. Nat. Rev. Gastroenterol. Hepatol. 2018;15:461–478. doi: 10.1038/s41575-018-0014-9. [DOI] [PubMed] [Google Scholar]
- 23.Connor AA, Gallinger S. Pancreatic cancer evolution and heterogeneity: integrating omics and clinical data. Nat. Rev. Cancer. 2022;22:131–142. doi: 10.1038/s41568-021-00418-1. [DOI] [PubMed] [Google Scholar]
- 24.Metwaly A, et al. Integrated microbiota and metabolite profiles link Crohn’s disease to sulfur metabolism. Nat. Commun. 2020;11:4322. doi: 10.1038/s41467-020-17956-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JWJ, et al. Multi-omics reveal microbial determinants impacting responses to biologic therapies in inflammatory bowel disease. Cell Host Microbe. 2021;29:1294–1304 e1294. doi: 10.1016/j.chom.2021.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao W, et al. Multi-faceted epigenetic dysregulation of gene expression promotes esophageal squamous cell carcinoma. Nat. Commun. 2020;11:3675. doi: 10.1038/s41467-020-17227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, et al. Multi-omics characterization of molecular features of gastric cancer correlated with response to neoadjuvant chemotherapy. Sci. Adv. 2020;6:eaay4211. doi: 10.1126/sciadv.aay4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busslinger GA, et al. Human gastrointestinal epithelia of the esophagus, stomach, and duodenum resolved at single-cell resolution. Cell Rep. 2021;34:108819. doi: 10.1016/j.celrep.2021.108819. [DOI] [PubMed] [Google Scholar]
- 29.May-Zhang AA, et al. Combinatorial transcriptional profiling of mouse and human enteric neurons identifies shared and disparate subtypes in situ. Gastroenterology. 2021;160:755–770 e726. doi: 10.1053/j.gastro.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruepunga N, et al. Anatomy of rodent and human livers: What are the differences? Biochim Biophys. Acta Mol. Basis Dis. 2019;1865:869–878. doi: 10.1016/j.bbadis.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, et al. Comparative analysis of cell lineage differentiation during hepatogenesis in humans and mice at the single-cell transcriptome level. Cell Res. 2020;30:1109–1126. doi: 10.1038/s41422-020-0378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monberg ME, et al. Occult polyclonality of preclinical pancreatic cancer models drives in vitro evolution. Nat. Commun. 2022;13:3652. doi: 10.1038/s41467-022-31376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-David U, et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560:325–330. doi: 10.1038/s41586-018-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, et al. 3D cell culture models: Drug pharmacokinetics, safety assessment, and regulatory consideration. Clin. Transl. Sci. 2021;14:1659–1680. doi: 10.1111/cts.13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beumer J, Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 2016;143:3639–3649. doi: 10.1242/dev.133132. [DOI] [PubMed] [Google Scholar]
- 36.Kim TH, Shivdasani RA. Stomach development, stem cells and disease. Development. 2016;143:554–565. doi: 10.1242/dev.124891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willet SG, Mills JC. Stomach organ and cell lineage differentiation: from embryogenesis to adult homeostasis. Cell Mol. Gastroenterol. Hepatol. 2016;2:546–559. doi: 10.1016/j.jcmgh.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, et al. The development and stem cells of the esophagus. Development. 2021;148:dev193839. doi: 10.1242/dev.193839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Funata M, et al. The promise of human organoids in the digestive system. Cell Death Differ. 2021;28:84–94. doi: 10.1038/s41418-020-00661-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puschhof J, Pleguezuelos-Manzano C, Clevers H. Organoids and organs-on-chips: Insights into human gut-microbe interactions. Cell Host Microbe. 2021;29:867–878. doi: 10.1016/j.chom.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Dedhia PH, Bertaux-Skeirik N, Zavros Y, Spence JR. Organoid models of human gastrointestinal development and disease. Gastroenterology. 2016;150:1098–1112. doi: 10.1053/j.gastro.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prior N, Inacio P, Huch M. Liver organoids: from basic research to therapeutic applications. Gut. 2019;68:2228–2237. doi: 10.1136/gutjnl-2019-319256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H, et al. Organoid model: A new hope for pancreatic cancer treatment? Biochim Biophys. Acta Rev. Cancer. 2021;1875:188466. doi: 10.1016/j.bbcan.2020.188466. [DOI] [PubMed] [Google Scholar]
- 44.Zhang M, Liu Y, Chen YG. Generation of 3D human gastrointestinal organoids: principle and applications. Cell Regen. 2020;9:6. doi: 10.1186/s13619-020-00040-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mithal A, et al. Generation of mesenchyme free intestinal organoids from human induced pluripotent stem cells. Nat. Commun. 2020;11:215. doi: 10.1038/s41467-019-13916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Georgakopoulos N, et al. Long-term expansion, genomic stability and in vivo safety of adult human pancreas organoids. BMC Dev. Biol. 2020;20:4. doi: 10.1186/s12861-020-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Artegiani B, et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat. Cell Biol. 2020;22:321–331. doi: 10.1038/s41556-020-0472-5. [DOI] [PubMed] [Google Scholar]
- 48.Beumer J, Clevers H. Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 2021;22:39–53. doi: 10.1038/s41580-020-0278-0. [DOI] [PubMed] [Google Scholar]
- 49.Mun SJ, et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 2019;71:970–985. doi: 10.1016/j.jhep.2019.06.030. [DOI] [PubMed] [Google Scholar]
- 50.Breunig M, et al. Modeling plasticity and dysplasia of pancreatic ductal organoids derived from human pluripotent stem cells. Cell. Stem Cell. 2021;28:1105–1124 e1119. doi: 10.1016/j.stem.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau HCH, Kranenburg O, Xiao H, Yu J. Organoid models of gastrointestinal cancers in basic and translational research. Nat. Rev. Gastroenterol. Hepatol. 2020;17:203–222. doi: 10.1038/s41575-019-0255-2. [DOI] [PubMed] [Google Scholar]
- 52.Verdu EF, Schuppan D. Co-factors, Microbes, and Immunogenetics in Celiac Disease to Guide Novel Approaches for Diagnosis and Treatment. Gastroenterology. 2021;161:1395–1411 e1394. doi: 10.1053/j.gastro.2021.08.016. [DOI] [PubMed] [Google Scholar]
- 53.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 54.Sayols S, et al. Signalling codes for the maintenance and lineage commitment of embryonic gastric epithelial progenitors. Development. 2020;147:dev188839. doi: 10.1242/dev.188839. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, et al. 3D modeling of esophageal development using Human PSC-derived basal progenitors reveals a critical role for notch signaling. Cell. Stem Cell. 2018;23:516–529 e515. doi: 10.1016/j.stem.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 57.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 58.Mahe MM, et al. Establishment of human epithelial enteroids and colonoids from whole tissue and biopsy. J. Vis. Exp. 2015;97:52483. doi: 10.3791/52483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huch M, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartfeld S, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–136 e126. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roos FJM, et al. Human branching cholangiocyte organoids recapitulate functional bile duct formation. Cell. Stem Cell. 2022;29:776–794 e713. doi: 10.1016/j.stem.2022.04.011. [DOI] [PubMed] [Google Scholar]
- 63.Zachos NC, et al. Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J. Biol. Chem. 2016;291:3759–3766. doi: 10.1074/jbc.R114.635995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munera JO, et al. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell. Stem Cell. 2017;21:51–64 e56. doi: 10.1016/j.stem.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Munera JO, et al. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell. Stem Cell. 2019;24:829. doi: 10.1016/j.stem.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCracken KW, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trisno SL, et al. Esophageal organoids from human pluripotent stem cells delineate Sox2 functions during esophageal specification. Cell. Stem Cell. 2018;23:501–515 e507. doi: 10.1016/j.stem.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takebe T, et al. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat. Protoc. 2014;9:396–409. doi: 10.1038/nprot.2014.020. [DOI] [PubMed] [Google Scholar]
- 70.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 71.Liu H, et al. A droplet microfluidic system to fabricate hybrid capsules enabling stem cell organoid engineering. Adv. Sci. (Weinh.). 2020;7:1903739. doi: 10.1002/advs.201903739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guye P, et al. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat. Commun. 2016;7:10243. doi: 10.1038/ncomms10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eicher AK, et al. Functional human gastrointestinal organoids can be engineered from three primary germ layers derived separately from pluripotent stem cells. Cell. Stem Cell. 2022;29:36–51 e36. doi: 10.1016/j.stem.2021.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Broda TR, McCracken KW, Wells JM. Generation of human antral and fundic gastric organoids from pluripotent stem cells. Nat. Protoc. 2019;14:28–50. doi: 10.1038/s41596-018-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frum T, Spence JR. hPSC-derived organoids: models of human development and disease. J. Mol. Med (Berl.). 2021;99:463–473. doi: 10.1007/s00109-020-01969-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fan, N. et al. Subculture and cryopreservation of esophageal adenocarcinoma organoids: pros and cons for single cell digestion. J. Vis. Exp.185, (2022). [DOI] [PubMed]
- 77.Boyle EC, Wunschel EJ, Grassl GA. Salmonella enterica Infection of Human and Mouse Colon Organoid-Derived Monolayers. Methods Mol. Biol. 2022;2427:149–163. doi: 10.1007/978-1-0716-1971-1_13. [DOI] [PubMed] [Google Scholar]
- 78.Kulkarni, G. et al. Combining human organoids and organ-on-a-chip technology to model intestinal region-specific functionality. J. Vis. Exp.183, (2022). [DOI] [PubMed]
- 79.Raggi C, Selleri S, M’Callum MA, Paganelli M. Generation of complex syngeneic liver organoids from induced pluripotent stem cells to model human liver pathophysiology. Curr. Protoc. 2022;2:e389. doi: 10.1002/cpz1.389. [DOI] [PubMed] [Google Scholar]
- 80.Pettinato G, Perelman LT, Fisher RA. Development of a scalable three-dimensional culture of human induced pluripotent stem cells-derived liver organoids. Methods Mol. Biol. 2022;2455:131–147. doi: 10.1007/978-1-0716-2128-8_12. [DOI] [PubMed] [Google Scholar]
- 81.Lin SC, Haga K, Zeng XL, Estes MK. Generation of CRISPR-Cas9-mediated genetic knockout human intestinal tissue-derived enteroid lines by lentivirus transduction and single-cell cloning. Nat. Protoc. 2022;17:1004–1027. doi: 10.1038/s41596-021-00669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gabriel, V. et al. Standardization and maintenance of 3D canine hepatic and intestinal organoid cultures for use in biomedical research. J. Vis. Exp.179, (2022). [DOI] [PubMed]
- 83.Breunig M, et al. Differentiation of human pluripotent stem cells into pancreatic duct-like organoids. STAR Protoc. 2021;2:100913. doi: 10.1016/j.xpro.2021.100913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Co JY, Margalef-Catala M, Monack DM, Amieva MR. Controlling the polarity of human gastrointestinal organoids to investigate epithelial biology and infectious diseases. Nat. Protoc. 2021;16:5171–5192. doi: 10.1038/s41596-021-00607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Puschhof J, et al. Intestinal organoid cocultures with microbes. Nat. Protoc. 2021;16:4633–4649. doi: 10.1038/s41596-021-00589-z. [DOI] [PubMed] [Google Scholar]
- 86.Qu, N., Daoud, A., Jeffcoat, B. & Munera, J. O. Generation, maintenance, and characterization of human pluripotent stem cell-derived intestinal and colonic organoids. J. Vis. Exp.173, (2021). [DOI] [PubMed]
- 87.Campbell SA, et al. Signalling pathways and transcriptional regulators orchestrating liver development and cancer. Development. 2021;148:dev199814. doi: 10.1242/dev.199814. [DOI] [PubMed] [Google Scholar]
- 88.Armandi A, Schattenberg JM. NAFLD between genes and environment: what drives fibrogenesis? Gut. 2021;70:815–816. doi: 10.1136/gutjnl-2020-321964. [DOI] [PubMed] [Google Scholar]
- 89.Kayali S, et al. Inverse association between Helicobacter pylori and inflammatory bowel disease: myth or fact? Acta Biomed. 2018;89:81–86. doi: 10.23750/abm.v89i9-S.7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCracken KW, et al. Wnt/beta-catenin promotes gastric fundus specification in mice and humans. Nature. 2017;541:182–187. doi: 10.1038/nature21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat. Rev. Genet. 2018;19:671–687. doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- 92.Schneeberger K, Roth S, Nieuwenhuis EES, Middendorp S. Intestinal epithelial cell polarity defects in disease: lessons from microvillus inclusion disease. Dis. Model Mech. 2018;11:dmm031088. doi: 10.1242/dmm.031088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou H, et al. Gene editing in pluripotent stem cells and their derived organoids. Stem Cells Int. 2021;2021:8130828. doi: 10.1155/2021/8130828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen C, Ji W, Niu Y. Primate organoids and gene-editing technologies toward next-generation biomedical research. Trends Biotechnol. 2021;39:1332–1342. doi: 10.1016/j.tibtech.2021.03.010. [DOI] [PubMed] [Google Scholar]
- 95.Idris M, et al. Intestinal multicellular organoids to study colorectal cancer. Biochim Biophys. Acta Rev. Cancer. 2021;1876:188586. doi: 10.1016/j.bbcan.2021.188586. [DOI] [PubMed] [Google Scholar]
- 96.Sugimoto S, et al. Reconstruction of the Human Colon Epithelium In Vivo. Cell. Stem Cell. 2018;22:171–176 e175. doi: 10.1016/j.stem.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 97.Kasendra M, et al. Intestinal organoids: roadmap to the clinic. Am. J. Physiol. Gastrointest. Liver Physiol. 2021;321:G1–G10. doi: 10.1152/ajpgi.00425.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Serra D, et al. Self-organization and symmetry breaking in intestinal organoid development. Nature. 2019;569:66–72. doi: 10.1038/s41586-019-1146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gjorevski N, et al. Tissue geometry drives deterministic organoid patterning. Science. 2022;375:eaaw9021. doi: 10.1126/science.aaw9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang J, Xu Z, Ren J. Small Intestinalization of Colon Using Ileum Organoids. Trends Cell Biol. 2021;31:517–519. doi: 10.1016/j.tcb.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 101.Leng, C., Rings, E., de Wildt, S. N. & van, I. S. C. D. Pharmacological and parenteral nutrition-based interventions in microvillus inclusion disease. J. Clin. Med. 10, 22 (2020). [DOI] [PMC free article] [PubMed]
- 102.Jayawardena, D., Alrefai, W. A., Dudeja, P. K. & Gill, R. K. Recent advances in understanding and managing malabsorption: focus on microvillus inclusion disease. F1000Res. 8, F1000 Faculty Rev-2061 (2019). [DOI] [PMC free article] [PubMed]
- 103.Dhekne HS, et al. MYO5B, STX3, and STXBP2 mutations reveal a common disease mechanism that unifies a subset of congenital diarrheal disorders: A mutation update. Hum. Mutat. 2018;39:333–344. doi: 10.1002/humu.23386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wiegerinck CL, et al. Loss of syntaxin 3 causes variant microvillus inclusion disease. Gastroenterology. 2014;147:65–68 e10. doi: 10.1053/j.gastro.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 105.Hess MW, et al. Advanced microscopy for liver and gut ultrastructural pathology in patients with MVID and PFIC caused by MYO5B mutations. J. Clin. Med. 2021;10:1901. doi: 10.3390/jcm10091901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaji I, et al. Cell differentiation is disrupted by MYO5B loss through Wnt/Notch imbalance. JCI Insight. 2021;6:e150416. doi: 10.1172/jci.insight.150416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Haas JT, et al. DGAT1 mutation is linked to a congenital diarrheal disorder. J. Clin. Invest. 2012;122:4680–4684. doi: 10.1172/JCI64873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang L, et al. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020;581:329–332. doi: 10.1038/s41586-020-2280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van Rijn JM, et al. Intestinal failure and aberrant lipid metabolism in patients with DGAT1 deficiency. Gastroenterology. 2018;155:130–143 e115. doi: 10.1053/j.gastro.2018.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mou W, et al. A novel homozygous TTC7A missense mutation results in familial multiple intestinal atresia and combined immunodeficiency. Front Immunol. 2021;12:759308. doi: 10.3389/fimmu.2021.759308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Govindarajan KK, Annamalai M. Multiple small bowel atresia: resection or conservation? J. Coll. Physicians Surg. Pak. 2021;30:740–742. doi: 10.29271/jcpsp.2021.06.740. [DOI] [PubMed] [Google Scholar]
- 112.Bigorgne AE, et al. TTC7A mutations disrupt intestinal epithelial apicobasal polarity. J. Clin. Invest. 2014;124:328–337. doi: 10.1172/JCI71471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lopez-Valdez JA, et al. Cystic fibrosis: current concepts. Bol. Med. Hosp. Infant Mex. 2021;78:584–596. doi: 10.24875/BMHIM.20000372. [DOI] [PubMed] [Google Scholar]
- 114.Dekkers JF, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 115.Ciciriello F, et al. Theratyping of the Rare CFTR Variants E193K and R334W in rectal organoid-derived epithelial monolayers. J. Pers. Med. 2022;12:632. doi: 10.3390/jpm12040632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.de Winter-de Groot KM, et al. Forskolin-induced swelling of intestinal organoids correlates with disease severity in adults with cystic fibrosis and homozygous F508del mutations. J. Cyst. Fibros. 2020;19:614–619. doi: 10.1016/j.jcf.2019.10.022. [DOI] [PubMed] [Google Scholar]
- 117.Li VSW. Modelling intestinal inflammation and infection using ‘mini-gut’ organoids. Nat. Rev. Gastroenterol. Hepatol. 2021;18:89–90. doi: 10.1038/s41575-020-00391-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee M, Chang EB. Inflammatory bowel diseases (IBD) and the microbiome-searching the crime scene for clues. Gastroenterology. 2021;160:524–537. doi: 10.1053/j.gastro.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Agrawal M, et al. Multiomics to elucidate inflammatory bowel disease risk factors and pathways. Nat. Rev. Gastroenterol. Hepatol. 2022;19:399–409. doi: 10.1038/s41575-022-00593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nanki K, et al. Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature. 2020;577:254–259. doi: 10.1038/s41586-019-1844-5. [DOI] [PubMed] [Google Scholar]
- 121.Sarvestani SK, et al. Induced organoids derived from patients with ulcerative colitis recapitulate colitic reactivity. Nat. Commun. 2021;12:262. doi: 10.1038/s41467-020-20351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rawat M, et al. IL1B Increases Intestinal Tight Junction Permeability by Up-regulation of MIR200C-3p, Which Degrades Occludin mRNA. Gastroenterology. 2020;159:1375–1389. doi: 10.1053/j.gastro.2020.06.038. [DOI] [PubMed] [Google Scholar]
- 123.Chakravarti D, et al. Telomere dysfunction instigates inflammation in inflammatory bowel disease. Proc. Natl Acad. Sci. USA. 2021;118:e2024853118. doi: 10.1073/pnas.2024853118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Howell KJ, et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology. 2018;154:585–598. doi: 10.1053/j.gastro.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Porpora M, et al. Inflammation is present, persistent and more sensitive to proinflammatory triggers in celiac disease enterocytes. Int. J. Mol. Sci. 2022;23:1973. doi: 10.3390/ijms23041973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kakiuchi N, et al. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature. 2020;577:260–265. doi: 10.1038/s41586-019-1856-1. [DOI] [PubMed] [Google Scholar]
- 127.Schnalzger TE, et al. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 2019;38:e100928. doi: 10.15252/embj.2018100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ooft SN, et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019;11:eaay2574. doi: 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- 129.Yao Y, et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell. Stem Cell. 2020;26:17–26 e16. doi: 10.1016/j.stem.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 130.Crespo M, et al. Corrigendum: Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat. Med. 2018;24:526. doi: 10.1038/nm0418-526a. [DOI] [PubMed] [Google Scholar]
- 131.JE IJ, Vermeulen L, Meijer GA, Dekker E. Serrated neoplasia-role in colorectal carcinogenesis and clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2015;12:401–409. doi: 10.1038/nrgastro.2015.73. [DOI] [PubMed] [Google Scholar]
- 132.Bae JM, Kim JH, Kang GH. Molecular subtypes of colorectal cancer and their clinicopathologic features, with an emphasis on the serrated neoplasia pathway. Arch. Pathol. Lab Med. 2016;140:406–412. doi: 10.5858/arpa.2015-0310-RA. [DOI] [PubMed] [Google Scholar]
- 133.Inamura K. Colorectal cancers: an update on their molecular pathology. Cancers. 2018;10:26. doi: 10.3390/cancers10010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Guinney J, et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ganesh K, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat. Med. 2019;25:1607–1614. doi: 10.1038/s41591-019-0584-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li H, et al. Modeling tumor development and metastasis using paired organoids derived from patients with colorectal cancer liver metastases. J. Hematol. Oncol. 2020;13:119. doi: 10.1186/s13045-020-00957-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Matano M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 138.Roper J, et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 2017;35:569–576. doi: 10.1038/nbt.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Drost J, et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 2017;358:234–238. doi: 10.1126/science.aao3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zheng X, et al. Single-cell transcriptomic profiling unravels the adenoma-initiation role of protein tyrosine kinases during colorectal tumorigenesis. Signal Transduct. Target Ther. 2022;7:60. doi: 10.1038/s41392-022-00881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bolhaqueiro ACF, et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 2019;51:824–834. doi: 10.1038/s41588-019-0399-6. [DOI] [PubMed] [Google Scholar]
- 142.Kawasaki K, et al. Chromosome engineering of human colon-derived organoids to develop a model of traditional serrated adenoma. Gastroenterology. 2020;158:638–651 e638. doi: 10.1053/j.gastro.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 143.Saenz JB, Mills JC. Acid and the basis for cellular plasticity and reprogramming in gastric repair and cancer. Nat. Rev. Gastroenterol. Hepatol. 2018;15:257–273. doi: 10.1038/nrgastro.2018.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rocken C. Molecular classification of gastric cancer. Expert Rev. Mol. Diagn. 2017;17:293–301. doi: 10.1080/14737159.2017.1286985. [DOI] [PubMed] [Google Scholar]
- 145.Suh YS, et al. Comprehensive molecular characterization of adenocarcinoma of the gastroesophageal junction between esophageal and gastric adenocarcinomas. Ann. Surg. 2022;275:706–717. doi: 10.1097/SLA.0000000000004303. [DOI] [PubMed] [Google Scholar]
- 146.Riquelme I, et al. Molecular classification of gastric cancer: Towards a pathway-driven targeted therapy. Oncotarget. 2015;6:24750–24779. doi: 10.18632/oncotarget.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Luan RR, Liu FF. Updates on the molecular classification of gastric cancer. Zhonghua Bing. Li Xue Za Zhi. 2021;50:1205–1209. doi: 10.3760/cma.j.cn112151-20210401-00252. [DOI] [PubMed] [Google Scholar]
- 148.Nanki K, et al. Divergent routes toward wnt and r-spondin niche independency during human gastric carcinogenesis. Cell. 2018;174:856–869 e817. doi: 10.1016/j.cell.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 149.Liabeuf D, Oshima M, Stange DE, Sigal M. Stem cells, helicobacter pylori, and mutational landscape: utility of preclinical models to understand carcinogenesis and to direct management of gastric cancer. Gastroenterology. 2022;162:1067–1087. doi: 10.1053/j.gastro.2021.12.252. [DOI] [PubMed] [Google Scholar]
- 150.Seidlitz T, et al. Human gastric cancer modelling using organoids. Gut. 2019;68:207–217. doi: 10.1136/gutjnl-2017-314549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zou J, et al. Construction of gastric cancer patient-derived organoids and their utilization in a comparative study of clinically used paclitaxel nanoformulations. J. Nanobiotechnology. 2022;20:233. doi: 10.1186/s12951-022-01431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yan HHN, et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell. Stem Cell. 2018;23:882–897 e811. doi: 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 153.Fatehullah A, et al. A tumour-resident Lgr5(+) stem-cell-like pool drives the establishment and progression of advanced gastric cancers. Nat. Cell Biol. 2021;23:1299–1313. doi: 10.1038/s41556-021-00793-9. [DOI] [PubMed] [Google Scholar]
- 154.Rosekrans SL, Baan B, Muncan V, van den Brink GR. Esophageal development and epithelial homeostasis. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;309:G216–G228. doi: 10.1152/ajpgi.00088.2015. [DOI] [PubMed] [Google Scholar]
- 155.Dong J, et al. Determining risk of barrett’s esophagus and esophageal adenocarcinoma based on epidemiologic factors and genetic variants. Gastroenterology. 2018;154:1273–1281 e1273. doi: 10.1053/j.gastro.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fabian T, Leung A. Epidemiology of Barrett’s Esophagus and Esophageal Carcinoma. Surg. Clin. North Am. 2021;101:381–389. doi: 10.1016/j.suc.2021.03.001. [DOI] [PubMed] [Google Scholar]
- 157.Nowicki-Osuch K, et al. Molecular phenotyping reveals the identity of Barrett’s esophagus and its malignant transition. Science. 2021;373:760–767. doi: 10.1126/science.abd1449. [DOI] [PubMed] [Google Scholar]
- 158.Lavery DL, et al. The stem cell organisation, and the proliferative and gene expression profile of Barrett’s epithelium, replicates pyloric-type gastric glands. Gut. 2014;63:1854–1863. doi: 10.1136/gutjnl-2013-306508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Harada, K. et al. Recent advances in treating oesophageal cancer. F1000Res. 9, F1000 Faculty Rev-1189 (2020).
- 160.Thrift AP. Global burden and epidemiology of Barrett oesophagus and oesophageal cancer. Nat. Rev. Gastroenterol. Hepatol. 2021;18:432–443. doi: 10.1038/s41575-021-00419-3. [DOI] [PubMed] [Google Scholar]
- 161.Li X, et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat. Commun. 2018;9:2983. doi: 10.1038/s41467-018-05190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Secrier M, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat. Genet. 2016;48:1131–1141. doi: 10.1038/ng.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 164.Kijima T, et al. Three-dimensional organoids reveal therapy resistance of esophageal and oropharyngeal squamous cell carcinoma cells. Cell Mol. Gastroenterol. Hepatol. 2019;7:73–91. doi: 10.1016/j.jcmgh.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ehrhardt, J. D. & Gomez, F. in StatPearls (2022).
- 166.Henry BM, et al. Development of the human pancreas and its vasculature - An integrated review covering anatomical, embryological, histological, and molecular aspects. Ann. Anat. 2019;221:115–124. doi: 10.1016/j.aanat.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 167.Zhou Q, Melton DA. Pancreas regeneration. Nature. 2018;557:351–358. doi: 10.1038/s41586-018-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Miyabayashi K, et al. Intraductal transplantation models of human pancreatic ductal adenocarcinoma reveal progressive transition of molecular subtypes. Cancer Discov. 2020;10:1566–1589. doi: 10.1158/2159-8290.CD-20-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Park W, Chawla A, O’Reilly EM. Pancreatic cancer: a review. JAMA. 2021;326:851–862. doi: 10.1001/jama.2021.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen X, Kang R, Kroemer G, Tang D. Targeting ferroptosis in pancreatic cancer: a double-edged sword. Trends Cancer. 2021;7:891–901. doi: 10.1016/j.trecan.2021.04.005. [DOI] [PubMed] [Google Scholar]
- 171.Watanabe S, et al. Establishment of patient-derived organoids and a characterization-based drug discovery platform for treatment of pancreatic cancer. BMC Cancer. 2022;22:489. doi: 10.1186/s12885-022-09619-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tiriac H, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8:1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Driehuis E, et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl Acad. Sci. USA. 2019;116:26580–26590. doi: 10.1073/pnas.1911273116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Seino T, et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell. Stem Cell. 2018;22:454–467 e456. doi: 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 175.Lee J, et al. Reconstituting development of pancreatic intraepithelial neoplasia from primary human pancreas duct cells. Nat. Commun. 2017;8:14686. doi: 10.1038/ncomms14686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Huang L, et al. Commitment and oncogene-induced plasticity of human stem cell-derived pancreatic acinar and ductal organoids. Cell. Stem Cell. 2021;28:1090–1104 e1096. doi: 10.1016/j.stem.2021.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Huppert SS, Iwafuchi-Doi M. Molecular regulation of mammalian hepatic architecture. Curr. Top. Dev. Biol. 2019;132:91–136. doi: 10.1016/bs.ctdb.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Guillaud O, et al. Long term results of liver transplantation for alpha-1 antitrypsin deficiency. Dig. Liver Dis. 2021;53:606–611. doi: 10.1016/j.dld.2020.10.016. [DOI] [PubMed] [Google Scholar]
- 179.Ray K. Liver: Purified A1AT holds promise as therapy for acute liver failure. Nat. Rev. Gastroenterol. Hepatol. 2014;11:140. doi: 10.1038/nrgastro.2014.16. [DOI] [PubMed] [Google Scholar]
- 180.Gomez-Mariano G, et al. Liver organoids reproduce alpha-1 antitrypsin deficiency-related liver disease. Hepatol. Int. 2020;14:127–137. doi: 10.1007/s12072-019-10007-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kamath BM, Spinner NB, Rosenblum ND. Renal involvement and the role of Notch signalling in Alagille syndrome. Nat. Rev. Nephrol. 2013;9:409–418. doi: 10.1038/nrneph.2013.102. [DOI] [PubMed] [Google Scholar]
- 182.Andersson ER, et al. Mouse model of alagille syndrome and mechanisms of Jagged1 missense mutations. Gastroenterology. 2018;154:1080–1095. doi: 10.1053/j.gastro.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Heeren J, Scheja L. Metabolic-associated fatty liver disease and lipoprotein metabolism. Mol. Metab. 2021;50:101238. doi: 10.1016/j.molmet.2021.101238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Samuel VT, Shulman GI. Nonalcoholic fatty liver disease as a nexus of metabolic and hepatic diseases. Cell Metab. 2018;27:22–41. doi: 10.1016/j.cmet.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang L, et al. Recapitulating lipid accumulation and related metabolic dysregulation in human liver-derived organoids. J. Mol. Med (Berl.). 2022;100:471–484. doi: 10.1007/s00109-021-02176-x. [DOI] [PubMed] [Google Scholar]
- 186.Pericleous M, et al. Wolman’s disease and cholesteryl ester storage disorder: the phenotypic spectrum of lysosomal acid lipase deficiency. Lancet Gastroenterol. Hepatol. 2017;2:670–679. doi: 10.1016/S2468-1253(17)30052-3. [DOI] [PubMed] [Google Scholar]
- 187.Ouchi R, et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 2019;30:374–384 e376. doi: 10.1016/j.cmet.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Leite SB, et al. Novel human hepatic organoid model enables testing of drug-induced liver fibrosis in vitro. Biomaterials. 2016;78:1–10. doi: 10.1016/j.biomaterials.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 190.Sato K, et al. Organoids and spheroids as models for studying cholestatic liver injury and cholangiocarcinoma. Hepatology. 2021;74:491–502. doi: 10.1002/hep.31653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Shi S, et al. RecapitUlating Cholangiopathy-associated Necroptotic Cell Death In Vitro Using Human Cholangiocyte Organoids. Cell Mol. Gastroenterol. Hepatol. 2022;13:541–564. doi: 10.1016/j.jcmgh.2021.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang S, et al. Human ESC-derived expandable hepatic organoids enable therapeutic liver repopulation and pathophysiological modeling of alcoholic liver injury. Cell Res. 2019;29:1009–1026. doi: 10.1038/s41422-019-0242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Woo HG, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70:3034–3041. doi: 10.1158/0008-5472.CAN-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Broutier L, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nuciforo S, et al. Organoid models of human liver cancers derived from tumor needle biopsies. Cell Rep. 2018;24:1363–1376. doi: 10.1016/j.celrep.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chan LH, et al. PRMT6 regulates RAS/RAF binding and MEK/ERK-Mediated cancer stemness activities in hepatocellular carcinoma through CRAF methylation. Cell Rep. 2018;25:690–701 e698. doi: 10.1016/j.celrep.2018.09.053. [DOI] [PubMed] [Google Scholar]
- 197.Artegiani B, et al. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell. Stem Cell. 2019;24:927–943 e926. doi: 10.1016/j.stem.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 198.Belenguer G, et al. RNF43/ZNRF3 loss predisposes to hepatocellular-carcinoma by impairing liver regeneration and altering the liver lipid metabolic ground-state. Nat. Commun. 2022;13:334. doi: 10.1038/s41467-021-27923-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wong CC, et al. The cholesterol uptake regulator PCSK9 promotes and is a therapeutic target in APC/KRAS-mutant colorectal cancer. Nat. Commun. 2022;13:3971. doi: 10.1038/s41467-022-31663-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Arnauts K, et al. Microbiota, not host origin drives ex vivo intestinal epithelial responses. Gut Microbes. 2022;14:2089003. doi: 10.1080/19490976.2022.2089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cheng WY, Wu CY, Yu J. The role of gut microbiota in cancer treatment: friend or foe? Gut. 2020;69:1867–1876. doi: 10.1136/gutjnl-2020-321153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Xavier JB, et al. The cancer microbiome: distinguishing direct and indirect effects requires a systemic view. Trends Cancer. 2020;6:192–204. doi: 10.1016/j.trecan.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhao L, Zhao N. Demonstration of causality: back to cultures. Nat. Rev. Gastroenterol. Hepatol. 2021;18:97–98. doi: 10.1038/s41575-020-00400-6. [DOI] [PubMed] [Google Scholar]
- 204.Matson V, Chervin CS, Gajewski TF. Cancer and the microbiome-influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology. 2021;160:600–613. doi: 10.1053/j.gastro.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Amoroso C, et al. The role of gut microbiota biomodulators on mucosal immunity and intestinal inflammation. Cells. 2020;9:1234. doi: 10.3390/cells9051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ferreira RM, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67:226–236. doi: 10.1136/gutjnl-2017-314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ma C, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360:eaan5931. doi: 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Singh V, et al. Dysregulated microbial fermentation of soluble fiber induces cholestatic liver cancer. Cell. 2018;175:679–694 e622. doi: 10.1016/j.cell.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nejman D, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kennedy EA, King KY, Baldridge MT. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol. 2018;9:1534. doi: 10.3389/fphys.2018.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Chaudhari SN, McCurry MD, Devlin AS. Chains of evidence from correlations to causal molecules in microbiome-linked diseases. Nat. Chem. Biol. 2021;17:1046–1056. doi: 10.1038/s41589-021-00861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bayer F, Ascher S, Pontarollo G, Reinhardt C. Antibiotic treatment protocols and germ-free mouse models in vascular research. Front Immunol. 2019;10:2174. doi: 10.3389/fimmu.2019.02174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhao L, et al. Parvimonas micra promotes colorectal tumorigenesis and is associated with prognosis of colorectal cancer patients. Oncogene. 2022;41:4200–4210. doi: 10.1038/s41388-022-02395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.De Palma G, et al. Histamine production by the gut microbiota induces visceral hyperalgesia through histamine 4 receptor signaling in mice. Sci. Transl. Med. 2022;14:eabj1895. doi: 10.1126/scitranslmed.abj1895. [DOI] [PubMed] [Google Scholar]
- 215.Aron-Wisnewsky J, Warmbrunn MV, Nieuwdorp M, Clement K. Metabolism and metabolic disorders and the microbiome: the intestinal microbiota associated with obesity, lipid metabolism, and metabolic health-pathophysiology and therapeutic strategies. Gastroenterology. 2021;160:573–599. doi: 10.1053/j.gastro.2020.10.057. [DOI] [PubMed] [Google Scholar]
- 216.Dutta D, Heo I, Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 217.Pleguezuelos-Manzano C, et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature. 2020;580:269–273. doi: 10.1038/s41586-020-2080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Co JY, et al. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep. 2019;26:2509–2520 e2504. doi: 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Giobbe GG, et al. SARS-CoV-2 infection and replication in human gastric organoids. Nat. Commun. 2021;12:6610. doi: 10.1038/s41467-021-26762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ettayebi K, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Schlaermann P, et al. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2016;65:202–213. doi: 10.1136/gutjnl-2014-307949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhang J, et al. Coculture of primary human colon monolayer with human gut bacteria. Nat. Protoc. 2021;16:3874–3900. doi: 10.1038/s41596-021-00562-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Neal JT, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988 e1916. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Nikolaev M, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. 2020;585:574–578. doi: 10.1038/s41586-020-2724-8. [DOI] [PubMed] [Google Scholar]
- 225.Shah P, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Park SE, Georgescu A, Huh D. Organoids-on-a-chip. Science. 2019;364:960–965. doi: 10.1126/science.aaw7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Verhulsel M, et al. Developing an advanced gut on chip model enabling the study of epithelial cell/fibroblast interactions. Lab Chip. 2021;21:365–377. doi: 10.1039/D0LC00672F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Schuster B, et al. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat. Commun. 2020;11:5271. doi: 10.1038/s41467-020-19058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Shin W, Kim HJ. 3D in vitro morphogenesis of human intestinal epithelium in a gut-on-a-chip or a hybrid chip with a cell culture insert. Nat. Protoc. 2022;17:910–939. doi: 10.1038/s41596-021-00674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sontheimer-Phelps A, et al. Human colon-on-a-chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cell Mol. Gastroenterol. Hepatol. 2020;9:507–526. doi: 10.1016/j.jcmgh.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wang Y, et al. Long-term culture captures injury-repair cycles of colonic stem cells. Cell. 2019;179:1144–1159 e1115. doi: 10.1016/j.cell.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.In J, et al. Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell Mol. Gastroenterol. Hepatol. 2016;2:48–62 e43. doi: 10.1016/j.jcmgh.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Karve SS, Pradhan S, Ward DV, Weiss AA. Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PLoS One. 2017;12:e0178966. doi: 10.1371/journal.pone.0178966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Pradhan S, et al. Tissue responses to shiga toxin in human intestinal organoids. Cell Mol. Gastroenterol. Hepatol. 2020;10:171–190. doi: 10.1016/j.jcmgh.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Allen J, et al. Colon Tumors in Enterotoxigenic Bacteroides fragilis (ETBF)-colonized mice do not display a unique mutational signature but instead possess host-dependent alterations in the APC gene. Microbiol Spectr. 2022;10:e0105522. doi: 10.1128/spectrum.01055-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hall AJ, et al. The roles of Clostridium difficile and norovirus among gastroenteritis-associated deaths in the United States, 1999–2007. Clin. Infect. Dis. 2012;55:216–223. doi: 10.1093/cid/cis386. [DOI] [PubMed] [Google Scholar]
- 237.Leslie JL, et al. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 2015;83:138–145. doi: 10.1128/IAI.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Engevik MA, et al. Human Clostridium difficile infection: altered mucus production and composition. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;308:G510–G524. doi: 10.1152/ajpgi.00091.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhou C, et al. Expansion of intestinal secretory cell population induced by listeria monocytogenes infection: accompanied with the inhibition of NOTCH pathway. Front Cell Infect. Microbiol. 2022;12:793335. doi: 10.3389/fcimb.2022.793335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhou C, et al. Involvement of CCN1 protein and TLR2/4 signaling pathways in intestinal epithelial cells response to listeria monocytogenes. Int. J. Mol. Sci. 2022;23:2739. doi: 10.3390/ijms23052739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Muhlberg E, et al. Renaissance of vancomycin: approaches for breaking antibiotic resistance in multidrug-resistant bacteria. Can. J. Microbiol. 2020;66:11–16. doi: 10.1139/cjm-2019-0309. [DOI] [PubMed] [Google Scholar]
- 242.Kock R, Cuny C. [Multidrug-resistant bacteria in animals and humans] Med Klin. Intensivmed. Notfmed. 2020;115:189–197. doi: 10.1007/s00063-018-0487-x. [DOI] [PubMed] [Google Scholar]
- 243.Xiong X, et al. Emerging enterococcus pore-forming toxins with MHC/HLA-I as receptors. Cell. 2022;185:1157–1171 e1122. doi: 10.1016/j.cell.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019;17:156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Rubinstein MR, et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1. EMBO Rep. 2019;20:e47638. doi: 10.15252/embr.201847638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hong J, et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. 2021;70:2123–2137. doi: 10.1136/gutjnl-2020-322780. [DOI] [PubMed] [Google Scholar]
- 247.Yang Y, et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152:851–866 e824. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Engevik MA, et al. Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. mBio. 2021;12:e02706-20. doi: 10.1128/mBio.02706-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Petersen L, et al. The essential role of Rac1 glucosylation in clostridioides difficile toxin B-Induced arrest of G1-S transition. Front Microbiol. 2022;13:846215. doi: 10.3389/fmicb.2022.846215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Amieva M, Peek RM., Jr. Pathobiology of helicobacter pylori-induced gastric cancer. Gastroenterology. 2016;150:64–78. doi: 10.1053/j.gastro.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Cover TL, Lacy DB, Ohi MD. The helicobacter pylori cag type IV secretion system. Trends Microbiol. 2020;28:682–695. doi: 10.1016/j.tim.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Alipour M. Molecular mechanism of helicobacter pylori-induced gastric cancer. J. Gastrointest. Cancer. 2021;52:23–30. doi: 10.1007/s12029-020-00518-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Morey P, et al. Helicobacter pylori depletes cholesterol in gastric glands to prevent interferon gamma signaling and escape the inflammatory response. Gastroenterology. 2018;154:1391–1404 e1399. doi: 10.1053/j.gastro.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 254.Maubach G, Vieth M, Boccellato F, Naumann M. Helicobacter pylori-induced NF-kappaB: trailblazer for gastric pathophysiology. Trends Mol. Med. 2022;28:210–222. doi: 10.1016/j.molmed.2021.12.005. [DOI] [PubMed] [Google Scholar]
- 255.Yao X, Smolka AJ. Gastric parietal cell physiology and helicobacter pylori-induced disease. Gastroenterology. 2019;156:2158–2173. doi: 10.1053/j.gastro.2019.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cao L, et al. Helicobacter pylori-induced RASAL2 through activation of nuclear factor-kappaB promotes gastric tumorigenesis via beta-catenin signaling axis. Gastroenterology. 2022;162:1716–1731 e1717. doi: 10.1053/j.gastro.2022.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Holokai L, et al. Increased programmed death-ligand 1 is an early epithelial cell response to helicobacter pylori infection. PLoS Pathog. 2019;15:e1007468. doi: 10.1371/journal.ppat.1007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Mun, S. J. et al. Effect of microbial short-chain fatty acids on CYP3A4-mediated metabolic activation of human pluripotent stem cell-derived liver organoids. Cells. 10, (2021). [DOI] [PMC free article] [PubMed]
- 259.Nakamoto N, et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat. Microbiol. 2019;4:492–503. doi: 10.1038/s41564-018-0333-1. [DOI] [PubMed] [Google Scholar]
- 260.Shen, M. et al. Three of a kind: control of the expression of liver-expressed antimicrobial peptide 2 (LEAP2) by the endocannabinoidome and the gut microbiome. Molecules. 27, (2021). [DOI] [PMC free article] [PubMed]
- 261.Munch NS, et al. High-fat diet accelerates carcinogenesis in a mouse model of barrett’s esophagus via interleukin 8 and alterations to the gut microbiome. Gastroenterology. 2019;157:492–506 e492. doi: 10.1053/j.gastro.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Brusilovsky M, et al. Host-microbiota interactions in the esophagus during homeostasis and allergic inflammation. Gastroenterology. 2022;162:521–534 e528. doi: 10.1053/j.gastro.2021.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Plat VD, et al. Esophageal microbiota composition and outcome of esophageal cancer treatment: a systematic review. Dis. Esophagus. 2021;35:doab076. doi: 10.1093/dote/doab076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Thomas RM, Jobin C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat. Rev. Gastroenterol. Hepatol. 2020;17:53–64. doi: 10.1038/s41575-019-0242-7. [DOI] [PubMed] [Google Scholar]
- 265.McAllister F, Khan MAW, Helmink B, Wargo JA. The tumor microbiome in pancreatic cancer: bacteria and beyond. Cancer Cell. 2019;36:577–579. doi: 10.1016/j.ccell.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 266.Aron-Wisnewsky J, Warmbrunn MV, Nieuwdorp M, Clement K. Nonalcoholic fatty liver disease: modulating gut microbiota to improve severity? Gastroenterology. 2020;158:1881–1898. doi: 10.1053/j.gastro.2020.01.049. [DOI] [PubMed] [Google Scholar]
- 267.Kolodziejczyk AA, et al. Acute liver failure is regulated by MYC- and microbiome-dependent programs. Nat. Med. 2020;26:1899–1911. doi: 10.1038/s41591-020-1102-2. [DOI] [PubMed] [Google Scholar]
- 268.Nishikawa, H. et al. Dysbiosis and liver diseases (Review). Int. J. Mol. Med. 48, (2021). [DOI] [PubMed]
- 269.Ettayebi K, et al. Antiviral activity of olanexidine-containing hand rub against human noroviruses. mBio. 2022;13:e0284821. doi: 10.1128/mbio.02848-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Liu D, et al. Fingerprinting of human noroviruses co-infections in a possible foodborne outbreak by metagenomics. Int J. Food Microbiol. 2020;333:108787. doi: 10.1016/j.ijfoodmicro.2020.108787. [DOI] [PubMed] [Google Scholar]
- 271.Ramani S, Atmar RL, Estes MK. Epidemiology of human noroviruses and updates on vaccine development. Curr. Opin. Gastroenterol. 2014;30:25–33. doi: 10.1097/MOG.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Mboko, W. P. et al. Advances in understanding of the innate immune response to human norovirus infection using organoid models. J. Gen. Virol. 103, (2022). [DOI] [PMC free article] [PubMed]
- 273.Costantini V, et al. Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg. Infect. Dis. 2018;24:1453–1464. doi: 10.3201/eid2408.180126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Guo M, Tao W, Flavell RA, Zhu S. Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat. Rev. Gastroenterol. Hepatol. 2021;18:269–283. doi: 10.1038/s41575-021-00416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Eyre DW, et al. Effect of Covid-19 vaccination on transmission of alpha and delta variants. N. Engl. J. Med. 2022;386:744–756. doi: 10.1056/NEJMoa2116597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Han Y, et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Salahudeen AA, et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature. 2020;588:670–675. doi: 10.1038/s41586-020-3014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Yang H, Rao Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021;19:685–700. doi: 10.1038/s41579-021-00630-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Chen H, et al. Acoustic bioprinting of patient-derived organoids for predicting cancer therapy responses. Adv. Healthc. Mater. 2022;11:e2102784. doi: 10.1002/adhm.202102784. [DOI] [PubMed] [Google Scholar]
- 282.Kruger J, et al. Drug inhibition of SARS-CoV-2 replication in human pluripotent stem cell-derived intestinal organoids. Cell Mol. Gastroenterol. Hepatol. 2021;11:935–948. doi: 10.1016/j.jcmgh.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Prelli Bozzo C, et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 2021;12:4584. doi: 10.1038/s41467-021-24817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Singh B, Kaur P, Maroules M. Autoimmune hepatitis-primary biliary cholangitis overlap syndrome triggered by COVID-19. Eur. J. Case Rep. Intern Med. 2021;8:002264. doi: 10.12890/2021_002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Klocperk, A. et al. Complex immunometabolic profiling reveals the activation of cellular immunity and biliary lesions in patients with severe COVID-19. J. Clin. Med. 9, (2020). [DOI] [PMC free article] [PubMed]
- 286.Faruqui S, et al. Cholangiopathy after severe COVID-19: clinical features and prognostic implications. Am. J. Gastroenterol. 2021;116:1414–1425. doi: 10.14309/ajg.0000000000001264. [DOI] [PubMed] [Google Scholar]
- 287.Zhao, Y. et al. Mechanistic insight of SARS-CoV-2 infection using human hepatobiliary organoids. Gut gutjnl-2021-326617 (2022). [DOI] [PMC free article] [PubMed]
- 288.D’Souza S, Lau KC, Coffin CS, Patel TR. Molecular mechanisms of viral hepatitis induced hepatocellular carcinoma. World J. Gastroenterol. 2020;26:5759–5783. doi: 10.3748/wjg.v26.i38.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Harputluoglu M, Carr BI. Hepatitis B before and after hepatocellular carcinoma. J. Gastrointest. Cancer. 2021;52:1206–1210. doi: 10.1007/s12029-021-00745-4. [DOI] [PubMed] [Google Scholar]
- 290.De Crignis, E. et al. Application of human liver organoids as a patient-derived primary model for HBV infection and related hepatocellular carcinoma. Elife. 10, (2021). [DOI] [PMC free article] [PubMed]
- 291.Wose Kinge, C. N., Bhoola, N. H. & Kramvis, A. In vitro systems for studying different Genotypes/Sub-genotypes of hepatitis B virus: Strengths and limitations. Viruses. 12, (2020). [DOI] [PMC free article] [PubMed]
- 292.Hossain T, Romal S, Mahmoudi T. Production of recombinant hepatitis B virus (HBV) and detection of HBV in infected human liver organoids. Bio Protoc. 2022;12:e4392. doi: 10.21769/BioProtoc.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Rao S, Hossain T, Mahmoudi T. 3D human liver organoids: An in vitro platform to investigate HBV infection, replication and liver tumorigenesis. Cancer Lett. 2021;506:35–44. doi: 10.1016/j.canlet.2021.02.024. [DOI] [PubMed] [Google Scholar]
- 294.Romal S, Hossain T, Mahmoudi T. Generation, maintenance and HBV infection of human liver organoids. Bio Protoc. 2022;12:e4358. doi: 10.21769/BioProtoc.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Nie YZ, et al. Recapitulation of hepatitis B virus-host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine. 2018;35:114–123. doi: 10.1016/j.ebiom.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Younossi ZM, et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 297.Yang J, et al. Epstein-Barr virus-associated gastric cancer: A distinct subtype. Cancer Lett. 2020;495:191–199. doi: 10.1016/j.canlet.2020.09.019. [DOI] [PubMed] [Google Scholar]
- 298.Okabe A, et al. Cross-species chromatin interactions drive transcriptional rewiring in Epstein-Barr virus-positive gastric adenocarcinoma. Nat. Genet. 2020;52:919–930. doi: 10.1038/s41588-020-0665-7. [DOI] [PubMed] [Google Scholar]
- 299.Wallaschek N, et al. Ephrin receptor A2, the epithelial receptor for Epstein-Barr virus entry, is not available for efficient infection in human gastric organoids. PLoS Pathog. 2021;17:e1009210. doi: 10.1371/journal.ppat.1009210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Santos-Ferreira, N., Van Dycke, J., Neyts, J. & Rocha-Pereira, J. Current and future antiviral strategies to tackle gastrointestinal viral infections. Microorganisms. 9, (2021). [DOI] [PMC free article] [PubMed]
- 301.Gasco, S. & Munoz-Fernandez, M. A. A review on the current knowledge on ZIKV infection and the interest of organoids and nanotechnology on development of effective therapies against Zika infection. Int. J. Mol. Sci. 22, (2020). [DOI] [PMC free article] [PubMed]
- 302.Saito Y, Nakaoka T, Saito H. A new molecular mechanism underlying the antitumor effect of DNA methylation inhibitors via an antiviral immune response. Adv. Protein Chem. Struct. Biol. 2017;106:227–242. doi: 10.1016/bs.apcsb.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 303.Fujii M, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell. Stem Cell. 2016;18:827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 304.Barresi V, Pedrazzani C. Colorectal signet ring cell carcinoma: advancing research in a rare cancer. Future Oncol. 2020;16:1161–1163. doi: 10.2217/fon-2020-0242. [DOI] [PubMed] [Google Scholar]
- 305.Lowenthal BM, et al. Gastric medullary carcinoma with sporadic mismatch repair deficiency and a TP53 R273C mutation: an unusual case with wild-type BRAF. Case Rep. Pathol. 2017;2017:3427343. doi: 10.1155/2017/3427343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Akbaba TH, Bekircan-Kurt CE, Balci-Peynircioglu B, Balci-Hayta B. Biologia Futura: the importance of 3D organoids-a new approach for research on neurological and rare diseases. Biol. Futur. 2021;72:281–290. doi: 10.1007/s42977-021-00070-8. [DOI] [PubMed] [Google Scholar]
- 307.Haugabook SJ, Ferrer M, Ottinger EA. In vitro and in vivo translational models for rare liver diseases. Biochim Biophys. Acta Mol. Basis Dis. 2019;1865:1003–1018. doi: 10.1016/j.bbadis.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 308.Kondo T. Current status and perspectives of patient-derived rare cancer models. Hum. Cell. 2020;33:919–929. doi: 10.1007/s13577-020-00391-1. [DOI] [PubMed] [Google Scholar]
- 309.Schork NJ. Personalized medicine: Time for one-person trials. Nature. 2015;520:609–611. doi: 10.1038/520609a. [DOI] [PubMed] [Google Scholar]
- 310.Tiriac H, Plenker D, Baker LA, Tuveson DA. Organoid models for translational pancreatic cancer research. Curr. Opin. Genet Dev. 2019;54:7–11. doi: 10.1016/j.gde.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Lee JA, et al. Gene therapy for cystic fibrosis: new tools for precision medicine. J. Transl. Med. 2021;19:452. doi: 10.1186/s12967-021-03099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Zhou Z, Cong L, Cong X. Patient-derived organoids in precision medicine: drug screening, organoid-on-a-chip and living organoid biobank. Front Oncol. 2021;11:762184. doi: 10.3389/fonc.2021.762184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Veninga V, Voest EE. Tumor organoids: Opportunities and challenges to guide precision medicine. Cancer Cell. 2021;39:1190–1201. doi: 10.1016/j.ccell.2021.07.020. [DOI] [PubMed] [Google Scholar]
- 314.Aboulkheyr Es,H, et al. Personalized cancer medicine: an organoid approach. Trends Biotechnol. 2018;36:358–371. doi: 10.1016/j.tibtech.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 315.LeSavage BL, et al. Next-generation cancer organoids. Nat. Mater. 2022;21:143–159. doi: 10.1038/s41563-021-01057-5. [DOI] [PubMed] [Google Scholar]
- 316.Sontheimer-Phelps A, Hassell BA, Ingber DE. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer. 2019;19:65–81. doi: 10.1038/s41568-018-0104-6. [DOI] [PubMed] [Google Scholar]
- 317.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 318.Huang, L. et al. PDX-derived organoids model in vivo drug response and secrete biomarkers. JCI Insight. 5, (2020). [DOI] [PMC free article] [PubMed]
- 319.Pauli C, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Li, L. et al. Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI Insight. 4, (2019). [DOI] [PMC free article] [PubMed]
- 321.Kraiczy J, et al. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut. 2019;68:49–61. doi: 10.1136/gutjnl-2017-314817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Wang R, et al. Systematic evaluation of colorectal cancer organoid system by single-cell RNA-Seq analysis. Genome Biol. 2022;23:106. doi: 10.1186/s13059-022-02673-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Vlachogiannis G, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Karakasheva TA, et al. Patient-derived organoids as a platform for modeling a patient’s response to chemoradiotherapy in esophageal cancer. Sci. Rep. 2021;11:21304. doi: 10.1038/s41598-021-00706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Weeber F, Ooft SN, Dijkstra KK, Voest EE. Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chem. Biol. 2017;24:1092–1100. doi: 10.1016/j.chembiol.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 327.Lin M, Gao M, Cavnar MJ, Kim J. Utilizing gastric cancer organoids to assess tumor biology and personalize medicine. World J. Gastrointest. Oncol. 2019;11:509–517. doi: 10.4251/wjgo.v11.i7.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Steele NG, et al. An organoid-based preclinical model of human gastric cancer. Cell Mol. Gastroenterol. Hepatol. 2019;7:161–184. doi: 10.1016/j.jcmgh.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Gao M, et al. Development of patient-derived gastric cancer organoids from endoscopic biopsies and surgical tissues. Ann. Surg. Oncol. 2018;25:2767–2775. doi: 10.1245/s10434-018-6662-8. [DOI] [PubMed] [Google Scholar]
- 330.Armstrong, A. et al. Multiplex patient-based drug response assay in pancreatic ductal adenocarcinoma. Biomedicines. 9, (2021). [DOI] [PMC free article] [PubMed]
- 331.Usui, T. et al. Hedgehog signals mediate anti-cancer drug resistance in three-dimensional primary colorectal cancer organoid culture. Int. J. Mol. Sci. 19, (2018). [DOI] [PMC free article] [PubMed]
- 332.Shen X, et al. KLF5 inhibition overcomes oxaliplatin resistance in patient-derived colorectal cancer organoids by restoring apoptotic response. Cell Death Dis. 2022;13:303. doi: 10.1038/s41419-022-04773-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Cho K, et al. YAP/TAZ suppress drug penetration into hepatocellular carcinoma through stromal activation. Hepatology. 2021;74:2605–2621. doi: 10.1002/hep.32000. [DOI] [PubMed] [Google Scholar]
- 334.Bruun J, et al. Patient-derived organoids from multiple colorectal cancer liver metastases reveal moderate intra-patient pharmacotranscriptomic heterogeneity. Clin. Cancer Res. 2020;26:4107–4119. doi: 10.1158/1078-0432.CCR-19-3637. [DOI] [PubMed] [Google Scholar]
- 335.Schutte M, et al. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat. Commun. 2017;8:14262. doi: 10.1038/ncomms14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Verissimo, C. S. et al. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. Elife. 5, (2016). [DOI] [PMC free article] [PubMed]
- 337.Rodrigues D, et al. Unravelling mechanisms of doxorubicin-induced toxicity in 3D human intestinal organoids. Int. J. Mol. Sci. 2022;23:1286. doi: 10.3390/ijms23031286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Rodrigues D, et al. A transcriptomic approach to elucidate the mechanisms of gefitinib-induced toxicity in healthy human intestinal organoids. Int. J. Mol. Sci. 2022;23:2213. doi: 10.3390/ijms23042213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Kim H, et al. Development of human pluripotent stem cell-derived hepatic organoids as an alternative model for drug safety assessment. Biomaterials. 2022;286:121575. doi: 10.1016/j.biomaterials.2022.121575. [DOI] [PubMed] [Google Scholar]
- 340.Rowe RG, Daley GQ. Induced pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Genet. 2019;20:377–388. doi: 10.1038/s41576-019-0100-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Savoia D. New antimicrobial approaches: reuse of old drugs. Curr. Drug Targets. 2016;17:731–738. doi: 10.2174/1389450116666150806124110. [DOI] [PubMed] [Google Scholar]
- 342.Jourdan JP, Bureau R, Rochais C, Dallemagne P. Drug repositioning: a brief overview. J. Pharm. Pharmacol. 2020;72:1145–1151. doi: 10.1111/jphp.13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Norkin M, Ordonez-Moran P, Huelsken J. High-content, targeted RNA-seq screening in organoids for drug discovery in colorectal cancer. Cell Rep. 2021;35:109026. doi: 10.1016/j.celrep.2021.109026. [DOI] [PubMed] [Google Scholar]
- 344.Li L, et al. Protein synthesis inhibitor omacetaxine is effective against hepatocellular carcinoma. JCI Insight. 2021;6:e138197. doi: 10.1172/jci.insight.138197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Chen B, et al. Fangchinoline inhibits non-small cell lung cancer metastasis by reversing epithelial-mesenchymal transition and suppressing the cytosolic ROS-related Akt-mTOR signaling pathway. Cancer Lett. 2022;543:215783. doi: 10.1016/j.canlet.2022.215783. [DOI] [PubMed] [Google Scholar]
- 346.Ghate NB, et al. VprBP directs epigenetic gene silencing through histone H2A phosphorylation in colon cancer. Mol. Oncol. 2021;15:2801–2817. doi: 10.1002/1878-0261.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Ilegems E, et al. HIF-1alpha inhibitor PX-478 preserves pancreatic beta cell function in diabetes. Sci. Transl. Med. 2022;14:eaba9112. doi: 10.1126/scitranslmed.aba9112. [DOI] [PubMed] [Google Scholar]
- 348.Loe AKH, et al. Uncovering the dosage-dependent roles of Arid1a in gastric tumorigenesis for combinatorial drug therapy. J. Exp. Med. 2021;218:e20200219. doi: 10.1084/jem.20200219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Han S, et al. Defining the identity and dynamics of adult gastric isthmus stem cells. Cell. Stem Cell. 2019;25:342–356 e347. doi: 10.1016/j.stem.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Qu M, et al. Establishment of intestinal organoid cultures modeling injury-associated epithelial regeneration. Cell Res. 2021;31:259–271. doi: 10.1038/s41422-020-00453-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Villablanca EJ, Selin K, Hedin CRH. Mechanisms of mucosal healing: treating inflammatory bowel disease without immunosuppression? Nat. Rev. Gastroenterol. Hepatol. 2022;19:493–507. doi: 10.1038/s41575-022-00604-y. [DOI] [PubMed] [Google Scholar]
- 352.Tan SH, et al. AQP5 enriches for stem cells and cancer origins in the distal stomach. Nature. 2020;578:437–443. doi: 10.1038/s41586-020-1973-x. [DOI] [PubMed] [Google Scholar]
- 353.Arnold K, et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell. Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Choi E, et al. Lrig1+ gastric isthmal progenitor cells restore normal gastric lineage cells during damage recovery in adult mouse stomach. Gut. 2018;67:1595–1605. doi: 10.1136/gutjnl-2017-313874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Mitoyan L, et al. A stem cell population at the anorectal junction maintains homeostasis and participates in tissue regeneration. Nat. Commun. 2021;12:2761. doi: 10.1038/s41467-021-23034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Chacon-Martinez CA, Koester J, Wickstrom SA. Signaling in the stem cell niche: regulating cell fate, function and plasticity. Development. 2018;145:dev165399. doi: 10.1242/dev.165399. [DOI] [PubMed] [Google Scholar]
- 358.Frey N, Schwank G. CRISPR-based screening in three-dimensional organoid cultures to identify TGF-beta pathway regulators. Methods Mol. Biol. 2022;2488:99–111. doi: 10.1007/978-1-0716-2277-3_8. [DOI] [PubMed] [Google Scholar]
- 359.Okamoto R, Watanabe M. Role of epithelial cells in the pathogenesis and treatment of inflammatory bowel disease. J. Gastroenterol. 2016;51:11–21. doi: 10.1007/s00535-015-1098-4. [DOI] [PubMed] [Google Scholar]
- 360.Sugimoto S, et al. An organoid-based organ-repurposing approach to treat short bowel syndrome. Nature. 2021;592:99–104. doi: 10.1038/s41586-021-03247-2. [DOI] [PubMed] [Google Scholar]
- 361.Karvellas CJ, Francoz C, Weiss E. Liver transplantation in acute-on-chronic liver failure. Transplantation. 2021;105:1471–1481. doi: 10.1097/TP.0000000000003550. [DOI] [PubMed] [Google Scholar]
- 362.Sarin SK, Choudhury A. Acute-on-chronic liver failure: terminology, mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2016;13:131–149. doi: 10.1038/nrgastro.2015.219. [DOI] [PubMed] [Google Scholar]
- 363.Messina, A., Luce, E., Hussein, M. & Dubart-Kupperschmitt, A. Pluripotent-stem-cell-derived hepatic cells: hepatocytes and organoids for liver therapy and regeneration. Cells. 9, (2020). [DOI] [PMC free article] [PubMed]
- 364.Schneemann SA, et al. Ethical challenges for pediatric liver organoid transplantation. Sci. Transl. Med. 2020;12:eaau8471. doi: 10.1126/scitranslmed.aau8471. [DOI] [PubMed] [Google Scholar]
- 365.Reza HA, Okabe R, Takebe T. Organoid transplant approaches for the liver. Transpl. Int. 2021;34:2031–2045. doi: 10.1111/tri.14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Tamai M, Adachi E, Kawase M, Tagawa YI. Syngeneic implantation of mouse hepatic progenitor cell-derived three-dimensional liver tissue with dense collagen fibrils. World J. Gastroenterol. 2022;28:1444–1454. doi: 10.3748/wjg.v28.i14.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Sampaziotis F, et al. Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science. 2021;371:839–846. doi: 10.1126/science.aaz6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 369.Vining KH, Mooney DJ. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017;18:728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Jasper H. Intestinal stem cell aging: origins and interventions. Annu Rev. Physiol. 2020;82:203–226. doi: 10.1146/annurev-physiol-021119-034359. [DOI] [PubMed] [Google Scholar]
- 371.Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 372.Shivdasani RA, Clevers H, de Sauvage FJ. Tissue regeneration: Reserve or reverse? Science. 2021;371:784–786. doi: 10.1126/science.abb6848. [DOI] [PubMed] [Google Scholar]
- 373.Bankaitis ED, Ha A, Kuo CJ, Magness ST. Reserve stem cells in intestinal homeostasis and injury. Gastroenterology. 2018;155:1348–1361. doi: 10.1053/j.gastro.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Higa T, et al. Spatiotemporal reprogramming of differentiated cells underlies regeneration and neoplasia in the intestinal epithelium. Nat. Commun. 2022;13:1500. doi: 10.1038/s41467-022-29165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Hageman JH, et al. Intestinal regeneration: regulation by the microenvironment. Dev. Cell. 2020;54:435–446. doi: 10.1016/j.devcel.2020.07.009. [DOI] [PubMed] [Google Scholar]
- 376.Liu Y, Chen YG. Intestinal epithelial plasticity and regeneration via cell dedifferentiation. Cell Regen. 2020;9:14. doi: 10.1186/s13619-020-00053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Gupta PB, et al. Phenotypic plasticity: driver of cancer initiation, progression, and therapy resistance. Cell. Stem Cell. 2019;24:65–78. doi: 10.1016/j.stem.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Tetteh PW, et al. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell. Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 379.Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci. Transl. Med. 2014;6:265sr266. doi: 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Galliot B, Crescenzi M, Jacinto A, Tajbakhsh S. Trends in tissue repair and regeneration. Development. 2017;144:357–364. doi: 10.1242/dev.144279. [DOI] [PubMed] [Google Scholar]
- 381.Sprangers J, Zaalberg IC, Maurice MM. Organoid-based modeling of intestinal development, regeneration, and repair. Cell Death Differ. 2021;28:95–107. doi: 10.1038/s41418-020-00665-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Guillermin O, et al. Wnt and Src signals converge on YAP-TEAD to drive intestinal regeneration. EMBO J. 2021;40:e105770. doi: 10.15252/embj.2020105770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Yang L, et al. CREPT is required for murine stem cell maintenance during intestinal regeneration. Nat. Commun. 2021;12:270. doi: 10.1038/s41467-020-20636-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Kim YH, et al. Evaluation of the radiation response and regenerative effects of mesenchymal stem cell-conditioned medium in an intestinal organoid system. Biotechnol. Bioeng. 2020;117:3639–3650. doi: 10.1002/bit.27543. [DOI] [PubMed] [Google Scholar]
- 385.Drost J, Clevers H. Translational applications of adult stem cell-derived organoids. Development. 2017;144:968–975. doi: 10.1242/dev.140566. [DOI] [PubMed] [Google Scholar]
- 386.Date S, Sato T. Mini-gut organoids: reconstitution of the stem cell niche. Annu Rev. Cell Dev. Biol. 2015;31:269–289. doi: 10.1146/annurev-cellbio-100814-125218. [DOI] [PubMed] [Google Scholar]
- 387.Banerjee A, et al. Targeting Wnt signaling through small molecules in governing stem cell fate and diseases. Endocr. Metab. Immune Disord. Drug Targets. 2019;19:233–246. doi: 10.2174/1871530319666190118103907. [DOI] [PubMed] [Google Scholar]
- 388.Um J, Lee JH, Jung DW, Williams DR. Re-education begins at home: an overview of the discovery of in vivo-active small molecule modulators of endogenous stem cells. Expert Opin. Drug Discov. 2018;13:307–326. doi: 10.1080/17460441.2018.1437140. [DOI] [PubMed] [Google Scholar]
- 389.Li W, Li K, Wei W, Ding S. Chemical approaches to stem cell biology and therapeutics. Cell. Stem Cell. 2013;13:270–283. doi: 10.1016/j.stem.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Wolffling S, et al. EGF and BMPs govern differentiation and patterning in human gastric glands. Gastroenterology. 2021;161:623–636 e616. doi: 10.1053/j.gastro.2021.04.062. [DOI] [PubMed] [Google Scholar]
- 391.Tsakmaki A, et al. ISX-9 manipulates endocrine progenitor fate revealing conserved intestinal lineages in mouse and human organoids. Mol. Metab. 2020;34:157–173. doi: 10.1016/j.molmet.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Villegas-Novoa C, Wang Y, Sims CE, Allbritton NL. Development of a primary human intestinal epithelium enriched in L-cells for assay of GLP-1 secretion. Anal. Chem. 2022;94:9648–9655. doi: 10.1021/acs.analchem.2c00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Hou Q, et al. Regulation of the Paneth cell niche by exogenous L-arginine couples the intestinal stem cell function. FASEB J. 2020;34:10299–10315. doi: 10.1096/fj.201902573RR. [DOI] [PubMed] [Google Scholar]
- 394.Mead BE, et al. Screening for modulators of the cellular composition of gut epithelia via organoid models of intestinal stem cell differentiation. Nat. Biomed. Eng. 2022;6:476–494. doi: 10.1038/s41551-022-00863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Sorrentino G, et al. Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology. 2020;159:956–968 e958. doi: 10.1053/j.gastro.2020.05.067. [DOI] [PubMed] [Google Scholar]
- 396.Veenvliet JV, Herrmann BG. Modeling mammalian trunk development in a dish. Dev. Biol. 2021;474:5–15. doi: 10.1016/j.ydbio.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 397.Zhou R, et al. The protein phosphatase PPM1A dephosphorylates and activates YAP to govern mammalian intestinal and liver regeneration. PLoS Biol. 2021;19:e3001122. doi: 10.1371/journal.pbio.3001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Cujba AM, et al. An HNF1alpha truncation associated with maturity-onset diabetes of the young impairs pancreatic progenitor differentiation by antagonizing HNF1beta function. Cell Rep. 2022;38:110425. doi: 10.1016/j.celrep.2022.110425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Yu W, et al. ACE2 contributes to the maintenance of mouse epithelial barrier function. Biochem Biophys. Res Commun. 2020;533:1276–1282. doi: 10.1016/j.bbrc.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Huang M, et al. Emc3 maintains intestinal homeostasis by preserving secretory lineages. Mucosal Immunol. 2021;14:873–886. doi: 10.1038/s41385-021-00399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Farin HF, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530:340–343. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- 402.Kabiri Z, et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 2014;141:2206–2215. doi: 10.1242/dev.104976. [DOI] [PubMed] [Google Scholar]
- 403.Biton M, et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell. 2018;175:1307–1320 e1322. doi: 10.1016/j.cell.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Shyer AE, et al. Bending gradients: how the intestinal stem cell gets its home. Cell. 2015;161:569–580. doi: 10.1016/j.cell.2015.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.von Furstenberg RJ, et al. Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G409–G417. doi: 10.1152/ajpgi.00453.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Wang F, et al. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology. 2013;145:383–395 e381-321. doi: 10.1053/j.gastro.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348:74–80. doi: 10.1126/science.aaa6204. [DOI] [PubMed] [Google Scholar]
- 408.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Hai J, et al. Generation of genetically engineered mouse lung organoid models for squamous cell lung cancers allows for the study of combinatorial immunotherapy. Clin. Cancer Res. 2020;26:3431–3442. doi: 10.1158/1078-0432.CCR-19-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Koh V, et al. Hedgehog transcriptional effector GLI mediates mTOR-Induced PD-L1 expression in gastric cancer organoids. Cancer Lett. 2021;518:59–71. doi: 10.1016/j.canlet.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Westcott PMK, et al. Low neoantigen expression and poor T-cell priming underlie early immune escape in colorectal cancer. Nat. Cancer. 2021;2:1071–1085. doi: 10.1038/s43018-021-00247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Zhao X, Yuan C, Wangmo D, Subramanian S. Tumor-secreted extracellular vesicles regulate T-cell costimulation and can be manipulated to induce tumor-specific T-cell responses. Gastroenterology. 2021;161:560–574 e511. doi: 10.1053/j.gastro.2021.04.036. [DOI] [PubMed] [Google Scholar]
- 413.Zou F, et al. The CD39(+) HBV surface protein-targeted CAR-T and personalized tumor-reactive CD8(+) T cells exhibit potent anti-HCC activity. Mol. Ther. 2021;29:1794–1807. doi: 10.1016/j.ymthe.2021.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Liu T, et al. High-affinity neoantigens correlate with better prognosis and trigger potent antihepatocellular carcinoma (HCC) activity by activating CD39(+)CD8(+) T cells. Gut. 2021;70:1965–1977. doi: 10.1136/gutjnl-2020-322196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Dijkstra KK, et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174:1586–1598 e1512. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Montel-Hagen A, et al. Organoid-induced differentiation of conventional T cells from human pluripotent stem cells. Cell. Stem Cell. 2019;24:376–389 e378. doi: 10.1016/j.stem.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Wang Z, et al. 3D-organoid culture supports differentiation of human CAR(+) iPSCs into highly functional CAR T cells. Cell. Stem Cell. 2022;29:515–527 e518. doi: 10.1016/j.stem.2022.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Cattaneo CM, et al. Tumor organoid-T-cell coculture systems. Nat. Protoc. 2020;15:15–39. doi: 10.1038/s41596-019-0232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Gronholm M, et al. Patient-derived organoids for precision cancer immunotherapy. Cancer Res. 2021;81:3149–3155. doi: 10.1158/0008-5472.CAN-20-4026. [DOI] [PubMed] [Google Scholar]
- 420.Shi Y, Inoue H, Wu JC, Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Yuki K, Cheng N, Nakano M, Kuo CJ. Organoid models of tumor immunology. Trends Immunol. 2020;41:652–664. doi: 10.1016/j.it.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Ye W, et al. Organoids to study immune functions, immunological diseases and immunotherapy. Cancer Lett. 2020;477:31–40. doi: 10.1016/j.canlet.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 423.Tsai S, et al. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer. 2018;18:335. doi: 10.1186/s12885-018-4238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Courau T, et al. Cocultures of human colorectal tumor spheroids with immune cells reveal the therapeutic potential of MICA/B and NKG2A targeting for cancer treatment. J. Immunother. Cancer. 2019;7:74. doi: 10.1186/s40425-019-0553-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Gwag T, Ma E, Zhou C, Wang S. Anti-CD47 antibody treatment attenuates liver inflammation and fibrosis in experimental non-alcoholic steatohepatitis models. Liver Int. 2022;42:829–841. doi: 10.1111/liv.15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Xiang Z, et al. Dexamethasone suppresses immune evasion by inducing GR/STAT3 mediated downregulation of PD-L1 and IDO1 pathways. Oncogene. 2021;40:5002–5012. doi: 10.1038/s41388-021-01897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Frey N, et al. Loss of Rnf31 and Vps4b sensitizes pancreatic cancer to T cell-mediated killing. Nat. Commun. 2022;13:1804. doi: 10.1038/s41467-022-29412-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Kong JCH, et al. Tumor-Infiltrating Lymphocyte Function Predicts Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. JCO Precis Oncol. 2018;2:1–15. doi: 10.1200/PO.18.00075. [DOI] [PubMed] [Google Scholar]
- 429.Deng J, et al. CDK4/6 inhibition augments antitumor immunity by enhancing T-cell activation. Cancer Discov. 2018;8:216–233. doi: 10.1158/2159-8290.CD-17-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Sermer D, Brentjens R. CAR T-cell therapy: Full speed ahead. Hematol. Oncol. 2019;37(Suppl 1):95–100. doi: 10.1002/hon.2591. [DOI] [PubMed] [Google Scholar]
- 431.Majzner RG, Mackall CL. Tumor antigen escape from CAR T-cell therapy. Cancer Discov. 2018;8:1219–1226. doi: 10.1158/2159-8290.CD-18-0442. [DOI] [PubMed] [Google Scholar]
- 432.Kaneko S. Successful organoid-mediated generation of iPSC-derived CAR-T cells. Cell. Stem Cell. 2022;29:493–495. doi: 10.1016/j.stem.2022.03.005. [DOI] [PubMed] [Google Scholar]
- 433.Kumari R, et al. Preclinical pharmacology modeling of chimeric antigen receptor T therapies. Curr. Opin. Pharmacol. 2021;61:49–61. doi: 10.1016/j.coph.2021.08.008. [DOI] [PubMed] [Google Scholar]
- 434.Michie J, et al. Antagonism of IAPs Enhances CAR T-cell Efficacy. Cancer Immunol. Res. 2019;7:183–192. doi: 10.1158/2326-6066.CIR-18-0428. [DOI] [PubMed] [Google Scholar]
- 435.Jing R, et al. EZH1 repression generates mature iPSC-derived CAR T cells with enhanced antitumor activity. Cell. Stem Cell. 2022;29:1181–1196 e1186. doi: 10.1016/j.stem.2022.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Sachs, N. et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 38, (2019). [DOI] [PMC free article] [PubMed]
- 437.Lane A, et al. Modeling and Rescue of RP2 Retinitis Pigmentosa Using iPSC-Derived Retinal Organoids. Stem Cell Rep. 2020;15:67–79. doi: 10.1016/j.stemcr.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell. Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 439.Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017;14:573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Gomes AC, Hoffmann C, Mota JF. The human gut microbiota: Metabolism and perspective in obesity. Gut Microbes. 2018;9:308–325. doi: 10.1080/19490976.2018.1465157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Bajaj JS. Alcohol, liver disease and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019;16:235–246. doi: 10.1038/s41575-018-0099-1. [DOI] [PubMed] [Google Scholar]
- 442.Sommer F, et al. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017;15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 443.Nishida A, et al. Can control of gut microbiota be a future therapeutic option for inflammatory bowel disease? World J. Gastroenterol. 2021;27:3317–3326. doi: 10.3748/wjg.v27.i23.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Vendrik KEW, et al. Fecal microbiota transplantation in neurological disorders. Front Cell Infect. Microbiol. 2020;10:98. doi: 10.3389/fcimb.2020.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Costello SP, et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 2019;321:156–164. doi: 10.1001/jama.2018.20046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Aron-Wisnewsky J, Clement K, Nieuwdorp M. Fecal microbiota transplantation: a future therapeutic option for obesity/diabetes? Curr. Diab Rep. 2019;19:51. doi: 10.1007/s11892-019-1180-z. [DOI] [PubMed] [Google Scholar]
- 447.Chen D, et al. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int J. Cancer. 2019;145:2021–2031. doi: 10.1002/ijc.32003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Margolis KG, Cryan JF, Mayer EA. The microbiota-gut-brain axis: from motility to mood. Gastroenterology. 2021;160:1486–1501. doi: 10.1053/j.gastro.2020.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Maruvada P, Leone V, Kaplan LM, Chang EB. The human microbiome and obesity: moving beyond associations. Cell Host Microbe. 2017;22:589–599. doi: 10.1016/j.chom.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 450.Cammarota G, et al. International consensus conference on stool banking for faecal microbiota transplantation in clinical practice. Gut. 2019;68:2111–2121. doi: 10.1136/gutjnl-2019-319548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Antushevich H. Fecal microbiota transplantation in disease therapy. Clin. Chim. Acta. 2020;503:90–98. doi: 10.1016/j.cca.2019.12.010. [DOI] [PubMed] [Google Scholar]
- 452.Malsagova K, et al. Biobanks-a platform for scientific and biomedical research. Diagnostics (Basel) 2020;10:485. doi: 10.3390/diagnostics10070485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Perrone F, Zilbauer M. Biobanking of human gut organoids for translational research. Exp. Mol. Med. 2021;53:1451–1458. doi: 10.1038/s12276-021-00606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.VanDussen KL, et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2015;64:911–920. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Geurts MH, et al. CRISPR-based adenine editors correct nonsense mutations in a cystic fibrosis organoid biobank. Cell. Stem Cell. 2020;26:503–510 e507. doi: 10.1016/j.stem.2020.01.019. [DOI] [PubMed] [Google Scholar]
- 457.Roerink SF, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018;556:457–462. doi: 10.1038/s41586-018-0024-3. [DOI] [PubMed] [Google Scholar]
- 458.Tiriac H, et al. Successful creation of pancreatic cancer organoids by means of EUS-guided fine-needle biopsy sampling for personalized cancer treatment. Gastrointest. Endosc. 2018;87:1474–1480. doi: 10.1016/j.gie.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Saito Y, et al. Induction of differentiation of intrahepatic cholangiocarcinoma cells to functional hepatocytes using an organoid culture system. Sci. Rep. 2018;8:2821. doi: 10.1038/s41598-018-21121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Sachs N, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386 e310. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 461.Dekkers JF, et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 2016;8:344ra384. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- 462.Chey WD, Kurlander J, Eswaran S. Irritable bowel syndrome: a clinical review. JAMA. 2015;313:949–958. doi: 10.1001/jama.2015.0954. [DOI] [PubMed] [Google Scholar]
- 463.Forsythe SD, et al. Organoid platform in preclinical investigation of personalized immunotherapy efficacy in appendiceal cancer: feasibility study. Clin. Cancer Res. 2021;27:5141–5150. doi: 10.1158/1078-0432.CCR-21-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Almeqdadi M, Mana MD, Roper J, Yilmaz OH. Gut organoids: mini-tissues in culture to study intestinal physiology and disease. Am. J. Physiol. Cell Physiol. 2019;317:C405–C419. doi: 10.1152/ajpcell.00300.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Beumer J, et al. High-resolution mRNA and secretome atlas of human enteroendocrine cells. Cell. 2020;181:1291–1306 e1219. doi: 10.1016/j.cell.2020.04.036. [DOI] [PubMed] [Google Scholar]
- 466.de Lau W, et al. Peyer’s patch M cells derived from Lgr5(+) stem cells require SpiB and are induced by RankL in cultured “miniguts”. Mol. Cell Biol. 2012;32:3639–3647. doi: 10.1128/MCB.00434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Uehara K, et al. Epithelial-derived factors induce muscularis mucosa of human induced pluripotent stem cell-derived gastric organoids. Stem Cell Rep. 2022;17:820–834. doi: 10.1016/j.stemcr.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Beumer J, et al. BMP gradient along the intestinal villus axis controls zonated enterocyte and goblet cell states. Cell Rep. 2022;38:110438. doi: 10.1016/j.celrep.2022.110438. [DOI] [PubMed] [Google Scholar]
- 469.Workman MJ, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 2017;23:49–59. doi: 10.1038/nm.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Read, E., Jowett, G. M., Coman, D. & Neves, J. F. Co-culture of murine small intestine epithelial organoids with innate lymphoid cells. J. Vis. Exp.181 (2022). [DOI] [PubMed]
- 471.Jung KB, et al. Interleukin-2 induces the in vitro maturation of human pluripotent stem cell-derived intestinal organoids. Nat. Commun. 2018;9:3039. doi: 10.1038/s41467-018-05450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Noel G, et al. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 2017;7:45270. doi: 10.1038/srep45270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Na YR, et al. Prostaglandin E2 receptor PTGER4-expressing macrophages promote intestinal epithelial barrier regeneration upon inflammation. Gut. 2021;70:2249–2260. doi: 10.1136/gutjnl-2020-322146. [DOI] [PubMed] [Google Scholar]
- 474.Votanopoulos KI, Skardal A. ASO author reflections: co-cultured lymph node and tumor organoids as a platform for the creation of adaptive immunity and predict response to immunotherapy. Ann. Surg. Oncol. 2020;27:1968–1969. doi: 10.1245/s10434-020-08351-7. [DOI] [PubMed] [Google Scholar]
- 475.Lahar N, et al. Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PLoS One. 2011;6:e26898. doi: 10.1371/journal.pone.0026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Stzepourginski I, et al. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl Acad. Sci. Usa. 2017;114:E506–E513. doi: 10.1073/pnas.1620059114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Jabaji Z, et al. Type I collagen as an extracellular matrix for the in vitro growth of human small intestinal epithelium. PLoS One. 2014;9:e107814. doi: 10.1371/journal.pone.0107814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Sigal M, et al. Stromal R-spondin orchestrates gastric epithelial stem cells and gland homeostasis. Nature. 2017;548:451–455. doi: 10.1038/nature23642. [DOI] [PubMed] [Google Scholar]
- 479.Cordero-Espinoza L, et al. Dynamic cell contacts between periportal mesenchyme and ductal epithelium act as a rheostat for liver cell proliferation. Cell. Stem Cell. 2021;28:1907–1921 e1908. doi: 10.1016/j.stem.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Hou Q, et al. Exogenous L-arginine increases intestinal stem cell function through CD90+ stromal cells producing mTORC1-induced Wnt2b. Commun. Biol. 2020;3:611. doi: 10.1038/s42003-020-01347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Lee JH, et al. Microfluidic co-culture of pancreatic tumor spheroids with stellate cells as a novel 3D model for investigation of stroma-mediated cell motility and drug resistance. J. Exp. Clin. Cancer Res. 2018;37:4. doi: 10.1186/s13046-017-0654-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Ohlund D, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Baghdadi MB, et al. Enteric glial cell heterogeneity regulates intestinal stem cell niches. Cell. Stem Cell. 2022;29:86–100 e106. doi: 10.1016/j.stem.2021.10.004. [DOI] [PubMed] [Google Scholar]
- 484.Pastula A, et al. Three-Dimensional Gastrointestinal Organoid Culture in Combination with Nerves or Fibroblasts: A Method to Characterize the Gastrointestinal Stem Cell Niche. Stem Cells Int. 2016;2016:3710836. doi: 10.1155/2016/3710836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Greicius G, et al. PDGFRalpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc. Natl Acad. Sci. Usa. 2018;115:E3173–E3181. doi: 10.1073/pnas.1713510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Takahashi Y, et al. Reciprocal inflammatory signaling between intestinal epithelial cells and adipocytes in the absence of immune cells. EBioMedicine. 2017;23:34–45. doi: 10.1016/j.ebiom.2017.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Watson CL, et al. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Gao Y, et al. Fusobacterium nucleatum enhances the efficacy of PD-L1 blockade in colorectal cancer. Signal Transduct. Target Ther. 2021;6:398. doi: 10.1038/s41392-021-00795-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Skardal A, et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017;7:8837. doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Koike H, et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature. 2019;574:112–116. doi: 10.1038/s41586-019-1598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Kakni, P., Truckenmuller, R., Habibovic, P. & Giselbrecht, S. Challenges to, and prospects for, reverse engineering the gastrointestinal tract using organoids. Trends Biotechnol. (2022). [DOI] [PubMed]
- 492.Silva AC, et al. Co-emergence of cardiac and gut tissues promotes cardiomyocyte maturation within human iPSC-derived organoids. Cell. Stem Cell. 2021;28:2137–2152 e2136. doi: 10.1016/j.stem.2021.11.007. [DOI] [PubMed] [Google Scholar]
- 493.Tao T, et al. Microengineered multi-organoid system from hiPSCs to recapitulate human liver-islet axis in normal and Type 2 diabetes. Adv. Sci. (Weinh.). 2022;9:e2103495. doi: 10.1002/advs.202103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol. 2005;15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 495.Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 496.Kozlowski MT, Crook CJ, Ku HT. Towards organoid culture without Matrigel. Commun. Biol. 2021;4:1387. doi: 10.1038/s42003-021-02910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Giobbe GG, et al. Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat. Commun. 2019;10:5658. doi: 10.1038/s41467-019-13605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Takezawa T, et al. Collagen vitrigel: a novel scaffold that can facilitate a three-dimensional culture for reconstructing organoids. Cell Transplant. 2004;13:463–473. doi: 10.3727/000000004783983882. [DOI] [PubMed] [Google Scholar]
- 499.Nguyen HT, Massino M, Keita C, Salmon JB. Microfluidic dialysis using photo-patterned hydrogel membranes in PDMS chips. Lab Chip. 2020;20:2383–2393. doi: 10.1039/D0LC00279H. [DOI] [PubMed] [Google Scholar]
- 500.Cruz-Acuna R, et al. PEG-4MAL hydrogels for human organoid generation, culture, and in vivo delivery. Nat. Protoc. 2018;13:2102–2119. doi: 10.1038/s41596-018-0036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Gjorevski N, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 503.Schneeberger K, et al. Converging biofabrication and organoid technologies: the next frontier in hepatic and intestinal tissue engineering? Biofabrication. 2017;9:013001. doi: 10.1088/1758-5090/aa6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Ashammakhi N, et al. Bioinks and bioprinting technologies to make heterogeneous and biomimetic tissue constructs. Mater. Today Bio. 2019;1:100008. doi: 10.1016/j.mtbio.2019.100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Yu F, Hunziker W, Choudhury D. Engineering microfluidic organoid-on-a-chip platforms. Micromachines (Basel) 2019;10:165. doi: 10.3390/mi10030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Staab JF, et al. Co-culture system of human enteroids/colonoids with innate immune cells. Curr. Protoc. Immunol. 2020;131:e113. doi: 10.1002/cpim.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Seppala TT, et al. Patient-derived organoid pharmacotyping is a clinically tractable strategy for precision medicine in pancreatic cancer. Ann. Surg. 2020;272:427–435. doi: 10.1097/SLA.0000000000004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Czerniecki SM, et al. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell. Stem Cell. 2018;22:929–940 e924. doi: 10.1016/j.stem.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Clevers HC. Organoids: avatars for personalized medicine. Keio J. Med. 2019;68:95. doi: 10.2302/kjm.68-006-ABST. [DOI] [PubMed] [Google Scholar]
- 510.Fujii M, et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell. Stem Cell. 2018;23:787–793 e786. doi: 10.1016/j.stem.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 511.Garcia-Rodriguez I, Sridhar A, Pajkrt D, Wolthers KC. Put some guts into it: intestinal organoid models to study viral infection. Viruses. 2020;12:1288. doi: 10.3390/v12111288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Gu W, et al. Rapid establishment of human colonic organoid knockout lines. STAR Protoc. 2022;3:101308. doi: 10.1016/j.xpro.2022.101308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Chiang HY, et al. IL-22 initiates an IL-18-dependent epithelial response circuit to enforce intestinal host defence. Nat. Commun. 2022;13:874. doi: 10.1038/s41467-022-28478-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Breau KA, et al. Efficient transgenesis and homology-directed gene targeting in monolayers of primary human small intestinal and colonic epithelial stem cells. Stem Cell Rep. 2022;17:1493–1506. doi: 10.1016/j.stemcr.2022.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Bollen Y, et al. Efficient and error-free fluorescent gene tagging in human organoids without double-strand DNA cleavage. PLoS Biol. 2022;20:e3001527. doi: 10.1371/journal.pbio.3001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Bues J, et al. Deterministic scRNA-seq captures variation in intestinal crypt and organoid composition. Nat. Methods. 2022;19:323–330. doi: 10.1038/s41592-021-01391-1. [DOI] [PubMed] [Google Scholar]
- 517.Grun D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]