Abstract
Collagen is the most abundant structural protein in humans, with dozens of sequence variants accounting for over 30% of the protein in an animal body. The fibrillar and hierarchical arrangements of collagen are critical in providing mechanical properties with high strength and toughness. Due to this ubiquitous role in human tissues, collagen-based biomaterials are commonly used for tissue repairs and regeneration, requiring chemical and thermal stability over a range of temperatures during materials preparation ex vivo and subsequent utility in vivo. Collagen unfolds from a triple helix to a random coil structure during a temperature interval in which the midpoint or Tm is used as a measure to evaluate the thermal stability of the molecules. However, finding a robust framework to facilitate the design of a specific collagen sequence to yield a specific Tm remains a challenge, including using conventional molecular dynamics modeling. Here we propose a de novo framework to provide a model that outputs the Tm values of input collagen sequences by incorporating deep learning trained on a large data set of collagen sequences and corresponding Tm values. By using this framework, we are able to quickly evaluate how mutations and order in the primary sequence affect the stability of collagen triple helices. Namely, we confirm that mutations to glycines, mutations in the middle of a sequence, and short sequence lengths cause the greatest drop in Tm values
Keywords: Collagen, machine learning, melting temperature, deep learning, long short-term memory artificial recurrent neural network
1. Introduction
Type I collagen is the most abundant protein in animals, forming the matrix of the skin, tendons, bone, and vasculature. Collagen consists of amino acid sequences, generally in the form of repeat units of glycine-X-Y where amino acids proline and hydroxyproline are especially abundant1. The fixed angles of proline or hydroxyproline enable collagen sequences to fold into a triple helical structure, or tropocollagen, which forms the basic structural unit of collagen. In this triple-helical arrangement, glycine is the only amino acid that can be incorporated in the helix without distortion and is, therefore, a strict requirement for fibril-forming collagens. This fundamental structural unit is typically 300 nm long, and 1.5 nm in diameter and can pack with other three-stranded collagen molecules to form hierarchical structures, including fibrils and fibers 2-8. This hierarchical structure enables significant mechanical tensile loading of collagen in physiological conditions 9-13; collagen fibrils can reach a tensile modulus of 0.2 to 0.86 GPa while maintaining elasticity in the human body 11,14-17.
With such remarkable structure and mechanics, as well as biocompatibility, collagen-based biomaterials are routinely sought for use in vivo tissue repairs. Collagen-based biomaterials have been successfully used in drug delivery systems, skin repair, and other biomedical applications 18,19. However, the use of collagen as biomaterial constructs remains limited in large part due to the inability to design and assemble collagen structures in vitro that emulate the structural hierarchy seen in vivo, reflective of triple-helical stability as well as higher order assembly. Such limits in the ability to recapitulate collagen triple helical structures in vitro, are reflected in reduced temperature stabilization and, in practice, in thus reduced mechanics and more rapid degradation in vivo. Such limitations remain as significant hurdles to the more widespread use of collagen in a broader range of tissue repairs. Thus, one metric to evaluate collagen’s mechanical integrity is from its melting point or Tm, defined as the midpoint during the temperature window in which the triple helix unfolds.
Several researchers have examined various biological species to discover or design collagen peptides with greater thermal stability 20-25. Others have tried to add various additives to increase Tm 21. A significant effort by Persikov et al. developed equations to predict Tm values based on local interactions between different amino acid chemistries in collagen tripeptides 26-29. While significant work has sought to understand the effects of variables including primary sequence and additives on the thermal stability of collagen, having a predictive framework that facilitates a priori design of collagen sequences with specific Tm values without prior knowledge of chemical interactions would enable researchers to more efficiently design and synthesize thermally stable collagen for desired applications. Such a framework for discovery, when combined with advancing concepts in controlled self-assembly, would propel the field of collagen-based biomaterials forward.
However, a major challenge to this goal has been to uncover sequence-structure-function relationships in collagen across its length scales 6,11,30,31. Most of the earlier studies have reported progress using atomistic-based methods, such as molecular dynamics or coarse-graining, however, these approaches tend to be computationally expensive and cannot easily explore vast variations of sequences and mutations. In this paper we propose to develop a method that overcomes some of these limitations to set the foundation for an effective machine-learning based approach towards identify sequence-structure-function relationships in collagen molecules. We hypothesize that machine learning can effectively relate complex sequences to effective physical, chemical and biological functions without knowledge of underlying biological interactions. Machine learning has emerged as a useful tool in the analysis of large datasets to help develop design principles of biological materials 32-36. Through careful design and application of neural networks and use of appropriate datasets, machine learning can provide useful information for predicting various behaviors of biological materials without prior knowledge of chemical interactions 33,34
This paper reports the development of a deep neural network machine learning model to predict Tm values of de novo collagen sequences. Through the application of a self-evolutionary algorithm, 1D convolution, bidirectional long short-term memory (LSTM), and dropout features, the model trains on a dataset of existing collagen sequences and Tm values derived from the literature. Using this framework, the effect of mutations in specific residues, sequence length, and order on Tm values is quickly analyzed to derive design principles about the thermal stability of collagen. The goal of this work is to twofold. First, this machine learning method applied to generate thermally stable de novo collagen sequences creates a general framework for the use of machine learning models in biomaterial applications. Second, the insights gained from rapid testing of collagen sequences contribute general design principles about the thermal stability of collagen, with implications not only in biomaterial designs, but also in understanding collagen-related mutations and associated disease states.
2. Materials and Methods
2.1. Collagen Dataset
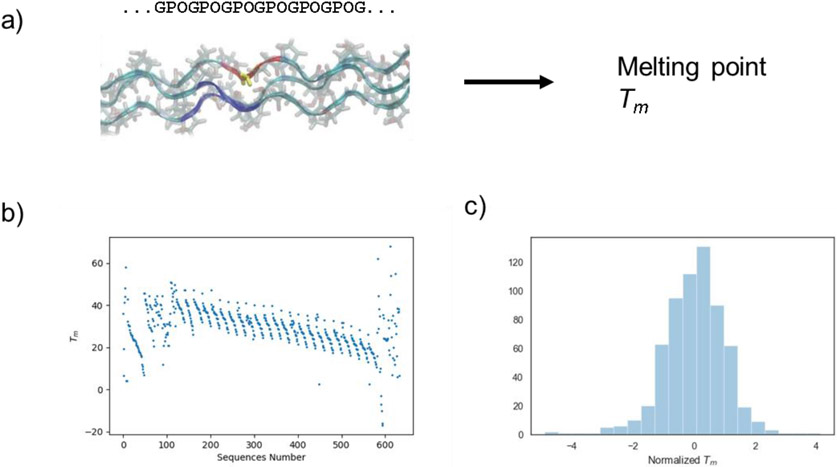
We collected 566 collagen sequences with reported Tm values from a survey of literature (see Table S1). These available melting temperatures were collected from the PubMed, Web of Science, Scopus, Directory of Open Access Journals (DOAJ) Google Scholar databases 26,27,45-54,37,55-57,38-44. Experimental thermal stability data sets for the observed Tm values (in °C) for the Gly-X-Y tripeptide units in triple helical collagens-like peptides are based on host-guest peptides and are integrated to produce an algorithm for predicting global melting temperatures. These experimental results are expanded further using predictions of Tm values of host-guest sequences from Persikov et. al.26 The distribution of the dataset is presented in Figure 1 where sequences have experimentally measured melting temperatures ranging from a few degrees C to 70°C with the mean at ~30°C. The data shows a normal distribution which is used in the machine learning model (Figure 1c). Outliers in the data, specifically the negative Tm value sequences, are generally from extrapolated experimental data.58
Fig. 1. Distribution of data from literature, based on experimental results.
a) Overview of the problem studied here, to predict the melting point Tm from the sequence of collagen molecules. b) Experimental melting temperatures collected. c) Normalized Tm value distribution. Thermal stability data sets for observed Tm values for Gly-X-Y tripeptide units in triple helical collagen-like peptides are integrated here to produce an algorithm for predicting global melting temperatures. Data from 26,27,45-54,37,55-57,38-44.
2.2. Development of the End-to-end Deep Learning Model
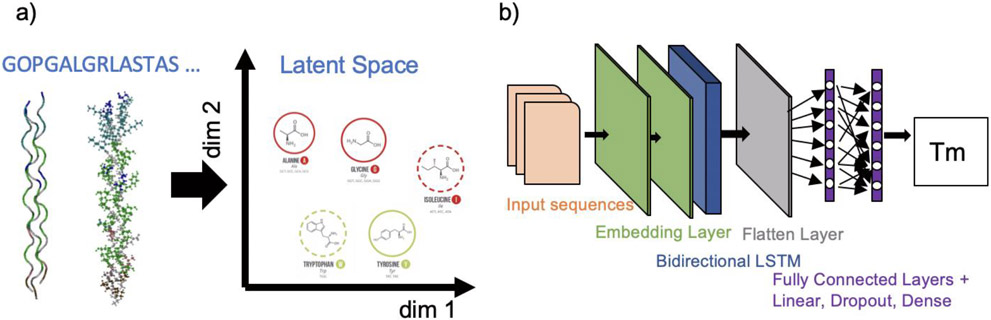
We provide a summary of the deep learning model reported here, as shown in Figure 2. This model is named ColGen to represent a collagen sequences generator which is capable of generating new type I collagen sequences with known Tm values. A natural language processing (NLP) model was adopted 59,60.
Fig. 2. Overview of machine learning model.
a) We design a deep learning network to discover hidden features of collagen sequences by introducing embedding layer. b) The structure of our deep learning model starts at an embedding layer, followed by two 1D convolution layers, then we flatten all the features and send them into a fully connected layer for regression to determine Tm value. Figures S1 and S2 provide details of the neural network model.
Tokenization is used, reflecting a common approach when processing the raw texts or sequences of symbols. Tokens can be considered as the fundamental building blocks of our data which was represented in single letter code for each amino acid along the main chin61,62. Tokenization is first applied to the collagen sequences before passing them through the deep learning model. Every collagen sequence was first encoded into a series of digital tokens, that is, every amino acid is treated as a unique number from 1 to 21 for all the 20 essential amino acids and one nonessential amino acid, hydroxyproline, found in collagen.
2.3. Neural net structure
The tokenized sequences were passed through an embedding layer which is able to recognize the relationship between tokens during the training process. After the embedding layer, data flows through a 1D convolutional layer to harness the internal features from the input. The data is then routed into bidirectional LSTM layers to learn all hidden features from each collagen sequences. Finally, a fully connected neural network - composed by two dense layers – is used to ultimately output the predicted Tm value as a scalar value. This flow of information provides an end-to-end model that relates a sequence of amino acids of varying lengths to its melting temperature.
Figures S1 and S2 provide detailed information into the architecture of the neural network used. The neural network features a total of 49,041 trainable parameters.
2.4. Model training
The dataset was randomly split into training dataset testing dataset, where 80% of the data is used for training and the other 20% is used for testing to examine the ability of prediction of our model.
The model is trained on a Xeon workstation with a GTX-3090 GPU, for 200 epochs. It is worth noting here, the well-trained model can be deployed in a laptop or desktop computer without further requirement of GPUs.
3. Results
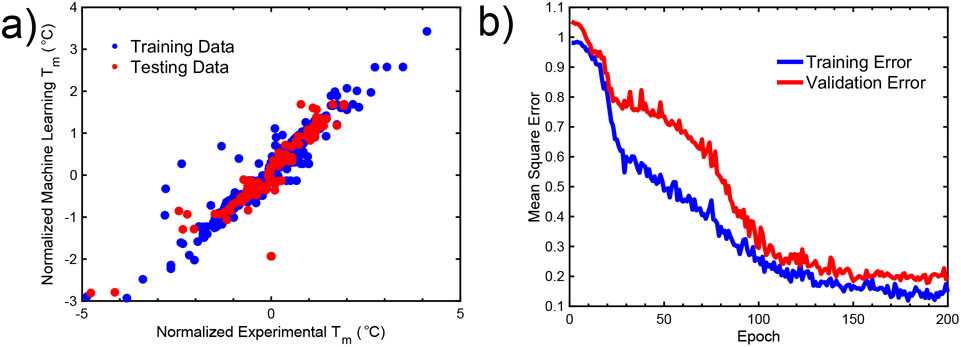
We begin the analysis by training the machine learning model. We find that ColGen demonstrates good predictive accuracy of Tm values in the testing set (Figure 3a). The data shows that testing data is generally well predicted, and a large range of temperatures can be predicted by the model. In addition, the training and validation error remain consistent at a mean squared error of ~0.2 after 120 epochs (Figure 3b).
Fig. 3. Predictive accuracy of ColGen, and training performance.
a) Data comparing training with test set demonstrates a 95% confidence interval. Plotting R2 of training / testing / generation. b) Training and validation error over epochs demonstrate a well fit model. The validation and training errors reach a plateau around 150 epochs.
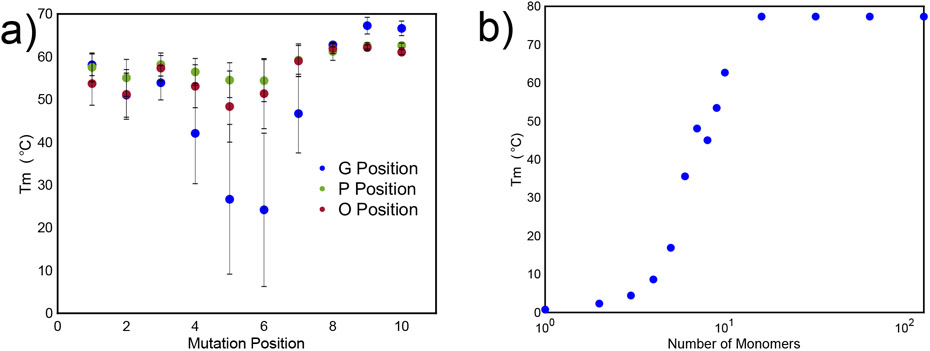
Because the ColGen model enables fast characterization of the Tm values of different collagen sequences, the model was used to understand how mutations and chain length affect Tm value. ColGen enables a rapid search of these effects. (GPO)10 was used as a standard comparison, referred to here as the “pristine sequence,” as it has the highest known thermal stability value in literature 20-56-63-64 due to stereoelectronic effects from hydroxyproline 65-67. Mutations were made in either the G, P, or O position, where either the G, P, or O positions were replaced with another amino acid, and this process is repeated over all amino acid substitutions. The resulting Tm values from each of these calculations are then averaged.
Based on the GolGen model, mutations in the middle of the pristine collagen sequence have the greatest destabilizing effect on collagen Tm values (Figure 4a). This suggests the presence of a critical transition location along the length of the sequence, potentially near the midpoint of the sequence or at least away from the chain ends depending on overall length, that is critical in holding the full chain length together. Further, mutations made along the first several residues at the N terminus of the collagen sequence are more destabilized and have a lower Tm value than mutations made along the last several residues at the C terminus. This indicates a directionality along the sequence and is validated by experimental data which shows that the N-terminal regime is required for the trimerization of other triple helical collagens 68,69. This is in contrast to other work on fibril-forming procollagens which suggests that type 1 collagen molecules in vivo have a C-terminal that is responsible for chain selection and trimerization 70-74. This difference between our model and procollagen fibrillar formation results is likely because several peptides included in this training data may have been folded in N-to-C direction with a nucleation domain at the N terminus 75,76.
Fig. 4. Characterization of the effects of various types of mutations, predicted by ColGen.
a) Tm values of mutations in G, P, or O position demonstrates that mutations in the middle of the sequence are the most destabilizing for Tm values. Mutations in the G position are the most destabilizing to the peptide. Error bars indicate standard deviation of all amino acids that were mutated. b) Thermal stability as a function of collagen sequence length, where length is number of repeat units (GOP) demonstrates that there is a critical length at which the Tm can no longer be increased significantly. This critical length is consistent with other studies.85
The effect of chain length on Tm was also measured. As shown in Figure 4b, the model faithfully captures that Tm values increase upon increasing number of amino acids (triplets) due to increasing hydrogen bonding between triplets. However, there is a limit to the increase in thermal stability that is also captured by the model. This leveling off is achieved at about 14 triplet repeats at around 80C and is consistent with experiments and a thermal stability prediction algorithm developed by Persikov et. al 26. Similar intrinsic strength limits have been found in hydrogen bonded alpha helix and beta sheet structures 77-79.
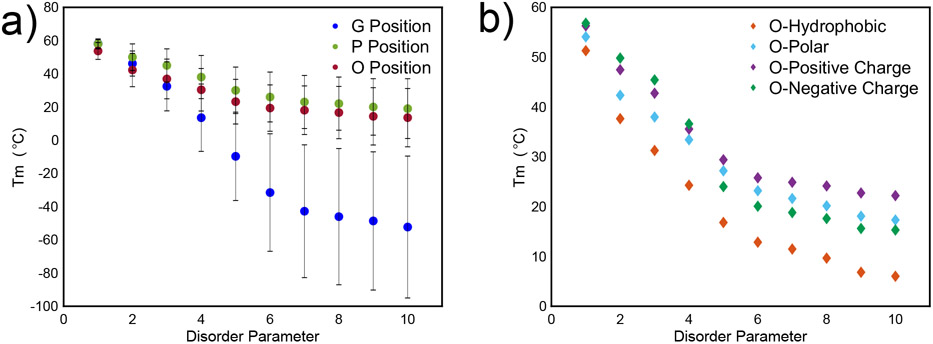
To quantify how the increasing number of mutations affects the thermal stability of collagen, we define the term “disorder parameter” which means the number of repeating triplets with mutations in the G, P, or O position compared to the pristine sequence. Thus, increasing disorder parameter increases deviation from the pristine sample. As expected, increasing disorder decreases the thermal stability of the pristine sequence consistent with experimental data (Figure 5a) 26. Further, disruptions in glycines are the most destabilizing. This is consistent with experimental findings that disruptions in glycine severely impact stability and often constitute disease states, though we cannot directly correlate the position of our glycine mutations to the position of naturally observed mutations due to the shorter sequence length we employ in this study 58,80-82.
Figure 5. Characterization of effect of disorder on Tm, as predicted by the model.
A) Tm values of disorder arranged by G, P, or O position confirm that increasing mutations along the chain decreases thermal stability of the triple helix. Error bars indicate standard deviation of all amino acids that were mutated. b) Tm values of disorder in the O position demonstrates that initial mutations to polar, positive charged, and negative charged amino acids confer the same degree of stability in the molecule. However, upon increasing mutations, polar amino acids are the least destabilizing to the triple helix, suggesting that they should be used for bacterial expression of collagen where expression of O is not possible.
This model could be useful in informing sequence design of bacterially-produced collagen, which enable larger production of tailored collagen sequences 23,50,83. Given that bacteria are unable to express hydroxyproline for bacterially-produced collagen, the machine learning model developed here could also serve as a tool to predict which amino acids may help as a replacement to hydroxyproline. Figure 5b demonstrates these results, showing that positively charged, negatively charged and polar amino acids are only slightly destabilizing compared to (GPO)10 up to a certain extent, but positively charged amino acids (R, H, K) are the least destabilizing compared to other types of amino acids if significant mutations are introduced in the O position.
4. Discussion and conclusions
We developed a platform that adopts a deep learning model trained with input sequences collected from the literature to predict Tm values and a self-evolutionary module to optimize Tm values. To evaluate the long-term interference within a cut-off, the size of kernel function is prescribed and the data flows through several fully connected neuron layers with non-linear activation functions. The deep learning model predicts Tm values of de novo sequences as the output.
Our model shows good prediction power in extrapolating Tm values of the testing set for collagen sequences not included in our training dataset. The trained deep learning model is able to predict Tm values within an acceptable error range considering the amount of experimental data. The machine learning algorithm also allows us to quickly determine how specific amino acids mutations, the amount of disorder in the sequence, and the sequence length affect the thermal stability of collagen. We determine that mutations in the middle of the sequence greatly affect stability and that there the maximum achievable temperature is already reached at a sequence length of 14 repeat units.
While the model enables a quick prediction of Tm values, there are some limitations that should be expanded upon in future work. These limitations primarily arise from the data set used to train the model, which could be further expanded to include a wider range of sequences and sequence lengths. For example, the current dataset has a maximum Tm range of 70C. While the collagen model is able to predict sequences up to 80C, the model’s predictive capacity beyond this temperature range is unclear. New sequences with higher Tm ranges would provide one way to validate the model. Further, the model is only trained on the standard amino acids and hydroxyproline. As such, it would be unable to predict the Tm values of other non-native amino acids and this problem too should be resolved by an expanded data set. Further, experimental melting temperature tests of the predictions from the collagen sequence mutation would help validate the model.
Despite these limitations, the application of this algorithm to collagen sequences more broadly would enable researchers and engineers to design specific collagen sequences with desired Tm values to match specific processing steps related to collagen materials formation and subsequent biomedical applications. More importantly, this approach would lead to more stable and thus mechanically more robust collagen biomaterials to meet new medical applications. Such design control would also provide a foundation for understanding collagen degradation rates in vivo, thus correlating Tm to rates of collagen-based biomaterial remodeling/regeneration in vivo. Tuning such rates would be a significant advance the field of regenerative medicine.
Many collagen-based diseases are based on mutations in the primary sequence, which relate to the models and predictive tools offered here. Thus, this new method would offer insight and new perspectives on disease states in the context of collagen stability, along with implications for possible interventions and regeneration/repair routes in the future. Further, this approach could be propagated up length scales to enable predictions of macroscale biomaterial features and assembly.
In addition to expanding the utility of thermally stable collagen sequences, this work represents a starting point future work on a priori design of protein sequences with specific properties without prior chemical knowledge, for instance through the use of genetic algorithms in conjunction with the machine learning model reported here – to solve the inverse problem 84 A more immediate application may be other fibrous proteins, such as silks, keratins, resilins, reflectins and elastins, which have repeating protein sequences and could be optimized with such an algorithm to maximize mechanics, stability and functions. The formulation reported here could find many other generalizable applications.
Supplementary Material
Acknowledgements:
This research was supported by the IBM-MIT Watson AI Lab, ARO (W911NF-17-1-0384), NOR, NIH P41EB027062 and U01 EB014976). MJB acknowledges the support from ONR (N000141612333 and N000141912375. We would like to thank the Google Cloud computing program (GCP) of the MIT-AI lab, NCREE and NTU joint AI center for providing computational resources. CHY acknowledges support from the Ministry of Science and Technology in Taiwan (MOST 109-2222-E-006 -005 -MY2 and MOST 110-2224-E-007 - 003 -). EK would like to acknowledge support from NSF GRFP. We acknowledge fruitful discussions with Barbara Brodsky.
Data availability:
The processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study. The raw data set is provided as Table S1
References
- 1.Lodish H, Berk A & Zipursky SL Molecular Cell Biology: The Fibrous Proteins of the Matrix. Molecular Cell Biology (W. H. Freeman, 2000). [Google Scholar]
- 2.Prockop DJ & Kivirikko KI Collagens: Molecular biology, diseases, and potentials for therapy. Annual Review of Biochemistry 64, 403–434 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Orgel JPRO et al. The in situ supermolecular structure of type I collagen. Structure 9, 1061–1069 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Ramachandran GN, G. K. Structure of collagen. Nature 593–595 (1955). [DOI] [PubMed] [Google Scholar]
- 5.Rich A, F. C. The structure of collagen. Nature 915–916 (1955). doi: 10.1021/ja01644a065 [DOI] [PubMed] [Google Scholar]
- 6.Buehler MJ & Wong SY Entropic elasticity controls nanomechanics of single tropocollagen molecules. Biophys. J 93, 37–43 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharjee A & Bansal M Collagen structure: The Madras triple helix and the current scenario. IUBMB Life 57, 161–172 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Puxkandl R et al. Viscoelastic properties of collagen: Synchrotron radiation investigations and structural model. Philos. Trans. R. Soc. B Biol. Sci 357, 191–197 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nalla RK, Kruzic JJ, Kinney JH & Ritchie RO Mechanistic aspects of fracture and R-curve behavior in human cortical bone. Biomaterials 26, 217–231 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Ritchie RO, Kruzic JJ, Muhlstein CL, Nalla RK & Stach EA Characteristic dimensions and the micro-mechanisms of fracture and fatigue in ‘nano’ and ‘bio’ materials. Int. J. Fract 128, 1–15 (2004). [Google Scholar]
- 11.Buehler MJ Nature designs tough collagen: Explaining the nanostructure of collagen fibrils. Proc. Natl. Acad. Sci. U. S. A 103, 12285–12290 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautieri A, Vesentini S, Redaelli A & Buehler MJ Viscoelastic properties of model segments of collagen molecules. Matrix Biol. 31, 141–149 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Yeo J et al. Multiscale modeling of keratin, collagen, elastin and related human diseases: Perspectives from atomistic to coarse-grained molecular dynamics simulations. Extrem. Mech. Lett 20, 112–124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen ZL, Dodge MR, Kahn H, Ballarini R & Eppell SJ Stress-strain experiments on individual collagen fibrils. Biophys. J 95, 3956–3963 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Der Rijt JAJ, Van Der Werf KO, Bennink ML, Dijkstra PJ & Feijen J Micromechanical testing of individual collagen fibrils. Macromol. Biosci 6, 697–702 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Yang L et al. Mechanical properties of native and cross-Linked type i collagen fibrils. Biophys. J 94, 2204–2211 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svensson RB, Hassenkam T, Grant CA & Magnusson SP Tensile properties of human collagen fibrils and fascicles are insensitive to environmental salts. Biophys. J 99, 4020–4027 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee CH, Singla A & Lee Y Biomedical applications of collagen. Int. J. Pharm 221, 1–22 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Parenteau-Bareil R, Gauvin R & Berthod F Collagen-based biomaterials for tissue engineering applications. Materials (Basel). 3, 1863–1887 (2010). [Google Scholar]
- 20.Burjanadze TV Hydroxyproline content and location in relation to collagen thermal stability. Biopolymers 18, 931–938 (1979). [DOI] [PubMed] [Google Scholar]
- 21.Gekko K & Koga S Increased thermal stability of collagen in the presence of sugars and polyols. J. Biochem 94, 199–205 (1983). [DOI] [PubMed] [Google Scholar]
- 22.Rigby BJ Amino-acid composition and thermal stability of the skin collagen of the antarctic ice-fish [19]. Nature 219, 166–167 (1968). [DOI] [PubMed] [Google Scholar]
- 23.Mohs A et al. Mechanism of stabilization of a bacterial collagen triple helix in the absence of hydroxyproline. J. Biol. Chem 282, 29757–29765 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Inouye K et al. Synthesis and physical properties of (hydroxyproline-proline-glycine)10: Hydroxyproline in the X-position decreases the melting temperature of the collagen triple helix. Arch. Biochem. Biophys 219, 198–203 (1982). [DOI] [PubMed] [Google Scholar]
- 25.Sakakibara S et al. Synthesis of (Pro-Hyp-Gly)n of defined molecular weights Evidence for the stabilization of collagen triple helix by hydroxypyroline. BBA - Protein Struct. 303, 198–202 (1973). [DOI] [PubMed] [Google Scholar]
- 26.Persikov AV, Ramshaw JAM & Brodsky B Prediction of collagen stability from amino acid sequence. J. Biol. Chem 280, 19343–19349 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Persikov AV, Ramshaw JAM, Kirkpatrick A & Brodsky B Amino acid propensities for the collagen triple-helix. Biochemistry 39, 14960–14967 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Persikov AV, Ramshaw JAM, Kirkpatrick A & Brodsky B Peptide investigations of pairwise interactions in the collagen triple-helix. J. Mol. Biol 316, 385–394 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Persikov AV, Ramshaw JAM, Kirkpatrick A & Brodsky B Electrostatic interactions involving lysine make major contributions to collagen triple-helix stability. Biochemistry 44, 1414–1422 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Gronau G et al. A review of combined experimental and computational procedures for assessing biopolymer structure-process-property relationships. Biomaterials 33, 8240–8255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buehler MJ Atomistic and continuum modeling of mechanical properties of collagen: Elasticity, fracture, and self-assembly. J. Mater. Res 21, 1947–1961 (2006). [Google Scholar]
- 32.Wang J, Cao H, Zhang JZH & Qi Y Computational Protein Design with Deep Learning Neural Networks. Sci. Rep 8, 6349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin Z et al. Artificial intelligence method to design and fold alpha-helical structural proteins from the primary amino acid sequence. Extrem. Mech. Lett 36, 100652 (2020). [Google Scholar]
- 34.Yu CH, Qin Z, Martin-Martinez FJ & Buehler MJ A Self-Consistent Sonification Method to Translate Amino Acid Sequences into Musical Compositions and Application in Protein Design Using Artificial Intelligence. ACS Nano 13, 7471–7482 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Al-Shahib A, Breitling R & Gilbert DR Predicting protein function by machine learning on amino acid sequences – a critical evaluation. BMC Genomics 8, 78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu GX, Chen CT, Richmond DJ & Buehler MJ Bioinspired hierarchical composite design using machine learning: Simulation, additive manufacturing, and experiment. Mater. Horizons 5, 939–945 (2018). [Google Scholar]
- 37.Bolboacă SD & L, J. Amino Acids Sequence Analysis on Collagen. Bull. USAMV-CN 64, 311–316 (2007). [Google Scholar]
- 38.Bretscher LE, Jenkins CL, Taylor KM, DeRider ML & Raines RT Conformational stability of collagen relies on a stereoelectronic effect [23]. J. Am. Chem. Soc 123, 777–778 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Brodsky B, Thiagarajan G, Madhan B & Kar K Triple-helical peptides: An approach to collagen conformation, stability, and self-association. Biopolymers 89, 345–353 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Brodsky B & Persikov AV Molecular structure of the collagen triple helix. Adv. Protein Chem 70, 301–339 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Fallas JA, Dong J, Tao YJ & Hartgerink JD Structural insights into charge pair interactions in triple helical collagen-like proteins. J. Biol. Chem 287, 8039–8047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fidler AL, Boudko SP, Rokas A & Hudson BG The triple helix of collagens - An ancient protein structure that enabled animal multicellularity and tissue evolution. J. Cell Sci 131, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Germann H-P & Heidemann E A synthetic model of collagen: An experimental investigation of the triple-helix stability. Biopolymers 27, 157–163 (1988). [DOI] [PubMed] [Google Scholar]
- 44.Goldberga I, Li R & Duer MJ Collagen Structure-Function Relationships from Solid-State NMR Spectroscopy. Acc. Chem. Res 51, 1621–1629 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Jenkins CL & Raines RT Insights on the conformational stability of collagen. Nat. Prod. Rep 19, 49–59 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Jenkins CL, Bretscher LE, Guzei IA & Raines RT Effect of 3-hydroxyproline residues on collagen stability. J. Am. Chem. Soc 125, 6422–6427 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Kar K et al. Aromatic interactions promote self-association of collagen triple-helical peptides to higher-order structures. Biochemistry 48, 7959–7968 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katti MV, Sami-Subbu R, Ranjekar PK & Gupta VS Amino acid repeat patterns in protein sequences: Their diversity and structural-functional implications. Protein Sci. 9, 1203–1209 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orgel JPRO, Irving TC, Miller A & Wess TJ Microfibrillar structure of type I collagen in situ. Proc. Natl. Acad. Sci 103, 9001–9005 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Z, An B, Ramshaw JAM & Brodsky B Bacterial collagen-like proteins that form triple-helical structures. J. Struct. Biol 186, 451–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker KT et al. Non-linearity of the collagen triple helix in solution and implications for collagen function. Biochem. J 474, 2203–2217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun M et al. Collagen V is a dominant regulator of collagen fibrillogenesis: Dysfunctional regulation of structure and function in a corneal-stroma-specific Col5a1-null mouse model. J. Cell Sci 124, 4096–4105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shoulders MD, Hodges JA & Raines RT Reciprocity of steric and stereoelectronic effects in the collagen triple helix. J. Am. Chem. Soc 128, 8112–8113 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shoulders MD & Raines RT Collagen structure and stability. Annual Review of Biochemistry 78, 929–958 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu Y et al. Collagen Gly missense mutations: Effect of residue identity on collagen structure and integrin binding. J. Struct. Biol 203, 255–262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Persikov AV, Ramshaw JAM & Brodsky B Collagen model peptides: Sequence dependence of triple-helix stability. Biopolym. -Pept. Sci. Sect 55, 436–450 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Persikov AV, Xu Y & Brodsky B Equilibrium thermal transitions of collagen model peptides. Protein Sci. 13, 893–902 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beck K et al. Destabilization of osteogenesis imperfecta collagen-like model peptides correlates with the identity of the residue replacing glycine. Proc. Natl. Acad. Sci. U. S. A 97, 4273–4278 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pennington J, Socher R & Manning C Glove: Global Vectors for Word Representation. in Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) 1532–1543 (Association for Computational Linguistics, 2014). doi: 10.3115/v1/D14-1162 [DOI] [Google Scholar]
- 60.Mirabello C & Wallner B rawMSA: proper Deep Learning makes protein sequence profiles and feature extraction obsolete. doi: 10.1101/394437 [DOI] [Google Scholar]
- 61.Qin L, Dong G & Peng J Chemical-protein Interaction Extraction via ChemicalBERT and Attention Guided Graph Convolutional Networks in Parallel. Proc. - 2020 IEEE Int. Conf. Bioinforma. Biomed. BIBM 2020 708–715 (2020). doi: 10.1109/BIBM49941.2020.9313234 [DOI] [Google Scholar]
- 62.Li X & Fourches D SMILES Pair Encoding: A Data-Driven Substructure Tokenization Algorithm for Deep Learning. J. Chem. Inf. Model 61, 1560–1569 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Buijanadze TV & Kisiriya EL Dependence of thermal stability on the number of hydrogen bonds in water-bridged collagen structure. Biopolymers 21, 1695–1701 (1982). [DOI] [PubMed] [Google Scholar]
- 64.Sakakibara S et al. Synthesis of (Pro-Hyp-Gly)n of defined molecular weights Evidence for the stabilization of collagen triple helix by hydroxypyroline. BBA - Protein Struct. 303, 198–202 (1973). [DOI] [PubMed] [Google Scholar]
- 65.Kotch FW, Guzei IA & Raines RT Stabilization of the collagen triple helix by O-methylation of hydroxyproline residues. J. Am. Chem. Soc 130, 2952–3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raines RT 2005 Emil Thomas Kaiser Award. Protein Sci. 15, 1219–25 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu S, Gu M, Wu K & Li G Unraveling the Role of Hydroxyproline in Maintaining the Thermal Stability of the Collagen Triple Helix Structure Using Simulation. J. Phys. Chem. B 123, 7754–7763 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Snellman A et al. A short sequence in the N-terminal region is required for the trimerization of type XIII collagen and is conserved in other collagenous transmembrane proteins. EMBO J. 19, 5051–5059 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Areida SK et al. Properties of the Collagen Type XVII Ectodomain. J. Biol. Chem 276, 1594–1601 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Bachinger HP, Bruckner P, Timpl R & Engel J The Role of Cis-Trans Isomerization of Peptide Bonds in the Coil + Triple Helix Conversion of Collagen. Eur. J. Biochem. YO (1978). [DOI] [PubMed] [Google Scholar]
- 71.Bachinger HP, Bruckner P, Timpl R, Prockop DJ & Engel J Folding Mechanism of the Triple Helix in Type-I11 Collagen and Type-I11 pN-Collagen Role of Disulfide Bridges and Peptide Bond Isomerization. Eur. J. Biochem 106, (1980). [DOI] [PubMed] [Google Scholar]
- 72.Buevich Alexei V., ‡,§, Qing-Hong Dai, ‡,§, Xiaoyan Liu, §,∥, Barbara Brodsky, ⊥ and & Jean Baum*, § Site-Specific NMR Monitoring of cis–trans Isomerization in the Folding of the Proline-Rich Collagen Triple Helix†·. (2000). doi: 10.1021/BI992584R [DOI] [PubMed] [Google Scholar]
- 73.Doege KJ & Fessler JH Folding of carboxyl domain and assembly of procollagen I. J. Biol. Chem 261, 8924–8935 (1986). [PubMed] [Google Scholar]
- 74.McLaughlin SH & Bulleid NJ Molecular recognition in procollagen chain assembly. Matrix Biol. 16, 369–377 (1998). [DOI] [PubMed] [Google Scholar]
- 75.Buevich AV, Silva T, Brodsky B & Baum J Transformation of the mechanism of triple-helix peptide folding in the absence of a C-terminal nucleation domain and its implications for mutations in collagen disorders. J. Biol. Chem 279, 46890–5 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Stultz CM The folding mechanism of collagen-like model peptides explored through detailed molecular simulations. Protein Sci. 15, 2166–77 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keten S & Buehler MJ Asymptotic strength limit of hydrogen-bond assemblies in proteins at vanishing pulling rates. Phys. Rev. Lett 100, 1–4 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Keten S & Buehler MJ Geometric confinement governs the rupture strength of h-bond assemblies at a critical length scale. Nano Lett. 8, 743–748 (2008). [DOI] [PubMed] [Google Scholar]
- 79.Ackbarow T, Chen X, Keten S & Buehler MJ Hierarchies, multiple energy barriers, and robustness govern the fracture mechanics of α-helical and β-sheet protein domains. Proc. Natl. Acad. Sci. U. S. A 104, 16410–16415 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Culbert AA et al. Substitutions of aspartic acid for glycine-220 and of arginine for glycine-664 in the triple helix of the proα1(I) chain of type I procollagen produce lethal osteogenesis imperfecta and disrupt the ability of collagen fibrils to incorporate crystalline hy. Biochem. J 311, 815–820 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bodian DL, Madhan B, Brodsky B & Klein TE Predicting the clinical lethality of osteogenesis imperfecta from collagen glycine mutations. Biochemistry 47, 5424–5432 (2008). [DOI] [PubMed] [Google Scholar]
- 82.Buevich AV, Silva T, Brodsky B & Baum J Transformation of the mechanism of triple-helix peptide folding in the absence of a C-terminal nucleation domain and its implications for mutations in collagen disorders. J. Biol. Chem 279, 46890–5 (2004). [DOI] [PubMed] [Google Scholar]
- 83.Cheng H et al. Location of glycine mutations within a bacterial collagen protein affects degree of disruption of triple-helix folding and conformation. J. Biol. Chem 286, 2041–2046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu CH, Qin Z & Buehler MJ Artificial intelligence design algorithm for nanocomposites optimized for shear crack resistance. Nano Futur. 3, (2019). [Google Scholar]
- 85.Persikov AV, Ramshaw JAM & Brodsky B Prediction of collagen stability from amino acid sequence. J. Biol. Chem 280, 19343–19349 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study. The raw data set is provided as Table S1