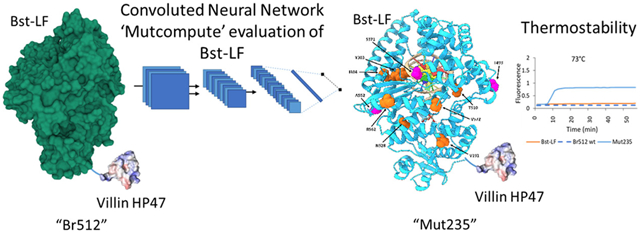
Abstract
The DNA polymerase I from Geobacillus stearothermophilus (also known as Bst DNAP) is widely used in isothermal amplification reactions, where its strand displacement ability is prized. More robust versions of this enzyme should be enabled for diagnostic applications, especially for carrying out higher temperature reactions that might proceed more quickly. To this end, we appended a short fusion domain from the actin-binding protein villin that improved both stability and purification of the enzyme. In parallel, we have developed a machine learning algorithm that assesses the relative fit of individual amino acids to their chemical microenvironments at any position in a protein and applied this algorithm to predict sequence substitutions in Bst DNAP. The top predicted variants had greatly improved thermotolerance (heating prior to assay), and upon combination, the mutations showed additive thermostability, with denaturation temperatures up to 2.5 °C higher than the parental enzyme. The increased thermostability of the enzyme allowed faster loop-mediated isothermal amplification assays to be carried out at 73 °C, where both Bst DNAP and its improved commercial counterpart Bst 2.0 are inactivated. Overall, this is one of the first examples of the application of machine learning approaches to the thermostabilization of an enzyme.
Graphical Abstract
INTRODUCTION
The DNA polymerase I from Bacillus stearothermphilus1 (now classified as a Geobacillus2) is uniquely useful for a variety of isothermal amplification reactions due to its robust strand displacement abilities. In particular, it is a favored polymerase for the implementation of loop-mediated isothermal amplification (LAMP).3 LAMP is a powerful and widely used diagnostic method, including for SARS-CoV-2 detection,4–6 that can rival PCR in sensitivity and speed. Since it does not require thermocycling and associated instrumentation, it is frequently found to be more convenient for both clinical and field use.7,8 While we have previously engineered LAMP for high surety diagnostics by integrating it with oligonucleotide strand displacement (OSD) probes that transduce only true amplicons into signals,9 further improvements, such as increasing the temperatures at which reactions are carried out, may enable faster detection, directly from biological samples without prior processing. The key to such improvements is the ability to successfully engineer the unique strand-displacing Bst DNA polymerase used in LAMP reactions. Despite the biotechnological, biomedical, and commercial importance of this enzyme, there have been relatively few studies that have attempted to engineer its biophysical or kinetic properties,10,11 with only a few active site residues having been mutated.12 The related enzyme from Geobacillus caldoxylosilyticus has also been purified and characterized, and two mutations that impacted substrate specificity and strand displacement were characterized.13 Further analysis of related enzymes and the incorporation of amino acid substitutions observed in phylogeny have also led to improvements in Bst DNAP strand displacement activity.14
In order to greatly expand engineering approaches to the understudied Bst DNAP, we have begun to apply machine learning approaches. We had previously adapted a convolutional neural network (CNN) developed by Torng and Altman,15 and that was trained on the large set of structural data, that is, the Protein Data Bank, to protein engineering applications.16 Our resultant algorithm, MutCompute, evaluates the relative steric and chemical suitabilities of 20 amino acids for a microenvironment represented by 20 Å cubes with 1 Å voxel resolution centered at the alpha-carbon of any given residue in any protein. While MutCompute typically repredicts wild-type residues across all proteins with upward of 70% accuracy, the remaining residues are potential candidates for mutation, and we have developed experimental validations of gain-of-function predictions for three model proteins amenable to quantitative high-throughput screening: blue fluorescence protein (BFP) (PDB: 3M24), phosphomannose isomerase (PDB: 1PMI), and TEM-1 β-lactamase (PDB: 1BTL).16 We have also found that combining individual machine learning mutations can have an additive impact on the phenotype and lead to engineered proteins with much higher activity. For example, multiple slightly improved variants of BFP could be combined to yield a variant with 5-fold greater fluorescence, while multiple slightly improved variants of a phosphomannose isomerase could be combined to yield a variant with 5-fold greater solubility.16
By applying MutCompute predictions to the moderately thermostable Bst DNA polymerase and screening for thermostability, we have been able to greatly improve its thermostability and, in consequence, improve its performance in isothermal amplification assays. To our knowledge, this is the first time that unsupervised machine learning approaches have been used for the thermostabilization of any protein and certainly for optimization of a complex enzyme such as a DNA polymerase.
METHODS
Chemicals and Reagents.
All chemicals were of analytical grade and were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise indicated. All commercially sourced enzymes and related buffers were purchased from New England Biolabs (NEB, Ipswich, MA, USA) unless otherwise indicated. All oligonucleotides and gene blocks (Table S1) were obtained from Integrated DNA Technologies (IDT, Coralville, IA, USA).
Br512 and Enzyme Variants Purification Protocol.
Br512 was cloned into an in-house E. coli expression vector under the control of a T7 RNA polymerase promoter (pKAR2). Full sequence and annotations of the pKAR2-Br512 plasmid (Addgene Plasmid #161875) are available in Table S2. The Br512 expression construct pKAR2-Br512 and its variants were then transformed into E. coli BL21(DE3) (NEB, C2527H). A single colony was seed-cultured overnight in 5 mL of superior broth (Athena Enzyme Systems, catalog number: 0105). The next day, 1 mL of seed culture was inoculated into 1 L of superior broth and grown at 37 °C until it reached an OD600 of 0.7–0.8. Enzyme expression was induced with 1 mM IPTG and 100 ng/mL of anhydrous tetracycline (aTc) at 18 °C for 18 h (or overnight). The induced cells were pelleted at 5000 × g for 10 min at 4 °C and resuspended in 30 mL of ice-cold lysis buffer (50 mM phosphate buffer, pH 7.5, 300 mM NaCl, 20 mM imidazole, 0.1% Igepal CO-630, 5 mM MgSO4, 1 mg/mL HEW lysozyme, 1× EDTA-free protease inhibitor tablet, Thermo Scientific, A32965). The samples were then sonicated (1 s ON, 4 s OFF) for a total time of 4 min with 40% amplitude. The lysate was centrifuged at 35,000×g for 30 min at 4 °C. The supernatant was transferred to a clean tube and filtered through a 0.2 μm filter. Protein from the supernatant was purified using metal affinity chromatography on a Ni-NTA column. Briefly, 1 mL of Ni-NTA agarose slurry was packed into a 10 mL disposable column and equilibrated with 20 column volumes (CVs) of equilibration buffer (50 mM phosphate buffer, pH 7.5, 300 mM NaCl, 20 mM imidazole). The sample lysate was loaded onto the column, and the column was developed by gravity flow. Following loading, the column was washed with 20 CVs of equilibration buffer and 5 CVs of wash buffer (50 mM phosphate buffer, pH 7.5, 300 mM NaCl, 50 mM imidazole). Br512 was eluted with 5 mL of elution buffer (50 mM phosphate buffer, pH 7.5, 300 mM NaCl, 250 mM imidazole). The eluate was dialyzed twice with 2 L of Ni-NTA dialysis buffer (40 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 mM DTT, 0.1% Igepal CO-630). The dialyzed eluate was further passed through an equilibrated 5 mL heparin column (HiTrap Heparin HP) on an FPLC (AKTA pure, GE healthcare) and eluted using a linear NaCl gradient generated from heparin buffers A and B (40 mM Tris–HCl, pH 7.5, 100 mM NaCl for buffer A; 2 M NaCl for buffer B, 0.1% Igepal CO-630). The collected final eluate was dialyzed first with 2 L of heparin dialysis buffer (50 mM Tris–HCl, pH 8.0, 50 mM KCl, 0.1% Tween-20) and second with 2 L of final dialysis buffer (50% glycerol, 50 mM Tris–HCl, pH 8.0, 50 mM KCl, 0.1% Tween-20, 0.1% Igepal CO-630, 1 mM DTT). The purified Br512 was quantified by the Bradford assay and SDS-PAGE/Coomassie gel staining alongside a bovine serum albumin standard.
Real-Time GAPDH LAMP-OSD.
LAMP-OSD reaction mixtures were prepared in 25 μL volume containing indicated amounts of human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA templates along with a final concentration of 1.6 μM each of BIP and FIP primers, 0.4 μM each of B3 and F3 primers, and 0.8 μM of the loop primer. Amplification was performed in 1× isothermal buffer (NEB) (20 mM Tris–HCl, 10 mM (NH4)2SO4, 50 mM KCl, 2 mM MgSO4, 0.1% Tween 20, pH 8.8 at 25 °C). The buffer was appended with 1 M betaine, 0.4 mM dNTPs, 2 mM additional MgSO4, and either Bst 2.0 DNA polymerase (16 units), Bst-LF DNA polymerase (20 pmol) or Br512 DNA polymerase (0.2, 2, 20, or 200 pmol). Assays read using OSD probes received 100 nM fluorophore-labeled OSD strands annealed with a 5-fold excess of the quencher-labeled OSD strands by incubation at 95 °C for 1 min followed by cooling at the rate of 0.1 °C/s to 25 °C. Assays read using intercalating dyes received 1× EvaGreen (Biotium, Freemont, CA, USA) instead of OSD probes. For real-time signal measurement, these LAMP reactions were transferred into a 96-well PCR plate, which was incubated in a LightCycler 96 real-time PCR machine (Roche, Basel, Switzerland) maintained at 65 °C for 90 min. Fluorescence signals were recorded every 3 min in the FAM channel and analyzed using the LightCycler 96 software. For assays read using EvaGreen, amplification was followed by a melt curve analysis on LightCycler 96 to distinguish target amplicons from spurious background.
Site-Directed Mutagenesis.
Site-directed mutagenesis was performed using the Q5 site-directed mutagenesis kit from NEB (E0554S) according to the manufacturer’s instructions. The pKAR2-Br512 plasmid was used as a template to introduce mutations suggested by the Mutcompute analysis. All primer sequences are listed in Table S1. The introduced mutations on the plasmids were confirmed by Sanger sequencing. The list of predictions by Mutcompute is available at https://www.mutcompute.com/polymerase/3tan.
Heat Challenge and High-Temperature LAMP.
LAMP reaction mixtures were prepared in 25 μL volume containing 10 pg of GAPDH DNA template plasmid along with a final concentration of 1.6 μM each of BIP and FIP primers, 0.4 μM each of B3 and F3 primers, and 0.8 μM of the loop primer. The reaction mixtures were preassembled on ice and aliquoted into PCR tubes. A total 20 pmol of enzyme variants were added to the wells. Amplification was performed in the following buffer [1× LAMP heat challenge buffer: 40 mM Tris–HCl, pH 8.0, 10 mM (NH4)2SO4, 80 mM KCl, 4 mM MgCl2] supplemented with 0.4 mM dNTP, 1× Evagreen Dye, and 0.4 M betaine unless otherwise indicated. For heat challenges, PCR tubes that contain the reaction mixtures and 20 pmol of Br512 enzyme variants were challenged on a PCR machine that was preheated to temperatures indicated in the figures. After the heat challenges, the tubes were immediately removed from the PCR machine and cooled on an ice-cooled metal rack for at least 5 min. LAMP assay was performed at 65 °C for 2 h unless otherwise indicated. Fluorescence signals were recorded every 4 min in the FAM channel provided by LightCycler 96 software preset.
Dye-Based Protein Thermal Shift Assay.
The Tm (transition midpoint; melting temperature) of the various enzyme variants were measured using Protein Thermal ShiftTM reagents (Thermo Fisher; catalog number: 4461146) according to the manufacturer’s instruction. Briefly, a total of 40 μg (5 μg/μL) of each enzyme variant in the final dialysis buffer (see Br512 purification protocol) was added into a reaction mixture (20 μL) containing 1× Protein Thermal Shift buffer and 1× Protein Thermal Shift dye. Fluorescence signals were measured in Texas Red channel provided by LightCycler 96 software preset. The red fluorescence change was measured from 37 to 95 °C with 0.1 °C/s ramp speed. The measured values (delta fluorescence/delta temperature) were plotted on the graph with a Tm calling tool provided by LightCycler 96 analytical software (Roche).
RESULTS
Adding a Fast-folding Domain to Bst DNAP as a Starting Point for Engineering.
In order to prepare Bst DNAP for the introduction of additional mutations that could potentially perturb, rather than hopefully enhance, the function, we first added a stabilizing fusion domain. Previously, fusion domains such as the DNA-binding domain Sso7d have been used in the construction of thermostable DNA polymerases (Phusion) that have improved properties, such as increased processivity and resistance to PCR inhibitors.17 In the current instance, we attached a novel fusion domain based on the terminal 47 amino acids of the villin headpiece (HP47)18–20 (Figure 1). This headpiece consists of three α-helices that form a highly conserved hydrophobic core21 and exhibits cotranslational, ultrafast, and autonomous folding properties that may circumvent kinetic traps during protein folding.22 The ultrafast folding property of the villin headpiece subdomain has made it a model for protein folding dynamics and simulation studies.23 In addition, the head displays thermostability with a transition midpoint (Tm) of 70 °C.21,22 It also contains clusters of positively charged amino acids that may facilitate interaction with DNA.
Figure 1.
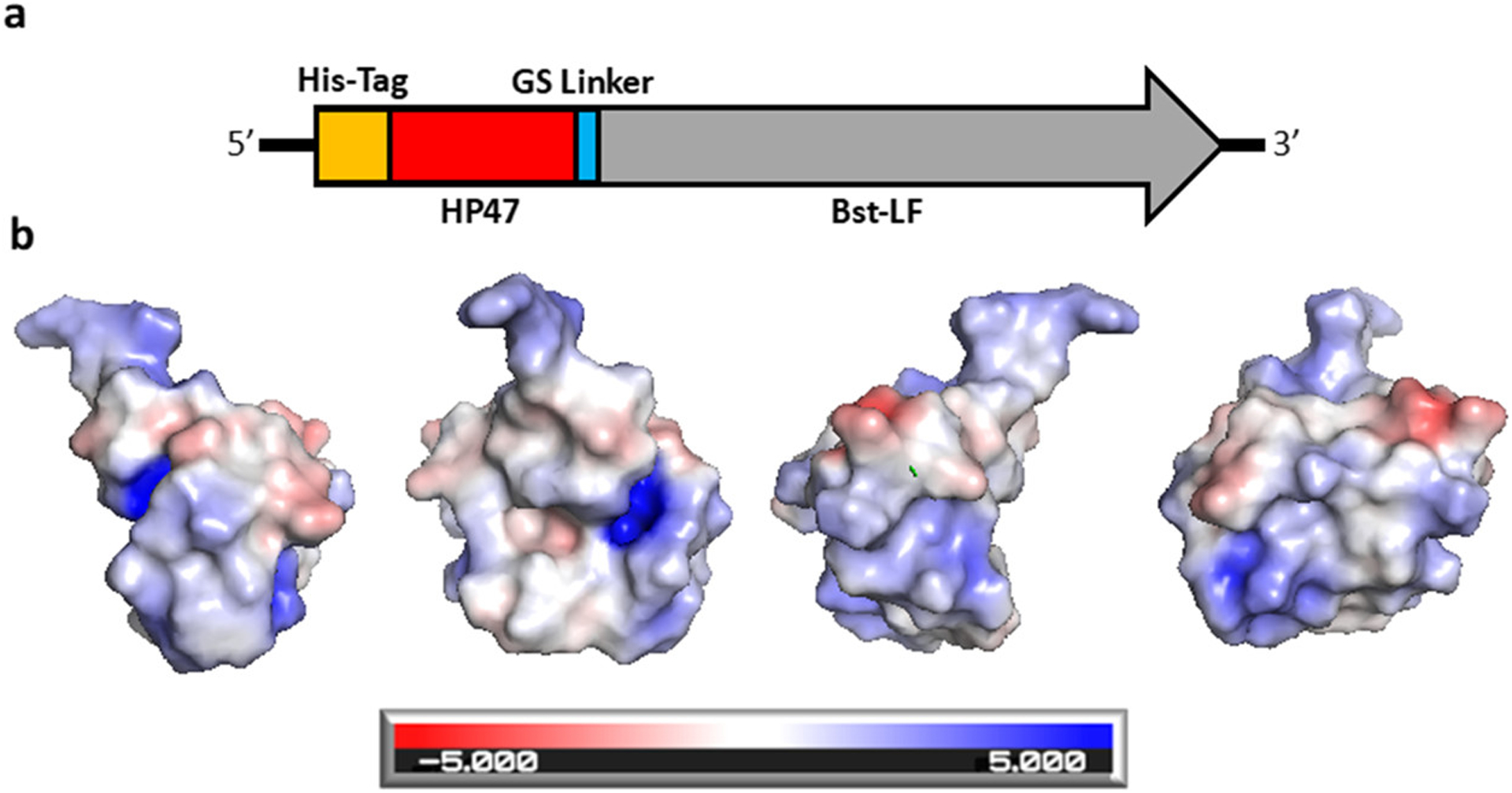
Graphical representation of Br512 and the electrostatic force map of HP47. (a) Br512 was constructed by fusing HP47 with a GS linker to the N-terminal of Bst-LF. An 8× His-Tag was added at the N-terminal of the new fusion protein to aid with purification. (b) Models of HP47 electrostatic force using an Adaptive Poisson–Boltzmann Solver to identify surface charge. The charge designations are referenced in the bar at the bottom. Each graphic is the same model with different orientations rotated on the Y-axis. Graphics were created in PyMol.
We centered our initial designs around the large fragment of DNA polymerase I (Pol I) from Geobacillus stearothermophilus (bst, GenBank L42111.1), which is frequently used for isothermal amplification reactions.8,24,25 This fragment (here-after Bst-LF) lacks a 310 amino acid N-terminal domain that is responsible for 5′ to 3′ exonuclease activity, leading to an increased efficiency of dNTP polymerization.26 The HP47 tag was added to the amino terminus of the large fragment of Bst-LF, leading to the enzyme we denote as Br512 (Figure 1). Br512 also contains a N-terminal 8× His-tag for immobilized metal affinity chromatography (IMAC; Ni-NTA).
Performance of Br512 in Isothermal Amplification Assays.
The development of isothermal amplification assays that are both sensitive and robust to sampling is key for continuing to mitigate the ongoing coronavirus pandemic.27 However, LAMP is well known to frequently produce spurious amplicons, even in the absence of a template, and thus colorimetric and other methods that do not use sequence-specific probes may be at risk for generating false positive results,9 and we therefore developed oligonucleotide strand displacement probes that are only triggered in the presence of specific amplicons. These probes are essentially the equivalent of TaqMan probes for qPCR and can work either in an endpoint or continuous fashion with LAMP.9 In their simplest form, OSD probes are hemiduplex DNAs composed of a long fluorophore-labeled strand annealed to a short complementary quencher-labeled strand. The single-stranded “toehold” in the hemiduplex can bind to its complement in the single-stranded LAMP amplicon loop and initiate strand displacement, leading to separation of the fluorophore and the quencher and a fluorescent signal (Figure S1). Base-pairing to the toehold region is extremely sensitive to mismatches, ensuring specificity, and the programmability of both primers and probes makes possible rapid adaptation to the evolution of new SARS-CoV-2 or other disease variants. We have also shown that higher-order molecular information processing is also possible, such as integration of signals from multiple amplicons.28
Our improved version of LAMP, which we term LAMP-OSD (Figure S1) is designed to be easy to use and interpret, and we have previously shown that it can sensitively and reliably detect SARS-CoV-2, including following direct dilution from saliva.28 Although we have largely mitigated nonspecific signaling of LAMP and made it more robust for point-of-need application, the limited choice and supply and concomitant expense of LAMP enzymes constitute a significant roadblock to widespread application of rapid LAMP-based diagnostics. Br512 presents a potential generally available solution to these issues.
To assess whether the folding domain introduced in Br512 had an impact on enzyme activity, we first compared the strand-displacing DNA polymerase activity of Br512 with that of the parental Bst-LF enzyme. We set up duplicate LAMP-OSD assays9 for the GAPDH gene using either 20 pmol of Bst-LF (a previously optimized amount) or 0.2, 2, 20, or 200 pmol of Br512. Real-time measurement of OSD probe fluorescence revealed that in the presence of 6000 template DNA copies, the DNA polymerase activity of 20 pmol of Br512 was comparable to that of 16 units of Bst 2.0 (Figure S2). The addition of more Br512 did not yield further improvements, although lower amounts reduced the amplification efficiency. In the absence of specific templates, none of the enzymes generated false OSD signals.
We then set up assays with different numbers of template copies and optimized enzyme amounts and found that Br512 had a faster time-to-result and similar detection limit compared to the parental Bst-LF enzyme (Figure 2). In fact, 20 pmol of Br512 performed comparably to 16 units of Bst 2.0 and Bst3.0 in terms of both speed and limit of detection (Figure S3). Similar results were observed via real-time measurements of amplification kinetics using the fluorescent intercalating dye, EvaGreen, in place of sequence-specific OSD probes (Figure S4). While LAMP reactions with Br512 and monitored with intercalating dyes revealed some spurious amplicons, these could be readily distinguished from true amplicons by their distinct melting temperatures (Figure S4). More importantly, these spurious amplicons did not produce any false signals in OSD-based LAMP assays (Figure S3). Spurious amplification is a common problem in LAMP assays, especially when using highly active polymerase variants. For instance, Bst 3.0, which was engineered for improved amplification speed compared to Bst 2.0, has been documented to frequently generate spurious amplicons.29 Taken together, these results demonstrate that the presence of the villin HP47 fusion domain in Br512 improves its speed of amplification relative to Bst-LF, bringing it on par with the DNA amplification abilities of some of the best available commercial enzymes.
Figure 2.
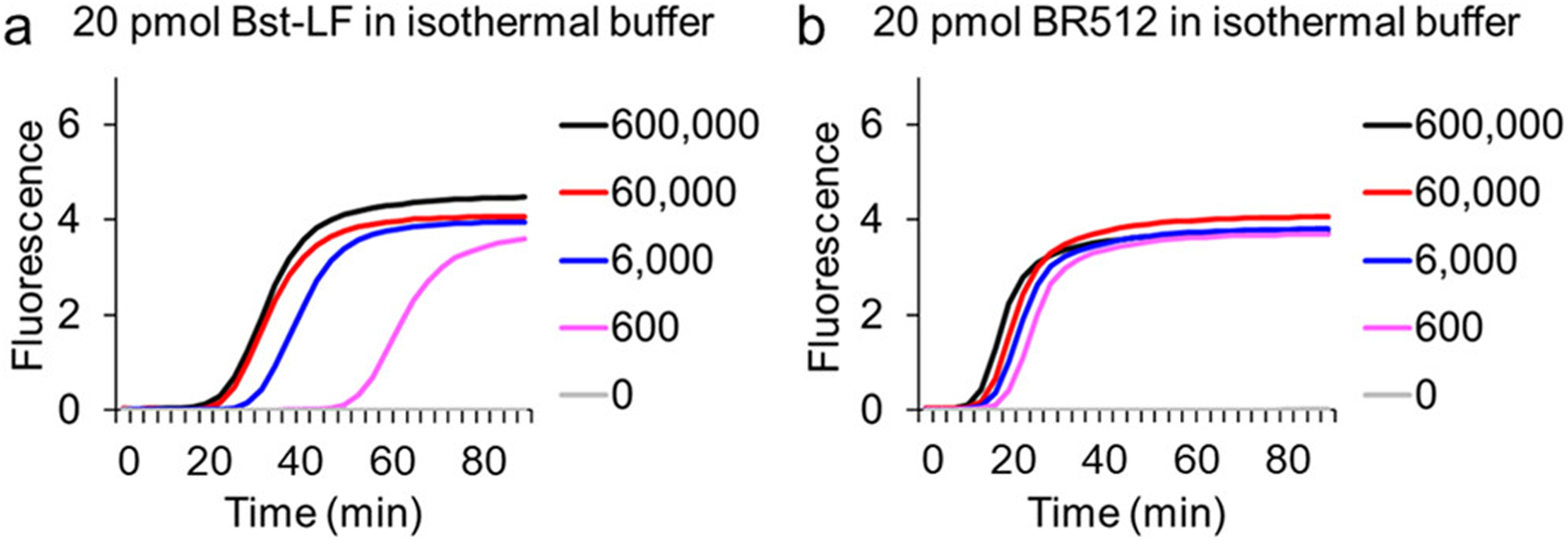
Comparison of Br512 and Bst-LF in LAMP-OSD assays of DNA templates. LAMP-OSD assays for the human GAPDH gene were carried out with 20 pmol of in-house purified Bst-LF (panel A) or 20 pmol of Br512 (panel B) in the indicated reaction buffer. Amplification curves were observed in real-time at 65 °C by measuring OSD fluorescence in reactions seeded with 600,000 (black traces), 60,000 (red traces), 6000 (blue traces), 600 (pink traces), and 0 (gray traces) copies of GAPDH plasmid templates.
Machine Learning Predictions Thermostabilize Bst DNAP.
The CNN model employed (MutCompute) has been previously published and is available to the community at www.mutcompute.com.16 Briefly, MutCompute is a self-supervised CNN that has been trained to predict the identities of individual amino acids based on their local chemical microenvironments (Figure S11). For each residue in a protein, MutCompute outputs a discrete probability distribution for a given position, spanning the 20 possible amino acids. The CNN model was trained on ~1.6M microenvironments sampled from ~19K diverse PDB structures and is capable of predicting wild-type residues with ~70% accuracy. We have previously hypothesized and found that positions, where the wild-type amino acid is not predicted by MutCompute, can frequently be substituted with another, more chemically congruent amino acid, and in consequence, gains in protein stabilities and other functionalities can be achieved. We are now further testing this hypothesis in the context of Bst DNAP. MutCompute evaluated the suitability of each residue in the Bst LF structure, with the exception of residues that were in the first contact shell with the cocrystallized DNA, as the algorithm does not yet incorporate nonprotein atoms (ligands/DNA/RNA). To identify residues prone for gain-of-function, residues were then sorted according to their wild-type probabilities, and those positions that were predicted to be “least fit” for the wild-type residue were experimentally prioritized (Figure 3b).
Figure 3.
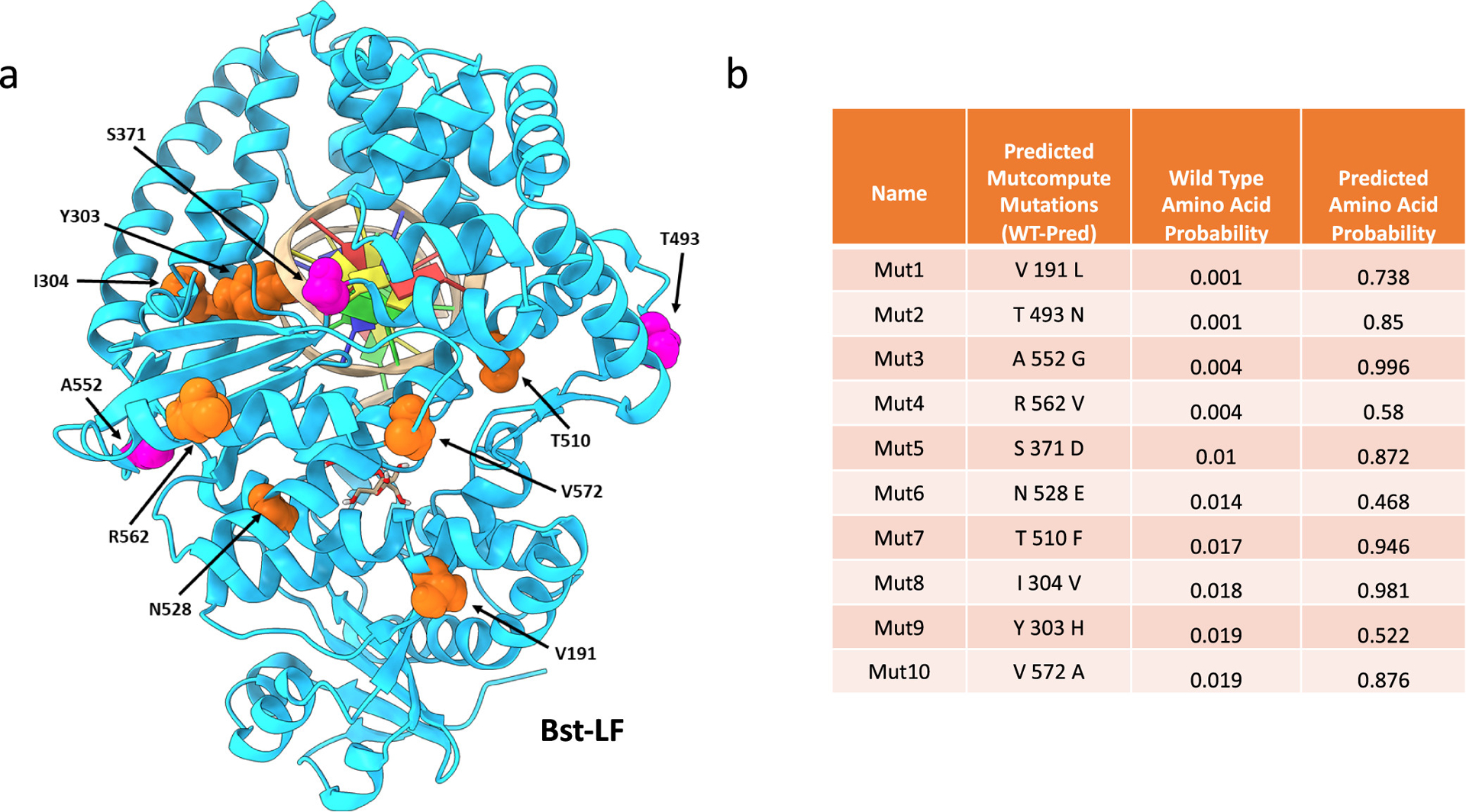
Mutcompute mutations in Br512. (a) Global landscape of the top 10 residues flagged for mutagenesis by MutCompute. Pink residues made it into the final variant and orange residues did not. (b) Table listing Br512 top 10 stabilizing amino acid substitutions predicted by MutCompute (PDB ID: 3TAN). They were designated as Mut1 to Mut10 according to their predicted priorities. The calculated probabilities of the wild-type and predicted amino acids at each position are indicated in columns 3 and 4, respectively.
Surprisingly, of the top 10 residues MutCompute flagged for mutagenesis, only two (Mut6 and Mut9) showed little or no activity in a standard LAMP assay targeting the gene for human GAPDH, while the top 5 (Mut1–5) showed activities as good as or better than the parent Br512 enzyme (Figure S5). Because we were hoping to introduce additive substitutions and achieve higher thermostability, we also adapted LAMP to serve as a simple screen for improved activity at higher temperatures. Initially, enzymes were challenged at temperatures above those typically used for LAMP (75 and 80 °C) before carrying out LAMP reactions at their normal temperature (65 °C). Mut1–5 were further assayed with a heat challenge to determine if they had imparted additional stability to the polymerase (Figure S6), and both Mut2 and Mut 3 were found to be more thermotolerant than the parental enzyme.
Combining Predicted Substitutions Yields Additive Thermotolerance.
We have previously had great success in combining individual mutations predicted via machine learning approaches to generate significantly improved proteins, such as an engineered BFP with 5-fold greater fluorescence and a 1PMI enzyme variant with 5-fold greater solubility.16 Therefore, we examined combinations of the point mutations that showed the greatest activity. We initially generated all possible double mutants of the Muts 1–4 (Mut12, 13, 14, 23, 24, and 34) and carried out LAMP assays and thermal challenges (Figure S7). Mut23 yielded the most robust activity, in keeping with the results of the initial thermal challenges.
We finally generated four additional triple mutant variants (Mut123, Mut124, Mut234, and Mut235) centered on Mut23 (Figure 3). All four triple mutant variants examined showed robust performance in the normal GAPDH LAMP assay (Figures 4a, S8), and the combined machine learning predicted mutations also displayed strong thermotolerance relative to the parental enzyme, which itself was already superior to Bst-LF (Figures 4b,c, S8). Mut235 showed the highest activity and was therefore used in further analysis. We also carried out a comparative LAMP-OSD assay between some of the top-performing variants (Mut23 and Mut235) and other commercially available Bst polymerases (Bst2.0 and Bst3.0). We observed comparable performances of our engineered variants to Bst2.0 and Bst3.0 in terms of both speed and limit of detection (Figure S3). Interestingly, Mut5 on its own has an inactive phenotype at higher temperatures and seems to serve as a potentiating mutation for additional substitutions.
Figure 4.
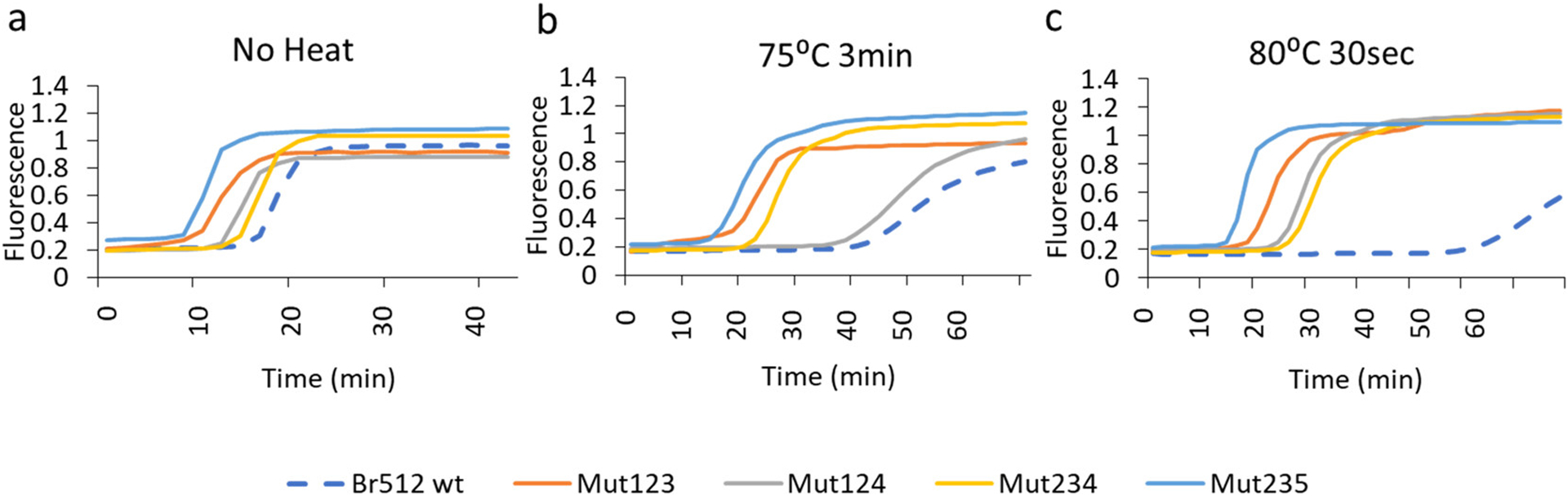
Effect of MutCompute mutations on Br512 enzyme thermal stability. (a–c) Effect of thermal challenge on wild-type and triple mutant MutCompute variants of Br512. Identical GAPDH LAMP assays assembled using the same amount of indicated enzymes were subjected to either no heat challenge (a), 3 min at 75 °C (b), or 30 s at 80 °C (c) prior to real-time measurement of GAPDH DNA amplification kinetics at 65 °C. Representative amplification curves generated by measuring increases in EvaGreen dye fluorescence (Y-axis) over time (X-axis; time in minutes) are depicted as dotted blue (Br512 wild-type), burnt orange (Mut123), gray (Mut124), yellow (Mut234), and blue (Mut235) traces.
Combined Substitutions can Carry out Faster LAMP Reactions at Higher Temperatures.
In addition to determining if the substitutions predicted by machine learning approaches would lead to greater thermotolerance, we attempted to carry out LAMP reactions at higher temperatures. Surprisingly, the Br512 domain not only improves the performance of Bst DNAP (Figure 2), improving the time to signal by 6 min relative to Bst-LF, but also provides thermostabilization in a LAMP reaction up to 72 °C (Figure 5a,b), where Bst-LF shows no activity. Further increases in performance are provided by the addition of the substitutions predicted by machine learning, with the enzyme now being stable in LAMP reactions up to 73 °C (Figure 5c) and the overall reaction proceeding 2–4 min more quickly than Br512 (Figure 5a,b). We also compared the thermostabilities of the engineered variants and commercially available enzymes (Bst2.0 and Bst3.0) in high-temperature LAMP-OSD assays seeded with 60,000, 6000, 600, or 0 copies of the GAPDH plasmid template. At 73 °C, Bst2.0 exhibited reduced activity and showed delayed LAMP amplification at only the highest template input. In contrast, the engineered variants, Mut23 and Mut235, and Bst.3.0 all showed robust activity at 73 °C and generated LAMP-OSD amplification signals at all three template amounts, suggesting a comparable thermotolerance for these enzymes (Figure S9).
Figure 5.
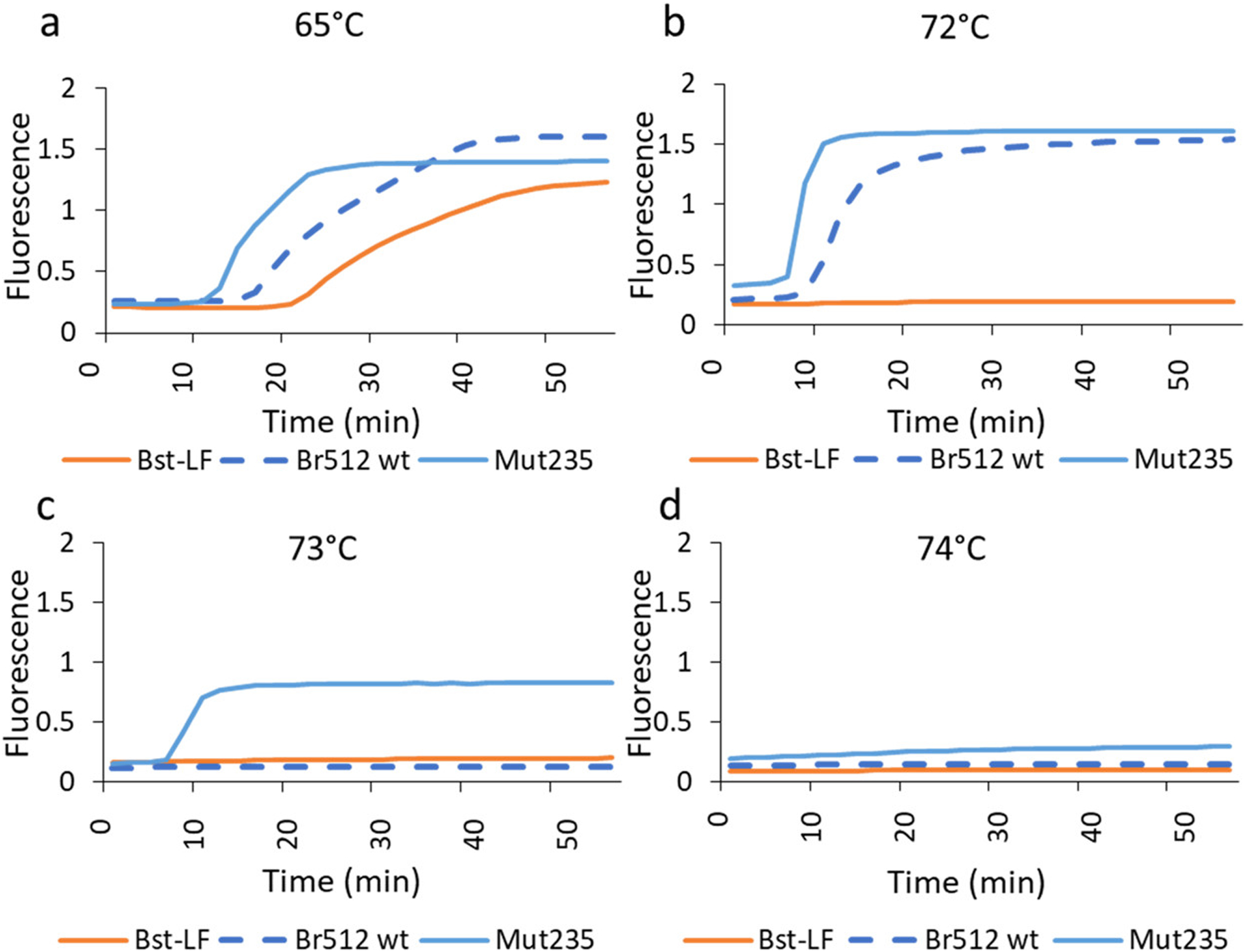
High-temperature GAPDH LAMP assay at 72, 73, and 73 °C. (a–d) GAPDH LAMP assays were assembled using 20 pmol of Bst-LF, wild-type Br512, and Mut235 variant. Amplification kinetics at (a) 65 °C, (b) 72 °C, (c) 73 °C, and (d) 74 °C were determined by measuring EvaGreen fluorescence. Representative amplification curves showing changes in fluorescence (Y-axis) over time (X-axis; minutes) are depicted as blue (Bst-LF), burnt orange (Br512 wild-type), and gray (Mut235) traces.
The increased thermostability of the engineered variants was strongly indicated by the thermal challenge assays. We also carried out a dye-based protein thermal shift assay (TSA) to determine the melting temperatures of the proteins (Figure S10). Br512 showed a slightly higher Tm value (76.1 °C) compared to the parental enzyme Bst-LF (75.5 °C), whereas Mut235 demonstrated a greatly improved Tm value (78.1 °C), further supporting that the computationally predicted substitutions enhanced the thermostability.
DISCUSSION
While there are a variety of DNA polymerases available commercially for isothermal amplification reactions (i.e., Pyrophage 3173 exo-DNA polymerase from Lucigen, IsoPol BST+ from ArcticZymes, Bsm DNA Pplymerase, large fragment from Thermo Fisher, and GspSSD LF DNA polymerase from OptiGene), the primary enzyme used for most diagnostic assays remains Bst DNAP. There have been a number of commercial improvements of the Bst DNAP, notably NEB’s Bst 2.0 and Bst 3.0 enzymes, but there is little scientific literature on the engineering involved in developing these enzymes (including in the patent descriptions themselves).
We therefore set out to develop a series of improvements in Bst DNAP, focusing on understanding the rationales by which this critical enzyme could be engineered. We initially encountered some difficulty purifying the parental Bst-LF enzyme, in part because of solubility issues. In consequence, we appended a portion of the villin headpiece to anchor folding and/or improve stability and solubility.21,22 Indeed, beyond demonstrating the increased thermotolerance relative to Bst LF (Figures 4, S9 and S10), the engineered Br512 derivative greatly improved yields from our purification protocol, ultimately producing up to 35 mg of homogenous protein per liter compared to only 10 mg/L of Bst-LF in a comparable preparation.
It is possible that a cluster of positively charged amino acids that are known to be crucial for the actin-binding activity of the headpiece domain30 may have provided another potential advantage to Br512 in carrying out LAMP reactions (Figure 1b), allowing productive interactions with nucleic acid templates, similar to how Phusion DNA polymerase relies on the Sso7d DNA binding protein from Sulfolobus solfactaricus for enhanced processivity.17 Similarly, a fusion of the Bst-like polymerase Gss-polymerase, and DNA polymerase I from Geobacillus sp. 777 with the DNA-binding domains from the DNA ligase of Pyrococcus abyssi or the Sto7d protein from Sulfolobus tokodaii yielded 3-fold increase in processivity and a 4-fold increase in DNA yield during whole genome amplifications.31
The villin fusion domain also imparted increased thermostability, consonant with its own known thermostability. While the use of the villin headpiece to increase a protein’s thermostability has not previously been attempted, the general opportunities for thermal enhancement via appending DNA-binding fusions and catalytic domains of polymerases has been remarked on,32 with the helix-hairpin-helix DNA-binding domain of topoisomerase V (Topo V) of Methanopyrus kandleri improving the thermal stability of a number of polymerases, including Bst DNAP.33 Interestingly, another DNA-binding domain, Sto7d, a counterpart of Sso7d that was used in the popular Phusion polymerase, did not impart increased thermostability to Bst DNAP,31 possibly indicating that key interactions between the DNA-binding and catalytic domains may be important for stabilization.
There have been a variety of machine learning approaches for understanding the protein sequence, structure, and function. For example, a random forest model has been used to predict solubility,34 support vector machines have been used to predict changes to enzyme stability upon mutagenesis,35,36 and K-nearest neighbor has been used to predict enzyme function (gene ontology).37 Recently, more advanced machine learning algorithms, such as natural language processing algorithms, have shown the ability to recapitulate known protein chemistry phenomena such as physicochemical similarities between the amino acids and secondary structural propensities in an unsupervised fashion.38,39
These algorithms have now begun to be used for engineering proteins, generally in concert with directed evolution approaches. For example, Frances Arnold’s group recently used machine learning approaches to guide the directed evolution of enantiodivergent Rhodothermus marinus nitric oxide dioxygenase variants capable of producing S- and R-enantiomers with 93 and 79% ee, respectively.40 Similarly, Gaussian process models have enabled the rapid evolution of a GFP into over 12 different variants with yellow fluorescence protein41 and of novel thermostable cytochrome p450s.42
While these previous studies demonstrate how machine learning can accelerate the directed evolution of proteins, they are also potentially limited by the innate need to generate a labeled dataset in order to train a supervised model, which is resource- and time-intensive and in many cases not possible (requiring large labeled datasets for sufficient training).43 To alleviate this bottleneck, we used a self-supervised CNN trained on large numbers of known protein structures. Self-supervision is a type of unsupervised learning that consists of generating an artificial label from the data in an automated fashion to guide learning, thus obviating human (and potentially biased) labeling of datasets (Figure S11). For MutCompute, the artificial label is the wild-type amino acid. Since every protein in the Protein Data Bank is the product of evolution, by initially using wild-type amino acid labels, we capture the signal available from evolution on protein stability and functionality and can use this signal for machine learning. By assessing individual microenvironments for every amino acid in a protein structure, MutCompute can identify particular positions that are primed for gain-of-function without the need to curate a genotype-phenotype dataset a priori. However, because our CNN model is not trained with phenotype data, the model’s mutational predictions of “fit” must be assessed experimentally for an actual, desired phenotype. Previously, we have found that such mutational predictions led to improved protein function and solubility;16 we now show that these predictions can lead to thermotolerance and thermostability as well.
Overall, by creating a fusion between Bst DNAP and the villin headpiece and subsequently using machine learning to precisely guide the introduction of strategic mutations, we have created a set of engineered enzymes that are superior not only to the parental Bst-LF DNA polymerase but also surpass the functional limits of one of the most widely used enzymes for isothermal amplification, Bst 2.0. The improved Br512 variants (i) are more robust to purification, generating more than 3-fold higher yields compared to Bst-LF, (ii) achieve time-to-signal and detection limits that are on par with Bst 2.0, and (iii) exhibit greater thermotolerance and thermostability allowing LAMP amplification at temperatures as high as 73 °C, where both Bst-LF and Bst 2.0 are inactivated. The combined impact of the engineered additions can ultimately speed the time-to-signal relative to the parental enzyme by upward of 10 min, allowing LAMP-OSD assays to be conducted in under 15 min. The combined enhancements not only convert the widely available Bst DNAP into a viable resource for conducting LAMP-based diagnostics, especially in resource-poor settings, but, with these studies, also yield a better understood, more robust Bst DNAP chassis for engineering further enzyme improvements.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Science Foundation (2027169), the National Institutes of Health (1R01EB027202-01A1 and 3R01EB027202-01A1S1), the Welch Foundation (F-1654), and the National Aeronautics and Space Administration (NNX15AF46G).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.1c00451.
Schematic diagram of LAMP-OSD; effect of varying amounts of Br512 on LAMP-OSD of DNA templates; comparison of Br512, Mut23, Mut235, Bst-LF, Bst2.0, and Bst3.0 in LAMP-OSD assays of DNA templates; comparison of Br512, Bst-LF, and Bst 2.0 in LAMP assays of DNA templates read using EvaGreen intercalating dye; initial evaluation of computationally predicted substitutions on Br512 (Bst-LF) activity; heat challenge LAMP assay with computationally predicted single amino acid substitutions; heat challenge LAMP assay with double mutation Br512 variants; Ct analysis of triple Mutcompute variants; comparison of Br512, Mut23, Mut235, Bst-LF, Bst2.0, and Bst3.0 in LAMP-OSD assays executed at 73 °C; protein TSA for engineered Bst-LF variants; oligonucleotide and template sequences used in the study; and full sequence of pKAR2-Br512 (PDF)
Accession Codes
Bst DNA polymerase I (Uniprot id: Q45458, PDB id: 3TAN); villin-1 (Uniprot id: P02640).
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.1c00451
The authors declare no competing financial interest.
Contributor Information
Inyup Paik, Department of Molecular Biosciences, College of Natural Sciences, the University of Texas at Austin, Austin, Texas 78712, United States; Center for Systems and Synthetic Biology, The University of Texas at Austin, Austin, Texas 78712, United States;.
Phuoc H. T. Ngo, Department of Molecular Biosciences, College of Natural Sciences, the University of Texas at Austin, Austin, Texas 78712, United States; Center for Systems and Synthetic Biology and Department of Chemistry, College of Natural Sciences, The University of Texas at Austin, Austin, Texas 78712, United States
Raghav Shroff, Department of Molecular Biosciences, College of Natural Sciences, the University of Texas at Austin, Austin, Texas 78712, United States; Center for Systems and Synthetic Biology, The University of Texas at Austin, Austin, Texas 78712, United States; CCDC Army Research Lab-South, Austin, Texas 78712, United States;.
Daniel J. Diaz, Center for Systems and Synthetic Biology and Department of Chemistry, College of Natural Sciences, The University of Texas at Austin, Austin, Texas 78712, United States
Andre C. Maranhao, Department of Molecular Biosciences, College of Natural Sciences, the University of Texas at Austin, Austin, Texas 78712, United States; Center for Systems and Synthetic Biology, The University of Texas at Austin, Austin, Texas 78712, United States
David J.F. Walker, Department of Molecular Biosciences, College of Natural Sciences, the University of Texas at Austin, Austin, Texas 78712, United States; Center for Systems and Synthetic Biology, The University of Texas at Austin, Austin, Texas 78712, United States
Sanchita Bhadra, Department of Molecular Biosciences, College of Natural Sciences, the University of Texas at Austin, Austin, Texas 78712, United States; Center for Systems and Synthetic Biology, The University of Texas at Austin, Austin, Texas 78712, United States;.
Andrew D. Ellington, Department of Molecular Biosciences, College of Natural Sciences, the University of Texas at Austin, Austin, Texas 78712, United States; Center for Systems and Synthetic Biology, The University of Texas at Austin, Austin, Texas 78712, United States;.
REFERENCES
- (1).Aliotta JM; Pelletier JJ; Ware JL; Moran LS; Benner JS; Kong H Thermostable Bst DNA polymerase I lacks a 3′ → 5′ proofreading exonuclease activity. Genet Anal 1996, 12, 185–195. [PubMed] [Google Scholar]
- (2).Nazina TN; Tourova TP; Poltaraus AB; Novikova EV; Grigoryan AA; Ivanova AE; Lysenko AM; Petrunyaka VV; Osipov GA; Belyaev SS; Ivanov MV Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int. J. Syst. Evol. Microbiol 2001, 51, 433–446. [DOI] [PubMed] [Google Scholar]
- (3).Panno S; Matic S; Tiberini A; Caruso AG; Bella P; Torta L; Stassi R; Davino AS Loop Mediated Isothermal Amplification: Principles and Applications in Plant Virology. Plants 2020, 9, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Park G-S; Ku K; Baek S-H; Kim S-J; Kim SI; Kim B-T; Maeng J-S Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mod. Dynam 2020, 22, 729–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Huang WE; Lim B; Hsu CC; Xiong D; Wu W; Yu Y; Jia H; Wang Y; Zeng Y; Ji M; Chang H; Zhang X; Wang H; Cui Z RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. J. Microb. Biotechnol 2020, 13, 950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yan C; Cui J; Huang L; Du B; Chen L; Xue G; Li S; Zhang W; Zhao L; Sun Y; Yao H; Li N; Zhao H; Feng Y; Liu S; Zhang Q; Liu D; Yuan J Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect 2020, 26, 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Thi VLD; Herbst K; Boerner K; Meurer M; Kremer LP; Kirrmaier D; Freistaedter A; Papagiannidis D; Galmozzi C; Stanifer ML; Boulant S; Klein S; Chlanda P; Khalid D; Barreto Miranda I; Schnitzler P; Krausslich HG; Knop M; Anders S A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med 2020, 12, No. eabc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Notomi T; Okayama H; Masubuchi H; Yonekawa T; Watanabe K; Amino N; Hase T Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, No. e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Jiang YS; Bhadra S; Li B; Wu YR; Milligan JN; Ellington AD Robust strand exchange reactions for the sequence-specific, real-time detection of nucleic acid amplicons. Anal. Chem 2015, 87, 3314–3320. [DOI] [PubMed] [Google Scholar]
- (10).Coulther TA; Stern HR; Beuning PJ Engineering Polymerases for New Functions. Trends Biotechnol. 2019, 37, 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Nikoomanzar A; Chim N; Yik EJ; Chaput JC Engineering polymerases for applications in synthetic biology. Q. Rev. Biophys 2020, 53, No. e8. [DOI] [PubMed] [Google Scholar]
- (12).Ma Y; Zhang B; Wang M; Ou Y; Wang J; Li S Enhancement of Polymerase Activity of the Large Fragment in DNA Polymerase I from Geobacillus stearothermophilus by Site-Directed Mutagenesis at the Active Site. BioMed Res. Int 2016, 2016, 2906484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Sandalli C; Singh K; Modak MJ; Ketkar A; Canakci S; Demir İ; Belduz AO A new DNA polymerase I from Geobacillus caldoxylosilyticus TK4: cloning, characterization, and mutational analysis of two aromatic residues. Appl. Microbiol. Biotechnol 2009, 84, 105–117. [DOI] [PubMed] [Google Scholar]
- (14).Piotrowski Y; Gurung MK; Larsen AN Characterization and engineering of a DNA polymerase reveals a single amino-acid substitution in the fingers subdomain to increase strand-displacement activity of A-family prokaryotic DNA polymerases. BMC Mol. Cell Biol 2019, 20, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Torng W; Altman RB 3D deep convolutional neural networks for amino acid environment similarity analysis. BMC Bioinf. 2017, 18, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Shroff R; Cole AW; Diaz DJ; Morrow BR; Donnell I; Annapareddy A; Gollihar J; Ellington AD; Thyer R Discovery of Novel Gain-of-Function Mutations Guided by Structure-Based Deep Learning. ACS Synth. Biol 2020, 9, 2927–2935. [DOI] [PubMed] [Google Scholar]
- (17).Wang Y; Prosen DE; Mei L; Sullivan JC; Finney M; Horn PBV A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res. 2004, 32, 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bazari WL; Matsudaira P; Wallek M; Smeal T; Jakes R; Ahmed Y Villin sequence and peptide map identify six homologous domains. Proc. Natl. Acad. Sci. U.S.A 1988, 85, 4986–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Paik I; Ngo PH; Shroff R; Maranhao AC, Walker DJ; Bhadra S; Ellington AD Multi-modal engineering of Bst DNA polymerase for thermostability in ultra-fast LAMP reactions, Cold Spring Harbor Laboratory, bioRxiv, 2021, DOI: 10.1101/2021.04.15.439918. [DOI] [Google Scholar]
- (20).Maranhao A; Bhadra S; Paik I; Walker D; Ellington AD An improved and readily available version of Bst DNA Polymerase for LAMP, and applications to COVID-19 diagnostics. MedRxiv 2020, DOI: 10.1101/2020.10.02.20203356. [DOI] [Google Scholar]
- (21).Chiu TK; Kubelka J; Herbst-Irmer R; Eaton WA; Hofrichter J; Davies DR High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein. Proc. Natl. Acad. Sci. U.S.A 2005, 102, 7517–7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).McKnight JC; Doering DS; Matsudaira PT; Kim PS A thermostable 35-residue subdomain within villin headpiece. J. Mol. Biol 1996, 260, 126–134. [DOI] [PubMed] [Google Scholar]
- (23).Lei H; Wu C; Liu H; Duan Y Folding free-energy landscape of villin headpiece subdomain from molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A 2007, 104, 4925–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Tanner NA; Evans TC Jr. Loop-mediated isothermal amplification for detection of nucleic acids. Curr. Protoc. Mol. Biol 2014, 105, 15.14. [DOI] [PubMed] [Google Scholar]
- (25).Hsieh K; Mage PL; Csordas AT; Eisenstein M; Tom Soh H Simultaneous elimination of carryover contamination and detection of DNA with uracil-DNA-glycosylase-supplemented loop-mediated isothermal amplification (UDG-LAMP). Chem. Commun 2014, 50, 3747–3749. [DOI] [PubMed] [Google Scholar]
- (26).Lawyer FC; Stoffel S; Saiki RK; Chang SY; Landre PA; Abramson RD; Gelfand DH High-level expression, purification, and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5’ to 3’ exonuclease activity. PCR Methods Appl. 1993, 2, 275–287. [DOI] [PubMed] [Google Scholar]
- (27).Esbin MN; Whitney ON; Chong S; Maurer A; Darzacq X; Tjian R Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Bhadra S; Riedel TE; Lakhotia S; Tran ND; Ellington AD High-Surety Isothermal Amplification and Detection of SARS-CoV-2. mSphere 2021, 6, 00911–00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Rolando JC; Jue E; Barlow JT; Ismagilov RF Real-time kinetics and high-resolution melt curves in single-molecule digital LAMP to differentiate and study specific and non-specific amplification. Nucleic Acids Res. 2020, 48, No. e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Friederich E; Vancompernolle K; Huet C; Goethals M; Finidori J; Vandekerckhove J; Louvard D An actin-binding site containing a conserved motif of charged amino acid residues is essential for the morphogenic effect of villin. Cell 1992, 70, 81–92. [DOI] [PubMed] [Google Scholar]
- (31).Oscorbin IP; Belousova EA; Boyarskikh UA; Zakabunin AI; Khrapov EA; Filipenko ML Derivatives of Bst-like Gsspolymerase with improved processivity and inhibitor tolerance. Nucleic Acids Res. 2017, 45, 9595–9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ishino S; Ishino Y DNA polymerases as useful reagents for biotechnology—the history of developmental research in the field. Front. Microbiol 2014, 5, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Pavlov AR; Pavlova NV; Kozyavkin SA; Slesarev AI Cooperation between Catalytic and DNA Binding Domains Enhances Thermostability and Supports DNA Synthesis at Higher Temperatures by Thermostable DNA Polymerases. Biochemistry 2012, 51, 2032–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Yang Y; Niroula A; Shen B; Vihinen M PON-Sol: prediction of effects of amino acid substitutions on protein solubility. Bioinformatics 2016, 32, 2032–2034. [DOI] [PubMed] [Google Scholar]
- (35).Teng S; Srivastava AK; Wang L Sequence feature-based prediction of protein stability changes upon amino acid substitutions. BMC Genom. 2010, 2, S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Folkman L; Stantic B; Sattar A; Zhou Y EASE-MM: Sequence-Based Prediction of Mutation-Induced Stability Changes with Feature-Based Multiple Models. J. Mol. Biol 2016, 428, 1394–1405. [DOI] [PubMed] [Google Scholar]
- (37).Koskinen P; Törönen P; Nokso-Koivisto J; Holm L PANNZER: high-throughput functional annotation of uncharacterized proteins in an error-prone environment. Bioinformatics 2015, 31, 1544–1552. [DOI] [PubMed] [Google Scholar]
- (38).Alley EC; Khimulya G; Biswas S; AlQuraishi M; Church GM Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 2019, 16, 1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Rao R; Bhattacharya N; Thomas N; Duan Y; Chen X; Canny J; Abbeel P; Song YS Evaluating Protein Transfer Learning with TAPE. Adv. Neural Inf. Process. Syst 2019, 32, 9689–9701. [PMC free article] [PubMed] [Google Scholar]
- (40).Wu Z; Kan SBJ; Lewis RD; Wittmann BJ; Arnold FH Machine learning-assisted directed protein evolution with combinatorial libraries. Proc. Natl. Acad. Sci. U.S.A 2019, 116, 8852–8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Saito Y; Oikawa M; Nakazawa H; Niide T; Kameda T; Tsuda K; Umetsu M Machine-Learning-Guided Mutagenesis for Directed Evolution of Fluorescent Proteins. ACS Synth. Biol 2018, 7, 2014–2022. [DOI] [PubMed] [Google Scholar]
- (42).Romero PA; Krause A; Arnold FH Navigating the protein fitness landscape with Gaussian processes. Proc. Natl. Acad. Sci. U.S.A 2013, 110, E193–E201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wittmann BJ; Johnston KE; Wu Z; Arnold FH Advances in machine learning for directed evolution. Curr. Opin. Struct. Biol 2021, 69, 11–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


