Abstract
We have successfully generated induced pluripotent stem cells (iPSC) from dermal fibroblasts and peripheral blood mononuclear cells from patients with a homozygous missense mutation in the gene encoding PSMB8. Biallelic loss of function mutations in this gene are responsible for the PSMB8 deficiency termed Chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature (CANDLE). The iPSC carrying the homozygous PSMB8 gene mutation (c.224C>T, T75M) are phenotypically normal and have the capacity to differentiate toward the three germ layers. These iPSC have great potential to study the role of PMSB8 in the regulation of immune responses and other cellular pathways.
Resource Utility
Human induced pluripotent stem cells (hiPSC) carrying homozygous PSMB8 gene mutations possess the potential of differentiating into variety of cell types including immune cells. The derivatives sustaining the mutation could be a powerful platform allowing for the investigation of the molecular mechanisms and identification of potential therapeutic targets in patients.
Resource Details
Chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature (CANDLE) syndrome, now also classified within the broader proteasome-associated autoinflammatory syndromes (PRAAS), is a rare, mendelian auto-inflammatory disease related to loss-of-function genetic mutations of proteasome components resulting in proteasomal dysfunction (Brehm et al., 2015, Jesus et al., 2019). With the cell’s protein homeostasis disrupted, it is believed the resulting intracellular stress from accumulating polyubiquinated proteins is one of the mechanisms that cause an inflammatory state seen in patients associated with a strong interferon response gene signature thought to be driven by type 1, and to a lesser extent type 2, interferon (Brehm et al., 2015). Clinical features of CANDLE syndrome patients vary, but includes sterile fevers, a pathognomonic erythematous rash, lipodystrophy, panniculitis, muscloskeletal involvement, metabolic derangements, central nervous system disorders, and vascular disorders such as systemic and pulmonary arterial hypertension (Brehm et al., 2015, Liu et al., 2012). Though it is now known that digenic mutations can occur with PRAAS phenotypes, initial reporting for CANDLE syndrome was with the homozygous PSMB8 mutation which encodes the B5i subunit containing the chemotrypsin-like enzymatic activity of the immuno-proteasome (Brehm et al., 2015, Liu et al., 2012). The NIH CANDLE/PRAAS cohort’s most common founder mutation is the homozygous p.T75M missense mutation seen in mostly Spanish, Portuguese, and Latin American patients (Liu et al., 2012). Specific treatment includes JAK-STAT 1/2 inhibitors with resulting improvement of the interferon response gene signature and clinical symptoms in most, but not all, patients (Jesus et al., 2019). The exact molecular mechanism of intracellular stress and inflammation has not truly been elucidated in CANDLE/PRAAS patients with the homozygous PSMB8 p.T75M mutation, both in hemopoietic cells and nonhemopoietic cell types (which do not constitutively express the immunoproteasome).
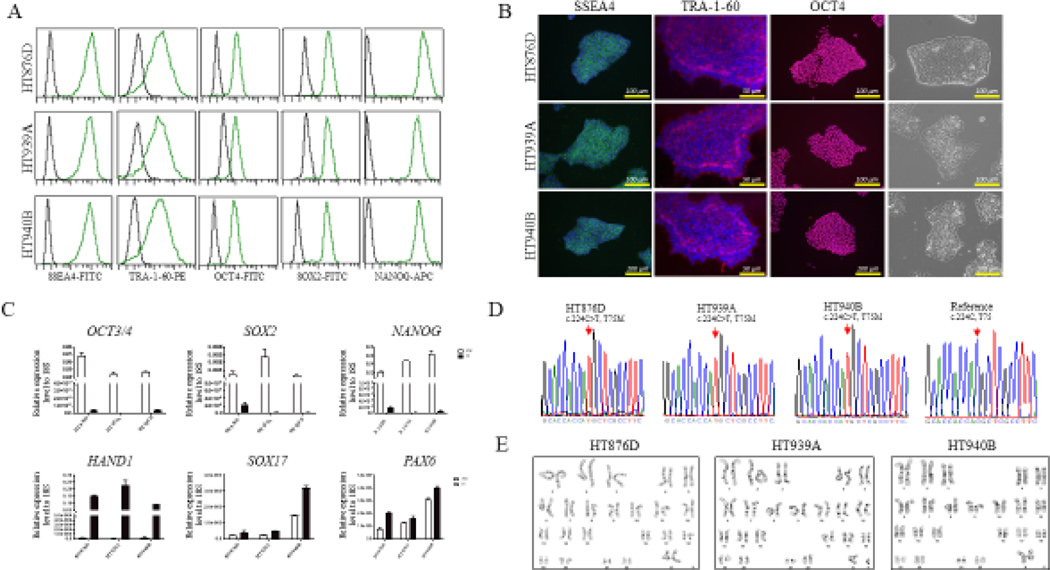
We identified three patients carrying the homozygous missense mutation in the PSMB8 gene by exome sequencing. The patients were enrolled into our NHLBI clinical protocols for further investigation. Information regarding clinical features of this patients were obtained using the standard clinical questionnaire (Table 1). Skin punch biopsy and peripheral blood samples were collected at the NIH Clinical Center. Using a Sendai-OKSM delivery system expressing four transcription factors (OCT4, SOX2, KLF4, and C-MYC), we successfully generated iPSC lines from skin fibroblasts or peripheral blood cells from each patient (HT876D, HT939A, and HT940B). Each iPSC line maintained the typical morphology of iPSCs and expressed the pluripotency markers OCT4, NANOG, TRA-160, SSEA4, and SOX2, as shown by FACS (Figure 1A), immunocytochemistry (Figure 1B), and/or real-time (RT)-qPCR (Figure 1C). The iPSC lines were free of Sendai virus confirmed by RT-PCR at the 15th passage (Supplement file 2). Genotyping of the generated iPSC lines confirmed the point mutation (c.224C>T, T75M) in the PSMB8 gene that were the same as their parental cells (Figure 1D). To test the differentiation potential of the iPSC lines, we performed a monolayer differentiation assay to drive the cells towards the three germ layers in vitro. We determined the marker gene expression for the mesoderm (HAND1, RUNX1), endoderm (SOX17, AFP), and ectoderm (PAX6, NESTIN) by RT-qPCR, which showed that all three iPSC lines are able to differentiate into mesoderm, endoderm, and ectoderm cells (Figure 1C, Supplement file 1). Short tandem repeat (STR) profiles indicated that the iPSC lines matched with its parental cells completely in 15 amplified STR loci. All cultures were routinely tested for Mycoplasma contamination and were found to be Mycoplasma free (Supplementary file 2). All three iPSC lines demonstrated chromosomal stability and a normal karyotype with G-banding (Figure 1E). Overall, the three patient derived iPSC lines exhibited the pluripotent potential for self-renew and differentiation, suggesting the successful generation of iPSCs from CANDLE patients.
Table 1.
Summary of three individuals with CANDLE patients
| iPSC line names | Abbreviation in figures | Gender | Current Age (years) | Ethnicity | Genotype of locus | Disease |
|---|---|---|---|---|---|---|
| NIHTVBi 016-A | HT876D | F | 22 | Hispanic/Latino | PSMB8, c.224C>T, p.T75M | CANDLE |
| NIHTVBi 017-A | HT939A | M | 6 | Not Hispanic | PSMB8, c.224C>T, p.T75M | CANDLE |
| NIHTVBi 018-A | HT940B | F | 41 | Hispanic/Latino | PSMB8, c.224C>T, p.T75M | CANDLE |
Fig 1.
Generation and characterization of human induced pluripotent stem cells (iPSCs) from three CANDLE syndrome patients carried a homozygous mutation in the PSMB8 gene.
Materials and Methods
Subjects and Derivation of fibroblasts
The fibroblasts were derived from skin punch biopsy samples obtained from the CANDLE patients with the homozygous PMSB8 gene mutation. Cells were propagated as previously described (Chen et al., 2019). This study was approved by the NHLBI’s institutional review board, and samples were collected after obtaining informed written consent.
Generation and culture of human iPSC from fibroblasts or peripheral mononuclear blood cells
Fibroblasts or peripheral mononuclear blood cells from the CANDLE patients were reprogrammed and expanded in a typical hESC/iPSC culture condition as previously described (Chen et al., 2019).
Immunofluorescent staining and Flow Cytometry Analysis (FACS)
iPSC colonies (passage 12) were fixed with 4% paraformaldehyde and stained for NANOG, SOX2, SSEA4, and TRA-1–60 (Table 3) following the previous protocol (Chen et al., 2019).
Table 3.
Reagents
| Antibodies used for immunocytochemistry and FACS | |||||
|---|---|---|---|---|---|
|
| |||||
| Antibody | Dilution | Company | Cat# | RRID | |
| Primary antibodies | Mouse anti-SSEA4 | 1:100 | MilliporeSigma | MAB4304 | AB_177629 |
| Mouse anti-TRA-1–60 | 1:150 | MilliporeSigma | MAB4360 | AB_2119183 | |
| Alexa Fluor 488 antiSSEA4 Antibody | 1:10 | BioLegend | 330412 | AB_1089198 | |
| PE anti-TRA-1–60 Antibody | 1:10 | BioLegend | 330610 | AB_2119065 | |
| Alexa Fluor 488 anti-SOX2 Antibody | 1:10 | BioLegend | 656110 | AB_2563957 | |
| Alexa Fluor 488 anti-OCT4 Antibody | 1:10 | BioLegend | 653708 | AB_2563184 | |
| Alexa Fluor 647 anti-NANOG Antibody | 1:10 | BioLegend | 674210 | AB_2650619 | |
| Secondary antibodies | Alexa Fluor 594 Donkey anti-rabbit | 1:300 | Life Technologies | A21207 | AB_141637 |
| Alexa Fluor 594 Donkey anti-mouse | 1:300 | Life Technologies | A21203 | AB_141633 | |
| Alexa Fluor 488 Donkey anti-mouse | 1:300 | Life Technologies | A21202 | AB_141607 | |
| Primers used for RT-qPCR and PCR | |
|---|---|
|
| |
| Target | Forward/reverse primer (5’−3’) |
| NANOG | AGG GAA ACA ACC CAC TTC T/CCT TCT GCG TCA CAC CAT T |
| SOX2 | CCC AGC AGA CTT CAC ATG T/CCT CCC ATT TCC CTC GTT TT |
| AFP | AGC TTG GTG GAT GAA AC/CCC TCT TCA GCA AAG CAG AC |
| NESTIN | GCG TTG GAA CAG AGG TTG GA/TGG GAG CAA AGA TCC AAG AC |
| RUNX1 | CTG CCC ATC GCT TTC AAG GT/GCC GAG TAG TTT TCA TTG CC |
| PMSB8 | CCT TCT CCC AAG CCA TTT CC /TCC CAG TAC TGA CAG TCT GC |
FACS data acquisition and analysis was performed on the iPSCs (passage 12) for the designed antibodies (Table 3) as previously described (Chen et al., 2019).
Monolayer differentiation assay
The in vitro assessment of iPSC differentiation ability at passage 12 was performed as per the previous protocol with harvesting of cells for analysis at day 7 (Chen et al., 2019).
Gene expression analysis by RT-PCR
Total RNA isolation, cDNA synthesis, and PCR using the primers indicated in Table 2 was performed as previously described (Chen et al., 2019).with the iPSC obtained at passage 5 serving as the positive control (Pos)(Supplement file 3).
Table 2.
Characterization and validation
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Phase-contrast microscope | Normal | Figure 1B |
| Phenotype | Qualitative analysis (immunofluorescence staining) | Expression of pluripotency markers: OCT4, NANOG, SSEA4 and TRA-1–60 | Figure 1B |
| Quantitative analysis (RT-qPCR) | Expression of pluripotency markers: SOX2 and NANOG | Figure 1C | |
| Qualitative analysis (FACS) | Expression of pluripotency markers: NANOG, SOX2, SSEA4, TRA-1–60, OCT4 | Figure 1A | |
| Genotype | Karyotype (G-banding) and resolution | 46, XY; resolution 450–500 bands | Figure 1E |
| Identity | Microsatellite PCR OR STR analysis | Not performed 15 sites tested, 100% match | N/A Supplementary file 2 |
| Mutation analysis (IF APPLICABLE) | DNA sequencing | Homozygous, PMSB8 gene point mutation | Figure 1D |
| Southern blot OR WGS | Not performed | N/A | |
| Microbiology and virology | Mycoplasma testing by luminescence | Negative | Supplementary file 3 |
| Differentiation potential | Monolayer differentiation assay | Differentiating cells are expression of RUNX1, AFP, and NESTIN; iPSC were able to differentiate into three germ layers | Figure 1C |
| Donor screening (OPTIONAL) | HIV1 + HIV2, hepatitis B virus, hepatitis C virus | Not performed | N/A |
| Genotype additional | Blood group genotyping | Not performed | N/A |
| info (OPTIONAL) | HLA tissue typing | Not performed | N/A |
After 15 passages, iPSC was tested for Sendai virus (SeV) residues as described (Alonso-Barroso et al., 2017).
Endogenous mRNA expression levels of OC4, NANOG, SOX2, AFP, SOX17, PAX6, NESTIN, HAND1 and RUNX1 were determined in iPSC (iPSC) and in differentiating cells at day 7 (differentiated) (Figure 1C). For this, RT-qPCR was performed as previously described (Chen et al., 2019) using the primers shown in Table 3.
Karyotyping assay
The karyotype of the iPSC was evaluated by the WiCell Research Institute using G-banding metaphase karyotype analysis.
DNA sequencing and STR
DNA sequencing and STR analysis was derived from previous methods (Chen et al., 2019) with exception being that PCR was performed with specific primers (Table 3) in order to amplify the corresponding deletion position in PMSB8 gene.
Mycoplasma detection
Mycoplasma testing was performed on the supernatant of iPSCs as per the previous protocol (Chen et al., 2019) using the MycoAlert™ Mycoplasma Detection Kit (Lonza, LT27–224) (Supplement file 2).
Supplementary Material
Resource Table
| Unique stem cell lines identifier | NIHTVBi016-A |
| NIHTVBi017-A | |
| NIHTVBi018-A | |
| Alternative names of stem cell lines | HT876D (NIHTVBi016-A) |
| HT939A (NIHTVBi017-A) | |
| HT940B (NIHTVBi018-A) | |
| Institution | National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH), Bethesda, Maryland, USA |
| Contact information of distributor | Manfred Boehm; boehmm@nhlbi.nih.gov |
| Type of cell lines | iPSC |
| Origin | Human |
| Cell Source | Dermal fibroblasts and peripheral blood cells |
| Clonality | Clonal cell lines |
| Method of reprogramming | Sendai-virus vectors containing the transcription factors Oct-4, Klf4, Sox2 and c-MYC |
| Multiline rationale | Lines derived from the individual |
| Gene modification | Yes |
| Type of modification | Hereditary |
| Assoicated disease | Chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature (CANDLE) syndrome |
| Gene/locus | PMSB8 |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | July 1, 2021 |
| Cell line repository/bank |
https://hpscreg.eu/cell-line/NIHTVBi016-A
https://hpscreg.eu/cell-line/NIHTVBi017-A https://hpscreg.eu/cell-line/NIHTVBi018-A |
| Ethical approval | NCT02974595 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brehm A, Liu Y, Sheikh A, Marrero B, Omoyinmi E, Zhou Q, et al. Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J Clin Invest. 2015; 125: 4196–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesus AA, Brehm A, VanTries R, Pillet P, Parentelli AS, Montealegre Sanchez GA, Deng Z, Paut IK, Goldbach-Mansky R, Krüger E. Novel proteasome assembly chaperone mutations in PSMG2/PAC2 cause the autoinflammatory interferonopathy CANDLE/PRAAS4. J Allergy Clin Immunol. 2019. May;143(5):1939–1943.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ramot Y, Torrelo A, Paller AS, Si N, Babay S, Kim PW, Sheikh A, Lee CC, Chen Y, Vera A, Zhang X, Goldbach-Mansky R, Zlotogorski A. Mutations in PSMB8 cause CANDLE syndrome with evidence of genetic and phenotypic heterogeneity. Arthritis Rheum. 2012;64(3):895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Jin H, Yu Z, Liu Y, Li Z, Navarengom K, Schwartzbeck R, Dmitrieva N, Cudrici C, Ferrante EA, Biesecker LG, Yang D, Boehm M. Generation of human induced pluripotent stem cells from individuals with a homozygous CCR5Δ32 mutation. Stem Cell Research, 38 (2019), 101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Barroso E, Brasil S, Briso-Montiano A, Navarrete R, Perez-Cerda C, Ugarte M, Perez B, Desviat LR, Richard E. Generation and characterization of a human iPSC line from a patient with propionic acidemia due to defects in the PCCA gene. Stem Cell Research, 23 (2017), 173–177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.