Abstract
Pseudomonas stutzeri has type IV pili for which the pilA gene (here termed pilAI) provides the structural protein and which are required for DNA uptake and natural genetic transformation. Downstream of pilAI we identified a gene, termed pilAII, coding for a deduced protein with a size similar to that of PilAI with 55% amino acid sequence identity and with a typical leader peptide including a leader peptidase cleavage site. Fusions to lacZ revealed that pilAII is expressed only about 10% compared to pilAI, although the genes are cotranscribed as shown by reverse transcription-PCR. Surprisingly, insertional inactivation of pilAII produced a hypertransformation phenotype giving about 16-fold-increased transformation frequencies. Hypertransformation also occurred in pilAI pilAII double mutants expressing heterologous pilA genes of nontransformable bacteria, like Pseudomonas aeruginosa or Dichelobacter nodosus. The overexpression of pilAII decreased transformation up to 5,000-fold compared to that of the pilAII mutant. However, neither inactivation of pilAII nor its overexpression affected the amounts of [3H]thymidine-labeled DNA that were competence-specifically bound and taken up by the cells. In the pilAII mutant, the transformation by purified single-stranded DNA (which depends on comA and exbB, as does transformation by duplex DNA) was also increased 17-fold. It is concluded that PilAII suppresses a step in transformation after the uptake of duplex DNA into the cell and perhaps before its translocation into the cytoplasm. The idea that the degree of the transformability of cells could be permanently adjusted by the expression level of an antagonistic protein is discussed.
The gram-negative bacterium Pseudomonas stutzeri is ubiquitous and lives in terrestrial habitats and freshwater and marine environments. It is among the species capable of natural genetic transformation (7, 21). This horizontal gene transfer mechanism involves the active uptake of free DNA by the cell from the environment and the heritable incorporation of its informational content into the genome. Transformation is thought to contribute to adaptation of populations to a changing environment and thereby to evolution and speciation (12, 21). Recently it was shown by electron microscopy that cells of P. stutzeri strain JM300 have type IV pili (15), which are found in a large number of diverse gram-negative bacteria (18, 41). Type IV pili can mediate the interaction of bacteria with surfaces, including those of epithelial cells, and may allow the flagellum-independent translocation of bacteria on solid surfaces, such as on agar medium (41, 45). The phenomenon was termed twitching motility and has been observed in about 20 species, including P. stutzeri (5, 18, 41, 45). Mutations preventing pilus formation abolished twitching motility in P. stutzeri (15). It was also found that the pili allow the infection of P. stutzeri by the pilus-specific phage PO4 (15), which was initially isolated with Pseudomonas aeruginosa as a host (4). Several genes involved in the pilus biogenesis of P. stutzeri were identified, including pilA, pilB, pilC, and pilD (15), and their putative proteins showed extensive amino acid sequence similarity to proteins which are required for pilus formation in P. aeruginosa and other bacteria (14, 24, 41). Among them, the pilA gene codes for prepilin, which is proteolytically processed and transported through the cytoplasm membrane and then forms the pilus by polymerization.
In P. stutzeri, inactivation of pilA not only prevented formation of type IV pili accompanied by loss of twitching motility and PO4 infection but also blocked natural transformation and strongly decreased DNA uptake, indicating a function of pili in the internalization of transforming DNA (15). All these defects were fully complemented by the presence of the gene for the structural protein of pili from other bacteria, including Dichelobacter nodosus and P. aeruginosa (15). In Neisseria gonorrhoeae and Legionella pneumophila, inactivation of the gene for the structural protein of type IV pili prevents pilus formation and also natural transformation (14, 40). During DNA sequencing near the pilABCD region of P. stutzeri we identified a gene downstream of pilA coding for a putative protein with high amino acid sequence identity to PilA. Here we report on this gene, which we termed pilAII, and show that it limits the natural level of transformation of the cells. Its normal expression keeps transformability at a low level, while the insertional inactivation produces a hypertransformation phenotype. Our studies provide evidence that the suppression of transformation is caused by interference of PilAII with a step subsequent to DNA uptake into the cell and before DNA translocation into the cytoplasm.
MATERIALS AND METHODS
Bacterial strains and microbiological methods
The bacterial strains and plasmids used are listed in Table 1. The broth media for P. stutzeri and Escherichia coli were Luria-Bertani (LB) agar plates or LB liquid medium (34). If necessary, LB medium was supplemented with ampicillin (1 g liter−1 for P. stutzeri and 100 mg liter−1 for E. coli), gentamicin (10 mg liter−1), kanamycin (60 mg liter−1), rifampin (20 mg liter−1), or nalidixic acid (50 mg liter−1). Strains with pSG plasmids were grown on media with 1 mM isopropylthiogalactoside (IPTG). The minimal medium for P. stutzeri was minimal pyruvate agar medium (20). Incubations were done at 37°C. β-Galactosidase activity was determined as described previously (27). One unit of activity corresponds to 1 nmol of o-nitrophenyl-β-d-galactoside hydrolyzed per min.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference(s) |
|---|---|---|
| Strains | ||
| E. coli K-12 | ||
| SF8 recA | recA56 | 16 |
| XL10 | recA Tcr | Stratagene |
| P. stutzeri | ||
| JM375 | Rifr Smr | 7 |
| JM302 | hisX1 | 7, 38 |
| LO15 | JM302 Rifr Nalr | 15 |
| Tf300 | LO15 pilAI::Gmr (insertion in BglII site) | 15 |
| Tf301 | LO15 pilAI::lacZ-Gmr (insertion in BglII site) | This study |
| Tf400 | LO15 pilAII::Gmr (insertion in SacII site) | This study |
| Tf401 | LO15 pilAII::lacZ-Gmr (insertion in SacII site) | This study |
| Tf500 | Tf400 pilAI::Kmr (insertion in BglII site) | This study |
| Plasmids | ||
| pST81 | About 40 kb of religated chromosomal DNA of Tf81 including inserted pSUP102-Gm::Tn5Bn20 | 15 |
| pUCP19, pUCP18 | oricolE1 oripRO1600 Apr | 36 |
| pUCGmr | Apr Gmr | 37 |
| pAB2001 | AprlacZ-Gmr cassette | 3 |
| pAW102-O | pUCP19 carrying pilA+ of P. aeruginosa PAO | 46 |
| pAW103-K | pUCP19 carrying pilA+ of P. aeruginosa PAK | 46 |
| pAW107-Dn | pUCP18 carrying fimA+ of D. nodosus | 46 |
| pSG | pBRR1 derivative containing tac promoter and lac1q | This study |
| pSGA2 | pSG carrying pilAII+ of LO15 under control of the tac promoter | This study |
| pUCA1 | pUCP19 carrying pilAI+ of LO15 | 15 |
| pUCA1/2 | pUCP19 carrying the pilAI+pilAII+ region of LO15 | This study |
| pUCA2lacZ-Gmr | pUCA1/2 carrying a lacZ-Gmr cassette in the SacII site of pilAII | This study |
| pBluescript KS+ | Apr | Stratagene |
| pKSA1/2 | pBluescript KS+ carrying the pilAI+pilAII+ region of LO15 | This study |
| pKSA2lacZ-Gmr | pKSA1/2 carrying a lacZ-Gmr cassette in the SphI site of pilAII | This study |
| pKSA1Kmr | pKSA1/2 carrying a Kmr gene in the BglII site of pilAI | This study |
| pKSA1lacZ-Gmr | pKSA1/2 carrying a lacZ-Gmr cassette in the BglII site of pilAI | This study |
| pUCA2Gmr | pUCA1/2 carrying a Gmr gene in the SacII site of pilAII | This study |
| pPM1 | pBluescript SK+ with the 2.18-kb PstI fragment covering hisX+ | P. Meier, C. Berndt, N. Weger, and W. Wackernagel |
DNA manipulations and plasmid and strain constructions.
Plasmids and genomic DNA were prepared using Qiagen (Hilden, Germany) columns according to the instructions of the manufacturer. Electrocompetent cells of P. stutzeri and E. coli were prepared according to the methods of Pemberton and Penfold (29) and Dower et al. (11), respectively. Electroporation conditions used with the Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.) were 12.5 kV cm−1, 25 μF, and 200 Ω. E. coli XL10 was used for transformation with derivatives of pBluescript, and strain E. coli SF8 recA was used for transformation with the other plasmids.
Chromosomal integration of mutant alleles was achieved by homologous recombination during plate transformation. The plasmids were linearized by restriction enzyme cleavage before transformation to avoid cointegrate formation by single-crossover events. Selection was performed on plates with gentamicin or kanamycin. The correct integration of the insert in the genomic DNA was verified by PCR analysis.
The pilA region from nucleotide positions 220 to 1233 (Fig. 1), including the putative promoter region and the pilAI+ and pilAII+ genes, was amplified by PCR from chromosomal DNA of LO15 (using the primers PilAPro4 [5′-CATGCCGGCATACTAGACAT-3′] and PilA5 [5′-TTAGAGGCATTTCCCCGGCTTATATCGG-3′]). The PCR product was purified using the PCR purification kit from Qiagen and cloned into the SmaI sites of pUCP19 and pBluescript KS(+) to yield pUCA1/2 and pKSA1/2, respectively (Table 1). For construction of strain Tf400, pUCA1/2 was treated with SacII (cutting in pilAII) and ligated to a gentamicin resistance gene amplified by PCR from pUCGmr DNA (36) using the primers Gm1 (5′-CAGCGGTGGTAACGGCGCAG-3′) and Gm2 (5′-TTTACCGAACAACTCCGCGG-3′). The pilAII::Gmr region present in the resulting plasmid, pUCA2Gmr, was integrated into the chromosome of LO15 by plate transformation to yield Tf400 (Table 1). For construction of Tf500, pKSA1/2 was treated with BglII (cutting in pilAI) and ligated to a kanamycin resistance gene amplified by PCR from pSKKmr using primers Km1 (5′-ATGGCGATAGCTAGACTGGG-3′) and Km2 (5′-TGGTCGGTCATTTCGAACCC-3′) to obtain pKSA1Kmr. The pilAI::Kmr region of pKSA1Kmr was integrated by plate transformation into the chromosome of Tf400, which already had a Gmr gene in pilAII, yielding Tf500 (Table 1). The pilAI::lacZ-Gmr allele and the two pilAII::lacZ-Gmr alleles were constructed by ligation of the lacZ-Gmr cassette (3) into the BglII or SphI site of pKSA1/2 or the SacII site of pUCA1/2 (Fig. 1), yielding pKSA1lacZ-Gmr, pKSA2lacZ-Gmr, or pUCA2lacZ-Gmr, respectively. These plasmids were used for plate transformation of LO15 to obtain Tf301 and Tf401. For cloning of pilAII+, the gene was amplified from chromosomal DNA of LO15 by PCR using the primers PilA6 (5′-tttaacggaattccgttaaaggaggatatattaATGAAGCCCGTGAAATCTTCGTGCC-3′) containing an EcoRI site (boldface type) and a ribosome binding site (italicized) and PilA5 (5′-gggaattcctcgagcatatgTTAGAGGCATTTCCCCGGCTTATATCGG-3′) containing an XhoI site, shown in boldface type (capital letters indicate DNA homologous to the pilAII+ gene). The PCR product was purified as described above, digested with EcoRI and XhoI, and ligated to EcoRI/XhoI-digested pSG vector DNA to obtain pSGA2. The pSG vector is a derivative of pBBR1 (2) carrying lacIq and the tac promoter upstream of the multiple-cloning site, so that in pSGA2 the pilAII+ gene is under the control of the IPTG-inducible tac promoter.
FIG. 1.
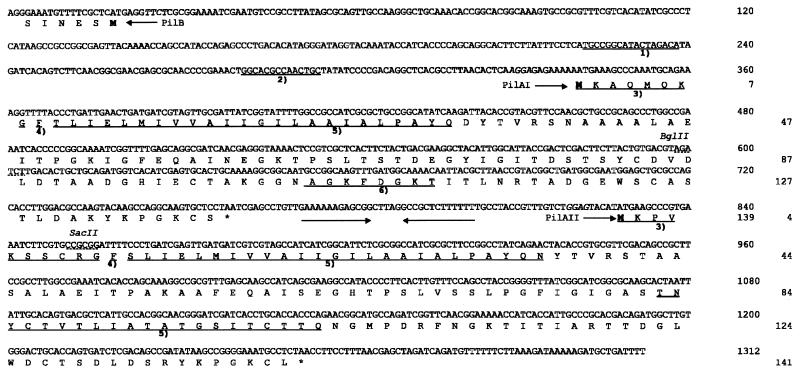
Nucleotide and protein sequences of the pilA region. The nucleotide sequence of the pilA region and the deduced amino acid sequences for PilB (partial), PilAI, and PilAII are shown in the one-letter code. The cloning sites for insertional inactivation of pilAI+ and pilAII+ are indicated (dashed lines), as are the names of the restriction endonucleases used. Putative ribosome binding sites are printed in italics. The putative start of a protein is indicated by a methionine printed in boldface type. ∗, stop codon. A putative rho-independent transcriptional terminator is indicated by two inverted arrows. 1), putative NifA-like recognition sequence; 2), putative ς54 promoter; 3), leader peptide; 4), first amino acid of the mature protein; 5), hydrophobic domain; 6), ATP/GTP binding site.
RT-PCR with total RNA.
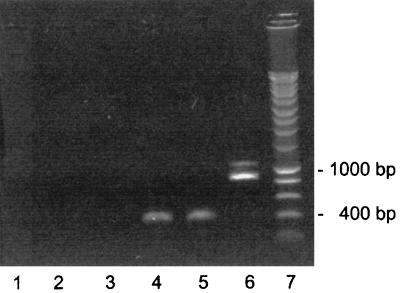
Total RNA was prepared from 2 × 109 late-logarithmic-phase LO15 cells using the S.N.A.P. total RNA isolation kit (Invitrogen, Groningen, The Netherlands). The procedure includes a DNase I treatment of RNA to remove DNA. Reverse transcription (RT) of RNA was performed with the cDNA cycle kit (Invitrogen) using the primer PilA11 (5′-ATGAAAGCCCAAATGCAGAA) at 0.5 μM. The PCR amplifications with material from the RT reaction (30 cycles of 1 min at 94°C, 2 min at 55°C, and 3 min at 72°C) employed the primers (each at 1 μM) PilA11 and PilA12 for pilAI (5′-TTAGGAGCACTTGCCTGGCT), PilA21 (5′-ATGAAGCCCGTGAAATCTTC) and PilA22 (5′-TTAGAGGAATTTCCCCGGCT) for pilAII, and PilA11 and PilA22 for the pilAI-pilAII region. The electrophoresis of PCR products in 0.8% agarose included samples of PCR amplifications which contained material from RT reactions run without reverse transcriptase as controls for the absence of genomic DNA.
DNA sequencing.
DNA sequencing was performed with the dideoxynucleotide chain termination method (35) as described previously (15). pST81 was used for sequencing of pilAII+ (Table 1).
Natural transformation of P. stutzeri by duplex and single-stranded DNA and DNA binding and uptake measurements.
Quantitative plate transformation of the histidine-auxotrophic strain LO15 (hisX1 [38]) and its derivatives by his+ DNA (isolated from strain JM375) was performed as described before (15). In these experiments 1 μg of DNA was included in 50 μl of a cell suspension and incubation was done for 24 h at 37°C on LB agar. Transformation with single-stranded DNA was performed by plate transformation (P. Meier, C. Berndt, N. Weger, and W. Wackernagel, unpublished data). The transformation frequency is the number of transformants per viable count. Competent cells from a stock suspension stored at −80°C were used in the DNA binding and uptake experiments, which were performed as described before (15). The specific radioactivity of the chromosomal P. stutzeri DNA labeled with [3H]thymidine was 6 × 106 to 8 × 106 cpm per μg.
Plating of phage PO4 and determination of twitching motility.
Plaque formation of PO4 on P. stutzeri was performed in a spot test as described by Bradley (4). Twitching motility was determined by inspecting single colonies of P. stutzeri for spreading zones on LB agar after incubation in a humid atmosphere at 37°C for 10 days.
Nucleotide sequence accession number.
The sequence reported here has the accession number AJ249743 in the EMBL database.
RESULTS
Identification of pilAII.
The pilA gene of P. stutzeri has its own promoter and is followed by a perfect inverted repeat of 13 bp (ΔG, −28.7 kcal mol−1) resembling a rho-independent transcription terminator, which suggests that pilA constitutes a single gene transcription unit (15). Upon further DNA sequencing downstream of pilA an open reading frame with its own ribosome binding site was found oriented in the same direction as pilA and coding for a putative protein of 141 amino acids (aa) (Fig. 1). The deduced amino acid sequence was 55% identical (and 65% similar) over a stretch of 132 aa to that of the previously identified pilA gene (Fig. 2). Therefore, the previously found gene was termed pilAI and the novel gene was termed pilAII. The amino acid sequence identity of PilA of P. aeruginosa with PilAI of P. stutzeri is 50%, and the amino acid sequence identity with PilAII is 51%. Two tyrosine residues at positions 21 and 24 after the putative prepilin cleavage site typical of group A of type IV pili (41) are present in PilAI and PilAII. The alignment of the amino acid sequences of PilAI and PilAII indicates an almost full identity in a hydrophobic region of 33 aa following the N-terminal hydrophilic leader peptide (Fig. 2). A similar localized identity was observed with and between the pilus proteins from several bacteria, including P. aeruginosa (28), D. nodosus (25), N. gonorrhoeae (26), Eikenella corrodens (44), Moraxella bovis (23), and Kingella denitrificans (47), and with proteins involved in extracellular protein secretion from various species (41). In contrast to PilAI, the PilAII protein has no motif for nucleotide binding and has a hydrophobic domain close to the C terminus (Fig. 1).
FIG. 2.
Amino acid sequence alignment of PilAI and PilAII prepilin proteins of P. stutzeri, generated by the CLUSTALW (1.73) program (42). Asterisks indicate identical residues and dots indicate related residues. Gaps introduced for alignment are represented by dashes. Amino acid numbers are listed on the right. The arrowhead marks the putative cleavage site of the leader peptidase, PilD, followed by a phenylalanine which is the first amino acid of the mature pilin. PilAI and PilAII showed 55% amino acid identity and 65% amino acid similarity using BLAST (1).
Phenotype of a pilAII::Gmr mutant.
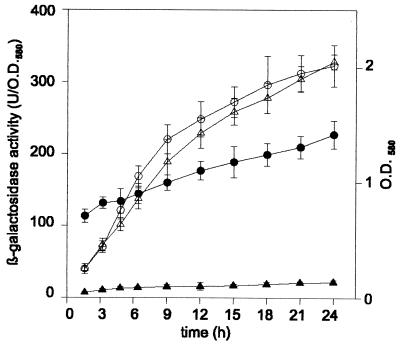
The pilAII gene was cloned after PCR amplification into the vector pUCP19, and then a gentamicin resistance cassette was ligated in the sense orientation into the SacII restriction site located in the pilAII open reading frame (Fig. 1). Transformation of naturally competent LO15 cells with the linearized plasmid DNA yielded strain Tf400 having a chromosomal pilAII::Gmr produced by allelic exchange, which was verified by PCR. Surprisingly, strain Tf400 showed a transformation frequency 16-fold higher than that of the parental strain LO15 (Table 2, entry 2). The hypertransformation phenotype suggested that pilAII is expressed in LO15 and that the gene exerts a suppressive effect on transformability. In order to determine the expression of pilAII and to compare it to that of pilAI, chromosomal lacZ fusions of pilAI and pilAII were created. A lacZ-Gmr cassette was used in a procedure similar to that described above for the creation of the pilAII::Gmr fusion. pilAII was expressed at a level about 10% of that of pilAI in logarithmic and early-stationary-phase cells (Fig. 1 and 3). The pilAI and pilAII genes are cotranscribed, which was determined by PCR amplification of reverse transcripts of mRNA covering both genes (Fig. 4). The normal twitching motility and PO4 sensitivity of the pilAII mutant indicated that these phenotypes, requiring functional type IV pili, were not dependent on pilAII (Table 2). The electron microscopic examination of cells revealed no difference of piliation between the wild type and the pilAII mutant.
TABLE 2.
Relative transformation frequencies, PO4 sensitivities and twitching motilities of the P. stutzeri wild type and various pilAI and pilAII mutantsa
| Strainb | Plasmid |
pilA allele on chromosome
|
Presence of pilA+ gene with indicated allele on plasmid
|
Relative transformation frequencyc | PO4 platingd | Twitching motilitye | ||
|---|---|---|---|---|---|---|---|---|
| AI | AII | AI | AII | |||||
| 1. LO15 | + | + | 1 | + | + | |||
| 2. Tf400 | + | − | 16.1 ± 2.3 | + | + | |||
| 3. Tf400 | pUCA1 | + | − | + | 20.3 ± 3.0 | + | + | |
| 4. Tf400 | pSGA2 | + | − | + | 0.02 ± 0.002 | + | + | |
| 5. Tf400 | pUCA1/2 | + | − | + | + | 3.1 ± 3.0 | + | + |
| 6. Tf300 | pUCA1/2 | − | + | + | + | 0.8 ± 0.1 | (+) | + |
| 7. LO15 | pUCA1/2 | + | + | + | + | 1.1 ± 0.1 | + | + |
| 8. LO15 | pSGA2 | + | + | + | 0.003 ± 0.0001 | + | + | |
| 9. Tf300 | − | + | ≤0.00019 | − | − | |||
| 10. Tf300 | pSGA2 | − | + | + | ≤0.0002 | − | − | |
| 11. Tf500 | − | − | ≤0.00012 | − | − | |||
| 12. Tf500 | pUCA1 | − | − | + | 14.2 ± 1.7 | (+) | + | |
| 13. Tf500 | pSGA2 | − | − | + | ≤0.00017 | − | − | |
Transformation frequencies were obtained by plate transformation and are expressed relative to the frequency of LO15 (2.0 × 10−5 ± 0.2 × 10−5).
As controls, the vectors pSG and pUCP19, corresponding to the plasmids with the pilA genes, were electroporated into LO15, Tf300, Tf400, and Tf500. These strains gave transformation frequencies, PO4 plating, and twitching motilities similar to those of the corresponding strains without vector plasmid.
Means of three experiments.
Phage titer was 108 ml−1; 0.02 ml was spotted. +, confluent lysis of cells; (+), single plaques; −, no visible plaques.
—, twitching motility was not observed within 10 days on an LB plate.
FIG. 3.
β-Galactosidase activities of strains with lacZ-Gmr fusions to pilAI (Tf301 [●]) and pilAII (Tf401 [▴]) during growth in LB medium. The growth of the cultures was determined as optical density at 580 nm (O.D.580) (open symbols). The activity is presented as units per O.D.580 of the culture (27). Data are given with standard deviations (n = 3).
FIG. 4.
Cotranscription of pilAI and pilAII. After RT-PCR of total RNA from LO15 cells, PCR amplification of the DNA was performed employing primer pairs specific for pilAI (420 bp), pilAII (426 bp), and the pilAI-pilAII region (913 bp). The amplified DNA obtained with the primer pairs and electrophoresed in a 0.8% agarose gel is shown. Lane 4, pilAI; lane 5, pilAII; lane 6, pilAI plus the intergenic region and pilAII. Parallel amplifications with material from RT-PCRs in which the reverse transcriptase was omitted show that no contaminating DNA was present (lane 1, pilAI; lane 2, pilAII; lane 3, pilAII). Lane 7 shows a molecular size marker DNA ladder.
Overexpression of pilAI+ cloned in the multicopy plasmid pUCP19 (36) did not further increase the high transformability of the pilAII mutant Tf400 (Table 2, entry 3) and increased the transformation of LO15 about twofold (data not shown).
Effect of pilAII overexpression.
The pilAII+ gene was cloned after PCR amplification into the multicopy vector pSG to obtain pSGA2. Overexpression of pilAII+ from this plasmid drastically decreased the high transformability of the pilAII strain (Table 2, entry 4), providing further evidence for a suppressive influence of PilAII on PilAI-promoted transformation. The 800-fold reduction of transformation by multicopy pilAII+ (Table 2, compare entries 2 and 4) exceeded the 16-fold reduction of transformation by the single-copy chromosomal pilAII+ gene in the parental strain (Table 2, compare entries 2 and 1).
The pilAI-pilAII region cloned on a multicopy plasmid (pUCA1/2) gave a somewhat increased transformation in a pilAII mutant background (Table 2, entry 5) and a normal transformation phenotype in a pilAI mutant background (Table 2, entry 6). This can be interpreted to mean that pilAI+ and pilAII+ are expressed in a certain ratio from their chromosomal loci and that this ratio determines the transformability of the cell. The ratio is the same when the pilAI-pilAII region is present once per chromosome (in LO15) or several times on a multicopy plasmid (pUCA1/2). This assumption was supported by the finding that the pilAI-pilAII region on a multicopy plasmid did not alter the transformability of parental strain LO15 (Table 2, entry 7). Accordingly, additionally produced PilAII would be expected to change the normal PilAI/PilAII ratio to a ratio giving low transformation. This was seen with strains Tf400(pSGA2) (Table 2, entry 4) and Tf300(pUCA1/2) (Table 2, entry 6) but most strongly with LO15(pSGA2) (Table 2, entry 8), in which the single chromosomal pilAI+ gene is faced with the pilAII+ genes on the multicopy plasmid plus pilAII+ on the chromosome. In this strain, transformation was more than 5,000-fold lower than in the strains with only a defective pilAII gene [Tf400 and Tf400(pUCA1)]. Strains having no intact pilAI gene were nontransformable, irrespective of the presence of pilAII+ in a single copy or in multiple copies or without pilAII+ (Table 2, entries 9 to 11). These mutants were also defective for twitching motility and PO4 plating, demonstrating the essential role of pilAI+ for these phenotypes. The fact that the pilAI pilAII double deletion mutant was not transformable suggests that a third pilA+ gene that could promote transformation and would also be functionally suppressed by pilAII is not expressed in P. stutzeri. The presence of pilAI+ on a plasmid in the double deletion mutant restored the hypertransformation phenotype, while the pilAII+ gene had no discernible effect in that background (Table 2, entries 12 and 13).
PilAII does not form pili.
The pilAI mutant cells have no pili visible in the electron microscope (15). The detection of the second pilus gene raised the question of why pilAII+ did not support formation of pili that would be visible in the pilAI mutant. Besides other reasons, the expression of pilAII+ may be too low (Fig. 3). The latter possibility was tested by electron microscopic examination of strains overexpressing pilAII+ from a plasmid (more than 100 cells inspected per strain). Overexpression did not change the piliation in the wild-type background [LO15(pSGA2)], suggesting that overexpression does not interfere with normal pilus formation. This is consistent with the normal twitching motiliy and PO4 infection of the strains (Table 2). PilAII overexpression in a pilAI mutant [Tf300(pSG2)] did not result in visible pili (and twitching motility plus PO4 infection), suggesting that PilAII itself does not form type IV pili.
DNA binding and uptake.
Since the pilAII mutant was hypertransformable, we investigated whether the cells would bind or take up more DNA than LO15. Cells of strain Tf400 in the late logarithmic growth phase, in which LO15 is on its competence peak, bound 144 ± 20 pg of DNA/5 × 108 cells within 90 min (mean ± standard deviation; n = 3), an amount similar to that found with LO15 cells (152 ± 41 pg/5 × 108 cells; n = 15). In the pilAII mutant cells roughly 35% of the bound DNA was taken up into a DNase I-resistant state, which is similar to what has been previously found with LO15 cells (15). It is concluded that the hypertransformability of the pilAII mutant does not result from enhanced binding and uptake of DNA. This was supported by the observation that pilAII overexpression which strongly suppressed transformation did not reduce binding and uptake of DNA (data not shown). Thus, per amount of taken-up DNA much more transformants are formed with the pilAII mutant than with wild-type cells.
Influence of pilAII on heterologous pilus gene-mediated transformation.
pilAI is essential for natural transformation of P. stutzeri. It can be functionally replaced by heterologous prepilin genes from nontransformable bacteria, including P. aeruginosa PAK, P. aeruginosa PAO, and D. nodosus, restoring pilus formation, twitching motility, PO4 infection, and transformability (15). The transformability of P. stutzeri pilAI was restored to 0.3 to 1.4 times the wild-type level. We wanted to see whether the negative effect exerted by the PilAII protein on transformation would also extend to the transformation mediated by heterologous pilin proteins. Table 3 shows that insertional inactivation of pilAII led to 7- to 28-fold increases of the transformability of strains with the heterologous pilA genes, while twitching motility and PO4 infection were normal. The same phenotypes of cells expressing heterologous pilA genes were also observed in a pilAI pilAII background, indicating that in fact pilAII+ suppressed the transformability mediated by pilins from the nontransformable species. The data in Table 3 confirm that the heterologous pilA genes can fully restore twitching motility and PO4 plating in the pilAI mutant (15) and show that inactivation of pilAII does not interfere with heterologous complementation.
TABLE 3.
Relative transformation frequencies, PO4 sensitivities, and twitching motilities of strains Tf400 and Tf500 complemented by heterologous pilA+ genesa
| Strainb | Plasmid |
pilA allele on chromsome
|
Foreign pilA+ gene on plasmid | Relative transformation frequencyc | Plaque formation with PO4d | Twitching motilitye | |
|---|---|---|---|---|---|---|---|
| AI | AII | ||||||
| LO15 | + | + | 1 | + | + | ||
| Tf400 | + | − | 16.1 ± 2.3 | + | + | ||
| Tf400 | pAW102-O | + | − | PAO | 28.4 ± 4.0 | + | + |
| Tf400 | pAW103-K | + | − | PAK | 12.9 ± 1.7 | + | + |
| Tf400 | pAW107-Dn | + | − | Dn | 7.2 ± 1.0 | + | + |
| Tf500 | − | − | ≤0.00019 | − | − | ||
| Tf500 | pAW102-O | − | − | PAO | 10.3 ± 1.6 | + | + |
| Tf500 | pAW103-K | − | − | PAK | 11.1 ± 1.9 | + | + |
| Tf500 | pAW107-Dn | − | − | Dn | 5.4 ± 0.9 | + | + |
Transformation frequencies were obtained by plate transformation and are expressed relative to the frequency in LO15 (2.0 × 10−5 ± 0.2 × 10−5); the heterologous pilA+ genes were from P. aeruginosa PAO(pAW102-O), from P. aeruginosa PAK(pAW103-K), and from D. nodosus(pAW107-Dn).
As controls the vector pUCP19, corresponding to the plasmids with the heterologous pilA+ genes, was introduced into LO15, Tf400, and Tf500. These strains gave transformation frequencies, PO4 plating, and twitching motilities similar to those of the corresponding strains without vector plasmid.
Means of three experiments.
Phage titer of 108 ml−1; 0.02 ml was spotted. +, confluent lysis of the cells.
+, twitching motility was observed with 10 days on an LB plate.
Increased transformation by single-stranded DNA.
Natural transformation of P. stutzeri LO15 (hisX1) by the purified 5′ strand of the hisX+ gene region has been demonstrated recently (Meier et al., unpublished). Transformation by single-stranded DNA was about 30-fold lower than with an equivalent amount of duplex DNA. It depended on functional pili, since it was absent in pilAI and pilC mutants (Table 4) (Meier et al., unpublished). In P. stutzeri a defect in comA abolished transformation with duplex DNA (17) and with single-stranded DNA (Meier et al., unpublished). For entering the cytoplasm, preformed single strands apparently depend on the DNA translocation machinery that has been identified in transformation studies with duplex DNA in several transformable organisms (12). As shown in Table 4, transformation of the pilAII strain by single-stranded DNA was increased 17-fold compared to that of LO15, which corresponds to the about 20-fold increase of duplex DNA transformation (Tables 2 and 4). Thus, PilAII also suppresses the transformation by preformed single strands.
TABLE 4.
Transformation of LO15 and pilAI and pilAII mutants with purified single-stranded and duplex DNAa
| Strain | Relevant genotype | Relative his+ transformation frequency
|
|
|---|---|---|---|
| Single-stranded DNA | Duplex DNA | ||
| LO15 | Wild type | 1b | 1c |
| Tf300 | pilAI | ≤0.003 | ≤0.0002 |
| Tf400 | pilAII | 17 | 22 |
The duplex DNA was pPM1 containing the hisX+ gene. The plasmid does not replicate in P. stutzeri (Meier et al., unpublished). The single-stranded DNA (5′ strand of the hisX+ region) was the plus strand of pPM1 isolated from purified fd phage (Meier et al., unpublished). The DNA concentrations were 1.6 μg per 50 μl of cell suspension. The data with the mutants are means of two independent determinations.
The transformation frequency was 2.4 × 10−7 (±1.1 × 10−7; n = 4).
The transformation frequency was 5.1 × 10−6 (±2.1 × 10−6; n = 4).
DISCUSSION
The soil bacterium P. stutzeri forms type IV pili, which are essential for natural genetic transformation and for which pilAI provides the structural pilin (15). Here we show that the cells have a second pilA-like gene (pilAII) downstream of pilAI coding for a protein with high amino acid sequence similarity to the pilAI protein of P. stutzeri and to corresponding proteins of other bacteria. The pilAII gene is expressed at about 1/10 the rate of pilAI during the logarithmic and early stationary growth phases, as determined by expression of lacZ fusions to these genes. Since pilAI and pilAII are cotranscribed (presumably from the putative ς54 promoter preceding pilAI) (15), the attenuation of pilAII expression relative to pilAI is probably caused by the perfect inverted repeat of 13 bp separated by four nucleotides which is located in the intergenic region between the two cistrons. Such elements function as intrinsic transcription terminators and can serve to decrease expression of genes within operons (33). Recently it has been shown that of the two tandemly located pilA1 and pilA2 genes of E. corrodens the downstream pilA2 gene is much less transcribed than pilA1 and that a region of 25 bp in the intergenic region can form a nearly perfect hairpin structure (44). In E. corrodens the pilA1 gene is the pilus structural gene (44). Although E. corrodens is naturally transformable (43), the roles of pilAI and pilAII in transformation have not yet been studied. In P. stutzeri, insertional inactivation of pilAI but not of pilAII abolished piliation, which argues against an important contribution of pilAII to pilus biogenesis. However, the loss of pilAII function had the unexpected result of making cells hypertransformable.
All of the known naturally transformable gram-negative bacteria with type IV pili, including N. gonorrhoeae, L. pneumophila, Acinetobacter sp. strain BD4, and P. stutzeri, have more than one gene coding for prepilin or prepilin orthologs (6, 31, 40, 51). Mutational inactivation of the type IV pilus prepilin gene resulted in the loss of transformability in N. gonorrhoeae (14), L. pneumophila (40), and P. stutzeri (15). The nonpiliated mutants of N. gonorrhoeae and P. stutzeri were unable to take up DNA (14, 15). However, the mere physical presence of pili is not sufficient for transformation. This was concluded from the observations that piliated strains of N. gonorrhoeae and P. stutzeri are nontransformable and are defective for DNA uptake if they carry a mutation in the pilT gene (49; S. Graupner and W. Wackernagel, unpublished data). The cytoplasmic PilT protein is assumed to be involved in the retraction or depolymerization of type IV pili (4, 5, 50), which is required for twitching motility in N. gonorrhoeae (49), P. aeruginosa (48), and P. stutzeri (Graupner and Wackernagel, unpublished) as well as for PO4 infection of P. aeruginosa (48) and P. stutzeri (Graupner and Wackernagel, unpublished). In N. gonorrhoeae, pilus retraction is possibly important for the infection of eukaryotic cells (32, 50). PilAII is not required for pilus formation, twitching motility, and PO4 infection. Thus, pilAII+ is dispensable not only for the biogenesis but also for the function of type IV pili in P. stutzeri.
Inactivation of secondary prepilin-coding genes abolished transformability by no longer allowing DNA binding in N. gonorrhoeae (51) and Acinetobacter sp. strain BD4, in which the pilus subunit gene has not yet been identified (6, 31). Similarly, the inactivation of any of the pilA-like genes in organisms having no pili, like Bacillus subtilis (comGC, comGD, comGE, and comGG [8]), Streptococcus pneumoniae (cglC and cglD [30]), or Streptococcus gordonii (comYC and comYD [22]), blocked transformation by preventing DNA binding. A deletion of the pilA gene from Haemophilus influenzae, which is not involved in pilus formation, also blocked transformation (10). These observations indicate that the PilA-like proteins by themselves or along with the type IV pilus subunit protein are essential for DNA binding and uptake. By contrast, inactivation of pilAII in P. stutzeri increased transformability. Such a situation has not been described before. Moreover, the detrimental effect of PilAII on transformation increased with overexpression of pilAII+ and resulted in an up to 5,000-fold reduction of transformation, although pilus formation and function remained unaffected.
At what stage could PilAII hinder the transformation process? A loss of pilAII+ function increased transformability about 20-fold, while binding and uptake of DNA were not increased. Thus, PilAII acts after DNA uptake and not in any interaction with PilAI of P. stutzeri during DNA uptake. This is in agreement with the increased transformability of the pilAII mutants in which the defective pilAI gene was complemented by the pilA genes of the nontransformable bacteria P. aeruginosa and D. nodosus. The uptake of DNA into P. stutzeri cells is a two-step process. After binding of duplex DNA to the cell, the DNA enters the periplasmic space in duplex conformation and thereby becomes DNase I resistant. The second step is the translocation of DNA from the periplasm into the cytoplasm. While the first step requires functional pili (15), the second step depends on comA and is strongly facilitated by exbB (17). The polytopic membrane protein ComA and its orthologs are key components of the DNA transmembrane translocation machinery of other transformable organisms, like N. gonorrhoeae (13), H. influenzae (9), B. subtilis (19), and S. pneumoniae (30). ComA protein is thought to form the pore through which a single strand derived from duplex transforming DNA by degradation of one strand by an unknown DNase reaches the cytoplasm (12). In P. stutzeri, ExbB, having two transmembrane-spanning domains, perhaps helps to energize the translocation (17). Since the P. stutzeri comA mutant was no longer transformable by single-stranded DNA (and the transformation of the exbB mutant was strongly decreased), it was concluded that ComA also translocates preformed single-stranded DNA into the cytoplasm (Meier et al., unpublished). PilAII suppresses transformation by duplex DNA as well as by preformed single-stranded DNA (Table 4), suggesting that PilAII interferes with a transport process of single- and double-stranded DNA within the cell following uptake. The typical leader peptide including the leader peptidase cleavage site of PilAII suggests its transport into the periplasm. Possibly the protein interferes with the DNA transport to the ComA pore or through it.
Our finding of a PilA ortholog being an antagonistic factor of genetic transformation may be linked to the different levels of transformability found in various isolates of naturally transformable species. It was suggested that some of the most studied strains having particularly high transformability, including B. subtilis 168 Marburg, Acinetobacter sp. strain BD4, and Vibrio sp. strain WJT-1C, might represent transformation overshoot mutants (21). The pilAII mutant generated in this study would be an example of such an overshoot strain, if such a mutant had been isolated from nature. Recent observations on the level of transformability of natural isolates of P. stutzeri from the environment suggest a broad difference of the transformation potential of individual isolates and even strong differences of closely related members of the same population (N. Teschner, J. Sikorski, and W. Wackernagel, unpublished data).
ACKNOWLEDGMENTS
We thank Cecilia Berndt, Petra Meier, and Nicole Weger for help with some of the experiments. The work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoine R, Locht C. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol Microbiol. 1992;6:1785–1799. doi: 10.1111/j.1365-2958.1992.tb01351.x. [DOI] [PubMed] [Google Scholar]
- 3.Becker A, Schmidt M, Jager W, Pühler A. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene. 1995;162:37–39. doi: 10.1016/0378-1119(95)00313-u. [DOI] [PubMed] [Google Scholar]
- 4.Bradley D E. A pilus-dependent Pseudomonas aeruginosa bacteriophage with a long noncontractile tail. Virology. 1973;51:489–492. doi: 10.1016/0042-6822(73)90448-0. [DOI] [PubMed] [Google Scholar]
- 5.Bradley D E. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol. 1980;26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 6.Busch S, Rosenplänter C, Averhoff B. Identification and characterization of ComE and ComF, two novel pilin-like competence factors involved in natural transformation of Acinetobacter sp. strain BD413. Appl Environ Microbiol. 1999;65:4568–4574. doi: 10.1128/aem.65.10.4568-4574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson C A, Pierson L S, Rosen J J, Ingraham J L. Pseudomonas stutzeri and related species undergo natural transformation. J Bacteriol. 1983;153:93–99. doi: 10.1128/jb.153.1.93-99.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung Y S, Dubnau D. All seven comG open reading frames are required for DNA binding during the transformation of competent Bacillus subtilis. J Bacteriol. 1998;180:41–45. doi: 10.1128/jb.180.1.41-45.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clifton S W, McCarthy D, Roe B A. Sequence of the rec-2 locus of Haemophilus influenzae: homologies to comE-ORF3 of Bacillus subtilis and msbA of Escherichia coli. Gene. 1994;146:95–100. doi: 10.1016/0378-1119(94)90840-0. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty B A, Smith H O. Identification of Haemophilus influenzae Rd transformation genes using cassette mutagenesis. Microbiology. 1999;145:401–409. doi: 10.1099/13500872-145-2-401. [DOI] [PubMed] [Google Scholar]
- 11.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 13.Facius D, Meyer T F. A novel determinant (comA) essential for natural transformation competence in Neisseria gonorrhoeae and the effect of a comA defect on pilin variation. Mol Microbiol. 1993;10:699–712. doi: 10.1111/j.1365-2958.1993.tb00942.x. [DOI] [PubMed] [Google Scholar]
- 14.Fussenegger M, Rudel T, Barten R, Ryll R, Meyer T F. Transformation competence and type IV pilus biogenesis in Neisseria gonorrhoeae—a review. Gene. 1997;192:125–134. doi: 10.1016/s0378-1119(97)00038-3. [DOI] [PubMed] [Google Scholar]
- 15.Graupner S, Frey V, Hashemi R, Lorenz M G, Brandes G, Wackernagel W. Type IV pilus genes pilA and pilC of Pseudomonas stutzeri are required for natural genetic transformation, and pilA can be replaced by corresponding genes from nontransformable species. J Bacteriol. 2000;182:2184–2190. doi: 10.1128/jb.182.8.2184-2190.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graupner S, Wackernagel W. Identification of multiple plasmids released from recombinant genomes of Hansenula polymorpha by transformation of Escherichia coli. Appl Environ Microbiol. 1996;62:1839–1841. doi: 10.1128/aem.62.5.1839-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graupner, S., and W. Wackernagel. Identification and characterization of novel competence genes comA and exbB involved in natural genetic transformation of Pseudomonas stutzeri. Res. Microbiol., in press. [DOI] [PubMed]
- 18.Henrichson J. Twitching motility. Annu Rev Microbiol. 1983;37:81–93. doi: 10.1146/annurev.mi.37.100183.000501. [DOI] [PubMed] [Google Scholar]
- 19.Inamine G S, Dubnau D. ComEA, a Bacillus subtilis integral membrane protein required for genetic transformation, is needed for both DNA binding and transport. J Bacteriol. 1995;177:3045–3051. doi: 10.1128/jb.177.11.3045-3051.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenz G M, Wackernagel W. High frequency of natural genetic transformation of Pseudomonas stutzeri in soil extract supplemented with a carbon/energy and phosphorus source. Appl Environ Microbiol. 1991;57:1246–1251. doi: 10.1128/aem.57.4.1246-1251.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenz G M, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lunsford R D, Roble A G. comYA, a gene similar to comGA of Bacillus subtilis, is essential for competence-factor-dependent DNA transformation in Streptococcus gordonii. J Bacteriol. 1997;179:3122–3126. doi: 10.1128/jb.179.10.3122-3126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marrs C F, Schoolnik G, Koomey J M, Hardy J, Rothbard J, Falkow S. Cloning and sequencing of a Moraxella bovis pilin gene. J Bacteriol. 1985;163:132–139. doi: 10.1128/jb.163.1.132-139.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattick J S, Whitchurch C B, Alm R A. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa—a review. Gene. 1996;179:147–155. doi: 10.1016/s0378-1119(96)00441-6. [DOI] [PubMed] [Google Scholar]
- 25.McKern N M, Stewart D J, Strike P M. Amino acid sequences of pilins from serologically distinct strains of Bacteroides nodosus. J Protein Chem. 1988;7:157–164. doi: 10.1007/BF01025245. [DOI] [PubMed] [Google Scholar]
- 26.Meyer T F, Billyard E, Haas R, Storzbach S, So M. Pilus genes of Neisseria gonorrhoeae: chromosomal organization and DNA sequence. Proc Natl Acad Sci USA. 1984;81:6110–6114. doi: 10.1073/pnas.81.19.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28.Paranchych W, Frost L S, Carpenter M. N-terminal amino acid sequence of pilin isolated from Pseudomonas aeruginosa. J Bacteriol. 1978;134:1179–1180. doi: 10.1128/jb.134.3.1179-1180.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pemberton J M, Penfold R J. High-frequency electroporation and maintenance of pUC- and pBR-based cloning vectors in Pseudomonas stutzeri. Curr Microbiol. 1992;25:25–29. doi: 10.1007/BF01570078. [DOI] [PubMed] [Google Scholar]
- 30.Pestova E V, Morrison D A. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J Bacteriol. 1998;180:2701–2710. doi: 10.1128/jb.180.10.2701-2710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porstendörfer D, Drotschmann U, Averhoff B. A novel competence gene, comP, is essential for natural transformation of Acinetobacter sp. strain BD413. Appl Environ Microbiol. 1997;63:4150–4157. doi: 10.1128/aem.63.11.4150-4157.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pujol C, Eugène E, Marceau M, Nassif X. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc Natl Acad Sci USA. 1999;96:4017–4022. doi: 10.1073/pnas.96.7.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson J P, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 822–848. [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 37.Schweizer H P. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- 38.Sikorski J, Graupner S, Lorenz M G, Wackernagel W. Natural transformation of Pseudomonas stutzeri in non-sterile soil. Microbiology. 1998;144:569–576. doi: 10.1099/00221287-144-2-569. [DOI] [PubMed] [Google Scholar]
- 39.Stone B J, Kwaik Y A. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun. 1998;66:1768–1775. doi: 10.1128/iai.66.4.1768-1775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stone B J, Kwaik Y A. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol. 1999;181:1395–1402. doi: 10.1128/jb.181.5.1395-1402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strom S M, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 42.Thompson J D, Higgins D G, Gibson T J. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tønjum T, Freitag N E, Namork E, Koomey M. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol Microbiol. 1995;16:451–464. doi: 10.1111/j.1365-2958.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 44.Villar T M, Helber J T, Hood B, Schaefer M R, Hirschberg R L. Eikenella corrodens phase variation involves a posttranslational event in pilus formation. J Bacteriol. 1999;181:4154–4160. doi: 10.1128/jb.181.14.4154-4160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 46.Watson A A, Mattick J S, Alm R A. Functional expression of heterologous type IV fimbriae in Pseudomonas aeruginosa. Gene. 1996;175:143–150. doi: 10.1016/0378-1119(96)00140-0. [DOI] [PubMed] [Google Scholar]
- 47.Weir S, Marrs C F. Identification of type IV pili in Kingella denitrificans. Infect Immun. 1992;60:3437–3441. doi: 10.1128/iai.60.8.3437-3441.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitchurch C B, Hobbs M, Livingston S P, Krishnapillai V, Mattick J S. Characterization of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- 49.Wolfgang M, Lauer P, Park H-S, Brossay L, Hébert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 50.Wolfgang M, Park H-S, Hayes A F, van Putten J P M, Koomey M. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1998;95:14973–14978. doi: 10.1073/pnas.95.25.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfgang M, van Putten J P M, Hayes S F, Koomey M. The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol Microbiol. 1999;31:1345–1357. doi: 10.1046/j.1365-2958.1999.01269.x. [DOI] [PubMed] [Google Scholar]