ABSTRACT
COVID-19 and influenza are both highly contagious respiratory diseases that have been serious threats to global public health. It is necessary to develop a bivalent vaccine to control these two infectious diseases simultaneously. In this study, we generated three attenuated replicating recombinant vesicular stomatitis virus (rVSV)-based vaccine candidates against both SARS-CoV-2 and influenza viruses. These rVSV-based vaccines coexpress SARS-CoV-2 Delta spike protein (SP) bearing the C-terminal 17 amino acid (aa) deletion (SPΔC) and I742A point mutation, or the SPΔC with a deletion of S2 domain, or the RBD domain, and a tandem repeat harboring four copies of the highly conserved influenza M2 ectodomain (M2e) that fused with the Ebola glycoprotein DC-targeting/activation domain. Animal immunization studies have shown that these rVSV bivalent vaccines induced efficient humoral and cellular immune responses against both SARS-CoV-2 SP and influenza M2 protein, including high levels of neutralizing antibodies against SARS-CoV-2 Delta and other variant SP-pseudovirus infections. Importantly, immunization of the rVSV bivalent vaccines effectively protected hamsters or mice against the challenges of SARS-CoV-2 Delta variant and lethal H1N1 and H3N2 influenza viruses and significantly reduced respiratory viral loads. Overall, this study provides convincing evidence for the high efficacy of this bivalent vaccine platform to be used and/or easily adapted to produce new vaccines against new or reemerging SARS-CoV-2 variants and influenza A virus infections.
IMPORTANCE Given that both COVID-19 and influenza are preferably transmitted through respiratory droplets during the same seasons, it is highly advantageous to develop a bivalent vaccine that could simultaneously protect against both COVID-19 and influenza. In this study, we generated the attenuated replicating recombinant vesicular stomatitis virus (rVSV)-based vaccine candidates that target both spike protein of SARS-Cov-2 Delta variant and the conserved influenza M2 domain. Importantly, these vaccine candidates effectively protected hamsters or mice against the challenges of SARS-CoV-2 Delta variant and lethal H1N1 and H3N2 influenza viruses and significantly reduced respiratory viral loads.
KEYWORDS: bivalent vaccine, VSV vector, SARS-CoV-2 Delta variant, influenza, neutralizing antibody, M2 protein, spike protein, ectodomain
INTRODUCTION
The ongoing pandemic of COVID-19 has been the most serious threat to global public health. Numbers of vaccines have been successfully developed and approved for the prevention of COVID-19 (1, 2). However, the continuing spread of some highly transmissible variants of concern (VOCs) and their ability to infect immunized people (breakthrough infections) have challenged the effectiveness of current vaccines which calls for the reformulation of COVID-19 vaccines.
SARS-CoV-2 is a member of the betacoronavirus subfamily that causes severe symptoms in respiratory, gastrointestinal, and neurological systems (3–6). In December 2020, the SARS-CoV-2 Delta variant (B.1.617.2) emerged in India and spread across the globe within a few months of its outbreak making it the dominant variant in 2021 (7). Then, another VOC, SARS-CoV-2 Omicron (B.1.1.529), was designated in November of 2021 and possesses an excessive number of mutations compared with other variants, especially with 32 amino acid residue changes in the spike protein (SP) (8, 9). Previous reports found that people infected by the Delta variant had viral loads that were over 1,000 times higher than those of individuals infected with the original strain in 2020. Also, the disease caused by the Delta variant was more severe with an increased risk of death than previous VOCs (10, 11). Our recent study has shown that the Delta SP has an enhanced fusogenic ability and modulates inflammatory cytokine production (12). Therefore, the Delta variant is an excellent model for an emerging strain with altered virulence and transmission and unique vaccine requirements.
Influenza is another contagious respiratory illness, caused mainly by the influenza A virus (IAV). Surprisingly, after 100 years of the IAV global outbreak resulting in about 50 million deaths globally (13, 14), seasonal influenza still poses a large threat to public health, with global annual mortality of over 300,000 (15). Although vaccination remains the most effective method to prevent influenza-associated illness, the effectiveness of the seasonal influenza vaccine is only approximately 10% to 60% because the vaccine strains may not be well matched to circulating strains (16, 17). Therefore, it is necessary to develop a universal vaccine that offers broad protection against different influenza strains. Recently, we demonstrated that the recombinant vesicular stomatitis virus (rVSV)-EΔM-tM2e vaccine containing the highly conserved extracellular domain of influenza matrix protein (M2e) could efficiently protect mice from H1N1 and H3N2 infection (18). Given that both COVID-19 and influenza are contagious respiratory diseases transmitted during the same seasons with an increasing threat to the globe, it is highly advantageous to develop a bivalent vaccine that could simultaneously protect against both COVID-19 and influenza.
The rVSV-based vaccine platform has been used as an attenuated replication-competent vaccine that lacks the VSV-glycoprotein (G) related pathogenesis because the virulent G is replaced with different designed immunogene(s). It induces a rapid and robust immune response to viral antigens after a single immunization and has been shown to protect against several pathogens, including the Ebola virus, Zika virus, HIV, and Nipah virus (19–23). An rVSV can robustly elicit mucosal and systemic immunity because its preexisting antibodies in the population are considerably low (24). In addition, rVSV can accommodate large insert and heterogenous genes in its genome. Specifically, the safe and effective rVSV-based Zaire Ebola glycoprotein (GP) vaccine (rVSV-ZEBOV-GP) was approved for medical use in the United States in 2019. Within the last 2 years, several VSV-based SARS-CoV-2 vaccines expressing the SP of SARS-CoV-2 (Wu-Han-1) have been reported (25–30). Interestingly, a recent report indicated that intranasal vaccination with VSV-SARS-CoV-2 resulted in protection in hamsters if administered within 10 days before the SARS-CoV-2 challenge, demonstrating that VSV-based vaccines are fast-acting vaccine candidates that are protective against COVID-19 (28).
We have previously established the EboGP Mucin-Like Domain (MLD) Replacement System (EboGP with MLD deleted, EboGPΔM) as vaccine platform, in which immunogens can be fused with EboGPΔM and inserted into VSV-ΔG vector, and the EboGPΔM mediate the VSV entry to cells especially DCs and macrophages (31). Based on above system, we have produced three rVSV bivalent vaccine candidates coexpressing an attenuated SARS-CoV2 Delta variant SP (full-length or S2-deleted) or RBD domain, and four copies of highly conserved influenza M2 ectodomain (M2e) fused with EboGPΔM. Here, we characterized the expressions of SARS-CoV-2 Delta variant spike protein and influenza M2e of these bivalent vaccine candidates and their abilities to induce immune responses against SARS-CoV-2 SP and influenza M2e. Furthermore, we demonstrated the protection of the bivalent VSV vaccines against the SARS-CoV-2 Delta variant and lethal H1N1 and H3N2 influenza infection in hamster and mouse models, respectively.
RESULTS
Generation of rVSV-based vaccines expressing both SARS-CoV-2 Delta-SP and influenza conserved M2e.
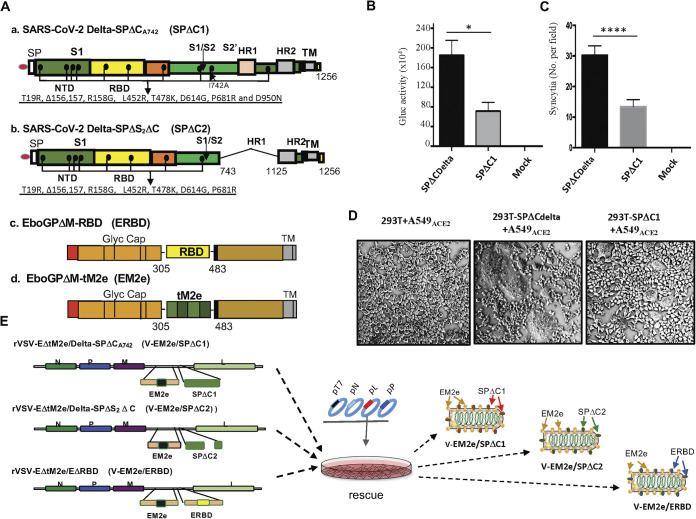
We first generated cDNAs encoding SARS-CoV-2 Delta-SP (SPDelta) containing a C-terminal 17 aa deletion and an I742A point mutation (SPDeltaΔCA742 or SPΔC1) (Fig. 1A, panel a). The C-terminus 17 aa deletion facilitates the transportation of SP to the plasma membrane and its assembly into the virus (32–34). Whereas the I742A point mutation in SPΔC1 significantly reduced pseudovirus infectivity and the SP-mediated syncytia formation compared to the wild-type SPΔCDelta in A549-ACE2 cells (Fig. 1B to D). Also, we generated SPDeltaΔS2ΔC (named SPΔC2), in which a 381-aa fragment in the S2 domain (744 to 1,124 aa) was deleted (Fig. 1A, panel b). Meanwhile, an RBD from SARS-CoV-2 (Wu-Han-1) SP was inserted into the Ebola GP (EboGPΔM) generating EboGPΔM-RBD (ERBD) (Fig. 1A, panel c). Finally, we inserted each cDNA encoding SPΔC1, SPΔC2 or ERBD into a recently reported rVSV-EM2e vaccine vector (Fig. 1A and D) (35) generating V-EM2e/SPΔC1, V-EM2e/SPΔC2, or V-EM2e/ERBD (Fig. 1E). This rVSV-EM2e vaccine vector contains an EboGPΔM fused with the tandem repeats of four copies of influenza M2 ectodomain (24 aa) polypeptide, in which two copies are from human flu strains, one from swine flu strain and one from avian flu strain (35).
FIG 1.
Construction and rescue of rVSV Delta SP and influenza M2e bivalent vaccines. (A) Schematic diagram of the Delta SPΔC and EboGPΔM-tM2e immunogens present in the bivalent vaccines. (a) SARS-CoV-2 Delta-SPΔCA742 (SPΔC1), containing a C-terminal 17 aa (DEDDSEPVLKGVKLHYT) deletion and an I742A mutation as indicated. The nine mutations in Delta SP are listed in the lower part. (b) Delta SPΔC2, containing the C-terminal 17 aa deletion and another 381 aa (encompassing aa744 to aa1124) deletion in the S2 domain. The eight mutations in SPΔC2, are listed in lower part. (c) EboGPΔM-RBD, the RBD of SARS-CoV-2 was used to replace the MLD domain in EboGP. (d) EboGPΔM-tM2e, four copies of influenza virus M2 ectodomain (24 aa) polypeptide (tM2e) replaced the MLD domain in EboGP. (B) The attenuated virus entry of SPΔC1. A549ACE2 cells were infected with equal amounts of SPΔCDelta-PVs or SPΔC1-PVs (adjusted by P24) carrying the Gluc gene, as indicated. At 48 h postinfection, the Gluc activity in the supernatant of different infected cultures was measured. Data represent the mean ± SD of two replicates from a representative experiment out of three performed. (C and D) The attenuated cell-to-cell fusion ability of SPΔCDelta- or SPΔC1-mediated syncytia formation was analyzed by coculturing the SPΔCDelta- or SPΔC1-expressing 293T cells with A549ACE2 cells. The amounts of syncytia were counted after 24 h in five different views of the microscope (C) and was also imaged under bright-field microscopy (D). (E) Schematic diagram of V-EM2e/SPΔC1, V-EM2e/SPΔC2 and V-EM2e/ERBD and the virus rescuing procedures. 293T and Vero E6 coculture cells were cotransfected with V-ΔG-EM2/SPΔC1, V-ΔG-EM2/SPΔC or V-ΔG-EM2/RBD and helping plasmids (T7, N, L, P plasmids). The supernatants containing V-EM2e/SPΔC1, V-EM2e/SPΔC2, and V-EM2e/ERBD viruses were used to infect Vero E6 cells to generate the rVSV stocks.
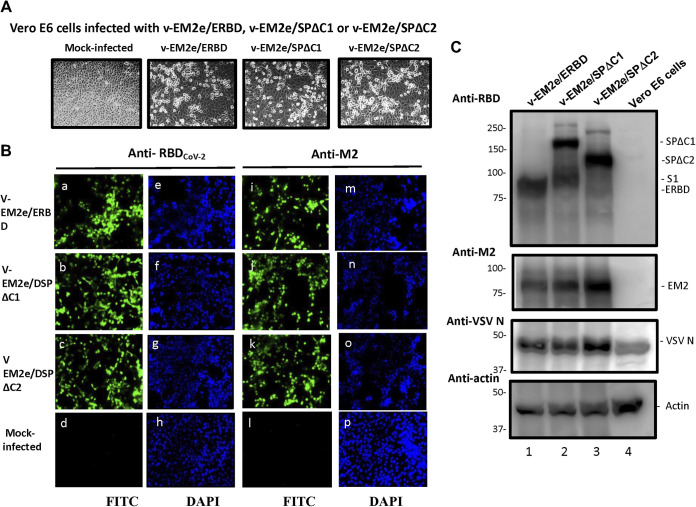
The V-EM2e/SPΔC1, V-EMe2/SPΔC2, and V-EM2e/ERBD were successfully rescued by using reverse genetics technology (36), as described in the Materials and Methods (Fig. 1E). The cytopathic effect was observed in above vaccine candidates infected Vero E6 (Fig. 2A) and the abundant expression of SPΔC1, SPΔC2, ERBD, and EM2e were detected in each corresponding rVSV-infected cells by immunofluorescence assay and the Western blotting (WB) (Fig. 2B and C, top two gels). The VSV nucleocapsid (N) protein was detected in all rVSV-infected cells (Fig. 2C, top third gel, lanes 1–3).
FIG 2.
Expression of V-EM2e/SPΔC1, V-EM2e/SPΔC2, or V-EM2e/ERBD in infected Vero E6 cells. (A) The infection of V-EM2/SPΔC1, V-EM2/SPΔC2 or V-EM2/ERBD in Vero E6 cells induced the cytopathic effects after 4 days postinfection. (B) Representative immunofluorescence images of Vero E6 cells infected with V-EM2e/SPΔC1, V-EM2e/SPΔC2, V-EM2e/ERBD, or mock-infected, stained with anti-SARS-CoV-2 RBD antibody (a to d) or anti-M2e antibody (i to l), and DAPI (e to h, m to p). (C) Vero E6 cells infected with the rescued V-EM2/SPΔC1, V-EM2/SPΔC, or V-EM2/ERBD were lysed and processed with SDS-PAGE followed by WB with a rabbit anti-SARS-CoV-2 RBD antibody (top panel), a mouse antibody against influenza M2e (top second gel), anti-VSV nucleocapsid (N) (top third gel), or anti-action (a cellular protein as an internal control) (low panel).
Replication attenuation and different cell tropisms of bivalent rVSV vaccine candidates compared with the wild-type VSV.
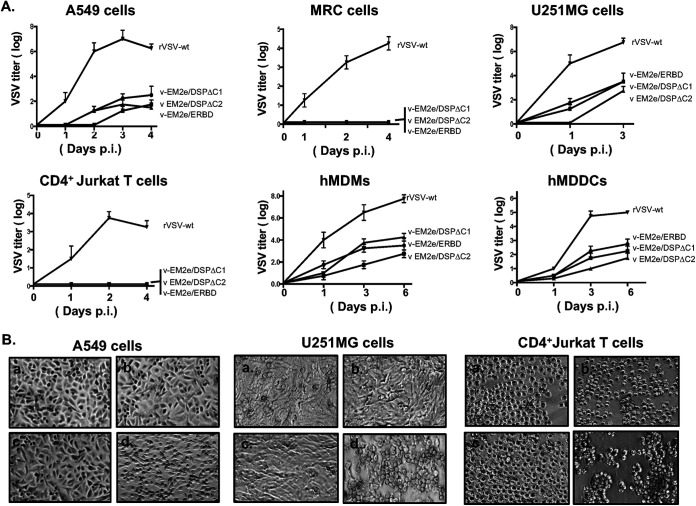
Given that rVSV is a replication-competent vector, the replication ability of the vaccine candidates was investigated using a dose of 100 TCID50 of each rVSV to infect the A549, a type II pulmonary epithelial cell (37); MRC-5, a human lung fibroblast cell (38); U251MG, a glioblastoma cells; CD4+ Jurkat T cells; human monocyte-derived macrophages (MDMs) and dendritic cells (DCs) (Fig. 3A). We assessed the cytopathic effect (CPE) of rVSV vaccine candidates and their replicating kinetics. As expected, wild-type VSV replicated efficiently in several cell lines, including A549, U251, and CD4+ Jurkat T cells, and induced typical CPEs, such as cell rounding detachment or forming cell clusters (Fig. 3A and B, panel d), while the rVSV vaccine candidates were unable to infect CD4+ Jurkat T cells and MRC-5 cells (Fig. 3A and B, panels a to c). In A549, U251, MDMs and MDDCs, three rVSV vaccine candidates displayed positive infection but in much slower replication kinetics and less CPE than wild-type VSV. These data showed that the rVSV vaccine candidates possessed significantly lower replication ability, less cytopathic effects, and altered cell tropism, compared to wild-type VSV.
FIG 3.
Characterization of the replication kinetics and the cell tropisms of bivalent rVSV vaccine candidates. (A) Each of the bivalent VSV vaccine candidates or the rVSV expressing VSV-G (rVSV-wt) was used to infect different cell lines, including A549, MRC-5, U251MG, CD4+ Jurkat T cells, human monocyte-derived macrophages (MDMs), and dendritic cells (MDDCs). Supernatants were collected at different time points postinfection as indicated and were titrated on Vero E6 cells. Data represent mean ± SD and were obtained from two replicates of a representative experiment out of two performed. (B) The ability of induced cytopathic effects in A549, U251MG and CD4+ Jurkat T cells, by each rVSV were observed after 4 days postinfection under microscopy. (a) V-EM2e/ERBD; (b) V-EM2e/SPΔC1; (c) V-EM2e/SPΔC2; (d) rVSVwt.
Evaluation of anti-SARS-CoV-2 and anti-influenza humoral immune responses induced by bivalent VSV vaccine candidates.
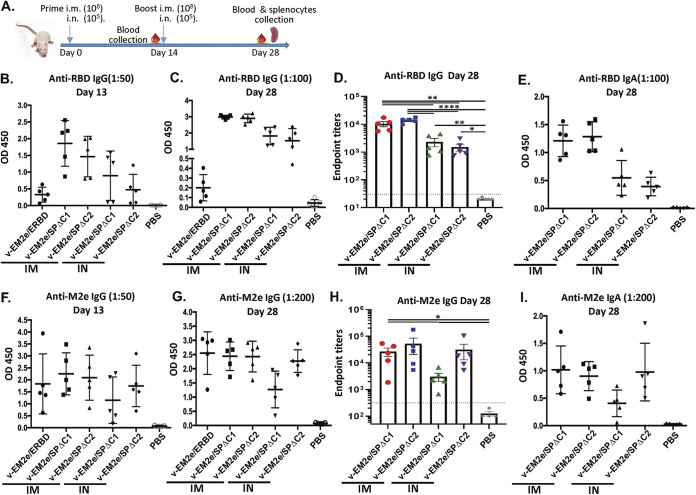
To test whether the above bivalent rVSV vaccine candidates could induce specific immune responses against SARS-CoV-2 SP and influenza M2, two doses of each vaccine (prime on day 0 and boost on day 14) were administered in BALB/c mice intramuscularly (IM; 1 × 108 TCID50) or intranasally (IN; 1 × 105 TCID50) (Fig. 4A). The potential adverse effects and body weight of mice were monitored daily for 1 week following vaccination and no changes were noticed (data not shown). Sera from immunized mice were collected on days 13 and 28 for measuring the anti-SARS-CoV-2 RBD and anti-M2 antibody levels using the corresponding antigen-coated ELISA. The results showed (i) IM immunization with V-EM2e/SPΔC1 or V-EM2e/SPΔC2 induced higher levels of circulating anti-SARS-CoV-2 RBD IgG and IgA antibodies than IN immunization (Fig. 4B to D); (ii) V-EM2e/ERBD IM administration induced much lower levels of anti-RBD IgG antibodies than the other two vaccines (Fig. 4B), indicating the SP-specific antibody responses induced by SPΔC1 and SPΔC2 immunogens; and (iii) all vaccine candidates elicited high levels of anti-M2-specific IgG and IgA antibodies regardless of the route of administration (Fig. 4E to G). All of these observations indicate that IM and IN immunizations with V-EM2/SPΔC1 and V-EM2/SPΔC2 induced efficient anti-SARS-CoV-2 RBD and anti-M2 immune responses.
FIG 4.
Anti-SARS-CoV-2 RBD and anti-influenza M2e immune responses induced by immunization with different bivalent VSV vaccine candidates. (A) Schematic of the bivalent rVSV vaccine candidate immunization protocol in the mouse. BALB/c mice were immunized with V-EM2e/SPΔC1, V-EM2e/SPΔC2, or V-EM2e/ERBD via intramuscular (IM) or intranasal (IN) routes, as indicated. The mice sera were collected on days 13 and 28 and were measured for anti-SARS-CoV-2 RBD IgG and IgA antibody levels (B to D) or measured for anti-M2e IgG and IgA antibody levels (F to H). (E and I) The anti-SARS-CoV-2 RBD and anti-M2e IgA antibody levels at 28 days. Data represent mean ± SD. Statistical significance was determined using an unpaired T-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
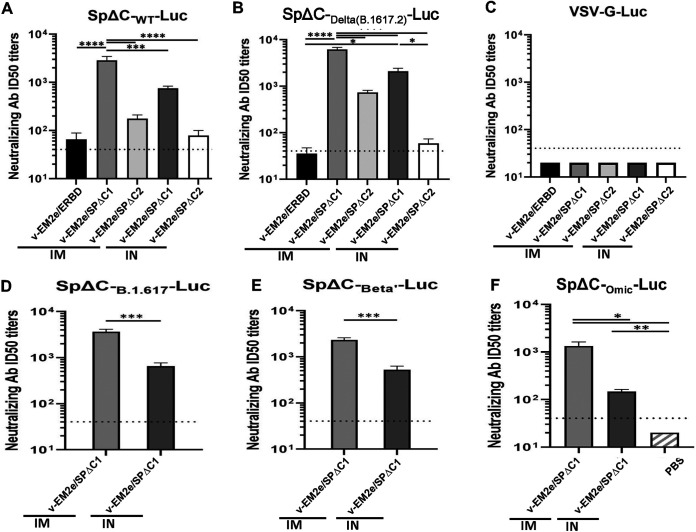
Vaccination with bivalent VSV vaccines induced potent neutralizing antibodies against different SARS-CoV-2 SP-pseudoviruses.
We tested whether the antibodies induced by bivalent rVSV-based vaccines could possess neutralizing activities. Various single-cycle SpΔC-pseudoviruses (PV) expressing firefly luciferase (Luc) were produced and used for the neutralization assay. Results showed that the V-EM2e/SPΔC1 immunized mice sera contained the highest levels of neutralizing antibodies against SpΔCWT- or SpΔCDelta-PV infections, while V-EM2e/ERBD immunization showed very low neutralizing activity (Fig. 5A and B), which was consistent with the low level of anti-RBD IgG antibody in the immunization sera (Fig. 4B). These results indicate that the full-length SP is the most efficient in producing neutralizing antibody (NAb). As expected, the V-EM2e/SPΔC1 and V-EM2e/SPΔC2 induced higher NAb titers against the Delta variant than the wild-type, SpΔCB.1.617, or SpΔCBeta’ (Fig. 5, compare B, D, E with A). Interestingly, the V-EM2e/SPΔC1 IM-immunized mice sera still retained over 103 titers of NAb against SpΔCOmic-PV infection even though it was significantly reduced compared with NAbs against SpΔCDelta-PV (Fig. 5F, bar 1). We also noticed that IM administration elicited significantly higher NAb titers than IN, probably because the immunization dose for the IM groups was 1,000-fold higher than that for the IN groups. As expected, all tested mice sera did not show any neutralization activity against VSV-G-PV infection (Fig. 5C). In summary, among three dual-action vaccine candidates, the V-EM2e/SPΔC1 elicited the highest titers of NAb against Delta SP-pseudovirus infection and, to a lesser extent, against SpΔCWT-, SpΔCBeta’-, and SpΔCOmic-PV infections in vitro.
FIG 5.
rVSV Delta SP vaccine candidates elicited neutralization antibodies. The neutralization titers (50% inhibition) in immunized mice sera against SpΔCWT-Luc-PVs (A), SpΔCDelta (B.1.617.2)-Luc-PVs (B), SpΔCBeta’-Luc-PVs (D), SpΔCB.1.617-Luc-PVs (E), and SpΔCOmic-Luc-PVs (F) infections. VSV-G-Luc-PVPs (C) were used as the negative control. The mouse serum of each immunization group collected on day 28 was pooled, 2× serially diluted, and incubated with different Luc-PVs (~104 RLU). Then, the mixtures were added in A549ACE2.cell cultures and the infection of PVs was determined by Luciferase assay at approximately 48 to 66 h postinfection. The percentage of infection was calculated compared with no serum control and neutralizing titers were calculated by using sigmoid 4PL interpolation with GraphPad Prism 9.0, as described in the Materials and Methods. Data represent mean ± SD and were obtained from over three independent experiments. Statistical significance was determined using an ordinary one-way ANOVA test and Turkey’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Induction of Th1/2 cytokines in splenocytes from the mice immunized by the bivalent VSV vaccine candidates.
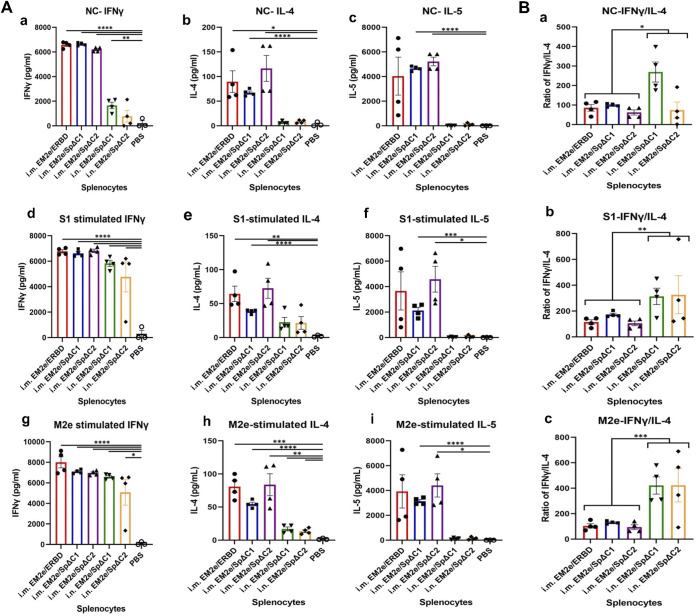
Effective vaccination involves the induction of T-helper cells that produce cytokines to shape subsequent humoral adaptive immune responses. To test the effect of vaccination-induced T cell responses, the splenocytes collected from the control and the immunized mice were cultured in the absence of any peptides (Fig. 6A, panels a to c), with SARS-CoV-2 SP subunit 1 (S1) overlapping peptide pool (Fig. 6A, panels d to f) or with influenza M2e peptides (Fig. 6A, panels c to i) (Fig. 6A, panels a to c). The levels of Th1 cytokines (IFN-γ) and Th2 cytokines (IL-4, IL-5) production in the medium were examined to determine whether T cells were stimulated in the immunized mice.
FIG 6.
T-cell cytokine response induced by bivalent VSV vaccine candidates. (A) Splenocytes isolated from immunized mice (as described in Fig. 4A) were cultured without peptide (no peptide control, NC) (a to c), or stimulated with SARS-CoV-2 SP subunit 1 (S1) peptide pool (d to f) or influenza M2e peptide (g to i) (1 μg/mL for each peptide). After 4 days of stimulation, supernatants were collected, and the release of IFN-γ, IL-4, and IL-5 cytokines in the supernatants was quantified with an MSD U-plex mouse cytokine immunoassay kit and counted in the MESO Quickplex SQ120 instrument. Each symbol indicated one individual mouse. (B) The ratios of Th1/Th2 cytokines from splenocytes of each mouse in the same culture condition were calculated, respectively, and the representative data (IFN-γ/IL-4) were shown. Statistical significance between the two groups was determined using an unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
As expected, we observed low/no levels of Th1 and Th2 cytokines in the splenocytes of PBS-treated mice, while variably high levels of Th cytokines were detected in the mice immunized with vaccine candidates. For the IN-immunized mouse splenocytes, the stimulation with S1 or M2e peptides markedly further elevated the secretion of IFN-γ and, to a lesser extent, IL-4 compared with PBS control (Fig. 6A, compare d, e, g, and h with a and b, bars 4 and 5), suggesting the S1/M2e-specific reactivation ability (memory) of these splenocytes. However, the presence of S1 or M2e peptides could not stimulate IL-5 production (Fig. 6A, compare f and g to c, bars 4 and 5). For the IM-immunized mice, significantly high levels of IFN-γ, IL-4 and IL-5 production were observed in the vaccinated splenocytes cultures in the absence of S1, M2e peptides stimulation (Fig. 6A, panels a to c), while stimulation of S1, M2e peptides did not further augment cytokine production (Fig. 6A, panels d to i, bar 1–3), suggesting the splenocytes after 14 days of IM-vaccination may be still in an activated status. Furthermore, the ratios of IFN-γ (Th1)/IL-4 (Th2) induced by the IN-immunized mice after peptide treatments (Fig. 6B, panels b to c, bars 4–5) were significantly higher than the ratios in IM-immunized mice (Fig. 6B, , panels b to c, bars 1–3), implying that IN immunization stimulated a very strong Th1-biased response, and IM immunization stimulated a more Th1/Th2-balanced cellular response with a little Th1-bias. Collectively, the above results suggested that our dual-action VSV vaccines have good immunogenicity to elicit strong T-cell immune responses.
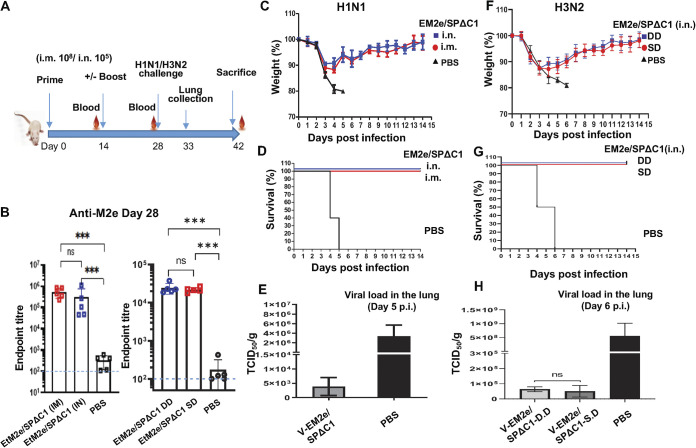
Immunization with V-EM2/SPΔC1 protects mice from lethal H1N1 and H3N2 influenza virus challenge.
The above-mentioned experiments have demonstrated the strong humoral and cellular immune responses induced by the bivalent VSV vaccine candidates. We next investigated whether V-EM2e/SPΔC1 immunization could protect against influenza virus infection. Briefly, groups of five mice were vaccinated with V-EM2/SPΔC1 via either IM or IN route and boosted on day 14, while control mice received only PBS (IN) (Fig. 7A). On day 28, all mice were challenged with a fatal dose of the A/Puerto Rico/8/34 H1N1 strain (2.1 × 103 TCID50/mouse) as previously described (35). For mouse-adapted H3N2 virus challenge experiments, mice were immunized with V-EM2/SPΔC1 via IN route, with (DD) or without (SD) boost on Day14 and intranasally challenged with H3N2 virus (1.4 × 104 TCID50/mouse) at day 28. Before the viral challenge, we confirmed that high levels of anti-M2e antibodies were induced in immunized mice in both vaccine delivery routes (Fig. 7B).
FIG 7.
Mice immunized with V-EM2/SPΔC1 were protected against the lethal challenge of H1N1 and H3N2 influenza viruses. (A) Schematic of the bivalent VSV vaccine candidate immunization and influenza virus challenge protocol used in the study. For the H1N1 challenge experiment, the BALB/c mice were immunized with 1 × 108 TCID50 (IM) or 1 × 105 TCID50 (IN) of V-EM2e/SPΔC1 or PBS on day 0 and day 14. On day 27, the blood samples were collected and measured for anti-influenza M2e antibody levels by ELISA (B). On day 28, all the mice were challenged with 2,100 PFU of H1N1 influenza virus. Weight loss (C) and survival rates (D) of the mice were monitored daily for 2 weeks. (E) Viral loads in the lung tissue of immunized mice and PBS group at day 5 post-H1N1 challenge were measured in MDCK cell line, as described in the Materials and Methods. For the H3N2 challenge experiment, the BALB/c mice were immunized with 1 × 105 TCID50 (IN) of V-EM2e/SPΔC1 or PBS on day 0 (single-dose, SD), or on day 0 and 14 (double-dose, DD). On day 28, all the mice were challenged with 1.4 × 104 PFU of H3N2. (F) Weight loss, (G) survive rates, and (H) viral loads in the lung tissue of immunized mice and PBS group at day 6 after H3N2 challenge.
Following challenge with either H1N1 or H3N2 strain, a high morbidity rate was observed among the control group mice, exhibiting over 20% weight loss (the humane endpoint for euthanasia) within 5 or 6 days (Fig. 7C and F). In contrast, V-EM2/SPΔC1-vaccinated mice via IM or IN routes showed moderate weight loss (~11%) until day 4 following a full recovery by days 8 to 11. The survival curve further indicated that both IM- and IN-immunized mice had 100% protection against H1N1 and H3N2 infections (Fig. 7D and G). We also monitored the H1N1 or H3N2 viral loads in the lungs of animals at 5- or 6-days postchallenge. The result showed that the virus titers of H1N1 or H3N2 in control-treated mice reached approximately 3 × 106 or 5 × 108 TCID50/gm of lung tissue, while the virus titers in the immunized mice were only 3 × 103 or 5 × 104 TCID50/gm of lung tissue, respectively, indicating that H1N1 and H3N2 virus replications were considerably suppressed in the lungs of immunized mice (Fig. 7E and H). Interestingly, the results also indicated that a single-dose administration via IN achieved similar protection efficiency as that of a double-dose administration (Fig. 7F to H). Overall, these results provide strong evidence that both IM and IN vaccination with the bivalent vaccine V-EM2e/SPΔC1 effectively protected mice from lethal H1N1 and H3N2 influenza challenges.
V-EM2/SPΔC1 and V-EM2/SPΔC2 protect Syrian hamsters from SARS-CoV-2 Delta virus infection.
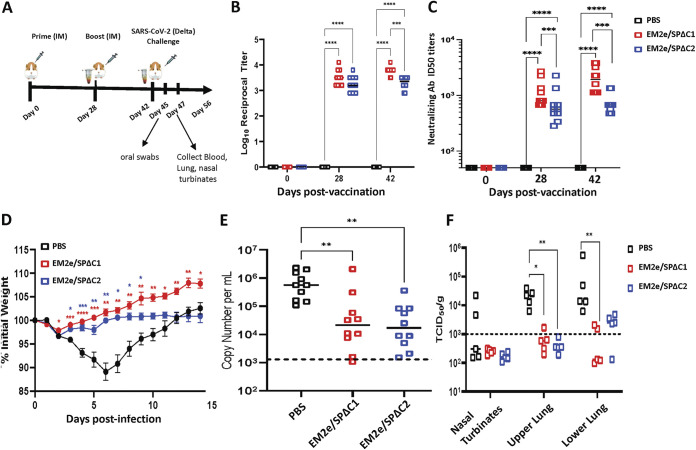
Next, we investigated whether immunization with V-EM2/SPΔC1 and V-EM2/SPΔC2 could protect Syrian hamsters from SARS-CoV-2 virus infection. Briefly, three groups of 10 hamsters were vaccinated with either V-EM2/SPΔC1 or V-EM2/SPΔC2 via an IM route and boosted on day 28, while control hamsters received only PBS (Fig. 8A). On days 28 and 42 (before virus challenge), we monitored anti-SARS-CoV-2 SP IgG titers and results showed that one dose (1 × 108 TCID50/hamster) of either vaccine induced a strong anti-SP antibody response at 28 days postvaccination (Fig. 8B). Interestingly, 2 weeks following the boost, both V-EM2/SPΔC1 and V-EM2/SPΔC2 vaccinated animals had small/no increase in antibody titer, suggesting that the levels of anti-SARS SP antibodies in mice have reached the peak after a single dose of rVSV vaccination. Notably, the serum from V-EM2/SPΔC1- and V-EM2/SPΔC2-vaccinated mice induced a strong nAb against the infection of SARS-CoV-2 Delta strain (Fig. 8C). Especially, the mice immunized with V-EM2/SPΔC1 had significantly higher nAb than the mice immunized with V-EM2/SPΔC2. Moreover, the nAb levels were not much different between single dose (day 28) and double dose (day 42).
FIG 8.
V-EM2/SPΔC1 and V-EM2/SPΔC2 provided protection against SARS-CoV-2 Delta infection in Syrian Hamsters. (A) Schematic of the bivalent VSV vaccine candidate immunization and SARS-CoV-2 Delta variant challenge protocol used in the study. (B) Total serum anti-SARS-CoV-2 spike IgG titers in hamsters following prime and boost vaccination. (C) The neutralizing antibody titers in immunized mice sera against SARS-CoV-2 Delta variant were measured and neutralizing titers were calculated by using sigmoid 4PL interpolation with GraphPad Prism 9.0, as described in the Materials and Methods. (D) Weight loss in the vaccinated or the control Syrian hamsters following infection with the SARS-CoV-2 Delta variant. (E) Viral RNA levels in oral swabs on day 3 following infection with SARS-CoV-2 Delta variant. (F) Infectious SARS-CoV-2 Delta virus titers in nasal turbinates and lung tissues on day 5 following infection with SARS-CoV-2 delta. n = 10 for B (each time point), n = 10 for C, 10 through day 28, and 10 at day 42; n = 10 for D (at day 3 postinfection) and n = 5 for E (from day 5 postinfection). Statistical significance was assessed by two-way analysis of variance with multiple comparisons (A), mixed effects analysis with multiple comparisons (B), and the Kruskal-Wallis test with multiple comparisons (C and D). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. For (B), colored asterisks indicate significant differences between the same colored group compared with the PBS group. Shown are medians for each group in A, C, and D, and mean + SEM in B.
Fourteen days following the second vaccine dose, all animals were challenged intranasally with the SARS-CoV-2 Delta variant. Animals in the PBS group continued to lose weight until maximal weight loss was seen on day 6, before recovering to the initial weight by day 12 (Fig. 8D). Vaccinated animals in all groups showed slight weight loss in the first 2 days but began to trend back toward their initial starting weights. The mean weight of the animals in both vaccinated groups remained significantly higher than the PBS group up to days 9 to 14. Interestingly, V-EM2/SPΔC2-vaccinated animals, while being protected from the weight loss seen in control animals, did not see the overall weight gain throughout infection seen in the V-EM2/SPΔC1 immunized animals.
Following infection, oral swabs were collected to examine viral shedding in all animals on day 3 and results showed that both V-EM2/SPΔC1 and V-EM2/SPΔC2 vaccinated groups had significantly reduced levels of viral RNA (Fig. 8E), indicating that vaccination may be capable of reducing viral shedding. On day 5, infectious viral titers were examined in the nasal turbinate, upper lung, and lower lung of five animals from each group. Both groups had significantly reduced viral titers in the upper lung, and V-EM2/SPΔC1 animals had reduced virus levels in the lower lung as well (Fig. 8F). These experiments provide strong evidence that V-EM2/SPΔC1 and V-EM2/SPΔC2 can protect with significantly reduced viral replication, leading to reduced disease and viral shedding.
DISCUSSION
COVID-19 and influenza are both highly contagious respiratory diseases with great global burdens. In this study, we have developed rVSV bivalent vaccines that specifically target the SARS-CoV-2 Delta variant and influenza viruses. The animal immunization studies have demonstrated that both V-EM2e/SPΔC1 and V-EM2e/SPΔC2 can induce robust humoral and cellular immune responses, including the NAbs against different SARS-CoV-2 SP-PV infections and effectively protect hamsters or mice against SARS-CoV-2 Delta variant and lethal H1N1 and H3N2 infections.
The SARS-CoV-2 Delta variant emerged in 2020 and quickly became the predominant circulating variant worldwide, showing increased potential for transmission and disease severity (39), and significant immune evasion ability (40–42). These indicate that variant-specific vaccine formulations could provide improved neutralization in immunized individuals. Therefore, several rVSV-based bivalent vaccines that specifically target the SARS-CoV-2 Delta variant have been generated in this study and the results clearly showed that the rVSV-based vaccines expressing either the full-length SP or S1 domain derived from the Delta variant provided efficient protection against SARS-CoV-2 Delta variant infection (Fig. 8). Interestingly, the immune response, including the nAb induced by V-EM2e/SPΔC2 expressing S1 and part of S2, tended to be weaker than that of V-EM2e/SPΔC1 expressing the full-length SPDelta (Fig. 4 and 8B to C). In line with this, the challenge experiment showed that although both vaccines protected infected hamsters from significant weight loss, the EM2e/SPΔC2-vaccinated animals recovered close to their initial weight without further weight gain. One explanation for the above difference in immune response and protection could be that the S2 domain of SP plays a yet undefined role in enhancing the induction of NAbs or antibody-dependent cellular cytotoxicity (ADCC). Indeed, a study reported that two segments, 884 to 891 and 1,116 to 1,123, located in SP-S2, are very effective in inducing host immune responses (43). Another interesting finding in this study was that the second dose of vaccine in hamsters did not significantly boost antibody titers (Fig. 8B), implying further study is required to demonstrate whether a single dose of VSV vaccination study is sufficient to protect against SARS-CoV-2 infection. Furthermore, our study revealed vaccination of either V-EM2e/SPΔC1 or V-EM2e/SPΔC2 significantly reduces viral shedding as measured by viral RNA load in oral swabs and viral titers (44) in the lungs. Additionally, no infectious virus was detected in any vaccinated animals in the nasal turbinate (Fig. 8E). This provides strong evidence that vaccination via the IM route can inhibit viral replication in the tissues of the upper and lower airways, suggesting that each vaccine candidate provides significant protection in the Syrian hamster models against clinical disease, viral shedding, and viral replication. This demonstrates that further development of this vaccine platform is warranted.
Notably, the continuing emergence of COVID-19 VOC has decreased the licensed vaccine’s effectiveness in terms of preventing infection (45–47). Particularly, the newly emerged Omicron possesses an extensive number of mutations compared with other variants (8) and the rapid spread of this VOC was facilitated by its high transmissibility and immune evasion ability (9, 48). Therefore, it is necessary to assess whether EM2e/SPΔC1 vaccine could induce broad NAbs against different SARS-CoV-2 variants. Our results indicated that the V-EM2e/SPΔC1 vaccination in mice elicited high titers of Nabs, specifically against Delta as well as wild-type, B.1.617, and Beta infections while exhibiting reduced neutralization against SpΔCOmic infection (Fig. 5F). Although the NAb titers against SpΔCOmic-PV could still reach a level (>103) that was likely comparable with the anti-2019-nCoV NAb titers detected in COVID-19 patients in a previous study (49), whether V-EM2e/SPΔC1 could provide in vivo effective protection from Omicron infection still requires further investigation.
Our bivalent vaccines have also been designed to target influenza A virus M2e domains from human, avian, and swine influenza virus strains (50) (Fig. 1A, panel d) in order to induce broad heterosubtypic immune responses to influenza viruses. Interestingly, immunization with the lead vaccine candidate V-EM2/SPΔC1 has elicited a high level of M2e-specific immune responses (Fig. 4) and effectively protected mice from lethal H1N1 and H3N2 influenza virus infection (Fig. 7C to H). Importantly, even a single IN immunization with V-EM2/SPΔC1 achieved equally efficient protection from H3N2 challenge compared with prime-boost IN immunization (Fig. 7F to H).
The rapid T cell response following vaccination is generally considered a key part of the immune response required to elicit effective protection. In this study, we observed high levels of secreted cytokines, including IFN-γ, IL-4, and IL-5, in splenocytes from the immunized mice (Fig. 6A), suggesting that the vaccine candidates could elicit both humoral and cellular responses in mice. Nevertheless, the lower cytokine levels, including IFN-γ, IL-4, and IL-5, were observed in IN route groups compared with IM route groups. The possible reasons are (i) the dose difference (1,000 times less in IN route groups) and (ii) the immunization via IN (a noninvasive administration) may stimulate more local mucosal immune response and less systemic immune response, compared with IM route (an invasive administration), so the splenocytes were significantly less activated by IN immunization. Interestingly, the S1/M2e-specific reactivation ability (memory) was only observed in IN-immunized mice splenocytes, but not in IM-immunized mice (Fig. 6A). In the IM group, the high proinflammatory cytokine productions had been there before antigen stimulation. This might be the rapid cellular response that has not waned to baseline after the boost. Some previous studies showed that VSV-EBOV-GP provided complete protection against lethal EBOV challenge in cynomolgus macaques at the early stage of immunization (3 days after vaccination, before antibody is detected) by robust innate antiviral immunity (51, 52), which included high expression of interferon (IFN)-stimulated genes (ISGs) and viral RNA sensors (53). Therefore, it is not surprising that a continued high proinflammatory cytokine response could be observed at 14 days after the boost in the IM group, probably due to the higher dose of rVSV (1 × 108 TCID50) used. Although the involvement of innate immunity in the protection of our bivalent vaccine for IM-administered animals remains to be elucidated, our IN experiments provide evidence to support the specific Th1-biased cellular response against influenza and SARS-CoV-2 virus infection that can benefit the removal of virus in the early stage (54, 55). Moreover, a previous study has shown that administration of VSV-expressing EboGP vector alone could not protect mice against influenza challenges (56).
The safety profile is also an important issue for vaccine development. A previous study revealed that, after mice were immunized intraperitoneally with VSV-ZEBOV-GP vaccine, no vaccine virus viremia was detected in tissues by either RT-PCR or virus isolation at different time points (1 to 7 days and 2, 3, 4 weeks) after immunization (57). Also, numbers of studies have been performed to test systemic/mucosal immunization or intrathalamic inoculation (in the brain) of VSV-ZEBOV-GP vaccine to mice (57), pigs (58), normal nonhuman primates (NHP) (59), or simian-HVI-infected NHPs (60), and verified that the immunization/inoculation with VSV-ZEBOV-GP did not result in neurological disease or histologic lesions, indicating a lack of neurovirulence of this vaccine. Our study has used several means to increase the safety profile of this bivalent vaccine platform, including the replacement of VSV-G with EM2 and introduction of I742A mutation into SARS-CoV-2 SP, to further attenuate the cytopathic effects. In addition, the concern for the impact of anti-VSV vector immune responses on vaccine efficacy remains, and the rVSV-EboGP-vaccine design and vaccination strategy still need to be optimized in future studies. Overall, these studies provided evidence for the high efficacy and the attenuated feature of this bivalent vaccine platform that can be used and easily adapted to produce new vaccines simultaneously against both emerging SARS-CoV-2 and influenza contagious respiratory infections.
MATERIALS AND METHODS
Ethics statement.
All animals were maintained in a specific-pathogen-free animal facility and used according to protocols approved by the Central Animal Care Facility, University of Manitoba (Protocol Approval No. 20-034) or by the Animal Care Committee at the Canadian Science Center for Human and Animal Health.
Plasmid constructions.
The gene encoding SPΔCDelta from our previously described study (12) was amplified and the I742A mutation was introduced by the mutagenesis technique. The amplified SPΔCDelta-I742A gene was cloned into the rVSV-EΔM-M2e vector (35) generating V-EM2e/SPΔC1. For V-EM2e/SPΔC2, a cDNA that carried an additional 381 aa deletion in the S2 region of SPΔCDelta (Fig. 1A and b), was cloned into the V-EΔM-M2e vector. To construct V-EM2e/ERBD, a cDNA encoding RBD of SARS-CoV-2 (Wuhan-Hu-1, GenBank accession No. MN908947) SP was amplified, inserted in pCAGGS-EboGPΔM at the MLD region (35) (Fig. 1A and D), and then cloned into the rVSV-EΔM-EM2e (Fig. 1B). For constructing pCAGGS-SPΔCBeta’ and pCAGGS-SPΔCB.1.617, the cDNAs for SPΔCBeta’ (harboring main mutations K417N, E484K, N501Y, D614G of Beta/B.1.351 but not others) and for SPΔCB.1.617 (the earliest lineage of Delta variant, that has mutations L452R, E484Q, and D614G) were introduced into the pCAGGS-nCoVSPΔC plasmid (12). The cDNA encoding SPΔCOmic, as described previously (8), was synthesized (Genescript) and cloned into the pCAGGS plasmid. All the inserted SPΔC transgenes in various plasmids were confirmed by sequencing.
Cells, antibodies, recombinant proteins, and viruses.
The HEK293T cells, human lung type II pulmonary epithelial cell line (A549), A549ACE2 cell line (12), human lung fibroblast cell line (MRC-5), human glioblastoma-derived cell line (U251GM), and Vero E6 and Madin-Darby canine kidney (MDCK) cell line were cultured in Dulbecco’s modified Eagle medium (DMEM) or DMEM/F-12 medium (21331-020, Gibco). CD4+ Jurkat cells were cultured in RPMI 1640 medium. MDMs and dendritic cells (MDDCs) were isolated from healthy donors as previously described (61). The antibodies used in this study included the rabbit polyclonal antibody against SARS-CoV-2 SP/RBD (Cat# 40150-R007, Sino Biological), anti-SARS-CoV-2 S-NTD (E-AB-V1030, Elabscience), anti-M2 (14C2: sc-32238, Santa Cruz Biotech.), and anti-VSV-Nucleoprotein, clone 10G4 (Cat. #MBAF2348, EMD Millipore Corp.). Recombinant proteins included SARS-CoV-2 RBD peptides (RayBiotech, Cat. #230-30162) and S1 peptide pool (JPT Peptide Technologies. Cat. #PM-WCPV-S-SU1-1). Influenza M2e peptide, mouse-adapted influenza A/Puerto Rico/8/34 (H1N1) and H3N2 strains were described previously (35).
rVSV rescue, virus growth kinetics experiments, and syncytia formation assay.
All three bivalent VSV vaccine candidates were rescued in 293T-Vero E6 cocultured cells, propagated and titrated as described previously (35). To examine the growth kinetics of bivalent rVSVs, different cell lines in a 24-well plate were infected with each of wild-type VSV (VSVwt) or vaccine candidate at a dose of 100 TCID50. After 2 h of infection, the cells were washed and the supernatants were collected at different times. The titers of rVSV were titrated in Vero E6 cells. To detect the expression of EM2, SPΔCDelta, and other viral proteins, rVSV-infected cells were lysed or fixed with 4% paraformaldehyde for 15 min and analyzed by WB or immunofluorescence assay with each corresponding antibody (12, 35). To test SPDelta-mediated syncytia formation, 293T cells transfected with various SPΔC plasmids were mixed with A549ACE2 cells at a 1:3 ratio and syncytium formation observed was imaged with an Axiovert 200 microscope (12).
SP pseudovirus production, infection, and neutralization assays.
Different SARS-CoV-2 SP pseudovirus expressing luciferase (Luc) or Gaussia luciferase (Gluc) were produced by cotransfecting 293T cells with each of the pCAGGS-SPΔC plasmids (33, 62), an HIV vector (pNL4-3-R-E-Luc) (63) or Gluc expressing HIV vector ΔRI/E/Gluc, as described previously (33). The SPΔC proteins are Wu-han-1 (wild-type), B.1.617.2 (Delta), B.1.617 (early lineage of Delta without mutation T478K), Beta’(with mutations K417N, E484K, N501Y of Beta/B.1.351 but not others) and B.1.1.529 (Omicron). The purified pseudoviruses were quantified by HIV-1 p24 amounts using an HIV-1 p24 ELISA (12). To measure the infection of SPΔC-pseudotyped VPs, equal amounts of each SPΔC-PV (as adjusted by p24 levels) were used to infect A549ACE2 and the viral infection levels were monitored by measuring Gluc activity in the supernatants. The neutralization assay was performed on A549/hACE2 cells according to previously reported methods with some modifications (64, 65). Briefly, each 2× serially diluted inactivated mouse sera were preincubated each SPΔC pseudovirus preparation for 1.5 h, then A549ACE2 (1.25 × 104 cells/well) were added in and were incubated at 37°C. After 48 h postinfection, cells were lysed followed by the luciferase RLU measurement. Neutralization assay was also performed using SARS-CoV-2 Delta virus. Briefly, the Delta virus was incubated with the serially diluted heat-inactivated mouse serum to a final concentration of 1 PFU/μL for 1 h at 37°C. The serum-virus mixture was then added to the Vero cells in 96-well plate and incubated for 5 days at 37°C. The wells containing infected cells were scored for CPE and the ID50 was calculated by using sigmoid 4PL interpolation with GraphPad Prism 9.0.
Mouse immunization and viral challenge.
Female BALB/c mice aged 6 to 8 weeks (five per group) were immunized IM (1 × 108 TCID50) or IN (1 × 105 TCID50) with rVSV vaccine candidates and boosted on day 14. Mice were sacrificed on day 28, and samples were collected for analyses on days 13 and 28.
For influenza virus challenge in mice, different groups of mice (five for each group) were IM or IN immunized with V-EM2e/SPΔC1 or PBS with or without boost on day 14. On day 28, all the mice were intranasally infected with a mouse-adapted strain A/Puerto Rico/8/34 (H1N1) (2.1 × 103 PFU/mouse) or H3N2 virus (1.4 × 104 PFU). The weight and survival rate of the mice were monitored daily for 2 weeks after the challenge. Moreover, 5 to 6 days postchallenge, the mice from the PBS group and vaccinated group were sacrificed, and the lungs were collected, homogenized, and used for viral titration in MDCK cells.
The SARS-CoV-2 challenge experiments were carried out at the National Microbiology Laboratory (NML). Different groups of 10 Syrian Golden hamsters were administered with 108 PFU of either V-EM2e/SPΔC1 or V-EM2e/SPΔC2 or PBS via IM injection and boosted after 28 days. Animals were monitored daily for any adverse signs following vaccine administration. After 14 days of the boost, hamsters were intranasally infected with 8.9 × 104 TCID50/100 μL of the SARS-CoV-2 delta variant (SARS-CoV-2; B.1.617.2; hCoV-19/Canada/ON-NML-63169/2021). After infection, animals were weighed and monitored daily for 14 days. On day 3 postinfection, oral swabs were performed on all animals. Groups of five animals (three male and two female) from each experimental group were euthanized on day 5 postinfection for examination of viral burden in the tissues.
Measurement of viral burden in the tissues.
For viral RNA detection in oral swabs, viral RNA was extracted with the QIAamp Viral RNA minikit (Qiagen) and the detection of the SARS-CoV-2 E gene was carried out on a QuantStudio 5 real-time PCR system (Applied Biosystems), as per manufacturer’s instructions. RNA was reverse transcribed and amplified using the primers reported by the WHO and, including E_Sarbeco_F1 (5′-ACAGGTACGTTAATAGTTAATAGCGT-3′) and E_Sarbeco_R2 (5′-ATATTGCAGCAGTACGCACACA-3′) and probe E_Sarbeco_P1 (5′-FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ-3′). A standard curve for each plate using synthesized DNA was used for the viral genome copy number quantification per mL of media.
For detection of infectious SARS-CoV-2 Delta virus and influenza H1N1 and H3N2 in tissues, thawed tissue samples were weighed and placed in 1 mL of DMEM and homogenized. Then, supernatants were serially diluted in 10-fold using in the same media and added to 96-well plates of 95% confluent Vero or MDCK cells containing 50 μL of the same medium. After 5 days, plates were scored for the presence of a cytopathic effect. TCID50 titers were calculated using the Reed and Muench method (66).
Measurement of vaccine-induced specific antibody and T cell responses.
Briefly, ELISA was performed as previously described using 0.75 μg/mL RBD recombinant protein or 0.5 μg/mL M2e peptide (35). To assess general T cell reactivity, mouse splenocytes from the sacrificed mice on day 28 were collected and plated in 48-well plates (2 × 106/200 μL per well) with SARS-CoV-2 S1 overlapping peptide pool, the influenza virus M2e peptide (1 μg/mL for each peptide), or RPMI (no peptide control). The measurement of the extracellular cytokines released from splenocytes was performed using the same previously described protocols (12).
Statistics.
Statistical analysis of antibody/cytokine levels was performed using the unpaired t test (considered significant at P ≥ 0.05) by GraphPad Prism 5.01 software. The statistical analysis of neutralizing antibodies was performed using the one-way ANOVA multiple-comparison tests followed by Tukey’s test by GraphPad Prism.
Data availability.
The data sets generated and/or analyzed during the current study are available from the corresponding author.
ACKNOWLEDGMENTS
We thank Kevin Coombs for kindly providing the U251 cell lines and William Peter Stefura for his technique support. T.A.O. is a recipient of the University Manitoba Graduate scholarship. This work was supported by the Canadian 2019 Novel Coronavirus (COVID-19) Rapid Research Funding (OV5-170710) by the Canadian Institutes of Health Research (CIHR, cihr-irsc.gc.ca/e/193.html) and Research Manitoba (researchmanitoba.ca), and CIHR COVID-19 Variant Supplement grant (VS1-175520 to X.Y., S.K., K.R.F., and D.K.). This work was also supported by the Manitoba Research Chair Award from the Research Manitoba (R.M.) to X.Y.
The experiments were conceived of and designed by X.Y., S.K., K.R.F., and D.K. The constructions and rescue of different rVSV viruses and the viral replication characterization were performed by Z.A. and X.Y. SARS-CoV-2 SP-pseudotyped virus infections and the neutralizing experiments were conducted by M.J.O. and Z.A. Animal immunization studies were performed by M.J.O., T.A.O., Z.A., and M.Z.; cytokine production analyses were performed by M.J.O., M.Z., and Z.A.; animal influenza challenge experiments were carried out by T.A.O. and M.J.O.; SARS-CoV-2 challenge experiments and different analyses were carried out by B.W., R.V., T.T., and C.M. The initial draft of the paper was written by Z.A., M.J.O., T.A.O., and B.W. All other authors contributed to editing the paper into its final form. The work was managed and supervised by S.K., K.R.F., D.K., and X.Y.
We declare we have no conflicts of interest.
Contributor Information
Darwyn Kobasa, Email: darwyn.kobasa@phac-aspc.gc.ca.
Xiaojian Yao, Email: xiao-jian.yao@umanitoba.ca.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Kim JH, Marks F, C JD. 2021. Looking beyond COVID-19 vaccine phase 3 trials. Nat Med 27:205–211. 10.1038/s41591-021-01230-y. [DOI] [PubMed] [Google Scholar]
- 2.Tregoning JS, Flight KE, Higham SL, Wang Z, Pierce BF. 2021. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat Rev Immunol 21:626–636. 10.1038/s41577-021-00592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Z, Kang H, Li S, Zhao X. 2020. Understanding the neurotropic characteristics of SARS-CoV-2: from neurological manifestations of COVID-19 to potential neurotropic mechanisms. J Neurol 267:2179–2184. 10.1007/s00415-020-09929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed MU, Hanif M, Ali MJ, Haider MA, Kherani D, Memon GM, Karim AH, Sattar A. 2020. Neurological Manifestations of COVID-19 (SARS-CoV-2): a Review. Front Neurol 11:518. 10.3389/fneur.2020.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim GU, Kim MJ, Ra SH, Lee J, Bae S, Jung J, Kim SH. 2020. Clinical characteristics of asymptomatic and symptomatic patients with mild COVID-19. Clin Microbiol Infect 26:948–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin L, Jiang X, Zhang Z, Huang S, Zhang Z, Fang Z, Gu Z, Gao L, Shi H, Mai L, Liu Y, Lin X, Lai R, Yan Z, Li X, Shan H. 2020. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 69:997–1001. 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 7.Dougherty K, Mannell M, Naqvi O, Matson D, Stone J. 2021. SARS-CoV-2 B.1.617.2 (Delta) variant COVID-19 outbreak associated with a gymnastics facility—Oklahoma, April–May 2021. Morbidity and Mortality Wkly Report 70:1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma W, Yang J, Fu H, Su C, Yu C, Wang Q, de Vasconcelos ATR, Bazykin GA, Bao Y, Li M. 2022. Genomic perspectives on the emerging SARS-CoV-2 omicron variant. Genomics Proteomics Bioinformatics. 10.1016/j.gpb.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, Duyvesteyn HME, Ginn HM, Mentzer AJ, Tuekprakhon A, Nutalai R, Wang B, Dijokaite A, Khan S, Avinoam O, Bahar M, Skelly D, Adele S, Johnson SA, Amini A, Ritter TG, Mason C, Dold C, Pan D, Assadi S, Bellass A, Omo-Dare N, Koeckerling D, Flaxman A, Jenkin D, Aley PK, Voysey M, Costa Clemens SA, Naveca FG, Nascimento V, Nascimento F, Fernandes da Costa C, Resende PC, Pauvolid-Correa A, Siqueira MM, Baillie V, Serafin N, Kwatra G, Da Silva K, Madhi SA, Nunes MC, Malik T, Openshaw PJM, Baillie JK, Semple MG, ISARIC4C Consortium., et al. 2022. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 185:467–484. 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Deng A, Li K, Hu Y, Li Z, Shi Y, Xiong Q, Liu Z, Guo Q, Zou L, Zhang H, Zhang M, Ouyang F, Su J, Su W, Xu J, Lin H, Sun J, Peng J, Jiang H, Zhou P, Hu T, Luo M, Zhang Y, Zheng H, Xiao J, Liu T, Tan M, Che R, Zeng H, Zheng Z, Huang Y, Yu J, Yi L, Wu J, Chen J, Zhong H, Deng X, Kang M, Pybus OG, Hall M, Lythgoe KA, Li Y, Yuan J, He J, Lu J. 2022. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat Commun 13:1–9. 10.1038/s41467-022-28089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisman DN, Tuite AR. 2021. Evaluation of the relative virulence of novel SARS-CoV-2 variants: a retrospective cohort study in Ontario, Canada. CMAJ 193:E1619–E1625. 10.1503/cmaj.211248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ao Z-j, Ouyang MJ, Olukitibi TA, Yao X-J. 2022. SARS-CoV-2 Delta spike protein enhances the viral fusogenicity and inflammatory cytokine production. iScience 25:104759. 10.1016/j.isci.2022.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nickol ME, Kindrachuk J. 2019. A year of terror and a century of reflection: perspectives on the great influenza pandemic of 1918–1919. BMC Infect Dis 19:117. 10.1186/s12879-019-3750-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kedzierska K, Van de Sandt C, Short KR. 2018. Back to the future: lessons learned from the 1918 influenza pandemic. Front Cell Infect Microbiol 8:343. 10.3389/fcimb.2018.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paget J, Spreeuwenberg P, Charu V, Taylor RJ, Iuliano AD, Bresee J, Simonsen L, Viboud C, Global Seasonal Influenza-Associated Mortality Collaborator Network and GLaMOR Collaborating Teams. 2019. Global mortality associated with seasonal influenza epidemics: new burden estimates and predictors from the GLaMOR Project. J Global Health 9:e020421-21. 10.7189/jogh.09.020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weir JP, Gruber MF. 2016. An overview of the regulation of influenza vaccines in the United States. Influenza Other Respir Viruses 10:354–360. 10.1111/irv.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention. 2021. Past seasons vaccine effectiveness estimates. Centers for Disease Control and Prevention. https://www.cdc.gov/flu/vaccines-work/past-seasons-estimates.html. [Google Scholar]
- 18.Olukitibi T, Ao Z-j, Azizi H, Mahmoudi M, Coombs K, Kobasa D, Kobinger G, Yao X-j. 2022. Development and characterization of influenza M2 ectodomain and/or HA stalk-based DC-targeting vaccines for different influenza infections. Front Micro 13:937192. 10.3389/fmicb.2022.937192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emanuel J, Callison J, Dowd KA, Pierson TF, Feldmann H, Marzi A. 2018. A VSV-based Zika virus vaccine protects mice from lethal challenge. Sci Rep 8:11043. 10.1038/s41598-018-29401-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose NF, Marx PA, Luckay A, Nixon DF, Moretto W, Donahoe SM, Montefiori D, Roberts A, Buonocore L, Rose JK. 2001. An effective AIDS vaccine based on live attenuated vesicularstomatitis virus recombinants. Cell 106:539–549. 10.1016/S0092-8674(01)00482-2. [DOI] [PubMed] [Google Scholar]
- 21.DeBuysscher BL, Scott D, Marzi A, Prescott J, Feldmann H. 2014. Single-dose live-attenuated Nipah virus vaccines confer complete protection by eliciting anti-bodies directed against surface glycoproteins. Vaccine 32:2637–2644. 10.1016/j.vaccine.2014.02.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marzi A, Robertson SJ, Haddock E, Feldmann F, Hanley PW, Scott DP, Strong JE, Kobinger G, Best SM, H HF. 2015. VSV-EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science 349:739–742. 10.1126/science.aab3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suder E, Furuyama W, Feldmann H, Marzi A, de Wit E. 2018. The vesicular stomatitis virus-based Ebola virus vaccine: from concept to clinical trials. Hum Vaccin Immunother 14:2107–2113. 10.1080/21645515.2018.1473698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts A, Buonocore L, Price R, Forman J, Rose JK. 1999. Attenuated vesicular stomatitis viruses as vaccine vectors. J Virol 73:3723–3732. 10.1128/JVI.73.5.3723-3732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Case JB, Rothlauf PW, Chen RE, Kafai NM, Fox JM, Smith BK, Shrihari S, McCune BT, Harvey IB, Keeler SP, Bloyet L-M, Zhao H, Ma M, Adams LJ, Winkler ES, Holtzman MJ, Fremont DH, Whelan SPJ, Diamond MS. 2020. Replication-competent vesicular stomatitis virus vaccine vector protects against SARS-CoV-2-mediated pathogenesis in mice. Cell Host Microbe 28:465–474. 10.1016/j.chom.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malherbe DC, Kurup D, Wirblich C, Ronk AJ, Mire C, Kuzmina N, Shaik N, Periasamy S, Hyde MA, Williams JM, Shi P-Y, Schnell MJ, Bukreyev A. 2021. A single dose of replication-competent VSV-vectored vaccine expressing SARS-CoV-2 S1 protects against virus replication in a hamster model of severe COVID-19. NPJ Vaccines 6. 10.1038/s41541-021-00352-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yahalom-Ronen Y, Tamir H, Melamed S, Politi B, Shifman O, Achdout H, Vitner EB, Israeli O, Milrot E, Stein D, Cohen-Gihon I, Lazar S, Gutman H, Glinert I, Cherry L, Vagima Y, Lazar S, Weiss S, Ben-Shmuel A, Avraham R, Puni R, Lupu E, Bar-David E, Sittner A, Erez N, Zichel R, Mamroud E, Mazor O, Levy H, Laskar O, Yitzhaki S, Shapira SC, Zvi A, Beth-Din A, Paran N, Israely T. 2020. A single dose of recombinant VSV-ΔG-spike vaccine provides protection against SARS-CoV-2 challenge. Nat Commun 11:6402. 10.1038/s41467-020-20228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Donnell KL, Clancy CS, Griffin AJ, Shifflett K, Gourdine T, Thomas T, Long CM, Furuyama W, Marzi A. 2021. Optimization of single-dose VSV-based COVID-19 vaccination in hamsters. Front Immunol 12:788235. 10.3389/fimmu.2021.788235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuyama W, Shifflett K, Pinski AN, Griffin AJ, Feldmann F, Okumura A, Gourdine T, Jankeel A, Lovaglio J, Hanley PW, Thomas T, Clancy CS, Messaoudi I, O’Donnell KL, Marzi A. 2022. Rapid protection from COVID-19 in nonhuman primates vaccinated intramuscularly but not intranasally with a single dose of a vesicular stomatitis virus-based vaccine. mBio 13:e03379-21. 10.1128/mbio.03379-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim GN, Choi JA, Wu K, Saeedian N, Yang E, Park H, Woo SJ, Lim G, Kim SG, Eo SK, Jeong HW, Kim T, Chang JH, Seo SH, Kim NH, Choi E, Choo S, Lee S, Winterborn A, Li Y, Parham K, Donovan JM, Fenton B, Dikeakos JD, Dekaban GA, Haeryfar SMM, Troyer RM, Arts EJ, Barr SD, Song M, Kang CY. 2021. A vesicular stomatitis virus-based prime-boost vaccination strategy induces potent and protective neutralizing antibodies against SARS-CoV-2. PLoS Pathog 17:e1010092. 10.1371/journal.ppat.1010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ao Z-j, Wang L-j, Azizi H, Olukitibi T, Mahmoudi M, Kobinger G, Yao X-j. 2021. Development and evaluation of an Ebola virus glycoprotein mucin-like domain replacement system as a new dendritic cell-targeting vaccine approach against HIV-1. J Virol 95:e0236820. 10.1128/JVI.02368-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petit CM, Melancon JM, Chouljenko VN, Colgrove R, Farzan M, Knipe DM, Kousoulas KG. 2005. Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion. Virology 341:215–230. 10.1016/j.virol.2005.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ao Z-j, Chan M, Ouyang M, Abiola T, Mahmoudi M, Kobasa D, Yao X-J. 2021. Identification and evaluation of the inhibitory effect of Prunella vulgaris extract on SARS-coronavirus 2 virus entry. PLoS One 16:e0251649. 10.1371/journal.pone.0251649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robison CS, Whitt MA. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J Virol 74:2239–2246. 10.1128/jvi.74.5.2239-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olukitibi T, Ao Z-j, Azizi H, Mahmoudi M, Coombs K, Kobasa D, Kobinger G, Yao X-j. 2021. Development and characterization of influenza M2 ectodomain and/or HA stalk-based DC-targeting vaccines for different influenza infections. BioRxiv. 10.1101/2021.10.07.463539. [DOI] [PMC free article] [PubMed]
- 36.Witko SE, Kotash CS, Nowak RM, Johnson JE, Boutilier LA, Melville KJ, Heron SG, Clarke DK, Abramovitz AS, Hendry RM, Sidhu MS, Udem SA, Parks CL. 2006. An efficient helper-virus-free method for rescue of recombinant paramyxoviruses and rhadoviruses from a cell line suitable for vaccine development. J Virol Methods 135:91–101. 10.1016/j.jviromet.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Foster KA, Oster CG, Mayer MM, Avery ML, Audus KL. 1998. Characterization of the A549 cell line as a type ii pulmonary epithelial cell model for drug metabolism. Exp Cell Res 243:359–366. 10.1006/excr.1998.4172. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto R, Lin LS, Lowe R, Warren MK, White TJ. 1990. The human lung fibroblast cell line, MRC-5, produces multiple factors involved with megakaryocytopoiesis. J Immunol 144:1808–1816. [PubMed] [Google Scholar]
- 39.Kirola L. 2021. Genetic emergence of B.1.617.2 in COVID-19. New Microbes New Infect 43:100929. 10.1016/j.nmni.2021.100929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis C, Logan N, Tyson G, Orton R, Harvey WT, Perkins JS, Mollett G, Blacow RM, The C-G, Peacock TP, Barclay WS, Cherepanov P, Palmarini M, Murcia PR, Patel AH, Robertson DL, Haughney J, Thomson EC, Willett BJ, on behalf of the C-DVCSi. 2021. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLoS Pathog 17:e1010022. 10.1371/journal.ppat.1010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O. 2021. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596:276–280. 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 42.Tian D, Sun Y, Zhou J, Ye Q. 2021. The global epidemic of the SARS-CoV-2 Delta variant, key spike mutations and immune escape. Front Immunol 12. 10.3389/fimmu.2021.751778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia X. 2021. Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design. Viruses 13:109. 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffin BD, Warner BM, Chan M, Valcourt E, Tailor N, Banadyga L, Leung A, He S, Boese AS, Audet J, Cao W, Moffat E, Garnett L, Tierney K, Tran KN, Albietz A, Manguiat K, Soule G, Bello A, Vendramelli R, Lin J, Deschambault Y, Zhu W, Wood H, Mubareka S, Safronetz D, Strong JE, Embury-Hyatt C, Kobasa D. 2021. Host parameters and mode of infection influence outcome in SARS-CoV-2-infected hamsters. iScience 24:103530. 10.1016/j.isci.2021.103530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dolgin E. 2021. COVID vaccine immunity is waning - how much does that matter? Nature 597:606–607. 10.1038/d41586-021-02532-4. [DOI] [PubMed] [Google Scholar]
- 46.Azzi L, Dalla Gasperina D, Veronesi G, Shallak M, Ietto G, Iovino D, Baj A, Gianfagna F, Maurino V, Focosi D, Maggi F, Ferrario MM, Dentali F, Carcano G, Tagliabue A, Maffioli LS, Accolla RS, Forlani G. 2022. Mucosal immune response in BNT162b2 COVID-19 vaccine recipients. EBioMedicine 75:103788. 10.1016/j.ebiom.2021.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Gier B, Andeweg S, Joosten RT, Schegget R, Smorenburg N, van de Kassteele J, Hahné SJ, van den Hof S, de Melker HE, Knol MJ, members of the RIVM COVID-19 surveillance and epidemiology team. 2021. Vaccine effectiveness against SARS-CoV-2 transmission and infections among household and other close contacts of confirmed cases, the Netherlands, February to May 2021. Euro Surveill 26:2100640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jalali N, Brustad HK, Frigessi A, MacDonald E, Meijerink H, Feruglio S, Nygård K, Rø GI, Madslien EH, De Blasio BF. 2022. Increased household transmission and immune escape of the SARS-CoV-2 Omicron variant compared to the Delta variant: evidence from Norwegian contact tracing and vaccination data. medRxiv. 10.1101/2022.02.07.22270437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dogan M, Kozhaya L, Placek L, Gunter C, Yigit M, Hardy R, Plassmeyer M, Coatney P, Lillard K, Bukhari Z, Kleinberg M, Hayes C, Arditi M, Klapper E, Merin N, Liang BT-T, Gupta R, Alpan O, Unutmaz D. 2021. SARS-CoV-2 specific antibody and neutralization assays reveal the wide range of the humoral immune response to virus. Commun Biol 4:129. 10.1038/s42003-021-01649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu W, Zou P, Ding J, Lu Y, Chen YH. 2005. Sequence comparison between the extracellular domain of M2 protein human and avian influenza A virus provides new information for bivalent influenza vaccine design. Microbes Infect 7:171–177. 10.1016/j.micinf.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 51.Tsuda Y, Safronetz D, Brown K, LaCasse R, Marzi A, Ebihara H, Feldmann H. 2011. Protective efficacy of a bivalent recombinant vesicular stomatitis virus vaccine in the Syrian hamster model of lethal Ebola virus infection. J Infect Dis 204 Suppl 3:S1090–7. 10.1093/infdis/jir379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marzi A, Hanley P, Haddock E, Martellaro C, G K, H F. 2016. Efficacy of vesicular stomatitis virus-Ebola virus postexposure treatment in Rhesus macaques infected with Ebola virus Makona. J Infect Dis 214:S360–S366. 10.1093/infdis/jiw218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Menicucci AR, Jankeel A, Feldmann H, Marzi A, Messaoudi I. 2019. Antiviral innate responses induced by VSV-EBOV vaccination contribute to rapid protection. mBio 10. 10.1128/mBio.00597-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aleebrahim-Dehkordi E, Molavi B, Mokhtari M, Deravi N, Fathi M, Fazel T, Mohebalizadeh M, Koochaki P, Shobeiri P, Hasanpour-Dehkordi A. 2022. T helper type (Th1/Th2) responses to SARS-CoV-2 and influenza A (H1N1) virus: from cytokines produced to immune responses. Transpl Immunol 70:101495. 10.1016/j.trim.2021.101495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pavel AB, Glickman JW, Michels JR, Kim-Schulze S, Miller RL, Guttman-Yassky E. 2021. Th2/Th1 cytokine imbalance is associated with higher COVID-19 risk mortality. Front Genet 12:706902. 10.3389/fgene.2021.706902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Furuyama W, Reynolds P, Haddock E, Meade-White K, Quynh Le M, Kawaoka Y, Feldmann H, Marzi A. 2020. A single dose of a vesicular stomatitis virus-based influenza vaccine confers rapid protection against H5 viruses from different clades. NPJ Vaccines 5:4. 10.1038/s41541-019-0155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones SM, Stroher U, Fernando L, Qiu X, Alimonti J, Melito P, Bray M, Klenk HD, Feldmann H. 2007. Assessment of a vesicular stomatitis virus-based vaccine by use of the mouse model of Ebola virus hemorrhagic fever. J Infect Dis 196 Suppl 2:S404–12. 10.1086/520591. [DOI] [PubMed] [Google Scholar]
- 58.de Wit E, Marzi A, Bushmaker T, Brining D, Scott D, Richt JA, Geisbert TW, Feldmann H. 2015. Safety of recombinant VSV-Ebola virus vaccine vector in pigs. Emerg Infect Dis 21:702–704. 10.3201/eid2104.142012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mire CE, Miller AD, Carville A, Westmoreland SV, Geisbert JB, Mansfield KG, Feldmann H, Hensley LE, Geisbert TW. 2012. Recombinant vesicular stomatitis virus vaccine vectors expressing filovirus glycoproteins lack neurovirulence in nonhuman primates. PLoS Negl Trop Dis 6:e1567. 10.1371/journal.pntd.0001567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geisbert TW, Daddario-Dicaprio KM, Lewis MG, Geisbert JB, Grolla A, Leung A, Paragas J, Matthias L, Smith MA, Jones SM, Hensley LE, Feldmann H, Jahrling PB. 2008. Vesicular stomatitis virus-based Ebola vaccine is well-tolerated and protects immunocompromised nonhuman primates. PLoS Pathog 4:e1000225. 10.1371/journal.ppat.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ao Z, Wang L, Mendoza EJ, Cheng K, Zhu W, Cohen EA, Fowke K, Qiu X, Kobinger G, Yao X. 2019. Incorporation of Ebola glycoprotein into HIV particles facilitates dendritic cell and macrophage targeting and enhances HIV-specific immune responses. PLoS One 14:e0216949. 10.1371/journal.pone.0216949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu J, Li Z, He X, Gebre MS, Bondzie EA, Wan H, Jacob-Dolan C, Martinez DR, Nkolola JP, Baric RS, Barouch DH. 2021. Deletion of the SARS-CoV-2 spike cytoplasmic tail increases infectivity in pseudovirus neutralization assays. J Virol 95. 10.1128/JVI.00044-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ao Z, Fowke KR, Cohen EA, Yao X. 2005. Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import. Retrovirology 2:62. 10.1186/1742-4690-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donofrio G, Franceschi V, Macchi F, Russo L, Rocci A, Marchica V, Costa F, Giuliani N, Ferrari C, Missale G. 2021. A simplified SARS-CoV-2 pseudovirus neutralization assay. Vaccines 9:389. 10.3390/vaccines9040389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu J, Gao Q, He C, Huang A, Tang N, Wang K. 2020. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis 7:551–557. 10.1016/j.gendis.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. American J Epidemiology 27:493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated and/or analyzed during the current study are available from the corresponding author.