Abstract
A novel gene, hetF, was identified as essential for heterocyst development in the filamentous cyanobacterium Nostoc punctiforme strain ATCC 29133. In the absence of combined nitrogen, hetF mutants were unable to differentiate heterocysts, whereas extra copies of hetF in trans induced the formation of clusters of heterocysts. Sequences hybridizing to a hetF probe were detected only in heterocyst-forming cyanobacteria. The inactivation and multicopy effects of hetF were similar to those of hetR, which encodes a self-degrading serine protease thought to be a central regulator of heterocyst development. Increased transcription of hetR begins in developing cells 3 to 6 h after deprivation for combined nitrogen (N step-down), and the HetR protein specifically accumulates in heterocysts. In the hetF mutant, this increase in hetR transcription was delayed, and a hetR promoter::green fluorescent protein (GFP) transcriptional reporter indicated that increased transcription of hetR occurred in all cells rather than only in developing heterocysts. When a fully functional HetR-GFP fusion protein was expressed in the hetF mutant from a multicopy plasmid, HetR-GFP accumulated nonspecifically in all cells under nitrogen-replete conditions; when expressed in the wild type, HetR-GFP was observed only in heterocysts after N step-down. HetF therefore appears to cooperate with HetR in a positive regulatory pathway and may be required for the increased transcription of hetR and localization of the HetR protein in differentiating heterocysts.
Diazotrophic cyanobacteria growing in the absence of combined nitrogen face a dilemma: nitrogenase, the enzyme complex that catalyzes the reduction of dinitrogen gas to ammonium, is extremely sensitive to oxygen, the primary product of the cyanobacterial oxygenic photoautotrophic life style. Certain filamentous forms, such as Anabaena and Nostoc, respond to the dilemma by differentiating heterocysts, which are highly specialized, nearly anoxic cells. Heterocysts maintain a low intercellular level of oxygen through several means. First, the presence of an envelope composed of glycolipid and polysacccharide retards the diffusion of atmospheric gases into the cell. Second, the oxygen-producing photosystem II complex is deactivated and the phycobiliprotein light-harvesting pigments are degraded. Finally, any residual oxygen is scavenged by an enhanced level of respiration (24).
Heterocyst development has been studied primarily in Anabaena sp. strain PCC 7120 and more recently in the symbiotically competent Nostoc punctiforme strain ATCC 29133 (16). In N. punctiforme, within 12 h of combined nitrogen limitation (N step-down), cells at regular intervals degrade phycobiliproteins as the first sign of development. By 24 to 36 h, mature heterocysts are observed singly at regular intervals, at a frequency of approximately 8% of the cell population. Although many genes have been identified as required for heterocyst function (23), only three, ntcA (21), hanA (13), and hetR (3), are known to be essential to the initiation of heterocyst development. Mutation of any one of these genes leads to a Het− phenotype in Anabaena strain 7120. By definition, in any Het− mutant, heterocyst development is blocked sufficiently early that the cell-specific degradation of phycobiliproteins does not occur; no morphological traits of heterocysts can be observed on prolonged nitrogen limitation and the cultures cannot grow in the absence of a source of combined nitrogen.
Of the three genes that are essential for the initiation of heterocyst development, only hetR appears to be involved specifically in the differentiation process. NtcA, which belongs to the Crp family of bacterial transcriptional activators, appears to function as a nitrogen-dependent global transcriptional regulator in all cyanobacteria (14, 22). Anabaena strain 7120 ntcA mutants fail to grow on nitrate, initiate heterocyst differentiation, or induce nitrogenase expression (9, 21). NtcA influences the expression of a number of heterocyst-specific genes, including indirectly hetR (9) and other genes expressed later in heterocyst development (15, 20, 21). HanA belongs to the bacterial histone-like HU protein family (13). Mutations in hanA resulted in a pleiotrophic Het− mutant that required secondary mutations to maintain viability, indicating that hanA is probably an essential regulator in other processes as well (13).
In contrast to ntcA mutants, Anabaena strain 7120 hetR mutants are Het− but can grow on nitrate (3). Thus, HetR is not a general nitrogen regulator and appears to be temporally the earliest known regulatory protein specific to heterocyst differentiation. When hetR is present in multiple copies, heterocysts differentiate at a higher frequency in strings of two or more (multiple contiguous heterocyst [Mch] phenotype) in the absence of combined nitrogen (3). hetR is transcribed at a low level in nitrate-grown cells and induced within 3.5 h following N step-down (2). The transcriptional induction is autocatalytic, requiring a functional HetR, and is confined to cells developing into heterocysts (2). Although no NtcA-binding site has been identified upstream of hetR, induction of hetR requires a functional copy of ntcA (9). HetR is a serine-type protease which degrades itself but accumulates in heterocysts (25, 26). HetR is also modified in an unknown fashion upon N step-down (25), but the exact mechanisms through which HetR promotes heterocyst development in cells remain unknown.
We report in this paper the identification of a gene, hetF, in N. punctiforme whose mutation and multicopy effects are similar to those of hetR and whose product directly or indirectly affects the specific accumulation of HetR in developing cells.
MATERIALS AND METHODS
Cultures and media.
Wild-type and mutant strains of N. punctiforme (Table 1) were grown in liquid Allen and Arnon medium diluted fourfold (AA/4) and on Noble agar-solidified AA medium plates as previously described (6). When necessary, the medium was supplemented with 2.5 mM NH4Cl buffered with 5 mM morpholineproparasulfonic acid (MOPS) (pH 7.8) (+N). Neomycin at 25 μg/ml, ampicillin at 10 μg/ml, and erythromycin at 15 μg/ml were used for the selection and maintenance of recombinant N. punctiforme strains. For N step-down time courses, 500-ml cultures of vegetative filaments were grown in AA/4+N to 2 to 3 μg of chlorophyll a/ml, collected by centrifugation at 1,000 × g for 10 min, washed three times by resuspension with AA/4 medium, and subjected to centrifugation. Samples containing approximately equal biomass from just before (time zero) and at 3, 6, 12, 24, and 36 h after the N step-down were collected by centrifugation and frozen in liquid nitrogen for RNA isolation. Mature heterocysts at typical frequencies were observed by light microscopy 36 h after N step-down.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| N. punctiforme | ||
| ATCC 29133 | Wild type | 16 |
| UCD 415 | hetR::Tn5-1063 | This study |
| UCD 401 | hetF::Tn5-1063 | This study |
| UCD 416 | hetF::Tn5-1063 | This study |
| UCD 445 | hetF::pRL271 | This study |
| UCD 444 | ntcA::Ω Nmr cassette | This study |
| UCD 483 | Wild type carrying pSCR93 | This study |
| UCD 484 | UCD 416 carrying pSCR93 | This study |
| UCD 485 | Wild type carrying pSCR90 | This study |
| UCD 486 | UCD 416 carrying pSCR90 | This study |
| UCD 487 | Wild type carrying pSCR60 | This study |
| UCD 488 | UCD 416 carrying pSCR60 | This study |
| UCD 489 | UCD 415 carrying pSCR60 | This study |
| E. coli | ||
| DH5α-MCR | Methylation-dependent restriction defective derivative of strain DH5α; used for general cloning | Gibco-BRL |
| Plasmids | ||
| pBluescript KS(+); pBS | Cloning vector; Apr | Stratagene |
| pRL271 | Conjugatable nonreplicating cyanobacterial vector containing SacB; Emr Cmr | 4 |
| pSCR202 | Multicopy replicating plasmid for use in N. punctiforme and E. coli; Apr | 19 |
| pGFPCR | Cloning vector for E. coli with GFPuv as reporter; modified restriction sites from pGFPuv (Clonetech); Apr | 7 |
| pSCR32 | 1.0-kb BspDI-BamHI fragment containing sequence internal to hetF upstream of the Tn5-1063 insertion site in UCD 401 cloned in pBS | This study |
| pSCR45 | Insert in pSCR32 cloned into XhoI and SstI sites of pRL271 | This study |
| pSCR58 | 2.8-kb EcoRV fragment containing hetF 3′ sequence subcloned from cosmid clone 15A8 into pBS | This study |
| pSCR60 | 3.5-kb NheI-BstEII fragment containing hetF as the only ORF in pSCR202 | This study |
| pSCR79 | 736-bp C-terminal coding sequence of hetR fused in frame with GFPuv in pGFPCR | This study |
| pSCR81 | 3.3-kb EcoRI-PvuII fragment containing hetR as the only ORF subcloned from cosmid clone 18C8 into pBS | This study |
| pSCR84 | 0.7-kb C-terminal coding sequence of hetR in pSCR81 replaced with the 1.5-kb hetR3′-gfp fusion from pSCR79 | This study |
| pSCR90 | 4.1-kb BssHI insert from pSCR84 cloned into pSCR202, containing the full: length hetR-gfp fusion driven by native promoters | This study |
| pSCR93 | 646-bp hetR upstream sequence and first 8 codons of hetR fused in frame with gfp, cloned in pSCR202 | This study |
All Escherichia coli strains (Table 1) were grown in Luria-Bertani broth supplemented with antibiotics at the following concentrations: kanamycin, 25 μg/ml; ampicillin, 100 μg/ml; and chloramphenicol, 30 μg/ml.
Isolation and characterization of the hetF mutant strains.
Methods for transposon mutagenesis of N. punctiforme with Tn5-1063 and the isolation and characterization of mutants unable to fix nitrogen in air (Fox−) were described previously (6). Het− mutants were distinguished from Fox− mutants by microscopic examination of strains during N step-down.
To identify the sequence of hetF, the transposon in strain UCD 416, together with 3.0 kbp of flanking DNA, was recovered from DraI-digested genomic DNA as a plasmid. The flanking DNA was found to contain approximately 1.8 kbp of the 5′ region of an open reading frame (ORF) and 1.2 kbp of upstream sequence. To recover the remainder of the ORF, a 1.0-kbp BspDI-BamHI fragment containing sequence internal to the ORF was used to probe a cosmid library consisting of random sheared fragments of the N. punctiforme genome (6). A 2.8-kbp EcoRV fragment of the cosmid clone 15A8 hybridized to the probe, and this fragment was cloned into pBluescript to form pSCR58. The insert in pSCR58 was sequenced and contains the remaining 3′ region of the ORF, now designated hetF. To complement the original mutant, a 3.5-kbp fragment containing hetF as the only ORF was isolated from NheI-BstEII-digested cosmid 15A8. The fragment was Klenow treated to blunt the ends and cloned into Ecl136II-digested pSCR202 (19) to form pSCR60. pSCR60 was then transformed by electroporation into the wild-type strain and strain UCD 416 by a previously described method (19). N. punctiforme maintains 15 copies of pSCR202-based plasmids per chromosome (19). To reconstruct the hetF mutation, the 1.0-kbp BspDI-BamHI fragment used above in cosmid hybridizations was cloned into the pBluescript KS(+) vector to form pSCR32. The fragment was excised by SstI and XhoI, ligated into the complementary site in pRL271 (4) to form pSCR45, and transferred by conjugation into wild-type N. punctiforme via triparental mating (6). Strain UCD 445, a Het− mutant whose hetF locus was disrupted by pSCR45 via a single recombination event, was identified as an Emr colony. Its genotype was verified by Southern analysis, and cultures were maintained in the presence of erythromycin. Bacterial luciferase assays were carried out as previously described (5).
Molecular biology procedures.
Preparations and manipulations of plasmid DNA from E. coli were done using standard methods (17). Large-scale plasmid purifications from E. coli were carried out using a commercial kit (Qiagen). DNA restriction and modification enzymes were obtained from New England Biolabs or Gibco-BRL and used as specified by the manufacturer. Total cyanobacterial genomic and plasmid DNAs were isolated as previously described (6). DNA sequencing was performed by contract using ABI Prism sequencers and the dye termination method. Total RNA was isolated as previously described (18). For Northern blots, total RNA was denatured with a mixture of 6% (vol/vol) formaldehyde and 50% (vol/vol) formamide and separated on a 1% agarose gel containing 2% (vol/vol) formaldehyde. For RNA slot blots, 3 to 5 μg of total RNA was loaded on a blot using a Bio-Rad Bio-dot SF apparatus. Hybridization probes were labeled with [α-32P]dCTP (Dupont NEN) by using a random-priming reaction kit from Gibco-BRL. All PCR amplifications were done using reagents from Amersham Scientific. DNA and RNA hybridization analyses were performed with GeneScreen Plus membrane in 50% formamide hybridization buffers as previously described (19). Blots were exposed to a Molecular Dynamics phosphorscreen, and hybridization signals were quantitated on a Storm PhosphorImager system using ImageQuant software (Molecular Dynamics). The hybridization probe for hetR was generated by PCR amplification of an 851-bp fragment from genomic DNA using primers P33-4 (5′-GCTGGACTTAGTGATATCTG-3′) and P34-6 (5′-CCAAGGAGAATCTATGCGTG-3′). The 2,842-bp probe used in Northern and Southern analyses for hetF was amplified from pSCR60 using primers P44-4 (5′-CCGAATCAGAAACATTCAGG-3′) and P58-4 (5′-CCTTTAGACAGCCTGAACC-3′).
Construction of GFP fusion reporters.
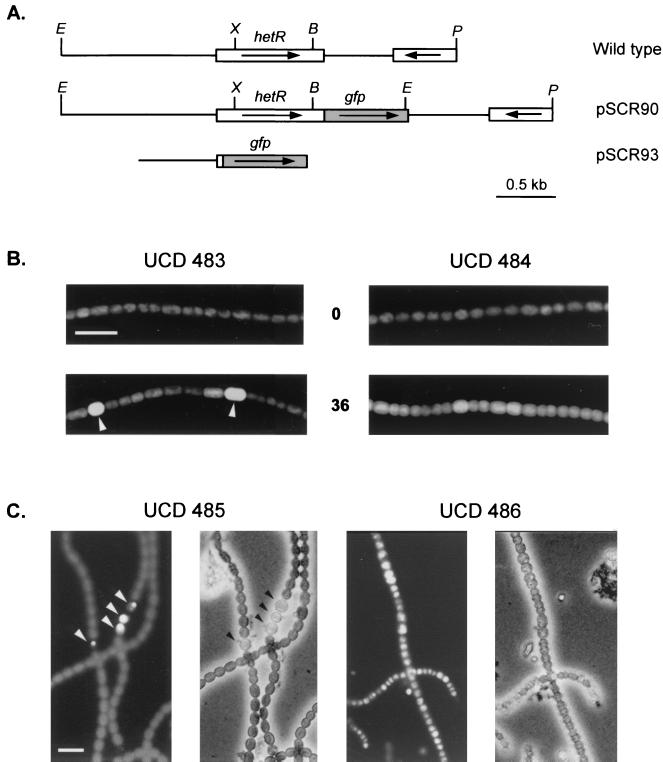
To construct the hetR promoter::gfp (PhetR::gfp) transcriptional reporter (pSCR93), a 670-bp fragment was amplified from the cosmid clone 18C8 using primers PhetR5-1 (5′-CGCGCCAGACCCTCAAGCCAACGGCTTATACGC-3′) and PhetR5-2 (5′-CGCATACCGGTATCAGATCTATGTCGTTACTC-3′). This fragment, which contains 646 bp upstream of the translational start and the first eight codons of hetR, was fused in frame with green fluoroscent protein (GFP) by digestion with XbaI and AgeI and cloned into the corresponding sites in vector pGFPCR (7) to form pSCR92. A 1.4-kbp XbaI-StuI fragment containing the PhetR::gfp fusion was cloned into Ecl136II-digested pSCR202 to form the plasmid pSCR93. To construct the full-length HetR-GFP fusion (pSCR90), a 736-bp fragment coding for the C-terminal region of HetR was amplified with primers PhetRGFP1 (5′-CCTATCTAGAGCAGGGACAAAACCTGCG-3′) and PhetRGFP2 (5′-GTCTTCTTTTTCACCAAAAACCGTCTG-3′). The fragment was end filled with Klenow and cloned into XbaI-digested, end-filled pGFPCR to form pSCR79. This step generated an in-frame fusion between the sequence coding for the C terminus of HetR and GFP. A 3.3-kbp EcoRI-PvuII fragment containing hetR was subcloned from cosmid clone 18C8 to form pSCR81. The sequence coding for the C-terminal HetR-GFP fusion protein in pSCR79 was excised by XbaI and StuI and ligated into XbaI-BsaBI-digested pSCR81, thereby replacing the wild-type 3′ hetR sequence with one fused to the GFP coding sequence to form pSCR84. The 4.1-kbp fragment insert in pSCR84 was cut out by BssHII, end filled, and ligated into Ecl136II-digested pSCR202 to form pSCR90 (see Fig. 5A). The coding fidelity of the hetR fusion was confirmed by sequence analysis and phenotypic complementation of a hetR mutant.
FIG. 5.
GFP reporter expression in wild-type and mutant cells. (A) Physical map of the GFP fusion constructs. The arrows show the transcriptional orientation of the ORFs. The top schematic shows a map of the 3.3-kb region containing hetR subcloned from cosmid clone 18C8. The middle schematic shows the insert in pSCR90 and is essentially the same as for the wild type, except that the gfp sequence is fused in frame with hetR at its 3′ end. The bottom schematic shows the insert in pSCR93 with the gfp sequence fused in frame with first 8 codon sequence of hetR. Restriction sites: B, BsaBI; E, EcoRI; P, PvuII; and X, XbaI. (B and C) Photomicrographs of N. punctiforme GFP-expressing strains. Bars, 10 μm. Heterocysts are indicated by arrowheads. (B) Blue-light-excited, GFP emission epifluorescence images of the wild-type strain and strain UCD 416 carrying plasmid pSCR93 (strains UCD 483 and UCD 484, respectively). Numbers in the center indicate hours after N step-down. (C) Blue-light-excited epifluorescent and phase-contrast images of the wild-type strain and strain UCD 416 carrying pSCR90 (strains UCD 485 and UCD 486, respectively). Strain UCD 485 was grown with nitrate, and strain UCD 486 was grown with ammonium.
Image acquisition and analysis.
Images of N. punctiforme strains in the N step-down time course and strains carrying pSCR93 were captured by a Hamamatsu C4742-95-12 digital charge-coupled device camera mounted on a Nikon Eclipse E600 microscope. Images were captured with Openlab 2.2 and edited with Adobe Photoshop 5.0. Fluorescence levels of original captured images were measured using NIH Image SXM 1.62 (http://reg.ssci.liv.ac.uk). Phase-contrast light microscopy and other epifluorescence images were taken with a Zeiss Universal III microscope fitted with an internal camera. The images were captured on film, scanned, and edited into Adobe Photoshop 5.0.
Sequence analyses.
Protein sequence alignments were carried out using the GAP program in SeqWeb v.1.2 (Genetics Computer Group, Inc.). Similarity searches were carried out using the BLAST program against the databases available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/blast) (1), the unfinished Anabaena strain 7120 sequence database at the Kasuza Institute website (http://www.kazusa.or.jp/cyano/anabaena/), and the unfinished N. punctiforme, Prochlorococcus marinus, and marine Synechococcus sp. genome sequence databases at the Joint Genome Institute website (http://www.jgi.doe.gov). Protein motif searches were carried out using ProfileScan at the Swiss Institute for Experimental Cancer Research website (http://www.isrec.isb-sib.ch/software/PFSCAN_form.html). Transmembrane domain prediction was based on results from the SOSUI program at the Tokyo University of Technology and Agriculture website (http://sosui.proteome.bio.tuat.ac.jp) and TMHMM v.2.0 at the Technical University of Denmark website (http://www.cbs.dtu.dk/services/TMHMM-2.0).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 4,975-bp region containing hetF and the 3,776-bp region containing hetR are AF288130 and AF318069, respectively.
RESULTS
Isolation of Het− mutants, and identification of hetF and the hetF mutant phenotype.
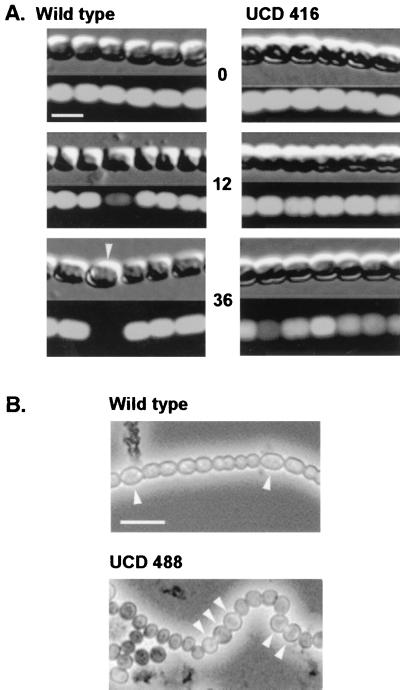
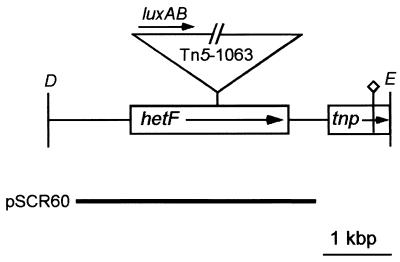
To identify genes essential to early heterocyst development in N. punctiforme, we generated 25,000 Nmr mutants by Tn5-1063 transposition, from which 69 Fox− mutants were identified. Six Het− mutants were identified by microscopically screening the Fox− mutants during N step-down. Het− mutants exhibited no patterned, cell-specific degradation of phycobiliproteins (bleaching) as seen by epifluorescence microscopy between 12 and 24 h during N step-down, and they showed no morphological signs of differentiation and displayed a loss of viability on prolonged incubation in nitrogen-limited medium (Fig. 1A). Mutant vegetative cells will eventually bleach as nitrogen reserves are mobilized, but the bleaching of vegetative cells does not appear in a specific pattern. Of the six mutants, sequence analysis revealed that one, strain UCD 415, has a Tn5-1063 insertion in an ORF encoding a protein with 92% similarity to HetR of Anabaena strain 7120. Its complete characterization will be published elsewhere. The remainder had insertions in an ORF with no significant similarity to genes in the current database. We designated the ORF hetF. Similar to strain UCD 415, hetF mutants are Het− (Fig. 1A). A 4,975-bp region surrounding the hetF insertion in strain UCD 416 was sequenced on both strands. hetF was 2.4 kbp in length, and there was no discernible ORF within 1.2 kbp upstream of the hetF presumptive ATG translational start site (Fig. 2). A gene encoding a protein with high homology to a transposase found in Synechocystis strain PCC 6803 is located downstream of hetF, although a premature stop codon is present 0.7-kbp after the presumptive ATG start site. To reconstruct the hetF mutation, an internal fragment of hetF was cloned into pRL271 and transferred into the wild type. The recombinant strain, UCD 445, exhibited the expected Het− phenotype upon N step-down, and Southern analysis verified the specific disruption of the hetF gene (data not shown).
FIG. 1.
Photomicrographs of N. punctiforme filaments. Arrowheads indicate heterocysts. (A) Typical filament of wild-type and hetF mutant strain UCD 416 in liquid culture undergoing N step-down. The top panel in each block shows Normarski interference images of filaments, while the bottom panel shows the same field of view in epifluorescence, showing phycobiliprotein (green light)-excited emission. Numbers in the center column indicate hours after N step-down. Bar, 5 μm. (B) Phase-contrast images of wild-type N. punctiforme and strain UCD 488 (strain UCD 416 carrying pSCR60) taken 36 to 48 h after N step-down. Bar, 10 μm. Images were processed and edited with Adobe Photoshop 5.0.
FIG. 2.
Physical map of the 4,975-bp sequence containing the hetF locus. The orientations of the ORFs are indicated by arrows. The inverted triangle shows the Tn5-1063 insertion and luxAB orientation in strain UCD 416. Incomplete tnp has similarity to Synechocystis strain PCC 6803 transposase; the diamond shows the premature stop codon. The heavy line indicates the fragment cloned into pSCR60. The DraI (D) and EcoRV (E) restriction sites are shown. Not all sites are shown.
When hetF on plasmid pSCR60 was transformed by electroporation into both the wild type and strain UCD 416 to generate strains UCD 487 and UCD 488, respectively, vegetative filament morphology and heterocyst development (Fig. 1B) were both affected. Normally straight vegetative filaments became irregular, due apparently to cell septation out of the normal plane of division. pSCR60, replicated in multiple copies, not only restored the ability of strain UCD 416 to differentiate heterocysts but also caused an increase in the frequency of heterocysts when cells were shifted into medium without combined nitrogen (Fig. 1B). Most heterocysts appeared as two or three contiguous cells, but single heterocysts were also observed. The heterocysts appear to be fragile, and the cultures do not survive prolonged incubation in medium without combined nitrogen. No heterocysts were observed when strains UCD 487 and UCD 488 were grown with ammonium or nitrate. To test whether the increased copy number of hetF can complement the mutation in hetR, pSCR60 was transformed into the hetR mutant background to form strain UCD 489. pSCR60 did not restore the ability to differentiate heterocysts, since strain UCD 489 exhibited the Het− phenotype regardless of the nitrogen source.
Transcriptional pattern of hetF.
In strain UCD 416, luxAB (encoding bacterial luciferase) formed a transcriptional reporter with hetF (Fig. 2). Strain UCD 416 did not show any significant change in luciferase activity within 24 h of N step-down, implying that hetF is transcribed constitutively (data not shown). Luciferase expression by strain UCD 416 is about half that of the constitutive level of the hetR::luxAB transcriptional reporter in ammonium-grown strain UCD 415. Similarly, when a probe specific for hetF was used in a slot blot experiment to measure the amount of hetF transcript in total RNA isolated from wild-type cells harvested during N step-down, no change in the level of the hetF hybridizing signal was observed up to the 24-h time point (data not shown). Attempts to visualize an intact hetF mRNA in Northern blot experiments were unsuccessful. Only a highly degraded hybridization signal smear could be seen when 50 μg of total RNA from the wild type was probed with a hetF-specific probe (data not shown). Distinct bands hybridizing to hetR and 16S rRNA probes could be visualized using the same RNA preparation in a smaller amount (10 μg).
Distribution of hetF in cyanobacteria.
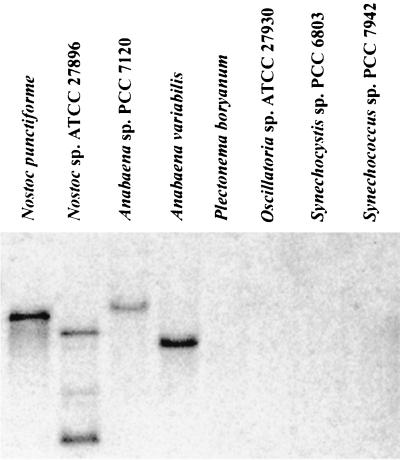
The deduced HetF protein sequence has no significant similarity to proteins in the GenBank databases by BLAST analysis. No protein motifs were found by scanning the sequence using the ProfileScan program. A transmembrane domain was predicted between amino acid residues 500 and 518 by both the SOSUI and TMHMM v.2.0 programs. A gene encoding a protein with 73% similarity to HetF was found in the unfinished Anabaena strain 7120 genomic sequence database, and, as expected, an identical hetF sequence was present in the N. punctiforme database. No significant sequence similarity to hetF was found in the Prochlorococcus marinus and marine Synechococcus sp. genome sequence databases. Genomic DNA isolated from various strains of cyanobacteria was analyzed by Southern hybridization with a probe specific for hetF; hybridizing bands were found only in heterocyst-forming strains (Fig. 3). No hybridizing band was found in the genomic DNA of unicellular strains or non-heterocyst-forming filamentous strains, including nitrogen-fixing Plectonema boryanum and non-nitrogen-fixing Oscillatoria sp. strain ATCC 27930.
FIG. 3.
Hybridization of hetF probe to genomic DNA from several morphological groups of cyanobacteria. A 1-μg (heterocyst-forming strains) or 5-μg (all others) portion of DNA was digested with EcoRI, separated in an agarose gel, and blotted onto a nylon membrane. All Nostoc and Anabaena strains are filamentous, heterocyst-forming strains. P. boryanum is a filamentous strain that fixes nitrogen under microoxic conditions, while Oscillatoria strain ATCC 29730 is a filamentous strain with no detectable nitrogenase activity; neither strain differentiates heterocysts. Synechocystis strain PCC 6803 and Synechococcus strain PCC 7942 are non-nitrogen-fixing, unicellular strains.
Early transcriptional induction of hetR is affected by the hetF mutation.
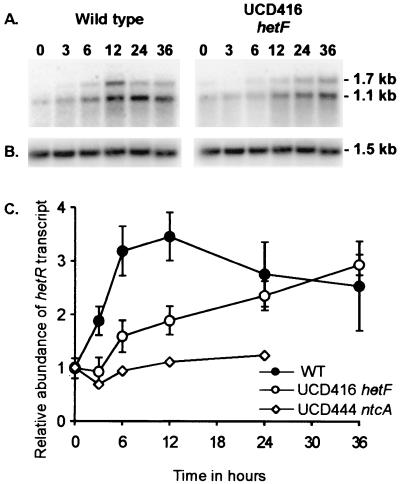
Since hetF inactivation and the presence of multigene copies yield similar phenotypes to those of hetR and both genes are essential to heterocyst development, we determined the epistatic relationship between the two genes by examining whether hetR transcription was altered in strain UCD 416. Total RNA was isolated from the wild-type strain and strain UCD 416 cultures at various time points following N step-down. Filaments in the wild-type culture contained both vegetative cells and morphologically mature heterocysts by 24 h. Total RNA samples across the time course were then analyzed with a probe specific for hetR. In wild-type N. punctiforme, a major hetR transcript of 1.1 kb and a less abundant 1.7-kb transcript were identified (Fig. 4A). Both transcripts were present in nitrogen-replete cultures (time zero), and their levels were elevated as early as 3 h after N step-down. The induction peaked at 12 h, after which both transcripts declined to a steady-state level that was clearly higher than that of time zero. In strain UCD 416, both hetR transcripts were present at time zero but the induction pattern differed from that of the wild type. Significant induction was not detected until between 6 and 12 h, although both transcripts appeared similar in intensity to the wild type by 24 to 36 h. To quantitatively confirm the apparent differences in the induction pattern of hetR between the wild-type strain and strain UCD 416, total RNAs isolated from cells from three independent N step-down time courses were analyzed with the hetR probe in RNA slot blot experiments. The hetR signals were quantified and normalized to 16S rRNA signal, and the results are compiled in Fig. 4C. In the wild type, hetR transcripts increased within 3 h of N step-down, reaching a level about 3-fold higher than that of nitrogen-replete cultures by 12 h, and then fell slightly to a level that was about 2.5-fold higher than that at time zero. In contrast, the early burst of hetR transcription was absent in the hetF mutant strain, although by 24 and 36 h the transcript levels reached approximately the same values as in the wild type. Similar to Anabaena strain 7120, hetR transcriptional induction in N. punctiforme was ntcA dependent since hetR transcripts in a ntcA mutant strain were present but remained constitutively low throughout the time course of nitrogen starvation (Fig. 4C).
FIG. 4.
Northern analysis of hetR transcripts during a time course study of heterocyst development in wild-type cells and the hetF mutant strain UCD 416. (A and B) Samples of total RNA (10 μg) from various time points (designated in hours [given across the top]) following the N step-down were fractionated, blotted, and hybridized with a hetR-specific probe (A) or a probe specific for the 16S rDNA (B). The same blot was used for both hybridizations. (C) Quantitation of the total hetR transcript level. A 2- to 3-μg portion of total RNA was used in slot blot experiments. The hetR transcript signal was normalized using the 16S rRNA signal, and then the transcript level was divided by the value for the wild type at 0 h of N step-down (arbitrarily set to 1.0). The values are the average of three independent time course experiments except for that of the ntcA mutant strain UCD 444. Error bars indicate standard error.
hetR transcript and product accumulate nonspecifically in the hetF mutant.
hetR transcription is induced in specific cells by 3.5 h after N step-down (2). Since hetR induction kinetics are altered in strain UCD 416, we examined whether the cell-specific increase in the level of hetR transcript and accumulation of HetR protein were disrupted. A multicopy plasmid carrying the PhetR::gfp transcriptional reporter (pSCR93) (Fig. 5A) was introduced into the wild-type strain and strain UCD 416, forming strains UCD 483 and UCD 484, respectively. Both strains expressed GFP fluorescence strongly and uniformly in all cells when growing with ammonium. By 36 h after N step-down, strain UCD 484 expressed on average about two- to threefold-higher GFP fluorescence in all cells as determined by quantitative image analysis (data not shown), and no heterocysts were observed. In contrast, under the same N step-down conditions, GFP fluorescence in strain UCD 483 was elevated 6- to 10-fold specifically in heterocysts (Fig. 5B). PhetR::gfp indicates only the level of hetR transcript in cells but does not reveal whether any posttranslational degradation of the gene product occurs. To examine that possibility, a full-length hetR-gfp fusion was constructed and cloned into a multicopy plasmid (pSCR90) (Fig. 5A). The ability of the hetR mutant to differentiate heterocysts was restored by pSCR90, indicating that the HetR-GFP fusion protein is functional (data not shown). pSCR90 was introduced into the wild-type strain and strain UCD 416, forming strains UCD 485 and UCD 486, respectively. In strain UCD 486 grown with ammonium, HetR-GFP protein accumulated in all cells even though no heterocysts were observed. The vegetative cells were morphologically altered, and cells gradually lost the HetR-GFP expression even in the presence of ampicillin selection. The presence of the HetR-GFP fusion protein did not promote heterocyst differentiation or allow growth of strain UCD 486 in the absence of combined nitrogen. In contrast, HetR-GFP protein was not observed in strain UCD 485 vegetative cells under all conditions. Strain UCD 485 differentiated less than 1% of its cells into heterocysts when growing with ammonium but differentiated single and multiple heterocysts to approximately 8% of the total cells when growing in nitrate. Under all growth conditions, HetR-GFP accumulated specifically in heterocysts (Fig. 5C).
DISCUSSION
We have identified a novel gene, hetF, in N. punctiforme with inactivation and multicopy phenotypic characteristics that are similar but not identical to those of hetR. hetF mutants display a Het− phenotype (Fig. 1A) and can grow in the presence of nitrate, similar to hetR mutants but unlike ntcA mutants. In the absence of combined nitrogen, wild-type N. punctiforme carrying multiple copies of hetF displays a Mch phenotype (Fig. 1B). This phenotype is similar to those of Anabaena strain 7120 (3) and N. punctiforme wild-type filaments carrying extra copies of hetR (Fig. 5C). These results imply that HetF plays a similar or ancillary role to HetR in the positive regulation of heterocyst development.
However, there are at least four distinct differences between hetF and hetR. First, strains carrying extra copies of hetR develop heterocysts in the presence of nitrate whereas strains carrying extra copies of hetF do not develop heterocysts unless they are incubated in the complete absence of combined nitrogen. Second, vegetative filaments of N. punctiforme strains carrying extra copies of hetF displayed an altered morphology (Fig. 1B) while those carrying extra copies of hetR exhibited normal vegetative cells morphology (Fig. 5C). This phenotype may be explained by the apparent membrane attachment of HetF, in contrast to the cytoplasmic localization of HetR. Overexpression of membrane-associated proteins is known to alter cell morphology (11). Third, by similarity searches and Southern analysis, we have shown that hetF is found only in heterocyst-forming strains, unlike hetR, which is present in all filamentous strains surveyed thus far (12), including P. boryanum, where hetR has been identified (3) but hetF is absent (Fig. 3). Fourth, hetF is transcribed constitutively at a low level regardless of the nitrogen status. Both luxAB reporter and RNA slot blot studies showed that the hetF transcript level was constant during nitrogen-replete conditions and up to 24 h after the N step-down. Northern analysis indicated that the hetF transcript was low in abundance and unstable. These results are in contrast to those for hetR, whose stable transcripts were present under nitrogen-replete conditions and elevated during N step-down (Fig. 4A).
Other than a putative transmembrane domain, no significant sequence similarity to proteins or motifs was found using HetF as a search sequence to the major databases. Thus, the primary sequence gives no clue to the function of HetF. Both the hetR mutant strain carrying supernumerary copies of hetF and the hetF mutant carrying extra copies of a functional hetR-gfp fusion display the Het− phenotype in the absence of combined nitrogen, indicating that the elevated copy number of one of the genes is insufficient to induce heterocyst differentiation in the absence of the other. We concluded that both hetF and hetR are essential to heterocyst development and may act in concert. As discussed below, we present evidence that hetR transcripts and protein accumulate aberrantly in the hetF mutant under nitrogen-limiting conditions, which may indicate a role for HetF in the regulation of heterocyst development.
In N. punctiforme, hetR is transcribed as two mRNAs, whose abundance increases at 3 h after N step-down to a maximum of threefold higher between 6 and 12 h (Fig. 4). This transcriptional induction pattern is similar to that of Anabaena strain 7120 hetR (3). Induction of the two N. punctiforme hetR transcripts is dependent on the presence of an intact copy of ntcA, again similar to Anabaena strain 7120 (9). Although both hetR transcripts are present and induced in hetF mutant strain UCD 416, the induction pattern is altered such that the 0- to 6-h burst is absent. The hetR transcripts in strain UCD 416 accumulate in an almost linear fashion and reach the wild-type level only at approximately 24 h after N step-down. Since hetR is induced in specific, developing cells as early as 3.5 h after N step-down (2), the delay seen in strain UCD 416 may indicate that the cell-specific elevation of hetR transcripts and consequently the accumulation of HetR protein was disrupted.
Results obtained with GFP transcriptional and functional fusion reporters confirmed that the hetR transcript and protein accumulated nonspecifically in the hetF mutant. Under nitrogen-limiting conditions, GFP expressed from a PhetR::gfp transcriptional reporter was elevated in all cells in hetF mutant strain UCD 484. HetR-GFP expressed in hetF mutant strain UCD 486 transiently accumulated in all cells to a high level under nitrogen-replete conditions, even though no cells resembling heterocysts were observed (Fig. 5). Overexpression of a functional HetR appears to be detrimental to a hetF mutant, as evidenced by the gradual loss of HetR-GFP expression in strain UCD 486 and by our inability to isolate a hetF mutant strain carrying wild-type hetR in trans (data not shown).
Our experimental results lend support to the hypothesis that HetR, while synthesized in all cells, autodegrades itself in vegetative cells and accumulates in heterocysts, and that this activity is important in regulating the intracellular amount of HetR (8). Emission from GFP expressed from the PhetR::gfp transcriptional reporter in hetF+ strain UCD 483 was sufficiently high to completely eclipse the background chlorophyll fluorescence in the vegetative cells, and the reporter GFP accumulated to an even higher level in heterocysts (Fig. 5B). These results confirm that hetR is transcribed in vegetative cells and that its level is elevated specifically in heterocysts. However, emission from the functional HetR-GFP fusion protein in hetF+ strain UCD 485 was not visible amid the red chlorophyll fluorescence in vegetative cells (Fig. 5C). In this construct, emission was clearly seen only from heterocysts, indicating that HetR does not accumulate in vegetative cells, irrespective of hetR transcription. Similar to the N. punctiforme hetF mutant, the original Anabaena strain 7120 hetR mutant, strain 216, which carries a hetR gene with a single loss-of-function mutation, accumulates HetR protein aberrantly in all cells. Upon N step-down, strain 216 expresses the mutant HetR protein to levels higher than those in the wild type (8). The mutant HetR protein in strain 216 has no autoproteolytic activity (8) and Haselkorn stated that strain 216 carrying a PhetR::gfp transcriptional reporter exhibited enhanced fluorescence in all cells under combined nitrogen-limiting condition (10). Collectively, these data imply that the autoproteolytic activity of HetR is essential for the differential accumulation of HetR in vegetative cells and heterocysts.
The mechanism by which HetR promotes heterocyst differentiation is unknown, but it appears that accumulation in specific cells is essential to its function. The role of HetF in the positive regulatory pathway may be to regulate the autoproteolytic activity of HetR, which subsequently enhances hetR transcriptional induction and cell specific accumulation of HetR.
We should like to note that the lack of transcriptional induction of hetF and the absence of known motifs in the protein sequence preclude its ever being identified as a cellular differentiation regulatory factor by any transcriptional assay or database analysis. We were able to discern a role for HetF in heterocyst differentiation only by mutation and phenotypic characterization.
ACKNOWLEDGMENTS
This work was supported by grant NRICGP 98-35305-6748 from the U.S. Department of Agriculture.
We thank Elsie Campbell, Kari Hagen, and John Ingraham for critical review of the manuscript. We thank our departmental colleague Kazuhiro Shiozaki for the use of the Nikon microscope and image-capturing equipment, and we thank Jeff Elhai for providing the pGFPCR plasmid.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Milller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black T A, Cai Y, Wolk C P. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol. 1993;9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 3.Buikema W J, Haselkorn R. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 1991;5:321–330. doi: 10.1101/gad.5.2.321. [DOI] [PubMed] [Google Scholar]
- 4.Cai Y P, Wolk C P. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol. 1990;172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen M F, Meeks J C. A hormogonium regulating locus, hrmUA, of the cyanobacterium Nostoc punctiforme strain ATCC 29133 and its response to an extract of a symbiotic plant partner Anthoceros punctatus. Mol Plant-Microbe Interact. 1997;10:280–289. doi: 10.1094/MPMI.1997.10.2.280. [DOI] [PubMed] [Google Scholar]
- 6.Cohen M F, Wallis J G, Campbell E L, Meeks J C. Transposon mutagenesis of Nostoc sp. strain ATCC 29133, a filamentous cyanobacterium with multiple cellular differentiation alternatives. Microbiology. 1994;140:3233–3240. doi: 10.1099/13500872-140-12-3233. [DOI] [PubMed] [Google Scholar]
- 7.Cormack, R. S., and I. E. Somssich. 28 May 1997, posting date. Cloning of PCR products using the green fluorescent protein. Elsevier Trends J. Tech. Tips 1:T01107. [www.bmn.com]
- 8.Dong Y, Huang X, Wu X Y, Zhao J. Identification of the active site of HetR protease and its requirement for heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 2000;182:1575–1579. doi: 10.1128/jb.182.6.1575-1579.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frias J E, Flores E, Herrero A. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 1994;14:823–832. doi: 10.1111/j.1365-2958.1994.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 10.Haselkorn R. How cyanobacteria count to 10. Science. 1998;282:891–892. doi: 10.1126/science.282.5390.891. [DOI] [PubMed] [Google Scholar]
- 11.Hay N A, Tipper D J, Gygi D, Hughes C. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol. 1999;181:2008–2016. doi: 10.1128/jb.181.7.2008-2016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janson S, Matveyev A, Bergman B. The presence and expression of hetR in the non-heterocystous cyanobacterium Symploca PCC 8002. FEMS Microbiol Lett. 1998;168:173–9. doi: 10.1111/j.1574-6968.1998.tb13270.x. [DOI] [PubMed] [Google Scholar]
- 13.Khudyakov I, Wolk C P. Evidence that the hanA gene coding for HU protein is essential for heterocyst differentiation in, and cyanophage A-4(L) sensitivity of, Anabaena sp. strain PCC 7120. J Bacteriol. 1996;178:3572–3577. doi: 10.1128/jb.178.12.3572-3577.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luque I, Flores E, Herrero A. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 1994;13:2862–2869. doi: 10.1002/j.1460-2075.1994.tb06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muro-Pastor A M, Valladares A, Flores E, Herrero A. The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J Bacteriol. 1999;181:6664–6669. doi: 10.1128/jb.181.21.6664-6669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rippka R, Herdman M. Pasteur culture collection of cyanobacteria in axenic culture. Paris, France: Institut Pasteur; 1992. pp. 44–57. [Google Scholar]
- 17.Sambrook J, Maniatis T, Fritsch E F. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 18.Schmidt-Goff C M, Federspiel N A. In vivo and in vitro footprinting of a light-regulated promoter in the cyanobacterium Fremyella diplosiphon. J Bacteriol. 1993;175:1806–1813. doi: 10.1128/jb.175.6.1806-1813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Summers M L, Wallis J G, Campbell E L, Meeks J C. Genetic evidence of a major role for glucose-6-phosphate dehydrogenase in nitrogen fixation and dark growth of the cyanobacterium Nostoc sp. strain ATCC 29133. J Bacteriol. 1995;177:6184–6194. doi: 10.1128/jb.177.21.6184-6194.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valladares A, Muro-Pastor A M, Fillat M F, Herrero A, Flores E. Constitutive and nitrogen-regulated promoters of the petH gene encoding ferredoxin:NADP+ reductase in the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 1999;449:159–164. doi: 10.1016/s0014-5793(99)00404-4. [DOI] [PubMed] [Google Scholar]
- 21.Wei T F, Ramasubramanian T S, Golden J W. Anabaena sp. strain PCC. 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol. 1994;176:4473–4482. doi: 10.1128/jb.176.15.4473-4482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei T F, Ramasubramanian T S, Pu F, Golden J W. Anabaena sp. strain PCC 7120 bifA gene encoding a sequence-specific DNA-binding protein cloned by in vivo transcriptional interference selection. J Bacteriol. 1993;175:4025–4035. doi: 10.1128/jb.175.13.4025-4035.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolk C P. Heterocyst formation in Anabaena. In: Brun Y V, Shimkets L J, editors. Prokaryotic development. Washington, D.C.: American Society for Microbiology; 2000. pp. 83–104. [Google Scholar]
- 24.Wolk C P. Heterocyst metabolism and development. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 769–823. [Google Scholar]
- 25.Zhou R, Cao Z, Zhao J. Characterization of HetR protein turnover in Anabaena sp. PCC 7120. Arch Microbiol. 1998;169:417–23. doi: 10.1007/s002030050592. [DOI] [PubMed] [Google Scholar]
- 26.Zhou R, Wei X, Jiang N, Li H, Dong Y, Hsi K L, Zhao J. Evidence that HetR protein is an unusual serine-type protease. Proc Natl Acad Sci USA. 1998;95:4959–4963. doi: 10.1073/pnas.95.9.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]