Summary
Molecular profiling studies have enabled discoveries for metastatic prostate cancer (MPC) but have predominantly occurred in academic medical institutions and involved non-representative patient populations. We established the Metastatic Prostate Cancer Project (MPCproject, mpcproject.org), a patient-partnered initiative to involve patients with MPC living anywhere in the US and Canada in molecular research. Here, we present results from our partnership with the first 706 MPCproject participants. While 41% of patient partners live in rural, physician-shortage, or medically underserved areas, the MPCproject has not yet achieved racial diversity, a disparity that demands new initiatives detailed herein. Among molecular data from 333 patient partners (572 samples), exome sequencing of 63 tumor and 19 cell-free DNA (cfDNA) samples recapitulated known findings in MPC, while inexpensive ultra-low-coverage sequencing of 318 cfDNA samples revealed clinically relevant AR amplifications. This study illustrates the power of a growing, longitudinal partnership with patients to generate a more representative understanding of MPC.
Keywords: cancer genomics, patient advocacy, health disparities, metastatic prostate cancer, precision medicine, patient-reported data, geographic analysis, clinicogenomic partnership, diversity, oncology
Graphical abstract
Highlights
-
•
MPCproject partners with metastatic prostate cancer patients for molecular research
-
•
Over 1,000 patient partners to date are from across the US and Canada
-
•
41% of patient partners are from rural or medically underserved areas
-
•
Remotely donated samples from real-world settings recapitulate genomic findings
Crowdis et al. describe the MPCproject (mpcproject.org), a decentralized initiative to partner with patients with metastatic prostate cancer in the US and Canada to accelerate molecular research. The authors describe clinicogenomic results from the first 706 geographically diverse patient partners and lay the foundation for sustained and inclusive partnership in this disease.
Introduction
Prostate cancer is the second most diagnosed cancer in men, with nearly 200,000 men diagnosed in 2020 alone in the US.1 Survival rates for localized disease are high, but the 5-year survival rate for the over 300,000 men currently living with metastatic prostate cancer (MPC) is only 31%, representing the third leading cause of death for men.1,2 Genomic sequencing studies have enabled new therapeutic targets for MPC, but obtaining large cohorts of tumor biopsies for molecular study has been difficult, as MPC often spreads to bone and requires technically challenging procedures to sample.3, 4, 5, 6 Because prostate cancer can shed cell-free DNA (cfDNA) into the bloodstream, blood biopsies that sample this circulating tumor DNA have proven to be a useful alternative for the study of MPC.7,8
Historically, quaternary care academic medical institutions have had the necessary infrastructure to lead clinically integrated MPC sequencing studies. However, the resulting clinical and genomic data is often siloed within these institutions, leading many to push for mandatory data sharing.9,10 These efforts, while important, do not directly improve access to molecular research programs and do not address underlying ethnic, socioeconomic, and geographic patient disparities in such studies, which threaten to bias findings and eventually care toward select patient populations.11, 12, 13, 14 Commercial sequencing options for prostate cancer are emerging but are often proprietary, only available with appropriate insurance, and regularly inaccessible for research use.15, 16, 17 Indeed, despite growing interest from patients with MPC in clinical and research-based genomic sequencing, there are only limited mechanisms for these patients to partner with the research community to accelerate discoveries.18, 19, 20
We hypothesized that a patient-partnered framework that empowers patients with MPC to share their biological samples, clinical histories, and lived experiences directly with researchers regardless of geographic location or hospital affiliation would lead to new clinicogenomic discoveries and begin to address demographic inequities and data-access barriers in molecular studies for this disease. Thus, we established the Metastatic Prostate Cancer Project (MPCproject, mpcproject.org), a research model that leverages patient advocacy and social media to enable patients with MPC to participate in genomic research remotely at no personal cost.
Results
Development of a patient-partnered MPC research model
Working with patients, loved ones, and advocates, we developed an MPCproject enrollment process for men living with MPC in the US and Canada (Figure 1A). The MPCproject outreach model is community centered and utilizes advocacy partnerships, social media campaigns, and educational initiatives to engage patients (Figure S1). To enroll, patient partners complete an online survey describing their experience with MPC, followed by signing electronic consent and release forms, which allow the MPCproject team to contact their hospitals to request medical records and optionally archival tumor tissue for research-grade genomic sequencing (Figure S2). Enrolled patient partners can also use a mailed kit to donate saliva and/or blood at routine blood draws at no cost, and these samples are sequenced to assess germline DNA and cfDNA, respectively (Figures S3 and S4).
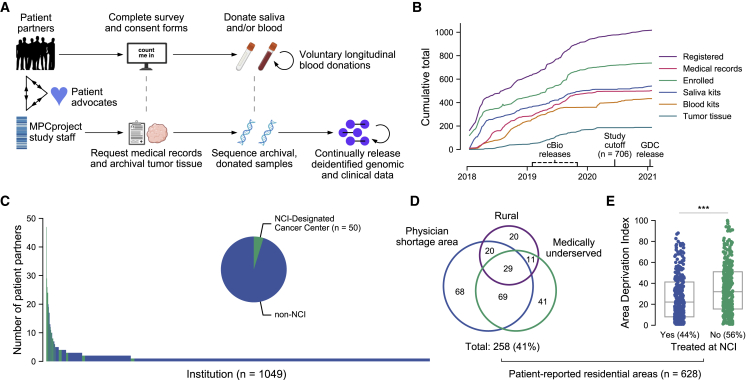
Figure 1.
Partnering with diverse patients to enhance our understanding of metastatic prostate cancer
(A) Summary of MPCproject enrollment process. Patients learn about the project primarily through outreach and partnered advocacy groups. If they register, patient partners complete online intake, consent, and medical release forms, then can opt into donating saliva via a mailed kit and/or blood at routine blood draws at no charge. In parallel, MPCproject staff request medical records and archival tumor samples from patients’ medical institutions, then abstract medical information from obtained records and sequence archival tumor tissue and/or donated blood and saliva (STAR Methods). Deidentified clinical, genomic, and patient-reported data are released on a continual, prepublication basis and deposited in public repositories.
(B) Enrollment statistics and timeline for the MPCproject. Depicted are the cumulative number of patients that began the registration process (registered), patients that completed the survey and consent forms (enrolled), patients with at least one medical record received (medical records), and blood kits, saliva kits, and archival tumor tissue received at the Broad Institute for sequencing (blood kits, saliva kits, and tumor tissue, respectively). 706 patient partners enrolled before “study cutoff,” June 1, 2020, and are included in this study’s analyses. cBioPortal (cbioportal.org) releases include summary abstracted medical, genomic, and patient-reported data; Genomic Data Commons (GDC) releases include raw sequencing files and demographic data.
(C) Represented medical institutions among patient partners living in the US and Canada. Shown are the 1,049 unique institutions (x axis) where patient partners report receiving care for their prostate cancer, with the number of distinct patient partners at each institution (y axis). NCI-designated cancer centers are shown in green. Patient partners that did not complete this survey question (n = 36) and institutions outside the US and Canada (n = 56) are not shown.
(D) Access to medical care among patient partners living in the US. Patient-reported data were used to identify residential census tracts that were overlapped with primary care health-physician-shortage areas (HPSAs), medically underserved population/areas (MUAs), and rural areas obtained from the Health Resources and Services Administration and US Census. Patient partners that live in Canada (n = 30) who did not provide residential data (n = 40) or who provided only a P.O. box (n = 8) are not shown.
(E) Patient partners living in more disadvantaged areas are less likely to attend NCI cancer centers. The Area Deprivation Index, a metric that assesses neighborhood disadvantage, was assessed for each residential census block group. Higher values indicate more disadvantage. The x axis reflects whether patient partners reported receiving care at an NCI-designated cancer center. ∗∗∗ p < 0.001 in a logistic regression model that adjusts for rural, MUA, and HPSA status.
Patient partners and advocates are involved in every step of the project’s design and execution—we respond directly to their feedback and keep them informed of our progress and findings (supplemental information; Figure S5). Patient advocates help design the website and all patient-facing enrollment material, lead patient information sessions about the project, and advise the project’s mission. We also work with patient partners who continue donating blood to help the research community understand the evolution of MPC, and we regularly release prepublication, deidentified genomic, patient-reported, and clinical data in public repositories for research use.
Partnering with a demographically distinct patient population
To date, the MPCproject has partnered with over 1,000 patients in the US and Canada and has orchestrated three public data releases (Figure 1B). The analyses presented here are based on the 706 men from the US and Canada who had enrolled (completed consent forms) as of June 1, 2020 (Figure S6).
Using patient-reported survey data, we assessed the geographical diversity of our patient partners. Hailing from 49 US states and six Canadian provinces, patient partners reported receiving care for their prostate cancer at over 1,000 distinct medical institutions, 91% of which were reported by two or fewer patients (Figure 1C). We found that 56% of patient partners have never received care at an NCI-designated cancer center, where genomic research is traditionally conducted (Table S1). These patient partners were three times less likely to report participating in a clinical trial (7% versus 20%, p = 1 × 10−6, Fisher’s exact test).
We then used patient-reported data to identify residential census tracts and their geographic characteristics (n = 628/706 participants had identifiable census tracts; STAR Methods). We found that 13% of patient partners live in rural areas defined by the USDA, a proportion consistent with patients with MPC in the US (11%).21 We additionally found that 30% of patient partners live in health-physician-shortage areas (HPSAs) and that 24% live in medically underserved areas (MUAs) as defined by the Health Resources and Services Administration (Figure 1D; STAR Methods).22 These proportions could not be compared with patients with MPC in the US or with other sequencing efforts due to a lack of published data but are significantly enriched compared with the US population (25% HPSAs, 5% MUAs, p = 0.03 and 1 × 10−82, respectively, Fisher’s exact test).23,24 While living in a rural area was associated with being in an MUA or HPSA, 28% of MPCproject patient partners live in urban primary care MUAs or HPSAs (p = 5.7 × 10−13, Fisher’s exact test). We additionally found that patient partners living in rural areas compared with urban areas lived a median of 160 km farther from institutions where they reported receiving treatment, suggesting that they may travel farther for cancer care (p < 10−11, Mann-Whitney U test; Figure S7).
We next examined the socioeconomic traits of patient partner residential areas using the national Area Deprivation Index (ADI), a 0–100 ranking that includes factors of income, education, employment, and housing quality, where 100 indicates the most disadvantage.25 The average ADI of patient partner residential areas was lower than the age- and race-matched national average (31 versus 46), which may reflect the relative success of patient partner engagement via social media outreach, the usage of which is correlated with socioeconomic status, compared with our community-driven efforts to date (Figure S7).26 Notably, we cannot compare this average with patient populations from existing sequencing studies due to a lack of published data. We also found that patient partners living in more disadvantaged areas were less likely to attend NCI cancer centers for treatment, even after controlling for rural, MUA, and HPSA status (ADI = 35 versus 27, NCI treated versus not, p < 0.001, logistic regression) (Figure 1E). We are cautious, however, in interpreting the results of these geographic analyses. Patient partners may not currently live in their reported locations, we do not directly survey their income or socioeconomic status, and their experiences may not be represented by their residential area. We did not observe significant associations in baseline clinical factors, therapies received, or likelihood to participate in a clinical trial with ADI or across patient partners in rural areas, MUAs, or HPSAs.
The combination of the MPCproject’s online enrollment and patient-centered outreach through advocacy partnerships enabled the creation of a geographically distinct prostate cancer research program. Despite the project’s geographical diversity, however, fewer than 10% of patient partners self-identify as non-White (Table S2). While similar to existing studies, this representation remains well below the proportion of minority patients with prostate cancer generally (20%).21 The lack of racial diversity in our study is a critical flaw that is thus far insufficient to accelerate research for communities of color, and it has spurred new, community-driven MPCproject initiatives to connect with these patients, as detailed in the limitations of the study.
Patient-reported data augment medical records to amplify patient stories
Through the patient-reported data, we sought to understand the real-world experiences of those living with MPC. 45% of patient partners report being diagnosed with de novo metastatic disease, with bone (48%) and lymph node (39%) lesions as the most common metastatic sites (Figures 2A and 2B). 48% of patient partners reported a family history of prostate or breast cancer, while 24% reported having at least one other cancer diagnosis in their lifetime, 30% of which was a non-skin form of cancer (Figures 2C and 2D). The average age at diagnosis was significantly younger than the national average (61 versus 65 years old, p < 10−39, t-test), and 24% of participants were diagnosed with early-onset prostate cancer (≤55 years at diagnosis; Table S2).27 We note that these characteristics of our patient partners are likely influenced by participation bias and may differ from other prostate cancer studies as a result.
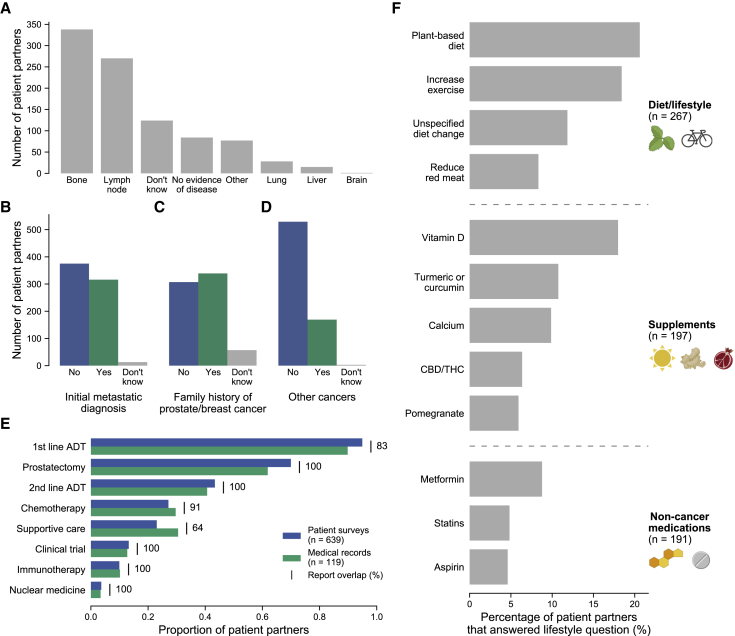
Figure 2.
Patient voices reveal the landscape of living with metastatic prostate cancer
(A–D) Self-reported data of 706 patient partners related to their prostate cancer.
(A) Patient partners were asked for the current location of their cancer. Participants were free to choose multiple if their cancer had metastasized to multiple locations.
(B–D) Responses were tabulated from questions asking patient partners if their initial prostate cancer diagnosis was metastatic (B), if they have a family history of prostate/breast cancer (C), or if they have ever had another cancer diagnosis (D). Patient partners who did not complete these questions (n < 5 for all questions) are not shown.
(E) Self-reported therapies show strong overlap with medical records. Therapy categories are shown on the y axis, with the proportion of patient partners from each data type (patient surveys and medical records) receiving therapies of that category shown on the x axis. In the online survey, patient partners selected therapies they received for their metastatic prostate cancer from a list. 639/706 patient partners reported at least one therapy and are shown. 119 of these participants also had abstracted therapy data from medical records. Report overlap refers to how often patient partners report receiving a therapy when their medical records show that they have received that therapy as a percentage. Only therapies available for selection in the patient survey were used in this comparison (Table S4).
(F) Landscape of lifestyle changes for patient partners. Participants were asked to list additional medications, alternative medications, or lifestyle changes since their diagnosis of prostate cancer. Free-text responses were manually abstracted and categorized into diet/lifestyle changes, supplements, and non-cancer medications. The y axis shows individual instances of diet/lifestyle changes, supplements, or medications. The x axis shows the percentage of patient partners with that lifestyle change or that were taking that supplement/therapy out of all patient partners that responded to the lifestyle question (n = 456). CBD/THC, cannabidiol/tetrahydrocannabinol (oils, medical marijuana, etc.).
We used the MPCproject’s comprehensive abstracted medical records together with patient-reported data to evaluate the treatments received in this real-world cohort (STAR Methods; Figure 2E). Patient partners reported taking an average of 2.8 therapies (range 1–13) to treat their prostate cancer. 119 (17%) patient partners had abstracted medical records at the time of writing, and there was 90% concordance between therapies noted in formal medical records and therapies reported by these patient partners. The overlap was lowest for treatments typically given earlier in the therapeutic timeline (first-line androgen deprivation therapy, 83%), supportive care therapies (64%), or treatments abandoned quickly due to side effects (Figure 2E).
We also used the patient-reported data to assess how living with prostate cancer has changed the daily lives of our patient partners. 56% of patient partners reported a lifestyle change because of living with their cancer, with the most common being a change in diet or exercise (Figure 2F). Common nutritional supplements reported include vitamin D and antioxidant-based supplements, while common non-cancer medications included metformin and statins.
Whole-exome sequencing of a real-world MPC patient cohort
To complement the demographic, patient-reported, and clinical data, we have completed molecular profiling of 572 samples from 333 patient partners to date, including ultra-low-pass whole-genome sequencing (ULP-WGS; average depth of 0.1×) of cfDNA from 318 donated blood samples; whole-exome sequencing (WES) of cfDNA from 47 of those blood samples; WES of 106 tumor samples; and WES of 148 germline samples from donated saliva or blood buffy coat. In total, 82 exome-sequenced samples (63 tumor and 19 cfDNA) from 79 patient partners enrolled before June 1, 2020, were included in downstream genomic analyses after assessment of sufficient tumor purity (≥10%) and coverage (STAR Methods).
Exome sequencing from the tumor and cfDNA samples recapitulated known genomic patterns in MPC (Figure 3A). TP53 and SPOP were recurrently altered, consistent with previous studies of both metastatic and primary prostate cancer (q < 0.1 via MutSig2CV).3,4,6 In primary tumor samples from this cohort, the mutation frequency of TP53 (29%) was more consistent with metastatic cohorts than those of primary prostate cancer.3,6 Twenty-four (38%) primary tumor samples were from men diagnosed with de novo metastatic disease, and samples from these patient partners were more likely to carry TP53 mutations (p = 0.04, Fisher’s exact test). We also observed known patterns of copy-number alteration in prostate cancer, including recurrent amplifications of androgen receptor (AR) and FOXA1, as well as recurrent deletions of PTEN (q < 0.1 via GISTIC2.0; Figure 3A).28 Whole-genome doubling was present in 6/63 tumor samples and 2/19 cfDNA samples, including in two tumor samples from patient partners initially diagnosed with localized prostate cancer. Both patient partners were diagnosed with metastatic disease within a few months of their initial diagnosis.
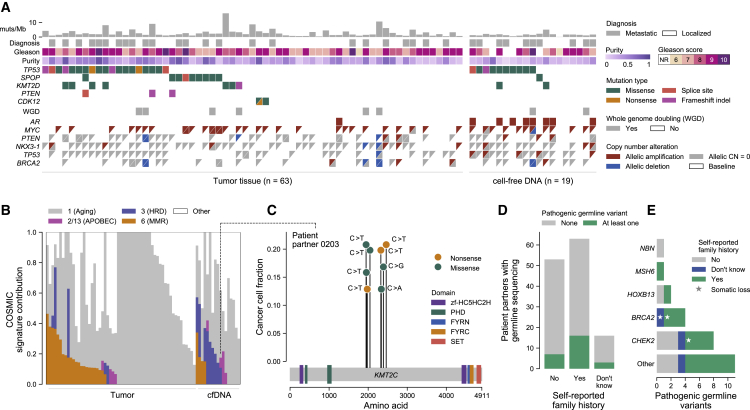
Figure 3.
Remotely donated tumor and cell-free DNA samples obtained through patient partnership recapitulate known genomic findings in metastatic prostate cancer
(A) Genomic and clinical landscape of 82 sequenced samples. Columns represent samples, separated into tumor (prostate, left) and cell-free DNA (cfDNA; donated blood, right) samples, while rows represent select clinical and genomic features. Gleason scores for tumor samples are taken from the pathology report received with the sample (n = 58) or the patient partner’s medical records (n = 5) if Gleason scores were not provided in the report. Gleason scores for cfDNA were taken from pathology reports in the medical record, with NR representing cases where a Gleason score was not reported in the medical record. Diagnosis refers to whether the initial diagnosis of prostate cancer was localized or metastatic. Multiple mutations in the same gene are represented as triangles. WGD refers to whole-genome doubling. Copy-number calls are allelic and defined with respect to baseline allelic ploidy (2 for samples with WGD, one for those without), with calls for the two alleles indicated by two triangles (except for AR, which has only one allele in men and so is shown as a single box). Allelic CN = 0 refers to complete allelic deletions. Allelic deletions that are not complete deletions are possible in samples with WGD. Figure created with CoMut.29
(B) Mutational signature analysis of sequenced samples. The relative contribution of select COSMIC v.2.0 mutational signatures are shown, separated by tumor and cfDNA (donated blood) sample type.30 APOBEC refers to signatures associated with activity of APOBEC family of cytidine deaminases (signatures 2 and 13); MMR to the signature associated with deficient DNA mismatch repair (signature 6); and HRD to the signature associated with homologous recombination deficiency (signature 3). To be denoted as present, a signature cutoff of 6% was used. Samples with too few mutations for signature analysis (<50 mutations, n = 5 samples) are not shown.
(C) Instance of localized hypermutation (kataegis) of KMT2C in cfDNA from a donated blood sample. The y axis shows the cancer cell fraction of each mutation, while the x axis shows their amino acid within KMT2C. Domains taken from Pfam.31 The dotted line connects to this sample’s mutational signature profile.
(D and E) Germline pathogenic alterations and their overlap with patient-reported family history. Pathogenic germline alterations (as annotated by ClinVar) in genes from a select panel of genes previously implicated in cancer heritability were detected in patient partners with sequenced saliva or blood buffy coat (n = 132) (STAR Methods; Tables S3 and S5).32 Survey responses to a question asking about a family history of prostate or breast cancer were tabulated and overlapped with this genomic data. Stars in (E) indicate instances where a somatic deletion also affected that gene in a tumor or cfDNA sample from that patient partner, suggesting biallelic inactivation.
To understand the mutational processes in this cohort’s exome-sequenced samples, we used a mutation-based method (deconstructSigs) to determine the contribution of COSMIC v.2.0 signatures to each sample30,33 (Figure 3B; STAR Methods). We detected the presence of aging-associated clock-like signature one in all samples and the presence of signature 3 (associated with homologous recombination deficiency [HRD]) and signature 6 (associated with mismatch repair deficiency [MMR]) in a subset of samples. These results are consistent with previous studies implicating these signatures in prostate cancer, although they likely overestimate the prevalence of signature six in tumor samples due to formalin-induced deamination artifacts.34,35 We found that the presence of signature three was enriched in metastasis-associated samples (cfDNA and primary tumors obtained in the metastatic setting) relative to tumor tissue from patient partners with strictly localized tumors at time of resection (p = 0.04, Fisher’s exact test). While some samples with signature three had at least one alteration in BRCA1 or BRCA2 (n = 9/16), this association was not statistically significant, highlighting the potential role of other homologous repair defects in the etiology of signature 3, as noted in prior studies of prostate and breast cancer.5,36, 37, 38, 39 All samples with signature 3, however, had at least one alteration in a DNA-repair pathway gene, and biallelic BRCA2 alterations were associated with copy-number-based estimations of HRD (STAR Methods; Figure S8).40
In 10% of samples (8/82), we observed contributions from COSMIC signatures 2 and 13, which are driven by APOBEC cytidine deaminases and are known to operate at a baseline level in prostate cancer.34,41 APOBEC-driven mutagenesis has been implicated in kataegis—rare, localized hypermutation in specific nucleotide contexts that is associated with genomic instability and increased Gleason score in prostate cancer.42,43 In one patient partner’s cfDNA sample, we detected eight distinct mutations within a 2-kB window in KMT2C, a known prostate cancer driver (Figure 3C).3 Six of these mutations were in a T(C>T)A nucleotide context, and this sample had a detectable contribution from COSMIC signature 13. We found that two pairs of the mutations, p.S1947F/p.S1954F and p.Q2325∗/p.S2337Y, were each present on individual sequencing reads, confirming that these mutations existed within the same cell and strongly implicating KMT2C disruption through kataegis (Figure S9).
Given the strong heritability of prostate cancer, we assessed inherited germline alterations and their overlap with patient-reported family history of cancer.44 We found that among the 132 patient partners (19%) with WES of donated saliva or blood buffy coat, 15 and 11 had pathogenic germline alterations in select genes implicated in prostate cancer and other cancers, respectively.45 Men that self-reported a family history of prostate or breast cancer were more likely to have a pathogenic germline alteration associated with cancer, although this difference was not statistically significant (25% versus 13%, p = 0.11, Fisher’s exact test; Figure 3D). The most mutated gene was CHEK2 (8 patient partners), followed by BRCA2 (4 patient partners). In three cases, we detected an accompanying somatic loss of a germline-mutated gene (Figures 3E and S10).
Longitudinal blood biopsies enable study of tumor evolution in a patient-partnered model
Ten patient partners had WES from both tumor tissue and cfDNA, and three patient partners had both samples pass quality-control metrics. Using the molecular data and abstracted medical records, we sought to explore the evolutionary relationships between these longitudinal samples in the context of patient clinical trajectories. Like most men with MPC, one participant, patient partner 0495, received a diverse range of treatments between biopsy timepoints (Figure 4A). After responding to first-line anti-androgen therapy (leuprolide + bicalutamide), they took second-generation anti-androgen inhibitors (abiraterone, enzalutamide), as well as experimental radiotherapy and immunotherapy. To explore the relationship between samples, we utilized PhylogicNDT, an algorithm that clusters mutations based on their prevalence in the tumor (cancer cell fraction) into evolutionarily related subclones (STAR Methods).46 In the cfDNA sample of patient partner 0495, but not the primary tumor, we observed two distinct frameshift mutations in ASXL2, a gene implicated in castration-resistant MPC, as well as an amplification of AR, a known resistance mechanism to abiraterone and enzalutamide.47,48 Patient partner 0093’s tumor had clonal mutations in TP53 and KMT2D but harbored an NF2 mutation solely in the cfDNA sample. Patient partner 0213’s tumor had a TP53 mutation and APOBEC-associated COSMIC signature 13 detected exclusively in the cfDNA sample.
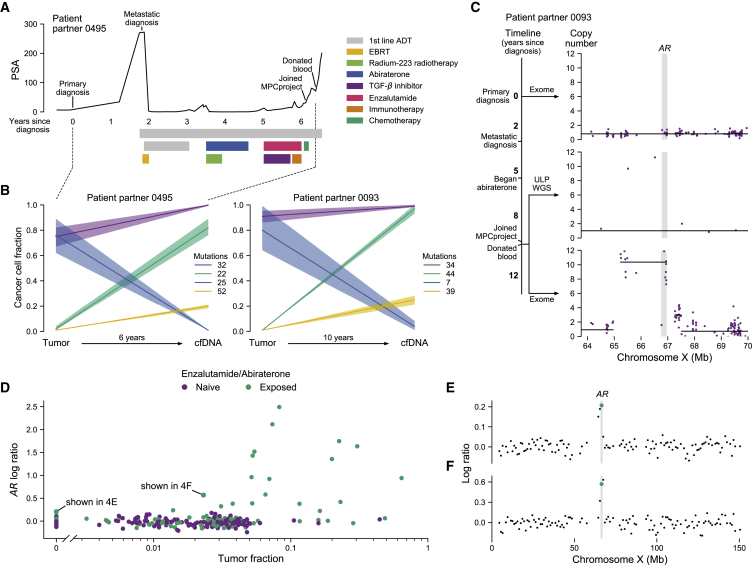
Figure 4.
cfDNA from donated blood reveals patterns of clonal dynamics and clinically relevant genomic changes
(A) Clinical trajectory of patient partner 0495. This patient partner’s prostate-specific antigen (PSA) trajectory is shown on the y axis, time in years since initial diagnosis is shown on the x axis, and bars denote the beginning and end of therapies. EBRT, external beam radiation therapy; first-line androgen deprivation therapy (ADT), leuprolide and bicalutamide; immunotherapy, nivolumab; chemotherapy, cisplatin and etoposide.
(B) Tumor evolution from primary tumor to metastatic cfDNA samples. The y axis shows the cancer cell fraction (CCF) of clonal clusters identified between tumor and cfDNA samples (x axis). Time between samples shown on the x axis. Colors indicate how many mutations were identified in each clone, with a 95% confidence interval around the estimated CCF. Purple represents the truncal/ancestral clone. Clusters with CCF <0.10 across all biopsies are omitted. The clinical trajectory of patient partner 0495 (left) is shown in (A), while the trajectory of patient partner 0093 (right) is shown in (C).
(C) Emergence of AR amplification in patient partner 0093 induced by anti-androgen therapy. The timeline depicts this patient’s clinical trajectory, while the plots show the absolute copy number (y axis) of the genomic region around AR (x axis, gene body shown in gray). The first plot depicts exome sequencing from the patient’s archival tumor tissue; the second and third plots depict ultra-low-pass whole-genome sequencing (ULP-WGS) and exome sequencing of cfDNA from the patient’s donated blood, respectively. Individual points represent copy number of target regions (exome) or copy number of 1 Mb genomic windows (ULP-WGS). Black lines represent discrete copy-number segments.
(D–F) ULP-WGS reveals clinically relevant AR amplifications even at low tumor fraction. In (D), tumor fraction of 318 cfDNA samples from donated blood of 300 patient partners with ULP-WGS sequencing is shown on the x axis, while the log copy ratio (logR) of the genomic interval containing AR is shown on the y axis. Points are colored by whether patient partners self-reported taking enzalutamide or abiraterone. 89 samples are shown with tumor fraction of 0 (undetectable), while 229 have non-zero tumor fractions. Two samples, one at a tumor fraction of 0 and another at a tumor fraction of 0.023, have chromosome X log copy ratio profiles shown in (E) and (F), respectively. The green points represent the values shown in (D), with the genomic interval containing AR highlighted in gray.
Two of these patient partners, 0495 and 0093, were initially diagnosed with primary prostate cancer (Gleason score 4 + 3 and 5 + 4, respectively), while patient partner 0213 was diagnosed with de novo metastatic disease. Their donated blood samples were separated from their primary tissue biopsies by a range of years (2–10 years). Despite these varied disease presentations, clinical trajectories, and biopsy timelines, we observed similar patterns of a “clonal switch” between the primary tumor and cfDNA, wherein different subclones were dominant in each sample (Figures 4B and S11). We did not, however, observe primary tumor-specific copy-number alterations, bolstering previous claims that subclonal diversification in MPC via mutations may happen after acquisition of ancestral copy-number alterations (Figure S12).49 Furthermore, we observed likely primary-tumor-specific mutations across all seven other patient partners with both tumor and cfDNA samples, although the samples had low purity (Figure S13). While we cannot account for the sampling bias of tumor biopsies, these results suggest that such clonal switches may be common in the development of metastatic disease.
In several cases, we detected the emergence of an amplification in the AR between the initial diagnosis and metastatic blood sample that was captured using ULP-WGS of cfDNA (example patient partner shown in Figure 4C). This led us to examine AR copy number using ULP-WGS of cfDNA samples across the entire cohort (n = 300 patient partners, 318 samples; Figures 4D and S14). We found that patient partners who reported taking enzalutamide or abiraterone had significantly higher AR log copy ratios across a range of tumor fractions (p < 0.001, linear regression). Men who had taken enzalutamide or abiraterone also had significantly higher tumor fractions, likely reflecting a more advanced disease state and subsequent higher tumor burden in blood (p < 0.001, Mann-Whitney U test).50 We observed that AR amplifications are often detectable in ULP-WGS of cfDNA even when the tumor fraction is below 0.03 (Figures 4E and 4F). For one patient partner, the tumor fraction within their donated blood was inferred as undetectable, but we nevertheless observed a clear AR amplification (Figure 4E). This highlights the potential efficacy of cfDNA to reveal clinically relevant changes in MPC, even in cases of very low or undetectable tumor burden. Attempts to identify other common copy-number changes were limited by tumor fraction (Figure S15). Broadly, these sequencing results illustrate the feasibility of identifying relevant genomic and evolutionary alterations from both archival tumor tissue and donated blood samples irrespective of geographical source site, enabling patient partners to participate in genomic research at no cost and with little effort.
Discussion
Here, we describe the MPCproject, a patient-driven framework for partnering with patients with MPC in the US and Canada to increase access to genomics research and strengthen our understanding of this disease. The online enrollment process was jointly created with patient partners and advocates to emphasize simplicity, requiring only the completion of online consent and survey forms, along with optional mailed saliva and blood kits. To our knowledge, no previous effort in MPC has used patient partnership to integrate demographic, clinical, patient-reported, and genomic data from patients at a national level.
To that end, we demonstrated the feasibility of working with over 700 patient partners, 41% of whom live in rural areas, MUAs, or HPSAs, a metric unreported in previous molecular profiling efforts. We found that 56% of our patient partners have never received care at an NCI-designated cancer center and that patient partners living in more disadvantaged areas were less likely to attend those institutions for treatment. Taken together with previous studies showing disparities in standard treatment and clinical trial outcomes by socioeconomic status, these results highlight existing barriers in access to care and sequencing studies.51, 52, 53 Furthermore, a recent study found that incomplete medical records are associated with shorter overall survival for patients with MPC, particularly for those with complicated clinical histories or whose care is fragmented between institutions.54 Our analysis of abstracted medical record data revealed a strong overlap between clinical histories represented in medical records and patient-reported data, even for patient partners with complex treatment trajectories or who had received treatment at multiple hospitals, supporting the use of patient surveys to improve care in this disease.
We also demonstrated that tumor tissue collected from archival samples and cfDNA from donated blood samples from across the US and Canada accurately recapitulate known genomic findings in MPC and place findings in the context of both patient-reported and abstracted medical record data. There has been substantial effort in the field to identify molecular features associated with selective response to therapies like PARP inhibition and immunotherapy, including the use of mutational signatures to assess targetable HRD, MMR, and APOBEC deficiencies in cases without a causative molecular alteration.36,55 Our results strengthen previous findings that such signatures can be detected using cfDNA and, combined with our ability to obtain cfDNA from participants nationwide, demonstrate the scalability of a patient-partnered approach to identify and validate such genomic findings within a real-world cohort in parallel to existing molecular approaches.56,57
Moreover, we used archival tumor tissue and cfDNA from donated blood to reconstruct tumor phylogenetic profiles, revealing polyclonality between primary and metastatic diagnosis. Despite well-known findings of heterogeneity in both primary and MPC, there is a paucity of matched primary-metastatic studies, owing mostly to the invasiveness and logistical challenges of longitudinal biopsy studies.34,58 Our project enables such studies paired with comprehensive clinical histories with minimal patient effort. To that end, we also found clinically relevant AR amplifications via low-pass WGS of cfDNA from donated blood, even at very low or undetectable tumor fractions. This result provides additional inexpensive utility to the suggested use of cfDNA tumor fraction as a clinically relevant biomarker in MPC.50,56 We are working with patient partners who continue to donate blood and have been able to collect multiple secondary blood biopsy kits for future longitudinal analysis.
New approaches in molecular cancer research are needed to address an increased desire from patients to actively participate in research and a pressing need for equity in the clinic. Paired with emerging open-access clinical trials, patient-driven studies hold great promise to achieve equity and accelerate discovery in genomic research.59 The MPCproject is part of a wider “Count Me In” patient-partnered initiative (joincountmein.org) that has already yielded new findings in angiosarcoma and has expanded to metastatic breast cancer and osteosarcoma, among others.60, 61, 62 The achievements of the MPCproject are based entirely on the courage and altruism of the men with whom we partner, who, in the words of one participant, hope that their “participation will help other men […] and lead eventually to a cure.”
Limitations of the study
Despite the geographic diversity of our patient partners, we acknowledge that they do not reflect the racial diversity of patients with MPC, a critical issue given substantial disparities in both cancer care and genomics research by race and ethnicity.11,63,64 These unmet disparities demand that we rethink our models of outreach and patient engagement, and our effort cannot be considered a success until sustained and equitable partnership is achieved.65 Recognizing that building trust in marginalized communities takes time, we must continue to work longitudinally with community-based advocacy organizations to partner with Black communities. Since the launch of our project, we have worked to build an engagement model that meets patients in their communities, including churches, barbershops, and fraternities. Using the longitudinal model of this study, we will continue to iteratively learn from community engagement successes and failures. We received feedback, for example, that Black patients and their cancer stories are rarely heard—in response, we are building a campaign to amplify the voices of Black patients with cancer and their lived experiences (www.BlackCancerVoices.org). Additionally, a common request is for the project to return clinically relevant sequencing results to patient partners and their physicians. We are working with regulatory, clinical, and sequencing experts to build the infrastructure necessary to fulfill this request.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Raw sequencing files | This paper | dbGaP study accession phs001939.v3.p1 |
| Raw sequencing files (processed by GDC) | This paper | https://portal.gdc.cancer.gov/projects/CMI-MPC |
| Processed and deidentified sequencing and clinical data | This paper | https://www.cbioportal.org/study/summary?id=mpcproject_broad_2021 |
| Processed and deidentified figure data and code | This paper | https://github.com/vanallenlab/mpcproject-paper |
| Study information and materials seen by patients | This paper | https://mpcproject.org/ |
| Rural-area continuum codes (2010) | USDA66 | https://www.ers.usda.gov/data-products/rural-urban-commuting-area-codes.aspx |
| Information on MPC patients nationwide (2018) | SEER21 | https://seer.cancer.gov/data-software/ |
| Medically underserved and health-physician shortage areas (accessed Dec 2021) | HRSA23 | https://data.hrsa.gov/tools/shortage-area |
| National Area Deprivation Index 2019 data | Kind and Buckingham, 201867 | https://www.neighborhoodatlas.medicine.wisc.edu/ |
| ClinVar (2019) | Landrum et al., 201832 | https://www.ncbi.nlm.nih.gov/clinvar/ |
| Variant Effect Predictor GRCh37 Cache | McLaren et al., 201668 | https://useast.ensembl.org/info/docs/tools/vep/script/vep_cache.html |
| COSMIC germline cancer census gene set v86 | Sondka et al., 201869 | https://cancer.sanger.ac.uk/census |
| Software and algorithms | ||
| Python 3.8 | Python Software Foundation, 202170 | https://www.python.org/ |
| R 3.5.1 | R Core Team, 202171 | https://www.r-project.org/ |
| BWA | Li and Durbin, 200972 | http://bio-bwa.sourceforge.net/ |
| GATK 3.7 | McKenna et al., 201073 | https://github.com/broadinstitute/gatk/releases |
| Sequence alignment and alteration calling (component algorithms detailed below) | The Getz Laboratory | https://portal.firecloud.org/#methods/getzlab/CGA_WES_Characterization_Pipeline_v0.1_Dec2018/ |
| Mutect v1.1.6 | Cibulskis et al., 201374 | http://archive.broadinstitute.org/cancer/cga/mutect |
| FilterByOrientationBias | McKenna et al., 201073 | https://gatk.broadinstitute.org/hc/en-us/articles/360037060232 |
| Strelka v2.8.0 | Saunders et al., 201275 | https://github.com/Illumina/strelka |
| Oncotator v1.9.9.0 | Ramos et al., 201576 | https://github.com/broadinstitute/oncotator |
| MutSig2CV | Lawrence et al., 201477 | https://github.com/getzlab/MutSig2CV |
| GATK 3.7 (CNV) | McKenna et al., 201073 | https://gatk.broadinstitute.org/hc/en-us/articles/360035531092 |
| ABSOLUTE v1.5 | Carter et al., 201278 | https://software.broadinstitute.org/cancer/cga/absolute_download |
| FACETS v0.6.2 | Shen and Seshan, 201679 | https://github.com/mskcc/facets |
| GISTIC2.0 v2.0.23 | Mermel et al., 201128 | https://github.com/broadinstitute/gistic2 |
| DeTiN v2.0.1 | Taylor-Weiner et al., 201880 | https://github.com/getzlab/deTiN |
| ContEst | Cibulskis et al., 201181 | https://software.broadinstitute.org/cancer/cga/contest |
| CrossCheckFingerprints (GATK 3.7) | McKenna et al., 201073 | https://gatk.broadinstitute.org/hc/en-us/articles/360037594711 |
| ichorCNA | Adalsteinsson et al., 201756 | https://github.com/broadinstitute/ichorCNA |
| deconstructSigs (COSMIC v2 signatures, v1.9.0) | Rosenthal et al., 201633 | https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-0893-4 |
| DeepVariant v0.8.0 | Poplin et al., 201882 | https://github.com/google/deepvariant |
| PhylogicNDT | Leshchiner et al., 201846 | https://github.com/broadinstitute/PhylogicNDT |
| Other | ||
| Repository for regenerating main study findings and figures of this paper | This paper | https://github.com/vanallenlab/mpcproject-paper, https://doi.org/10.5281/zenodo.6816267 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Eliezer M. Van Allen (Eliezerm_vanallen@dfci.harvard.edu).
Materials availability
This study did not generate any new unique reagents.
Experimental model and subject details
Patients who chose to enroll in this research study provided informed consent using a web-based consent form approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board (DF/HCC Protocol 15-057B). Patient partners can exit the study at any time. All patient partners were male, with age and other features detailed in Table S2. If patient partners consented, FFPE exomes were requested from hospitals where they received treatment. Germline DNA was collected using mailed saliva collection kits. cfDNA from blood biopsies was collected through blood draws by medical providers or Quest Diagnostics (with a complimentary voucher), received by mail (Method details).
Method details
MPCproject website
The MPCproject utilizes a website (https://mpcproject.org/) to enroll patients through an online consent and release form. The website provides information about the project and advocacy groups that have partnered with the study. The website design, messaging, and workflow were developed with direct input from patient partners and advocates.
Informed consent
A link to the electronic informed consent document for formal enrollment in the study (https://mpcproject.org/ConsentAndRelease.pdf) was sent to registrant emails, and upon signing, a copy of the completed form was shared. At minimum, informed consent enabled study staff to request and abstract medical records, send a saliva kit directly to patients, perform sequencing on any returned saliva samples, and release de-identified integrated clinical, genomic, and patient-reported data for research use. Patient partners had the additional option to consent to study staff obtaining a portion of archived tumor tissue and/or a blood sample for further sequencing analysis.
Patient-reported data
After registering, patient partners completed a 17-question survey asking them about themselves and their disease (https://mpcproject.org/AboutYouSurvey.pdf). All questions were optional. Information on how question responses were standardized and categorized can be found in the supplemental methods.
Acquisition of medical records
Medical records were obtained for patient partners from the U.S. and Canada who completed the consent and medical release forms. Later in project development, a donated saliva or blood sample was also required. Study staff submitted medical record requests to all institutions and physician offices at which the patient reported receiving clinical care for their prostate cancer. A detailed medical record request form, along with the consent and release forms, were electronically faxed to each facility listed in a patient’s release form. Medical records were returned to the project via mail, fax, or secure online portals. If a record request was not fulfilled in six months, study staff called the hospital, and a second request was submitted, with up to three requests made. Patient partners that communicated with study staff about changes in their treatment could request a medical record update, in which case their current hospital was again contacted for medical records. All medical records were saved in an electronic format to a secure drive at the Broad Institute.
Acquisition of patient samples
All consented patient partners living in the United States or Canada were mailed saliva kits with appropriate instructions, a sample tube labeled with a unique barcode, and a prepaid return box to send back the saliva sample. Samples were returned to the Broad Institute Genomics Platform, logged, and stored at room temperature (25 °C) until further sequencing.
If a consented patient partner opted into the blood biopsy component of the study, they were sent a blood kit with instructions (https://mpcproject.org/BloodSampleInstructions.pdf, Figure S4). Participants could take this kit to their next blood draw and request a courtesy draw by their medical provider; if a courtesy draw was not possible, patient partners could go to Quest Diagnostics with a complimentary voucher to have their blood drawn. Blood kits were returned free of charge to the Broad Institute Genomics Platform where they were fractionated into plasma and buffy coats and stored at −80 °C. If a patient partner did not provide a saliva sample, buffy coats were used to extract germline DNA for WES. Plasma samples continued to WES if ultra-low pass WGS detected a tumor fraction of circulating tumor DNA greater than 0.03. Some patient partners were selected to provide additional blood samples and were sent a new consent form. If they agreed to submit another blood sample, a new blood kit was shipped.
For patient partners that provided a germline sample and consented to the acquisition of some of their archival tumor tissue, study staff reviewed each patient’s medical records and identified available tissue supplemental methods). Patient partners were screened by the study staff to determine if they had metastatic or advanced prostate cancer based on the definition by our study. If a patient partner had a sample that met the project’s strict requesting criteria, study staff coordinated with that hospital’s pathology department to fax a request for one H&E-stained slide as well as either 5–20 5-μm unstained slides or one formalin-fixed paraffin-embedded tissue block. Requests explicitly asked that the pathology department should not exhaust a sample to fulfill the request. Samples were sent to the MPCproject by mail. Tissue samples received as slides were labeled with unique barcode identifiers and submitted for whole exome sequencing. Tissue samples received as blocks were cut into three 30-μm scrolls per block, labeled with unique barcode identifiers, and then submitted for whole exome sequencing.
Medical record abstraction
A data dictionary comprising 60 clinical fields with possible options was curated by trained study staff working with prostate oncologists. Electronic health records were converted to searchable PDF files using the Optical Character Recognition (OCR) engine known as Tesseract.83 Three study staff abstractors were involved in the abstraction and QC process for each record (supplemental methods). If a field had lack of concordance between abstractors or there were outstanding questions, a prostate cancer oncologist reviewed the content. Whenever possible, clinical data was abstracted directly from the records. For information that’s not found, it was abstracted as 'NOT FOUND IN RECORD'. In instances where ambiguity or incomplete data was present, inferences were made considering the whole narrative of the medical record. Incomplete dates missing the day or month are abstracted as the first day of the month or first month of the year, respectively. While all medical records will eventually be abstracted, medical records from patient partners that received molecular sequencing of some form were prioritized for this study, resulting in 125 patient partners with medical record abstractions, 119 of which had at least one therapy noted. In examining the overlap between patient surveys and medical record therapies, we only considered therapies that were given for metastatic prostate cancer at least one week before the patient enrolled.
Geographic analysis
Using patient-reported data and secure Census Bureau geocoding, we identified residential census tracts for 628/706 patient partners.84 To identify patient partners living in rural areas, this information was overlapped with rural-area continuum (RUCA) codes from the United States Department of Agriculture (USDA).66 Census tracts with a secondary RUCA code greater than 3 were designated as rural. For comparison, the proportion of metastatic prostate cancer patients within each RUCA code from 2004 – 2017 was taken from Surveillance, Epidemiology, and End Results (SEER) using SEER∗stat with the following selection table: {Site and Morphology.Site recode ICD-O-3/WHO 2008} = 'Prostate' AND {Stage - Summary/Historic.SEER Combined Summary Stage 2000 (2004-2017)} != 'In situ', 'Localized only', 'Not applicable', 'Unknown/unstaged/unspecified/DCO', 'Blank(s)'.21 To identify patient partners living in medical shortage areas, census tracts were overlapped with primary care health physician shortage areas (HPSA) and medically underserved areas (MUA) defined by the Health Resources and Services Administration (HRSA).23 Census tracts were labelled as existing within a MUA or HPSA if they were designated as within a medically underserved area/population or within a primary care HPSA, respectively. Published geographic datasets of cancer patients (e.g., SEER, NPCR) do not contain census-tract resolved data or summary results of MUA/HPSA status, so for comparison we instead used the total U.S. population living in HPSAs and MUAs, taken from HRSA, divided by the entire U.S. population taken from the U.S. Census.23,24 To calculate appointment distances, we calculated the round-trip Haversine distances between residential zip codes and the zip code of reported institutions. To assess socioeconomic advantage, we used secure Census Bureau geocoding to identify residential census block groups (12 digit FIPS codes) and cross-referenced them with a publicly available dataset of Area Deprivation Index (https://www.neighborhoodatlas.medicine.wisc.edu/download).67 We used the National ADI, which ranks neighborhoods by percentiles (1–100), with 100 indicating the highest level of disadvantage.
To protect privacy, geographic locations in the graphical abstract do not represent real patient partner residential areas. Random counties from the state of each reported residential area are shown instead.
Whole exome sequencing analysis
Whole exome sequences were captured using Illumina technology and the sequence data processing and analysis was performed using Picard and FireCloud pipelines on Terra (https://terra.bio/) (supplemental methods). The Picard pipeline (http://picard.sourceforge.net) was used to produce a BAM file with aligned reads. This includes alignment to the GRCh37 human reference sequence using BWA72 and estimation and recalibration of base quality score with the Genome Analysis Toolkit (GATK).73 Somatic alterations for tumor samples were called using a customized version of the Getz Lab CGA WES Characterization pipeline (https://portal.firecloud.org/#methods/getzlab/CGA_WES_Characterization_Pipeline_v0.1_Dec2018/) developed at the Broad Institute. Briefly, MuTect v1.1.6 algorithm was used to identify somatic mutations.74 Somatic mutation calls were filtered using a panel of normals (PoN), oxoG filter and an FFPE filter to remove artifacts introduced during the sequencing or formalin fixation process.85 Small somatic insertions and deletions were detected using the Strelka algorithm.75 Somatic mutations were annotated using Oncotator.76 Recurrently altered mutations were identified using MutSig2CV.77 To define somatic copy ratio profiles, we used GATK CNV.73 To generate allele-specific copy number profiles and assess tumor purity and ploidy, we used ABSOLUTE and FACETS.78,79 Final segmentation calls were taken from ABSOLUTE, except for the X chromosome, which was taken from FACETS. We utilized GISTIC2.0 to identify significantly recurrent amplification and deletion peaks.28 For determining allele-specific copy number alterations, we assessed the absolute allelic copy numbers of the segment containing each gene. Mutation burden was calculated as the total number of mutations (non-synonymous + synonymous) detected for a given sample divided by the length of the total genomic target region captured with appropriate coverage from whole exome sequencing.
Whole exome sequencing quality control
Samples with average coverage below 55x in the tumor sample or below 30x in the normal sample were excluded. Samples with purity <0.10 from both ABSOLUTE and FACETS were excluded. DeTiN was applied to samples to estimate the amount of tumor contamination in the normal samples; samples with TiN (tumor in normal) > 0.25 were excluded.80 ContEst was applied to measure the amount of cross-sample contamination in samples; samples with contamination >0.04 were excluded.81 The Picard task CrossCheckFingerprints was applied to determine sample mixups; samples with Fingerprints LOD value <0 were excluded.86 Two FFPE samples that failed sequence processing and were noted to have extensive segment fragmentation and allelic imbalance were also excluded due to suspicion of poor sequencing. A table of samples with quality control metrics for each sample can be found in the Supplementary Data. Samples which passed quality control were submitted to cBioPortal and GDC.
Ultra-low pass whole genome sequencing analysis
ichorCNA was used to assess the tumor fraction in cfDNA samples that completed ultra-low pass whole genome sequencing.56 The log copy ratio of AR was assessed by the log copy ratio of the genomic interval containing AR. This value could not consistently be converted to absolute copy number due to the low tumor fractions of many samples.
Mutational signature analysis and kataegis
Mutational processes in our cohort were determined using deconstructSigs with default parameters applying COSMIC v2 signatures as the reference with a maximum number of signatures of 629,30. A signature was assessed as present if the signature contribution was greater than 6%. Because tumor samples were formalin-fixed and paraffin embedded (FFPE), a process known to introduce stranded mutational artifacts in specific nucleotide contexts, we used a filter to remove likely FFPE artifacts according to nucleotide context and strand bias before using deconstructSigs.87 We also tried to assess the colocalization of the kataegis event with structural variant breakpoints but were limited by targeted sequencing in exomes and low coverage in ULP-WGS. KMT2C and its surrounding region were not copy number altered in the sample with kataegis. Kataegis was not identified in any other sample.
Germline variant discovery
To call short germline single-nucleotide polymorphisms, insertions, and deletions from germline WES data, we used DeepVariant (v0.8.0).82,88 Specifically, we used the publicly-released WES model (https://console.cloud.google.com/storage/browser/deepvariant/models/DeepVariant/0.8.0/DeepVariant-inception_v3-0.8.0+data-wes_standard/) to generate single-sample germline variant call files using the human genome reference GRCh37(b37). We filtered variants with bcftools v1.9 to only keep high-quality variants annotated as “PASS” in the “FILTER” column. The high-quality variants were merged into single-sample Variant Call Format (VCF) files using CombineVariants from GATK 3.7 (https://github.com/broadinstitute/gatk/releases). To decompose multiallelic variants and normalize variants, we used the computational package vt v3.13 (https://github.com/atks/vt). Lastly, germline variants were annotated using the VEP v92 with the publicly-released GRCh37 cache file (https://github.com/Ensembl/ensembl-vep).68 An alteration was also considered if there was a pathogenic germline alteration, denoted by “Pathogenic”, “Pathogenic/Likely_pathogenic”, “Likely_pathogenic”, “_risk_factor”, or “Conflicting_interpretations_of_pathogenicity” (if at least one expert source indicated “Likely_pathogenic” or “Pathogenic”) in ClinVar (Dec 2019 version).32 An alteration was also considered if it had an “HIGH” predicted impact on protein function and had a maximum allele fraction of <0.01 in all populations. The germline cancer predisposition genes were selected based on the level of evidence supporting their Mendelian disease susceptibility. This is composed of the well-curated COSMIC germline cancer census gene set (v86; http://cancer.sanger.ac.uk/census) and the germline cancer gene set listed in Huang et al. 2018 and Rahman 2014.30,69,89,90
Association of DNA-repair alterations and presence of signature 3
Alterations in a select list of genes previously implicated in DNA-repair were examined (Table S3). An alteration was considered if there was a somatic single-copy deletion, double deletion, nonsense mutation, missense mutation, frameshift indel, or splice site mutation. An alteration was also considered if there was a pathogenic germline alteration. An alteration was considered biallelic for Figure S7 if there was a double somatic deletion, a pathogenic germline/protein-altering somatic variant plus a somatic loss, or more than one mutation in the same gene, although we cannot confirm the biallelic nature of multiple mutations.
Phylogenetic analysis
To compare mutations between distinct samples (tumor and cfDNA) from the same patient, we used a previously described method designed to recover evidence for mutations called in one sample in all other samples derived from the same individual.91 In brief, the ‘force-calling’ method uses the strong prior of the mutation being present in at least one sample in the patient to detect and recover mutations that might otherwise be missed. A mutation was deemed tumor/cfDNA specific if there were no force-called reads that supported the mutation in the other sample, although this process underestimates the proportion of shared mutations in low purity tumors. The cancer cell fraction (CCF) of mutations were defined using ABSOLUTE, which calculates the CCF based on variant allele frequency, purity, and local allelic copy number.78 To reconstruct tumor phylogenies, we used PhylogicNDT, which clusters mutations into subclones across multiple samples based on their underlying similar CCFs.46
Quantification and statistical analysis
Statistical analysis
Except where otherwise specified, analysis and data visualization were performed with Python 3.8, SciPy v.1.5.2, Matplotlib v.3.3.2, seaborn v.0.11.0 and R v.3.5.1.90,91 The code used to generate most main figures, analyses, and supplementary figures can be found at https://github.com/vanallenlab/mpcproject-paper or Zenodo: https://doi.org/10.5281/zenodo.6816267, except for figures and analyses requiring sample-level germline data. Between-group comparisons of continuous variables were performed with the Mann-Whitney U test (Wilcoxon rank sum test) or Student’s t-test. Contingency table tests were performed with Fisher’s exact test. All tests were two-sided.
Additional resources
MPCproject website: https://mpcproject.org/.
Acknowledgments
We thank our patient partners, caregivers, loved ones, project advisory council, and advocacy partners, without whom this project would not be possible. We would like to pay our respects to the late Jack Whelan, a patient with MPC and advocate, who was instrumental in developing the MPCproject. We also thank the staff of the MPCproject, the engineering team from the Data Sciences Platform at the Broad Institute (A. Zimmer, E. Baker, S. Maiwald, P. Taheri, D. Kaplan, J. Lapan, S. Sutherland), and all members of Count Me In who work daily to ensure that all patients with MPC can participate in research. Finally, we would like to express our gratitude to the Broad Institute Cancer Program, the Broad Institute Genomics Platform, Broad Institute Communications and Development teams, and the compliance team at the Broad Institute for their support of the project. Figure 1A and parts of Figure 2F were created with BioRender.com. This work was funded by the following: Count Me In, Inc., Fund for Innovation in Cancer Informatics (E.M.V.A.); PCF-Movember Challenge Award (E.M.V.A.); NIH R01CA227388 and U01CA233100 (E.M.V.A.); Mark Foundation Emerging Leader Award (E.M.V.A.); Participant Engagement and Cancer Genome Sequencing (U2CCA252974); US Department of Defense (W81XWH-21-1-0084 and PC200150, S.H.A.); Prostate Cancer Foundation (S.H.A.); Conquer Cancer Foundation of the American Society of Clinical Oncology (S.H.A.); National Science Foundation (GRFP DGE1144152, M.X.H.); and National Institutes of Health (T32 GM008313).
Author contributions
N.W., C.A.P., and E.M.V.A. conceived and designed the MPCproject with support from E.S.L. J.C., S.B., L.S., and E.M.V.A. designed and prepared the study and interpreted the data. J.C. wrote the manuscript and performed the analyses. S.B. and L.S. led study operations including tumor sample and medical record acquisition, sample sequencing, and patient coordination. L.S., B.S.T., M.D., E.A., S.S., A.L.D., R.R., D.M.S., and I.K.S. oversaw medical record abstraction. S.Y.C. provided feedback on various analyses of the study and completed germline variant calling with oversight from S.H.A. S.B., L.S., J.C., B.N.T., M.D., M.M., and P.S.C. coordinated data releases. M.M., P.S.C., A.D., and B.Z. led recent project operations. M.D. supervised early project operations. C.M.N. and E.A. led patient advocacy and outreach efforts. A.T.M.C. and S.W. oversaw early project sequencing analyses. M.X.H. provided feedback of study analyses. A.K.T. provided feedback on medical record abstractions and tissue sample collection. D.K. enabled electronic medical record searching. J.N., J.M., I.H.G., and B.O. contributed to survey design, project development, assessment of patient criteria, and outreach strategy.
Declaration of interests
M.X.H. has been a consultant to Amplify Medicines and Ikena Oncology and is a current employee of Genentech/Roche. E.S.L. is currently in the process of divesting any relevant holdings. N.W. reports advisory relationships and consulting with Eli Lilly and Co.; advising and stockholding interest in Relay Therapeutics; and grant support from Puma Biotechnology. E.M.V.A. reports advisory relationships and consulting with Tango Therapeutics, Genome Medical, Invitae, Illumina, Enara Bio, Mani-fold Bio, and Janssen; research support from Novartis and BMS; equity in Tango Therapeutics, Genome Medical, Syapse, Mani-fold Bio, and Enara Bio; and travel reimbursement from Roche and Genentech, outside the submitted work.
Published: August 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2022.100169.
Supplemental information
Data and code availability
The MPCproject releases deidentified clinical, patient-reported and research-grade genomic data into public repositories, such as cBioPortal: mpcproject_broad_2021 (https://www.cbioportal.org/study/summary?id=mpcproject_broad_2021), the Genomic Data Commons: CMI-MPC (https://portal.gdc.cancer.gov/projects/CMI-MPC), and dbGaP: phs001939.v3.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001939.v3.p1) at regular intervals and prepublication. Data is processed and formatted as required by each repository’s guidelines. All patient identifiers are stripped prior to data deposition to protect patient privacy. On the MPCproject data release webpage (https://mpcproject.org/data-release), patients can access project data, additional information about the data, a list of common terms used in research, methods used to generate the data, and an e-mail address for any additional data-related questions. All other data used in this paper are from publicly available resources. The code used to generate most main figures, central analyses, and supplementary figures can be found at can be found at https://github.com/vanallenlab/mpcproject-paper, except for figures and analyses requiring sample-level germline data. An unchanging version of the code at time of publication is also available at Zenodo: https://doi.org/10.5281/zenodo.6816267. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA. Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Litwin M.S., Tan H.-J. The diagnosis and treatment of prostate cancer: a review. JAMA. 2017;317:2532–2542. doi: 10.1001/jama.2017.7248. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armenia J., Wankowicz S.A.M., Liu D., Gao J., Kundra R., Reznik E., Chatila W.K., Chakravarty D., Han G.C., Coleman I., et al. The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 2018;50:645–651. doi: 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bono J., Mateo J., Fizazi K., Saad F., Shore N., Sandhu S., Chi K.N., Sartor O., Agarwal N., Olmos D., et al. Olaparib for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 6.Abida W., Cyrta J., Heller G., Prandi D., Armenia J., Coleman I., Cieslik M., Benelli M., Robinson D., Van Allen E.M., et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. USA. 2019;116:11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Annala M., Vandekerkhove G., Khalaf D., Taavitsainen S., Beja K., Warner E.W., Sunderland K., Kollmannsberger C., Eigl B.J., Finch D., et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8:444–457. doi: 10.1158/2159-8290.CD-17-0937. [DOI] [PubMed] [Google Scholar]
- 8.Sonpavde G., Agarwal N., Pond G.R., Nagy R.J., Nussenzveig R.H., Hahn A.W., Sartor O., Gourdin T.S., Nandagopal L., Ledet E.M., et al. Circulating tumor DNA alterations in patients with metastatic castration-resistant prostate cancer. Cancer. 2019;125:1459–1469. doi: 10.1002/cncr.31959. [DOI] [PubMed] [Google Scholar]
- 9.Siu L.L., Lawler M., Haussler D., Knoppers B.M., Lewin J., Vis D.J., Liao R.G., Andre F., Banks I., Barrett J.C., et al. Facilitating a culture of responsible and effective sharing of cancer genome data. Nat. Med. 2016;22:464–471. doi: 10.1038/nm.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joly Y., Dove E.S., Knoppers B.M., Bobrow M., Chalmers D. Data sharing in the post-genomic world: the experience of the international cancer genome consortium (ICGC) data access compliance office (DACO) PLoS Comput. Biol. 2012;8:e1002549. doi: 10.1371/journal.pcbi.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spratt D.E., Chan T., Waldron L., Speers C., Feng F.Y., Ogunwobi O.O., Osborne J.R. Racial/ethnic disparities in genomic sequencing. JAMA Oncol. 2016;2:1070–1074. doi: 10.1001/jamaoncol.2016.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feyman Y., Provenzano F., David F.S. Disparities in clinical trial access across US urban areas. JAMA Netw. Open. 2020;3:e200172. doi: 10.1001/jamanetworkopen.2020.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huey R.W., Hawk E., Offodile A.C. Mind the Gap: precision Oncology and its potential to widen disparities. J. Oncol. Pract. 2019;15:301–304. doi: 10.1200/JOP.19.00102. [DOI] [PubMed] [Google Scholar]
- 14.Mamun A., Nsiah N.Y., Srinivasan M., Chaturvedula A., Basha R., Cross D., Jones H.P., Nandy K., Vishwanatha J.K. Diversity in the era of precision medicine - from bench to bedside implementation. Ethn. Dis. 2019;29:517–524. doi: 10.18865/ed.29.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Messner D.A., Al Naber J., Koay P., Cook-Deegan R., Majumder M., Javitt G., Deverka P., Dvoskin R., Bollinger J., Curnutte M., et al. Barriers to clinical adoption of next generation sequencing: perspectives of a policy Delphi panel. Appl. Transl. Genom. 2016;10:19–24. doi: 10.1016/j.atg.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.American Cancer Society Cancer Action Network . American Cancer Society Cancer Action Network; 2020. Payer Coverage Policies of Tumor Biomarker Testing. [Google Scholar]
- 17.Chakradhar S. Tumor sequencing takes off, but insurance reimbursement lags. Nat. Med. 2014;20:1220–1221. doi: 10.1038/nm1114-1220. [DOI] [PubMed] [Google Scholar]
- 18.McGuire A.L., Oliver J.M., Slashinski M.J., Graves J.L., Wang T., Kelly P.A., Fisher W., Lau C.C., Goss J., Okcu M., et al. To share or not to share: a randomized trial of consent for data sharing in genome research. Genet. Med. 2011;13:948–955. doi: 10.1097/GIM.0b013e3182227589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Husedzinovic A., Ose D., Schickhardt C., Fröhling S., Winkler E.C. Stakeholders’ perspectives on biobank-based genomic research: systematic review of the literature. Eur. J. Hum. Genet. 2015;23:1607–1614. doi: 10.1038/ejhg.2015.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.2022. Sequence me - demand genomic & DNA testing for cancer treatments.https://sequenceme.org/ [Google Scholar]
- 21.Surveillance Research Program . National Cancer Institute SEER∗Stat Software. National Cancer Institute; 2022. [Google Scholar]
- 22.Economic Research Service . U.S. Department of Agriculture; 2013. Rural-Urban Continuum Codes - Documentation. [Google Scholar]
- 23.https://data.hrsa.gov/tools/shortage-area; 2022.
- 24.Population Clock, (2022). https://www.census.gov/popclock/.
- 25.Maroko A.R., Doan T.M., Arno P.S., Hubel M., Yi S., Viola D. Integrating social determinants of health with treatment and prevention: a new tool to assess local area deprivation. Prev. Chronic Dis. 2016;13:E128. doi: 10.5888/pcd13.160221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson M., Perrin A. Pew Research Center Internet Science Tech; 2017. Technology Use Among Seniors.https://www.pewresearch.org/internet/2017/05/17/technology-use-among-seniors/ [Google Scholar]
- 27.Rawla P. Epidemiology of prostate cancer. World J. Oncol. 2019;10:63–89. doi: 10.14740/wjon1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crowdis J., He M.X., Reardon B., Van Allen E.M. CoMut: visualizing integrated molecular information with comutation plots. Bioinformatics. 2020;36:4348–4349. doi: 10.1093/bioinformatics/btaa554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E., et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenthal R., McGranahan N., Herrero J., Taylor B.S., Swanton C. deconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerhauser C., Favero F., Risch T., Simon R., Feuerbach L., Assenov Y., Heckmann D., Sidiropoulos N., Waszak S.M., Hübschmann D., et al. Molecular evolution of early-onset prostate cancer identifies molecular risk markers and clinical trajectories. Cancer Cell. 2018;34:996–1011.e8. doi: 10.1016/j.ccell.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., Nava Rodrigues D., Robinson D., Omlin A., Tunariu N., et al. DNA-repair defects and Olaparib in metastatic prostate cancer. N. Engl. J. Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pritchard C.C., Mateo J., Walsh M.F., De Sarkar N., Abida W., Beltran H., Garofalo A., Gulati R., Carreira S., Eeles R., et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N. Engl. J. Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polak P., Kim J., Braunstein L.Z., Karlic R., Haradhavala N.J., Tiao G., Rosebrock D., Livitz D., Kübler K., Mouw K.W., et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet. 2017;49:1476–1486. doi: 10.1038/ng.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sztupinszki Z., Diossy M., Krzystanek M., Borcsok J., Pomerantz M.M., Tisza V., Spisak S., Rusz O., Csabai I., Freedman M.L., Szallasi Z. Detection of molecular signatures of homologous recombination deficiency in prostate cancer with or without BRCA1/2 mutations. Clin. Cancer Res. 2020;26:2673–2680. doi: 10.1158/1078-0432.CCR-19-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sztupinszki Z., Diossy M., Krzystanek M., Reiniger L., Csabai I., Favero F., Birkbak N.J., Eklund A.C., Syed A., Szallasi Z. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. Npj Breast Cancer. 2018;4:16. doi: 10.1038/s41523-018-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanton C., McGranahan N., Starrett G.J., Harris R.S. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015;5:704–712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A., et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraser M., Sabelnykova V.Y., Yamaguchi T.N., Heisler L.E., Livingstone J., Huang V., Shiah Y.-J., Yousif F., Lin X., Masella A.P., et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature. 2017;541:359–364. doi: 10.1038/nature20788. [DOI] [PubMed] [Google Scholar]
- 44.Mucci L.A., Hjelmborg J.B., Harris J.R., Czene K., Havelick D.J., Scheike T., Graff R.E., Holst K., Möller S., Unger R.H., et al. Familial risk and heritability of cancer among twins in nordic countries. JAMA. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.AlDubayan S.H. Considerations of multigene test findings among men with prostate cancer - knowns and unknowns. Can. J. Urol. 2019;26:14–16. [PubMed] [Google Scholar]
- 46.Leshchiner I., Livitz D., Gainor J.F., Rosebrock D., Spiro O., Martinez A., Mroz E., Lin J.J., Stewart C., Kim J., et al. Comprehensive analysis of tumour initiation, spatial and temporal progression under multiple lines of treatment. bioRxiv. 2018:508127. doi: 10.1101/508127. Preprint at. [DOI] [Google Scholar]
- 47.Grasso C.S., Wu Y.-M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C., et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tucci M., Zichi C., Buttigliero C., Vignani F., Scagliotti G.V., Di Maio M. Enzalutamide-resistant castration-resistant prostate cancer: challenges and solutions. OncoTargets Ther. 2018;11:7353–7368. doi: 10.2147/OTT.S153764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Espiritu S.M.G., Liu L.Y., Rubanova Y., Bhandari V., Holgersen E.M., Szyca L.M., Fox N.S., Chua M.L.K., Yamaguchi T.N., Heisler L.E., et al. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell. 2018;173:1003–1013.e15. doi: 10.1016/j.cell.2018.03.029. [DOI] [PubMed] [Google Scholar]
- 50.Choudhury A.D., Werner L., Francini E., Wei X.X., Ha G., Freeman S.S., Rhoades J., Reed S.C., Gydush G., Rotem D., et al. Tumor fraction in cell-free DNA as a biomarker in prostate cancer. JCI Insight. 2018;3:e122109. doi: 10.1172/jci.insight.122109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Onega T., Duell E.J., Shi X., Demidenko E., Goodman D. Determinants of NCI Cancer Center attendance in Medicare patients with lung, breast, colorectal, or prostate cancer. J. Gen. Intern. Med. 2009;24:205–210. doi: 10.1007/s11606-008-0863-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Unger J.M., Moseley A.B., Cheung C.K., Osarogiagbon R.U., Symington B., Ramsey S.D., Hershman D.L. Persistent disparity: socioeconomic deprivation and cancer outcomes in patients treated in clinical trials. J. Clin. Oncol. 2021;39:1339–1348. doi: 10.1200/JCO.20.02602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh G.K., Jemal A. Socioeconomic and racial/ethnic disparities in cancer mortality, incidence, and survival in the United States, 1950-2014: over six decades of changing patterns and widening inequalities. J. Environ. Public Health. 2017;2017:2819372. doi: 10.1155/2017/2819372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang D.X., Khera R., Miccio J.A., Jairam V., Chang E., Yu J.B., Park H.S., Krumholz H.M., Aneja S. Prevalence of missing data in the national cancer database and association with overall survival. JAMA Netw. Open. 2021;4:e211793. doi: 10.1001/jamanetworkopen.2021.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abida W., Campbell D., Patnaik A., Shapiro J.D., Sautois B., Vogelzang N.J., Voog E.G., Bryce A.H., McDermott R., Ricci F., et al. Non-BRCA DNA damage repair gene alterations and response to the PARP inhibitor rucaparib in metastatic castration-resistant prostate cancer: analysis from the phase II TRITON2 study. Clin. Cancer Res. 2020;26:2487–2496. doi: 10.1158/1078-0432.CCR-20-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adalsteinsson V.A., Ha G., Freeman S.S., Choudhury A.D., Stover D.G., Parsons H.A., Gydush G., Reed S.C., Rotem D., Rhoades J., et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017;8:1324. doi: 10.1038/s41467-017-00965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritch E., Fu S.Y.F., Herberts C., Wang G., Warner E.W., Schönlau E., Taavitsainen S., Murtha A.J., Vandekerkhove G., Beja K., et al. Identification of hypermutation and defective mismatch repair in ctDNA from metastatic prostate cancer. Clin. Cancer Res. 2020;26:1114–1125. doi: 10.1158/1078-0432.CCR-19-1623. [DOI] [PubMed] [Google Scholar]
- 58.Gundem G., Van Loo P., Kremeyer B., Alexandrov L.B., Tubio J.M.C., Papaemmanuil E., Brewer D.S., Kallio H.M.L., Högnäs G., Annala M., et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKay R.R., Gold T., Zarif J.C., Chowdhury-Paulino I.M., Friedant A., Gerke T., Grant M., Hawthorne K., Heath E., Huang F.W., et al. Tackling diversity in prostate cancer clinical trials: a report from the diversity working group of the ironman registry. JCO Glob. Oncol. 2021;7:495–505. doi: 10.1200/GO.20.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Painter C.A., Jain E., Tomson B.N., Dunphy M., Stoddard R.E., Thomas B.S., Damon A.L., Shah S., Kim D., Gómez Tejeda Zañudo J., et al. The Angiosarcoma Project: enabling genomic and clinical discoveries in a rare cancer through patient-partnered research. Nat. Med. 2020;26:181–187. doi: 10.1038/s41591-019-0749-z. [DOI] [PubMed] [Google Scholar]
- 61.Count Me In. Count Me In. (2022). https://joincountmein.org/.
- 62.Wagle N., Painter C., Krevalin M., Oh C., Anderka K., Larkin K., Lennon N., Dillon D., Frank E., Winer E.P., et al. The Metastatic Breast Cancer Project: a national direct-to-patient initiative to accelerate genomics research. J. Clin. Oncol. 2016;34:LBA1519. doi: 10.1200/JCO.2016.34.18_suppl.LBA1519. [DOI] [Google Scholar]
- 63.Ward E., Jemal A., Cokkinides V., Singh G.K., Cardinez C., Ghafoor A., Thun M. Cancer disparities by race/ethnicity and socioeconomic status. CA. Cancer J. Clin. 2004;54:78–93. doi: 10.3322/canjclin.54.2.78. [DOI] [PubMed] [Google Scholar]
- 64.Rebbeck T.R. Prostate cancer disparities by race and ethnicity: from nucleotide to neighborhood. Cold Spring Harb. Perspect. Med. 2018;8:a030387. doi: 10.1101/cshperspect.a030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bailey Z.D., Krieger N., Agénor M., Graves J., Linos N., Bassett M.T. Structural racism and health inequities in the USA: evidence and interventions. Lancet. 2017;389:1453–1463. doi: 10.1016/S0140-6736(17)30569-X. [DOI] [PubMed] [Google Scholar]
- 66.USDA ERS - Documentation. (2020). https://www.ers.usda.gov/data-products/rural-urban-commuting-area-codes/documentation/.
- 67.Kind A.J.H., Buckingham W.R. Making neighborhood-disadvantage metrics accessible — the neighborhood atlas. N. Engl. J. Med. 2018;378:2456–2458. doi: 10.1056/NEJMp1802313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sondka Z., Bamford S., Cole C.G., Ward S.A., Dunham I., Forbes S.A. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018;18:696–705. doi: 10.1038/s41568-018-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Python Software Foundation. Vol. 3. Python Software Foundation; 2022. (Python). [Google Scholar]
- 71.R Core Team . R Foundation for Statistical Computing; 2021. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 72.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saunders C.T., Wong W.S.W., Swamy S., Becq J., Murray L.J., Cheetham R.K. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 76.Ramos A.H., Lichtenstein L., Gupta M., Lawrence M.S., Pugh T.J., Saksena G., Meyerson M., Getz G. Oncotator: cancer variant annotation tool. Hum. Mutat. 2015;36:E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P.W., Onofrio R.C., Winckler W., Weir B.A., et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shen R., Seshan V.E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taylor-Weiner A., Stewart C., Giordano T., Miller M., Rosenberg M., Macbeth A., Lennon N., Rheinbay E., Landau D.-A., Wu C.J., Getz G. DeTiN: overcoming tumor-in-normal contamination. Nat. Methods. 2018;15:531–534. doi: 10.1038/s41592-018-0036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cibulskis K., McKenna A., Fennell T., Banks E., DePristo M., Getz G. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poplin R., Chang P.-C., Alexander D., Schwartz S., Colthurst T., Ku A., Newburger D., Dijamco J., Nguyen N., Afshar P.T., et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018;36:983–987. doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 83.Tesseract OCR. 2022. https://github.com/tesseract-ocr/tesseract
- 84.Geocoder - U.S. Census Bureau. (2022). https://geocoding.geo.census.gov/.
- 85.FilterByOrientationBias GATK. 2019. https://gatk.broadinstitute.org/hc/en-us/articles/360037060232-FilterByOrientationBias-EXPERIMENTAL-
- 86.CrosscheckFingerprints (Picard) – GATK. 2021. https://gatk.broadinstitute.org/hc/en-us/articles/360037594711-CrosscheckFingerprints-Picard-
- 87.Prentice L.M., Miller R.R., Knaggs J., Mazloomian A., Aguirre Hernandez R., Franchini P., Parsa K., Tessier-Cloutier B., Lapuk A., Huntsman D., et al. Formalin fixation increases deamination mutation signature but should not lead to false positive mutations in clinical practice. PLoS One. 2018;13:e0196434. doi: 10.1371/journal.pone.0196434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.AlDubayan S.H., Conway J.R., Camp S.Y., Witkowski L., Kofman E., Reardon B., Han S., Moore N., Elmarakeby H., Salari K., et al. Detection of pathogenic variants with germline genetic testing using deep learning vs standard methods in patients with prostate cancer and melanoma. JAMA. 2020;324:1957–1969. doi: 10.1001/jama.2020.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang K.-L., Mashl R.J., Wu Y., Ritter D.I., Wang J., Oh C., Paczkowska M., Reynolds S., Wyczalkowski M.A., Oak N., et al. Pathogenic germline variants in 10, 389 adult cancers. Cell. 2018;173:355–370.e14. doi: 10.1016/j.cell.2018.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rahman N. Realizing the promise of cancer predisposition genes. Nature. 2014;505:302–308. doi: 10.1038/nature12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stachler M.D., Taylor-Weiner A., Peng S., McKenna A., Agoston A.T., Odze R.D., Davison J.M., Nason K.S., Loda M., Leshchiner I., et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat. Genet. 2015;47:1047–1055. doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MPCproject releases deidentified clinical, patient-reported and research-grade genomic data into public repositories, such as cBioPortal: mpcproject_broad_2021 (https://www.cbioportal.org/study/summary?id=mpcproject_broad_2021), the Genomic Data Commons: CMI-MPC (https://portal.gdc.cancer.gov/projects/CMI-MPC), and dbGaP: phs001939.v3.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001939.v3.p1) at regular intervals and prepublication. Data is processed and formatted as required by each repository’s guidelines. All patient identifiers are stripped prior to data deposition to protect patient privacy. On the MPCproject data release webpage (https://mpcproject.org/data-release), patients can access project data, additional information about the data, a list of common terms used in research, methods used to generate the data, and an e-mail address for any additional data-related questions. All other data used in this paper are from publicly available resources. The code used to generate most main figures, central analyses, and supplementary figures can be found at can be found at https://github.com/vanallenlab/mpcproject-paper, except for figures and analyses requiring sample-level germline data. An unchanging version of the code at time of publication is also available at Zenodo: https://doi.org/10.5281/zenodo.6816267. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.