Abstract
The bacterial fatty acid pathway is essential for membrane synthesis and a range of other metabolic and cellular functions. The β-ketoacyl-ACP synthases carry out the initial elongation reaction of this pathway, utilizing acetyl-CoA as a primer to elongate malonyl-ACP by two carbons, and subsequent elongation of the fatty acyl-ACP substrate by two carbons. Here we describe the structures of the β-ketoacyl-ACP synthase I from Brucella melitensis in complex with platencin, 7-hydroxycoumarin, and (5-thiophen-2-ylisoxazol-3-yl)methanol. The enzyme is a dimer and based on structural and sequence conservation, harbors the same active site configuration as other β-ketoacyl-ACP synthases. The platencin binding site overlaps with the fatty acyl compound supplied by ACP, while 7-hydroxyl-coumarin and (5-thiophen-2-ylisoxazol-3-yl)methanol bind at the secondary fatty acyl binding site. These high-resolution structures, ranging between 1.25 and 1.70 Å resolution, provide a basis for in silico inhibitor screening and optimization, and can aid in rational drug design by revealing the high-resolution binding interfaces of molecules at the malonyl-ACP and acyl-ACP active sites.
Keywords: drug design, fatty acid synthesis, structure
1 |. INTRODUCTION
Fatty acid synthesis is an essential and ubiquitous process required for membrane synthesis, cell signaling, energy storage, and a wide range of other cellular functions. The reaction pathway required for fatty acid synthesis is similar in both prokaryotes and eukaryotes; however, key differences exist in the enzyme architecture that catalyze these reactions. Mammals and fungi utilize the type I fatty acid synthase (FAS) complex, a multifunctional enzyme which catalyzes all the reactions required for fatty acid elongation, while bacteria utilize a type II FAS system, where the enzymes are discrete and monofunctional.1–4 Differences between bacterial and eukaryotic FAS enzymes are becoming thoroughly described through the impressive library of high-resolution structures,5–11 and represent attractive targets for the development of novel antibacterial inhibitors. Antibiotics targeting the type II FAS pathway are currently in use, and the mechanisms of inhibition have been resolved using structural methods.12-16
The canonical bacterial type II FAS pathway, based on Escherichia coli fatty acid biosynthesis, utilize three β-ketoacyl-acyl carrier protein (ACP) synthases, KASI, KASII, and KASIII (also known as FabB, FabF, and FabH, respectively). β-Ketoacyl-ACP synthase III (FabH) carries out the initial elongation reaction of the type II FAS pathway, typically utilizing acetyl-CoA as a primer to elongate malonyl-ACP by two carbons via Claisen condensation.12,13 Subsequent elongation of the acyl-ACP substrate by two carbons is performed by either β-ketoacyl-ACP synthase I (FabB) or II (FabF), by a similar catalytic mechanism. The β-ketoacyl-ACP synthases I and II contain a His-His-Cys catalytic triad at the base of their substrate binding pockets, while the β-ketoacyl-ACP synthase III contains a His-Asn-Cys catalytic triad.17–19 In E. coli, β-ketoacyl-ACP synthase I and II mediate the elongation of both saturated and unsaturated fatty acids.
Inhibitors thiolactomycin and cerulenin have been shown to bind the active site of β-ketoacyl-ACP synthase I, blocking the malonyl-ACP and acyl-ACP substrate binding pockets, respectively.12,13 The β-ketoacyl-ACP synthase inhibitors platencin and platensimycin have demonstrated antimicrobial activity against Streptococcus pneumoniae, extensively drug resistant Mycobacterium tuberculosis, and methicillin-resistant Staphylococcus aureus.20–22 Based on the structures of E. coli β-ketoacyl-ACP synthase II in complex with platencin (PDB ID: 3HO2) and platensimycin (PDB ID: 2GFX), these inhibitors block the malonyl subsite of β-ketoacyl-ACP synthases to prevent elongation of the fatty acyl-ACPs. Using these examples as a template for successful inhibition, new compounds can be screened to target β-ketoacyl-ACP synthases.
Brucella spp. are the causative agents of brucellosis, a common zoonotic disease that has a nearly worldwide distribution, with particularly high incidence in the Middle East and central Asia.23,24 Those working closely with livestock and animal products, such as milk and meat, are the most vulnerable.25 Three species, Brucella melitensis, B. abortus, and B. suis, are classified as select agents, pathogens that present a substantial threat to human health, animal health, and animal products by both the Centers of Disease Control and Prevention and US Department of Agriculture (https://www.selectagents.gov/SelectAgentsandToxinsList.html.). B. melitensis is the most likely species to cause zoonotic infections, and also has the most acute symptoms.26,27 The vaccine for B. melitensis, while able to limit incidence of infection, has not been sufficiently attenuated for human use.25 The most effective treatment has been a combination of doxycycline and streptomycin,28,29 although the parenteral route of administration for streptomycin complicates this regimen. Given the widespread distribution, lack of human vaccine, and need for multidrug treatment, the discovery of new treatment options is crucial.
The structural determination of the B. melitensis β-ketoacyl-ACP synthase I and complexes with bound molecules at the active site will provide new information on an essential protein of a select agent, and a platform for in silico drug design. Here we describe the structures of B. melitensis β-ketoacyl-ACP synthase I in complex with the compounds platencin, 7-hydroxycoumarin, and (5-thiophen-2-ylisoxazol-3-yl)methanol. Two of these compounds, platencin and (5-thiophen-2-ylisoxazol-3-yl)methanol, have been investigated for antibiotic properties with other bacterial targets.20,30,31 The third compound, 7-hydroxycoumarin, is known to interact with a bacterial reductase.32 These structures will aid in rational drug design by revealing the high-resolution binding interfaces of molecules at the malonyl-ACP and acyl-ACP active sites.
2 |. MATERIALS AND METHODS
2.1 |. Cloning, expression, and purification of β-ketoacyl-ACP synthase I
The gene for BrabA.00113.a was amplified from genomic DNA and cloned into the expression vector pBG1861 using ligation-independent cloning.33 The expression vector provides a 3C-cleavable N-terminal His6-tag (SSGCID target ID BrabA.00113.a, SSGCID construct ID BrabA.00113.a.A1, SSGCID batch BrabA.00113.a.A1. PW25441). BrabA.00113.a was expressed in E. coli Rosetta BL21 (DE3)R3 following standard SSGCID protocols, as described previously.34 Purification was performed using Ni-NTA affinity and size exclusion chromatography following standard SSCID protocols.35 The His-tag was not cleaved. The purified protein was concentrated to 22.94 mg/mL in its final buffer (25 mM HEPES pH 7.0, 500 mM NaCl, 5% glycerol, 2 mM DTT, 0.025% NaN3), flash frozen in liquid nitrogen and stored at −80°C.
2.2 |. Crystallization, data collection, and structure determination
Crystallization set ups were done with apo protein at 22.94 mg/mL, using 96-well XJR crystallization trays (Rigaku Reagents) with 0.4 μL protein mixed with 0.4 μL reservoir, equilibrating against 80 μL reservoir solution. Crystallization conditions were searched for with sparse matrix screens JCSG+ (Rigaku Reagents), CrystalScreen HT, Index HT (Hampton Research), and PACT (Molecular Dimensions). Crystallization trays were incubated at 285 K. Crystals were observed in all trays. A crystal from condition PACT H12 (20% PEG 3350, 100 mM BisTris propane pH 8.5, 200 mM sodium malonate) was cryoprotected with a solution of reservoir with 25% ethylene glycerol, and vitrified in liquid nitrogen. Diffraction data were collected in-house on a Rigaku FR-E+ SuperBright rotating anode equipped with Rigaku VariMax optics and a Saturn 944+ detector, using CuKα X-rays. All data sets were reduced with the XDS package.36 Diffraction data are summarized in Table 1.
TABLE 1.
Crystallization statistics for the structures determined in this study
| Crystal parameters Sample | apo | (1-methyl-1h-indazol-3-yl) methanol | 7-hydroxycoumarin | Platencin |
|---|---|---|---|---|
| Space group | C2 | C2 | C2 | C2 |
| Cell dimensions | 77.48, 83.27, 72.87 | 78.07, 83.62, 73.64 | 78.56, 83.25, 73.81 | 78.16, 84.48, 74.02 |
| a = b = c (Å), α = β = γ | 90, 120.97, 90 | 90, 121.37, 90 | 90, 121.20, 90 | 90, 120.81, 90 |
| Data set | ||||
| X-ray Source | Rigaku F-RE+ SuperBright | ALS beamline 5.0.1 | APS beamline 21-ID-G | Rigaku F-RE+ SuperBright |
| Wavelength (Å) | 1.5418 | 0.9774 | 0.97856 | 1.5418 |
| Resolution (Å) | 50–1.60 (1.64–1.60) | 41–1.25 (1.28–1.25) | 39–1.25 (1.28–1.25) | 50–1.70 (1.74–1.70) |
| R merge | 0.039 (0.327) | 0.091 (0.404) | 0.061 (0.350) | 0.033 (0.187) |
| I/sigma (I) | 34.2 (4.9) | 12.1 (4.0) | 15.3 (3.6) | 47.8 (7.5) |
| Completeness (%) | 98.1 (91.8) | 99.7 (99.1) | 97.8 (93.0) | 98.5 (96.8) |
| # reflections, unique | 51 414 (3537) | 111 226 (8269) | 109 578 (7661) | 44 869 (3278) |
| Multiplicity | 8.1 (4.8) | 5.4 (5.2) | 4.5 (3.6) | 11.2 (4.3) |
| Refinement statistics | ||||
| R work | 0.129 | 0.113 | 0.122 | 0.142 |
| R free | 0.158 | 0.133 | 0.134 | 0.169 |
| RMSD bond lengths (Å) | 0.015 | 0.018 | 0.008 | 0.015 |
| RMSD bond angles (°) | 1.47 | 1.69 | 1.32 | 1.40 |
| Ramachandran: | - | |||
| favored (%) | 96.79 | 96.79 | 97.28 | 96.52 |
| allowed (%) | 2.72 | 2.72 | 3.21 | 4.11 |
| Outliers (%) | 0.49 | 0.49 | 0.49 | 0.63 |
| Molprobity clash score | 1.14 | 3.00 | 3.65 | 1.32 |
| Molprobity score | 1.02 | 1.29 | 1.29 | 1.09 |
| B factors | ||||
| overall | 12.9 | 10.7 | 10.3 | 16.9 |
| protein | 10.9 | 8.7 | 8.4 | 15.3 |
| ligands | 8.75 | 10.4 | 18.4 | 24.2 |
| solvent | 25.2 | 24.3 | 21.1 | 28.2 |
| PDB code | 3lrf | 3mqd | 3u0f | 4jv3 |
The structure of apo BrabA.00113.a was solved using the program PHASER37 from the CCP4 package,38 with PDB entry 1DD8 modified with the CCP4 program CHAINSAW39 as the search model. The initial model was extended with ARP/wARP.40 Manual model building was performed using Coot,41 and the structure was refined in reciprocal space with Refmac5.42 The coordinates and structure factors of the apo structure were deposited in the PDB with accession code 3LRF.
For fragment studies, crystals were grown in condition PACT H12. 2.5 μL of 50 mM compound stock solution in methanol were dried in the well of a crystallization tray. The dried fragment was overlaid with 2.5 μL of a soaking solution (100 mM MES pH 6.5, 30% PEG 3350, 10% glycerol), and crystals were transferred into the soaking drop. Crystals were incubated overnight, and vitrified by plunging them into liquid nitrogen. Diffraction data were collected at ALS beamline 5.0.1 on an ADSC Q310r CCD detector (3MQD, 3U0E), or at APS beamline 21-ID-G on a Rayonix MX-300 CCD detector (3U0F).
For the platencin co-crystal structure, BrabA.00113.a was crystallized in the presence of 2.5 mM platencin (Bioaustralis) in PACT condition H2 (20% PEG 3350, 100 mM BisTris propane pH 8.5, 200 mM sodium bromide). The crystal was cryoprotected with 20% ethylene glycol, and vitrified in liquid nitrogen. All ligand-bound structures were refined as described for the non-ligand bound structure.
3 |. RESULTS
3.1 |. Structure of the β-ketoacyl-ACP synthase I from Brucella melitensis
Bacterial β-ketoacyl-ACP synthases catalyze the elongation reactions of the type II FAS pathway. These enzymes perform essential functions and exhibit structural differences to eukaryote β-ketoacyl-ACP synthases, making them promising targets for antibacterial drug design. To facilitate the design of new inhibitory molecules, we set out to determine the high-resolution crystallographic structures of a β-ketoacyl-ACP synthase from B. melitensis in the absence and presence of platencin and fragment screen molecules.43 The β-ketoacyl-ACP synthase from B. melitensis was expressed recombinantly, and purified by affinity and size exclusion chromatography. Crystals belonging to the C2 space group with unit cell parameters a = 77.48 Å, b = 83.27 Å, c = 72.87 Å, β = 120.97° diffracted to 1.6 Å resolution. Molecular replacement using the E. coli structure of β-ketoacyl-ACP synthase I, PDB entry 1DD8 as the search model was used to solve the structure, which together with model building and refinement, resulted in a high-quality structural model with Rwork and Rfree values of 0.129 and 0.158, respectively. A summary of the crystallographic and refinement statistics is presented in Table 1.
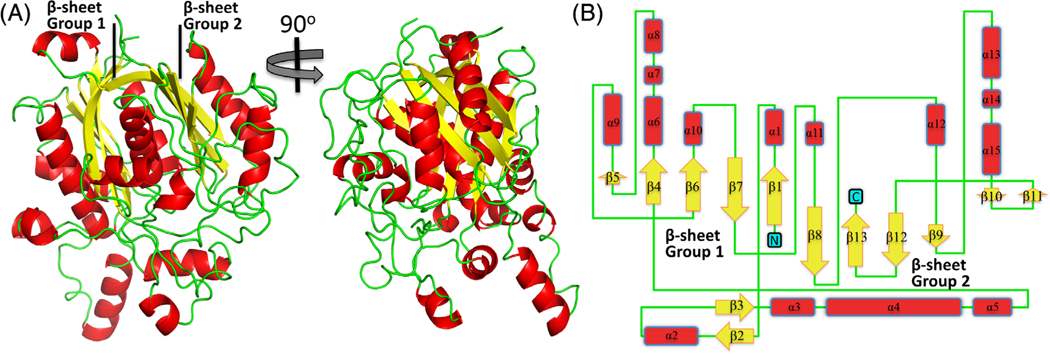
The β-ketoacyl-ACP synthase protomer is comprised of 15 α-helices, 13 β-strands, and two mixed β-sheets (Figures 1 and 2). This typical thiolase-like fold arrangement is conserved in β-ketoacyl-ACP synthases. A DALI search44 revealed three similar structures deposited to the PDB, all of which were β-ketoacyl-ACP synthase I molecules (FabB): Vibrio cholera (r.m.s.d 0.8 Å, sequence identity 64% over 403 aligned residues, PDB ID: 4XOX), E. coli (r.m.s.d 0.8 Å, sequence identity 61% over 401 aligned residues, PDB ID: 1G5X), and Yersinia pestis (r.m.s.d 0.8 Å, sequence identity 56% over 404 aligned residues, PDB ID: 3OYT). A clear separation was observed for the next class of structurally related β-ketoacyl-ACP synthases, all of which were β-ketoacyl-ACP synthase II molecules (FabF): Burkholderia vietnamiensis (r.m.s.d 1.4 Å, sequence identity 39% over 401 aligned residues, PDB ID: 4DDO), Streptococcus pneumoniae (r.m.s.d 1.4 Å sequence identity 38% over 396 aligned residues, PDB ID: 2RJT), and B. melitensis (r.m.s.d 1.5 Å sequence identity 41%, over 400 aligned residues PDB ID: 3KZU). No reasonable degree of sequence or structural identity to β-ketoacyl-ACP synthase III molecules (FabH) was observed. These results suggest that the B. melitensis β-ketoacyl-ACP synthase reported here is most likely to be a β-ketoacyl-ACP synthase I.
FIGURE 1.
The tertiary structure of the β-ketoacyl-ACP synthase I from Brucella melitensis. A, A cartoon representation of β-ketoacyl-ACP synthase I showing the two β-sheets. B, Topology arrangement of secondary structural elements of β-ketoacyl-ACP synthase I [Color figure can be viewed at wileyonlinelibrary.com]
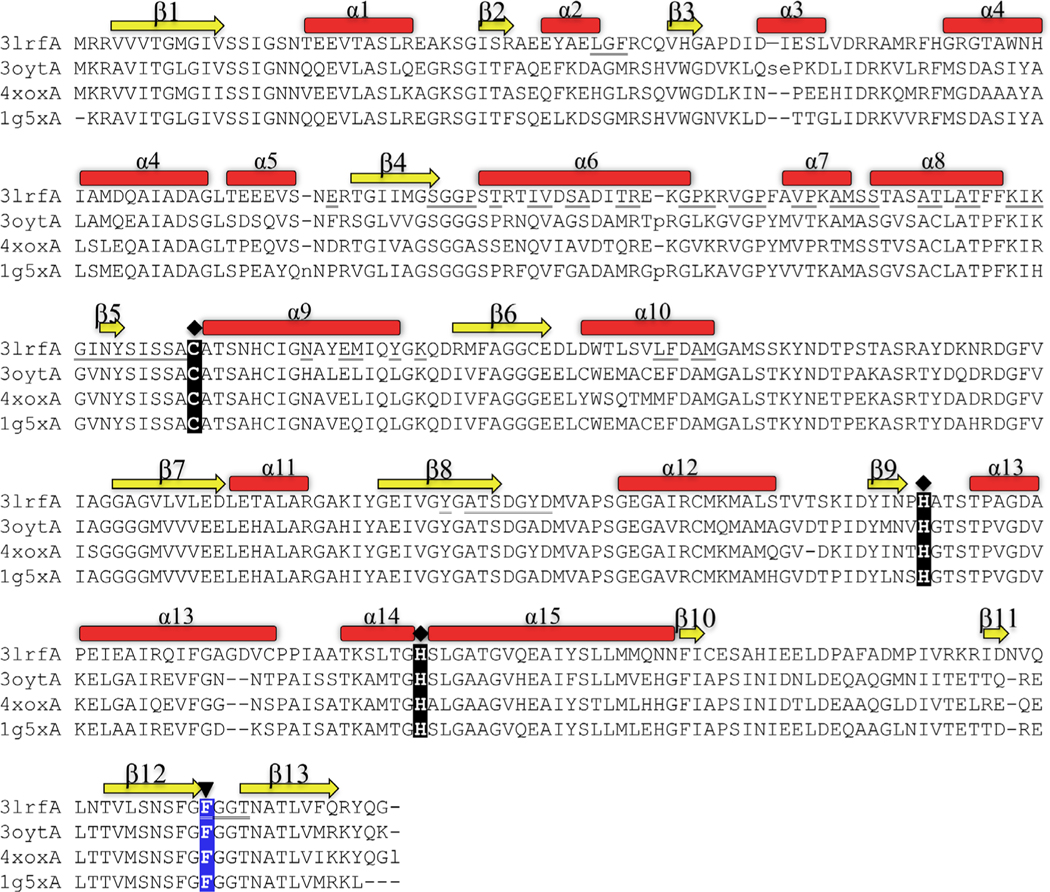
FIGURE 2.
Structural alignment of the most closely related structures in the PDB. 3lrfA is the β-ketoacyl-ACP synthase I from Brucella melitensis in this study. 3oytA is the β-ketoacyl-ACP synthase I from Yersinia pestis (r.m.s.d 0.8 Å, sequence identity 56%). 4xoxA is the β-ketoacyl-ACP synthase I from Vibrio cholera (r.m.s.d 0.8 Å, sequence identity 64%) and 1g5x is the β-ketoacyl-ACP synthase I from Escherichia coli (r.m.s.d 0.8 Å, sequence identity 61%. The catalytic active site residues (Cys, His, His) are highlighted with a diamond-shaped symbol, and the highly conserved Phe residue at the end of β12 (highlighted with an upside-down triangle) mediates substrate specificity and entry of an acyl chain to the binding cavity. β-Sheets are displayed yellow; α-helices are displayed red. Underlined residues indicate residues at the dimer interface (see also Figure 3B) [Color figure can be viewed at wileyonlinelibrary.com]
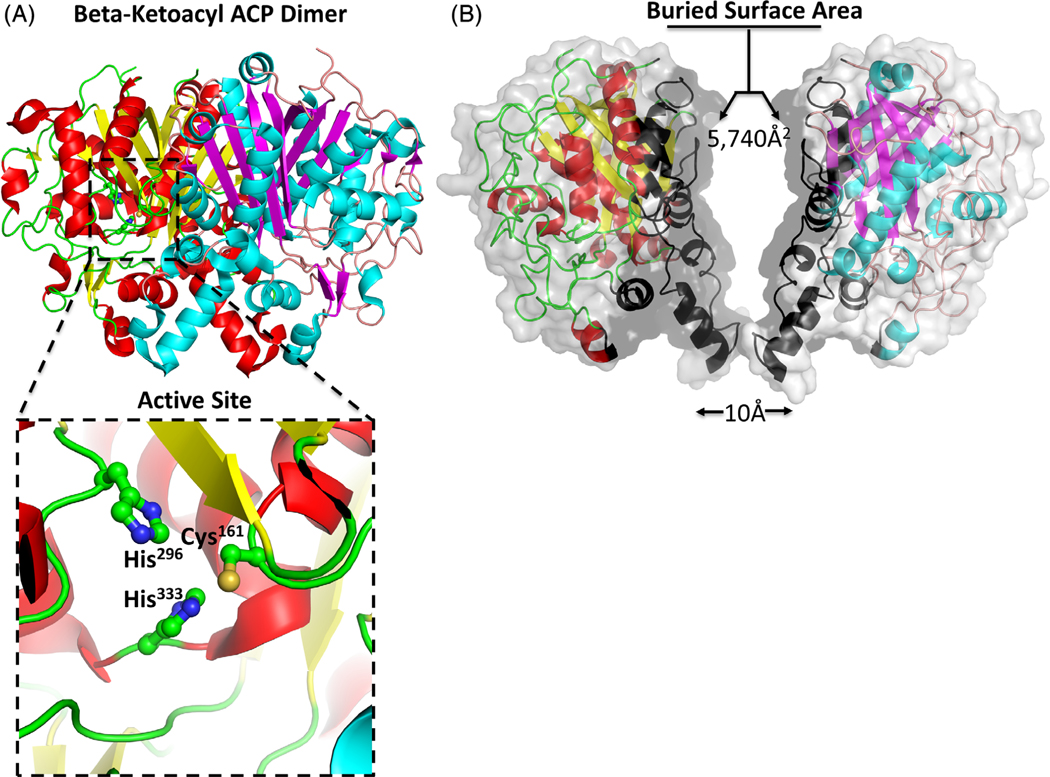
While the crystal asymmetric unit contained a single protomer, an extensive surface area of 5740Å2 was buried between two symmetry related protomers, suggesting the enzyme is likely to exist as a dimer (Figure 3). Analysis of the protein interfaces using PISA45 indicates that the dimer is stable in solution. Moreover, the same biological assemblies were formed from closely related homologues identified in the DALI search described above. The dimer interface for the β-ketoacyl-ACP synthase in this structure is mediated by 30 hydrogen bonds and 10 salt bridges (see Table 2 for a full description of the interactions). The majority of the dimer interactions are located on secondary structure elements, with α-helices 6 to 10 and β-strand 8 involved in dimer formation. Other interactions are localized to the loop regions connecting α2-β3 (LGF44), α5-β4 (E96), β4-α6 (GGP107), α6-α7 (PK125, VGP129), α7-α8 (S137), α8-β5 (KIKG152), β5-α9 (SISSA160), α9-β6 (K179), β8-α12 (DGYD266), and β12-β13 (FGG395) (Figure 2).
FIGURE 3.
The β-ketoacyl-ACP synthase I from Brucella melitensis forms a dimer. A, The two monomers are shown in cartoon form, with one monomer colored red (α-helices) and yellow (β-sheets), and the second monomer colored blue (α-helices) and purple (β-sheets). The insert shows a zoom of the conserved active site residues Cys161, His296, His333. B, The two monomers bury a surface area 5740 Å2. The contacting interface residues are shaded black, and each the monomers have been translated 10 Å2 to illustrate the extent of the buried interface [Color figure can be viewed at wileyonlinelibrary.com]
TABLE 2.
Hydrogen bond and salt bridge interactions at the dimer interface
| Hydrogen bond interactions | |||
|---|---|---|---|
| 1 | A:ARG 120[ NH1] | 2.72 | A:ASP 117[ OD2] |
| 2 | A:ARG 120[ NH2] | 2.97 | A:ASP 117[ OD1] |
| 3 | A:SER 137[ N ] | 2.74 | A:GLY 105[ O ] |
| 4 | A:SER 137[ OG ] | 2.89 | A:SER 137[ OG ] |
| 5 | A:ASN 154[ ND2] | 2.89 | A:GLY 394[ O ] |
| 6 | A:ASN 154[ ND2] | 3.22 | A:SER 262[ OG ] |
| 7 | A:SER 156[ N ] | 2.93 | A:SER 158[ O ] |
| 8 | A:SER 158[ N ] | 2.84 | A:SER 156[ O ] |
| 9 | A:SER 158[ OG ] | 3.55 | A:SER 158[ OG ] |
| 10 | A:ALA 160[ N ] | 3.54 | A:SER 138[ OG ] |
| 11 | A:LYS 179[ NZ ] | 2.66 | A:GLU 173[ OE2] |
| 12 | A:SER 262[ N ] | 3.16 | A:LYS 151[ O ] |
| 13 | A:SER 262[ N ] | 2.88 | A:GLY 152[ O ] |
| 14 | A:SER 262[ OG ] | 2.65 | A:ILE 150[ O ] |
| 15 | A:ARG 277[ NH1] | 2.74 | A:LYS 151[ O ] |
| 16 | A:THR 396[ OG1] | 2.63 | A:ASN 154[ O ] |
| 17 | A:ASP 117[ OD2] | 2.72 | A:ARG 120[ NH1] |
| 18 | A:ASP 117[ OD1] | 2.97 | A:ARG 120[ NH2] |
| 19 | A:GLY 105[ O ] | 2.74 | A:SER 137[ N ] |
| 20 | A:SER 262[ OG ] | 3.22 | A:ASN 154[ ND2] |
| 21 | A:GLY 394[ O ] | 2.89 | A:ASN 154[ ND2] |
| 22 | A:SER 158[ O ] | 2.93 | A:SER 156[ N ] |
| 23 | A:SER 156[ O ] | 2.84 | A:SER 158[ N ] |
| 24 | A:SER 138[ OG ] | 3.54 | A:ALA 160[ N ] |
| 25 | A:GLU 173[ OE2] | 2.66 | A:LYS 179[ NZ ] |
| 26 | A:GLY 152[ O ] | 2.88 | A:SER 262[ N ] |
| 27 | A:LYS 151[ O ] | 3.16 | A:SER 262[ N ] |
| 28 | A:ILE 150[ O ] | 2.65 | A:SER 262[ OG ] |
| 29 | A:LYS 151[ O ] | 2.74 | A:ARG 277[ NH1] |
| 30 | A:ASN 154[ O ] | 2.63 | A:THR 396[ OG1] |
| Salt bridges | |||
| 1 | A:ARG 120[ NH1] | 3.62 | A:ASP 117[ OD1] |
| 2 | A:ARG 120[ NH1] | 2.72 | A:ASP 117[ OD2] |
| 3 | A:ARG 120[ NH2] | 2.97 | A:ASP 117[ OD1] |
| 4 | A:ARG 120[ NH2] | 3.50 | A:ASP 117[ OD2] |
| 5 | A:LYS 179[ NZ ] | 2.66 | A:GLU 173[ OE2] |
| 6 | A:ASP 117[ OD1] | 3.62 | A:ARG 120[ NH1] |
| 7 | A:ASP 117[ OD2] | 2.72 | A:ARG 120[ NH1] |
| 8 | A:ASP 117[ OD1] | 2.97 | A:ARG 120[ NH2] |
| 9 | A:ASP 117[ OD2] | 3.50 | A:ARG 120[ NH2] |
| 10 | A:GLU 173[ OE2] | 2.66 | A:LYS 179[ NZ ] |
3.2 |. Active site and bound inhibitors
The active site of β-ketoacyl-ACP synthases is well established.5–8,11,12,46 A Cys residue (Cys161 in this structure) positioned at the N-terminus of α9 is activated by the strong helix dipole moment, and accepts the acyl-group from acyl-ACP. Upon binding malonyl-ACP, two His residues positioned at the end of β9 and α14 (His296 and His333 in this structure) initiate a second nucleophilic attack and stabilize formation of an enol intermediate. A highly conserved Phe residue at the end of β12 (Phe393 in this structure) is involved in substrate specificity and guiding the entry of an acyl chain to the binding cavity. Overall, the structure of the β-ketoacyl-ACP synthase in this study supports the highly conserved and well-described active site (Figures 2 and 3).
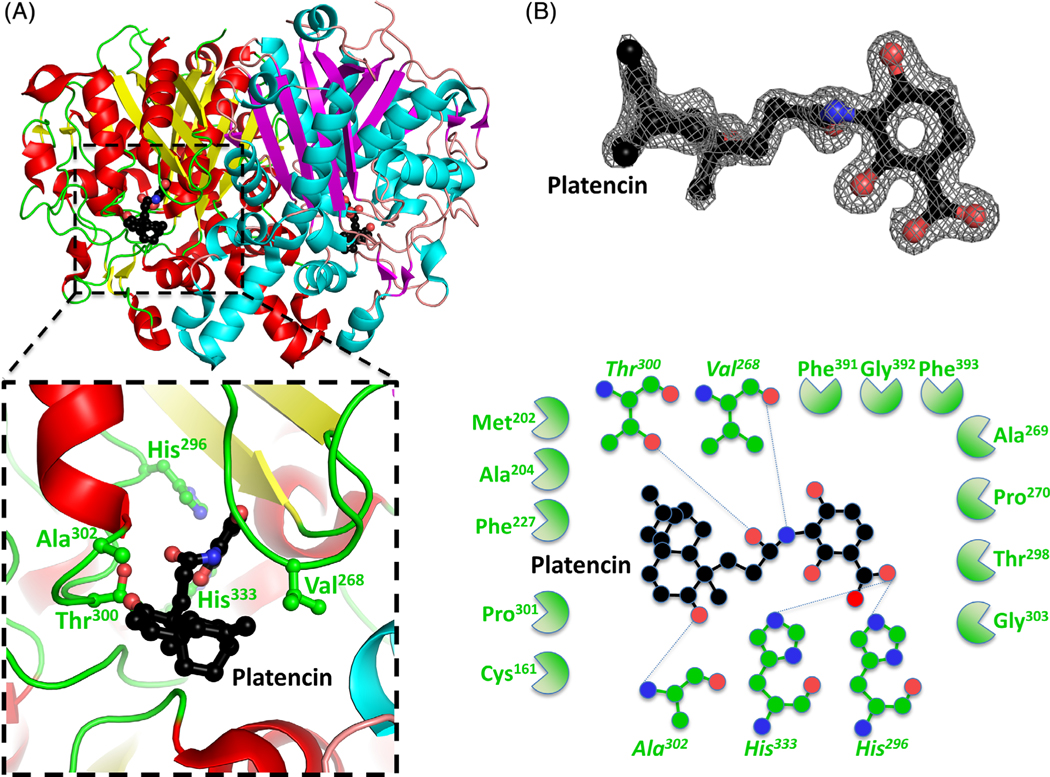
To understand the binding mechanism of inhibitors and fragment screen molecules at the active site, we crystallized the enzyme in the presence of both platencin, and fragment screens. Platencin is a potent broad-spectrum inhibitor, with activity against β-ketoacyl-ACP synthase III and β-ketoacyl-ACP synthase II,20,21 however, structural characterization of platencin bound to a β-ketoacyl-ACP synthase I is yet to be reported. Crystals formed in the presence of platencin displayed the same space group as the unbound β-ketoacyl-ACP synthase (Table 1), with well-ordered density for platencin (Figure 4). The binding interface for platencin is 385Å2, and is mediated by five hydrogen bonds and a range of hydrophobic interactions. Key hydrogen bonds include active site residues His296 and His333, and non-catalytic residues Val268, Thr300, and Ala302 (Figure 4).
FIGURE 4.
Structure of the β-ketoacyl-ACP synthase I from Brucella melitensis bound to platencin. A, The two monomers are shown in cartoon form, with one monomer colored red (α-helices) and yellow (β-sheets), and the second monomer colored blue (α-helices) and purple (β-sheets). Platencin is colored black (carbon), red (oxygen), and blue (nitrogen). The insert shows the binding pocket and interface residues. B, The location of platencin was supported by good electron density. The electron density map (2Fo-Fc) is contoured to 2 sigma, and colored gray. The full binding pocket and interacting residues are presented in cartoon form. The blue dashed lines indicate hydrogen bonds. The green shaped amino acids indicate hydrophobic interactions [Color figure can be viewed at wileyonlinelibrary.com]
There are currently two other structures of platencin bound to β-ketoacyl-ACP synthases in the Protein Data Bank; E. coli (PDB ID: 3HO2), and B. vietnamiensis (PDB ID: 4F32). The binding is similar across all three structures, with the E. coli counterpart exhibiting an interface of 423Å2 and four hydrogen bonds mediated by His340 (equivalent to His333), as well as Thr307 (equivalent to Thr300), Ala309 (equivalent to Ala302), and an additional Thr270. Similarly, the B. vietnamiensis binding interface is 402Å2, and mediated by six hydrogen bonds, including the active site residues His313 (equivalent to His296), His349 (equivalent to His333), Thr317 (equivalent to His300), Val319 (equivalent to Ala302), while additional interactions with Thr280, and Gly282 were not observed in our structure.
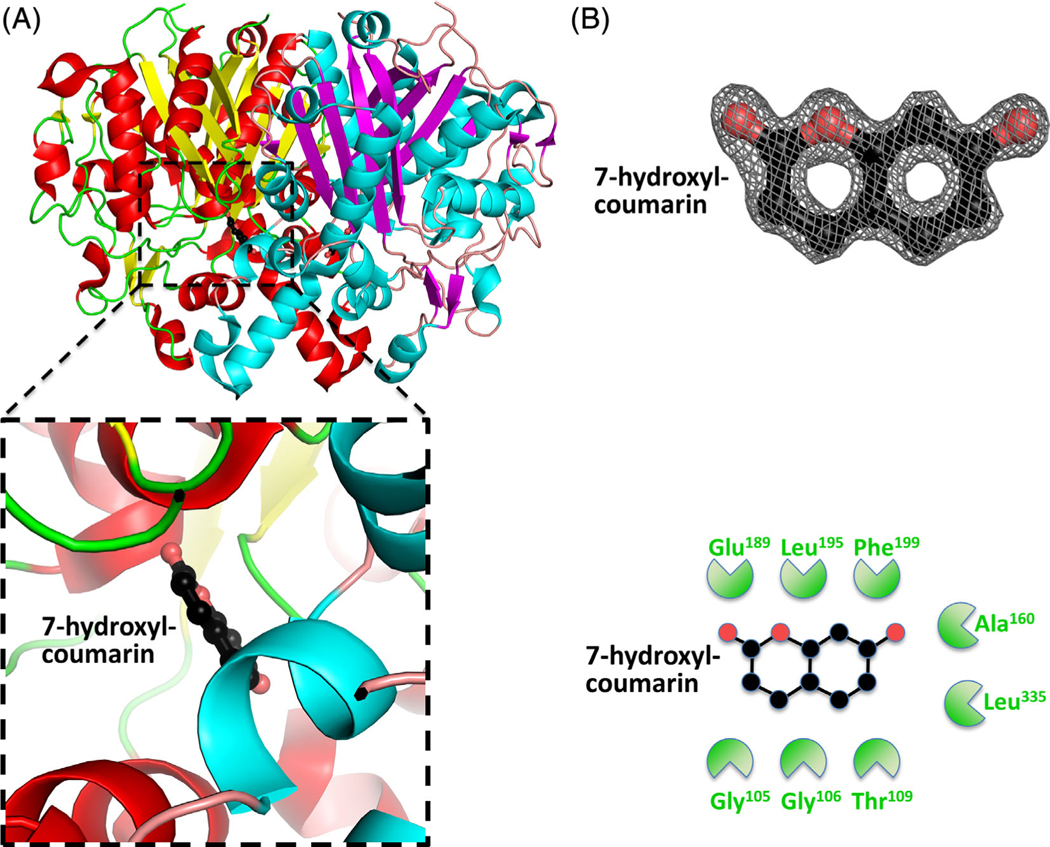
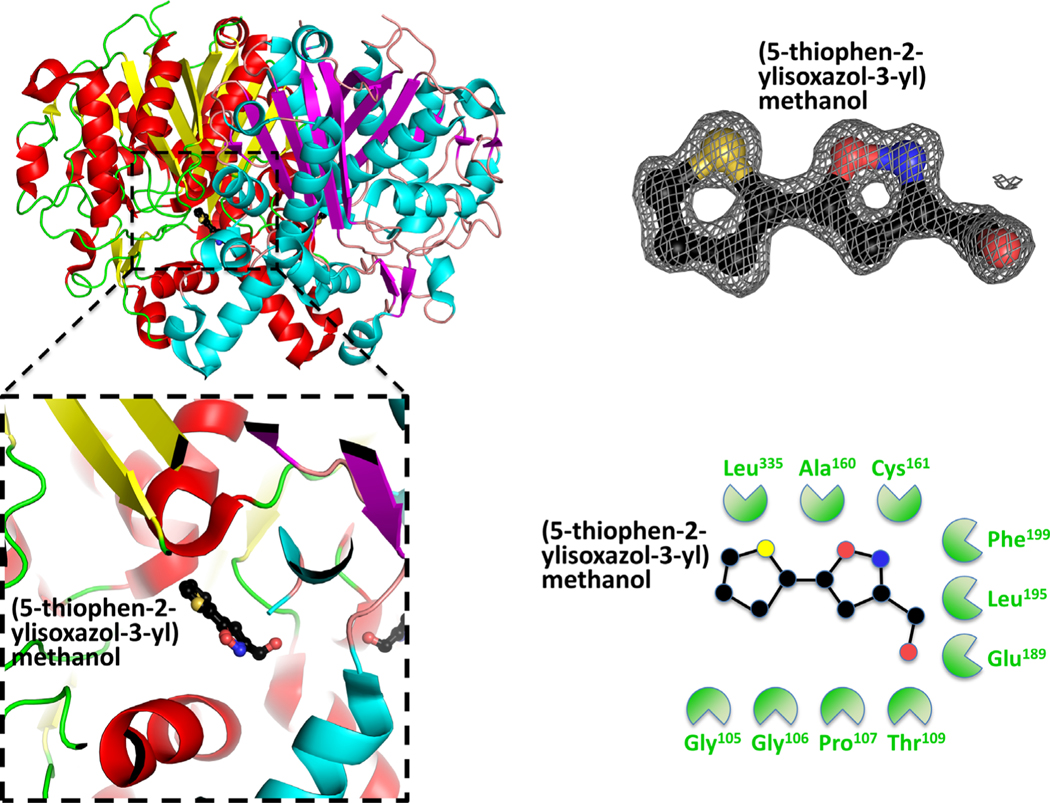
To assist with inhibitor screening and in silico drug design, we also crystallized the β-ketoacyl-ACP synthase in the presence of small molecules from fragment screens. We identified two molecules bound in close proximity to the β-ketoacyl-ACP synthase active site (Table 1). Fragments 7-hydroxyl-coumarin (PDB ID: 3U0F) (Figure 5), and (5-thiophen-2-ylisoxazol-3-yl)methanol (PDB ID: 3MQD) (Figure 6), were bound within >4 Å of the active site Cys161, but at discrete sites from platencin (closest atoms are 5.8 Å apart). The 7-hydroxyl-coumarin bound through hydrophobic interactions involving Gly105, Gly106, Thr109, Ala160, Glu189, Leu195, Phe199, and Leu335. The (5-thiophen-2-ylisoxazol-3-yl)methanol bound through Gly105, Gly106, Pro107, Thr109, Ala160, Cys161, Glu189, Leu195, Phe199, and Leu335.
FIGURE 5.
Structure of the β-ketoacyl-ACP synthase I from Brucella melitensis bound to 7-hydroxyl-coumarin. A, The two monomers are shown in cartoon form, with one monomer colored red (α-helices) and yellow (β-sheets), and the second monomer colored blue (α-helices) and purple (β-sheets). 7-Hydroxyl-coumarin is colored black (carbon), red (oxygen), and blue (nitrogen). The insert shows the binding pocket and interface residues. B, The location of 7-hydroxyl-coumarin was supported by good electron density. The electron density map (2Fo-Fc) is contoured to 2 sigma, and colored gray. The full binding pocket and interacting residues are presented in cartoon form. The green shaped amino acids indicate hydrophobic interactions [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 6.
Structure of the β-ketoacyl-ACP synthase I from Brucella melitensis bound to (5-thiophen-2-ylisoxazol-3-yl)methanol. A, The two monomers are shown in cartoon form, with one monomer colored red (α-helices) and yellow (β-sheets), and the second monomer colored blue (α-helices) and purple (β-sheets). (5-thiophen-2-ylisoxazol-3-yl)methanol is colored black (carbon), red (oxygen) and blue (nitrogen). The insert shows the binding pocket and interface residues. B, The location of (5-thiophen-2-ylisoxazol-3-yl)methanol was supported by good electron density. The electron density map (2Fo-Fc) is contoured to 2 sigma, and colored gray. The full binding pocket and interacting residues are presented in cartoon form. The green shaped amino acids indicate hydrophobic interactions [Color figure can be viewed at wileyonlinelibrary.com]
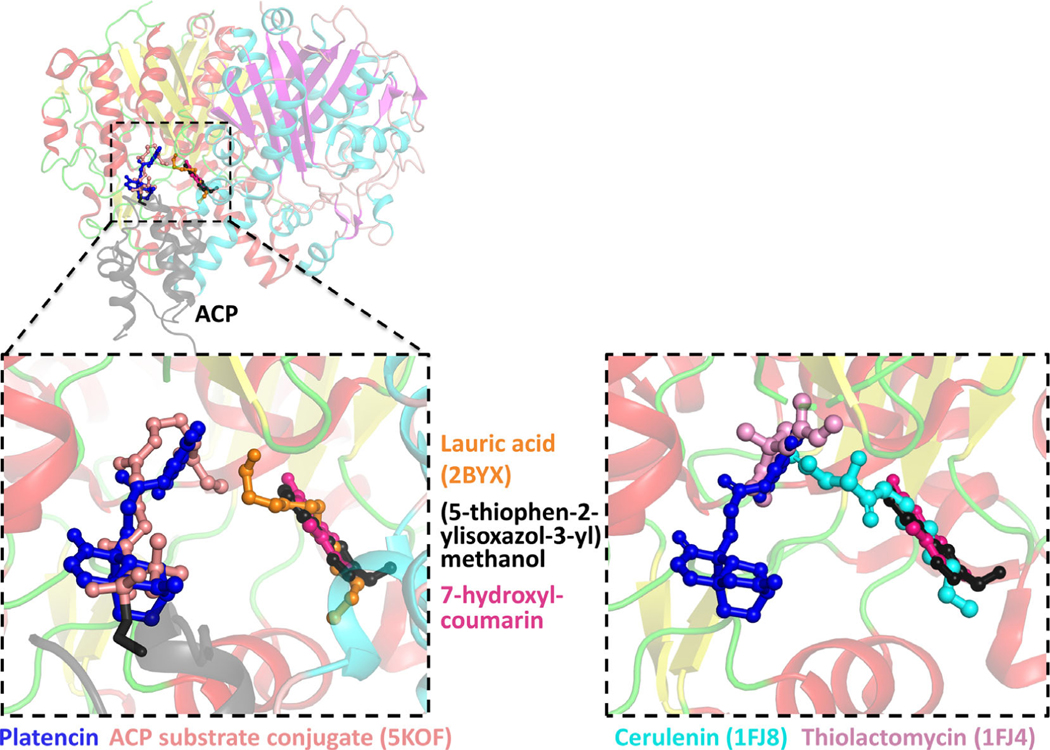
To assess the relative binding site of platencin, 7-hydroxyl-coumarin, and (5-thiophen-2-ylisoxazol-3-yl)methanol to the substrate binding sites, we superimposed various substrate bound structures. These structures included lauric acid bound synthase to E. coli β-ketoacyl-ACP synthase I11 (PDB ID: 2BYX), and an unpublished structure of a β-ketoacyl-ACP synthase I bound to ACP and a reaction mimic (unpublished, PDB ID: 5KOF). Platencin overlaps directly with the ACP linked acyl-compound, while the 7-hydroxyl-coumarin and (5-thiophen-2-ylisoxazol-3-yl)methanol bind at the second fatty acid site (Figure 7). These structural analyses suggest these compounds bind at the two distinct fatty acid substrate binding sites, and represent good platforms for in silico drug design.
FIGURE 7.
Left panel: the structure of the ACP substrate conjugate (5KOF) (colored light pink) and lauric acid (2BYX) (colored orange) bound to β-ketoacyl-ACP synthase I were superimposed onto the structures determined in this study. Platencin (dark blue) bound in the same site as the ACP substrate conjugate, while 7-hydroxyl-coumarin (dark pink) and (5-thiophen-2-ylisoxazol-3-yl)methanol (black) superimposed with lauric acid. Right panel: Cerulenin (cyan) (1FJ8) and thiolactomycin (pink) (1FJ4) also superimposed with the ligands determined in this study
4 |. CONCLUSION
In this article, we describe the structure of β-ketoacyl-ACP synthase from B. melitensis in the presence of platencin, 7-hydroxyl-coumarin and (5-thiophen-2-ylisoxazol-3-yl)methanol bound at two substrate binding sites. Based on the structure, the enzyme is most closely related to β-ketoacyl-ACP synthase I (FabB). The enzyme harbors the same dimer and active site configuration as other β-ketoacyl-ACP synthases. Platencin binding overlaps with the same binding site as the fatty acyl compound supplied by ACP, while 7-hydroxyl-coumarin and (5-thiophen-2-ylisoxazol-3-yl)methanol bind at the fatty acyl binding site. These high-resolution structures of the β-ketoacyl-ACP synthase I from B. melitensis bound to molecules that block substrate binding, range between 1.25 and 1.70 Å resolution, providing a basis for in silico inhibitor screening and optimization.
ACKNOWLEDGMENTS
We thank the SSGCID Cloning, Protein Production and Crystal Core groups and Banu Sankaran at the Advanced Light Source at Lawrence Berkeley National Laboratory. This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases (contract nos. HHSN272201700059C, HHSN27220120002 5C, and HHSN272200700057C to P.J.M.).
Funding information
National Institutes of Health/National Institute of Allergy and Infectious Diseases, Grant/Award Numbers: HHSN272200700057C, HHSN272201200025C, HHSN272201700059C
REFERENCES
- 1.Herbst D, Townsend C, Maier T The architectures of iterative type I PKS and FAS. Nat Prod Rep. 2018;35:1046–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grininger M Perspectives on the evolution, assembly and conformational dynamics of fatty acid synthase type I (FAS I) systems. Curr Opin Struct Biol. 2014;25:49–56. [DOI] [PubMed] [Google Scholar]
- 3.White S, Zheng J, Zhang Y-M, Rock C The structural biology of type II fatty acid biosynthesis. Biochemistry-US. 2005;74:791–831. [DOI] [PubMed] [Google Scholar]
- 4.Beld J, Lee J, Burkart M Fatty acid biosynthesis revisited: structure elucidation and metabolic engineering. Mol Biosyst. 2014;11:38–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsen J, Kadziola A, von Wettstein-Knowles P, Siggaard-Andersen M, Lindquist Y, Larsen S The X-ray crystal structure of β-ketoacyl [acyl carrier protein] synthase I. FEBS Lett. 1999;460:46–52. [DOI] [PubMed] [Google Scholar]
- 6.Price A, Rock C, White S The 1.3-angstrom-resolution crystal structure of β-Ketoacyl-acyl carrier protein synthase II from Streptococcus pneumoniae. J Bacteriol. 2003;185:4136–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang W, Jia J, Edwards P, Dehesh K, Schneider G, Lindquist Y Crystal structure of β-ketoacyl-acyl carrier protein synthase II from E. coli reveals the molecular architecture of condensing enzymes. EMBO J. 1998;17:1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies C, Heath R, White S, Rock C The 1.8 Å crystal structure and active-site architecture of β-ketoacyl-acyl carrier protein synthase III (FabH) from Escherichia coli. Structure. 2000;8:185–195. [DOI] [PubMed] [Google Scholar]
- 9.Nanson J, Himiari Z, Swarbrick C, Forwood J Structural characterisation of the Beta-Ketoacyl-acyl carrier protein synthases, FabF and FabH, of Yersinia pestis. Sci Rep-UK. 2015;5(14):797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Costa T, Nanson J, Forwood J Structural characterisation of the fatty acid biosynthesis enzyme FabF from the pathogen Listeria monocytogenes. Sci Rep-UK. 2017;7:srep39277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Wettstein-Knowles P, Olsen J, McGuire K, Henriksen A Fatty acid synthesis. FEBS J. 2006;273:695–710. [DOI] [PubMed] [Google Scholar]
- 12.Price A, Choi K, Heath RJ, Li Z, White SW, Rock CO Inhibition of β-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin structure and mechanism. J Biol Chem. 2001;276:6551–6559. [DOI] [PubMed] [Google Scholar]
- 13.Luckner S, Machutta C, Tonge P, Kisker C Crystal structures of Mycobacterium tuberculosis KasA show mode of action within cell wall biosynthesis and its inhibition by thiolactomycin. Structure. 2009;17: 1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart M, Parikh S, Xiao G, Tonge P, Kisker C Structural basis and mechanism of enoyl reductase inhibition by triclosan. J Mol Biol. 1999;290:859–865. [DOI] [PubMed] [Google Scholar]
- 15.Dessen A, Quémard A, Blanchard JS, Jacobs WR, Sacchettini JC Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science. 1995;267:1638–1641. [DOI] [PubMed] [Google Scholar]
- 16.Chollet A, Mourey L, Lherbet C, et al. Crystal structure of the enoyl-ACP reductase of Mycobacterium tuberculosis (InhA) in the apo-form and in complex with the active metabolite of isoniazid pre-formed by a biomimetic approach. J Struct Biol. 2015;190:328–337. [DOI] [PubMed] [Google Scholar]
- 17.Garwin JL, Klages AL, Cronan JE Structural, enzymatic, and genetic studies of beta-ketoacyl-acyl carrier protein synthases I and II of Escherichia coli. J Biol Chem. 1980;255(11):949–956. [PubMed] [Google Scholar]
- 18.De Mendoza D, Ulrich K, Cronan JE Thermal regulation of membrane fluidity in Escherichia coli. Effects of overproduction of beta-ketoacyl-acyl carrier protein synthase I. J Biol Chem. 1983;258: 2098–2101. [PubMed] [Google Scholar]
- 19.Feng Y, Cronan J Escherichia coli unsaturated fatty acid synthesis complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J Biol Chem. 2009;284: 29 526–29 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Kodali S, Lee SH, et al. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc Natl Acad Sci. 2007;104:7612–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Soisson SM, Young K, et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441: 358–361. [DOI] [PubMed] [Google Scholar]
- 22.Moustafa G, Nojima S, Yamano Y, et al. Potent growth inhibitory activity of (±)-platencin towards multi-drug-resistant and extensively drug-resistant Mycobacterium tuberculosis. MedChemComm. 2013;4: 720–723. [Google Scholar]
- 23.Pappas G, Akritidis N, Bosilkovski M, Tsianos E Brucellosis. New Engl J Med. 2005;352:2325–2336. [DOI] [PubMed] [Google Scholar]
- 24.Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos E The new global map of human brucellosis. Lancet Infect Dis. 2006;6:91–99. [DOI] [PubMed] [Google Scholar]
- 25.Seleem M, Boyle S, Sriranganathan N Brucellosis: a re-emerging zoonosis. Vet Microbiol. 2010;140:392–398. [DOI] [PubMed] [Google Scholar]
- 26.Mantur BG, Amarnath SK, Shinde RS Review of clinical and laboratory features of human brucellosis. Indian J Med Microbiol. 2007;25: 188–202. [DOI] [PubMed] [Google Scholar]
- 27.Reguera JM, Alarcon A, Miralles F, Pachon J, Juarez C, Colmenero JD Brucella endocarditis: clinical, diagnostic, and therapeutic approach. Eur J Clin Microbiol Infect Dis. 2003;22:647–650. [DOI] [PubMed] [Google Scholar]
- 28.Solera J, Rodriguez-Zapata M, Geijo P, et al. Doxycycline-rifampin vs doxycycline-streptomycin in treatment of human brucellosis due to Brucella melitensis. The GECMEI group. Grupo de Estudio de Castilla-la Mancha de Enfermedades Infecciosas. Antimicrob Agents Chem. 1995; 39:2061–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seleem M, Jain N, Pothayee N, Ranjan A, Riffle JS, Sriranganathan N Targeting Brucella melitensis with polymeric nanoparticles containing streptomycin and doxycycline. FEMS Microbiol Lett. 2009;294:24–31. [DOI] [PubMed] [Google Scholar]
- 30.Baugh L, Gallagher LA, Patrapuvich R, et al. Combining functional and structural genomics to sample the essential Burkholderia Structome. PLoS One. 2013;8:e53851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moynié L, Leckie SM, McMahon SA, et al. Structural insights into the mechanism and inhibition of the β-hydroxydecanoyl-acyl carrier protein dehydratase from Pseudomonas aeruginosa. J Mol Biol. 2013; 425:365–377. [DOI] [PubMed] [Google Scholar]
- 32.Werther T et al. Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase. Nat Commun. 2017;8:ncomms16084. [Google Scholar]
- 33.Aslanidis C, Jong P d. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 1990;18:6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi R et al. Immobilized metal-affinity chromatography protein-recovery screening is predictive of crystallographic structure success. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:998–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bryan C, Bhandari J, Napuli AJ, et al. High-throughput protein production and purification at the Seattle structural genomics Center for Infectious Disease. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch W XDS. Acta Crystallogr Sect D Biol Crystallogr. 2010;66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy A, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ Phaser crystallographic software. J Appl Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Project N The CCP4 suite: programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. [DOI] [PubMed] [Google Scholar]
- 39.Stein N CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J Appl Crystallogr. 2008;41:641–643. [Google Scholar]
- 40.Langer G, Cohen S, Lamzin V, Perrakis A Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 42.Vagin AA, Steiner RA, Lebedev AA, et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2184–2195. [DOI] [PubMed] [Google Scholar]
- 43.Davies D Structural genomics and drug discovery: methods and protocols. Methods Mol Biol Clifton N J. 2014;1140:315–323. [Google Scholar]
- 44.Holm L, Laakso L Dali server update. Nucleic Acids Res. 2016;44: W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krissinel E, Henrick K Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y-M, Hurlbert J, White SW, Rock CO Roles of the active site water, Histidine 303, and phenylalanine 396 in the catalytic mechanism of the elongation condensing enzyme of Streptococcus pneumoniae. J Biol Chem. 2006;281:17 390–17 399. [DOI] [PubMed] [Google Scholar]