Abstract
Microtubule-associated protein 2 (MAP2) is an important neuronal target of extracellular signal–regulated kinase 2 (ERK2) involved in Raf signaling pathways, but mechanistic details of MAP2 phosphorylation are unclear. Here, we used NMR spectroscopy to quantitatively describe the kinetics of phosphorylation of individual serines and threonines in the embryonic MAP2 variant MAP2c. We carried out real-time monitoring of phosphorylation to discover major phosphorylation sites that were not identified in previous studies relying on specific antibodies. Our comparison with the phosphorylation of MAP2c by a model cyclin-dependent kinase CDK2 and with phosphorylation of the MAP2c homolog Tau revealed differences in phosphorylation profiles that explain specificity of regulation of biological functions of MAP2c and Tau. To probe the molecular basis of the regulatory effect of ERK2, we investigated the interactions of phosphorylated and unphosphorylated MAP2c by NMR with single-residue resolution. As ERK2 phosphorylates mostly outside the regions binding microtubules, we studied the binding of proteins other than tubulin, namely regulatory subunit RIIα of cAMP-dependent PKA, adapter protein Grb2, Src homology domain 3 of tyrosine kinases Fyn and Abl, and ERK2 itself. We found ERK2 phosphorylation interfered mostly with binding to proline-rich regions of MAP2c. Furthermore, our NMR experiments in SH-SY5Y neuroblastoma cell lysates showed that the kinetics of dephosphorylation are compatible with in-cell NMR studies and that residues targeted by ERK2 and PKA are efficiently phosphorylated in the cell lysates. Taken together, our results provide a deeper characterization of MAP2c phosphorylation and its effects on interactions with other proteins.
Keywords: microtubule-associated protein, extracellular signal–regulated kinase, cyclin-dependent kinase, Src homology 3 domain, PKA, NMR, growth factor receptor-bound protein 2 (GRB2)
Abbreviations: AKAP, A-kinase anchoring protein; CDK, cyclin-dependent kinase; ERK2, extracellular signal–regulated kinase 2; GSK, glycogen synthase kinase; MAP, microtubule-associated protein; MARK, MAP/microtubule affinity-regulating kinase; MST, microscale thermophoresis; MTBD, microtubule-binding domain; PRR, proline-rich region; VCR, variable central region
Cytoskeletal microtubule-associated proteins (MAPs) are proteins of critical importance for regulating the stability and dynamics of microtubules (1). MAP2 and Tau represent MAP subfamilies expressed in neurons, with MAP2 localized in dendrites, whereas Tau is found mainly in axons (2). Tau and MAP2 belong to the class of intrinsically disordered proteins (IDPs), which lack a unique structure and which exist in multiple, quickly interconverting conformations (3, 4, 5, 6, 7).
Tau and MAP2 differ in their N-terminal projection domains, containing the N-terminal region, the variable central region (VCR), and proline-rich regions (PRRs), whereas the C-terminal part, containing microtubule-binding domain (MTBD) and the C-terminal region, are homologous (8, 9).
The MAP2 family is composed of two high-molecular weight proteins, MAP2a and MAP2b, and two low-molecular weight proteins MAP2c and MAP2d, consisting of 467 and 498 amino-acids, respectively. The MAP2 isoforms differ mainly in the projection domain (9), with MAP2c being the shortest functional isoform. MAP2c is mainly expressed perinatally (2). Postnatally, its expression is restricted to regions exhibiting postnatal plasticity, such as the olfactory bulb (10), suggesting a role in neuronal development.
MAP2 is highly phosphorylated in vivo, and its phosphorylation is regulated during development (11) by a precise balance of activation of specific kinases and phosphatases (for review see (12)). Phosphorylation of MAPs regulates their binding to microtubules (13, 14, 15, 16, 17, 18) and consequently microtubule dynamics (19) and is implied in neuronal development and plasticity (12). MAP2 phosphorylation can also regulate binding of MAP2 to binding partners (20, 21). Serine/threonine kinases phosphorylating MAPs in nonproliferating neurons of the adult brain include cAMP-dependent PKA, extracellular signal–regulated kinases (ERKs), cyclin-dependent kinase 5 (CDK5), PKC, MAP/microtubule affinity-regulating kinase (MARK), and glycogen synthase kinase 3β (GSK3β) (17, 18, 22, 23, 24). MARKs phosphorylate Tau and MAP2 in the MTBD, decreasing their affinity for the microtubules (24), whereas PKA phosphorylates Tau but not MAP2 in the MTBD, inhibiting the microtubule-stabilizing activity of Tau but not MAP2 (21, 25, 26, 27).
Abnormal hyperphosphorylation of Tau by proline-directed kinases has been found in paired helical filaments and neurofibrillary tangles formed in brains of patients suffering from the Alzheimer disease (28, 29). Therefore, phosphorylation of Tau by proline-directed kinases has been studied in detail. The results showed that CDK2 and ERK2 phosphorylate Tau in the PRR, outside of the MTBD, and that the phosphorylation induces conformational changes affecting the ability of Tau to aggregate but its effect on microtubule binding is less clear (12, 30, 31, 32, 33, 34, 35).
Phosphorylation of MAP2 by proline-directed kinases has been less extensively studied and does not have an effect on binding to microtubules but may interfere with microtubule nucleation (36, 37). Only a few phosphorylated residues have been identified, including (using numbering of rat MAP2c) Ser136, Thr259, Thr262, Thr289, Ser293. These sites have been identified using antibodies specific for phosphorylated residues, not providing information about possible phosphorylation sites not probed by the antibodies. MAP2 phosphorylation sites by PKA have been investigated using mass spectrometry (21, 38).
In this study, we used NMR spectroscopy to identify all Ser/Thr-Pro sites of MAP2c phosphorylated by proline-directed kinases ERK2 and CDK2 and to quantitatively describe differences in the phosphorylation kinetics of individual sites. NMR has been used previously to study phosphorylation of MAP2c by PKA (21), phosphorylation of Tau by PKA and ERK2 (27, 34), as well as phosphorylation of other proteins such as histone H3 and p53, both in vitro and in cell extracts (39, 40, 41) and proved to be a reliable method to probe the individual phosphorylation sites. As the level of phosphorylation in vivo is a result of a balance between the actions of kinases and phosphatases, we complemented the study by real-time monitoring of dephosphorylation of individual residues by phosphatases in the SH-SY5Y neuroblastoma cell extract. Finally, we showed that collocation of SH3-binding sites and target sites of proline-directed kinases results in regulation of the interactions of MAP2c with SH3 domains and RIIα regulatory subunit of PKA (RIIα-PKA) by Ser/Thr-Pro phosphorylation.
Results
Interaction of MAP2c with ERK2
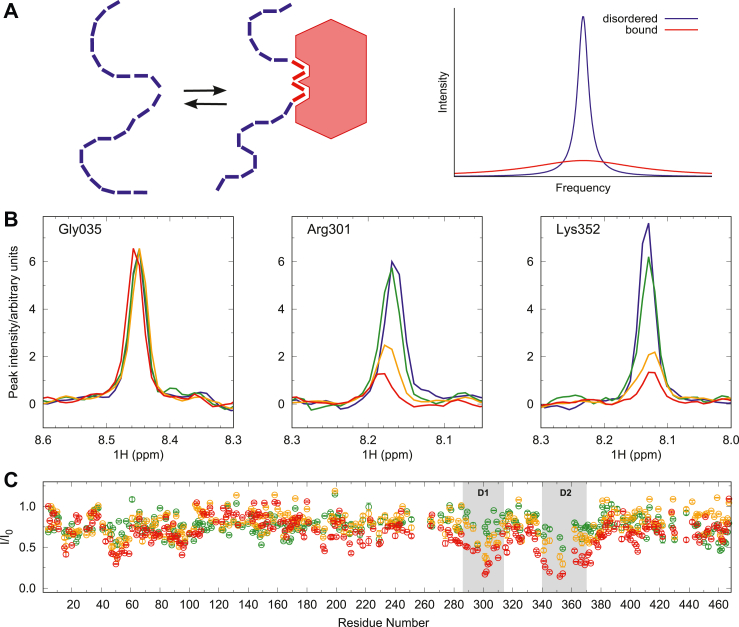
As a first step in our study of the effect of ERK2 on MAP2c, we investigated the physical interaction of the proteins. We took advantage of the fact that heights of NMR peaks of intrinsically disordered proteins decrease when interactions with other proteins result in formation of well-ordered complexes due to the faster NMR relaxation in large, well-ordered proteins (Fig. 1, A and B). To determine the docking site of ERK2 on MAP2c, we measured 3D HNCO spectra of the complex ERK2:[15N,13C]-MAP2c at 0.3:1, 0.6:1, and 1:1 molar ratios and compared the intensity of the peaks to the intensity of the peaks of free MAP2c (Fig. 1C). We observed a gradual decrease in intensity in the MTBD upon addition of ERK2, in regions 290 to 310 and 340 to 360, containing two potential canonical ERK D-box docking sites: D1, 298KQLRL302 and D2, 336TKKIDL341 (42). The peak intensities decrease simultaneously in both D-docking sites, showing that ERK2 binds both sites with a comparable affinity. The dissociation constant of the ERK2:MAP2c complex, determined by microscale thermophoresis (MST), was (0.8 ± 0.4) μM (Fig. S1 in Supplementary material). In addition, the peak intensities decrease also in the N-terminal region, at residues 10 to 30 and 40 to 120, and in the VCR, between residues 180 and 230. These regions do not contain a D-box docking site and could therefore correspond to a secondary binding site.
Figure 1.
Interaction of MAP2c with ERK2.A, schematic representation of a decrease of NMR peak height of residues of IDPs bound to a well-ordered large protein. B, one-dimensional traces of peaks of Gly305, not interacting with ERK2, and of Arg301 and Lys352 located in the D-box D1 and D2, respectively, of MAP2c. The traces are plotted for free MAP2c (blue) and for the MAP2c:ERK2 ratios 1:0.3 (green), 1:0.6 (orange), and 1:1 (red). C, heights of peaks in HNCO spectra of [13C,15N]-MAP2c with 0.3 (green), 0.6 (orange), and 1 (red) molar equivalents of ERK2, compared to free MAP2c. The identified D-docking sites D1 and D2 are indicated by shaded boxes. ERK2, extracellular signal–regulated kinase 2; IDPs, intrinsically disordered proteins; MAP, microtubule-associated protein.
Assignment of NMR spectra
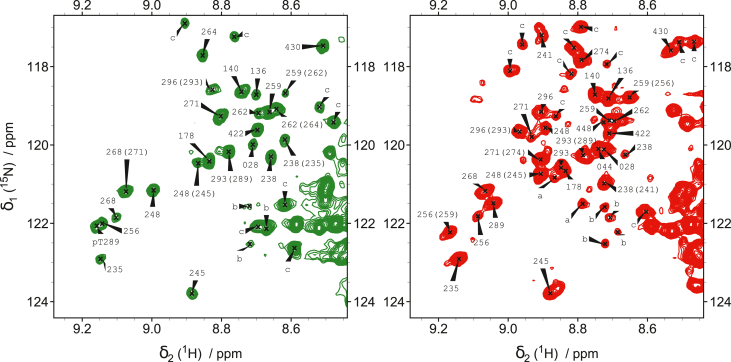
MAP2c phosphorylation was studied by NMR spectroscopy. Series of 1H,15N-HSQC spectra of [15N]-MAP2c were measured after addition of ERK2 or CDK2 for 20 h. Within 4 h of phosphorylation at our conditions, several new intense peaks appeared at proton frequencies downfield from 8.5 ppm, corresponding to a chemical shift of NH groups influenced by phosphorylation. More than 20 peaks of phosphorylated residues could be seen in the spectra of MAP2c phosphorylated by ERK2 and CDK2 (Fig. 2), which is a number expected for the 19 Ser/Thr residues followed by proline (43).
Figure 2.
1H,15N-HSQC spectra of the region of phosphoserines and phosphothreonines of [15N]-MAP2c phosphorylated by CDK2 (green) and ERK2 (red) after 20 h of phosphorylation. The well-resolved peaks of phosphorylated Ser/Thr are labeled with numbers. For phosphorylated residues clustered in the sequence, phosphorylated neighbor residues that influence the position of the peak are indicated in parenthesis. Label a indicates Ser/Thr residues that could not be assigned unambiguously, b designates residues preceded by proline in cis-conformation, and c indicates other unassigned residues. The peaks that are not labeled are already present in nonphosphorylated MAP2c. CDK, cyclin-dependent kinase; ERK2, extracellular signal–regulated kinase 2; MAP, microtubule-associated protein.
In order to assign NMR peaks of phosphorylated Ser/Thr, the type of the preceding residue was first identified based on frequencies in 3D CBCA(CO)NH spectra. Unambiguous assignment was obtained by recording 1H,15N-HSQC spectra of four shorter MAP2c fragments (1–159, 159–254, 250–347, and 300–467) and of MAP2c mutants prepared by site-directed mutagenesis of serines and threonines followed by proline (mutants S28D, S136D, S140D, S178D, T245E, T259E, T262E, S264D, S274D, S422D, MAP2c-159–254/T238E, MAP2c-250–347/T268E, MAP2c-250–347/T271E, MAP2c-250–347/T289E, MAP2c-250–347/S293A). In the case of MAP2c phosphorylated by ERK2, assignment of Thr248 and unexpected phosphorylation at Ser274 were confirmed by following connectivities in 2D FOSY NMR spectra (44). The chemical shifts of the phosphorylated Ser/Thr are listed in Table S1 in Supplementary material.
We assigned peaks of all serines and threonines phosphorylated efficiently by ERK2 including twelve Pro-Xxx-Ser/Thr-Pro ERK consensus sequences and seven other Ser/Thr-Pro motifs present in MAP2c. Phosphorylation of some serines not followed by prolines was also detected, most notably of Ser274 by ERK2. On the other hand, CDK2 but not ERK2 phosphorylates Ser264. Slow phosphorylation of other residues (Ser44, Thr80, Ser241) was also observed. Eighteen of the ERK2 sites were phosphorylated by CDK2 as well. Ser274 and Ser448 were phosphorylated by ERK2 but not by CDK2 under our conditions. In addition, peaks of several residues immediately following the phosphorylated Ser/Thr-Pro sites appeared in the downfield region of the 1H,15N-HSQC spectra (Ala142, Gly240, Gly261, Lys273, Lys292, Ala295). Chemical shifts of Cβ observed in CBCA(CO)NH spectra (45) allowed us to distinguish peaks of residues following cis- and trans-prolines in the Ser/Thr-Pro sites.
The variable central region of MAP2c contains three important phosphorylation sites, Ser136, Ser140, and Ser178. Phosphorylated Ser136 is recognized by the antibody AP18 and its biological role has been discussed in the literature (36).
Twelve target motifs for proline-directed kinases are located in the PRR of MAP2c, forming five clusters of two or three Ser/Thr-Pro sites. Starting from Thr256, the MAP2c sequence is homologous with that of Tau. Thus, three clusters of Ser/Thr-Pro sites in MAP2c PRR can be aligned with the Tau epitopes AT8, AT100, and AT180.
The cluster 256TPGTPGTPS264 (epitope 305) is homologous with the Tau epitope AT8 (199SPGSPGTPGS208). Phosphorylation at Ser202 and Thr205 in AT8 induces formation of a turn-like structure (32), which is disrupted by further phosphorylation at Ser208, resulting in Tau aggregation (33). Interestingly, CDK2 produces a MAP2c motif with a distribution of phosphates resembling fully phosphorylated AT8 of Tau. The important difference between Tau and MAP2c is insertion of Gly207 and presence of two arginine residues in the Tau sequence, critical for the formation of the turn-like motif (32).
The cluster 268TPGTPKSGIL277 in MAP2c substantially differs from the homologous Tau epitope AT100 (212TPSLPTPPTR221). The major PKA phosphorylation site Ser214 and the class-I SH3-binding site 216PTPPTR221 of Tau are missing in the MAP2c sequence. Interestingly, Ser274 in MAP2c is phosphorylated by ERK2, but not by CDK2. ERK2 thus creates distribution of phosphates more similar to the Tau epitope AT100, consisting of phosphorylated Thr212, Ser214, and Thr217.
The MAP2c cluster 288RTPPKSPATP297 aligns with the Tau sequence 230RTPPKSPSSA239, containing the major in vitro ERK2 phosphorylation site Ser235 (34). The Tau sequence is recognized by the antibody AT180 when phosphorylated at Thr231 and Ser235. Phosphorylation of the epitope interferes with Tau binding to microtubules (46). Phosphorylation of Ser235, Ser237, and Ser238 in Tau shifts the conformational equilibrium from polyproline II to α-helical structure (47). The discussed sequences of both Tau and MAP2c contain the class I SH3 recognition sites RTPPKSP.
The Ser/Thr-Pro sites occur also in the terminal regions of MAP2c and Tau. The N-terminal sequences of MAP2c and Tau differ substantially and direct comparison of the phosphorylation sites is not possible. In the case of MAP2c, the only N-terminal Ser/Thr-Pro site is Ser28 in the sequence 26PHSP29. Three Ser-Pro motifs are present in the highly homologous C-terminal regions of MAP2c and Tau. Yet, some differences between the MAPs exist. ERK2 phosphorylates Ser396 slowly in Tau (34) but rapidly the homologous Ser422 in MAP2c. The other two serines are phosphorylated efficiently in both MAPs. Phosphorylation of Ser404 primes Tau for phosphorylation by GSK3β at Ser400 and subsequently at Ser396, forming the epitope PHF-1 (48). Ser430 of MAP2c is in proximity to the major PKA phosphorylation site Ser435, whereas the homologous Ser409 in Tau is phosphorylated only slowly by PKA. Phosphorylation of Ser422 in Tau may control cleavage in the immediately preceding caspase-3 recognition sequence that is missing in MAP2c (49, 50, 51). A biological role of phosphorylation at Ser178 or Ser430 is not known, but proximity to Ser184 and Ser435, rapidly phosphorylated by PKA, suggests a possible cross-talk between the kinases (see below).
Kinetics of ERK2 and CDK2 phosphorylation
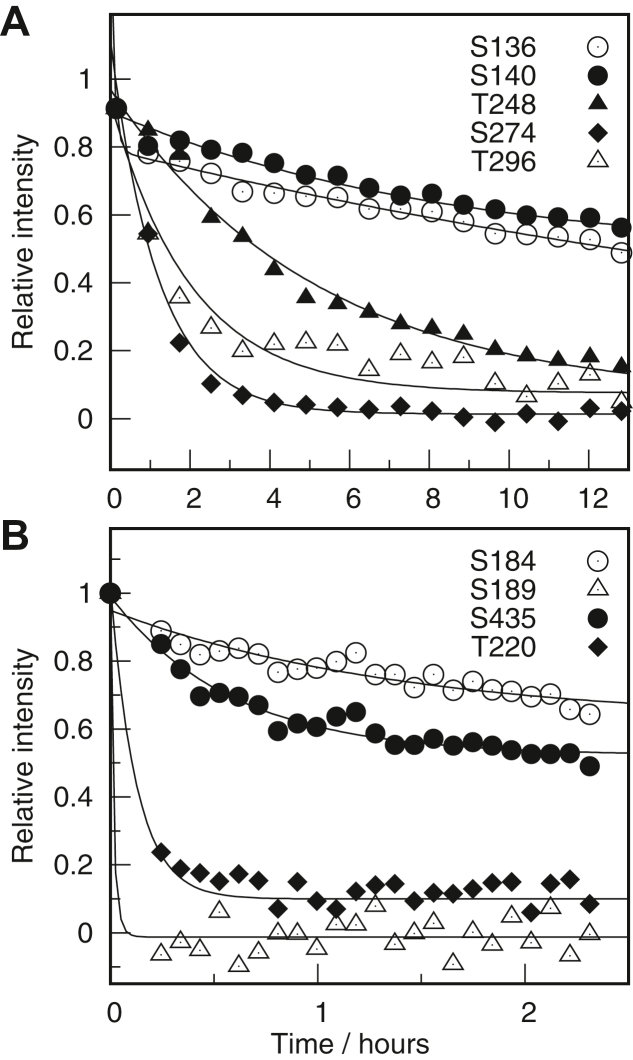
We studied the kinetics of phosphorylation of MAP2c by analyzing peak intensities in the acquired 1H,15N-HSQC spectra. The 2D 1H,15N-SOFAST-HMQC experiment, providing higher time resolution but worse frequency resolution, was used to track peaks of Ser178, which was phosphorylated most rapidly by CDK2. Concentrations of kinases were chosen such that the highest phosphorylation rates were comparable.
A large number of serines and threonines competing for the enzyme and potentially interfering mutually constitute a very complex kinetic system. Therefore, exact determination of individual kinetic constants is not feasible. However, phosphorylation of individual sites can be compared. For this purpose, we used an empirical parameter τ1/2 corresponding to the time needed to phosphorylate half of the given residue under the given conditions (see Experimental procedures). The comparison is presented in Table 1.
Table 1.
Values of τ1/2 (in hours) for phosphorylation of individual MAP2c residues by ERK2 and CDK2
| Residue | ERK2 | CDK2 |
|---|---|---|
| Ser28 | 0.96 ± 0.04 | 9.6 ± 2.9 |
| Ser136 | 0.29 ± 0.02 | 7.1 ± 2.4 |
| Ser140 | 0.30 ± 0.02 | 3.76 ± 0.18 |
| Ser178 | (6.32 ± 0.21)a | 0.27 ± 0.03 |
| Thr235 | 1.09 ± 0.04 | 17 ± 10 |
| Thr238 | 1.21 ± 0.12 | 2.94 ± 0.19 |
| Thr245 | 1.09 ± 0.05 | 8.2 ± 3.2 |
| Thr248 | 0.95 ± 0.09 | 0.96 ± 0.04 |
| Thr256 | 1.18 ± 0.04 | peak overlap |
| Thr259 | peak overlap | 0.93 ± 0.09 |
| Thr262 | peak overlap | 2.34 ± 0.20 |
| Ser264 | not phosphorylated | 16.9 ± 5.2 |
| Thr268 | 0.38 ± 0.19 | 1.90 ± 0.19 |
| Thr271 | 1.04 ± 0.09 | 0.44 ± 0.08 |
| Ser274 | 0.96 ± 0.01 | not phosphorylated |
| Thr289 | 3.69 ± 0.14 | peak overlap |
| Ser293 | (≈1.1)b | 0.53 ± 0.08 |
| Thr296 | 1.18 ± 0.07 | 0.94 ± 0.05 |
| Ser422 | 0.90 ± 0.03 | 0.99 ± 0.04 |
| Ser430 | 0.74 ± 0.01 | 0.93 ± 0.02 |
| Ser448 | (0.87 ± 0.23)c | not phosphorylated |
determined for the MAP2c fragment 1 to 159.
estimated for the MAP2c fragment 250 to 347.
determined for the MAP2c fragment 300 to 467.
Five Ser-Pro sites (with Ser28, Ser178, Ser422, Ser430, and Ser448) are isolated in the MAP2c sequence and analysis of their phosphorylation is straightforward. By contrast, most Ser/Thr-Pro sites are clustered in the MAP2c sequence. For serines and threonines in such clusters, several peaks with variable phosphorylations of the Ser/Thr neighbors appeared in the course of the reaction. In most cases, the resolution of the spectra allowed us to estimate the parameter τ1/2 from the sum of intensities of peaks corresponding to the same phosphoserine/phosphothreonine with differently phosphorylated neighbors (Fig. S2 in Supplementary material). Peak overlaps were observed for Thr256 and Thr289. In the case of ERK2, peaks of Ser178 and Ser293 are not well resolved, and peaks of Ser448 overlapped with peaks of Thr259 and Thr262. As discussed below, similar phosphorylation rates are observed for full-length MAP2c and for its shorter fragments. Therefore, τ1/2 values determined from phosphorylation of the fragments serve as good descriptors of phosphorylation efficiency of residues not resolved in the full-length spectra (Table 1).
ERK2 and CDK2 substantially differ in the phosphorylation kinetics. The difference in kinetics can be explained by different consensus sequences of the kinases: ERK2 prefers proline at position -2, whereas CDK2 prefers a positive residue at position +3 and ideally also proline at -2, glycine at +2, and a positive residue at +4 (52).
Our results show that CDK2 is more specific. Only residues followed by proline are phosphorylated by CDK2, with a noteworthy exception of Ser264. Ser178 (with Lys181 and Arg282 at positions +3 and +4, respectively) is phosphorylated much faster than any other residue by CDK2, and phosphorylation of Ser422 and Ser430, with arginines at position +3, is also relatively fast. On the other hand, phosphorylation of Ser28 is very slow and Ser448 remains unphosphorylated under our conditions. The first residues in the tandem sites Ser136/Ser140, Thr235/Thr238, and Thr245/Thr248 are also phosphorylated very slowly. Phosphorylation of the second residues is somewhat faster. Slower phosphorylation of Thr235, Thr238, (and possibly of unresolved Thr256) can be explained by the fact that threonines/serines in the other Ser/Thr-Pro sites in PRR have proline at the position -2 or positive charge at the position +3. The positive charge of Arg267 also explains why Ser264, not followed by proline, is phosphorylated. Phosphorylation rates of residues clustered in more complex sites are close to those of Ser422 and Ser430, with the exception of Thr262 and Thr268. Interestingly, sequences 256TPGTPGTP263 and 268TPGT271 are bridged by the aforementioned Ser264.
ERK2 is more promiscuous. More peaks of residues not followed by proline appear in the spectra and rates of phosphorylation of the Ser/Thr-Pro sites are more uniform. Ser136 and Ser140 are phosphorylated most rapidly. It is expected as both serines in the sequence 134PPSPPPSP141 represent ideal ERK2 targets. Phosphorylated Ser136 constitutes the epitope AP18 (36), but Ser140, not contributing to the epitope AP18 (36), is phosphorylated at a similar rate. This confirms that ERK2 efficiently creates the AP18 epitope but also suggests that phosphorylation of Ser140 may contribute to physiological effects attributed to phospho-Ser136. Phosphorylation rates of Thr235 and Thr245 are comparable with those of other threonines in the clustered sites. Differences between serines/threonines with and without proline at position -2 are marginal. The same applies to Ser422, Ser430, and Ser448 in the C-terminal region, which are efficiently phosphorylated by ERK2 despite the lack of proline at position -2. The differences between phosphorylation by ERK2 and CDK2 can be thus mostly attributed to a more specific consensus sequence of the latter kinase. However, proline at -2 may contribute to the observed efficient ERK2 phosphorylation of Ser274 that is not followed by proline. Phosphorylation of Ser264 by ERK2 is negligible. Slow phosphorylation of other residues (Ser44, Thr80, Ser241) was also observed.
It is well described for Tau (53) that phosphorylation by some kinases depends on prior phosphorylation by other kinases. Therefore, we probed if phosphorylation by ERK2 is primed by PKA, CDK2, or GSK3β (kinetics for residues with well-resolved peaks is presented in Fig. S3 in Supplementary material). Prior phosphorylation by PKA did not have any effect on the kinetics of phosphorylation by ERK2. Prior phosphorylation by CDK2 moderately increased the rate of ERK2 phosphorylation, most notably for Thr235, Thr238, Thr245, and Thr274. This suggests that ERK2 phosphorylates faster when other residues in clusters of ERK2 target sites (in our case Thr238, Thr235, Thr248, Thr271, respectively) are already phosphorylated by CDK2. A similar effect was observed for GSK3β, but peaks of residues phosphorylated by GSK3β are yet to be assigned. We also compared phosphorylation kinetics of full-length MAP2c with MAP2c fragments 1 to 159 and 159 to 254, lacking the ERK D-box docking sites. Phosphorylation rates were comparable for all three samples (Fig. S4 in Supplementary material). This indicates that at our conditions, ERK2 is saturated with the substrates (MAP2c fragments) even if they lack the docking sites and thus the interactions of ERK2 with the docking sites do not influence phosphorylation kinetics significantly.
Effect of ERK2 phosphorylation on long-range interactions of MAP2c
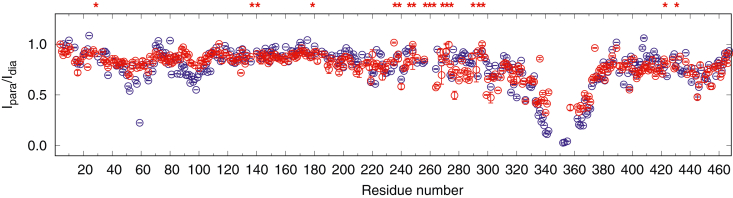
MAP2c exhibits long-range intramolecular and intermolecular interactions between the MTBD and the N-terminal region (40–60, 80–120) and the C-terminal region (440–467) (8). We have shown previously that phosphorylation of MAP2c by PKA strongly reduces the intermolecular interactions (8). To determine if phosphorylation of MAP2c by proline-directed kinases modifies the long-range interaction, we labeled [15N,13C]-MAP2c on its single cysteine C348 with the paramagnetic tag MTSL and compared the intensity of MAP2c residues with those in MAP2c tagged with diamagnetic MTS, at 300 μM concentration (Fig. 3).
Figure 3.
Intensities of the peaks of the paramagnetic [15N,13C]-MAP2c in HNCO spectra, compared to the intensities of the peaks of diamagnetic [15N,13C]-MAP2c at 300 μM concentration, unphosphorylated (blue) and phosphorylated by ERK2 (red). The ERK2 phosphorylation sites are marked by asterisks. ERK2, extracellular signal–regulated kinase 2.
ERK2 had a similar effect as PKA on the intermolecular interactions involving the N-terminal region of MAP2c. Paramagnetic relaxation enhancement between residues 40 and 110 was strongly reduced upon phosphorylation. On the other hand, the intensity ratio decreased upon phosphorylation by ERK2 in the region between residues 270 to 300, showing a stronger interaction between the MTBD and this region after ERK2 phosphorylation. The region 270 to 300 contains several phosphorylation sites (Thr268, Thr271, Ser274, Thr289, Ser293, and Thr296). These data suggest that MAP2c phosphorylated by ERK2 forms a more compact core between the PRR and the MTBD, whereas the intermolecular and/or intramolecular interaction between the N-terminus and the MTBD decrease.
Effect of ERK2 phosphorylation on interactions with PKA
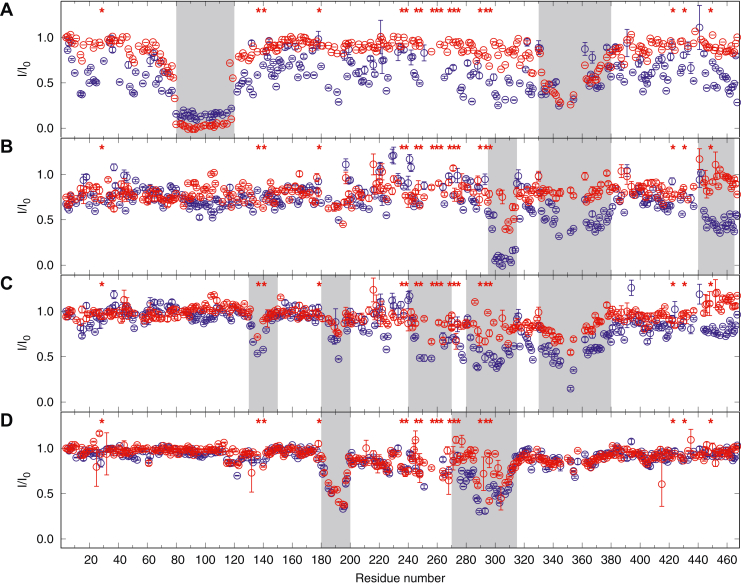
The putative RIIα-PKA–binding site, in the region 80 to 120 (54), is close to two main ERK2 phosphorylation sites, Ser136 and Ser140. Phosphorylation of Ser136 (detected by the antibody AP18) has been suggested to affect binding of the RIIα-PKA (36). We determined the binding site of the RIIα-PKA using NMR (Fig. 4A). In the presence of RIIα-PKA, a strong decrease of peak intensity is seen in the region 80 to 120. However, intensity decrease is observed for many residues in all regions of MAP2c, suggesting that binding RIIα-PKA is not a local effect, limited to the recognition site between Thr80 and Pro120. In MAP2c phosphorylated by ERK2, the peak intensity decreases only in the region 80 to 120 or in the MTBD. This result shows that ERK2 phosphorylation does not affect the major binding site of RIIα-PKA but interferes with interactions in other regions of the MAP2c complex with RIIα-PKA.
Figure 4.
Interactions of MAP2c with various binding partners. Intensities (I) of the peaks in [15N,13C]-MAP2c HNCO spectra upon addition of RIIα-PKA (A), Fyn-SH3 (B), Abl-SH3 (C), and Grb2 (D) compared with intensities of the peaks in free MAP2c (I0). Blue color is used for unphosphorylated MAP2c, red color for MAP2c phosphorylated by ERK2. Asterisks above the plot indicate the phosphorylated residues. The region of MAP2c interacting with the different partners are shaded. ERK2, extracellular signal–regulated kinase 2; MAP, microtubule-associated protein.
After finding that Ser136 does not influence the binding of RIIα-PKA to MAP2c, we also checked if phosphorylation by ERK2 affects the catalytic activity of PKA. We prepared MAP2c phosphorylated by ERK2 and measured rates of a subsequent phosphorylation by PKA (Fig. S5 in Supplementary material). No effect was observed for Ser189, and phosphorylation rate of Ser184 was reduced only moderately. However, the major site targeted by PKA, Ser435, was phosphorylated substantially slower when MAP2c was phosphorylated by ERK2 prior to PKA. This indicates that the phosphate group at Ser430, introduced by ERK2, interferes with otherwise very fast phosphorylation of Ser435 by PKA.
Effect of ERK2 phosphorylation on interactions with SH3 domains
MAP2c contains 13 potential SH3 domain binding motifs (PXXP) (55), eight of them located in the PRR. All these sites are phosphorylated by ERK2 and could therefore modulate the interaction of SH3 domains with MAP2c. We therefore investigated the interaction of the classical SH3 domain of the tyrosine kinases Fyn (Fyn-SH3), Abl (Abl-SH3), and of the adapter protein Grb2, containing two SH3 domains, N- and C-terminal (56).
The intensity of the peaks in 3D HNCO spectra in presence of Fyn-SH3, Abl-SH3, or Grb2 was compared to those in the spectrum of free MAP2c. The same experiments were repeated with [15N, 13C]-MAP2c phosphorylated by ERK2. The results are shown in Figure 4, B–D.
In the presence of an equimolar concentration of Fyn-SH3, we observed a strong (almost 100%) decrease of intensities of the peaks in the region 295 to 315 (immediately following the class I SH3-binding site 288RTPPKSP294) in the PRR and some (about 50%) decrease in the MTBD and in the C-terminal region of MAP2c. Phosphorylation by ERK2 reduced the strong decrease in the peak intensities of residues 295 to 305 to approximately 50% and almost completely suppressed the signs of binding in other regions.
In the presence of equimolar amount of Abl-SH3, intensities of the MAP2c peaks decreased to approximately 50% in the region 240 to 300 in PRR, in MTBD, but also for residues 130 to 150 and 180 to 200 in the variable central region. Phosphorylation of MAP2c by ERK2 resulted in a weaker effect of binding of Abl-SH3 on MAP2c spectra, indicating that phosphorylation in the PRR disrupts the interaction with Abl-SH3.
These results show that Fyn-SH3 and Abl-SH3 interact with the SH3-binding motifs in the PRR and VCR, and that phosphorylation of the SH3 motifs inhibits the interaction. The difference in the effect of the binding is in agreement with different affinities of the SH3 domains (Fig. S6 in Supplementary material). The equimolar concentration of Fyn-SH3, binding with nanomolar affinity (dissociation constant of the MAP2c:Fyn-SH3 determined by MST was (22 ± 5) nM), completely saturates MAP2c. On the other hand, similar amounts of free and bound MAP2c are present at 100 μM concentrations of MAP2c and Abl-SH3, in agreement with the dissociation constant of (68 ± 10) μM estimated by MST. In addition, Fyn-SH3 and Abl-SH3 interact with the MTBD, where ERK2 does not phosphorylate. The reduction of the effects of the interactions upon ERK2 phosphorylation observed also in the MTBD indicates an indirect effect.
In the presence of equimolar Grb2, we observed a 50% decrease in intensities of residues 180 to 210 and 280 to 310, suggesting that the two regions are the main Grb2-interacting sites. Intensities of the peaks of residues 220 to 270 also decreased, suggesting that this region is also involved in Grb2 binding. After ERK2 phosphorylation of MAP2c, the intensity in the region 220 to 270 is close to the intensity of free MAP2c, showing that phosphorylation by ERK2 interferes with interactions of this region with Grb2. On the other hand, the interaction with the MTBD decreases only partially and the interaction with the N-terminal residues 180 to 210 is not affected by phosphorylation.
MAP2c dephosphorylation in neuroblastoma cell extract
The actual level of MAP2 phosphorylation in the cell is a result of a balance between the activities of kinases and phosphatases. As the roles of individual phosphatases are characterized much less than those of the kinases, we probed dephosphorylation in the extracts of neuroblastoma SH-SY5Y cells.
We incubated [15N]-MAP2c phosphorylated by ERK2 in SH-SY5Y neuroblastoma cell extract and followed its phosphorylation state with 20-min 1H,15N-SOFAST-HMQC spectra (Figs. 5A and S7 in Supplementary material). The spectra showed that Ser274, Thr271, and Thr245 are dephosphorylated most rapidly, followed by Thr235, Thr238, Thr259, Thr262, and Thr248. Other residues (Ser136, Ser140, Thr256, Thr268, Ser422, and Ser430) were dephosphorylated more slowly.
Figure 5.
Dephosphorylation of phosphorylated MAP2c in SH-SY5Y neuroblastoma cell extracts. Relative decrease of the intensities of the peaks of selected phosphorylated serines and threonines in 1H,15N-SOFAST-HMQC spectra of [15N]-MAP2c phosphorylated by ERK2 (A) and PKA (B) in SH-SY5Y neuroblastoma cell extracts. The relative intensities of the phosphorylated peaks were normalized to the intensity of the Trp14 side chain peak. ERK2, extracellular signal–regulated kinase 2; MAP, microtubule-associated protein.
In order to compare dephosphorylation rates of residues phosphorylated by ERK2 with nonproline–dependent kinases, we repeated the experiments with [15N]-MAP2c phosphorylated by PKA (Fig. 5B). A series of 5-min SOFAST-HMQC spectra revealed that dephosphorylation of Ser189 and Thr220 is almost complete after 20 min, showing that these two sites are highly sensitive to phosphatases. This is in agreement with a previous report of fast dephosphorylation of phospho-Thr220 (57). Dephosphorylation of Ser435 is much slower and phospho-Ser184 is insensitive to phosphatases.
These results show that the residues phosphorylated most rapidly by ERK2 or PKA remain phosphorylated for a long time in the presence of uninhibited mixture of kinases and phosphatases in the neuroblastoma cell lysate.
MAP2c phosphorylation in neuroblastoma cell extract
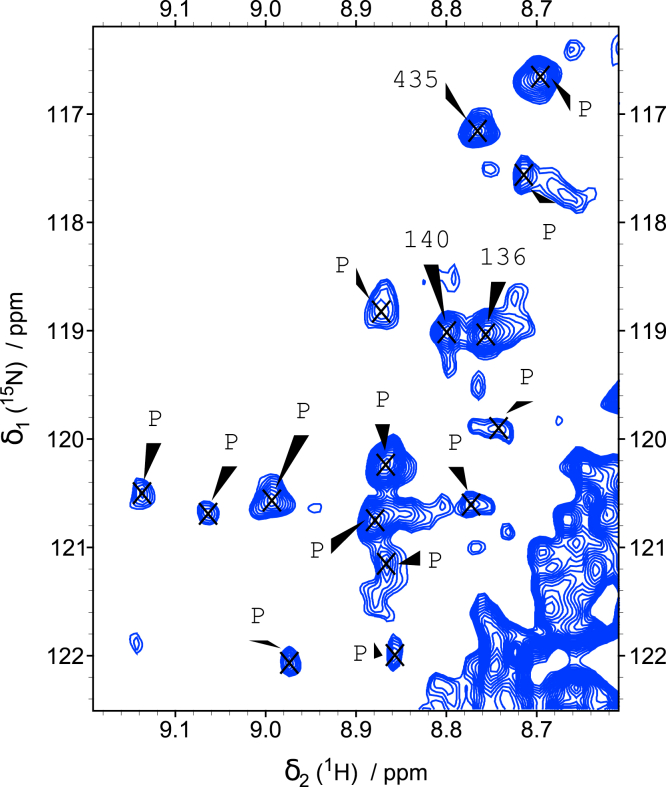
We also checked the ability of the neuroblastoma cell extract to phosphorylate MAP2c. Upon incubation of unphosphorylated [15N]-MAP2c in SH-SY5Y cell extract supplemented with nonspecific phosphatase inhibitors, several signals appeared in the region of the 1H,15N-SOFAST-HMQC spectra corresponding to phosphothreonines and phosphoserines (Fig. 6). By overlaying the 1H,15N-SOFAST-HMQC spectra of MAP2c after phosphorylation in cell extract with the 1H,15N-SOFAST-HMQC spectra of MAP2c phosphorylated by ERK2 or PKA, some of the signals could be assigned to Ser435, phosphorylated by PKA, to Ser178, phosphorylated most quickly by CDK2, and to major phosphorylation sites of ERKs, Ser136 and Ser140. To confirm our assignment, we repeated the phosphorylation in cell extract using the mutants [15N]-MAP2c-S435D and [15N]-MAP2c-S140D. In these mutants, the peak corresponding to putative phosphorylated Ser435 and Ser140, respectively, disappear, confirming that Ser435 and Ser140 are phosphorylated in the cell extract (Fig. S8 in Supplementary material). These results document that MAP2c is the target of PKA and proline-directed kinases in the SH-SY5Y cell, as well as of other kinases that remain to be characterized.
Figure 6.
Phosphorylation of [15]N-MAP2c in SH-SY5Y neuroblastoma cell extract.1H,15N- SOFAST-HMQC spectrum of the region of phosphorylated serines and threonines after 17 h of incubation in the cell extract. Peaks of phosphorylated Ser/Thr are labeled by P. The assigned phosphorylated residues (Ser140, Ser136, and Ser435) are labeled with numbers. MAP, microtubule-associated protein.
Discussion
ERK2 is a mitogen-activated protein kinase involved in Raf signaling pathways triggered by various growth factors. ERK2 is strongly expressed in the brain and associated with microtubules (58). Phosphorylation of Tau by ERK2 has been associated with the progression of the Alzheimer disease (59) and studied in detail in vitro (34), but MAP2 is also an important target of ERK2 under normal physiological conditions (12, 36, 60, 61). Therefore, we employed NMR spectroscopy to fully characterize phosphorylation of Ser/Thr-Pro sites in MAP2c by ERK2 in vitro, including quantitative comparison of kinetics of phosphorylation at individual residues.
In parallel, we investigated phosphorylation of MAP2c by another proline-directed kinase, CDK2. Although CDK2 is not active in the adult brain, Sibille et al. (62) successfully used CDK2 to mimic a combined action of CDK5 and GSK3β on Tau. In the case of MAP2c, expressed prenatally when the brain develops, investigation of CDK2, active through most phases of the cell cycle, is also physiologically relevant. Last but not least, higher specificity of CDK2 helped us to confirm assignment of the NMR peaks of MAP2c phosphorylated by ERK2.
We identified 20 ERK2 phosphorylation sites in MAP2c. Our results cover epitopes of frequently used antibodies AT18 (36), 305 (63), CP9 (64), and MC6 (65, 66) but show that other residues may be efficiently phosphorylated as well. As reported for Tau, the ERK2 phosphorylation sites are not confined to the docking sites of ERK2 on MAP2c (Fig. 1C). But in contrast to the ERK2–Tau interactions, ERK2 strongly binds to the N-terminal domain of MAP2c. However, the docking sites do not seem to drive the phosphorylation to specific sites, as MAP2c fragments lacking the docking sites show the same kinetics.
The phosphorylation by ERK2 influences conformational ensemble of MAP2c, the main difference being the formation of a more compact core between the MTBD and the PRR upon ERK2 phosphorylation (Fig. 3). On the other hand, the intermolecular and/or intramolecular interaction between the N-terminus and the MTBD decrease, leading to an opening of the paper clip conformation, similar to what was observed upon PKA phosphorylation (8).
It should be emphasized that phosphorylations observed in vitro are not necessarily present in the cell. Experiments in the lysates of SH-SY5Y cells document that the exact phosphorylation state of MAP2c is determined by kinetics of phosphorylation but also dephosphorylation. We assume that residues with the highest phosphorylation rates are most likely to be biologically relevant. The major phosphorylation sites of ERK2, Ser136, and Ser140, together with the major phosphorylation site of PKA, Ser435, are efficiently phosphorylated by the lysate of SH-SY5Y neuroblastoma cells. On the other hand, phosphates at residues of PRR are hydrolyzed faster, but the relationship of phosphorylation and dephosphorylation kinetics is complex. For example, Ser184, phosphorylated more slowly by PKA than Ser435, is most resistant to hydrolysis, whereas dephosphorylation of Thr271, relatively rapidly phosphorylated by ERK2, is fast.
How does the phosphorylation by ERK2 control the biological role(s) of MAP2c? In order to answer this question, we first compare ERK2 with other kinases active in nerve cells. Distribution of phosphorylation sites targeted by different kinases strongly correlates with the fact that particular regions of MAPs play different biological roles. MAP2c and Tau are known primarily as proteins stabilizing microtubules. Sites recognized by MARKs are localized in the middle of microtubule-binding repeats of Tau and MAP2 (67). PKA phosphorylates Tau and MAP2 very differently. It targets proline-rich region P2 and microtubule-binding repeat 3 in Tau, but not the homologous regions in MAP2c (27). On the other hand, PKA phosphorylates most rapidly Ser435 in the C-terminal region of MAP2c and also a proline-rich region encoded by exon 14, unique for MAP2 (21). In this study, we observed that prior phosphorylation by ERK2 slows down phosphorylation of Ser435 by PKA. This finding may serve as an example of a cross-talk between two signaling pathways (ERK2 affecting the major target of PKA in MAP2c).
Proline-directed kinases, investigated in this study, do not phosphorylate MTBDs of Tau or MAP2c. Interactions with microtubules may be influenced by phosphorylation in PRR (12, 68), but the major ERK2 phosphorylation sites of MAP2c have no impact on microtubule binding (36). Yet, various physiological activities have been related to phosphorylation in the variable central region and in the C-terminal region of MAP2. Phosphorylation at Ser136 in rat olfactory bulb is modified by olfactory experience (69) and has been linked to contextual memory (70). It has been shown recently that MAP2 is involved in activity-dependent synaptic plasticity in mature hippocampal networks, and that this function involves ERK (71). Phosphorylation of the epitope PHF-1 in the C-terminal region of Tau has been reported to suppress interaction of Tau with cholinergic muscarinic receptors (72). Ser1782 (corresponding to Ser422 of rat MAP2c) in a homologous region of MAP2 is hyperphosphorylated in schizophrenic patients, while phosphorylation of residues corresponding to Thr259 and Thr262 in PRR is reduced (73, 74).
Importance of other interactions than microtubule binding is also evident at the molecular level. MAP2c and Tau interact with different proteins in cells, in addition to tubulin. In this study, we investigated ability of MAP2c to bind proteins that potentially link phosphorylation of MAP2c by ERK2 with other phosphorylation cascades. In particular, we studied interactions with a regulatory subunit of PKA, with SH3 domains of two tyrosine kinases, and with an adapter protein Grb2.
The dimeric RIIα regulatory subunit (RIIα-PKA) localizes PKA in the cell by recognizing helical motifs of various A-kinase anchoring proteins (AKAPs). PKA is activated by cAMP, which releases RIIα-PKA from the inactive complex and liberates the active catalytic subunit of PKA that then phosphorylates target proteins in the proximity of AKAP. Despite its disordered nature, MAP2c was identified as one of the AKAPs. Residues 80 to 120 in the N-terminal domain of MAP2c bind RIIα-PKA (54) and phosphorylation of Ser136 has been suggested to interfere with its binding (36). Our direct monitoring of RIIα-PKA binding to MAP2c revealed that RIIα-PKA is indeed recognized by residues 80 to 120 of MAP2c, but motions of many residues throughout the whole MAP2c sequence are restricted when the complex is formed (blue circles in Fig. 4A). On the contrary, backbone flexibility is strongly reduced by the RIIα binding only for residues 80 to 120 and in MTBD of MAP2c phosphorylated by ERK2 (red circles in Fig. 4A). It indicates that multiple residues of MAP2c form secondary contacts directly with bound RIIα-PKA or with regions of MAP2c interacting with RIIα-PKA. Phosphorylation by ERK2 weakens such secondary interactions but does not disrupt strong binding of RIIα-PKA to the recognition site (residues 80–120) and interactions involving the unphosphorylated MTBD. Our results thus do not support the hypothesis that phosphorylation of Ser136, efficiently targeted by ERK2, suppresses the interaction of RIIα-PKA with the AKAP motif of MAP2c.
SH3 domains are typical recognition structures of various tyrosine kinases, including Fyn and Abl. Fyn plays a fundamental role in cytoskeletal remodeling (for review, see (75)), and MAP2c interacts with Fyn-SH3 in vivo and in vitro (20, 76). Interaction of Abl-SH3 with MAP2c has not been shown yet, however, they colocalize in dendrites, and Abl is necessary for MAP2c segregation in dendrites, suggesting a possible interaction of Abl and MAP2 in vivo (77). Our data revealed that Fyn-SH3 and Abl-SH3 bind to MAP2c with different affinities, explaining why Fyn, but not Abl, was reported to interact with MAP2c (20, 76). Resolution of the NMR spectra allowed us to identify the actual binding sites of MAP2c. Tau and MAP2c contain 7 and 13 minimal interaction sites for SH3 domains (Pro-Xxx-Xxx-Pro), respectively. PRR of Tau contains one classical class I and one classical class II SH3 consensus site, whereas only one class I motif is present in MAP2c (286IIRTPPKSPATPK298) (43) that overlaps with a cluster of residues phosphorylated by ERK2. Indeed, MAP2c binds Fyn-SH3 and Abl-SH3 in the vicinity of the class I motif and the interaction is strongly reduced by ERK2 phosphorylation (Fig. 4, B and C). Phosphorylation by ERK2 also suppresses weaker interactions of MAP2c with Fyn-SH3 and Abl-SH3. Interestingly, phosphorylation at Ser136 and Ser140 seems to directly interfere with the Abl-SH3 binding to the 134PPSPPPSP141 motif. This region is similar to the C-terminal region of p41 peptide (APSYSPPPP), a high affinity ligand of Abl-SH3 (78) binding the hydrophobic surface of Abl-SH3. In a similar manner, phosphorylation of Ser422, Ser430, and Ser448 inhibits interaction of Fyn-SH3 with the C-terminal domain. In summary, our results suggest that ERK2 regulates interactions of MAP2c with Fyn (and Abl, if biologically relevant).
The last examined interaction was binding of MAP2c to the adapter protein Grb2 linking tyrosine kinases with the Ras signaling pathway (Fig. 4D). Binding of Grb2 to MAP2 has been reported (20, 79), but complete residue-specific mapping of the interactions have not been performed yet. We observed that Grb2, containing two SH3 domains, binds to the class I SH3-binding site of MAP2c, but also to residues 180 to 210, not targeted by proline-directed kinases but phosphorylated by PKA (21). Phosphorylation by ERK2 thus reduces only the interaction with the class I motif. In addition, it has been described (79) that Grb2 binds to Tyr67 phosphorylated by Fyn. Therefore, it seems that Grb2 can interact with several MAP2c sites and each interaction may be controlled by a different kinase.
In summary, in vitro NMR investigation of phosphorylation of MAP2c by ERK2 provided a detailed picture of phosphorylated sites and of their kinetics. Among 20 residues targeted by ERK2, serines 136 and 140 are phosphorylated most rapidly. Phosphorylation of Ser136 and Ser140 was also confirmed in neuroblastoma cell lysate and relatively slow hydrolysis of these phosphoserines in the lysate supports their potential biological function. The phosphorylation by ERK2 changes compaction of MAP2c. A strong suppression of phosphorylation of Ser435 by PKA in MAP2c phosphorylated by ERK2 was the most evident sign of cross-talk between kinases tested in this study. Binding studies with four proteins involved in various signaling pathways reveals that phosphorylation by ERK2 reduces binding of SH3 domains to their class I recognition motif in the proline-rich region of MAP2c but does not influence interactions of RIIα-PKA with the N-terminal MAP2c domain and interactions of Grb2 with MAP2c residues 180 to 210. It documents that ERK2 is able to specifically influence interactions of MAP2c with other regulatory proteins. Whereas some ERK2 phosphorylation sites in MAP2c have been reported in the literature (12, 36, 37, 61, 63, 76), kinetics of the phosphorylation has not been studied systematically. Our work advances the field by providing quantitative description of the phosphorylation of individual residues of MAP2c by ERK2 and by describing its biologically relevant consequences (regulation of interactions with proteins involved in signaling pathways and interference with phosphorylation by PKA).
Experimental procedures
DNA constructs
DNA encoding the fragments MAP2c-1–467, MAP2c-1–159, MAP2c-159–254, MAP2c-250–347, and MAP2c-300–467 was amplified by PCR using MAP2c gene in pET3d vector as template and cloned in pETM11 (MAP2c-1–467, MAP2c-1–159, MAP2c-159–254, MAP2c-250–347) or pET28 (MAP2c-300–467) by restriction cutting using NcoI and BamHI and ligation. Single point mutants of MAP2c (S28D, S136D, S140D, S178D, T245E, T259E, T262E, S264D, S274D, S422D), MAP2c-159–254 (T238E), MAP2c-250–347 (T268E, T271E, T289E, S293A) were produced following the protocol from (80) using MAP2c in the pET3d vector, MAP2c-159–254 or MAP2c-250–347 in pETM11 as a template. The primers used are listed in Table S2 in Supplementary material. MAP2c/S435D mutant was prepared previously (21). The result of mutations was confirmed by sequencing.
The plasmids coding for ERK2, MEKR, CDK2, and CycA3 were a kind gift from Isabelle Landrieu (Université de Lille). The plasmids coding for human RIIα-PKA was a kind gift from Enno Klussmann (Max Delbrück Centrum für Molekulare Medizin). pET28-His-Grb2 (C32S, C198A) was used for its better stability. The parental plasmid for the expression of His-tagged WT human Grb2 protein (NP 002077.1) was a gift by JCD Houtman (81). C32S and C198A mutations for the reduction of Grb2 dimerization were introduced based on the work of Yuzawa et al. (82) The mutations were introduced using a QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent) with Grb2 C32S 5′–cctcaaggttttgaacgaagaaagtgatcagaactgg– 3′ and Grb2 C198A 5′–ctggtggaaaggagctgcccacgggcagac–3′ as mutagenesis primers.
The pGEX-2T-Abl SH3 plasmid for the expression of the GST-tagged SH3 domain (aa 64–120) of human Abl (NP 005148.2) has been described in (83).
The parental plasmid for expression of Fyn-SH3 was a gift from Filippo Giancotti (Addgene plasmid nbr 16032, (84)). The SH3 domain of Fyn (aa 85–141) was cloned in pETM11 vector using the primers described in Table S2 in Supplementary material.
Preparation of recombinant proteins
MAP2c expression and purification was performed as described earlier (1, 85). The same protocol was used for the expression and purification of the phosphorylation mutants and MAP2c-300–467. MAP2c-1–467 with N-terminal histidine tag (H6-MAP2c), MAP2c-1–159, MAP2c-159–254, and MAP2c-250–347 were expressed in Escherichia coli BL21(DE3)RIPL strain overnight at 20 °C and bound to a HisTrap column (GE Healthcare), in 20 mM phosphate buffer, 500 mM NaCl, 20 mM imidazole, and eluted with a 0 to 300 mM imidazole gradient. After cutting of the Histidine tag with TEV, MAP2c-159–254 and MAP2c-250–347 were purified on HiTrap SP HP column (GE Healthcare) in 50 mM acetate, 1 mM MgCl2, 1 mM EDTA and eluted with a gradient of 0 to 500 mM NaCl. MAP2c-1–159 was further purified on HiTrap Q column (GE Healthcare) in 20 mM Tris, pH 8 and eluted with a step gradient of 0 to 500 mM NaCl. After purification on His-Trap column, H6-MAP2c was dialyzed in 50 mM MOPS, purified on HiTrap SP HP column in 50 mM MOPS, 1 mM EDTA, and eluted with a gradient of 0 to 500 mM NaCl. All proteins were dialyzed against NMR buffer consisting of 50 mM MOPS, pH 6.9, 100 mM NaCl, 0.5 mM TCEP. [15N]-MAP2c and [15N,13C]-MAP2c and the fragments were expressed in M9 medium containing [15N]H4Cl and/or [13C6]glucose. ERK2, MEKR, and CDK2/CycA3 were purified as described previously (86, 87).
The SH3 domain of human Abl fused to GST (GST-Abl-SH3) was expressed in E. coli BL21(DE3) overnight at 20 °C. GST-Abl-SH3 was purified on Protino-agarose-4B (Macherey-Nagel) in 20 mM Tris–HCl pH 7.5, 150 mM NaCl, and eluted with 10 mM GSH. The SH3 domain of Fyn fused to hexahistidine tag (Fyn-SH3) was expressed in E. coli BL21(DE3) overnight at 22 °C and purified on HisTrap HP column in 50 mM Tris–HCl, 500 mM NaCl, 20 mM imidazole, and eluted with a 0 to 300 mM imidazole gradient. The eluted fractions were dialyzed in 20 mM Tris–HCl, pH 8 and further purified on HiTrap Q column (GE Healthcare) in 20 mM Tris, pH 8 and eluted with a step gradient of 0 to 500 mM NaCl.
Grb2-C32S/C198A was expressed in BL21(DE3)RIPL overnight at 20 °C and purified on HisTrap HP column in 50 mM Tris–HCl, 500 mM NaCl, 25 mM imidazole, and eluted with a 0 to 500 mM imidazole gradient. The eluted fractions were dialyzed in 20 mM Tris–HCl, 150 mM NaCl, pH 8 and cleaved with thrombin (0.5 U/mg Grb2) overnight at room temperature. Thrombin was removed by purification on benzamidine column (GE Healthcare). Grb2 was further purified on HiTrap Q column in 20 mM Tris–HCl, pH 8.0 and eluted with a 0 to 500 mM NaCl gradient. RIIα-PKA was expressed in BL21(DE3)RIPL overnight at 20 °C and purified on HisTrap HP column in 20 mM sodium phosphate, pH 7.5, 500 mM NaCl, 20 mM imidazole, and eluted with a 0 to 300 mM imidazole gradient. The eluted fractions were dialyzed in 20 mM Tris–HCl, pH 8 overnight at 4 °C. RIIα-PKA was further purified on HiTrap Q column in 20 mM Tris–HCl, pH 8.0 and eluted with a 0 to 500 mM NaCl gradient. ERK2, Fyn-SH3, GST-Abl-SH3, Grb2, and RIIα-PKA were dialyzed in NMR buffer and concentrated using Amicon devices (Millipore) before NMR measurements.
MAP2c phosphorylation
MAP2c was phosphorylated by ERK2 by mixing MAP2c, ERK2, and MEKR at a mass ratio 50:1:1. For CDK2 phosphorylation, 5 μg of CDK2/CycA3 was used per mg of MAP2c. For PKA phosphorylation, 2.5 μg of PKA was used. All phosphorylation reactions were done in NMR buffer containing 10 mM ATP, 10 mM MgCl2, 0.1 mM EDTA, at 27 °C for 20 h. For HNCO and CBCA(CO)NH measurements, MAP2c was purified on HiTrap SP column after phosphorylation and dialyzed in NMR buffer.
NMR spectroscopy
NMR experiments were acquired using 850 and 950 MHz Bruker Avance III spectrometer equipped with TCI cryogenic probe heads with z-axis gradients. All experiments were performed at 27 °C with the temperature calibrated according to the chemical shift differences of pure methanol peaks. The indirect dimensions in 3D experiments were acquired in a nonuniformly sampled manner. On-grid Poisson disk sampling with a Gaussian probability distribution (88) was applied.
The 1H,15N-HSQC (89, 90) spectra were recorded with spectral widths set to 11,904 Hz in the direct dimension and to 2240 Hz in the indirect dimension. Two thousand forty eight and two hundred fifty six complex points were acquired in the direct and the indirect dimensions, respectively.
To follow real-time kinetics, 1H,15N-SOFAST-HMQC (91) spectra were recorded with spectral widths set to 13297 Hz in the direct dimension and to 2500 Hz in the indirect dimension. For phosphorylation of Ser178 by CDK2, 2048 and 128 complex points were acquired in the direct and the indirect dimensions respectively. Sixteen scans with the recycle delay set to 228 ms were recorded in each experiment. For dephosphorylation of [15N]-MAP2c phosphorylated by PKA, 2048 and 64 complex points were acquired in the direct and the indirect dimensions, respectively, and 16 scans were recorded in each experiment. For dephosphorylation of [15N]-MAP2c phosphorylated by ERK2 in the cell extract and [15N]-MAP2c phosphorylation in cell extract, 2048 and 256 complex points were acquired in the direct and the indirect dimensions respectively. Thirty two and sixty four scans were recorded in each experiment for dephosphorylation of MAP2c phosphorylated by ERK and phosphorylation of MAP2c in the cell extract, respectively.
The 3D HNCO (92) spectra were acquired with spectral widths set to 18939 (aq) × 2000 (15N) × 2000 (13C′) Hz and maximal evolution times of 120 ms for 15N and 80 ms for 13C′ indirectly detected dimensions. The overall number of 2048 complex points was acquired in the acquisition dimension and 2000 hypercomplex points were randomly distributed over the indirectly detected dimensions. The 3D CBCA(CO)NH experiment (93) was acquired with spectral widths set to 18939 (aq) × 2500 (15N) × 17921 (13C′) Hz. The number of recorded complex points was 2048, 40, and 128 for 1H, 15N, and 13C dimensions respectively.
2D hnco(CA)NH and hncoCA(N)H FOSY spectra (44) with selective excitation of the probed frequencies were acquired with spectral widths set to 18939 (aq) × 2500 (15N) × 17921 (13C′) Hz. The number of recorded complex points was 2048, 40, and 128 for 1H, 15N, and 13C dimensions respectively.
Uniformly sampled data processing and direct dimension processing of non-uniformly sampled data was done using NMRPipe software (94). The Multidimensional Fourier Transform with iterative algorithm for artifact suppression (95) was employed to process indirect dimensions in three-dimensional experiments. Spectra analysis was done using software Sparky 3.115 (T.D. Goddard and D. G. Kneller, University of California). Peak intensities were extracted from 3D HNCO spectra using NMR pipe.
Phosphorylation kinetics
Real-time phosphorylation by ERK2 and CDK2 was followed by recording 1H,15N-HSQC spectra at 27 °C in 45-min intervals for 20 h after addition of kinase. [15N]-MAP2c, [15N]-MAP2c-1–159, or [15N]-MAP2c-159–254 at 0.1 mM concentration in 50 mM MOPS, pH 6.9, 100 mM NaCl, 0.5 mM TCEP, 10 mM MgCl2, 10 mM ATP, 0.1 mM EDTA were mixed with 77 μg of ERK2 and MEKR and 20 μg CDK2, respectively, directly in the NMR tube. The final volume was 600 μl. The first 1H,15N-HSQC experiment was started 3 min after addition of kinase. Relative concentrations of individual phosphorylation sites were assumed to be proportional to the corresponding peak volumes and kinases were assumed to be saturated by ATP and magnesium. To determine the kinetics of Ser178 phosphorylation by CDK2, we measured 1H,15N-SOFAST-HMQC spectra at 27 °C in 11-min intervals for CDK2 for a total of 15 h. The sum of peak intensities (heights) for a given residue y during fastest phosphorylation was fitted to the equation y = a(e− ln 2(t−Δ)/(τ1/2−Δ)), where the parameter τ1/2 is time when half of the given serine/threonine is phosphorylated.
Priming of MAP2c with CDK2, GSK3β, and PKA
Ten milligrams of [15N]-MAP2c was incubated with 50 μg CDK2, 1 mg GSK3β, and 5 μg PKA in 50 mM MOPS, 100 mM NaCl, 10 mM ATP, 0.1 mM EDTA, 1 mM PMSF, overnight at 27 °C. Phosphorylated [15N]-MAP2c was then purified on HiTrap SP column, dialyzed in NMR buffer and concentrated to 100 μM. Kinetics of phosphorylation by ERK2 was measured as described, using 20-min 1H,15N-SOFAST-HMQC experiments. For comparison of the phosphorylation rates, the kinetics of phosphorylation of [15N]-MAP2c WT was repeated using the same conditions and 20-min 1H,15N-SOFAST-HMQC experiments.
Kinetics of phosphorylation by PKA of MAP2c primed by ERK2
Three milligrams of [15N]-MAP2c was incubated with 60 μg ERK2 and 60 μg MEKR in 50 mM MOPS, 100 mM NaCl, 10 mM ATP, 0.1 mM EDTA, 1 mM PMSF, overnight at 27 °C. Phosphorylated [15N]-MAP2c ([15N]-MAP2c-ERK) was then purified on HiTrap SP column, dialyzed in NMR buffer, and concentrated to 100 μM. Kinetics of phosphorylation of 100 μM [15N]-MAP2c or [15N]-MAP2c-ERK by PKA was measured using 2.5 μg PKA in 50 mM MOPS, 100 mM NaCl, 10 mM ATP, 0.1 mM EDTA at 27 °C using 20-min 1H,15N-SOFAST-HMQC experiments.
Paramagnetic labeling of MAP2c
After purification and phosphorylation, the cysteines were reduced by incubating [15N,13C]-MAP2c with 0.5 mM TCEP. For spin labeling, the buffer was changed to 20 mM potassium phosphate, 150 mM potassium chloride, pH 7.4 using size-exclusion chromatography on PD–10 desalting column (GE Healthcare) and the protein was eluted in a 10-fold molar excess of MTSL or MTS (Toronto Research Chemicals) dissolved in DMSO, and MTSL or MTS was allowed to bind overnight at 4 °C. The protein was then dialyzed into 50 mM MOPS, 150 mM sodium chloride, pH 6.9 before measurement. 3D HNCO spectra were measured for the paramagnetic and diamagnetic MAP2c mutants. The paramagnetic relaxation enhancement profiles were obtained by calculating the ratio of peak intensities of the paramagnetic and diamagnetic samples.
Interaction of MAP2c with ERK2, GST-Abl-SH3, Fyn-SH3, and Grb2 and RIIα-PKA
The interactions of [15N,13C]-MAP2c and [15N,13C]-MAP2c phosphorylated by ERK2 were measured by NMR spectroscopy using 3D HNCO spectra. All proteins were dialyzed in NMR buffer. Titration of ERK2 with MAP2c was done by mixing 100 μM [13C,15N]-MAP2c with 30, 60 and 100 μM ERK2. For interaction studies, 100 μM [15N,13C]-MAP2c was mixed with 200 μM Fyn-SH3, 200 μM GST-Abl-SH3, 100 μM Grb2, or 200 μM RIIα-PKA. The samples containing Grb2 were supplemented with PMSF at the final concentration of 1 mM to prevent cleavage of MAP2c by residual thrombin. The error was calculated from noise.
Microscale thermophoresis
Binding affinities between MAP2c and ERK2, Fyn-SH3, and Abl-SH3 were measured by MST at 27 °C. H6-MAP2c was fluorescently labeled on the histidine tag with RED-tris-NTA (Nanotemper). Titration experiments were performed in a buffer containing 50 mM MOPS, 100 mg/ml NaCl, pH 6.9, 0.5 mg/ml bovine serum albumin, and 0.05% Tween-20. The concentration of H6-MAP2c was 5 nM. Concentration of ERK2, Fyn-SH3, and Abl-SH3 ranges from 1.3 nM to 45 μM, 0.046 nM to 1.5 μM, and 1.8 μM to 533 μM, respectively. Binding studies were performed at 40% laser power for ERK2 and Fyn-SH3 and 20% laser power for Abl-SH3, with a Monolith NT.115 Pico device (NanoTemper Technologies) at medium MST power. The mean values ± SD for each concentration point were calculated from triplicate measurements. The data were fitted to the following model:
where Fnorm = F/Fmax is the normalized fluorescence, c is the total concentration of MAP2c, x is the total concentration of ligand, and KD’ is the apparent dissociation constant.
Dephosphorylation and phosphorylation in SH-SY5Y cell extract
SH-SY5Y human neuroblastoma cells (CRL-2266, ATCC) were grown at 37 °C in humidified 5% (v/v) CO2 incubators in 80% Dulbecco’s modified Eagle’s medium, 20% fetal bovine serum, and 50 μg/ml gentamycin. Cells were split at 70 to 80% confluence and not passaged more than 6 to 12 times prior to NMR experiments. All media, supplements, and PBS were purchased from PAA Laboratories. Cell lines were routinely tested for mycoplasma contaminations and were mycoplasma free. For each NMR experiment, (4–10) × 106 cells were harvested 24 h after final feeding at 70 to 80% confluence. Cells were harvested by scraping them on ice into lysis buffer (20 mM Tris–HCl pH 7.5, 1.5 mM EDTA, 100 mM NaCl, 1% (v/v) Triton-X100, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 2X EDTA-free protease inhibitor cocktail (Cat. nb. 04693159001, Roche), 1.4 μg/ml pepstatin A (Cat. nb. P4265, Sigma Aldrich), and 0.2 mM PMSF). In this buffer, whole cell lysates were vortexed for 5 s and mutated on ice for 30 min followed by centrifugation at 21, 000g for 30 min at 4 °C. The supernatant was separated and total protein concentration was determined by Bradford assay. Aliquots were snap-frozen in liquid nitrogen and stored at −80 °C.
Thirty microliters of 1.1 mM [15N]-MAP2c phosphorylated by PKA was diluted to 100 μM in 141 μl of cell extract at 5.53 mg/ml protein and 114 μl lysis buffer with 5% D2O. Sixty five microliters of [15N]-MAP2c phosphorylated by ERK2 (465 μM) was diluted to 100 μM in 220 μl of cell extract at 3.4 mg/ml protein with 5% D2O. Intensities of the peaks were normalized to intensity of Trp14 side chain and calculated as the decrease of relative intensities. The dephosphorylation rates were modeled using the equation y = a0 + y0e−kt, where y0 is the intensity at t = 0 and a0 is final intensity.
For the phosphorylation in the cell extract, 50 μl [15N]-MAP2c (1.5 mM) was diluted to 200 μM in 220 μl cell extract at 1.89 mg/ml–containing phosphatase inhibitors at doubled concentration (PhosSTOP, Roche), 5 mM ATP, 10 mM MgCl2 and 5% D2O. For reference, we measured the spectra of the phosphorylated or unphosphorylated MAP2c at the same concentration in lysis buffer. The decrease of intensity of Leu467 shows the extent of MAP2c proteolysis over time.
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Petr Padrta, Radka Dopitová, Michal Rečlo, and Veronika Vundrová for their technical help. This work was supported by the Czech Science Foundation, grant GA20-12669S. The authors thank Prof. Isabelle Landrieu (Université de Lille, France) and Dr Enno Klussmann (Max Delbrück Centrum Für Molekulare Medizin, Berlin, Germany) for kindly providing plasmids with the genes of kinases used in this study. CIISB, Instruct-CZ Centre of Instruct-ERIC EU consortium, funded by MEYS CR infrastructure project LM2018127, is gratefully acknowledged for the financial support of the measurements at the Josef Dadok National NMR Centre and at the Proteomics and Biomolecular Interactions and Crystallization CEITEC core facilities.
Author contributions
J. P., S. J., S. N., A. L., and L. Z. conceptualization; J. P., S. J., S. N., A. L., and L. Z. methodology; J. P., S. J., S. N., A. L., M. L., S. M. F., and L. Z. investigation; J. P., S. J., and L. Z. writing–original draft; J. P., S. J., S. N., A. L., M. L., S. M. F., and L. Z. writing–review and editing.
Funding and additional information
S. F. is supported by the DFG RTG 2467 “Intrinsically disordered proteins” (project number 391498659).
Edited by Donita Brady
Supporting information
References
- 1.Gamblin T.C., Nachmanoff K., Halpain S., Williams R.C. Recombinant microtubule-associated protein 2c reduces the dynamic instability of individual mi-crotubules. Biochemistry. 1996;35:12576–12586. doi: 10.1021/bi961135d. [DOI] [PubMed] [Google Scholar]
- 2.Jalava N.S., Lopez-Picon F.R., Kukko-Lukjanov T.K., Holopainen I.E. Changes in microtubule-associated protein-2 (MAP2) expression during development and after status epilepticus in the immature rat hippocampus. Int. J. Dev. Neurosci. 2007;25:121–131. doi: 10.1016/j.ijdevneu.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Dunker A.K., Obradovic Z., Romero P., Garner E.C., Brown C.J. Intrinsic protein disorder in complete genomes, genome informatics. Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 4.Dunker A.K., Oldfield C.J., Meng J., Romero P., Yang J.Y., Chen J.W., et al. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9:S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 6.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 7.Fink A.L. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Melkova K., Zapletal V., Jansen S., Nomilner E., Zachrdla M., Hritz J., et al. Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics. J. Biol. Chem. 2018;293:13297–13309. doi: 10.1074/jbc.RA118.001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dehmelt L., Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2005;6:204. doi: 10.1186/gb-2004-6-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viereck C., Tucker R.P., Matus A. The adult rat olfactory system expresses microtubule-associated proteins found in the developing brain. J. Neurosci. 1989;9:3547–3557. doi: 10.1523/JNEUROSCI.09-10-03547.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Díaz-Nido J., Serrano L., Hernández M.A., Avila J. Phosphorylation of microtubule proteins in rat brain at different developmental stages: comparison with that found in neuronal cultures. J. Neurochem. 1990;54:211–222. doi: 10.1111/j.1471-4159.1990.tb13303.x. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez C., Díaz-Nido J., Avila J. Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function. Prog. Neurobiol. 2000;61:133–168. doi: 10.1016/s0301-0082(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 13.Vallee R. Structure and phosphorylation of microtubule-associated protein 2 (MAP2) Proc. Natl. Acad. Sci. U. S. A. 1980;77:3206–3210. doi: 10.1073/pnas.77.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamauchi T., Fujisawa H. Disassembly of microtubules by the action of calmodulin-dependent protein kinase (kinase II) which occurs only in the brain tissues. Biochem. Biophys. Res. Commun. 1983;110:287–291. doi: 10.1016/0006-291x(83)91293-7. [DOI] [PubMed] [Google Scholar]
- 15.Burns R.G., Islam K., Chapman R. The multiple phosphorylation of the microtubule-associated protein MAP2 controls the MAP2:tubulin interaction. Eur. J. Biochem. 1984;141:609–615. doi: 10.1111/j.1432-1033.1984.tb08236.x. [DOI] [PubMed] [Google Scholar]
- 16.Hoshi M., Akiyama T., Shinohara Y., Miyata Y., Ogawara H., Nishida E., et al. Protein-kinase-C-catalyzed phosphorylation of the microtubule-binding domain of microtubule-associated protein 2 inhibits its ability to induce tubulin polymerization. Eur. J. Biochem. 1988;174:225–230. doi: 10.1111/j.1432-1033.1988.tb14086.x. [DOI] [PubMed] [Google Scholar]
- 17.Ainsztein A.M., Purich D.L. Stimulation of tubulin polymerization by MAP-2. Control by protein kinase C-mediated phosphorylation at specific sites in the microtubule-binding region. J. Biol. Chem. 1994;269:28465–28471. [PubMed] [Google Scholar]
- 18.Illenberger S., Drewes G., Trinczek B., Biernat J., Meyer H.E., Olmsted J.B., et al. Phosphorylation of microtubule-associated proteins MAP2 and MAP4 by the protein kinase p110mark. Phosphorylation sites and regulation of microtubule dynamics. J. Biol. Chem. 1996;271:10834–10843. doi: 10.1074/jbc.271.18.10834. [DOI] [PubMed] [Google Scholar]
- 19.Drewes G., Ebneth A., Mandelkow E.M. MAPs, MARKs and microtubule dynamics. Trends Biochem. Sci. 1998;23:307–311. doi: 10.1016/s0968-0004(98)01245-6. [DOI] [PubMed] [Google Scholar]
- 20.Lim R., Halpain S. Regulated association of microtubule-associated protein 2 (MAP2) with Src and Grb2: evidence for MAP2 as a scaffolding protein. J. Biol. Chem. 2000;275:20578–20587. doi: 10.1074/jbc.M001887200. [DOI] [PubMed] [Google Scholar]
- 21.Jansen S., Melková K., Trošanová Z., Hanáková K., Zachrdla M., Nováček J., et al. Quantitative mapping of microtubule-associated protein 2c (MAP2c) phosphorylation and regulatory protein 14-3-3ζ-binding sites reveals key differences between MAP2c and its homolog Tau. J. Biol. Chem. 2017;292:6715–6727. doi: 10.1074/jbc.M116.771097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Illenberger S., Zheng-Fischhöfer Q., Preuss U., Stamer K., Baumann K., Trinczek B., et al. The endogenous and cell cycle-dependent phosphorylation of tau protein in living cells: implications for Alzheimer’s disease. Mol. Biol. Cell. 1998;9:1495–1512. doi: 10.1091/mbc.9.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avila J., Domínguez J., Díaz-Nido J. Regulation of microtubule dynamics by microtubule-associated protein expression and phosphorylation during neuronal development. Int. J. Dev. Biol. 1994;38:13–25. [PubMed] [Google Scholar]
- 24.Drewes G., Ebneth A., Preuss U., Mandelkow E.-M., Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 25.Brandt R., Lee G., Teplow D.B., Shalloway D., Abdel-Ghany M. Differential effect of phosphorylation and substrate modulation on tau’s ability to promote microtubule growth and nucleation. J. Biol. Chem. 1994;269:11776–11782. [PubMed] [Google Scholar]
- 26.Itoh T.J., Hisanaga S., Hosoi T., Kishimoto T., Hotani H. Phosphorylation states of microtubule-associated protein 2 (MAP2) determine the regulatory role of MAP2 in microtubule dynamics. Biochemistry. 1997;36:12574–12582. doi: 10.1021/bi962606z. [DOI] [PubMed] [Google Scholar]
- 27.Landrieu I., Lacosse L., Leroy A., Wieruszeski J.M., Trivelli X., Sillen A., et al. NMR analysis of a Tau phosphorylation pattern. J. Am. Chem. Soc. 2006;128:3575–3583. doi: 10.1021/ja054656+. [DOI] [PubMed] [Google Scholar]
- 28.Drewes G., Lichtenberg-Kraag B., Döring F., Mandelkow E.M., Biernat J., Goris J., et al. Mitogen activated protein (MAP) kinase transforms tau protein into an Alzheimer-like state. EMBO J. 1992;11:2131–2138. doi: 10.1002/j.1460-2075.1992.tb05272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumann K., Mandelkow E.-M., Biernat J., Piwnica-Worms H., Mandelkow E. Abnormal alzheimer-like phosphorylation of tau-protein by cyclin-dependent kinases cdk2 and cdk5. FEBS Lett. 1993;336:417–424. doi: 10.1016/0014-5793(93)80849-p. [DOI] [PubMed] [Google Scholar]
- 30.Biernat J., Mandelkow E.M., Schröter C., Lichtenberg-Kraag B., Steiner B., Berling B., et al. The switch of tau protein to an Alzheimer-like state includes the phosphorylation of two serine-proline motifs upstream of the microtubule binding region. EMBO J. 1992;11:1593–1597. doi: 10.1002/j.1460-2075.1992.tb05204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bielska A.A., Zondlo N.J. Hyperphosphorylation of tau induces local polyproline II helix. Biochemistry. 2006;45:5527–5537. doi: 10.1021/bi052662c. [DOI] [PubMed] [Google Scholar]
- 32.Gandhi N.S., Landrieu I., Byrne C., Kukic P., Amniai L., Cantrelle F.-X., et al. A phosphorylation-induced turn defines the Alzheimer’s disease AT8 antibody epitope on the tau protein. Angew. Chem. Int. Ed. Engl. 2015;54:6819–6823. doi: 10.1002/anie.201501898. [DOI] [PubMed] [Google Scholar]
- 33.Despres C., Byrne C., Qi H., Cantrelle F.-X., Huvent I., Chambraud B., et al. Identification of the Tau phosphorylation pattern that drives its aggregation. Proc. Natl. Acad. Sci. U. S. A. 2017;114:9080–9085. doi: 10.1073/pnas.1708448114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi H., Prabakaran S., Cantrelle F.-X., Chambraud B., Gunawardena J., Lippens G., et al. Characterization of neuronal tau protein as a target of extracellular signal-regulated kinase. J. Biol. Chem. 2016;291:7742–7753. doi: 10.1074/jbc.M115.700914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amniai L., Barbier P., Sillen A., Wieruszeski J.-M., Peyrot V., Lippens G., et al. Alzheimer disease specific phosphoepitopes of Tau interfere with as-sembly of tubulin but not binding to microtubules. FASEB J. 2009;23:1146–1152. doi: 10.1096/fj.08-121590. [DOI] [PubMed] [Google Scholar]
- 36.Berling B., Wille H., Röll B., Mandelkow E.M., Garner C., Mandelkow E. Phosphorylation of microtubule-associated proteins MAP2a,b and MAP2c at Ser136 by proline-directed kinases in vivo and in vitro. Eur. J. Cell Biol. 1994;64:120–130. [PubMed] [Google Scholar]
- 37.Hoshi M., Ohta K., Gotoh Y., Mori A., Murofushi H., Sakai H., et al. Mitogen-activated-protein-kinase-catalyzed phosphorylation of microtubule-associated proteins, microtubule-associated protein 2 and microtubule-associated protein 4, induces an alteration in their function. Eur. J. Biochem. 1992;203:43–52. doi: 10.1111/j.1432-1033.1992.tb19825.x. [DOI] [PubMed] [Google Scholar]
- 38.Ozer R.S., Halpain S. Phosphorylation-dependent localization of microtubule-associated protein MAP2c to the actin cytoskeleton. Mol. Biol. Cell. 2000;11:3573–3587. doi: 10.1091/mbc.11.10.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theillet F.-X., Smet-Nocca C., Liokatis S., Thongwichian R., Kosten J., Yoon M.-K., et al. Cell signaling, post-translational protein modifications and nmr spectroscopy. J. Biomol. NMR. 2012;54:217–236. doi: 10.1007/s10858-012-9674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theillet F.-X., Rose H.M., Liokatis S., Binolfi A., Thongwichian R., Stuiver M., et al. Site-specific NMR mapping and time-resolved monitoring of serine and threonine phosphorylation in reconstituted kinase reactions and mammalian cell extracts. Nat. Protoc. 2013;8:1416–1432. doi: 10.1038/nprot.2013.083. [DOI] [PubMed] [Google Scholar]
- 41.Rose H.M., Stuiver M., Thongwichian R., Theillet F.-X., Feller S.M., Selenko P. Quantitative nmr analysis of erk activity and inhibition by u0126 in a panel of patient-derived colorectal cancer cell lines. Biochim. Biophys. Acta. 2013;1834:1396–1401. doi: 10.1016/j.bbapap.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 42.Bardwell A.J., Bardwell L. Two hydrophobic residues can determine the specificity of mitogen-activated protein kinase docking interactions. J. Biol. Chem. 2015;290:26661–26674. doi: 10.1074/jbc.M115.691436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melková K., Zapletal V., Narasimhan S., Jansen S., Hritz J., Škrabana R., et al. Structure and func-tions of microtubule associated proteins tau and MAP2c: similarities and differ-ences. Biomolecules. 2019;9:105. doi: 10.3390/biom9030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lesovoy D.M., Georgoulia P.S., Diercks T., Matečko-Burmann I., Burmann B.M., Bocharov E.V., et al. Unambiguous tracking of protein phosphorylation by fast high-resolution FOSY NMR. Angew. Chem. Int. Ed. Engl. 2021;60:23540–23544. doi: 10.1002/anie.202102758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alderson T.R., Benesch J.L.P., Baldwin A.J. Proline isomerization in the C-terminal region of HSP27. Cell Stress Chaperones. 2017;22:639–651. doi: 10.1007/s12192-017-0791-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kadavath H., Hofele R.V., Biernat J., Kumar S., Tepper K., Urlaub H., et al. Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc. Natl. Acad. Sci. U. S. A. 2015;112:7501–7506. doi: 10.1073/pnas.1504081112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwalbe M., Kadavath H., Biernat J., Ozenne V., Blackledge M., Mandelkow E., et al. Structural impact of tau phosphorylation at threonine 231. Structure. 2015;23:1448–1458. doi: 10.1016/j.str.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Li T., Paudel H.K. Glycogen synthase kinase 3β phosphorylates Alzheimer’s disease-specific Ser396 of microtubule-associated protein tau by a sequential mechanism. Biochemistry. 2006;45:3125–3133. doi: 10.1021/bi051634r. [DOI] [PubMed] [Google Scholar]
- 49.Sandhu P., Naeem M.M., Lu C., Kumarathasan P., Gomes J., Basak A. Ser 422 phosphorylation blocks human Tau cleavage by caspase-3: biochemical implications to Alzheimer’s disease. Bioorg. Med. Chem. Lett. 2017;27:642–652. doi: 10.1016/j.bmcl.2016.11.087. [DOI] [PubMed] [Google Scholar]
- 50.Fifre A., Sponne I., Koziel V., Kriem B., Potin F.T.Y., Bihain B.E., et al. Microtubule-associated protein MAP1A, MAP1B, and MAP2 proteolysis during soluble amyloid β-peptide-induced neuronal apoptosis: synergistic involvement of calpain and caspase-3. J. Biol. Chem. 2006;281:229–240. doi: 10.1074/jbc.M507378200. [DOI] [PubMed] [Google Scholar]
- 51.Berry R.W., Abraha A., Lagalwar S., LaPointe N., Gamblin T.C., Cryns V.L., et al. Inhibition of tau polymerization by its carboxy-terminal caspase cleavage fragment. Biochemistry. 2003;42:8325–8331. doi: 10.1021/bi027348m. [DOI] [PubMed] [Google Scholar]
- 52.Safaei J., Maňuch J., Gupta A., Stacho L., Pelech S. Prediction of 492 human protein kinase substrate specificities. Proteome Sci. 2011;9:1–13. doi: 10.1186/1477-5956-9-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hashiguchi M., Hashiguchi T. Kinase-kinase interaction and modulation of tau phosphorylation. Int. Rev. Cell Mol. Biol. 2013;300:121–160. doi: 10.1016/B978-0-12-405210-9.00004-7. [DOI] [PubMed] [Google Scholar]
- 54.Rubino H.M., Dammerman B., Shafit-Zagardo B., J E. Localization and characterization of the binding site for the regulatory subunit of type II cAMP-dependent protein kinase on MAP2. Neuron. 1998;3:631–638. doi: 10.1016/0896-6273(89)90273-0. [DOI] [PubMed] [Google Scholar]
- 55.Ren R., Mayer B.J., Cicchetti P., Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 56.Chardin P., Cussac D., Maignan S., Ducruix A. The Grb2 adaptor. FEBS Lett. 1995;369:47–51. doi: 10.1016/0014-5793(95)00578-w. [DOI] [PubMed] [Google Scholar]
- 57.Alexa A., Schmidt G., Tompa P., Ogueta S., Vázquez J., Kulcsár P., et al. The phosphorylation state of threonine-220, a uniquely phosphatase-sensitive protein kinase A site in microtubule-associated pro-tein MAP2c, regulates microtubule binding and stability. Biochemistry. 1992;41:12427–12435. doi: 10.1021/bi025916s. [DOI] [PubMed] [Google Scholar]
- 58.Reszka A.A., Seger R., Diltz C.D., Krebs E.G., Fischer E.H. Association of mitogen-activated protein kinase with the microtubule cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8881–8885. doi: 10.1073/pnas.92.19.8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pei J.-J., Braak H., An W.-L., Winblad B., Cowburn R.F., Iqbal K., et al. Up-regulation of mitogen-activated protein kinases ERK1/2 and MEK1/2 is associated with the progression of neurofibrillary degeneration in Alzheimer’s disease. Brain Res. Mol. Brain Res. 2002;109:45–55. doi: 10.1016/s0169-328x(02)00488-6. [DOI] [PubMed] [Google Scholar]
- 60.Bhat K. Cell-cell signaling during neurogenesis: some answers and many ques-tions. Int. J. Dev. Biol. 1998;42:127–139. [PubMed] [Google Scholar]
- 61.Edbauer D., Cheng D., Batterton M.N., Wang C.-F., Duong D.M., Yaffe M.B., et al. Identification and characterization of neuronal mitogen-activated protein kinase substrates using a specific phosphomotif antibody. Mol. Cell. Proteomics. 2009;8:681–695. doi: 10.1074/mcp.M800233-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sibille N., Huvent I., Fauquant C., Verdegem D., Amniai L., Leroy A., et al. Structural characterization by nuclear magnetic resonance of the impact of phosphorylation in the proline-rich region of the disordered tau protein. Proteins: Struct. Funct. Bioinform. 2012;80:454–462. doi: 10.1002/prot.23210. [DOI] [PubMed] [Google Scholar]
- 63.Sánchez C., Tompa P., Szücs K., Friedrich P., Avila J. Phosphorylation and dephosphorylation in the proline-rich c-terminal domain of microtubule-associated protein 2. Eur. J. Biochem. 1996;241:765–771. doi: 10.1111/j.1432-1033.1996.00765.x. [DOI] [PubMed] [Google Scholar]
- 64.Lu P.J., Wulf G., Zhou X.Z., Davies P., Lu K.P. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- 65.Vincent I., Rosado M., Davies P. Mitotic mechanisms in Alzheimer’s disease? J. Cell Biol. 1996;132:413–425. doi: 10.1083/jcb.132.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jicha G.A., Lane E., Vincent I., Otvos L., Jr., Hoffmann R., Davies P. A conformation- and phosphorylation-dependent antibody recognizing the paired he-lical filaments of Alzheimer’s disease. J. Neurochem. 1997;69:2087–2095. doi: 10.1046/j.1471-4159.1997.69052087.x. [DOI] [PubMed] [Google Scholar]
- 67.Ebneth A., Drewes G., Mandelkow E.M., Mandelkow E. Phosphorylation of MAP2c and MAP4 by MARK kinases leads to the destabilization of microtubules in cells. Cell Motil. Cytoskeleton. 1999;44:209–224. doi: 10.1002/(SICI)1097-0169(199911)44:3<209::AID-CM6>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 68.Sánchez C., Pérez M., Avila J. GSK3 beta-mediated phosphorylation of the microtubule-associated protein 2C (MAP2C) prevents microtubule bundling. Eur. J. Cell Biol. 2000;79:252–260. doi: 10.1078/s0171-9335(04)70028-x. [DOI] [PubMed] [Google Scholar]
- 69.Philpot B.D., Lim J.H., Halpain S., Brunjes P.C. Experience-dependent modifications in MAP2 phosphorylation in rat olfactory bulb. J. Neurosci. 1997;17:9596–9604. doi: 10.1523/JNEUROSCI.17-24-09596.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woolf N.J., Zinnerman M.D., Johnson G.V.W. Hippocampal microtubule-associated protein-2 alterations with contextual memory. Brain Res. 1999;821:241–249. doi: 10.1016/s0006-8993(99)01064-1. [DOI] [PubMed] [Google Scholar]
- 71.Kim Y., Jang Y.-N., Kim J.-Y., Kim N., Noh S., Kim H., et al. Microtubule-associated protein 2 mediates induction of long-term potentiation in hippocampal neurons. FASEB J. 2020;34:6965–6983. doi: 10.1096/fj.201902122RR. [DOI] [PubMed] [Google Scholar]
- 72.Díaz-Hernández M., Gómez-Ramos A., Rubio A., Gómez-Villafuertes R., Naranjo J.R., Miras-Portugal M.T., et al. Tissue-nonspecific alkaline phos-phatase promotes the neurotoxicity effect of extracellular tau. J. Biol. Chem. 2010;285:32539–32548. doi: 10.1074/jbc.M110.145003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cotter D., Kerwin R., Doshi B., Martin C.S., Everall I.P. Alterations in hippocampal non-phosphorylated MAP2 protein expression in schizophrenia. Brain Res. 1997;765:238–246. doi: 10.1016/s0006-8993(97)00575-1. [DOI] [PubMed] [Google Scholar]
- 74.Grubisha M.J., Sun X., MacDonald M.L., Garver M., Sun Z., Paris K.A., et al. MAP2 is differentially phosphorylated in schizophrenia, altering its function. Mol. Psych. 2021;26:5371–5388. doi: 10.1038/s41380-021-01034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richter-Landsberg C. The cytoskeleton in oligodendrocytes. J. Mol. Neurosci. 2008;35:55–63. doi: 10.1007/s12031-007-9017-7. [DOI] [PubMed] [Google Scholar]
- 76.Zamora-Leon S.P., Lee G., Davies P., Shafit-Zagardo B. Binding of Fyn to MAP-2c through an SH3 binding domain. Regulation of the interaction by ERK2. J. Biol. Chem. 2001;276:39950–39958. doi: 10.1074/jbc.M107807200. [DOI] [PubMed] [Google Scholar]
- 77.Jones S.B., Lu H.Y., Lu Q. Abl tyrosine kinase promotes dendrogenesis by inducing actin cytoskeletal rearrangements in cooperation with rho family small gtpases in hippocampal neurons. J. Neurosci. 2004;24:8510–8521. doi: 10.1523/JNEUROSCI.1264-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pisabarro M.T., Serrano L., Wilmanns M. Crystal structure of the Abl-SH3 domain complexed with a designed high-affinity peptide ligand: implications for SH3-ligand interactions. J. Mol. Biol. 1998;281:513–521. doi: 10.1006/jmbi.1998.1932. [DOI] [PubMed] [Google Scholar]
- 79.Zamora-Leon S.P., Bresnick A., Backer J.M., Shafit-Zagardo B. Fyn phos-phorylates human map-2c on tyrosine 67. J. Biol. Chem. 2005;280:1962–1970. doi: 10.1074/jbc.M411380200. [DOI] [PubMed] [Google Scholar]
- 80.Zheng L., Baumann U., Reymond J.-L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucl. Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Houtman J.C.D., Yamaguchi H., Barda-Saad M., Braiman A., Bowden B., Ap-pella E., et al. Oligomerization of signaling complexes by the multipoint binding of GRB2 to both LAT and SOS1. Nat. Struct. Mol. Biol. 2006;13:798–805. doi: 10.1038/nsmb1133. [DOI] [PubMed] [Google Scholar]
- 82.Yuzawa S., Yokochi M., Hatanaka H., Ogura K., Kataoka M., Miura K.-i., et al. Solution structure of Grb2 reveals extensive flexibility necessary for target recognition. J. Mol. Biol. 2001;306:527–537. doi: 10.1006/jmbi.2000.4396. [DOI] [PubMed] [Google Scholar]
- 83.Wenzel J., Sanzenbacher R., Ghadimi M., Lewitzky M., Zhou Q., Kaplan D.R., et al. Multiple interactions of the cytosolic polyproline region of the CD95 ligand: hints for the reverse signal transduction capacity of a death factor. FEBS Lett. 2001;509:255–262. doi: 10.1016/s0014-5793(01)03174-x. [DOI] [PubMed] [Google Scholar]
- 84.Mariotti A., Kedeshian P.A., Dans M., Curatola A.M., Gagnoux-Palacios L., Giancotti F.G. Egf-r signaling through fyn kinase disrupts the function of integrin α6β4 at hemidesmosomes: role in epithelial cell migration and carcinoma invasion. J. Cell Biol. 2001;155:447–458. doi: 10.1083/jcb.200105017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nováček J., Janda L., Dopitová R., Žídek L., Sklenář V. Efficient protocol for backbone and side-chain assignments of large, intrinsically disordered proteins: transient secondary structure analysis of 49.2 kDa microtubule associated protein 2c. J. Biomol. NMR. 2013;56:291–301. doi: 10.1007/s10858-013-9761-7. [DOI] [PubMed] [Google Scholar]
- 86.Qi H., Despres C., Prabakaran S., Cantrelle F.-X., Chambraud B., Gunawardena J., et al. The study of posttranslational mod-ifications of tau protein by nuclear magnetic resonance spectroscopy: phospho-rylation of tau protein by ERK2 recombinant kinase and rat brain extract, and acetylation by recombinant creb-binding protein. Met. Mol. Biol. 2017;1523:179–213. doi: 10.1007/978-1-4939-6598-4_11. [DOI] [PubMed] [Google Scholar]
- 87.Welburn J., Endicott J. Methods for preparation of proteins and protein complexes that regulate the eukaryotic cell cycle for structural studies. Met. Mol. Biol. 2005;296:219–235. doi: 10.1385/1-59259-857-9:219. [DOI] [PubMed] [Google Scholar]
- 88.Kazimierczuk K., Zawadzka A., Koźmiński W. Optimization of random time domain sampling in multidimensional NMR. J. Magn. Reson. 2008;192:123–130. doi: 10.1016/j.jmr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 89.Bodenhausen G., Ruben D.J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980;69:185–189. [Google Scholar]
- 90.Sklenar V., Piotto M., Leppik R., Saudek V. Gradient-tailored water suppres-sion for 1H-15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. 1993;102:241–245. [Google Scholar]
- 91.Schanda P., Brutscher B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of second. J. Am. Chem. Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 92.Kay L.E., Ikura M., Tschudin R., Bax A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J. Magn. Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 93.Grzesiek S., Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 1992;114:6291–6293. [Google Scholar]
- 94.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 95.Stanek J., Koźmiński W. Iterative algorithm of discrete Fourier transform for processing randomly sampled NMR data sets. J. Biomol. NMR. 2010;47:65–77. doi: 10.1007/s10858-010-9411-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.