Abstract
Nucleophosmin 1 (NPM1) is a nucleus-cytoplasmic shuttling protein which is predominantly located in the nucleolus and exerts multiple functions, including regulation of centrosome duplication, ribosome biogenesis and export, histone assembly, maintenance of genomic stability and response to nucleolar stress. NPM1 mutations are the most common genetic alteration in acute myeloid leukemia (AML), detected in about 30–35% of adult AML and more than 50% of AML with normal karyotype. Because of its peculiar molecular and clinico-pathological features, including aberrant cytoplasmic dislocation of the NPM1 mutant and wild-type proteins, lack of involvement in driving clonal hematopoiesis, mutual exclusion with recurrent cytogenetic abnormalities, association with unique gene expression and micro-RNA profiles and high stability at relapse, NPM1-mutated AML is regarded as a distinct genetic entity in the World Health Organization (WHO) classification of hematopoietic malignancies. Starting from the structure and functions of NPM1, we provide an overview of the potential targeted therapies against NPM1-mutated AML and discuss strategies aimed at interfering with the oligomerization (compound NSC348884) and the abnormal traffic of NPM1 (avrainvillamide, XPO1 inhibitors) as well as at inducing selective NPM1-mutant protein degradation (ATRA/ATO, deguelin, (-)-epigallocatechin-3-gallate, imidazoquinoxaline derivatives) and at targeting the integrity of nucleolar structure (actinomycin D). We also discuss the current therapeutic results obtained in NPM1-mutated AML with the BCL-2 inhibitor venetoclax and the preliminary clinical results using menin inhibitors targeting HOX/MEIS1 expression. Finally, we review various immunotherapeutic approaches in NPM1-mutated AML, including immune check-point inhibitors, CAR and TCR T-cell-based therapies against neoantigens created by the NPM1 mutations.
Subject terms: Acute myeloid leukaemia, Drug development
Introduction
Nucleophosmin 1 (NPM1) is a ubiquitous nucleus-cytoplasmic shuttling protein [1] predominantly resident in the nucleolus whose functions are pivotal for many cellular processes including histone assembly, centrosome duplication, ribosome biogenesis and export, maintenance of genomic stability and response to nucleolar stress [2].
Mutations of NPM1 gene are the most frequent genetic lesion in acute myeloid leukemia (AML), being detectable in about one-third of adult AML and 50–60% of AML with normal karyotype [3, 4]. These mutations are a driver genetic lesion and AML defining event that occurs in the context of clonal hematopoiesis, frequently promoted by genes such as DNMT3A and TET2 [4]. Distinctive features of NPM1-mutated AML include the mutual exclusion with recurrent cytogenetic abnormalities, the association with specific gene expression [5] and microRNA [6] profiles and the high stability of NPM1 mutations at relapse [7]. Another characteristic of NPM1-mutated AML is the aberrant cytoplasmic dislocation of the NPM1 mutant and NPM1 wild-type proteins (through heterodimerization) [3, 8, 9]. Moreover, the mutant NPM1 is directly involved in promoting high expression of homeobox (HOX) genes [10] which are necessary for maintaining the undifferentiated state of leukemic cells. Notably, this function is closely dependent on the cytoplasmic localization of the mutant. However, the mechanisms underlying leukemogenesis in NPM1-mutated AML still remain largely unknown [4]. According to the 5th edition of the World Health Organization (WHO) of hematolymphoid tumors, NPM1-mutated AML can be diagnosed irrespective of the percentage of blasts, based on previous observations that cases classified as MDS or MDS/MPN with NPM1 mutations quickly progressed to AML [11]. Because of the above unique features, NPM1-mutated AML is recognized as a distinct entity, within the category of AML with recurrent genetic abnormalities of the WHO classification [11]. The main characteristics of NPM1-mutated AML are summarized in Table 1.
Table 1.
Clinical, pathological and molecular features of NPM1-mutated AML.
| Main characteristics of NPM1-mutated AML |
|---|
| About 30–35% of adult AML (50–60% of AML with normal karyotype). Female predominance |
| Markedly hypercellular bone marrow. Rare fibrosis. Frequent myelomonocytic (FAB M4) or monocytic (FAB M5) appearance but other FAB categories (except M7) can be represented |
| Frequent multilineage involvement, as shown by IHC (cytoplasmic NPM1) |
| Diagnosis can be done irrespective of the percentage of blast cellsa |
| Low/moderate WBC count in the absence of FLT3-ITD. Progressively increase in WBC when concomitant FLT3-ITD and/or DNMT3A mutations are present |
| Frequent extramedullary involvement, especially skin (easily detectable by IHC) |
| No/low expression of CD34 in the bulk leukemic population. The rare CD34+ leukemic stem cells harbor the NPM1 mutation |
| Excellent response to induction chemotherapy |
| Relatively good outcome in the absence of FLT3-ITD. Prognosis may vary depending upon concomitant mutations |
| Amenable for MRD monitoring by qRT-PCR for NPM1 mutant transcripts |
“AML with cytoplasmic nucleophosmin” (NPM1c) has been also used as a synonym of NPM1-mutated AML.
IHC immunohistochemistry, WBC white blood cell count, MRD measurable residual disease, qRT-PCR quantitative reverse transcription polymerase chain reaction.
aAccording to the 5th edition of WHO classification [11].
The standard therapy of NPM1-mutated AML in young adults is based upon induction chemotherapy (±FLT3 inhibitors) and consolidation cycles with high/intermediate dose of cytarabine (ARA-C) ± allogeneic hematopoietic stem cell transplantation (HSCT) in first complete remission (CR), depending on the status of the FLT3 gene and measurable residual disease-MRD [12, 13]. However, despite the remarkable advances in the treatment of NPM1-mutated AML, about 50% of patients still died of progressive disease. Thus, there is a need for new therapeutic opportunities. Whole genomic approaches have unraveled the molecular heterogeneity of AML [14, 15] and given a great input to the development of small molecules aimed to target specific genetic abnormalities [16, 17]. Because of their small-size (<500 Da), these compounds can easily penetrate the cell membrane and exert their activity on intracellular proteins involved in cell signaling mechanisms (e.g., kinases) that promote the tumor growth [18]. In the past 5 years, several small molecules have been approved by the Food and Drug Administration (FDA) for AML treatment, including FLT3, IDH1/2 and BCL-2 inhibitors [19, 20]. Contemporarily, the use of immunotherapy as an additional therapeutic strategy has been explored.
Here, we provide an overview of the current status and future perspectives of targeted therapies in NPM1-mutated AML.
Molecular therapeutic targeting of NPM1-mutated AML
Targeting the structure of NPM1
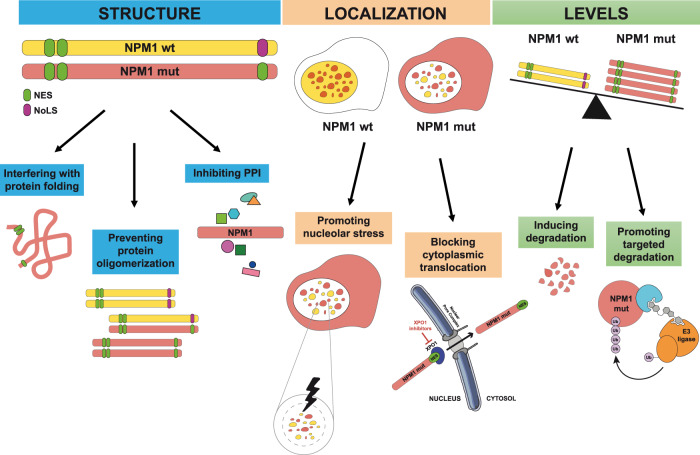
Small molecules can be potentially designed to bind to any portion of a given protein to modulate its function [21]. The multi-domain conformation of NPM1 makes it an appealing target. In fact, NPM1 consists of three domains: the amino-terminal core region (N-term) similar to the members of the nucleophosmin family; the central region involved in the histone binding and the carbossi-terminal region (C-term) unique for NPM1 protein, that is important for its nucleolar localization [2, 22].
The N-term domain contains two nuclear export signals (NES) that promote the shuttling of NPM1 from the nucleus to the cytoplasm through the interaction with the nuclear exporter XPO1 (CRM1, Exportin-1) [9]. The N-term region has also chaperone activity which is crucial for ribosome maturation and oligomerization of NPM1 with other partners and itself [23]. The three-dimensional structure of the N-term consists of eight antiparallel β-strands forming a β-barrel with jelly-roll topology [24]. NPM1 monomers are unfold and intrinsically unstable [23]. However, they are stabilized through formation of a crown-shaped pentamer, with two pentamers interacting in a head-to-head fashion to form a decamer [25]. Association into pentamers is strictly regulated and it is critical for NPM1 functions.
Therefore, disruption of NPM1 oligomers may promote unfolding of the protein and alter NPM1 structure and functions. Targeting NPM1 oligomerization can be achieved using NSC348884, a water-insoluble compound that interacts with the high hydrophobic NPM1 dimerization surface [26]. NSC348884 was more effective in disrupting NPM1 oligomerization and inducing apoptosis in NPM1-mutated OCI-AML3 cells than in NPM1 wild-type HL-60 and OCI-AML2 cells [27] (Fig. 1). This is somewhat surprising because NPM1 mutations target the C-terminal domain whilst NSC348884 targets the N-terminal domain.
Fig. 1. Targeting the structure, localization and levels of the wild-type and mutant NPM1 proteins.
Mechanisms of targeting include: interference with protein folding, prevention of NPM1 oligomerization, inhibition of protein–protein interactions (PPIs), promotion of nucleolar stress, block of nuclear export, and induction of protein degradation.
The N-term region is critical for the interaction of NPM1 with its partners through positively charged surface. These molecular interactions are critical to build-up the structure of the nucleolus [28, 29]. In fact, the nucleolus is the result a “liquid-liquid” phase separation leading to the segregation of NPM1 and other components of the nucleolus from the surrounding nucleoplasm [30, 31], similarly to how oil and water separate from each other when mixed. This process is mediated by the interaction of the native pentameric NPM1 molecule with proteins containing R-motifs, i.e., multivalent arginine-rich linear motifs (sharing features with nucleolar localization signals) and nascent ribosomal RNA (rRNA) [29, 32]. Distruption of NPM1 oligomers can interfere with this process.
Many NPM1 interactors that are thought to be involved in leukemogenesis have been claimed to be dislocated in the cytoplasm by the NPM1 mutant [9], although this event has been never conclusively demonstrated in primary NPM1-mutated AML cells [4]. Thus, small molecules targeting NPM1 protein–protein interactions could also help preventing downstream effects on important molecules potentially implicated in AML pathogenesis.
The NPM1 C-terminus consists of a three-helix bundle [33] stabilized by strictly conserved aromatic residues (Phe268, Phe276, Trp288, Trp290) [25], and contains the nucleolar localization signal (NoLS) [25]. This aromatic-rich NoLS seems to be rather specific for NPM1. Mutations of tryptophans 288 and 290 (or 290 only) cause unfolding of the three-helix bundle and loss of NPM1 NoLS [9]. This event together with the insertion of a new NES motif, are responsible for the aberrant delocalization of NPM1 in the cytoplasm of AML cells carrying NPM1 mutations [8, 9].
The C-term of NPM1 mutated protein is unfolded and therefore difficult to target [24, 25]. The only small molecule capable of directly binding this region is avrainvillamide. This natural alkaloid was found to form tight complexes with the NPM1 protein by S-alkylation of cysteine residues [34, 35] and to induce partial nuclear relocalization of mutant NPM1 in OCI-AML3 cells and primary AML cells. This effect was secondary to the ability of avrainvillamide to alkylate specifically Cys275 (in helix H2 of the three-helix bundle at C-terminus) of certain NPM1 mutants [34] and to inhibit the nuclear export of XPO1 cargo proteins, including NPM1 mutants [34]. Thus, avrainvillamide can relocate cytoplasmic NPM1, acting as a substitute for NoLS [34] rather than inducing refolding of the C-terminus three-helix structure of the mutant [34] (Fig. 1). Avrainvillamide was more active against NPM1-mutated than wild-type AML cells [36], probably due to the unfolded structure of the C-terminus three-helix bundle, and caused proteasomal degradation of NPM1 mutant and differentiation of OCI-AML3 cells [36]. Moreover, it demonstrated strong anti-proliferative activity against a PDX model of NPM1-mutated AML [36].
Targeting the nucleolus of NPM1-mutated cells
NPM1 acts as a nucleolar stress sensor [37]. The nucleolus of NPM1-mutated AML cells may be particularly vulnerable to stress because it is NPM1 depleted due to both haploinsufficiency and cytoplasmic delocalization [38]. Therefore, inducing nucleolar stress represents a therapeutic option in NPM1-mutated AML [39] (Fig. 1). This prompted us to evaluate actinomycin D which, at low dosage, is a selective inhibitor of RNA polymerase I and ribosome biogenesis. This drug induced nucleolar stress in NPM1-mutated cells [40, 41] and CR in relapsed/refractory NPM1-mutated AML [38, 40]. Other drugs causing nucleolar stress through inhibition of ribosome biogenesis [42] should be investigated in NPM1-mutated AML.
However, the mechanism of action of actinomycin D in NPM1-mutated AML may be even more complex. In fact, NPM1 mutants can impede the formation of acute promyelocytic leukemia (APL) nuclear bodies (NBs) [43], which in turn are regulators of mitochondria fitness and key senescence effectors [44]. Thus, NPM1-mutated AML cells are characterized by defective mitochondrial function [44]. Actinomycin D acts on NPM1 mutant-primed mitochondria by releasing mitochondrial DNA, activating cyclic GMP-AMP synthase signaling, producing reactive-oxygen species (ROS) that restores NB formation to drive TP53 activation and senescence of NPM1-mutated AML cells. Interestingly, actinomycin D appears to potentiate the activity of venetoclax on mitochondrial function and apoptosis [44].
Targeting NPM1 protein levels
Targeting the levels of mutant NPM1 represents another therapeutic opportunity (Fig. 1). Combining arsenic trioxide (ATO) with all-trans retinoic acid (ATRA) resulted into proteasome-dependent degradation of NPM1 oncoprotein and death in AML cell lines and primary AML cells carrying NPM1 mutations [43, 45]. Treatment of the NPM1-mutated AML cell lines OCI-AML3 with deguelin, a rotenoid isolated from several plant species [46, 47], and IMS-M2 with (-)-epigallocatechin-3-gallate (ECGT) [48], a major catechin found in green tea, were effective in reducing the NPM1 mutant but not the wild-type protein and in inducing apoptosis. Moreover, the imidazoquinoxaline derivative EAPB0503 induced a selective proteasome-mediated degradation of NPM1 mutant protein through EAPB0503-mediated SUMOylation and ubiquitylation [49]. This event was followed by restoration of NPM1 wild-type protein in the nucleolus [50], apoptosis (through selective downregulation of HDM2 and activation of p53 [49]) and reduction of leukemia burden in NPM1-mutated AML xenografts [49, 50]. Moreover, introducing NPM1-mutation into cells normally bearing wild-type NPM1 sensitized them to EAPB0503, leading to their growth arrest [50].
Mutant NPM1 can be also targeted through proteolysis targeting chimera (PROTAC) which allows ubiquitination and proteasome-mediated degradation of the target protein [51] (Fig. 1). PROTAC promoted degradation of fused oncoproteins in MLL leukemia subtypes [52] and its use could be extended to NPM1-mutated AML. In this regard, selective degradation of mutant NPM1 through degron-tag, invariably induced differentiation, and growth arrest of NPM1-mutated cell lines [10], confirming the validity of such approach.
Targeting NPM1 localization
The multilayered nucleolar structure (the fibrillar core, surrounded by the dense fibrillar component and the granular component) is dictated by the “liquid-liquid” phase separation involving nucleolar proteins, primarily native NPM1 and fibrillarin that segregate in the granular component and dense fibrillar component, respectively [30]. NPM1 is predominantly localized in the nucleolus, and shuttles to the cytoplasm as a consequence of the interaction of the two N-terminal NES with XPO1. XPO1 recognizes and binds also other NES-containing molecules including tumor-suppressor proteins [53, 54], promoting their export from the nucleus to the cytoplasm.
Altered nuclear-cytoplasmic shuttling is a peculiar characteristic of NPM1-mutated AML [9]. Thus, targeting nuclear export is an appealing strategy in this pathological condition (Fig. 1). The natural compound leptomycin B shows strong XPO1 inhibitory activity in vitro [55] but its irreversible binding to XPO1 is associated with severe toxicity [56]. More recently, inhibitors that bind reversibly to XPO1 have become available. These drugs, collectively known as selective inhibitors of nuclear export (SINEs), include KPT-185, KPT-249, KPT-251, KPT-276, KPT-330, and KPT-335 [57] (Table 2). Among them, KPT-330 (selinexor) is the most well-known SINE compound that has been also tested in AML. [58] However, selinexor was scarcely active towards NPM1-mutated AML patients in early-phase clinical trials [59–61]. This is likely due to the once or twice/week administration schedule of selinexor (due to its toxicity profile) which is not sufficient to stably inhibit the interaction between mutated NPM1 and XPO1 (Pianigiani et al. BioRxiv 2021). An attractive alternative to selinexor is the second-generation SINE compound KPT-8602 (eltanexor) that crosses at lower extent the blood-brain barrier resulting in better tolerability. More importantly, the prolonged and frequent dosing schedule of eltanexor (i.e., 5 days/week) led to a greater anti-leukemic efficacy in preclinical animal models of hematological malignancies [62, 63]. This 5 days/week eltanexor schedule resulted into more robust anti-leukemic activity than selinexor alone in models of NPM1-mutated AML cells in vitro and in vivo (unpublished data). Interestingly, eltanexor synergizes with a BCL-2 inhibitor by increasing apoptosis in primary AML cells [64].
Table 2.
Summary of in vitro and in vivo data of given compounds and related clinical trials.
| Drug | Target | Mechanism of action | In vivo and in vivo efficacya | Clinical trial (ClinicalTrials.gov Identifier) | Phase; Patients; Recruitment Status | Combination therapies | Clinical outcome in NPM1-mut AML pts (CR/CRi) |
|---|---|---|---|---|---|---|---|
| Leptomycin B (LMB) | XPO1 inhibitor natural compound (irreversible) | NPM1 export inhibition | OCI-AML2, OCI-AML3, HL-60, KG1, MV4-11; RPMI-8226 and NCI-H929 (MM cell lines). | / | / | / | / |
| CBS9106 | XPO1 degradation (reversible) | NPM1 export inhibition | MM.1R, MM.1S, RPMI-8226, and ARH-77 (MM cell lines). RPMI-8226 xenograft mice. | / | / | / | / |
| KPT-185 | XPO1 inhibitors (reversible) SINE, 1st generation | NPM1 export inhibition | HL-60, Kasumi-1, KG1a, MOLM13, MV4-11, OCI-AML3, and THP-1 cell lines; NCI-H929 and RPMI-8226 (MM cell lines). AML primary patient samples with NPM1 and FLT3 -ITD mutations. Primary CLL cells. | / | / | / | / |
| KPT-249 | HL-60; APL cell lines; NCI-H929 and RPMI-8226 (MM cell lines). | / | / | / | / | ||
| KPT-251 | Primary CLL cells; MV4-11 xenograft mice. | / | / | / | / | ||
| KPT-276 | HL-60, MV4-11; NCI-H929 and RPMI-8226 (MM cell lines). MV4-11 xenograft mice. | / | / | / | / | ||
| KPT-330 (Selinexor) | HL-60, K562, KG-1, IMS-M2, MOLM13, MOLM16, MOLT-4, MV4-11, NB4, OCI-AML3, OCI-AML5, THP-1 and U937 cell lines; NCI-H929 and RPMI-8226 (MM cell lines). MV4-11 xenograft mice and others AML-derived xenografts (including CN and FLT3 -ITD). | NCT02093403 | Phase 1; 25 patients; Enrollment completed | Decitabine | Evaluating efficacy | ||
| NCT02249091 | Phase 2; 42 patients; Enrollment completed | Ara-C and Idarubicin | Evaluating efficacy | ||||
| NCT02088541 | Phase 2; 317 patients; Enrollment completed | Hydroxyurea and Ara-C | Evaluating efficacy | ||||
| NCT01607892 | Phase 1; 286 patients (95 AML patients); Enrollment completed | / | Evaluating efficacy | ||||
| NCT03955783 | Phase 1b; 78 patients; Recruiting | Venetoclax | Evaluating efficacy | ||||
| KPT-335 | Jurkat (T-cell leukemia); OCI-Ly3 and OCI-Ly10 (B-cell lymphoma) cell lines; CLBL1 (canine lymphoma) cell line; primary DLBCL cells. | / | / | / | Evaluating efficacy | ||
| KPT-8602 (eltanexor) | XPO1 inhibitors (reversible) SINE, 2nd generation | NPM1 export inhibition | K562, Kasumi-1, KG1, MOLM13, MOLM16, Mono-Mac-1, MV4-11, NB4, OCI-AML2, OCI-AML3, SKM1, and U937. MV4-11 xenograft mice. | NCT02649790 | Phase 1/2; 119 patients;Recruiting | ASTX727 and Dexamethasone | Evaluating efficacy |
| VTX (Venetoclax) | BCL-2 inhibitor | Apoptosis | GDM1, HL-60, KG1, K562, ME1, ML2, MOLM13, MOLM16, Mono-Mac-6, MV4-11, OCI-AML2, OCI-AML3, OCI-M1, NB4, NOMO1, THP-1 and U937. AML primary patient samples with NPM1, FLT3-ITD mutations. MV4-11 xenograft mice and others AML-PDX models harboring different mutations including NPM1, DNMT3A, FLT3, IDH1, PML-RARα, CEBPA. | NCT04867928 | Phase 2; 35 patients; Recruiting | Azacitidine | Evaluating efficacy |
| NCT02203773 | Phase 1b; 212 patient Active, not recruiting | Azacitidine or Decitabine |
91.5% (N = 23, 21/23) |
||||
| NCT02993523 | Phase 3; 400 patients; Active, not recruiting | Azacitidine |
66.7% (N = 27, 18/27) |
||||
| NCT02287233 | Phase 1/2; 94 patients; Enrollment completed | LDAC |
89% (N = 9, 8/9) |
||||
| NCT03069352 | Phase 3; 211 patients; Active, not recruiting | LDAC |
78% (N = 18, 14/18) |
||||
| ACTRN12616000445471b | Phase 1b; 48 patients; Recruiting | 5 + 2 (cytarabine + idarubicin) |
80% (N = 10, 8/10) |
||||
| NCT03214562 | Phase 1b/2; 116 patients; Recruiting | FLAG + IDA |
100% (N = 8, 8/8) |
||||
| MI-2-2 | MLL-Menin protein–protein interaction inhibition | HOX genes and MEIS1 downregulation | HL-60, KOPN-8, ML-2, MOLM13, MV4-11 and OCI-AML3 cell lines; MLL-AF9, MLL-AF6 and MLL-AF1p. OCI-AML3 xenograft mice. | / | / | / | / |
| MI-503 | MV4-11 and OCI-AML3 cell lines. MLL-AF9 BMCs from patients. MOLM13, MV4-11 and OCI-AML3 xenograft mice. | / | / | / | / | ||
| MI-3454 | K562, KOPN-8, MOLM13, MV4-11, SET2 and U937. RS4-11 and SEM (B-cell leukemia) cell lines. AML primary patient samples with NPM1 mutations or MLL1 translocations. MOLM13 and MV4-11 xenograft mice. | / | / | / | / | ||
| KO-539 | MOLM13, MV4-11, OCI-AML3 cell lines. AML primary patient samples with NPM1, KMT2A and FLT3-TKD mutations. PDX models with MLL-FP or NPM1 mutations. | NCT04067336 (KOMET-1) | Phase 1/2; 60 patients; Recruiting | FLT3 inhibitors | Evaluating efficacy | ||
| VTP-50469 | MLL-Menin protein–protein interaction inhibition | HOX genes and MEIS1 downregulation | HL-60, K562, ML2, MOLM13, MV4-11, NOMO1, OCI-AML3, THP1 cell lines. RS4-11, KOPN-8 and HB11-19 (B-cell leukemia) cell lines. Mouse MOZ-TIF2 cells and MLL-AF9 cells. MV4-11 xenograft mice and others AML-PDX models harboring different mutations including NPM1, FLT3, DNMT3A and IDH1. | / | / | / | / |
| SNDX-5613 | MOLM13, MV4-11 and OCI-AML3. MOLM13 xenograft mice and others AML-PDX models harboring different mutations including NPM1, DNMT3A, FLT3, IDH1, WT1, KMT2C. | NCT04065399 (AUGMENT-101) | Phase 1/2; 186 patients; Recruiting | / |
30% (N = 10, 3/10) |
||
| JNJ-75276617 | AML cell lines (not specified). AML patient samples with NPM1 and KMT2Ar mutations. | NCT04811560 | Phase 1; 110 patients; Recruiting | FLT3 inhibitors | Evaluating efficacy | ||
| DS-1594b | Menin inhibitor | HOX genes and MEIS1 downregulation | AML and ALL cells with MLLr (not specified). | NCT04752163 | Phase 1/2; 122 patients; Recruiting | Azacitidine, Venetoclax, or mini-HCVD | Evaluating efficacy |
| BMF-219 | Menin inhibitor (irreversible) | HOX genes and MEIS1 downregulation | MOLM13 cell line. CLL and MM cell lines (not specified). AML patient samples with NPM1 and MLLr mutations. | NCT05153330 (COVALENT-101) | Phase 1; 100 patients; Recruiting | / | Evaluating efficacy |
CR complete response, CRi complete response with incomplete count recovery, CN cytogenetically normal, SINE selective inhibitor of nuclear export, MM Multiple myeloma, DLBCL diffuse large B-cell lymphoma, HMAs hypomethylating agents, LDAC Low-Dose Cytarabine, mini-HCVD mini–hyper fractionated cyclophosphamide, vincristine and dexamethasone, FLAG-IDA fludarabine, cytarabine, granulocyte colony-stimulating factor (G-CSF) and idarubicin.
aData from the most important studies in AML cells are reported, unless otherwise indicated.
bAustralian Clinical Trials.
Targeting of other NPM1 pathways
Targeting apoptosis pathway
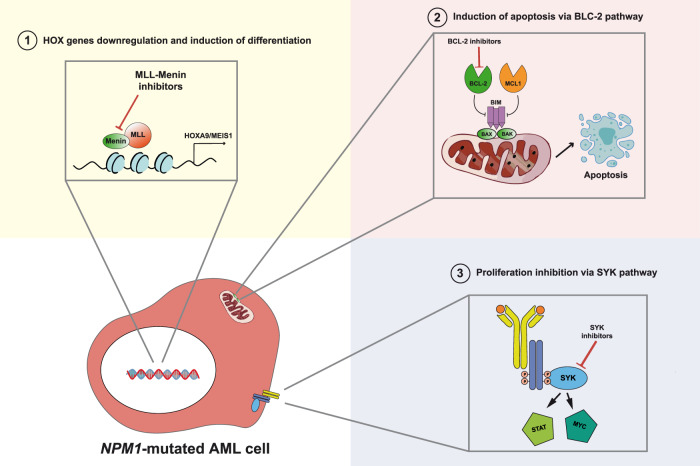
The BCL-2 family of proteins play a key role in the intrinsic mitochondrial apoptotic response and BCL-2 is a key survival factor in AML. The anti-apoptotic proteins BCL-2 and MCL1 inhibit apoptosis by sequestering the pro-apoptotic protein BIM, which is required for activation of BAX/BAK and the subsequent induction of mitochondrial outer membrane permeabilization. Venetoclax, a selective BCL-2 inhibitor, when combined with hypomethylating agents (e.g., 5-azacytidine) or low-dose cytarabine (LDAC), shows anti-leukemic activity in 60-70% of AML patients [65]. By downregulating MCL1 and inducing the expression of the pro-death proteins NOXA and PUMA, azacytidine inhibits synergistically the pro-survival proteins MCL1 and BCL-XL, increasing the dependence of leukemia cells on BCL-2. Moreover, venetoclax (with 5-azacytidine) induces leukemic stem cells toxicity by decreasing amino acid uptake, which is essential for oxidative phosphorylation and survival [66, 67]. Thus, venetoclax + 5-azacytidine (or decitabine) and venetoclax + LDAC have become the standard treatment for newly diagnosed older or unfit AML patients [12, 65]. In keeping with preclinical studies [68], NPM1-mutated AML patients appear to be particularly sensitive to venetoclax (Fig. 2). Whether this efficacy is related to the high expression of HOX genes [5], that in turn are linked to BCL-2 inhibitor sensitivity and responsiveness [69], remains to be defined. NPM1 mutant-primed defect in mitochondrial function may also be responsible for the higher sensitivity to venetoclax [44].
Fig. 2. Other approaches to target NPM1-mutated AML.
NPM1-mutated AML can be targeted with selective inhibitors of menin to downregulate HOX/MEIS, with BCL-2 (venetoclax) to induce apoptosis and with inhibitors of the SYK pathway (entospletinib).
NPM1-mutated AML particularly benefits from venetoclax-based regimens, both at first diagnosis [70, 71] (independently by the FLT3 status [72]) and in the relapsed/refractory setting [73–75] (Table 2). In the phase 3 clinical trial (NCT02993523) of venetoclax plus azacytidine, 66.7% of NPM1-mutated AML patients achieved CR + CRi [65], a majority of them being negative for measurable residual disease (MRD). Similar good results have been reported in real-world [76]. One-year OS for elderly patients with NPM1-mutated AML exceeded 80%, with an estimated 2-year OS of 70% [70]. Usually, the response was reached with 1–2 cycles and a good safety profile [65]. In trials NCT02287233 and NCT03069352, venetoclax plus LDAC resulted into CR + CRi of 89% and 78%, respectively, in NPM1-mutated AML patients [77, 78]. CR and CRi of 80% and 100%, were also achieved in NPM1-mutated AML patients, when venetoclax was combined with 5 + 2 (cytarabine + idarubicin) or FLAG + IDA (fludarabine, cytarabine, granulocyte colony-stimulating factor, and idarubicin) (ACTRN12616000445471, NCT03214562) [79, 80].
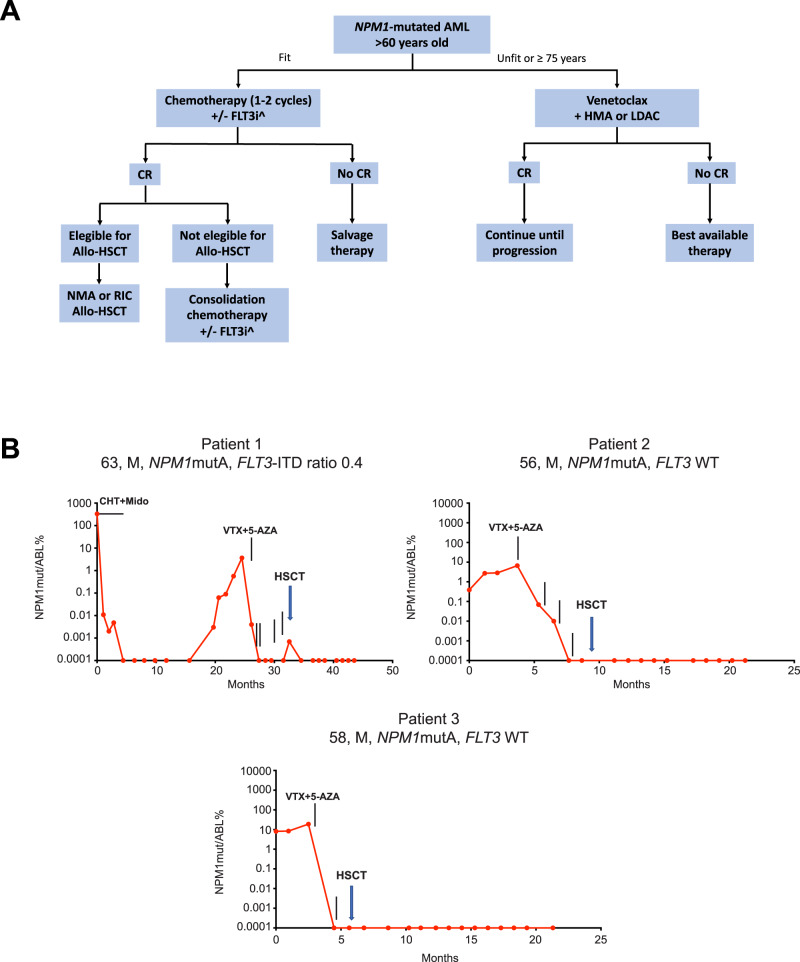
CR rates in refractory/relapsed NPM1-mutated AML are lower, ranging between 46% and 66.8%. In general, venetoclax plus 5-azacitidine was associated with better results than venetoclax plus decitabine or LDAC. Therapy was usually administered continuously until progression. However, about half of AML patients carrying NPM1 and/or IDH2 mutations and/or achieving a molecular CR after at least 12 months of a venetoclax-based regimen, experienced a long treatment-free remission, after therapy cessation [81]. Venetoclax-based regimens have been also used pre-emptively to treat 12 NPM1-mutated AML patients with persistent or relapsed/progressed MRD [82]. All five patients with persistent MRD and 6/7 patients with relapsed/progressed MRD, achieved durable molecular CR, after 1–2 cycles of venetoclax, with <17% grade >3 non-hematological toxicities [82]. Thus, venetoclax-based therapies have the potential to be used for NPM1-mutated MRD-positive patients, as bridge to allotransplant (Fig. 3A, B).
Fig. 3. Venetoclax in NPM1-mutated AML.
A Algorithm for the treatment of NPM1-mutated AML patients older than 60 years. ^Based on the presence or absence of FLT3 mutations. CR complete remission, FLT3i FLT3 inhibitors, HMA hypomethylating agents, LDAC low-dose cytarabine, allo-HSCT allogeneic hematopoietic stem cell transplantation. B Examples of preemptive therapy with venetoclax as bridging to allo-HSCT. All three patients achieved molecular CR (negativity for NPM1 mutant transcripts) before allo-HSCT. VTX venetoclax, 5-AZA 5-azacytidine, CHT chemotherapy.
Unfortunately, most patients treated with these regimens develop resistance over time and eventually relapse. To circumvent this problem, new combinations are currently tested, including venetoclax plus XPO1 [64] or menin [83, 84] inhibitors. In mouse models of NPM1-mutated/FLT3-ITD AML, venetoclax plus menin inhibitor was superior to the menin inhibitor alone, in eliminating leukemic cells (including leukemia stem/progenitor cells), decreasing Bcl-2 and Bcl-xL levels [83], and significantly prolonging mice OS [83]. ATO can potentiate the activity of venetoclax by attenuating Mcl-1 upregulation induced by venetoclax in AML cells. ATO plus venetoclax synergistically induced apoptosis in OCI-AML3 cells in vitro and were active in two R/R NPM1-mutated AML patients [85].
Targeting the MLL-menin complex
The Menin (MEN1) gene is located on chromosome 11q13 and encodes for a protein which regulates tissue-specific gene expression [86]. In human, germline MEN1 mutations cause the multiple endocrine neoplasia type-1 syndrome [87]. In mice, knock-out of the Men1 gene leads to reduced expression of Hoxc6 and Hoxc8 genes during embryogenesis [88]. Menin is a co-factor of the histone-lysine-N-methyltransferase 2A (KMT2A) that induces the trimethylation of lysine 4 on histone 3, a histone mark that correlates with active transcription of HOX genes and their co-factor MEIS1 (HOX/MEIS). HOXA and HOXB gene clusters are highly expressed in normal adult hematopoietic stem/progenitor cells but are physiologically silenced in mature blood cells, suggesting their role in self-renewal [89]. High expression of HOXA and HOXB cluster genes and their co-factor MEIS1 contributes to the gene signature typical of NPM1-mutated AML [4, 5]. Interestingly, the HOXA and HOXB expression levels in NPM1-mutated AML overlap with those of normal hematopoietic stem/progenitor cells, pointing to common mechanisms regulating HOX expression and to persistent expression rather than upregulation of HOX genes in NPM1-mutated AML [90].
More recently, we proved that HOX expression is directly dependent upon the aberrant cytoplasmic dislocation of NPM1 mutants [10]. Indeed, nuclear relocalization or targeted degradation of cytoplasmic mutated NPM1 resulted into the rapid loss of HOX expression, followed by differentiation and growth arrest [10]. However, the link between cellular localization of NPM1 mutants and HOX expression remains unclear. A possible model implies that NPM1 directly binds nucleolar putative factors that repress HOX genes required for proper differentiation and displaces them from the nucleus to the cytoplasm [4]. This would be in keeping with the finding that nuclear relocalization of NPM1 mutants from the cytoplasm to the nucleus restore the normal activity of these factors, leading to downregulation of HOX genes [4]. Alternatively, recruitment of NPM1 mutants to HOX loci through their interaction with chromatin-bound XPO1 [91] could be responsible for HOX expression maintenance, cytoplasmic delocalization of NPM1 representing a mere epiphenomenon. We previously suggested that NPM1 mutants may induce leukemia by acting both at the chromatin level and by delocalizing NPM1-interacting partners in the cytoplasm [4].
Dependence of NPM1-mutated AML cells on epigenetic machinery for HOX regulation [92] provides the rationale for using inhibitors of KMT2A-menin protein interaction [93, 94] (Fig. 2; Table 2). The VTP-50469 inhibitor showed strong in vitro and in vivo anti-leukemic activity on NPM1-mutated AML cells, causing a dose-dependent reduction in cell proliferation, a significant downregulation of HOXA/B clusters and MEIS1 gene expression, a marked differentiation of leukemic cells, a reduction of AML engraftment and a prolonged survival in mice PDX models [92, 95, 96]. This occurred especially through rapid repression of important co-factors of HOX genes (MEIS1 and PBX3), the effect on expression of HOXA and HOXB genes being not relevant [97]. The MI-3454 inhibitor was very effective in inhibiting cell proliferation and differentiation of NPM1-mutated AML cells, independently from the coexistence of other mutations in patients’ samples, and in reducing blast infiltration of organs and expression of MEIS1 and FLT3 in PDX NPM1-mutated mouse models [95].
Menin inhibitors were reported to prevent the transformation of Npm1-mutated mouse hematopoietic progenitors into leukemic cells, implying that they could also be effective in Npm1-mutated preleukemia [96]. However, we believe that this concept is difficult to translate into clinic because NPM1 mutations in patients do not associate with a preleukemic state [4]. Moreover, myelodysplasia with NPM1 mutations is very rare and cases with these characteristics usually represent already early-stage AML [12]. Thus, we expect that the value of menin inhibitors in clinic will be mainly limited to the therapy of frank NPM1-mutated AML.
Menin inhibitors was also combined with inhibitors of FLT3 (mutated in about 40% of NPM1-mutated AML [3]), demonstrating a synergistic effect that resulted in stronger cell growth inhibition, apoptosis and differentiation of AML blast cells [98] and induction of long-lasting CR in PDX mice models of NPM1-mutated/FLT3-ITD AML [99]. Menin inhibitors have been also combined with XPO1 inhibitors. The rationale for this association is that nuclear relocation of the NPM1 mutant by the XPO1 inhibitors is associated with downregulation of HOX genes [10]. Thus, the two compounds could exert a cumulative effect on downregulation of HOX genes through different mechanisms. The utility of combining menin inhibitors with venetoclax has been already mentioned [83]. All these associations could help preventing the rapid development of resistance to targeted monotherapies.
Menin inhibitors, including KO-539, SNDX-5613, JNJ-75276617 and DS-1594b, are currently evaluated in clinical trials [100, 101]. KO-539 induced CR in 2/6 patients with R/R AML who were evaluable for efficacy analysis. One of them had an NPM1-mutated AML co-mutated for DNMT3A and KMT2D who received KO-539 at 200 mg/die, as the eight line of therapy, achieving an MRD-negative CR [101]. This trial continues to enroll patients with NPM1-mutated and KTM2A-rearranged AML on the doses of 200 and 600 mg (NCT04067336). SNDX-5613 (an analog of VTP-50469) was very active in PDX mouse models of NPM1-mutated AML, including animals remaining in CR 1 year after cessation of therapy [96, 102]. The safety and efficacy of SNDX-5613 in adult patients with R/R NPM1-mutated or KTM2A-rearranged AML is being evaluated in the AUGMENT-101 phase I/II trial (NCT04065399). Preliminary results of this study were released by Syndax in April 2021. By March 2021, the trial had enrolled in the phase 1 cohort, 43 patients (median age 54 years) who had received a median of 3 previous lines of treatment. The most common side effects (>5%) included QT prolongation (14%), differentiation syndrome (5%) and anemia (5%). The overall response rate in the 7 patients with NPM1-mutated AML was 29% (2/7). In keeping with the proposed mechanism of menin inhibitors, RNA-Seq analysis of the bone marrow samples from responding patients exhibited downregulation of MEIS and HOXA9 genes and upregulation of the differentiation antigens CD11b, CD14 and CD13. The phase 2 part of the trial is ongoing. JNJ-75276617 is a potent inhibitor of the binding between menin and KTM2A. Its safety and activity are being tested in NCT04811560 that enrolls AML patients harboring NPM1 mutations or KMT2A rearrangements. However, no data have been released so far. The safety and efficacy of DS-1594b menin inhibitor will be evaluated as single drug or in combination with azacytidine and venetoclax regimens in a phase 1/2 clinical trial (NCT04752163). To maximize the depth and durability of clinical response, the Biomea Fusion, Inc. has recently developed BMF-219, an orally bioavailable, potent and selective irreversible covalent menin inhibitor. Clinical trial with this compound is ongoing (NCT05153330).
Targeting SYK signaling
In a phase 1b/2 trial (NCT02343939), entospletinib, a selective oral inhibitor of the spleen tyrosine kinase (SYK) which is constitutively activated in AML promoting survival and proliferation [103], when combined with chemotherapy, was more active in patients with HOXA9/MEIS1 signature (as in NPM1-mutated AML) than in the whole patient population [104] (Fig. 2). These results suggest that the increased expression and activity of SYK protein is strictly dependent upon the deregulation of HOXA and MEIS genes [105]. Based on this evidence, FDA approved a phase 3 trial to assess the efficacy and safety of entospletinib in combination with chemotherapy in adult patients with newly diagnosed NPM1-mutated AML (NCT05020665).
Immunotherapy of NPM1-mutated AML
Ideally, any target antigen for AML immunotherapy should be expressed at high levels in the whole leukemic population, including leukemic stem cells, and to be absent or low expressed in normal hematopoietic cells and other tissues. Leukemia-associated antigens (e.g., CD33 and CD123) are usually strongly expressed in AML cells (especially in NPM1-mutated AML cells [106–108]) but can also be detected in normal hematopoietic stem cells and in extramedullary tissues (e.g., CD123 in endothelial cells). This limits their use as target antigens for immunotherapy because of potential off-target effects.
Conversely, leukemia-specific antigens deriving from altered proteins encoded by leukemogenic mutations (e.g., NPM1), are specifically expressed in malignant clones and therefore represent ideal targets. In particular, the NPM1 mutant neoantigen can be considered an ideal AML target for a number of reasons. First, NPM1 mutations are common driver, gate-keeper events [109], very stable at relapse [4], specific for AML and absent in normal tissues [110]. Second, the NPM1 mutated proteins are detectable in chemoresistant leukemic stem cells [108], making them possibly vulnerable to immune surveillance and eradication. Third, although >50 NPM1 mutations have been identified, the 4 bp frameshift insertion occurring in NPM1 mutant A is responsible for almost 80-85% of all mutations and more rare NPM1 mutations lead to the same amino acidic changes at NPM1 C-terminus. Fourth, the newly acquired amino acid C-terminus sequence of NPM1 mutant proteins is highly immunogenic in animals, eliciting specific antibodies. Fifth, the aberrant localization of the NPM1 mutant proteins in the cytoplasm of leukemic cells may favor their processing by the Human Leukocyte Antigen (HLA) MHC class I degradation pathway leading to HLA presentation and anti-cancer immune response. Indeed, using in silico analysis, we predicted that several peptides could bind to specific HLA class I molecules [111]. Sixth, specific autologous cytotoxic T-cell responses against NPM1 mutant peptides could be detected in NPM1-mutated AML patients [112–115]. These immune responses associated with molecular CR [116] and may explain the relatively favorable outcome of NPM1-mutated AML [117]. interestingly, NPM1-mutated AML sensitivity to T-cell immunity has been observed not only in the autologous but also in allogeneic setting. Although NPM1-mutated AML patients without FLT3-ITD has a good prognosis, those who underwent allogeneic HSCT showed a particularly long-term disease control [118], probably due to specific graft-versus leukemia effect. Moreover, polyspecific T-cell anti-leukemic responses, even against NPM1-mutated peptides, have been observed following preemptive donor lymphocyte infusions (DLIs) at molecular relapse after allogeneic HSCT [116]. Finally, the importance of eradicating the NPM1-mutated clone to achieve cure of AML is exemplified by the clinical observation of patients with NPM1-DNMT3A double-mutated AML after cessation of therapy. These cases, when achieve long-term molecular (MRD-negative) remission, are likely to be cured, even though the persistence of detectable copies of the DNMT3A mutant (indicating persistent clonal hemopoiesis) may expose them to a low risk of developing a second AML [4].
Antibodies against CD33 and CD123
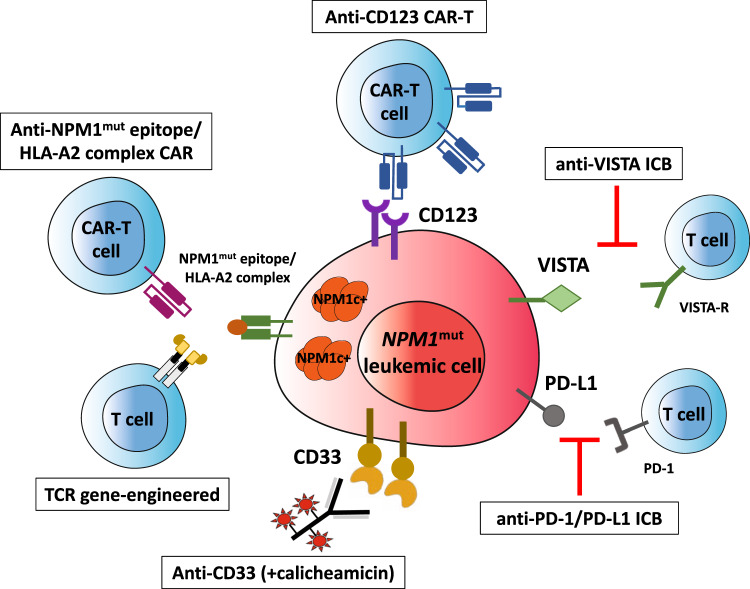
CD33 is expressed in all stages of myeloid differentiation [119] and it is detectable in most cases of AML, the expression levels being high in NPM1-mutated AML [106]. Thus, CD33 is an useful target for immunotherapy with an anti-CD33 monoclonal antibody conjugated with a DNA-damaging calicheamicin derivative (Gemtuzumab Ozogamicin-GO) (Fig. 4). In a metanalysis study [120], adding GO to chemotherapy showed a survival benefit for intermediate-risk cytogenetics and NPM1-mutated AML patients because of a reduced relapse risk. Similar results were reported by the ALFA0701 trial that also demonstrated the impact of GO in reducing NPM1-mut transcripts level [121]. Higher reduction of NPM1-mut transcript levels were also observed in the GO arm of the AMLSG study that translated into a lower cumulative incidence of relapse [122]. However, the AMLSG 09-09 Phase III Study failed to meet the early primary end point (event-free survival) due to higher early mortality in the GO arm [123]. Nevertheless, a significant clinical benefit was observed in females older than 70 years with NPM1-mutated/FLT3 wild-type genotype [123] in terms of both event-free-survival and cumulative incidence of relapse. Collectively, the above studies support the incorporation of GO into the frontline treatment of NPM1-mutated AML.
Fig. 4. Immunotherapeutic approaches to NPM1-mutated AML.
NPM1-mutated AML can be targeted using antibody-drug conjugates (e.g., gentuzumab ozogamicin, anti-CD33), immune check-point inhibitors, CAR and TCR-based adoptive T-cell therapies directed against NPM1 mutated epitope/HLA complex. CAR chimeric antigen receptor, TCR T-cell receptor.
CD123 is highly expressed in NPM1-mutated AML both at diagnosis and relapse (Fig. 4), the highest expression being observed in CD34+ CD38− leukemic cells [107]. Moreover, CD123 expression was enhanced by FLT3 mutations, suggesting that the subset of NPM1/FLT3 double-mutated AML patients could particularly benefit from anti-CD123 targeted therapies [107]. So far, tagraxofusp (SL401) which is formed by the fusion of IL-3 with diphtheria toxin and the CD123-directed chimeric antigen receptor (CAR) T cells (MB-102) developed by Mustang Bio Inc. are approved or received the Orphan drug designation (MB-102) for the treatment of blastocytic plasmacytoid dendritic cell neoplasm [124] which strongly expresses CD123. Only scarce information is available in CD123-positive R/R AML. Main limitation of these products is myelotoxicity [124].
Antibodies against PD-1 and PD-L1
AML is a poorly immunogenic and highly immune-suppressive hematological malignancy. High expression of PD-L1 has been found in NPM1-mutated AML patients, especially in the leukemic progenitors/stem cell compartment (CD34+ CD38−) [125]. High PD-L1 expression in blasts of AML with NPM1-mutated/FLT3-ITD genotype predicted inferior survival [41]. Moreover, in a comprehensive immunogenomic analysis of AML, mutations of NPM1 and FLT3 preferentially associated with low T-cell cytolytic activity and a reduced expression of HLA-II (and/or related genetic determinants of HLA-II expression, as CIITA) [126]. Interestingly, CIITA methylation may limit antigen presentation by primary NPM1mutIDH1mut AML blasts through downregulation of MHC-II, thereby inducing immune evasion [126]. Immune evasion in NPM1-mutated AML is also contributed by VISTA (V-domain Ig suppressor of T-cell activation) and ULBP1 (NKG2 ligand) immunoregulatory circuitries that are both significantly upregulated in NPM1-mutated AML patients [126] (Fig. 4). VISTA-Ig, which shows a partial homology with other B7 family members, is predominantly expressed in hematopoietic cells of myeloid lineage. This circuitry in mainly involved in suppressing proliferation of T cells and blunting the production of T-cell cytokines, making it a potential target for immune check-point blockade combinations [127].
More recently, the anti-PD-1 antibody, nivolumab, was found to increase leukemia-associated antigen-stimulated cytotoxic T cells and cytotoxicity against stem cell-like cells, especially those carrying NPM1 mutations [128]. These findings provide a rationale for the treatment of NPM1-mutated AML, combining anti-PD-1 and anti NPM1-mutation specific immunotherapy (see below). Moreover, targeted immune gene expression and multiplexed digital spatial profiling showed distinct AML immune microenvironments [129]. The immune-infiltrated microenvironment that was characterized by severe immune suppression (high expression of PD-L1, CTLA4, IDO1 and BTLA), higher dependence from IFN-γ driven adaptive immune responses, high T-cell infiltration and expression of major histocompatibility complex, closely clustered with the adverse-risk genetic AML categories (specifically, TP53 and RUNX1 mutated AML) [129]. Conversely, the NPM1-mutated cases (with or without FLT3-ITD) more frequently showed an immune-depleted microenvironment [129]. Finally, NPM1-mutated AML harboring concomitant clonal hematopoiesis driven mutations (e.g., DNMT3A, TET2) showed an enriched tumor inflammation signature score, predicting a clinical benefit from anti-PD-1 treatment [130]. This finding is in keeping with the observation linking in a mouse model a persistent immune stimulation to an accelerated NPM1mut myeloproliferative phenotype in vivo [131].
Although the above findings suggest that NPM1-mutated AML may be a potential candidate for immune check-point inhibition (Fig. 4), the few studies performed so far with anti-PD-L1 antibodies in AML patients have shown only modest clinical activity. Their impact has been evaluated also in combination with hypomethylating agents [132–134], since they induce the expression of several immune-related genes, including HLA-I and HLA-II, leukemia-associated antigens (e.g., PRAME, WT1) [135], PD-1 and PD-L1 [136]. Despite most studies did not specifically evaluate NPM1-mutated AML, they showed that patients who might benefit more from these drug combinations are those who are naïve for hypomethylating agents or have <20% blasts and a higher pre-therapy infiltration of bone marrow by CD3+, CD4+ Teff, and CD8+ T cells [132]. The clinical trial NCT03769532 is currently evaluating the safety/efficacy of pembrolizumab plus 5-azacitidine in NPM1-mutated AML patients. The impact of pembrolizumab 200 mg (i.v. on day 14) has been assessed also in association with high-dose cytarabine in 37 R/R AML patients (9/37, 24%, bearing NPM1 mutations) [137]. The overall response rate, composite CR rate and median OS were 46%, 38% and 11.1 months, respectively. Responding patients exhibited a higher percentage of progenitor exhausted TCF1+ CD8+ T cells and an increased diversity of the T-cell receptor at baseline [137].
As previously mentioned, venetoclax is very active in NPM1-mutated AML [70]. This effect is also contributed by the immunomodulatory effect of the drug that enhances the T-cell-mediated anti-leukemic response by increasing reactive-oxygen species (ROS) production [138] and increases the PD1+ T-effector memory cells and anti-tumor efficacy in combination with immune check-point blockade [139]. The NCT02397720 trial is assessing the combination of nivolumab, azacytidine and venetoclax in frontline and R/R AML, while the NCT04284787 trial is assessing the impact of pembrolizumab plus azacytidine and venetoclax in newly diagnosed AML patients unfit for conventional chemotherapy.
CAR and TCR engineered T cell therapy
CAR T cells or T-cell receptor (TCR) gene therapy could be promising approaches against NPM1-mutated AML. Immune targeting can be distinguished into: (1) HLA-dependent therapies relying on the presentation of NPM1 neoantigen [113]; or (2) HLA-independent therapies identifying molecules differentially expressed on leukemic cells relative to normal cells (tumor-associated antigens).
Searching for HLA class I ligandome of primary AMLs, multiple ΔNPM1-derived immunogenic peptides, have been identified, including AIQDLCLAV, AIQDLCVAV, CLAVEEVSL, LAVEEVSLR, AVEEVSLRK 9-mer, and CLAVEEVSLRK 11-mer, representative of the more common NPM1 mutation types. These peptides are able to efficiently bind to at least most common HLA types (A*02:01, A*03:01) which are often detected in the Caucasian population [111–115, 140–142]. Using yeast surface display, a human single-chain variable fragment (scFv) that specifically identifies the NPM1 mutant epitope/HLA-A2 complex but not HLA-A2 or HLA-A2 loaded with control peptides was generated and used to construct CAR T cells (Fig. 4). These engineered cells showed strong in vitro and in vivo activity against preclinical models of NPM1-mutated AML cells carrying NPM1-mutant/HLA-A2 complex but not against NPM1 wild-type/HLA-A2+ AML cells or HLA-A2 negative tumor cells [140]. More recently, memory-like NK cells armed with the same neoepitope-specific CAR showed strong activity against NPM1-mutated AML in absence of toxicity [143].
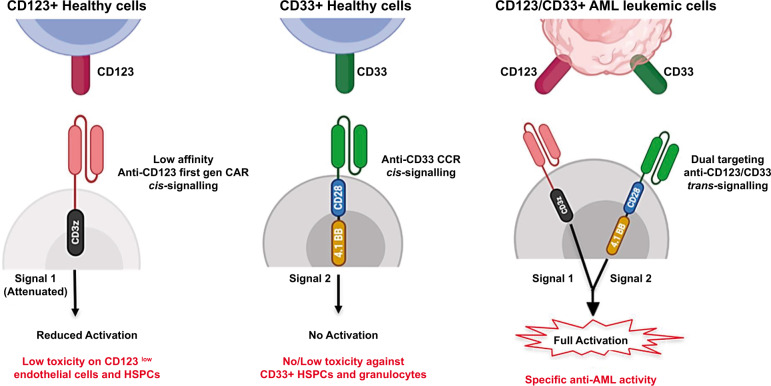
CD123 and CD33 are strongly expressed both in NPM1-mutated AML cells and healthy tissue. Thus, aiming to improve selectivity for leukemic cells while minimizing toxicity towards normal cells, a dual targeting model was exploited through Cytokine Induced Killer (CIK) cells co-expressing a first-generation low affinity anti-CD123 and an anti-CD33 as costimulatory receptor without activation signaling domains. This trans-signaling strategy could allow: i) low toxicity profile against CD123+ endothelial cells and HSPC, due to a reduced cell activation given by the suboptimal first-generation CAR signal; ii) no or low myelotoxicity against CD33+ HSPC cells, due to absence of CIK cell activation upon the sole costimulatory signal engagement; and iii) full CAR-CIK activation only against double expressing CD123+/CD33+ leukemic cells [144] (Fig. 5).
Fig. 5. Dual CAR targeting of CD33 and CD123.
The rationale of CD123/CD33 dual targeting trans-signaling strategy is to induce a full cell activation against only CD123/CD33+ leukemic cells while reducing cell stimulation against CD33+ HSPCs and CD123+ endothelial cells [144].
Specific T cells for HLA-A*02:01-binding CLAVEEVSL have been searched in healthy individuals using peptide-HLA tetramers. Tetramer-positive CD8+ T cells were isolated and their activity towards primary AMLs investigated. The TCR was then isolated from a clone with high anti-leukemic reactivity and its capability to specifically recognize and lyse HLA-A*02:01-positive ΔNPM1 AML demonstrated after retroviral transduction of CD8+ and CD4+ T cells [145]. Moreover, T cells transduced with TCR for HLA-A*02:01-binding CLAVEEVSL efficiently killed AML cells and prolonged OS of NSG mice engrafted with HLA-A*02:01-positive NPM1-mutated OCI-AML3 human cells [145] (Fig. 4). Thus, CLAVEEVSL is a neoantigen that can be efficiently targeted on AML by ΔNPM1 TCR gene transfer. While such TCR gene-engineered T-cell therapy prove to be potent and safe, it must match the TCR haplotype restriction to HLA-A*02:01-positive patients, representing around 40% of Caucasian population. Moreover, HLA class I downregulation or loss of neoantigen expression could be a possible mechanism of immune escape from NPM1-mutated TCR gene-engineered T-cell therapy.
DLI using T cells derived from healthy donors and specifically directed against the NPM1-mutated neoantigen with the aim to elicit graft-versus-leukemia may be a therapeutic option in patients experiencing molecular relapse following allogeneic HSCT. Eliciting endogenous immune responses through vaccination with NPM1 neoantigens is unlikely to be effective in patients with high-burden newly diagnosed or relapsed AML but could be of benefit, possibly in combination with immune check-point inhibitors, to treat pre-emptively NPM1-mutated AML with persistent NPM1 transcripts or in molecular relapse.
Future perspectives
Molecular mechanisms underlying NPM1-mutated AML are still poorly understood. Endogenous tagging of wild-type and mutated NPM1 proteins followed by mass spectrometry may unravel their interactions with other partners and functions in the cytoplasm. How NPM1 mutants deregulate the HOX program remains also to be better defined. Moreover, all current experimental data are based upon the analysis of OCI-AML3 and IMS-M2 human AML cell lines that both carry the most frequent NPM1 mutation (i.e., mutation A). Whether similar results can be extended also to the rarer NPM1 variants remains to be determined. Clarifying these issues may lead to the development of new targeted therapeutic strategies.
Unfit NPM1-mutated AML patients relapsing after venetoclax-based regimens represent a medical need. The mechanisms of resistance to venetoclax and its use in combination with other drugs to prevent relapse should be better investigated. Menin inhibitors are emerging as the most promising agents for targeted therapy of NPM1-mutated AML. The ongoing trials will tell us which is the real impact of these compounds in NPM1-mutated AML and suggest which are the best combinations to maximize the clinical benefit. Menin inhibitors, alone or in combination with venetoclax or other agents, could be incorporated in the treatment algorithm, as that shown in Fig. 3A, to reduce or eradicate NPM1-related MRD, possibly as bridge to allo-HSCT, in eligible patients. Moreover, menin and XPO1 inhibitors have the potential to be used alone or in combination (e.g., with FLT3 inhibitors), at initial diagnosis, especially in patients who are older or unfit for intensive chemotherapy.
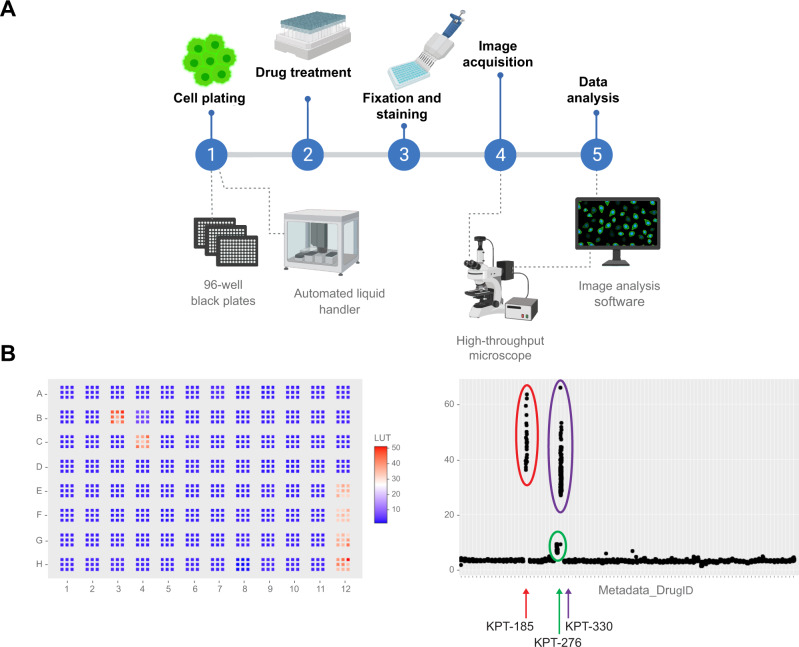
Identifying novel drugs for NPM1-mutated AML using synthetic lethality approaches are under way [146]. High-throughput screening technology [147] allows to screen a large number of compound libraries at a rate that may exceed a few thousand compounds per day or per week [148, 149]. Searching for new molecules able to re-localize or reduce the expression of the NPM1 mutated protein, we have established a microscopy-based screening strategy suitable to analyze hundreds selected drugs and compounds using high-throughput microscope technique and image analysis (Fig. 6). Among other compounds, we identified inhibitors with known re-localizing activity on NPM1 mutated protein, thus confirming the value of our experimental strategy (unpublished data).
Fig. 6. High-throughput screening for novel drugs in NPM1-mutated AML.
A Workflow of microscopy-based screening strategy (created with BioRender.com). B Example of 96 well plate subjected to image and data processing (generated with ShinyHTM software). Arrows indicate results obtained with XPO1 inhibitors (KPT-185, KPT-276 and KPT-330). The higher number of points for selinexor (KPT-330) results from its use as a positive control.
Acknowledgements
This work has been supported by the European Research Council (ERC Adv grant 2016 n. 740230 to BF and ERC Cons grant 2016 n. 725725 to MPM) and the Italian Association for Cancer research (AIRC grant n. 23604 to BF and grant Start-Up 2019 n. 22895 to LB).
Author contributions
BF had the original idea and wrote and supervised the work together with MPM and LB. RR, GP, SP, FM and IG contributed to the drug screening and to wrote the section of molecular therapeutic targeting of NPM1-mutated AML. SS take care of NPM1-mutated AML patients treated with venetoclax. VC followed the trial on actinomycin D in NPM1-mutated AML. VMP and AM contributed to write the section on immunotherapy of NPM1-mutated AML.
Competing interests
MPM declares honoraria from Rasna Therapeutics, Inc for scientific advisor activities and serves as consultant for scientific advisory boards of Abbvie, Amgen, Celgene, Janssen, Novartis, Pfizer and Jazz Pharmaceuticals. LB declares consultancy at scientific advisory boards for Abbvie and Amgen. BF licensed a patent on NPM1 mutants (n. 102004901256449) and declares honoraria from Rasna Therapeutics, Inc for scientific advisor activities. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Roberta Ranieri, Giulia Pianigiani.
These authors jointly supervised this work: Maria Paola Martelli, Brunangelo Falini, Lorenzo Brunetti.
References
- 1.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–90. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 2.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 3.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–66. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 4.Falini B, Brunetti L, Sportoletti P, Martelli MP. NPM1-mutated acute myeloid leukemia: from bench to bedside. Blood. 2020;136:1707–21. doi: 10.1182/blood.2019004226. [DOI] [PubMed] [Google Scholar]
- 5.Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106:899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 6.Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105:3945–50. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivey A, Hills RK, Simpson MA, Jovanovic JV, Gilkes A, Grech A, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374:422–33. doi: 10.1056/NEJMoa1507471. [DOI] [PubMed] [Google Scholar]
- 8.Falini B, Bolli N, Shan J, Martelli MP, Liso A, Pucciarini A, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107:4514–23. doi: 10.1182/blood-2005-11-4745. [DOI] [PubMed] [Google Scholar]
- 9.Falini B, Bolli N, Liso A, Martelli MP, Mannucci R, Pileri S, et al. Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia. 2009;23:1731–43. doi: 10.1038/leu.2009.124. [DOI] [PubMed] [Google Scholar]
- 10.Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, et al. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell. 2018;34:499–512.e. doi: 10.1016/j.ccell.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: myeloid and histiocytic /dendritic neoplasms. Leukemia. 2022;36:1703–19. doi: 10.1038/s41375-022-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falini B, Brunetti L, Martelli MP. How I diagnose and treat NPM1-mutated AML. Blood. 2021;137:589–99. doi: 10.1182/blood.2020008211. [DOI] [PubMed] [Google Scholar]
- 13.Falini B, Sciabolacci S, Falini L, Brunetti L, Martelli MP. Diagnostic and therapeutic pitfalls in NPM1-mutated AML: notes from the field. Leukemia. 2021;35:3113–26. doi: 10.1038/s41375-021-01222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal S. Targeted cancer therapies. Nat Rev Drug Discov. 2010;9:427–8. doi: 10.1038/nrd3186. [DOI] [PubMed] [Google Scholar]
- 17.Spotlight on cancer genomics. Nat Cancer. 2020;1:265–6. [DOI] [PubMed]
- 18.Zhong L, Li Y, Xiong L, Wang W, Wu M, Yuan T, et al. Small molecules in targeted cancer therapy: advances, challenges, and future perspectives. Signal Transduct Target Ther. 2021;6:201. doi: 10.1038/s41392-021-00572-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai C, Doucette K, Norsworthy K. Recent drug approvals for acute myeloid leukemia. J Hematol Oncol. 2019;12:100. doi: 10.1186/s13045-019-0774-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estey E, Karp JE, Emadi A, Othus M, Gale RP. Recent drug approvals for newly diagnosed acute myeloid leukemia: gifts or a Trojan horse? Leukemia. 2020;34:671–81. doi: 10.1038/s41375-019-0704-5. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 22.Cela I, Di Matteo A, Federici L. Nucleophosmin in its interaction with ligands. Int J Mol Sci. 2020;21:4885. doi: 10.3390/ijms21144885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitrea DM, Grace CR, Buljan M, Yun MK, Pytel NJ, Satumba J, et al. Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc Natl Acad Sci USA. 2014;111:4466–71. doi: 10.1073/pnas.1321007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Matteo A, Franceschini M, Chiarella S, Rocchio S, Travaglini-Allocatelli C, Federici L. Molecules that target nucleophosmin for cancer treatment: an update. Oncotarget. 2016;7:44821–40. doi: 10.18632/oncotarget.8599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Federici L, Falini B. Nucleophosmin mutations in acute myeloid leukemia: a tale of protein unfolding and mislocalization. Protein Sci. 2013;22:545–56. doi: 10.1002/pro.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi W, Shakalya K, Stejskal A, Goldman A, Beeck S, Cooke L, et al. NSC348884, a nucleophosmin inhibitor disrupts oligomer formation and induces apoptosis in human cancer cells. Oncogene. 2008;27:4210–20. doi: 10.1038/onc.2008.54. [DOI] [PubMed] [Google Scholar]
- 27.Balusu R, Fiskus W, Rao R, Chong DG, Nalluri S, Mudunuru U, et al. Targeting levels or oligomerization of nucleophosmin 1 induces differentiation and loss of survival of human AML cells with mutant NPM1. Blood. 2011;118:3096–106. doi: 10.1182/blood-2010-09-309674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emmott E, Hiscox JA. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10:231–8. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, et al. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife. 2016;5:e13571. doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–97. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitrea DM, Cika JA, Stanley CB, Nourse A, Onuchic PL, Banerjee PR, et al. Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat Commun. 2018;9:842. doi: 10.1038/s41467-018-03255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, et al. Composition-dependent thermodynamics of intracellular phase separation. Nature. 2020;581:209–14. doi: 10.1038/s41586-020-2256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grummitt CG, Townsley FM, Johnson CM, Warren AJ, Bycroft M. Structural consequences of nucleophosmin mutations in acute myeloid leukemia. J Biol Chem. 2008;283:23326–32. doi: 10.1074/jbc.M801706200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee H, Chan KP, Andresen V, Hanley ML, Gjertsen BT, Myers AG. Interactions of the natural product (+)-avrainvillamide with nucleophosmin and exportin-1 Mediate the cellular localization of nucleophosmin and its AML-associated mutants. ACS Chem Biol. 2015;10:855–63. doi: 10.1021/cb500872g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wulff JE, Siegrist R, Myers AG. The natural product avrainvillamide binds to the oncoprotein nucleophosmin. J Am Chem Soc. 2007;129:14444–51. doi: 10.1021/ja075327f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andresen V, Erikstein BS, Mukherjee H, Sulen A, Popa M, Sornes S, et al. Anti-proliferative activity of the NPM1 interacting natural product avrainvillamide in acute myeloid leukemia. Cell Death Dis. 2016;7:e2497. doi: 10.1038/cddis.2016.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang K, Wang M, Zhao Y, Sun X, Yang Y, Li X, et al. A redox mechanism underlying nucleolar stress sensing by nucleophosmin. Nat Commun. 2016;7:13599. doi: 10.1038/ncomms13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falini B, Brunetti L, Martelli MP. Dactinomycin in NPM1-mutated acute myeloid leukemia. N Engl J Med. 2015;373:1180–2. doi: 10.1056/NEJMc1509584. [DOI] [PubMed] [Google Scholar]
- 39.Falini B, Gionfriddo I, Cecchetti F, Ballanti S, Pettirossi V, Martelli MP. Acute myeloid leukemia with mutated nucleophosmin (NPM1): any hope for a targeted therapy? Blood Rev. 2011;25:247–54. doi: 10.1016/j.blre.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Gionfriddo I, Brunetti L, Mezzasoma F, Milano F, Cardinali V, Ranieri R, et al. Dactinomycin induces complete remission associated with nucleolar stress response in relapsed/refractory NPM1-mutated AML. Leukemia. 2021;35:2552–62. doi: 10.1038/s41375-021-01192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brodska B, Otevrelova P, Salek C, Fuchs O, Gasova Z, Kuzelova K. High PD-L1 expression predicts for worse outcome of leukemia patients with concomitant NPM1 and FLT3 mutations. Int J Mol Sci. 2019;20:2823. doi: 10.3390/ijms20112823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burger K, Muhl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem. 2010;285:12416–25. doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martelli MP, Gionfriddo I, Mezzasoma F, Milano F, Pierangeli S, Mulas F, et al. Arsenic trioxide and all-trans retinoic acid target NPM1 mutant oncoprotein levels and induce apoptosis in NPM1-mutated AML cells. Blood. 2015;125:3455–65. doi: 10.1182/blood-2014-11-611459. [DOI] [PubMed] [Google Scholar]
- 44.Wu HC, Rerolle D, Berthier C, Hleihel R, Sakamoto T, Quentin S, et al. Actinomycin D targets NPM1c-primed mitochondria to restore PML-driven senescence in AML therapy. Cancer Discov. 2021;11:3198–213. doi: 10.1158/2159-8290.CD-21-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Hajj H, Dassouki Z, Berthier C, Raffoux E, Ades L, Legrand O, et al. Retinoic acid and arsenic trioxide trigger degradation of mutated NPM1, resulting in apoptosis of AML cells. Blood. 2015;125:3447–54. doi: 10.1182/blood-2014-11-612416. [DOI] [PubMed] [Google Scholar]
- 46.Yi S, Wen L, He J, Wang Y, Zhao F, Zhao J, et al. Deguelin, a selective silencer of the NPM1 mutant, potentiates apoptosis and induces differentiation in AML cells carrying the NPM1 mutation. Ann Hematol. 2015;94:201–10. doi: 10.1007/s00277-014-2206-x. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Zhao Z, Yi S, Wen L, He J, Hu J, et al. Deguelin induced differentiation of mutated NPM1 acute myeloid leukemia in vivo and in vitro. Anticancer Drugs. 2017;28:723–38. doi: 10.1097/CAD.0000000000000494. [DOI] [PubMed] [Google Scholar]
- 48.Chi HT, Ly BT, Vu HA, Sato Y, Dung PC, Xinh PT. Down-regulated expression of NPM1 in IMS-M2 cell line by (-)-epigallocatechin-3-gallate. Asian Pac J Trop Biomed. 2014;4:570–4. doi: 10.12980/APJTB.4.2014APJTB-2014-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skayneh H, Jishi B, Hleihel R, Hamie M, El Hajj R, Deleuze-Masquefa C, et al. EAPB0503, an imidazoquinoxaline derivative modulates SENP3/ARF mediated SUMOylation, and induces NPM1c degradation in NPM1 mutant AML. Int J Mol Sci. 2022;23:3421. doi: 10.3390/ijms23073421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nabbouh AI, Hleihel RS, Saliba JL, Karam MM, Hamie MH, Wu HJM, et al. Imidazoquinoxaline derivative EAPB0503: a promising drug targeting mutant nucleophosmin 1 in acute myeloid leukemia. Cancer. 2017;123:1662–73. doi: 10.1002/cncr.30515. [DOI] [PubMed] [Google Scholar]
- 51.Sun X, Gao H, Yang Y, He M, Wu Y, Song Y, et al. PROTACs: great opportunities for academia and industry. Signal Transduct Target Ther. 2019;4:64. doi: 10.1038/s41392-019-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He Y, Khan S, Huo Z, Lv D, Zhang X, Liu X, et al. Proteolysis targeting chimeras (PROTACs) are emerging therapeutics for hematologic malignancies. J Hematol Oncol. 2020;13:103. doi: 10.1186/s13045-020-00924-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–60. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 54.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–11. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 55.Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, et al. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–7. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 56.Newlands ES, Rustin GJ, Brampton MH. Phase I trial of elactocin. Br J Cancer. 1996;74:648–9. doi: 10.1038/bjc.1996.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Q, Chen X, Zhou Q, Burstein E, Yang S, Jia D. Inhibiting cancer cell hallmark features through nuclear export inhibition. Signal Transduct Target Ther. 2016;1:16010. doi: 10.1038/sigtrans.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garzon R, Savona M, Baz R, Andreeff M, Gabrail N, Gutierrez M, et al. A phase 1 clinical trial of single-agent selinexor in acute myeloid leukemia. Blood. 2017;129:3165–74. doi: 10.1182/blood-2016-11-750158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhatnagar B, Zhao Q, Mims AS, Vasu S, Behbehani GK, Larkin K, et al. Selinexor in combination with decitabine in patients with acute myeloid leukemia: results from a phase 1 study. Leuk Lymphoma. 2020;61:387–96. doi: 10.1080/10428194.2019.1665664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fiedler W, Chromik J, Amberg S, Kebenko M, Thol F, Schlipfenbacher V, et al. A Phase II study of selinexor plus cytarabine and idarubicin in patients with relapsed/refractory acute myeloid leukaemia. Br J Haematol. 2020;190:e169–73. doi: 10.1111/bjh.16804. [DOI] [PubMed] [Google Scholar]
- 61.Sweet K, Bhatnagar B, Dohner H, Donnellan W, Frankfurt O, Heuser M, et al. A 2:1 randomized, open-label, phase II study of selinexor vs. physician’s choice in older patients with relapsed or refractory acute myeloid leukemia. Leuk Lymphoma. 2021;62:3192–203. doi: 10.1080/10428194.2021.1950706. [DOI] [PubMed] [Google Scholar]
- 62.Hing ZA, Fung HY, Ranganathan P, Mitchell S, El-Gamal D, Woyach JA, et al. Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies. Leukemia. 2016;30:2364–72. doi: 10.1038/leu.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Etchin J, Berezovskaya A, Conway AS, Galinsky IA, Stone RM, Baloglu E, et al. KPT-8602, a second-generation inhibitor of XPO1-mediated nuclear export, is well tolerated and highly active against AML blasts and leukemia-initiating cells. Leukemia. 2017;31:143–50. doi: 10.1038/leu.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischer MA, Friedlander SY, Arrate MP, Chang H, Gorska AE, Fuller LD, et al. Venetoclax response is enhanced by selective inhibitor of nuclear export compounds in hematologic malignancies. Blood Adv. 2020;4:586–98. doi: 10.1182/bloodadvances.2019000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DiNardo CD, Jonas BA, Pullarkat V, Thirman MJ, Garcia JS, Wei AH, et al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N Engl J Med. 2020;383:617–29. doi: 10.1056/NEJMoa2012971. [DOI] [PubMed] [Google Scholar]
- 66.Jones CL, Stevens BM, D’Alessandro A, Reisz JA, Culp-Hill R, Nemkov T, et al. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell. 2018;34:724–40. doi: 10.1016/j.ccell.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin M, et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med. 2018;24:1859–66. doi: 10.1038/s41591-018-0233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bisaillon R, Moison C, Thiollier C, Krosl J, Bordeleau ME, Lehnertz B, et al. Genetic characterization of ABT-199 sensitivity in human AML. Leukemia. 2020;34:63–74. doi: 10.1038/s41375-019-0485-x. [DOI] [PubMed] [Google Scholar]
- 69.Kontro M, Kumar A, Majumder MM, Eldfors S, Parsons A, Pemovska T, et al. HOX gene expression predicts response to BCL-2 inhibition in acute myeloid leukemia. Leukemia. 2017;31:301–9. doi: 10.1038/leu.2016.222. [DOI] [PubMed] [Google Scholar]
- 70.Lachowiez CA, Loghavi S, Kadia TM, Daver N, Borthakur G, Pemmaraju N, et al. Outcomes of older patients with NPM1-mutated AML: current treatments and the promise of venetoclax-based regimens. Blood Adv. 2020;4:1311–20. doi: 10.1182/bloodadvances.2019001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DiNardo CD, Tiong IS, Quaglieri A, MacRaild S, Loghavi S, Brown FC, et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood. 2020;135:791–803. doi: 10.1182/blood.2019003988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Konopleva M, Thirman MJ, Pratz KW, Garcia JS, Recher C, Pullarkat V, et al. Impact of F LT3 mutation on outcomes after venetoclax and azacitidine for patients with treatment-naive acute myeloid leukemia. Clin Cancer Res. 2022;28:2744–52. doi: 10.1158/1078-0432.CCR-21-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stahl M, Menghrajani K, Derkach A, Chan A, Xiao W, Glass J, et al. Clinical and molecular predictors of response and survival following venetoclax therapy in relapsed/refractory AML. Blood Adv. 2021;5:1552–64. doi: 10.1182/bloodadvances.2020003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Labrador J, Saiz-Rodriguez M, de Miguel D, de Laiglesia A, Rodriguez-Medina C, Vidriales MB, et al. Use or venetoclax in patients with relapsed or refractory acute myeloid leukemia: the PETHEMA registry experience. Cancers. 2022;14:1734. doi: 10.3390/cancers14071734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang YW, Tsai CH, Lin CC, Tien FM, Chen YW, Lin HY, et al. Cytogenetics and mutations could predict outcome in relapsed and refractory acute myeloid leukemia patients receiving BCL-2 inhibitor venetoclax. Ann Hematol. 2020;99:501–11. doi: 10.1007/s00277-020-03911-z. [DOI] [PubMed] [Google Scholar]
- 76.Winters AC, Gutman JA, Purev E, Nakic M, Tobin J, Chase S, et al. Real-world experience of venetoclax with azacitidine for untreated patients with acute myeloid leukemia. Blood Adv. 2019;3:2911–9. doi: 10.1182/bloodadvances.2019000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei AH, Strickland SA, Jr., Hou JZ, Fiedler W, Lin TL, Walter RB, et al. Venetoclax combined with low-dose cytarabine for previously untreated patients with acute myeloid leukemia: results from a phase Ib/II Study. J Clin Oncol. 2019;37:1277–84. doi: 10.1200/JCO.18.01600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wei AH, Montesinos P, Ivanov V, DiNardo CD, Novak J, Laribi K, et al. Venetoclax plus LDAC for newly diagnosed AML ineligible for intensive chemotherapy: a phase 3 randomized placebo-controlled trial. Blood. 2020;135:2137–45. doi: 10.1182/blood.2020004856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chua CC, Roberts AW, Reynolds J, Fong CY, Ting SB, Salmon JM, et al. Chemotherapy and venetoclax in elderly acute myeloid leukemia trial (CAVEAT): a Phase Ib dose-escalation study of venetoclax combined with modified intensive chemotherapy. J Clin Oncol. 2020;38:3506–17. doi: 10.1200/JCO.20.00572. [DOI] [PubMed] [Google Scholar]
- 80.DiNardo CD, Lachowiez CA, Takahashi K, Loghavi S, Xiao L, Kadia T, et al. Venetoclax combined with FLAG-IDA induction and consolidation in newly diagnosed and relapsed or refractory acute myeloid leukemia. J Clin Oncol. 2021;39:2768–78. doi: 10.1200/JCO.20.03736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chua CC, Hammond D, Kent A, Tiong IS, Konopleva MY, Pollyea DA, et al. Treatment-free remission after ceasing venetoclax-based therapy in patients with acute myeloid leukemia. Blood Adv. 2022;6:3879–83. doi: 10.1182/bloodadvances.2022007083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tiong IS, Dillon R, Ivey A, Teh TC, Nguyen P, Cummings N, et al. Venetoclax induces rapid elimination of NPM1 mutant measurable residual disease in combination with low-intensity chemotherapy in acute myeloid leukaemia. Br J Haematol. 2021;192:1026–30. doi: 10.1111/bjh.16722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carter BZ, Tao W, Mak PY, Ostermann LB, Mak D, McGeehan G, et al. Menin inhibition decreases Bcl-2 and synergizes with venetoclax in NPM1/FLT3-mutated AML. Blood. 2021;138:1637–41. doi: 10.1182/blood.2021011917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fiskus W, Boettcher S, Daver N, Mill CP, Sasaki K, Birdwell CE, et al. Effective Menin inhibitor-based combinations against AML with MLL rearrangement or NPM1 mutation (NPM1c) Blood Cancer J. 2022;12:5. doi: 10.1038/s41408-021-00603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu HH, Qian JJ, Sun WJ, You LS, Wang QQ, Naranmandura H, et al. Venetoclax and arsenic showed synergistic anti-leukemia activity in vitro and in vivo for acute myeloid leukemia with the NPM1 mutation. Am J Hematol. 2020;95:E55–7. doi: 10.1002/ajh.25719. [DOI] [PubMed] [Google Scholar]
- 86.Matkar S, Thiel A, Hua X. Menin: a scaffold protein that controls gene expression and cell signaling. Trends Biochem Sci. 2013;38:394–402. doi: 10.1016/j.tibs.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert-Buck MR, et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276:404–7. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 88.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–97. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 89.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–76. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 90.Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O’Laughlin M, et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia. 2015;29:1279–89. doi: 10.1038/leu.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oka M, Mura S, Otani M, Miyamoto Y, Nogami J, Maehara K, et al. Chromatin-bound CRM1 recruits SET-Nup214 and NPM1c onto HOX clusters causing aberrant HOX expression in leukemia cells. Elife. 2019;8:e46667. doi: 10.7554/eLife.46667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuhn MW, Song E, Feng Z, Sinha A, Chen CW, Deshpande AJ, et al. Targeting chromatin regulators inhibits leukemogenic geneeExpression in NPM1 mutant leukemia. Cancer Discov. 2016;6:1166–81. doi: 10.1158/2159-8290.CD-16-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8:277–84. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ozyerli-Goknar E, Nizamuddin S, Timmers HTM. A box of chemistry to inhibit the MEN1 tumor suppressor gene promoting leukemia. ChemMedChem. 2021;16:1391–402. doi: 10.1002/cmdc.202000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klossowski S, Miao H, Kempinska K, Wu T, Purohit T, Kim E, et al. Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia. J Clin Investig. 2020;130:981–97. doi: 10.1172/JCI129126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Uckelmann HJ, Kim SM, Wong EM, Hatton C, Giovinazzo H, Gadrey JY, et al. Therapeutic targeting of preleukemia cells in a mouse model of NPM1 mutant acute myeloid leukemia. Science. 2020;367:586–90. doi: 10.1126/science.aax5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gundry MC, Goodell MA, Brunetti L. It’s all about MEis: Menin-MLL inhibition eradicates NPM1-mutated and MLL-rearranged acute leukemias in mice. Cancer Cell. 2020;37:267–9. doi: 10.1016/j.ccell.2020.02.011. [DOI] [PubMed] [Google Scholar]
- 98.Dzama MM, Steiner M, Rausch J, Sasca D, Schonfeld J, Kunz K, et al. Synergistic targeting of FLT3 mutations in AML via combined menin-MLL and FLT3 inhibition. Blood. 2020;136:2442–56. doi: 10.1182/blood.2020005037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miao H, Kim E, Chen D, Purohit T, Kempinska K, Ropa J, et al. Combinatorial treatment with menin and FLT3 inhibitors induces complete remission in AML models with activating FLT3 mutations. Blood. 2020;136:2958–63. doi: 10.1182/blood.2020006575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Issa GC, Ravandi F, DiNardo CD, Jabbour E, Kantarjian HM, Andreeff M. Therapeutic implications of menin inhibition in acute leukemias. Leukemia. 2021;35:2482–95. doi: 10.1038/s41375-021-01309-y. [DOI] [PubMed] [Google Scholar]
- 101.Swaminathan M, Bourgeois W, Armstrong SA, Wang ES. Menin inhibitors in acute myeloid leukemia-What does the future hold? Cancer J. 2022;28:62–6. doi: 10.1097/PPO.0000000000000571. [DOI] [PubMed] [Google Scholar]
- 102.Krivtsov AV, Evans K, Gadrey JY, Eschle BK, Hatton C, Uckelmann HJ, et al. A Menin-MLL inhibitor induces specific chromatin changes and eradicates disease in models of MLL-rearranged leukemia. Cancer Cell. 2019;36:660–73.e11. doi: 10.1016/j.ccell.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cremer A, Ellegast JM, Alexe G, Frank ES, Ross L, Chu SH, et al. Resistance mechanisms to SYK inhibition in acute myeloid leukemia. Cancer Discov. 2020;10:214–31. doi: 10.1158/2159-8290.CD-19-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Walker AR, Byrd JC, Blachly JS, Bhatnagar B, Mims AS, Orwick S, et al. Entospletinib in combination with induction chemotherapy in previously ontreated acute myeloid leukemia: response and predictive significance of HOXA9 and MEIS1 expression. Clin Cancer Res. 2020;26:5852–9. doi: 10.1158/1078-0432.CCR-20-1064. [DOI] [PubMed] [Google Scholar]
- 105.Mohr S, Doebele C, Comoglio F, Berg T, Beck J, Bohnenberger H, et al. Hoxa9 and Meis1 cooperatively induce addiction to Syk signaling by suppressing miR-146a in acute myeloid leukemia. Cancer Cell. 2017;31:549–62.e11. doi: 10.1016/j.ccell.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.De Propris MS, Raponi S, Diverio D, Milani ML, Meloni G, Falini B, et al. High CD33 expression levels in acute myeloid leukemia cells carrying the nucleophosmin (NPM1) mutation. Haematologica. 2011;96:1548–51. doi: 10.3324/haematol.2011.043786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perriello VM, Gionfriddo I, Rossi R, Milano F, Mezzasoma F, Marra A, et al. CD123 Is consistently expressed on NPM1-mutated AML Cells. Cancers. 2021;13:496. doi: 10.3390/cancers13030496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martelli MP, Pettirossi V, Thiede C, Bonifacio E, Mezzasoma F, Cecchini D, et al. CD34+ cells from AML with mutated NPM1 harbor cytoplasmic mutated nucleophosmin and generate leukemia in immunocompromised mice. Blood. 2010;116:3907–22. doi: 10.1182/blood-2009-08-238899. [DOI] [PubMed] [Google Scholar]
- 109.Sportoletti P, Varasano E, Rossi R, Mupo A, Tiacci E, Vassiliou G, et al. Mouse models of NPM1-mutated acute myeloid leukemia: biological and clinical implications. Leukemia. 2015;29:269–78. doi: 10.1038/leu.2014.257. [DOI] [PubMed] [Google Scholar]
- 110.Liso A, Bogliolo A, Freschi V, Martelli MP, Pileri SA, Santodirocco M, et al. In human genome, generation of a nuclear export signal through duplication appears unique to nucleophosmin (NPM1) mutations and is restricted to AML. Leukemia. 2008;22:1285–9. doi: 10.1038/sj.leu.2405045. [DOI] [PubMed] [Google Scholar]
- 111.Liso A, Colau D, Benmaamar R, De Groot A, Martin W, Benedetti R, et al. Nucleophosmin leukaemic mutants contain C-terminus peptides that bind HLA class I molecules. Leukemia. 2008;22:424–6. doi: 10.1038/sj.leu.2404887. [DOI] [PubMed] [Google Scholar]
- 112.Narayan R, Olsson N, Wagar LE, Medeiros BC, Meyer E, Czerwinski D, et al. Acute myeloid leukemia immunopeptidome reveals HLA presentation of mutated nucleophosmin. PLoS ONE. 2019;14:e0219547. doi: 10.1371/journal.pone.0219547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van der Lee DI, Reijmers RM, Honders MW, Hagedoorn RS, de Jong RC, Kester MG, et al. Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia. J Clin Investig. 2019;129:774–85. doi: 10.1172/JCI97482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kuzelova K, Brodska B, Fuchs O, Dobrovolna M, Soukup P, Cetkovsky P. Altered HLA class I profile associated with type A/D nucleophosmin mutation points to possible anti-nucleophosmin immune response in acute myeloid leukemia. PLoS ONE. 2015;10:e0127637. doi: 10.1371/journal.pone.0127637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Forghieri F, Riva G, Lagreca I, Barozzi P, Vallerini D, Morselli M, et al. Characterization and dynamics of specific T cells against nucleophosmin-1 (NPM1)-mutated peptides in patients with NPM1-mutated acute myeloid leukemia. Oncotarget. 2019;10:869–82. doi: 10.18632/oncotarget.26617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hofmann S, Gotz M, Schneider V, Guillaume P, Bunjes D, Dohner H, et al. Donor lymphocyte infusion induces polyspecific CD8(+) T-cell responses with concurrent molecular remission in acute myeloid leukemia with NPM1 mutation. J Clin Oncol. 2013;31:e44–7. doi: 10.1200/JCO.2011.41.1116. [DOI] [PubMed] [Google Scholar]
- 117.Greiner J, Schneider V, Schmitt M, Gotz M, Dohner K, Wiesneth M, et al. Immune responses against the mutated region of cytoplasmatic NPM1 might contribute to the favorable clinical outcome of AML patients with NPM1 mutations (NPM1mut) Blood. 2013;122:1087–8. doi: 10.1182/blood-2013-04-496844. [DOI] [PubMed] [Google Scholar]
- 118.Aldoss I, Nakamura R, Yang D, Salhotra A, Stein AS, Pullarkat V, et al. Favorable outcomes for allogeneic hematopoietic cell transplantation in elderly patients with NPM1-mutated and FLT3-ITD-negative acute myeloid leukemia. Bone Marrow Transplant. 2020;55:473–5. doi: 10.1038/s41409-019-0553-x. [DOI] [PubMed] [Google Scholar]
- 119.Laszlo GS, Estey EH, Walter RB. The past and future of CD33 as therapeutic target in acute myeloid leukemia. Blood Rev. 2014;28:143–53. doi: 10.1016/j.blre.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 120.Hills RK, Castaigne S, Appelbaum FR, Delaunay J, Petersdorf S, Othus M, et al. Addition of gemtuzumab ozogamicin to induction chemotherapy in adult patients with acute myeloid leukaemia: a meta-analysis of individual patient data from randomised controlled trials. Lancet Oncol. 2014;15:986–96. doi: 10.1016/S1470-2045(14)70281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fournier E, Duployez N, Ducourneau B, Raffoux E, Turlure P, Caillot D, et al. Mutational profile and benefit of gemtuzumab ozogamicin in acute myeloid leukemia. Blood. 2020;135:542–6. doi: 10.1182/blood.2019003471. [DOI] [PubMed] [Google Scholar]
- 122.Kapp-Schwoerer S, Weber D, Corbacioglu A, Gaidzik VI, Paschka P, Kronke J, et al. Impact of gemtuzumab ozogamicin on MRD and relapse risk in patients with NPM1-mutated AML: results from the AMLSG 09-09 trial. Blood. 2020;136:3041–50. doi: 10.1182/blood.2020005998. [DOI] [PubMed] [Google Scholar]
- 123.Schlenk RF, Paschka P, Krzykalla J, Weber D, Kapp-Schwoerer S, Gaidzik VI, et al. Gemtuzumab Ozogamicin in NPM1-mutated acute myeloid leukemia: early results from the prospective randomized AMLSG 09-09 Phase III study. J Clin Oncol. 2020;38:623–32. doi: 10.1200/JCO.19.01406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pemmaraju N, Konopleva M. Approval of tagraxofusp-erzs for blastic plasmacytoid dendritic cell neoplasm. Blood Adv. 2020;4:4020–7. doi: 10.1182/bloodadvances.2019000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Greiner J, Hofmann S, Schmitt M, Gotz M, Wiesneth M, Schrezenmeier H, et al. Acute myeloid leukemia with mutated nucleophosmin 1: an immunogenic acute myeloid leukemia subtype and potential candidate for immune checkpoint inhibition. Haematologica. 2017;102:e499–501. doi: 10.3324/haematol.2017.176461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dufva O, Polonen P, Bruck O, Keranen MAI, Klievink J, Mehtonen J, et al. Immunogenomic landscape of hematological malignancies. Cancer Cell. 2020;38:424–8. doi: 10.1016/j.ccell.2020.08.019. [DOI] [PubMed] [Google Scholar]
- 127.Lines JL, Pantazi E, Mak J, Sempere LF, Wang L, O’Connell S, et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014;74:1924–32. doi: 10.1158/0008-5472.CAN-13-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Greiner J, Goetz M, Schuler PJ, Bulach C, Hofmann S, Schrezenmeier H, et al. Enhanced stimulation of antigen-specific immune responses against nucleophosmin 1 mutated acute myeloid leukaemia by an anti-programmed death 1 antibody. 2022. Br J Haematol. 10.1111/bjh.18326. [DOI] [PubMed]
- 129.Vadakekolathu J, Minden MD, Hood T, Church SE, Reeder S, Altmann H, et al. Immune landscapes predict chemotherapy resistance and immunotherapy response in acute myeloid leukemia. Sci Transl Med. 2020;12:eaaz0463. doi: 10.1126/scitranslmed.aaz0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Damotte D, Warren S, Arrondeau J, Boudou-Rouquette P, Mansuet-Lupo A, Biton J, et al. The tumor inflammation signature (TIS) is associated with anti-PD-1 treatment benefit in the CERTIM pan-cancer cohort. J Transl Med. 2019;17:357. doi: 10.1186/s12967-019-2100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tripodo C, Burocchi A, Piccaluga PP, Chiodoni C, Portararo P, Cappetti B, et al. Persistent immune stimulation exacerbates genetically driven myeloproliferative disorders via stromal remodeling. Cancer Res. 2017;77:3685–99. doi: 10.1158/0008-5472.CAN-17-1098. [DOI] [PubMed] [Google Scholar]
- 132.Daver N, Garcia-Manero G, Basu S, Boddu PC, Alfayez M, Cortes JE, et al. Efficacy, safety, and biomarkers of response to azacitidine and nivolumab in relapsed/refractory acute myeloid leukemia: a nonrandomized, open-label, Phase II study. Cancer Discov. 2019;9:370–83. doi: 10.1158/2159-8290.CD-18-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Goswami M, Gui G, Dillon LW, Lindblad KE, Thompson J, Valdez J, et al. Pembrolizumab and decitabine for refractory or relapsed acute myeloid leukemia. J Immunother Cancer. 2022;10:e003392. doi: 10.1136/jitc-2021-003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saxena K, Herbrich SM, Pemmaraju N, Kadia TM, DiNardo CD, Borthakur G, et al. A phase 1b/2 study of azacitidine with PD-L1 antibody avelumab in relapsed/refractory acute myeloid leukemia. Cancer. 2021;127:3761–71. doi: 10.1002/cncr.33690. [DOI] [PubMed] [Google Scholar]
- 135.Vago L, Gojo I. Immune escape and immunotherapy of acute myeloid leukemia. J Clin Invest. 2020;130:1552–64. doi: 10.1172/JCI129204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen X, Pan X, Zhang W, Guo H, Cheng S, He Q, et al. Epigenetic strategies synergize with PD-L1/PD-1 targeted cancer immunotherapies to enhance antitumor responses. Acta Pharm Sin B. 2020;10:723–33. doi: 10.1016/j.apsb.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zeidner JF, Vincent BG, Ivanova A, Moore D, McKinnon KP, Wilkinson AD, et al. Phase II trial of pembrolizumab after high-dose cytarabine in relapsed/refractory acute myeloid leukemia. Blood Cancer Discov. 2021;2:616–29. doi: 10.1158/2643-3230.BCD-21-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee JB, Khan DH, Hurren R, Xu M, Na Y, Kang H, et al. Venetoclax enhances T cell-mediated antileukemic activity by increasing ROS production. Blood. 2021;138:234–45. doi: 10.1182/blood.2020009081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kohlhapp FJ, Haribhai D, Mathew R, Duggan R, Ellis PA, Wang R, et al. Venetoclax increases intratumoral effector T cells and antitumor efficacy in combination with immune checkpoint blockade. Cancer Discov. 2021;11:68–79. doi: 10.1158/2159-8290.CD-19-0759. [DOI] [PubMed] [Google Scholar]
- 140.Xie G, Ivica NA, Jia B, Li Y, Dong H, Liang Y, et al. CAR-T cells targeting a nucleophosmin neoepitope exhibit potent specific activity in mouse models of acute myeloid leukaemia. Nat Biomed Eng. 2021;5:399–413. doi: 10.1038/s41551-020-00625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Greiner J, Ono Y, Hofmann S, Schmitt A, Mehring E, Gotz M, et al. Mutated regions of nucleophosmin 1 elicit both CD4(+) and CD8(+) T-cell responses in patients with acute myeloid leukemia. Blood. 2012;120:1282–9. doi: 10.1182/blood-2011-11-394395. [DOI] [PubMed] [Google Scholar]
- 142.Kuzelova K, Brodska B, Schetelig J, Rollig C, Racil Z, Walz JS, et al. Association of HLA class I type with prevalence and outcome of patients with acute myeloid leukemia and mutated nucleophosmin. PLoS ONE. 2018;13:e0204290. doi: 10.1371/journal.pone.0204290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dong H, Ham JD, Hu G, Xie G, Vergara J, Liang Y, et al. Memory-like NK cells armed with a neoepitope-specific CAR exhibit potent activity against NPM1 mutated acute myeloid leukemia. Proc Natl Acad Sci USA. 2022;119:e2122379119. doi: 10.1073/pnas.2122379119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Perriello VM, Rotiroti RM, Pisani I, Alberti G, Pianigiani G, Rossi R, et al. CD123 and CD33 co-targeting by balanced signaling on CAR-CIK cells reduces potential off-target toxicity while preserving the anti-leukemic activity of acute myeloid leukemia. Blood. 2021;138:1699. [Google Scholar]
- 145.van der Lee DI, Koutsoumpli G, Reijmers RM, Honders MW, de Jong RCM, Remst DFG, et al. An HLA-A*11:01-binding neoantigen from mutated NPM1 as target for TCR gene therapy in AML. Cancers. 2021;13:5390. doi: 10.3390/cancers13215390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang B, Tang C, Yao Y, Chen X, Zhou C, Wei Z, et al. The tumor therapy landscape of synthetic lethality. Nat Commun. 2021;12:1275. doi: 10.1038/s41467-021-21544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Carnero A. High throughput screening in drug discovery. Clin Transl Oncol. 2006;8:482–90. doi: 10.1007/s12094-006-0048-2. [DOI] [PubMed] [Google Scholar]
- 148.Hoelder S, Clarke PA, Workman P. Discovery of small molecule cancer drugs: successes, challenges and opportunities. Mol Oncol. 2012;6:155–76. doi: 10.1016/j.molonc.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Coussens NP, Braisted JC, Peryea T, Sittampalam GS, Simeonov A, Hall MD. Small-molecule screens: a gateway to cancer therapeutic agents with case studies of Food and Drug Administration-approved drugs. Pharm Rev. 2017;69:479–96. doi: 10.1124/pr.117.013755. [DOI] [PMC free article] [PubMed] [Google Scholar]