Abstract
Plant pathogenic Gram-negative bacteria evade the host plant immune system by secreting Type III (T3E) and Type IV effector (T4E) proteins into the plant cytoplasm. Mostly T3Es are secreted into the plant cells to establish pathogenicity by affecting the vital plant process viz. metabolic pathways, signal transduction and hormonal regulation. Ubiquitin-26S proteasome system (UPS) exists as one of the important pathways in plants to control plant immunity and various cellular processes by employing several enzymes and enzyme components. Pathogenic and non-pathogenic bacteria are found to secrete effectors into plants with structural and/or functional similarity to UPS pathway components like ubiquitin E3 ligases, F-box domains, cysteine proteases, inhibitor of host UPS or its components, etc. The bacterial effectors mimic UPS components and target plant resistance proteins for degradation by proteasomes, thereby taking control over the host cellular activities as a strategy to exert virulence. Thus, the bacterial effectors circumvent plant cellular pathways leading to infection and disease development. This review highlights known bacterial T3E and T4E proteins that function and interfere with the ubiquitination pathway to regulate the immune system of plants.
Keywords: Plant immunity, Effectors, T3E, Ubiquitin-proteasome system, E3 ubiquitin ligases, Pathogen-associated molecular patterns
1. Introduction
Plants confront beneficial and pathogenic microorganisms throughout their life cycle. The ability of plant pathogenic bacteria to invade a host depends on its secretions, virulence, and pathogenicity. To avert and defend against pathogens, plants have developed sophisticated immune systems besides their inherent physical barriers. The first line of host defense or the basal defense is triggered instantaneously upon the detection of conserved pathogen- or microbe-associated molecular patterns (P/MAMPs). The recognition of different PAMPs by specific pattern recognition receptors (PRRs) initiates PAMP-triggered immunity (PTI) (Shan et al., 2008; Zipfel, 2014). P/MAMPs are compounds produced or present only in microorganisms like conserved cell surface structures, including bacterial flagellin, elongation factor Tu (EF-Tu), lipopolysaccharides, and peptidoglycan, or fungal cell wall components like chitin or glucan (Torres, 2010; Zipfel et al., 2006; Torres, 2010; Boller and Felix, 2009). The PTI produces cellular responses that include production of reactive oxygen species (ROS), ion fluxes and activation of defense-related genes, resulting in callose deposition at the cell wall, defense hormone synthesis, stomatal closure (Zipfel, 2014) and localized cell death. Adapted plant pathogens overcome PTI by delivering type III secretory proteins or effector proteins (T3Es) directly into the host cell cytoplasm through type III secretory system (T3SS) and induce effector-triggered susceptibility1 (ETS1) and promote bacterial virulence.
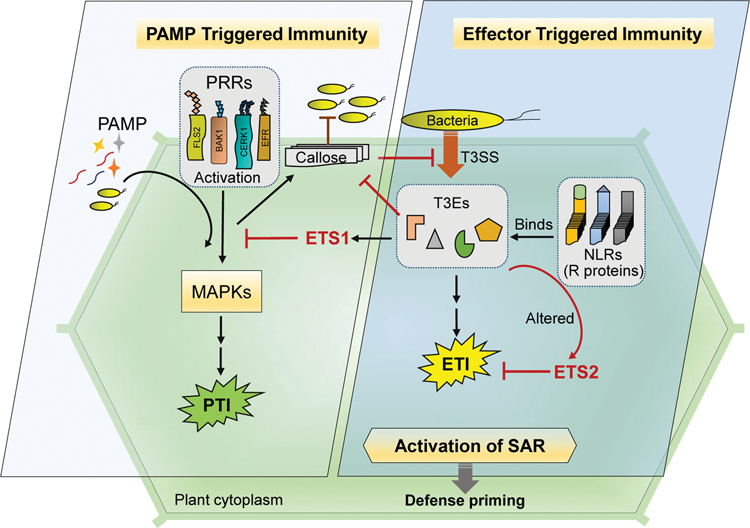
To counter attack the pathogen-secreted effectors, plants have evolved a second line of immune response. This response acts primarily inside the plant cell and relies upon the ability to detect effector intrusion through intracellular immune receptors called nucleotide-binding, leucine-rich repeat receptors (NLRs) or R proteins, resulting in effector triggered immunity (ETI) (Jones and Dangl, 2006). ETI is an amplified and accelerated PTI response resulting in disease resistance (Jones and Dangl, 2006). ETI is typically accompanied by the hypersensitive response (HR), a form of localized programmed cell death (PCD) at the primary infection site that restricts pathogen spread within infected tissue (Hofius et al., 2009). Loss or modification of R proteins due to effector action can lead to effector-triggered susceptibility2 (ETS2), which inhibits the ETI. When the R protein takes over during localized pathogen attack, increased resistance toward secondary infection is observed in uninfected parts of plants. This resistance is referred to as systemic acquired resistance (SAR) (Craig et al., 2009; Fu and Dong, 2013). SAR-induced plants are said to be sensitized or primed to respond more rapidly and effectively to secondary infection (Fig. 1). The arms race continues depending on the ability of the pathogenic effectors or the plant immune system to co-evolve during the process of invasion and resistance.
Fig. 1.
Activation of plant immune system. A two-tiered immune system comprising pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) occur in plants. The instant immune response is mediated by PRRs that triggers PTI whereas NLR activate ETI upon translocation of T3E into the cytoplasm via. T3SS. Signalling events by PRRs and NLRs leads to overlapping downstream cellular responses, including defense-gene expression, production of ROS and callose deposition. The primary plant immune receptors viz. PRRs and NLRs function synergistically to ensure a robust level of key immune components during ETI. SAR is induced when the ETI takes over the pathogenic effector and plants are able to resist secondary infection in uninfected parts of plants.
Recent studies unite two major immune signaling cascades in plants viz., PTI and ETI, and find PTI to be an integral component of ETI. Production of ROS as an early response during PTI is a critical early signaling event that play a major role in establishing the synergy between PRR- and NLR-mediated immunity, and also the PRR mediated downstream receptor signaling following the perception of pathogen is indispensable for producing an effective ETI response (Yuan et al., 2021). Activation of both PTI and ETI immune systems is necessary for mediating strong defenses against pathogens and either of them alone is insufficient for establishing robust immune response and resistance in plants (Ngou et al., 2021). Effective defense against pathogens relies on mutual potentiation of both PTI and ETI components (Pruitt et al., 2021). These studies indicate the importance of both immune responses for establishing a strong defense system.
Bacterial effectors or virulence factors, the main players for pathogen survival are delivered by specialized and sophisticated protein secretion systems, which are grouped into at least six different classes, designated type I to type VI (Costa et al., 2015; Hayes et al., 2010). Among the secretion systems, type I, type III, and type IV are involved in virulence and are highly conserved among the pathogens of plants and humans (Guttman, 2004). T3E proteins interfere and modulate many plant regulatory processes viz., basal PTI defense, signal transduction pathways, proteasome-dependent protein degradation, phytohormone signaling, plant gene expression and the plant cytoskeleton (Buttner, 2016). T3SS are present in plant- and animal-pathogenic bacteria viz., Aeromonas, Bordetella, Burkholderia, Chlamydia, Erwinia, Escherichia, Pseudomonas, Ralstonia, Rhizobium, Salmonella, Shigella, Vibrio and Xanthomonas spp. (Troisfontaines and Cornelis, 2005) as well as in the non-pathogenic symbiotic bacterium Rhizobium (Dai et al., 2008). Bacteria have developed strategies to invade plants using plant-like proteins, including E3 ligases, F-box proteins, etc., to increase pathogenicity. T3Es from pathogenic and non-pathogenic bacteria have been reported to modulate the ubiquitin-26S proteasome system (UPS) and other cellular processes (Angot et al., 2007; Ashida et al., 2014; Üstün et al., 2014; Weßling et al., 2014; Buttner, 2016). In addition, plant viruses are found to regulate host UPS (Chen et al., 2020; Gao and Luo, 2006; Gustin et al., 2011), and eukaryotic plant-parasitic nematodes are identified that secrete Ub extension proteins to interfere with the UPS in plant cells (Tytgat et al., 2004). The effectors mimicking UPS from animal and human pathogens have been well-reviewed (Angot et al., 2007; Ashida et al., 2014; Kim et al., 2014; Lin and Machner, 2017). However, plant pathogenic bacterial effectors regulating UPS are less explored despite numerous plant pathogenic bacteria. With this perspective, the current review bestows an overview of the effectors from pathogenic and non-pathogenic bacteria that are associated with ubiquitination and/or the UPS pathway to remodel the immune response in plants.
2. Ubiquitin-26S proteasome system
The UPS and autophagy are the major protein degradation pathways that occur in eukaryotes. Regulated protein turnover by UPS and autophagy controls many facets of plant immunity, including recognition of pathogens, immune receptor accumulation, and downstream defense signaling (Langin et al., 2020; Leary et al., 2019; Üstün et al., 2017, 2018). Autophagy was considered as a non-specific recycling process that occurs during nutrient limitation; however, several reports have shown that autophagy can act selectively to degrade protein aggregates, organelles, and specific proteins (Kraft et al., 2009; Randow and Youle, 2014; Zaffagnini and Martens, 2016).
In autophagy, double-membrane vesicles termed autophagosomes form that sequester and deliver cytoplasmic cargo to vacuoles for breakdown and recycling. In this pathway, protein aggregates, specific proteins, macromolecules like ribosomes and proteasomes, fragments of endoplasmic reticulum (ER) or nuclei, whole organelles like mitochondria, peroxisomes, and chloroplasts, and possibly even invading pathogens, can be selectively eliminated (Marshall and Vierstra, 2018). Viral effectors are reported to target the autophagic pathway (Yang et al., 2018; Hafren et al., 2017, Hafren, 2018). However, bacterial effectors targeting autophagy-mediated degradation has been reported only in a few bacteria (Üstün et al., 2018; Leong et al., 2021). More research is needed to identify bacterial effectors that target autophagic pathway (Langin et al., 2020).
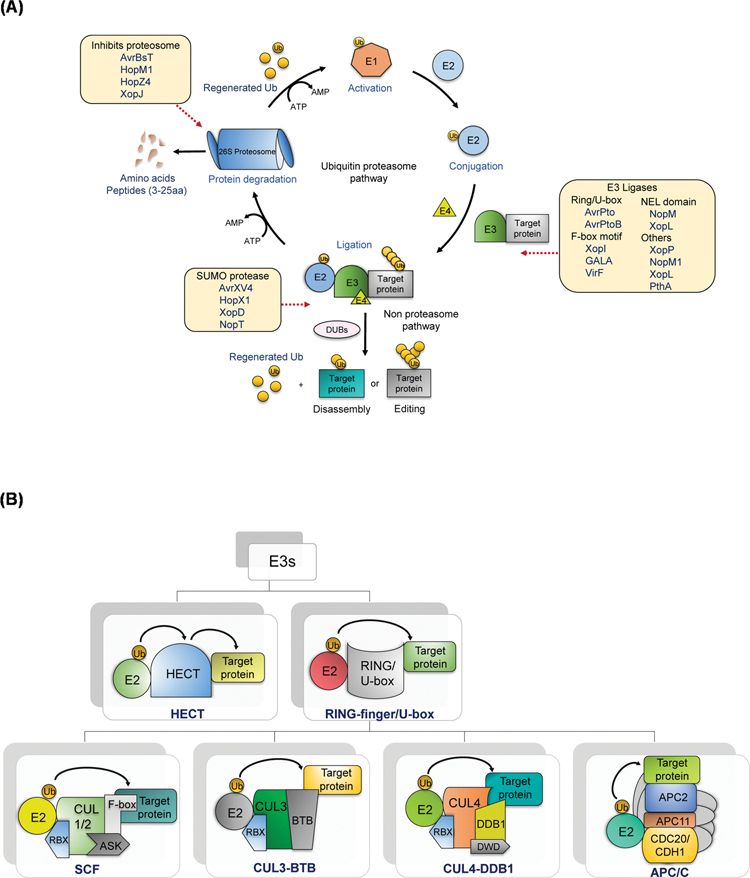
In contrast to autophagy where bulk protein, organelles or pathogens are degraded, UPS mainly degrades short-lived proteins or mis-folded proteins through a process called ubiquitination. Ubiquitination is a post-translational modification (PTM) of proteins that controls a variety of processes in eukaryotic cells. Ubiquitination can target a protein for degradation (turnover) by 26S proteasomes resulting in the removal of rate-limiting enzymes in regulation and mis-folded proteins in protein quality control as well as the maintenance of amino acid pools during growth and starvation. Ubiquitination also regulates hormonal signaling (Wang and Deng, 2011; Kim et al., 2016), stress response (Shu and Yang, 2017; Stone, 2014), defense response (Luo et al., 2010; Lin et al., 2008), programmed cell death (Lin et al., 2008), vesicle trafficking (Xu et al., 2009; Glickman and Ciechanover, 2002; Foot et al., 2017; Pickart and Eddins, 2004), cell cycle (Petroski and Deshaies, 2005), DNA repair (Chen and Sun, 2009) etc. Ubiquitination targets immune receptors and downstream signaling components to maintain homeostasis in the absence of pathogen (Copeland and Li, 2019). Genome-wide studies predict nearly 6 % of the Arabidopsis thaliana genome encodes UPS components (Vierstra, 2009). In this pathway, ubiquitin (Ub), a small regulatory protein, serves as a reusable tag that targets proteins for selective turnover as well as alterations in protein-protein interactions, localization and enzyme activity. Monomeric units or polymeric chains of Ub are formed as covalent attachments on protein targets through a three-step cascade composed of three major enzymes: E1 (ubiquitin-activation enzyme), E2 (ubiquitin-conjugating enzyme) and E3 (ubiquitin ligase) (Thrower et al., 2000; Weissman et al., 2011) (Fig. 2A). The pathway originates with the activation of ubiquitin that requires an ATP and the formation of a thioester linkage between the carboxy-terminal glycine of ubiquitin and a specific cysteine residue of the E1. The activated ubiquitin is transferred to the E2 protein, after which the E3 ligase mediates the transfer of ubiquitin from E2 to a lysine residue on the substrate. Efficient poly ubiquitination is facilitated by multi-ubiquitin chain assembly factors, E4, to transfer additional ubiquitin moieties (Koegl et al., 1999). Ubiquitin has eight linking options to form the polymeric Ub chains, including seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) and its N-terminal methionine residue (M1) (Swatek and Komander, 2016). Ubiquitin linkages on Ser, Thr and Cys residues of target proteins are also reported (McClellan et al., 2019). Target proteins with more than four Ubs attached in K48 linkages are typically degraded by 26S proteasomes (Thrower et al., 2000).
Fig. 2.
(A). Ubiquitination-26S proteasome pathway.
Ubiquitin is first activated by ubiquitin-activating enzyme (E1), at the expenditure of ATP. Then, the ubiquitin molecule is passed on to the second enzyme, ubiquitin-conjugating enzyme (E2). The final enzyme, ubiquitin ligase (E3), recognizes target substrate and binds and labels it with the ubiquitin. The poly-ubiquitination is facilitated by E4, which transfers additional ubiquitin moieties. Proteins modified by sequential linkage of multiple ubiquitin residues of at least four via ubiquitin degradation are targeted by the 26S proteasome. In the non-proteasomal pathway, deubiquitinase (DUBs) catalyze the disassembly and editing of the Ub moieties attached to protein substrates. The various plant effectors that interfere with the UPS are indicated along the pathway.
(B). Plant E3 ubiquitin ligases.
E3 can be divided into HECT and RING/U-box domain-containing E3s based on their mode of transfer of Ub. RING E3s catalyze the transfer of Ub, whereas in the HECT, the E3 forms an intermediate and transfers to the target. The RING/U-box E3s are the multi-subunit complex of E3s viz., SCF, CUL3-BTB, CUL4- DDB1 and APC/C.
The process of ubiquitination requires variety of enzymes and enzyme components, and the substrate specificity of this system depends on multiple E2 and E3 combinations. Plants usually encode for only one or two E1 proteins that activate the Ub through adenylation and thioester intermediate formation (Callis et al., 1995). The E2 Ub conjugating enzymes that serve to transfer the Ub from the E1 to the E3s or substrate are more numerous and clustered into 12 groups (Callis and Vierstra, 2000). The E3 Ub ligases that transfer the Ub from an E2-Ub thioester to the substrate protein are the most diverse class of enzymes that define the substrate specificity of ubiquitination. More than 1000 E3s occur in plants and are found to function as a single component or multi-subunit complex (Chen and Hellmann, 2013). E2s share many well-conserved catalytic domains, but E3s share only a few conserved motifs. E3 enzymes are classified into two broad structural classes: i) Homologous to E6-associated protein Carboxyl Terminus (HECT) and ii) Really Interesting New Gene (RING)/U-box domain. The RING-type E3 enzymes simply catalyze Ub transfer without being ubiquitinated, whilst the HECT-type E3s form an intermediate thioester complex and transfer Ub to the target (Vierstra, 2009). RING-type E3s function as monomers, dimers, or as multi-subunit complexes and are divided into three broad categories: i) S-phase kinase-associated protein 1 (SKP1)-cullin 1 (CUL1)-F-box (SCF), ii) Cullin3-Bric-a-BracTramtrackBroad-complex (CUL3-BTB), and iii) Cullin4-DNA-Damage Binding 1 (CUL4-DDB1). The multi-component E3s contain: i) a catalytic RING domain, e.g., RING-box 1 (RBX1) or Anaphase Promoting Complex 11(APC11), ii) an assembly platform, e.g., CUL1–4 or APC2, and iii) a substrate recognition domain, e.g., F-box, BTB, or DDB1. RING E3s can act independently or as part of a multi-subunit complex such as SCF. The type of cullin subunit determines which recognition protein is to be incorporated into the complex. A fourth multi-subunit E3 is named APC/C (APC/cyclosome) that consists of 11 subunits (Zhang et al., 2014) (Fig. 2B, Supplementary Figs. S1 and S2). Ubiquitin-like proteins (UBLs/Ulps), Nedd8 (or Rub1) and small ubiquitin-related modifier proteins (SUMOs) have also been identified that can be covalently attached to target proteins similar to Ub (Vierstra, 2012).
The opposing action of ubiquitination is carried out by the deubiquitinase (DUBs), that act on the ubiquitinated substrates and either edit or disassemble the Ub linked target, leading to an alternative pathway (Nijman et al., 2005). DUBs can release Ub from an inactive precursors that are translated from Ub gene fusions or can cleave Ub moieties that are attached post-translationally to the target protein (Hepowit et al., 2012). DUBs are the second most diverse group of UPS components with around 64 members, as reported in A. thaliana (Vierstra, 2009).
Proteins modified with K48 linked poly-ubiquitin chains are substrates for the 26S proteasome, a 2.5 MDa ATP-dependent protease complex that is present in both the cytoplasm and nucleus. The 26S proteasome of yeast, mammals and some plant (Arabidopsis and rice) are quite similar and comprised of 31 subunits divided into two sub-complexes, the 20S core protease (CP) and 19S regulatory particle (RP). However plants have more advanced 26S proteasome with assembled multiple isoforms (Smalle and Vierstra, 2004). The opening to this chamber is sufficiently narrow to restrict entry to only those substrates that are unfolded and threaded inside. The regulatory particle (RP) is found on either or both ends of the CP. The RP is involved in ubiquitin-conjugate recognition, Ub recycling, unfolding the target protein, transporting the target protein into the CP chamber and presumably releasing the breakdown products. More specifically, a ring of six RP triple-A (AAA+) ATPases (RPTs 1–6) covers the opening to the CP and assists in target unfolding. The RP non-ATPases (RPNs) 10 and 13 are Ub receptors, and RPN11 is a deubiquitylating enzyme (DUB) that helps to release bound ubiquitins. The functions of many of the RP subunits have not been elucidated. However, several RP subunits in A. thaliana had substrate-specific functions. RPN10 and RPN12a participate in ABA and cytokinin signalling, respectively. RPN1 and RPN5 are essential for embryogenesis and RPT2a participates in root development (Vierstra, 2009).
Owing to the importance of the UPS in various plant cellular processes and host defense, this system has become a major target for pathogenic microbes to subvert plant immunity and gain control over plants to enable pathogen survival. Important plant pathogenic bacteria and non-pathogenic bacteria, like Rhizobium spp., produce effectors that have similarities to plant proteins participating in the UPS pathway, thereby interfering with the host cellular immunity. The ‘arms race’ between pathogens and host plants might have led to the evolution of numerous types of effectors in microbial pathogens that converge and have high degree of interaction on common host plant proteins and defense-related or R proteins in plants (Rovenich et al., 2016; Weßling et al., 2014 Langin et al., 2020).
3. Pseudomonas syringae: RING/U-box domain like-effectors and proteasome inhibitors
The pathogen P. syringae pv. tomato (Pst) that causes bacterial speck disease in tomato is one of the best-studied pathosystems for plant-microbe interaction. Pst effectors viz., AvrPto and AvrPtoB are the pioneer effectors in the T3E studies to describe the modulation, evolution and interplay of bacterial effectors in plants. Pst delivers about 30 effectors through T3SS into the plant cell, and a subset of eight effectors are shown to be sufficient to establish full virulence in one of its plant hosts (Cunnac et al., 2011). Among this subset, AvrPtoB (HopAB2) is an T3E of Pst that targets the plant UPS and acts with priority in suppressing immunity in the modulation, evolution and interplay of the bacterial pathogen with its host. AvrPtoB triggers the HR and immunity in plant during pathogenesis (Guo et al., 2009). The tomato gene conferring resistance to bacterial speck disease is ‘Pto’ (resistance to P. syringae pv. tomato) which encodes for the kinase protein Pto. The Pto-mediated ETI in tomato is the best example of effector evolution and the R-protein mediated defense to counteract the effectors. The interplay between pathogen and plant is well studied, and at least 25 genes in the Pto-mediated ETI are reported (Oh and Martin, 2011). In tomato genotypes that are immune to speck disease, the Pst effectors AvrPtoB as well as AvrPto were recognized by the tomato R protein Pto, to initiate HR and resistance leading to ETI. Pto interacts with another R protein Prf and, upon binding of AvrPtoB induces resistance, via programmed cell death (PCD), thereby limiting the pathogen growth (Oh and Martin, 2011; Pedley and Martin, 2003) (Fig. 3). Fen is evolved in plants to activate defense signaling. To counteract Fen recognition and suppress the basal defense mechanism induced in tomato plants, the N-terminal domain of AvrPtoB from Pst had acquired a C-terminal E3 ligase domain to mediate the Fen degradation (Rosebrock et al., 2007). Finally, the Pto was evolved to restore resistance response to suppress AvrPtoB ubiquitination activity. Pto possibly phosphorylates AvrPtoB near its E3 ubiquitin ligase domain to prevent the targeted ubiquitination and degradation of plant defense proteins (Mathieu et al., 2014).
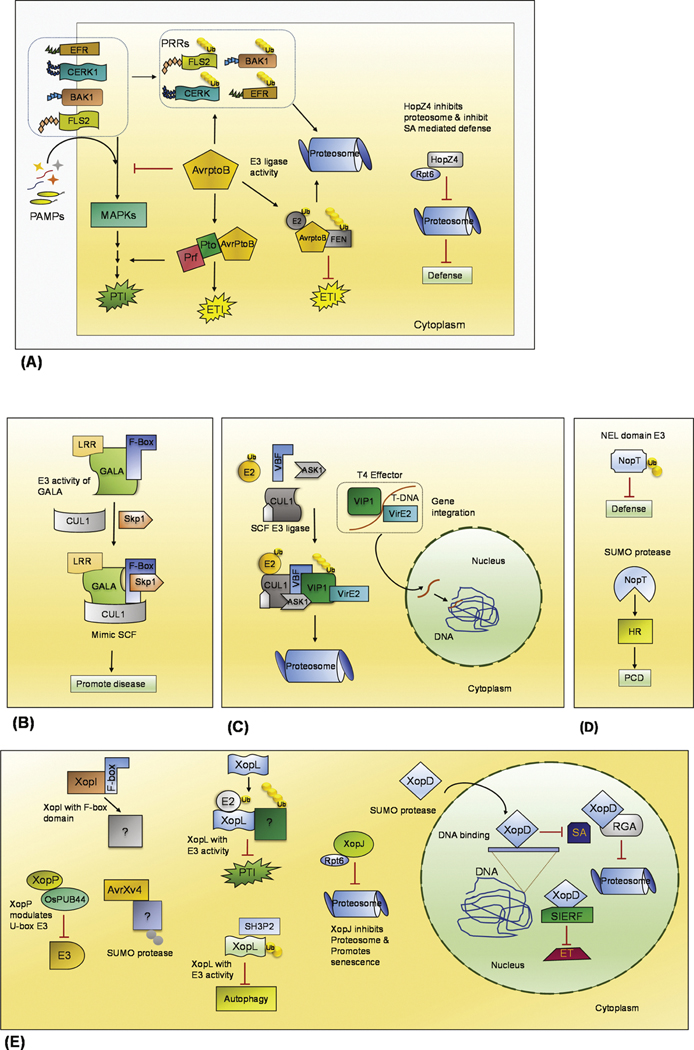
Fig. 3.
Illustration of bacterial effectors that manipulate the plant ubiquitination pathway. The action of effectors delivered from Gram-negative bacteria into the plant cell cytoplasm through the T3SS or T4SS is depicted. The effectors exert their action by acting as E3 ubiquitin ligase with domains RING/U-box or F-box of SCF or NEL, SUMO or cysteine protease, that binds with proteasome subunit and interfere with signaling pathway. (A) The P. syringae T3E AvrPtoB has U-box/RING- E3 ubiquitin ligase activity that mimics plant E3. It has both kinase and E3 ligase activity that enable functions in the ubiquitination and degradation of PRRs (FLS2, BAK, CERK, EFR) and R proteins (Pto, Fen, Prf). This prevents the downstream MAPK cascade and inhibits PTI and degradation of R protein that leads to prevention of ETI. HopZ4 acts on the 26S proteasome subunit component Rpt6 and thereby preventing SA-mediated defense. (B) The R. solanacearum has GALA protein that has LRR and an F-box domain that mimic plant E3, thereby promoting disease. (C) A. tumefaciens VirF has F-box domain and it mimics SCF E3 ligase that targets the VIP1 and VirE2 proteolysis thereby enabling integration of the DNA to promote crown gall. (D) The Rhizobium sp. NopM is a NEL family E3 and it reduce the ROS production and NopT effector induces PCD. (E) The Xanthomonas sp. harbors many effectors that has been characterized from few species. The T3E XopP from Xoo has RING/U-box domain with E3 activity. XcvXopD has SUMO protease activity that promotes senescence in plants, XccXopD alters host components RGA and interferes with signaling to promote infection. Similarly, Xcv AvrXv4 possess SUMO protease activity, Xcv 85–10 XopI with F-box domain was identified, and XcvXopL with NEL domain effector and E3 activity has been reported.
AvrPtoB is a bifunctional and bipartite effector with its N-terminal domain possessing the structural features that control hypersensitive cell death suppression and the C-terminal domain exhibiting in vitro E3 Ub ligase activity (Janjusevic et al., 2006; Abramovitch et al., 2006). The C-terminal domain of AvrPtoB resembles RING/U-box E3 ligases (Supplementary Fig. S3). In the suppression of PTI, AvrPtoB functions in the ubiquitination of PRRs, like flagellin-sensitive 2 (FLS2) receptor, chitin elicitor receptor kinase 1 (CERK1), and elongation factor thermo unstable receptor (EF-Tu receptor/EFR) (Gimenez-Ibanez et al., 2014; Göhre et al., 2008; Langin et al., 2020; Shan et al., 2008). AvrPtoB also interacts with the PRR-associated proteins BRI1-Associated Kinase1 (BAK1) and Botrytis-Induced Kinase1 (BIK1) and prevents the MAPK (Mitogen-Activated Protein Kinase) cascade (Shan et al., 2008). AvrPtoB also binds the host R proteins, including the protein kinases Pto and Fen as well as the nucleotide-binding site-leucine-rich repeat protein Prf leading to their ubiquitination and degradation (Mathieu et al., 2014; Munkvold and Martin, 2009; Rosebrock et al., 2007). In addition, AvrPtoB interacts with the plasma membrane-anchored protein RIN4 (RPM1-resistance to P. syringae pv. maculicola 1 interacting protein 4), an important regulator of plant immunity (Luo et al., 2009; Ray et al., 2019). Through this interaction, AvrPtoB targets RIN4 for degradation and activates PTI in the presence of Pto and Prf (Luo et al., 2009).
Another effector from P. syringae is HopM1, which promotes the ubiquitination of the immunity-associated protein AtMIN7 (BIG5) to inhibit the induced defense responses (Nomura et al., 2006). AtMIN7 functions in PTI, ETI, and salicylic acid (SA)-regulated immunity in A. thaliana, with homologs widespread in plants. AtMIN7 is an ADP ribosylation factor-guanine nucleotide exchange factor that localizes to the trans-Golgi network/early endosome, where it regulates the endocytic cycling of proteins in the plasma membrane (Nomura et al., 2011; Tanaka et al., 2009). The bacterial HopM1 facilitates the ubiquitination and the destruction of AtMIN7 via the host UPS (Nomura et al., 2006). The 712 amino acid HopM1 is translocated via the bacterial T3SS into the host cell and acts in a host endomembrane compartment or compartments to promote disease (Nomura et al., 2011). HopM1 recruits AtMIN7, or an AtMIN7-containing complex, via its N terminus and promotes the subsequent ubiquitination and degradation of AtMIN7 via the host proteasome (Nomura et al., 2006).
Jasmonate ZIM-domain (JAZ) family proteins are key regulators of jasmonate (JA) signaling in the immunity and development of plants that can be altered by plant pathogens. JAs are essential phytohormones in plant growth and development. JAZ proteins negatively regulate the transcriptional activation of these JA responses. To overcome this repression, JA triggers the degradation of JAZ proteins through the SCF (COI1) E3 ubiquitin ligase associated UPS (Chini et al., 2007; Thines et al., 2007). Pathogenic P. syringae secretes T3Es and several toxins such as coronatine (COR), which mimic the plant hormone jasmonate-isoleucine (JA-Ile). Like JA-Ile, COR functions as an SA antagonist to promote virulence via suppression of host defenses. Only a few P. syringae pathovars produce COR, and this toxin can act synergistically with bacterial T3Es to induce the JA pathway. JA-Ile and its COR mimic activation of plant responses by promoting physical interaction between the E3 ligase COI1 and JAZ repressors. This interaction leads to JAZ ubiquitination and subsequent degradation by 26S proteasomes, releasing transcription factors from repression. A major regulatory mechanism of JAZs in the presence of JA or COR is through COI-dependent UPS degradation of host proteins. COI1 is an F-box protein that determines the substrate specificity of a SCF E3 ubiquitin ligase-SCFCOI1. COR can also suppress host defense responses by triggering JA signaling in a COI1-dependent manner, thus, altering the normal progression of plant immunity and development (Katsir et al., 2008).
Other P. syringae plant pathogens that do not produce COR are still found to alter the host JAZ repressors. For example, the P. syringae pv. tabaci 11528, which does not produce COR, activates the JA pathway to promote susceptibility by degrading JAZ repressors through its effector HopX1. HopX1 acts as a cysteine protease that associates with JAZ proteins through its central ZIM domain and JAZ degradation occurs in a proteasome- and COI1- independent manner. HopX1 adopts different strategies to target similar host components as an alternative evolutionary solution to COR with similar physiological outcomes. JAZ proteins are direct targets of HopX1 to promote the activation of JA-induced defenses and susceptibility in Arabidopsis (Gimenez-Ibanez et al., 2014). P. syringae T3E HopZ1a, which belongs to the widely distributed YopJ family of cysteine proteases/acetyl transferases produced by plant and animal bacterial pathogens, is also found to interact with and modify the JAZ proteins in plant hosts. HopZ1a activates JA signaling and promotes bacterial multiplication in Arabidopsis. HopZ1a subverts host immunity by directly targeting the receptor complex of a defense-associated hormone in plants (Jiang et al., 2013).
Proteasome activity is intensely induced during basal defense in plants, and proteasomal mutations render plants susceptible to increased growth of virulent strains of P. syringae (Üstün et al., 2016). Bacteria appear to have taken advantage of this susceptibility by secreting effector proteins that inhibit the plant proteasomes. The P. syringae pv lachrymans HopZ4, a member of the YopJ family of T3Es, interacts with the host Rpt6, an AAA ATPase subunit of the 19S RP of 26S proteasomes, and inhibits proteasome activity during infection (Üstün et al., 2014). In effect, this results in decreased turnover of the salicylic acid (SA) master regulator Non-expressor of Pathogenesis Related genes1 (NPR1) and the attenuation of SA-dependent defense responses. The Pst effectors were screened to find potential T3Ss that may interfere with proteasome activity, resulting in the identification of HopM1, HopAO1, HopA1, and HopG1 as putative proteasome inhibitors. HopM1 was identified to interact with several E3 ubiquitin ligases and proteasome subunits, suggesting it may have an additional role beyond promoting the ubiquitination and degradation of MIN7 proteins (Üstün et al., 2016).
Bacterial effectors target the autophagy pathway for pathogen survival in host cells. The P. syringae pv. tomato DC3000 (PstDC3000) activates autophagy in a HopM1 dependent manner and stimulates the autophagic removal of proteasomes that suppress immune responses in plants and supports bacterial proliferation. PstDC3000 induces autophagy and increases autophagic flux upon delivery of the HopM1 (Üstün et al., 2018). While the PstDC3000 induced autophagy promotes bacterial proliferation, processes associated with the selective autophagy cargo receptor NBR1 counteract this proliferation by suppressing the formation of HopM1 induced water-soaked lesions (aqueous extracellular space). Distinct autophagy pathways are also shown to contribute to host immunity and bacterial pathogenesis during PstDC3000 infection with intimate crosstalk between proteasomes and autophagy. Thus, autophagic pathways with opposing pro- and antimicrobial functions appear to act in parallel during PstDC3000 infection, which is predicted to be a consequence of the long-lasting co-evolution of plants and their associated bacterial pathogens (Üstün et al., 2018).
4. Ralstonia solanacearum: F-box domain effector
R. solanacearum is a gram-negative soil-borne plant pathogenic bacterium that causes bacterial wilt in more than 450 plant species (Cunnac et al., 2004; Remigi et al., 2011). Analysis of the R. solanacearum genome reveals a large repertoire of up to 80 candidates of putative T3SS effectors, among which seven are homologs of plant-specific leucine-rich repeat (LRR) proteins. These seven T3SS effectors were found to have a conserved GAxALA motif in their LRR and, thus, are referred to as “GALA” proteins. These GALA proteins contain not only a leucine-rich repeat region but also an F-box domain related to plants (Angot et al., 2006). F-box proteins typically interact with Skp1 and Cullin1 to form SCF E3 Ub ligases that control ubiquitination. The bacterial GALA proteins are able to mimic plant F-box proteins by interacting with a subset of the 19 different Arabidopsis Skp1-like proteins. Deletion or mutation of the seven GALA effector genes of R. solanacearum results in the loss of pathogenesis on Arabidopsis and reduced virulence on tomato. Of these, the R. solanacearum GALA7 is found to be a host specificity factor in Medicago truncatula that requires its F-box domain for virulence function (Angot et al., 2006). Thus the T3E from R. solanacearum, GALA7, appears to hijack the host SCF-type E3 Ub ligases and affect the host UPS to promote disease.
5. Agrobacterium tumefaciens: type IV effector VirF contains F-box domain
A. tumefaciens is known to cause crown gall disease in many plants by its virulence (vir) region-encoded transport system (Schrammeijer et al., 2001). Agrobacterium genetically transforms plant hosts using a DNA-protein complex (T-complex) composed of a single T-DNA strand and the bacterial protein effectors VirD2, VirE2, and VirE3 (Li et al., 2018). The T-DNA is transported into the plant nucleus and subsequently integrated into the host genome. T-DNA nuclear import is mediated by the bacterial protein effectors, along with the host protein interactors AtKAP-a5 and VirE2 interacting protein 1 (VIP1), and integrated by host cell proteins. A. tumefaciens encodes and translocates VirF, to facilitate infection and suppress host immune system. VirF is an F-box protein that localizes into the plant cell nucleus and interacts with the nuclear protein VIP1. The VirF-containing SCF E3 ligase complex targets the VIP1 and VirE2 for proteolysis, leading to the release of T-DNA from the DNA-protein complex. Upon release, the T-DNA is integrated into the host chromatin (Tzfira et al., 2004). Some plant species do not require the bacterial VirF for transformation. In these latter cases, A. tumefaciens can induce the expression of a plant F-box protein, VIP1-binding F-box protein (VBF), that can functionally replace VirF. VBF interacts with the plant SKP1-like component (ASK1) in the SCF-VBF complex. VBF also binds VIP1 and its associated VirE2 to form a ternary VBF-VIP1-VirE2 complex. VBF then turns to destabilize both VIP1 and VirE2 via the SCF-VBF pathway leading to its proteolytic degradation (Zaltsman et al., 2010) (Fig. 3).
6. Xanthomonas spp.: E3 and SUMO proteases
Approximately 27 species of Xanthomonas are known to infect a wide range of economically important crop plants, such as rice, citrus, banana, cabbage, tomato, pepper, bean etc. (Ryan et al., 2011). T3Es that interfere with UPS have been studied from X. oryzae pv. oryzae (Xoo) that causes bacterial leaf blight, X. campestris pv.vesicatoria (Xcv) that causes bacterial spot of pepper and tomato, X. axonopodis pv. citri (Xac) that causes citrus canker, and X. axonopodis pv. manihotis (Xam) that causes cassava bacterial blight (CBB) (Üstün and Börnke, 2014). From a few of these species, the T3Es are characterized using a sequence-based approach (Furutani et al., 2009; Darrasse et al., 2013; Roux et al., 2015). In Xoo MAFF311018, about 16 effectors are found to be translocated via T3SS and among them, nine are homologs to known effectors in other plant-pathogenic bacteria (Furutani et al., 2009). The T3Es are generally conserved among Xanthomonas spp. with some specific to Xoo (Furutani et al., 2009). X. fuscans subsp. fuscans (Xff) that causes bacterial blight of bean was predicted to encode 29 T3Es (Darrasse et al., 2013). Comparative genome sequence analysis of pathovar strains of X. campestris revealed XopP, XopF1 and XopAL1 formed a common core of putative T3Es among pathovar incanae, pathovar raphani and a pathovar formerly named barbareae (Roux et al., 2015). Of the numerous T3Es reported, few have been characterized for their interference with UPS and the immune system of plants (Table 1; Fig. 3).
Table 1.
Effectors identified from bacteria that interact with plant UPS pathway are listed along with the functional domain and host targets.
| Bacteria | Effector | Interacting protein | Domain | Activity | Test plant | Reference |
|---|---|---|---|---|---|---|
|
| ||||||
| P. syringae pv. tomato | AvrPto/AvrPtoB | Fen, CERK1, FLS2, BAK1 | RING/ U-box | E3 Ubiquitin ligase | Tomato, Arabidopsis, Tobacco | Abramovitch et al., 2006; Janjusevic et al., 2006 |
| P. syringae pv. tomato DC3000 | HopM1 | AtMIN7 | ? | Promotes ubiquitination of AtMIN7, Inhibit proteasome subunit | Arabidopsis, Tobacco | Nomura et al., 2006; Üstün et al., 2016 |
| P. syringae pv. tabaci11528 | HopX1 | JAZ | – | Cystein protease | Tobacco, Pepper | Gimenez-Ibanez et al., 2014 |
| P. syringae pv. lachrymans | HopZ4 | RPT6 | – | Inhibit proteasome activity | Nicotiana benthamiana | Üstün et al., 2014 |
| A. tumefaciens | VirF | VIP1, ASK1 | F-box motif | F-box (protein protein interaction) | Tobacco | Tzfira et al., 2004; Zaltsman et al., 2010 |
| X. campestris pv. vesicatoria 85–10 | XopI | ? | F-box motif | ? | Pepper | Schulze et al., 2012 |
| R. solanacearum | GALA7 | Skp1 | F-box domain | SCF-type E3s | Arabidopsis, Tomato, Medicago truncatula, Eggplant | Angot et al., 2006; Remigi et al., 2011 |
| Rhizobium sp. strain NGR234 | NopM | ? | NELs | E3 Ubiquitin ligase | N. benthamiana | Xin et al., 2012 |
| Rhizobium sp. strain NGR234 | NopT | – | catalytic triad of protease | Cystein protease | Arabidopsis, Tobacco | Dai et al., 2008 |
| X. oryzae pv. oryzae | XopP | OsPUB44 | ? | Inhibits the host E3 ligase activity | Rice | Ishikawa et al., 2014 |
| X. campestris pv. vesicatoria 85–10 | XopL | E2 | XL-box domain | E3 Ubiquitin ligase | Tomato and pepper | Singer et al., 2013 |
| X. axonopodis pv. citri | PthA | E2 enzyme complex Ubc13&Uev | Central domain of tandem repeats | Transcriptional activators in the plant cell nucleus | Citrus sinensis and N. benthamiana | Domingues et al., 2010 |
| X. campestris pv. campestris 8004 | XopD | DELLA protein RGA | SUMO proteases | DeSUMOylating activity | Tomato | Hotson et al., 2003; Tan et al., 2014 |
| Xanthomonas sp. | XopD | SlERF4 | Cysteine proteases | SUMO protease | Tomato | Kim et al., 2008, 2013 |
| X. campestris | AvrXv4 | ? | Cysteine proteases | SUMO protease | N. benthamiana | Roden et al., 2004 |
| X. campestris pv. vesicatoria75–3 | AvrBsT | SnRK1 or Rpn8 | Cysteine proteases | Disrupt proteasome-mediated protein turnover | Arabidopsis, Tobacco | Szczesny et al., 2010 |
| X. campestris pv. vesicatoria 85–10 | XopJ | RPT6 | Cysteine proteases | Compete 26S proteasome subunit | Pepper | Bartetzko et al., 2009; Üstün et al., 2013; 2014; Üstün and Börnke, 2015 |
In rice, the OsPUB44 encoding E3 Ub ligase positively regulated the immune response against Xoo pathogen, while the XopP effector released by the rice pathogen Xoo strongly suppressed peptidoglycan (PGN) and chitin-triggered resistance (Ishikawa et al., 2014). This was effected by targeted binding of XopP to U-box domain of OsPUB44 via two amino-acid residues, thereby reducing the E3 ligase activity (Ishikawa et al., 2014).
The T3E XopD of Xcv is a cysteine protease with plant-specific SUMO substrate specificity. XopD is a member of the ubiquitin-like protease family that includes the yeast Ulp1 demonstrated to cleave SUMO/SMT3-conjugated proteins, thus, removing the SUMO tag. During Xcv pathogenesis, XopD is translocated to subnuclear loci of the host cell where it hydrolyzes SUMO-conjugated proteins. XopD interferes with the regulation of host proteins during Xcv infection by mimicking endogenous plant SUMO isopeptidases. The C-terminal domain (amino acids 322–520) is the portion of XopD related to the C48 family of cysteine peptidases with amino acids 309–481 homologous to the C-terminal catalytic domain of Ulp1. The XopD H409, D421 and C470 residues are predicted to form the catalytic core based on their relationship to the active site residues of the Ulp1 cysteine protease. Substrate recognition residues are also conserved. The target proteins affected by XopD proteolysis appear to be specific for plant SUMO isoforms, as XcvXopD cleaves a C-terminal hemagglutinin (HA) tag from plant SUMO but not mammalian SUMO isoforms. Thus, XopD is hypothesized to alter SUMO protein targets in the plant nucleus to control plant susceptibility and/or plant defense (Hotson et al., 2003).
The XcvXopD, not only cleaves SUMO-conjugates but also alters host transcription. Structure-function studies of XopD performed in vitro and in planta suggest that XopD mimicks the function of some plant transcriptional regulators (Kim et al., 2013, 2008). XopD alters host gene transcription by modulating senescence- and defense-associated mRNA levels. A putative helix-loop-helix region spanning amino acids 113–131 in the N-terminal domain of XopD appears to participate in DNA binding; however, this binding appears non-specific to DNA sequence. By contrast, the transcriptional repression activity of XopD appears specific and dependent upon two tandem EAR (ERF-associated amphiphilic repression) motifs in a region distinct from the cysteine protease active site. These EAR motifs are important for XopD to repress SA- and JA-induced gene transcription in planta. Thus, XopD appears multifaceted in its ability to cleave SUMO-conjugates and repress transcription to mediate virulence. In this virulence pathway, XopD is associated with promoting Xcv growth and suppressing host defense and pathogen-induced cell death responses (Kim et al., 2013, 2008).
The XopD homolog of Xcc8004 (XopDXcc8004) also appears to alter host components in initiating disease tolerance and enhancing bacterial survival. Through its EAR motif region, XopDXcc8004 interacts with the DELLA domain of the host protein RGA (repressor of ga1–3) (Tan et al., 2014). DELLA domain proteins, such as RGA, repress gibberellin (GA) signaling and promote host tolerance during plant stress. GA is important in signaling plant development processes and induces degradation of DELLA proteins by the UPS (Feng et al., 2008). By binding RGA, XopDXcc8004 apparently interferes with the GA-induced binding of the GA receptor GID1 to RGA. This interference appears to partially stabilize DELLA proteins in the nucleus and delay their GA-induced degradation via. the UPS. XopDXcc8004 cannot deubiquitinate the ubiquitination of RGA and does not appear to alter the RGA transcript levels. Thus, the protein: protein interaction of XopDXcc8004 with RGA is proposed to lead to disease suppression and enhanced bacterial survival (Tan et al., 2014).
Four XcvT3Es (AvrXv4, AvrBsT, AvrRxv, and XopJ) are identified to be members of the YopJ/AvrRxv family (Lewis et al., 2011). YopJ family T3Es are proposed to have three basic functional domains: translocation, interaction, and catalytic, including cysteine protease and/or acetyltransferase activity (Whalen et al., 2008). Of the XcvYopJ family proteins, AvrXv4 functions as an apparent cysteine protease that cleaves SUMO-conjugates during Xcv-plant interactions, as its expression in planta leads to a reduced abundance of SUMO-conjugates (Roden et al., 2004). The localization of the Ulp1-related XopD to the plant nucleus (Hotson et al., 2003) and AvrXv4 to the plant cytoplasm (Roden et al., 2004) suggests that these R proteins have evolved to recognize bacterial SUMO proteases in distinct cellular compartments. Like AvrXv4, the Xcv AvrBsT is an apparent cysteine protease as it exhibits a weak protease activity in vitro that is dependent upon its conserved catalytic cysteine residue. In vivo, AvrBsT localizes to the cytoplasm and nucleus of plant cells (Roden et al., 2004). In the cytoplasm, AvrBsT suppresses the induction of the AvrBs1-specific HR apparently via interaction with the SNF1-related kinase (SnRK1) in pepper plants (Szczesny et al., 2010). The YopJ-like XopJ is targeted to the host plasma membrane when expressed in plant cells (Noël et al., 2003; Thieme et al., 2007). In addition to its homology to YopJ, XopJ has a conserved N-myristoylation motif that is required for plasma membrane localization (Thieme et al., 2007). Thus, targeting XopJ to the host membrane is thought to occur through N-myristoylation. XopJ also interacts with the AAA ATPase proteasomal subunit RPT6 in yeast and in planta, and this appears to inhibit proteasome activity. Proteasome inhibition prevents the accumulation of the defense phytohormone SA and attenuates SA mediated symptom development as well as pathogen-induced senescence (Bartetzko et al., 2009; Üstün et al., 2013; 2014, Üstün and Börnke, 2015). AvrRxv has a cysteine protease catalytic core important in eliciting the HR and inhibiting bacterial growth in resistant plants (Bonshtien et al., 2005). In host cells, AvrRxv localizes predominantly to the cytoplasm and may associate with plasma and nuclear membranes (Bonshtien et al., 2005). AvrRxv binds the host 14–3-3 protein called AvrRxv Interactor 1 (ARI1), which is associated with HR-inducing activity in tomato (Whalen et al., 2008). 14–3-3 proteins are involved in a range of protein-protein interaction (PPI) functions in plants (Ryan et al., 2011; Sehnke et al., 2002).
In pursuit of new T3Es in the Xcv model strain 85–10, the F-box motif protein XopI was identified. A plant-inducible promoter (PIP) box, upstream of the encoding gene, suggests transcriptional expression of xopI is regulated by HrpX (Schulze et al., 2012). HrpX is an AraC-type transcriptional activator that binds to PIP box motifs and controls the expression of T3Es in plant pathogens (Koebnik et al., 2006). Translocation of XopI into plant cells is dependent on the T3SS chaperone HpaB (Schulze et al., 2012). While F-box proteins are typically one of three components of SCF complexes that mediate protein ubiquitination for degradation by the UPS, the function of the F-box in XopI is not yet elucidated.
A new family of E3 Ub ligases with a novel structural domain (termed NEL for Novel E3 Ligase) distinct from either the RING or HECT domains is identified in select bacteria. This family is exemplified by the Shigella spp. IpaH9.8 and Salmonella spH1 proteins which display E3 Ub ligase activity despite lacking sequence similarity to any known E3 ligases (Singer et al., 2008). NEL E3 ligases comprise a large family of bacterial effector proteins encoded by a subset of pathogenic bacteria (Hicks and Galán, 2010; Rohde et al., 2007; Zhu et al., 2008). From plant pathogenic bacteria X. campestris pv. vesicatoria (Xcv), the effector XopL is identified to have a novel E3 ligase domain that induces plant cell death and suppresses PTI (Singer et al., 2013). XopL is encoded by a gene that has a PIP (pathogen-inducible promoter) box in its promoter region that contributes to virulence and suggests the co-regulation of this gene with the T3SS. The C-terminal domain of XopL catalyzes E3 ligase activity and explicitly interacts with the plant E2 to form K11-linked poly-ubiquitin chains. The crystal structure of the XopL C-terminal domain reveals a novel fold, termed the XL-box. The N-terminal region of XopL confirmed the presence of a LRR domain and may form a protein-protein interaction module for ubiquitination target recognition and suppression of PAMP responses while the E3 ligase activity-induced plant cell death (Singer et al., 2013).
The X. axonopodis pv. citri T3E PthA modulates host transcription to promote citrus canker. PthA belongs to the AvrBs3/PthA protein family and has a central domain of tandem 34 amino acid repeats that mediate protein-protein and protein-DNA interactions (Domingues et al., 2010). PthA proteins are encoded as multiple variants in a single bacterial strain. These PthA variants localize in the nucleus of plant cells and form homo- and heterodimers. The PthA heterodimers are hypothesized to include interactions with distinct host targets. PthA2 and PthA3 appear to interact with the citrus cyclophilin and TDX (tetratricopeptide domain-containing thioredoxin) proteins. In addition, PthA2 and PthA3 associate with the E2 complex of Ubc13 and Uev (Ub conjugating E2 variant), required for K63-linked ubiquitination and DNA repair. The citrus Ubc13 and Uev proteins complement the DNA repair phenotype of the yeast, indicating that they are also involved in K63-linked ubiquitination and DNA repair. PthA2 inhibits K63-linked ubiquitination required for DNA repair (Domingues et al., 2010).
Similar to effectors altering UPS, microbes have evolved mechanisms to modulate autophagy through their T3Es. One such effector is XopL secreted by Xcv that acts as E3 ligase and exerts its role in a novel way by self-modifying T3Es to trap host cellular degradation pathways. A sub proportion of XopL undergoes self-ubiquitination, subsequently triggering its own degradation by the NBR1/Joka2-mediated autophagy to act as a bait to trap host cellular degradation machineries to boost virulence of Xcv by attenuating autophagic degradation. XopL inhibits autophagy by interacting and subsequently degrading the autophagic component SH3P2 via. its E3 ligase activity. By degrading the SH3P2, it partially escapes its own degradation and blocks autophagy (Leong et al., 2021).
7. Rhizobium sp.: NEL domain effectors and cysteine protease
The NopM (nodulation outer protein M) effector of the nitrogen-fixing Sinorhizobium sp. (Rhizobium sp.) strain NGR234 is an IpaH family effector with NEL domain. NopM is similar to IpaH family effectors and consists of a variable N-terminal domain composed of LRR domain and a conserved C-terminal NEL domain. In vitro, NopM has E3 Ub ligase activity, including the formation of unanchored poly Ub chains and auto-ubiquitination activity (Xu et al., 2018). In vivo, NopM, but not the active site variant NopMC338A, promotes symbiotic nodulation of the host legume Lablab purpureus (Xin et al., 2012). When expressed in yeast, NopM inhibits mating pheromone signaling, a MAPK pathway. Consistent with this finding, NopM can be phosphorylated in vitro by the tobacco MAPK, the SA-induced protein kinase (NtSIPK). In addition, NopM is phosphorylated at Ser2 in planta (Xu et al., 2018). NopM inhibits the plant defense response in Nicotiana benthamiana through reduced ROS production in response to the flagellin peptide flg22 and defense gene expression. These results suggest NopM as a functional NEL domain E3 Ub ligase that is serine phosphorylated and can interact with itself, with Ub, and with MAPKs.
The novel rhizobial T3E (named NopT for nodulation outer protein T) is secreted via., the T3SS in Rhizobium sp. NGR234 causing chlorotic and necrotic symptoms in Arabidopsis plants (Dai et al., 2008; Kambara et al., 2009). Tobacco plants expressing nopT elicit an HR response, a form of programmed cell death that would block the further attack (Kambara et al., 2009). The NopT of NGR234 is related in sequence to the NopT of Bradyrhizobium japonicum USDA110 and the T3Es AvrPphB of the pathogen P. syringae pv. phaseolicola, YopT from Yersinia spp., and LopT from P. luminescens. NopT contains the conserved active site residues required for proteolytic activity of cysteine proteases (C/H/D residues; invariant catalytic triad in proteins belonging to the YopT-AvrPphB cysteine protease family) (Shao et al., 2002) and induces an R protein-mediated defense response in non-host plants (Kambara et al., 2009). By acting as cysteine protease, NopT elicits HR, a localized PCD response in tobacco and induces cytotoxic effects in Arabidopsis (Dai et al., 2008).
8. Conclusion and future perspective
Bacterial effectors that alter the plant UPS pathway and thereby affect the immune system of plants have been characterized in several important plant pathogenic and non-pathogenic bacteria. The P. syringae AvrPtoB effector is a good example of how bacterial and plant proteins co-evolve in an arms race to develop immunity. R. solancarum promotes itself through its mimicry of F-box proteins and establishes inside plants. A. tumificiencs uses T4Es to genetically transform plants, revealing how bacterial genes can be integrated into the genome of plant systems to enable efficient colonization. The genus Xanthomonas includes several important pathogens and encodes a set of effectors that mimic E3s or its subunits to alter plant responses. Some of the Xanthomonas effectors act as SUMO and/or cysteine proteases and others target proteins for degradation by the UPS to alter the immune system of plants in favor of the pathogen. Interestingly, T3Es from the non-pathogenic Rhizobium spp. include those that function as components of E3s and cysteine proteases in non-host plants. At present, approximately 126 whole-genome sequences of pathogenic bacteria are available (Xu and Wang, 2019). Screening for potential E3 subunits or other UPS components from these genomes might reveal new bacterial proteins that mimic plant proteins in pathogenesis. The main strategy adopted by bacterial pathogens in altering the UPS and immunity of plants is through structural mimicry, binding, modifying, inhibiting, or degradation of target proteins of the plant host. Finding ways to circumvent the effector invasion inside plants and identifying plant targets would benefit plant defense. The process underlining protein co-evolution in plants and microbe for its survival during the plant-microbe interaction is yet another aspect that is less understood. Yet, whole-genome mapping of plant and pathogen may help to identify new novel variants in R and effector gene; and understanding its target role may widen the opportunities to develop resistant cultivars through molecular or mutational breeding and genome editing. Besides targeting the effector binding sequences and susceptible genes in plants for increased immunity to prevent disease (Joshi et al., 2020) as a promising strategy, the plant UPS mimicking effectors can also be targeted for resistance in the future.
Supplementary Material
Acknowledgements
The research was supported by the Ministry of Human Resource Development (MHRD-FAST CoE) (F.No.5–6/2013-TS-VII), from the Government of India under grant number F.No.5–6/2013-TSVII sanctioned to SU. This work was also supported by awards to JMF through the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences and Biosciences, Physical Biosciences Program (DOE DE-FG02–05ER15650) to advance microbial biocatalysts and the National Institutes of Health (NIH R01 GM57498) to understand ubiquitin-proteasome systems in disease. The authors also thank DBT-BIOCARe for the financial support through project grant No. BT/PR18134/BIC/101/795/2016 sanctioned to PR and Core funding from the Department of Agriculture, Tamil Nadu Government through University-PDF support to BJ and PR by Tamil Nadu Agricultural University, Coimbatore.
Abbreviations:
- APC/C
Anaphase Promoting Complex/cyclosome
- ASK1
Apoptosis signal-regulating kinase 1
- AvrPto
avirulentPto
- BIK1
Botrytis-induced kinase1
- BAK1
BRI1-Associated Kinase1
- BTB
broad complex/tramtrack/bric-a-brac
- CDC20
cell division cycle protein 20
- CERK1
chitin elicitor receptor kinase 1
- COI1
coronatine insensitive
- CUL
cullin
- DUBs
deubiquitinase
- DDB1
DNA-Damage Binding 1
- E1
ubiquitin-activation enzyme
- E2
ubiquitin-conjugating enzyme
- E3
ubiquitin ligase
- EAR
ERF-associated amphiphilic repression
- ETI
effector-triggered immunity
- EFR
EF-Tu receptor
- EF-Tu
elongation factor Tu
- ET
ethylene
- FLS2
flagellin-sensitive 2 receptor
- GA
gibberellins
- HR
hypersensitive response
- JA
jasmonate
- JAZ
jasmonate ZIM-domain, JA-ZIM domain (JAZ) repressor proteins
- LRR
leucine-rich repeat
- MAPK
mitogen-activated protein kinase
- NEDD
neddylation
- NPR1
nonexpressor of pathogenesis related genes 1
- NEL
novel E3 ubiquitin ligase
- P/MAMPs
pathogen- or microbe-associated molecular patterns
- PRRs
pattern recognition receptors
- PTM
post-translational modification
- PCD
programmed cell death
- Protein Pto
resistance to Pseudomonas syringaepathovar (pv.) tomato
- ROS
reactive oxygen species
- RPNs
regulatory particle non-ATPases
- R proteins
resistance proteins
- RGA
repressor of ga1–3
- RIN4
RPM1- resistance to P. syringaepv. maculicola 1 interacting protein 4
- RBX1
RING-box 1
- Rub
rubylation
- SA
salicylic acid
- SKP1
S-Phase Kinase Associated Protein 1
- SUMOs
small ubiquitin related modifier proteins
- SCF
S-phase kinase-associated protein 1 (SKP1)-cullin 1 (CUL1)–F-box
- SAR
systemic acquired resistance
- T3SS
type III secretory system
- Ub
ubiquitin
- UPS
ubiquitin 26S proteasome system
- UBLs/Ulps
ubiquitin-like proteins
- VBF
VIP1-binding F-box protein
- VIP1
VirE2 interacting protein 1
- ZIM
zinc-finger inflorescence meristem
Footnotes
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.micres.2021.126810.
References
- Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB, 2006. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. U. S. A. 103 (8), 2851–2856. 10.1073/pnas.0507892103%J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angot A, Peeters N, Lechner E, Vailleau F, Baud C, Gentzbittel L, et al. , 2006. Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. U. S. A. 103 (39), 14620–14625. 10.1073/pnas.0509393103%J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angot A, Vergunst A, Genin S, Peeters N, 2007. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 3 (1), e3. 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Kim M, Sasakawa C, 2014. Exploitation of the host ubiquitin system by human bacterial pathogens. Nat. Rev. Microbiol. 12 (6), 399–413. 10.1038/nrmicro3259. [DOI] [PubMed] [Google Scholar]
- Bartetzko V, Sonnewald S, Vogel F, Hartner K, Stadler R, Hammes UZ, Börnke F, 2009. The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: evidence for interference with cell wall-associated defense responses. Mol. Plant Microbe Interact. 22 (6), 655–664. 10.1094/mpmi-22-6-0655. [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G, 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Bonshtien A, Lev A, Gibly A, Debbie P, Avni A, Sessa G, 2005. Molecular properties of the Xanthomonas AvrRxv effector and global transcriptional changes determined by its expression in resistant tomato plants. Mol. Plant Microbe Interact. 18 (4), 300–310. 10.1094/mpmi-18-0300. [DOI] [PubMed] [Google Scholar]
- Buttner D, 2016. Behind the lines-actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 40 (6), 894–937. 10.1093/femsre/fuw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J, Vierstra RD, 2000. Protein degradation in signaling. Curr. Opin. Plant Biol. 3 (5), 381–386. 10.1016/s1369-5266(00)00100-x. [DOI] [PubMed] [Google Scholar]
- Callis J, Carpenter T, Sun CW, Vierstra RD, 1995. Structure and evolution of genes encoding poly-ubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics 139 (2), 921–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hellmann H, 2013. Plant E3 ligases: flexible enzymes in a sessile world. Mol. Plant 6 (5), 1388–1404. 10.1093/mp/sst005. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ, 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286. [DOI] [PubMed] [Google Scholar]
- Chen ZQ, Zhao JH, Chen Q, Zhang ZH, Li J, Guo Z, et al. , 2020. DNA geminivirus infection induces an imprinted E3 ligase gene to epigenetically activate viral gene transcription. Plant Cell. 10.1105/tpc.20.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, et al. , 2007. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 (7154), 666–6671.. 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Copeland C, Li X, 2019. Regulation of plant immunity by the proteasome. Int. Rev. Cell Mol. Biol. 343, 37–63. 10.1016/bs.ircmb.2018.06.004. [DOI] [PubMed] [Google Scholar]
- Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G, 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13 (6), 343–359. 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- Craig A, Ewan R, Mesmar J, Gudipati V, Sadanandom A, 2009. E3 ubiquitin ligases and plant innate immunity. J. Exp. Bot. 60, 1123–1132. 10.1093/jxb/erp059. [DOI] [PubMed] [Google Scholar]
- Cunnac S, Boucher C, Genin S, 2004. Characterization of the cis-acting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. J. Bacteriol. 186 (8), 2309–2318. 10.1128/jb.186.8.2309-2318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A, 2011. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc. Natl. Acad. Sci. U. S. A. 108 (7), 2975–2980. 10.1073/pnas.1013031108%J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W-J, Zeng Y, Xie Z-P, Staehelin C, 2008. Symbiosis-promoting and deleterious effects of NopT, a novel type 3 effector of Rhizobium sp. Strain NGR234. J. Bacteriol. 190 (14), 5101–5110. 10.1128/jb.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrasse A, Carrère S, Barbe V, Boureau T, Arrieta-Ortiz ML, Bonneau S, et al. , 2013. Genome sequence of Xanthomonas fuscans subsp. Fuscansstrain 4834-R reveals that flagellar motility is not a general feature of Xanthomonads. BMC Genomics 14 (1), 761. 10.1186/1471-2164-14-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues MN, De Souza TA, Cernadas RA, de Oliveira ML, Docena C, Farah CS, Benedetti CE, 2010. The Xanthomonas citri effector protein PthA interacts with citrus proteins involved in nuclear transport, protein folding and ubiquitination associated with DNA repair. Mol. Plant Pathol. 11 (5), 663–675. 10.1111/j.1364-3703.2010.00636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foot N, Henshall T, Kumar S, 2017. Ubiquitination and the regulation of membrane proteins. Physiol. Rev. 97, 253–281. [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X, 2013. Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- Furutani A, Takaoka M, Sanada H, Noguchi Y, Oku T, Tsuno K, et al. , 2009. Identification of novel type III secretion effectors in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 22 (1), 96–106. 10.1094/mpmi-22-1-0096. [DOI] [PubMed] [Google Scholar]
- Gao G, Luo H, 2006. The ubiquitin–proteasome pathway in viral infections. Special Issue, entitled Young Investigator’s Forum. Can. J. Physiol. Pharmacol. 84 (1), 5–14. 10.1139/y05-144. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Boter M, Fernández-Barbero G, Chini A, Rathjen JP, Solano R, 2014. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 12 (2) 10.1371/journal.pbio.1001792 e1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A, 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428. [DOI] [PubMed] [Google Scholar]
- Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, et al. , 2008. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18 (23), 1824–1832.. 10.1016/j.cub.2008.10.063. [DOI] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, Alfano JR, 2009. The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol. Plant Microbe Interact. 22 (9), 1069–1080. 10.1094/MPMI-22-9-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin JK, Moses AV, Früh K, Douglas JL, 2011. Viral takeover of the host ubiquitin system. Front. Microbiol. 2, 161. 10.3389/fmicb.2011.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman DS, 2004. Plants as models for the study of human pathogenesis. Biotechnol. Adv. 22 (5), 363–382. 10.1016/j.biotechadv.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Hafren A, et al. , 2017. Selective autophagy limits cauliflower mosaic virus infection by NBR1 –mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. U. S. A. 114, E2026–E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafren A, et al. , 2018. Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HC pro. Plant Physiol. 176, 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Aoki SK, Low DA, 2010. Bacterial contact-dependent delivery systems. Annu. Rev. Genet. 44, 71–90. 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- Hepowit NL, Uthandi S, Miranda HV, Toniutti M, Prunetti L, Olivarez O, et al. , 2012. Archaeal JAB1/MPN/MOV34 metalloenzyme (HvJAMM1) cleaves ubiquitin-like small archaeal modifier proteins (SAMPs) from protein-conjugates. Mol. Microbiol. 86 (4), 971–987. 10.1111/mmi.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SW, Galán JE, 2010. Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr. Opin. Microbiol. 13 (1), 41–46. 10.1016/j.mib.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, et al. , 2009. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137, 773–783. [DOI] [PubMed] [Google Scholar]
- Hotson A, Chosed R, Shu H, Orth K, Mudgett MB, 2003. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50 (2), 377–389. 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Yamaguchi K, Sakamoto K, Yoshimura S, Inoue K, Tsuge S, et al. , 2014. Bacterial effector modulation of host E3 ligase activity suppresses PAMP-triggered immunity in rice. Nat. Commun. 5 (1), 5430. 10.1038/ncomms6430. [DOI] [PubMed] [Google Scholar]
- Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE, 2006. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311 (5758), 222–226. 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- Jiang S, Yao J, Ma K-W, Zhou H, Song J, He SY, Ma W, 2013. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 9 (10) 10.1371/journal.ppat.1003715 e1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL, 2006. The plant immune system. Nature 444 (7117), 323–329. 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Joshi JB, Arul L, Ramalingam J, Uthandi S, 2020. Advances in the Xoo-rice pathosystem interaction and its exploitation in disease management. J. Biosci. 112 10.1007/s12038-020-00085-8. [DOI] [PubMed] [Google Scholar]
- Kambara K, Ardissone S, Kobayashi H, Saad MM, Schumpp O, Broughton WJ, Deakin WJ, 2009. Rhizobia utilize pathogen-like effector proteins during symbiosis. Mol. Microbiol. 71 (1), 92–106. 10.1111/j.1365-2958.2008.06507.x. [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA, 2008. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U. S. A. 105 (19), 7100–7105. 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Taylor KW, Hotson A, Keegan M, Schmelz EA, Mudgett MB, 2008. XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas-infected tomato leaves. Plant Cell 20 (7), 1915–1929. 10.1105/tpc.108.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Stork W, Mudgett MB, 2013. Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 13 (2), 143–154. 10.1016/j.chom.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Otsubo R, Morikawa H, Nishide A, Takagi K, Sasakawa C, Mizushima T, 2014. Bacterial effectors and their functions in the ubiquitin-proteasome system: insight from the modes of substrate recognition. Cells. 3 (3), 848–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Jang I-C, Seo HS, 2016. COP1 controls abiotic stress responses by modulating AtSIZ1 function through its E3 ubiquitin ligase activity. Front. Plant Sci. 7, 1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik R, Krüger A, Thieme F, Urban A, Bonas U, 2006. Specific binding of the Xanthomonas campestris pv. vesicatoria AraC-type transcriptional activator HrpX to plant-inducible promoter boxes. J. Bacteriol. 188 (21), 7652–7660. 10.1128/jb.00795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S, 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96 (5), 635–644. 10.1016/S0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Kraft C, Reggiori F, Peter M, 2009. Selective types of autophagy in yeast. Biochim. Biophys. Acta 1793 (9), 1404–1412. [DOI] [PubMed] [Google Scholar]
- Langin G, Gouguet P, Üstün S, 2020. Microbial effector proteins – a journey through the proteolytic landscape. Trends Microbiol. 28 (7), 523–535. 10.1016/j.tim.2020.02.010. [DOI] [PubMed] [Google Scholar]
- Leary AY, et al. , 2019. Contrasting and emerging roles of autophagy in plant immunity. Curr. Opin. Plant Biol. 52, 46–53. [DOI] [PubMed] [Google Scholar]
- Leong JX, Raffeiner M, Spinti, et al. , 2021. Self-ubiquitination of a pathogen type-III effector traps and blocks the autophagy machinery to promote disease. bioRxiv. 10.1101/2021.03.17.435853, 2021.2003.2017.435853. [DOI] [Google Scholar]
- Lewis JD, Lee A, Ma W, Zhou H, Guttman DS, Desveaux D, 2011. The YopJ superfamily in plant-associated bacteria. Mol. Plant Pathol. 12 (9), 928–937. 10.1111/j.1364-3703.2011.00719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tu H, Pan SQ, 2018. Agrobacterium delivers anchorage protein VirE3 for companion VirE2 to aggregate at host entry sites for T-DNA protection. Cell Rep. 25 (2), 302–311. 10.1016/j.celrep.2018.09.023 e306. [DOI] [PubMed] [Google Scholar]
- Lin YH, Machner MP, 2017. Exploitation of the host cell ubiquitin machinery by microbial effector proteins. J. Cell. Sci. 130 (12), 1985–1996. 10.1242/jcs.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SS, Martin R, Mongrand S, Vandenabeele S, Chen KC, Jang IC, Chua NH, 2008. RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. Plant J. 56, 550–561. [DOI] [PubMed] [Google Scholar]
- Luo Y, Caldwell KS, Wroblewski T, Wright ME, Michelmore RW, 2009. Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell 21 (8), 2458–2472. 10.1105/tpc.107.056044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Laluk K, Lai Z, Veronese P, Song F, Mengiste T, 2010. The Arabidopsis botrytis susceptible1 interactor defines a subclass of RING E3 ligases that regulate pathogen and stress responses. Plant Physiol. 154, 1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD, 2018. Autophagy: the master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208, 16. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Schwizer S, Martin GB, 2014. Pto kinase binds two domains of AvrPtoB and its proximity to the effector E3 ligase determines if it evades degradation and activates plant immunity. PLoS Pathog. 10 (7) 10.1371/journal.ppat.1004227 e1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Laugesen SH, Ellgaard L, 2019. Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open Biol. 9 (9), 190147 10.1098/rsob.190147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold KR, Martin GB, 2009. Advances in experimental methods for the elucidation of Pseudomonas syringae effector function with a focus on AvrPtoB. Mol. Plant Pathol. 10 (6), 777–793. 10.1111/j.1364-3703.2009.00586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou BPM, Ahn HK, Ding P, et al. , 2021. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. 10.1038/s41586-021-03315-7. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R, 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123 (5), 773–786. 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Noël L, Thieme F, Gäbler J, Büttner D, Bonas U, 2003. XopC and XopJ, two novel type III effector proteinsfrom Xanthomonas campestris pv.vesicatoria. J. Bacteriol. 185 (24), 7092–7102. 10.1128/JB.185.24.7092-7102.2003%J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY, 2006. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313 (5784), 220–223. 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- Nomura K, Mecey C, Lee YN, Imboden LA, Chang JH, He SY, 2011. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 108 (26), 10774–10779. 10.1073/pnas.1103338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Martin GB, 2011. Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci. 16 (3), 132–140. 10.1016/j.tplants.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB, 2003. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41, 215–243. 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ, 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ, 2004. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72. [DOI] [PubMed] [Google Scholar]
- Pruitt RN, Gust AA, Nürnberger T, 2021. Plant immunity unified. Nat. Plants 382–383. 10.1038/s41477-021-00903-3. [DOI] [PubMed] [Google Scholar]
- Randow F, Youle RJ, 2014. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe 15 (4), 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray SK, Macoy DM, Kim W-Y, Lee SY, Kim MG, 2019. Role of RIN4 in regulating PAMP-Triggered immunity and effector-triggered immunity: current status and future perspectives. Mol. Cells 42 (7), 503–511. 10.14348/molcells.2019.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigi P, Anisimova M, Guidot A, Genin S, Peeters N, 2011. Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 192 (4), 976–987. 10.1111/j.1469-8137.2011.03854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden J, Eardley L, Hotson A, Cao Y, Mudgett MB, 2004. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant Microbe Interact. 17 (6), 633–643. 10.1094/mpmi.2004.17.6.633. [DOI] [PubMed] [Google Scholar]
- Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C, 2007. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1 (1), 77–83. 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB, 2007. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448 (7151), 370–374. 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B, Bolot S, Guy E, Denancé N, Lautier M, Jardinaud M-F, et al. , 2015. Genomics and transcriptomics of Xanthomonas campestris species challenge the concept of core type III effectome. BMC Genomics 16 (1), 975. 10.1186/s12864-015-2190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovenich H, Zuccaro A, Thomma BPHJ, 2016. Convergent evolution of filamentous microbes towards evasion of glycan-triggered immunity. New Phytol. 212 (4), 896–901. 10.1111/nph.14064. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Vorhölter F-J, Potnis N, Jones JB, Sluys M-AV, Bogdanove AJ, Dow JM, 2011. Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nat. Rev. Microbiol. 9, 344–355. [DOI] [PubMed] [Google Scholar]
- Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuïnk TJ, Crosby WL, Hooykaas PJ, 2001. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr. Biol. 11 (4), 258–262. 10.1016/s0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- Schulze S, Kay S, Büttner D, Egler M, Eschen-Lippold L, Hause G, et al. , 2012. Analysis of new type III effectors from Xanthomonas uncovers XopB and XopS as suppressors of plant immunity. New Phytol. 195 (4), 894–911. 10.1111/j.1469-8137.2012.04210.x. [DOI] [PubMed] [Google Scholar]
- Sehnke PC, Rosenquist M, Alsterfjord M, DeLille J, Sommarin M, Larsson C, Ferl RJ, 2002. Evolution and isoform specificity of plant 14–3-3 proteins. Plant Mol. Biol. 50 (6), 1011–1018. 10.1023/a:1021289127519. [DOI] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, et al. , 2008. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4 (1), 17–27. 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao ZQ, Innes RW, Dixon JE, 2002. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109 (5), 575–588. 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- Shu K, Yang W, 2017. E3 ubiquitin ligases: ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 58, 1461–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer AU, Rohde JR, Lam R, Skarina T, Kagan O, Dileo R, et al. , 2008. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat. Struct. Mol. Biol. 15 (12), 1293–1301. 10.1038/nsmb.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD, 2004. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590. [DOI] [PubMed] [Google Scholar]
- Stone SL, 2014. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 5, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek KN, Komander D, 2016. Ubiquitin modifications. Cell Res. 26 (4), 399–422. 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny R, Büttner D, Escolar L, Schulze S, Seiferth A, Bonas U, 2010. Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol. 187 (4), 1058–1074. 10.1111/j.1469-8137.2010.03346.x. [DOI] [PubMed] [Google Scholar]
- Tan L, Rong W, Luo H, Chen Y, He C, 2014. The Xanthomonas campestris effector protein XopDXcc8004 triggers plant disease tolerance by targeting DELLA proteins. New Phytol. 204 (3), 595–608. 10.1111/nph.12918. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Kitakura S, De Rycke R, De Groodt R, Friml J, 2009. Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19 (5), 391–397. 10.1016/j.cub.2009.01.057. [DOI] [PubMed] [Google Scholar]
- Thieme F, Szczesny R, Urban A, Kirchner O, Hause G, Bonas U, 2007. New type III effectors from Xanthomonas campestris pv. vesicatoria trigger plant reactions dependent on a conserved N-myristoylation motif. Mol. Plant Microbe Interact. 20 (10), 1250–1261. 10.1094/mpmi-20-10-1250. [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, et al. , 2007. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448 (7154), 661–6665.. 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM, 2000. Recognition of the poly-ubiquitin proteolytic signal. EMBO J. 19 (1), 94–102. 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, 2010. ROS in biotic interactions. Physiol. Plant. 138 (4), 414–429. 10.1111/j.1399-3054.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- Troisfontaines P, Cornelis GR, 2005. Type III secretion: more systems than you think. Physiology (Bethesda) 20, 326–339. 10.1152/physiol.00011.2005. [DOI] [PubMed] [Google Scholar]
- Tytgat T, Vanholme B, De Meutter J, Claeys M, Couvreur M, Vanhoutte I, et al. , 2004. A new class of ubiquitin extension proteins secreted by the dorsal pharyngeal gland in plant parasitic cyst nematodes. Mol. Plant Microbe Interact. 17 (8), 846–8852.. 10.1094/mpmi.2004.17.8.846. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V, 2004. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431 (7004), 87–92. 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- Üstün S, Börnke F, 2014. Interactions of Xanthomonas type-III effector proteins with the plant ubiquitin and ubiquitin-like pathways. Front. Plant Sci. 5, 736. 10.3389/fpls.2014.00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S, Börnke F, 2015. The Xanthomonas campestris type III effector XopJ proteolytically degrades proteasome subunit RPT6. Plant Physiol. 168 (1), 107–119. 10.1104/pp.15.00132%J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S, Bartetzko V, Börnke F, 2013. The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic-acid mediated plant defence. PLoS Pathog. 9 (6) 10.1371/journal.ppat.1003427 e1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S, König P, Guttman DS, Börnke F, 2014. HopZ4 from Pseudomonas syringae, a member of the HopZ type III effector family from the YopJ superfamily, inhibits the proteasome in plants. Mol. Plant Microbe Interact. 27 (7), 611–623. 10.1094/mpmi-12-13-0363-r. [DOI] [PubMed] [Google Scholar]
- Üstün S, Sheikh A, Gimenez-Ibanez S, Jones A, Ntoukakis V, Börnke F, 2016. The proteasome acts as a hub for plant immunity and is targeted by Pseudomonas type III effectors. Plant Physiol. 172 (3), 1941–1958. 10.1104/pp.16.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S, et al. , 2017. Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol 40, 122–130. [DOI] [PubMed] [Google Scholar]
- Üstün S, et al. , 2018. Bacteria exploit autophagy for proteasome degradation and enhanced virulence in plants. Plant Cell 30, 668–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD, 2009. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10 (6), 385–397. 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- Vierstra RD, 2012. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 160 (1), 2–14. 10.1104/pp.112.200667%J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Deng XW, 2011. Plant ubiquitin-proteasome pathway and its role in gibberellin signaling. Cell Res. 21, 1286–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM, Shabek N, Ciechanover A, 2011. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 12 (9), 605–620. 10.1038/nrm3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling R, Epple P, Altmann S, He Y, Yang L, Henz SR, et al. , 2014. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16 (3), 364–375.. 10.1016/j.chom.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen M, Richter T, Zakhareyvich K, Yoshikawa M, Al-Azzeh D, Adefioye A, et al. , 2008. Identification of a host 14–3-3 Protein that interacts with Xanthomonas effector AvrRxv. Physiol. Mol. Plant Pathol. 72 (1–3), 46–55. 10.1016/j.pmpp.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin D-W, Liao S, Xie Z-P, Hann DR, Steinle L, Boller T, Staehelin C, 2012. Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathog. 8 (5) 10.1371/journal.ppat.1002707 e1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang N, 2019. Where are we going with genomics in plant pathogenic bacteria? Genomics 111 (4), 729–736. 10.1016/j.ygeno.2018.04.011. [DOI] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J, 2009. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CC, Zhang D, Hann DR, Xie ZP, Staehelin C, 2018. Biochemical properties and in planta effects of NopM, a rhizobial E3 ubiquitin ligase. J. Biol. Chem. 293 (39), 15304–15315. 10.1074/jbc.RA118.004444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, et al. , 2018. Barley stripe mosaic virus gamma b protein subverts autophagy to promote viral infection by disrupting the ATG7–ATG8 interaction. Plant Cell 30, 1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi G, et al. , 2021. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. 10.1038/s41586-021-03316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini G, Martens S, 2016. Mechanisms of selective autophagy. J. Mol. Biol. 428, 1714–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Krichevsky A, Loyter A, Citovsky V, 2010. Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe 7 (3), 197–209. 10.1016/j.chom.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wan L, Dai X, Sun Y, Wei W, 2014. Functional characterization of Anaphase Promoting Complex/Cyclosome (APC/C) E3 ubiquitin ligases in tumorigenesis. Biochim. Biophys. Acta 1845 (2), 277–293. 10.1016/j.bbcan.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li H, Hu L, Wang J, Zhou Y, Pang Z, Liu L, Shao F, 2008. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat. Struct. Mol. Biol. 15 (12), 1302–1308. 10.1038/nsmb.1517. [DOI] [PubMed] [Google Scholar]
- Zipfel C, 2014. Plant pattern-recognition receptors. Trends Immunol. 35 (7), 345–351. 10.1016/j.it.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G, 2006. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts agrobacterium-mediated transformation. Cell 125 (4), 749–760. 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.