Abstract
Mesorhizobium sp. strain N33 (Oxytropis arctobia), a rhizobial strain isolated in arctic Canada, is able to fix nitrogen at very low temperatures in association with a few arctic legume species belonging to the genera Astragalus, Onobrychis, and Oxytropis. Using mass spectrometry and nuclear magnetic resonance spectroscopy, we have determined the structure of N33 Nod factors, which are major determinants of nodulation. They are pentameric lipochito-oligosaccharides 6-O sulfated at the reducing end and exhibit other original substitutions: 6-O acetylation of the glucosamine residue next to the nonreducing terminal glucosamine and N acylation of the nonreducing terminal glucosamine by methyl-branched acyl chains of the iso series, some of which are α,β unsaturated. These unusual substitutions may contribute to the peculiar host range of N33. Analysis of N33 whole-cell fatty acids indicated that synthesis of the methyl-branched fatty acids depended on the induction of bacteria by plant flavonoids, suggesting a specific role for these fatty acids in the signaling process between the plant and the bacteria. Synthesis of the methyl-branched α,β-unsaturated fatty acids required a functional nodE gene.
Rhizobia are soil bacteria, now classified in several genera (e.g., Rhizobium, Sinorhizobium, Mesorhizobium, Bradyrhizobium, Azorhizobium), which form a symbiotic association with legume plants. The interaction between bacteria and plants results in the formation of nodules on the host plant roots, in which rhizobia fix atmospheric nitrogen. The association between rhizobia and legumes is specific: each rhizobial strain has a defined host range. For example, Mesorhizobium sp. strain N33, isolated from Oxytropis arctobia, also nodulates Astragalus alpinus and Onobrychis viciifolia (19, 26). In contrast, Sinorhizobium meliloti efficiently nodulates alfalfa (Medicago sativa) as well as Melilotus and Trigonella species. Mesorhizobium sp. strain N33 was isolated in the Canadian high arctic and is able to grow and fix nitrogen at temperatures as low as 5°C. In addition, it was shown that arctic rhizobia promoted better growth of O. viciifolia at low temperatures than did temperate strains (27). Thus, it might be valuable to extend the host range of arctic rhizobia to agronomically important legumes, in order to improve nitrogen fixation by these plants at low temperatures. It is therefore important to understand the molecular mechanisms controlling the nodulation specificity of arctic rhizobia.
Earlier work has shown that nodulation and host specificity in rhizobium-legume symbiosis are determined by signal exchanges between the bacteria and the host plant (21). Flavonoids excreted by the plant roots induce the expression of rhizobial nodulation (nod) genes. Most of these genes are involved in the biosynthesis and secretion of bacterial signals, the Nod factors, that can specifically induce symbiotic responses of the host plants. These responses include root hair deformation, division of root cortical cells and, in some instances, nodule formation (8, 32). The structures of Nod factors produced by several rhizobial species have been characterized. They are all lipochito-oligosaccharides (LCOs) consisting of β-1,4-linked oligomers of three to five N-acetylglucosamine residues with an amide-linked acyl chain on the nonreducing terminal residue (8, 10, 25). In addition, the glucosamine residues can carry various substitutions. Nod factor specificity is determined by the nature of these substitutions and of the N-acyl chain.
A number of nodulation genes have been identified in Mesorhizobium sp. strain N33 (2, 3, 4). The nodABC genes, which are found in all rhizobial species, are responsible for the synthesis of the lipo-oligosaccharide core common to all Nod factors. In contrast to most rhizobial species, where nodABC belong to one operon, in Mesorhizobium sp. strain N33 nodA and nodBC belong to two different operons (2, 4). In addition to nodABC, several nod genes likely to be involved in Nod factor substitutions have been characterized (3, 4). The nodHPQ genes, which are also found in S. meliloti and Rhizobium tropici, have been shown to specify O sulfation of the Nod factor reducing end in these two species (11, 18, 29). The nodFE genes, which are also present in S. meliloti and R. leguminosarum, determine in these two species the synthesis of various polyunsaturated fatty acids with one or several double bounds conjugated to the carbonyl group (7, 34, 39). Therefore, one might expect the Mesorhizobium sp. strain N33 Nod factors to be sulfated and substituted by a fatty acid with conjugated double bonds. However, given the peculiar host range of this species, the Mesorhizobium sp. strain N33 Nod factors are likely to carry novel substitutions. In this paper, we describe the structure of the Mesorhizobium sp. strain N33 Nod factors. We show that they have original structures with substitutions never described before, and we study several features of their biosynthesis.
MATERIALS AND METHODS
Microbiological techniques.
Bacterial strains and plasmids are described in Table 1. Mesorhizobium sp. strain N33 was grown in TY medium (33). The transfer of IncP and IncQ plasmids to Mesorhizobium sp. strain N33 was performed by triparental mating as described previously (1). Large-scale cultures were grown in Vincent medium (33) supplemented with 0.2% sodium glutamate and 0.1% sodium succinate as nitrogen and carbon sources, biotin at 0.5 μg ml−1, and other vitamins (Sigma MS vitamins) at 0.1 g liter−1. Formononetin (50 μM) or p-coumaric acid (100 μM) was used for the induction of nod gene expression.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Rhizobial strains | ||
| Mesorhizobium sp. strain N33 | Wild type; Nod+ on O. arctobia | 26 |
| JC253 | N33 nodC::Tn5; Smr Nmr | 2 |
| JC72 | N33 nodE::Tn5; Smr Nmr | 4 |
| JC20 | N33 nodG::Tn5; Smr Nmr | 4 |
| JC62 | N33 orfZ::Tn5; Smr Nmr | 4 |
| GMI5886 | S. meliloti 2011 ΔnodFEG; Nmr Phlr | 5 |
| GMI3202 | S. meliloti 2011 ΔnodA ΔnodFEG; Nmr Phlr | This work |
| Plasmids | ||
| pJC372 | 4.8-kb insert containing N33 nodAFEG in pRK7813 | This work |
| pA28 | pRK7813 derivative carrying NGR234 nodD1 | 28 |
| pML122 | Cloning vector; IncQ; Gmr | 17 |
| pJQ92 | pJQ200KS derivative with a nodA deletion; Gmr | 6 |
| pRK2013 | Helper plasmid for mobilization of IncP and IncQ plasmids; Kmr | 9 |
| LITMUS 28 | Cloning vector; ColE1; Apr | New England Biolabs |
| pRK7813 | Cloning vector; IncP; Tcr | 14 |
| pNGRD | 4-kb insert with NGR234 nodD1 in pML122 | This work |
Smr, streptomycin resistant; Nmr, neomycin resistant; Phlr, phleomycin resistant; Gmr, gentamicin resistant; Kmr, kanamycin resistant; Apr, ampicillin resistant; Tcr, tetracycline resistant.
DNA techniques.
Plasmid pJC372 was constructed as follows. A lambda EMBL3 genomic bank from strain N33 (2) was screened using a N33 nodAFEG probe (4), allowing the isolation of phage 235-1. From this phage, we isolated a 4.8-kb BamHI-BglII fragment containing nodAFEG with 1.3 and 0.9 kb of 5′- and 3′-flanking sequences, respectively, including the nod box. This fragment was cloned in BamHI-BglII-digested LITMUS 28, amplified, and cloned in BamHI-digested pRK7813.
In order to construct a plasmid carrying Rhizobium NGR234 nodD1 compatible with IncP plasmid pJC372, a 4-kb SalI fragment from pA28 (28) containing NGR234 nodD1 was cloned in SalI-digested IncQ plasmid pML122 (17), yielding pNGRD. S. meliloti GMI3202 with a deletion of the nodAFEG genes was constructed as described earlier (6) by introducing a deletion in nodA nonpolar on nodBC into S. meliloti GMI5886, which has a deletion of the nodFEG genes.
Nod factor purification.
Nod factors were extracted from filtered culture supernatants by butanol extraction (30). Purification was initially performed by high-performance liquid chromatography (HPLC) with a semipreparative C18 reverse-phase column (7.5 by 250 mm; Spherisorb ODS2; 5 μm) for 10 min in isocratic solvent A (water-acetonitrile, 80:20 [vol/vol]), followed by a linear gradient from solvent A to solvent B (100% acetonitrile) for 40 min at a flow rate of 1 ml min−1; the UV absorption at 206 and 220 nm was monitored. The fraction eluting between 36 and 62% acetonitrile was collected. More accurate purification was achieved with the same column and the same flow rate but with a 20-min linear gradient running from water-acetonitrile (70:30 [vol/vol]) containing 50 mM ammonium acetate (solvent A) to water-acetonitrile (50:50 [vol/vol]) containing 50 mM ammonium acetate (solvent B). Ammonium acetate and solvents from the collected fractions were removed by two successive lyophilizations.
Analytical methods. (i) Alkali hydrolysis of Nod factors and of bacteria.
Nod factors were hydrolyzed with a 5 N KOH solution at 100°C for 2 h. The solution was acidified with 6 N HCl, and fatty acids were extracted with diethyl ether. Lyophilized bacteria were suspended in a 5 N KOH solution and stirred at 110°C for 4 h. After acidification, the solution was extracted with diethyl ether. The acidic components were extracted from the ether layer with a 2 N KOH solution. The aqueous phase was acidified, and the fatty acids were extracted with diethyl ether.
(ii) Derivatization of fatty acids. (a) Pentafluorobenzyl ester derivatives.
Free fatty acids (less than 100 μg) were dissolved in a solution of 10 μl of anhydrous methanol and 50 μl of anhydrous acetonitrile. Two microliters of diisopropylamine and 2 μl of pentafluorobenzyl bromide were added. The reaction was complete after 1 h at room temperature. After total evaporation under nitrogen flux, the fatty acid derivatives were redissolved in dichloromethane.
(b) Methyl ester derivatives.
Fatty acids dissolved in dichloromethane were methylated in situ by coinjecting them with 1 μl of trimethylsulfonium hydroxide (Macherey-Nagel, Hoerdt, Germany) into the gas chromatograph injector kept at 250°C.
(iii) Mass spectrometry (MS).
An AUTOSPEC 6F sector instrument (Micromass, Altrincham, United Kingdom) with the EBE-EBE configuration was fitted with a cesium ion gun and a liquid secondary-ion MS (LSIMS) ion source. For sensitivity reasons, only the first half of the instrument was used. The energy of the Cs+ ions was 25 keV, and the accelerating voltage of the instrument was set to 8 kV. The 1:1 metanitrobenzyl alcohol-glycerol matrix was acidified with 1% trichloroacetic acid in water. Product ions resulting from decompositions in the first field free region were recorded in the constant B/E-linked scan mode.
The same instrument fitted with a chemical ionization or electron impact (EI) ion sources was used for gas chromatography (GC)-MS and GC-MS-MS studies. In the EI positive-ion mode, the electron energy was 70 eV. In the negative-ion electron capture mode, the reactant gas was ammonia bombarded by electrons at 70 eV. Carboxylate anions were dissociated by collision with helium gas in the cell located just after the magnet, and product ions were collected by scanning the second electrostatic analyzer monitored ion kinetic energy spectrometry. As soon as the different fatty acids were eluted from the GC column, the magnet field was adjusted manually to transmit the mass of the corresponding carboxylate. The helium pressure in the collision cell was adjusted to reduce the intensity of the ion beam by 50% of its original value.
(iv) NMR.
Nuclear magnetic resonance (NMR) experiments were done with a Brüker AMX 500 spectrometer operating at an observation frequency of 500 MHz for 1H. Data were recorded at 303 K. Samples were dissolved in a 3:7 D2O-CD3OD mixture. Methanol was used as an internal reference for chemical shift (δ) determinations. Phase-sensitive 1H-1H-correlated spectroscopy (COSY) (2J, 3J, and long-range coupling) was recorded.
RESULTS
Mesorhizobium sp. strain N33 Nod factors are pentameric LCOs 6-O sulfated at the reducing end.
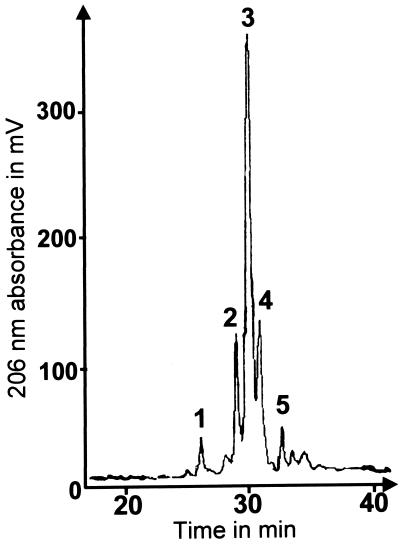
Previous work had shown that the Mesorhizobium sp. strain N33 nodulation genes were induced by compounds such as formononetin, an isoflavanone, and p-coumaric acid, a phenylpropane derivative (2). In order to prepare Nod factors, 5 liters of Mesorhizobium sp. strain N33 was grown in the presence of formononetin. The culture supernatant was extracted with n-butanol, and the LCOs in the extract were purified as described in Materials and Methods. Final purification was performed by HPLC on a C18 column with a water-acetonitrile gradient in the presence of ammonium acetate (Fig. 1), which allows better purification of sulfated species by reverse-phase HPLC. A total of 2.5 mg of Nod factors was obtained from a 5-liter culture. Nod factors were also extracted from a culture of Mesorhizobium sp. strain N33 induced by p-coumaric acid. Compounds identical to those obtained after induction by formononetin were obtained, as judged by the HPLC profile and MS analysis (data not shown).
FIG. 1.
HPLC profile of a Nod factor-enriched extract from a formononetin-induced N33 culture supernatant. Separation was performed in the presence of ammonium acetate as described in Materials and Methods, with monitoring of the absorption at 206 nm. See Table 2 for a description of the compounds found in fractions 1 to 4. No Nod factors were found in fraction 5.
Five HPLC fractions from a formononetin-induced culture were collected (Fig. 1) and analyzed by LSIMS. The positive-ion spectra of compounds in fractions 1 to 4 exhibited a complex series of protonated and alkali ion-attached molecules assigned to [M + Na]+, [M + K]+, [M − H + 2Na]+, [M − H + Na + K]+, and [M − H + 2K]+ (Fig. 2a). Confirmation of the molecular weights of the various molecules was performed by negative-ion spectrometry, which showed only [M − H]− ions. Ions appearing 80 mass units (u) lower than each MH+ ion in the positive-ion spectra suggested the presence of a sulfate substituent at the Nod factor reducing end. This suggestion was confirmed by product ion scanning of each MH+ ion after collision with helium, which showed an extensive loss of 80 units together with fragments of the chito-oligosaccharide backbone (Fig. 2b). Two-dimensional NMR (COSY) analysis of the Nod factor mixture indicated that the sulfate was located at position 6. The resonances of protons linked to the O-sulfated glucosaminyl residue were similar to those observed previously for synthetic Nod factors 6-O sulfated at the reducing end (36): H6′,—4.24 ppm, dd, J = 9 and 3.5 Hz; H6—4.16 ppm, dd, J = 9 and 2 Hz; and H5—4.05 ppm, m.
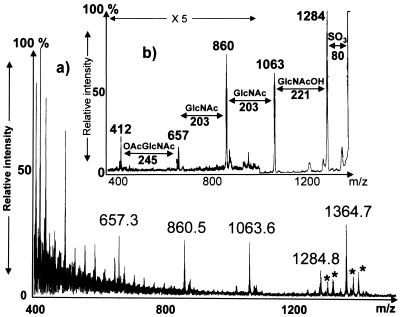
FIG. 2.
LSIMS analysis of N33 Nod factors. (a) Spectrum of HPLC fraction 4, showing an MH+ ion at m/z 1,364.7 for the Nod factor identified as NodMN33 V (iso-C17:1, Ac, S). See Table 2 for Nod factor nomenclature. Sodium and potassium salt adducts are indicated by asterisks. (b) Product ion spectrum from dissociation of the ion at m/z 1,364 (constant B/E-linked scans). Intervals of 203 u between the fragment ions indicate N-acetylglucosamine residues, while the interval of 245 u indicates additional O acetylation of the glucosamine residue next to the nonreducing end. The section of panel b that was magnified (magnification of ×5) is indicated.
Collision-induced decomposition of MH+ ions also induces the cleavage of each interglycosidic bond. Analysis of the collision-induced dissociation (CID)–MS patterns obtained for the different HPLC fractions showed that two types of Nod factors were present. The first one possessed a chitopentameric backbone substituted on both ends, with a sulfate on the reducing end and various N-acyl groups on the nonreducing end. The second one had a molecular weight 42 u higher, suggesting the presence of an additional acetyl group (see below).
Mesorhizobium sp. strain N33 Nod factors are partly 6-O acetylated on the glucosamine residue next to the non reducing terminal glucosamine.
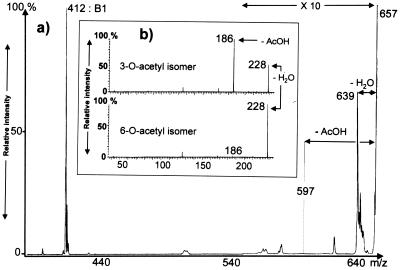
The position of the acetyl group on the chito-oligosaccharide backbone was determined by measuring the mass intervals between consecutive ions of the B series (oxonium ions); these intervals corresponded to 203 u (N-acetylglucosamine residues), except for that between B1 and B2, which was equal to 245 u (Fig. 2). These results indicated that the acetyl group is located on the glucosamine residue next to the nonreducing terminal glucosamine. Location of the O-acetyl group on this residue was attempted by measuring the spontaneous decomposition of the B2 ion. Either O-3 or O-6 acetylation is possible on an internal 1,4-linked N-acetylglucosamine residue. Studies performed previously on model compounds showed that an O-acetyl substitution at position 6 leads mostly to the elimination of water, whereas an O-acetyl substitution at position 3 is characterized by the loss of acetic acid (Fig. 3b) (38). The major decomposition product of the B2 ion at m/z 657 was the B1 ion at m/z 412, but the spectrum also showed the ion at m/z 639 resulting from the loss of water, whereas the elimination of acetic acid was undetectable (Fig. 3). Therefore, the acetyl group in N33 Nod factors is located at position 6. This assessment was confirmed by two-dimensional NMR (COSY) analysis (data not shown). The proton resonances were close to those observed previously for S. meliloti and R. leguminosarum bv. viciae Nod factors, which are both 6-O acetylated, although on another glucosaminyl residue (15, 26): H6′—4.43 ppm, dd, J = 12 and 2.5 Hz; H6—4.36 ppm, dd, J = 12 and 1.5 Hz; H5—4.16 ppm, m.
FIG. 3.
Location on O-6 (and not O-3) of the acetyl substituent in N33 Nod factors. (a) LSIMS of products from spontaneous dissociation (constant B/E-linked scans) of the B2 ion at m/z 657, generated in the LSIMS spectrum of the Nod factor shown in Fig. 2. Note the presence of an ion at m/z 639 resulting from the loss of water and the absence of an ion at m/z 597. The section of panel a that was magnified (magnification of ×10) is indicated. (b) Decomposition of oxonium ions (B ions) at m/z 246 from synthetic 3-O- and 6-O-acetyl-N-acetylglucosamine isomers, shown for comparison. Arrows indicate characteristic fragmentations.
A putative gene, called orfZ, encoding a protein homologous to microbial O-acetyltransferases, was previously found upstream of the N33 nodA gene (4). In order to test whether this gene is involved in the O acetylation of N33 Nod factors, LCOs were purified from the culture supernatant of an N33 derivative carrying a Tn5 insertion in orfZ. LSIMS analysis of HPLC-purified fractions indicated that compounds with m/z values corresponding to those of O-acetylated Nod factors were still present (data not shown).
Original saturated and α,β-unsaturated methyl-branched acyl chain substitutions in N33 Nod factors.
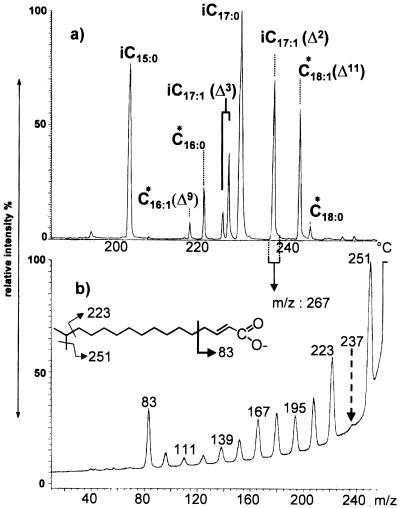
Calculations based on the masses of the B1 fragment ions suggested that the terminal nonreducing glucosamine of N33 Nod factors was substituted either by an acyl chain with an uneven number of carbons (C15 or C17) or by a methyl group and an acyl chain with an uneven number of carbons (C14 and C16). To identify the acyl substituents, the Nod factor mixture was submitted to alkali hydrolysis. The liberated fatty acids were derivatized as perfluorobenzyl esters and analyzed by capillary GC-MS in the dissociative electron capture ionization mode associated with high-energy CID of the carboxylate anions (20) (Fig. 4). The major components were identified as saturated and α,β-unsaturated methyl-branched fatty acids of the iso series. The presence of a methyl branch on the carbon atom next to the methyl end, characteristic of the iso series, was deduced from the greatly reduced loss of the C-2 neutral fragment (Fig. 4). The main saturated methyl-branched fatty acids were characterized as 13-methyl-tetradecanoic (iso-C15:0) and 15-methyl-hexadecanoic (iso-C17:0) acids. Monounsaturated methyl-branched fatty acids were also identified, the most abundant being α,β-unsaturated 15-methyl-2-hexadecenoic acid (iso- C17:1 Δ2). The position of the double bond in this compound was deduced from enhanced cleavage of the allylic bond, giving the ion at m/z 83 but not lower-mass fragments (20, 29). This main component was accompanied by two 15-methyl-3-hexadecenoic acids (assigned to the E and Z isomers), which probably resulted from alkali-induced isomerization of 15-methyl-2-hexadecenoic acid. Indeed, 1H NMR studies of the whole mixture of Nod factors confirmed the above assumption, as no signal due to a double bond at position 3 could be detected. The iso-methyl group appeared as a doublet at 0.87 ppm, with J = 6.7 Hz, correlating with a CH proton at 1.53 ppm. The conjugated double bond in the E configuration induced three characteristic resonances: H2 at 5.99 ppm, dt, J = 15 and 1.5 Hz; H3 at 6.85 ppm, dt, J = 15 and 7 Hz; and H4 at 2.12 ppm, m.
FIG. 4.
Characterization of fatty acids from a crude preparation of N33 Nod factors by negative-ion GC-MS of their perfluorobenzyl derivatives. (a) Total ion current (GC-MS profile). Peaks labeled by asterisks corresponded to fatty acids not characterized as Nod factor substituents by examination of the LSIMS mass spectra. This profile was recorded in the negative-ion electron capture mode. i, iso. (b) MS-MS profile (CID monitored ion kinetic energy spectrometry) of the carboxylate ion at m/z 267 from 15-methyl-2-hexadecenoic acid (iso-C17:1 Δ2). The low abundance of the ion at m/z 237 (loss of ethane) demonstrated the presence of a methyl group on C15. The higher abundance of the ion at m/z 83 (allylic cleavage), associated with a lack of ions at lower m/z values, was characteristic of Δ2 unsaturations.
Synthesis of methyl-branched fatty acids by N33 is flavonoid dependent.
Fatty acids of the iso series have never been described as components of Nod factors. However, they are important components of lipids in many bacteria (15, 16). We wondered whether the presence of such fatty acids in N33 Nod factors reflected the general fatty acid composition of lipids in this bacterium or whether the iso fatty acids were specific for the symbiosis signals. To address this question, we first characterized the fatty acid composition of N33 cells grown in the presence or in the absence of a flavonoid inducer. Whole N33 cell pellets were therefore submitted to alkaline hydrolysis, and the liberated fatty acids were identified by capillary GC coupled with electron impact MS (Fig. 5). For N33 cells grown in the presence of a flavonoid, vaccenic acid (C18:1) was identified as the most abundant component by far, and palmitic (C16:0) and stearic (C18:0) acids were also found (Fig. 5a). These are the fatty acids most commonly found in rhizobial lipids (37). In addition, small peaks attributed to iso-C15 and iso-C17 fatty acids were seen.
FIG. 5.
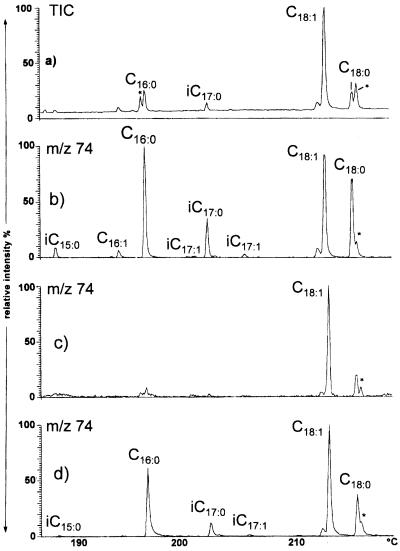
Characterization of fatty acids released by alkali hydrolysis of N33 cells grown in the presence or in the absence of flavonoids. The GC-MS profile of methyl esters was recorded in the electron impact ionization mode. Asterisks indicate nonacidic contamination. (a) Total ion current (TIC) from formononetin-induced cells. (b) Reconstructed chromatogram of the ion at m/z 74 from the same sample. This ion is the base peak in the spectra of methyl esters of saturated fatty acids. This profile was used to improve the signal-over-noise detection of saturated fatty acids, indicating the presence of iso-C15 and iso-C17 saturated fatty acids. (c) Reconstructed chromatogram of the ion at m/z 74 ion from a similar preparation from noninduced N33 cells. (d) Reconstructed chromatogram of the ion at m/z 74 from a similar preparation of formononetin-induced cells from the N33 nodC::Tn5 mutant. i, iso.
For better detection of the iso-C15 and iso-C17 fatty acids relative to the major fatty acid, vaccenic acid (C18:1), enhancement of the detection of saturated fatty acids versus unsaturated ones was performed by reconstructing the chromatogram of the ion at m/z 74, which is the base peak in the spectra of saturated fatty acids of the straight-chain or iso series. Fatty acids of the iso series, both saturated (iso-C15:0 and iso-C17:0) and α,β unsaturated (iso-C17:1), were clearly identified in flavonoid-induced cells (Fig. 5b) by this method. In contrast, they were not detected in the chromatogram obtained for noninduced cells (Fig. 5c). It is noteworthy that palmitic acid (C16:0), although not detected as a component of secreted Nod factors, also seemed more abundant in lipids of flavonoid-induced cells. The observation that iso-series fatty acids are found only in flavonoid-induced cells suggests that these compounds are specifically involved in the signaling process between plants and bacteria.
It was shown for R. leguminosarum that once synthesized, a portion of the Nod factors is secreted in the external medium, while another portion of them remains in the cells (22, 24). Thus, a portion of the iso-series fatty acids that we found in N33 cell pellets are probably substituents of Nod factors. In order to establish whether iso-series fatty acids might be components of other lipids, we studied the fatty acid composition of a nodC mutant of N33. nodC encodes the oligochitin synthase responsible for the synthesis of the Nod factor oligosaccharide backbone. nodC mutants are therefore unable to produce chitin oligomers. Fatty acids released by alkaline hydrolysis of the cells of a nodC mutant grown with or without a flavonoid were analyzed as before. Fatty acids of the methyl-branched iso series were still detected in cells grown with the flavonoid, although in smaller proportions than in the flavonoid-induced wild-type strain (Fig. 5d). Thus, it is likely that when an oligochitin acceptor is not available, methyl-branched fatty acids can be substituents of other lipids.
The Mesorhizobium sp. strain N33 nodE gene is required for the synthesis of Nod factors with α,β-unsaturated fatty acids.
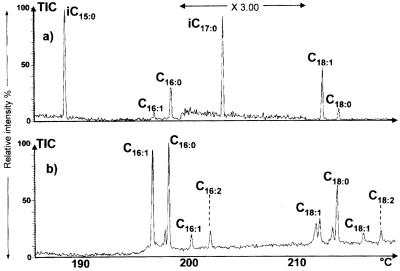
The presence of Nod factors with fatty acids bearing carbonyl-conjugated double bonds was expected, since the strain under study possesses nodFE genes. These genes are required for the synthesis of Nod factors with α,β-unsaturated acyl substituents in R. leguminosarum and S. meliloti (7, 34). We thus studied Nod factors produced by an N33 strain carrying a Tn5 insertion in the nodE gene. Nod factors were purified from the culture medium of the mutant grown in the presence of flavonoids and were hydrolyzed with alkali. The released fatty acids were analyzed by capillary GC coupled with MS as methyl ester derivatives. Unsaturated fatty acids of the iso series could not be detected, whereas saturated fatty acids of the iso series were still present, with a high proportion of the iso-C15 component (Fig. 6a). Fatty acids in whole-cell lipids were also characterized by the same method; again, no unsaturated fatty acids of the iso series were detected (data not shown).
FIG. 6.
Role of the nodE gene in the synthesis of N33 α,β-unsaturated Nod factors. (a) GC-MS profile (electron impact ionization mode) of methyl ester derivatives of fatty acids released by hydrolysis of Nod factors secreted by the N33 nodE::Tn5 mutant. Note the absence of the iso-C17:1 fatty acid. The section of panel a that was magnified (magnification of ×3) is indicated. i, iso. (b) GC-MS profile (electron impact ionization mode) of methyl ester derivatives of fatty acids in Nod factors secreted by the S. meliloti ΔnodAFEG (pN33nodAFEG) hybrid strain. TIC, total ion current.
In order to check that the defect in Nod factor synthesis observed for the nodE mutant was not due to a polar effect of the mutation on downstream genes, we characterized the acyl chain in Nod factors produced by a strain with a mutation in nodG, located just downstream of nodE. Unsaturated fatty acids of the iso series (iso-C17:1) were detected (data not shown). Therefore, nodG is not required for the synthesis of these fatty acids, whereas nodE is.
An intriguing observation is that the only fatty acid with a carbonyl-conjugated double bond is the methyl-branched C17 fatty acid of the iso series, while most of the fatty acids produced by Mesorhizobium sp. strain N33 are straight-chain fatty acids. In other rhizobia that produce straight-chain polyunsaturated Nod factors, it is hypothesized that NodFE proteins act on the same short-chain precursors as for the synthesis of common fatty acids (vaccenic and stearic acids). Is the selectivity for branched chains observed here due to a strict specificity of N33 NodFE or NodA (acyltransferase) for structures of the iso series? To address this question, we constructed an S. meliloti strain in which the nodAFEG genes were replaced by those of N33. We first constructed an S. meliloti strain carrying two deletions eliminating nodA and nodFEG. A plasmid carrying the N33 nodAFEG genes under the control of their nod box was then transferred to the deletion strain. Finally, a plasmid carrying the Rhizobium sp. strain NGR234 nodD1 gene was introduced into the transconjugant in order to overexpress the other nod genes and produce sufficient amounts of Nod factors for characterization. Nod factors were purified from the culture supernatant, and the Nod factor acyl chains were analyzed by GC-MS as described above. In the profile of fatty acids released from the Nod factors, a variety of straight-chain fatty acids with zero, one, and two double bonds were detected. Their structures were similar to those already identified for Nod factors from wild-type S. meliloti strains. Thus, the NodFE and NodA proteins from N33 are able to use straight-chain precursors as substrates.
DISCUSSION
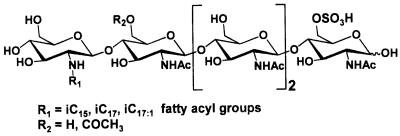
The Nod factors of Mesorhizobium sp strain N33 (O. arctobia) are chitopentamers of N-acetylglucosamine, 6-O sulfated at the reducing end, partially 6-O acetylated at the glucosamine residue proximal to the nonreducing terminal glucosamine, and N acylated at the nonreducing end by saturated and α,β-monounsaturated methyl-branched fatty acids (Fig. 7 and Table 2). In comparison with the previously described Nod factor structures, these molecules have novel features. These molecules are 6-O acetylated at the glucosamine residue proximal to the nonreducing terminal glucosamine. Recently, substitution of this residue was described for Mesorhizobium loti NZP2213, where a fucose substituent was present on O-3 (23), and for R. galegae, where an acetyl substitution was found on O-3 (40). The biological importance of these substituents is currently unknown, as are the genes encoding the substitutions. O acetylation of Nod factors on the terminal nonreducing and reducing glucosamine residues was described previously, and two different genes encoding the corresponding O-acetyltransferases were found in rhizobia: nodL and nodX, which encode 6-O-acetyltransferases specific for the nonreducing and reducing glucosaminyl ends, respectively (10, 25). Sequencing of the N33 DNA upstream of nodA revealed the presence of orfZ (4), which is homologous to bacterial O-acetyl transferase genes. However, a strain mutated in orfZ still produced acetylated Nod factors, suggesting that either this gene is not involved in the acetylation of N33 Nod factors or other N33 genes can replace it in this function.
FIG. 7.
General structure of Mesorhizobium sp. strain N33 (O. arctobia) Nod factors. i, iso.
TABLE 2.
Summary of the main Nod factors from Mesorhizobium sp. strain N33 (O. arctobia)
| Nod factor structurea | Molecular mass (Da) | HPLC fraction |
|---|---|---|
| NodMN33 V (iso-C15:0, Ac, S) | 1,337.7 | 1 |
| NodMN33 V (iso-C17:1, S) | 1,321.7 | 2 |
| NodMN33 V (iso-C17:1, Ac, S) | 1,363.7 | 3 |
| NodMN33 V (iso-C17:0, Ac, S) | 1,365.7 | 4 |
The roman numeral V indicates the number of glucosamine residues. The substitutions are listed in the parentheses as follows: Ac, acetate; S, sulfate.
The other substitution peculiar to N33 Nod factors is the fatty acyl chain. Methyl-branched fatty acids of the iso series, one of them α,β unsaturated, are substituents of N33 Nod factors. All of the Nod factor acyl chains described so far in other rhizobia are unbranched, as are the major fatty acid components of rhizobial lipids. However, in many bacteria, methyl-branched fatty acids are important components of cellular lipids. They play a role in the adaptation to cold temperatures, since they decrease the phase transition temperature of the lipids in which they are incorporated (16). We therefore wondered whether methyl-branched fatty acids could be major components of lipids in arctic strain N33 and thus could replace linear fatty acids in Nod factors. We found that in bacteria grown at 20°C in the absence of flavonoids, methyl-branched fatty acids could not be detected; in addition, they were quantitatively minor components of lipids in bacteria cultivated in the presence of flavonoids. Synthesis of methyl-branched fatty acids thus seems specific for the signal exchange between N33 and its host plant, and as such, it could contribute to the specificity of this symbiotic interaction. However, we cannot exclude the possibility that synthesis of the methyl-branched fatty acids is also induced when Mesorhizobium sp. strain N33 is grown at low temperatures. If this were the case then methyl-branched fatty acids could also be involved in a general adaptation of N33 to life at low temperatures.
In addition to LCOs, methyl-branched fatty acids could be substituents of other N33 lipids. This idea is suggested by our observation that an N33 strain mutated in nodC and therefore unable to synthesize the chito-oligosaccharide backbone of Nod factors still contained methyl-branched fatty acids. Such a substitution of lipids different from LCOs by symbiosis-specific fatty acids was described previously by Geiger et al. (12, 13). These authors showed that nodFE-dependent polyunsaturated fatty acids could be substituents of R. leguminosarum bv. viciae phospholipids. However, it is not known whether these nodFE-dependent phospholipids play a role in the symbiotic process.
The presence of methyl-branched fatty acyl chains in N33 LCOs raises the question of the biosynthesis of the corresponding fatty acids in N33. These fatty acids are important components of membrane lipids in bacteria such as Bacillus, where their biosynthetic pathways have been studied. Differences in the biosynthesis of methyl-branched fatty acids and straight-chain fatty acids lie in the primers that are used in the initial condensation reaction catalyzed by the fatty acid synthase (16). For branched-chain fatty acids of the iso series, such as iso-C15:0 or iso-C17:0, the primers are derivatives of the methyl-branched amino acid leucine, most likely α-ketoisocaproyl- or isovaleryl-coenzyme A. Synthesis of such primers or their conversion to acyl carrier protein derivatives by a fatty acid synthase complex might be flavonoid dependent in N33. This idea would explain why flavonoids are required for the production of lipids with branched-chain fatty acids.
In addition to being methyl branched, one of the fatty acyl components of N33 Nod factors is α,β unsaturated. In N33, acyl groups with more than one conjugated double bond have not been detected, while in other rhizobia synthesizing Nod factors with α,β-unsaturated fatty acyl groups (R. leguminosarum bv. viciae, R. leguminosarum bv. trifolii, M. huakuii, R. galegae, and S. meliloti), up to three carbonyl-conjugated double bonds have been characterized (20, 31, 34, 35, 40). In addition, in the latter four species, contrary to N33, fatty acid chains are linear and generally carry a cis double bond at the ω7 position. As in R. leguminosarum and S. meliloti, the nodE gene is required in N33 for the synthesis of Nod factors with α,β-unsaturated fatty acids. It was proposed earlier (12) that the acyl carrier protein NodF and the putative condensing enzyme NodE could modify normal fatty acid biosynthesis in such a way that α,β-unsaturated fatty acid derivatives would not undergo the step of reduction of the enoyl intermediate. Such a model could also apply to the biosynthesis of N33 Nod factor acyl chains, if it is postulated that N33 NodFE proteins have a higher affinity for branched-chain intermediates than for unbranched ones. However, this preference would not be strict, since when the nodAFEG genes of N33 were transferred to an S. meliloti strain with a deletion of its nodAFEG genes, Nod factors possessing linear α,β-unsaturated fatty acyl chains were produced. It is likely that in S. meliloti, branched precursors are not synthesized, so that N33 NodFE proteins instead use available linear precursors, such as acetyl- and malonyl-acyl carrier proteins.
In a recent work (40), it was proposed that a cluster of phylogenetically related legumes are nodulated by rhizobia producing Nod factors with α,β-unsaturated fatty acids. This cluster includes the Trifolieae, Vicieae, and Galegeae tribes. Strain N33, which produces Nod factors with α,β-unsaturated fatty acids, was isolated from O. arctobia, a member of the Galegeae tribe. It also nodulates O. viciifolia, a member of the Hedysareae tribe, which is closely related to the Galegeae tribe. Therefore, the present work confirms the earlier proposal and extends the cluster of related legumes nodulated by rhizobia producing Nod factors with α,β-unsaturated fatty acids to members of the Hedysareae tribe. We hypothesized that in this group of plants, a new mechanism allowing the recognition of structural features of the Nod factor acyl chain evolved. This led to new possibilities for determining the specificity of rhizobium-plant interactions. Previously, two sources of variation among rhizobial LCO α,β-unsaturated fatty acids were known, the length of the acyl chain and the number of conjugated double bonds. With N33, another source of variation was explored, the branching of the acyl chain; the latter might contribute to the peculiar host range of this species, together with the other substitutions. Methyl-branched fatty acids could also play a role in the adaptation of signaling mechanisms to low temperatures. It would be interesting to determine the structure of the acyl chain in Nod factors produced by other arctic rhizobia.
ACKNOWLEDGMENT
We are grateful to P. Roche for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Ardourel M, Demont N, Debellé F, Maillet F, De Billy F, Promé J C, Dénarié J, Truchet G. Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cloutier J, Laberge S, Prevost D, Antoun H. Sequence and mutational analysis of the common nodBCIJ region of Rhizobium sp. (Oxytropis arctobia) strain N33, a nitrogen-fixing microsymbiont of both arctic and temperate legumes. Mol Plant-Microbe interact. 1996;9:523–531. doi: 10.1094/mpmi-9-0523. [DOI] [PubMed] [Google Scholar]
- 3.Cloutier J, Laberge S, Castonguay Y, Antoun H. Characterization and mutational analysis of nodHPQ genes of Rhizobium sp. strain N33. Mol Plant-Microbe Interact. 1996;9:720–728. doi: 10.1094/mpmi-9-0720. [DOI] [PubMed] [Google Scholar]
- 4.Cloutier J, Laberge S, Antoun H. Sequence and mutational analysis of the 6.7-kb region containing nodAFEG genes of Rhizobium sp. strain N33: evidence of DNA rearrangements. Mol Plant-Microbe Interact. 1997;10:401–406. doi: 10.1094/MPMI.1997.10.3.401. [DOI] [PubMed] [Google Scholar]
- 5.Debellé F, Maillet F, Vasse J, Rosenberg C, de Billy F, Truchet G, Dénarié J, Ausubel F M. Interference between Rhizobium meliloti and Rhizobium trifolii nodulation genes: genetic basis of R. meliloti dominance. J Bacteriol. 1988;170:5718–5727. doi: 10.1128/jb.170.12.5718-5727.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debellé F, Plazanet C, Roche P, Pujol C, Savagnac A, Rosenberg C, Promé J C, Dénarié J. The NodA proteins of Rhizobium meliloti and Rhizobium tropici specify the N-acylation of Nod factors by different fatty acids. Mol Microbiol. 1996;22:303–314. doi: 10.1046/j.1365-2958.1996.00069.x. [DOI] [PubMed] [Google Scholar]
- 7.Demont N, Debellé F, Aurelle H, Dénarié J, Promé J C. Role of the Rhizobium meliloti nodF and nodE genes in the biosynthesis of lipo-oligosaccharidic nodulation factors. J Biol Chem. 1993;268:20134–20142. [PubMed] [Google Scholar]
- 8.Dénarié J, Debellé F, Promé J C. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 9.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Downie J A. Functions of rhizobial nodulation genes. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Molecular biology of model plant-associated bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 387–402. [Google Scholar]
- 11.Folch-Mallol J L, Marroqui S, Sousa C, Manyani H, Lopez-Lara I M, van der Drift K M, Haverkamp J, Quinto C, Gil-Serrano A, Thomas-Oates J, Spaink H P, Megias M. Characterization of Rhizobium tropici CIAT899 nodulation factors: the role of nodH and nodPQ genes in their sulfation. Mol Plant-Microbe Interact. 1996;9:151–163. doi: 10.1094/mpmi-9-0151. [DOI] [PubMed] [Google Scholar]
- 12.Geiger O, Thomas-Oates J E, Glushka J, Spaink H P, Lugtenberg B J J. Phospholipids of Rhizobium contain nodE-determined highly unsaturated fatty acid moieties. J Biol Chem. 1994;269:11090–11097. [PubMed] [Google Scholar]
- 13.Geiger O, Glushka J, Lugtenberg B J, Spaink H P, Thomas-Oates J E. NodFE-dependent fatty acids that lack an alpha-beta unsaturation are subject to differential transfer, leading to novel phospholipids. Mol Plant-Microbe Interact. 1998;11:33–44. doi: 10.1094/MPMI.1998.11.1.33. [DOI] [PubMed] [Google Scholar]
- 14.Jones J D G, Gutterson N. An efficient mobilizable cosmid vector, pRK7813, and its use in a rapid method for marker exchange in Pseudomonas fluorescens strain HV37a. Gene. 1987;61:299–306. doi: 10.1016/0378-1119(87)90193-4. [DOI] [PubMed] [Google Scholar]
- 15.Kaneda T. Fatty acids of the genus Bacillus: an example of branched-chain preference. Bacteriol Rev. 1977;41:391–418. doi: 10.1128/br.41.2.391-418.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Rev. 1991;55:288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labes M, Puhler A, Simon R. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene. 1990;89:37–46. doi: 10.1016/0378-1119(90)90203-4. [DOI] [PubMed] [Google Scholar]
- 18.Laeremans T, Caluwaerts I, Verreth C, Rogel M A, Vanderleyden J, Martinez-Romero E. Isolation and characterization of Rhizobium tropici Nod factor sulfation genes. Mol Plant-Microbe Interact. 1996;9:492–500. doi: 10.1094/mpmi-9-0492. [DOI] [PubMed] [Google Scholar]
- 19.Laguerre G, van Berkum P, Amarger N, Prevost D. Genetic diversity of rhizobial symbionts isolated from legume species within the genera Astragalus, Oxytropis, and Onobrychis. Appl Environ Microbiol. 1997;63:4748–4758. doi: 10.1128/aem.63.12.4748-4758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé J C, Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 21.Long S R. Rhizobium symbiosis: nod factors in perspective. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKay I A, Djordjevic M A. Production and excretion of Nod metabolites by Rhizobium leguminosarum bv. trifolii are disrupted by the same environmental factors that reduce nodulation in the field. Appl Environ Microbiol. 1993;59:3385–3392. doi: 10.1128/aem.59.10.3385-3392.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsthoorn M M A, Lopez-Lara I M, Petersen B O, Bock K, Haverkamp J, Spaink H P, Thomas-Oates J E. Novel branched nod factor structure results from alpha-(1→3) fucosyl transferase activity: the major lipo-chitin oligosaccharides from Mesorhizobium loti strain NZP2213 bear an alpha-(1→3) fucosyl substituent on a nonterminal backbone residue. Biochemistry. 1998;37:9024–9032. doi: 10.1021/bi972937r. [DOI] [PubMed] [Google Scholar]
- 24.Orgambide G G, Lee J, Hollingsworth R I, Dazzo F B. Structurally diverse chitolipooligosaccharide nod factors accumulate primarily in membranes of wild type Rhizobium leguminosarum biovar trifolii. Biochemistry. 1995;34:3832–3840. doi: 10.1021/bi00011a041. [DOI] [PubMed] [Google Scholar]
- 25.Perret X, Staehelin C, Broughton W J. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prévost D, Bordeleau L M, Caudry-Reznick S, Schulman H M, Antoun H. Characteristics of rhizobia isolated from three legumes indigenous to the Canadian high arctic: Astragalus alpinus, Oxytropis maydelliana, and Oxytropis arctobia. Plant Soil. 1987;98:313–324. [Google Scholar]
- 27.Prévost D, Bordeleau L M, Michaud R, Lafrenière C, Waddington J, Biederbeck V O. Nitrogen fixation efficiency of cold-adapted rhizobia on sainfoin (Onobrychis viciifolia): laboratory and field evaluation. In: Graham P H, Sadowsky M J, Vance C P, editors. Symbiotic nitrogen fixation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 171–176. [Google Scholar]
- 28.Price N P, Relic B, Talmont F, Lewin A, Promé D, Pueppke S G, Maillet F, Dénarié J, Promé J C, Broughton W J. Broad-host-range Rhizobium species strain NGR234 secretes a family of carbamoylated, and fucosylated, nodulation signals that are O-acetylated or sulphated. Mol Microbiol. 1992;6:3575–3584. doi: 10.1111/j.1365-2958.1992.tb01793.x. [DOI] [PubMed] [Google Scholar]
- 29.Roche P, Debellé F, Maillet F, Lerouge P, Faucher C, Truchet G, Dénarié J, Promé J C. Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell. 1991;67:1131–1143. doi: 10.1016/0092-8674(91)90290-f. [DOI] [PubMed] [Google Scholar]
- 30.Roche P, Lerouge P, Ponthus C, Promé J C. Structural determination of bacterial nodulation factors involved in the Rhizobium meliloti-alfalfa symbiosis. J Biol Chem. 1991;266:10933–10940. [PubMed] [Google Scholar]
- 31.Schultze M, Quiclet-Sire B, Kondorosi E, Virelizer H, Glushka J N, Endre G, Gero S D, Kondorosi A. Rhizobium meliloti produces a family of sulfated lipooligosaccharides exhibiting different degrees of plant host specificity. Proc Natl Acad Sci USA. 1992;89:192–196. doi: 10.1073/pnas.89.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultze M, Kondorosi A. Regulation of symbiotic root nodule development. Annu Rev Genet. 1998;32:33–57. doi: 10.1146/annurev.genet.32.1.33. [DOI] [PubMed] [Google Scholar]
- 33.Somasegaran P, Hoben H J. Handbook for rhizobia. Berlin, Germany: Springer-Verlag KG; 1994. [Google Scholar]
- 34.Spaink H P, Sheeley D M, van Brussel A A, Glushka J, York W S, Tak T, Geiger O, Kennedy E P, Reinhold V N, Lugtenberg B J. A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature. 1991;354:125–130. doi: 10.1038/354125a0. [DOI] [PubMed] [Google Scholar]
- 35.Spaink H P, Bloemberg G V, van Brussel A A N, Lugtenberg B J J, van der Drift K M G M, Haverkamp J, Thomas-Oates J E. Host specificity of Rhizobium leguminosarum is determined by the hydrophobicity of highly unsaturated fatty acyl moieties of the nodulation factors. Mol Plant-Microbe Interact. 1995;8:155–164. [Google Scholar]
- 36.Tailler D, Jacquinet J C, Beau J M. Total synthesis of Nod-Rm-IV(S): a sulfated lipotetrasaccharide symbiotic signal from Rhizobium meliloti. J Chem Soc Chem Commun. 1994;16:1827–1828. [Google Scholar]
- 37.Tighe S W, de Lajudie P, Dipietro K, Lindstrom K, Nick G, Jarvis B D. Analysis of cellular fatty acids and phenotypic relationships of Agrobacterium, Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium species using the Sherlock Microbial Identification System. Int J Syst Evol Microbiol. 2000;50:787–801. doi: 10.1099/00207713-50-2-787. [DOI] [PubMed] [Google Scholar]
- 38.Treilhou M, Ferro M, Monteiro C, Poinsot V, Jabbouri S, Kanony C, Promé D, Promé J C. Differentiation of O-acetyl and O-carbamoyl esters of N-acetyl-glucosamine by decomposition of their oxonium ions. Application to the structure of the nonreducing terminal residue of Nod factors. J Am Soc Mass Spectrom. 2000;11:301–311. doi: 10.1016/s1044-0305(99)00152-x. [DOI] [PubMed] [Google Scholar]
- 39.van der Drift K M, Spaink H P, Bloemberg G V, van Brussel A A, Lugtenberg B J, Haverkamp J, Thomas-Oates J E. Rhizobium leguminosarum bv. trifolii produces lipo-chitin oligosaccharides with nodE-dependent highly unsaturated fatty acyl moieties. An electrospray ionization and collision-induced dissociation tandem mass spectrometric study. J Biol Chem. 1996;271:22563–22569. doi: 10.1074/jbc.271.37.22563. [DOI] [PubMed] [Google Scholar]
- 40.Yang G-P, Debellé F, Savagnac A, Ferro M, Schiltz O, Maillet F, Promé D, Treilhou M, Vialas C, Lindstrom K, Dénarié J, Promé J C. Structure of the Mesorhizobium huakuii and Rhizobium galegae Nod factors: a cluster of phylogenetically related legumes are nodulated by rhizobia producing α,β-unsaturated fatty acids. Mol Microbiol. 1999;34:227–237. doi: 10.1046/j.1365-2958.1999.01582.x. [DOI] [PubMed] [Google Scholar]