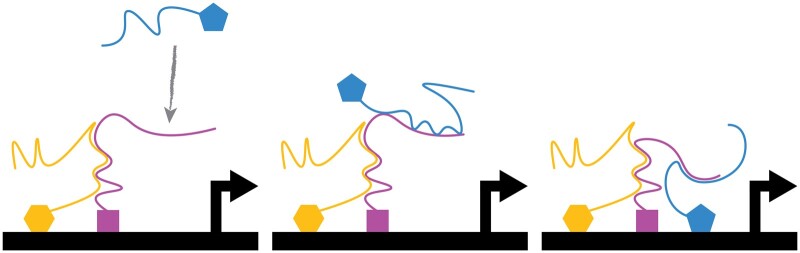
In the traditional view of transcription factors, the DNA binding domain bears sole responsibility for identifying target loci in the genome; however, recent results contradict this view. Staller highlights those results and proposes that transcription factors search the nucleus for target loci with a combination of protein-protein interactions and protein-DNA interactions. This two-step nuclear search explains both old and new results.
Keywords: transcription factor, intrinsically disordered region, DNA-binding domain, transcriptional condensate, liquid–liquid phase separation, transcription, gene regulation, transcriptional activation domain, transcription initiation
Abstract
Transcription factors regulate gene expression by binding to regulatory DNA and recruiting regulatory protein complexes. The DNA-binding and protein-binding functions of transcription factors are traditionally described as independent functions performed by modular protein domains. Here, I argue that genome binding can be a 2-part process with both DNA-binding and protein-binding steps, enabling transcription factors to perform a 2-step search of the nucleus to find their appropriate binding sites in a eukaryotic genome. I support this hypothesis with new and old results in the literature, discuss how this hypothesis parsimoniously resolves outstanding problems, and present testable predictions.
Graphical abstract
Graphical abstract.
Transcription factors have 2 jobs: binding DNA and regulating transcription. Site-specific transcription factors bind short DNA sequences, called motifs, with DNA-binding domains. Eukaryotic transcription factors regulate transcription with effector domains that bind to regulatory complexes: repression domains bind corepressors and activation domains bind coactivators. Transcription factors have other functions, but most of their other domains (e.g. dimerization domains, degrons, and ligand-binding domains) modulate DNA binding or coregulator binding. In this review, I argue that the standard model is incomplete and that some transcription factors search the nucleus in a 2-step process. These transcription factors use protein–protein interactions to perform a global search of the nucleus to find a “protein cloud” and then use DNA-binding domains to perform a local search of the DNA within that protein cloud. This expanded model is motivated by examples where deleting the DNA-binding domain does not prevent transcription factors from localizing to the correct promoters (Brodsky et al. 2020; Gera et al. 2022), which I discuss in detail below. The global search with protein–protein interactions localizes the transcription factor to the right region of the nucleus, and then the DNA-binding domain scans the DNA in that region and dwells on the cognate motif. Critically, the protein–protein interactions that perform the global search for the protein cloud require protein sequences outside the DNA-binding domain.
I have chosen the term “protein cloud” to emphasize that this idea is still cloudy. I am picturing a nonstoichiometric cluster of transcription factors engaged in both homotypic interactions between multiple copies of the same transcription factor and heterotypic interactions between different transcription factors. This cluster may or may not include coactivator proteins, which could, in principle, bridge multiple TF molecules (Tuttle et al. 2018; Sanborn et al. 2021). I am not invoking a large, energetically stable liquid–liquid phase-separated droplet, but something more dynamic, in line with the original definition of a condensate or with a transcription factor hub (Shin and Brangwynne 2017; Chong et al. 2018). I am picturing dozens of molecules, not hundreds. In plants, the AUXIN RESPONSE FACTOR (ARF)7, ARF19, and EARLY FLOWERING3 (ELF3) transcription factors each become inactive when they enter a condensate (Powers et al. 2019; Jung et al. 2020). In human cell culture, much of the attention on transcriptional condensates has focused on transcriptional activation. Although I assume a rather explicit mechanism for transcriptional activation (see below), this hypothesis is not about activation. Instead, it addresses the problem of selecting active regions of the genome. It is related to the problem of identifying where transcriptional condensates or hubs form, which is the same as the old problem of why a region of the genome is an active enhancer in 1 cell type and inert in another.
In transcription factor biology, we know a lot more about DNA-binding domains than we know about the rest of the protein. DNA-binding domains are structured, conserved, and predictable based on protein sequences (Latchman 2008; El-Gebali et al. 2019). DNA-binding domains are the basis for transcription factor family organization schemes (Lambert et al. 2018). There are many methods for measuring protein–DNA interactions in vitro and in vivo (Stormo 2013). Outside of the DNA-binding domain, transcription factors are primarily composed of long intrinsically disordered regions (IDRs) that do not fold into a single 3D structure and instead exhibit multiple conformations (Liu et al. 2006; van der Lee et al. 2014). The sequence of an IDR controls whether these ensembles are expanded, collapsed, or form hairpins (Das and Pappu 2013). The nomenclature in the literature is confusing: some IDRs have been called low-complexity domains because they contain only a few types of amino acids (Chong et al. 2018; Cascarina et al. 2020). The terms activation domain, transactivation domain, or activator domain have been used to refer to everything outside of the DNA-binding domain or to minimized, highly active regions (Latchman 2008; Staller et al. 2018; Tycko et al. 2020). Here, I use the term activation domain to refer to short, highly active regions that directly contact coactivators, and I use the term IDR to refer to extended regions outside of DNA-binding domains and other folded domains.
Classically, it was argued that DNA-binding domains and activation domains were independent, modular components, but this idea is approaching the end of its usefulness. In the few cases that have been carefully examined, activation domains can modulate DNA affinity, increase specificity for cognate motifs, or increase affinity for random DNA (Liu et al. 2008; Krois et al. 2018; Baughman et al. 2022). For the remainder of this piece, I assume that true modularity is rare. All activation domains are disordered in solution, and many fold upon binding to partners (Dyson and Wright 2016). The one known exception is IRF3, which is natively folded (Qin et al. 2003). There are a handful of well-studied repression domains, notably the KRAB and POZ/BTB domains, but aside from these 2 types, there are no good predictors of repression domains (Bintu et al. 2016; Soto et al. 2022). There is a rich body of work examining activation domain coactivator interactions with NMR; for example, p53, RelA, the ETV family, Hif1a, and CITED2 (Dyson and Wright 2016; Raj and Attardi 2017; Currie et al. 2017; Berlow et al. 2022) in human and Gcn4 and Gal4 in yeast (Brzovic et al. 2011; Hahn and Young 2011; Tuttle et al. 2021). There has been some progress predicting acidic activation domains from protein sequence in yeast and human proteomes (Ravarani et al. 2018; Erijman et al. 2020; Sanborn et al. 2021; Staller et al. 2022), but it has been difficult to distill the features of other classes, such as proline-rich or glutamine-rich activation domains (Latchman 2008). In recent work, I argued that the critical sequence feature of acidic activation domains is the balance between acidic residues and aromatic and leucine residues (Staller et al. 2022).
This 2-step nuclear search hypothesis is motivated by a result from Barkai and colleagues showing how IDRs of Msn2p and Yap1p are necessary and sufficient for targeting a transcription factor to the correct promoter in yeast (Brodsky et al. 2020; Gera et al. 2022). This hypothesis is further influenced by single-molecule imaging of transcription factor dynamics in living nuclei, where the IDRs of Hif1α and Hif2α are necessary and sufficient to control the fraction of molecules bound to chromatin and the diffusion rates of mobile molecules (Chen et al. 2021). However, this hypothesis can also explain several puzzling results from genomics over the last 2 decades and reemphasizes outstanding questions. In the following sections, I develop this hypothesis, contrast it with several models in the literature, and discuss testable predictions.
Assumptions
Implicit in the 2-step nuclear search hypothesis are several assumptions about how transcription factors work together to activate transcription. First, I assume a thermodynamic framework, where protein–protein interactions and transcription factor–DNA interactions occur quickly enough to come to equilibrium. Protein clouds can nucleate anywhere, but they preferentially accumulate at genomic sites with many transcription factor-binding sites. Traditionally, the thermodynamic framework assumed constant microscopic on-rates and slower off-rates at cognate sites, but there is accumulating evidence that DNA sequence modulates transcription factor–DNA on rates (Marklund et al. 2022). Second, I assume a key feature of transcriptional regulation is enhancer occupancy, or the total fraction of time an enhancer is bound by transcription factors (and not the residence times of individual molecules, which are generally less than 15 s) (Sherman and Cohen 2012; Stormo 2013; Chen et al. 2014, 2021; Hansen et al. 2018). Genome specificity is achieved thermodynamically by equilibrium binding of transcription factors. Third, I assume that all transcriptional regulation is combinatorial: namely, that multiple transcription factors must simultaneously achieve high occupancy to activate transcription. It is not yet clear whether each transcription factor brings in a different coactivator or if multiple transcription factor molecules together recruit 1 coactivator (e.g. a p53 tetramer binding 4 domains of p300; Ferreon et al. 2009). Fourth, I assume that an enhancer acts as a scaffold to bring together the multiple biochemical activities necessary to progress through the steps of the transcription cycle (e.g. opening chromatin, assembling the basal transcriptional machinery, forming the polymerase initiation complex, initiating polymerase and releasing paused polymerase) (Fuda et al. 2009). While it is clear that there is more than 1 step in transcription, it is not clear how many of these steps are near rate limiting at a given gene. For a thorough and highly accessible discussion of kinetic control of transcription, see Scholes et al. (2017). Fifth, I assume that multivalent-binding “cycles” that bridge multiple molecules are a critical feature: transcription factors simultaneously bind DNA and other proteins and simultaneous release of all contacts is rare, slowing transcription factor escape from a protein cloud (Deeds et al. 2012; Sanborn et al. 2021). Sixth, I will assume that histone modifications are the time integral of recent transcription factor-binding activity, serving as a short-term memory for occupancy (Long et al. 2016).
A new phenomenon requires a new model
The crucial new data motivating the 2-step nuclear search hypothesis are the recent work from Barkai and colleagues (Brodsky et al. 2020) showing that long IDRs are necessary and sufficient to target Msn2p and Yap1p to the correct promoters in yeast. Critically, the DNA-binding domain is dispensable for targeting to the correct promoter: transcription factors lacking the DNA-binding domain lost the sharp peak in binding signal over the DNA motif, but they retained substantial binding throughout the promoter. The integral of the binding signal over the full promoter was largely unchanged between full length Msn2p and the DNA-binding domain deletion. In contrast, the Msn2p DNA-binding domain alone bound some, but not all, of the same promoters and bound to new promoters. For promoters that retained binding of the DNA-binding domain only, the integral of the binding signal was reduced and the remaining binding shifted to motifs in the nucleosome free region (the ∼100-bp upstream of a transcription start site). For Msn2p, the binding signal over the promoter decreased as the IDR was shortened. Notably, the annotated activation domains were dispensable for proper promoter targeting. One important coactivator subunit, Med15p, was also dispensable for proper promoter targeting. In reciprocal chimeras that exchanged the IDRs and DNA-binding domains of Msn2p and Nrg2p, the IDR dominated promoter selection. This result upends the classical picture of a modular transcription factor where the DNA-binding domain is solely responsible for localization to the correct genomic locations.
The 2-step nuclear search hypothesis can explain this result: the IDR localizes the transcription factor to the protein cloud at the correct target promoters and the DNA-binding domain scans this promoter and binds to its cognate motif. AD–coactivator interactions may contribute to localizing a transcription factor to the right protein cloud, but they are neither necessary nor sufficient (Brodsky et al. 2020). Targeting the transcription factor to the protein cloud requires additional protein-protein interactions. I anticipate these interactions will include both homotypic interactions between multiple copies of the same transcription factor and heterotypic interactions between different transcription factors. There is direct evidence for homotypic clusters of Sp1, Mig1p, and Msn2p (Su et al. 1991; Wollman et al. 2017; Chong et al. 2018). This IDR-mediated nuclear search is primarily used to find existing protein clouds at specific genomic locations, not nucleate new ones. I discuss below how these protein clouds nucleate at specific genomic regions.
Importantly, Brodsky et al. could not detect this phenomenon with traditional ChIP-seq and required a more sensitive method, ChEC-seq (Brodsky et al. 2020). Independent work using Calling Cards, an orthogonal method, found that for 2 paralogous yeast transcription factors, regions outside the DNA-binding domain control targeting to the correct promoters (Shively et al. 2019). Gera et al. examined 30 pairs of transcription factor paralogs and showed that for 18 pairs, genomic localization is determined primarily by regions outside the DNA-binding domain (Gera et al. 2022). The remaining 12 behaved like traditional transcription factors, with the DNA-binding domain determining promoter selection.
It is likely that Chen et al. are observing the same phenomenon as Brodsky et al. and Gera et al. at the single-molecule level (Chen et al. 2021). By comparing chimeras of 2 paralogous transcription factors, they have shown that the fraction of molecules immobilized on the chromatin and the diffusion rate of mobile proteins are determined primarily by the IDR and not the DNA-binding domain. The different diffusion rates of the mobile fractions can be explained by the IDRs orchestrating distinct constellations of protein–protein interactions, namely distinct clusters that wander the nucleus at different rates. The changes in the fraction of molecules bound to chromatin are hard to rationalize without something akin to the 2-step nuclear search hypothesis. The 2-step nuclear search explains both of these single-molecule phenomena.
A 2-step search solves old problems
Invoking a 2-step nuclear search solves 3 old problems: (1) Why do only a minority of residues in transcription factors have known functions? (2) Why are only a tiny fraction of transcription factor motifs in a metazoan genome bound in vivo? (3) Why do many genome regions detected by ChIP-seq assays not contain motifs for the precipitated transcription factor?
First, the known functional domains in most transcription factors cover only a minority of residues (Lambert et al. 2018; Soto et al. 2022). Most eukaryotic transcription factors have a short, structured, and conserved DNA-binding domain, while the majority of the protein is intrinsically disordered and poorly conserved. Even in well-characterized transcription factors, the known activation domains, repression domains, ligand-binding domains, dimerization domains, and other Pfam domains cover only the minority of residues (Soto et al. 2022). What is the rest of the protein doing? Some of these residues are flexible linkers between activation domains and are necessary for multivalent, fuzzy binding to coactivators (Harmon et al. 2017; Tuttle et al. 2018). However, we should be skeptical of the idea that the majority of residues in a transcription factor are linkers. We must also grant that most effector domains are not yet annotated, but known examples are short, with a median length of 91 residues (Soto et al. 2022). Under the 2-step nuclear search hypothesis, some of these long IDRs bind other IDRs to localize transcription factors to a protein cloud at target promoters. Metazoan transcription factors have expanded IDRs (Liu et al. 2006; Jana et al. 2021), which may result from neutral drift (Lynch et al. 2016) but may enable the expansion of protein–protein interactions that accompanied multicellularity (Dunker et al. 2015). There is evidence that long IDRs can mediate homotypic and heterotypic interactions that cause clustering in the nucleus (Chong et al. 2018; Boija et al. 2018). Under the 2-step nuclear search hypothesis, the unannotated regions of IDRs perform the global search.
Second, how do transcription factors avoid getting lost in the genome? Only a tiny fraction of predicted transcription factor-binding sites in a metazoan genome are bound by a transcription factor: there are millions of predicted motifs, thousands of which are bound in ChIP-seq assays and a subset of which are active in reporter gene assays. What distinguishes the bound sites from the unbound sites? This problem has enthralled genomicists for over 20 years (Harbison et al. 2004; Harrison et al. 2011; White et al. 2013). For a thorough review of the specificity problem see Brodsky et al. (2021). This problem has been formalized with information theory: metazoan genomes are large and transcription factor motifs are short, so there is not enough information in a single motif occurrence to uniquely define genomic addresses (Wunderlich and Mirny 2009). In the human genome, a cluster of 10–15 sites are necessary to uniquely encode a 500–1,000 bp genomic location. In the 2-step nuclear search hypothesis, the IDR performs the global search, contributing additional information to find the right loci. Once the transcription factor is in the protein cloud, the DNA-binding domain is only responsible for the local search of a much smaller amount of DNA. The local search then becomes efficient, leading to high occupancy and sharp peaks over cognate motifs in ChEC-seq (Brodsky et al. 2020). The 2-step search similarly explains how large clusters of Ultrabithorax protein can accumulate at low-affinity transcription factor-binding sites that control development of bristles in fly (Crocker et al. 2015). A protein cloud with dozens of members, each with an expanded IDR, also offers a larger search target than a single DNA-binding site.
Third, genome-wide ChIP-seq data contain a second paradox: many peaks do not contain a DNA motif for the precipitated transcription factor. By some estimates 30–70% of called ChIP-seq peaks do not contain a motif for the precipitated transcription factor (Harrison et al. 2011; Spitz and Furlong 2012; reviewed in Jana et al. 2021). There are at least 3 classes of peaks without motifs: (1) “Hyperchipable” regions caused by DNA/RNA hybrids, high expression, and other fixation artifacts (Teytelman et al. 2013); (2) highly occupied target (HOT) regions of highly open chromatin that are bound by practically every transcription factor and are sometimes computationally removed as an artifact (Kvon et al. 2012); and (3) true enhancers bound by partner transcription factors. The third class motivated the transcription factor collective model: active enhancers are bound by a group of cell-type-specific transcription factors that together activate expression (Spitz and Furlong 2012). Any given enhancer has binding sites for most but not all transcription factors in this group. Under the 2-step nuclear search hypothesis, a transcription factor will spend significant time in all compatible protein clouds, not just those with cognate-binding sites, and these clouds will provide ChIP-seq signal. Some will consider this 2-step nuclear search hypothesis to be a restatement of the transcription factor collective model, but I argue below that this hypothesis makes several more precise predictions.
Additional support from the literature
Further support of the 2-step nuclear search hypothesis comes from ChIP-exo and single-particle tracking experiments on transcription factor mutants that remove the IDR or mutate the DNA-binding domain (Chen et al. 2014). Compared to the full-length protein, the Sox2 DNA-binding domain alone spent less time in 3D diffusion and had double the number of ChIP-exo peaks, and its mean dwell time on chromatin was shorter. This result was interpreted as more binding to “pseudotargets” with lower quality motifs (more ChIP-exo peaks and shorter binding times to these lower-quality motifs). The reciprocal perturbation, a mutation disrupting the Sox2 DNA-binding domain, still bound ∼26% of original genomic loci, showing that the IDR is sufficient for genomic localization, similar to Msn2p in yeast (Brodsky et al. 2020). Compared to the full protein, the DNA-binding domain-inactivating mutant spent more time in 3D diffusion and had a lower fraction of immobilized molecules, and these immobile molecules had longer dwell times. These results imply that the IDR is reducing binding to incorrect genomic loci, either by increasing time spent in protein clouds at the correct loci or by other means (like directly competing with the DNA-binding domain; Krois et al. 2018). The results are confusing but can be interpreted as follows: the DNA-binding domain contributes both short-lived binding at random DNA and medium-lived binding at motifs, while the IDR contributes long-lived binding to protein clouds. The WT protein is a convolution of these 3 binding modes. Reciprocally, WT 3D search can be interrupted by DNA binding to a true motif, nonspecific DNA binding to random open DNA, or IDR binding to a protein cloud. Under the 2-step nuclear search hypothesis, the interpretation of these data is that the protein–protein interactions that retain transcription factors in protein clouds have slower off-rates (longer dwell times) than DNA-binding interactions at low-quality motifs. Also, consistent with the 2-step nuclear search, single-particle tracking of the glucocorticoid receptor observed low mobility (confined) and chromatin-bound states (Garcia et al. 2021). Deleting the glucocorticoid receptor IDR caused a loss of the confined state and the majority of ChIP-seq peaks.
Relationship to other models
The 2-step nuclear search hypothesis is a reimagining of the Transcription Factor Funnel Model where the funnel is protein–protein interactions instead of DNA (Castellanos et al. 2020). In the DNA funnel model, partial transcription factor-binding sites near a “real” transcription factor-binding site can slow down a DNA-binding domain during 1D scanning of DNA, effectively concentrating the transcription factor near the real binding sites (Wunderlich and Mirny 2008). The transcription factor funnel model has always been hard to rationalize with eukaryotic chromatin and its short regions of naked DNA between histones. The observed partial sites can just as easily be the product of binding site turnover (Ludwig et al. 2000; Hare et al. 2008). By contrast, the 2-step nuclear search hypothesis uses protein–protein interactions rather than DNA-binding domain–DNA interactions to concentrate protein at active enhancers.
The 2-step nuclear search hypothesis is compatible with the original formulation of the pioneer factor hypothesis. Pioneer factors are transcription factors with specialized DNA-binding domains and specialized activation domains that bind closed chromatin and open it up for other transcription factors, defining the active enhancer landscape and specifying cell types (Zaret 2020). This function is analogous to nucleating and localizing the protein clouds. The 2-step nuclear search hypothesis is more useful for explaining global gene regulation if only some transcription factors follow it: some transcription factors define the locations of the protein clouds with DNA-binding domains and others are followers with IDRs. For example, on long time scales, developmental master regulator transcription factors would localize the protein clouds at cell-type-specific enhancers, and then fast acting, signaling effector transcription factors could simply join these clouds (e.g. Glucocorticoid receptor; Barolo and Posakony 2002; Vockley et al. 2016). The IDR-dominated Msn2p, Hif1α, and Hif2α are stress response transcription factors (Brodsky et al. 2020; Chen et al. 2021).
However, the 2-step nuclear search hypothesis is equally compatible with the Collaborative Competition Model, where transcription factors work together to evict nucleosomes and open chromatin (Polach and Widom 1996; Mirny 2010). Once formed, a protein cloud has many DNA-binding domains that together outcompete nucleosomes. In the Collaborative Competition Model, DNA-binding domains have quantitatively different affinities for DNA rather than specialized subclasses.
It bears noting that Brodsky and colleagues offer 2 other explanations for their observed phenomena (Brodsky et al. 2020, 2021; Jana et al. 2021; Gera et al. 2022). They propose that the IDR-mediated nuclear localization could be driven by condensates. More intriguingly, they propose that the IDR can directly bind to specific DNA sequences in a highly distributed manner. In vitro experiments may be necessary to distinguish these 2 models or the 2-step nuclear search.
Do transcription factors hunt the genome for binding sites in packs or as lone wolves?
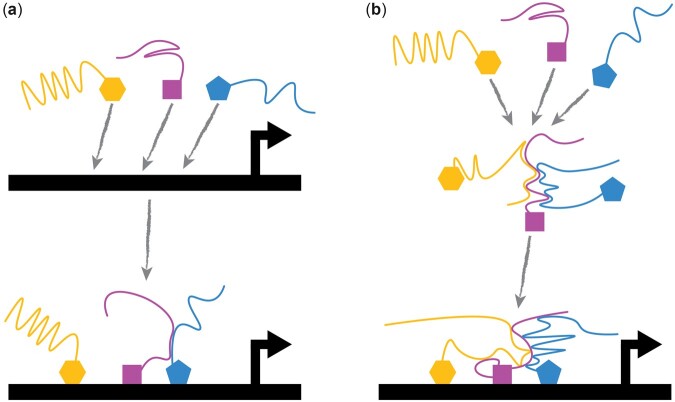
Most cartoons of transcription factor function depict a single protein molecule diffusing through the nucleoplasm searching for its cognate-binding site. The implicit assumption is that transcription factors are lone wolves that search for their binding sites by themselves (Fig. 1a).
Fig. 1.
Transcription factors could hunt the genome for binding sites in wolf packs. a) In the traditional model, transcription factors arrive at a promoter independently, hunting for binding sites like lone wolves. They often bind cooperatively on arrival (e.g. interface between purple and blue IDRs). b) Some transcription factors can form clusters in the nucleoplasm and search for promoters as a group, hunting the genome like a wolf pack. Solid shapes are folded DNA binding domains and the tails are IDRs.
A corollary to the 2-step nuclear search hypothesis is that clusters of transcription factors could search the nucleoplasm together as a single unit, collaboratively hunting for binding sites, like a wolf pack (Fig. 1b). This cluster of transcription factors, or nascent protein cloud, would have many DNA-binding domains that together contain enough motif information to uniquely specify regions of the genome. A heterotypic cluster of transcription factors matches the clusters of heterotypic binding sites in an enhancer. These transcription factor wolf packs would have variable sizes, which can explain why some transcription factors have a broad range of apparent diffusion constants in single-particle tracking experiments (Heckert et al. 2021; Chen et al. 2021). For some transcription factors, like Mig1p and Msn2p, the functional unit is likely a small cluster (Wollman et al. 2017). Notably, a wolf pack would complicate some models of cooperative activation of transcription (Estrada et al. 2016; DePace AH, personal communication).
It is not clear if a wolf pack would speed up or slow down nuclear search kinetics. More DNA-binding domains would increase the number of nonspecific DNA-binding events, which could slow the search. More DNA-binding domains would also slow the off-rate at real target sites, ensuring that more collisions with real targets are productive. Under the assumption of a thermodynamic framework here, the wolf pack aids in the selection of correct genomic locations. The transcription factors that establish the protein cloud could search the nucleus as a wolf pack and signal response effector transcription factors would join the clouds by performing the 2-step nuclear search.
Am I kicking the can down the road?
The biggest weakness with the 2-step nuclear search hypothesis is the lingering question of specificity. How does the protein cloud form at or localize to the right parts of the genome? This weakness is a restatement of other important problems: what distinguishes active enhancers in a cell? or what nucleates transcriptional condensates? One answer comes from the thermodynamic framework, where all euchromatin is sampled with approximately the same on-rate, and slower off-rates at clusters of binding sites nucleate the protein clouds. Protein clouds emerge at clusters of binding sites by equilibrium binding of transcription factors to DNA. Protein–protein interactions between the transcription factors stabilize the clouds in a feed-forward manner. In the wolf pack framework, master regulator transcription factors bind each other in the nucleoplasm and search the genome as a unit. Once they find a cognate transcription factor-binding site cluster, they would have an extended dwell time. Individual molecules would still have short residence times, but the protein cloud would have a longer dwell time, resulting in higher DNA occupancy (Sanborn et al. 2021).
A parallel problem with the 2-step nuclear search hypothesis is the issue of protein cloud diversity. Do individual transcription factors join multiple types of protein clouds? Are all the clouds similar? It is safe to assume that many different clouds will eventually activate transcription by recruiting coactivators like p300/CBP, Mediator, SAGA, and TFIID (Latchman 2008). Do these transcription factor-coactivator interactions occur before or after a cloud settles on a genomic locus? It follows that transcription factor–coactivator interactions are poor candidates for protein–protein interactions to nucleate protein clouds because coactivators must be able to activate many (sometimes all) genes and must be able to enter potentially all protein clouds. For example, in yeast, it has been argued that mediator is necessary for transcription of virtually all genes (Petrenko et al. 2017), but degrading mediator with degrons changes the expression of only 6% of genes (Warfield et al. 2021). Degrading mediator in human cells has similarly modest effects (El Khattabi et al. 2019). If instead, the dominant force creating protein clouds is transcription factor-transcription factor interactions (homotypic or heterotypic), then it is easy to create diverse protein clouds.
Combining DNA-binding domain-driven and IDR-driven nuclear search—allowing for a diversity of transcription factors
So far, I have drawn a strong contrast between traditional DNA-binding domain-driven nuclear search and a 2-step, IDR-driven nuclear search, but biology rarely works in absolutes. We can imagine a continuum between a DNA-binding domain-only mode and an IDR-only mode of genomic site selection. This continuum is anchored by Max, which contains only a DNA-binding domain, and the Notch Intracellular Domain, which has no DNA-binding domain (Grandori et al. 2000; Hori et al. 2013). The Notch signaling protein is cleaved in response to extracellular signals, allowing the Notch Intracellular Domain to enter the nucleus and bind to CSL (also known as Suppressor of Hairless in flies or Lag1 in worms), displacing corepressors and recruiting mastermind and other coactivators (Hori et al. 2013). Notch lacks a DNA-binding domain and performs the global search using its IDR. Other transcription factors would lie on this continuum between Max/Max dimers and the Notch intracellular domain. For each transcription factor, genomic site selection would be the combination of the DNA-binding domain contribution and the IDR contribution. This combination may or may not be a simple sum. The transcription factors that are DNA-binding domain-dependent would establish the protein clouds while the transcription factors that are IDR dependent would go to the existing clouds. The 2-step nuclear search is more useful for gene regulation if some transcription factors set up the protein clouds and others follow.
Moreover, it is formally possible that the same transcription factor might find different binding sites in the genome with different mixtures of the 2 parts of the 2-step search: that some genomic sites will be selected by the DNA-binding domain and other genomic sites will be selected by the IDR. New work from the Barkai group found that 12 pairs of transcription factor paralogs had largely overlapping genomic localization. For 12 other pairs, the IDR dominated promoter localization; for the remaining 6 pairs, both the IDR and the DNA-binding domain contributed (Gera et al. 2022). This blend of genomic site selection parallels the recent argument that transcription factors can have pioneering activity at specific genomic sites (Hansen et al. 2022). A transcription factor might help establish a protein cloud at the genomic locations with high-quality motifs for its DNA-binding domain and be a follower with its IDR at other genomic locations.
Testing the 2-step nuclear search model
A model is most useful when it can make testable predictions. The 2-step nuclear search hypothesis predicts that more transcription factors will behave like the Brodsky et al. data: transcription factors without DNA-binding domains (or with mutant DNA-binding domains) will continue to localize to the correct enhancers and promoters but lose the focal peaks above motifs. Truncating transcription factor IDRs will gradually shift genome binding from endogenous targets toward DNA-binding domain-only targets, which will be more enriched for motifs and general open chromatin.
There are 3 more predictions. First, there will be regions of the protein that are responsible for genomic localization outside of the activation domains and DBDs. They will be necessary and sufficient for the global search. Brodsky et al. have demonstrated this prediction genome wide and Chen et al. have demonstrated it for single molecules. All that remains is to find more examples and exceptions. Second, the reciprocal prediction is that if we cut out the internal “inert” regions of transcription factors, there will be genome localization defects, i.e. a minimal transcription factor with all the known minimal activation domains and DNA-binding domain will not bind to and activate endogenous targets. Third, chimeras that swap DNA-binding domains and IDRs between pairs of transcription factors could reveal more cases where the IDR or the DNA-binding domain dominates genome binding. These experiments are now feasible.
Conclusion
I have proposed that transcription factors search the nucleus for binding sites with a combination of a global search with protein–protein interactions mediated by the IDR and a local search with protein–DNA interactions mediated by the DNA-binding domain. This 2-step nuclear search hypothesis can explain several long-standing irregularities in the literature. It follows that these protein–protein interactions may initiate off of the DNA, yielding small wolf packs of transcription factors that together hunt the nucleus for binding sites. So far, I have discussed this idea only in the context of active euchromatin. If the tight meshwork of heterochromatin (Ou et al. 2017) precludes transcription factor wolf packs from entering, this would further ensure a tight off state, reduce the genomic search space, and speed up nuclear searches.
Acknowledgments
The author would like thank Alex Holehouse, Zeba Wunderlich, Vincent Fan, Yu Chen, Ben Vincent, Thomas Graham, Angela DePace, Ryan Friedman, Xavier Darzacq, Clarice Kit Hong, Robert Tjian, Michael Gabriel Hayes, Jordan Stefani, Abrar Abidi, Michael White, Nicholas Morffy, and members of the Tjian-Darzacq labs for helpful discussions and comments on the manuscript. The author would also like to thank Jasper Rine for editorial feedback.
Funding
This work was supported by the Burroughs Wellcome Fund Postdoctoral Enrichment Program grant 1017384, NSF grant 2112057, and startup funding from UC Berkeley.
Conflicts of Interest
None declared.
Data Availability
No data were generated for this article.
Literature cited
- Barolo S, Posakony JW.. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16(10):1167–1181. [DOI] [PubMed] [Google Scholar]
- Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin M, Gunther TR, Wolynes PG, Komives EA. An intrinsically disordered transcription activation domain alters the DNA binding affinity and specificity of NFκB p50/RelA. bioRxiv 2022.04.11.487922, 2022, accessed 10 July 2022. 10.1101/2022.04.11.487922 [DOI] [PMC free article] [PubMed]
- Berlow RB, Dyson HJ, Wright PE.. Multivalency enables unidirectional switch-like competition between intrinsically disordered proteins. Proc Natl Acad Sci U S A. 2022;119(3). e2117338119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Yong J, Antebi YE, McCue K, Kazuki Y, Uno N, Oshimura M, Elowitz MB.. Dynamics of epigenetic regulation at the single-cell level. Science. 2016;351(6274):720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175(7):1842–1855.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky S, Jana T, Barkai N.. Order through disorder: the role of intrinsically disordered regions in transcription factor binding specificity. Curr Opin Struct Biol. 2021;71:110–115. [DOI] [PubMed] [Google Scholar]
- Brodsky S, Jana T, Mittelman K, Chapal M, Kumar DK, Carmi M, Barkai N.. Intrinsically disordered regions direct transcription factor in vivo binding specificity. Mol Cell. 2020;79(3):459–471. e4. [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, Warfield L, Littlefield P, Baker D, Klevit RE, et al. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol Cell. 2011;44(6):942–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascarina SM, Elder MR, Ross ED.. Atypical structural tendencies among low-complexity domains in the Protein Data Bank proteome. PLoS Comput Biol. 2020;16(1):e1007487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos M, Mothi N, Muñoz V.. Eukaryotic transcription factors can track and control their target genes using DNA antennas. Nat Commun. 2020;11(1):540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen B-C, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156(6):1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cattoglio C, Dailey G, Zhu Q, Tjian R, Darzacq X. Mechanisms governing target search and binding dynamics of hypoxia-inducible factors. bioRxiv 2021.10.27.466110, 2021, accessed 10 July 2022. doi: 10.1101/2021.10.27.466110 [DOI] [PMC free article] [PubMed]
- Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361(6400). 10.1126/science.aar2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F, et al. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell. 2015;160(1–2):191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie SL, Doane JJ, Evans KS, Bhachech N, Madison BJ, Lau DKW, McIntosh LP, Skalicky JJ, Clark KA, Graves BJ, et al. ETV4 and AP1 transcription factors form multivalent interactions with three sites on the MED25 activator-interacting domain. J Mol Biol. 2017;429(20):2975–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das RK, Pappu RV.. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc Natl Acad Sci U S A. 2013;110(33):13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeds EJ, Bachman JA, Fontana W.. Optimizing ring assembly reveals the strength of weak interactions. Proc Natl Acad Sci U S A. 2012;109(7):2348–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Bondos SE, Huang F, Oldfield CJ.. Intrinsically disordered proteins and multicellular organisms. Semin Cell Dev Biol. 2015;37:44–55. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE.. Role of intrinsic protein disorder in the function and interactions of the transcriptional coactivators CREB-binding protein (CBP) and p300. J Biol Chem. 2016;291(13):6714–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khattabi L, Zhao H, Kalchschmidt J, Young N, Jung S, Van Blerkom P, Kieffer-Kwon P, Kieffer-Kwon K-R, Park S, Wang X, et al. A pliable mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell. 2019;178(5):1145–1158. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erijman A, Kozlowski L, Sohrabi-Jahromi S, Fishburn J, Warfield L, Schreiber J, Noble WS, Söding J, Hahn S.. A high-throughput screen for transcription activation domains reveals their sequence features and permits prediction by deep learning. Mol Cell. 2020;78(5):890–902. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada J, Wong F, DePace A, Gunawardena J.. Information integration and energy expenditure in gene regulation. Cell. 2016;166(1):234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon JC, Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE.. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci U S A. 2009;106(16):6591–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT.. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461(7261):186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DA, Johnson TA, Presman DM, Fettweis G, Wagh K, Rinaldi L, Stavreva DA, Paakinaho V, Jensen RAM, Mandrup S, et al. An intrinsically disordered region-mediated confinement state contributes to the dynamics and function of transcription factors. Mol Cell. 2021;81(7):1484–1498. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gera T, Jonas F, More R, Barkai N.. Evolution of binding preferences among whole-genome duplicated transcription factors. eLife. 2022;11: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN.. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. [DOI] [PubMed] [Google Scholar]
- Hahn S, Young ET.. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189(3):705–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Woringer M, Grimm JB, Lavis LD, Tjian R, Darzacq X.. Robust model-based analysis of single-particle tracking experiments with Spot-On. eLife. 2018;7. 10.7554/eLife.33125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JL, Loell KJ, Cohen BA.. The pioneer factor hypothesis is not necessary to explain ectopic liver gene activation. eLife. 2022;11. 10.7554/eLife.73358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne J-B, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431(7004):99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB.. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4(6):e1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon TS, Holehouse AS, Rosen MK, Pappu RV.. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. eLife. 2017;6. 10.7554/eLife.30294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li X-Y, Kaplan T, Botchan MR, Eisen MB.. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7(10):e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckert A, Dahal L, Tjian R, Darzacq X. Recovering mixtures of fast diffusing states from short single particle trajectories. bioRxiv 2021.05.03.442482, 2021, accessed 10 July 2022. doi: 10.1101/2021.05.03.442482 [DOI] [PMC free article] [PubMed]
- Hori K, Sen A, Artavanis-Tsakonas S.. Notch signaling at a glance. J. Cell Sci. 2013;126:2135–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana T, Brodsky S, Barkai N.. Speed–specificity trade-offs in the transcription factors search for their genomic binding sites. Trends Genet. 2021;37(5):421–432. [DOI] [PubMed] [Google Scholar]
- Jung J-H, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F, et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature. 2020;585(7824):256–260. [DOI] [PubMed] [Google Scholar]
- Krois AS, Dyson HJ, Wright PE.. Long-range regulation of p53 DNA binding by its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci U S A. 2018;115(48):E11302–E11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Stampfel G, Yáñez-Cuna JO, Dickson BJ, Stark A.. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev. 2012;26(9):908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT, et al. The human transcription factors. Cell. 2018;175(2):598–599. [DOI] [PubMed] [Google Scholar]
- Latchman DS. Eukaryotic Transcription Factors. San Diego: Elsevier Science & TechnologyOxford University Press; 2008. [Google Scholar]
- Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK.. Intrinsic disorder in transcription factors. Biochemistry. 2006;45(22):6873–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Matthews KS, Bondos SE.. Multiple intrinsically disordered sequences alter DNA binding by the homeodomain of the Drosophila hox protein ultrabithorax. J Biol Chem. 2008;283(30):20874–20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J.. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167(5):1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M.. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403(6769):564–567. [DOI] [PubMed] [Google Scholar]
- Lynch M, Ackerman MS, Gout J-F, Long H, Sung W, Thomas WK, Foster PL.. Genetic drift, selection and the evolution of the mutation rate. Nat Rev Genet. 2016;17(11):704–714. [DOI] [PubMed] [Google Scholar]
- Marklund E, Mao G, Yuan J, Zikrin S, Abdurakhmanov E, Deindl S, Elf J.. Sequence specificity in DNA binding is mainly governed by association. Science. 2022;375(6579):442–445. [DOI] [PubMed] [Google Scholar]
- Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Proc Natl Acad Sci U S A. 2010;107(52):22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC.. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357(6349). 10.1126/science.aag0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko N, Jin Y, Wong KH, Struhl K.. Evidence that mediator is essential for Pol II transcription, but is not a required component of the preinitiation complex in vivo. eLife. 2017;6. 10.7554/eLife.28447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polach KJ, Widom J.. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol. 1996;258(5):800–812. [DOI] [PubMed] [Google Scholar]
- Powers SK, Holehouse AS, Korasick DA, Schreiber KH, Clark NM, Jing H, Emenecker R, Han S, Tycksen E, Hwang I, et al. Nucleo-cytoplasmic partitioning of ARF proteins controls auxin responses in Arabidopsis thaliana. Mol Cell. 2019;76(1):177–190. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin BY, Liu C, Lam SS, Srinath H, Delston R, Correia JJ, Derynck R, Lin K.. Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat Struct Biol. 2003;10(11):913–921. [DOI] [PubMed] [Google Scholar]
- Raj N, Attardi LD.. The transactivation domains of the p53 protein. Cold Spring Harb Perspect Med. 2017;7(1):a026047. 10.1101/cshperspect.a026047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravarani CN, Erkina TY, De Baets G, Dudman DC, Erkine AM, Babu MM.. High-throughput discovery of functional disordered regions: investigation of transactivation domains. Mol Syst Biol. 2018;14(5):e8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Yeh BT, Feigerle JT, Hao CV, Townshend RJ, Lieberman Aiden E, Dror RO, Kornberg RD.. Simple biochemical features underlie transcriptional activation domain diversity and dynamic, fuzzy binding to Mediator. eLife. 2021;10. 10.7554/eLife.68068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes C, DePace AH, Sánchez Á.. Combinatorial gene regulation through kinetic control of the transcription cycle. Cell Syst. 2017;4(1):97–108. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MS, Cohen BA.. Thermodynamic state ensemble models of cis-regulation. PLoS Comput Biol. 2012;8(3):e1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP.. Liquid phase condensation in cell physiology and disease. Science. 2017;357(6357). 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- Shively CA, Liu J, Chen X, Loell K, Mitra RD.. Homotypic cooperativity and collective binding are determinants of bHLH specificity and function. Proc Natl Acad Sci U S A. 2019;116(32):16143–16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto LF, Li Z, Santoso CS, Berenson A, Ho I, Shen VX, Yuan S, Fuxman Bass JI.. Compendium of human transcription factor effector domains. Mol Cell. 2022;82(3):514–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM.. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613–626. [DOI] [PubMed] [Google Scholar]
- Staller MV, Holehouse AS, Swain-Lenz D, Das RK, Pappu RV, Cohen BA.. A high-throughput mutational scan of an intrinsically disordered acidic transcriptional activation domain. Cell Syst. 2018;6(4):444–455. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller MV, Ramirez E, Kotha SR, Holehouse AS, Pappu RV, Cohen BA.. Directed mutational scanning reveals a balance between acidic and hydrophobic residues in strong human activation domains. Cell Syst. 2022;13(4):334–345. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD. Introduction to protein-DNA Interactions: Structure, Thermodynamics, and Bioinformatics. New York: Cold Spring Harbor Laboratory Press; 2013. [Google Scholar]
- Su W, Jackson S, Tjian R, Echols H.. DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 1991;5(5):820–826. [DOI] [PubMed] [Google Scholar]
- Teytelman L, Thurtle DM, Rine J, van Oudenaarden A.. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci U S A. 2013;110(46):18602–18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle LM, Pacheco D, Warfield L, Luo J, Ranish J, Hahn S, Klevit RE.. Gcn4-mediator specificity is mediated by a large and dynamic fuzzy protein-protein complex. Cell Rep. 2018;22(12):3251–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle LM, Pacheco D, Warfield L, Wilburn DB, Hahn S, et al. Mediator subunit Med15 dictates the conserved “fuzzy” binding mechanism of yeast transcription activators Gal4 and Gcn4. Nat. Commun. 2021;12:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko J, DelRosso N, Hess GT, Banerjee A, Mukund A, Van MV, Ego BK, Yao D, Spees K, Suzuki P, et al. High-throughput discovery and characterization of human transcriptional effectors. Cell. 2020;183(7):2020–2035. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114(13):6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vockley CM, D'Ippolito AM, McDowell IC, Majoros WH, Safi A, Song L, Crawford GE, Reddy TE.. Direct GR binding sites potentiate clusters of TF binding across the human genome. Cell. 2016;166(5):1269–1281. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield L, Donczew R, Mahendrawada L, Hahn S. Mediator is broadly recruited to gene promoters via a Tail-independent mechanism. bioRxiv 2021.12.21.473728, 2021, accessed 10 July 2022. doi: 10.1101/2021.12.21.473728 [DOI] [PMC free article] [PubMed]
- White MA, Myers CA, Corbo JC, Cohen BA.. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc Natl Acad Sci U S A. 2013;110(29):11952–11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman AJ, Shashkova S, Hedlund EG, Friemann R, Hohmann S, Leake MC.. Transcription factor clusters regulate genes in eukaryotic cells. eLife. 2017;6. 10.7554/eLife.27451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich Z, Mirny LA.. Spatial effects on the speed and reliability of protein–DNA search. Nucleic Acids Res. 2008;36(11):3570–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich Z, Mirny LA.. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 2009;25(10):434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS. Pioneer transcription factors initiating gene network changes. Annu Rev Genet. 2020;54:367–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were generated for this article.