ABSTRACT
Humans often show variable responses to dietary, prebiotic, and probiotic interventions. Emerging evidence indicates that the gut microbiota is a key determinant for this population heterogeneity. Here, we provide an overview of some of the major computational and experimental tools being applied to critical questions of microbiota-mediated personalized nutrition and health. First, we discuss the latest advances in in silico modeling of the microbiota-nutrition-health axis, including the application of statistical, mechanistic, and hybrid artificial intelligence models. Second, we address high-throughput in vitro techniques for assessing interindividual heterogeneity, from ex vivo batch culturing of stool and continuous culturing in anaerobic bioreactors, to more sophisticated organ-on-a-chip models that integrate both host and microbial compartments. Third, we explore in vivo approaches for better understanding of personalized, microbiota-mediated responses to diet, prebiotics, and probiotics, from nonhuman animal models and human observational studies, to human feeding trials and crossover interventions. We highlight examples of existing, consumer-facing precision nutrition platforms that are currently leveraging the gut microbiota. Furthermore, we discuss how the integration of a broader set of the tools and techniques described in this piece can generate the data necessary to support a greater diversity of precision nutrition strategies. Finally, we present a vision of a precision nutrition and healthcare future, which leverages the gut microbiota to design effective, individual-specific interventions.
Keywords: prebiotic, probiotic, diet, microbiome, microbiota, personalized nutrition, personalized healthcare, precision nutrition, precision healthcare
Statement of Significance: Humans often show variable responses to dietary, prebiotic, and probiotic interventions. Here, we provide an overview of some of the major computational and experimental tools being applied to critical questions of microbiota-mediated personalized nutrition and health.
Introduction
The gut microbiota aids in digestion, including the degradation of complex fibers and phytochemicals, the production and absorption of vitamins, and the conversion of primary bile acids, xenobiotics, and other bioactive compounds into metabolites that can be readily absorbed by the host (1–6). Thus, the metabolic activity of commensal microbes is closely intertwined with human physiology, digestive health, and the nutritive value of diet. Just as individuals each have a distinct genotype, each carries a unique set of commensal microbiota, with individual-specific functional capacities (7, 8). This person-specific gut ecology has been associated with human population-wide heterogeneity in responses to dietary, lifestyle, and pharmacological interventions (9–13). However, there is limited understanding of exactly how individual variation in the ecology of the gut microbiota modulates the biological impact of dietary, prebiotic (i.e., a substrate that is selectively utilized by host-associated microorganisms, conferring a health benefit) (14), or probiotic (i.e., live microorganisms that when administered in adequate amounts confer a health benefit on the host) (15) interventions on human health and nutrition (16). It is, therefore, one of the critical goals of 21st century healthcare and nutrition to design personalized, predictive frameworks and viable intervention strategies that target and exploit each person's unique gut microbiota and its specific capabilities to optimize human health (7). This article represents the end-product of an International Life Sciences Institute (ILSI) Europe probiotics task force initiative on predicting individual responses to nutritional interventions, which focused, in particular, on how the human gut microbiota (i.e., the set of microorganisms that reside within the gastrointestinal tract) can mediate these personalized responses.
Developing predictive frameworks for individualized responses to dietary, prebiotic, and probiotic interventions will rely on iterative approaches that can span the full translational axis, integrating feedback between computational models of microbe–host interactions, in vitro approaches for testing these model predictions, and ultimately in vivo studies on the impact of these interventions on host physiology and health in both nonhuman animals and in humans. In this Perspective, we briefly outline state-of-the-art in silico, in vitro, and in vivo approaches for the rational design and testing of personalized dietary, prebiotic, and probiotic interventions (Table 1).
TABLE 1.
A nonexhaustive summary of in silico, in vitro, and in vivo approaches to exploring how the commensal gut microbiota drive individual-specific responses to dietary, prebiotic, and probiotic interventions
| Model | Advantages | Challenges | References | |
|---|---|---|---|---|
| In silico | Machine learning | ● Strong predictions● Data-driven | ● Predictions specific to training cohort● Mechanistically opaque | Zeevi et al., 2015 (9)Ben-Yacov et al., 2021 (18) |
| Metabolic modeling | ● Mechanistic● No training data● Predicts function● N-of-1 enabling● Computationally tractable | ● Lacks dynamics● Limited by model database● Cannot capture nonmetabolic phenomena | Magnúsdóttir et al., 2017 (32)Diener et al., 2020 (35)Thiele et al., 2020 (36) | |
| Dynamical modeling | ● Capture dynamics● Mechanistic● Predictive | ● Computationally intractable for complex ecosystems● Mismatches between sampling timescales and dynamics | Harcombe et al., 2014 (25)Bucci et al., 2016 (26)Mainali et al., 2019 (27) | |
| In vitro | Batch culture | ● Cost-effective● Easy to implement● Well suited to high-throughput screening● Ability to monitor metabolite production | ● Composition of the medium changes through time● No host absorption or interactions● Difficulty in culturing certain commensals | Liu et al., 2020 (44)Gurry et al., 2021 (45) |
| Continuous culture | ● Can adjust or maintain medium composition through time● Well suited to comparing steady states before and after treatment● Ability to monitor metabolite production | ● Lack of host tissue interaction models● Difficulty in culturing certain commensals | Salgaço et al., 2021 (53)Walton et al., 2012 (47) | |
| Gut on a chip | ● Captures host-tissue interactions | ● Experimentally complex | Pimenta et al., 2021 (61)Xiang et al., 2020 (60) | |
| In vivo | Invertebrates | ● A complete host–microbe system● Highly experimentally tractable● High degree of replication● Low cost | ● Divergent anatomy and physiology from vertebrates● Smaller size can limit the types of possible interventions | Hashmi et al., 2013 (67)Marsh and May, 2012 (68) |
| Vertebrates | ● Address systemic responses within the context of digestion and absorption● Control over microbial community● Access to host tissues of interest● Control over genetic background and diet | ● Nonhuman anatomy (e.g., hindgut fermenters)● Nonhuman physiology● Microbiota specific to each species● Lack of background genetic diversity within many model species | Kim et al., 2021 (69)Kemis et al., 2019 (70)Christoforidou et al., 2019 (73) | |
| Humans | ● Address systemic responses within the context of digestion and absorption● Directly applicable to human outcomes | ● Limited access to host tissues of interest● Controlling diet for long-term studies is challenging and expensive● Limited experimental tractability | Lichtenstein et al., 2021 (75)Kane et al., 2021 (77)Nogal et al., 2021 (78)Lancaster et al., 2022 (79) |
In Silico Approaches
There have been a number of recent advances in the application of computational models to large human cohorts to make personalized predictions for how interventions will influence health-relevant outcomes. Modeling approaches are highly flexible and enable arbitrary degrees of resolution for exploring parameter space or spatiotemporal dynamics. These approaches fall into 2 broad categories, statistical modeling and mechanistic modeling, which we discuss below.
Classic univariate statistical modeling has been applied to human cohorts undergoing dietary or lifestyle interventions, and has identified individual taxonomic (e.g., Prevotella dominance) and functional (e.g., bacterial amylase gene frequencies) features of the gut microbiome (i.e., the taxonomic and/or functional composition of the gut microbiota, inferred from either DNA or RNA sequencing) significantly associated with “responders” and “nonresponders” to weight loss interventions (10, 11, 17). Additionally, cross-sectional correlation-based analyses have been used to identify consistent associations between gut bacterial taxon abundances and personalized blood lipid profiles, in response to variation in diet (12).
In addition to these simpler, univariate approaches, personalized glycemic responses to variation in diet have been successfully predicted using machine learning (ML) models trained on multivariate phenotypic input data from individuals, including data on the composition of the gut microbiome (9). These personalized dietary predictions were recently shown to outperform the standard Mediterranean diet in managing blood glucose concentrations in prediabetes (18). Others are working to build ML models for predicting personalized responses to drugs or cancer immunotherapies based on the baseline composition of the gut microbiota (13, 19–21). Although these advances are exciting and provide an important proof-of-concept for personalized, microbiota-based interventions, they rely upon the manual integration of univariate model outputs or on complex multivariate models (e.g., random forest regression or convolutional neural networks), which can be somewhat opaque to mechanistic interpretation and are highly reliant on training data. Statistical models can fail to perform well on cohorts that are not well represented in the training data. For example, most microbiome research has been conducted on individuals in the United States and Europe, which means that precision-intervention ML models trained on these populations might not perform as well on populations from other parts of the world (22). For a more detailed review of ML and its applications to microbiome-mediated precision interventions, please see the review articles in references 20, 23, and 24.
Models that can incorporate both longitudinal and cross-sectional information are also important, but they can become computationally intractable if the number of parameters is too large, requiring long, high-density time series for parameter fitting and for validating predictions (25–27). Furthermore, the timescales of dynamics must match the timescales of sampling in these dynamical models, which is often not the case in the human gut, where bacterial growth rates are much faster than the defecation rate (28). Just as with the purely cross-sectional ML models described above, the input data sets for these kinds of models are often enriched in healthy individuals from more affluent populations in developed countries, which likely limits their application to unhealthy individuals, to individuals in the developing world, or to indigenous societies, where significant differences in gut ecology have been observed (29–31). Thus, effective models that require fitting to training data are often limited by a lack of mechanistic interpretability and by a reliance on incomplete training sets.
Alternatively, mechanistic models provide detailed insights into the potentially causal associations between enteric microbiota and human health. These kinds of models do not require a training data set, because they rely upon a network of validated, causal interactions. Thus, mechanistic models can perform more robustly than statistical models when applied across diverse populations. However, these models are limited by information that is available in existing knowledge bases. For instance, we do not yet have a large enough knowledge base on immune–commensal interactions to build a reliable mechanistic model for this particular interface. Genome-scale metabolic models are currently the most promising kinds of mechanistic models in host–microbe systems due to the vast amount of pre-existing knowledge on human and gut bacterial metabolism (32, 33). Prior work has demonstrated that baseline microbial metabolic gene profiles in the gut can be used to predict the probability of engraftment of a probiotic strain, indicating that the probiotic is unable to engraft when its metabolic niche is occupied by other established commensals (34).
Recent progress has been made in integrating multiple human gut bacterial commensals into a computationally tractable metagenome-scale metabolic model, which enables mapping of ecosystem structure to metabolic outputs and allows for the personalized simulation of metabolic responses to dietary, prebiotic, and probiotic interventions (35). Additionally, significant progress has been made in the last few years in the integration of host and gut bacterial metabolism into whole-body metabolic models (36). These types of models enable the simulation of gut commensal metabolism in the context of host metabolic variation, such as phenylketonuria or the loss of lactase expression in adulthood (8, 36). Indeed, ecosystem-scale metabolic modeling is a powerful tool for constructing a “digital twin” (i.e., a computational model of an individual person that can be used to assess the impacts of potential interventions in silico) for designing and testing prebiotic, probiotic, and dietary interventions in silico (8, 37). For a more comprehensive review of microbiome metabolic modeling and precision medicine, see reference 8. However, metabolism is only one piece of a broader picture, and integration of these models with models of the immune system, nervous system, endocrine system, and behavior are still needed (37). The accumulation of high-quality, longitudinal data, collected using standardized approaches from human populations, will be crucial to the development and validation of these mechanistic host–microbiota models.
Overall, both statistical and mechanistic approaches are required to make progress when it comes to precision engineering of the gut microbiota to optimize health. Data-intensive statistical methods are optimal for making predictions for host–microbiota interfaces that are poorly understood, but these models are often, by their nature, somewhat opaque to interpretation. Mechanistic models, on the other hand, leverage existing and emerging knowledge bases and personalized constraints on microbiome composition and diet to make personalized, N-of-1 predictions that do not require training data and that have more transparent, biologically interpretable outputs. These kinds of N-of-1, or “digital twin,” personalized models that enable in silico precision intervention response predictions are becoming increasingly popular in other fields (e.g., in the treatment of cancers or infectious diseases) (38, 39).
In Vitro Approaches
Developing a predictive framework for leveraging the gut microbiota requires a quantitative and dynamic understanding of the properties we wish to predict within the system. However, time-resolved and continuous data are difficult to obtain in vivo. Thus, in vitro approaches offer a compromise between accuracy and flexibility. Environmental variables or microbial community composition can be precisely controlled, and sampling timescales can be arbitrarily tuned, making in vitro models ideal for mechanistically dissecting variable responses to prebiotic and probiotic interventions (40), testing specific hypotheses about host–diet–microbe interactions (41, 42), or providing in silico models with detailed training or validation data (36).
Model complexity can vary widely in vitro (43). The simplest and most versatile approaches involve short-term (i.e., <24–48-h) batch culturing of microbial communities. Temperature, pH, oxygen, carbon dioxide, and medium/nutrient composition can be tightly controlled, at modest cost. Batch culturing methods are ideal for high-throughput screening purposes, such as testing the impact of specific nutrient challenges on a given stool homogenate sample. Over short incubation windows, changes in metabolite abundances can be used to assess production and consumption fluxes of these molecules (i.e., fluxomics) in batch culture. For example, this approach has been used to study the effects of dietary fibers isolated from sweet potato on microbiome composition (44), or to study the production of SCFAs from the same fiber inputs to different fecal microbial communities from human volunteers (45). In the latter example, stark differences in SCFA production found in vitro were well correlated with significant differences in fecal SCFA concentrations from human volunteers fed the same fibers (46), indicating some degree of accuracy in reproducing community-level functional properties of an in vivo gut microbiota. However, a key weakness of these models lies in the absence of absorption or transformation of microbially produced metabolites by host tissue, resulting in their accumulation over time beyond normal physiological concentrations.
Continuous culturing methods introduce a layer of complexity by maintaining the medium contents at a steady state, and allow for longer experimental timescales (i.e., weeks to months) or repeat-dosing experiments. For example, the 3-stage Macfarlane and Gibson continuous culturing model, built to reflect different ecological niches along the intestinal tract, was shown to reproduce the bifidogenic effects of galacto-oligosaccharides observed in a human trial and provided spatially resolved predictions for where specific fermentative processes were likely occurring in the gut (47, 48). Even more sophisticated methods include the Simulator of the Human Intestinal Microbial Ecosystem (SHIME), a computer-controlled set of reactors connected by peristaltic pumps, or the TNO Gastro-Intestinal Model (49–51), which consists of 2 compartments simulating the upper and lower gastrointestinal tracts. These systems have been used in numerous nutrient and microbial community-challenge studies (50, 52–57). Although these models have incorporated some elements of host absorption, they suffer from certain drawbacks, notably a lack of host tissue-specific interactions.
In vitro systems can also be leveraged to study the effect of a microbiota on specific host tissues (50). In one study, supernatants from batch cultures were used in cell challenge assays with peripheral blood mononuclear cells (PBMCs) isolated from human volunteers to study cytokine production, finding higher IL-10 production in supernatants derived from batch cultures supplemented with inulin and trans-galacto-oligosaccharides, both prebiotic fibers, resulting in a higher production of the anti-inflammatory SCFA butyrate (58). However, one limitation of this study was that these supernatants were applied directly to PBMCs without first passing through intestinal and liver tissues, which would degrade or transform many microbially derived metabolites before they could pass into general circulation. Similarly, mucin-coated beads can be added to the culture medium to simulate both the luminal and mucus-associated microbial community in the colonic reactors, as has been demonstrated in the SHIME system (54).
Direct contact with host tissues can be achieved in vitro in organoid or organ-on-a-chip models (59, 60), which can be connected microfluidically with peristaltic pumps (61). Advances in coupling microfluidic and cell culture approaches with 3D organ-on-a-chip systems have allowed for novel experimental possibilities and have greatly increased in vitro model complexity (62). In recent work, leveraging a Caco-2 cell model, researchers investigated gut epithelial barrier integrity in the presence of different human commensal strains, highlighting the power of these more advanced in vitro systems (63).
Overall, in vitro methods have aided greatly in determining which interventions to take forward into animal models and human trials. However, they are limited by the fact that many human commensal species remain difficult to culture, making it hard to experimentally reproduce the human digestive system and compromising the validity of any underlying microbial community dynamics. Furthermore, the lack of complex multitissue interactions and the buildup of metabolites give rise to an artificial environment for the microbiota, which leads to “bottle effects” where the ecology of the in vitro community begins to diverge from what it would look like in vivo (64). Nonetheless, significant progress in culturing and isolation methods, host cell and tissue culturing, and the release of multiple gut microbiota strain banks that contain representative commensal strain diversity from large and diverse human populations, such as the Broad Institute-OpenBiome Microbiome Library (65) and the Global Microbiome Conservancy (66), in combination with the development of more robust experimental systems that include host tissues (60, 61), continue to widen the scope of applicability of in vitro models.
In Vivo Approaches
Predicting personalized responses to dietary, prebiotic, and probiotic interventions in vivo requires adequate variation in the study population, careful collection of relevant metadata and biological measures that serve as covariates, and robust approaches for characterizing the response or outcome. Whether it is a preclinical animal model or a human population, sampling across sufficient genetic, phenotypic, and environmental variation is important to develop personalized prediction models.
Invertebrate models, like Caenorhabditis elegans, provide the highest degree of experimental tractability and replication, but are not the most biologically relevant models for translation to humans (67, 68). Inbred rodent models, on the other hand, are more translationally relevant and have been an integral component of nutrition research, enhancing our understanding of physiological and pathological processes and providing mechanistic insights into causality. The phenotypic spectrum available across various mouse strains allows for discovery of host genetic features related to intervention responses. However, to date, most studies have been conducted within a small number of genetically homogeneous mouse strains. More recently, development of multiparent advanced generation intercross populations (i.e., collaborative cross mice) has expanded the assessment of host genetics on biological traits, as well as response to dietary interventions (69). Further, it has provided an approach to characterizing mechanisms underlying the effects of host genetics on gut microbiota composition and function (70). Use of germ-free mice and mice treated with antibiotic cocktails, colonized with human fecal communities, further extends the examination of microbe-driven differences in responses to dietary inputs (e.g., dietary fiber sources) (71, 72). Finally, pig studies can provide a more physiologically relevant model of the human body, while allowing samples to be readily obtained from multiple locations along the gastrointestinal tract at any degree of longitudinal resolution. Overall, nonhuman animal models provide detailed spatiotemporal information on digestion, immune function, and physiology following dietary or probiotic interventions (73). Universal concerns related to relevance of exposures and doses, blinding, use of appropriate comparators, and bias related to housing conditions, handling, and overall relevance to human biology, are important considerations in the rigor of these experiments and supporting translation from nonhuman animal models to humans (74). In addition, food companies are increasingly reluctant to use nonhuman animal models for human research due to rising consumer concerns for animal welfare.
Traditionally, nutrition studies in humans range from observational studies in prospective cohorts to randomized controlled trials (RCTs). RCTs are considered the gold standard for establishing causal relations between interventions and biomarkers or outcomes in humans. However, the choice of study design (e.g., parallel, crossover, factorial, cluster), the duration of intervention, and the sampling timescales are critical to addressing the relevant biological mechanisms and timescales on which intervention-induced changes might occur (75). Prior work in a healthy human cohort found that 5–9 longitudinal samples, taken 2–3 d apart from one another, were optimal for estimating the average population sizes of commensal gut bacteria within an individual's gut (65, 76). Additionally, interindividual heterogeneity in microbiome composition makes traditional randomized trials difficult to interpret, which suggests that N-of-1 trial designs and crossover trials, where individuals serve as their own controls, are best when considering personalized microbiota–intervention interactions (77–79). Duration of interventions to study gut microbiota-related outcomes can vary widely, and longer duration interventions make crossover trials impractical (77). Some aspects of gut microbiota activity and composition respond rapidly to short-term interventions, if the particular microbes and pathways targeted by the intervention are already present in the system (80, 81). Immediate diet–microbiota interactions can be evident from looking at metabolomic or physiological biomarkers within hours of intake (9). However, if the goal is to substantially change gut community composition in order to alter its functional outputs, then longer-term interventions might be needed (82–84). Planning dietary interventions in free-living humans poses additional challenges related to timing of intake, adherence, and sampling (85).
Consideration and collection of appropriate metadata, including relevant host factors and environmental exposures, is critical to crafting robust, covariate-adjusted prediction models using data from observational studies, as well as from RCTs. Multiple host factors affecting microbial and host phenotypes include: dietary habits, genetics, age, adiposity, gut permeability, bile acids, mucus production, glycosylation and fucosylation, macrophysiology (e.g., gut transit; stomach and intestinal pH), and clinical factors. Environmental factors, both direct and indirect, that affect diet–microbiota interactions include: pollution, stress/anxiety, medication use, physical activity, smoking, etc. Often studies are hampered in their capacity to capture this information accurately due to limitations of participant recall, inadequate databases, and financial constraints. Technologies that enable cost-effective, accurate, and comprehensive systems-scale data collection on human populations, such as the expanding diversity and availability of wearable devices and smartphone-based applications (86–88), will provide greater insight into the causal mechanisms that underlie how the microbiota mediates personalized responses to dietary, prebiotic, and probiotic interventions. Finally, it is important to decrease the costs and logistical hurdles of these precision approaches to increase the representation of indigenous, nonindustrialized, and rural populations in microbiome research so that the societal benefits of precision nutrition and healthcare are more equitably distributed (22, 66, 89).
Synthesis
It has become clear that the human gut microbiota influences how humans respond to dietary, prebiotic, and probiotic interventions (9–12, 17, 34, 90). In the specific case of postprandial blood glucose responses, these insights have been commercialized by DayTwo, Inc, by integrating microbiome data, clinical data, and blood glucose responses to standardized meals into predictive models that can be leveraged to design optimal personalized interventions that perform better than the current standard-of-care (9, 18). However, many current commercial precision dietary, prebiotic, and probiotic interventions remain nascent, often overpromising on their predictive capabilities in a highly underregulated market. In this context, it is incumbent upon researchers to collect more evidence from well-designed, hypothesis-generating human observational studies and hypothesis-testing experimental intervention trials where dense phenotypic, clinical, and behavioral information is combined with gut microbiome profiling. Furthermore, it is unclear whether or not precision nutrition models trained on relatively affluent developed-world cohorts are broadly applicable to the rest of the world, necessitating a sharper focus on running observational and interventional trials in indigenous, nonindustrialized, and rural populations (22, 66, 89).
Large, diverse, densely phenotyped human cohorts can be leveraged to build statistical learning models that enable personalized predictions of phenotypic responses to interventions (9). However, these statistical modeling approaches are limited in that they do not necessarily provide detailed mechanistic insights and they rely upon a training cohort. If new data come from individuals who are sufficiently different from the training cohort, predictions will begin to break down. Thus, our growing knowledge bases should be leveraged to build better mechanistic models, which enable robust personalized predictions based on causally validated host–microbe and microbe–microbe interactions. Recent advances in host-microbiota metabolic modeling enable high-throughput, personalized predictions of responses to dietary, prebiotic, or probiotic interventions (8, 35, 36). In order to properly validate these in silico model predictions, it is necessary to have controlled in vitro models that allow for high-resolution, spatially resolved, longitudinal sampling. For instance, metabolic model predictions for personalized SCFA production in response to specific fiber amendments could be directly validated using fluxomics data collected from in vitro batch or continuous culture systems (45). In addition to validating existing statistical or mechanistic models, sophisticated in vitro models and in vivo nonhuman animal models can be directly leveraged to explore uncharacterized microbe–host associations (2, 60, 73). Feedback from these experiments can be exploited to build upon our knowledge bases and eventually to serve as a foundation for better mechanistic modeling. Finally, human observational and intervention trials are critical components to the final validation and potential regulatory approval of personalized dietary, prebiotic, and probiotic interventions. In order to achieve regulatory approval, it will be important to have quantitatively validated outcomes, predicted by in silico, in vitro, and in vivo models, that leverage the composition of the gut microbiota to optimize human health, nutrition, and overall wellness.
A Vision of the Future: Microbiome-Informed Precision Nutrition and Healthcare
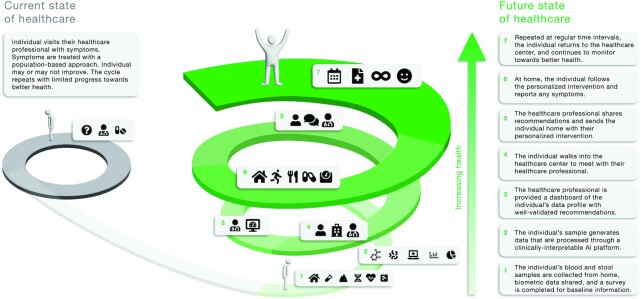
Historically, healthcare and interactions with healthcare professionals (HCPs) have been triggered by patients reporting disease symptoms, visiting practitioners, and receiving standard-of-care, population-based treatments (Figure 1). Preventative, personalized interventions have not been widely integrated into our current health systems, and public health messages remain largely untargeted to specific populations. It is possible to imagine a not-too-distant future where a relatively healthy individual walks into a healthcare center and receives an effective, personalized, microbiome-informed intervention (Figure 1). In our example, the individual is known to be prediabetic. Before their visit, the individual would have provided both blood and stool samples for data generation on blood proteins, blood metabolites, whole genome sequencing, gut metagenomic sequencing, live stool homogenates for in vitro stool assays, and questionnaire data on dietary intake, lifestyle, medication use, and health history. The individual's data are processed through a clinically interpretable artificial intelligence platform, which provides individual-specific predictions based on ML models trained on a large reference population, and also includes mechanistic model predictions for thousands of host–microbiota–diet metabolic interactions. The HCP or dietitian is given a dashboard of the individual's data profile, which includes a number of well-validated recommendations (e.g., genomically inferred disease risk or pharmacogenomic recommendations on drug usage) and a high-level summary of the data. The individual is overweight, has elevated hemoglobin A1c, shows a relatively low amylase gene copy number in their genome, and has a Prevotella copri–enriched gut microbiome. The data dashboard informs the HCP or dietitian that individuals with low salivary amylase and a Prevotella-enriched gut microbiome are known to lose weight readily in response to a high-fiber dietary intervention (11). Therefore, based on this data profile, the healthcare provider can see that the individual should respond well to a healthy lifestyle intervention, including a high-fiber diet and a 1-h walk in the evenings after dinner, and should not need a pharmaceutical intervention to achieve and sustain weight loss and improved cardiometabolic health. In addition, the individual shows elevated levels of systemic inflammation and reports constipation. The dietary and exercise interventions should help, in part, to alleviate these issues. However, to further reduce constipation the healthcare provider prescribes supplementation with prebiotic inulin, which has been shown to increase bowel movement frequency (91). Furthermore, inulin has been associated with increased fat oxidation and the promotion of SCFA production (92). Finally, the in silico metabolic simulations predict that a diet rich in pectin is likely to greatly increase butyrate production in this person's gut, which should further reduce systemic inflammation and help with maintaining weight loss. To validate this personalized prediction, the HCP orders the hospital lab to run the individual's stool homogenate through a panel of dietary fibers on a gut-on-a-chip bioreactor system prior to their visit, measuring SCFA fluxes. Indeed, of the dozens of dietary compounds tested, pectin elicited the greatest increase in butyrate production in vitro, and the HCP prescribes more pectin-rich foods, like carrots, tangerines, and apples to the individual. The healthcare provider sends the individual home with their personalized, microbiome-informed intervention regime. The individual is given a wearable activity tracker and a cell phone application with a patient-facing data-health dashboard, which provides healthy lifestyle advice, allows the individual to report adverse symptoms to the HCP in real time, and schedules a follow-up appointment in 6 mo for another blood draw and stool sample. This data-driven, personalized, participatory process involves feedback between an individual and their HCP and helps to drive better compliance and sustained progress towards individually defined, goal-oriented, precision health outcomes (93). Additionally, the information gained from tracking individual-specific responses to interventions across many people can be used to design impactful public health messages targeting specific, at-risk subpopulations. As such, this approach would result in a reduced risk of many chronic diseases associated with obesity, systemic inflammation, and poor metabolic health, potentially extending healthspan and lifespan (18, 94, 95).
FIGURE 1.
Conceptual schematic of the current nonpersonalized state of healthcare that focuses on population-based approaches to treating symptoms, and a vision for a future state of healthcare that leverages personalized data (e.g., microbiomes, dietary intake, genomes, blood analytes, etc.) to inform precision interventions (e.g., combinations of prebiotic, probiotic, lifestyle, dietary, or clinical regimes) aimed at addressing the root causes of illness to improve health and well-being.
Growing evidence supports a vision of personalized, participatory nutrition and healthcare that is achievable in the near term (96). Indeed, nascent microbiome-mediated precision nutrition and health interventions, like those implemented by the companies DayTwo or Zoë for optimizing blood glucose responses and blood lipid profiles, respectively (9, 12, 18), are already available to consumers. However, there are many other quantitative nutritional and health-related targets, beyond blood glucose and blood lipids, such as increasing colonic SCFA production to reduce systemic inflammation levels (97), reducing toxic bacterial metabolites (e.g., trimethylamine N-oxide or imidazole propionate) in the bloodstream (98, 99), or weight loss (10), which will require new kinds of precision intervention strategies. The existing tools and techniques discussed in this Perspective bring us closer to an even wider array of personalized dietary, prebiotic, and probiotic interventions that leverage the unique ecological capacities of our gut microbes to improve our individual lives and reduce the societal burden of chronic disease. However, an enormous engineering and optimization challenge lies ahead of us, where we must learn to harness these tools to achieve specific, quantitative outcomes across a range of diverse contexts. It is unlikely that we will find a general-purpose algorithm, so we must move forward, one targeted application at a time, to build up an ecosystem of precision intervention tools. Thus, we will need to generate more and more dense phenotypic data from small-to-large-scale longitudinal human trials to help refine and optimize our targeted models, where individual-specific responses to prebiotic, probiotic, and/or dietary inputs are tracked. Finally, we will need to develop a rigorous set of standards and best practices for designing and assessing the efficacy of personalized interventions (100). Although the pace of progress in this field is accelerating, the ultimate timescales for translating microbiome science into personalized nutrition and healthcare will be limited by the availability of funding, by the size of the scientific workforce, by access to diverse human cohorts, and ultimately, by buy-in from healthcare systems.
Glossary
Batch culture: A closed system in which cells are grown in a fixed volume of growth medium under controlled environmental conditions for a limited period of time.
Continuous culture: An open system in which cells are grown on balanced inflow and outflow of growth medium, under specific environmental conditions, over a potentially indefinite period of time, with long-term growth conditions approaching a steady-state.
Digital twin: A computational model of an individual that can be used to assess the impacts of potential interventions in silico.
Ex vivo: Research conducted or produced by means of taking biological material out of its in vivo context into an in vitro context.
Fluxomics: A variety of techniques that aim to determine the rates of metabolic reactions within a cell, a tissue, an organism, a microbial community, or any other kind of biological system.
Gut microbiome: The genomic/metagenomic/metatrans-criptomic content of the gut microbiota, as inferred from sequencing nucleic acids (i.e., DNA or RNA).
Gut microbiota: The microorganisms that reside in the gastrointestinal tracts of nonhuman animals and humans.
In silico: Research conducted or produced by means of computer modeling or computer simulation.
In vitro: Research conducted or produced in the laboratory, outside the context of an intact, living host organism.
In vivo: Research conducted or produced within a living host organism.
Machine learning: A broad range of statistical modeling techniques that are designed to draw inferences from data and improve performance on a set of user-specified tasks (i.e., to “learn” from data).
N-of-1 study design: Longitudinal studies that leverage individuals as their own controls to assess individual-specific responses to interventions. For example, crossover trials apply a sequence of interventions (e.g., alternating between placebo and active treatment several times) to the same individual, with the ordering of these interventions depending on randomized assignment to a cross-sectional group, to quantify both individual-specific and cross-sectional responses.
Organ-on-a-chip: A continuous-culture, multichannel, 3D, microfluidic system that is designed to simulate the structure and physiology of an entire organ or organ system.
Prebiotic: A substrate that is selectively utilized by host microorganisms, conferring a health benefit.
Probiotic: Live microorganisms that when administered in adequate amounts confer a health benefit on the host.
Training data: Data used to train a machine learning algorithm to make user-specified inferences.
Validation data: A collection of data that is independent of the training data set, which is used to test the performance of a machine learning algorithm.
ACKNOWLEDGEMENTS
Figure 1 was designed by Allison Kudla. Thanks to Thea Swanson for copy-editing.
This work was conducted by an expert group (EG) of ILSI Europe. According to ILSI Europe policies, the EG is composed of ≥50% external nonindustry members. This publication was coordinated by the Probiotics Task Force and the Prebiotics Task Force. Industry members of this task force and the composition of the EG are listed on the ILSI Europe website at https://ilsi.eu/scientific-activities/nutrition/probiotics/ and https://ilsi.eu/scientific-activities/nutrition/prebiotics/. Experts are not paid for the time spent on this work; however, the nonindustry members within the EG were offered small compensatory sums (honoraria) with the option to decline. The EG carried out the work, that is, collecting/analyzing data/information and writing the scientific paper separate from other activities of the task forces. The research reported is the result of a scientific evaluation in line with ILSI Europe's framework to provide a precompetitive setting for public-private partnership. ILSI Europe facilitated scientific meetings and coordinated the overall project management and administrative tasks relating to the completion of this work. For further information about ILSI Europe, please email info@ilsieurope.be or call +32 2 771 00 14.
The authors’ responsibilities were as follows—All authors: were responsible for the design of the manuscript; SMG, JWL, TG: wrote the first draft of the manuscript; and all authors: contributed to editing and revising this draft and approving the final content, and read and approved the final version.
Notes
SMG was supported by the Washington Research Foundation Distinguished Investigator Award and startup funds from the Institute for Systems Biology. AE is research associate at FRS-FNRS (Fonds de la Recherche Scientifique) and recipient of grants from FNRS (WELBIO-CR-2019S-03, WELBIO-CR-2019S-03R, CDR-convention: J.0075.22). The opinions expressed herein and the conclusions of this publication are those of the authors and do not necessarily represent the views of International Life Sciences Institute (ILSI) Europe nor those of its member companies.
Author disclosures: AC is an employee of Cargill. AE is inventor on patent applications dealing with the use of gut microbes in the treatment of metabolic and food intake disorders. AMV is a consultant for CPKelco and Zoe Global Ltd. BP is employed by Yakult Europe BV. GG is employee of Reckitt/Mead Johnson Nutrition. JAL is an employee of Health and Happiness Group. JM is an employee of IFF Health & Biosciences. TG is an employee and shareholder of Myota GmbH. VD is an employee of Sensus BV (Royal Cosun). YZ is an employee of BENEO-Institute/Südzucker Group, Germany. The authors report no other conflicts of interest.
SMG, TG, and JWL contributed equally to this work.
Perspective articles allow authors to take a position on a topic of current major importance or controversy in the field of nutrition. As such, these articles could include statements based on author opinions or point of view. Opinions expressed in Perspective articles are those of the author and are not attributable to the funder(s) or the sponsor(s) or the publisher, Editor, or Editorial Board of Advances in Nutrition. Individuals with different positions on the topic of a Perspective are invited to submit their comments in the form of a Perspectives article or in a Letter to the Editor.
Abbreviations used: EG, expert group; HCP, healthcare professional; ILSI, International Life Sciences Institute; ML, machine learning; PBMC, peripheral blood mononuclear cell; RCT, randomized controlled trial; SHIME, Simulator of the Human Intestinal Microbial Ecosystem.
Contributor Information
Sean M Gibbons, Institute for Systems Biology, Seattle, WA, USA; Department of Bioengineering, University of Washington, Seattle, WA, USA.
Thomas Gurry, Pharmaceutical Biochemistry group, School of Pharmaceutical Sciences, University of Geneva, Geneva, Switzerland; Institute of Pharmaceutical Sciences of Western Switzerland (PSI-WS), University of Geneva/University of Lausanne, Geneva, Switzerland.
Johanna W Lampe, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Anirikh Chakrabarti, Cargill R&D Center Europe, Vilvoorde, Belgium.
Veerle Dam, Sensus BV (Royal Cosun), Roosendaal, The Netherlands.
Amandine Everard, Metabolism and Nutrition Research Group, Louvain Drug Research Institute, Walloon Excellence in Life Sciences and BIOtechnology (WELBIO), UCLouvain, Université Catholique de Louvain, Brussels, Belgium.
Almudena Goas, Department of Food, Nutrition, and Exercise Sciences, University of Surrey, Guildford, United Kingdom.
Gabriele Gross, Medical and Scientific Affairs, Reckitt| Mead Johnson Nutrition Institute, Nijmegen, The Netherlands.
Michiel Kleerebezem, Host Microbe Interactomics Group, Wageningen University & Research, Wageningen, The Netherlands.
Jonathan Lane, Health and Happiness Group, H&H Research, Cork, Ireland.
Johanna Maukonen, IFF Health & Biosciences, Kantvik, Finland.
Ana Lucia Barretto Penna, Department of Food Engineering and Technology, São Paulo State University, São José do Rio Preto, Brazil.
Bruno Pot, Yakult Europe BV, Almere, The Netherlands.
Ana M Valdes, Nottingham NIHR Biomedical Research Centre at the School of Medicine, University of Nottingham, Nottingham, United Kingdom.
Gemma Walton, Food and Nutritional Sciences, University of Reading, Reading, United Kingdom.
Adrienne Weiss, Yili Innovation Center Europe, Wageningen, The Netherlands.
Yoghatama Cindya Zanzer, BENEO-Institute/Südzucker Group, Obrigheim/Pfalz, Germany.
Naomi V Venlet, International Life Sciences Institute, European Branch, Brussels, Belgium.
Michela Miani, International Life Sciences Institute, European Branch, Brussels, Belgium.
References
- 1. Yadav M, Verma MK, Chauhan NS. A review of metabolic potential of human gut microbiome in human nutrition. Arch Microbiol. 2018;200(2):203–17. [DOI] [PubMed] [Google Scholar]
- 2. Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570(7762):462–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Javdan B, Lopez JG, Chankhamjon P, Lee Y-CJ, Hull R, Wu Qet al. . Personalized mapping of drug metabolism by the human gut microbiome. Cell. 2020;181(7):1661–79.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Das P, Babaei P, Nielsen J. Metagenomic analysis of microbe-mediated vitamin metabolism in the human gut microbiome. BMC Genomics. 2019;20(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tian Y, Gui W, Koo I, Smith PB, Allman EL, Nichols RGet al. . The microbiome modulating activity of bile acids. Gut Microbes. 2020;11(4):979–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collins SL, Patterson AD. The gut microbiome: an orchestrator of xenobiotic metabolism. Acta Pharm Sin B. 2020;10(1):19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilmanski T, Rappaport N, Diener C, Gibbons SM, Price ND. From taxonomy to metabolic output: what factors define gut microbiome health?. Gut Microbes. 2021;13(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heinken A, Basile A, Hertel J, Thinnes C, Thiele I. Genome-scale metabolic modeling of the human microbiome in the era of personalized medicine. Annu Rev Microbiol. 2021;75(1):199–222. [DOI] [PubMed] [Google Scholar]
- 9. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger Aet al. . Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–94. [DOI] [PubMed] [Google Scholar]
- 10. Diener C, Qin S, Zhou Y, Patwardhan S, Tang L, Lovejoy JCet al. . Baseline gut metagenomic functional gene signature associated with variable weight loss responses following a healthy lifestyle intervention in humans. Msystems. 2021;6(5):e0096421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hjorth MF, Christensen L, Larsen TM, Roager HM, Krych L, Kot Wet al. . Pretreatment Prevotella-to-Bacteroides ratio and salivary amylase gene copy number as prognostic markers for dietary weight loss. Am J Clin Nutr. 2020;111(5):1079–86. [DOI] [PubMed] [Google Scholar]
- 12. Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DAet al. . Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021;27(2):321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilmanski T, Kornilov SA, Diener C, Conomos MP, Lovejoy JC, Sebastiani Pet al. . Heterogeneity in statin responses explained by variation in the human gut microbiome. Med (N Y). 2022;3(6):388–405.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJet al. . Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14(8):491–502. [DOI] [PubMed] [Google Scholar]
- 15. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot Bet al. . The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–14. [DOI] [PubMed] [Google Scholar]
- 16. Salehi B, Dimitrijević M, Aleksić A, Neffe-Skocińska K, Zielińska D, Kołożyn-Krajewska Det al. . Human microbiome and homeostasis: insights into the key role of prebiotics, probiotics, and symbiotics. Crit Rev Food Sci Nutr. 2021;61:1415–28. [DOI] [PubMed] [Google Scholar]
- 17. Christensen L, Vuholm S, Roager HM, Nielsen DS, Krych L, Kristensen Met al. . Prevotella abundance predicts weight loss success in healthy, overweight adults consuming a whole-grain diet ad libitum: a post hoc analysis of a 6-wk randomized controlled trial. J Nutr. 2019;149(12):2174–81. [DOI] [PubMed] [Google Scholar]
- 18. Ben-Yacov O, Godneva A, Rein M, Shilo S, Kolobkov D, Koren Net al. . Personalized postprandial glucose response-targeting diet versus Mediterranean diet for glycemic control in prediabetes. Diabetes Care. 2021;44(9):1980–91. [DOI] [PubMed] [Google Scholar]
- 19. McCoubrey LE, Gaisford S, Orlu M, Basit AW. Predicting drug-microbiome interactions with machine learning. Biotechnol Adv. 2022;54:107797. [DOI] [PubMed] [Google Scholar]
- 20. Cammarota G, Ianiro G, Ahern A, Carbone C, Temko A, Claesson MJet al. . Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat Rev Gastroenterol Hepatol. 2020;17:635–48. [DOI] [PubMed] [Google Scholar]
- 21. Limeta A, Ji B, Levin M, Gatto F, Nielsen J. Meta-analysis of the gut microbiota in predicting response to cancer immunotherapy in metastatic melanoma. JCI Insight. 2020;5(23):e140940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abdill RJ, Adamowicz EM, Blekhman R. Public human microbiome data are dominated by highly developed countries. PLoS Biol. 2022;20(2):e3001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCoubrey LE, Elbadawi M, Orlu M, Gaisford S, Basit AW. Harnessing machine learning for development of microbiome therapeutics. Gut Microbes. 2021;13(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bashiardes S, Godneva A, Elinav E, Segal E. Towards utilization of the human genome and microbiome for personalized nutrition. Curr Opin Biotechnol. 2018;51:57–63. [DOI] [PubMed] [Google Scholar]
- 25. Harcombe WR, Riehl WJ, Dukovski I, Granger BR, Betts A, Lang AHet al. . Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014;7(4):1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bucci V, Tzen B, Li N, Simmons M, Tanoue T, Bogart Eet al. . MDSINE: Microbial Dynamical Systems INference Engine for microbiome time-series analyses. Genome Biol. 2016;17(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mainali K, Bewick S, Vecchio-Pagan B, Karig D, Fagan WF. Detecting interaction networks in the human microbiome with conditional Granger causality. PLoS Comput Biol. 2019;15(5):e1007037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carr A, Diener C, Baliga NS, Gibbons SM. Use and abuse of correlation analyses in microbial ecology. ISME J. 2019;13(11):2647–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Groussin M, Poyet M, Sistiaga A, Kearney SM, Moniz K, Noel Met al. . Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell. 2021;184(8):2053–67.e18. [DOI] [PubMed] [Google Scholar]
- 30. Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid Get al. . Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science. 2017;357(6353):802–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duvallet C, Gibbons S, Gurry T, Irizarry R, Alm E. Meta analysis of microbiome studies identifies shared and disease-specific patterns. Nat Commun. 2017;8(1):1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Magnúsdóttir S, Heinken A, Kutt L, Ravcheev DA, Bauer E, Noronha Aet al. . Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol. 2017;35(1):81–9. [DOI] [PubMed] [Google Scholar]
- 33. Brunk E, Sahoo S, Zielinski DC, Altunkaya A, Dräger A, Mih Net al. . Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat Biotechnol. 2018;36(3):272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maldonado-Gómez MX, Martínez I, Bottacini F, O'Callaghan A, Ventura M, van Sinderen Det al. . Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe. 2016;20(4):515–26. [DOI] [PubMed] [Google Scholar]
- 35. Diener C, Gibbons SM, Resendis-Antonio O. MICOM: metagenome-scale modeling to infer metabolic interactions in the gut microbiota. Msystems [Internet]. 2020;5(1). doi: 10.1128/mSystems.00606-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thiele I, Sahoo S, Heinken A, Hertel J, Heirendt L, Aurich MKet al. . Personalized whole-body models integrate metabolism, physiology, and the gut microbiome. Mol Syst Biol. 2020;16(5):e8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gkouskou K, Vlastos I, Karkalousos P, Chaniotis D, Sanoudou D, Eliopoulos AG. The “virtual digital twins” concept in precision nutrition. Adv Nutr. 2020;11(6):1405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hernandez-Boussard T, Macklin P, Greenspan EJ, Gryshuk AL, Stahlberg E, Syeda-Mahmood Tet al. . Digital twins for predictive oncology will be a paradigm shift for precision cancer care. Nat Med. 2021;27(12):2065–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laubenbacher R, Sluka JP, Glazier JA. Using digital twins in viral infection. Science. 2021;371(6534):1105–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pham VT, Mohajeri MH. The application of in vitro human intestinal models on the screening and development of pre- and probiotics. Benef Microbes. 2018;9(5):725–42. [DOI] [PubMed] [Google Scholar]
- 41. Pearce SC, Coia HG, Karl JP, Pantoja-Feliciano IG, Zachos NC, Racicot K. Intestinal in vitro and ex vivo models to study host-microbiome interactions and acute stressors. Front Physiol. 2018;9:1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nissen L, Casciano F, Gianotti A. Intestinal fermentation in vitro models to study food-induced gut microbiota shift: an updated review. FEMS Microbiol Lett. 2020;367(12):fnaa097. [DOI] [PubMed] [Google Scholar]
- 43. Williams CF, Walton GE, Jiang L, Plummer S, Garaiova I, Gibson GR. Comparative analysis of intestinal tract models. Ann Rev Food Sci Technol. 2015;6(1):329–50. [DOI] [PubMed] [Google Scholar]
- 44. Liu M, Li X, Zhou S, Wang TTY, Zhou S, Yang Ket al. . Dietary fiber isolated from sweet potato residues promotes a healthy gut microbiome profile. Food Funct. 2020;11(1):689–99. [DOI] [PubMed] [Google Scholar]
- 45. Gurry T, Nguyen LTT, Yu X, Alm EJ. Functional heterogeneity in the fermentation capabilities of the healthy human gut microbiota. PLoS One. 2021;16(7):e0254004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio. 2019;10(1):e02566–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Walton GE, van den Heuvel E, Kosters MHW, Rastall RA, Tuohy KM, Gibson GR. A randomised crossover study investigating the effects of galacto-oligosaccharides on the faecal microbiota in men and women over 50 years of age. Br J Nutr. 2012;107(10):1466–75. [DOI] [PubMed] [Google Scholar]
- 48. Macfarlane GT, Gibson GR. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine. In: Mackie RI, White BA, editors. Gastrointestinal microbiology. Boston (MA): Springer;. 1997. p.; 269–318. [Google Scholar]
- 49. Molly K, Vande Woestyne M, De Smet I. Validation of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) reactor using microorganism-associated activities. Microb Ecol Health Dis. 1994;7:191–200. [Google Scholar]
- 50. Marzorati M, Van de Wiele T. An advanced in vitro technology platform to study the mechanism of action of prebiotics and probiotics in the gastrointestinal tract. J Clin Gastroenterol. 2016;50(Suppl 2):S124–5. [DOI] [PubMed] [Google Scholar]
- 51. Minekus M. Chapter 5: The TNO gastro-intestinal model (TIM). In: Verhoeckx K, Cotter P, López-Expósito I, Kleiveland C, Lea T, Mackie Aet al. editors. The impact of food bioactives on health: in vitro and ex vivo models. Cham (Switzerland): Springer; 2018. 1–19. [Google Scholar]
- 52. Hage RE, El Hage R, Hernandez-Sanabria E, Arroyo MC, Props R, Van de Wiele T. Propionate-producing consortium restores antibiotic-induced dysbiosis in a dynamic in vitro model of the human intestinal microbial ecosystem. Front Microbiol [Internet]. 2019;10:1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Salgaço MK, Perina NP, Tomé TM, Mosquera EMB, Lazarini T, Sartoratto Aet al. . Probiotic infant cereal improves children's gut microbiota: insights using the Simulator of Human Intestinal Microbial Ecosystem (SHIME®). Food Res Int. 2021;143:110292. [DOI] [PubMed] [Google Scholar]
- 54. Duysburgh C, Van den Abbeele P, Kamil A, Fleige L, De Chavez PJ, Chu Yet al. . In vitro–in vivo validation of stimulatory effect of oat ingredients on lactobacilli. Pathogens. 2021;10(2):235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oddi S, Huber P, Rocha Faria Duque AL, Vinderola G, Sivieri K. Breast-milk derived potential probiotics as strategy for the management of childhood obesity. Food Res Int. 2020;137:109673. [DOI] [PubMed] [Google Scholar]
- 56. da Cruz Rodrigues VC, Duque ALR, de Carvalho Fino L, Simabuco FM, Sartoratto A, Cabral Let al. . Modulation of the intestinal microbiota and the metabolites produced by the administration of ice cream and a dietary supplement containing the same probiotics. Br J Nutr. 2020;124(1):57–68. [DOI] [PubMed] [Google Scholar]
- 57. Helbig A, Silletti E, van Aken GA, Oosterveld A, Minekus M, Hamer RJet al. . Lipid digestion of protein stabilized emulsions investigated in a dynamic in vitro gastro-intestinal model system. Food Dig. 2013;4(2-3):58–68. [Google Scholar]
- 58. Liu Y, Gibson GR, Walton GE. An in vitro approach to study effects of prebiotics and probiotics on the faecal microbiota and selected immune parameters relevant to the elderly. PLoS One. 2016;11(9):e0162604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol. 2014;12(4):207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xiang Y, Wen H, Yu Y, Li M, Fu X, Huang S. Gut-on-chip: recreating human intestine in vitro. J Tissue Eng. 2020;11:204173142096531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pimenta J, Ribeiro R, Almeida R, Costa PF, da Silva MA, Pereira B. Organ-on-chip approaches for intestinal 3D in vitro modelling. Cell Mol Gastroenterol Hepatol. 2022;13(2):351–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fedi A, Vitale C, Ponschin G, Ayehunie S, Fato M, Scaglione S. In vitro models replicating the human intestinal epithelium for absorption and metabolism studies: a systematic review. J Controlled Release. 2021;335:247–68. [DOI] [PubMed] [Google Scholar]
- 63. Zhang J, Hernandez-Gordillo V, Trapecar M, Wright C, Taketani M, Schneider Ket al. . Coculture of primary human colon monolayer with human gut bacteria. Nat Protoc. 2021;16(8):3874–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Coenye T, Nelis HJ. In vitro and in vivo model systems to study microbial biofilm formation. J Microbiol Methods. 2010;83(2):89–105. [DOI] [PubMed] [Google Scholar]
- 65. Poyet M, Groussin M, Gibbons SM, Avila-Pacheco J, Jiang X, Kearney SMet al. . A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat Med. 2019;25(9):1442–52. [DOI] [PubMed] [Google Scholar]
- 66. Rabesandratana T. Microbiome conservancy stores global fecal samples. Science. 2018;362(6414):510–11. [DOI] [PubMed] [Google Scholar]
- 67. Hashmi S, Wang Y, Parhar RS, Collison KS, Conca W, Al-Mohanna Fet al. . A C. elegans model to study human metabolic regulation. Nutr Metab. 2013;10(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Marsh EK, May RC. Caenorhabditis elegans, a model organism for investigating immunity. Appl Environ Microbiol. 2012;78(7):2075–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim M, Huda MN, O'Connor A, Albright J, Durbin-Johnson B, Bennett BJ. Hepatic transcriptional profile reveals the role of diet and genetic backgrounds on metabolic traits in female progenitor strains of the collaborative cross. Physiol Genomics. 2021;53(5):173–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kemis JH, Linke V, Barrett KL, Boehm FJ, Traeger LL, Keller MPet al. . Genetic determinants of gut microbiota composition and bile acid profiles in mice. PLos Genet. 2019;15(8):e1008073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Murga-Garrido SM, Hong Q, Cross T-WL, Hutchison ER, Han J, Thomas SPet al. . Gut microbiome variation modulates the effects of dietary fiber on host metabolism. Microbiome. 2021;9(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rodriguez J, Hiel S, Neyrinck AM, Le Roy T, Pötgens SA, Leyrolle Qet al. . Discovery of the gut microbial signature driving the efficacy of prebiotic intervention in obese patients. Gut. 2020;69(11):1975–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Christoforidou Z, Mora Ortiz M, Poveda C, Abbas M, Walton G, Bailey Met al. . Sexual dimorphism in immune development and in response to nutritional intervention in neonatal piglets. Front Immunol. 2019;10:2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Walter J, Armet AM, Finlay BB, Shanahan F. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell. 2020;180(2):221–32. [DOI] [PubMed] [Google Scholar]
- 75. Lichtenstein AH, Petersen K, Barger K, Hansen KE, Anderson CAM, Baer DJet al. . Perspective: design and conduct of human nutrition randomized controlled trials. Adv Nutr. 2021;12(1):4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gibbons SM, Kearney SM, Smillie CS, Alm EJ. Two dynamic regimes in the human gut microbiome. PLoS Comput Biol. 2017;13(2):e1005364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kane PB, Bittlinger M, Kimmelman J. Individualized therapy trials: navigating patient care, research goals and ethics. Nat Med. 2021;27(10):1679–86. [DOI] [PubMed] [Google Scholar]
- 78. Nogal B, Blumberg JB, Blander G, Jorge M. Gut microbiota-informed precision nutrition in the generally healthy individual: are we there yet?. Curr Dev Nutr. 2021;5(9):nzab107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lancaster SM, Lee-McMullen B, Abbott CW, Quijada JV, Hornburg D, Park Het al. . Global, distinctive, and personal changes in molecular and microbial profiles by specific fibers in humans. Cell Host Microbe. 2022;30(6):848–62.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gurry T, HST Microbiome Consortium, Gibbons SM, Nguyen LTT, Kearney SM, Ananthakrishnan Aet al. . Predictability and persistence of prebiotic dietary supplementation in a healthy human cohort. Sci Rep. 2018;8:12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BEet al. . Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SAet al. . Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bolte LA, Vich Vila A, Imhann F, Collij V, Gacesa R, Peters Vet al. . Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut. 2021;70(7):1287–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BMet al. . US immigration westernizes the human gut microbiome. Cell. 2018;175(4):962–72.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RRet al. . Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe. 2019;25(6):789–802.e5. [DOI] [PubMed] [Google Scholar]
- 86. Dunn J, Runge R, Snyder M. Wearables and the medical revolution. Per Med. 2018;15(5):429–48. [DOI] [PubMed] [Google Scholar]
- 87. Kellogg RA, Dunn J, Snyder MP. Personal omics for precision health. Circ Res. 2018;122(9):1169–71. [DOI] [PubMed] [Google Scholar]
- 88. Schembre SM, Liao Y, O'Connor SG, Hingle MD, Shen S-E, Hamoy KGet al. . Mobile ecological momentary diet assessment methods for behavioral research: systematic review. JMIR Mhealth Uhealth. 2018;6(11):e11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Foxx AJ, Franco Meléndez KP, Hariharan J, Kozik AJ, Wattenburger CJ, Godoy-Vitorino Fet al. . Advancing equity and inclusion in microbiome research and training. Msystems. 2021;6(5):e0115121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EOet al. . Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65(3):426–36. [DOI] [PubMed] [Google Scholar]
- 91. Watson AW, Houghton D, Avery PJ, Stewart C, Vaughan EE, Meyer PDet al. . Changes in stool frequency following chicory inulin consumption, and effects on stool consistency, quality of life and composition of gut microbiota. Food Hydrocolloids. 2019;96:688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. van der Beek CM, Canfora EE, Kip AM, Gorissen SHM, Olde Damink SWM, van Eijk HMet al. . The prebiotic inulin improves substrate metabolism and promotes short-chain fatty acid production in overweight to obese men. Metabolism. 2018;87:25–35. [DOI] [PubMed] [Google Scholar]
- 93. Olsen JM, Nesbitt BJ. Health coaching to improve healthy lifestyle behaviors: an integrative review. Am J Health Promot. 2010;25(1):e1–e12. [DOI] [PubMed] [Google Scholar]
- 94. Bailey JM, Bartlem KM, Wiggers JH, Wye PM, Stockings EAL, Hodder RKet al. . Systematic review and meta-analysis of the provision of preventive care for modifiable chronic disease risk behaviours by mental health services. Prev Med Rep. 2019;16:100969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Slawson DL, Fitzgerald N, Morgan KT. Position of the Academy of Nutrition and Dietetics: the role of nutrition in health promotion and chronic disease prevention. J Acad Nutr Diet. 2013;113(7):972–9. [DOI] [PubMed] [Google Scholar]
- 96. Flores M, Glusman G, Brogaard K, Price ND, Hood L. P4 medicine: how systems medicine will transform the healthcare sector and society. Per Med. 2013;10(6):565–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ratajczak W, Rył A, Mizerski A, Walczakiewicz K, Sipak O, Laszczyńska M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim Pol. 2019;66:1–12. [DOI] [PubMed] [Google Scholar]
- 98. Dannenberg L, Zikeli D, Benkhoff M, Ahlbrecht S, Kelm M, Levkau Bet al. . Targeting the human microbiome and its metabolite TMAO in cardiovascular prevention and therapy. Pharmacol Ther. 2020;213:107584. [DOI] [PubMed] [Google Scholar]
- 99. Molinaro A, Bel Lassen P, Henricsson M, Wu H, Adriouch S, Belda Eet al. . Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat Commun. 2020;11(1):5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Davidson KW, Silverstein M, Cheung K, Paluch RA, Epstein LH. Experimental designs to optimize treatments for individuals: personalized N-of-1 trials. JAMA Pediatr. 2021;175(4):404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]