Abstract
Escherichia coli responds to oxidative stress by activating sets of coregulated genes that help the cell to maintain homeostasis. Identified previously by genetic and biochemical approaches, the soxRS system mediates the induction of 18 of these redox-inducible genes (including the soxS gene itself). An overlapping set of genes is activated by an assortment of structurally unrelated molecules with antibiotic activities; many genes in this response are controlled by the marRAB system. The activation of either the soxRS or the marRAB system results in enhanced resistance to both superoxide-generating agents and multiple antibiotics. In order to probe the extent of these regulatory networks, we have measured whole-genome transcriptional profiles of the E. coli response to the superoxide-generating agent paraquat (PQ), an inducer of the soxRS system, and to the weak acid salt sodium salicylate (NaSal), an inducer of the marRA system. A total of 112 genes was modulated in response to PQ, while 134 genes were modulated in response to NaSal. We have also obtained transcriptional profiles of the SoxS and MarA regulons in the absence of global stress, in order to establish the regulatory hierarchies within the global responses. Several previously unrelated genes were shown to be under SoxS or MarA control. The genetic responses to both environmental insults revealed several common themes, including the activation of genes coding for functions that replenish reducing potential; regulate iron transport and storage; and participate in sugar and amino acid transport, detoxification, protein modification, osmotic protection, and peptidoglycan synthesis. A large number of PQ- and NaSal-responsive genes have no known function, suggesting that many adaptive metabolic changes that ensue after stress remain uncharacterized.
Escherichia coli responds to oxidative stress by modifying the expression of many genes. Early studies using two-dimensional gels to analyze variations in protein expression have shown that the synthesis of more than 80 proteins is activated in response to oxidative stress (19). Some of these induced proteins were identified as possessing fundamental antioxidant functions, e.g., superoxide dismutase and catalase. The search for mutants with altered antioxidant defenses led to the isolation and characterization of pleiotropic regulators that operate as redox-regulated genetic switches (3, 20, 42, 43, 45). The best-characterized pleiotropic regulators of the antioxidant responses are the OxyR and SoxR proteins (36). Both proteins have the remarkable ability of directly transducing oxidative signals to genetic regulation. Both proteins are expressed constitutively in an inactive state and are transiently activated in cells under specific types of oxidative stress. The activation of the OxyR and SoxR proteins results in the transcriptional enhancement of sets of genes (regulons) whose products relieve the stress by eliminating oxidants and preventing or repairing oxidative damage (36).
SoxR is a member of the MerR family of metal-binding transcription factors, and it exists in solution as a homodimer, with each subunit containing a [2Fe-2S] cluster. In nonactivated SoxR, these clusters are in the reduced state and their oxidation activates SoxR as a powerful transcription factor (12, 16). Interestingly, SoxR can also be activated by nitric oxide (NO) by direct nitrosylation of the iron-sulfur clusters (11). The active (oxidized or nitrosylated) form of SoxR activates transcription of the soxS gene up to 100-fold. The soxS gene product, SoxS protein, belongs to the AraC/XylS family of DNA-binding transcription factors (3), but its activity seems to be regulated solely at the level of expression. Conventional analysis using limited proteomics and genetic approaches showed that SoxS activates the expression of 17 genes or operons. The known SoxS-activated genes are sodA (encoding Mn-superoxide dismutase), fpr (NADPH-ferredoxin oxidoreductase), micF (antisense RNA, repressor of OmpF translation), ribA (cyclic GMP hydrolase), inaA (unknown function), fldA and fldB (flavodoxins A and B), nfo (endonuclease IV), marRAB (multiple-antibiotic-resistance operon), nfsA (also called mdaA, a nitroreductase), zwf (glucose-6-phosphate dehydrogenase), fur (an iron-binding repressor of iron uptake), fumC (fumarase C), acnA (aconitase), tolC (outer membrane protein), acrAB (drug efflux pump), and rimK (a modifier of ribosomal protein S6). Activating tolC, acrAB, micF, and rimK alters the sensitivity of E. coli and Salmonella enterica to a broad range of antibiotics (9, 10, 27, 33). SoxS is also a repressor of the soxS gene (34) and thus limits its own synthesis.
The diversity of genes activated by OxyR and SoxR illustrates the variety of cellular defense mechanisms against oxidative stress. Antioxidant mechanisms include the scavenging of reactive species (sodA, ahpCF), synthesis of reducing species (acnA, zwf), repair of oxidative damage (nfo, fpr), drug efflux (acrAB, tolC), reduction of cell permeability (micF), and replacement of redox-sensitive isozymes by redox-resistant isozymes (fumC). This variety is hardly surprising, given the large number of targets for oxidative damage; virtually all biological macromolecules can be damaged by oxidants. Particularly sensitive are electron-rich moieties, such as metal centers in proteins, unsaturated bonds in phospholipids, aromatic amino acids, and the double bonds of bases in nucleic acids (40). Oxidative stress has other, indirect effects, such as depletion of reducing power by consumption of NADH and NADPH in antioxidant reactions (40).
Interestingly, the induction of the marRAB regulon also results in enhanced resistance to oxidative agents and multiple antibiotics (3, 18). The genes of the marRAB regulon overlap significantly with those of the soxRS regulon (2, 5). This overlap evidently results from the structural similarity between the MarA and SoxS proteins and their respective DNA-binding sites (29, 30). Thus, two overlapping sets of genes are modulated by different signals sensed by different regulatory circuits. While the soxS gene is under the redox-regulated, positive control of SoxR, marA is under negative control by MarR, a repressor whose DNA-binding activity is regulated by the binding of small molecules with toxic effects (1, 31, 39). Among marRAB inducers are sodium salicylate (NaSal), the naphthoquinones menadione and plumbagin, and dinitrophenol. Although NaSal does not activate the soxRS regulon and although paraquat (PQ), a superoxide-generating agent, only marginally activates the marA regulon (30), the naphthoquinones activate both regulons (33). In contrast to PQ, menadione is a natural plant product. This fact has led to the hypothesis that the early evolution of the soxRS and marA regulons was shaped by the environmental stress mediated by naphthoquinones and other noxious xenobiotics from natural sources (33).
Although a complete profile of the cellular responses to oxidative stress has been lacking, the availability of the complete sequence of the E. coli genome now provides important tools for analyzing gene expression. The analysis of the variations of the “transcriptome,” or transcriptional profiling, has already yielded abundant biological information in several organisms, including E. coli (4, 37, 44), Bacillus subtilis (14), Caulobacter crescentus (25), and Saccharomyces cerevisiae (15, 23).
Here we have determined genome-wide transcriptional profiles of E. coli cells exposed to the superoxide-producing agent PQ or to NaSal. We have also begun to dissect regulatory hierarchies within these responses by expressing, in the absence of stress, individual transcription factors that respond to environmental insult.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strain GC4468 (K-12 ΔlacU169 rpsL) (19) was used as the wild type in all experiments. The GC4468 ΔsoxRS derivative DJ901 (20) transformed with pJP105 (placZ::soxS lacI Apr) (35) was used in the SoxS expression experiment. The GC4468 ΔmarRAB derivative MB106 was constructed for this study by transduction of the kanamycin-tagged 1.24-kb deletion spanning the marRAB operon from strain AG100/Kan (28) and was transformed with pMB102 (placZ::marA lacI Ap). Plasmid pMB102 was constructed by replacing the robA gene in plasmid pMB101 (6) with a PCR product containing the marA gene. The sequence of the cloned marA gene was confirmed at the Molecular Biology Core Facility of the Dana-Farber Cancer Center.
Culture growth and RNA isolation.
Overnight cultures were diluted 1/100 in 15 ml of Luria broth contained in 125-ml Erlenmeyer flasks and were grown at 37°C and 250 rpm to an optical density at 600 nm (OD600) of 0.5. At this point, the cultures were either left untreated or exposed to the different inducers for 45 min. The cells were harvested by centrifugation of 1.5-ml aliquots in a microcentrifuge for 30 s. An RNAeasy kit (Qiagen) was used to lyse the cells and extract total RNA. The RNA preparation obtained from the RNAeasy columns was extracted with phenol (pH 5.0) at 60°C, followed by extraction with a mixture of acidic phenol and chloroform and finally with chloroform alone. The RNA was precipitated with 90% ethanol and 3 M sodium acetate overnight at −20°C and was recovered by centrifugation for 10 min at 4°C. The RNA pellet was washed twice with cold 70% ethanol, dried in a Speed-Vac, and resuspended in diethylpyrocarbonate-treated water. The RNA was quantified by light absorption at 260/280 nm. Samples of the RNA were separated by electrophoresis in agarose gels and stained with ethidium bromide to verify the purity and integrity of the RNA preparation, using the 16S and 23S ribosomal bands as indicators. The absence of chromosomal DNA was verified by using the RNA preparations as the template in PCR amplifications. Briefly, ∼100 ng of total RNA or ∼20 ng of chromosomal DNA was incubated in PCR cocktails containing sodB-specific primers. While the reactions containing chromosomal DNA consistently yielded a DNA fragment of the expected size, the reactions containing RNA preparations always failed to yield a product.
cDNA synthesis.
The RNA preparations from each sample were used as the template for cDNA synthesis by employing a commercial set of 4,290 open reading frame (ORF)-specific oligonucleotides (Sigma-Genosys) as primers. A total of 1 μg of total RNA was incubated in reaction buffer with the ORF-specific oligonucleotides, dTTP, dATP, and dGTP, at 90°C for 2 min and was then cooled to 42°C at a rate of 2.4°C/min. Two hundred units of avian myeloma virus reverse transcriptase (Sigma-Genosys) was added to each reaction, together with 20 μCi of [33P]dCTP (3,000 Ci/mmol). The mixture was incubated at 42°C for 2 h. Labeled products were purified on gel filtration columns (Sephadex G-50).
Hybridization.
The Panorama gene arrays (Sigma-Genosys) are positively charged nylon membranes onto which ORF-specific PCR products have been spotted in duplicate. The arrays represent the complete set of known and predicted ORFs as deduced from the complete genomic sequence of the E. coli K-12 strain MG1655 (7). For all experiments, a pair of membranes was used: one was hybridized with cDNA synthesized from the untreated cell culture, and the other was hybridized with the cDNA synthesized from the treated cell culture. Hybridization and washing steps were carried out following the manufacturer's instructions. The nylon filters were prehybridized at 65°C for 1 h in cylindrical tubes containing 5 ml of hybridization solution (5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA {pH 7.7}], 2% sodium dodecyl sulfate, 1× Denhardt's reagent, and 100 μg of sonicated salmon testes DNA/ml). The whole cDNA preparation was first denatured at 100°C in 3 ml of hybridization solution, and the prehybridization solution was discarded from the tubes and replaced by the mix containing the labeled cDNA. The filters were hybridized for ∼16 h at 65°C in a rotary oven. After this, the filters were rinsed with washing solution (0.5× SSPE, 0.2% sodium dodecyl sulfate) twice at room temperature and three more times at 60°C. The filters were then air dried and wrapped in clear plastic film. Two series of experiments were performed using two different pairs of membranes. For each experimental series, the hybridized membranes were stripped and reprobed up to four times, following the procedures recommended by the manufacturer.
Array imaging and analysis.
Phosphorimaging screens were exposed to the hybridized filters for 48 to 72 h at room temperature. The screens were scanned in a Storm 840 PhosphorImager (Molecular Dynamics) at a 50-μm resolution. The resulting files were analyzed by determination of pixel density using Arrayvision software, which determined the intensity of each duplicate spot, measured in arbitrary units. The background signal was determined for each filter by averaging the intensities of 42 spots that did not contain DNA. This average background was then subtracted from the intensity at each DNA spot, and the corrected intensity of each spot was expressed as a percentage of the sum of all intensities. This treatment allowed comparison between filters independently of total hybridization intensity. The corrected intensities of duplicate spots were averaged, and the untreated and treated intensities from two independent experiments were averaged. The expression ratio for each gene was calculated as the treated/untreated intensities. Thus, an expression ratio of 1 indicated an invariable level of a transcript, whereas expression ratios larger or smaller than 1 indicated up- or down-regulated levels of mRNA, respectively.
Two stringency criteria were applied to each data set. First, only those genes were further analyzed that had an expression level equal to the average background plus 3 standard deviations in at least one of the culture conditions in both duplicate experiments. This minimum expression threshold helped to discard genes with very low expression in control or experimental samples with a confidence of 99.9%. Second, only those genes for which the log of the expression ratio was equal to the mean plus or minus 2.5 standard deviations were considered activated or down-regulated, respectively. This statistical threshold provides ratios that are significantly different from the mean with confidence higher than 99%.
Gene annotation.
The expression values and ratios for each gene were transferred to Excel spreadsheets for statistical analysis and integration into updated, annotated databases (38), accessible online (http://genprotec.mbl.edu).
Web access.
The complete data sets for all experiments are available online (http://www.hsph.harvard.edu/demplelab/genomics).
Gene probe synthesis and Northern blot analysis.
The RNA samples used in the gene array experiments were also used in Northern blot experiments as an independent way to assess the quantitative validity of transcriptional profiling. Probes for specific genes were generated by PCR amplification using chromosomal DNA from strain GC4468 as template and gene-specific primers (ORF-mers) obtained from Sigma-Genosys. Typically, PCR amplifications were carried out in 30 cycles of annealing at 60°C (45 s), elongation at 72°C (1 min), and denaturation at 94°C (30 s). The PCR products were resolved by electrophoresis in 1.25% agarose gels, recovered by excision from the gel, and purified using Qiaquick DNA-binding microspin columns (Qiagen). The DNA fragments were labeled by nick translation using a Klenow DNA polymerase fragment, random hexamers (Gibco BRL), and [32P]dCTP (3,000 Ci/mmol) plus unlabeled dATP, dGTP, and dTTP. The labeled probe was purified by gel filtration in Sephadex G-25 columns (Pharmacia). For the Northern blot experiments, 2 μg of total RNA was run per lane in 1.25% agarose gels containing formaldehyde and was transferred to Nytran membranes using a Turboblotter setup (Schleicher & Schuell). The RNA was cross-linked to the membrane by UV irradiation, and the membranes were then hybridized at 65°C with radioactively labeled DNA fragments in cylindrical tubes using QuickHyb solution (Stratagene). The membranes were washed according to the instructions from the manufacturer. X-ray films were exposed to the membranes at −70°C and were developed using a Fuji automatic developer. The radioactive signals were measured using an Applied Biosystems phosphorimager.
RESULTS
A transcriptional profile of E. coli exposed to PQ.
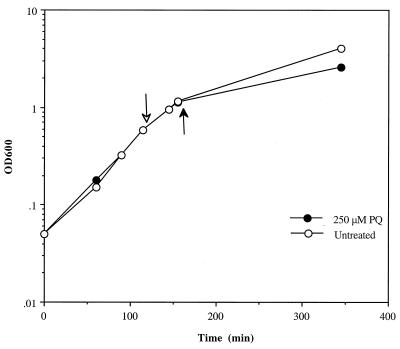
The genomic transcriptional profile of E. coli growing on Luria broth under superoxide stress was determined by comparison of cultures in which one sample was left untreated while the other was exposed to 250 μM PQ for 45 min. This concentration of PQ induced the genes of the soxRS regulon (35) but failed to inhibit growth rate in any substantial way during the 45-min exposure. We expected that these exposure conditions would prevent the activation of “general stress” pathways that generally accompanies growth inhibition (22) and would thus maximize the chance to identify specific, oxidative stress-responsive genes. Figure 1 shows the growth of the untreated and PQ-treated cultures and demonstrates the lack of effect of the chosen PQ concentration on growth during the 45-min exposure.
FIG. 1.
Growth of E. coli strain GC4468. OD600 as a function of time for strain GC4468 was measured. The cultures were either left untreated (empty circles) or treated with 250 μM PQ (filled circles). PQ was added to log-phase cultures (empty arrow), and cells were harvested after 45 min (filled arrow). For detailed culture conditions, see Materials and Methods.
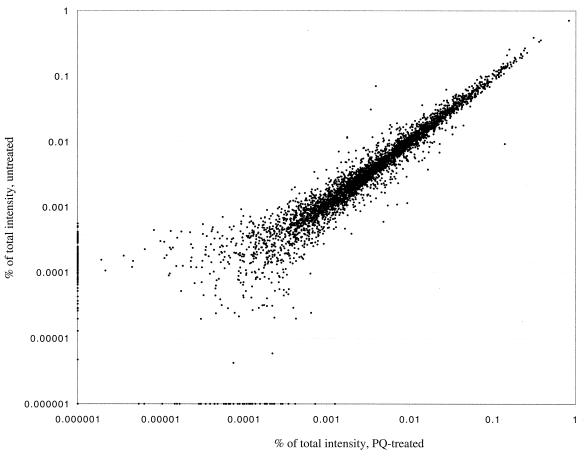
Total RNA was extracted from treated and untreated cultures and was used to synthesize cDNA, which was hybridized to gene arrays. The radioactive signal from each spot in the arrays served as a measure of the expression level of each gene and was used to calculate the expression ratio between the PQ-treated and untreated cultures for all genes in duplicate experiments (see Materials and Methods). Figure 2 shows the correlation between the treated and untreated samples of the averaged expression levels for the 4,290 genes and control spots represented in the arrays for one experiment. It is clear that the vast majority of the mRNAs did not vary significantly with exposure to PQ, an observation substantiated by a correlation coefficient of 0.987 between the expression values for the untreated and PQ-treated cells. Table 1 shows the genes that were activated or down-regulated significantly after exposure of growing cells to PQ. The complete data set for the genome-wide expression ratios is available online (http://www.hsph.harvard.edu/demplelab/genomics). In total, 112 genes were modulated by PQ beyond threshold levels, with 66 genes activated and 46 down-regulated. Of the 16 SoxS-activated genes represented in the array, 9 were detected. Of the remaining seven SoxS-activated genes, five (nfo, rimK, tolC, fldB, and mdaA) were activated with values below threshold while one (ribA) appeared slightly repressed. The micF gene codes for an untranslated RNA and therefore is not represented in the array. However, the down-regulation of ompF, the effect of micF induction, was registered (Table 1).
FIG. 2.
Scatter plot of expression levels for the E. coli genome: untreated and PQ-treated cells. The percent total intensity for each gene represented in the arrays is plotted on a log scale. A relatively small number of genes had associated signal intensities that were below average background levels. The subtraction of the background value from these signals resulted in a negative corrected value and therefore a negative percentage of total signal. These genes were given an arbitrary value of 0.000001% of total intensity.
TABLE 1.
PQ-regulated genesa
| Gene type | No. | Gene | Ratio | Description |
|---|---|---|---|---|
| PQ-activated | b0463 | acrA | 2.0 | AcrAB efflux system effects Mar multiple resistance |
| b0605 | ahpC | 2.8 | Alkyl hydroperoxide reductase small subunit | |
| b1415 | aldA | 2.3 | Aldehyde dehydrogenase, NAD linked | |
| b0863 | artI | 1.9 | Periplasmic binding protein of Arg transport system | |
| b0864 | artP | 1.9 | Arg periplasmic transport system | |
| b0710 | b0710 | 2.0 | Function unknown | |
| b1378 | b1378 | 1.7 | Putative pyruvate-flavodoxin oxidoreductase | |
| b1452 | b1452 | 1.9 | Function unknown | |
| b2351 | b2351 | 1.7 | CPS-53 prophage; putative bactoprenol glucosyl transferase | |
| b2962 | b2962 | 2.4 | Function unknown | |
| b4131 | cadA | 1.8 | Lysine decarboxylase | |
| b4133 | cadC | 2.4 | Regulatory gene | |
| b2198 | ccmD | 1.7 | Cytochrome c-related | |
| b0429 | cyoD | 1.7 | Cytochrome o oxidase subunit IV | |
| b2752 | cysD | 1.7 | Sulfate adenylyltransferase | |
| b2414 | cysK | 1.9 | Cysteine synthase A | |
| b1190 | dadX | 1.7 | Alanine racemase | |
| b4383 | deoB | 1.9 | Deoxyribouratase, phosphopentomutase | |
| b0184 | dnaE | 2.0 | DNA polymerase III, alpha subunit | |
| b0812 | dps | 1.7 | Stress response DNA-binding protein | |
| b0684 | fldA | 4.7 | Flavodoxin | |
| b3288 | fmt | 1.8 | Methionyl-tRNA formyltransferase | |
| b3924 | fpr | 3.6 | Ferredoxin NADP+ reductase; anaerobic | |
| b1611 | fumC | 6.9 | Fumarase C | |
| b0683 | fur | 2.2 | Ferric iron uptake, negative regulatory gene | |
| b2094 | gatA | 2.9 | Galactitol-specific enzyme IIA of PTSb | |
| b2093 | gatB | 3.7 | Galactitol-specific enzyme IIB of PTS | |
| b2091 | gatD | 1.7 | Galactitol-1-phosphate dehydrogenase | |
| b0450 | glnK | 1.8 | Regulated through NRI/NRII two-component regulatory system | |
| b0720 | gltA | 3.1 | Citrate synthase | |
| b2552 | hmpA | 1.8 | Dihydropteridine reductase 2 and nitric oxide dioxygenase NAD/FAD activity | |
| b0440 | hupB | 1.7 | Histone-like protein HU-alpha, HU-1 | |
| b2237 | inaA | 2.9 | Function unknown | |
| b4036 | lamB | 4.8 | Maltose high-affinity uptake | |
| b0096 | lpxC | 2.0 | Cell envelope and cell separation | |
| b4034 | malE | 2.1 | Maltose-binding protein, periplasmic; transport and chemotaxis | |
| b4035 | malK | 1.9 | Maltose transport complex, ATP-binding subunit | |
| b0168 | map | 1.7 | Methionine aminopeptidase | |
| b1531 | marA | 2.6 | Transcription activator of multiple-antibiotic-resistance system | |
| b0086 | murF | 1.7 | d-Alanyl:d-alanine-adding enzyme | |
| b0578 | nfnB | 2.1 | Resistance to nitrofurantoin; a nitroreductase | |
| b2281 | nuoI | 1.8 | NADH dehydrogenase I subunit | |
| b2279 | nuoK | 1.9 | NADH dehydrogenase I subunit | |
| b0113 | pdhR | 1.9 | Pyruvate-dehydrogenase repressor | |
| b4025 | pgi | 2.1 | Glucose phosphate isomerase | |
| b1101 | ptsG | 3.3 | PTS family enzyme IIC, glucose-specific | |
| b1658 | purR | 1.9 | Purine repressor | |
| b3317 | rplB | 1.9 | 50S ribosomal subunit protein L2 | |
| b3319 | rplD | 2.5 | 50S ribosomal subunit protein L4 | |
| b3305 | rplF | 1.7 | 50S ribosomal subunit protein L6 | |
| b3985 | rplJ | 1.7 | 50S ribosomal subunit protein L10 | |
| b3318 | rplW | 1.8 | 50S ribosomal subunit protein L23 | |
| b3312 | rpmC | 1.7 | 50S ribosomal subunit protein L29 | |
| b3314 | rpsC | 1.7 | 30S ribosomal subunit protein S3 | |
| b3321 | rpsJ | 2.0 | 30S ribosomal subunit protein S10 | |
| b3316 | rpsS | 1.7 | 30S ribosomal subunit protein S19 | |
| b0724 | sdhB | 1.8 | Succinate dehydrogenase iron-sulfur protein | |
| b3908 | sodA | 12.3 | Superoxide dismutase, Mn | |
| b4062 | soxS | 2.3 | Regulatory protein of soxRS regulon | |
| b0729 | sucD | 1.9 | Succinyl-coenzyme A synthetase alpha subunit | |
| b3708 | tnaA | 2.4 | Tryptophanase | |
| b0850 | ybjC | 1.7 | Function unknown | |
| b2523 | yfhI | 1.9 | Peptidase B; aminopeptidase | |
| b3520 | yhjB | 3.4 | Putative transcriptional regulator (LuxR/UhpA family) | |
| b4217 | ytfK | 2.3 | Function unknown | |
| b1852 | zwf | 2.7 | Glucose-6-phosphate dehydrogenase | |
| PQ-down-regulated | b3237 | argR | 0.6 | Repressor of Arg regulon |
| b4139 | aspA | 0.5 | Aspartate ammonia-lyase (aspartase) | |
| b0218 | b0218 | 0.3 | Function unknown | |
| b0271 | b0271 | 0.5 | Function unknown | |
| b0938 | b0938 | 0.6 | Function unknown | |
| b1029 | b1029 | 0.4 | Function unknown | |
| b1567 | b1567 | 0.6 | Qin prophage | |
| b1903 | b1903 | 0.5 | Function unknown | |
| b2102 | b2102 | 0.5 | Function unknown | |
| b2355 | b2355 | 0.5 | Function unknown | |
| b2629 | b2629 | 0.4 | Function unknown | |
| b2768 | b2768 | 0.6 | Function unknown | |
| b0032 | carA | 0.5 | Carbamoylphosphate synthase (glutamine-hydrolyzing) light subunit | |
| b0620 | criR | 0.5 | Response regulator in two-component regulatory system with DpiB | |
| b1575 | dicB | 0.5 | Control of cell division. Activates MinC | |
| b2369 | evgA | 0.6 | Multicopy on plasmid in strain with a deletion of envZ induces ompC expression | |
| b2323 | fabB | 0.4 | Beta-ketoacyl-acyl carrier protein synthase I | |
| b1925 | fliS | 0.5 | Flagellar synthesis; flagellar regulon member | |
| b2025 | hisF | 0.5 | Cyclase component of IGP synthase complex | |
| b3580 | lyxK | 0.5 | l-Xylulose kinase, cryptic | |
| b0923 | mukE | 0.5 | Involved in chromosome partitioning | |
| b1224 | narG | 0.3 | Nitrate reductase alpha subunit | |
| b3480 | nikE | 0.6 | Formate hydrogen-lyase activity | |
| b0929 | ompF | 0.2 | Outer membrane protein, porin | |
| b4245 | pyrB | 0.3 | Aspartate transcarbamylase, catalytic subunit | |
| b0945 | pyrD | 0.5 | Dihydroorotate oxidase | |
| b4387 | smp | 0.6 | Membrane protein | |
| b1656 | sodB | 0.5 | Superoxide dismutase, Fe | |
| b1646 | sodC | 0.6 | Superoxide dismutase, Cu, Zn | |
| b2497 | uraA | 0.4 | Uracil concentration dependence of pyr mutants; Ura ABC transporter | |
| b0193 | yaeF | 0.4 | Function unknown | |
| b0315 | yahA | 0.6 | Putative transcriptional repressor (LuxR/UhpA family) | |
| b1347 | ydaC | 0.6 | Rac prophage | |
| b2597 | yfiA | 0.5 | Ribosome-associated factor, stabilizes ribosomes against dissociation | |
| b3087 | ygjR | 0.4 | Putative NADP-binding dehydrogenase | |
| b3088 | ygjT | 0.5 | Putative transmembrane protein | |
| b3112 | yhaQ | 0.4 | Anaerobic pathway, l-serine deaminase, l-serine dehydratase | |
| b3516 | yhiX | 0.5 | Putative transcriptional regulator (AraC/XylS family) | |
| b3582 | yiaR | 0.5 | Putative hexulose-6-phosphate isomerase | |
| b3879 | yihR | 0.6 | Putative aldose-1-epimerase | |
| b3888 | yiiD | 0.5 | Putative acyltransferase | |
| b4048 | yjbM | 0.5 | Function unknown | |
| b4333 | yjiK | 0.6 | Putative outer membrane protein | |
| b4337 | yjiO | 0.4 | Putative transport protein | |
| b4380 | yjjI | 0.6 | Function unknown | |
| b4394 | yjjX | 0.4 | Function unknown |
No., unique identifier. Boldface denotes genes modulated by either exposure to PQ or expression of SoxS. Underlining denotes previously characterized soxRS genes.
PTS, phosphotransferase system.
The hmpA gene, which encodes a hemoglobin-like protein and is induced by PQ in a soxRS-independent manner (32), was also detected by the gene array experiments. Only two OxyR-regulated genes (ahpC and dps) were activated above the statistical threshold, indicating that the levels of hydrogen peroxide generated under our conditions were relatively low. Finally, 7 of the activated genes and 22 of the down-regulated genes have no function known at this time. The absence of induction of any heat shock genes or genes for stress-responsive sigma factors was consistent with the lack of growth inhibition by PQ. Thus, the observed transcriptional profile of the response to PQ bears most of the hallmarks of the known responses to superoxide stress while providing abundant new information on putative antioxidant functions.
Validation by Northern blotting of expression ratios from transcriptional profiling.
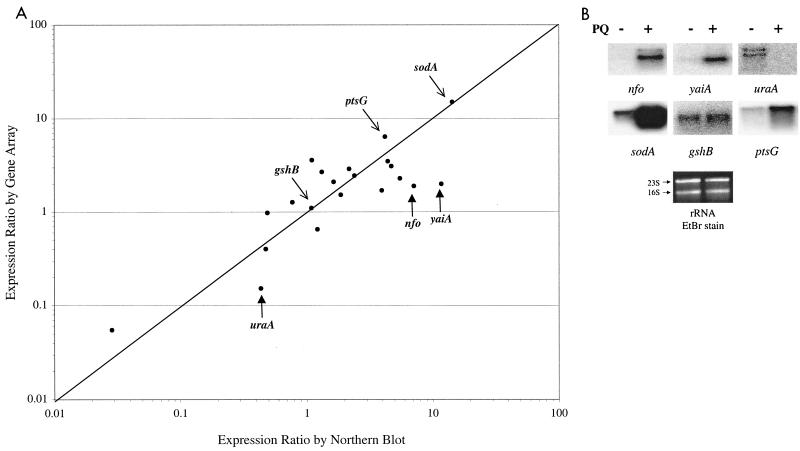
In order to test independently the values for the expression ratios obtained from the gene arrays, we performed Northern blot analysis for 21 genes. The same RNA samples used as templates in one of the PQ-treatment experiments were run on an agarose gel, transferred to nylon membranes, and hybridized with labeled gene probes (see Materials and Methods). The intensity of each signal was measured by phosphorimaging, and the expression ratio (treated/untreated intensities) was calculated for each gene. Figure 3A shows the comparison of the expression ratios obtained from gene array experiments and conventional Northern blots. The 21 genes selected for the comparison included 11 genes that scored as activated in the gene array experiment, 7 genes that scored as unmodified (expression ratios between 0.5 and 2), and 3 genes that scored as down-regulated. In general, the results of the two methods were similar across at least 2 orders of magnitude. However, the correlation between the Northern blot and the array values was weaker for genes with basal (or down-regulated) expression levels that were close to background. Figure 3B shows three examples of genes (nfo, uraA, and yaiA) with comparatively large differences between the expression ratios obtained by transcriptional profiling and Northern blotting. In all three cases, the basal or down-regulated level of mRNA was extremely low as estimated by Northern blotting. In contrast, genes with detectable basal levels (sodA, gshB, and ptsG) show good correlation in their expression ratios measured by the two methods (Fig. 3A).
FIG. 3.
Comparison of expression ratios from transcriptional profiling and Northern blots. (A) Total RNA was extracted from untreated or PQ-treated cultures (250 μM for 45 min). Aliquots from these preparations were used as a template for cDNA synthesis and hybridization with gene arrays or were run in agarose gels, transferred to Nytran membranes, and probed with labeled gene-specific PCR fragments. The expression ratio of 21 genes is shown for Northern blotting (horizontal axis) or cDNA synthesis and hybridization to gene arrays (vertical axis). The genes tested were cyoD, cysK, dnaE, gshB, inaA, lpxC, nadE, nfo, nupC, pdhR, ptsG, pyrB, rpsS, sodA, speE, uraA, yadJ, ybjC, yhiM, and zwf. See Materials and Methods for detailed protocols. (B) Northern blots of the nfo, yaiA, uraA, sodA, gshB, and ptsG genes. −, absence of PQ; +, presence of PQ. A replicate gel was run and stained with ethidium bromide (EtBr), revealing the 16S and 23S rRNA, which serves as loading control.
A search for novel SoxS-regulated genes.
Although transcriptional profiling provides information about relative RNA levels, it does not establish regulatory hierarchies among genes. In an effort to begin dissecting the regulatory cascade within the response to PQ, we expressed SoxS protein in the absence of oxidative stress. The plasmid pJP105 expresses the SoxS protein from an isopropyl-β-d-thiogalactopyranoside (IPTG)-regulated promoter and has been used to identify SoxS-regulated operon fusions (35). The activity of the SoxS protein is regulated exclusively by its intracellular concentration, and thus the artificial induction of soxS expression from pJP105 should be sufficient to modulate all genes of the soxRS regulon. The E. coli strain DJ901 (ΔsoxRS) was transformed to Apr with plasmid pJP105, and cultures were either left untreated or were treated with IPTG. The labeled cDNA from these cultures was hybridized to Panorama gene arrays, and the blots were analyzed using the same stringency conditions applied to the PQ experiment. Again, the vast majority of genes was unaffected, with only 95 genes modulated beyond the statistical threshold: 37 genes were activated and 58 genes were down-regulated. The activated genes included 11 out of the 16 known SoxS-activated genes represented in the gene array (data not shown). The indirect down-regulation of ompF was also registered. Of the 37 SoxS-activated genes revealed by transcriptional profiling, 14 were also registered in the PQ exposure experiments. The genes that were activated both by PQ and by expression of SoxS are indicated in Table 1 in boldface. The complete results from the SoxS-expression experiment can be obtained online (http://www.harvard.hsph.edu/demplelab/genomics).
A transcriptional profile of the E. coli response to NaSal.
NaSal is a natural product with a role in signal transduction in the response to plant infection (8). In the millimolar concentration range, NaSal dissipates the proton gradient across the inner membrane, chelates iron, inhibits growth, and induces the heat shock and marA regulons (41), the latter by binding to the repressor of the marRAB operon (31, 39). In order to characterize the global response to NaSal and to identify novel regulatory overlaps with the response to PQ, the transcriptional profile of growing cultures of E. coli treated with NaSal was determined.
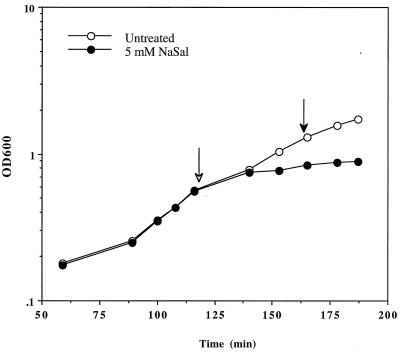
The concentration of NaSal used (5 mM) was chosen because it has been used experimentally to induce the marA regulon (30). Figure 4 shows that this concentration of NaSal inhibited growth; therefore, we expected to observe both a NaSal-specific and a “general stress” response.
FIG. 4.
Growth of E. coli GC4468 exposed to NaSal. OD600 as a function of time for strain GC4468 was determined. The cultures were either left untreated (empty circles) or treated with 5 mM NaSal (filled circles). NaSal was added to log-phase cultures (empty arrow), and cells were harvested after 45 min (filled arrow). For detailed culture conditions, see Materials and Methods.
Figure 5 depicts the correlation of averaged expression levels for the 4,290 genes and control spots between the untreated and NaSal-treated samples, which shows that the expression of most genes was unaffected by the treatment. Table 2 shows the genes that were significantly activated or down-regulated after the NaSal exposure. The complete data set for the genome-wide expression ratios is available online (http://www.hsph.harvard.edu/demplelab/genomics). In total, 144 genes were modulated beyond threshold levels in response to NaSal, with 84 genes activated and 60 down-regulated.
FIG. 5.
Scatter plot of expression levels for the E. coli genome: untreated and NaSal-treated cells. The percent total intensity for each gene represented in the arrays is plotted on a log scale. A relatively small number of genes had associated signal intensities that were below average background levels. The subtraction of the background value from these signals resulted in a negative corrected value and therefore a negative percentage of total signal. These genes were given an arbitrary value of 0.000001% of total intensity.
TABLE 2.
NaSal-regulated genesa
| Gene type | No. | Gene | Ratio | Description |
|---|---|---|---|---|
| NaSal-activated | b4014 | aceB | 2.3 | Malate synthase A |
| b1276 | acnA | 2.3 | Aconitase A | |
| b1241 | adhE | 2.2 | Acetaldehyde-coenzyme A dehydrogenase | |
| b2252 | ais | 3.8 | Aluminum-inducible protein; function unknown | |
| b0864 | artP | 2.5 | Arg periplasmic transport system | |
| b0485 | b0485 | 2.7 | Putative glutaminase | |
| b0881 | b0881 | 2.1 | Function unknown | |
| b1112 | b1112 | 9.0 | Function unknown | |
| b1164 | b1164 | 3.4 | Function unknown | |
| b1165 | b1165 | 3.2 | Function unknown | |
| b1200 | b1200 | 5.9 | Putative dihydroxyacetone kinase | |
| b1450 | b1450 | 2.3 | Putative transcriptional regulator (GntR family) | |
| b1452 | b1452 | 3.0 | Function unknown | |
| b1643 | b1643 | 2.1 | Function unknown | |
| b1795 | b1795 | 2.9 | Function unknown | |
| b2174 | b2174 | 2.4 | Putative permease | |
| b2266 | b2266 | 2.9 | Function unknown | |
| b2672 | b2672 | 2.6 | Function unknown | |
| b3004 | b3004 | 2.5 | Function unknown | |
| b3024 | b3024 | 2.3 | Function unknown | |
| b3238 | b3238 | 27.6 | Function unknown | |
| b3242 | b3242 | 6.9 | Function unknown | |
| b1661 | cfa | 3.7 | Cyclopropane fatty acid synthase | |
| b3806 | cyaA | 2.4 | Adenylate cyclase | |
| b2414 | cysK | 2.4 | Cysteine synthase | |
| b4382 | deoA | 2.4 | Thymidine phosphorylase | |
| b4383 | deoB | 2.1 | Deoxyribouratase, phosphopentomutase | |
| b0014 | dnaK | 2.6 | HSP-70-type molecular chaperone | |
| b0812 | dps | 7.0 | Stress response DNA-binding protein | |
| b0593 | entC | 2.2 | Isochorismate synthetase | |
| b3289 | fmu | 2.4 | 16S rRNA m5C967 methyltransferase, S-adenosyl-l-methionine dependent | |
| b1611 | fumC | 8.4 | Fumarase C, aerobic; member of soxRS regulon | |
| b3517 | gadA | 2.7 | Glutamate decarboxylase | |
| b1493 | gadB | 2.7 | Glutamate decarboxylase | |
| b1779 | gapA | 2.6 | GAPDH A | |
| b2094 | gatA | 5.3 | Galactitol-specific enzyme IIA of PTSb | |
| b2093 | gatB | 5.2 | Galactitol-specific enzyme IIB of PTS | |
| b2092 | gatC | 2.5 | Galactitol-specific enzyme IIC of PTS | |
| b2091 | gatD | 3.8 | Galactitol-1-phosphate dehydrogenase | |
| b2090 | gatR_2 | 2.3 | Split transcriptional repressor of galactitol utilization, fragment 2 (DeoR family) | |
| b2095 | gatZ | 2.8 | Tagatose 6-phosphate aldolase 2, subunit with GatY | |
| b0720 | gltA | 2.6 | Citrate synthase | |
| b3212 | gltB | 2.8 | Glutamate synthase, large subunit | |
| b2947 | gshB | 3.0 | Glutathione synthetase | |
| b3510 | hdeA | 9.9 | Periplasmic, unknown function, has sigma S-dependent promoter | |
| b3509 | hdeB | 9.2 | Periplasmic, unknown function, has sigma S-dependent promoter | |
| b2237 | inaA | 3.9 | Function unknown | |
| b1732 | katE | 2.1 | Catalase hydroperoxidase III | |
| b3604 | lctR | 4.9 | Regulatory gene for lld operon | |
| b3603 | lldP | 2.7 | l-Lactate permease | |
| b4129 | lysU | 2.1 | Lysyl tRNA synthetase, inducible | |
| b1817 | manX | 2.3 | PTS family, mannose-specific enzyme IIA component | |
| b1818 | manY | 2.4 | Mannose PTS, EIIC component | |
| b1531 | marA | 11.3 | Transcription activator of multiple-antibiotic-resistance system | |
| b1532 | marB | 9.0 | Regulatory gene for mar | |
| b1530 | marR | 20.7 | Repressor of mar operon | |
| b3028 | mdaB | 2.2 | Modulator of drug activity | |
| b3601 | mtlR | 2.5 | Mannitol repressor | |
| b0578 | nfnB | 3.4 | Resistance to nitrofurantoin; a nitroreductase | |
| b1482 | osmC | 4.4 | Osmotically inducible protein C; nonessential gene | |
| b4376 | osmY | 2.9 | Periplasmic, sigma S-dependent protein (stationary phase) | |
| b1897 | otsB | 2.2 | Trehalose phosphate phosphatase | |
| b0932 | pepN | 2.1 | Aminopeptidase N | |
| b0903 | pflB | 2.1 | Pyruvate formate lyase I; induced anaerobically | |
| b1014 | putA | 3.2 | Proline dehydrogenase | |
| b3506 | slp | 2.1 | C starvation and stationary phase-inducible; outer membrane lipoprotein | |
| b3284 | smg | 2.1 | Function unknown | |
| b3908 | sodA | 2.3 | Member of soxRS regulon; superoxide dismutase, Mn | |
| b2703 | srlA_2 | 2.7 | PTS family, glucitol/sorbitol-specific enzyme IIB component | |
| b0958 | sulA | 2.9 | Inhibits cell division and ftsZ ring formation | |
| b1004 | wrbA | 2.1 | Affects association between Trp repressor and operators in stationary phase | |
| b0707 | ybgA | 2.1 | Function unknown | |
| b0719 | ybgD | 2.2 | Putative fimbria-like protein | |
| b0850 | ybjC | 2.7 | Function unknown | |
| b1003 | yccJ | 2.6 | Function unknown | |
| b1198 | ycgC | 2.3 | Function unknown | |
| b2597 | yfiA | 3.4 | Ribosome-associated factor, stabilizes ribosomes against dissociation | |
| b2946 | yggJ | 2.0 | Function unknown | |
| b3160 | yhbW | 2.9 | Putative monooxygenase | |
| b3516 | yhiX | 2.3 | Putative transcriptional regulator (AraC/XylS family) | |
| b3555 | yiaG | 3.3 | Putative transcriptional regulator | |
| b4367 | yjjS | 3.9 | Ferric hydroxamate transport | |
| b4378 | yjjV | 2.2 | Putative hydrolase | |
| b3098 | yqjD | 2.8 | Function unknown | |
| NaSal-down-regulated | b3734 | atpA | 0.4 | Membrane-bound ATP synthase, F1 sector, alpha subunit |
| b3731 | atpC | 0.4 | Membrane-bound ATP synthase, F1 sector, epsilon subunit | |
| b3736 | atpF | 0.3 | Membrane-bound ATP synthase, F0 sector, subunit b | |
| b3735 | atpH | 0.3 | Membrane-bound ATP synthase, F1 sector, delta subunit | |
| b3739 | atpI | 0.4 | Membrane-bound ATP synthase subunit, F1-F0-type proton-ATPase | |
| b0295 | b0295 | 0.4 | Function unknown | |
| b3261 | fis | 0.2 | Transcriptional activator for rRNA operons | |
| b3340 | fusA | 0.4 | Fusidic acid resistance; protein chain elongation factor G | |
| b2215 | ompC | 0.5 | Outer membrane protein, porin | |
| b0565 | ompT | 0.3 | Outer membrane protein, protease VII | |
| b4201 | priB | 0.4 | Primosomal protein N | |
| b3259 | prmA | 0.3 | Methyltransferase for 50S subunit L11 protein modification | |
| b4245 | pyrB | 0.4 | Aspartate transcarbamylase, catalytic subunit | |
| b3984 | rplA | 0.3 | 50S ribosomal subunit protein L1 | |
| b3317 | rplB | 0.3 | 50S ribosomal subunit protein L2 | |
| b3320 | rplC | 0.5 | 50S ribosomal subunit protein L3 | |
| b3319 | rplD | 0.3 | 50S ribosomal subunit protein L4 | |
| b3308 | rplE | 0.4 | 50S ribosomal subunit protein L5 | |
| b3305 | rplF | 0.3 | 50S ribosomal subunit protein L6 | |
| b4203 | rplI | 0.2 | 50S ribosomal subunit protein L9 | |
| b3985 | rplJ | 0.3 | 50S ribosomal subunit protein L10 | |
| b3983 | rplK | 0.4 | 50S ribosomal subunit protein L11 | |
| b3986 | rplL | 0.3 | 50S ribosomal subunit protein L7/L12 | |
| b3231 | rplM | 0.4 | 50S ribosomal subunit protein L13 | |
| b3310 | rplN | 0.3 | 50S ribosomal subunit protein L14 | |
| b3301 | rplO | 0.4 | 50S ribosomal subunit protein L15 | |
| b3313 | rplP | 0.2 | 50S ribosomal subunit protein L16 | |
| b3294 | rplQ | 0.3 | 50S ribosomal subunit protein L17 | |
| b3304 | rplR | 0.4 | 50S ribosomal subunit protein L18 | |
| b3186 | rplU | 0.3 | 50S ribosomal subunit protein L21 | |
| b3315 | rplV | 0.1 | 50S ribosomal subunit protein L22 | |
| b3318 | rplW | 0.4 | 50S ribosomal subunit protein L23 | |
| b3309 | rplX | 0.3 | 50S ribosomal subunit protein L24 | |
| b3185 | rpmA | 0.4 | 50S ribosomal subunit protein L27 | |
| b3637 | rpmB | 0.4 | 50S ribosomal subunit protein L28 | |
| b3312 | rpmC | 0.3 | 50S ribosomal subunit protein L29 | |
| b3302 | rpmD | 0.3 | 50S ribosomal subunit protein L30 | |
| b1717 | rpmI | 0.4 | 50S ribosomal subunit protein A (L35) | |
| b3299 | rpmJ | 0.3 | 50S ribosomal subunit protein X (L36) | |
| b3295 | rpoA | 0.4 | RNA polymerase, alpha subunit | |
| b0169 | rpsB | 0.4 | 30S ribosomal subunit protein S2 | |
| b3314 | rpsC | 0.4 | 30S ribosomal subunit protein S3 | |
| b3296 | rpsD | 0.4 | 30S ribosomal subunit protein S4 | |
| b3303 | rpsE | 0.3 | 30S ribosomal subunit protein S5 | |
| b3306 | rpsH | 0.3 | 30S ribosomal subunit protein S8 | |
| b3230 | rpsI | 0.3 | 30S ribosomal subunit protein S9 | |
| b3321 | rpsJ | 0.3 | 30S ribosomal subunit protein S10 | |
| b3297 | rpsK | 0.5 | 30S ribosomal subunit protein S11 | |
| b3307 | rpsN | 0.3 | 30S ribosomal subunit protein S14 | |
| b2609 | rpsP | 0.3 | 30S ribosomal subunit protein S16 | |
| b3311 | rpsQ | 0.3 | 30S ribosomal subunit protein S17 | |
| b3316 | rpsS | 0.2 | 30S ribosomal subunit protein S19 | |
| b0023 | rpsT | 0.4 | 30S ribosomal subunit protein S20 | |
| b0121 | speE | 0.4 | Spermidine synthase | |
| b4240 | treB | 0.4 | IITre, translocation system, Tre-specific PTS enzyme II | |
| b2607 | trmD | 0.3 | tRNA (guanine-7)-methyltransferase | |
| b3384 | trpS | 0.5 | Tryptophanyl-tRNA synthetase | |
| b0170 | tsf | 0.5 | EF-Ts, elongation factor for transcription, stable | |
| b3339 | tufA | 0.5 | Duplicate gene for EF-Tu subunit, elongation factor, unstable | |
| b2014 | yeeF | 0.4 | Putative amino acid transport protein |
No., unique identifier. Boldface denotes genes modulated by either exposure to NaSal or MarA expression.
PTS, phosphotransferase system.
Of the 62 genes postulated to be activated by MarA (5), 19 were detected in our NaSal experiments and an additional 22 were modulated following the previously reported trend but below our statistical threshold. As expected from its inhibitory effect over growth and from previous observations (41), NaSal activated a set of genes associated with general cell stress and damage. These included genes coding for proteins involved in heat shock (dnaK), an inhibitor of cell division (sulA), adenylate cyclase (cyaA), and several rpoS-activated genes: the DNA-binding iron chelator (dps), two periplasmic proteins (hdeAB), and a catalase (katE). The genes encoding the global regulators ςS (rpoS) and ςE (rpoE) were also activated, albeit under threshold levels. Exposure to NaSal also produced a down-regulation of genes coding for translation machinery elements (e.g., ribosomal proteins and elongation factors) and ATP synthase subunits. Thus, the response to NaSal exposure bears the hallmarks of the activation of marA-regulated genes, plus a substantial number of the characteristics of a growth-limited culture.
A search for MarA-regulated genes.
To begin dissecting the regulatory cascade of the response to NaSal, we expressed MarA in the absence of exogenous toxic agents. The plasmid pMB102 harbors the marA gene under the control of an IPTG-regulated promoter. As with SoxS, the activity of the MarA protein is regulated exclusively by its intracellular concentration (2). Thus, the artificial induction of marA from pMB102 should be sufficient to modulate all MarA target genes. Cultures of an E. coli ΔmarRAB strain containing plasmid pMB102 were grown and either left untreated or treated with IPTG. After 45 min, total RNA was extracted from both cultures and analyzed by hybridization to gene arrays. As for the previous experiments, background-corrected expression values were determined and used to calculate the expression ratios for each gene and the same significance thresholds were applied. In total, 88 genes were modulated by MarA expression; 67 genes were activated and 21 genes were down-regulated. The activated genes include 21 of the 62 postulated MarA-regulated genes (5) (data not shown). An additional 19 genes had similar expression trends as reported but fell below our statistical threshold. Those genes activated both by exposure to NaSal and by expression of MarA, 20 in total, are indicated in boldface in Table 2. The complete results from the MarA-expression experiment can be obtained online (http://www.hsph.harvard.edu/demplelab/genomics).
The overlap between the PQ and NaSal stimulons.
Sixteen genes were modulated with the same trend by both PQ and NaSal (Table 3). Some genes were known from standard genetic studies to be regulatory targets for SoxS and MarA (fumC, inaA, marA, and sodA). In addition, the PQ- or SoxS-responsive genes gatABD, gltA, nfnB, and ybjC were recently shown to be activated by constitutive expression of MarA (5). The ybjC gene was also activated by expression of SoxS in our experiments (Table 1). This study identified five additional genes activated by both PQ and NaSal. These genes code for products involved in arginine transport (artP), cysteine synthesis (cysK), protection of DNA from iron-mediated oxidative damage (dps), and salvage of nucleotides (deoB). One other gene, activated by both PQ and NaSal, has no known function or extensive homology to any other gene of known function (b1452). Only one gene was down-regulated by both treatments: pyrB, which codes for aspartate transcarbamylase, involved in pyrimidine biosynthesis.
TABLE 3.
List of genesa
| No. | Gene | PQ ratio | NaSal ratio | Description |
|---|---|---|---|---|
| b0864 | artP | 1.9 | 2.5 | Arg periplasmic transport system |
| b1452 | b1452 | 1.9 | 3.0 | Function unknown |
| b2414 | cysK | 1.9 | 2.4 | Cysteine synthase A |
| b4383 | deoB | 1.9 | 2.1 | Deoxyribouratase, phosphopentomutase |
| b0812 | dps | 1.7 | 7.0 | Stress response DNA-binding protein |
| b1611 | fumC | 6.9 | 8.4 | Fumarase C, aerobic; member of soxRS regulon |
| b2094 | gatA | 2.9 | 5.3 | Galactitol-specific enzyme IIA of PTSb |
| b2093 | gatB | 3.7 | 5.2 | Galactitol-specific enzyme IIB of PTS |
| b2091 | gatD | 1.7 | 3.8 | Galactitol-1-phosphate dehydrogenase |
| b0720 | gltA | 3.1 | 2.6 | Citrate synthase |
| b2237 | inaA | 2.9 | 3.9 | Function unknown |
| b1531 | marA | 2.6 | 11.3 | Transcription activator of multiple-antibiotic-resistance system |
| b0578 | nfnB | 2.1 | 3.4 | Resistance to nitrofurantoin; a nitroreductase |
| b4245 | pyrB | 0.3 | 0.4 | Aspartate transcarbamylase, catalytic subunit |
| b3908 | sodA | 12.3 | 2.3 | Member of soxRS regulon; superoxide dismutase, Mn |
| b0850 | ybjC | 1.7 | 2.7 | Function unknown |
| b3317 | rplB | 1.9 | 0.3 | 50S ribosomal subunit protein L2 |
| b3319 | rplD | 2.5 | 0.3 | 50S ribosomal subunit protein L4 |
| b3305 | rplF | 1.7 | 0.3 | 50S ribosomal subunit protein L6 |
| b3318 | rplW | 1.8 | 0.4 | 50S ribosomal subunit protein L23 |
| b3312 | rpmC | 1.7 | 0.3 | 50S ribosomal subunit protein L29 |
| b3314 | rpsC | 1.7 | 0.4 | 30S ribosomal subunit protein S3 |
| b3321 | rpsJ | 2.0 | 0.3 | 30S ribosomal subunit protein S10 |
| b3316 | rpsS | 1.7 | 0.2 | 30S ribosomal subunit protein S19 |
| b2597 | yfiA | 0.5 | 3.4 | Ribosome-associated factor, stabilizes ribosomes against dissociation |
| b3516 | yhiX | 0.5 | 2.3 | Putative transcriptional regulator (AraC/XylS family) |
No., unique identifier. Underlining denotes previously characterized soxRS genes.
PTS, phosphotransferase system.
A second group of 10 genes was commonly regulated by exposure to PQ or NaSal but with opposite trends (Table 3). Eight of these genes code for ribosomal proteins, one codes for a ribosome-associated factor, and the last one codes for a putative member of the AraC/XylS family of transcriptional regulators.
DISCUSSION
We have used a functional genomics approach to identify novel genes that respond to oxidative stress. In all the experiments, our results not only bear the hallmarks of the cellular responses that we intended to evoke but also provide new insights in the physiology of oxidative stress. The resulting tally of the genes involved in these responses should be considered an underestimate for reasons that are inherent to the method used. First, commercially available gene arrays are a fixed platform that do not admit modifications. The arrays used excluded untranslated RNAs, which are clearly involved in responses to changing redox conditions (46–48). Second, transcriptional profiling reveals only comparative, steady-state levels of mRNAs, without any information about posttranscriptional processes or actual protein expression. Third, despite the almost complete coverage of the genome, transcriptional profiling experiments consistently fail to detect changes in genes known to be modulated by the stimuli of interest. For example, the use of gene arrays to analyze the heat shock response revealed only 23 of the 51 known genes (37). Finally, the results can vary significantly between transcriptional profiling experiments. We have tried to address this problem by averaging replicate experiments and by establishing statistical thresholds for the expression ratios beyond 99% confidence. For those experiments involving induction of the SoxS and MarA transcription factors, we performed a single experiment. However, we included in our analysis only those genes that were also modulated in the duplicate experiment involving the corresponding global inducer.
PQ activates genes involved in pathways that reconstitute NADH and NADPH pools.
PQ is reduced intracellularly at the expense of NADPH in a reaction catalyzed by at least three oxidoreductases (26). PQ reduced by one electron is oxidized by O2 to form superoxide, resulting in a redox cycle that produces a flux of superoxide. Thus, the cell faces a double threat under exposure to PQ and to other redox-cycling agents: the deleterious effects of superoxide itself and the decreased level of NADPH that limits biosynthetic capabilities. Equilibration of NADPH with NADH would generalize this limitation of cellular reducing power.
Previous observations suggested that treatment of growing cells with PQ induces pathways that replenish reducing power. First, PQ activates the expression of glucose-6-phosphate dehydrogenase, the first enzyme of the pentose phosphate pathway (19, 24). This pathway generates NADPH and is required for resistance to redox-cycling agents (17a). Second, PQ also activates the expression of two enzymes of the tricarboxylic acid cycle, fumarase C and aconitase. These enzymes contribute to the reduction of NAD+. The coordinated activation of G6PD, fumarase C, and aconitase by PQ occurs at the transcriptional level in a soxRS-dependent manner.
In our studies, exposure to PQ activated the expression of additional genes involved in pathways that contribute to replenish reducing power. These genes code for proteins involved in sugar transport (ptsG, gatABD, malEK, lamB), glycolysis (pgi), amino acid transport and degradation (artIP, tnaA, dadX), and the tricarboxylic acid cycle (gltA, sdhB, sucD). A similar redirection of carbon metabolism to pathways that reconstitute NADPH was observed in S. cerevisiae after treatment with hydrogen peroxide and proteome analysis by two-dimensional gels (17). Thus, regenerating NADPH may be a fundamental and general aspect of cellular responses to oxidative stress.
Evidence for adaptation and repair pathways under oxidative stress.
Challenge of growing E. coli cells with PQ induces the genes coding for nine ribosomal proteins (Table 1). This increase in ribosomal building blocks was not predicted, in that the bacterial growth rate was not significantly affected during the 45-min treatment with PQ. Another gene coding for a translational regulator, fmt, is also induced by PQ. The product of this gene, methionyl-tRNA formyltransferase, is an important factor in translational initiation (21). One possibility is that increased translational capacity counterbalances a faster turnover of proteins due to oxidative damage and increased degradation. In this scenario, enhanced synthesis would be required to maintain the high growth rate. The possibility of an increased metabolic rate under superoxide stress is consistent with the activation of genes coding for products involved in crucial anabolic and catabolic pathways. These genes include nuoI and nuoK coding for subunits of NADPH dehydrogenase, the first electron acceptor of the respiratory chain. Interestingly, 11 out of the 12 genes coding for NADPH dehydrogenase subunits showed some degree of activation in both PQ exposure experiments, albeit below the statistical threshold levels.
Recently, a direct regulatory connection between oxidative stress and iron metabolism was shown by Zheng et al., who demonstrated the transcriptional activation of fur by SoxS and OxyR (49). Our work confirmed the activation of fur by superoxide stress. In addition to the increase in fur expression, superoxide stress resulted in the down-regulation of sodB, a Fur-activated gene (13). Collectively, these changes in gene expression are consistent with a phenotypic iron deficiency in PQ-treated cells, but how this deficiency might be caused by oxidative stress is unknown. It has been suggested that the Fur-iron complex is sensitive to oxidative damage and that this damage leads to the eventual loss of repressor function (49). An alternative hypothesis is based on the observation that Fe3+ does not seem to function as corepressor. It is possible that under oxidative stress the Fe2+ associated with Fur is oxidized to Fe3+, which leads to the derepression of Fur-repressed genes. While observing that fur is not activated by treatment with NaSal, it is important to note that modulation by Fur does not account for all the observed regulation of iron uptake.
The activation of the dadX and murF genes, coding for proteins involved in peptidoglycan synthesis, and of the lpxC gene, coding for an enzyme involved in lipopolysaccharide synthesis, suggests that the repair mechanisms triggered by oxidative stress extend to extracytoplasmic structures.
Common themes in the responses to PQ and NaSal.
The list of genes activated by treatment of E. coli with PQ or NaSal (Table 3) is a first approximation to the common solutions to the physiological challenges posed by these two compounds. In addition to the common genes listed in Table 3, many other genes activated by the individual stresses have comparable functions. For example, the induction of genes involved in sugar transport occurs in both situations, albeit with specificity for different sugars. Exposure to both PQ and NaSal results in activation of the galactitol (gat) operon, while exposure to PQ activates the additional glucose and maltose transport genes (Table 1) and exposure to NaSal activates genes for the transport of sorbitol and mannose (Table 2).
At first sight, our results might appear to be inconsistent with those recently published by Barbosa and Levy (5), who also employed gene arrays from the same supplier. Despite this similarity, we detect only about one-third of the MarA-regulated genes that they proposed (5), while identifying 67 others. The actual disagreement may be less dramatic, however. Firstly, and noted earlier, gene arrays can miss large fractions of a coregulated group, as in the case of the heat shock regulon (37). Moreover, two methodological differences might have contributed to the observed discrepancies. Barbosa and Levy based their genomic analysis on a strain that expresses MarA constitutively, as opposed to our inducible MarA expression system. The constitutive expression of a gene regulator might provoke both direct and indirect gene modulation, thereby producing a different set of modulated genes. The statistical treatment of the data also differed between the two studies at several levels. Barbosa and Levy used an expression level cutoff equal to twice the average background level, while we set the threshold at the mean background plus 3 standard deviations. Barbosa and Levy also chose to treat both replicate experiments separately, admitting genes with the arbitrary expression ratio of at least 2 in one experiment and the same trend (≥1.2) in the other. We have averaged our two replicates and included only genes above a statistical cutoff of 2.5 standard deviations from the mean of the log ratios. In spite of these differences, both studies coincide on many novel genes, providing independent evidence for novel regulatory connections between MarA and its target genes. Finally, this example of independent studies addressing similar questions underscores the need for the availability of complete genomic data sets that can be compared beyond the particular statistical treatment selected by the original researchers.
From statistical significance to biological relevance.
Transcriptional profiling can measure changes in only the steady-state levels of mRNA, and the significance of these changes can be estimated by statistical analysis. However, the biological relevance of the detected variations in mRNA levels must be substantiated by genetic and biochemical analysis of the functions. We have constructed deletion mutants of the genes cysK and b2962 and measured the sensitivity of these strains to PQ and other oxidants. Preliminary results show that both deletion mutants are hypersensitive to PQ but not to H2O2 or to tert-butyl hydroperoxide (data not shown). These results suggest that even relatively small increases in gene expression, as in the cases of cysK and b2962, can point to genes with important roles in defense against oxidants.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant CA37831.
We thank R. Bennet for helping with computer programming and the members of G. Church's laboratory for helping with the use of the Storm PhosphorImager. S. Jelinsky was extremely helpful and generous with his knowledge of genomic databases. E. Lin, D. Fraenkel, and W. Wong read our manuscript and provided valuable suggestions.
P.J.P. and M.H.J.B contributed equally to this publication, and thus both should be considered first authors.
REFERENCES
- 1.Alekshun M N, Levy S B. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol. 1999;181:4669–4672. doi: 10.1128/jb.181.15.4669-4672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amabile-Cuevas C F, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arfin S M, Long A D, Ito E T, Tolleri L, Riehle M M, Paegle E S, Hatfield G W. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J Biol Chem. 2000;275:29672–29684. doi: 10.1074/jbc.M002247200. [DOI] [PubMed] [Google Scholar]
- 5.Barbosa T M, Levy S B. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol. 2000;182:3467–3474. doi: 10.1128/jb.182.12.3467-3474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennik M H, Pomposiello P J, Thorne D F, Demple B. Defining a rob regulon in Escherichia coli by using transposon mutagenesis. J Bacteriol. 2000;182:3794–3801. doi: 10.1128/jb.182.13.3794-3801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, Silva H, Klessig D F. Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science. 1993;262:1883–1886. doi: 10.1126/science.8266079. [DOI] [PubMed] [Google Scholar]
- 9.Chou J H, Greenberg J T, Demple B. Posttranscriptional repression of Escherichia coli OmpF protein in response to redox stress: positive control of the micF antisense RNA by the soxRS locus. J Bacteriol. 1993;175:1026–1031. doi: 10.1128/jb.175.4.1026-1031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen S P, McMurry L M, Levy S B. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding H, Demple B. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc Natl Acad Sci USA. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding H, Hidalgo E, Demple B. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J Biol Chem. 1996;271:33173–33175. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 13.Dubrac S, Touati D. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J Bacteriol. 2000;182:3802–3808. doi: 10.1128/jb.182.13.3802-3808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fawcett P, Eichenberger P, Losick R, Youngman P. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasch A P, Spellman P T, Kao C M, Carmel-Harel O, Eisen M B, Storz G, Botstein D, Brown P O. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci USA. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godon C, Lagniel G, Lee J, Buhler J M, Kieffer S, Perrot M, Boucherie H, Toledano M B, Labarre J. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- 17a.Greenberg, J. T. Ph.D. thesis. Harvard University, Cambridge, Mass.
- 18.Greenberg J T, Chou J H, Monach P A, Demple B. Activation of oxidative stress genes by mutations at the soxQ/cfxB/marA locus of Escherichia coli. J Bacteriol. 1991;173:4433–4439. doi: 10.1128/jb.173.14.4433-4439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg J T, Demple B. A global response induced in Escherichia coli by redox-cycling agents overlaps with that induced by peroxide stress J. Bacteriol. 1989;171:3933–3939. doi: 10.1128/jb.171.7.3933-3939.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg J T, Monach P, Chou J H, Josephy P D, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillon J M, Mechulam Y, Schmitter J M, Blanquet S, Fayat G. Disruption of the gene for Met-tRNA(fMet) formyltransferase severely impairs growth of Escherichia coli. J Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengge-Aronis R. The general stress response in Escherichia coli. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 161–178. [Google Scholar]
- 23.Jelinsky S A, Samson L D. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao S M, Hassan H M. Biochemical characterization of a paraquat-tolerant mutant of Escherichia coli. J Biol Chem. 1985;260:10478–10481. [PubMed] [Google Scholar]
- 25.Laub M T, McAdams H H, Feldblyum T, Fraser C M, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 26.Liochev S I, Hausladen A, Beyer W F, Jr, Fridovich I. NADPH:ferredoxin oxidoreductase acts as a paraquat diaphorase and is a member of the soxRS regulon. Proc Natl Acad Sci USA. 1994;91:1328–1331. doi: 10.1073/pnas.91.4.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 28.Maneewannakul K, Levy S B. Identification for mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:1695–1698. doi: 10.1128/aac.40.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin R G, Gillette W K, Rhee S, Rosner J L. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol Microbiol. 1999;34:431–441. doi: 10.1046/j.1365-2958.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- 30.Martin R G, Jair K W, Wolf R E, Jr, Rosner J L. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J Bacteriol. 1996;178:2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin R G, Rosner J L. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Membrillo-Hernández J, Kim S O, Cook G M, Poole R K. Paraquat regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12 is SoxRS independent but modulated by ςs. J Bacteriol. 1997;179:3164–3170. doi: 10.1128/jb.179.10.3164-3170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller P F, Sulavik M C. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol Microbiol. 1996;21:441–448. doi: 10.1111/j.1365-2958.1996.tb02553.x. [DOI] [PubMed] [Google Scholar]
- 34.Nunoshiba T, Hidalgo E, Li Z, Demple B. Negative autoregulation by the Escherichia coli SoxS protein: a dampening mechanism for the soxRS redox stress response. J Bacteriol. 1993;175:7492–7494. doi: 10.1128/jb.175.22.7492-7494.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pomposiello P J, Demple B. Identification of SoxS-regulated genes in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:23–29. doi: 10.1128/jb.182.1.23-29.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pomposiello P J, Demple B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 2001;19:109–114. doi: 10.1016/s0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 37.Richmond C S, Glasner J D, Mau R, Jin H, Blattner F R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riley M. Genes and proteins of Escherichia coli K-12. Nucleic Acids Res. 1998;26:54. doi: 10.1093/nar/26.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seoane A S, Levy S B. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J Bacteriol. 1995;177:3414–3419. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sies H. Oxidative stress: introduction, p. xv–xxii. In: Sies H, editor. Oxidative stress: oxidants and antioxidants. London, England: Academic Press; 1991. [Google Scholar]
- 41.Slonczewski J L, Foster J W. pH-regulated genes and survival at extreme pH. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1539–1549. [Google Scholar]
- 42.Storz G, Christman M F, Sies H, Ames B N. Spontaneous mutagenesis and oxidative damage to DNA in Salmonella typhimurium. Proc Natl Acad Sci USA. 1987;84:8917–8921. doi: 10.1073/pnas.84.24.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storz G, Tartaglia L A, Ames B N. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990;248:189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 44.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsaneva I R, Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wassarman K M, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 47.Wassarman K M, Zhang A, Storz G. Small RNAs in Escherichia coli. Trends Microbiol. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- 48.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng M, Doan B, Schneider T D, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]