Abstract
Antibiotic resistance is a major cause of treatment failure and leads to increased use of broad spectrum agents, which begets further resistance. This vicious cycle is epitomized by uncomplicated urinary tract infection (UTI), which affects 1 in 2 women during their life and is associated with increasing antibiotic resistance and high rates of prescription for broad spectrum second-line agents. To address this, we developed machine learning models to predict antibiotic susceptibility using electronic health record data, and built a decision algorithm for recommending the narrowest possible antibiotic to which a specimen is susceptible. When applied to a test cohort of 3,629 patients presenting between 2014 and 2016, the algorithm achieved a 67% reduction in the use of second-line antibiotics relative to clinicians. At the same time, it reduced inappropriate antibiotic therapy, defined as the choice of a treatment to which a specimen is resistant, by 18% relative to clinicians. For specimens where clinicians chose a second-line drug but the algorithm chose a first-line drug, 92% (1066/1157) of decisions ended up being susceptible to the first-line drug. When clinicians chose an inappropriate first-line drug, the algorithm chose an appropriate first-line drug 47% (183/392) of the time. Our machine learning decision algorithm provides antibiotic stewardship for a common infectious syndrome by maximizing reductions in broad spectrum antibiotic use while maintaining optimal treatment outcomes. Further work is necessary to improve generalizability by training models in more diverse populations.
ONE SENTENCE SUMMARY
Machine learning models for predicting antibiotic resistance could reduce the use of broad spectrum antibiotics in urinary tract infection.
INTRODUCTION
Uncomplicated urinary tract infection (UTI) refers to bacterial infection of a structurally normal lower urinary tract in a healthy female. It is an extremely common diagnosis that affects more than 1 in 2 women in their lifetime(1) and accounts for over 13 million outpatient and emergency room visits(2). It is the third most common indication for antibiotic treatment in the United States(3) resulting in 4.7 million prescriptions annually(4). Fluoroquinolone antibiotics such as ciprofloxacin and levofloxacin are the most commonly prescribed antibiotic class for uncomplicated UTI, despite being second-line agents(4). This may reflect the impact of increasing antibiotic resistance(5–7), which leads clinicians to choose broad spectrum therapies in order to minimize the risk of treatment failure.
Reducing the unnecessary use of fluoroquinolones has been a target for antimicrobial stewardship programs due to the well documented risks of serious adverse events that include secondary infection with Clostridioides difficile(8), selection of multidrug resistant organisms(9), tendinopathies(10), and aortic dissection(11). National practice guidelines published by the Infectious Diseases Society of America (IDSA)(12) provide a treatment algorithm that avoids the use of fluoroquinolones for uncomplicated UTI, but adherence is low(13–15), partly because they are designed to be broadly applicable across many populations, leaving the task of personalizing treatment decisions to the clinician(16, 17). Thus, a data-driven clinical decision support tool to identify candidates for the first-line therapies nitrofurantoin and trimethoprim-sulfamethoxazole (TMP-SMX) could greatly reduce harm to patients by avoiding exposure to fluoroquinolones, while still maintaining optimal treatment outcomes.
Computer algorithms have been employed for clinical decision support in the management of infectious diseases since the 1970s(18). More recently, machine learning has been used to predict antibiotic resistance in bloodstream infections(19–21), UTI(22), and from pathogen genomic data(23). These approaches can provide new insights into clinical phenomena(24, 25) but have not yet been widely adopted due to their difficulty integrating into clinical workflows, lack of interpretability, and an absence of evidence proving their generalizability and their utility in actual clinical settings.
Here, we use the syndrome of uncomplicated UTI to propose a solution to the challenge of antibiotic prescription in the era of resistance. We applied machine learning to data in the electronic health record (EHR) to predict the probability of antibiotic resistance to first- and second-line therapies. We then developed a decision algorithm that translates probabilities into recommendations designed to select the antibiotic of the narrowest possible spectrum while still achieving clinical cure, and benchmarked its performance relative to clinicians and a best-case adaptation of the national practice guidelines. We structured our algorithm with the intention of its deployment as an interpretable and personalized decision support tool embedded in the EHR to provide robust antimicrobial stewardship for a common outpatient diagnosis. Future efforts will focus on increasing the diversity of our sample to ensure robust recommendations across diverse race/ethnicities and socioeconomic strata.
RESULTS
Design and evaluation of a machine learning decision algorithm
We conducted this study in three parts. The first part consisted of building machine learning models to predict the probability of non-susceptibility to antibiotics used to treat uncomplicated UTI. Models were trained on data from 10,053 patients (11,865 specimens) with uncomplicated UTI presenting between January 1 2007 to December 31 2013 to Massachusetts General Hospital and Brigham & Women’s Hospital. We then developed a decision algorithm that translated probabilities into susceptibility phenotypes and chose the treatment of the narrowest spectrum among those that were susceptible. Last, we retrospectively evaluated the performance of our algorithm versus clinicians on a test set consisting of 3,629 patients (3,941 specimens) with uncomplicated UTI who presented to the same hospitals between January 1 2014 to December 31 2016. We also compared performance against an adaptation of the national practice guidelines designed to allow clinicians the use of second line antibiotics in a given percentage of decisions. Figure 1 outlines the analytic protocol.
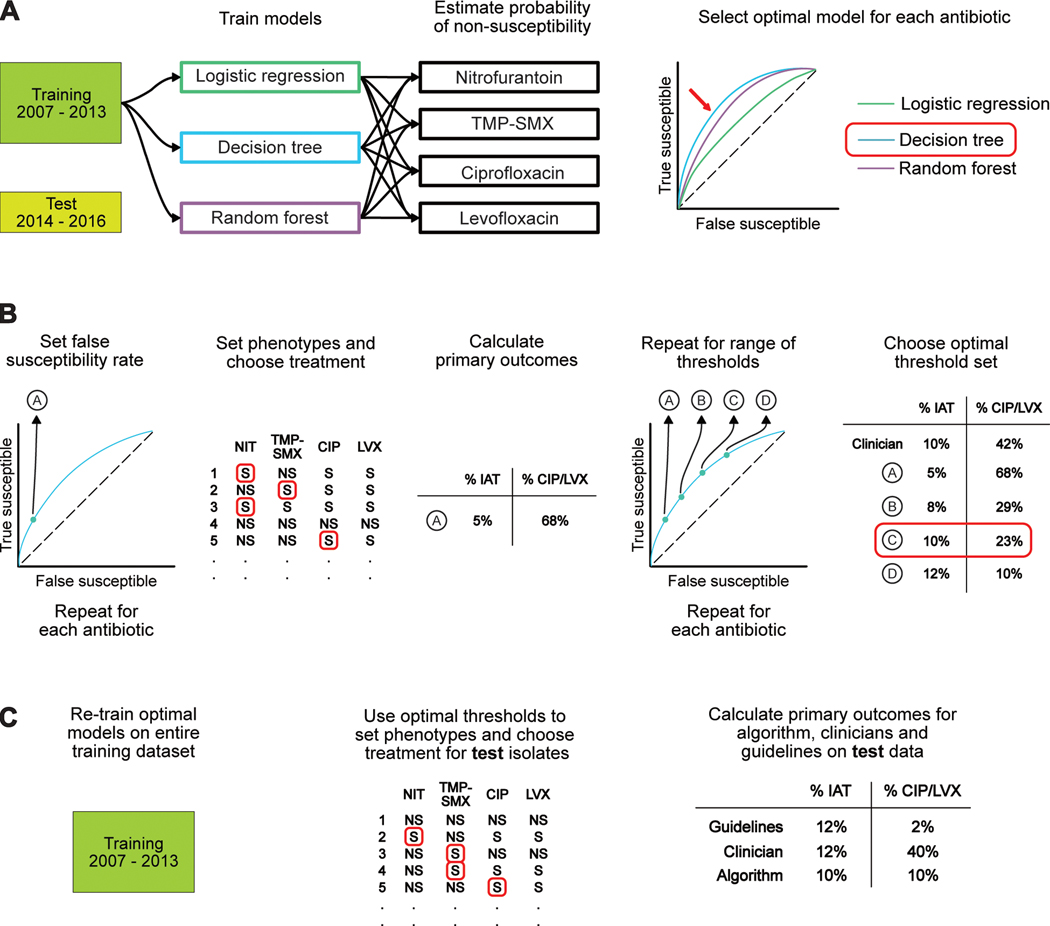
Figure 1: Schematic of analytic protocol.
(A) We trained decision tree, logistic regression, and random forest models to predict non-susceptibility to nitrofurantoin (NIT), TMP-SMX, ciprofloxacin (CIP), and levofloxacin (LVX). We selected the logistic regression models for use on our test cohort, which consisted of patients presenting between 2014 and 2016. (B) We set a false negativity rate and identified the corresponding probability threshold for a given antibiotic. Isolates with the predicted probabilities of non-susceptibility (NS) greater than this value were categorized as NS while those with probabilities below this threshold were categorized as ‘susceptible’ (S). This was repeated for all four antibiotics to yield a set of probability thresholds that could be used to bin predicted probabilities into phenotypes for each specimen. We then chose the antibiotic of the narrowest spectrum among those considered susceptible and calculated our two primary outcomes. This process was repeated for 1,331 sets of thresholds. The optimal threshold set was selected to meet a pre-specified target of minimizing IAT to the greatest extent possible while not exceeding a second-line antibiotic usage rate of 10%. (C) We evaluated our algorithm by retraining our chosen prediction models from part A on the entire training cohort and then performing prediction on the test cohort. Treatment decisions were made using the optimal threshold set from part B and the resulting primary outcomes were compared to the performance of clinicians and a best-case guideline-based policy.
Few patients with uncomplicated UTI have known risk factors for antibiotic resistance
Baseline characteristics for the combined, training and test cohorts are shown in Table 1. Mean age was 34 years (SD 10.9 years) and 64.2% of patients self-identified as white. Patients in the test set presented more frequently in the emergency room and had a higher prevalence of resistance to ciprofloxacin and levofloxacin. Test set patients were more likely to receive treatment with nitrofurantoin, reflecting a shift in prescribing patterns after the dissemination of the updated IDSA guidelines in 2010. Time trends for major features are located in fig. S1. The prevalence of resistance in our cohort to fluoroquinolones was lower than national estimates taken from a cross-sectional survey performed in 2012 (5.8% versus 10.6%) (5). Conversely, the prevalence of resistance to nitrofurantoin in our sample was higher (12.1% versus 3.3%). Among patients with antibiotic resistance, the majority had no observed risk factors for drug resistance (75.5%, 2,747 specimens in the training set; 80.5%, 1,000 specimens in the test set), defined as a prior resistant organism or antibiotic exposure in the previous 90 days.
Table 1:
Demographics, location of specimen collection, microbiologic and treatment characteristics for patients with uncomplicated UTI.
| Entire cohort (2007 – 2016) | Training set (2007 – 2013) | Test set (2014 – 2016) | P | |
|---|---|---|---|---|
| n (patients) | 13,682 | 10,053 | 3,629 | |
| n (specimens) | 15,806 | 11,865 | 3,941 | |
| Demographics | ||||
| Age, mean (SD) | 34.0 (10.9) | 34.1 (10.8) | 33.6 (11.1) | 0.007 |
| Race, n (%) | ||||
| White | 8,784 (64.2) | 6,497 (64.6) | 2,287 (63.0) | 0.083 |
| Non-white | 4,898 (35.8) | 3,556 (35.4) | 1,342 (37.0) | |
| Location, n (%) | ||||
| Outpatient | 11,639 (85.1) | 8,655 (86.1) | 2,984 (82.2) | <0.001 |
| Emergency room | 1,607 (11.7) | 1,074 (10.7) | 533 (14.7) | <0.001 |
| General inpatient | 534 (3.9) | 403 (4.0) | 131 (3.6) | 0.287 |
| Intensive care unit | 17 (0.1) | 13 (0.1) | 4 (0.1) | 0.580 |
| Organism, n (%) | ||||
| Escherichia coli | 11,901 (87.0) | 8,809 (87.6) | 3,092 (85.2) | <0.001 |
| Coagulase-negative Staphylococcus spp | 670 (4.9) | 448 (4.5) | 222 (6.1) | <0.001 |
| Klebsiella pneumoniae | 667 (4.9) | 503 (5.0) | 164 (4.5) | 0.246 |
| Enterococcus spp | 56 (0.4) | 28 (0.3) | 28 (0.8) | <0.001 |
| Staphylococcus aureus | 53 (0.4) | 32 (0.3) | 21 (0.6) | 0.028 |
| Other spp | 838 (6.1) | 628 (6.2) | 210 (5.8) | 0.322 |
| Current resistance, n (%) | ||||
| Nitrofurantoin | 1,654 (12.1) | 1,236 (12.3) | 418 (11.5) | 0.219 |
| TMP-SMX | 2,885 (21.1) | 2,147 (21.4) | 738 (20.3) | 0.196 |
| Ciprofloxacin | 792 (5.8) | 554 (5.5) | 238 (6.6) | 0.020 |
| Levofloxacin | 772 (5.6) | 533 (5.3) | 239 (6.6) | 0.004 |
| Prior resistance in the past 90 days, n (%) | ||||
| Nitrofurantoin | 98 (0.7) | 83 (0.8) | 15 (0.4) | 0.012 |
| TMP-SMX | 153 (1.1) | 153 (1.1) | 25 (0.7)) | 0.004 |
| Ciprofloxacin | 83 (0.6) | 65 (0.6) | 18 (0.5) | 0.260 |
| Levofloxacin | 85 (0.6) | 67 (0.7) | 18 (0.5) | 0.218 |
| Empiric therapy decision, n (%) | ||||
| Nitrofurantoin | 3,044 (22.2) | 1,745 (17.4) | 1,299 (35.8) | <0.001 |
| TMP-SMX | 5,676 (41.5) | 4,459 (44.4) | 1,217 (33.5) | <0.001 |
| Ciprofloxacin | 5,478 (40.0) | 4,247 (42.2) | 1,231 (33.9) | <0.001 |
| Levofloxacin | 418 (3.1) | 377 (3.8) | 41 (1.1) | <0.001 |
Patients in the training set presented to Massachusetts General Hospital and Brigham and Women’s Hospital between 2007 and 2013 and those in the test set presented between 2014 and 2016. P-values are for differences between the training and test sets and were calculated using two-sample t-tests for normally distributed variables and a non-parametric randomization test derived from random permutations of the dataset labels for non-normally distributed variables.
Accuracy of prediction models for antibiotic susceptibility is influenced by prior antibiotic exposure and prior antibiotic resistance
For each patient in our cohort, we constructed a feature vector containing demographics, microbiology, antibiotic exposures, comorbidities, procedures, and basic laboratory values. We additionally constructed two population-level features: colonization pressure, which is the population-level prevalence of resistance in urine specimens to an antibiotic in the 90 days preceding specimen submission, and hospital-wide antibiotic consumption. Major features except for colonization pressure were summarized 7, 14, 30, 90, and 180 days prior to the date of collection for a urine specimen. Additional features were added to indicate the presence of antibiotic non-susceptibility or antibiotic exposure at any previous point in time. We excluded any data that would not be present at the time of an empiric treatment decision. We then trained logistic regression, decision tree, and random forest models to predict the probability of non-susceptibility, defined as the likelihood an isolate would be called ‘intermediate’ or ‘resistant’ by the clinical microbiology laboratory, to first- and second-line treatments. The models were trained on patients presenting between 2007 and 2013 and tested on patients presenting between 2014 and 2016.
Logistic regression was selected for prediction of non-susceptibility to all four antibiotics, based on validation performance and interpretability. In the held-out cohort, the area under the receiver operator curve (AUROC) for nitrofurantoin and TMP-SMX were 0.56 (95% CI, 0.53–0.59) and 0.59 (95% CI, 0.57–0.62), respectively. For ciprofloxacin and levofloxacin the AUROCs were identical at 0.64 (95% CI, 0.60–0.68). Limiting prediction to the subset of patients with prior antibiotic resistance or antibiotic exposure in the past 6 months improved all AUROCs but had the greatest impact for the fluoroquinolones (Table 2). Model hyperparameters, ROC curves and calibration plots are located in table S1, fig. S2 and S3, respectively.
Table 2:
AUROCs for prediction of antibiotic non-susceptibility in patients presenting with uncomplicated UTI between 2014 and 2016.
| AUROC (95% CI)a | ||
|---|---|---|
| Drugb | Full test cohort | Prior antibiotic resistance or exposure |
| Nitrofurantoin | 0.56 (0.53, 0.59) | 0.61 (0.55, 0.66) |
| TMP-SMX | 0.59 (0.57, 0.62) | 0.67 (0.64, 0.71) |
| Ciprofloxacin | 0.64 (0.60, 0.68) | 0.76 (0.71, 0.80) |
| Levofloxacin | 0.64 (0.60, 0.68) | 0.77 (0.71, 0.82) |
AUROC and 95% CI calculated using 1000 bootstrapped samples taken from the test set.
Logistic regression was chosen for prediction for all four antibiotics.
A decision algorithm is able to reduce use of second-line therapies relative to clinicians and matches a best-case implementation of the treatment guidelines
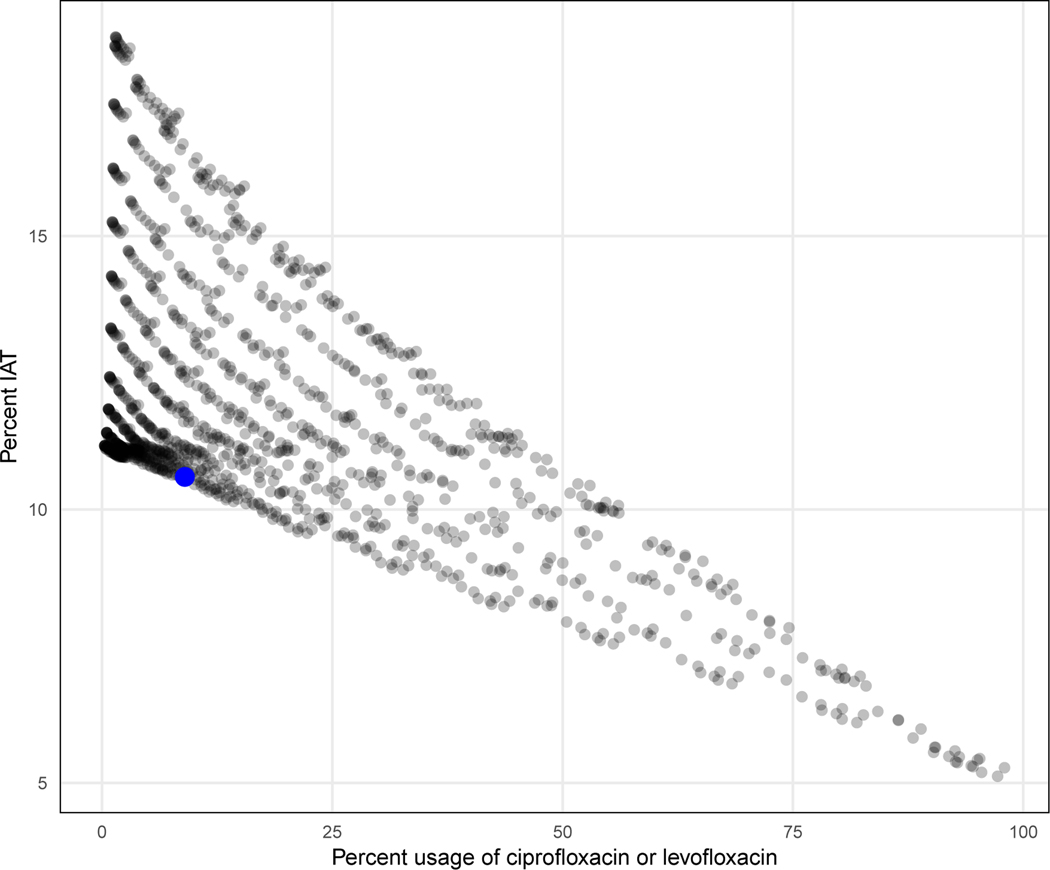
We next translated probabilities of non-susceptibility into phenotypes that fed into a decision algorithm designed to select the narrowest possible effective antibiotic. We achieved this by setting a threshold above which probabilities were phenotypically classified as ‘non-susceptible’ and below which they were phenotypically classified as ‘susceptible’. For each specimen, a set of four distinct thresholds was used on each of the four treatment choices and the algorithm subsequently recommended the antibiotic of narrowest spectrum among those predicted to be susceptible as the optimal treatment, making no decision if the specimen was predicted to be non-susceptible to all treatments. We then calculated our primary outcomes, which were the proportion of recommendations for second-line antibiotics and the proportion of recommendations that resulted in inappropriate antibiotic therapy (IAT), defined as the use of an antibiotic to which the organism has in vitro resistance. We repeated this analysis sequence for 1,331 threshold sets in total, and chose a final set that met a prespecified target of minimizing IAT while allowing second line usage in 10% of decisions in the validation set, which represented a realistic lower bound for clinicians in real-world settings (Fig. 2).
Figure 2: Threshold sensitivity analysis.
Primary outcomes for 1,331 unique threshold sets. The final threshold set (indicated by the blue dot) had the lowest IAT rate among the 1,331 threshold sets that had less than 10 percent usage of ciprofloxacin and levofloxacin on the validation set.
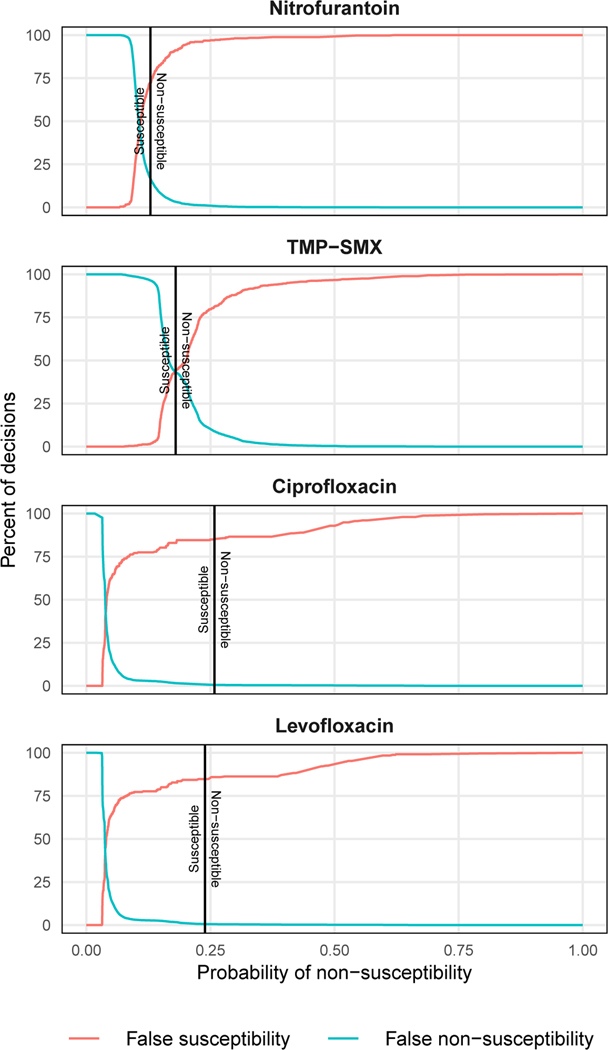
Using this process, we determined that the optimal thresholds for achieving our prespecified target were predicted probabilities of non-susceptibility of >13% for nitrofurantoin, >18% for TMP-SMX, >26% for ciprofloxacin, and >24% for levofloxacin (Fig. 3). A decision algorithm applied to the test data utilizing these thresholds was able to make a recommendation in 99% of specimens and chose ciprofloxacin or levofloxacin for 11.0% (95% CI 10.0% - 12.0%) of specimens. This was a 67% reduction in the selection of these antibiotics relative to clinicians (33.6%, 95% CI 32.1% - 35.0%). Algorithm recommendations resulted in IAT in 9.8% (95% CI 8.9% - 10.8%) of test specimens, an 18% reduction compared to clinicians (11.9%, 95% CI 10.9% - 12.9%). The algorithm had similar rates of IAT to a best-case implementation of the national treatment guidelines (10.7%, 95% CI 9.7% - 11.7%), where second line usage was capped at 10% (Table 3). See Retrospective Evaluation in Materials and Methods for a detailed description of the benchmark comparison.
Figure 3: False susceptibility and non-susceptibility rates.
Rates of false susceptibility and false non-susceptibility for prediction models derived from the training data. Final thresholds used on the test data are indicated by a black vertical line. Isolates with probabilities of susceptibility below the threshold were categorized as susceptible and those above were categorized as non-susceptible.
Table 3:
Comparison of primary outcomes for algorithm, clinicians and best-case guideline-based policy in patients presenting with uncomplicated UTI between 2014 and 2016.
| % (95% CI)a | |||
|---|---|---|---|
| Algorithm | Clinicians | Best-case guidelines | |
| Use of second-line therapyb | |||
| Recommendation cohortc (n =3911) | 10.8 (9.8–11.8) | 33.5 (32.1.−35.0) | 9.5 (8.6–10.4) |
| Full cohortd (n = 3941) | 11.0 (10.0–12.0) | 33.6 (32.1 – 35.0) | 9.7 (8.8–10.7) |
| Use of inappropriate antibiotic treatment | |||
| Recommendation cohortc (n = 3911) | 9.7 (8.8–10.6) | 11.8 (10.8–12.8) | 10.6 (9.6–11.5) |
| Full cohortd (n = 3941) | 9.8 (8.9–10.8) | 11.9 (10.9 – 12.9) | 10.7 (9.7–11.7) |
95% CI was calculated using a normal approximation to binomial distribution.
Second-line therapy refers to use of ciprofloxacin or levofloxacin.
Recommendation cohort refers to the 99% of specimens for which the algorithm made a treatment recommendation.
Full cohort refers to all specimens in the test cohort, including the 1% of patients where the algorithm was unable to make a recommendation. In these cases, the evaluation defaulted to the decision of the clinician.
The decision algorithm better differentiates non-susceptibility to first-line therapies relative to clinicians
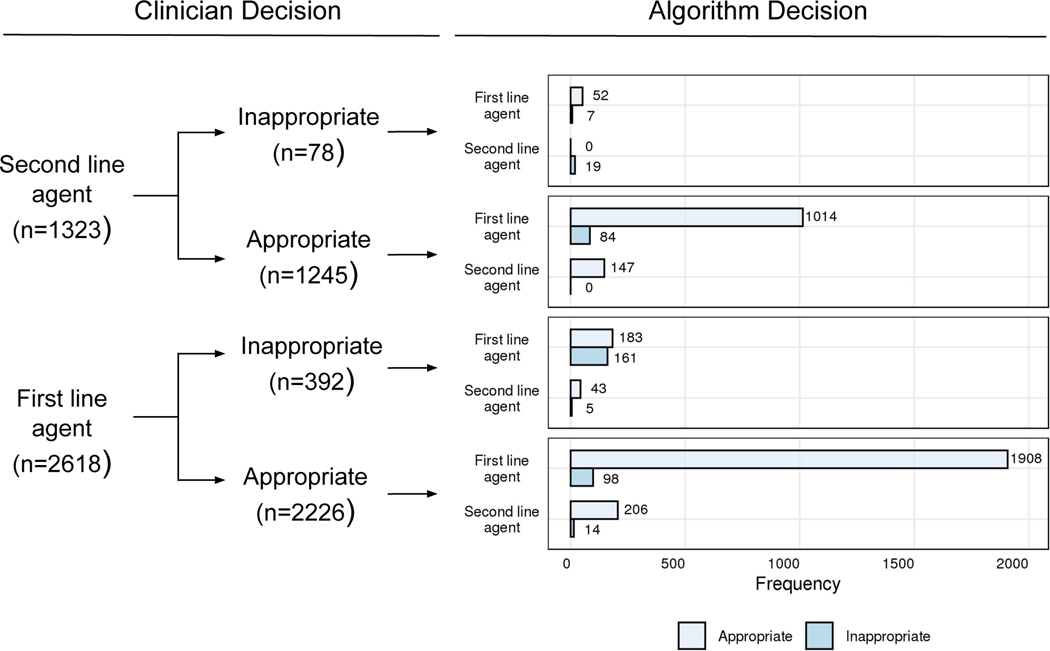
We sought to better understand the factors driving algorithmic and clinical decisions through a post-hoc analysis of the test results. The algorithm was able to discern non-susceptibility to first line agents better than clinicians. This difference was driven by the proportion of IAT with nitrofurantoin and TMP-SMX by clinicians (11.1% and 19.1%, respectively) relative to the algorithm (9.6% and 14.3%, respectively) (table S2). For cases where clinicians chose a second-line therapy but the algorithm chose a first-line agent, 92% (1066/1157) of decisions ended up being susceptible to the first-line agent. For cases where clinicians chose an inappropriate first-line therapy, the algorithm correctly chose the appropriate first-line agent 47% (183/392) of the time (Fig. 4). We performed a manual review of 18 randomly selected charts where the algorithm (but not the clinician) chose the proper first-line agent and found that 10 patients (56%) had no prior antibiotic resistance or exposure to first-line therapies, 1 patient (6%) had complicated UTI or pyelonephritis, and 2 patients (11%) had no clinical documentation. Using regularized logistic regression, we observed that the top 5 features predicting use of fluoroquinolones by clinicians were prior fluoroquinolone use, being of a white race, and being seen in the ER, suggesting that provider preferences rather than patient risk factors for resistance may be driving use. Being seen in an outpatient clinic and prior resistance to ciprofloxacin were negatively associated with fluoroquinolone prescription (table S3).
Figure 4: Post hoc analysis of clinician vs algorithm therapy decisions and appropriateness in patients with uncomplicated UTI presenting between 2014 and 2016.
Appropriate therapy was defined as the choice of an empiric antibiotic that has in vitro activity against the pathogen, whereas inappropriate therapy was defined as the choice of an empiric antibiotic that has no in vitro activity against the pathogen.
Algorithm recommendations would be actionable in actual clinical practice
As our model is not able to account for all factors used in treatment decisions, we anticipated that a percentage of recommendations would be ignored by clinicians due to contraindications. We sought to estimate that percentage and the reasons for contraindication through an additional manual review of 20 randomly selected charts. For the scenario where clinicians chose a second-line agent when the algorithm correctly recommended a first-line agent, 15/20 (75%) of recommendations were actionable and 3/20 (15%) were contraindicated due to suspicion of pyelonephritis or the presence of multiple infectious syndromes. The actionability of the algorithm could not be determined in 2/20 (10%) due to a lack of clinical documentation. Based on this chart review, we performed a conservative sensitivity analysis that assumed only 75% of algorithm recommendations would be actionable. Second-line use in the test set was 47% lower for the algorithm than for clinicians (17.8% versus 33.6%, respectively), while the proportion of IAT was nearly equal at 11.3% versus 11.8% for the algorithm and clinicians, respectively.
Antibiotic susceptibility to first-line therapies is influenced by prior resistance and antibiotic exposures
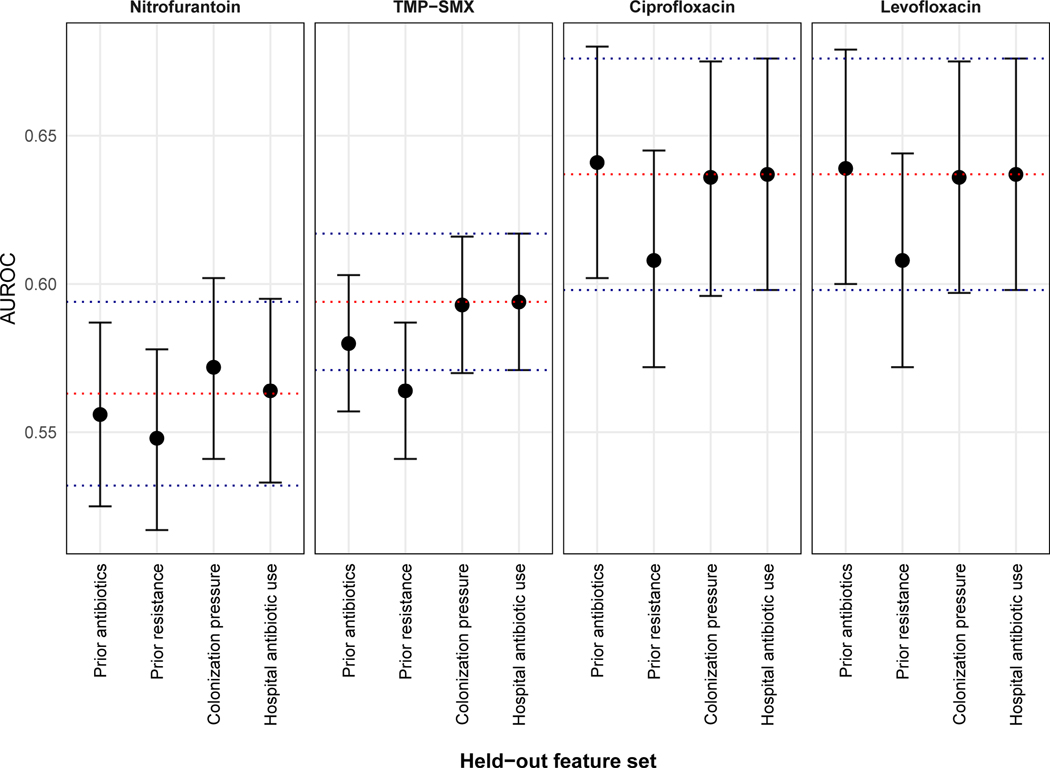
Lastly, we characterized the types of features predictive of antibiotic non-susceptibility in patients with uncomplicated UTI. We first grouped features into sets corresponding to risk factor domains known to be associated with resistance. We then inferred the importance of each feature group by estimating the decline in predictive performance when that set was left out of a regularized logistic regression model. Prior antibiotic resistance was the most important feature set predicting non-susceptibility to nitrofurantoin and fluoroquinolones, whereas both prior antibiotic exposures and resistance were most important for predicting non-susceptibility to TMP-SMX. None of the changes in AUROCs reached statistical significance (Fig. 5). A description of the ten most important individual features for predicting non-susceptibility to each antibiotic is located in table S4.
Figure 5: Feature importance characterization.
Data points represent the AUROC of logistic regression models that left out the feature set indicated on the x-axis. The red dotted line represents the AUROC for the full model, which contains all feature sets. The blue dotted lines represent the 95% CI for the full model. Error bars represent 95% CIs for the individual models.
DISCUSSION
In this study of patients presenting with uncomplicated UTI, we show how data-driven prescription strategies can help resolve the tension between maintaining optimal patient outcomes and reducing broad spectrum antibiotic use. Using only information passively collected in the electronic health record, our decision algorithm was able to reduce prescription of second-line agents by 67% while at the same time, also reduce inappropriate antibiotic treatment by 18% relative to clinicians. The implementation of this algorithm as a point-of-care clinical decision support tool could be a valuable addition to outpatient antimicrobial stewardship programs.
Machine learning applied to observational health data was used to predict antibiotic resistance in a large cohort of Israeli patients with UTI in a study by Yelin et al(22). Unlike the present study, which sought to balance the two competing objectives of reducing IAT and reducing broad spectrum antibiotic use, their algorithm had the single objective of reducing IAT. Their study also included males, pregnant females, and the elderly, further precluding a direct comparison. They achieved a retrospective reduction in IAT by 30% to 40% relative to clinicians by always selecting the antibiotic with the highest probability of susceptibility. However, since broad spectrum drugs have the lowest rates of resistance, the final model had a high rate of selection for fluoroquinolones. In contrast, our work focuses on using machine learning to fill an unmet need for antimicrobial stewardship. Therefore, our goal was to penalize the use of fluoroquinolones and evaluate model utility under conditions that mimic a real-world clinical scenario to the greatest extent possible. This motivated our use of strict inclusion criteria for uncomplicated UTI as it was essential to performing a fair evaluation that accounts for factors driving clinical decisions.
Across the United States, fluoroquinolones are prescribed in 42% of treatment decisions in uncomplicated UTI(4). This is a much higher proportion than what would be expected based on criteria set forth in the IDSA treatment guidelines. While the guidelines are intended to be a tool to reduce unnecessary use of fluoroquinolones, adherence has been poor. A major reason is their lack of personalization to the patient(13, 15) and because there is significant variability in tolerating treatment failure between physicians, regardless of prior risk of resistance(26). We noted that one in three patients in the test cohort were prescribed a fluoroquinolone and the primary drivers for this choice were presentation in the emergency room, white race and prior treatment with fluoroquinolones. One possible explanation for this is a lower tolerance for the risk of IAT among clinicians practicing in certain clinics or encountering specific patient populations. Our algorithm achieved antimicrobial stewardship targets that would be expected under a best-case scenario where guideline adherence leads to a second line usage of just 10%. Unlike the guidelines, it is able to do this in a manner that provides interpretable recommendations derived from models trained on data from the local population. Further research is necessary to determine whether these aspects would be sufficient to influence prescribing practices in settings where guidelines have been ineffective and tolerance for treatment failure is low.
From model conception through execution and evaluation, our intent was to promote generalizability by mimicking the clinical context in which we envisioned the algorithm would be deployed. This impacted our analysis in three ways. The first was our choice to eschew more sophisticated modeling approaches in favor of model classes that have greater interpretability and computational tractability. Second, we elected to use a time-based train/test split despite the secular trends in our covariates, as this would best recapitulate the implementation of our algorithm in clinical practice. Last, our thresholding-based method to translate continuous probabilities into categorical decisions represents a value judgment to minimize the use of second-line antibiotics at the cost of inappropriate therapy in a subset of patients. We believe this approach can be easily translated to identify empiric treatments for other infectious syndromes such as hospital acquired pneumonia and bloodstream infection by simply adjusting the tradeoff between the two outcomes.
There remain several outstanding questions regarding generalizability. First, as our cohort consisted of mostly Caucasians, it is possible that predictions will be biased when applied to more diverse populations. We have tried to minimize this by using nationally adopted criteria for uncomplicated UTI. Second, given that antibiotic resistance is an important predictor, we expect that the model would predict non-susceptibility more often in environments where the prevalence of antibiotic resistance is higher than what is seen in our training data. This is most pertinent for nitrofurantoin and TMP-SMX as our decision algorithm is heavily weighted to favor first-line agents. Increases in the risk of resistance to the fluoroquinolones may also indirectly negatively impact model performance because it is likely that settings with a high prevalence of resistance to fluoroquinolones also have a high prevalence of resistance to first line agents. The impact of antibiotic exposure and prior resistance on current antibiotic resistance is well established but quantifying the impact of each has been challenging (27). In this study of healthy patients with uncomplicated UTI, confounding by indication is unlikely to be an issue and we have assessed temporality by using longitudinal data.
In 25% of decisions, algorithm recommendations were non-actionable due to contraindications that would be known to clinicians but not to the model. Even in a worst-case scenario where we ignore 25% of recommendations, the algorithm still maintained a rate of IAT that was no worse than clinicians, and maintained a 47% reduction in second-line agent use. We anticipate that in clinical practice, the majority of non-actionable recommendations will be due to triggering of the decision support tool in patients with pyelonephritis and a minority due to allergies or antibiotic intolerance. Only 3% of patients are estimated to have an allergy or intolerance to TMP-SMX(28, 29), although that may be an underestimate given that documentation of allergies in medical charts is poor. We suggest that implementation of the algorithm be accompanied by a means for clinicians to provide feedback on contraindications when rejecting recommendations, thereby providing a mechanism for reducing inappropriate deployment. Although not impacting predictive performance, future work could also incorporate urinalysis results to further restrict deployment for only those patients with pyuria.
In summary, we have developed a decision algorithm for reducing unnecessary broad spectrum empiric treatment for patients with uncomplicated UTI. Further work is necessary to develop the algorithm into a clinical decision support tool that integrates seamlessly into clinical workflows and draws from a continually re-trained machine learning model to provide interpretable recommendations with measures of uncertainty in real-time. A randomized controlled trial designed to prove the efficacy of such a tool for antimicrobial stewardship is critical for adoption into routine practice.
MATERIALS AND METHODS
Study design
This study was designed as a retrospective analysis of 13,682 patients (15,806 specimens) with uncomplicated UTI who presented between 2007 and 2016 to the Massachusetts General Hospital and the Brigham & Women’s Hospital in Boston, MA. The cohort was split into a training dataset with 10,053 patients (11,865 specimens) presenting between January 1 2007 to December 31 2013 and a test set consisting of 3,629 patients (3,941 specimens) who presented between January 1 2014 to December 31 2016. Uncomplicated UTI was defined as infection in the lower urinary tract of a non-pregnant female between the ages of 18 and 55 years with no abnormalities of the genitourinary tract and no major comorbidities. All patients provided urine cultures with an organism burden sufficient to warrant antibiotic susceptibility testing (>50,000 colony forming units / ml with at most 2 organisms present) and received empiric antibiotic treatment with one of the first-line agents, nitrofurantoin or TMP-SMX, or one of the second-line agents, ciprofloxacin or levofloxacin. The empiric treatment window was defined as 48 hours prior to and up to 24 hours after specimen collection. We excluded patients with pyelonephritis and did not predict for fosfomycin as only 3.4% of specimens underwent susceptibility testing. We excluded specimens that did not undergo susceptibility testing for all four target antibiotics and any specimens that were prescribed multiple antibiotics during the empiric treatment window, as we would not be able to make a clean comparison between our algorithm’s recommendation and clinicians. We also excluded from the test set any specimens from patients who also submitted specimens in the training set (4.4% of all specimens) to prevent data leakage. The selection of patients is shown in fig. S4. This study was approved by the Institutional Review Board of Massachusetts General Hospital with a waived requirement for informed consent.
Description of model features
Our data were derived from the Boston Infectious Diseases Cohort, a database of 271,827 patients who submitted a specimen to the microbiology laboratories of Massachusetts General Hospital and Brigham & Women’s Hospital between 2000 and 2016. For our prediction models, we extracted patient-level microbiology, demographics, antibiotic exposures, comorbidities(30), procedures, and basic laboratory values. Microbiologic data included the hospital location of specimen collection and antibiotic susceptibility profiles. Breakpoints were applied to the raw susceptibility data as defined in the 27th edition of the M100 document published by the Clinical and Laboratory Standards Institute(31) to provide uniform interpretations over the course of the study. Specimens coming from the same body site and growing the same organism within a 14-day period were dropped. Antibiotic exposures, prior resistance, prior organism, laboratories, comorbidities, and prior hospitalizations were summarized over the 7, 14, 30, 90, and 180 days before the date of a microbiologic specimen. Antibiotic exposures and prior resistance were also summarized across all available patient history to capture the 35% of patients in our training set with a history of resistance or medication outside of the preceding 180 days. We did not have access to dose or duration of antibiotic therapy, urinalysis results, drug allergies, or data for patient encounters occurring outside of our two centers.
We incorporated two population-level features into our models. The first was an adaptation of colonization pressure(32), which we defined as the prevalence of resistance among urinary specimens to an antibiotic over a predefined service area in the previous 90 days. We calculated this metric for three hierarchies of service areas, a) the ward (separate prevalence for each outpatient clinic or inpatient ward), b) the facility (separate prevalence for 5 categories: hospital, general inpatient ward, intensive care unit, emergency room, or outpatient) and c) overall (a single prevalence across both hospitals). The second incorporated feature was cumulative antibiotic usage across both hospitals normalized by total patient volume per quarter. A detailed description of features and the analytic protocol is located in table S5 and fig. S5.
Machine learning architecture
We trained logistic regression (LR), decision tree (DT), and random forest (RF) models to predict the probability a specimen would be called non-susceptible to nitrofurantoin, TMP-SMX, ciprofloxacin, or levofloxacin. The term non-susceptible included both intermediate and resistant phenotypes. Models were trained on data from patients who submitted urine specimens between January 1 2007 and December 31 2013 (the training set). Hyperparameters for each combination of model class (LR, DT, RF) and antibiotic were tuned by training on 70% of the training data and evaluating the AUROC on the remaining 30% of the training data, referred to as the ‘validation set’. For each model class we chose the hyperparameter set that produced the highest mean AUROC, which was generated by averaging across five 70/30 splits of the training data. Using these hyperparameters, we then trained each of the three model classes on 20 new 70/30 splits of the training data and evaluated the mean AUROC on the validation set. LR performed best for nitrofurantoin and TMP-SMX. While RF performed marginally better than LR for ciprofloxacin and levofloxacin, LR was chosen for all subsequent analyses for reasons of interpretability.
Decision algorithm
We next translated probabilities output by our predictive models into susceptibility phenotypes by performing a sensitivity analysis with various false negative rates (FNRs) for each antibiotic. In this context, a false negative corresponds to falsely predicting susceptibility, also referred to as a ‘very major error’. We utilized only logistic regression and excluded decision trees and random forests based on their poor validation set performance, as well as their relative lack of interpretability.
Using the 20 70% splits of the training data noted above, we set an FNR value and identified the probability of non-susceptibility that would result in that value. We then used this probability as a threshold and applied it to the corresponding validation dataset to bin probabilities into susceptible or non-susceptible phenotypes. A ‘threshold set’ was generated by repeating this process for each antibiotic. For each specimen, we next selected the antibiotic of narrowest spectrum (nitrofurantoin < TMP-SMX < ciprofloxacin < levofloxacin) among those that were considered susceptible as the final treatment recommendation. If no antibiotic was considered susceptible, the decision algorithm made no choice. Using this set of recommendations, we calculated our primary outcomes, the IAT and second-line antibiotic usage rates, in that particular validation dataset. For any specimens where the algorithm was unable to make a treatment recommendation, we defaulted to the decision made by the clinician at evaluation time.
Due to the extensive time it takes to evaluate the performance of each FNR combination and the high correlation between resistance to ciprofloxacin and levofloxacin, we constrained the search space to combinations in which both second-line antibiotics had the same FNR. In total, we calculated reductions in IAT and second-line usage over 11 FNR values (0.001, 0.015, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9), one model class (logistic regression) and 3 antibiotics (nitrofurantoin, TMP-SMX and the fluoroquinolones combined), yielding 1,331 different combinations. The optimal threshold set was defined to be the one which minimized the mean IAT rate while not exceeding a mean second line antibiotic usage rate of 10% on the validation set.
Retrospective evaluation
We estimated the performance of the decision algorithm on a held-out test set of patients presenting with uncomplicated UTI between 2014 and 2016. We retrained our best performing models with tuned hyperparameters on 100% of the training data, applied the optimal threshold set (identified through our sensitivity analysis on the training data) to recommend empiric antibiotic treatments, and then calculated primary outcomes over the entire test dataset. For specimens where the model was unable to make a recommendation, the evaluation defaulted to the choice of the clinician. As our primary benchmark, we compared algorithm performance in the test dataset to the empiric treatment decisions made by clinicians.
We also sought to compare our performance to a conservative interpretation of the IDSA guidelines that preferentially chose the first-line agent to which the patient did not have prior antibiotic exposure and resistance in the prior 90 days. It avoided TMP-SMX if local rates of resistance were ≥20% and chose a fluoroquinolone only if the patient had exposure or resistance to both first-line agents. In our dataset, local rates of resistance to TMP-SMX exceeded 20% every year, leading the guidelines to always favor nitrofurantoin over TMP-SMX. This implementation of the guidelines yielded a 3.2% rate of broad spectrum usage in our validation cohort, which we deemed to be an unrealistic benchmark compared to real-world antibiotic prescribing practices(3, 4). It also ignores drug allergies or intolerance and prior treatment failures. Thus, we adjusted guideline recommendations to use second line agents 10% of the time as this represents a more realistic target for antimicrobial stewardship programs. This was done by defaulting to broad spectrum therapy in 18% of decisions where the conservative guidelines, but not the clinicians, chose a first line agent. This adjusted guideline-based policy represents a best-case scenario for implementation of the guidelines in actual clinical practice.
We identified factors driving decisions for clinicians through a post-hoc analysis using regularized logistic regression and manual chart review. Models included all of the covariates in our prediction models and were fit using the entire test dataset.
Feature importance characterization
A secondary aim of our study was to identify the features predictive of non-susceptibility. Based on the known risk factors for antibiotic resistance, we grouped features into 4 mutually exclusive sets, a) prior antibiotic exposure, b) prior antibiotic resistance and organism, c) colonization pressure, and d) hospital-wide antibiotic consumption. To estimate the impact of a given feature set, we compared the predictive performance between a full model and one where that set was held out. All models were trained in the same fashion as the prediction models and all contained demographics, comorbidities, laboratory values and hospital encounters.
Statistical analyses
P-values for comparisons of patient characteristics between train and test sets were calculated using two sample t-tests when a Shapiro-Wilk test failed to reject normality of the test statistic at a significance level of 0.05. Otherwise, p-values were computed using a non-parametric randomization test using 100,000 random permutations of the dataset labels (i.e, train vs. test set) for each characteristic. We report p-values for descriptive purposes, but do not assess statistical significance of these comparisons. Means and standard deviations for training set AUROCs were obtained by averaging over 5 70/30 splits for hyperparameter tuning and over 20 70/30 splits for model selection. For each of the 1,331 threshold sets, we calculated means for primary outcomes over the same 20 70/30 splits generated for the training step. For the retrospective evaluation, we calculated mean AUROCs using 1000 bootstrapped samples drawn with replacement from the test set. Each sample contained the same number of specimens as the full test set. 95% CIs for the reported AUROCs were calculated as follows:
where and Stdev(AUROCb) are the mean and standard deviation of the AUROC across the 1000 bootstrapped samples, respectively, and is the quantile function of the normal distribution, approximately equal to 1.96. The 95% CIs for our primary outcomes in the test set were calculated using a normal approximation to the binomial distribution, also known as a Wald interval. Given the value of a primary outcome , the 95% CI was given by:
where n is the sample size of the entire test set. For 95% CIs where sample sizes were <20, we computed the Jeffreys interval. All analyses were performed using Python version 3.6 and R version 3.5.0 (R Project for Statistical Computing).
Supplementary Material
Figure S1: Secular trends for specimen location and empiric treatment choices
Figure S2: Test set ROCs
Figure S3: Calibration plots
Figure S4: Cohort selection
Figure S5: Detailed methods schematic
Table S1: Model hyperparameters
Table S2: Detailed breakdown of primary outcomes
Table S3: Features predicting use of fluoroquinolones
Table S4: Top 10 features for prediction of non-susceptibility
Table S5: Description of the Boston Infectious Diseases Cohort
Data file S1: Data for all graphs
ACKNOWLEDGEMENTS
The authors would like to thank Yonatan Grad and Michael Klompas for their close review of the manuscript.
FUNDING
This work was supported by a Massachusetts General Hospital – Massachusetts Institute of Technology Grand Challenges Award (S.K., M.O., S.B., H.Z), a Harvard Catalyst CMeRIT grant (S.K.), and a National Science Foundation CAREER award (S.B., D.S.).
S.K. receives grant support from PhAST Diagnostics. D.S. is paid consultant for ASAPP, Memorial Sloan Kettering Cancer Center, Curai, and GNS Healthcare. H.Z. is a part-time student researcher at Google Health.
Footnotes
COMPETING INTERESTS
All other authors declare that they have no competing interests.
OVERLINE: ANTIBIOTICS
DATA AND MATERIALS AVAILABILITY
All data associated with this study are in the paper or supplementary materials. The code necessary to reproduce the main analyses from this paper is available at https://doi.org/10.5281/zenodo.3497335.
REFERENCES
- 1.Schappert SM, Rechtsteiner EA, Ambulatory medical care utilization estimates for 2007, Vital Health Stat 13, 1–38 (2011). [PubMed] [Google Scholar]
- 2.Niska R, Bhuiya F, Xu J, National Hospital Ambulatory Medical Care Survey: 2007 emergency department summary, Natl Health Stat Report, 1–31 (2010). [PubMed] [Google Scholar]
- 3.Shapiro DJ, Hicks LA, Pavia AT, Hersh AL, Antibiotic prescribing for adults in ambulatory care in the USA, 2007–09, J Antimicrob Chemother 69, 234–240 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Kabbani S, Hersh AL, Shapiro DJ, Fleming-Dutra KE, Pavia AT, Hicks LA, Opportunities to Improve Fluoroquinolone Prescribing in the United States for Adult Ambulatory Care Visits, Clin Infect Dis 67, 134–136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez GV, Babiker A, Master RN, Luu T, Mathur A, Bordon J, Antibiotic Resistance among Urinary Isolates from Female Outpatients in the United States in 2003 and 2012, Antimicrob. Agents Chemother 60, 2680–2683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez GV, Master RN, Karlowsky JA, Bordon JM, In vitro antimicrobial resistance of urinary Escherichia coli isolates among U.S. outpatients from 2000 to 2010, Antimicrob. Agents Chemother 56, 2181–2183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson L, Sabel A, Burman WJ, Everhart RM, Rome M, MacKenzie TD, Rozwadowski J, Mehler PS, Price CS, Emergence of fluoroquinolone resistance in outpatient urinary Escherichia coli isolates, Am J Med 121, 876–884 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Dingle KE, Didelot X, Quan TP, Effects of control interventions on Clostridium difficile infection in England: an observational study, Lancet Infectious Diseases 17, 411–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couderc C, Jolivet S, Thiébaut ACM, Ligier C, Remy L, Alvarez A-S, Lawrence C, Salomon J, Herrmann J-L, Guillemot D, Antibiotic Use and Staphylococcus aureus Resistant to Antibiotics (ASAR) Study Group, Fluoroquinolone use is a risk factor for methicillin-resistant Staphylococcus aureus acquisition in long-term care facilities: a nested case-case-control study, Clin Infect Dis 59, 206–215 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Stephenson AL, Wu W, Cortes D, Rochon PA, Tendon Injury and Fluoroquinolone Use: A Systematic Review, Drug Saf 36, 709–721 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Lee C-C, Lee M-TG, Chen Y-S, Lee S-H, Chen Y-S, Chen S-C, Chang S-C, Risk of Aortic Dissection and Aortic Aneurysm in Patients Taking Oral Fluoroquinolone, JAMA Intern Med 175, 1839–1847 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, Moran GJ, Nicolle LE, Raz R, Schaeffer AJ, Soper DE, Infectious Diseases Society of America, European Society for Microbiology and Infectious Diseases, International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis 52, e103–20 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Grigoryan L, Zoorob R, Wang H, Trautner BW, Low Concordance With Guidelines for Treatment of Acute Cystitis in Primary Care, Open Forum Infectious Diseases 2, ofv159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taur Y, Smith MA, Adherence to the Infectious Diseases Society of America guidelines in the treatment of uncomplicated urinary tract infection, Clin Infect Dis 44, 769–774 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Durkin MJ, Keller M, Butler AM, Kwon JH, Dubberke ER, Miller AC, Polgreen PM, Olsen MA, An Assessment of Inappropriate Antibiotic Use and Guideline Adherence for Uncomplicated Urinary Tract Infections, Open Forum Infectious Diseases 5, ofy198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livorsi D, Comer AR, Matthias MS, Perencevich EN, Bair MJ, Barriers to guideline-concordant antibiotic use among inpatient physicians: A case vignette qualitative study, J. Hosp. Med 11, 174–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez GV, Roberts RM, Albert AP, Johnson DD, Hicks LA, Effects of knowledge, attitudes, and practices of primary care providers on antibiotic selection, United States, Emerg. Infect. Dis 20, 2041–2047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchanan BG, Shortliffe EH, Rule-based expert systems (Addison Wesley Publishing Company, 1984). [Google Scholar]
- 19.Oonsivilai M, Mo Y, Luangasanatip N, Lubell Y, Miliya T, Tan P, Loeuk L, Turner P, Cooper BS, Using machine learning to guide targeted and locally-tailored empiric antibiotic prescribing in a children’s hospital in Cambodia, Wellcome Open Res 3, 131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman KE, Lessler J, Cosgrove SE, Harris AD, Lautenbach E, Han JH, Milstone AM, Massey CJ, Tamma PD, A Clinical Decision Tree to Predict Whether a Bacteremic Patient Is Infected With an Extended-Spectrum β-Lactamase–Producing Organism, Clin Infect Dis 63, 896–903 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan T, Ichikawa O, Dudley J, Li L, Aberg J, The Rapid Prediction of Carbapenem Resistance in Patients With Klebsiella pneumoniae Bacteremia Using Electronic Medical Record Data, Open Forum Infectious Diseases 5, ofy091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yelin I, Snitser O, Novich G, Katz R, Tal O, Parizade M, Chodick G, Koren G, Shalev V, Kishony R, Personal clinical history predicts antibiotic resistance of urinary tract infections, Nat Med 25, 1–23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen M, Long SW, McDermott PF, Olsen RJ, Olson R, Stevens RL, Tyson GH, Zhao S, Davis JJ, Using Machine Learning To Predict Antimicrobial MICs and Associated Genomic Features for Nontyphoidal Salmonella, J Clin Micro 57, e0120–18 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghassemi M, Celi LA, Stone DJ, State of the art review: the data revolution in critical care, Crit Care 19, 118–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiens J, Shenoy ES, Machine Learning for Healthcare: On the Verge of a Major Shift in Healthcare Epidemiology, Clin Infect Dis 66, 149–153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cressman AM, MacFadden DR, Verma AA, Razak F, Daneman N, Empiric Antibiotic Treatment Thresholds for Serious Bacterial Infections: A Scenario-Based Survey Study, Clin Infect Dis 69, 930–937 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Schechner V, Temkin E, Harbath S, Carmeli Y, Schwaber MJ, Epidemiological interpretation of studies examining the effect of antibiotic usage on resistance. Clin Microbiol Rev. 26, 289–307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macy E, Poon K-Y T, Self-reported antibiotic allergy incidence and prevalence: age and sex effects, Am J Med 122, 778.e1–7 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Jick H, Adverse reactions to trimethoprim-sulfamethoxazole in hospitalized patients, Rev Infect Dis 4, 426–428 (1982). [DOI] [PubMed] [Google Scholar]
- 30.Elixhauser A, Steiner C, Harris DR, Coffey RM, Comorbidity measures for use with administrative data, Med Care 36, 8–27 (1998). [DOI] [PubMed] [Google Scholar]
- 31.CLSI, M100 – 29th edition - 2019 (Clinical and Laboratory Standards Institute, 2018), pp. 1–320. [Google Scholar]
- 32.Bonten MJ, Slaughter S, Ambergen AW, Hayden MK, van Voorhis J, Nathan C, Weinstein RA, The role of “colonization pressure” in the spread of vancomycin-resistant enterococci: an important infection control variable, Arch Intern Med 158, 1127–1132 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Secular trends for specimen location and empiric treatment choices
Figure S2: Test set ROCs
Figure S3: Calibration plots
Figure S4: Cohort selection
Figure S5: Detailed methods schematic
Table S1: Model hyperparameters
Table S2: Detailed breakdown of primary outcomes
Table S3: Features predicting use of fluoroquinolones
Table S4: Top 10 features for prediction of non-susceptibility
Table S5: Description of the Boston Infectious Diseases Cohort
Data file S1: Data for all graphs
Data Availability Statement
All data associated with this study are in the paper or supplementary materials. The code necessary to reproduce the main analyses from this paper is available at https://doi.org/10.5281/zenodo.3497335.