Abstract
Monkeypox is a viral zoonotic disease that is caused by the monkeypox virus (MPXV) and is mainly transmitted to human through close contact with an infected person, animal, or fomites which is contaminated by the virus. In the present research work, reverse vaccinology and several other bioinformatics and immunoinformatics tools were utilized to design multi-epitopes-based vaccine against MPXV by exploring three probable antigenic extracellular proteins: cupin domain-containing protein, ABC transporter ATP-binding protein and DUF192 domain-containing protein. Both cellular and humoral immunity induction were the main concerning qualities of the vaccine construct, hence from selected proteins both B and T-cells epitopes were predicted. Antigenicity, allergenicity, toxicity, and water solubility of the predicted epitopes were assessed and only probable antigenic, non-allergic, non-toxic and good water-soluble epitopes were used in the multi-epitopes vaccine construct. The developed vaccine was found to be potentially effective against MPXV and to be highly immunogenic, cytokine-producing, antigenic, non-toxic, non-allergenic, and stable. Additionally, to increase stability and expression efficiency in the host E. coli, disulfide engineering, codon adaptation, and in silico cloning were employed. Molecular docking and other biophysical approaches were utilized to evaluate the binding mode and dynamic behavior of the vaccine construct with TLR-2, TLR-4, and TLR-8. The outcomes of the immune simulation demonstrated that both B and T cells responded more strongly to the vaccination component. The detailed in silico analysis concludes that the proposed vaccine will induce a strong immune response against MPXV infection, making it a promising target for additional experimental trials.
Communicated by Ramaswamy H. Sarma
Keywords: Monkeypox, molecular docking, molecular dynamic simulation, binding free energies calculation
1. Introduction
Monkeypox is a viral zoonotic disease with symptoms similar to those seen in previous cases of smallpox, though it is physiologically less severe. It is caused by the monkeypox virus (MPXV), a member of the Orthopoxvirus genus in the Poxviridae family. While humanity is still coping with the COVID-19 pandemic, a case of MPXV was reported to the WHO on May 7, 2022 (Bunge et al., 2022; Beer & Rao, 2019; Chastel, 2009; Ullah et al., 2021). Since the worldwide eradication of smallpox in 1977, much attention has been focused on monkeypox as a smallpox-like disease and potential bioterrorism agent because it is clinically almost identical to conventional smallpox. This virus drew a lot of attention when it first appeared in the Western Hemisphere in the spring of 2003, causing several cases in the United StatesMidwest. Despite this, there are approximately a hundred confirmed monkeypox cases in the United Kingdom, United States, and several other European countries as of June 4, 2022 (Weinstein et al., 2005; Cunha, 2004).
Monkeypox has a wide range of clinical symptoms, including fever, flu-like symptoms, back pain, malaise, distinctive rash, and headache (Weinstein et al., 2005). The MPXV cause smallpox like clinical symptoms in human such as fever, weight loss, lesion development, and death. The pathogenesis of MPXV is very similar to orthopoxviruses, like variola virus, and counters the response against this infectious disease. The exposure of non-human primates to MPXV may occur through inhalation of the aerosolized virus (Zaucha et al., 2001), nasal route (Saijo et al., 2006), tracheal route (Stittelaar et al., 2005; Stittelaar et al., 2006), and intravenous route. Special respiratory protective equipment and facilities have been demanding against aerosols transmission route whereas the earliest known route is intravenous inoculation which successfully asses the immunogenicity (Johnson et al., 2011; Hooper et al., 2004). Vaccination against smallpox is known to be effective against monkeypox virus infection in non-human primates (Hooper et al., 2004; Heraud et al., 2006; Control & C.f.D. & Prevention, 2001).
MPXV can be detected using real-time polymerase chain reaction (PCR) on samples collected via dry swabs of unroofed lesions or ulcers (Li et al., 2006; Li et al., 2010; Saijo et al., 2008). According to data, smallpox vaccination may have a protective effect against the MPXV and may ameliorate the clinical symptoms of the illness. The US Strategic National Stockpile (SNS) currently contains three smallpox vaccines: JYNNEOSTM (also known as IMVANEX, IMVAMUNE, MVA-BN), ACAM2000®, and the Aventis Pasteur Smallpox Vaccine (APSV), which might be used for smallpox under an IND (investigational new drug) protocol (Heymann et al., 1998; Hammarlund et al., 2005; Rizk et al., 2022). JYNNEOSTM is an attenuated, non-replicating orthopoxvirus vaccine created from the modified vaccinia Ankara-Bavarian Nordic (MVA-BN strain) (Petersen et al., 2022). According to historical data, smallpox vaccine with the vaccinia virus was about 85% effective against MPXV (Fine et al., 1988). The vaccine is licensed for smallpox in Europe as IMVANEX®, but the UK has been using it off-label to treat MPXV (Kupferschmidt, 2022). ACAM2000® contains live vaccinia virus as well. It was approved by the FDA in August 2007, replacing the previously licensed orthopoxvirus vaccine Dryvax®, whose maker had withdrawn it (Nalca & Zumbrun, 2010). During an outbreak, the CDC's emergency access IND protocol permits the use of ACAM2000® to treat non-variola orthopoxvirus infections (e.g. MPXV) (Rizk et al., 2022). When the licensed vaccines are not accessible or are not advised, APSV, a replication-competent vaccinia vaccine, may be used under an IND or Emergency Use Authorization (EUA). However, it is unknown if this vaccine can prevent MPXV (Rizk et al., 2022; Parrino & Graham, 2006).
The fundamental tenet of all vaccinations is that the vaccine can elicit an immune response more quickly than the virus can. Although animals immunized with traditional vaccines elicited powerful neutralizing and protective antibodies, these vaccines are allergic, costly, and time-consuming, and they involve the in vitro cultivation of dangerous viruses, raising significant safety concerns (Lo et al., 2013; Li et al., 2014). Hence, a safe and effective vaccine should be developed for the prevention of MPXV. In comparison to traditional vaccinations, peptide-based vaccine production is extremely safe and cost-effective. One approach that has been thought about in this regard is the use of epitope vaccines that use immunogenic epitopes specific to CD8+ and CD4+ cells and stimulate the immune system against these epitopes simultaneously and completely. Through the use of artificial peptide epitopes, the immune system can identify foreign invaders and respond to any viral or microbial contamination (Amer et al., 2018; Tahir Ul Qamar et al., 2020). A comprehensive map of virus epitopes and their immunogenicity is necessary to develop an MPXV virus disease vaccine. Furthermore, due to the presence of T cell and B cell epitopes, a multi-epitope vaccine significantly stimulates both humoral and cellular immune responses. Adjuvants are used in the construction of multi-epitope vaccines, and as a result, high immunogenicity and long-lasting immune responses are anticipated (Dey et al., 2022). Consequently, the goal of this study was to develop a multi-epitope vaccine from MPXV proteins using immunoinformatic and molecular docking studies.
2. Materials and methods
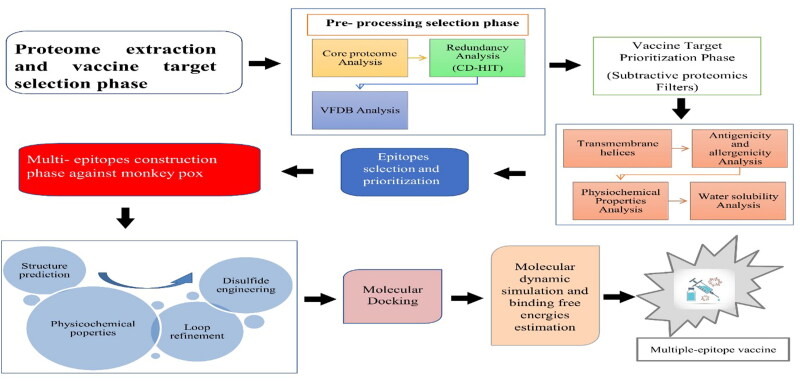
The overall methodology followed for the designing of multi-epitopes-based vaccine construct was described in the following Figure 1.
Figure 1.
Schematic representation of research methodology used herein.
2.1. Proteome extraction and vaccine target selection
This research work commenced with the retrieval of 39 fully sequenced genomes of monkeypox from NCBI Genbank (https://www.ncbi.nlm.nih.gov/genbank/) followed by core proteome analysis (Benson et al., 1993). In core proteome analysis, only core proteome was retrieved and processed for redundancy analysis in which redundant proteins were discarded and non-redundant proteins were selected using a CD-hit online webserver with a cut-off value of 0.8 (80%) (http://weizhong-lab.ucsd.edu/cd-hit/) (Huang et al., 2010; Ismail et al., 2021). Subsequently, antigenicity analysis was performed to determine the antigenicity of the sequences using VaxiJen 2.0 at the 0.4 threshold(http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) (Doytchinova & Flower, 2007). The ability to predict an antigenicity in a protein sequence is crucial because it tells us whether a viral protein sequence is likely to be recognized by immune cells in the human body. The antigenic sequences were further processed for physiochemical properties analysis using protparam expassy (https://web.expasy.org/protparam/) (Garg et al., 2016). Further, transmembrane helices were checked using TMHMM −2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (Krogh et al., 2001). Subcellular localization analysis was also done through CELLO2GO online webserver (http://cello.life.nctu.edu.tw/cello2go/) (Yu et al., 2014).
2.2. Epitopes selection phase
In the epitope selection phase, both B and T-cells epitopes were predicted using an online IEDB webserver (Vita et al., 2015). The B-cell epitopes with a score of 0.5 in the Bepipred linear epitope prediction method were selected. The T-cell epitopes both in MHC-I and MHC-II were opted based on the lowest percentile score. Similarly, a reference set of alleles were considered for predicting T-cell epitopes. Only a common set of epitopes were processed for additional analysis. The predicted epitopes were further checked for antigenicity, allergenicity, toxicity, and water solubility through vaxijen.2.0 (Doytchinova & Flower, 2007), AllerTOP v. 2.0 (Dimitrov et al., 2013), ToxinPred (Gupta et al., 2013), and INNOVAGEN online web servers, respectively and based on acceptable results, epitopes for multi-epitopes vaccine designing were selected.
2.3. Population coverage
Population coverage of predicted epitopes was conducted using the web-based population coverage analysis tool from Immune Epitope Database (IEDB) (http://tools.immuneepitope.org/tools/population/) (Bui et al., 2006; Dhanda et al., 2019). This tool provides human leukocytes antigens (HLA) allele frequencies of 78 population groups in 11 different geographical areas.
2.4. Vaccine construction and refinement
Epitopes are frequently non-immunogenic when used alone as a vaccine because of their small size (Li et al., 2014). They require a carrier that has strong immunostimulatory adjuvants to activate the innate and adaptive immune systems. Linkers are essential in replicating the vaccine construct’s ability to function as an independent immunogen and generate higher antibody concentrations than a single immunogen (Dong et al., 2020). The adjuvant was attached to the vaccine’s N terminal end using an EAAAK linker. The EAAAK linker promotes the separation of the domains of a bifunctional fusion protein, which is utilized to incorporate the first CTL epitope and the adjuvant. Linking two epitopes together is necessary for the epitope to function successfully. GPGPG linker was employed to integrate HTL and CTL epitopes, respectively, to efficiently detect epitopes inside the vaccine (Nezafat et al., 2014). Different studies have used these linkers and adjuvants for the designing of multi-epitope vaccine (Noor et al., 2022; Mahmood et al., 2021). The designed vaccine construct was subjected to the 3Dpro tool available in the SCRATCH suite for 3 D structure prediction (https://scratch.proteomics.ics.uci.edu/) (Cheng et al., 2005). 3Dpro is currently a de novo method. The amino acid sequence of the vaccine is given as input. The structure was further subjected to loops refinement using an online galaxy server (https://usegalaxy.org/) (Cheung & Sundram, 2017).
2.5. Structural and physiochemical properties
The structure of a vaccine’s potential to cause allergies and allergic reactions can be predicted by an assessment of its allergenicity. Hence, the AllerTop server was employed (https://www.ddg-pharmfac.net/AllerTOP/) (Garg et al., 2016). Using VaxiJen v2.0, structural antigenicity was evaluated (http://www.ddg-pharmfac.net/vaxijen/VaxiJen). The SOLpro server available in the SCRATCH suite was used to predict the solubility of the vaccine construct (https://scratch.proteomics.ics.uci.edu/) (Magnan et al., 2010). Stereochemical properties were predicted using the ProtParam server (https://web.expasy.org/protparam/) to check the nature and stability of the vaccine (Garg et al., 2016). SOPMA server was used to identify the secondary structure of the vaccine construct (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) (Geourjon & Deleage, 1995).
2.6. Molecular docking analysis
Molecular docking is a type of bioinformatics modeling that involves the interaction of two or more molecules to produce a stable adduct. It predicts the three-dimensional structure of any complex based on the binding characteristics of the ligand and target (Dar & Mir, 2017; Borkotoky et al., 2021; Rajkumari et al., 2018; Jain et al., 2021). To assess the binding potency of vaccine construct with immune cell receptors docking analysis was performed. Cluspro 2.0 online webserver (https://cluspro.bu.edu/login.php) was utilized for molecular docking (Desta et al., 2020). Toll-like receptors (TLR) play a key role in viral particle identification and innate immune system activation. TLRs could be a promising target for early disease control and vaccine development (Khanmohammadi & Rezaei, 2021). Different immune mediators toll-like receptors (TLR-2, TLR4- and TLR-8) were used as receptors molecules to determine the binding affinity of the vaccine construct with target immune cells receptors as it is crucial for immune cell activation. The three receptors are important in generating protectiveimmunity against the virus. Docked complexes having high binding affinities were further subjected to molecular dynamics simulations.
2.7. Molecular dynamic simulation and free binding energies estimation
Biophysical approach, a molecular dynamic simulation of 200 ns was performed to evaluate the dynamic behavior of the docked complexes (Ismail et al., 2021). Pre-processed and parameters generation was accomplished using an antechamber module of AMBER 20 (https://ambermd.org/AmberMD.php) (Salomon-Ferrer et al., 2013). Each docked complex was placed in a solvation box of 12 Å through a built-in Leap module. Protein force field Fd14SB was used to define both vaccine construct and receptors. Subsequently, Na + ions were added to the system to neutralize the charge density. Moreover, energy minimization, hydrogen atoms energy minimization for 500 steps, water molecules energy minimization for 1000 steps with restraining of 200 kcal/mol – Å2 on the remaining system, 1000 steps of energy minimization of all atoms exception to 5 kcal/mol – Å2 restrain on alpha carbon atoms and 300 steps energy minimization on non-heavy atoms with the restrain of 100 kcal/mol – Å2 on rest of the system, was performed during preprocessing. Systems were heated to 300 K (NVT ensemble), the temperature was maintained and hydrogen bond restriction was done using Langevin dynamics and SHAKE algorithm, respectively (Pastor et al., 1988; Kräutler et al., 2001). The complex systems were equilibrated for 1000ps and the system was compressed with an NPT ensemble constraining Cα atoms of 5 kcal/mol energy Å2. Lastly the production of 200 ns was executed and trajectory analysis was done using the CPPTRAJ module (Roe & Cheatham, 2013)
MMPBSA.py module of AMBER 20 was used to estimate the binding free energy and solvation free energies for the vaccine, receptors, and complexes. Estimation of binding energy for all three components was made either by Generalized Born (MMGBSA) or Poisson Boltzman (MMPBSA) equation (Genheden & Ryde, 2015). Mathematical interpretation of binding energy can be done as:
2.8. In silico cloning and codon optimization
The higher expression rates may be the result of a unique codon adaptation technique that was developed for E. coli K12, the most sequenced prokaryotic organism. The method was used in the host organism E. coli K12 to boost the expression of the subunit vaccination protein’s primary sequence, which was then sent to the JAVA Codon Adaptation Tool (http://www.prodoric.de/JCat) (Grote et al., 2005). RHO independent transcription termination, the prokaryote ribosome binding site, and the restriction enzyme cleavage site were all avoided. The altered nucleotide sequences of the created multi-subunit vaccine were further cloned into the E. coli pET-28a (+) vector between restriction sites using the restriction cloning module of the SnapGene program (https://www.snapgene.com/). SnapGene provides a simple and safe method for visualizing, planning, and documenting routine molecular biology processes. The software allows for the viewing of DNA sequences, sequence editing, sequence annotation, protein visualization, cloning, and simulation of typical cloning techniques, all with a simple user interface (Biotech, 2020).
2.9. Disulfide engineering
Disulfide engineering is a unique method of introducing disulfide bonds into the target protein structure. Disulfide bonds are covalent interactions that contribute to the enhancement of protein stability and the study of protein interactions and dynamics (Khatoon et al., 2017; Pandey et al., 2018). The vaccine structure was submitted to the Disulfide by Design (DbD) 2.12 tool (http://cptweb.cpt.wayne.edu/DbD2/) to increase stability (Craig & Dombkowski, 2013). A set of parameters (87 to + 97 chi3 value and 2.2 energy value) was used to select potential residue pairings, which were then mutated with a cysteine residue.
2.10. Immune simulation
Using the C-ImmSim server (https://150.146.2.1/C-IMMSIM/index.php), in silico immune simulations were carried out to assess the immunogenicity and immune response of the MPXV vaccine (Rapin et al., 2010). This immune trigger uses machine learning techniques and a position-specific scoring matrix (PSSM) to estimate immune interactivities and epitope prediction, respectively. The server’s prediction includes the simulation of three unique anatomical locations associated with immune responses in mammals: the thymus, the bone marrow, and a tertiary lymphatic organ such as a tonsil, lymph node, or spleen (Rapin et al., 2010).
Clinically, a month is the minimum amount of time that is advised between two doses of a vaccine (Jain et al., 2021). The immune simulation was carried out using the same protocol as that described in earlier studies (Chauhan & Singh, 2020; Tahir Ul Qamar et al., 2020). As one-time step is equivalent to 8 hours of daily life, 1, 84, and 168-time steps variables were prepared for a total of three inoculations, which were given with the recommended time intervals of 1 month. The default values for all other triggering parameters were maintained.
3. Results and discussion
3.1. Complete proteome extraction and selection of vaccine target
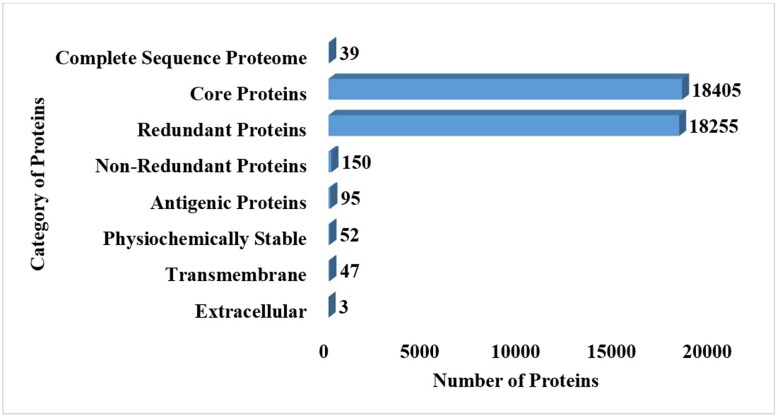
In the study, 39 fully sequence proteomes were extracted (Table S1), and these proteomes were subjected to core proteins analysis in which 18405 core proteins were predicted. Core proteins are mainly involved in several major structures and functions for example important for virion assembly, interaction between several host proteins (Maxwell & Frappier, 2007). Core proteins were further considered for redundancy analysis in which 18255 redundant and 150 non-redundant proteins were predicted. Redundant proteins are very highly similar to existing in the same species while the non-redundant proteins were present a single time in the complete proteome (Sikic & Carugo, 2010). Hence, non-redundant proteins were considered for further analysis. Among 150 non-redundant proteins, 95 proteins were found as probable antigenic with a threshold 0.4. Antigenic proteins are mostly involved in the pathogenicity of the pathogens and can induce the immune system properly. In previous studies, the researcher also considered antigenic proteins as good vaccine candidates and processed them for epitope prediction (Ullah et al., 2021). Further, the antigenic proteins were assessed for physiochemical properties analysis in which 43 proteins were found as unstable having an in-stability index greater than 40 (Guruprasad et al., 1990) . So these unstable proteins were discarded and 52 stable proteins were further examined for transmembrane helices. Too many helices in the structure can make the structure unstable (Noor et al., 2022). In 52 proteins 3 proteins were predicted to have more than one transmembrane helices so those 3 proteins were discarded and the remaining 47 proteins were processed for extracellular membrane analysis. In extracellular membrane analysis, only 3 proteins were predicted in extracellular region. Extracellular membrane proteins are present on the surface and can mostly interact with host immune cells as the interaction is vital for inducing an immune response. So these 3 extracellular proteins are considered a good candidates for vaccines. Extracellular and membrane proteins were preferred to choose epitopes and create a successful vaccine because of their role in pathogenic adhesion and virulence to host cells. The details of 3 selected proteins are described in Table 1. The overall number and categories of subtractive proteins are described in following Figure 2.
Table 1.
Final selected antigenic proteins with their transmembrane helices and subcellular localization.
| Name | Antigenicity | Transmembrane helices | Subcellular localization |
|---|---|---|---|
| Cupin domain-containing protein | 0.48 | 0 | Extracellular |
| ABC transporter ATP-binding protein | 0.63 | 0 | |
| DUF192 domain-containing protein | 0.62 | 0 |
Figure 2.
The number of proteins obtained at each step of subtractive proteomics.
3.2. Epitopes prediction and prioritization phase
Both B and T-cells epitopes were predicted from target proteins. B- cells epitopes were predicted to generate humoral immune response while for cellular immune responses, T-cells epitopes were predicted (Zheng et al., 2017). The predicted B-cells epitopes are tabulated in the following Table 2.
Table 2.
Predicted B-cells epitopes from three surface proteins to design MPXV vaccine.
| Proteins number/Accession number | Predicted B-cells epitopes |
|---|---|
| >core/2187/2/Org2_Gene165 (cupin domain-containing protein) (Accession ID: WP_038760017) |
SPQTSKKIGDD |
| SRNNT | |
| NYTKDKISYDSPYDD | |
| LTAGDA | |
| TNDTDKVDYE | |
| STPETISEKPEDIDNSNCSSVFEIATPEPITDNVEDHTDTVTYTSDSINTVNASSGESTTDETPEPITDKEEDHT VTDTVSYTTVSTSS | |
| TKSTTDDADLYDTYNDNDTVPPTTVGGSTTSISNYKTK | |
| KHPRKYKTE | |
| >core/85/1/Org1_Gene78 (ABC transporter ATP-binding protein) (Accession ID: WP_004184889) |
EQEANASAQT |
| FYIRQNHG | |
| SADADA | |
| LTPEQKAY | |
| TAALNIQTSVNTVVRDFENYVKQTCN | |
| CYGAPG | |
| KATTQIAPRQVAGTG | |
| KENVH | |
| YGNIKEFNATHAAFEYSKSIGGTPALDRRVQDVNDTISDVKQK | |
| >core/139/1/Org1_Gene141 (DUF192 domain-containing protein) (Accession ID: WP_038780458) |
AEVGPNNTISIRKFNTMRQC |
| TFSDVIN | |
| IAPNINNTE | |
| TFSDVIN |
In T-cells epitopes selection both MHC-I and MHC-II epitopes were predicted, and the predicted epitopes were prioritized on the base of least percentile score. MHC-I and MHC-II predicted epitopes with their least percentile score are tabulated in the following Table 3.
Table 3.
Predicted T-cells (MHC-I and MHC-II) epitopes from MPXV proteins with their corresponding percentile rank.
| MHC-I | Percentile rank | MHC-II | Percentile rank |
|---|---|---|---|
| NYTKDKISY | 0.86 | NYTKDKISYDSPYDD | 5.8 |
| ISYDSPYDD | 10 | SSVFEIATPEPITD | 0.95 |
| FEIATPEPI | 0.18 | ||
| DIDNSNCSSV | 3.4 | SEKPEDIDNSNCSSV | 36 |
| SEKPEDIDNS | 6.4 | ||
| VEDHTDTVTY | 0.15 | NVEDHTDTVTYTSDS | 30 |
| NTVNASSGE | 6.2 | INTVNASSGESTTDE | 8.2 |
| ASSGESTTDE | 14 | ||
| ITDKEEDHTV | 1.4 | PITDKEEDHTVTDTV | 27 |
| TVTDTVSYT | 0.52 | ||
| SYTTVSTSS | 6.7 | TVTDTVSYTTVSTSS | 14 |
| TTVGGSTTSI | 0.26 | PPTTVGGSTTSISN | 2.9 |
| DTYNDNDTV | 0.09 | DDADLYDTYNDNDTV | 17 |
| DDADLYDTY | 0.92 | TKSTTDDADLYDTY | 8.8 |
| STTDDADLY | 0.02 | TAALNIQTSVNTVV | 1.8 |
| AALNIQTSV | 0.2 | TVVRDFENYVKQTCN | 6.2 |
| TVVRDFENY | 0.06 | KATTQIAPRQVAGT | 12 |
| ENYVKQTCN | 29 | GNIKEFNATHAAFE | 1.9 |
| KATTQIAPR | 0.16 | YSKSIGGTPALDRRV | 12 |
| KEFNATHAAF | 0.09 | RVQDVNDTISDVKQK | 16 |
| KSIGGTPAL | 0.2 | AEVGPNNTISIRKF | 2.6 |
| DTISDVKQK | 0.03 | NTISIRKFNTMRQC | 2.7 |
3.3. Epitopes targeting phase
In epitopes targeting phase the predicted MHC-I and MHC-II epitopes were subjected for DRB*0101 binding, water-solubility, toxicity and allergenicity analysis, and good DRB*0101 binder, good water-soluble, non-toxic and non-allergic proteins were shortlisted for vaccine construction Table 4. Toxicity analysis was checked in order to remove toxic proteins, as the toxic proteins may cause toxic response in host.
Table 4.
Physicochemical properties of selected epitopes.
| Selected epitopes | DRB*0101, Predicted IC50 Value (nM) |
Water solubility | Toxicity | Allergenicity |
|---|---|---|---|---|
| NYTKDKISY | 10.26 | Good water soluble | Non-toxic | Non-allergic |
| ISYDSPYDD | 36.64 | |||
| KEFNATHAA | 55.85 | |||
| KSIGGTPAL | 19.82 |
3.4. Multi-epitopes vaccine construction and processing phase
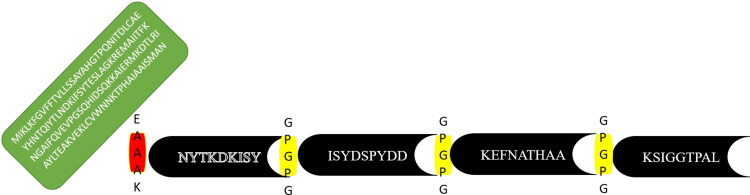
Humoral and cellular immune responses play important role in tackling the pathogens, as multi-epitopes based vaccine construct consist of several different types of B and T-cells epitopes which can induce both cellular and humoral immunity in host (Zhang, 2018). Multi-epitopes based vaccine construct was designed using the shortlisted epitopes, the epitopes were joined with each other via ‘GPGPG’ linkers and joined with cholera-toxin B subunit adjuvant through another EAAAK linker, as mentioned in Figure 3. The nontoxic cholera toxin B subunit is an effective adjuvant for DC-targeted vaccination. Humans can safely use the nontoxic cholera B subunit (CTB), which has the potential to activate CD4+ T cell responses (Antonio-Herrera et al., 2018).
Figure 3.
Schematic view of MPXV vaccine construct that shows 180 amino acids long and containing selected epitopes and adjuvant linked in the construct via EAAAK linker at N- terminal.
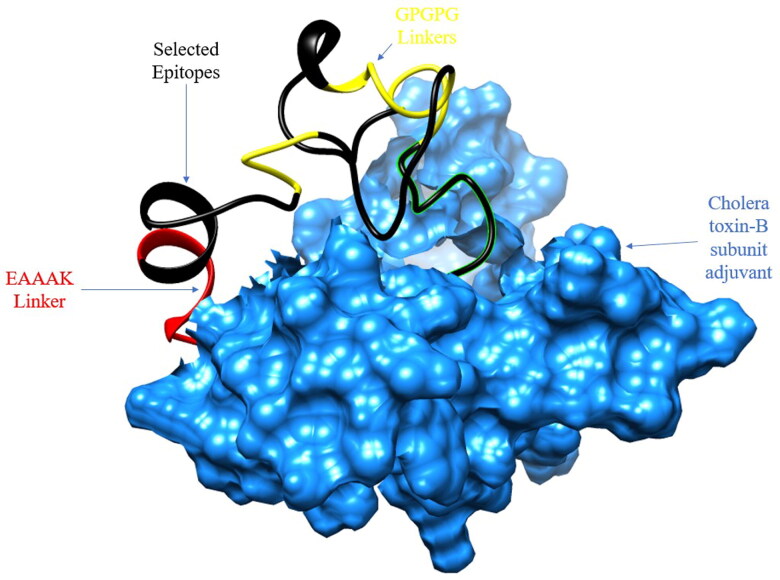
Three-dimensional structure of the designed vaccine construct was predicted by the 3DPro tool and is represented in the following Figure 4. To retain the structural stability, all the loops were refined using the galaxyweb server.
Figure 4.
3D structure of MPXV vaccine construct. Black color represents selected epitopes, red color represents EAAAK linker, yellow color represents GPGPG linkers and blue color represents adjuvant.
3.5. Population coverage analysis
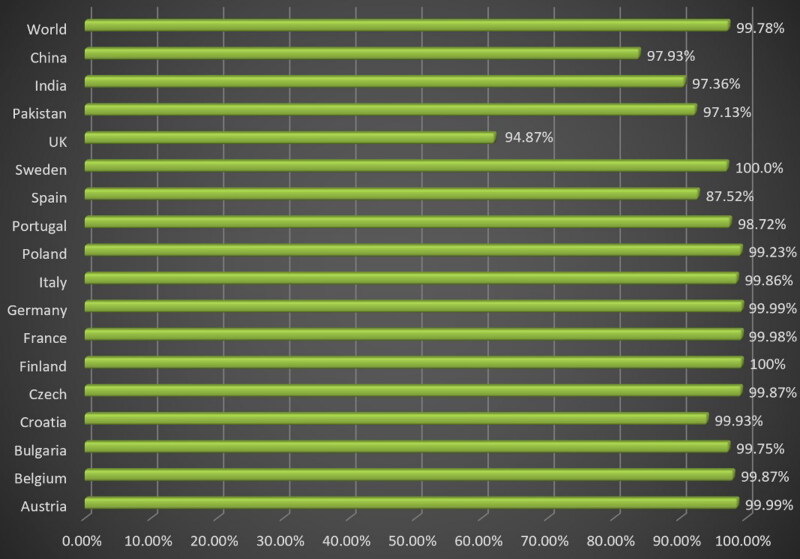
The Population coverage analysis was done on selected epitopes that have been used in designing of multi-epitopes vaccine constructs with their both MHC-I and MHC-II. The results predicted that the selected epitopes cover 99.78% world population followed by 97.93% Chinese population and 97.36% Indian population. Overall the different countries wise populations are mentioned in the following Figure 5.
Figure 5.
Population coverage map of MPXV vaccine covering 99.78% of the world population.
3.6. Physiochemical properties and water solubility analysis
Physiochemical properties analysis is important to deliver data for experimentalists to assist them in the experimental testing of the vaccine. Examining the stereo-chemical properties of the designed structure with the ProtParam program revealed that the vaccine structure has a molecular weight of 19450.09 Daltons and an isoelectric point of 8.43, indicating the basic nature of the designed vaccine structure. There are 17 negatively charged amino acids (aspartic acid, glutamic acid) and 19 positively charged amino acids (lysine, arginine). The aliphatic index is 72.78, and the instability of the designed structure is 31.76, indicating that the vaccine structure designed in the host is stable. The GRAVY index was −0.351, and it was reported that a negative value indicates that the vaccine structure is hydrophilic and could interact well with water molecules. This vaccine construct’s half-life was predicted to be 30 hours in mammals (in vivo), more than 20 hours in yeast (in vivo), and more than 10 hours in E. coli (in vivo). Based on the SOLpro server results, the designed vaccine structure was predicted soluble with a probability of 0.9, ensuring easy access to the host. Vaccine structures were also predicted to be non-toxic, non-allergenic, and antigenic. According to AntigenPro, the result of its antigenicity was 0.890876.
3.7. Molecular docking analysis
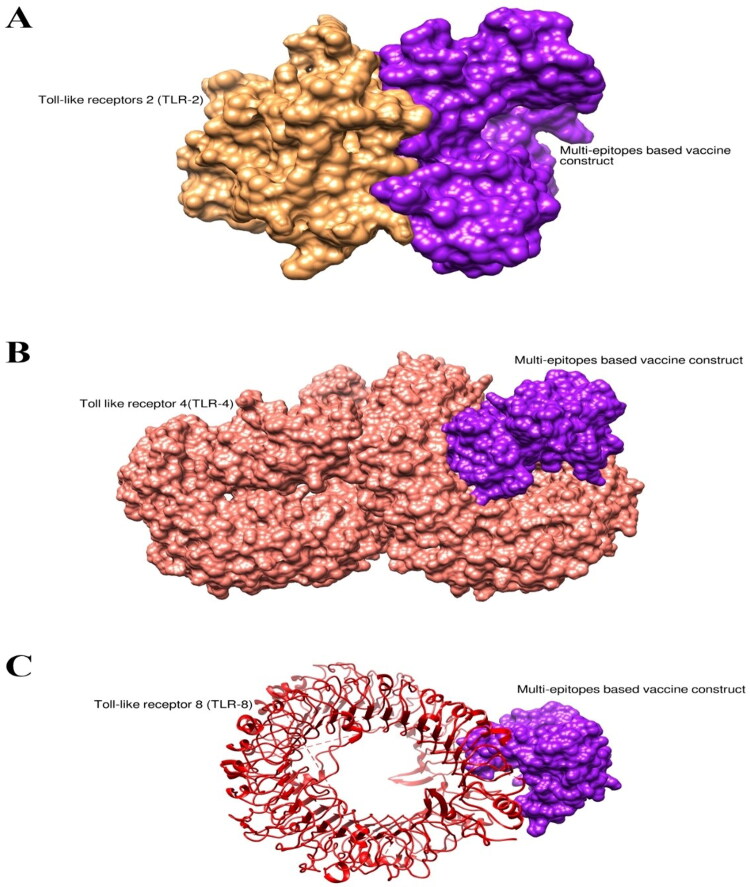
Interaction of vaccine with immune cell receptors is important to induce an immune response in the host body. The binding affinity of the designed vaccine molecules with selected immune cell receptors (TLR-2, 4, and 8) was analyzed through molecular docking. The server- generated docked complexes on the base of the complementary score, interface area, and atomic contact energy. Based on the lowest binding energy Top-1 complexes in the case of each receptor were subjected to molecular dynamic simulation analysis. The binding energy score of the Top-1 docked complex in the case of Vaccine-TLR-2, Vaccine-TLR-4 and Vaccine-TLR-8 is tabulated in the following Table 5 and is represented in Figure 6A–C.
Table 5.
Weighted score of the MPXV vaccine docked with immune cells receptors.
| Docked compounds | Representative | Weighted Score |
|---|---|---|
| Vaccine-TLR-2 | Center | −669.8 |
| Lowest energy | −829.5 | |
| Vaccine-TLR-4 | Center | −701.2 |
| Lowest energy | −869.1 | |
| Vaccine-TLR-8 | Center | −634.0 |
| Lowest energy | −735.1 |
Figure 6.
Docked confirmation of immune cells receptors with vaccine. (A) Docked structure of TLR-2 with MPXV, (B) Docked structure of TLR-4 with MPXV, (C) Docked structure of TLR-8 with MPXV. Purple color represent MPXV vaccine, while brown, pink, and red color represents TLR-2, TLR-4, and TLR-8, respectively.
3.8. Molecular dynamic simulation
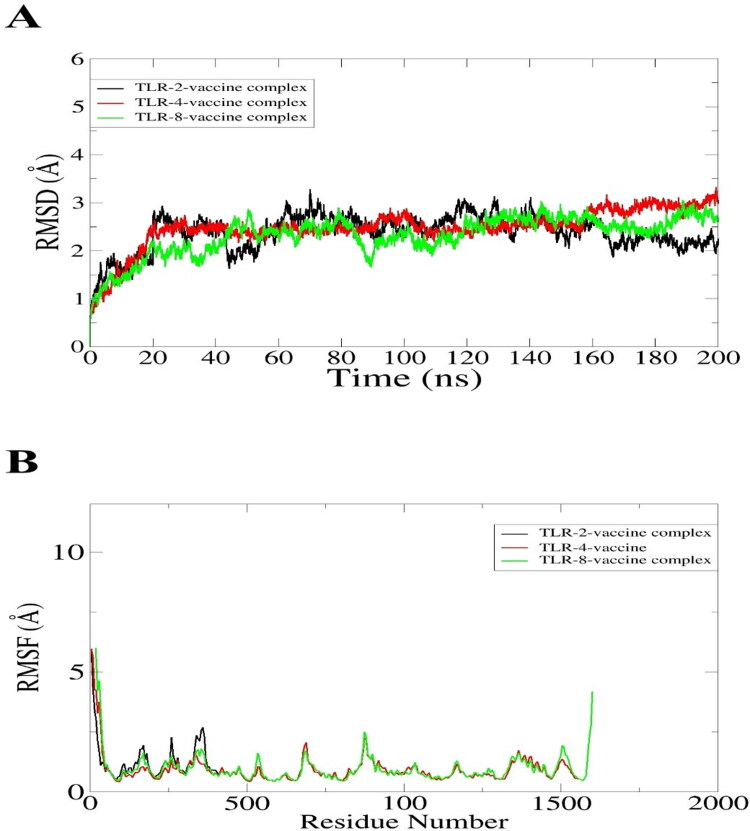
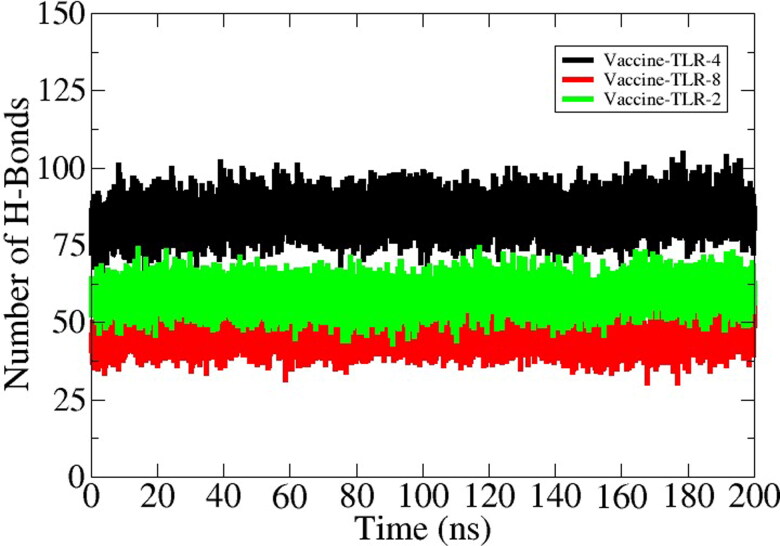
Molecular dynamic simulation analysis is based on computer simulation which is mainly assessed the dynamic behavior of the docked molecules in a specific time. For analysis of dynamic behavior and structural stability moleuar dynamic simulation of 200 nano second was conducted. The MD simulation was performed for TLR-2 – vaccine, TLR-4 – vaccine and TLR-8 – vaccine complexes. The trajectories of MD simulation was consisted of root mean square deviation and (RMSD) and root mean square fluctuation (RMSF). The RMSD backbone atom of the TLR-2 docked complex revealed that the initial deviation was between 0.2- 0.3 nm around upto 70 nano secnond but after 160 nano second the plot of RMSD got stabalized as presenetd in Figure 7A by black color. Followed by the TLR-2 vaccine TLR-4 showed proper stability as shown by the green color line in the RMSD graph. The maximum devaition is observed in the TLR-8 vaccine complex but that was due to loops present in the structure. The RMSD analysis found that the interaction between docked complexes isstabilized . Consequently, root mean square fluctuation (RMSF) for the atom of the backbone of the residues, the average fluctuation residues to be 0.05 nm. The RMSF analysis found lower fluctuation between docked complexes as presented in the following Figure 7B. Overall in the whole period of simulation, there are no drastic changes were observed in docked complexes. In order to ensure complexes stability, the simulation is repeated with different initial velocity and found to have stable dynamics reported in the first run (S-Fig.1). The number of hydrogen bonds in each complex is given in Figure 8. As can be seen in the figure that the vaccine formed robust hydrogen bonds with the receptors demonstrating stable complexes formation and stronger affinity of the vaccine for the receptors. In case of vaccine-TLR-4 complex, ∼ 75 hydrogen bonds were formed in each snapshot of the simulation trajectories. Likewise, the average number of hydrogen bonds for vaccine-TLR-2 and vaccine-TLR-8 is 50 and 45, respectively. In all three complexes, the hydrogen bond network is consistent throughout the simulation time. This analysis demonstrated that the vaccine showed strong binding affinity with the receptors and ensure efficient representation of the vaccine to host immune cells for activation of prompt immune resposnes.
Figure 7.
Simulation graphs of vaccine with immune cells receptors. (A) RMSD (B) RMSF.
Figure 8.
Number of hydrogen bonds formed in each frame of the MD simulation.
3.9. Binding free energies estimation
For further validation of docking results, we carried out binding free energy calculation. The ‘molecular mechanics the generalized born model and solvent accessibility MMGBSA’ method was adopted for energy calculation. The delta total net free binding energy for receptors TLR-2 vaccine construct was −328.29 kcal/mol, while with the TLR-4 and vaccine construct −397.39 kcal/mol and TLR-8 vaccine construct −404.06 kcal/mol, overall the VDWAALS energies was the most favorable region in the construct formation. Overall, the energy parameters and their value are presented in the following Table 6.
Table 6.
Binding free energies of MPXV vaccine docked with immune cell receptors.
| Energy parameter | TLR-2-vaccine complex | TLR-4-vaccine complex | TLR-8-vaccine complex |
|---|---|---|---|
| MM-GBSA | |||
| VDWAALS | −211.52 | −309.67 | −325.00 |
| EEL | −150.00 | −105.33 | −111.65 |
| EGB | 52.36 | 38.82 | 51.59 |
| ESURF | −19.13 | −21.21 | −19.00 |
| Delta G gas | −361.52 | −415 | −436.65 |
| Delta G solv | 33.23 | 17.61 | 32.59 |
| Delta Total | −328.29 | −397.39 | −404.06 |
3.10. In-silico cloning and codon optimization
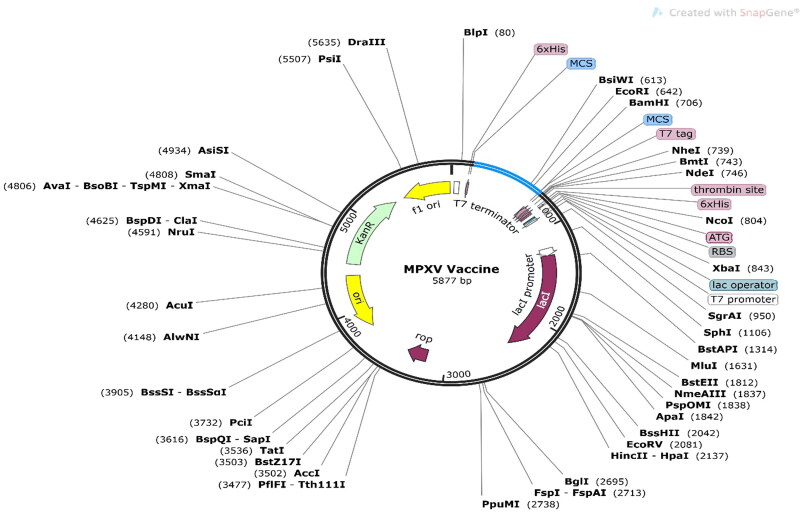
Codon adaptation is the best method to increase translational efficiency because variations in codon use limit the translation of foreign genes. The JCat tool was used to enhance the codon use of our developed vaccine concerning the E. coli K12 strain. The optimized sequence had a codon adaptation index (CAI) of 1, indicating effective expression in the E. coli host with a GC content of 50.18%. Following adaptation, the prokaryotic ribosome binding site, restriction enzyme cleavable sites, and rho independent transcription terminators were all eliminated, according to the results. The modified codon sequence from the vaccine construct was then inserted between the XhoI and BamHI restriction sites of the E. coli expression vector PET28a (+), as shown in Figure 9. The size of the clone was 5877 bp.
Figure 9.
In silico restriction cloning of the final multi-epitope vaccine using the pET28a (+) expression vector. The black circle represents the vector, and the blue portion is where the vaccine is inserted.
3.11. Disulfide engineering
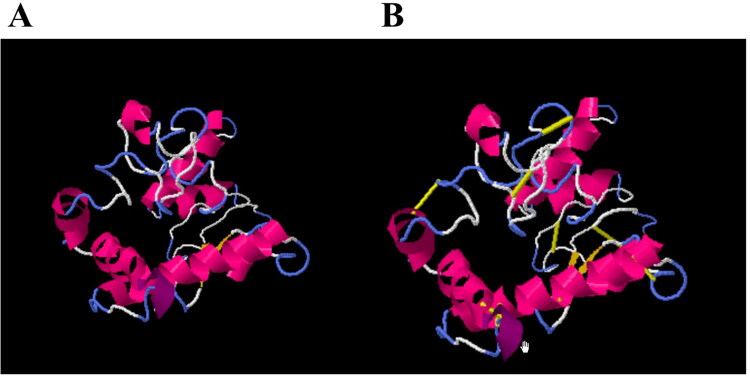
Disulfide by Design 2.13 tool was used to analyze the vaccine sequence, and a total of 10 potential residue pairings (Tyr39-Ile45, Ser51-Ala59, Gly66-Leu93, Ala67-Lys90, Ser76-Gln82, Ile95- Ala119, Val108-Ala116, Thr132-Gly141, Ser145-Pro154 and Gly168-Pro171) that could form a disulfide bond were discovered. Four pairs of residues were chosen after accounting for the bond energy and the χ3 parameters because their results met the requirements outlined in previous studies, which stipulated that the bond energy should be less than 2.2 kcal/mol and the χ3 angle should be between 87° and +97°. The mutated residues were described in the following Figure 10B by a yellow color stick while the original structure was presented in Figure 10A.
Figure 10.
(A)Wild and (B) mutated structure of designed vaccine. Yellow color represents the mutated residues.
3.12. Immune simulation
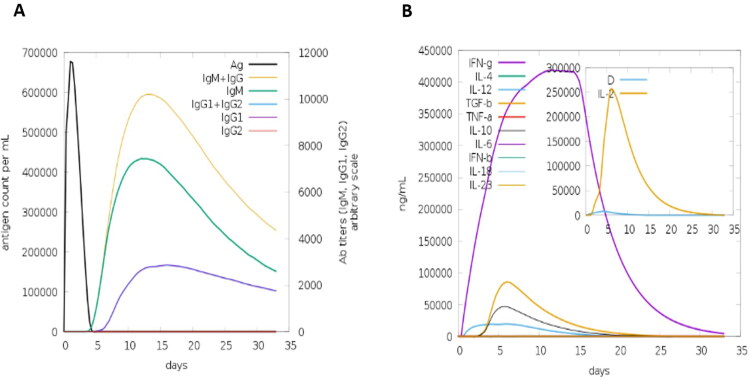
The C-IMMSIM server was used to obtain the immunogenic profile of the MPXV vaccine candidates. The results of the simulation showed that high IgM concentrations were detected during the initial response. The typical elevated levels of immunoglobulin activities (i.e. IgG1 + IgG2, IgM, and IgG + IgM antibodies) were noticeable in both secondary and tertiary responses with corresponding antigen depletion (Figure 11A). High levels of cytokines, such as IFN- and IL-2, which are important for suppressing viral replication and cellular immunity, were also discovered (Figure 11B). The immune elicit characteristics mentioned above ensured that the MPXV vaccine would be effective in human subjects.
Figure 11.
Immune simulation of the vaccine Construct. A. Immunoglobulin concentrations in relation to antigen concentration. B. Simpson index for cytokine and interleukin production.
MPXV vaccine has excellent features that make it superior to traditional vaccinations. Different multi-epitope vaccines have reported in previous studies (Dey, Mahapatra, Raj et al., 2022; Mahapatra et al., 2021; Dey et al., 2022; Dey et al., 2022; Dey et al., 2021; Chatterjee et al., 2021; Narang et al., 2021; Narang et al., 2021; Mahapatra et al., 2021). MPXV vaccine comprises B-cell and T-cell epitopes, and may thus be capable of eliciting humoral or cellular immunity within the host. Because it is connected to adjuvant, this vaccine can provide long-term protection to hosts. When administered orally, intranasally, or sublingually, MPXV vaccine may induce mucosal immunological responses, preventing pathogen entry into the host by stimulating the production of host-defensive B and T lymphocytes in the systemic and mucosal environments. The MPXV vaccine developed in this study will pave the way for future vaccinology research. Since the current work relies on an integrated computation pipeline; additional laboratory testing is required to prove the MPXV vaccine’s safety and efficacy
4. Conclusions
The main objective of the current work was to design and create a multi-epitope vaccine that can trigger an immune response against MPXV using three probable antigenic extracellular proteins as a target in order to create a novel vaccine that is risk-free and almost symptom-free. The most promising epitopes were subsequently chosen through a meticulous process and used for vaccine development. However, experimental validation and immunological base tests are strongly recommended to approve and validate the suitability of proposed constructed vaccine against MPXV. Hence, our research will support the creation of suitable therapeutics and encourage the creation of potential MPXV vaccines, which could mark a significant advancement in the development of an antiviral vaccine against MPXV.
Supplementary Material
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Amer, H., Alqahtani, A. S., Alaklobi, F., Altayeb, J., & Memish, Z. A. (2018). Healthcare worker exposure to Middle East respiratory syndrome coronavirus (MERS-CoV): Revision of screening strategies urgently needed. International Journal of Infectious Diseases: IJID: Official Publication of the International Society for Infectious Diseases, 71, 113–116. 10.1016/j.ijid.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio-Herrera, L., Badillo-Godinez, O., Medina-Contreras, O., Tepale-Segura, A., García-Lozano, A., Gutierrez-Xicotencatl, L., Soldevila, G., Esquivel-Guadarrama, F. R., Idoyaga, J., & Bonifaz, L. C. (2018). The nontoxic cholera B subunit is a potent adjuvant for intradermal DC-targeted vaccination. Frontiers in Immunology, 9, 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer, E. M., & Rao, V. B. (2019). A systematic review of the epidemiology of human monkeypox outbreaks and implications for outbreak strategy. PLoS Neglected Tropical Diseases, 13(10), e0007791. 10.1371/journal.pntd.0007791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D., Lipman, D. J., & Ostell, J. (1993). GenBank. Nucleic Acids Research, 21(13), 2963–2965. and 10.1093/nar/21.13.2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biotech, G. (2020). Snapgene viewer. Glick editor, 3(3).
- Borkotoky, S., Banerjee, M., Modi, G. P., & Dubey, V. K. (2021). Identification of high affinity and low molecular alternatives of boceprevir against SARS-CoV-2 main protease: A virtual screening approach. Chemical Physics Letters, 770, 138446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui, H.-H., Sidney, J., Dinh, K., Southwood, S., Newman, M. J., & Sette, A. (2006). Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics, 7(1), 153–155. 10.1186/1471-2105-7-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge, E. M., Hoet, B., Chen, L., Lienert, F., Weidenthaler, H., Baer, L. R., & Steffen, R. (2022). The changing epidemiology of human monkeypox—A potential threat? A systematic review. PLoS Neglected Tropical Diseases, 16(2), e0010141. 10.1371/journal.pntd.0010141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastel, C. (2009). Human monkeypox. Pathologie-biologie, 57(2), 175–183. 10.1016/j.patbio.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, R., Sahoo, P., Mahapatra, S. R., Dey, J., Ghosh, M., Kushwaha, G. S., Misra, N., Suar, M., Raina, V., & Son, Y.-O. (2021). Development of a conserved chimeric vaccine for induction of strong immune response against Staphylococcus aureus using immunoinformatics approaches. Vaccines, 9(9), 1038. 10.3390/vaccines9091038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, V., & Singh, M. P. (2020). Immuno-informatics approach to design a multi-epitope vaccine to combat cytomegalovirus infection. European Journal of Pharmaceutical Sciences: Official Journal of the European Federation for Pharmaceutical Sciences, 147, 105279. [DOI] [PubMed] [Google Scholar]
- Cheng, J., Randall, A. Z., Sweredoski, M. J., & Baldi, P. (2005). SCRATCH: A protein structure and structural feature prediction server. Nucleic Acids Research, 33(Web Server issue), W72–W76. 10.1093/nar/gki396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, G., & Sundram, F. (2017). Understanding the progression from physical illness to suicidal behavior: A case study based on a newly developed conceptual model. Clinical Gerontologist, 40(2), 124–129. 10.1080/07317115.2016.1217962 [DOI] [PubMed] [Google Scholar]
- Control, C.f.D. and Prevention . (2001). Vaccinia (smallpox) vaccine, recommendations of the Advisory Committee on Immunization Practices (ACIP), 2001. Morbidity and Mortality Weekly Report, 50, 1–25. [PubMed] [Google Scholar]
- Craig, D. B., & Dombkowski, A. A. (2013). Disulfide by Design 2.0: A web-based tool for disulfide engineering in proteins. BMC Bioinformatics, 14(1), 346–347. 10.1186/1471-2105-14-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha, B. E. (2004). Monkeypox in the United States: An occupational health look at the first cases. AAOHN Journal, 52(4), 164–168. 10.1177/216507990405200422 [DOI] [PubMed] [Google Scholar]
- Dar, A. M., & Mir, S. (2017). Molecular docking: Approaches, types, applications and basic challenges. Journal of Analytical & Bioanalytical Techniques, 08(02), 1–3. 10.4172/2155-9872.1000356 [DOI] [Google Scholar]
- Desta, I. T., Porter, K. A., Xia, B., Kozakov, D., & Vajda, S. (2020). Performance and its limits in rigid body protein-protein docking. Structure (London, England: 1993), 28(9), 1071–1081. e3. 10.1016/j.str.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey, J., Mahapatra, S. R., Lata, S., Patro, S., Misra, N., & Suar, M. (2022). Exploring Klebsiella pneumoniae capsule polysaccharide proteins to design multiepitope subunit vaccine to fight against pneumonia. Expert Review of Vaccines, 21(4), 569–587. 10.1080/14760584.2022.2021882 [DOI] [PubMed] [Google Scholar]
- Dey, J., Mahapatra, S. R., Patnaik, S., Lata, S., Kushwaha, G. S., Panda, R. K., Misra, N., & Suar, M. (2022). Molecular characterization and designing of a novel multiepitope vaccine construct against Pseudomonas aeruginosa. International Journal of Peptide Research and Therapeutics, 28(2), 1–19. 10.1007/s10989-021-10356-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey, J., Mahapatra, S. R., Raj, T. K., Kaur, T., Jain, P., Tiwari, A., Patro, S., Misra, N., & Suar, M. (2022). Designing a novel multi-epitope vaccine to evoke a robust immune response against pathogenic multidrug-resistant Enterococcus faecium bacterium. Gut Pathogens, 14(1), 1–20. 10.1186/s13099-022-00495-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey, J., Mahapatra, S. R., Singh, P., Patro, S., Kushwaha, G. S., Misra, N., & Suar, M. (2021). B and T cell epitope-based peptides predicted from clumping factor protein of Staphylococcus aureus as vaccine targets. Microbial Pathogenesis, 160, 105171. 10.1016/j.micpath.2021.105171 [DOI] [PubMed] [Google Scholar]
- Dhanda, S. K., Mahajan, S., Paul, S., Yan, Z., Kim, H., Jespersen, M. C., Jurtz, V., Andreatta, M., Greenbaum, J. A., Marcatili, P., Sette, A., Nielsen, M., & Peters, B. (2019). IEDB-AR: Immune epitope database—Analysis resource in 2019. Nucleic Acids Research, 47(W1), W502–W506. 10.1093/nar/gkz452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov, I., Flower, D. R., & Doytchinova, I. (2013). AllerTOP—A server for in silico prediction of allergens. In BMC bioinformatics. BioMed Central. 10.1186/1471-2105-14-S6-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, R., Chu, Z., Yu, F., & Zha, Y. (2020). Contriving multi-epitope subunit of vaccine for COVID-19: Immunoinformatics approaches. Frontiers in Immunology, 11, 1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova, I. A., & Flower, D. R. (2007). VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics, 8(1), 4. 10.1186/1471-2105-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine, P. E., Jezek, Z., Grab, B., & Dixon, H. (1988). The transmission potential of monkeypox virus in human populations. International Journal of Epidemiology, 17(3), 643–650. 10.1093/ije/17.3.643 [DOI] [PubMed] [Google Scholar]
- Garg, V. K., Avashthi, H., Tiwari, A., Jain, P. A., Ramkete, P. W., Kayastha, A. M., & Singh, V. K. (2016). MFPPI–multi FASTA ProtParam interface. Bioinformation, 12(2), 74–77. 10.6026/97320630012074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden, S., & Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery, 10(5), 449–461. 10.1517/17460441.2015.1032936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geourjon, C., & Deleage, G. (1995). SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Computer Applications in the Biosciences: CABIOS, 11(6), 681–684. 10.1093/bioinformatics/11.6.681 [DOI] [PubMed] [Google Scholar]
- Grote, A., Hiller, K., Scheer, M., Münch, R., Nörtemann, B., Hempel, D. C., & Jahn, D. (2005). JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Research, 33(Web Server issue), W526–W531. 10.1093/nar/gki376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Kapoor, P., Chaudhary, K., Gautam, A., Kumar, R., & Raghava, G. P. S. (2013). In silico approach for predicting toxicity of peptides and proteins. PLoS One. 8(9), e73957. 10.1371/journal.pone.0073957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruprasad, K., Reddy, B. B., & Pandit, M. W. (1990). Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Engineering, 4(2), 155–161. 10.1093/protein/4.2.155 [DOI] [PubMed] [Google Scholar]
- Hammarlund, E., Lewis, M. W., Carter, S. V., Amanna, I., Hansen, S. G., Strelow, L. I., Wong, S. W., Yoshihara, P., Hanifin, J. M., & Slifka, M. K. (2005). Multiple diagnostic techniques identify previously vaccinated individuals with protective immunity against monkeypox. Nature Medicine, 11(9), 1005–1011. 10.1038/nm1273 [DOI] [PubMed] [Google Scholar]
- Heraud, J.-M., Edghill-Smith, Y., Ayala, V., Kalisz, I., Parrino, J., Kalyanaraman, V. S., Manischewitz, J., King, L. R., Hryniewicz, A., Trindade, C. J., Hassett, M., Tsai, W.-P., Venzon, D., Nalca, A., Vaccari, M., Silvera, P., Bray, M., Graham, B. S., Golding, H., Hooper, J. W., & Franchini, G. (2006). Subunit recombinant vaccine protects against monkeypox. Journal of Immunology (Baltimore, Md.: 1950), 177(4), 2552–2564. 10.4049/jimmunol.177.4.2552 [DOI] [PubMed] [Google Scholar]
- Heymann, D. L., Szczeniowski, M., & Esteves, K. (1998). Re-emergence of monkeypox in Africa: A review of the past six years. British Medical Bulletin, 54(3), 693–702. 10.1093/oxfordjournals.bmb.a011720 [DOI] [PubMed] [Google Scholar]
- Hooper, J. W., Thompson, E., Wilhelmsen, C., Zimmerman, M., Ichou, M. A., Steffen, S. E., Schmaljohn, C. S., Schmaljohn, A. L., & Jahrling, P. B. (2004). Smallpox DNA vaccine protects nonhuman primates against lethal monkeypox. Journal of Virology, 78(9), 4433–4443. 10.1128/jvi.78.9.4433-4443.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Niu, B., Gao, Y., Fu, L., & Li, W. (2010). CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics (Oxford, England), 26(5), 680–682. 10.1093/bioinformatics/btq003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail, S., Shahid, F., Khan, A., Bhatti, S., Ahmad, S., Naz, A., Almatroudi, A., & Tahir Ul Qamar, M. (2021). Pan-vaccinomics approach towards a universal vaccine candidate against WHO priority pathogens to address growing global antibiotic resistance. Computers in Biology and Medicine, 136, 104705. [DOI] [PubMed] [Google Scholar]
- Jain, P., Joshi, A., Akhtar, N., Krishnan, S., & Kaushik, V. (2021). An immunoinformatics study: Designing multivalent T-cell epitope vaccine against canine circovirus. Journal of Genetic Engineering and Biotechnology, 19(1), 1–11. 10.1186/s43141-021-00220-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R. F., Dyall, J., Ragland, D. R., Huzella, L., Byrum, R., Jett, C., St Claire, M., Smith, A. L., Paragas, J., Blaney, J. E., & Jahrling, P. B. (2011). Comparative analysis of monkeypox virus infection of cynomolgus macaques by the intravenous or intrabronchial inoculation route. Journal of Virology, 85(5), 2112–2125. 10.1128/JVI.01931-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanmohammadi, S., & Rezaei, N. (2021). Role of Toll‐like receptors in the pathogenesis of COVID‐19. Journal of Medical Virology, 93(5), 2735–2739. 10.1002/jmv.26826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatoon, N., Pandey, R. K., & Prajapati, V. K. (2017). Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Scientific Reports, 7(1), 1–12. 10.1038/s41598-017-08842-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräutler, V., van Gunsteren, W. F., & Hünenberger, P. H. (2001). A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. Journal of Computational Chemistry, 22(5), 501–508. [DOI] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., & Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. Journal of Molecular Biology, 305(3), 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kupferschmidt, K. (2022). As monkeypox threat grows, scientists debate best vaccine strategy. Science, 376(6598)[ Mismatch [Google Scholar]
- Li, W., Joshi, M. D., Singhania, S., Ramsey, K. H., & Murthy, A. K. (2014). Peptide vaccine: Progress and challenges. Vaccines, 2(3), 515–536. 10.3390/vaccines2030515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Olson, V. A., Laue, T., Laker, M. T., & Damon, I. K. (2006). Detection of monkeypox virus with real-time PCR assays. Journal of Clinical Virology: The Official Publication of the Pan American Society for Clinical Virology, 36(3), 194–203. 10.1016/j.jcv.2006.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Zhao, H., Wilkins, K., Hughes, C., & Damon, I. K. (2010). Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. Journal of Virological Methods, 169(1), 223–227. 10.1016/j.jviromet.2010.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, Y.-T., Pai, T.-W., Wu, W.-K., & Chang, H.-T. (2013). Prediction of conformational epitopes with the use of a knowledge-based energy function and geometrically related neighboring residue characteristics. BMC Bioinformatics, 14(S4), 1–10. 10.1186/1471-2105-14-S4-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnan, C. N., Zeller, M., Kayala, M. A., Vigil, A., Randall, A., Felgner, P. L., & Baldi, P. (2010). High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics (Oxford, England), 26(23), 2936–2943. 10.1093/bioinformatics/btq551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra, S. R., Dey, J., Kaur, T., Sarangi, R., Bajoria, A. A., Kushwaha, G. S., Misra, N., & Suar, M. (2021). Immunoinformatics and molecular docking studies reveal a novel multi-epitope peptide vaccine against pneumonia infection. Vaccine, 39(42), 6221–6237. 10.1016/j.vaccine.2021.09.025 [DOI] [PubMed] [Google Scholar]
- Mahapatra, S. R., Dey, J., Kushwaha, G. S., Puhan, P., Mohakud, N. K., Panda, S. K., Lata, S., Misra, N., & Suar, M. (2021). Immunoinformatic approach employing modeling and simulation to design a novel vaccine construct targeting MDR efflux pumps to confer wide protection against typhoidal Salmonella serovars. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2021.1964600 [DOI] [PubMed] [Google Scholar]
- Mahmood, M., Javaid, A., Shahid, F., & Ashfaq, U. A. (2021). Rational design of multimeric based subunit vaccine against Mycoplasma pneumonia: Subtractive proteomics with immunoinformatics framework. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 91, 104795. [DOI] [PubMed] [Google Scholar]
- Maxwell, K. L., & Frappier, L. (2007). Viral proteomics. Microbiology and Molecular Biology Reviews: MMBR, 71(2), 398–411. 10.1128/MMBR.00042-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalca, A., & Zumbrun, E. E. (2010). ACAM2000™: The new smallpox vaccine for United States Strategic National Stockpile. Drug Design, Development and Therapy, 4, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang, P. K., Dey, J., Mahapatra, S. R., Ghosh, M., Misra, N., Suar, M., Kumar, V., & Raina, V. (2021). Functional annotation and sequence-structure characterization of a hypothetical protein putatively involved in carotenoid biosynthesis in microalgae. South African Journal of Botany, 141, 219–226. 10.1016/j.sajb.2021.04.014 [DOI] [Google Scholar]
- Narang, P. K., Dey, J., Mahapatra, S. R., Roy, R., Kushwaha, G. S., Misra, N., Suar, M., & Raina, V. (2021). Genome-based identification and comparative analysis of enzymes for carotenoid biosynthesis in microalgae. World Journal of Microbiology & Biotechnology, 38(1), 8–22. 10.1007/s11274-021-03188-y [DOI] [PubMed] [Google Scholar]
- Nezafat, N., Ghasemi, Y., Javadi, G., Khoshnoud, M. J., & Omidinia, E. (2014). A novel multi-epitope peptide vaccine against cancer: An in silico approach. Journal of Theoretical Biology, 349, 121–134. 10.1016/j.jtbi.2014.01.018 [DOI] [PubMed] [Google Scholar]
- Noor, F., Ahmad, S., Saleem, M., Alshaya, H., Qasim, M., Rehman, A., Ehsan, H., Talib, N., Saleem, H., Bin Jardan, Y. A., & Aslam, S. (2022). Designing a multi-epitope vaccine against Chlamydia pneumoniae by integrating the core proteomics, subtractive proteomics and reverse vaccinology-based immunoinformatics approaches. Computers in Biology and Medicine, 145, 105507. [DOI] [PubMed] [Google Scholar]
- Pandey, R. K., Ojha, R., Aathmanathan, V. S., Krishnan, M., & Prajapati, V. K. (2018). Immunoinformatics approaches to design a novel multi-epitope subunit vaccine against HIV infection. Vaccine, 36(17), 2262–2272. 10.1016/j.vaccine.2018.03.042 [DOI] [PubMed] [Google Scholar]
- Parrino, J., & Graham, B. S. (2006). Smallpox vaccines: Past, present, and future. The Journal of Allergy and Clinical Immunology, 118(6), 1320–1326. 10.1016/j.jaci.2006.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor, R. W., Brooks, B. R., & Szabo, A. (1988). An analysis of the accuracy of Langevin and molecular dynamics algorithms. Molecular Physics, 65(6), 1409–1419. 10.1080/00268978800101881 [DOI] [Google Scholar]
- Petersen, E., Zumla, A., Hui, D. S., Blumberg, L., Valdoleiros, S. R., Amao, L., Ntoumi, F., Asogun, D., Simonsen, L., Haider, N., Traore, T., Kapata, N., Dar, O., Nachega, J., Abbara, A., Al Balushi, A., Kock, R., Maeurer, M., Lee, S. S., … Koopmans, M. (2022). Vaccination for monkeypox prevention in persons with high-risk sexual behaviours to control on-going outbreak of monkeypox virus clade 3. International Journal of Infectious Diseases: IJID: Official Publication of the International Society for Infectious Diseases, 122, 569–571. 10.1016/j.ijid.2022.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumari, J., Borkotoky, S., Murali, A., & Busi, S. (2018). Anti-quorum sensing activity of Syzygium jambos (L.) Alston against Pseudomonas aeruginosa PAO1 and identification of its bioactive components. South African Journal of Botany, 118, 151–157. 10.1016/j.sajb.2018.07.004 [DOI] [Google Scholar]
- Rapin, N., Lund, O., Bernaschi, M., & Castiglione, F. (2010). Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PloS One, 5(4), e9862. 10.1371/journal.pone.0009862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk, J. G., Lippi, G., Henry, B. M., Forthal, D. N., & Rizk, Y. (2022). Prevention and treatment of monkeypox. Drugs, 82, 1–7. 10.1007/s40265-022-01767-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe, D. R., & Cheatham, T. E. (2013). Cheatham 3rd TE. 2013. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation, 9(7), 3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Saijo, M., Ami, Y., Suzaki, Y., Nagata, N., Iwata, N., Hasegawa, H., Ogata, M., Fukushi, S., Mizutani, T., Sata, T., Kurata, T., Kurane, I., & Morikawa, S. (2006). LC16m8, a highly attenuated vaccinia virus vaccine lacking expression of the membrane protein B5R, protects monkeys from monkeypox. Journal of Virology, 80(11), 5179–5188. 10.1128/JVI.02642-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo, M., Ami, Y., Suzaki, Y., Nagata, N., Iwata, N., Hasegawa, H., Ogata, M., Fukushi, S., Mizutani, T., Iizuka, I., Sakai, K., Sata, T., Kurata, T., Kurane, I., & Morikawa, S. (2008). Diagnosis and assessment of monkeypox virus (MPXV) infection by quantitative PCR assay: Differentiation of Congo Basin and West African MPXV strains. Japanese Journal of Infectious Diseases, 61(2), 140–142. [PubMed] [Google Scholar]
- Salomon-Ferrer, R., Case, D. A., & Walker, R. C. (2013). An overview of the Amber biomolecular simulation package. Wiley Interdisciplinary Reviews: Computational Molecular Science, 3(2), 198–210. 10.1002/wcms.1121 [DOI] [Google Scholar]
- Sikic, K., & Carugo, O. (2010). Protein sequence redundancy reduction: Comparison of various method. Bioinformation, 5(6), 234–239. 10.6026/97320630005234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stittelaar, K. J., Neyts, J., Naesens, L., van Amerongen, G., van Lavieren, R. F., Holý, A., De Clercq, E., Niesters, H. G. M., Fries, E., Maas, C., Mulder, P. G. H., van der Zeijst, B. A. M., & Osterhaus, A. D. M. E. (2006). Antiviral treatment is more effective than smallpox vaccination upon lethal monkeypox virus infection. Nature, 439(7077), 745–748. 10.1038/nature04295 [DOI] [PubMed] [Google Scholar]
- Stittelaar, K. J., van Amerongen, G., Kondova, I., Kuiken, T., van Lavieren, R. F., Pistoor, F. H. M., Niesters, H. G. M., van Doornum, G., van der Zeijst, B. A. M., Mateo, L., Chaplin, P. J., & Osterhaus, A. D. M. E. (2005). Modified vaccinia virus Ankara protects macaques against respiratory challenge with monkeypox virus. Journal of Virology, 79(12), 7845–7851. 10.1128/JVI.79.12.7845-7851.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir Ul Qamar, M., Rehman, A., Tusleem, K., Ashfaq, U. A., Qasim, M., Zhu, X., Fatima, I., Shahid, F., & Chen, L.-L. (2020). Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: Immunoinformatics and in silico approaches. PloS One, 15(12), e0244176. 10.1371/journal.pone.0244176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir ul Qamar, M., Shahid, F., Aslam, S., Ashfaq, U. A., Aslam, S., Fatima, I., Fareed, M. M., Zohaib, A., & Chen, L.-L. (2020). Reverse vaccinology assisted designing of multiepitope-based subunit vaccine against SARS-CoV-2. Infectious Diseases of Poverty, 9(1), 1–14. 10.1186/s40249-020-00752-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, A., Ahmad, S., Ismail, S., Afsheen, Z., Khurram, M., Tahir ul Qamar, M., AlSuhaymi, N., Alsugoor, M. H., & Allemailem, K. S. (2021). Towards a novel multi-epitopes chimeric vaccine for simulating strong immune responses and protection against morganella morganii. International Journal of Environmental Research and Public Health, 18(20), 10961. 10.3390/ijerph182010961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita, R., Overton, J. A., Greenbaum, J. A., Ponomarenko, J., Clark, J. D., Cantrell, J. R., Wheeler, D. K., Gabbard, J. L., Hix, D., Sette, A., & Peters, B. (2015). The immune epitope database (IEDB) 3.0. Nucleic Acids Research, 43(Database issue), D405–D412. 10.1093/nar/gku938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein, R. A., Nalca, A., Rimoin, A. W., Bavari, S., & Whitehouse, C. A. (2005). Reemergence of monkeypox: Prevalence, diagnostics, and countermeasures. Clinical Infectious Diseases, 41(12), 1765–1771. 10.1086/498155 [DOI] [PubMed] [Google Scholar]
- Yu, C.-S., Cheng, C.-W., Su, W.-C., Chang, K.-C., Huang, S.-W., Hwang, J.-K., & Lu, C.-H. (2014). CELLO2GO: A web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PloS One, 9(6), e99368. 10.1371/journal.pone.0099368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaucha, G. M., Jahrling, P. B., Geisbert, T. W., Swearengen, J. R., & Hensley, L. (2001). The pathology of experimental aerosolized monkeypox virus infection in cynomolgus monkeys (Macaca fascicularis). Laboratory Investigation; A Journal of Technical Methods and Pathology, 81(12), 1581–1600. 10.1038/labinvest.3780373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. (2018). Multi-epitope vaccines: A promising strategy against tumors and viral infections. Cellular & Molecular Immunology, 15(2), 182–184. 10.1038/cmi.2017.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J., Lin, X., Wang, X., Zheng, L., Lan, S., Jin, S., Ou, Z., & Wu, J. (2017). In silico analysis of epitope-based vaccine candidates against hepatitis B virus polymerase protein. Viruses, 9(5), 112. 10.3390/v9050112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.