Abstract
Immunotherapy offers the potential for durable clinical benefit but calls into question the association between tumor size and outcome that currently forms the basis for imaging-guided treatment. Artificial intelligence (AI) and radiomics allow for discovery of novel patterns in medical images that can increase radiology’s role in management of patients with cancer, although methodological issues in the literature limit its clinical application. Using keywords related to immunotherapy and radiomics, we performed a literature review of MEDLINE, CENTRAL, and Embase from database inception through February 2022. We removed all duplicates, non-English language reports, abstracts, reviews, editorials, perspectives, case reports, book chapters, and non-relevant studies. From the remaining articles, the following information was extracted: publication information, sample size, primary tumor site, imaging modality, primary and secondary study objectives, data collection strategy (retrospective vs prospective, single center vs multicenter), radiomic signature validation strategy, signature performance, and metrics for calculation of a Radiomics Quality Score (RQS). We identified 351 studies, of which 87 were unique reports relevant to our research question. The median (IQR) of cohort sizes was 101 (57–180). Primary stated goals for radiomics model development were prognostication (n=29, 33.3%), treatment response prediction (n=24, 27.6%), and characterization of tumor phenotype (n=14, 16.1%) or immune environment (n=13, 14.9%). Most studies were retrospective (n=75, 86.2%) and recruited patients from a single center (n=57, 65.5%). For studies with available information on model testing, most (n=54, 65.9%) used a validation set or better. Performance metrics were generally highest for radiomics signatures predicting treatment response or tumor phenotype, as opposed to immune environment and overall prognosis. Out of a possible maximum of 36 points, the median (IQR) of RQS was 12 (10–16). While a rapidly increasing number of promising results offer proof of concept that AI and radiomics could drive precision medicine approaches for a wide range of indications, standardizing the data collection as well as optimizing the methodological quality and rigor are necessary before these results can be translated into clinical practice.
Keywords: tumor biomarkers, translational medical research, review, immunotherapy, immunologic surveillance
Introduction
Immunotherapy, a treatment strategy that harnesses a patient’s own immune system, can improve outcomes in several types of cancer. Durable objective responses and improved overall survival (OS) have been achieved in patients with a wide range of malignancies through treatment with intratumoral oncolytic virus and chimeric antigen receptor T cells, like those against CD19 and B-cell maturation antigen, as well as antagonistic monoclonal antibodies directed against T-cell checkpoint molecules (eg, programmed cell death protein-1 (PD-1), programmed cell death-ligand 1 (PD-L1), cytotoxic T-lymphocytes-associated protein 4, and more recently Lymphocyte-activation gene 3 (LAG-3)).1 However, the unique mechanisms of action of these novel treatments has led to the emergence of new, atypical response patterns including delayed response, pseudoprogression, and mixed response, all of which confound the size-based criteria typically used to guide clinical decision-making.2 3 Moreover, these treatments have been associated with immune-related adverse events (irAEs) and potentially hyperprogression, which can be life-threatening and require identification and prediction by imaging.4–6 Adapting to these novel phenomena demands innovation in imaging, a long-standing cornerstone for the evaluation of cancer treatment response. Artificial intelligence (AI), including deep learning and radiomics, offers a potential solution to the growing complexity of response assessment and represents a pivotal upgrade to the role of imaging in immunotherapy, but many challenges must be overcome before a bench-to-bedside transition can be implemented. The present review will first introduce the fundamentals of this exciting technology before describing results from a literature survey covering ‘AI in immunotherapy’, synthesizing current trends in the field, and identifying broad characteristics of immunotherapy-related radiomics research.
Fundamentals of AI
AI broadly refers to algorithms created to perform tasks previously only achievable by human intelligence. Machine learning is a subfield of AI which involves algorithms that can modify themselves, or ‘learn’, to produce a desired output using the data available. The theory is that computer vision can distinguish characteristic and distinct phenotypes produced on imaging by underlying biological processes.7
Radiomics offers one method to convert images into statistically interpretable and quantifiable data.8 Traditionally, radiomics characterizes images using hand-crafted quantitative features and mathematical formulas which can describe the relationships of image pixels in a meaningful way. Domain expertize is required to choose these features, such as intensity heterogeneity, edge sharpness, and shape irregularity, that describe characteristics known to be associated with disease (eg, the pattern of central necrosis in an aggressive malignancy). A machine learning algorithm can then learn to adjust the importance of each feature or pattern, as well as their interactions, in order to combine radiomic features into a predictive signature. With the rise of open-source software packages to extract radiomic features from images, researchers around the world have built signatures for diverse clinical applications. These signatures have been correlated with distant metastasis, pathological response, cancer recurrence after radiation therapy, disease-free survival, and even with certain genotypes in one or more cancer types.9 10%5D
Deep learning is another type of machine learning that minimizes human input, instead seeking to discover patterns algorithmically. Each pixel, or set of pixels, serves as an initial data point for an algorithm commonly known as a neural network. Through training, the neural network learns to progressively combine information to automatically discover patterns, starting with simple characteristics such as a line or a circle before proceeding to more complex representations. There is much excitement about the potential for deep learning to discover previously unknown relationships in data and perform almost any complex mapping given the correct training. These algorithms have thus far produced remarkable advancements in the field of medical imaging and, in particular, have achieved impressive results in cancer detection, characterization, and monitoring.11 12
Radiomics and deep learning each offer advantages. The strength of radiomics lies in its intuitive operation, which affords more easily interpretable results. Using features engineered by domain experts provides reassurance that the algorithm is learning in the correct direction and is basing its decision-making on proven imaging patterns. Radiomics also typically requires a smaller data set to reach the threshold of learning, making it more feasible for experiments with sparse data. Lastly, radiomic features reflect fundamental properties of the images themselves, such as those directly observed by radiologists (contrast, shape, heterogeneity) as well as novel characteristics. These are more easily explained to other physicians, a fact that may help alleviate the historical apprehension towards this new technology.
On the other hand, deep learning holds a unique advantage in that the algorithm is allowed to create its own ‘features’, setting the stage for significant scientific progress; In 2012, AlexNet, one of the first modern convolutional neural networks (CNN), revolutionized the field of image recognition by winning the ImageNet competition with a substantial increase in accuracy over its competitors.13 Deep learning has no theoretical limit to what it can learn and can, with appropriate training data, continue to increase its performance, even learning to identify unexpected new associations between imaging and disease. Incorporating both deep learning and radiomics has been shown to produce even further performance improvements, potentially due to the analysis of previously unseen relationships that are uncovered by deep learning.14 15
Challenges of AI
The main barrier to integration of AI technology into clinical practice is the need for clinical validation of initial proof of concept studies. As preliminary reports are frequently prone to overfitting due to data set limitations, validation requires appropriate experimental design in order to properly assess generalizability.12 Several organizations have published recommendations for appropriate validation, which the Food and Drug Administration has incorporated in its guidance for Software as a Medical Device. It is recommended that validation is performed on data with high technical and demographic diversity, which may be best provided by multicenter data sets. A recent study evaluated design characteristics of AI medical image research and found that only 6.0% of the over 500 included articles qualified as validation studies. They concluded that the literature may not produce results that are robust enough for clinical translation.16
Another barrier, especially prominent in radiomics, is the issue of reproducibility. Sources of noise and variation are introduced at multiple steps of the radiomic pathway. Differences in scanner type, reconstruction parameters, and acquisition techniques have all been shown to affect radiomic feature reproducibility and model performance.7 17 18 Until 2020, no standardized definition had been published for even the most common imaging biomarkers.19 To establish a set of suggested research practices, such as the proper description of imaging protocols and providing open-source data and methods, a Radiomics Quality Score (RQS) was developed by Lambin et al.20 Studies that achieve higher scores using this metric could more easily facilitate translation of radiomic results into clinical practice.
Deep learning poses its own obstacles. These models can forgo several of the steps necessary for radiomics, including segmentation and feature extraction, and may be more robust to noise variability in image acquisition. However, deep learning requires a sufficiently large and high-quality data set for neural network training. The ImageNet challenge in 2012, which first established the breakthrough performance of CNNs, had a training data set of over 1.2 million images.13 Obtaining similarly large training data sets of medical images has historically been difficult due to high cost and concerns about patient confidentiality. Promising efforts towards resolving this problem have been made such as The Cancer Imaging Archive and the National Lung Cancer Screening Trial.21 22
Other issues with the deep learning strategy include a somewhat ‘black box’ decision-making process, unexplainable selected features, and a higher risk for overfitting when using small training sets. All of these may contribute to a lack of confidence from physicians that, in turn, impedes implementation in routine clinical settings. Current neural networks can have millions of parameters, leading to extreme model complexity. Repeat training on the same data set may generate networks that have significantly different visual appearances, yet still attain a similar result. It can be unclear what patterns the algorithm is learning and which parts of the image the algorithm is attaching importance to. As a result, there is a risk that algorithms may be prioritizing noise over true signal, prompting a need for extensive validation.23 To overcome this challenge, several breakthroughs have been made in visualizing neural networks and providing insight into the mechanism of their decision-making. SHAP and Grad-Cam are some of the technologies used to visualize the importance of certain pixels in input images and how they are related to the output, effectively allowing users to understand what the model ‘sees’ by using a heat map.24 25
Material and methods
Literature search strategy and study selection
In order to review the published literature relating to the use of radiomics in immunotherapy, we queried three databases, MEDLINE (PubMed), CENTRAL (Cochrane Central Register of Controlled Trials), and Embase, from their inception through February 26, 2022. This scoping review included the following key search terms: (immunotherapy) AND (CT OR MRI OR PET) AND (radiomics OR texture OR deep learning OR artificial intelligence). One additional relevant study that did not appear in this search but that was identified in literature references was added.26 Titles and abstracts of the articles were screened to determine eligibility and were included if they (1) involved immunotherapeutic treatment of human cancers or murine models of human cancers and (2) employed radiomics with positron emission tomography (PET), CT, or MRI imaging (or any combination of the three). Case reports, systematic reviews or meta-analyses, perspectives, editorials, book chapters, workshop reports, and conference abstracts were excluded from the analysis, as well as duplicate or non-English studies and publications including only an abstract.
Data extraction
Relevant data were extracted from each eligible publication using a standardized form recording the following information: (1) general publication information (date, PubMed reference number (PMID)/ Document Object Identifier (doi)), (2) sample size, (3) location of primary tumor, (4) imaging modality, (5) primary and secondary study objectives, (6) data collection strategy (retrospective vs prospective, single center vs multicenter), (7) radiomic signature validation strategy, (8) signature performance, and (9) metrics for calculation of an RQS.
Sample size
Sample sizes were calculated by addition of each cohort utilized in the study, including all training, validation, and test sets. Studies were grouped into sample size buckets (<50, 50–99, 100–199, 200–299, 300–399, 400–499, 500–599, 600–699, ≥700) for analysis. The sample size 0–99 group was split in half for higher resolution in order to better distinguish the large number of studies fitting this criterion.
Imaging modality
The imaging modality used to generate each study’s radiomics signature was identified. This was either MRI, CT, (18F)-fluorodeoxyglucose (FDG) PET nuclear imaging information alone (PET), a combination of (18F)-FDG PET and CT images (PET/CT), or a combination of MRI and CT (MRI/CT).
Tumor type
Primary tumor location of the cancer investigated in each study was identified. For simplicity and due to low numbers of individual studies, reports examining rectal, colon, gastric, and esophageal tumors were grouped together under ‘gastrointestinal tract’. Those investigating a variety of different cancers with one radiomics model (eg, ‘solid tumors’) were categorized as ‘mixed’.
Primary task
For identifying the primary task of each study, we established five broad categories: prognosis, treatment response, general classification, classification by immune environment, and classification by tumor phenotype. Measures of ‘prognosis’ were defined as OS, progression-free survival (time from treatment initiation to clinical or radiological disease progression), and durable clinical benefit (progression-free survival past a predetermined time point, eg, 6 months). ‘Treatment response’ included studies utilizing a primary endpoint of disease response as defined by Response Evaluation Criteria in Solid Tumors (RECIST) V.1.1 criteria. ‘General classification’ was comprised of studies performing other categorization-based tasks (eg, serious sequelae and adverse events from immunotherapy or adjuvant treatment). ‘Immune environment’ included reports examining immune cell (eg, CD8+, CD4+, CD3, T-helper 1/2, B-cells, natural killer cells, among others) infiltration of primary tumors, while ‘tumor phenotype’ covered studies focused on tumor PD-L1 expression or microsatellite instability.
Validation strategy
We examined how each radiomics model was validated. In the interest of clarity when describing model validation, we created the following definitions which were subsequently used to classify studies. ‘Cross validation’ was listed as the strategy when simple cross validation was used without a separate validation set. ‘Validation set’ indicated one of two things that: (1) a study’s original cohort was split into two distinct groups, the first used to train and refine the radiomics signature (the training set) and the second kept separate from training in order to be used for testing (the validation set) or (2) a training set and a validation set were recruited separately, but from the same institution using similar criteria (ie, they were not part of an aggregate original cohort that was split, but they had similar patient distributions and characteristics). The validation set thus acted as an ‘extension’ of the training set, drawn from the same or a similar patient base, but to which the model had not been previously exposed. In a similar fashion, ‘tuning; validation set’ meant that a study’s original cohort was split into three groups: a training, tuning (for adjustment of model parameters after training), and validation set. A test set was identified when the cohort used to evaluate model performance consisted of patients that were independent of the training/validation sets, that is, they were drawn from a different database/institution/clinical trial, received a different type of treatment (chemotherapy vs immunotherapy), or were collected prospectively following initial training of the radiomics model. Test sets were used by studies either directly after training, denoted by ‘test set’, or following training and the use of a validation set, which we labeled as ‘validation set; test set’. Studies were thus classified by their validation strategies as: ‘none’, ‘cross validation’, ‘validation set’, ‘tuning; validation set’, ‘test set’, or ‘validation set; test set’.
Temporal trend
We grouped studies by individual year of publication, except for those published in 2021 and 2022. For plotting purposes, these were combined as ‘2021+’ due to the relatively few studies published since the beginning of 2022.
Methodology
Each report was examined for investigational methodology in two domains: how the data was collected (retrospectively or prospectively) and where the data was collected (single or multiple institutions). If a study utilized an external data set (eg, The Cancer Genome Atlas Project) in addition to single-center cohort recruitment, the location was classified as ‘multiple institutions’.
Performance
Performance evaluation of radiomic signatures generally involves sequential application to each cohort available in the study—moving through the training set, validation set, and test sets, depending on study design—with a different performance metric (PM) reported at each stage. We extracted PMs from the most robust cohort applications reported in each individual study, such that metrics were used from the test set whenever present, the validation set if one was present but no test set was available, and the training set if neither test nor validation set was available. The highest reported performance was utilized if multiple metrics were reported at a given level (ie, the investigators tested their radiomics model on two validation sets).
RQS
An RQS was calculated for each eligible study. The articles were individually reviewed and data was extracted based on adherence to a 16 component scoring system originally defined by Lambin et al.20 Higher scores indicate higher quality studies, and the highest possible score is 36 points. This system was developed to provide a measure of standardization for the assessment of radiomics studies, assigning point values for different features of the scientific protocol (eg, points for if multiple segmentation or measures against overfitting were performed, if the study was prospective, if the model was validated and how, etc). The score does not take into account sample size or how the model actually performs, but instead represents an evaluation of both how rigorous model development is and how impactful the study may be to the field. A full summary of what features were evaluated, as well as the associated point values, is available in online supplemental table S1.
jitc-2022-005292supp001.pdf (1.4MB, pdf)
Tabulation and analysis
During our review, all eligible studies were recorded using Google Sheets (V.2022, Google, USA, 2022). Figures were created using Adobe Illustrator (V.2022, Adobe, USA, 2022).
Results
Identification and selection of studies
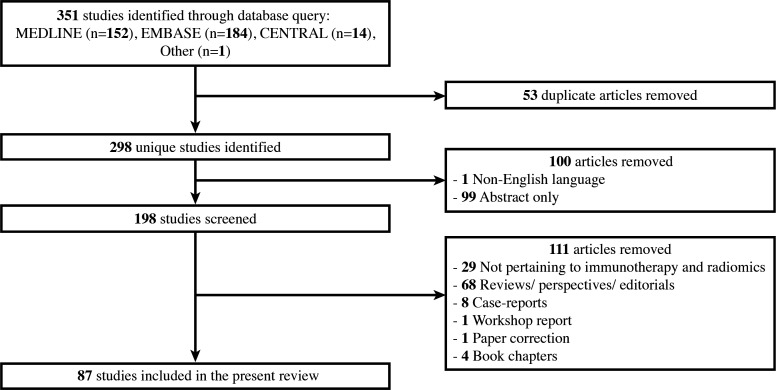
Our query of MEDLINE, CENTRAL, and Embase identified 350 studies reporting on the use of radiomics in immunotherapy. Among these, 53 studies were removed for being duplicates, 1 for not being in English, and 99 for only having an abstract available (including symposium/conference abstracts). The remaining 198 studies were screened to remove reviews, perspectives, and editorials (n=68), studies not pertaining to immunotherapy or radiomics (n=29), case reports (n=8), workshop reports (n=1), book chapters (n=4), and paper corrections (n=1). One additional study not identified in the search but previously known in the literature was added. Our selection process is outlined in figure 1 and yielded a total of 87 studies that were included in the final report. Their details are summarized in table 1 and figure 2.
Figure 1.
Visualization of our literature survey and study selection.
Table 1.
Literature review of MEDLINE, CENTRAL, and Embase from database inception through February 2022.
| Sample size | Tumor type | Modality | Task | Secondary task | Data collection | Multicenter | Validation strategy | RQS | Year | PMID/doi | Citation |
| 939 | Lung | CT | Classification (tumor phenotype) | Retrospective | No | Tuning; validation set | 13 | 2021 | 33500713 | 49 | |
| 697 | Lung | PET/CT | Classification (tumor phenotype) | Prognosis | Retrospective | Yes | Validation set; test set | 23 | 2021 | 34135101 | 39 |
| 575 | Melanoma | CT | Prognosis | Prospective | Yes | Validation set | 20 | 2022 | 35050320 | 34 | |
| 491 | Mix | CT | Classification (immune environment) | Prognosis; treatment response | Retrospective | Yes | Test set | 18 | 2018 | 30120041 | 30 |
| 491 | GI | MRI | Classification (tumor phenotype) | Retrospective | No | Validation set | 14 | 2021 | 33963688 | 50 | |
| 450 | Lung | CT | Classification | Prognosis; treatment response | Retrospective | No | Tuning; validation set | 15 | 2020 | 32636239 | 28 |
| 399 | Lung | PET/CT | Classification (tumor phenotype) | Retrospective | No | Validation set | 12 | 2020 | 31147234 | 51 | |
| 390 | Lung | CT | Classification (tumor phenotype) | Retrospective | No | Validation set | 2020 | 32176676 | 52 | ||
| 290 | Lung | CT | Prognosis | Classification | Retrospective | No | Validation set | 12 | 2018 | 29386574 | 53 |
| 255 | Lung | PET/CT | Classification (tumor phenotype) | Retrospective | No | Validation set | 15 | 2021 | 34976829 | 54 | |
| 224 | Lung | CT | Treatment response | Prognosis | Retrospective | Yes | Validation set | 15 | 2022 | 35096486 | 55 |
| 210 | Lung | PET/CT | Classification | Prognosis | Retrospective | Yes | Validation set; test set | 16 | 2021 | 33828255 | 56 |
| 207 | Liver | MRI | Classification (immune environment) | Retrospective | No | Validation set | 17 | 2019 | 30666445 | 57 | |
| 203 | Mix | CT | Treatment response | Prognosis | Retrospective | Yes | Validation set; test set | 12 | 2019 | 30895304 | 58 |
| 200 | Lung | CT | Treatment response | Prognosis | Retrospective | No | Cross validation | 12 | 2021 | 33594323 | 32 |
| 198 | GI | CT | Classification (tumor phenotype) | Retrospective | No | Validation set | 11 | 2019 | 31250180 | 59 | |
| 197 | Lung | CT | Treatment response | Classification (tumor phenotype) | Retrospective | Yes | Validation set | 18 | 2021 | 33816314 | 60 |
| 194 | Lung | PET/CT | Prognosis | Retrospective | No | Validation set; test set | 24 | 2019 | 31807885 | 38 | |
| 194 | Lung | PET/CT | Classification | Retrospective | No | Validation set; test set | 24 | 2020 | 33937811 | 40 | |
| 188 | Lung | CT | Treatment response | Prospective | Yes | Validation set | 25 | 2020 | 32198149 | 26 | |
| 184 | Pancreas | CT | Classification (immune environment) | Retrospective | No | Validation set | 17 | 2021 | 34094971 | 61 | |
| 181 | Lung | PET | Classification (immune environment) | Treatment response | Retrospective | Yes | Test set | 15 | 2020 | 32929383 | 62 |
| 179 | Head and neck | CT | Classification (tumor phenotype) | Retrospective | Yes | Validation set | 15 | 2021 | 34890937 | 63 | |
| 172 | Breast | MRI | Classification (immune environment) | Retrospective | No | Validation set | 16 | 2020 | 33795199 | 64 | |
| 166 | Brain | MRI | Prognosis | Retrospective | No | None | 4 | 2020 | 32065261 | 65 | |
| 165 | GI | CT | Classification (immune environment) | Prognosis | Retrospective | Yes | Validation set; test set | 16 | 2020 | 32395513 | 66 |
| 160 | Lung | CT | Prognosis | Retrospective | No | Validation set | 11 | 2020 | 33333202 | 67 | |
| 153 | Lung | CT | Classification (tumor phenotype) | Retrospective | No | Cross validation | 12 | 2020 | 32043309 | 68 | |
| 152 | Lung | CT | Prognosis | Retrospective | No | Validation set | 12 | 2021 | 33 738 253 | 35 | |
| 151 | Brain | MRI | Prognosis | Classification (immune environment) | Retrospective | Yes | Cross validation | 13 | 2021 | 10.1016/j.neucom.2020. 10.117 |
69 |
| 149 | Lung | CT | Classification (immune environment) | Retrospective | No | Test set | 16 | 2020 | 32251447 | 70 | |
| 140 | Melanoma | CT | Treatment response | Prognosis | Retrospective | No | Validation set | 13 | 2021 | 34795006 | 71 |
| 139 | Lung | CT | Treatment response | Prognosis | Retrospective | Yes | Validation set; test set | 15 | 2019 | 31719058 | 72 |
| 132 | Lymphoma | PET | Prognosis | Retrospective | No | Validation set | 12 | 2020 | 32248365 | 73 | |
| 131 | Lung | CT | Treatment response | Retrospective | Yes | Validation set | 16 | 2022 | 35083133 | 74 | |
| 127 | Lung | CT | Classification (tumor phenotype) | Retrospective | No | Cross validation | 11 | 2020 | 32953730 | 75 | |
| 125 | Brain | MRI | Prognosis | Retrospective | Yes | Test set | 13 | 2021 | 33761371 | 76 | |
| 120 | Lung | CT | Classification (tumor phenotype) | Retrospective | No | Validation set | 15 | 2021 | 34422625 | 77 | |
| 114 | Pancreas | MRI | Classification (immune environment) | Retrospective | No | Validation set | 17 | 2021 | 34355834 | 78 | |
| 109 | Lung | CT | Treatment response | Retrospective | Yes | Validation set | 16 | 2020 | 33051342 | 79 | |
| 103 | Lung | PET/CT | Classification (immune environment) | Retrospective | No | Validation set | 17 | 2021 | 34868999 | 80 | |
| 103 | Melanoma | CT | Prognosis | Retrospective | No | Validation set | 10 | 2019 | 31704599 | 81 | |
| 103 | Liver | MRI | Classification (tumor phenotype) | Retrospective | No | Cross validation | 10 | 2021 | 34679573 | 82 | |
| 101 | Lung | CT | Treatment response | Retrospective | Unknown | Validation set | 2022 | 35137628 | 83 | ||
| 100 | Lung | CT | Classification (immune environment) | Prognosis | Retrospective | No | Validation set | 13 | 2020 | 32548224 | 84 |
| 97 | Lung | CT | Classification (immune environment) | Retrospective | No | Validation set | 11 | 2021 | 34611410 | 85 | |
| 94 | Mix | CT | Treatment response | Classification (immune environment) | Retrospective | Yes | Test set | 13 | 2020 | 33188037 | 86 |
| 92 | Lung | CT | Prognosis | Retrospective | No | Validation set | 15 | 2021 | 34183009 | 87 | |
| 90 | GI | MRI | Classification (tumor phenotype) | Prospective | No | Validation set | 22 | 2021 | 34307164 | 88 | |
| 87 | GI | CT | Treatment response | Retrospective | No | Cross validation | 8 | 2020 | 33077705 | 89 | |
| 87 | GI | CT | Treatment response | Prognosis | Retrospective | No | Validation set | 17 | 2022 | 35073519 | 90 |
| 85 | Head and neck | CT | Prognosis | Retrospective | No | Validation set | 13 | 2021 | 34071518 | 91 | |
| 85 | Lung | CT | Classification | Retrospective | No | Cross validation | 9 | 2021 | 10.3390/cancers13040652 | 92 | |
| 75 | Ovary | CT | Prognosis | Retrospective | No | Cross validation | 10 | 2019 | 32914033 | 93 | |
| 74 | Lung | CT | Prognosis | Retrospective | Yes | Cross validation | 8 | 2021 | 34439148 | 94 | |
| 74 | Bladder | CT | Prognosis | Retrospective | No | Validation set | 13 | 2021 | 33889546 | 95 | |
| 73 | Lung | CT | Classification | Retrospective | No | Validation set | 12 | 2022 | 35026041 | 96 | |
| 71 | Head and neck | CT | Classification (immune environment) | Retrospective | Unknown | 12 | 2021 | 34519454 | 97 | ||
| 68 | Mix | CT | Prognosis | Retrospective | No | None | 12 | 2020 | 32569799 | 31 | |
| 64 | GI | CT | Treatment response | Prospective | Yes | Validation set | 23 | 2021 | 34717582 | 98 | |
| 63 | Lung | CT | Treatment response | Retrospective | No | None | 8 | 2021 | 33968716 | 99 | |
| 62 | Bladder | CT | Prognosis | Treatment response | Retrospective | No | Validation set | 17 | 2020 | 32394281 | 100 |
| 60 | Melanoma | CT | Treatment response | Retrospective | No | Cross validation | 10 | 2022 | 34666945 | 101 | |
| 59 | Lung | CT | Prognosis | Retrospective | Yes | Test set | 11 | 2019 | 10.3892/ol.2019.11220 | 102 | |
| 57 | Lung | PET | Prognosis | Treatment response | Retrospective | No | None | 9 | 2020 | 32380754 | 37 |
| 57 | Mix | CT | Treatment response | Retrospective | No | Cross validation | 9 | 2021 | 33849924 | 103 | |
| 56 | Melanoma | PET | Prognosis | Retrospective | Yes | Cross validation | 7 | 2022 | 35204479 | 104 | |
| 52 | Melanoma | PET/CT | Prognosis | Classification | Retrospective | No | None | 6 | 2021 | 33811161 | 105 |
| 50 | Melanoma | CT | Treatment response | Retrospective | No | Test set | 12 | 2020 | 32984000 | 106 | |
| 48 | Liver | MRI | Classification (immune environment) | Prognosis | Retrospective | No | None | 9 | 2020 | 32086577 | 107 |
| 48 | Kidney | CT | Prognosis | Retrospective | Unknown | 11 | 2021 | 34338919 | 108 | ||
| 46 | Lung | CT | Prognosis | Treatment response | Retrospective | No | Cross validation | 9 | 2021 | 33718125 | 109 |
| 42 | Lung | CT | Treatment response | Retrospective | Unknown | 2021 | 34034473 | 110 | |||
| 42 | Bladder | CT | Prognosis | Retrospective | No | Validation set | 10 | 2021 | 33849811 | 111 | |
| 41 | Bladder | CT | Treatment response | Retrospective | No | Validation set | 12 | 2020 | 34460530 | 112 | |
| 33 | Lung | CT | Treatment response | Retrospective | No | None | 10 | 2022 | 35053513 | 113 | |
| 32 | Melanoma | CT | Prognosis | Retrospective | No | Cross validation | 11 | 2021 | 34692481 | 114 | |
| 31 | Lung | PET | Classification (tumor phenotype) | Retrospective | No | None | 6 | 2020 | 32894535 | 115 | |
| 30 | Lung | PET | Prognosis | Prospective | No | Cross validation | 18 | 2020 | 32726293 | 36 | |
| 30 | Blood | CT | Treatment response | Experimental | Unknown | 2020 | 32832602 | 116 | |||
| 26 | Melanoma | PET/CT | Treatment response | Retrospective | Unknown | 2020 | 32259852 | 117 | |||
| 21 | Lung | CT | Prognosis | Treatment response | Prospective | No | None | 16 | 2021 | 33758304 | 118 |
| 16 | Pancreas | MRI | Prognosis | Treatment response | Experimental | Unknown | Cross validation | 2020 | 32039734 | 119 | |
| 15 | Lung | MRI/CT | Classification | Experimental | Unknown | Validation set | 2021 | 34428218 | 120 | ||
| 8 | Pancreas | MRI | Prognosis | Experimental | Unknown | Cross validation | 2021 | 32499156 | 121 | ||
| 7 | Pancreas | CT | Classification | Prospective | No | Cross validation | 15 | 2021 | 34261044 | 122 | |
| 7 | Brain | MRI | Treatment response | Prospective | No | None | 12 | 2020 | 32152626 | 123 |
CT, Computed Tomography; DOI, Document Object Identifier; GI, gastrointestinal; MRi, Magnetic Resonance Imaging; PET, positron emission tomography; PET, Positron Emission Tomography; PMID, PubMed reference number; RQS, Radiomics Quality Score.
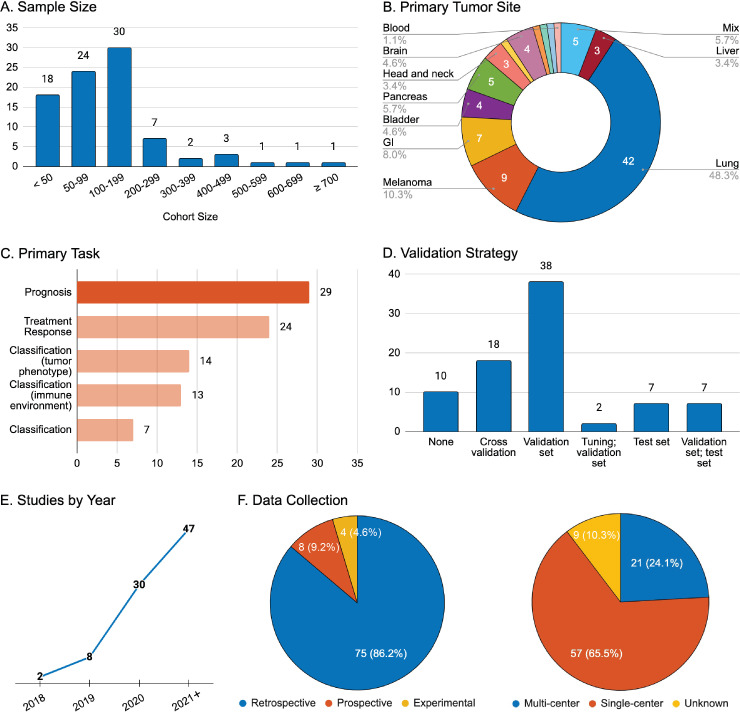
Figure 2.
General overview of study characteristics for reports involving radiomics and immunotherapy. (A) Aggregate number of patients included in the study for all purposes; (B) Primary tumor site of the disease investigated; (C) Stated task of the research: prognosis (overall survival, progression-free survival, durable clinical benefit), treatment response (defined by Response Evaluation Criteria in Solid Tumors (RECIST v1.1), tumor phenotype (programmed cell death-ligand 1 expression, microsatellite instability), immune environment (tumor immune cell infiltration), general classification (serious sequelae and adverse events from immunotherapy or adjuvant treatment); (D) Strategy for radiomics model performance validation; (E) Year of publication; (F) Data collection strategy and data source. GI, gastrointestinal.
Sample size
The vast majority of studies in this review had total sample sizes above 50 patients (n=69, 79.3%), with a mean of 146 patients. The median (IQR) of cohort sizes was 101 (57–180) (figure 2A). In studies with validation sets, many investigators split the original study cohort into training/testing divisions utilizing a ratio of at least 2:1, respectively. Although this placed an emphasis on large samples for signature development, evaluation of performance metrics was limited due to smaller validation cohorts.
Imaging modality
Radiomics was primarily applied to the evaluation of CT images in isolation (n=58, 66.7%), MRI alone (n=13, 14.9%), and PET/CT studies (n=9, 10.3%). There were few studies (n=6, 6.9%) building radiomics signatures based on PET alone and only one study (1.1%) utilizing an MRI/CT combination.
Tumor type
The site of primary malignancy for the studies reviewed was: lung (n=42, 48.3%), melanoma (n=9, 10.3%), gastrointestinal tract (n=7, 8.0%), pancreas (n=5, 5.7%), brain (n=4, 4.6%), bladder (n=4, 4.6%), head and neck (n=3, 3.4%), liver (n=3, 3.4%), ovary (n=1, 1.1%), breast (n=1, 1.1%), kidney (n=1, 1.1%), lymphatic system (n=1, 1.1%), and blood (study targeting myeloid-derived suppressor cells, n=1, 1.1%) (figure 2B). Five studies (5.7%) employed mixed primary malignancy sites to develop predictive models for solid tumors in general.
Primary task
Radiomic signatures were primarily employed to develop predictive models of prognosis (n=29, 33.3%) or treatment response (n=24, 27.6%) (figure 2C). The remaining studies focused on classification, describing either tumor phenotype (n=14, 16.1%), tumor immune microenvironment (n=13, 14.9%), or other general characteristics of the investigated disease (n=7, 8.0%). Secondary objectives were defined in 24 studies (27.6%), most of which were prognostic (n=11, 12.6%) or described treatment response (n=6, 6.9%).
Validation strategy
Assessment of validation strategy was possible for 82 studies (94.3%). Of these, the most popular strategy for model testing involved the use of a validation set (n=38, 46.3%). Other reports utilized both a tuning and a validation set (n=2, 2.4%), an independent test set alone (n=7, 8.5%), or a validation set followed by an independent test (n=7, 8.5%). The remaining studies either did not perform validation testing (n=10, 12.2%) or relied on less rigorous validation protocols, such as cross-validation on training set data alone (n=18, 22.0%).
Of the test sets identified (n=14, 17.0%), more than half were collected retrospectively from an external institution or database (n=9), while most of the others were derived from cohorts at the same institution as the training/validation sets but differing in the treatment regimen (n=2) or consisting of data collected prospectively after model training (n=2). Only one study (1.2%) followed what is considered the most thorough strategy for model development, progression through training and validation sets followed by performance evaluation on a prospective, external cohort. This latter workflow is illustrated in online supplemental figure S2.
Temporal trend
The literature search returned 87 relevant articles from 2018 through late February 2022. It is evident that interest in this particular application of radiomics is steadily increasing, with the number of published articles rising nearly linearly year on year (figure 2E). In fact, just over half of the reports detailed herein were published in the 13 months since January 2021 (n=47, 54.0%). Additionally, no MRI or PET/CT studies were identified before 2019.
Methodology
The data collection strategy for the vast majority of studies was retrospective in nature (n=75, 86.2%), with only a small percentage using prospective data sets (n=8, 9.2%) (figure 2F). Four (4.6%) relevant experimental studies have been published since 2018, employing murine models of human cancers to investigate radiomic predictors of serious sequelae, treatment response, or prognosis.
The majority of studies used patient cohorts recruited from a single institution (n=57, 65.5%) rather than multiple institutions (n=21, 24.1%) (figure 2F). Cohort origin was not applicable for the four experimental studies (4.6%), and could not be identified in five (5.7%) other publications.
Performance
A discrete measure of radiomic signature PM was identified in most studies (n=68, 78.2%) as either an area under the receiver operating curve or as a concordance index. Nine of these reports described performances of two separate models. Performance was either not reported or was addressed using a separate metric than those listed above (eg, HR) in 19 studies (21.8%).
Most (n=69) radiomics signatures could be placed into four broad categories for prediction of the following measures: prognosis, treatment response, immune environment, and tumor phenotype. The remaining eight radiomics models that could not thus be categorized involved a variety of classification goals, such as serious sequelae or immunotherapy side effect prediction. PMs, delineated by predictive aim, are detailed in online supplemental table S3. Within each category, we also clarified how PMs were validated, that is, whether only cross-validation on the training set was performed or if a validation set/independent test set was employed after model training. Signatures describing prognosis (n=26) had a mean PM of 0.787, with a median (IQR) of 0.771 (0.711–0.875). For signatures of treatment response (n=20), mean performance was higher at 0.808 with a median (IQR) of 0.810 (0.785–0.860). PMs reported on the validation sets within this category were higher than those for the training and independent test sets, although this could be an artifact from low numbers of the latter two study types. When describing immune environment (n=10) and tumor phenotype (n=13), models had PMs with means/medians (IQRs) of 0.787/0.760 (0.727–0.848) and 0.816/0.834 (0.790–0.848), respectively. The highest overall mean performances were seen in the radiomics signatures for predicting treatment response and describing tumor phenotype, with a larger number of reports contributing to the former.
RQS
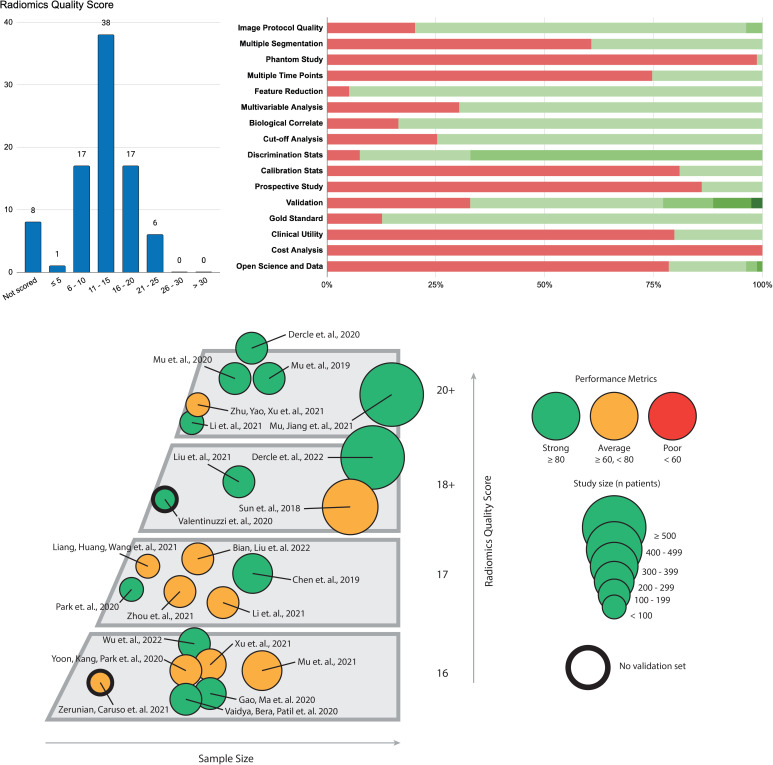
We computed an RQS for the studies identified in our survey based on the metrics detailed in online supplemental table S1. Distribution of radiomics scores as well as a visualization of study adherence to the different components of the RQS can be seen in figure 3. The four experimental studies were excluded from this analysis, and an additional four articles could not be assessed due to incomplete data. Out of a possible total score of 36 points, the median (IQR) RQS observed in the 79 studies reviewed was 12 (10–16). The vast majority of studies (n=55, 63.2%) fell within the range of 11–20, while only one study (1.3%) achieved a score greater than or equal to 25. The RQS categories with the highest adherence, that is, those for which over 75% of studies had at least one point, were image protocol quality (n=63, 79.7%), feature reduction (n=75, 94.9%), biological correlates (n=66, 83.5%), discrimination statistics (n=73, 92.4%), and comparison to a gold standard method (n=69, 87.3%). Most studies did not report calibration statistics (n=64, 81.0%) or clinical utility in the form of a decision curve (n=63, 79.7%) and most did not have or did not use open source data (n=62, 78.5%) or prospective cohorts (n=68, 86.1%). Notably, only one (1.3%) article reported a phantom study and none performed cost-analysis including a report of quality-adjusted life-years.
Figure 3.
Top: (Left) Histogram of radiomics quality scores assigned to the 87 studies included in this review. Scores have been placed in bins of five with the exception of the highest bracket, which ranges from 30 (inclusive) to the highest theoretical score of 36. (Right) Breakdown of individual radiomics score components in the studies surveyed. The red bar indicates the proportion of studies (out of 79 with scorable data) that did not receive a point in that category. The green bar overall represents the portion of studies which received at least a point in the category, with darker shades indicating serially increasing point values (eg, validation: lightest shade of green is one point with successively darker shades indicating additional points for more robust validation methods, as defined in the methods section and online supplemental table S1). Bottom: Comparison of key studies with a radiomics quality score >15 (n=23, 26.4%). The articles are organized by sample size (x-axis) and radiomics quality score (y-axis) and are represented by icons denoting studied sample size and reported performance metrics (area under the receiver operating curve, concordance index, etc). Studies without formal validation or test sets are demarcated by a black outline.
Discussion
Integration of AI to guide the care of patients with a diagnosis of cancer treated with immunotherapy holds great promise. Per our review, several studies have successfully trained models to perform specific tasks for guidance in patient management, including prediction of response to treatment (n=24, 27.6%) or prognosis (eg, survival, remission time) following immunotherapy (n=29, 33.3%). However, the field is undoubtedly still in its infancy. The earliest of the 87 studies which met our search criteria was published in 2018. The median cohort size of 101 suggests a high risk of overfitted results, further demonstrated by the fact that only 54 studies (out of 82 for which data was available, 65.8%) included a validation or independent testing data set. Out of a theoretical maximum RQS of 36 points, the median (IQR) was 12 (10–16), indicating a need for the field to adopt more robust methodology for radiomics model development and application.20
Moving forward, establishing a set of best practices for AI model development will involve addressing a few key points. Primarily, there is a need to precisely define the outcomes to be predicted by AI. For immunotherapy specifically, reference standards for treatment response and disease progression will need to account for atypical patterns of response including pseudoprogression and hyperprogression. Additionally, in order to direct the focus of future model development, the most interesting and predictive imaging biomarkers, such as those involved in radiogenomics and radiotranscriptomics (eg, PD-1/PD-L1 expression, tumor mutational burden, etc), must be identified and described using standardized definitions. With these addressed, a new gold-standard strategy for model development should be recognized that involves adherence to a set of systematic processes or measures, as described previously.9 20 To avoid overfitting, studies should divide initial patient cohorts into two groups: one for training and the second for model validation on a patient population similar to that of the training set but to which the model has not been previously exposed. The models must then be externally validated on multicenter data and prospective cohorts in order to best simulate real-world environments.16 Lastly, model development will need to implement stress testing to address the issue of underspecification, or failing to capture the inner logic of an underlying system due to confounding factors in data distributions.27 To ensure broad generalizability, these stress tests should become standard practice, just as crash tests are fundamental to the automotive industry.
Our survey revealed a dearth of truly robust cohort testing, limiting the widespread applicability of reported models and overall indicating that the use of AI for medical imaging in immunotherapy remains in a preliminary stage. The level of evidence and standardization will need to progress before the technology can be applied to clinical practice.
Notable studies and biomarkers
Notwithstanding the challenges with validation previously discussed, AI has so far achieved some success in the baseline assessment of tumors prior to therapy. He et al utilized deep learning features to distinguish the level of tumor mutational burden, which resulted in groups with distinctly different overall and progression-free survival after treatment with immune checkpoint inhibitors.28 Prior evidence also demonstrated the correlation between tumor infiltration by immune CD8+ cells and response to immunotherapy, especially with treatment specifically targeting anti-PD-1 and anti-PD-L1.29 Separately, a CT-based radiomic signature was created to predict the tumor immune environment and discriminate between tumor-inflamed and tumor-desert phenotypes.30 High baseline scores on this signature (and therefore greater CD8+ T cell infiltration) were associated with a higher chance of achieving objective response at 3 and 6 months as well as improved OS. This same radiomics signature was subsequently used to assess response after stereotactic body radiation therapy and pembrolizumab in metastatic treatment-refractory adult solid tumors, once again demonstrating a significant association with progression free survival.31 Additionally, a deep learning network incorporating multimodal data sources was trained on the multiomics of imaging, laboratory, and clinical data of patients with non-small cell lung cancer (NSCLC) who received anti-PD-1/PD-L1 agents.32 This model was capable of distinguishing responders from non-responders and predicting survival benefit to therapy in certain patients with stable disease.
Post-immunotherapy assessment is another promising application of AI. In a retrospective analysis of the CheckMate trials, a radiomics signature was developed that performed quantitative analysis of early tumor changes between baseline and first on-treatment assessment.26 It was found that treatment insensitivity and shorter OS were associated with an exponential increase in radiomic features assessing tumor volume, invasion of tumor boundaries, and tumor spatial heterogeneity. Using a similar concept in metastatic colorectal cancer treated with chemotherapy and other targeted therapies, the group demonstrated improved performance in prognostic classification over standard RECIST V.1.1 criteria.33 Another radiomics signature applied to metastatic melanoma also outperformed RECIST V.1.1 in estimating OS and was better able to distinguish between pseudoprogression and true progression at 3 months.34 Lastly, a neural network identified morphological changes on pretreatment and post-treatment chest CTs to predict 1-year survival in patients with NSCLC who received nivolumab.35 Here, the use of visualization heat mapping revealed the importance of gross morphologic changes, including those in nodal features, lung and bone metastases, pleural effusions, atelectasis, and consolidations.
AI has also found success in metabolic imaging. Certain PET features were found to be associated with OS and disease progression, predicting response of patients with NSCLC to immunotherapy.36 37 PET, CT, and PET/CT fusion features were extracted from patient with pretreatment NSCLC images to create a multiparametric radiomics signature that was able to accurately identify durable clinical benefit resulting from checkpoint blockade immunotherapy.38 These results were subsequently validated in both retrospective and prospective test cohorts. The same group then applied deep learning techniques to achieve similar results in the same data sets.39
While immunotherapy is a cornerstone of treatment in patients with advanced cancer, irAEs due to unbridled T-cell activation have emerged as a concern requiring specific detection and support. Several studies have applied radiomics to predict toxicity and irAE related to immunotherapy. One report utilized AI to distinguish between pituitary metastasis and hypophysitis by incorporating both MRI imaging and clinical features.5 In another study, PET/CT radiomics predicted development of severe irAEs in patients with NSCLC treated with immunotherapy.40 This model was validated on a prospective cohort in addition to standard training and testing data sets.
Finally, the application of AI has extended beyond radiographic and functional imaging alone towards the incorporation of histopathological slides. This approach has been studied for predicting diagnosis and prognosis, forecasting response to immune checkpoint blockers, and characterizing the tumor immune microenvironment through determination of genomic class, but is beyond the scope of this study.41–43 For further details, refer to several reviews published on these topics.41 44
Clinical application
The adoption of AI in clinical practice will require developing transparent machine-learning models in which the underlying logic of the learning process can be revealed to and understood by humans. Knowing how the model arrived at prognostic and predictive outputs allows rational application to real-world clinical scenarios. On review, we observed that the majority of relevant radiomics research using CT, MRI, and PET rely on the combination of a limited subset of imaging biomarkers that capture specific macroscopic, anatomical, and functional characteristics. The macroscopic features in particular are of great clinical use for assessing prognosis. For example, overall tumor burden and organ-specific localization of metastatic disease were found to be associated with survival, with liver metastases indicating the shortest survival and greatest acceleration in tumor growth.45 Additionally, skeletal muscle index—a surrogate marker of sarcopenia that is derived from the analysis of muscle surface area on a single CT scan slice at the level of the lumbar vertebra L3—was significantly associated with OS in patients after initiation of immunotherapy.45 Among more functional classifiers, increased bone marrow glucose metabolism was significantly and positively associated with transcriptomic profiles of regulatory T-cells, and spleen metabolism was associated with immunosuppressive environment and poor outcome.46 47
The goal of the RQS discussed in this review is to provide some measure of standardization for the assessment of studies in the field. The scoring system considers robustness in scientific protocol, including corrections for overfitting, consideration of temporal variations, cut-off analyses, etc, as well as an evaluation of the relevance of results in real-world settings, including identification of biological correlates, comparison to gold-standard methods, extent of validation, etc. The RQS thus assesses many of the features required to instill confidence in the technology for clinical practice, allowing physicians to transparently view how a biomarker was selected, how and on what cohorts the algorithm was validated, and overall how repeatable the entire sequence of testing would be given a different environment. However, the score overlooks some study components that may be useful in defining generalizability and in the field of oncology. Ideally, a more comprehensive score could incorporate the total number of lesions included in the model as well as how the actual lesions were segmented (two-dimensional vs three-dimensional). Additionally, quantification of how the examined immunotherapies were documented and identification of possible confounding variables (eg, local vs radiation therapy, intratumoral immunotherapy) would elucidate the applicability of a particular radiomic signature to other tumor types and therapies. Finally, inter-evaluator reproducibility may be a concern for RQS assignment, as demonstrated by Sanduleanu et al.48 More clear descriptions of individual score components might help the next iteration reduce the role of subjective evaluation in the RQS.
Conclusion
The results for AI application to medical imaging of patients on immunotherapy are promising but preliminary. We can envision a future in which AI-derived clinical decision support algorithms assist clinicians in distinguishing between the myriad of immune response patterns to achieve earlier and better identification of responders, non-responders, and those likely to develop adverse events. However, routine clinical implementation will require further evaluation, as the current evidence is still lacking in external validation and evidence of generalizability. Once these challenges of AI implementation have been overcome, three areas of application offer potentially transformative outcomes. First, there will be an opportunity to better define the diagnosis and character of a patient’s cancer with the help of AI to analyze pathology slides, blood biomarkers, and radiologic images. Paired with its ever improving predictive value through data accumulation, this deployment of AI will enable increased personalization of therapeutic strategies. Second, by using AI tools at baseline and throughout treatment, clinicians will have the opportunity to dynamically adapt their treatments earlier and more reliably than is currently possible, reacting to disease evolution at the first sign of change. Last but not least, AI will provide the field of immuno-oncology with new tools for deciphering the tumor immune environment by recognizing patterns on medical images that are associated with therapeutically actionable pathways and subsequently guiding theranostic approaches.
Footnotes
Twitter: @laurentdercle
LD and JM contributed equally.
Contributors: LD and JM contributed equally. The manuscript was written through contributions of all authors.
Funding: JM was funded by the Kirschstein-NRSA #5T35HL007616-42.
Competing interests: None declared.
Provenance and peer review: Commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Not applicable.
References
- 1.Vercellino L, de Jong D, di Blasi R, et al. Current and future role of medical imaging in guiding the management of patients with relapsed and refractory non-Hodgkin lymphoma treated with CAR T-cell therapy. Front Oncol 2021;11:1918. 10.3389/fonc.2021.664688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiou VL, Burotto M. Pseudoprogression and immune-related response in solid tumors. J Clin Oncol 2015;33:3541–3. 10.1200/JCO.2015.61.6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dercle L, Mokrane F-Z, Schiano de Colella JM, et al. Unconventional immune-related phenomena observed using 18F-FDG PET/CT in Hodgkin lymphoma treated with anti PD-1 monoclonal antibodies. Eur J Nucl Med Mol Imaging 2019;46:1391–2. 10.1007/s00259-019-04310-x [DOI] [PubMed] [Google Scholar]
- 4.Mekki A, Dercle L, Lichtenstein P, et al. Detection of immune-related adverse events by medical imaging in patients treated with anti-programmed cell death 1. Eur J Cancer 2018;96:91–104. 10.1016/j.ejca.2018.03.006 [DOI] [PubMed] [Google Scholar]
- 5.Mekki A, Dercle L, Lichtenstein P, et al. Machine learning defined diagnostic criteria for differentiating pituitary metastasis from autoimmune hypophysitis in patients undergoing immune checkpoint blockade therapy. Eur J Cancer 2019;119:44–56. 10.1016/j.ejca.2019.06.020 [DOI] [PubMed] [Google Scholar]
- 6.Champiat S, Dercle L, Ammari S, et al. Hyperprogressive disease is a new pattern of progression in cancer patients treated by anti-PD-1/PD-L1. Clin Cancer Res 2017;23:1920–8. 10.1158/1078-0432.CCR-16-1741 [DOI] [PubMed] [Google Scholar]
- 7.Zhao B. Understanding sources of variation to improve the reproducibility of Radiomics. Front Oncol 2021;11:633176. 10.3389/fonc.2021.633176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563–77. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dercle L, Henry T, Carré A, et al. Reinventing radiation therapy with machine learning and imaging bio-markers (radiomics): state-of-the-art, challenges and perspectives. Methods 2021;188:44–60. 10.1016/j.ymeth.2020.07.003 [DOI] [PubMed] [Google Scholar]
- 10.Limkin EJ, Sun R, Dercle L, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol 2017;28:1191–206. 10.1093/annonc/mdx034 [DOI] [PubMed] [Google Scholar]
- 11.Lee J-G, Jun S, Cho Y-W, et al. Deep learning in medical imaging: general overview. Korean J Radiol 2017;18:570–84. 10.3348/kjr.2017.18.4.570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol 2017;10:257–73. 10.1007/s12194-017-0406-5 [DOI] [PubMed] [Google Scholar]
- 13.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 2012;25 https://proceedings.neurips.cc/paper/2012/hash/c399862d3b9d6b76c8436e924a68c45b-Abstract.html [Google Scholar]
- 14.Wang X, Zhang L, Yang X, et al. Deep learning combined with radiomics may optimize the prediction in differentiating high-grade lung adenocarcinomas in ground glass opacity lesions on CT scans. Eur J Radiol 2020;129:109150. 10.1016/j.ejrad.2020.109150 [DOI] [PubMed] [Google Scholar]
- 15.Chen L, Zhou Z, Sher D, et al. Combining many-objective radiomics and 3D convolutional neural network through evidential Reasoning to predict lymph node metastasis in head and neck cancer. Phys Med Biol 2019;64:075011. 10.1088/1361-6560/ab083a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DW, Jang HY, Kim KW, et al. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol 2019;20:405–10. 10.3348/kjr.2019.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He L, Huang Y, Ma Z, et al. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep 2016;6:34921. 10.1038/srep34921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Lu L, Xiao M, et al. Ct slice thickness and convolution kernel affect performance of a radiomic model for predicting EGFR status in non-small cell lung cancer: a preliminary study. Sci Rep 2018;8:17913. 10.1038/s41598-018-36421-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative Radiomics for high-throughput image-based phenotyping. Radiology 2020;295:328–38. 10.1148/radiol.2020191145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749–62. 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 21.Clark K, Vendt B, Smith K, et al. The cancer imaging Archive (TCIA): maintaining and operating a public information Repository. J Digit Imaging 2013;26:1045–57. 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22., Aberle DR, Berg CD, et al. The National lung screening trial: overview and study design. Radiology 2011;258:243–53. 10.1148/radiol.10091808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zech JR, Badgeley MA, Liu M, et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med 2018;15:e1002683. 10.1371/journal.pmed.1002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selvaraju RR, Cogswell M, Das A. Grad-cam: visual explanations from deep networks via gradient-based localization. Proceedings of the IEEE international conference on computer vision, 2017:618–26. [Google Scholar]
- 25.Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst 2017;30 https://proceedings.neurips.cc/paper/2017/hash/8a20a8621978632d76c43dfd28b67767-Abstract.html [Google Scholar]
- 26.Dercle L, Fronheiser M, Lu L, et al. Identification of non-small cell lung cancer sensitive to systemic cancer therapies using Radiomics. Clin Cancer Res 2020;26:2151–62. 10.1158/1078-0432.CCR-19-2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eche T, Schwartz LH, Mokrane F-Z, et al. Toward generalizability in the deployment of artificial intelligence in radiology: role of computation stress testing to overcome Underspecification. Radiol Artif Intell 2021;3:e210097. 10.1148/ryai.2021210097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He B, Dong D, She Y, et al. Predicting response to immunotherapy in advanced non-small-cell lung cancer using tumor mutational burden radiomic biomarker. J Immunother Cancer 2020;8:e000550. 10.1136/jitc-2020-000550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321–30. 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 30.Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19:1180–91. 10.1016/S1470-2045(18)30413-3 [DOI] [PubMed] [Google Scholar]
- 31.Korpics MC, Polley M-Y, Bhave SR, et al. A validated T cell Radiomics score is associated with clinical outcomes following multisite SBRT and pembrolizumab. Int J Radiat Oncol Biol Phys 2020;108:189–95. 10.1016/j.ijrobp.2020.06.026 [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Yang J, Shen L, et al. A multi-omics-based serial deep learning approach to predict clinical outcomes of single-agent anti-PD-1/PD-L1 immunotherapy in advanced stage non-small-cell lung cancer. Am J Transl Res 2021;13:743–56. [PMC free article] [PubMed] [Google Scholar]
- 33.Dercle L, Zhao B, Gönen M, et al. An imaging signature to predict outcome in metastatic colorectal cancer using routine computed tomography scans. Eur J Cancer 2022;161:138–47. 10.1016/j.ejca.2021.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dercle L, Zhao B, Gönen M, et al. Early readout on overall survival of patients with melanoma treated with immunotherapy using a novel imaging analysis. JAMA Oncol 2022;8:385–92. 10.1001/jamaoncol.2021.6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trebeschi S, Bodalal Z, Boellaard TN, et al. Prognostic value of deep Learning-Mediated treatment monitoring in lung cancer patients receiving immunotherapy. Front Oncol 2021;11:609054. 10.3389/fonc.2021.609054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valentinuzzi D, Vrankar M, Boc N, et al. [18F]FDG PET immunotherapy radiomics signature (iRADIOMICS) predicts response of non-small-cell lung cancer patients treated with pembrolizumab. Radiol Oncol 2020;54:285–94. 10.2478/raon-2020-0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polverari G, Ceci F, Bertaglia V, et al. 18F-FDG Pet Parameters and Radiomics Features Analysis in Advanced Nsclc Treated with Immunotherapy as Predictors of Therapy Response and Survival. Cancers 2020;12:1163. 10.3390/cancers12051163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mu W, Tunali I, Gray JE, et al. Radiomics of 18F-FDG PET/CT images predicts clinical benefit of advanced NSCLC patients to checkpoint blockade immunotherapy. Eur J Nucl Med Mol Imaging 2020;47:1168–82. 10.1007/s00259-019-04625-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mu W, Jiang L, Shi Y, et al. Non-Invasive measurement of PD-L1 status and prediction of immunotherapy response using deep learning of PET/CT images. J Immunother Cancer 2021;9:e002118. 10.1136/jitc-2020-002118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mu W, Tunali I, Qi J, et al. Radiomics of 18F Fluorodeoxyglucose PET/CT Images Predicts Severe Immune-related Adverse Events in Patients with NSCLC. Radiol Artif Intell 2020;2:e190063. 10.1148/ryai.2019190063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bera K, Schalper KA, Rimm DL, et al. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 2019;16:703–15. 10.1038/s41571-019-0252-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, Cui C, Yang W, et al. Using deep learning to predict anti-PD-1 response in melanoma and lung cancer patients from histopathology images. Transl Oncol 2021;14:100921. 10.1016/j.tranon.2020.100921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie F, Zhang J, Wang J, et al. Multifactorial deep learning reveals pan-cancer genomic tumor clusters with distinct Immunogenomic landscape and response to immunotherapy. Clin Cancer Res 2020;26:2908–20. 10.1158/1078-0432.CCR-19-1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Y, Su G-H, Ma D, et al. Technological advances in cancer immunity: from immunogenomics to single-cell analysis and artificial intelligence. Signal Transduct Target Ther 2021;6:312. 10.1038/s41392-021-00729-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dercle L, Ammari S, Champiat S, et al. Rapid and objective CT scan prognostic scoring identifies metastatic patients with long-term clinical benefit on anti-PD-1/-L1 therapy. Eur J Cancer 2016;65:33–42. 10.1016/j.ejca.2016.05.031 [DOI] [PubMed] [Google Scholar]
- 46.Seban R-D, Nemer JS, Marabelle A, et al. Prognostic and theranostic 18F-FDG PET biomarkers for anti-PD1 immunotherapy in metastatic melanoma: association with outcome and transcriptomics. Eur J Nucl Med Mol Imaging 2019;46:2298–310. 10.1007/s00259-019-04411-7 [DOI] [PubMed] [Google Scholar]
- 47.Seban R-D, Synn S, Muneer I, et al. Spleen Glucose Metabolism on [18F]-FDG PET/CT for Cancer Drug Discovery and Development cannot be Overlooked. Curr Cancer Drug Targets 2021;21:944–52. 10.2174/1568009621666210720143826 [DOI] [PubMed] [Google Scholar]
- 48.Sanduleanu S, Woodruff HC, de Jong EEC, et al. Tracking tumor biology with radiomics: a systematic review utilizing a radiomics quality score. Radiother Oncol 2018;127:349–60. 10.1016/j.radonc.2018.03.033 [DOI] [PubMed] [Google Scholar]
- 49.Tian P, He B, Mu W, et al. Assessing PD-L1 expression in non-small cell lung cancer and predicting responses to immune checkpoint inhibitors using deep learning on computed tomography images. Theranostics 2021;11:2098–107. 10.7150/thno.48027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang W, Yin H, Huang Z, et al. Development and validation of MRI-based deep learning models for prediction of microsatellite instability in rectal cancer. Cancer Med 2021;10:4164–73. 10.1002/cam4.3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang M, Sun D, Guo Y, et al. Assessing PD-L1 expression level by radiomic features from PET/CT in nonsmall cell lung cancer patients: an initial result. Acad Radiol 2020;27:171–9. 10.1016/j.acra.2019.04.016 [DOI] [PubMed] [Google Scholar]
- 52.Sun Z, Hu S, Ge Y, et al. Radiomics study for predicting the expression of PD-L1 in non-small cell lung cancer based on CT images and clinicopathologic features. J Xray Sci Technol 2020;28:449–59. 10.3233/XST-200642 [DOI] [PubMed] [Google Scholar]
- 53.Tang C, Hobbs B, Amer A, et al. Development of an Immune-Pathology informed Radiomics model for non-small cell lung cancer. Sci Rep 2018;8:1922. 10.1038/s41598-018-20471-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J, Ge S, Sang S, et al. Evaluation of PD-L1 Expression Level in Patients With Non-Small Cell Lung Cancer by 18F-FDG PET/CT Radiomics and Clinicopathological Characteristics. Front Oncol 2021;11:789014. 10.3389/fonc.2021.789014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gong J, Bao X, Wang T, et al. A short-term follow-up CT based radiomics approach to predict response to immunotherapy in advanced non-small-cell lung cancer. Oncoimmunology 2022;11:2028962. 10.1080/2162402X.2022.2028962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mu W, Katsoulakis E, Whelan CJ, et al. Radiomics predicts risk of cachexia in advanced NSCLC patients treated with immune checkpoint inhibitors. Br J Cancer 2021;125:229–39. 10.1038/s41416-021-01375-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen S, Feng S, Wei J, et al. Pretreatment prediction of immunoscore in hepatocellular cancer: a radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur Radiol 2019;29:4177–87. 10.1007/s00330-018-5986-x [DOI] [PubMed] [Google Scholar]
- 58.Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol 2019;30:998–1004. 10.1093/annonc/mdz108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Golia Pernicka JS, Gagniere J, Chakraborty J, et al. Radiomics-based prediction of microsatellite instability in colorectal cancer at initial computed tomography evaluation. Abdom Radiol 2019;44:3755–63. 10.1007/s00261-019-02117-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y, Wu M, Zhang Y, et al. Imaging biomarkers to predict and evaluate the effectiveness of immunotherapy in advanced non-small-cell lung cancer. Front Oncol 2021;11:657615. 10.3389/fonc.2021.657615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J, Shi Z, Liu F, et al. XGBoost Classifier Based on Computed Tomography Radiomics for Prediction of Tumor-Infiltrating CD8+ T-Cells in Patients With Pancreatic Ductal Adenocarcinoma. Front Oncol 2021;11:671333. 10.3389/fonc.2021.671333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park C, Na KJ, Choi H, et al. Tumor immune profiles noninvasively estimated by FDG PET with deep learning correlate with immunotherapy response in lung adenocarcinoma. Theranostics 2020;10:10838–48. 10.7150/thno.50283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng Y-M, Zhan J-F, Yuan M-G, et al. A CT-based radiomics signature for preoperative discrimination between high and low expression of programmed death ligand 1 in head and neck squamous cell carcinoma. Eur J Radiol 2022;146:110093. 10.1016/j.ejrad.2021.110093 [DOI] [PubMed] [Google Scholar]
- 64.Xu N, Zhou J, He X, et al. Radiomics model for evaluating the level of tumor-infiltrating lymphocytes in breast cancer based on dynamic contrast-enhanced MRI. Clin Breast Cancer 2021;21:440–9. 10.1016/j.clbc.2020.12.008 [DOI] [PubMed] [Google Scholar]
- 65.Lin P, Peng Y-T, Gao R-Z, et al. Radiomic profiles in diffuse glioma reveal distinct subtypes with prognostic value. J Cancer Res Clin Oncol 2020;146:1253–62. 10.1007/s00432-020-03153-6 [DOI] [PubMed] [Google Scholar]
- 66.Gao X, Ma T, Bai S, et al. A CT-based radiomics signature for evaluating tumor infiltrating Treg cells and outcome prediction of gastric cancer. Ann Transl Med 2020;8:469. 10.21037/atm.2020.03.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiao Z, Li H, Xiao Y, et al. Integration of risk survival measures estimated from pre- and posttreatment computed tomography scans improves stratification of patients with early-stage non-small cell lung cancer treated with stereotactic body radiation therapy. Int J Radiat Oncol Biol Phys 2021;109:1647–56. 10.1016/j.ijrobp.2020.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoon J, Suh YJ, Han K, et al. Utility of CT radiomics for prediction of PD-L1 expression in advanced lung adenocarcinomas. Thorac Cancer 2020;11:993–1004. 10.1111/1759-7714.13352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaddad A, Daniel P, Zhang M, et al. Deep radiomic signature with immune cell markers predicts the survival of glioma patients. Neurocomputing 2022;469:366–75. 10.1016/j.neucom.2020.10.117 [DOI] [Google Scholar]
- 70.Yoon HJ, Kang J, Park H, et al. Deciphering the tumor microenvironment through radiomics in non-small cell lung cancer: correlation with immune profiles. PLoS One 2020;15:e0231227. 10.1371/journal.pone.0231227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brendlin AS, Peisen F, Almansour H, et al. A machine learning model trained on dual-energy CT radiomics significantly improves immunotherapy response prediction for patients with stage IV melanoma. J Immunother Cancer []. 2021;9. 10.1136/jitc-2021-003261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khorrami M, Prasanna P, Gupta A, et al. Changes in CT radiomic features associated with lymphocyte distribution predict overall survival and response to immunotherapy in non-small cell lung cancer. Cancer Immunol Res 2020;8:108–19. 10.1158/2326-6066.CIR-19-0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aide N, Fruchart C, Nganoa C, et al. Baseline 18F-FDG PET radiomic features as predictors of 2-year event-free survival in diffuse large B cell lymphomas treated with immunochemotherapy. Eur Radiol 2020;30:4623–32. 10.1007/s00330-020-06815-8 [DOI] [PubMed] [Google Scholar]
- 74.Wu M, Zhang Y, Zhang J, et al. A Combined-Radiomics approach of CT images to predict response to anti-PD-1 immunotherapy in NSCLC: a retrospective multicenter study. Front Oncol 2021;11:688679. 10.3389/fonc.2021.688679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu Y, Liu Y-L, Feng Y, et al. A CT-derived deep neural network predicts for programmed death ligand-1 expression status in advanced lung adenocarcinomas. Ann Transl Med 2020;8:930. 10.21037/atm-19-4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Z-Z, Liu P-F, An T-T, et al. Construction of a prognostic immune signature for lower grade glioma that can be recognized by MRI radiomics features to predict survival in LGG patients. Transl Oncol 2021;14:101065. 10.1016/j.tranon.2021.101065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wen Q, Yang Z, Dai H, et al. Radiomics study for predicting the expression of PD-L1 and tumor mutation burden in non-small cell lung cancer based on CT images and clinicopathological features. Front Oncol 2021;11:620246. 10.3389/fonc.2021.620246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bian Y, Liu C, Li Q, et al. Preoperative Radiomics Approach to Evaluating Tumor-Infiltrating CD8+ T Cells in Patients With Pancreatic Ductal Adenocarcinoma Using Noncontrast Magnetic Resonance Imaging. J Magn Reson Imaging 2022;55:803–14. 10.1002/jmri.27871 [DOI] [PubMed] [Google Scholar]
- 79.Vaidya P, Bera K, Patil PD, et al. Novel, non-invasive imaging approach to identify patients with advanced non-small cell lung cancer at risk of hyperprogressive disease with immune checkpoint blockade. J Immunother Cancer 2020;8:e001343. 10.1136/jitc-2020-001343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou J, Zou S, Kuang D, et al. A novel approach using FDG-PET/CT-Based Radiomics to assess tumor immune phenotypes in patients with non-small cell lung cancer. Front Oncol 2021;11:769272. 10.3389/fonc.2021.769272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schraag A, Klumpp B, Afat S, et al. Baseline clinical and imaging predictors of treatment response and overall survival of patients with metastatic melanoma undergoing immunotherapy. Eur J Radiol 2019;121:108688. 10.1016/j.ejrad.2019.108688 [DOI] [PubMed] [Google Scholar]
- 82.Tian Y, Komolafe TE, Zheng J, et al. Assessing PD-L1 expression level via preoperative MRI in HCC based on integrating deep learning and Radiomics features. Diagnostics 2021;11:1875. 10.3390/diagnostics11101875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin X, Wang H, Chen J, et al. The value of CT-based radiomics in early assessment of chemotherapeutic effect in patients with advanced lung adenocarcinoma: a preliminary study. Acta Radiol 2022:2841851221078290. 10.1177/02841851221078290 [DOI] [PubMed] [Google Scholar]
- 84.Mazzaschi G, Milanese G, Pagano P, et al. Dataset on the identification of a prognostic radio-immune signature in surgically resected non small cell lung cancer. Data Brief 2020;31:105781. 10.1016/j.dib.2020.105781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Min J, Dong F, Wu P, et al. A Radiomic Approach to Access Tumor Immune Status by CD8+TRMs on Surgically Resected Non-Small-Cell Lung Cancer. Onco Targets Ther 2021;14:4921–31. 10.2147/OTT.S316994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun R, Sundahl N, Hecht M, et al. Radiomics to predict outcomes and abscopal response of patients with cancer treated with immunotherapy combined with radiotherapy using a validated signature of CD8 cells. J Immunother Cancer 2020;8:e001429. 10.1136/jitc-2020-001429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang B, Zhou L, Zhong J, et al. Combination of computed tomography imaging-based radiomics and clinicopathological characteristics for predicting the clinical benefits of immune checkpoint inhibitors in lung cancer. Respir Res 2021;22:189. 10.1186/s12931-021-01780-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li Z, Dai H, Liu Y, et al. Radiomics analysis of Multi-Sequence Mr images for predicting microsatellite instability status preoperatively in rectal cancer. Front Oncol 2021;11:697497. 10.3389/fonc.2021.697497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ji Z, Cui Y, Peng Z, et al. Use of Radiomics to predict response to immunotherapy of malignant tumors of the digestive system. Med Sci Monit 2020;26:e924671. 10.12659/MSM.924671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liang Z, Huang A, Wang L, et al. A radiomics model predicts the response of patients with advanced gastric cancer to PD-1 inhibitor treatment. Aging 2022;14:907–22. 10.18632/aging.203850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Corino VDA, Bologna M, Calareso G, et al. A CT-based radiomic signature can be prognostic for 10-Months overall survival in metastatic tumors treated with nivolumab: an exploratory study. Diagnostics 2021;11:979. 10.3390/diagnostics11060979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mallio CA, Napolitano A, Castiello G, et al. Deep learning algorithm trained with COVID-19 pneumonia also identifies immune checkpoint inhibitor therapy-related pneumonitis. Cancers 2021;13:652. 10.3390/cancers13040652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Himoto Y, Veeraraghavan H, Zheng J, et al. Computed Tomography-Derived radiomic metrics can identify responders to immunotherapy in ovarian cancer. JCO Precis Oncol []. 2019;3. 10.1200/PO.19.00038. [Epub ahead of print: 15 08 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Granata V, Fusco R, Costa M, et al. Preliminary report on computed tomography Radiomics features as biomarkers to immunotherapy selection in lung adenocarcinoma patients. Cancers 2021;13:3992. 10.3390/cancers13163992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Trebeschi S, Bodalal Z, van Dijk N, et al. Development of a prognostic AI-Monitor for metastatic urothelial cancer patients receiving immunotherapy. Front Oncol 2021;11:637804. 10.3389/fonc.2021.637804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheng J, Pan Y, Huang W, et al. Differentiation between immune checkpoint inhibitor-related and radiation pneumonitis in lung cancer by CT radiomics and machine learning. Med Phys 2022;49:1547–58. 10.1002/mp.15451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang CY, Ginat DT. Preliminary computed tomography Radiomics model for predicting pretreatment CD8+ T-cell infiltration status for primary head and neck squamous cell carcinoma. J Comput Assist Tomogr 2021;45:629–36. 10.1097/RCT.0000000000001149 [DOI] [PubMed] [Google Scholar]
- 98.Zhu Y, Yao W, Xu B-C, et al. Predicting response to immunotherapy plus chemotherapy in patients with esophageal squamous cell carcinoma using non-invasive radiomic biomarkers. BMC Cancer 2021;21:1167. 10.1186/s12885-021-08899-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shen L, Fu H, Tao G, et al. Pre-Immunotherapy contrast-enhanced CT Texture-Based classification: a useful approach to non-small cell lung cancer immunotherapy efficacy prediction. Front Oncol 2021;11:591106. 10.3389/fonc.2021.591106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park KJ, Lee J-L, Yoon S-K, et al. Radiomics-based prediction model for outcomes of PD-1/PD-L1 immunotherapy in metastatic urothelial carcinoma. Eur Radiol 2020;30:5392–403. 10.1007/s00330-020-06847-0 [DOI] [PubMed] [Google Scholar]
- 101.Bonnin A, Durot C, Barat M, et al. Ct texture analysis as a predictor of favorable response to anti-PD1 monoclonal antibodies in metastatic skin melanoma. Diagn Interv Imaging 2022;103:97–102. 10.1016/j.diii.2021.09.009 [DOI] [PubMed] [Google Scholar]
- 102.Nardone V, Tini P, Pastina P, et al. Radiomics predicts survival of patients with advanced non-small cell lung cancer undergoing PD-1 blockade using nivolumab. Oncol Lett 2020;19:1559–66. 10.3892/ol.2019.11220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Colen RR, Rolfo C, Ak M, et al. Radiomics analysis for predicting pembrolizumab response in patients with advanced rare cancers. J Immunother Cancer 2021;9:e001752. 10.1136/jitc-2020-001752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Flaus A, Habouzit V, de Leiris N, et al. Outcome prediction at patient level derived from pre-treatment 18F-FDG PET due to machine learning in metastatic melanoma treated with anti-PD1 treatment. Diagnostics 2022;12:388. 10.3390/diagnostics12020388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aoude LG, Wong BZY, Bonazzi VF, et al. Radiomics biomarkers correlate with CD8 expression and predict immune signatures in melanoma patients. Mol Cancer Res 2021;19:950–6. 10.1158/1541-7786.MCR-20-1038 [DOI] [PubMed] [Google Scholar]
- 106.Wang Z-L, Mao L-L, Zhou Z-G, et al. Pilot study of CT-based Radiomics model for early evaluation of response to immunotherapy in patients with metastatic melanoma. Front Oncol 2020;10:1524. 10.3389/fonc.2020.01524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hectors SJ, Lewis S, Besa C, et al. Mri radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur Radiol 2020;30:3759–69. 10.1007/s00330-020-06675-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khene Z-E, Kokorian R, Mathieu R, et al. Metastatic clear cell renal cell carcinoma: computed tomography texture analysis as predictive biomarkers of survival in patients treated with nivolumab. Int J Clin Oncol 2021;26:2087–93. 10.1007/s10147-021-02003-w [DOI] [PubMed] [Google Scholar]
- 109.Liu C, Gong J, Yu H, et al. A CT-based Radiomics approach to predict nivolumab response in advanced non-small-cell lung cancer. Front Oncol 2021;11:544339. 10.3389/fonc.2021.544339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shen LL, Tao GY, Fu HC, et al. [Predicting response to non-small cell lung cancer immunotherapy using pre-treatment contrast-enhanced CT texture-based classification]. Zhonghua Zhong Liu Za Zhi 2021;43:541–5. 10.3760/cma.j.cn112152-20190725-00468 [DOI] [PubMed] [Google Scholar]
- 111.Rundo F, Bersanelli M, Urzia V, et al. Three-Dimensional deep noninvasive Radiomics for the prediction of disease control in patients with metastatic urothelial carcinoma treated with immunotherapy. Clin Genitourin Cancer 2021;19:396–404. 10.1016/j.clgc.2021.03.012 [DOI] [PubMed] [Google Scholar]
- 112.Rundo F, Banna GL, Prezzavento L, et al. 3D Non-Local neural network: a non-invasive biomarker for immunotherapy treatment outcome prediction. case-study: metastatic urothelial carcinoma. J Imaging 2020;6:133. 10.3390/jimaging6120133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barabino E, Rossi G, Pamparino S, et al. Exploring response to immunotherapy in non-small cell lung cancer using Delta-Radiomics. Cancers 2022;14:350. 10.3390/cancers14020350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guerrisi A, Russillo M, Loi E, et al. Exploring CT texture parameters as predictive and response imaging biomarkers of survival in patients with metastatic melanoma treated with PD-1 inhibitor nivolumab: a pilot study using a Delta-Radiomics approach. Front Oncol 2021;11:704607. 10.3389/fonc.2021.704607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim BS, Kang J, Jun S, et al. Association between immunotherapy biomarkers and glucose metabolism from F-18 FDG PET. Eur Rev Med Pharmacol Sci 2020;24:8288–95. 10.26355/eurrev_202008_22625 [DOI] [PubMed] [Google Scholar]
- 116.Devkota L, Starosolski Z, Rivas CH, et al. Detection of response to tumor microenvironment-targeted cellular immunotherapy using nano-radiomics. Sci Adv 2020;6:eaba6156. 10.1126/sciadv.aba6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dittrich D, Pyka T, Scheidhauer K, et al. Textural features in FDG-PET/CT can predict outcome in melanoma patients to treatment with vemurafenib and Ipililumab. Nuklearmedizin 2020;59:228–34. 10.1055/a-1140-5458 [DOI] [PubMed] [Google Scholar]
- 118.Zerunian M, Caruso D, Zucchelli A, et al. Ct based radiomic approach on first line pembrolizumab in lung cancer. Sci Rep 2021;11:6633. 10.1038/s41598-021-86113-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eresen A, Yang J, Shangguan J, et al. Mri radiomics for early prediction of response to vaccine therapy in a transgenic mouse model of pancreatic ductal adenocarcinoma. J Transl Med 2020;18:61. 10.1186/s12967-020-02246-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mihaylov IB, Totiger TM, Giret TM, et al. Toward prediction of abscopal effect in radioimmunotherapy: pre-clinical investigation. PLoS One 2021;16:e0255923. 10.1371/journal.pone.0255923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Eresen A, Yang J, Shangguan J, et al. Detection of immunotherapeutic response in a transgenic mouse model of pancreatic ductal adenocarcinoma using multiparametric MRI Radiomics: a preliminary investigation. Acad Radiol 2021;28:e147–54. 10.1016/j.acra.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vincent P, Maeder ME, Hunt B, et al. Ct radiomic features of photodynamic priming in clinical pancreatic adenocarcinoma treatment. Phys Med Biol 2021;66:175006. 10.1088/1361-6560/ac1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khatua S, Cooper LJN, Sandberg DI, et al. Phase I study of intraventricular infusions of autologous ex vivo expanded NK cells in children with recurrent medulloblastoma and ependymoma. Neuro Oncol 2020;22:1214–25. 10.1093/neuonc/noaa047 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005292supp001.pdf (1.4MB, pdf)