Abstract
Control of tropical theileriosis, caused by the apicomplexan Theileria annulata, depends on the use of a single drug, buparvaquone, the efficacy of which is compromised by the emergence of resistance. The present study was undertaken to improve understanding of the role of mutations conferring buparvaquone resistance in T. annulata, and the effects of selection pressures on their emergence and spread. First, we investigated genetic characteristics of the cytochrome b locus associated with buparvaquone resistance in 10 susceptible and 7 resistant T. annulata isolates. The 129G (GGC) mutation was found in the Q01 binding pocket and 253S (TCT) and 262S (TCA) mutations were identified within the Q02 binding pocket. Next, we examined field isolates and identified cytochrome b mutations 129G (GGC), 253S (TCT) and 262S (TCA) in 21/75 buffalo-derived and 19/119 cattle-derived T. annulata isolates, providing evidence of positive selection pressure. Both hard and soft selective sweeps were identified, with striking differences between isolates. For example, 19 buffalo-derived and 7 cattle-derived isolates contained 129G (GGC) and 253S (TCT) resistance haplotypes at a high frequency, implying the emergence of resistance by a single mutation. Two buffalo-derived and 12 cattle-derived isolates contained equally high frequencies of 129G (GGC), 253S (TCT), 129G (GGC)/253S (TCT) and 262S (TCA) resistance haplotypes, implying the emergence of resistance by pre-existing or recurrent mutations. Phylogenetic analysis further revealed that 9 and 21 unique haplotypes in buffalo and cattle-derived isolates were present in a single lineage, suggesting a single origin. We propose that animal migration between farms is an important factor in the spread of buparvaquone resistance in endemic regions of Pakistan. The overall outcomes will be useful in understanding how drug resistance emerges and spreads, and this information will help design strategies to optimise the use and lifespan of the single most drug use to control tropical theileriosis.
Keywords: Buparvaquone, Theilieria annulata, resistance, Soft selective sweep, Hard selective sweep
Graphical abstract

Highlights
-
•
First link between genetic mutations and buparvaquone drug resistance in T. annulata.
-
•
The data will help to design strategies to optimise the use and lifespan of the single most drug use to control tropical theileriosis.
1. Introduction
Tropical theileriosis, caused by Theileria annulata and transmitted by Hyalomma ticks, is one of the most important livestock diseases in Asia and North Africa. The disease is highly pathogenic, infecting mononuclear cells of mammalian hosts (Nene et al., 2016). T. annulata has a major impact on food production in low- and middle-income countries throughout South Asia, the Middle East and North Africa, where efficient agriculture and livestock production is a priority under the UN Sustainable Development Goals. Tropical theileriosis is less severe in indigenous cattle (20%) than in cross-breed or exotic animals (80%) in Pakistan. The disease become more substantial when programs were launched to increase milk production in this country. Mostly, the disease occurs in subclinical form, leading to significant economic losses. The disease causes severe economic losses due to high mortality and morbidity of livestock, having a significant impact on meat and milk production. The disease has been treated with the use of antiprotozoal drugs such as buparvaquone. The drug was introduced in the early 1990s, to exert its antiprotozoal actions against T. annulata (Jabbar et al., 2015).
Hydroxynaphthoquinone drugs, including buparvaquone and atovaquone compounds, are widely used for the treatment of protozoan parasites of livestock and humans, respectively. These drugs bind to parasite cytochrome b, inhibiting mitochondrial respiration (Fry and Pudney, 1992; McHardy et al., 1983, 1985; McHardy and Morgan, 1985). Different studies have shown associations between resistance to atovaquone in Plasmodium and Toxoplasma species and mutations in cytochrome b catalytic Q01 (codon 129–148), and oxidative Q02 (codon 244–266) binding pockets (Kessl et al., 2005, 2006, 2007; Korsinczky et al., 2000; Schwobel et al., 2003; Song et al., 2015; Srivastava et al., 1999). Atovaquone resistance-associated cytochrome b mutations have been reported at codon 268 in Plasmodium falciparum (David et al., 2003; Farnert et al., 2003; Fivelman et al., 2002; Musset et al., 2006; Schwartz et al., 2003); mutations at codon 133, 144 and 284 in Plasmodium berghei (Siregar et al., 2008; Syafruddin et al., 1999); and mutations at codon 129 and 254 in Toxoplasma gondii (McFadden et al., 2000).
The control of theileriosis heavily depends on the use of buparvaquone; it is the only effective commercially available drug for use in livestock all over the world including Pakistan. Clinical cases of buparvaquone resistance have been reported and represent a serious threat to efficient livestock production in the recent years (Cui et al., 2015; Mhadhbi et al., 2010; Muraguri et al., 2006). Current understanding of the mechanism of buparvaquone resistance in T. annulata is limited, albeit links between mutations in cytochrome b and the resistance phenotype have been demonstrated (Mhadhbi et al., 2015). Moreover, a few studies have demonstrated buparvaquone resistance-associated mutations at codons 129, 253 and 262 in T. annulata drug-treated isolates collected from Iran (Sharifiyazdi et al., 2012), Tunisia (Mhadhbi et al., 2015) and Sudan (Chatanga et al., 2019).
Selective sweep models could potentially explain how resistance mutations emerge in parasites; whereby the use of antiparasitic drugs provides positive selection pressure for adaptive mutations in the resistance candidate loci of the parasite (Chaudhry and Gilleard, 2015). A hard selective sweep is characterised by a single resistance haplotype rising from a recent mutation at a high frequency in each parasite isolate (Chaudhry et al., 2015, 2020; Shaukat et al., 2019). A soft selective sweep is characterised by the presence of multiple resistance haplotypes at a high frequency in each isolate, derived from either recurrent mutations appearing after the onset of selection, or pre-existing mutations before the onset of selection (Chaudhry et al., 2016a, 2020; Shaukat et al., 2019). Understanding adaptive mutations in response to selection can help to determine their origin and spread (Chaudhry and Gilleard, 2015). For example, phylogenetic models might imply that new resistance mutations could arise from a single origin, become fixed by selection and then spread through parasites by migration. This scenario would likely be a consequence of the gene flow of drug resistance alleles through host movement. In contrast, resistance mutations could repeatedly arise from multiple origins, become fixed by selection and migrate between parasites as a result of host movement (Chaudhry et al., 2015, 2016a; Shaukat et al., 2019, 2021).
High throughput deep amplicon sequencing using the Illumina Mi-Seq platform is an accurate and relatively low-cost method for the identification of amplicon sequence variants in parasites. The method has transformed the study of benzimidazole resistance in nematode parasites (Ali et al., 2019; Sargison et al., 2019), and pyrimethamine resistance in Plasmodium (Shaukat et al., 2019, 2021) and has the potential to open new areas of research to improve understanding of buparvaquone resistance in Theileria. The method can generate sequence reads up to 600 bp in length by targeting primer binding to conserved sites. Moreover, with the use of barcoded primers, up to 384 samples can be pooled and sequenced in a single Mi-Seq run.
The overall aim of the present study was to develop a genetic approach to improve understanding of the T. annulata cytochrome b locus in buparvaquone resistance. This allows the analysis of alleles conferring resistance and susceptibility to investigate different models of emergence and the spread of associated mutations. The specific objectives were: (i) To characterise single nucleotide polymorphism (SNPs) in the cytochrome b locus and their genetic role in buparvaquone resistance in T. annulata. (ii) To investigate the consequences of selection pressure on the emergence and spread of T. annulata cytochrome b resistance-conferring mutations in the field isolates. Achieving these objectives will lead to clearer understanding of buparvaquone resistance in an important tropical livestock pathogen, with the potential to inform enhanced animal health, improved global food security, and poverty alleviation through reduced production losses.
2. Materials and methods
2.1. Buparvaquone susceptible and resistant T. annulata isolates
We employed four infected cell lines (TA-ank, TA-but, TA-has, TA-mor) produced by the infection of blood mononuclear cells with laboratory-maintained stocks of T. annulata originally isolated in Turkey, India (Katzer et al., 1994), Tunisia and Morocco. The stock was selected to represent buparvaquone susceptible isolates. To extract the gDNA, 50 μl of each isolate was transferred into a fresh tube and centrifuged for 5 min, before removing the supernatant and mixing with 25 μl Direct PCR lysis buffer (Viagen), Proteinase K (Qiagen), and 1M dithiothreitol (DTT) (New England BioLabs) and dithiothreitol (DTT).
Six known buparvaquone susceptible isolates (674, Battan C, Battan P4, Chargui P5, Jed4, Jed4 P10) were selected from different endemic regions in northeast Tunisia. The Battan C, Battan P4, Jed4 P10 and Jed4 isolates were chosen based on previous studies confirming the susceptibility of these stocks to buparvaquone (Darghouth et al., 1996). The Chergui P5 and 674 isolates were chosen from the animals cured after the first injection of buparvaquone. Seven known buparvaquone-resistant isolates (ST2/13, ST2/19, 739, 881III, 5911, 8307, BC2T) were selected from different endemic regions in northeast Tunisia. Two buparvaquone-resistant isolates (ST2/13, ST2/19) were taken from the previous study by Mhadhbi et al. (2015). The resistant isolates were collected at different times after treatment as follows: isolate ST2/13 was taken 24 h after the first treatment and the ST2/19 isolate was taken 48 h after the third treatment (Mhadhbi et al., 2015). Five stocks (739, 881III, 5911, 8307, BC2T) were initially isolated at various times points from tropical theileriosis clinical cases of treatment failure based on the absence of clinical improvement despite the early initiation of treatment with the conventional dose of 2.5 mg/kg of buparvaquone, and the persistence of a high parasitaemia after at least three doses of buparvaquone. All isolates were collected by lymph node punctures or from whole blood and cultured in a complete RPMI 1640 medium. They had been used at the low passage to avoid in-vitro pressure selection (Mhadhbi et al., 2015). Genomic DNA was extracted using a Promega Wizard DNA Purification kit (Madison, WI, USA) according to the manufacturer's instructions.
2.2. Theileria annulata field isolates
The study was conducted in nine known tropical theileriosis endemically regions (Lahore, Gujranwala, Chakwal, Qadirabad, Okara, Sahiwal, Hafizabad, Bahawalpur, Vehari) of the Punjab province of Pakistan. We chose this region because animals are treated sporadically often with generic buparvaquone drugs of unknown quality. Buparvaquone has been widely used for the therapy of tropical theileriosis because they are relatively inexpensive (as per comm of Kiran Afshan).
Field isolates were taken from suspected piroplasm-infected cattle and buffalo and presented at veterinary clinics across the endemic regions of Pakistan between 2020 and 2021. Piroplasm-negative cattle blood samples were provided by Dr Tim Connelly, Roslin Institute, University of Edinburgh. The samples were collected by para-veterinary staff under the supervision of local veterinarians following consent from the animal owners. The study was approved by the Institutional Review Board of the Quaid-i-Azam University Islamabad Pakistan (No. #BEC-FBS-QAU2017). Peripheral blood smears were prepared and stained with 4% Giemsa and examined microscopically to detect piroplasms. Genomic DNA was isolated from positive samples by lysis with GS buffer containing proteinase K as described in the TIANamp Blood DNA Kit (TIANGEN Biotech Co. Ltd, Beijing) and stored at −20 °C. ‘Haemoprotobiome’ high-throughput sequencing was performed on piroplasm-positive blood samples to confirm the presence of T. annulata as previously described (Chaudhry et al., 2019). Briefly, 194-piroplasm positive field samples [buffalo (n = 75), cattle (n = 119)] were examined to identify the species of T. annulata involved in the infections.
2.3. PCR amplification, Illumina Mi-Seq run and bioinformatics data handling of cytochrome b locus
A 516 bp fragment of the cytochrome b locus of T. annulata the buparvaquone susceptible and resistant isolates and field isolates was amplified. The primer sets, adapter/barcoded PCR amplification conditions and magnetic bead purification were previously described by Chaudhry et al. (2021) (Supplementary Tables S1 and S2). Ten μl of each barcoded bead purified PCR product were combined to make a pooled library which was subject to agarose gel electrophoresis. Cytochrome b products were excised and purified from the gel using a commercial kit (QIAquick Gel Extraction Kit, Qiagen, Germany), followed by purification of eluted DNA using AMPure XP Magnetic Beads (1X) (Beckman Coulter, Inc.). Purified products were then combined into a single purified DNA pool library. The library was quantified with qPCR library quantification kit (KAPA Biosystems, USA) and then run on an Illumina MiSeq Sequencer using a 600-cycle pair-end reagent kit (MiSeq Reagent Kits v2, MS-103-2003) at a concentration of 15 nM with the addition of 15% Phix Control v3 (Illumina, FC-11-2003) described by Chaudhry et al. (2021).
The Illumina Mi-Seq post-run processing uses the barcoded indices to separate all sequences by sample and generate FASTQ files. The FASTQ files of the buparvaquone susceptible and resistant T. annulata isolates and T. annulata field isolates have been made freely available through the Mendeley database (https://doi.org/10.17632/n6xpdcntvf.1). These files were analysed using Mothur v1.39.5 software (Kozich et al., 2013; Schloss et al., 2009) with modifications in the standard operating procedures of Illumina Mi-Seq in the Command Prompt pipeline described by Chaudhry et al. (2021).
2.4. Allele frequency of buparvaquone resistance-associated SNPs in the cytochrome b locus
The calculation of the relative allele frequencies of T. annulata cytochrome b resistance-associated mutations identified in the buparvaquone susceptible and resistant isolates and field isolates was performed by dividing the number of sequence reads of each isolate by the total number of reads (package ggplot2).
2.5. Genetic models of buparvaquone resistance-associated SNPs in the cytochrome b locus
The sequence reads of the T. annulata cytochrome b locus from each isolate were imported into the FaBox 1.5 tool (birc.au.dk) to collapse the sequences that showed 100% base pair similarity after corrections into a single haplotype (freely available through the Mendeley database https://doi.org/10.17632/n6xpdcntvf.1). The haplotypes of cytochrome b were then aligned using the MUSCLE alignment tool in Geneious v10.2.5 software (Biomatters Ltd, New Zealand) for the analysis of the emergence of buparvaquone resistance-associated mutations in the cytochrome b locus. The haplotypes of the T. annulata cytochrome b locus were selected for neutrality analysis. Tests for selective neutrality were analysed to determine whether the observed frequency distribution of sequence polymorphism in T. annulata cytochrome b locus departed from neutral expectations using the DnaSP 5.10 software package (Librado and Rozas, 2009). The neutrality tests included Tajima's D (Tajima, 1989) and Fay & Wu's H (Fay and Wu, 2000) methods.
A split tree of cytochrome b haplotypes was created in the SplitTrees4 software (bio-soft.net) using the neighbour-joining method and the JukesCantor model of substitution for the analysis of the origin of buparvaquone resistance-associated mutations in the cytochrome b locus. The appropriate model of nucleotide substitutions for neighbour-joining analysis was selected by using the jModeltest 12.2.0 program (Posada, 2008). The network tree of T. annulata cytochrome b haplotypes was produced based on the neighbour-joining algorithm built on a sparse network and the epsilon parameter is set to zero default in Network 4.6.1 software (Fluxus Technology Ltd.) for the analysis of the spread of buparvaquone resistance-associated mutations in the cytochrome b locus. All unnecessary median vectors and links were removed with the star contractions. The number of mutations separating adjacent sequence nodes was indicated along connecting branches and the length of the lines connecting the haplotypes is proportional to the number of nucleotide changes.
3. Results
3.1. Buparvaquone resistance-associated SNPs in susceptible and resistant T. annulata isolates
A 516 bp region of the cytochrome b locus of the 10 susceptible (674, Battan C, Battan P4, Chargui P5, Jed4, Jes4P10, TA-ANK, TA-BUT, TA-HAS, TA-MARR) and 7 resistant (739, 881III, 5911, 8307, BC2T, ST2/13, ST2/19) T. annulata isolates were compared. Eleven point mutations in the T. annulata cytochrome b locus were found at codons A133V (GCT/GTT), V135F (GTC/TTC), I136L (ATA/CTA), G138V (GGT/GTT), L139F (TTG/TTT), L140F (TTG/TTT), K141N (AAA/AAC), F143L (GGA/GTA), G144V (GGA/GTA) and T146A (ACT/GCT), within the cytochrome b Q01 binding pocket. These SNPs were present at similar frequencies across the buparvaquone susceptible and resistant T. annulata isolates (Table 1). One point mutation was found in the Q01 binding pocket at codon 129G (GGC) in resistant isolates compared to S129 (AGC) in susceptible isolates (Table 1). Similarly, two-point mutations were found within the Q02 binding pocket at codons 253S (TCT) and 262S (TCA) in resistant isolates compared to P253 (CCT) and L262 (TTA) in susceptible isolates (Table 1). Overall, the results provide the analysis of cytochrome b mutations potentially involved in the development of buparvaquone resistance and the first high throughput genetic link between mutations and drug resistance in T. annulata.
Table 1.
Cytochrome b mutations identified in buparvaquone susceptible T. annulata isolates collected from an endemic region of Turkey, India, Tunisia, and Morocco and resistant T. annulata isolates collected from an endemic region of Tunisia. Comparison shows the buparvaquone susceptible and resistant isolates. Nonsynonymous mutations at positions S129G (AGC/GGC), P253S (CCT/TCT) and L262S (TTA/TCA) in the binding pockets Q01 and Q02 of the cytochrome b locus differ between susceptible and resistant T. annulata isolates.
| Q01 (130–148) |
Q02 (244–266) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide |
385 (AGC/GGC) |
398 (GCT/GTT) |
403 (GTC/TTC) |
406 (ATA/CTA) |
413 (GGT/GTT) |
418 (TTG/TTT) |
420 (TTG/TTT |
423 (AAA/AAC) |
427 (GGA/GTA) |
431 (GGA/GTA) |
436 (ACT/GCT) |
757 (CCT/TCT) |
785 (TTA/TCA) |
|
| Codon | S129G | A133V | V135F | I136L | G138V | L139F | L140F | K141N | F143L | G144V | T146A | P253S | L262S | |
| Susceptible isolates | 674 | S | A | V | I | G/V | L/F | L/F | K | F | G/V | T | P | L |
| Battan C | S | A | V/F | I | G/V | L/F | L/F | K/N | F/L | G/V | T | P | L | |
| Battan P4 | S | A/V | V/F | I/L | G/V | L/F | L/F | K/N | F | G/V | T | P | L | |
| Chargui P5 | S | A/V | V/F | I/L | G | L/F | L/F | K/N | F/L | G/V | T | P | L | |
| Jed4 | S | A | V | I | G | L | L/F | K | F/L | G/V | T | P | L | |
| Jed4P10 | S | A | V/F | I/L | G/V | L/F | L/F | K | F/L | G/V | T | P | L | |
| TA-ANK | S | A | V | I | G | L | L | K | F | G | T/A | P | L | |
| TA-BUT | S | A | V | I | G | L | L | K | F | G | T/A | P | L | |
| TA-HAS | S | A | V | I | G | L | L | K | F | G | T | P | L | |
| TA-MARR | S | A | V | I | G | L | L | K | F | G | T/A | P | L | |
| Resistant isolates | 739 | S/G | A | V/F | I | G/V | L/F | L/F | K | F | G/V | T | P/S | L/S |
| 881III | S/G | A | V | I | G/V | L | L/F | K | F | G/V | T | P/S | L | |
| 5911 | S/G | A | V/F | I | G | L/F | L/F | K | F | G/V | T/A | P/S | L | |
| 8307 | S/G | A | V/F | I | G/V | L | L/F | K | F | G/V | T | P/S | L | |
| BC2T | S/G | A | V | I | G/V | L | L/F | K | F | G/V | T | P/S | L | |
| ST2/13 | S | A | V/F | I | G/V | L/F | L/F | K/N | F | G/V | T/A | P/S | L/S | |
| ST2/19 | S/G | A/V | V/F | I/L | G/V | L/F | L/F | K | F | G/V | T | P/S | L/S | |
3.2. Buparvaquone resistance-associated SNPs in buffalo- and cattle-derived T. annulata field isolates and their impact on positive selection pressure
A 516 bp fragment of the T. annulata cytochrome b locus was amplified from 75 buffalo-derived and 119 cattle-derived isolates across the endemic regions of Pakistan (Supplementary Table S3). The relative allele frequencies of three mutations S129G (AGC/GGC), P253S (CCT/TCT) and L262S (TTA/TCA) correspond to the buparvaquone binding pockets were determined. Buparvaquone resistance-associated mutations 129G (GGC), 253S (TCT) and 129G (GGC)/253S (TCT) were present in 21/75 buffalo-derived T. annulata (Fig. 1, Table 2). The 129G (GGC) mutation was present at frequencies of 4.3–100%. The 253S (TCT) mutation was present at frequencies of 19.2–27.3%. Conversely, 129G (GGC)/253S (TCT) double mutations were detected at frequencies of 1.3–4.3%. 262S (TCA) was not detected in any buffalo-derived T. annulata isolates (Fig. 1, Table 2). In contrast, buparvaquone resistance-associated mutations 129G (GGC), 253S (TCT) and 262S (TCA) were present in 19/119 cattle-derived T. annulata isolates (Fig. 1, Table 2). The 129G (GGC) mutation was present at frequencies of 1.4–100%. 253S (TCT) mutation was present at frequencies between 3.9 and 43%. 262S (TCA) mutation was detected at frequencies of 2.6–13.7%. 129G (GGC)/253S (TCT) was not detected in any cattle-derived T. annulata (Fig. 1, Table 2). Overall, the results indicated the positive selection pressure on the cytochrome b locus of 21 buffalo-derived and 19 cattle-derived T. annulata isolates.
Fig. 1.
Relative allele frequencies of cytochrome b resistance-associated mutations in 75 buffalo-derived and 119 cattle-derived T. annulata isolates. The susceptible SNP frequency - S129 (AG), P253 (CCT) and L262 (TTA) - is shown in red and resistance-associated SNP frequency - 129G (GGC), 253S (TCT), 262S (TCA) and 129G (GGC)/253S (TCT) - is shown in light green, dark green, blue and pink. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Relative allele frequencies of the cytochrome b mutations in 21 buffalo-derived and 19 cattle-derived T. annulata resistant isolates collected from the endemic region of Pakistan. Each isolate was characterised by susceptible [S129 (AGC), P253 (CCT) and L262 (TTA)] and resistance mutations [129G (GGC), 253S (TCT), 262S (TCA) and 129G (GGC)/253S (TCT)].
| Field isolates | Total no of Illumina MiSeq reads | Susceptible reads | Resistance reads | Susceptible reads % [S129(AGC), P253 (CCT), L262(TTA)] | Resistant reads % [129G(GGC)] | Resistant reads % [253S(TCT)] | Resistant reads % [262S(TCA)] | Resistant reads % [129G(GGC)/253S(TCT)] | Location |
|---|---|---|---|---|---|---|---|---|---|
| Buffalo | |||||||||
| CY110–B | 18868 | 13699 | 5169 | 72.6 | 27.3 | Gujranwala | |||
| CY123–B | 46438 | 42178 | 4260 | 90.8 | 9.17 | Okara | |||
| CY59–B | 19801 | 14110 | 5691 | 71.2 | 28.7 | Gujranwala | |||
| CY49–B | 12313 | 12313 | 100 | Gujranwala | |||||
| CY5–B | 12341 | 12341 | 100 | Gujranwala | |||||
| CY48–B | 16542 | 16542 | 100 | Gujranwala | |||||
| CY4–B | 21578 | 16127 | 5451 | 74.7 | 25.2 | Gujranwala | |||
| CY60–B | 15433 | 15433 | 100 | Gujranwala | |||||
| CY47–B | 17642 | 17642 | 100 | Gujranwala | |||||
| CY254–B | 12256 | 12256 | 100 | Bahawalpur | |||||
| CY256–B | 12892 | 12892 | 100 | Bahawalpur | |||||
| CY257–B | 47942 | 47942 | 100 | Bahawalpur | |||||
| CY260–B | 13314 | 13314 | 100 | Vehari | |||||
| CY266–B | 21823 | 21823 | 100 | Vehari | |||||
| CY269–B | 12378 | 12378 | 100 | Vehari | |||||
| CY278–B | 21004 | 21004 | 100 | Vehari | |||||
| CY317–B | 14988 | 14988 | 100 | Gujranwala | |||||
| CY320–B | 23324 | 23324 | 100 | Gujranwala | |||||
| CY321–B | 24581 | 16842 | 7739 | 68.5 | 7.8 | 19.2 | 4.3 | Gujranwala | |
| CY322–B | 102021 | 73077 | 28944 | 71.6 | 4.3 | 22.7 | 1.3 | Gujranwala | |
| CY323–B | 12175 | 100 | Gujranwala | ||||||
| Cattle | |||||||||
| CY90–C | 65051 | 48644 | 16407 | 74.7 | 16.4 | 6.0 | 2.6 | Gujranwala | |
| CY136–C | 23906 | 18706 | 5200 | 78.2 | 21.7 | Okara | |||
| CY150–C | 28039 | 14904 | 13135 | 53.1 | 12.9 | 33.9 | Okara | ||
| CY6–C | 21001 | 17589 | 3412 | 83.75 | 16.2 | Gujranwala | |||
| CY26–C | 28672 | 16326 | 12346 | 56.9 | 43.0 | Lahore | |||
| CY38–C | 35600 | 2343 | 33257 | 6.5 | 93.4 | Qadirabad | |||
| CY39–C | 45065 | 1999 | 43066 | 4.4 | 95.5 | Qadirabad | |||
| CY40–C | 29245 | 1113 | 28132 | 3.8 | 96.1 | Qadirabad | |||
| CY41–C | 80153 | 2393 | 77760 | 2.9 | 97.0 | Qadirabad | |||
| CY42–C | 58645 | 1402 | 57243 | 2.3 | 97.6 | Qadirabad | |||
| CY43–C | 12398 | 12398 | 100 | Qadirabad | |||||
| CY277–C | 57086 | 51667 | 5419 | 90.5 | 2.1 | 7.3 | Vehari | ||
| CY272–C | 65893 | 60825 | 5068 | 92.3 | 7.6 | Vehari | |||
| CY235–C | 60586 | 47949 | 12637 | 79.1 | 7.1 | 13.7 | Lodhran | ||
| CY268–C | 4483 | 4483 | 76.9 | 23.0 | Vehari | ||||
| CY212–C | 39057 | 1240 | 37817 | 3.1 | 96.8 | Gujranwala | |||
| CY252–C | 5173 | 5173 | 100 | Bahawalpur | |||||
| CY270–C | 79794 | 78497 | 4297 | 98.3 | 1.4 | 3.9 | Vehari | ||
| CY258–C | 11512 | 11512 | 100 | Bahawalpur | |||||
3.3. Nature of selective sweeps associated with the emergence of cytochrome b resistance-associated SNPs in buffalo- and cattle-derived T. annulata field isolates
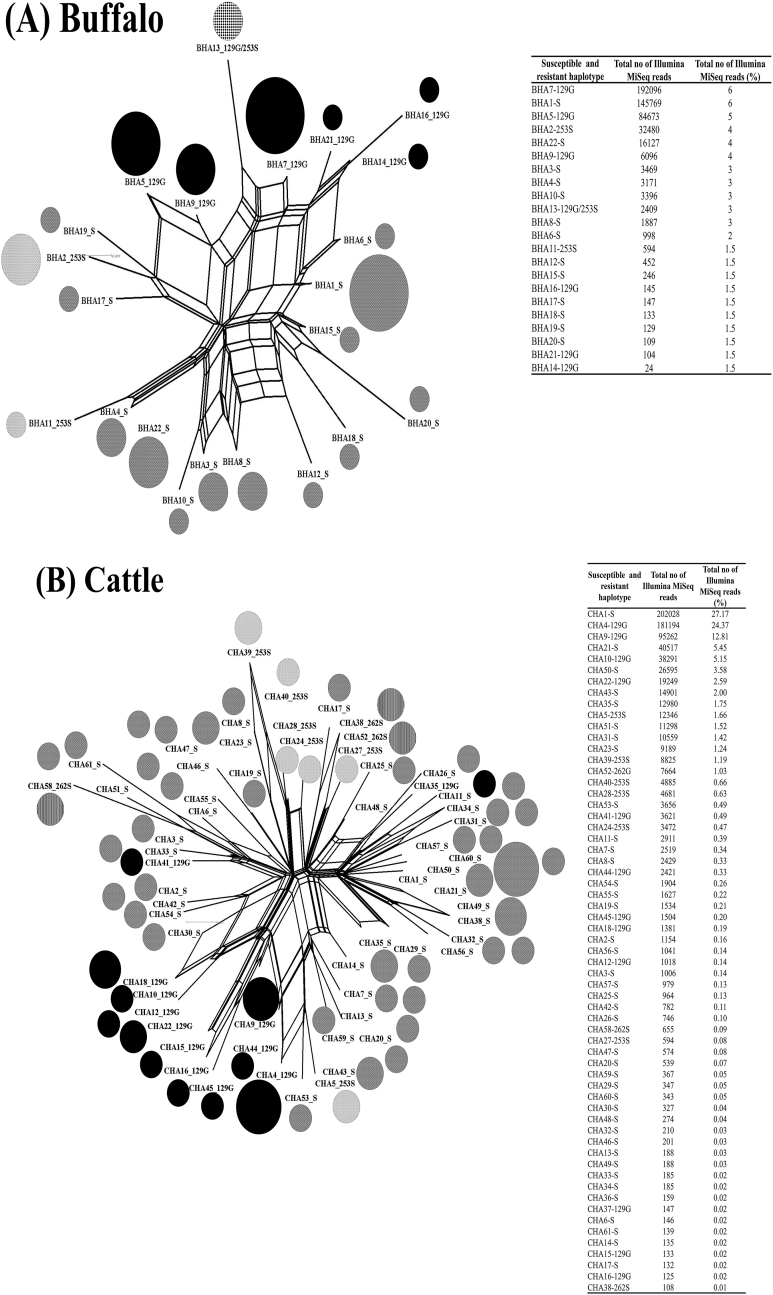
The analysis of 21 buffalo-derived T. annulata revealed that 18 isolates contained the 129G (GGC) resistance haplotype and one isolate (CY110–B) contained the 253S (TCT) resistance haplotype at a high frequency of 100% (Fig. 2A, Table 3), providing evidence of hard selective sweep patterns. In contrast, the CY321–B isolate contained resistance haplotypes of 129G (GGC) at a frequency of 24.5%, 253S (TCT) at a frequency of 59.9% and 129G (GGC)/253S (TCT) at a frequency of 13.8% (Fig. 2A, Table 3). The CY322–B isolate contained resistance haplotypes of 129G (GGC) at a frequency of 14.4%, 253S (TCT) at a frequency of 79.3% and 129G (GGC)/253S (TCT) at a frequency of 4.6% (Fig. 2A, Table 3). These two isolates had equally high frequencies of cytochrome b resistance-conferring haplotypes demonstrating evidence of soft selective sweep patterns. Neutrality analysis further revealed a significant departure of neutrality in those 2 isolates providing evidence of a signature of selection at the T. annulata cytochrome b locus (Table 3).
Fig. 2.
The 9/22 buffalo-derived (2A) and 21/61 cattle-derived (2B) resistance haplotypes (BHA and CHA) are represented by a different colour for the individual isolates (CY). Susceptible haplotypes are not indicated in the figure but are shown in Table 3. The haplotype distribution and the frequency based on the sequence reads generated per T. annulata isolates are shown in the insert table. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 3.
Haplotype diversity and signature of selection at the T. annulata cytochrome b locus. A total number of susceptible and resistant haplotypes from 21 buffalo-derived and 19 cattle-derived T. annulata isolates showed the haplotype diversity and signature of selection at the cytochrome b locus.
| Haplotype distribution |
Neutrality tests |
|||||||
|---|---|---|---|---|---|---|---|---|
| Field isolates | Total haplotypes | Susceptible haplotypes | 129G(GGC) resistance haplotype | 253S(TCT) resistance haplotype | 262S(TCA) resistance haplotype | 129G(GGC)/253S(TCT) resistance haplotype | Fay and Wu ‘s (H) | Tajima (D) |
| Buffalo | ||||||||
| CY110–B | 3 | 2 | 1 | 1.61917 | 1.95807 | |||
| CY123–B | 4 | 3 | 1 | 1.56652 | 1.95807 | |||
| CY59–B | 2 | 1 | 1 | 1.70958 | 2.46226 | |||
| CY49–B | 1 | 1 | N/A | |||||
| CY5–B | 1 | 1 | N/A | |||||
| CY48–B | 1 | 1 | N/A | |||||
| CY4–B | 2 | 1 | 1 | 1.55322 | 2.23140 | |||
| CY60–B | 1 | 1 | N/A | |||||
| CY47–B | 1 | 1 | N/A | |||||
| CY254–B | 1 | 1 | N/A | |||||
| CY256–B | 1 | 1 | N/A | |||||
| CY257–B | 1 | 1 | N/A | |||||
| CY260–B | 1 | 1 | N/A | |||||
| CY266–B | 1 | 1 | N/A | |||||
| CY269–B | 1 | 1 | N/A | |||||
| CY278–B | 1 | 1 | N/A | |||||
| CY317–B | 1 | 1 | N/A | |||||
| CY320–B | 1 | 1 | N/A | |||||
| CY321–B | 9 | 4 | 2 | 2 | 1 | 1.68770* | 1.75932* | |
| CY322–B | 15 | 9 | 3 | 2 | 1 | 1.10119* | 0.88492* | |
| CY323–B | 1 | 1 | N/A | |||||
| Total | 22 | 13 | 6 | 2 | 1 | |||
| Cattle | ||||||||
| CY90–C | 20 | 14 | 2 | 2 | 2 | 1.02457* | −0.58541* | |
| CY136–C | 6 | 5 | 1 | 1.26900 | 0.82865 | |||
| CY150–C | 6 | 3 | 1 | 2 | 1.39349 | 0.75819 | ||
| CY6–C | 4 | 3 | 1 | |||||
| CY26–C | 5 | 4 | 1 | |||||
| CY38–C | 4 | 1 | 3 | 1.54706* | 1.44162 | |||
| CY39–C | 8 | 3 | 5 | 1.74256* | 2.00448* | |||
| CY40–C | 5 | 2 | 3 | 1.69795* | 1.64710 | |||
| CY41–C | 4 | 1 | 3 | 1.62083* | 1.98644 | |||
| CY42–C | 6 | 3 | 3 | 1.53179* | 1.28502 | |||
| CY43–C | 1 | 1 | N/A | |||||
| CY277–C | 9 | 7 | 1 | 1 | 1.04126 | −0.29438 | ||
| CY272–C | 7 | 4 | 3 | 1.57241* | 1.52030 | |||
| CY235–C | 13 | 10 | 1 | 2 | 1.39659 | 0.51934 | ||
| CY268–C | 2 | 1 | 1 | 1.67822* | 2.34668 | |||
| CY212–C | 2 | 1 | 1 | |||||
| CY252–C | 1 | 1 | N/A | |||||
| CY270–C | 5 | 3 | 1 | 1 | 1.38874* | 1.01660 | ||
| CY258–C | 1 | 1 | N/A | |||||
| Total | 61 | 40 | 12 | 6 | 3 | |||
Statistically significant departure from neutrality determined with the use of simulations of the coalescent at p < 0.05*.
The analysis of 19 cattle-derived T. annulata revealed that 6 (CY212–C, 252-C, 258-C, 6-C, 43-C, 136-C) isolates contained 129G (GGC) resistance haplotypes and one isolate (CY26–C) contained 253S (TCT) resistance haplotype at a high-frequency of 100% (Fig. 2B, Table 3), demonstrating evidence of hard selective sweep patterns. In contrast, 9 isolates (CY268–C, 270-C, 272-C, 277-C, 38-C, 39-C, 40-C, 41-C, 42-C) contained 129G (GGC) resistance haplotypes and one isolate (CY150–C) contained 253S (TCT) resistance haplotypes at frequencies ranging between 16 and 84% (Fig. 2B, Table 3). The CY235–C isolate contained resistance haplotypes of 129G (GGC) at a frequency of 34.2% and 262S (TCA) at a frequency of 60.6%. The CY90–C isolate contained resistance haplotypes of 129G (GGC) at a frequency of 69.8%, 253S (TCT) at a frequency of 22.6% and 262S (TCA) at a frequency of 0.07% (Fig. 2B, Table 3). These 12 isolates had equally high frequencies of cytochrome b resistance-conferring haplotypes demonstrating evidence of soft selective sweep patterns. Neutrality analysis further revealed a significant departure of neutrality in 9 out of 12 isolates providing evidence of a signature of selection at the T. annulata cytochrome b locus (Table 3).
3.4. Phylogeny of cytochrome b resistance-associated SNPs in buffalo- and cattle-derived T. annulata field isolates and their impact on the origin and spread of buparvaquone resistance
The analysis of 21 buffalo-derived T. annulata revealed 22 unique cytochrome b haplotypes. Six haplotypes encoded 129G (GGC) resistance mutations, 2 haplotypes encoded 253S (TCT) resistance mutations, one haplotype encoded 129G (GGC)/253S (TCT) double resistance mutations and 13 haplotypes encoded S129 (AGC), P253 (CCT) and L262 (TTA) susceptible mutations (Fig. 3A, Fig. 4A, Table 3). A split tree analysis showed that 129G (GGC) resistance mutants and 129G (GGC)/253S (TCT) resistance mutants carried 7 haplotypes and 253S (TCT) resistance mutant carried 2 haplotypes were located in a single lineage demonstrating evidence of a single origin. (Fig. 3A). The network analysis showed that the BHA5-129G haplotype was present at a high frequency in eight different isolates collected from the livestock farms in the Okara and Gujranwala regions. The BHA7-129G haplotype was present in ten different isolates collected from the livestock farms in the Okara, Gujranwala, Vehari and Bahawalpur regions. BHA9-129G and BHA13-129G/253S haplotypes were present in two isolates collected from livestock farms in the Gujranwala region. BHA2-253S haplotype was present in three isolates and BHA11-253S haplotype was present in two isolates collected from the livestock farms in the Gujranwala region (Fig. 4A). The data provide evidence of a high level of gene flow predicted to occur due to the unregulated animal movement. However, three haplotypes (BHA14-129G, BHA16-129G and BHA21-129G) were present at a frequency of 100% in a single isolate collected from the livestock farms in the Gujranwala region (Fig. 4A), indicating that the resistance associated with those haplotypes had not yet spread.
Fig. 3.
A split tree of 22 buffalo (3A) and 61 cattle-derived (3B) haplotypes of cytochrome b locus was generated in SplitsTrees4 software (Huson and Bryant, 2006). The pie chart circles represent the different haplotypes and the size of each circle is proportional to the number of sequence reads and frequency generated in that haplotype as indicated in the inserted table. The mutations in T. annulata haplotypes are shaded as follows: susceptible haplotypes are shown by grey shading; 129G (GGC) resistant haplotypes are shaded black; 129G (GGC)/253S (TCT) double mutant resistant haplotypes are shown by dots shading; 253S (TCT) resistant haplotypes are shown by white shading and 262S (TCA) resistant haplotypes are shown by black line shading.
Fig. 4.
A network tree of 22 buffalo-derived (4A) and 61 cattle-derived (4A) haplotypes of cytochrome b locus was generated in Network 4.6.1 software (Fluxus Technology Ltd). The size of each pie chart circle representing the haplotype was proportional to the number of sequences generated from different isolates. The colours in the pie chart circles replicate the haplotype frequency and their distribution as indicated in the inserted table. The mutation-carrying T. annulata haplotypes in different livestock farms are shown in the inserted map. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The analysis of 19 cattle-derived T. annulata revealed 61 unique cytochrome b haplotypes. 12 haplotypes encoded 129G (GGC) resistance mutations, 6 haplotypes encoded 253S (TCT) resistance mutations, 3 haplotypes encoded 262S (TCA) resistance mutations and 40 haplotypes encoded S129 (AGC), P253 (CCT) and L262 (TTA) susceptible mutations (Figs. 3B and 4B, Table 3). A split tree analysis showed that 129G (GGC), 253S (TCT) and 262S (TCA) resistance mutants carried 10, 5 and 2 haplotypes were located in a single lineage (Fig. 3B) demonstrating evidence of a single origin (Fig. 3B). The network tree analysis showed that CHA4-129G haplotype was present at a high frequency in 7 different isolates collected from the livestock farms in the Gujranwala and Qadirabad regions. The CHA9-129G haplotype was present in 9 different isolates collected from the livestock farms in the Bahawalpur, Qadirabad, Vehari and Lodhran regions. CHA10-129G haplotype was present in 3 isolates, CHA12-129G in 2 isolates, CHA22-129G in 5 isolates at high frequency collected from the livestock farms in the Gujranwala, Qadirabad, Vehari and Okara regions. The analysis further revealed that CHA28-253S haplotype was present in two different isolates collected from the livestock farms in the Gujranwala and Vehari regions. CHA39-253S haplotype was present in three different isolates collected from the livestock farms in the Vehari and Okara regions (Fig. 4B). The data of those 7 haplotypes demonstrate the evidence of a high level of gene flow predicted to occur due to unregulated animal movement. Seven haplotypes (CHA15-129G, CHA16-129G, CHA18-129G, CHA37-129G, CHA41-129G, CHA44-129G, CHA45-129G) were present at a frequency of 100% in five isolates collected from the livestock farms in the Gujranwala, Qadirabad, Vehari and Okara regions (Fig. 4B). Four haplotypes (CHA5-253S, CHA24-253S, CHA27-253S, CHA39-253S) were present at a frequency of 100% in single isolates collected from the livestock farms in the Gujranwala and Okara regions (Fig. 4B). CHA38-262S, CHA52-262G, CHA58-262S haplotypes were present at a frequency of 100% in single isolates collected from the livestock farms in the Okara and Lodhran regions (Fig. 4B). These data demonstrated that the resistance conferred by these haplotypes has not yet spread elsewhere.
4. Discussion
The present study was designed to investigate the genetic characteristics of the cytochrome b locus and their impact on buparvaquone resistance in ruminants. The study further describes genetic models to understand the emergence and spread of buparvaquone resistance mutations at the cytochrome b locus of T. annulata.
Previous studies using low throughput sequencing methods have shown mutations in the cytochrome b locus of T. annulata isolates collected from clinical cases of treatment failure in Iran (Sharifiyazdi et al., 2012), Tunisia (Mhadhbi et al., 2015) and Sudan (Chatanga et al., 2019). In the present study, high-throughput deep amplicon sequencing was performed to identify 14 mutations in the cytochrome b catalytic Q01 and oxidative Q02 binding pockets of T. annulata collected from clinical cases of treatment failure in the endemic region of Tunisia. Eleven mutations were present at similar frequencies across the buparvaquone susceptible and resistant T. annulata isolates. One mutation at codon 129G (GGC) in resistant isolates and S129 (AGC) in susceptible isolates was found in the Q01 binding pocket. Similarly, two mutations at codons 253S (TCT) and 262S (TCA) in resistant isolates and P253 (CCT) and L262 (TTA) in susceptible isolates were found within the Q02 binding pocket. Overall, these data provide a high throughput sequencing analysis of cytochrome b loci involved in the development of buparvaquone resistance and the first genetic link between mutations and buparvaquone drug in T. annulata.
The present study used deep amplicon sequencing for the first time to explore the cytochrome b locus of buffalo- and cattle-derived T. annulata field isolates collected from the endemic regions of Pakistan. Buparvaquone resistance-associated mutations 129G (GGC), 253S (TCT) and 129G (GGC)/253S (TCT) were identified in 21 buffalo-derived and 129G (GGC), 253S (TCT) and 262S (TCA) were identified in 19 cattle-derived T. annulata isolates. This represents the first report of buparvaquone resistance-associated SNPs in T. annulata field isolates and their impact on positive selection pressure. A possible explanation for the differences is that were observed in the frequency of buparvaquone-conferring mutations with positive selection pressure may be variable drug doses; for example, if the 262S (TCA), 253S (TCT) and 129G (GGC)/253S (TCT) resistance mutations may be selected at low doses of buparvaquone, while 129G (GGC) may occur at higher doses. Overall, the data provide novel information for screening field samples for the detection of buparvaquone resistance mutations in multiple isolates across different geographical regions. This information could aid in understanding how resistance mutations differ between geographical locations with implications for decision-making tools for farmers, farm advisers and veterinarians to design parasite control regimes.
Several studies have assessed the selective sweep patterns in pyrimethamine resistance mutations of P. falciparum at the dihydrofolate reductase locus from different geographical regions. These emphasise a reduction in polymorphism around the dihydrofolate reductase locus with a single resistance haplotype indicative of hard selective sweeps (McCollum et al., 2007; Nair et al., 2003; Pearce et al., 2005; Roper et al., 2003). A few studies have examined the presence of multiple haplotypes in P. falciparum indicative of soft selective sweeps (McCollum et al., 2006, 2008). In contrast, there are few reports of selective advantage to parasites bearing resistance-conferring mutations at the P. vivax dihydrofolate reductase, providing evidence for a high level of polymorphism around the locus with multiple resistance haplotypes indicative of soft selective sweeps (Hawkins et al., 2008; Shaukat et al., 2019). There is currently no understanding of the nature of selective sweeps associated with the emergence of buparvaquone resistance mutations in T. annulata. Both hard and soft selective sweep patterns have been identified in the present study for the buparvaquone resistance mutations in T. annulata. A single resistance haplotype at a high frequency was detected in the 19 buffalo-derived and 7 cattle-derived isolates. The selective sweeps on these individual isolates were effectively hard with no evidence of a genetic footprint of selection detected by significant departures from the neutrality test. In contrast, multiple resistance haplotypes were detected at high frequencies in the 2 buffalo-derived and 12 cattle-derived isolates. The selective sweeps on these isolates were effectively soft and a genetic footprint of selection was also detected by significant departures from the neutrality test. Overall, the data provide novel information on single and or multiple emergences of resistance mutations in this group of parasites that may have implications for targeted selective treatment, or use of different drug combinations, including new drugs or the modification of current compounds in resistance mitigation strategies.
It can be challenging to determine the emergence of resistance mutations with multiple haplotypes, either due to pre-existing mutations before the onset of positive selection or due to recurrent mutations after the onset of positive selection (Chaudhry and Gilleard, 2015). If pre-existing mutations were the only source for the emergence of resistance, similar haplotypes would be present in the isolates (Chaudhry et al., 2016b). For example, in the present study, 2 buffalo-derived isolates (CY321–B and CY322–B) carried four similar resistance haplotypes (BHA2-253S, BHA9-129G, BHA11-253S, BHA13-129G/253S). This striking similarity between the isolates provides evidence for the emergence of resistance by pre-existing mutations. If the recurrent mutations were the only source for the emergence of resistance mutations, different haplotypes would be present in the isolates (Chaudhry et al., 2016b). For example, in the present study, 11 cattle-derived isolates have dramatically carried different haplotypes. This striking difference between the isolates provides evidence for the emergence of resistance by recurrent mutations.
There are several studies demonstrating the evolutionary origin of P. vivax and P. falciparum dihydrofolate reductase resistance mutants (Hawkins et al., 2008; Mita, 2010; Mita et al., 2007, 2009; Shaukat et al., 2021). A high level of genetic diversity may confer genetic adaptability, enabling the origin of pyrimethamine resistance mutations (Alam et al., 2007; Hong et al., 2016). In the present study, 9 unique buffalo-derived haplotypes of the 129G (GGC), 253S (TCT) resistance mutant and 129G (GGC)/253S (TCT) double resistance mutant were present in a single lineage. Similarly, 21 unique cattle-derived haplotypes of the 129G (GGC), 253S (TCT) and 262S (TCA) resistance mutants were present in a single lineage. These data demonstrate evidence of a single origin of the mutations in the T. annulata isolates examined.
Several studies indicate that there is a large amount of animal movement in the endemic region of Pakistan (Chaudhry et al., 2021), hence migration plays an important role in the spread of benzimidazole resistance mutations in livestock (Ali et al., 2019; Chaudhry et al., 2016a). The present study suggests that animal migration between farms is also important in the spread of buparvaquone resistance in the endemic region. The 129G (GGC), 253S (TCT) and 129G (GGC)/253S (TCT) and 262S (TCA) mutations were present in buffalo- and cattle-derived T. annulata collected from different livestock farms in the endemic regions and their phylogenetic relationship suggests that there is a high level of gene flow; possibly occurring due to unregulated animal movement. Notably, the two resistance haplotypes of the 129G (GGC) mutation predominate in 8 and 10 buffalo-derived T. annulata isolates and the other four resistance haplotypes of 129G (GGC), 253S (TCT) and 129G (GGC)/253S (TCT) mutations were present in 2 and 3 buffalo- and cattle-derived T. annulata isolates respectively. In cattle-derived T. annulata, six resistance haplotypes of the 129G (GGC) mutation predominate between 7 and 10 different isolates and the other two resistance haplotypes of 129G (GGC) and 253S (TCT) mutations were present in 2 and 3 different isolates respectively. Overall, the animal movement has likely contributed to the spread of buparvaquone resistance haplotypes in the endemic region of Pakistan. The spread of resistance mutations may require a degree of reproductive isolation of the hosts and parasites with resistant alleles (Chaudhry et al., 2021).
5. Conclusion
The data obtained in the present study provide the analysis of whether cytochrome b loci are involved in the development of buparvaquone resistance in T. annulata of ruminant livestock under natural field conditions. The results afford the first link between genetic mutations and buparvaquone drug resistance in T. annulata. The data then obtained in the present study provide information on single and, or, multiple emergences of resistance mutations, albeit it is still unclear how these spread in parasite populations under natural field conditions. Genetic models based on these results could help to inform novel buparvaquone resistance mitigation strategies.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgements
The study was financially supported by the Carnegie Trust Scotland and Biotechnology and Biological Sciences Research Council (BBSRC). Work at the Qaid-i-Azam University Pakistan and the University of Agriculture, Dera Ismail Khan Pakistan uses facilities funded by the Higher Education Commission of Pakistan.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2022.08.004.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Alam M.T., Agarwal R., Sharma Y.D. Extensive heterozygosity at four microsatellite loci flanking Plasmodium vivax dihydrofolate reductase gene. Mol. Biochem. Parasitol. 2007;153:178–185. doi: 10.1016/j.molbiopara.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Ali Q., Rashid I., Shabbir M.Z., Aziz Ul R., Shahzad K., Ashraf K., Sargison N.D., Chaudhry U. Emergence and the spread of the F200Y benzimidazole resistance mutation in Haemonchus contortus and Haemonchus placei from buffalo and cattle. Vet. Parasitol. 2019;265:48–54. doi: 10.1016/j.vetpar.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Chatanga E., Mosssad E., Abdo Abubaker H., Amin Alnour S., Katakura K., Nakao R., Salim B. Evidence of multiple point mutations in Theileria annulata cytochrome b gene incriminated in buparvaquone treatment failure. Acta Trop. 2019;191:128–132. doi: 10.1016/j.actatropica.2018.12.041. [DOI] [PubMed] [Google Scholar]
- Chaudhry U., Ali Q., Rashid I., Shabbir M.Z., Ijaz M., Abbas M., Evans M., Ashraf K., Morrison I., Morrison L., Sargison N.D. Development of a deep amplicon sequencing method to determine the species composition of piroplasm haemoprotozoa. Ticks Tick Borne Dis. 2019;10 doi: 10.1016/j.ttbdis.2019.101276. [DOI] [PubMed] [Google Scholar]
- Chaudhry U., Ali Q., Zheng L., Rashid I., Shabbir M.Z., Numan M., Ashraf K., Evans M., Rafiq S., Oneeb M., Morrison L.J., Ivan Morrison W., Sargison N.D. Contrasting population genetics of co-endemic cattle- and buffalo- derived Theileria annulata. Ticks Tick Borne Dis. 2021;12 doi: 10.1016/j.ttbdis.2020.101595. [DOI] [PubMed] [Google Scholar]
- Chaudhry U., Gilleard J. 2015. Molecular Analysis of Benzimidazole Resistance in Haemonchus contortus and Haemonchus Placei. PhD Thesis. [Google Scholar]
- Chaudhry U., Redman E., Ashraf K., Shabbir M.Z., Rashid M.I., Ashraf S., Gilleard J.S. Microsatellite marker analysis of Haemonchus contortus populations from Pakistan suggests that frequent benzimidazole drug treatment does not result in a reduction of overall genetic diversity. Parasites Vectors. 2016;9:349. doi: 10.1186/s13071-016-1624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry U., Redman E.M., Ashraf K., Shabbir M.Z., Rashid M.I., Ashraf S., Gilleard J.S. Microsatellite marker analysis of Haemonchus contortus populations from Pakistan suggests that frequent benzimidazole drug treatment does not result in a reduction of overall genetic diversity. Parasites Vectors. 2016;9:349. doi: 10.1186/s13071-016-1624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry U., Redman E.M., Kaplan R., Yazwinski T., Sargison N., Gilleard J.S. Contrasting patterns of isotype-1 β-tubulin allelic diversity in Haemonchus contortus and Haemonchus placei in the southern USA are consistent with a model of localised emergence of benzimidazole resistance. Vet. Parasitol. 2020;286 doi: 10.1016/j.vetpar.2020.109240. [DOI] [PubMed] [Google Scholar]
- Chaudhry U., Redman E.M., Raman M., Gilleard J.S. Genetic evidence for the spread of a benzimidazole resistance mutation across southern India from a single origin in the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2015;721:1016. doi: 10.1016/j.ijpara.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Cui L., Mharakurwa S., Ndiaye D., Rathod P.K., Rosenthal P.J. Antimalarial drug resistance: literature review and activities and findings of the ICEMR network. Am. J. Trop. Med. Hyg. 2015;93:57–68. doi: 10.4269/ajtmh.15-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darghouth M.A., Ben Miled L., Bouattour A., Melrose T.R., Brown C.G., Kilani M. A preliminary study on the attenuation of Tunisian schizont-infected cell lines of Theileria annulata. Parasitol. Res. 1996;82:647–655. doi: 10.1007/s004360050179. [DOI] [PubMed] [Google Scholar]
- David K.P., Alifrangis M., Salanti A., Vestergaard L.S., Ronn A., Bygbjerg I.B. Atovaquone/proguanil resistance in Africa: a case report. Scand. J. Infect. Dis. 2003;35:897–898. [PubMed] [Google Scholar]
- Farnert A., Lindberg J., Gil P., Swedberg G., Berqvist Y., Thapar M.M., Lindegardh N., Berezcky S., Bjorkman A. Evidence of Plasmodium falciparum malaria resistant to atovaquone and proguanil hydrochloride: case reports. Bmj. 2003;326:628–629. doi: 10.1136/bmj.326.7390.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J.C., Wu C.I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fivelman Q.L., Butcher G.A., Adagu I.S., Warhurst D.C., Pasvol G. Malarone treatment failure and in vitro confirmation of resistance of Plasmodium falciparum isolate from Lagos, Nigeria. Malar. J. 2002;1:1. doi: 10.1186/1475-2875-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry M., Pudney M. Site of action of the antimalarial hydroxynaphthoquinone, 2-[trans-4-(4'-chlorophenyl) cyclohexyl]-3-hydroxy-1,4-naphthoquinone (566C80) Biochem. Pharmacol. 1992;43:1545–1553. doi: 10.1016/0006-2952(92)90213-3. [DOI] [PubMed] [Google Scholar]
- Hawkins V.N., Auliff A., Prajapati S.K., Rungsihirunrat K., Hapuarachchi H.C., Maestre A., O'Neil M.T., Cheng Q., Joshi H., Na-Bangchang K., Sibley C.H. Multiple origins of resistance-conferring mutations in Plasmodium vivax dihydrofolate reductase. Malar. J. 2008;7:72. doi: 10.1186/1475-2875-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong N.V., Delgado-Ratto C., Thanh P.V., Van den Eede P., Guetens P., Binh N.T., Phuc B.Q., Duong T.T., Van Geertruyden J.P., D'Alessandro U., Erhart A., Rosanas-Urgell A. Population genetics of Plasmodium vivax in four rural communities in central Vietnam. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jabbar A., Abbas T., Sandhu Z.U., Saddiqi H.A., Qamar M.F., Gasser R.B. Tick-borne diseases of bovines in Pakistan: major scope for future research and improved control. Parasites Vectors. 2015;8:283. doi: 10.1186/s13071-015-0894-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzer F., Carrington M., Knight P., Williamson S., Tait A., Morrison I.W., Hall R. Polymorphism of SPAG-1, a candidate antigen for inclusion in a sub-unit vaccine against Theileria annulata. Mol. Biochem. Parasitol. 1994;67:1–10. doi: 10.1016/0166-6851(94)90090-6. [DOI] [PubMed] [Google Scholar]
- Kessl J.J., Ha K.H., Merritt A.K., Lange B.B., Hill P., Meunier B., Meshnick S.R., Trumpower B.L. Cytochrome b mutations that modify the ubiquinol-binding pocket of the cytochrome bc1 complex and confer anti-malarial drug resistance in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:17142–17148. doi: 10.1074/jbc.M500388200. [DOI] [PubMed] [Google Scholar]
- Kessl J.J., Ha K.H., Merritt A.K., Meshnick S.R., Trumpower B.L. Molecular basis of Toxoplasma gondii atovaquone resistance modeled in Saccharomyces cerevisiae. Mol. Biochem. Parasitol. 2006;146:255–258. doi: 10.1016/j.molbiopara.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Kessl J.J., Meshnick S.R., Trumpower B.L. Modeling the molecular basis of atovaquone resistance in parasites and pathogenic fungi. Trends Parasitol. 2007;23:494–501. doi: 10.1016/j.pt.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Korsinczky M., Chen N., Kotecka B., Saul A., Rieckmann K., Cheng Q. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob. Agents Chemother. 2000;44:2100–2108. doi: 10.1128/aac.44.8.2100-2108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- McCollum A.M., Basco L.K., Tahar R., Udhayakumar V., Escalante A.A. Hitchhiking and selective sweeps of Plasmodium falciparum sulfadoxine and pyrimethamine resistance alleles in a population from central Africa. Antimicrob. Agents Chemother. 2008;52:4089. doi: 10.1128/AAC.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum A.M., Mueller K., Villegas L., Udhayakumar V., Escalante A.A. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob. Agents Chemother. 2007;51:2085–2091. doi: 10.1128/AAC.01228-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum A.M., Poe A.C., Hamel M., Huber C., Zhou Z., Shi Y.P., Ouma P., Vulule J., Bloland P., Slutsker L., Barnwell J.W., Udhayakumar V., Escalante A.A. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J. Infect. Dis. 2006;194:189–197. doi: 10.1086/504687. [DOI] [PubMed] [Google Scholar]
- McFadden D.C., Tomavo S., Berry E.A., Boothroyd J.C. Characterization of cytochrome b from Toxoplasma gondii and Q(o) domain mutations as a mechanism of atovaquone-resistance. Mol. Biochem. Parasitol. 2000;108:1–12. doi: 10.1016/s0166-6851(00)00184-5. [DOI] [PubMed] [Google Scholar]
- McHardy N., Hudson A.T., Morgan D.W., Rae D.G., Dolan T.T. Activity of 10 naphthoquinones, including parvaquone (993C) and menoctone, in cattle artificially infected with Theileria parva. Res. Vet. Sci. 1983;35:347–352. [PubMed] [Google Scholar]
- McHardy N., Morgan D.W. Treatment of Theileria annulata infection in calves with parvaquone. Res. Vet. Sci. 1985;39:1–4. [PubMed] [Google Scholar]
- McHardy N., Wekesa L.S., Hudson A.T., Randall A.W. Antitheilerial activity of BW720C (buparvaquone): a comparison with parvaquone. Res. Vet. Sci. 1985;39:29–33. [PubMed] [Google Scholar]
- Mhadhbi M., Chaouch M., Ajroud K., Darghouth M.A., BenAbderrazak S. Sequence polymorphism of cytochrome b gene in Theileria annulata Tunisian isolates and its association with buparvaquone treatment failure. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhadhbi M., Naouach A., Boumiza A., Chaabani M.F., BenAbderazzak S., Darghouth M.A. In vivo evidence for the resistance of Theileria annulata to buparvaquone. Vet. Parasitol. 2010;169:241–247. doi: 10.1016/j.vetpar.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Mita T. Origins and spread of pfdhfr mutant alleles in Plasmodium falciparum. Acta Trop. 2010;114:166–170. doi: 10.1016/j.actatropica.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Mita T., Tanabe K., Kita K. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol. Int. 2009;58:201–209. doi: 10.1016/j.parint.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Mita T., Tanabe K., Takahashi N., Tsukahara T., Eto H., Dysoley L., Ohmae H., Kita K., Krudsood S., Looareesuwan S., Kaneko A., Björkman A., Kobayakawa T. Independent evolution of pyrimethamine resistance in Plasmodium falciparum isolates in Melanesia. Antimicrob. Agents Chemother. 2007;51:1071–1077. doi: 10.1128/AAC.01186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraguri G.R., Ngumi P.N., Wesonga D., Ndungu S.G., Wanjohi J.M., Bang K., Fox A., Dunne J., McHardy N. Clinical efficacy and plasma concentrations of two formulations of buparvaquone in cattle infected with East Coast fever (Theileria parva infection) Res. Vet. Sci. 2006;81:119–126. doi: 10.1016/j.rvsc.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Musset L., Bouchaud O., Matheron S., Massias L., Le Bras J. Clinical atovaquone-proguanil resistance of Plasmodium falciparum associated with cytochrome b codon 268 mutations. Microb. Infect. 2006;8:2599–2604. doi: 10.1016/j.micinf.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Nair S., Williams J.T., Brockman A., Paiphun L., Mayxay M., Newton P.N., Guthmann J.P., Smithuis F.M., Hien T.T., White N.J., Nosten F., Anderson T.J. A selective sweep driven by pyrimethamine treatment in southeast asian malaria parasites. Mol. Biol. Evol. 2003;20:1526–1536. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- Nene V., Kiara H., Lacasta A., Pelle R., Svitek N., Steinaa L. The biology of Theileria parva and control of East Coast fever - current status and future trends. Ticks Tick Borne Dis. 2016;7:549–564. doi: 10.1016/j.ttbdis.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Pearce R., Malisa A., Kachur S.P., Barnes K., Sharp B., Roper C. Reduced variation around drug-resistant dhfr alleles in African Plasmodium falciparum. Mol. Biol. Evol. 2005;22:1834–1844. doi: 10.1093/molbev/msi177. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Roper C., Pearce R., Bredenkamp B., Gumede J., Drakeley C., Mosha F., Chandramohan D., Sharp B. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet. 2003;361:1174–1181. doi: 10.1016/S0140-6736(03)12951-0. [DOI] [PubMed] [Google Scholar]
- Sargison N.D., MacLeay M., Morrison A.A., Bartley D.J., Evans M., Chaudhry U. Development of amplicon sequencing for the analysis of benzimidazole resistance allele frequencies in field populations of gastrointestinal nematodes. Int. J. Parasitol. Drugs Drug Resist. 2019;10:92–100. doi: 10.1016/j.ijpddr.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E., Bujanover S., Kain K.C. Genetic confirmation of atovaquone-proguanil-resistant Plasmodium falciparum malaria acquired by a nonimmune traveler to East Africa. Clin. Infect. Dis. 2003;37:450–451. doi: 10.1086/375599. [DOI] [PubMed] [Google Scholar]
- Schwobel B., Alifrangis M., Salanti A., Jelinek T. Different mutation patterns of atovaquone resistance to Plasmodium falciparum in vitro and in vivo: rapid detection of codon 268 polymorphisms in the cytochrome b as potential in vivo resistance marker. Malar. J. 2003;2:5. doi: 10.1186/1475-2875-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifiyazdi H., Namazi F., Oryan A., Shahriari R., Razavi M. Point mutations in the Theileria annulata cytochrome b gene is associated with buparvaquone treatment failure. Vet. Parasitol. 2012;187:431–435. doi: 10.1016/j.vetpar.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Shaukat A., Ali Q., Connelley T., Khan M.A.U., Saleem M.A., Evans M., Rashid I., Sargison N.D., Chaudhry U. Selective sweep and phylogenetic models for the emergence and spread of pyrimethamine resistance mutations in Plasmodium vivax. Infect. Genet. Evol. 2019;68:221–230. doi: 10.1016/j.meegid.2018.12.032. [DOI] [PubMed] [Google Scholar]
- Shaukat A., Ali Q., Raud L., Wahab A., Khan T.A., Rashid I., Rashid M., Hussain M., Saleem M.A., Sargison N.D., Chaudhry U. Phylogenetic analysis suggests single and multiple origins of dihydrofolate reductase mutations in Plasmodium vivax. Acta Trop. 2021;215 doi: 10.1016/j.actatropica.2020.105821. [DOI] [PubMed] [Google Scholar]
- Siregar J.E., Syafruddin D., Matsuoka H., Kita K., Marzuki S. Mutation underlying resistance of Plasmodium berghei to atovaquone in the quinone binding domain 2 (Qo(2)) of the cytochrome b gene. Parasitol. Int. 2008;57:229–232. doi: 10.1016/j.parint.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Song Z., Clain J., Iorga B.I., Yi Z., Fisher N., Meunier B. Saccharomyces cerevisiae-based mutational analysis of the bc1 complex Qo site residue 279 to study the trade-off between atovaquone resistance and function. Antimicrob. Agents Chemother. 2015;59:4053–4058. doi: 10.1128/AAC.00710-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava I.K., Morrisey J.M., Darrouzet E., Daldal F., Vaidya A.B. Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol. Microbiol. 1999;33:704–711. doi: 10.1046/j.1365-2958.1999.01515.x. [DOI] [PubMed] [Google Scholar]
- Syafruddin D., Siregar J.E., Marzuki S. Mutations in the cytochrome b gene of Plasmodium berghei conferring resistance to atovaquone. Mol. Biochem. Parasitol. 1999;104:185–194. doi: 10.1016/s0166-6851(99)00148-6. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.