Abstract
We designed an expression and export system that enabled the targeting of a reporter protein (the staphylococcal nuclease Nuc) to specific locations in Lactococcus lactis cells, i.e., cytoplasm, cell wall, or medium. Optimization of protein secretion and of protein cell wall anchoring was performed with L. lactis cells by modifying the signals located at the N and C termini, respectively, of the reporter protein. Efficient translocation of precursor (∼95%) is obtained using the signal peptide from the lactococcal Usp45 protein and provided that the mature protein is fused to overall anionic amino acids at its N terminus; those residues prevented interactions of Nuc with the cell envelope. Nuc could be covalently anchored to the peptidoglycan by using the cell wall anchor motif of the Streptococcus pyogenes M6 protein. However, the anchoring step proved to not be totally efficient in L. lactis, as considerable amounts of protein remained membrane associated. Our results may suggest that the defect is due to limiting sortase in the cell. The optimized expression and export vectors also allowed secretion and cell wall anchoring of Nuc in food-fermenting and commensal strains of Lactobacillus. In all strains tested, both secreted and cell wall-anchored Nuc was enzymatically active, suggesting proper enzyme folding in the different locations. These results provide the first report of a targeting system in lactic acid bacteria in which the final location of a protein is controlled and biological activity is maintained.
Lactic acid bacteria (LAB) are gram-positive bacteria that include lactococci, streptococci, and lactobacilli, all of which have long been used as starters for food fermentations (6). As such, live LAB are regularly and widely consumed by humans and animals. Although numerous in vivo beneficial effects (so-called probiotic effects) of LAB have been claimed, only a few have been unambiguously demonstrated experimentally (24, 34). Nevertheless, during their frequent passage through the gastrointestinal tract (GIT), LAB may produce bioactive molecules that accumulate in vivo. To study the potential probiotic capacity of LAB, our strategy is to specifically enhance a given phenotype of LAB and to analyze whether these bacteria, given orally to animals, induce the expected in vivo effect. These studies are aimed at enhancing a natural product of LAB (such as a vitamin or enzyme) or conferring expression of a new molecule, such as an antigen that would be produced in the gut of humans and animals. One advantage of live vaccines is that mucosal immunity induced in the GIT may prevent multiplication of the infectious agents via this common entry point (16). Also, as LAB may survive transiently in the GIT, they could be used to deliver digestive enzymes to supplement pancreatic deficiency in humans or animals. In this regard, some LAB already exert a positive action in lactose-intolerant consumers by providing lactase in the gut (14).
We are developing protein delivery systems in LAB for which the protein of interest is targeted to a defined cell location, i.e., the cytoplasm, the cell wall, or the medium. To this end, we constructed expression and export vectors based on (i) Sec-dependent machinery for protein translocation across the membrane and (ii) sortase-dependent machinery for protein anchorage to the cell wall. The Sec machinery is a ubiquitous secretion system comprised of a set of proteins that mediate translocation of a precursor protein (the mature protein plus an N-terminal signal peptide) across the cytoplasmic membrane (39). Upon translocation across the membrane, the signal peptide is cleaved off by signal peptidase, thus releasing the protein to the medium (39). The sortase machinery has been characterized for Staphylococcus aureus (45); it covalently anchors proteins by their C terminus to the peptidoglycan (36). Cell surface-anchored proteins are first synthesized as a preproprotein containing an N-terminal signal peptide and a C-terminal cell wall anchor (CWA) domain. The ∼30-amino-acid CWA consists of a conserved LPXTG motif, a transmembrane fragment, and a charged C terminus. Upon translocation of the precursor across the membrane, the sortase machinery, presumably membrane localized, catalyzes a transpeptidation reaction consisting of (i) cleavage of the amide bond between the threonine and the glycine residues of the LPXTG motif and (ii) covalent linkage of the C-terminal threonine to an amino acid of the peptide cross bridge in the peptidoglycan. Although sortase has been characterized for S. aureus, homologs are present in many gram-positive bacteria, including LAB. Indeed, the same C-terminal structure is present on many cell surface-located proteins from gram-positive bacteria, including some from LAB (10). In addition, anchoring of heterologous proteins using the CWA of proteins A and M6 from S. aureus and Streptococcus pyogenes, respectively, was demonstrated with various gram-positive hosts, including LAB (28, 29, 41). The combination of secretion and anchoring systems may provide the needed versatility for protein targeting in various LAB for use as delivery vectors.
We report the design and optimization of vectors for targeted heterologous gene expression in LAB. The gene products are addressed to the cytoplasm, the cell wall, or the extracellular medium. We describe the effects of different parameters on the expression and targeting efficiencies of the different vectors in various LAB strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Most plasmid constructions described below were performed with Escherichia coli TG1, DH5α, JM101, and JM105 strains or with their recA and pcnB derivatives (Table 1). E. coli cells were grown in Luria broth (35) supplemented with thymine at 37°C with shaking. Lactococcus lactis cells were grown in M17 medium (44) or brain heart infusion media (Difco, Detroit, Mich.) at 30°C without shaking. Lactobacillus sakei, Lactobacillus casei, and Lactobacillus plantarum lactobacilli were grown in MRS medium (9) at 30°C without shaking. Lactobacillus reuteri cells were grown in MRS medium or APTG10 (32) at 37°C in anaerobic jars. Where appropriate, antibiotics were added as follows: for E. coli, erythromycin (150 μg/ml) and ampicillin (100 μg/ml); for L. lactis, L. casei, L. sake, and L. plantarum, erythromycin (5 μg/ml) and chloramphenicol (10 μg/ml); and for L. reuteri, erythromycin (30 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strain | ||
| E. coli | ||
| TG1 | 35 | |
| TG1 pcnb | Kanr; ΔpcnB::kan derivative of strain TG1 | B. Michelb |
| DH5α | 35 | |
| JM101recA | Tetr; Δ(recA-srl)::Tn10 derivative of strain JM101 (35) | B. Michel |
| JM105recA | Tetr; Δ(recA-srl)::Tn10 derivative of strain JM105 (35) | B. Michel |
| LAB | ||
| L. lactis MG1363 | Plasmid-free strain | 15 |
| L. plantarum 37 A | Plasmid-free strain | R. Jimenez-Diazc |
| L. sake 23K | Plasmid-free strain | 2 |
| L. casei Sac7 | Plasmid-free strain | R. Rayad |
| L. reuteri BP83 | Plasmid-free strain | 12 |
| Plasmid | ||
| pBSII-SK+ | Apr; ColE1; 2.9 kb | Stratagene |
| pBSII-KS+ | Apr; ColE1; 2.9 kb | Stratagene |
| pUC18 | Apr; ColE1; 2.7 kb | 49 |
| pUC19 | Apr; ColE1; 2.7 kb | 49 |
| pIL252 | Emr; pAMβ1; 4.6 kb | 40 |
| pLEM7 | Eryr; 3.5 kb | 11 |
| pVE3588 | Apr; 4.6 kb; pBSII-SK+::usp45 | Y. Le Loire |
| pVE5201 | Apr; ColE1; 4.5 kb; pBSII-SK+::emm6 | 28 |
| pVE5207 | Apr; ColE1; 4.5 kb; pBSII-SK+::P59::emm6 | 28 |
| pVE5216 | Apr; ColE1; 2.8 kb; pUC19::P59 (48) | This work |
| pVE5218 | Apr; ColE1; 4.3 kb; pUC19::emm6 | This work |
| pVE5220 | Apr; ColE1; 2.8 kb; pUC18::P59 (48) | This work |
| pVE5221 | Apr; ColE1; 3.5 kb; pBSII-SK+::cwaM6 specifying the 140 C-terminal aa of the M6 protein (the CWA) | This work |
| pVE5222 | Apr; ColE1; 2.9 kb; pUC19::spM6 specifying the 47 N-terminal aa (signal peptide) of the M6 preprotein | This work |
| pVE5224 | Apr; ColE1; 2.8 kb; pUC18::P59 (48) | This work |
| pVE5228 | Apr; ColE1; 2.8 kb; pUC19::spM6::cwaM6 | This work |
| pVE5229 | Apr; ColE1; 3.4 kb; pBSII-SK+::spM6::cwaM6 | This work |
| pVE5231 | Apr; ColE1; 3.6 kb; pBSII-SK+::P59::spM6::cwaM6 | This work |
| pVE5232 | Apr; ColE1; 3.4 kb; pBSII-KS+::nucA (22) | This work |
| pVE5233 | Apr; ColE1; 4.0 kb; pBSII-SK+::P59::spM6::nucA::cwaM6 | This work |
| pVE5236 | Apr; Emr; ColE1 pAMβ1; 8.7 kb; pBS::pIL252::P59::spM6::nucA::cwaM6 | This work |
| pVE5239 | Apr; ColE1; 3.1 kb; pBSII-SK+::t1t2 (27) | This work |
| pVE5240 | Apr; ColE1; 3.7 kb; pBSII-SK+::P59::spM6::nucA::t1t2 | This work |
| pVE5243 | Apr Emr; ColE1 pAMβ1; 8.4 kb; pBS::pIL252::P59::spM6::nucA::t1t2 | This work |
| pVE5247 | Apr; ColE1; 3.1 kb; pBSII-KS+::spUsp45 specifying RBS and 32 N-terminal aa (signal peptide) of Usp45 preprotein (46) | This work |
| pVE5249 | Apr; ColE1; 4.0 kb; pBSII-SK+::spUsp45::nucA::cwaM6 | This work |
| pVE5250 | Apr; ColE1; 4.1 kb; pBSII-SK+::spUsp45::nucA::cwaM6::t1t2 | This work |
| pVE5251 | Apr; ColE1; 4.2 kb; pVE5250 with additional EcoRV site between nucA and cwaM6 | This work |
| pVE5252 | Apr; ColE1; 3.6 kb; pBSII-SK+::spUsp45::nucA::t1t2 | This work |
| pVE5253 | Apr; ColE1; 2.9 kb; pBSII-KS+::ttrpA::P59 | This work |
| pVE5253SX | Apr; ColE1; 2.9 kb; pVE5253 with deleted SalI and XhoI sites | This work |
| pVE5506 | Emr; pAMβ1; 4.7 kb; pIL252 XbaI digested and self-ligated (40) | This work |
| pVE5512 | Emr; pAMβ1; 4.7 kb; pIL252 SalI digested, filled in, and self-ligated | This work |
| pVE5514 | Emr; pAMβ1; 5.7 kb; pIL252::P59::spM6::nucA::t1t2 | This work |
| pVE5516 | Emr; pAMβ1; 5.5 kb; pIL252::P59::nucA::t1t2 | This work |
| pVE5517 | Emr; pAMβ1; 5.5 kb; pIL252::spUsp45::nucA::t1t2 | This work |
| pVE5518 | Emr; pAMβ1; 6.0 kb; pIL252::spUsp45::nucA::cwaM6::t1t2 | This work |
| pVE5523 | Apr Emr; ColE1 pAMβ1; pBS::pIL252::ttrpA::P59::spUsp45::nucA::t1t2 | This work |
| pVE5524 | Apr Emr; ColE1 pAMβ1; pBS::pIL252::ttrpA::P59::spUsp45::nucA::cwaM6::t1t2 | This work |
| pVE5529 | Apr Emr; ColE1 pAMβ1; pBS::pIL252::ttrpA::P59::nucA::t1t2 | This work |
| pVE5537 | Emr; 4.5 kb; pLEM7::ttrpA::P59::spUsp45::nucA::t1t2 | This work |
| pVE5546 | Apr Emr; ColE1 pAMβ1; pBS::pIL252::ttrpA::P59::spUsp45::nucA::cwaM6Δ105::t1t2 | This work |
| pVE8001 | Apr; ColE1; 3.0 kb; pBSII-KS+::ttrpA (8) | 30 |
ColE1 and pAMβ1 refer to the replicon. aa, amino acids; RBS, ribosome-binding site.
Laboratoire de Génétique Microbienne, INRA, Jouy-en-Josas, France.
Instituto de la Grassa y sus Derivados, Seville, Spain.
CERELA, S. M. de Tucuman, Argentina.
Unité de Recherches Laitières et Génétique Appliquée, INRA, Jouy-en-Josas, France.
DNA manipulation and transformation procedures.
General molecular biology techniques were performed essentially as methods described previously (35). Plasmid DNA was extracted from E. coli as described previously (3), and plasmid DNA was extracted from L. lactis, L. sakei, and L. reuteri as described previously (28). The protocol for plasmid extraction from L. casei and L. plantarum was similar to that used for L. sakei. Plasmids were established by electroporation as described for L. lactis and S. thermophilus (20), L. reuteri (4), and L. casei and L. sakei (2). For L. plantarum, the following protocol was used (R. Jimenez-Diaz, personal communication). An overnight preculture was prepared in MRS with 1% glucose and 0.1% glycine. The preculture was diluted 1:40 (vol/vol) in the same medium, grown to an optical density at 600 nm (OD600) of 0.6, and chilled on ice. Bacteria were harvested by a 10-min centrifugation at 6,800 × g and were washed successively in 1 culture volume of ice-cold 1 mM MgCl2 solution and in 1 culture volume of 30% polyethylene glycol 3000 plus 10% glycerol. The cells were resuspended in a 1:100 culture volume of 30% polyethylene glycol 3000 plus 10% glycerol. Electroporation was performed using 50 μl of competent cells and a maximum of 5 μl of plasmid DNA in 0.2-cm-wide electroporation cuvettes. Settings of 1.5 kV, 25 μF, and 400 Ω were used with the Gene Pulser (Bio-Rad, Hercules, Calif.). Electroporated cells were transferred into 0.5 ml of MRS containing 0.5 M sucrose plus 0.1 M MgCl2 and incubated 2 h at 30°C before plating on selective media.
Nuclease plate activity assays.
Nuc activity was detected using the metachromatic agar-diffusion method with toluidine blue-DNA agar (TB-D agar [18]). To identify Nuc+ clones, plates were overlaid with TB-D agar and analyzed for pink colonies. To detect Nuc in the cytoplasmic compartment of LAB, cells were lysed under mild conditions; the bacterial pellet was resuspended in TES (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 25% sucrose) plus 5 mg of lysozyme/ml and incubated at 37°C for 30 min. The resulting protoplasts were treated with 0.1% sodium dodecyl sulfate (SDS) and boiled for 10 min (note that nuclease is readily renatured after this treatment). Nuc activity in lysates was analyzed by spotting aliquots on TB-D agar and checking for pink halos after 37°C incubation. For detection of Nuc activity in culture supernatants, bacterial cultures were centrifuged for 5 min at 5,700 × g, and supernatants were filtered using 0.2-μm-pore-size filters (low-protein-retention filters; Millipore, Molsheim, France). Filtrates were then spotted on TB-D agar and observed for pink halos after a 37°C incubation.
Cell fractionation, protein extraction, and Western blot analysis.
Medium, cell wall, and protoplast fractionation and protein extractions were performed as previously described (28). For further fractionation between membranes and cytoplasm, protoplasts were washed with TES and resuspended in 500 μl of water. The suspension was subjected to five freeze-thawing cycles. Membranes were pelleted by a 45-min centrifugation at 21,000 × g at 4°C and resuspended in 100 μl of TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) containing 1% SDS per OD600 unit. Cytoplasmic proteins were recovered by trichloroacetic acid precipitation and resuspended in 100 μl of TE per OD600 unit. Equal volumes of 2× loading buffer were added to all samples. Extracts were run on SDS-polyacrylamide gel electrophoresis (PAGE) (12% acrylamide) gels (19). Electroblotting on polyvinylidene difluoride membranes (Millipore) and antibody reactions and detection (enhanced chemiluminescence) were performed according to the manufacturer's recommendations. Quantitation of Nuc was performed by scanning Western blots and comparing signals to those of known amounts of a purified commercial NucA (Sigma). Amounts are presented as milligrams per liter of culture corrected to an OD600 of 1.
Pulse-chase analysis.
Pulse and chase conditions (22) and immunoprecipitation (25) were performed as previously described. Extracts were run on SDS-PAGE (16% acrylamide) gels.
Cloning strategy.
Fusion genes using the nuc reporter (Fig. 1) were constructed via different intermediate plasmids (Table 1). The general strategy was to construct individual cassettes, each carrying promoter, terminators, export signals, or reporter genes, and to join them in different combinations; this strategy generates a flexible system in which a given cassette can be replaced to modify the nature of or the level of a given phenotype. Note that oligonucleotides discussed below, which were named “a” through “j,” are described at the end of this section.
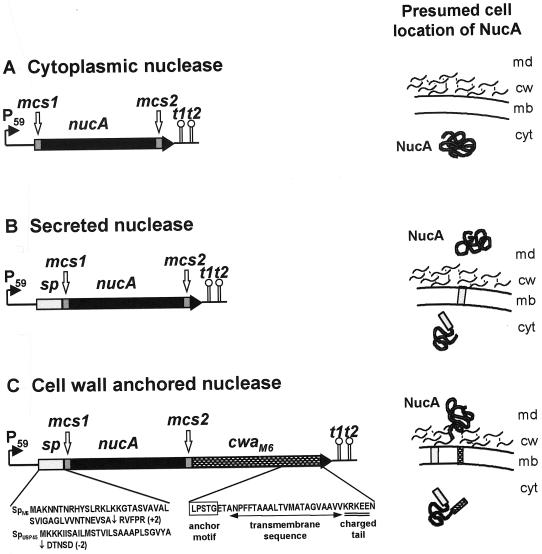
FIG. 1.
Fusion gene constructs with the Nuc reporter and expected cell location of final products. P59, lactococcal promoter; sp, signal peptide from the M6 preproprotein (spM6) or from the Usp45 preprotein (spUsp45); nucA, structural gene for staphylococcal NucA; cwaM6, sequence specifying the CWA domain from the M6 preproprotein; t1t2, transcriptional terminators; mcs1 and mcs2, multiple cloning sites; cyt, cytoplasm; mb, membrane; cw, cell wall; md, medium. The vertical black arrows in the amino acid sequences of SPM6 and of SPUsp45 indicate the signal peptidase cleavage sites; numbers in parentheses indicate the global net charge of the five amino acids following the cleavage sites.
Cloning of the various cassettes.
The P59 promoter cassette (48) was obtained by cloning the 130-bp BamHI-KpnI fragment of pVE5207 (28) into BamHI-KpnI-treated pUC18; the resulting pVE5220 plasmid was then digested by BamHI-PstI to delete unwanted restriction sites, treated with Klenow and T4 DNA polymerase, and self-ligated to yield pVE5224 (Table 1). Plasmid pVE5216 was obtained by cloning the 130-bp BamHI-KpnI fragment of pVE5207 (28) into BamHI-KpnI-treated pUC19.
Two cassettes specifying the M6 (spM6) and Usp45 (spUsp45) signal peptide were constructed. (i) The sequence encoding the M6 signal peptide was obtained by cloning the 1.6-kb EcoRI-BamHI fragment (containing the emm6 gene) from pVE5201 (28) into EcoRI-BamHI-treated pUC19 to yield pVE5218. The latter plasmid was further digested by StyI-XbaI, and the 2.8-kb fragment was self-ligated; the resulting pVE5222 plasmid, carrying spM6, was ClaI digested, blunted with Klenow, and self-ligated so that an NruI site was created 5′ of spM6 in the resulting pVE5228 plasmid. (ii) The spUsp45 cassette was obtained by PCR amplification from the usp45 gene, using oligonucleotides a and b and pVE3588 as the template. The PCR product was blunted using Klenow and T4 DNA polymerase and cloned into SmaI pBluescript II KS(+) (pBSII-KS+) to yield pVE5247.
The cassette containing the nucA reporter gene was obtained by PCR amplification from pBS::P59::nuc (22) using oligonucleotides c and d. The 0.5-kb PCR product was digested by SalI and ClaI and cloned into SalI-ClaI-linearized pBSII-KS+ to yield pVE5232.
The cassette corresponding to the CWA signal (cwaM6) was obtained by deleting the 5′ HindIII fragment of emm6 from pVE5201 (28); the HindIII ends were blunted using Klenow and self-ligated to yield pVE5221, which contained the 3′ end of emm6 encoding the 140 C-terminal amino acids of M6 proprotein.
Two cassettes specifying transcriptional terminators were used. One was present on pVE8001 and contained the sequence specifying the trpA operon terminator of E. coli (8), and it was a gift from I. Poquet (30). The T1T2 terminators from the rrnB operon of E. coli (27) (a gift from F. Chedin; see reference 5 for a similar construct) were cloned as an XbaI fragment into pBSII-SK+ to yield pVE5239.
Assembling of the cassettes.
The cassettes described above were assembled as follows. spM6::cwaM6 (present on pVE5229) was obtained by ligation of the ScaI-SalI 2.4-kb fragment of pVE5221 to the corresponding 1.0-kb fragment of pVE5228. P59::spM6::cwaM6 (present on pVE5231) was obtained by fusing the ScaI-EcoRI 1-kb fragment of pVE5224 to its 2.5-kb counterpart of pVE5229. P59::spM6::nucA::cwaM6 (present on pVE5233) was constructed by cloning the 0.5-kb SalI-ClaI nucA fragment of pVE5232 into SalI-ClaI-digested pVE5231. P59::spM6::nucA::t1t2 (present on pVE5240) was obtained by ligation of the 1.7-kb ClaI-BstXI fragment of pVE5233 to the ClaI-BstXI 2.0-kb fragment of pVE5239. A fusion of pVE5233 and pVE5240 to a pAMβ1-derived plasmid, pIL252, was formed by ligation at their BamHI site to yield pVE5236 and pVE5243, respectively; it was established in both E. coli and L. lactis MG1363. P59::nucA::t1t2 (present on pVE5516) was formed by first deleting the pBS part of pVE5243 by a SphI-BstXI digestion. It was treated with T4 DNA polymerase and self-ligated before introduction into L. lactis. The resulting pVE5514 plasmid was further digested with MslI and SalI, treated with T4 DNA polymerase, and self-ligated to yield pVE5516. This treatment removes the cassette corresponding to spM6 and introduces a TTG start codon recognized in L. lactis (47). ttrpA::P59 (present on pVE5253SX) was constructed by fusing the ScaI-BamHI-Klenow-treated 1.0-kb fragment of pVE5216 to the 1.8-kb ScaI-EcoRV fragment of pVE8001. The SalI site was removed from the resulting pVE5253 by a SalI-XhoI digestion and self-ligation to yield pVE5253SX. spUsp45::nucA::cwaM6 (present on pVE5249) was obtained by ligation of the ApoI-SalI 0.18-kb fragment of pVE5247 to the 3.8-kb ApoI-SalI fragment of pVE5233. spUsp45::nucA-::cwaM6::t1t2 was present on pVE5250 and pVE5251. pVE5250 was obtained by cloning t1t2 carrying a 0.17-kb XbaI fragment of pVE5239 into XbaI-linearized pVE5249. An EcoRV restriction site was inserted between nucA and cwaM6 by cloning annealed oligonucleotides e and f into NheI-ClaI-digested pVE5250 to yield pVE5251. spUsp45::nucA::t1t2 (present on pVE5252) was obtained by an NheI-SpeI digestion of pVE5251, followed by self-ligation of the 3.6-kb fragment; this deleted cwaM6 and introduced an in-frame stop codon between nucA and t1t2.
spUsp45::nucA::t1t2 and spUsp45::nucA::cwaM6::t1t2 fusions were recovered by ApoI-HpaI digestion of pVE5252 and pVE5251, respectively. They were inserted into SmaI-EcoRI pVE5512 (a pIL252-derived plasmid; Table 1) to yield pVE5517 and pVE5518, respectively. ttrpA::P59::spUsp45::nucA::t1t2 (present on pVE5523) and ttrpA::P59::spUsp45::nucA::cwaM6-t1t2 (present on pVE5524) were obtained by fusing pVE5253SX to pVE5517 and pVE5518, respectively, at their EcoRI sites. The functional orientation for transcription was detected by nuclease-positive phenotypes of the transformants. ttrpA::P59::spUsp45::nucA::cwaM6Δ105::t1t2 was present on pVE5546. pVE5524 was digested by EcoRV-BsgI, and the 7.6-kb fragment was recovered. It was ligated to the EcoRV-BsgI-digested PCR product obtained with oligonucleotides i and j, using pVE5524 as the template, to yield pVE5546. ttrpA::P59::nucA::t1t2 was present on pVE5529. A fragment specifying ttrpA::P59 was obtained by PCR amplification using oligonucleotides g and h and pVE5523 as the template; this fragment was digested by BspLU11I-SalI and cloned into BspLU11I-SalI-digested pVE5523. For the cloning of ttrpA::P59::spUsp45::nucA::t1t2 in L. reuteri cells, the ApaI-BamHI 1-kb fragment of pVE5523 was cloned into ApaI-BamHI-digested pLEM7 to yield pVE5537.
Oligonucleotides (relevant enzyme restriction sites [in bold] and restriction enzymes [in parentheses] are given) were as follows: a, 5′-AGTCGCGAAACCGAACTTAATGGGAGG-3′ (NruI); b, 5′-TAGTCGACCGCATCTTGTTTAGCAATATCTGAG-3′ (SalI); c, 5′-GTCGACCCATGGTCAACTAAAAAATTACATAAAGAACC-3′ (SalI); d, 5′-ATCGATTTGACCTGAATCAGCGTTGTCTTC-3′ (ClaI); e, 5′-ATCGATCTCGAGATATCGCTAGC-3′ (ClaI, EcoRV, NheI); f, 5′-GCTAGCGATATCTCGAGATCGAT-3′ (NheI, EcoRV, ClaI); g, 5′-CTGGACGACATGTGACGGTATCGATAGC-3′ (ClaI); h, 5′-CGGCCAGGTCGACCATAACTGTTCTTTTTTAATT-3′ (SalI); i, 5′-GCTCGATATCGTTACCATCAACAGGTGAAA-3′ (EcoRV); j, 5′-GCCAGTTTCGTCGTTAAATGCCCTTT-3′.
RESULTS
Targeting Nuc in L. lactis
We examined whether a heterologous enzyme could be targeted to various cell compartments of L. lactis. For this purpose, the S. aureus nucA open reading frame was fused to either (i) the strong P59 lactococcal promoter (plasmid pVE5516) for cytoplasmic localization of Nuc (Fig. 1A), (ii) the P59 promoter plus the M6 signal peptide (plasmid pVE5514) for extracellular secreted Nuc (Fig. 1B), or (iii) the promoter and signal peptide described above (ii) with an additional CWA domain from the M6 proprotein (plasmid pVE5236) for cell wall-anchored Nuc (Fig. 1C). These fusions were established in L. lactis cells on the low-copy-number replicon pIL252 and analyzed phenotypically and biochemically. Nuc production by bacterial colonies was examined (Fig. 2). As expected, no Nuc was detected for colonies harboring nucA (pVE5516), consistent with its cytoplasmic location (Fig. 2A). In contrast, upon bacterial lysis under mild conditions, strong Nuc activity was detected, indicating that it is indeed produced but not exported to the external medium (not shown). Colonies of L. lactis harboring spM6::nucA (on pVE5514) yielded pink halos on a plate assay, as expected if Nuc is exported (Fig. 2B); activity was detected in the culture supernatant, indicating that Nuc is indeed both exported and secreted (not shown). Colonies of L. lactis bearing spM6::nucA::cwaM6 (on pVE5236) also produced pink halos on TB-D agar (Fig. 2C), but only a little activity was detected in the culture supernatant (not shown), indicating that Nuc is exported to the cell surface of L. lactis but is not released in the supernatant. The halos formed by strains expressing spM6::nucA::cwaM6 (on pVE5236) were smaller than those formed by strains expressing spM6::nucA (on pVE5514). This is expected since, in the former, Nuc is cell wall bound and therefore does not diffuse in the medium, while in the latter it is secreted.
FIG. 2.
Nuclease activity detection in clones of L. lactis MG1363 expressing nucA in cytoplasm (nucA on pVE5516) (A), supernatant (spM6::nucA on pVE5514) (B), or cell wall (spM6::nucA::cwaM6 on pVE5236) (C). For an explanation, see the text.
Nuc distribution between protoplast, cell wall, and supernatant fractions was analyzed by Western blotting using NucA-specific antibodies (Fig. 3). In clones harboring nucA (on pVE5516), the majority of Nuc was detected in protoplasts, which corroborates results obtained by phenotypic analysis. However, a significant fraction was also present in cell walls; use of a cytoplasmic marker, PepX (7), confirmed that the above observation was not due to cell lysis. As Nuc activity was not detected upon plating of the clones on TB-D agar (Fig. 2A) and as no potential sequence acting as a signal peptide could be detected in the Nuc amino acid sequence, we cannot at present explain detection of the enzyme in the cell wall fraction.
FIG. 3.
Expression and targeting of Nuc in L. lactis. Western blottings using anti-NucA polyclonal antibodies were performed on fractionated protein extracts from L. lactis MG1363 cells expressing the fusion genes indicated on top. The strains contained plasmids pVE5516, pVE5514, and pVE5236, which express nucA, spM6::nucA, and spM6::nucA::cwaM6, respectively. P, protoplasts; CW, cell wall; SN, supernatant. Purified NucA was used as a reference, and the positions of molecular mass standards (in kilodaltons) are indicated.
Analysis of Nuc distribution in L. lactis expressing spM6::nucA (on pVE5514) showed that about half of the signal is present as two bands in the supernatant fraction. In this construct, processing of the signal peptide should give rise to a NucA protein containing additional amino acids at the N and C termini (Fig. 1; see Materials and Methods). We consider it likely that these tails are susceptible to proteolysis, giving rise to the NucA-size protein as seen in Fig. 3. The protoplast fraction of cells expressing spM6::nucA (pVE5514) also produced two bands: a high-molecular-mass band corresponding in size to the Nuc precursor (calculated molecular mass of 22.8 kDa) and the other corresponding to Nuc. We believe that the latter may have resulted from degradation of the precursor. A band of strong intensity and corresponding in size to NucA was also visible in the cell wall fraction. Given that no anchoring signal was present in the analyzed fusion, we wondered whether Nuc association to the cell wall could be due to a noncovalent linkage. In the above fusions, the molecular environment of the prepro-M6 signal peptidase cleavage site was conserved by maintaining the five N-terminal amino acids (RVFPR) of the mature M6 protein (Fig. 1C); since these amino acids provide a positive net charge of +2 at the N terminus of Nuc, we asked whether the interaction observed between Nuc and the cell wall with the spM6::nucA construct (on pVE5514) could be due to electrostatic interactions. We reasoned that salt addition could alleviate such an interaction and therefore cultivated the spM6::nucA-expressing strain in medium containing 170 or 340 mM NaCl. Cell fractions were analyzed by Western blotting. Protein expression levels were comparable in the presence or absence of salt in the medium. However, Nuc association with the cell wall was abolished when salt was present during growth (not shown), suggesting that this association is electrostatic. Interestingly, the apparent Nuc degradation product (cf. above) was reduced in 170 mM NaCl and nearly absent at 340 mM NaCl. This may suggest that the proteinase activity involved in the degradation of Nuc is inhibited by salt or that Nuc folding is accelerated when salt is present.
For cells expressing nucA with a C-terminal fusion containing 105 amino acids plus the M6 CWA domain (spM6::nucA::cwaM6 [on pVE5236]), no Nuc was detected in the supernatant fraction. Multiple bands migrating between 30 and 35 kDa were detected in the cell wall fraction, as previously reported for cell wall-anchored proteins. This is attributed to the presence of peptidoglycan fragments of various sizes that are covalently linked to the surface protein after digestion with a muramidase (37). These bands could also correspond to degradation products. A faint band corresponding in size to NucA was also visible in the cell wall fraction. In the protoplast fraction, an ∼38-kDa major band corresponded in size to the precursor protein. For this construct, ∼20% of precursor was processed and anchored to the cell wall.
To summarize, the constructed vectors enabled Nuc to be targeted to the desired cell compartment. Nevertheless, precursor processing was incomplete in the case of secreted and cell wall-anchored fusions and some Nuc was unexpectedly detected in the cell wall fraction of cytoplasmic and secreted fusions.
Signal peptide of Usp45 followed by anionic amino acids at N terminus of NucA improves protein export and prevents interaction of secreted Nuc with cell wall.
Protein targeting in the experiment described above was limited by incomplete protein export and interactions between secreted Nuc and cell wall. We therefore exchanged the signal peptide of prepro-M6 with that of pre-Usp45. Five additional amino acids (DTNSD) at the N terminus of mature Usp45 resulted in the creation of a negative net charge of −2 at the NucA N terminus (Fig. 1C), which was recently shown to improve secretion efficiency in L. lactis (22). Targeting efficiency of the cytoplasmic control (containing the N-terminal DTNSD residues), cell wall, and secreted constructions was examined by Western analysis (Fig. 4). Expression of nucA (from plasmid pVE5529) in L. lactis resulted in cytoplasmic Nuc accumulation; unlike the results above, which were obtained with pVE5516, no Nuc was detected in the cell wall fraction (compare Fig. 3 and 4).
FIG. 4.
Targeting Nuc to cytoplasm (nucA on pVE5529), supernatant (spUsp45::nucA on pVE5523), or cell wall (spUsp45::nucA::cwaM6 on pVE5524) in L. lactis. Protein extracts were analyzed by Western blotting using anti-NucA antibodies. Forms not indicated by arrows are presumably protein degradation products. P, protoplasts; CW, cell wall; SN, supernatant. The positions of molecular mass standards (in kilodaltons) are indicated.
Secretion efficiency of Nuc in L. lactis expressing spUsp45::nucA (from pVE5523) was assessed. The protoplast fraction contained a faint band corresponding to the size of the precursor (Fig. 4). The majority (95%) of the protein was in the supernatant fraction. Thus, secretion of this protein is highly efficient compared to that observed from the fusion with the M6 signal peptide (compare Fig. 3 and 4). This suggests that precursor processing is more efficient with the signal peptide and N-terminal environment of Usp45 than with that of M6. Also, all of the exported Nuc was detected in the supernatant fraction and not in the cell wall fraction; no interactions were detectable between the Nuc moiety and cell wall. Interestingly, compared with previous results (Fig. 3), Nuc secreted by this vector underwent only a little proteolytic cleavage.
Analysis of spUsp45::nucA::cwaM6 (on pVE5524) revealed improved targeting of the fusion to the cell wall fraction compared to that of spM6::nucA::cwaM6 (on pVE5236; compare Fig. 3 and 4). Export efficiency is enhanced to ca. 40%. As seen above (Fig. 3), NucA antibodies reacted with several bands in the cell wall fraction. A multiple banding pattern was also observed in the protoplast fraction; this point is addressed below.
Taken together, these results show that efficient export and proper localization of Nuc can be achieved by the use of the Usp45 signal peptide followed by anionic amino acids.
What is the limiting step in the processing of cell wall-anchored proteins?
The results above suggest that the signal peptide of Usp45 followed by anionic residues, which markedly improves secretion of Nuc, is less effective in exporting NucA-CWAM6. We first hypothesized that the 105-amino-acid sequence upstream of the C-terminal anchoring signal itself could impair precursor translocation. This sequence is indeed rich in glycine and proline residues that are known to be turn-promoting amino acids in proteins. We postulated that such a structure could be poorly translocated across the membrane. A fusion, spUsp45::nucA::cwaΔ105, in which this sequence was deleted was constructed and expressed in L. lactis. Comparison of the profiles obtained with spUsp45::nucA::cwa and spUsp45::nucA::cwaΔ105 showed no significant difference in the proportion of precursor processed and exported to the cell wall, i.e., about 40% Nuc was anchored to the cell wall while 60% remained unprocessed in the protoplasts (Fig. 5). However, the construct NucA-CWAΔ105 appeared as one major band in the cell wall fraction. This may suggest that the multiple-banding pattern observed with whole CWA was due, in our case, to degradation rather than to the linkage of Nuc to peptidoglycan fragments as was previously suggested (37). In the SPUsp45-NucA-CWAΔ105 construct, the absence of the glycine- and proline-rich 105-amino-acid peptide might have resulted in a better protein folding and thereby lower susceptibility to proteolysis. Alternatively, Nuc anchored to the cell wall with CWAΔ105 was presumably located closer to the cell envelope and could therefore be more protected from proteolysis than its counterpart anchored with whole CWA. It is noteworthy that both Nuc constructions were enzymatically active (not shown). However, for NucA-CWAΔ105, the appearance of pink halos on TB-D agar took longer, possibly because of reduced access of the enzyme to its DNA substrate.
FIG. 5.
A protein containing the complete CWA region CWAM6 is susceptible to degradation. Fusions spUsp45::nucA::cwaM6 (on pVE5524) and spUsp45::nucA::cwaM6Δ105 (on pVE5546) were expressed in L. lactis cells, and protein extracts were analyzed by Western blotting, using anti-NucA antibodies. P, protoplasts; CW, cell wall; SN, supernatant. The positions of molecular mass standards (in kilodaltons) are indicated. ∗, Nonmatured Nuc may correspond to SPUsp45-NucA-CWAM6 or to NucA-CWAM6. Forms not indicated by arrows are presumably protein degradation products.
The nature of the Nuc species (Fig. 5, nonmatured Nuc) remaining in the protoplast fraction in cells expressing spUsp45::nucA::cwaM6 was examined by fractionating the protoplast between membranes and cytoplasm. Western analysis shows that nonmatured Nuc species are exclusively in the membrane and are not in the cytoplasm (Fig. 6). We therefore believe that the membrane-bound species corresponds to NucA-CWAM6 rather than to SPUsp45-NucA-CWAM6, which we would expect to also be present in the cytoplasm fraction. Following this reasoning, we propose that cell Sec machinery does efficiently process the precursor and that sortase processing is limiting for protein maturation.
FIG. 6.
The fusion product of spUsp45::nucA::cwaM6 accumulates in the membrane fraction and not in the cytoplasm. Cells containing pVE5524 were fractionated into cytoplasm (CYT), membrane (MB), and cell wall (CW), and samples were analyzed by Western blotting, using anti-NucA antibodies. Refer to the text for the nature of nonmatured Nuc.
To examine this hypothesis, pulse-chase experiments were conducted with L. lactis cells expressing spUsp45::nucA::cwaM6. Pulse-labeled precursor SPUsp45-NucA-CWAM6 was present for 2 min after the chase (Fig. 7). In contrast, the NucA-CWAM6 form was only slowly processed into anchored Nuc, as it was present for at least 20 min after the chase. Note also that a single anchored species was detected, in keeping with the proposal that the multiple-banding pattern may correspond to degradation products. These observations reinforce the hypothesis that SPUsp45-NucA-CWAM6 is processed and translocated efficiently across the membrane while sortase processing of NucA-CWAM6 is slow and incomplete.
FIG. 7.
Anchoring is a limiting step for cell wall sorting of SPUsp45-NucA-CWAM6. Cells containing pVE5524 were pulse labeled for 2 min using [35S]methionine and chased with nonradioactive methionine. One minute before the chase and at time intervals 1, 2, 3, 5, and 20 min after the chase, an aliquot of culture was trichloroacetic acid precipitated and immunoprecipitated with anti-NucA antibodies. The immunoprecipitates were run on an SDS-PAGE (16% acrylamide) gel, and the gel was autoradiographed.
Nuc can be secreted and cell wall anchored in lactobacilli.
Lactobacilli are of great potential interest as vectors of secretion or for surface display of heterologous proteins. The export capacities of commensal lactobacilli, e.g., L. casei and L. reuteri, and food-fermenting lactobacilli, e.g., L. sakei and L. plantarum, were tested with the fusions described above. Plasmids pVE5523 (spUsp45::nucA) and pVE5524 (spUsp45::nucA::cwaM6) were used for expression in L. casei, L. sakei, and L. plantarum. The spUsp45::nucA fusions was recloned into pLEM7 for expression in L. reuteri cells. All strains expressed nucA, as shown by the appearance of pink halos around colonies (not shown). In L. casei, L. sakei, and L. plantarum cultures, the majority of Nuc was present in the supernatant as a single band (Fig. 8a, b, and c). Nuc export was very efficient in L. casei and L. sakei cells, while in L. plantarum cells, a portion of precursor (∼15%) remained unprocessed in protoplasts. In L. reuteri cells, the secreted spUsp45::nucA fusion was efficiently processed. However, the resulting Nuc was poorly released and the majority remained cell wall associated (Fig. 8d).
FIG. 8.
Secretion and anchoring of Nuc in lactobacilli. Fusions spUsp45::nucA and spUsp45::nucA::cwaM6 were expressed using pVE5523 and pVE5524, respectively, for L. casei, L. sakei, and L. plantarum. Fusion spUsp45::nucA was expressed in L. reuteri by using pVE5537. Protein extracts were analyzed by Western blotting using NucA antibodies. P, protoplasts; CW, cell wall; SN, supernatant. The positions of molecular mass standards (in kilodaltons) are indicated.
The capacity to anchor Nuc to the cell wall was demonstrated in L. casei, L. sakei, and L. plantarum cells. The presence of a single cell wall-associated species in these extracts suggests that less degradation may occur at the surface of lactobacilli than of L. lactis cells. A general estimation of expression levels was made by scanning Western blots and comparing the signals to those of internal standards of purified commercial NucA. In the case of exported fusions (i.e., both cell wall-anchored and secreted forms), Nuc was present at a 2 to 4 mg/liter range in all LAB strains; amounts were lower, i.e., 1 to 2 mg/liter, in the case of cytoplasmic Nuc (not shown). Taken together, the above results show that heterologous protein fusions can be successfully targeted in different locations in several Lactobacillus strains.
DISCUSSION
Secretion and cell wall anchoring of proteins (herein referred as protein export) are key functions in the interactions of LAB with their environment. Optimization of protein export is valuable in using these food-grade microorganisms in biotechnological applications both in vitro and in vivo in humans and animals. Our goal in this work was to design a system to target proteins in the desired location in different LAB species for use in numerous applications in food manufacturing and in health improvement. We therefore designed vectors that could target a model protein (Nuc) to the cytoplasm, cell wall, or medium by using L. lactis as a LAB model.
Nuc as a reporter to monitor protein targeting and conformation state.
The staphylococcal Nuc meets several required criteria for an efficient export reporter protein: (i) it is easily detected by simple activity tests on solid medium (18) or by immunoassays on Western blots (17), (ii) it is a convenient protein for biochemical analyses in that it is stable up to the boiling point and can be readily renatured after treatment by SDS and organic solvents, and (iii) it is inactive in the cytoplasm. The nuc gene encodes a preproprotein. The active 168-amino-acid proprotein, called NucB, is matured by several bacteria to form active NucA (17, 22, 23, 25, 38). Proprotein processing of NucB to NucA is mediated by the cell surface housekeeping proteinase HtrA in L. lactis (31). In our studies, only the nucA gene was used, thus reducing susceptibility of a fusion protein to proteolytic degradation. Using this reporter, we showed that Nuc remained active when it was anchored to cell wall of both lactococci and lactobacilli, even if the 105-amino-acid peptide spacer between Nuc and the anchor motif was removed. This indicates that proteins with a globular structure, such as Nuc (1), can fold properly, even when they are in close proximity to the peptidoglycan. This capacity is important for all applications of LAB involving surface display of proteins where spacial structure is crucial (e.g., for enzymes or conformational epitopes).
Signal peptide and N-terminal environment of Usp45 allows optimal translocation of precursors.
Based on the hypothesis that the unusually long M6 signal peptide might be necessary for protein sorting to the cell wall, we first used it to drive the export of both secreted and anchored Nuc. It appeared that it was a poor export signal in both cases, as much precursor remained unprocessed in the protoplast. The change of SPM6 to SPUsp45 greatly improved the situation: almost all precursor was processed and exported in the case of secreted Nuc, and the proportion of exported protein that was cell wall anchored was doubled. Protein Usp45 is the major secreted protein in L. lactis (46). We show here that its signal peptide plus first five amino acids of the mature protein are very efficient to drive secretion of heterologous proteins; this may reflect sequence or conformational features of Usp45 that are well recognized by the Sec machinery of L. lactis.
Nature of protein N terminus affects protein release in medium.
Efficient protein translocation across the membrane requires that the N terminus of a mature protein is less cationic than the N terminus of its signal peptide (51). This “positive inside rule” assures that the most cationic region of the precursor, i.e., the N terminus of the signal peptide, reacts with the anionic phospholipid heads on the inner side of membrane and determines the proper orientation of the signal peptide in the membrane (51). We propose here an additional effect of the charge at the protein N terminus. A fusion in which the five amino acids just after the signal peptide have a net positive charge remains (at about 30%) associated to the cell wall instead of being released in the medium (Fig. 3). This was observed using SPM6 as well as SPUsp45 (not shown). Addition of NaCl abolished this association, thus suggesting its electrostatic nature. Similar observations were made with Corynebacterium glutamicum (23). When the N-terminal amino acids were changed to five amino acids exhibiting a global net charge of −2, translocated Nuc was then entirely released in the medium (Fig. 5). The presence of anionic teichoic acids at the cell surface of gram-positive bacteria may interfere with protein targeting via nonspecific association of proteins with cationic tails, as exemplified here (26). Thus, an anionic spacer at the N terminus of mature proteins would not only improve protein translocation (22) but may also prevent nonspecific association with cell walls and possible further degradation. An alternative explanation to the observed phenomenon is that the short amino terminus of the two Nuc variants influences the overall protein folding, which in turn affects protein association with the cell wall. In either case, the choice of N-terminal amino acids may be important in protein localization when designing export systems for gram-positive bacteria.
Further improvements of the system.
In several experiments, we observed degradation products in all compartments, i.e., cytoplasm, cell wall, and supernatant. The extent of degradation varied according to the proteinaceous construct, i.e., we observed significant degradation of a Nuc derivative secreted using SPM6 and little degradation when Nuc was secreted using SPUsp45. The observation that NucA itself is not degraded is consistent with its stable globular structure (1). However, amino acid tails that were added at the N and C termini of NucA seemed to be highly susceptible to proteolysis, especially in SPM6-NucA. Degradation also varied according to the host strain tested, i.e., more degradation was observed in lactococci than in lactobacilli. We consider that in lactococci, the cell surface housekeeping proteinase HtrA involved in the processing of NucB into NucA is probably responsible for the degradation of amino acids tails fused at the ends of NucA (31). This proteolytic activity was not detected when salt was added to the culture medium. This may be due to HtrA inhibition or to a more rapid refolding and release of the potential HtrA target substrate. In either case, since these salt concentrations did not affect the L. lactis growth rate, use of these conditions could be a means of reducing proteolytic degradation when using L. lactis for biotechnological purposes.
An alternative way to improve protein yield will be to modify LAB host factors. Our studies suggest that, at the expression level used, sortase components may be limiting for complete and efficient anchoring and may even affect precursor maturation. Using the vectors for efficient protein targeting developed here, it will be of interest to overcome bottlenecks of export and anchoring. For example, (i) components of the sortase machinery could be overexpressed in order to achieve a better export and anchorage of cell surface proteins, (ii) lower proteolytic degradation could be achieved by disrupting the cell surface housekeeping proteinase htrA and/or its cytoplasmic counterpart clpP (13), and (iii) other genes that encode chaperone proteins involved in protein export and stability could be overexpressed (39).
Protein targeting in various LAB hosts opens the door to many applications.
We determined that the food-fermenting and commensal lactobacilli L. sakei, L. plantarum, and L. casei secreted and anchored Nuc as efficiently as L. lactis. Exported Nuc was estimated to be present at about 4 mg/liter of culture in all LAB strains tested; based on an average of 40% of Nuc anchored to the cell wall of those LAB per liter of culture at an OD600 of 1, the number of Nuc molecules associated with the cell wall corresponds to about 104 per cell. This is in agreement with levels reported with entire M6 in L. lactis and with the lipase anchored to the cell wall of Staphylococcus carnosus using the CWA domain of the fibronectin-binding protein of S. aureus (28, 43). The ability to target heterologous proteins to the cytoplasm, the cell wall, or the external medium is interesting for many applications. For example, live vaccine development using LAB as the vehicle has been proposed (33, 52). In this field, the ability to present antigens in various cell compartments of LAB will allow the study of the preferred antigen localization for mucosal immune response in the GIT (21, 50). Another application is the use of LAB as nutraceuticals and probiotics to provide enzymes, vitamins, or cytokines (42). In this field, the versatility of the system described here can be used to define the most appropriate mode of delivery in humans or animals.
ACKNOWLEDGMENTS
We are most grateful to several colleagues who provided us with precious material and tools: Michel Fons for L. reuteri strains and pLEM7 plasmid, Rufino Jimenez-Diaz for an L. plantarum strain and for sharing the L. plantarum transformation protocol before publication, Bénédicte Michel for several E. coli strains, James Miller for specific anti-NucA antibodies, Isabelle Poquet for pVE8001, Raul Raya for the L. casei strain, and Monique Zagorec for the L. sakei strain. We also thank Vincent Juillard for his help in the assay of PepX and Patrick Regent for the photography. We are grateful to our colleagues Philippe Langella, Yves Le Loir, and Isabelle Poquet for valuable discussions concerning this work.
REFERENCES
- 1.Alexandrescu A T, Hinck A P, Markley J L. Coupling between local structure and global stability of a protein: mutants of staphylococcal nuclease. Biochemistry. 1990;29:4516–4525. doi: 10.1021/bi00471a003. [DOI] [PubMed] [Google Scholar]
- 2.Berthier F, Zagorec M, Champommier-Vergès M, Ehrlich S D, Morel-Delville F. Efficient transformation of Lactobacillus sake by electroporation. Microbiology. 1996;142:12173–12179. doi: 10.1099/13500872-142-5-1273. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bringel F, Frey L, Hubert J-C. Characterization, cloning, curing, and distribution in lactic acid bacteria of pLP1, a plasmid from Lactobacillus plantarum CCM 1904 and its use in shuttle vector construction. Plasmid. 1989;22:193–202. doi: 10.1016/0147-619x(89)90002-4. [DOI] [PubMed] [Google Scholar]
- 5.Bruand C, Ehrlich S D. Transcription-driven DNA replication of plasmid pAMβ1 in Bacillus subtilis. Mol Microbiol. 1998;30:135–145. doi: 10.1046/j.1365-2958.1998.01044.x. [DOI] [PubMed] [Google Scholar]
- 6.Caplice E, Fitzgerald G F. Food fermentations: role of microorganisms in food production and preservation. Int J Food Microbiol. 1999;50:131–149. doi: 10.1016/s0168-1605(99)00082-3. [DOI] [PubMed] [Google Scholar]
- 7.Chich J-F, Chapot-Chartier M-P, Ribadeau-Dumas B, Gripon J-C. Identification of the active site serine of the X-prolyl dipeptyl aminopeptidase from Lactococcus lactis. FEBS Lett. 1992;314:139–142. doi: 10.1016/0014-5793(92)80960-o. [DOI] [PubMed] [Google Scholar]
- 8.Christie G E, Farnham P J, Platt T. Synthetic sites for transcription termination and a functional comparison with tryptophan operon termination sites in vitro. Proc Natl Acad Sci USA. 1981;78:4180–4184. doi: 10.1073/pnas.78.7.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Man J C, Rogosa M, Sharpe M T. A medium for cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 10.Fischetti V A, Pancoli V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from Gram-positive cocci. Mol Microbiol. 1990;4:1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 11.Fons M, Hege T, Ladire M, Raibaud P, Ducluzeau R, Maguin E. Isolation and characterization of a plasmid from Lactobacillus fermentum conferring erythromycin resistance. Plasmid. 1997;37:199–203. doi: 10.1006/plas.1997.1290. [DOI] [PubMed] [Google Scholar]
- 12.Fons M, Roels A, Ladire M, Raibaud P, Ducluzeau R. Recherche de gènes conférant un désavantage. In: Grimont P A D, editor. Nouvelles tendances en microbiologie anaérobie. Paris, France: Société Française de Microbiologie; 1994. p. 27. [Google Scholar]
- 13.Frees D, Ingmer H. ClpP participates in the degradation of misfolded proteins in Lactococcus lactis. Mol Microbiol. 1999;31:79–87. doi: 10.1046/j.1365-2958.1999.01149.x. [DOI] [PubMed] [Google Scholar]
- 14.Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66:365–378. [PubMed] [Google Scholar]
- 15.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmgren J, Czerkinsky C, Lycke N, Svennerholm A M. Mucosal immunity: implications for vaccine development. Immunobiology. 1992;184:157–179. doi: 10.1016/S0171-2985(11)80473-0. [DOI] [PubMed] [Google Scholar]
- 17.Kovacevic S, Veal L E, Hsiung H M, Miller J R. Secretion of staphylococcal nuclease by Bacillus subtilis. J Bacteriol. 1985;162:521–528. doi: 10.1128/jb.162.2.521-528.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lachica R V F, Genigeorgis C, Hoeprich P D. Metachromatic agar-diffusion methods for detecting staphylococcal nuclease activity. Appl Microbiol. 1971;21:585–587. doi: 10.1128/am.21.4.585-587.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Langella P, Le Loir Y S, Ehrlich D, Gruss A. Efficient plasmid mobilization by pIP501 in Lactococcus lactis subsp. lactis. J Bacteriol. 1993;175:5806–5813. doi: 10.1128/jb.175.18.5806-5813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leclerc C, Charbit A, Martineau P, Deriaud E, Hofnung M. The cellular location of a foreign B cell epitope expressed by recombinant bacteria determines its T cell-independent or T cell-dependent characteristics. J Immunol. 1991;147:3545–3552. [PubMed] [Google Scholar]
- 22.Le Loir Y, Gruss A, Ehrlich S D, Langella P. A 9-residue synthetic propeptide enhances secretion efficiency of heterologous proteins in Lactococcus lactis. J Bacteriol. 1998;180:1895–1903. doi: 10.1128/jb.180.7.1895-1903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liebl W, Sinskey A J, Schleifer K-H. Expression, secretion, and processing of staphylococcal nuclease by Corynebacterium glutamicum. J Bacteriol. 1992;174:1854–1861. doi: 10.1128/jb.174.6.1854-1861.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marteau P, Rambaud J-C. Potential of using lactic acid bacteria for therapy and immunomodulation in man. FEMS Microbiol Rev. 1993;12:207–220. doi: 10.1111/j.1574-6976.1993.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 25.Miller J R, Kovacevic S, Veal L E. Secretion and processing of staphylococcal nuclease by Bacillus subtilis. J Bacteriol. 1987;169:3508–3514. doi: 10.1128/jb.169.8.3508-3514.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perego M, Glasser P, Minutello A, Strauch M A, Leopold K, Fischer W. Incorporation of D-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. J Biol Chem. 1995;270:15598–15606. doi: 10.1074/jbc.270.26.15598. [DOI] [PubMed] [Google Scholar]
- 27.Peschke U, Beuck V, Bujard H, Gentz R, Le Grice S. Efficient utilization of Escherichia coli transcriptional signals in Bacillus subtilis. J Mol Biol. 1985;186:547–555. doi: 10.1016/0022-2836(85)90129-9. [DOI] [PubMed] [Google Scholar]
- 28.Piard J C, Hautefort I, Fischetti V A, Ehrlich S D, Fons M, Gruss A. The M6 protein from Streptococcus pyogenes: cloning and expression of its emm6 structural gene and cell wall anchoring efficiency in various lactic acid bacteria. J Bacteriol. 1997;179:3068–3072. doi: 10.1128/jb.179.9.3068-3072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piard J-C, Jimenez-Diaz R, Fischetti V A, Ehrlich S D, Gruss A. The M6 protein of Streptococcus pyogenes and its potential as a tool to anchor biologically active molecules at the surface of lactic acid bacteria. Adv Exp Med Biol. 1997;418:545–550. doi: 10.1007/978-1-4899-1825-3_126. [DOI] [PubMed] [Google Scholar]
- 30.Poquet I, Ehrlich S D, Gruss A. Identification of Lactococcus lactis exported proteins by use of a new reporter, the staphylococcal nuclease. J Bacteriol. 1998;180:1904–1912. doi: 10.1128/jb.180.7.1904-1912.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poquet I, Saint V, Seznec E, Simoes N, Bolotin A, Gruss A. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol Microbiol. 2000;35:1042–1051. doi: 10.1046/j.1365-2958.2000.01757.x. [DOI] [PubMed] [Google Scholar]
- 32.Raibaud P, Galpin J V G, Ducluzeau R, Mocquot G, Olivier G. Le genre Lactobacillus dans le tube digestif de rat. Ann Inst Pasteur Microbiol. 1973;124A:83–109. [PubMed] [Google Scholar]
- 33.Robinson K, Chamberlain L M, Schofield K M, Wells J M, Le Page R W F. Oral vaccination of mice against tetanus with recombinant Lactococcus lactis. Nat Biotechnol. 1997;15:653–657. doi: 10.1038/nbt0797-653. [DOI] [PubMed] [Google Scholar]
- 34.Salminen S, Bouley C, Boutron-Ruault M C, Cummings J H, Franck A, Gibson G R, Isolauri E, Moreau M C, Roberfroid M, Rowland I. Functional food science and gastrointestinal physiology and function. Br J Nutr. 1998;80(Suppl. 1):147–171. doi: 10.1079/bjn19980108. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Schneewind O, Model P, Fischetti V A. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 37.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983;22:181–189. doi: 10.1016/0378-1119(83)90102-6. [DOI] [PubMed] [Google Scholar]
- 39.Silmonen M, Palva I. Protein secretion in Bacillus species. Microbiol Rev. 1993;57:109–137. doi: 10.1128/mr.57.1.109-137.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simons D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 41.Stähl S, Uhlén M. Bacterial surface display: trends and progress. Trends Biotechnol. 1997;15:185–192. doi: 10.1016/S0167-7799(97)01034-2. [DOI] [PubMed] [Google Scholar]
- 42.Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, Fiers W, Remaut E. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–1355. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 43.Strauss A, Götz F. In vivo immobilizatiuon of enzymatically active polypeptides on the cell surface of Staphylococcus carnosus. Mol Microbiol. 1996;21:491–500. doi: 10.1111/j.1365-2958.1996.tb02558.x. [DOI] [PubMed] [Google Scholar]
- 44.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Environ Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ton-That H, Liu G, Mazmanian S K, Faull K F, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Asseldonk M, Rutten G, Oteman M, Siezen R J, de Vos W M, Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene. 1990;95:155–160. doi: 10.1016/0378-1119(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 47.Van de Gughte M, Kok J, Venema G. Gene expression in Lactococcus lactis. FEMS Microbiol Rev. 1992;88:73–92. doi: 10.1111/j.1574-6968.1992.tb04958.x. [DOI] [PubMed] [Google Scholar]
- 48.Van der Vossen J M B M, van der Lelie D, Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987;53:2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 50.Viret J-F, Favre D, Wegmüller B, Herzog C, Que J U, Cryz S J, Lang A B. Mucosal and systemic immune responses in humans after primary and booster immunizations with orally administered invasive and noninvasive live attenuated bacteria. Infect Immun. 1999;67:3680–3685. doi: 10.1128/iai.67.7.3680-3685.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Von Heijne G. Net N-C charge imbalance may be important for signal sequences function in bacteria. J Mol Biol. 1986;192:287–290. doi: 10.1016/0022-2836(86)90365-7. [DOI] [PubMed] [Google Scholar]
- 52.Wells J M, Robinson K, Chamberlain L M, Schofield K M, Le Page R W F. Lactic acid bacteria as vaccine delivery vehicles. Antonie Leeuwenhoek. 1996;70:317–330. doi: 10.1007/BF00395939. [DOI] [PubMed] [Google Scholar]