Preclinical studies demonstrate that relatlimab specifically blocks the interaction between LAG-3 and its ligands. The data provide a biological rationale for combining relatlimab with the PD-1 antibody nivolumab as an effective cancer immunotherapeutic strategy.
Abstract
Novel therapeutic approaches combining immune-checkpoint inhibitors are needed to improve clinical outcomes for patients with cancer. Lymphocyte-activation gene 3 (LAG-3) is an immune-checkpoint molecule that inhibits T-cell activity and antitumor immune responses, acting through an independent mechanism from that of programmed death-1 (PD-1) and cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4). Here, we describe the development and preclinical characterization of relatlimab, a human antibody that binds to human LAG-3 with high affinity and specificity to block the interaction of LAG-3 with the ligands MHC II and fibrinogen-like protein-1, and to reverse LAG-3–mediated inhibition of T-cell function in vitro. Consistent with previous reports, in mouse models, the combined blockade of LAG-3 and PD-1 with surrogate antibodies resulted in enhanced antitumor activity greater than the individual blockade of either receptor. In toxicity studies in cynomolgus monkeys, relatlimab was generally well tolerated when combined with nivolumab. These results are consistent with findings from the RELATIVITY-047 phase II/III trial showing that relatlimab combined with nivolumab is a well-tolerated regimen that demonstrates superior progression-free survival compared with nivolumab monotherapy in patients with unresectable or metastatic melanoma.
Introduction
Immune-checkpoint blockade (ICB) has revolutionized treatment options for patients with cancer, improving the survival of patients with a range of different malignancies (1). An effective immune response against cancer relies on immune surveillance of tumor antigens expressed on cancer cells, which ultimately results in an adaptive immune response and cancer cell death (2–4). Tumor immune escape, facilitated in part by the expression of inhibitory ligands, can be reversed by the blockade of receptors such as programmed death-1 (PD-1) and cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4), but there remains a need for additional novel combinations to improve patient outcomes (5–7).
Lymphocyte-activation gene 3 (LAG-3) is an inhibitory receptor on T cells frequently upregulated on the surface of tumor-infiltrating lymphocytes (TIL), including regulatory T cells, which contributes to T-cell exhaustion in the tumor microenvironment (TME) and limits antitumor T-cell responses (8–11). The mechanism by which LAG-3 exerts its immune-suppressive effects is not completely understood but includes engagement by major histocompatibility complex II (MHC II) expressed on antigen-presenting cells (APC) within the TME and on some tumor cells, as well as potential engagement by other reported ligands, including fibrinogen-like protein-1 (FGL1), LSECtin, α-synuclein fibrils, and galectin-3 (5, 12–15).
Previous studies have demonstrated that LAG-3 and PD-1 act in a nonredundant manner to suppress T-cell stimulation. In an in vitro antigen-specific T-cell stimulation system, the peptide responsiveness of T cells transduced to express both LAG-3 and programmed death ligand 1 (PD-L1) shows lower levels of interleukin-2 (IL2) secretion in coculture with APCs expressing PD-L1 and MHC II compared with the responses of T cells expressing either receptor alone (16). Preclinical data have demonstrated a synergistic relationship between the inhibitory receptors LAG-3 and PD-1 in regulating immune homeostasis, preventing autoimmunity, and enforcing tumor-induced tolerance (9, 16). Importantly, in mice, antibody blockade of both receptors results in more robust immune responses compared with blockade of either individual receptor (17–19).
Here, we describe the development of relatlimab, a human LAG-3 (hLAG-3)-blocking antibody, and preclinical analyses that demonstrate the binding affinity, specificity, functional activity, and safety of this novel immune-checkpoint inhibitor. In vitro assay results were consistent with published in vivo data showing that LAG-3 blockade combined synergistically with PD-1 blockade to achieve enhanced antitumor and immunomodulatory activity. Aside from MHC II as the canonical ligand of LAG-3, the literature on other reported LAG-3 ligands and their biology as a potential driver of LAG-3–mediated T-cell exhaustion in cancer is limited but represents an evolving area of scientific research. Nonetheless, data on FGL1 as a LAG-3 ligand led to the evaluation of the interaction between FGL1 and LAG-3, and of its modulation by relatlimab as part of the current study (12). We observed a weak, but measurable, interaction in vitro between FGL1 and LAG-3 and confirmed both the inhibitory potential of this interaction and the ability of relatlimab to block it. Our overall findings are consistent with the results from the RELATIVITY-047 study (NCT03470922), the first phase II/III trial evaluating dual administration of relatlimab and nivolumab in patients with previously untreated or unresectable melanoma. In this trial, the combined blockade of LAG-3 and PD-1 demonstrated superior progression-free survival (PFS) compared with the blockade of PD-1 alone (20, 21).
Materials and Methods
Mice
Generation of relatlimab was done using proprietary transgenic mice bred at Medarex, Inc., comprising germline configuration human immunoglobulin (Ig) miniloci in an endogenous IgH and Igκ knockout background (22, 23). For in vivo tumor efficacy studies, female C57BL/6 mice were obtained from Charles River Laboratories and female A/J mice were obtained from Harlan. All animals were provided chow (Prolab Isopro; Dean's Animal Feeds) and drinking water ad libitum. The Animal Use Protocols used for the antibody generation work and tumor efficacy studies were approved by the Medarex Institutional Animal Care and Use Committee prior to the initiation of animal treatments, and all animal husbandry was performed according to Medarex Standard Operating Procedures. The Medarex Animal Facility was accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
Cell lines
MC38 colon adenocarcinoma and SA1N fibrosarcoma tumor cells, obtained from the Bristol-Myers Squibb (BMS) Master Cell Bank, were maintained in Dulbecco's modified Eagle medium (DMEM; Corning/Cellgro) supplemented with 10% fetal bovine serum (FBS; Hyclone). P3×63Ag8.653 myeloma cells were obtained from the ATCC and maintained in RPMI-1640 (Corning/Cellgro) with 10% FBS (Gemini Bio-Products). Expi-293F cells were obtained from Invitrogen/Thermo Fisher Scientific and maintained in Expi293 expression media (Invitrogen). Human Daudi and Raji B-cell lymphoma lines expressing endogenous MHC II were obtained from the ATCC and maintained in RPMI-1640 and 10% FBS. The RJ225 MHC II-low variant of the Raji cell line was obtained from Dr. Matija Peterlin (University of California San Francisco, San Francisco) and maintained in RPMI-1640 and 10% FBS without selection (24). 3A9-hLAG-3 cells overexpressing full-length human LAG-3 and the LK35.2 APC line were obtained from Dr. Dario Vignali (University of Pittsburgh, Pittsburgh) and were maintained in RPMI-1640 with 10% FBS without selection.
The LK35-FGL1MT cell line was generated at BMS by electroporation of parental LK35.2 cells with a construct fusing FGL1 to the intracellular domain of Type II transmembrane protein human FIBCD1 following the example of Wang and colleagues (12) using the manufacturer-recommended protocol (Nucleofector 2b, 2 × 106 cells, 5 μg DNA, Solution V, Program L-013; Lonza). This cell line was archived in the BMS repository and maintained in RPMI-1640 medium (Corning/Cellgro) supplemented with 10% FBS (Gemini Bio-Products).
Chinese hamster ovary suspension (CHO-S) cells were obtained from Invitrogen and maintained in CD-CHO media (Invitrogen) containing 8 mmol/L glutamine (Invitrogen) and 1× HT supplement (Invitrogen). CHO-S cells expressing human LAG-3 (CHO-huLAG-3) were generated by electroporation of parental CHO-S cells with proprietary plasmid constructs for expression of full-length human LAG-3 (Genbank Accession code, X51985.3) using the manufacturer-recommended protocol (Nucleofector 2b, 1.0E+06 cells, 5 μg DNA, Solution V, Program U-024; Lonza). This cell line was archived in the BMS repository and maintained in CD-CHO media (Invitrogen) containing 8 mmol/L glutamine (Invitrogen) and 1× HT supplement (Invitrogen) and supplemented with 0.5 mg/mL G418 (Corning/Cellgro).
Cell lines were validated to be free of mycoplasma by PCR (PromoKine PCR Mycoplasma Test; VWR). The cell lines were maintained in culture for no more than 3 weeks prior to use in the appropriate in vitro or in vivo studies. The cell lines were not authenticated at the time of use. Cell lines used for in vivo studies were validated to be free of adventitious agents by PCR (Idexx Bioanalytics).
Relatlimab generation and characterization
Generation of human monoclonal anti–LAG-3 (relatlimab)
Proprietary transgenic mice, bred at Medarex (Medarex, Inc.), comprising germline configuration human Ig miniloci in an endogenous IgH and Igκ knockout background (22, 23), were immunized at 14-day intervals by i.p./s.c. administration with 10 μg recombinant human LAG-3–Fc protein (hLAG-3–hFc; R&D Systems), consisting of the extracellular domain of LAG-3 (Leu23-Leu450) fused to the Fc portion of human IgG1, together with Ribi adjuvant (Ribi lmmunoChemical Research). Spleens were harvested from immunized mice 2 weeks after the final immunization of antigen, and splenocytes were fused with P3×63Ag8.653 myeloma cells (ATCC) and screened for hybridomas producing human monoclonal antibodies (mAb) reactive to hLAG-3–hFc by enzyme-linked immunosorbent assay (ELISA), as described below. Purified antibodies specific for hLAG-3, including clones 25F7, 1C2, 23C9, and 10G7, were analyzed by flow cytometry, as described below, for the potency of binding to transfected CHO-S cells overexpressing hLAG-3 (BMS), but not to parental CHO cells. The variable region sequences of clone 25F7 were cloned and subsequently grafted onto human κ and IgG4 constant region sequences for recombinant CHO cell expression of the resulting derived antibodies, LAG3.1-G4P and LAG3.5-G4P (relatlimab; US patent US9505839B2).
Hybridoma screen via ELISA
Hybridomas derived from the fused spleens of hLAG-3-Fc–immunized mice were screened by ELISA for the production of human antibodies specific for hLAG-3. Hybridoma supernatants, including irrelevant supernatants from cultures of the myeloma fusion partner as control, were screened for binding activity to hLAG-3 by ELISA. Briefly, microtiter plates (Corning/Costar; catalog CLS9018) were coated with recombinant hLAG-3–Fc (R&D Systems) at 1 μg/mL in phosphate-buffered saline (PBS), 50 μL/well, incubated at 4°C overnight, and then blocked with 1% bovine serum albumin (BSA) in PBS buffer, pH7.4. Hybridoma supernatants were added neat and incubated for 1 hour. The plates were washed with PBS containing 0.05% Tween 20 (Sigma-Aldrich) and incubated with goat-anti-human kappa light chain conjugated with horseradish peroxidase (Bethyl Laboratories), diluted 1:25,000 in 1% BSA in PBS buffer, pH7.4, for 1 hour. After 3× washing, the plates were developed with ABTS substrate (Moss) and analyzed on a SpectraMax Plus 384 spectrophotometer (Molecular Devices) at optical density (OD) 405 nm. To rule out any nonspecific binding, an irrelevant human Fc-fusion protein control was used to test the hybridoma supernatants.
Epitope characterization
The relatlimab-binding epitope on LAG-3 was determined by several methods, including peptide library binding, binding to LAG-3 truncation mutants, differential chemical labeling, and X-ray crystallography. Detailed descriptions of the methods used are presented in the Supplementary Methods included in the Supplementary Data.
Octet biolayer interferometry characterization of relatlimab blockade of LAG-3/MHC II and LAG-3/FGL1 interactions
LAG-3/MHC II interaction and blockade by relatlimab
Recombinant human leukocyte antigen-DR isotype (HLA-DR) with a C-terminal biotin modification was generated at BMS using human HLA-DRA (accession code P01903, residues 26–207) and HLA-DRB1 (accession code P01911, residues 31–221) genes cloned into pTT5 expression plasmids (GenScript). HLA-DRA contains a C-terminal His6 tag and BirA recognition sequence, and HLA-DRB1 has a C-terminal Flag tag and an N-terminal peptide with a GS linker (SGPKYVKQNTLKLATGSGGGSLVPRGSGGGGSG; ref. 25). HLA-DRA and HLA-DRB1 were subsequently coexpressed in Expi293 cells, purified from clarified supernatant using Ni Sepharose Excel (GE Healthcare), and then run on a Superdex 200 16/60 column in PBS. The purified heterodimer was subsequently biotinylated using a BirA biotin-protein ligase bulk reaction kit according to the manufacturer's instructions using the kit-provided buffers (Avidity), followed by buffer exchange via dialysis into PBS without calcium and magnesium (Corning/Cellgro, catalog 21-040-CM) for further experiments. A recombinant hLAG-3 (D1–D4)–hFc fusion protein was produced by cloning hLAG-3 (D1–D4) into a pTT5 expression plasmid (GenScript) with an osteonectin signal peptide on the N-terminus and a C-terminal hFcG1 tag. Clarified supernatants were purified using MabSelect SuRe LX resin (GE Healthcare), washed with PBS, eluted in 100 mmol/L (pH 3.6) sodium citrate buffer, and neutralized with 1 mol/L (pH 8.0) Tris buffer (Corning/Cellgro). hLAG-3 (D1–D4) was further purified on a Superdex 200 column into 1× PBS. The binding of HLA-DR1 to the LAG-3 and evaluation of the effect of various antibodies on this interaction were investigated on an Octet HTX instrument with streptavidin capture (SA) biosensors (ForteBio/Sartorius). HLA-DR1 was captured at 10 μg/mL for 180 seconds in PBS + 0.05% BSA, with 0.05% TWEEN20, pH 7.4 (Sigma-Aldrich; PBST-BSA), followed by a 60-second wash with PBST-BSA. hLAG-3 (D1–D4)–hFc (1 μmol/L) alone or in the presence of 2.5-fold excess antibody was tested for binding to the captured HLA-DR protein over 300 seconds. For all biolayer interferometry (BLI) assays, the temperature was 30°C with a shake speed of 1,000 RPM. Data were visualized on ForteBio data analysis software.
LAG-3/FGL1 interaction and blockade by relatlimab
Recombinant human fibrinogen-like protein-1 (FGL1)–mFc fusion protein and recombinant human FGL1-FD-mFc fusion protein [consisting of only the fibrinogen domain of human FGL1 (residues 74–312)] were produced by BMS. Briefly, these were produced by cloning into a pTT5 expression plasmid (GenScript) with an osteonectin signal peptide followed by an mFcG1 tag on the N-terminus. These were expressed in Expi293 cells and clarified supernatants were purified using MabSelect SuRe LX resin (GE Healthcare), washed with PBS, eluted in 100 mmol/L (pH 3.6) sodium citrate buffer, and neutralized with 1 mol/L (pH 8.0) Tris buffer (Corning/Cellgro) and 1 mol/L (pH 8.5) arginine buffer. These were finally run on a Superdex 200 16/60 column. The binding of recombinant FGL1–mFc and FGL1-FD–mFc to recombinant hLAG-3–hFc was investigated on an Octet HTX instrument with anti-human IgG Fc capture (AHC) biosensors (ForteBio). LAG-3–hFc was captured at 10 μg/mL for 300 seconds in PBST-BSA, followed by a 60-second wash with PBST-BSA. Serial dilutions of FGL1–mFc and FGL1-FD–mFc were tested for binding to the captured LAG-3–hFc fusion protein. To assess the blockade of LAG-3/FGL1 engagement by relatlimab, LAG-3–hFc was first captured on AHC biosensors at 10 μg/mL for 600 seconds in PBST-BSA, followed by a 30-second wash with PBST-BSA. The biosensor was next quenched with a cocktail of keyhole limpet hemocyanin (KLH) human IgG1 (hIgG1), KLH-hIgG2, and KLH-hIgG4 antibodies (each at 15 μg/mL, produced at BMS) for 600 seconds in PBST-BSA to block the surface. This was followed by a 30-second wash with PBST-BSA, exposure to buffer alone or relatlimab (200 nmol/L) in PBST-BSA for 1,000 seconds, and a 10-second wash with PBST-BSA. Finally, for biosensors exposed to buffer alone, FGL1-FD–mFc (2 μmol/L) in the absence of relatlimab was tested for binding. For biosensors previously exposed to relatlimab, FGL1-FD–mFc (2 μmol/L) in the presence of excess relatlimab (200 nmol/L) was tested for binding. For all biolayer interferometry assays, the temperature was 30°C with a shake speed of 1,000 RPM. Data were visualized on ForteBio data analysis software.
LAG-3 deletion variants binding to relatlimab
Recombinant human LAG-3 (D1-D2), LAG-3 (D1-D2) ∆relatlimab, and LAG-3 (D1-D2) ∆loop insertion variants (deletion designs in the D1 domain described in Supplementary Fig. S9C) with C-terminal His6 tags were generated at BMS by cloning each into pTT5 expression plasmids (GenScript), which were subsequently coexpressed in Expi293 cells, purified from clarified supernatant using Ni Sepharose Excel (GE Healthcare), and polished on a Superdex 200 16/60 column in PBS. Binding of relatlimab to the LAG-3 variants was investigated on an Octet HTX instrument with AHC biosensors (ForteBio). Relatlimab was captured at 2 μg/mL for 180 seconds in PBS 0.05% BSA with 0.05% TWEEN20 pH 7.4 (Sigma-Aldrich), followed by a 60-second wash with PBST-BSA. The hLAG-3 variants at 100 nmol/L and 200 nmol/L were tested for binding to the captured relatlimab over 300 seconds. For all BLI assays, the temperature was 30°C with a shake speed of 1,000 RPM. Data were visualized on ForteBio data analysis software.
IHC analysis in normal human tissues
IHC analyses were performed in a selected panel of normal human tissues. Fresh frozen normal human tissues were purchased from commercial tissue networks/vendors (Analytical Biological Services, Inc.; Asterand, Inc.; and Cooperative Human Tissue Network). Tissues evaluated (n = 1 unless otherwise indicated) were human spleen, thymus, pancreas, cerebrum, cerebellum, heart, liver, lung, kidney, pituitary (n = 3), and tonsil (n = 2). Stained slides were evaluated using a DMLB brightfield microscope (Leica).
To detect tissue binding, the fluoresceinated forms of relatlimab, LAG3.1-G4P, and isotype control antibody were prepared and designated as relatlimab-FITC, LAG3.1-G4P-FITC, and hIgG4-FITC, respectively. Briefly, relatlimab (BMS), LAG-3.1-G4P (BMS), and hIgG4 (US Biological) were conjugated to fluorescein-5-isothiocyanate (FITC, Invitrogen). FITC was dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich) at 2 mg/mL, and the pH of the antibody solutions was adjusted to 9.2–9.4 using 0.1 mol/L Na2CO3 (Sigma-Aldrich). 40 μg of FITC was used per mg of antibody and incubated for 2.5 hours at room temperature. FITC-conjugated antibody and free FITC were separated using Econo-Pac 10DG desalting column (Bio-Rad). The conjugated antibody concentration and fluor/protein (F/P) conjugation ratio were determined by measuring OD at A280 and A495 on a Spectramax 384 Plus spectrophotometer (Molecular Devices). The F/P ratio ranged from 2.2 to 2.8.
Cryostat sections at 5 μm were fixed with acetone and then washed twice with PBS at room temperature. LAG-3–expressing lymphocytes and LAG-3-negative elements in tonsil sections were used as positive and negative control tissues, respectively. Cryostat sections of CHO-S–LAG-3 cells and CHO-S cells were also used as positive and negative controls. Endogenous peroxidase activity was blocked by incubation with peroxidase block supplied in the Dako EnVision System (Agilent; catalog K4011). Slides were washed in PBS and then incubated with Dako protein block supplemented with 0.5% human γ globulins (Sigma-Aldrich) to block nonspecific binding sites. Subsequently, primary antibodies (relatlimab-FITC or LAG-3.1-G4P-FITC) or isotype control (hIgG4-FITC) were applied onto sections and incubated for 1 hour. After washing, slides were subjected to serial 30-minute incubations separated by repeat washes, with rabbit anti-FITC, peroxidase-conjugated anti-rabbit IgG polymer, and finally 6 minutes with diaminobenzidine (DAB) substrate-chromogen solution (Dako EnVision System; Agilent). Slides were washed with deionized water, counterstained with Mayer's hematoxylin, dehydrated, cleared, and cover-slipped with Permount following routine histologic procedures. Stained slides were evaluated by light microscopy as described above. Grading criteria were as noted in Supplementary Table S4.
To verify the cell type within the pituitary gland to which relatlimab bound, double immunofluorescent staining of LAG-3.1-G4P-FITC with antibodies specific for five pituitary hormones was performed on cryostat sections. Hormone-specific antibodies used were the following: mouse anti-adrenocorticotropic hormone (Dako), antiluteinizing hormone (Dako), and anti-thyroid stimulating hormone (Abcam), or rabbit polyclonal anti-growth hormone (Dako), and rabbit anti-prolactin (Dako). In the same manner as above, slides were incubated with Dako protein block supplemented with 1% human γ globulins to block nonspecific binding sites. To reduce the background further, the PBS used in immunofluorescence contained high salt and low detergent (300 mmol/L NaCl and 0.01% TWEEN20). Subsequently, LAG-3.1-G4P-FITC with each of the above five hormone antibodies were simultaneously applied onto sections. Human IgG4-FITC, with either mouse IgG1 or rabbit IgG, was used as isotype control. After washing, slides were incubated with secondary antibodies: Alexa Fluor 488–conjugated goat anti-FITC/Oregon-Green (Invitrogen), Cy3-labeled donkey anti-mouse (Jackson ImmunoResearch), and Cy3-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch). After washing, sections were counterstained with Hoechst 33342 (Invitrogen) mounting with ProLong-Gold Antifade reagent (Invitrogen). To assess the specificity of the LAG-3.1-G4P-FITC staining, preabsorption of the primary antibody with human LAG-3 fusion protein was performed by preincubating LAG-3.1-G4P-FITC with 5- or 10-fold excess of human LAG-3 for 2 hours at room temperature before applying to the sections from two pituitary samples, following respective protocols, for immunoperoxidase and immunofluorescent methods. PBS supplemented with 0.5% human γ globulins and 0.5% BSA was used as a diluent for both primary and secondary antibodies. For more stringent nonspecific blocking conditions, Dako protein block supplemented with 1% human γ globulins or PBS supplemented with 1% human γ globulins was used as a blocking buffer or diluent, respectively. Both the staining intensity and frequency were evaluated using an Imager D1 fluorescence microscope (Zeiss).
Antitumor activity of anti–LAG-3 and anti–PD-1 in mouse models
Syngeneic tumor models
MC38 colon adenocarcinoma and SA1N fibrosarcoma tumor cells from the BMS Master Cell Bank were maintained in DMEM (Corning/Cellgro) supplemented with 10% FBS (Gemini Bio-Products). Cell lines used for in vivo studies were validated to be free of adventitious agents by PCR (Idexx Bioanalytics). Cells displayed a doubling time of 24 hours and were harvested near 80% confluence, washed in FBS-free medium three times, and resuspended in DMEM to provide subcutaneous injections of 1.0×106 cells (0.1 mL) into the right flank of each study animal using a 1-cc syringe (Becton Dickinson) and 25-gauge half-inch needle. Tumors were measured in 3 dimensions (l × w × h/2) with a Fowler Electronic Digital Caliper (Model 62379 531; Fred V. Fowler Co.) and recorded. Animals were randomized into study groups with similar tumor size ranges. Mice with palpable tumors on day 7 after tumor implantation (average volume 75–100 mm3) were administered with monoclonal antibodies interperitoneally, as appropriate by group, on days 7, 10, and 14 at a dose of 10 mg/kg, unless otherwise noted. Tumor size and body weight of study animals were monitored and recorded twice weekly until tumors reached endpoint (≤1,500 mm3).
Surrogate antibodies used in these studies included anti-mouse PD-1 monoclonal clone 4H2 (6), anti-mouse LAG-3 monoclonal clone C9B7W (4), anti-mouse LAG-3 monoclonal clone 19C7 [BMS, proprietary chimeric antibody raised against mouse LAG-3 antigen (R&D Systems) with rat IgG variable regions grafted onto mouse IgG1 Fc region incorporating a D265A mutation for reduced Fc receptor engagement, produced in a transfected CHO cell line], and purified mouse IgG1 (MOPC-21) isotype control antibody was obtained from Bio X Cell.
Toxicity studies in cynomolgus monkey model
Study design
As part of a 4-week multidose toxicity study to evaluate the potential toxicity of relatlimab alone and in combination with nivolumab when administered intravenously, cynomolgus monkeys (Macaca fascicularis) were administered a single intramuscular bolus of KLH antigen (Thermo Fisher/Pierce) on day 1 and were dosed once weekly (q.w.) for five total doses with relatlimab alone or in combination with nivolumab (both antibodies produced at BMS). KLH, a protein purified from the mollusk Megathura crenulata, can stimulate a T cell–dependent antibody response (TDAR) upon immunization. Antibody formation to KLH can be used to monitor the effect of drug candidates on the humoral immune response. In this study, blood samples were collected at preselected time points to measure systemic exposures to relatlimab and nivolumab, for analysis of TDAR to KLH (IgG, IgM, and IgA collectively) by ELISA, and for analysis of T-lymphocyte subsets by flow cytometry (see Supplementary Methods). At scheduled necropsies following the final antibody dose and following a 6-week recovery period, unfixed spleen sections were collected for analysis of splenic T-lymphocyte subsets by flow cytometry. The study procedures were approved by the Institutional Animal Care and Use Committee, and the monkeys were monitored daily for clinical signs. The clinical evaluation parameters and dosing strategy for this study are described in Supplementary Table S1.
The potential toxicity of relatlimab was also assessed in a 3-month, multidose toxicity study in monkeys administered drug intravenously q.w. for 3 months. The clinical evaluation parameters and dosing strategy for this study are described in Supplementary Table S2, and full details of the toxicity studies are provided in the Supplementary Methods included in Supplementary Data.
Statistical analyses
Statistical analyses of the peripheral blood lymphocyte phenotyping, KLH-specific antibodies, and splenic T-lymphocyte subset phenotyping data were performed by Global Biometric Sciences Nonclinical Biostatistics. Because the sample sizes within each sex were small, males and females were combined to provide sample sizes of about n = 6/group on day 30, and about n = 4/group on day 72. Homogeneity of group variances was assessed at the 5% significance level using Levene's test. Normality of the group distributions was assessed graphically. If significant heterogeneity of variance was observed (P ≤ 0.05), or if nonnormality was observed, an appropriate transformation of data was utilized. A one-way analysis of variance (ANOVA) model was utilized to analyze the percentage of splenic lymphocytes for each subset population; the Dunnett multiple-comparison t test procedure was utilized to compare the means for the vehicle-control group (group 1) to the means of each of the other groups (groups 2–5). Two-sided test statistics were calculated at the 5% significance level. Analyses were performed using SAS version 9.1.2. Graphs were produced using GraphPad Prism software.
Additional methods
Additional details on methods and experiments relating to relatlimab binding to human and cynomolgus monkey LAG-3, binding affinity, cell-based bio- and blocking assays, antibody-dependent cellular cytotoxicity (ADCC), PCR analysis, splenic T-lymphocyte subset phenotyping, ex vivo responses to KLH-specific antibodies, antibodies used, cell cultures, and procurement of human tissue samples can be found in the Supplementary Methods.
Data availability
The data generated in these studies are available within the article. The X-ray diffraction coordinates and structure factors are publicly available from the Research Collaboratory for Structural Bioinformatics Protein Data Bank under PDB ID 7UM3. The raw data underlying the included surface plasmon resonance (SPR) results were generated by a core facility and may not be available.
References are included with respect to animal models and reagents used in the study as appropriate. Relatlimab is commercially available as a therapeutic agent in the United States. Other anti–LAG-3 antibodies mentioned in the article can be made available upon reasonable request to the lead author and review by BMS, in compliance with BMS compound, technology, and data sharing policies (further details can be found at https://www.bms.com/researchers-and-partners/independent-research/compound-and-technology-requests.html and https://www.bms.com/researchers-and-partners/independent-research/data-sharing-request-process.html).
Results
Relatlimab antibody generation
A panel of human anti–hLAG-3 mAbs, including clones 25F7, 10G7, 23C9, and 1C2, was generated by immunization of transgenic mice bearing human Ig miniloci with recombinant LAG-3 protein. Hybridoma clone 25F7 was selected for expansion and further characterization based on its activity profile in biochemical and cell-based binding and blocking assays, as well as in functional T-cell activation assays. The variable region sequences of this antibody were cloned and subsequently grafted onto human κ and IgG4 constant region sequences for reduced Fc receptor engagement and a reduced potential for effector T cell–mediated cytotoxicity. The resulting antibody LAG3.1-G4P also incorporated an S228P stabilizing hinge mutation to prevent in vivo and in vitro IgG4 Fab-arm exchange (26).
The sequence of antibody LAG-3.1-G4P contained two potential deamidation sites in the CDR2 region of the heavy chain at residues N54 and N56, which were confirmed by biophysical analysis under forced deamidation conditions. These potential sequence liabilities were addressed by mutating these sites (N54R and N56S), with no deleterious effect observed on the functional activity of the resulting antibody designated LAG-3.5, later renamed BMS-986016 (relatlimab).
Relatlimab-binding specificity
Relatlimab displayed saturable and selective binding to immobilized hLAG-3–hFc by ELISA, with a half-maximal effective concentration (EC50) of 0.49 nmol/L compared with an EC50 of 1.46 nmol/L for binding to CHO cells expressing hLAG-3 (Supplementary Fig. S1A). To confirm that relatlimab recognized native LAG-3, the binding of relatlimab to primary activated human and cynomolgus CD4+ T cells was measured and found to be substantially higher for binding to human (mean EC50, 0.11 nmol/L) than monkey LAG-3 (mean EC50, 29.11 nmol/L; Fig. 1A and B). The parent hybridoma of relatlimab (clone 25F7) did not bind to cells expressing full-length murine LAG-3 (Supplementary Fig. S1B); by extension, relatlimab is also expected to not bind murine LAG-3.
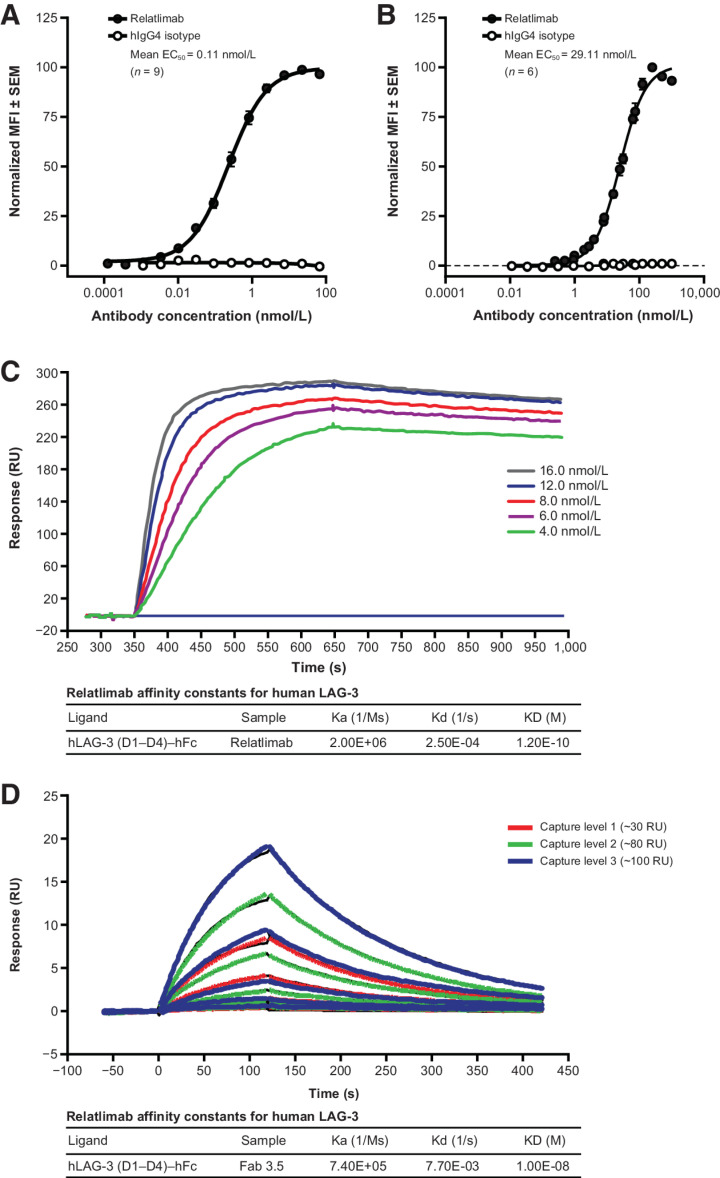
Figure 1.
Characterization of LAG-3 binding by relatlimab. A, Normalized flow cytometric binding of relatlimab to activated human CD4+ T cells from a total of nine donor samples over three experiments. B, Normalized flow cytometric binding of relatlimab to activated cynomolgus CD4+ T cells from a total of six donor samples over three experiments. Nonlinear regression analysis is shown with sample MFI values normalized to the highest (Y-Max) sample in each sample cohort as 100%. Error bars indicate SEM of normalized data across the combined cohort of donors in each panel (A, B). C, SPR sensogram depicting binding of bivalent relatlimab to LAG-3 under neutral pH conditions. Concentrations of flowed relatlimab are indicated. D, SPR sensogram depicting binding of monomeric relatlimab Fab under neutral pH conditions. Capture levels of hLAG-3–hFc ligand are indicated. Representative examples of kinetic analysis from at least three independent tests are shown (C, D). hIgG4, human IgG4; KD, dissociation constant; MFI, mean fluorescence intensity; RU, resonance units; SEM, standard error of the mean.
The kinetics of relatlimab binding to LAG-3 were determined by SPR for both intact bivalent relatlimab and its Fab fragment. The apparent affinity of the bivalent antibody was found to be 0.12 nmol/L at pH 7.4 under the experimental conditions detailed in the Supplementary Methods. The monovalent affinity of the Fab fragment measured with SPR was 10 nmol/L (Fig. 1C and D). Binding at pH 6, acidic conditions that likely exist in the TME (27), showed faster dissociation kinetics with a modest effect on affinity, indicating that relatlimab would still bind under these conditions (Supplementary Fig. S1C).
Disruption of LAG-3 receptor/ligand interactions by relatlimab
The functional potency of relatlimab in blocking the interaction of LAG-3 with MHC II and FGL1 was examined in a series of biochemical and cell-based assays.
MHC II binding
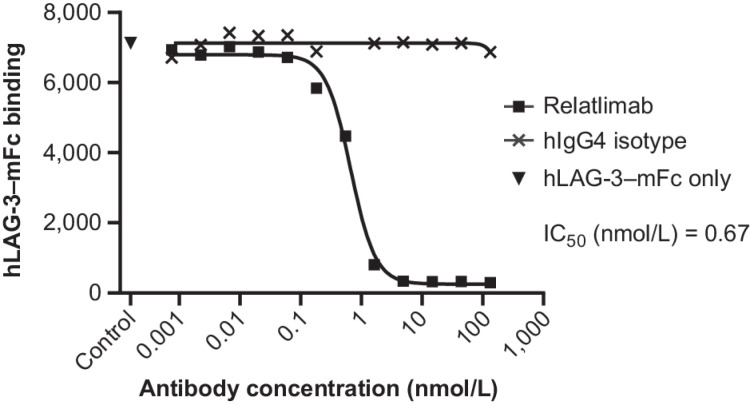
The interaction of LAG-3 with MHC II was confirmed by comparing the binding of mouse and hLAG-3–hFc to the wild-type Raji B lymphoid cell line and to an MHC II-low variant, RJ225 (24), with substantially less binding observed to the mutant cell line compared with the wild-type line (Supplementary Fig. S2). Relatlimab completely blocked detectable binding of hLAG-3–mFc to MHC II+ Daudi B lymphoid cells, exhibiting a half-maximal blockade (IC50) of 0.67 nmol/L compared with isotype control antibody (Fig. 2). Blockade was confirmed by BLI measurement of the binding of LAG-3–Fc to HLA-DR1 in the presence of either bivalent relatlimab or a Fab fragment of relatlimab (Fab 3.5). The interaction was equivalently blocked by both relatlimab and its Fab, but not by other antibody clones that bind within the D3–D4 extracellular domains (Supplementary Fig. S3).
Figure 2.
Flow-cytometric characterization of the blockade by relatlimab, compared with hIgG4 isotype control mAb, of hLAG-3–mFc binding to Daudi B lymphoid cells expressing endogenous MHC II. Representative data from three independent tests with nonlinear regression analysis is shown based sample MFI values. MFI, mean fluorescence intensity; RU, resonance units.
FGL1 binding
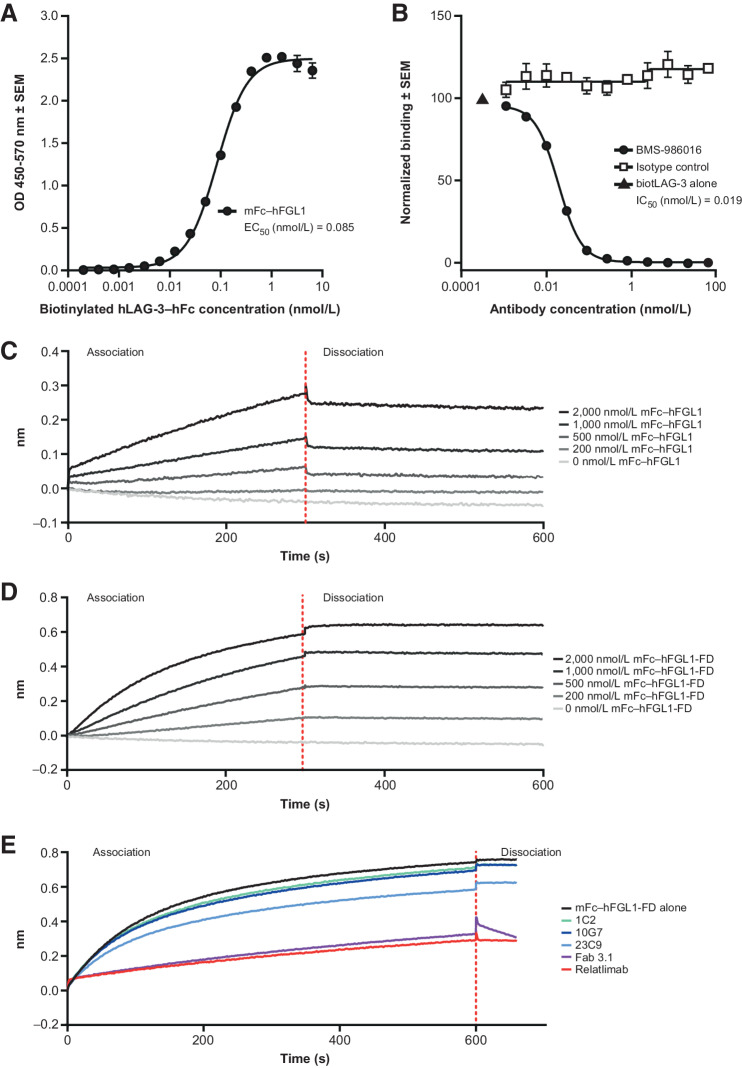
We next evaluated whether relatlimab can block the interaction between LAG-3 and the recently identified ligand, FGL1, using ELISA. Recombinant hLAG-3–hFc fusion protein bound immobilized (mFc–hFGL1) protein (3 μg/mL), with an EC50 of 0.085 nmol/L (Fig. 3A) and was blocked by relatlimab, with an IC50 of 0.019 nmol/L, but not by the isotype control antibody (Fig. 3A and B).
Figure 3.
Measurement of LAG-3/FGL1 interaction and blockade. A, Binding of hLAG-3–hFc to immobilized mFc–hFGL1 in ELISA. B, Blockade by relatlimab of hLAG-3–hFc binding to immobilized mFc–hFGL1 in ELISA. Representative examples of two independent tests with nonlinear regression analysis are shown; error bars indicate SEM of intra-assay technical replicates (N = 3; A, B). C–E, Characterization of LAG-3/FGL1 interactions by biolayer interferometry. Representative examples of at least three independent tests. C, Binding of serially diluted mFc–hFGL1 (full length) to captured hLAG-3–hFc. D, Binding of serially diluted mFc–hFGL1-FD (fibrinogen domain alone) to captured hLAG-3–hFc. E, Blockade of mFc–hFGL1-FD (fibrinogen domain alone) engagement with LAG-3 by relatlimab or by its Fab but not by antibodies specific for LAG-3 domains D3 or D4 (1C2, 10G7, and 23C9). biotLAG-3, biotinylated LAG-3. SEM, standard error of the mean.
Similar studies were conducted using BLI, which confirmed that both full-length FGL1 (mFc–hFGL1), as well as the fibrinogen domain alone (mFc–hFGL1-FD), bound hLAG-3–hFc in a dose-dependent, albeit nonsaturable, manner up to 2,000 nmol/L FGL1. Notably, the binding measurement of dimerized FGL1 to captured LAG-3-hFc is reflective of an avidity-driven interaction. Our data, as well as that from Wang and colleagues in supplement (12), indicated that the interaction between FGL1 and LAG-3 was relatively weak (>1 μmol/L; Fig. 3C and D). This interaction was inhibited by relatlimab and its Fab (Fab 3.5), but not by hLAG-3 antibodies specific to epitopes within the D3 or D4 domains of LAG-3 (clones 23C9, 1C2, and 10G7; Fig. 3E; Supplementary Fig. S3B). Although the mechanism by which soluble FGL1 may productively engage LAG-3 to drive T-cell inhibition is unknown, these results corroborate those of Wang and colleagues, suggesting that the FGL1 fibrinogen domain alone is sufficient to bind LAG-3 (12).
Functional blockade of ligand interaction with LAG-3
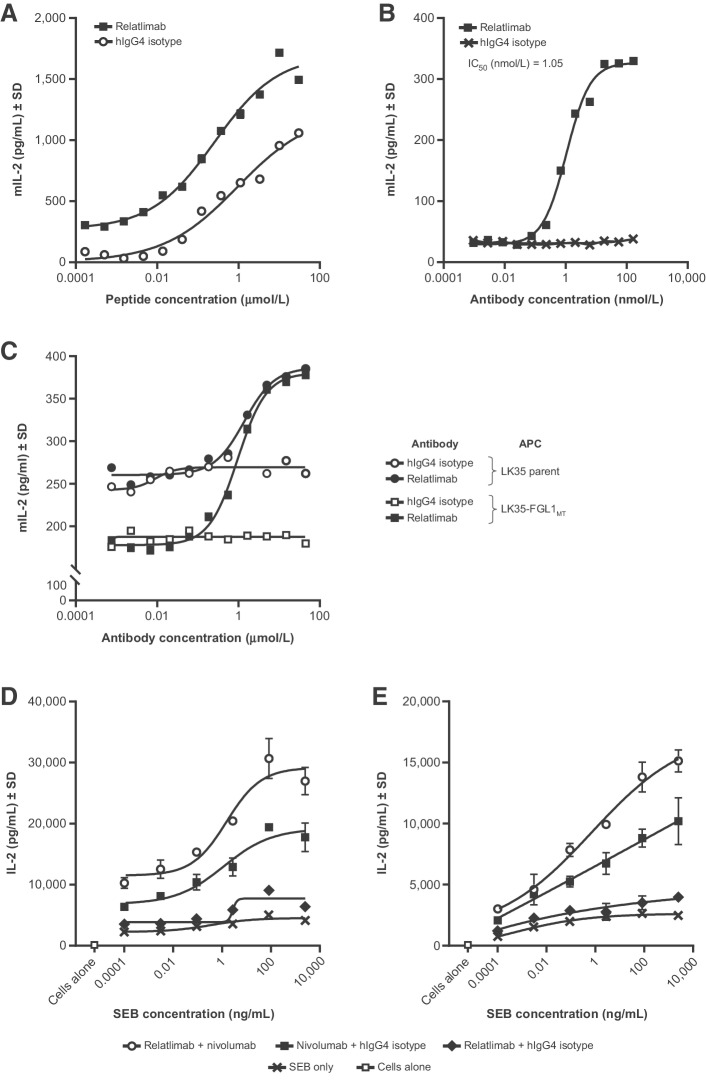
The ability of hLAG-3 to inhibit T-cell responses was studied using an antigen-specific T-cell hybridoma, 3A9, specific for hen egg lysozyme peptide (HEL48-62), presented by the MHC II-matched antigen-presenting mouse cell line LK35.2 (see Supplementary Methods: Functional cell-based bioassay of LAG-3/MHC II interaction and blockade by relatlimab; ref. 28). The expression of full-length hLAG-3 on the T cells (3A9-hLAG-3; Supplementary Fig. S4) resulted in attenuated T-cell peptide responsiveness, as demonstrated by lower murine IL2 secretion when cocultured with LK35.2 cells that could be enhanced by relatlimab in a dose-dependent manner (Fig. 4A).
Figure 4.
Characterization of relatlimab in vitro functional activity. A–E, Evaluation of the indicated cytokine responses by ELISA analysis of assay culture supernatants. A, Enhancement of the peptide responsiveness of T-cell hybridoma 3A9-hLAG-3 by relatlimab compared with control across a range of peptide concentrations. B, Dose-dependent reversal of LAG-3–mediated inhibition of T-cell hybridoma 3A9-hLAG-3 by relatlimab under conditions of suboptimal peptide stimulation. C, Measurement of LAG-3/FGL1 interaction and the blockade by relatlimab of T-cell inhibition mediated by APCs expressing MHC II alone (parent) or both FGL1 and MHC II together (FGL1MT) in the 3A9-hLAG-3 T-cell hybridoma assay. D and E, Enhanced activation of superantigen-stimulated human PBMC cultures from donor 1 (D) and donor 2 (E) by relatlimab alone or in combination with nivolumab. Nonlinear regression analysis is shown; error bars indicate SD of intra-assay technical replicates (N = 3; A–E). APC, antigen-presenting cell. mIL-2, mouse interleukin 2; MT, membrane-tethered; SEB, Staphylococcal enterotoxin B. SD, standard deviation.
Using an alternate format of the assay where cells were stimulated with a suboptimal concentration of HEL48-62 peptide in the presence of titrated mAb, relatlimab displayed potent blockade of LAG-3–mediated inhibition (IC50, 1.05 nmol/L) compared with the isotype control antibody (Fig. 4B). Collectively, these results suggest that the observed functional T-cell inhibition seen in coculture with LK35.2 cells and antigen, which is likely mediated by murine MHC II present on the APCs interacting with hLAG-3 on the T cells, can be reversed by relatlimab in a dose-dependent manner.
In contrast to published results (12), we observed only modest T-cell inhibitory activity by soluble mFc–hFGL1 that had been purified to ensure a homogeneous dimeric preparation (Supplementary Fig. S5) and, consequently, we next evaluated whether higher order oligomers of FGL1 could elicit enhanced inhibition. We generated an ordered oligomer of FGL1 by fusion to hexamer-forming hFc (E345R/E430G/S440Y; FGL1-RGY; refs. 29, 30). Although increased T-cell inhibition was observed with this variant relative to the wild-type Fc fusion protein alone, the impact on activity was still modest (Supplementary Fig. S5). For this reason, we sought to determine whether the expression of a membrane-tethered version of FGL1 on the cell surface could allow for a higher avidity interaction of FGL1 with LAG-3, thereby revealing functional inhibitory engagement. A membrane-tethered version of FGL1 (FGL1MT) was generated by fusing the fibrinogen domain of FGL1 with the transmembrane and intracellular domain of type II membrane fibrinogen C domain-containing protein 1 (12), and was expressed on LK35.2 cells (LK35-FGL1MT) (Supplementary Fig. S4). 3A9-hLAG-3 cells were more attenuated in their peptide responsiveness in coculture with LK35-FGL1MT cells compared with parental LK35.2 cells expressing MHC II alone (Fig. 4C). This suggests that FGL1-mediated inhibition in this setting is additive to the inhibition resulting from MHC II/LAG-3 engagement. Relatlimab reversed the T-cell inhibition resulting from coculture with either parental LK35.2 or LK35-FGL1MT cells with similar potency (IC50 of 1.39 nmol/L and 0.95 nmol/L, respectively) and to equivalent maximal levels of cytokine production, suggesting that relatlimab can effectively block the simultaneous engagement of LAG-3 by both MHC II and FGL1 (Fig. 4C; Supplementary Fig. S4).
The functional activity of relatlimab in the context of primary T cells was evaluated in human healthy donor peripheral blood mononuclear cell (PBMC) cultures stimulated with superantigen Staphylococcal enterotoxin B. Cultures from 15 of 18 donors showed enhanced IL2 secretion in the presence of relatlimab alone compared with the isotype control and, in most instances, the stimulation was less than that observed for treatment with nivolumab. The combination of relatlimab and nivolumab resulted in higher levels of stimulation compared with a combination of nivolumab and isotype control (Fig. 4D and E). Finally, relatlimab binding to activated T cells did not mediate significant levels of ADCC compared with the isotype control, whereas binding with positive control nonfucosylated human IgG1 anti-CD30 resulted in robust cell lysis (Supplementary Fig. S6).
These results are consistent with the synergistic in vivo antitumor activity observed from the combination antibody blockade of mouse LAG-3 and PD-1 in syngeneic tumor models. Similar to previously published reports (9), the antitumor activity of anti-mouse LAG-3 mAbs in combination with anti-mouse PD-1 monoclonal antibody 4H2 showed enhanced efficacy compared with LAG-3 or PD-1 single-agent blockade in both MC38 colon carcinoma tumors (Supplementary Fig. S7) and in Sa1N fibrosarcoma tumors (Supplementary Fig. S8). Substantial antitumor activity from the blockade of LAG-3 alone was observed only in the immunogenic fibrosarcoma Sa1N model using the C9B7W mAb. In the MC38 colon carcinoma model, enhanced efficacy was observed for all doses of anti-mouse LAG-3 mAb 19C7 > 1 mg/kg when combined with 4H2 compared with 4H2 alone.
Characterization of the relatlimab-binding site
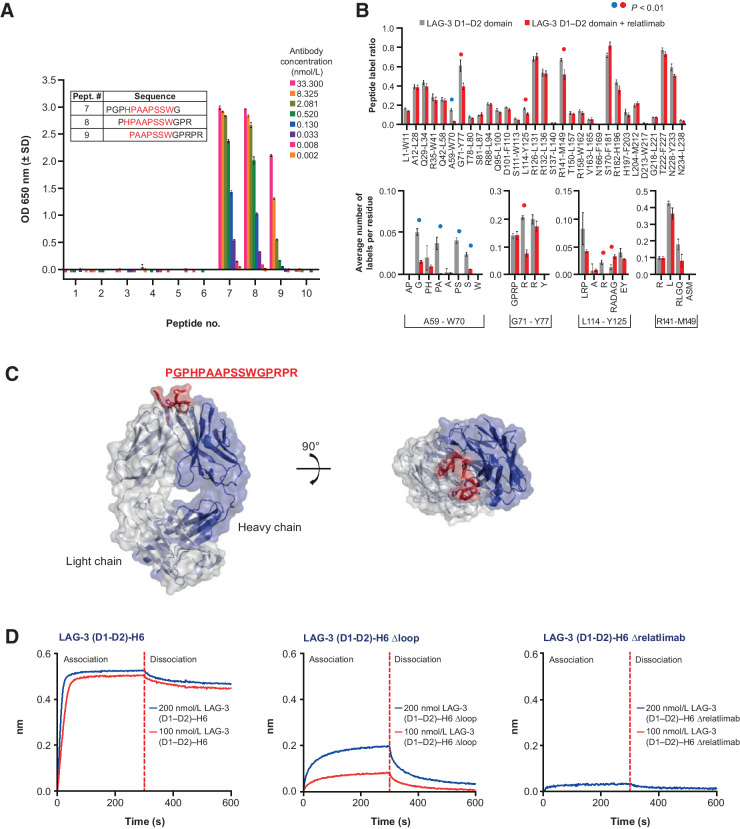
Previous studies by Triebel and colleagues (31) demonstrated that an extra insertion-loop sequence in the N-terminal extracellular D1 of LAG-3 potentially mediates the interaction with MHC II (32). Our initial analysis of hybridoma clones revealed that the most potent antibodies, including the parent clone for relatlimab (LAG3.1-G4P), were bound within the N-terminal D1–D2 domain region of LAG-3 (Supplementary Fig. S3C). A peptide consisting of the full-length D1 insertion-loop sequence (P60GPHPAAPSSWGPRPR75) was synthesized to test for antibody reactivity and LAG3.1-G4P was observed to bind strongly to this peptide by ELISA, with an EC50 of 0.44 nmol/L (Supplementary Fig. S9A and S9B). Next, LAG3.1-G4P was assessed by ELISA for binding a set of overlapping peptides spanning the 30 residues of the LAG-3 insertion loop (Supplementary Fig. S9A) and indicated that the antibody binds residues within the peptide H63PAAPSSW70 (Fig. 5A). Carbene chemical footprinting of a complex consisting of the LAG-3 D1–D2 domains and relatlimab Fab (Fab 3.5) confirmed that the epitope was contained within the peptide A59-W70 because this peptide showed the largest decrease in chemical labeling following complex formation compared with LAG-3 D1–D2 alone (Fig. 5B, top). Residue-level labeling of peptide A59-W70 identified multiple amino acids undergoing protection following complex formation contained in the region H63–W70 (Fig. 5B, bottom). Additional peptides were observed with labeling protection that was localized to individual amino acids in the D1 domain. Because the protein structure of LAG-3 with the insertion loop has not been determined, we speculate that these residues are likely to be structurally located near the A59-W70 peptide. Comparison of the insertion-loop sequences of human and cynomolgus monkey LAG-3 proteins showed 75% sequence identity for the epitope (H63–W70), likely accounting for the lower affinity of relatlimab binding to nonhuman primate LAG-3 (Supplementary Fig. S9C).
Figure 5.
Relatlimab epitope characterization. A, ELISA evaluation of relatlimab binding to immobilized 12-mer peptides with 10 residue overlaps spanning the LAG-3 insertion-loop sequence. Representative example of at least three independent tests with error bars indicating SD of intra-assay technical replicates (N = 3). B, Mapping of relatlimab epitope by carbene chemical footprinting. Blue circles denote the binding site peptide A59-W70. Red circles denote nearby residues in the D1 domain with some protection but outside the physical epitope. C, X-ray structure of the LAG3.1-G4P Fab (Fab 3.1) in complex with the binding epitope peptide. Density is observed for the underlined sequence. D, Binding of hLAG-3 loop deletion variants to captured relatlimab by BLI.
Structural characterization of relatlimab in complex with the LAG-3 insertion loop
To further characterize relatlimab interactions with LAG-3, the structure of the Fab of LAG3.1-G4P (Fab 3.1) in complex with a 16-residue peptide (Ac-PGPHPAAPSSWGPRPR-amide) derived from the insertion-loop sequence of the LAG-3 D1 domain was determined using X-ray crystallography (Supplementary Table S3). The three-dimensional structure, captured at 2.4 Å resolution, revealed distinct density for 12 of the 16 peptide residues (called out sequence in Fig. 5C) in the Fab 3.1 binding groove, with CDR-H3 and CDR-L3 pushed apart (Fig. 5C). Half of the accessible surface area of the peptide was buried by Fab 3.1, with light chain interactions more prevalent over heavy chain interactions (380 Å2 vs. 160 Å2, respectively). Roughly half (680 Å2) of the total accessible surface area of the peptide (1,350 Å2) was buried by Fab 3.1. Protein-binding assays using LAG-3 domain variants further confirmed this core peptide sequence as essential for the interaction between relatlimab and LAG-3, with deletion of the H63–W70 sequence resulting in the abolishment of relatlimab binding (Fig. 5D; Supplementary Fig. S9D). Overall, the peptide binding, BLI, and carbene chemical footprinting data are in good agreement and were corroborated by the X-ray crystal structure data and LAG-3 domain variant-binding data to collectively identify a linear epitope of relatlimab centered on residues H63–W70 of the insertion loop.
Relatlimab tissue-binding properties in normal human tissues
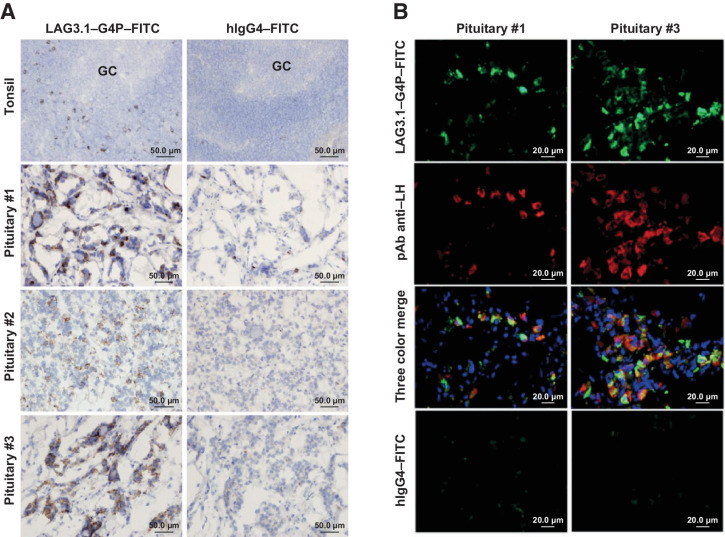
To confirm LAG-3 expression in immune cells, and to assess any unexpected tissue binding, tissue cross-reactivity of relatlimab and the parent clone of relatlimab, LAG3.1-G4P, was determined in a selected panel of normal human tissues by IHC (Supplementary Table S4). The tissue-binding patterns by relatlimab and LAG3.1-G4P were very similar. In hyperplastic tonsil tissue, strong positive staining was revealed in a small subset of lymphocytes primarily distributed in the interfollicular area (T-cell region), with few in the mantle zone, and only extremely rarely in the germinal center (Fig. 6A). This lymphocyte staining was expected and consistent with published observations (33). In pituitary tissue, rare to occasional moderate/strong immunoreactivity was displayed in the adenohypophysis. No specific staining was observed in the other tissues examined (Supplementary Table S4). Dual-color immunofluorescence analysis with LAG3.1-G4P confirmed the specificity of this staining localization of LAG-3 in pituitary gonadotroph cells, which included follicular-stimulating hormone- and luteinizing hormone (LH)-producing cells. Pituitary expression of LAG-3 was also confirmed by PCR (Fig. 6A and B; Supplementary Fig. S10 and S11; Supplementary Tables S5A and S5B).
Figure 6.
Binding of LAG3.1-G4P in the human pituitary gland. A, IHC of FITC-conjugated LAG-3.1-G4P (left) in positive control tissue (hyperplasia tonsil, n = 2) and normal human pituitary samples (n = 3). FITC-conjugated human IgG4 was used as isotype control (right). Counterstained with Mayer's hematoxylin. B, Colocalized binding of anti–LAG-3 antibody in LH-producing gonadotroph cells. Double immunofluorescent staining of FITC-conjugated LAG-3.1-G4P (green) and mouse mAb anti-LH (red) on cryostat sections from two normal human pituitary samples. FITC-conjugated human IgG4 and mouse IgG1 (not shown) were used as isotype controls (bottom) while nuclei were counterstained using Hoechst 33342. FITC, fluorescein isothiocyanate; GC, germinal center of the tonsil; LH, luteinizing hormone; mAb, monoclonal antibody.
Preclinical toxicity assessment in cynomolgus monkeys
Because relatlimab was shown to bind to cynomolgus LAG-3, we evaluated the potential toxicity of relatlimab ± nivolumab in cynomolgus monkeys. Additionally, splenic T-cell phenotyping, T-cell phenotyping, and TDARs were analyzed (Supplementary Methods).
Repeat-dose toxicity
In a 4-week repeat-dose toxicity study (Supplementary Table S1), relatlimab was clinically well tolerated by cynomolgus monkeys when administered intravenously q.w. at 30 or 100 mg/kg, with no adverse findings. Relatlimab, when administered at 100 mg/kg in combination with nivolumab at 50 mg/kg, was generally well tolerated in eight out of nine monkeys, with no adverse clinical signs; the exception being moribundity in one male monkey attributed to central nervous system (CNS) vasculitis (Supplementary Fig. S12A). Additional minimal histopathologic findings in the combination group were likely the result of enhanced immunostimulatory effects of nivolumab in combination with relatlimab because no treatment-related histopathologic changes were noted with relatlimab alone (Supplementary Table S6). Systemic exposures of relatlimab were evaluated and showed circulating half-lives of 490, 460, and 740 hours for the (relatlimab/nivolumab) 30/0, 100/0, and 100/50 mg/kg dose groups, respectively (Supplementary Table S7; Supplementary Fig. S12B). The CNS vascular findings may have been a result of a loss of tolerance to self-antigens based on the synergistic role of PD-1 and LAG-3 in maintaining self-tolerance. Given the long half-lives of relatlimab and nivolumab, the irreversibility of relatlimab plus nivolumab-related findings is likely a result of continued exposure to the test articles throughout the duration of the recovery period. In a 3-month toxicity study (Supplementary Table S2), relatlimab was generally well-tolerated by mature cynomolgus monkeys (4–7 years old) when administered intravenously q.w. up to 100 mg/kg.
In vivo pharmacodynamic effects of relatlimab administration
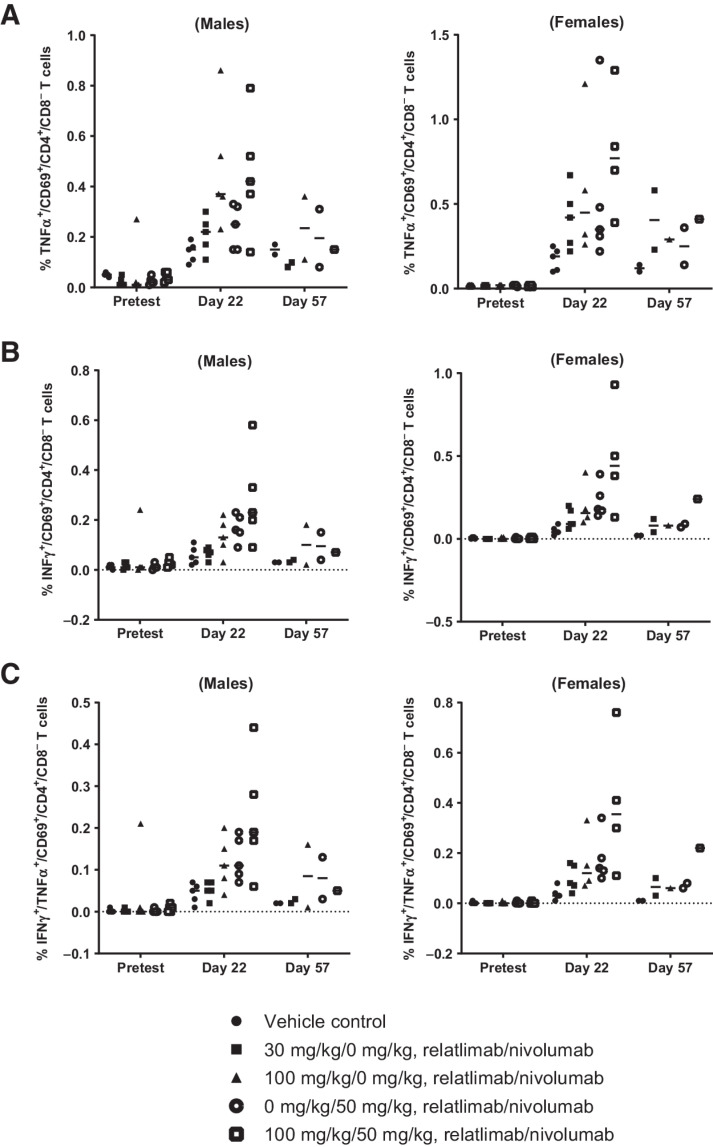
As part of the 4-week repeat-dose toxicity study of relatlimab and nivolumab in monkeys, all animals were immunized intramuscularly on day 1 of the study with 10 mg of KLH to permit assessment of in vivo antibody responses, ex vivo recall responses to KLH, and immunophenotypic analyses of peripheral blood and splenic T-lymphocyte subsets. There were no observed relatlimab- or nivolumab-related changes in TDARs to KLH among any of the study groups, and no measurable ex vivo T-cell recall responses to KLH. The comparatively low affinity of relatlimab for cynomolgus LAG-3 may have resulted in incomplete receptor blockade, leading to attenuated lymphocyte responses, including TDARs. Nevertheless, drug-related changes in ex vivo T-cell recall responses to KLH were indicative of an enhanced antigen-specific response. Increases in cytokine responses, especially IFNγ, were measured at day 22 in the mean percentage of both double-positive (CD69+TNFα+; CD69+IFNγ+) and triple-positive (CD69+TNFα+IFNγ+) CD4+CD8– splenic T cells among both male and female study groups, which received nivolumab alone, high-dose relatlimab (100 mg/kg) alone or in combination with nivolumab. Although the differences among these groups did not reach statistical significance, it was nevertheless interesting that the highest increases in both double-positive (Fig. 7A and B) and triple-positive (Fig. 7C) responding CD4+ T cells were observed in the relatlimab plus nivolumab combination group compared with the groups receiving either drug alone. These increases, which waned by day 57 across all groups, are consistent with the pharmacologic mechanisms of action of relatlimab and nivolumab.
Figure 7.
Flow-cytometric intracellular cytokine staining analysis of splenic CD4+ T-cell populations in male and female monkeys (n = 5/group, gender) from a 1-month toxicity study treated with a single dose of KLH at study initiation and 4 weekly doses of relatlimab ± nivolumab as indicated. A, TNFα+CD69+CD4+CD8–; B, IFNγ+CD69+CD4+CD8–; and C, triple-positive TNFα+IFNγ+CD69+CD4+CD8– T cells. Data points represent individual animal results as a percentage of CD4+CD8– cells (gated on live CD45+CD16–CD3+ CD69+CD4+ CD8–). Horizontal bars represent group means.
Phenotypic analysis was performed for splenic and peripheral blood memory T-cell subsets and for peripheral blood total lymphocytes (T, B, and NK cells). Overall, comparing pretest, day 15, and day 30 phenotypic profiles did not show identifiable trends in drug-related lymphocyte population changes across time points and female and male groups. Statistically significant differences among splenic memory T-cell subsets relative to the vehicle-control group were observed in CD4+ T regulatory cells in the nivolumab and combination groups (Supplementary Fig. S13; Supplementary Table S9). Among peripheral memory T-cell subsets, notable differences were observed mainly in naïve and central memory CD4+ and CD8+ T cells in the combination study group animals, as well as in activated CD25+CD4+ T cells among males, and in naïve and memory CD8+ T cells among females (Supplementary Table S10). No substantial changes were observed in total T, B, and NK cell numbers (Supplementary Table S11).
Discussion
Continuous exposure of T cells to cognate antigen leads to an exhausted, hyporesponsive phenotype characterized, in part, by the expression of multiple inhibitory receptors including PD-1, CTLA-4, LAG-3, and T-cell immunoglobulin and mucin domain-3 (34). LAG-3 has been shown to be expressed in TILs of several tumor types, including melanoma, hepatocellular carcinoma (HCC), and non–small cell lung cancer (NSCLC), often in parallel with increased PD-1 (35–37). Combination immunotherapies can result in improved clinical benefit compared with single checkpoint blockade, as exemplified by the combination of anti–PD-1, nivolumab, and anti–CTLA-4 (ipilimumab) approved in the United States and other countries for multiple indications, including melanoma, NSCLC, renal cell carcinoma, HCC, colorectal cancer, and pleural mesothelioma (38, 39). Targeting additional checkpoints, such as LAG-3, is a promising approach for overcoming resistance to ICB by targeting multiple inhibitory targets. LAG-3 acts in a nonredundant manner from that of PD-1 to suppress T-cell stimulation and presents an exciting new opportunity for combination ICB that may improve clinical responses.
Preclinical data presented in recent years illustrate a clear synergy between the inhibitory receptors LAG-3 and PD-1 in controlling immune homeostasis, preventing autoimmunity, and enforcing tumor-induced tolerance (9, 16). Although the focus of this report is the characterization of relatlimab itself, our in vivo results corroborate a large body of existing literature that demonstrates the combined treatment of mice with blocking antibodies against LAG-3 and PD-1 receptors results in more robust immune responses than either single-agent treatment (9, 40). There is a well-developed understanding that the coblockade of LAG-3 and PD-1 acts to reinvigorate exhausted T cells more robustly than single ICB and results in enhanced polyfunctionality (IFNγ and TNFα production and cytotoxic killing) to potentiate antitumor activity (9, 11, 17–19, 41–43). Similarly, in the in vitro primary T-cell superantigen stimulation assays reported here, only modest activity was observed from LAG-3 single-agent blockade with relatlimab compared with the substantially enhanced responsiveness in the context of coblockade of LAG-3 and PD-1 with relatlimab and nivolumab, respectively. Although our studies did not specifically interrogate the individual contributions of CD4+ and CD8+ T cells to the overall T-cell responses observed, superantigens have been demonstrated to stimulate both CD4+ and CD8+ T cells, and our data suggest that combined PD-1 and LAG-3 blockade likely potentiates both subsets (44). Moreover, it has previously been demonstrated that LAG-3 blockade, particularly in combination with PD-1 blockade, can enhance tumor killing by CD8+ T cells in vitro (17). As expected for an IgG4 isotype antibody null for Fc receptor engagement, there was no measurable ADCC mediated by relatlimab. Toxicologic assessment of relatlimab in cynomolgus monkeys showed it to be generally well tolerated, alone and in combination with nivolumab. Relatlimab displayed no unexpected normal tissue cross-reactivity by IHC, except for observed on-target binding in the pituitary. The role of LAG-3 in the pituitary has not been determined.
Herein, we have demonstrated in vitro functional binding and blocking activity of relatlimab. The antibody binds with a higher affinity to activated human T cells than to activated cynomolgus monkey T cells, most likely due to species differences in the sequence of the D1 domain that is targeted by the mAb. Relatlimab reversed the functional inhibition of T-cell activation by LAG-3 in an in vitro antigen-specific T-cell hybridoma assay, demonstrating that the antibody can functionally block the inhibition of T cells in the context of LAG-3 engagement by peptide-loaded MHC II.
Wang and colleagues recently reported the identification of FGL1 as a new putative ligand of LAG-3, presenting compelling evidence for its functional role in LAG-3 signaling and its potential clinical relevance in certain cancers (12). In our investigation of FGL1-mediated T-cell suppression, we showed that the engineered coexpression of a cell membrane-tethered FGL1 in the context of a mouse MHC II–positive APC resulted in more potent inhibition of T-cell responsiveness compared with the engagement of the receptor by MHC II alone. In biochemical assays, relatlimab blocked the interaction of LAG-3 with both MHC II and FGL1. Consistent with these observations, relatlimab also strongly blocked the enhanced T-cell suppression observed in cocultures with MHC II/FGL1 coexpressing APCs. In these assays, relatlimab restored T-cell cytokine production to a level equivalent to that observed for antibody treatment of T cells in coculture with APCs expressing MHC II alone. Additional work is needed to fully understand the nature of the interactions between these three molecules, but it is possible that a ternary complex of LAG-3, MHC II, and FGL1 could exist that may promote potent T-cell inhibition. Our data indicate that the FGL1/LAG-3 interaction is relatively weak (>1 μmol/L), but in the artificial context of APCs engineered for surface expression of FGL1, the increased valency likely fosters an avidity-driven enhancement of FGL1-mediated T-cell inhibition, as we observed. The mechanism by which soluble FGL1 can functionally engage LAG-3 in the periphery or intratumorally to inhibit T-cell responses remains unclear and we note a recent report that contradicts several findings from Wang and colleagues (45). FGL1 is produced by the liver and secreted into the bloodstream, where it plays a role in hepatocyte regeneration and metabolism to suppress environmentally induced inflammation (12, 46–49). FGL1 has been reported to promote invasion and metastasis in gastric cancer and to mediate drug resistance in lung and liver cancers (50–52). It has been suggested that FGL1 may associate with extracellular matrix components to facilitate LAG-3 interactions, similar perhaps to the interactions of the latent transforming growth factor (TGF)-β complex with αv integrins and the subsequent release of active TGFβ (53), but this has yet to be demonstrated. There is some evidence that FGL1 may form high-molecular-weight oligomers with FGL2 (54), but it is unclear whether FGL1 is able to assemble into homogeneous high-molecular-weight complexes alone. Dimeric Fc-tagged FGL1 protein, carefully processed to ensure a homogeneous dimer preparation, failed to inhibit in the 3A9 T-cell hybridoma assay as previously reported (12), but we observed some evidence of 3A9 cell inhibition when cells were treated with FGL1 that contains an Fc tag containing mutations to promote a hexamer. These differences in results may reflect differences in the biochemical properties of the purified proteins used in the different assays. It remains to be determined what the physical oligomerization state of FGL1 is in the periphery and the TME, and it remains unclear what effect soluble LAG-3 in either compartment may have on the ability of tumor-expressed FGL1 to functionally engage LAG-3 on T cells.
Biochemical and biophysical analyses demonstrated that the epitope of relatlimab resides in the insertion loop of the D1 domain of LAG-3 and is centered on the peptide H63–W70. Wang and colleagues showed evidence that the Y77F mutation in the C’ strand of D1, which has been shown to abrogate MHC II interaction with LAG-3, does not perturb the interaction with FGL1 (12). These results support the hypothesis that FGL1 and MHC II interaction sites on LAG-3 are independent of one another. Although the mechanism of soluble FGL1-mediated inhibition remains to be determined, our data demonstrate that relatlimab can block its interaction with LAG-3, supporting the potential utility of relatlimab in cancer indications where FGL1 expression is high (e.g., HCC), or where its expression correlates with poor prognosis (e.g., lung adenocarcinoma; refs. 12, 52).
The efficacy and manageable safety profile of relatlimab combined with nivolumab has been demonstrated in the phase II/III clinical trial RELATIVITY-047, where prolonged PFS benefit was observed with relatlimab combined with nivolumab compared with nivolumab monotherapy in patients with previously untreated metastatic or unresectable melanoma (20, 21). Collectively, these data support the development of a combination of relatlimab plus nivolumab as a promising therapeutic strategy in clinical oncology that has the potential to enhance antitumor responses and broaden the range of responding cancer types compared with nivolumab monotherapy.
Supplementary Material
Acknowledgments
This study was funded by Bristol-Myers Squibb. Medical writing support and editorial assistance were provided by Katerina Pipili, PhD, and Matthew Weddig of Spark Medica Inc., funded by Bristol Myers Squibb, according to Good Publication Practice guidelines. Jun Zhang contributed to antibody generation efforts. Christine Bee and Mohan Srinivasan contributed to SPR studies with additional support and review from Gavin Dollinger and Andrew Drake. Winse Morishige assisted with SPR data visualization. Jennifer Price provided PCR study support and Olufemi Adelakun contributed to IHC studies. Rangan Vangipuram managed antibody purification work, and Sharon Viajar aided in protein purification. Peter S. Lee provided guidance on structural characterization, and Arvind Rajpal provided input on biophysical studies. In support of the 1-month monkey toxicity study, Jagannatha Mysore provided pathology analysis, Carol Gleason provided biostatistical analysis, and Gautham Rao provided input on study design and analyses of pharmacodynamic endpoints. Bonnie Wang generated pharmacokinetic data as part of the repeat dose toxicity study in cynomolgus monkeys. The authors particularly thank Drew Pardoll and Dario Vignali for discussions, feedback, and support as subject-matter experts of LAG-3 biology, as well as Dario Vignali for providing the 3A9 hybridoma cell lines and C9B7W antibody used in these studies.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
K. Thudium reports personal fees from BMS outside the submitted work; in addition, K. Thudium has a patent for US 11,001,630 issued to BMS. M. Selby reports a patent for US11236164B2 issued. J.A. Zorn reports personal fees from BMS outside the submitted work; and Dr Zorn is an employee and shareholder of BMS. X. Wang reports personal fees from BMS outside the submitted work. R.T. Bunch reports being an employee of BMS and owns stock in the company. J.M. Hogan reports personal fees from BMS outside the submitted work. P. Strop reports personal fees from BMS during the conduct of the study; personal fees from BMS outside the submitted work. A.J. Korman reports other support from BMS outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
K. Thudium: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. M. Selby: Conceptualization, data curation, formal analysis, supervision, investigation, methodology, project administration, writing–review and editing. J.A. Zorn: Formal analysis, investigation, visualization, methodology, writing–review and editing. G. Rak: Investigation, writing–original draft, writing–review and editing. X.-T. Wang: Formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. R.T. Bunch: Conceptualization, resources, supervision, writing–original draft, writing–review and editing. J.M. Hogan: Investigation, visualization, writing–original draft, writing–review and editing. P. Strop: Conceptualization, supervision, project administration, writing–review and editing. A.J. Korman: Conceptualization, supervision, writing–review and editing.
References
- 1. Hargadon KM, Johnson CE, Williams CJ. Immune-checkpoint blockade therapy for cancer: an overview of FDA-approved immune-checkpoint inhibitors. Int Immunopharmacol 2018;62:29–39. [DOI] [PubMed] [Google Scholar]
- 2. Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol 2002;3:991–8. [DOI] [PubMed] [Google Scholar]
- 3. Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol 2006;6:715–27. [DOI] [PubMed] [Google Scholar]
- 4. Pardoll D. Does the immune system see tumors as foreign or self? Annu Rev Immunol 2003;21:807–39. [DOI] [PubMed] [Google Scholar]
- 5. Long L, Zhang X, Chen F, Pan Q, Phiphatwatchara P, Zeng Y, et al. The promising immune checkpoint LAG-3: from tumor microenvironment to cancer immunotherapy. Genes Cancer 2018;9:176–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao X, Subramanian S. Intrinsic resistance of solid tumors to immune checkpoint blockade therapy. Cancer Res 2017;77:817–22. [DOI] [PubMed] [Google Scholar]
- 7. Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined nivolumab and ipilimumab or monotherapy in previously untreated melanoma. N Engl J Med 2015;373:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol 2009;10:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Woo S-R, Turnis ME, Goldberg MV, Bankoti J, Selby M, Nirschl CJ, et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res 2012;72:917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang Z-Z, Kim HJ, Villasboas JC, Chen Y-P, Price-Troska T, Jalali S, et al. Expression of LAG-3 defines exhaustion of intratumoral PD-1+ T cells and correlates with poor outcome in follicular lymphoma. Oncotarget 2017;8:61425–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gandhi MK, Lambley E, Duraiswamy J, Dua U, Smith C, Elliott S, et al. Expression of LAG-3 by tumor-infiltrating lymphocytes is coincident with the suppression of latent membrane antigen–specific CD8+ T-cell function in Hodgkin lymphoma patients. Blood 2006;108:2280–9. [DOI] [PubMed] [Google Scholar]
- 12. Wang J, Sanmamed MF, Datar I, Su TT, Ji L, Sun J, et al. Fibrinogen-like protein 1 is a major immune inhibitory ligand of LAG-3. Cell 2019;176:334–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu F, Liu J, Liu D, Liu B, Wang M, Hu Z, et al. LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T-cell responses. Cancer Res 2014;74:3418–28. [DOI] [PubMed] [Google Scholar]
- 14. Mao X, Ou MT, Karuppagounder SS, Kam T-I, Yin X, Xiong Y, et al. Pathological α-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science 2016;353:aah3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kouo T, Huang L, Pucsek AB, Cao M, Solt S, Armstrong T, et al. Galectin-3 shapes antitumor immune responses by suppressing CD8+ T cells via LAG-3 and inhibiting expansion of plasmacytoid dendritic cells. Cancer Immunol Res 2015;3:412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okazaki T, Okazaki I-mWang J, Sugiura D, Nakaki F, Yoshida T, et al. PD-1 and LAG-3 inhibitory co-receptors act synergistically to prevent autoimmunity in mice. J Exp Med 2011;208:395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gestermann N, Saugy D, Martignier C, Tillé L, Fuertes Marraco SA, Zettl M, et al. LAG-3 and PD-1+LAG-3 inhibition promote anti-tumor immune responses in human autologous melanoma/T cell co-cultures. Oncoimmunology 2020;9:1736792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lichtenegger FS, Rothe M, Schnorfeil FM, Deiser K, Krupka C, Augsberger C, et al. Targeting LAG-3 and PD-1 to enhance T cell activation by antigen-presenting cells. Front Immunol 2018;9:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zelba H, Bedke J, Hennenlotter J, Mostböck S, Zettl M, Zichner T, et al. PD-1 and LAG-3 dominate checkpoint receptor–mediated T-cell Inhibition in renal cell carcinoma. Cancer Immunol Res 2019;7:1891–9. [DOI] [PubMed] [Google Scholar]
- 20. Lipson EJ, Tawbi HA-H, Schadendorf D, Ascierto PA, Matamala L, Gutiérrez EC, et al. (Relatlimab RELA) plus nivolumab (NIVO) versus NIVO in first-line advanced melanoma: Primary phase III results from RELATIVITY-047 (CA224-047). J Clin Oncol 2021;39:Abstract 9503. [Google Scholar]
- 21. Tawbi HA, Schadendorf D, Lipson EJ, Ascierto PA, Matamala L, Castillo Gutiérrez E, et al. Relatlimab and nivolumab versus nivolumab in untreated advanced melanoma. N Engl J Med 2022;386:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lonberg N, Taylor LD, Harding FA, Trounstine M, Higgins KM, Schramm SR, et al. Antigen-specific human antibodies from mice comprising four distinct genetic modifications. Nature 1994;368:856–9. [DOI] [PubMed] [Google Scholar]
- 23. Fishwild DM, O'Donnell SL, Bengoechea T, Hudson DV, Harding F, Bernhard SL, et al. High-avidity human IgG kappa monoclonal antibodies from a novel strain of minilocus transgenic mice. Nat Biotechnol 1996;14:845–51. [DOI] [PubMed] [Google Scholar]
- 24. Spilianakis C, Kretsovali A, Agalioti T, Makatounakis T, Thanos D, Papamatheakis J. CIITA regulates transcription onset via Ser5-phosphorylation of RNA Pol II. EMBO J 2003;22:5125–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yin Y, Wang XX, Mariuzza RA. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc Natl Acad Sci U S A 2012;109:5405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Silva J-P, Vetterlein O, Jose J, Peters S, Kirby H. The S228P mutation prevents in vivo and in vitro IgG4 Fab-arm exchange as demonstrated using a combination of novel quantitative immunoassays and physiological matrix preparation. J Biol Chem 2015;290:5462–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang X, Lin Y, Gillies RJ. Tumor pH and its measurement. J Nucl Med 2010;51:1167–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Workman CJ, Rice DS, Dugger KJ, Kurschner C, Vignali DAA. Phenotypic analysis of the murine CD4-related glycoprotein, CD223 (LAG-3). Eur J Immunol 2002;32:2255–63. [DOI] [PubMed] [Google Scholar]
- 29. de Jong RN, Beurskens FJ, Verploegen S, Strumane K, van Kampen MD, Voorhorst M, et al. A novel platform for the potentiation of therapeutic antibodies based on antigen-dependent formation of IgG hexamers at the cell surface. PLoS Biol 2016;14:e1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Diebolder CA, Beurskens FJ, de Jong RN, Koning RI, Strumane K, Lindorfer MA, et al. Complement is activated by IgG hexamers assembled at the cell surface. Science 2014;343:1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Triebel F, Jitsukawa S, Baixeras E, Roman-Roman S, Genevee C, Viegas-Pequignot E, et al. LAG-3, a novel lymphocyte activation gene closely related to CD4. J Exp Med 1990;171:1393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huard B, Mastrangeli R, Prigent P, Bruniquel D, Donini S, El-Tayar N, et al. Characterization of the major histocompatibility complex class II binding site on LAG-3 protein. Proc Natl Acad Sci U S A 1997;94:5744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huard B, Gaulard P, Faure F, Hercend T, Triebel F. Cellular expression and tissue distribution of the human LAG-3-encoded protein, an MHC class II ligand. Immunogenetics 1994;39:213–7. [DOI] [PubMed] [Google Scholar]
- 34. Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He Y, Yu H, Rozeboom L, Rivard CJ, Ellison K, Dziadziuszko R, et al. LAG-3 protein expression in non-small cell lung cancer and its relationship with PD-1/PD-L1 and tumor-infiltrating lymphocytes. J Thorac Oncol 2017;12:814–23. [DOI] [PubMed] [Google Scholar]
- 36. Maruhashi T, Sugiura D, Okazaki I-mOkazaki T. LAG-3: from molecular functions to clinical applications. J Immunother Cancer 2020;8:e001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guo M, Yuan F, Qi F, Sun J, Rao Q, Zhao Z, et al. Expression and clinical significance of LAG-3, FGL1, PD-L1 and CD8+T cells in hepatocellular carcinoma using multiplex quantitative analysis. J Transl Med 2020;18:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bristol Myers Squibb; Princeton NJ. 2022. Yervoy® (ipilimumab) [package insert]. Available from: https://packageinserts.bms.com/pi/pi_yervoy.pdf. Accessed 2022 March 17.
- 39. Bristol Myers Squibb; Princeton NJ. 2022. OPDIVO® (nivolumab) [package insert]. Available fromhttps://packageinserts.bms.com/pi/pi_opdivo.pdf. Accessed 2022 June 10.
- 40. Burova E, Hermann A, Dai J, Ullman E, Halasz G, Potocky T, et al. Preclinical development of the anti-LAG-3 antibody REGN3767: characterization and activity in combination with the anti–PD-1 antibody cemiplimab in human PD-1xLAG-3–knockin mice. Mol Cancer Ther 2019;18:2051–62. [DOI] [PubMed] [Google Scholar]
- 41. Mimura K, Kua L-F, Xiao J-F, Asuncion BR, Nakayama Y, Syn N, et al. Combined inhibition of PD-1/PD-L1, Lag-3, and Tim-3 axes augments antitumor immunity in gastric cancer-T cell coculture models. Gastric Cancer 2021;24:611–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harris-Bookman S, Mathios D, Martin AM, Xia Y, Kim E, Xu H, et al. Expression of LAG-3 and efficacy of combination treatment with anti-LAG-3 and anti-PD-1 monoclonal antibodies in glioblastoma. Int J Cancer 2018;143:3201–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schöffski P, Tan DSW, Martín M, Ochoa-de-Olza M, Sarantopoulos J, Carvajal RD, et al. Phase I/II study of the LAG-3 inhibitor ieramilimab (LAG525) ± anti-PD-1 spartalizumab (PDR001) in patients with advanced malignancies. J Immunother Cancer 2022;10:e003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Coppola MA, Blackman MA. Bacterial superantigens reactivate antigen-specific CD8+ memory T cells. Int Immunol 1997;9:1393–403. [DOI] [PubMed] [Google Scholar]
- 45. Maruhashi T, Sugiura D, Okazaki IM, Shimizu K, Maeda TK, Ikubo J, et al. Binding of LAG-3 to stable peptide-MHC class II limits T cell function and suppresses autoimmunity and anti-cancer immunity. Immunity 2022;55:912–24. [DOI] [PubMed] [Google Scholar]
- 46. Demchev V, Malana G, Vangala D, Stoll J, Desai A, Kang HW, et al. Targeted deletion of fibrinogen-like protein 1 reveals a novel role in energy substrate utilization. PLoS One 2013;8:e58084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hara H, Yoshimura H, Uchida S, Toyoda Y, Aoki M, Sakai Y, et al. Molecular cloning and functional expression analysis of a cDNA for human hepassocin, a liver-specific protein with hepatocyte mitogenic activity. Biochim Biophys Acta 2001;1520:45–53. [DOI] [PubMed] [Google Scholar]
- 48. Li C-Y, Cao C-Z, Xu W-X, Cao M-M, Yang F, Dong L, et al. Recombinant human hepassocin stimulates proliferation of hepatocytes in vivo and improves survival in rats with fulminant hepatic failure. Gut 2010;59:817–26. [DOI] [PubMed] [Google Scholar]
- 49. Liu Z, Ukomadu C. Fibrinogen-like protein 1, a hepatocyte-derived protein is an acute phase reactant. Biochem Biophys Res Commun 2008;365:729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Y, Qiao H-X, Zhou Y-T, Hong L, Chen J-H. Fibrinogen‑like‑protein 1 promotes the invasion and metastasis of gastric cancer and is associated with poor prognosis. Mol Med Rep 2018;18:1465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sun C, Gao W, Liu J, Cheng H, Hao J. FGL1 regulates acquired resistance to Gefitinib by inhibiting apoptosis in non-small cell lung cancer. Respir Res 2020;21:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Son Y, Shin N-R, Kim S-H, Park S-C, Lee H-J. Fibrinogen-like protein 1 modulates sorafenib resistance in human hepatocellular carcinoma cells. Int J Mol Sci 2021;22:5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shi M, Zhu J, Wang R, Chen X, Mi L, Walz T, et al. Latent TGF-β structure and activation. Nature 2011;474:343–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nagdas SK, Winfrey VP, Olson GE. Two fibrinogen-like proteins, FGL1 and FGL2 are disulfide-linked subunits of oligomers that specifically bind nonviable spermatozoa. Int J Biochem Cell Biol 2016;80:163–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in these studies are available within the article. The X-ray diffraction coordinates and structure factors are publicly available from the Research Collaboratory for Structural Bioinformatics Protein Data Bank under PDB ID 7UM3. The raw data underlying the included surface plasmon resonance (SPR) results were generated by a core facility and may not be available.
References are included with respect to animal models and reagents used in the study as appropriate. Relatlimab is commercially available as a therapeutic agent in the United States. Other anti–LAG-3 antibodies mentioned in the article can be made available upon reasonable request to the lead author and review by BMS, in compliance with BMS compound, technology, and data sharing policies (further details can be found at https://www.bms.com/researchers-and-partners/independent-research/compound-and-technology-requests.html and https://www.bms.com/researchers-and-partners/independent-research/data-sharing-request-process.html).