Abstract
Stony corals (order: Scleractinia) differ in growth form and structure. While stony corals have gained the ability to form their aragonite skeleton once in their evolution, the suite of proteins involved in skeletogenesis is different for different coral species. This led to the conclusion that the organic portion of their skeleton can undergo rapid evolutionary changes by independently evolving new biomineralization-related proteins. Here, we used liquid chromatography-tandem mass spectrometry to sequence skeletogenic proteins extracted from the encrusting temperate coral Oculina patagonica. We compare it to the previously published skeletal proteome of the branching subtropical corals Stylophora pistillata as both are regarded as highly resilient to environmental changes. We further characterized the skeletal organic matrix (OM) composition of both taxa and tested their effects on the mineral formation using a series of overgrowth experiments on calcite seeds. We found that each species utilizes a different set of proteins containing different amino acid compositions and achieve a different morphology modification capacity on calcite overgrowth. Our results further support the hypothesis that the different coral taxa utilize a species-specific protein set comprised of independent gene co-option to construct their own unique organic matrix framework. While the protein set differs between species, the specific predicted roles of the whole set appear to underline similar functional roles. They include assisting in forming the extracellular matrix, nucleation of the mineral and cell signaling. Nevertheless, the different composition might be the reason for the varying organization of the mineral growth in the presence of a particular skeletal OM, ultimately forming their distinct morphologies.
Subject terms: Molecular evolution, Computational biology and bioinformatics, Marine biology, Protein sequencing, Mass spectrometry
Introduction
Stony corals produce huge masses of calcium carbonate structures in the ocean in the form of aragonite and are among the oldest biomineralizing Metazoa1,2. In addition, they exhibit high variance in growth forms within and between taxa, ranging from simple encrusting to tree-like branching forms3 and mediating the formation of massive reefs in tropical, sub-tropical and cold-water seas. These structures then provide a complex three-dimensional ecosystem that offers shelter and sanctuary to a range of marine life4,5. These complex ecosystems are facilitated by depositing an aragonitic skeleton with species-specific macromorphology that can vary along environmental gradients and locations6.
At the macro scale, the skeletons of scleractinian corals are morphologically diverse3; for example, massive as in Favia, branching as in Stylophora, encrusting as in Oculina, or table-like similarly to Turbinaria. However, this diversity is not evident at the micro-scale, where stony coral skeletons reveal textural similarities, displaying needle-like single aragonite crystal fibers radiating from centers of calcification (CoCs)7–9 and forming superstructures that are reported to have a spherulitic organization10.
Besides the aragonitic mineral, coral skeletons are composed of up to 2.5 w% water associated with organics molecules10. Those biomolecules, dubbed the skeletal organic matrix (OM), are secreted through the calicoblastic cell layer (the cell layer that deposits the coral skeleton). The skeletal OM is composed of polysaccharides, lipids, and proteins, where proteins are the most studied fraction, having critical roles in skeleton formation2,11,12 It has been suggested that corals' ability to control skeleton formation is exercised through the skeletal OM, whose biosynthesis is orchestrated in space and time by the activity of the calicoblastic cells11.
However, the exact function of only a few skeletal OM proteins is known to date13. Efforts to sequence stony coral skeletal OM proteins have revealed many seemingly unique proteins at both the family and species levels14–19. While many of these biomolecules differ between taxa, explorations of the evolutionary history of coral skeletal OM proteins from divergent coral genera have found that a minor portion of proteins is conserved across species, referred to as the "core molecular biomineralization toolkit19". Nevertheless, approximately half of the skeletal OM proteins were independently co-opted from ancestors shared with other phyla, some containing no extant skeleton-forming taxa19. While it appears that the specific elements to form a skeleton are diverse between species, there might be a conserved genetic basis for some of the shared microstructural aspects of coral skeleton formation13, as it was found in vivo and in vitro that the skeletal OM will bias the mineral polymorph toward aragonite20,21.
These features may influence the ongoing viability of the different species to withstand the combined challenge of global warming and ocean acidification. Therefore, it is of primary importance to study the formation of the skeleton in the extent of coral species, which exhibit different population organization and resilience to environmental stresses, like ocean acidification and increased temperature.
In this context, we investigated the sequences of the skeletal OM of an encrusting temperate coral, Oculina patagonica, which is endemic to the Mediterranean Sea22. We first used liquid chromatography-tandem mass spectrometry to characterize the protein composition of skeletal proteins extracted from the temperate coral O. patagonica. We then compared our results with previously published skeletal OM data of the branching sub-tropic corals: Stylophora pistillata14,17, Acropora digitifera16 and A. millepora15. The corals O. patagonica and S. pistillata belong to the Robusta coral clade, while both A. digitifera and A. millepora belong to the Complexa clade23. In addition, we compared the sequence composition of the two Robusta coral representatives, O. patagonica and S. pistillata and tested the effect of their soluble skeletal OM fraction (SOM) on the precipitation of CaCO3 on calcite seeds in vitro. We found that each Robusta species utilizes a different set of proteins, contains different amino acid compositions, and has a different morphology modification capacity on calcite overgrowth. Our results further support the hypothesis that the different coral species utilize a species-specific protein set of independent gene co-option to construct their own unique organic matrix framework.
Materials and methods
Materials
All chemicals were obtained from Merck©, were of analytical grade and were used without further purification. All glassware was cleaned in ethanol and rinsed with distilled water before being air-dried.
Coral samples collection and preparation for protein extraction
Colonies of O. patagonica were collected in the Israeli Mediterranean Sea at Sdot-Yam (32° 49 N 34° 88 E) from 1 to 3 m depth. In addition, S. pistillata colonies were collected in the Israeli Red Sea, in front of the H. Steinitz Marine Biology Laboratory, Eilat (29° 30 N, 34° 56 E), from 3 to 5 m depth. Samples were collected under permit number 42410/2019 from the Israeli Natural Parks Authority.
Removal of organics tissue from the skeleton was done following the modified methods of Stoll et al.17,24. First, coral colonies were fragmented and oxidized with 20 ml 1:1 of 30% H2O2 and 3% NaClO solution while adding 1.5 ml of 3% NaClO every 20 min. After overnight incubation at room temperature, the solution was removed by washing the fragments five times for one minute with ultra-pure water and drying overnight at 60 °C. To ensure that no organic residue remained, we crushed the fragments to ≤ 63 µm in diameter with a mortar and pestle, oxidized, and washed them in ultra-pure water three more times. Between each cycle, both solutions were removed from the skeletal powder by centrifugation at 5000×g for 3 min at 4 °C and dried overnight at 60 °C.
Extraction and purification of skeletal proteins
Approximately 1.5 g of cleaned skeleton powder was used from each sample to extract the coral skeletal proteins, using the "CF4" method described in Peled et al.17. In brief, samples were decalcified in 0.5 M acetic acid for three hours at room temperature in Falcon tubes. Next, the samples were centrifuged at 5000×g for 5 min at 4 °C, and the supernatant was stored at 4 °C. The undissolved pellets were further treated until decalcification was completed, around three rounds, and until the measured pH in the solution was ~ 6. Next, the samples were frozen overnight at − 80 °C, lyophilized until dry and later merged by resuspension in 10 ml of ultra-pure water. The merged samples were frozen overnight at − 80 °C and lyophilized until dry. Next, the lyophilized pellet was resuspended in 12 ml ultra-pure water and centrifuged on a 3 kDa cutoff Amicon® Ultra 15 centrifugal filter units (Merck©) at 5000×g (4 °C) to a 0.5 ml final volume of desalted and concentrated skeletal OM. This process was repeated twice (three rounds in total), and the solution was separated into soluble and insoluble fractions by centrifugation at 5000×g for 5 min at 4 °C. The skeletal OM from each species was characterized by amino acid analysis and Fourier-transform infrared spectroscopy (FTIR) (see SI). The skeletal OM concentration (µg/ml) was expressed as the amount of protein from the amino acid analysis25.
O. patagonica proteomic analysis
Protein sequencing
O. patagonica skeletal protein samples were sequenced using the S-trap method26, where the resulting peptides were analyzed using a nanoflow liquid chromatography (nanoAcquity) coupled with a mass spectrometer (Fusion Lumos) (see SI). The Byonic search engine (Protein Metrics Inc.) was used to examine the resulting data against the predicted proteins from a de novo transcriptome of O. patagonica27 and a common contaminants database. First, no false discovery rate (FDR) filtering was implemented in the examination to generate a focused database for a second search. Next, the FDR was set to 1%, allowing fixed carbamidomethylation on C and variable oxidation on molecular weight, deamidation on NQ and protein N-terminal acetylation.
Data sorting
We used the predicted proteins O. patagonica27 as a reference peptide database for the MS analysis. We also included a common contaminants database. Only proteins with at least two significant peptides or at least one significant peptide with at least ten spectra and an identification score of 250 or greater were retained. To further filter out potential human proteins inadvertently introduced during sample preparation, we used the filtering criteria in Peled et al.17. In brief, all sequences were BLASTed against the 'Primates' nr database in NCBI using the Blast + command line (2.10.1)28. Lastly, we identified and removed from our analysis the BLAST sequence alignments of scleractinian versus Homo sapiens proteins with e-values lower than e−50 and percent mean similarity greater than 50% and sequences with e-values lower than e−100.
Characterization and annotation of SOM protein sequences
O. patagonica's protein sequences identified through the proteomic analysis were annotated using the Trinotate pipeline29.
The orthologous relationship between the species was determined using OrthoFinder 2.5.230,31. As sufficient species sampling is required to infer orthologous relationships between species, we sampled 12 species from the scleractinian order with an annotated genome supplemented with a combination of diverse metazoans from public databases (Table SI3). OrthoFinder generates orthology groups (Orthogroups) based on normalized reciprocal best BLAST hits' bit scores and then estimates orthologues genes pairs within Orthogroups. We then selected all pairs of O. patagonica skeletal OM sequences orthologous to S. pistillata SOM sequences (1:1, 1:many, many:many relationships). It is noteworthy that the use of de novo transcriptomes in inferring orthology is not recommended. However, the combination of a transcriptomic database and the species proteome is robust for accurately identifying proteins.
Nevertheless, another concern might be identifying multiple isoforms and transcripts of the same fragmented gene. To overcome this ambiguity, we have manually reviewed each rooted gene tree produced by OrthoFinder and their respective multiple sequence alignments (MSA) where skeletal OM proteins were identified. We examined all terminal nodes resulting from predicted duplication events to identify multiple isoforms classified as different skeletal OM proteins. In cases where the sequences aligned at over a 90% similarity, we considered them redundant and only kept the isoform with better MS evidence. In cases where the transcripts did not align with each other, the closest sequence (derived from a speciation event) was used as a scaffold to align the transcript as they were considered to be fragments of the same protein. A presence-absence (PA) matrix was generated based on orthologous sequences identified between at least two species out of O. patagonica, S. pistillata, A. digitifera And A. millepora (Table SI5). Non-metric multidimensional scaling (NMDS) was calculated based on the Jaccard distance matrix using the PA matrix.
Skeletal OM protein sequences from both O. patagonica (this study) and S. pistillata14,17 were analyzed using the InterProScan 5.50 platform32 in order to find conserved functional domains (Pfam)33 and gene ontology (GO) terms34. Using the InterPro predictive information, each skeletal OM protein domain and sequence were categorized into functional categories, representing their predictive role in the mineral formation. The non-redundant GO term sets were visualized using Revigo35. Intrinsic disorder regions (IDR) were predicted by an in silico analysis using flDPnn36, where a score above 0.3 is predicted for the region to be disordered.
In vitro calcification experiment in the presence of SOM
A 30 cm diameter desiccator was utilized for CaCO3 synthesis. It contained one glass beaker (50 ml) with crushed (NH4)2CO3 powder covered with parafilm, punched with three-needle holes and a Petri dish containing 5 g of anhydrous CaCl2. They were put at the bottom of the desiccator in advance. Cellular culture microplates containing a round glass coverslip in each well were used. In each well, 750 μl of 10 mM CaCl2 solution was poured. In the same solution, different amounts of SOM were added to investigate its effect on CaCO3 formation. When CaCO3 overgrowth experiments were performed, the bare round glass coverslip was replaced with a round glass coverslip with its surface covered mainly by rhombohedral calcite crystals (Fig. 5). A complete description of the experimental procedures is reported in the SI.
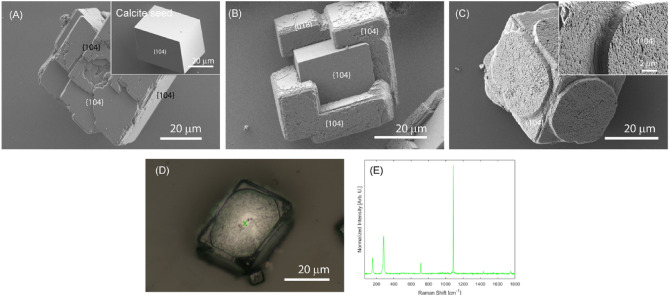
Figure 5.
SEM images of CaCO3 products obtained from overgrowth experiments. (A) Control experiment, i.e., overgrowth experiment in the absence of soluble fraction of the skeletal OM. (B) Overgrowth experiment in the presence of 13.3 µg/mL of SOM extracted from O. patagonica. (C) overgrowth experiment in the presence of 33.3 µg/mL SOM extracted from S. pistillata. The insets show details of the overgrowth of CaCO3. The Miller index of the crystalline faces is reported. (D) Optical microscope image of an overgrown disc-like mineral on the surface of a calcite seed. (E) Raman spectrum collected at the point indicated by the green cross in (D), the vibrational absorption bands observed indicate only the presence of calcite.
After a four-day crystallization time, the glass coverslips were lightly rinsed with Milli-Q water, dried, and examined using an optical microscope, FTIR, Raman spectroscopy and X-ray powder diffraction (see SI). Next, the formed crystals were coated in gold and examined with a scanning electron microscope (see SI).
A preliminary set of calcium carbonate precipitation experiments were performed to define the optimal SOM concentration for the overgrowth experiments. Different SOM concentrations of 66.7 µg/ml, 33.3 µg/ml and 13.3 µg/ml were tested (see SI). The optimal one was selected based on the trade-off of having the most evident effect on calcite morphology modification compared to the control (absence of SOM) and the inhibition of the crystal growth/aggregation of the SOM (Fig. SI1–4). A concentration of 13.3 µg/ml of O. patagonica and 33.3 µg/ml of S. pistillata were selected using the data from these experiments.
Results
Identification of coral skeletal OM proteins
We identified 73 skeletal OM proteins in O. patagonica using a de novo transcriptome database27. These proteins were compared to the published S. pistillata, A. digitifera and A. millepora skeletal OM proteins14–17. The NMDS analysis indicates a strong separation between the Robusta and Complexa species along the first axis (Fig. 1). Based on this result, we further focus our analysis on S. pistillata, the most similar of these species to O. patagonica.
Figure 1.
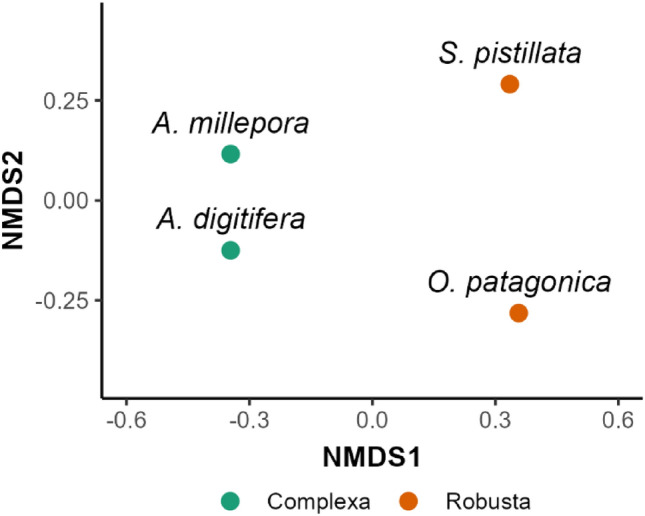
Dissimilarity in composition (Jaccard computed on generic presence-absence) between scleractinian species based on skeletal OM orthologous sequences.
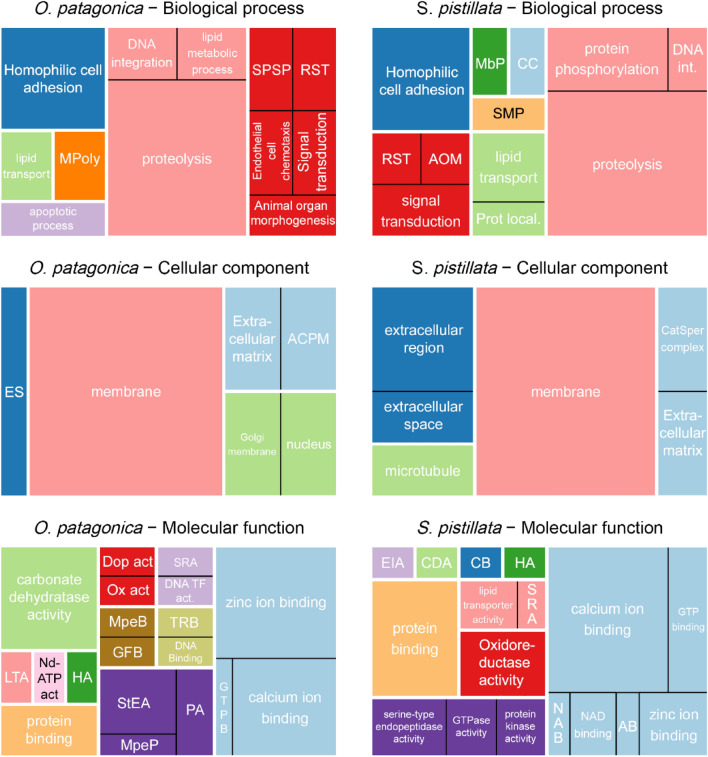
Further gene ontology (GO) analysis of the sequence composition between O. patagonica (Op-OMP) and S. pistillata (Sp-OMP) assigned 38 and 43 sequences to GO terms, respectively (Table SI4). The GO terms of the skeletal OM proteins data set are represented mainly by metabolic processes (including proteolysis and DNA integration), adhesion, cell communication, binding (including metal ion binding and protein binding) and catalytic activity (typically hydrolysis) (Fig. 2). Furthermore, the ontologies suggest key representations of proteins in the cell membrane and the extracellular space (Fig. 2). While the overall representation of the skeletal OM proteins' GO terms appear to be similar between species, the specific proteins that comprise the individual OMs are not the same. Furthermore, orthology analysis identified that 9 Op-OMPs share an orthologous relationship to Sp-OMPs, covering seven orthogroups (Tables SI4 and SI5). The shared orthogroups contain sets of MAM domain-containing proteins, cadherin proteins, acidic skeletal organic matrix proteins, ferroxidase proteins, carbonic anhydrase proteins, stereocilin proteins, and hemicentin proteins that, in turn, can be classified into three groups: (1) adhesion, (2) enzymic and (3) acidic proteins.
Figure 2.
Comparison of GO term lists visualized as a treemap. Each rectangle is a single cluster representative. The representatives are joined into "superclusters" of loosely related terms, visualized with different colors. The rectangles' sizes were adjusted to reflect the frequency of representation for each species of the GO term. Abbreviations and cluster information can be found in Table SI6.
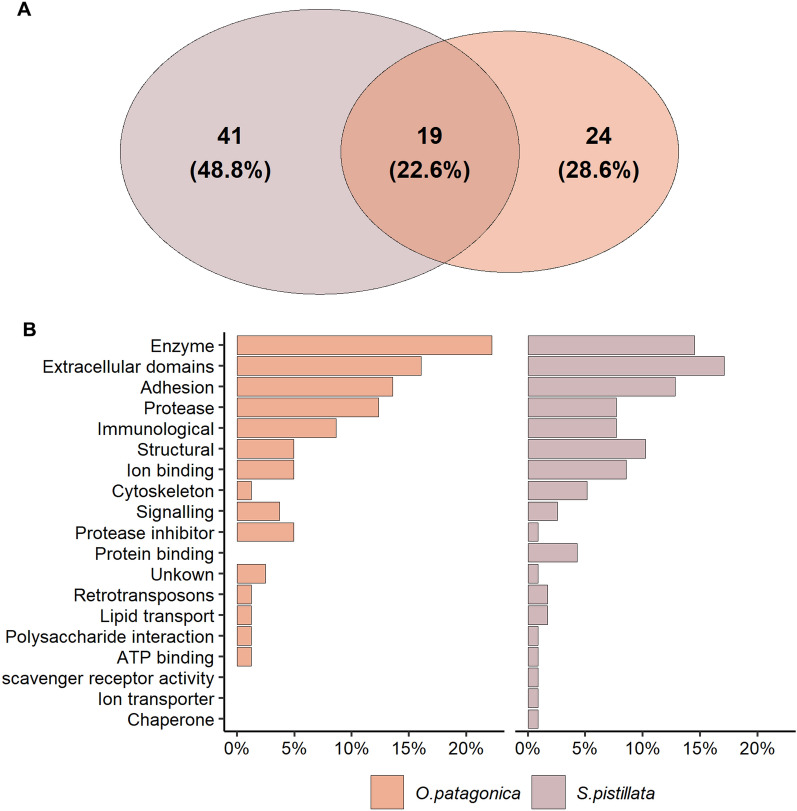
We aimed to identify functional domains in the skeletal OM protein sequences and found 19 shared domains between both species (Fig. 3A and Table SI4). The shared domains were classified to participate in adhesion, ion binding and lipid transport. Furthermore, some appear to have an enzymic role, including proteolysis and hydrolysis, and some were annotated to be involved in the immunological response and have extracellular domains. Furthermore, while most functional domains found were species-specific (41 and 24 for S. pistillata and O. patagonica, respectively) (Fig. 3A), the domains' overall predicted roles were found to be similarly represented between the species (Fig. 3B). The most common roles of the functional domains include enzymic, proteolysis, extracellular domains, adhesive, ion binding and Immunological representing 75% and 68% of the total identified domains for O. patagonica and S. pistillata, respectively. However, certain predicted roles were identified as S. pistillata specific, including protein binding, scavenger receptor activity, ion transporter and chaperones.
Figure 3.
(A) Comparison of functional domain numbers according to the PFAM analysis. (B) Distribution of PFAM domain role. The values represent the percentage of each domain sharing a functional role compared to the total number of functional roles for each species.
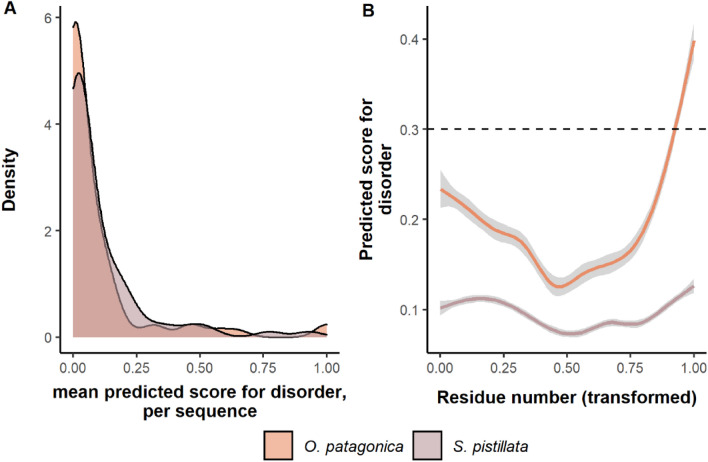
The IDR analysis revealed that both species have a mean disorder score per residue of 0.1, which signifies that they are not predicted to be intrinsically disordered. Furthermore, only two O. patagonica and a single S. pistillata skeletal OM proteins were completely intrinsically disordered. While the overall predictions appear similar between species, a Wilcoxon signed-rank test showed significantly higher IDR residues in S. pistillata (p value = 0.02). Furthermore, 80% of S. pistillata skeletal OM proteins are predicted to have at least a single IDR, while O. patagonica's prediction is 50%. Lastly, we identify that the IDRs are predominantly found at the sequences' start and end (up to ~ 25% and from ~ 75%, respectively, of its length) for both species (Fig. 4).
Figure 4.
Prediction of intrinsically disordered regions. (A) A density plot displaying the sequences’ disorder mean predicted score per species. (B) The average (± standard error in grey) of the disordered score per residue across SOM proteins with at least a single region predicted to be disordered. The residue number is represented as the percentage of the entire sequence. Regions with a score above the dotted line are predicted to be disordered.
Skeletal OM proteins composition
The proteins present in the skeletal OM from O. patagonica have a content of Asp and Glu (52.1 mol%) higher than that present in the SOM from S. pistillata (30.1 mol%). The latter, however, has a higher Ser content (14.4 mol% compared to 4.5 mol% in O. patagonica) (Table SI1).
In vitro overgrowth experiment
Next, we wished to test the effect of the different soluble fractions of the skeletal OMs on mineral formation. Therefore, a series of overgrowth experiments on calcite seeds were performed using the vapor diffusion method37 (results summarized in Table SI2).
The calcite seed crystals showed the typical {104} rhombohedral faces (Fig. 5A, inset). Overgrowth control experiments were also performed without SOM as control experiments. The overgrowth from the 10 mM CaCl2 solution induced secondary nucleation events and the deposition of additional calcite {104} layers on the calcite seeds (Fig. 5A). The effect of SOM on the CaCO3 overgrowth process on calcite crystal seeds was evaluated after a preliminary screening in the absence of seeds (Figs. SI1–4). The SOM extracted from O. patagonica and S. pistillata were used in the overgrowth experiments with concentrations equal to 13.3 µg/mL and 33.3 µg/mL, respectively. The products of these experiments were characterized, and each species included distinctive characteristics and mineral patterns of the overgrowth crystals. In the presence of SOM from O. patagonica, regular calcite crystals overgrew and covered some of the crystal edges (Fig. 5B). Their surface was irregular compared with the calcite seed's surface, showing many irregular pits. In addition, it showed {018} faces on their edges36. In the presence of S. pistillata SOM, the outcome was different. The formation of disk-like shapes centered on each {104} face of the calcite seed was observed. They uncovered the crystal edges (Fig. 5C inset) and showed the surface to be rich in irregular pits. The Raman microscopy analysis revealed that although differently shaped, this structure was calcite (Fig. 5D,E).
Discussion
In the Scleractinia order, many species have evolved which differ in population structure, trophic strategy, macro-scale morphology, and micro-scale texture38,39. For each species, the skeleton's structural characteristics are a direct result of genetic control on the production of skeletal OM macromolecules13. These then control the calcification process, which is affected by the environment in which the coral has adapted to live40. To date, mineralization-associated proteins found in the skeletal OM of scleractinians were identified only in three tropical species, S. pistillata, Acropora digitifera and A. millepora. While those species diverged over 400 million years ago1,6 and belong to the Robusta and Complexa scleractinian clades, they share a similar branching growth pattern. Here we study the skeletal OM protein sequence compositions of the encrusting temperate coral O. patagonica belonging to the Robusta clade and compare it to previously published skeletal OM proteomes14–17.
Previous skeletal OM proteome analysis identified distinct sets of proteins, with only six ortholog groups across all species, termed the “core biomineralization toolkit”19. These proteins are predicted to have a role in the mineral nucleation, cell adhesion and structure of the organic matrix13. Indeed, the same core proteins are still present when cross-comparing the three tropical species with a temperate one that further differentiates in its growth pattern (Table SI5). Furthermore, our results suggest that the biomineralization mechanism is partly a function of phylogenetic proximity among species (Fig. 1). However, it is important to point out that this analysis is on a small dataset that only includes four species with high variability in their evolutionary distance (family compared to coral clades). As such, the characterization of the skeletal OM proteins of other scleractinian species from a wide array of lineages would allow for a better understanding of the disparity of proteins involved in the broader scleractinian skeleton formation.
Despite the importance of the proposed “core biomineralization toolkit”, most skeletal OM proteomes differ between the species. It was proposed that integral key molecular pathways used by skeleton-forming organisms evolved through the co-option of proteins that previously served other biological functions41. Interestingly, as mineralization-associated protein datasets have increased, it was found that this co-option evolution was not achieved by a common ancestor of the skeleton-forming organisms but independently within and across lineages. These findings further suggest evolutionary plasticity in which their underlying functions can be performed using a different set of proteins by different organisms, regardless of their evolutionary distance42.
As our results support a separation between the two coral clades (Fig. 1), we further focused our comparison on O. patagonica’s closest species in this dataset, S. pistillata. While both species belong to the Robusta coral clade, they grow under a wide range of environmental conditions43–45 and exhibit different growth pattern3.
Not all proteins present in the skeletal OM are involved in CaCO3 nucleation and growth processes. While random incorporation within the mineral fraction cannot be ignored, it was reported that part of the skeletal OM proteins might also regulate cell communication and the formation of the extracellular matrix46,47. Previous studies roughly divided the mineralization-associated proteins in metazoans into different categories. These categories include broad functional ones such as matrix formers, nucleation assisters, communicators, remodelers46, and functional domains such as low-complexity regions, extracellular domains, adhesion, immunological, polysaccharide interactions, enzymes, protease inhibitors and others47. By analyzing the specific functional annotation of both species, we identify a low rate of overlap (Fig. 3A). Nonetheless, the domains' main functionalities were similar and related to extracellular domains, immunological, enzymes and protease inhibitors (Fig. 3B). Furthermore, we found that the shared broad functional categories identified in both species are similar and relate to communication (adhesion proteins), nucleation assisters (acidic proteins) and remodelers (proteases) (Fig. 2). Regarding extracellular domains and adhesion, the most prevalent class of proteins is von Willebrand factor (vWF) proteins, which are suggested to take part in the initial mineralization process48,49. vWF proteins also appear to have a role in the structural organization of the organic matrix and the mineral, similar to collagens50. Enzymes, especially proteases and protease inhibitors, have a vital role in the remodeling of the matrix environment. One well-studied set of proteins is the carbonic anhydrases which allow for the rapid conversion of carbon dioxide to bicarbonate51. A sequence feature highly regarded in the protein–protein matrix and aragonitic assembly is regions of intrinsic disorder52 as the free energy needed to bind IDR proteins to precursor mineral is low53. In this study, we found a variation between species in the extent of IDR proteins (Fig. 4A) that might be associated with the difference in the quality of the skeletal OM proteome referenced sequences54. Yet, most skeletal OM proteins in both species were identified to have at least a single IDR, with an overall trend of these regions being found at the start and the end of the sequences (Fig. 4B). This further emphasizes that while the specific building blocks to create a skeleton are different, the overall elements that provide functionality are conserved.
The soluble fraction of skeletal OMs effect on CaCO3 formation was performed by in vitro homogeneous and overgrowth experiments on calcite seeds, as coral calcification starts in proximity to the larvae settlement on the substrate55,56. Furthermore, the best-known biochemical signals arise from mineralized crustose coralline algae that deposit calcitic structures57,58. The in vitro homogeneous precipitation of CaCO3 showed that a higher concentration of skeletal OM from S. pistillata is required to have a nonspecific morphological modification of calcite particles that resembles what is observed for the O. patagonica SOM (see SI). This different interaction capability of the SOMs with growing CaCO3 crystals has already been observed for other coral species having different characteristics12,59. A possible explanation can come from the different amino acid compositions of the two skeletal OMs. One key difference between species regards the aspartic and glutamic concentration, which was 1.7 times higher in O. patagonica than in S. pistillata SOM (SI). Usually, molecules with a high content of charged functional groups and missing a conformation interact with the growing CaCO3 nuclei modifying their morphology in a nonspecific way25,60. According to this consideration, we can suppose that the SOM macromolecules from O. patagonica and S. pistillata in a homogeneous solution do not assume a conformation. Indeed, several studies have reported that the skeletal OM molecules are intrinsically disordered in solution53. These observations may support the different nanoscale filling mechanisms suggested for spherulitic growth61. Furthermore, the authors reported a higher abundance of "sprinklers" like particles at the O. patagonica skeleton compared to S. pistillata62, suggesting that those "sprinklers" are the first nucleation seeds of each crystalline fiber.
The overgrowth experiments on calcite seeds confirmed the differences in mineralizers between the two Robusta SOMs. The presence of calcite seeds modifies the interaction between the SOM and the growing CaCO3 crystals concerning the precipitation in a homogeneous solution63. The results show that in the presence of the O. patagonica SOM, calcite crystallization occurs mainly on the edges of the seeds, which surfaces with the highest energy are found64. In contrast, the overgrowth of calcite in the presence of SOM from S. pistillata occurs at the center of {104} faces of the calcite seeds generating disk-like structures. The presence of seeds reduces the supersaturation from nucleation65, and the effects of the SOMs' macromolecules, or other additives, can be more evident over the growth process, which usually produces a change in crystal morphology25,60,66. In this context, the effect of the SOM molecules from S. pistillata as crystal morphology modifiers seems more specific, favoring the formation of overgrown calcite crystals in which a completely different organization (disk-like) is observed compared to those detected in the presence of the SOM from O. patagonica or in the control experiment in the absence of SOM molecules. However, to extensively understand the interaction between skeletal OM molecules and CaCO3 crystals, a study on the single molecules of the skeletal OM and their combination is necessary. This requires the not-so-easy task of their purification or biochemical expression. The information available on CARPs50,67 indicates that they do not assume a conformation and probably are intrinsically disordered similar to other families of highly acidic proteins68.
In conclusion, the observations discussed above can be contextualized in the different micro-texture of the skeleton of the two coral species. Furthermore, the knowledge gained both from the proteomic analysis and these in vitro experiments on the calcification process in coral can be integrated with the already available information on the molecular toolkit that controls the calcification process13,50 and on the role of single proteins69–72 in addressing the pathway of the mineralization process. Although the proposed "core biomineralization toolkit"19 and the skeletal OM proteome set of functional categories are conserved across scleractinians, this study emphasizes that each species utilizes its own specific elements to achieve the conserved functional category. This suggests that the early diversification1,6 allowed species-specific adaptations to diverse environmental conditions. Therefore, with an eye on better understanding the ability of stony corals to calcify under diverse environmental conditions, we need to better explore the differences in those categories and across a wide range of species.
Supplementary Information
Acknowledgements
We thank Dr. David Morgenstern at the de Botton Institute for Protein Profiling at Nancy and Stephen Grand Israel National Center for Personalized Medicine for proteomic sequencing and analysis at the Weizmann Institute of Science. We thank Dr. Jeana L. Drake from UCLA for her valuable inputs and English proofing. In addition, we thank Yanai Peled and Dr. Ricardo Almuly at the University of Haifa for laboratory assistance. This research was supported by the Israeli Binational Science Foundation (BSF 2016321) and the Ministry of Innovation, Science & Technology, Israel. Computations presented in this work were performed on the Hive computer cluster at the University of Haifa, partly funded by ISF Grant 2155/15. I.P. is the incumbent of the Sharon Zuckerman research fellow chair.
Author contributions
T.Z., T.M. and G.F. wrote the main manuscript text. T.Z. and G.F. prepared the figures. A-P.D-B and G.F. performed the in vitro overgrowth. I.P. performed the Raman measurements. T.Z. preprocessed and analyzed the sequencing data. All authors reviewed the manuscript.
Data availability
All alignments, trees, and protein sequences used for orthology inference are available on GitHub (https://github.com/Mass-Lab/Zaquin_Op_Sp_SOM_comparison.git) and are publicly available. The datasets generated during the current study are publicly available in the ProteomeXchange repository under file number PXD034601 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD034601).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tal Zaquin, Email: tzaquin@campus.haifa.ac.il.
Giuseppe Falini, Email: giuseppe.falini@unibo.it.
Tali Mass, Email: tmass@univ.haifa.ac.il.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-20744-0.
References
- 1.Stolarski J, et al. The ancient evolutionary origins of Scleractinia revealed by azooxanthellate corals. BMC Evol. Biol. 2011;11:316. doi: 10.1186/1471-2148-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drake JL, et al. How corals made rocks through the ages. Glob. Change Biol. 2020;26:31–53. doi: 10.1111/gcb.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veron, J. E. N. Corals of the world (2000).
- 4.Veron JEN, et al. The coral reef crisis: The critical importance of <350 ppm CO2. Mar. Pollut. Bull. 2009;58:1428–1436. doi: 10.1016/j.marpolbul.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Roberts JM, Wheeler A, Freiwald A, Cairns S. Cold-Water Corals: The Biology and Geology of Deep-Sea Coral Habitats. Cambridge University Press; 2009. [Google Scholar]
- 6.McFadden CS, et al. Phylogenomics, origin, and diversification of anthozoans (phylum Cnidaria) Syst. Biol. 2021;70:635–647. doi: 10.1093/sysbio/syaa103. [DOI] [PubMed] [Google Scholar]
- 7.Barnes DJ. Coral skeletons: An explanation of their growth and structure. Science. 1970;170:1305–1308. doi: 10.1126/science.170.3964.1305. [DOI] [PubMed] [Google Scholar]
- 8.Cuif J-P, Lecointre G, Perrin C, Tillier A, Tillier S. Patterns of septal biomineralization in Scleractinia compared with their 28S rRNA phylogeny: A dual approach for a new taxonomic framework. Zool. Scr. 2003;32:459–473. doi: 10.1046/j.1463-6409.2003.00133.x. [DOI] [Google Scholar]
- 9.Cuif J-P, Dauphin Y. The two-step mode of growth in the scleractinian coral skeletons from the micrometre to the overall scale. J. Struct. Biol. 2005;150:319–331. doi: 10.1016/j.jsb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Cuif J-P, Dauphin Y, Berthet P, Jegoudez J. Associated water and organic compounds in coral skeletons: Quantitative thermogravimetry coupled to infrared absorption spectrometry. Geochem. Geophys. Geosyst. 2004;5:Q11011. doi: 10.1029/2004GC000783. [DOI] [Google Scholar]
- 11.Tambutté S, et al. Coral biomineralization: from the gene to the environment. J. Exp. Mar. Biol. Ecol. 2011;408:58–78. doi: 10.1016/j.jembe.2011.07.026. [DOI] [Google Scholar]
- 12.Falini G, Fermani S, Goffredo S. Coral biomineralization: A focus on intra-skeletal organic matrix and calcification. Semin. Cell Dev. Biol. 2015;46:17–26. doi: 10.1016/j.semcdb.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Drake JL, Varsano N, Mass T. Genetic basis of stony coral biomineralization: History, trends and future prospects. J. Struct. Biol. 2021;213:107782. doi: 10.1016/j.jsb.2021.107782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drake JL, et al. Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc. Natl. Acad. Sci. 2013;110:3788–3793. doi: 10.1073/pnas.1301419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramos-Silva P, et al. The skeletal proteome of the coral Acropora millepora: The evolution of calcification by co-option and domain shuffling. Mol. Biol. Evol. 2013;30:2099–2112. doi: 10.1093/molbev/mst109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi T, Yamada L, Shinzato C, Sawada H, Satoh N. Stepwise evolution of coral biomineralization revealed with genome-wide proteomics and transcriptomics. PLoS ONE. 2016;11:e0156424. doi: 10.1371/journal.pone.0156424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peled Y, et al. Optimization of skeletal protein preparation for LC–MS/MS sequencing yields additional coral skeletal proteins in Stylophora pistillata. BMC Mater. 2020;2:8. doi: 10.1186/s42833-020-00014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, et al. The evolution of calcification in reef-building corals. Mol. Biol. Evol. 2021;38:3543–3555. doi: 10.1093/molbev/msab103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaquin T, Malik A, Drake JL, Putnam HM, Mass T. Evolution of protein-mediated biomineralization in scleractinian corals. Front. Genet. 2021;12:52. doi: 10.3389/fgene.2021.618517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuyama I, Higuchi T. Differential gene expression in skeletal organic matrix proteins of scleractinian corals associated with mixed aragonite/calcite skeletons under low mMg/Ca conditions. PeerJ. 2019;7:e7241. doi: 10.7717/peerj.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sancho-Tomás M, Fermani S, Gómez-Morales J, Falini G, García-Ruiz JM. Calcium carbonate bio-precipitation in counter-diffusion systems using the soluble organic matrix from nacre and sea-urchin spine. Eur. J. Mineral. 2014;26:523–535. doi: 10.1127/0935-1221/2014/0026-2389. [DOI] [Google Scholar]
- 22.Leydet KP, Hellberg ME. The invasive coral Oculina patagonica has not been recently introduced to the Mediterranean from the Western Atlantic. BMC Evol. Biol. 2015;15:79. doi: 10.1186/s12862-015-0356-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitahara MV, Cairns SD, Stolarski J, Blair D, Miller DJ. A comprehensive phylogenetic analysis of the Scleractinia (Cnidaria, Anthozoa) based on mitochondrial CO1 sequence data. PLoS ONE. 2010;5:e11490. doi: 10.1371/journal.pone.0011490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoll HM, et al. A first look at paleotemperature prospects from Mg in coccolith carbonate: Cleaning techniques and culture measurements. Geochem. Geophys. Geosyst. 2001;2:1047-14. doi: 10.1029/2000GC000144. [DOI] [Google Scholar]
- 25.Albeck S, Aizenberg J, Addadi L, Weiner S. Interactions of various skeletal intracrystalline components with calcite crystals. J. Am. Chem. Soc. 1993;115:11691–11697. doi: 10.1021/ja00078a005. [DOI] [Google Scholar]
- 26.HaileMariam M, et al. S-Trap, an ultrafast sample-preparation approach for shotgun proteomics. J. Proteome Res. 2018;17:2917–2924. doi: 10.1021/acs.jproteome.8b00505. [DOI] [PubMed] [Google Scholar]
- 27.Zaquin T, Zaslansky P, Pinkas I, Mass T. Simulating bleaching: Long-term adaptation to the dark reveals phenotypic plasticity of the Mediterranean Sea coral Oculina patagonica. Front. Mar. Sci. 2019;6:662. doi: 10.3389/fmars.2019.00662. [DOI] [Google Scholar]
- 28.Camacho C, et al. BLAST Command Line Applications User Manual, BLAST® Help. Natl. Cent. Biotechnol. Inf. (US); 2008. [Google Scholar]
- 29.Bryant DM, et al. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017;18:762–776. doi: 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emms DM, Kelly S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emms DM, Kelly S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones P, et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburner M, et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu G, et al. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 2021;12:4438. doi: 10.1038/s41467-021-24773-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ihli J, Bots P, Kulak A, Benning LG, Meldrum FC. Elucidating mechanisms of diffusion-based calcium carbonate synthesis leads to controlled mesocrystal formation. Adv. Funct. Mater. 2013;23:1965–1973. doi: 10.1002/adfm.201201742. [DOI] [Google Scholar]
- 38.Stolarski J. Three-dimensional micro- and nanostructural characteristics of the scleractinian coral skeleton: A biocalcification proxy. Acta Palaeontol. Pol. 2003;48:497–530. [Google Scholar]
- 39.Cuif J-P. Calcification in the Cnidaria through time: An overview of their skeletal patterns from individual to evolutionary viewpoints. In: Goffredo S, Dubinsky Z, editors. The Cnidaria, Past, Present and Future. Springer; 2016. pp. 163–179. [Google Scholar]
- 40.Todd PA. Morphological plasticity in scleractinian corals. Biol. Rev. 2008;83:315–337. doi: 10.1111/j.1469-185X.2008.00045.x. [DOI] [PubMed] [Google Scholar]
- 41.Lowenstam HA, Margulis L. Evolutionary prerequisites for early Phanerozoic calcareous skeletons. Biosystems. 1980;12:27–41. doi: 10.1016/0303-2647(80)90036-2. [DOI] [PubMed] [Google Scholar]
- 42.Murdock DJE. The ‘biomineralization toolkit’ and the origin of animal skeletons. Biol. Rev. 2020;95:1372. doi: 10.1111/brv.12614. [DOI] [PubMed] [Google Scholar]
- 43.Fine M, Zibrowius H, Loya Y. Oculina patagonica: A non-Lessepsian scleractinian coral invading the Mediterranean Sea. Mar. Biol. 2001;138:1195–1203. doi: 10.1007/s002270100539. [DOI] [Google Scholar]
- 44.Mass T, et al. Photoacclimation of Stylophora pistillata to light extremes: Metabolism and calcification. Mar. Ecol. Prog. Ser. 2007;334:93–102. doi: 10.3354/meps334093. [DOI] [Google Scholar]
- 45.Malik A, et al. Molecular and skeletal fingerprints of scleractinian coral biomineralization: From the sea surface to mesophotic depths. Acta Biomater. 2020 doi: 10.1016/j.actbio.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Evans JS. The biomineralization proteome: Protein complexity for a complex bioceramic assembly process. Proteomics. 2019;19:1900036. doi: 10.1002/pmic.201900036. [DOI] [PubMed] [Google Scholar]
- 47.Liu C, Zhang R. Biomineral proteomics: A tool for multiple disciplinary studies. J. Proteomics. 2021;238:104171. doi: 10.1016/j.jprot.2021.104171. [DOI] [PubMed] [Google Scholar]
- 48.Hayward DC, et al. Differential gene expression at coral settlement and metamorphosis—A subtractive hybridization study. PLoS ONE. 2011;6:e26411. doi: 10.1371/journal.pone.0026411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holcomb M, Cohen AL, Gabitov RI, Hutter JL. Compositional and morphological features of aragonite precipitated experimentally from seawater and biogenically by corals. Geochim. Cosmochim. Acta. 2009;73:4166–4179. doi: 10.1016/j.gca.2009.04.015. [DOI] [Google Scholar]
- 50.Mummadisetti MP, Drake JL, Falkowski PG. The spatial network of skeletal proteins in a stony coral. J. R. Soc. Interface. 2021;18:20200859. doi: 10.1098/rsif.2020.0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertucci A, et al. Carbonic anhydrases in anthozoan corals—A review. Bioorg. Med. Chem. 2013;21:1437–1450. doi: 10.1016/j.bmc.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 52.Evans JS. Aragonite-associated biomineralization proteins are disordered and contain interactive motifs. Bioinformatics. 2012;28:3182–3185. doi: 10.1093/bioinformatics/bts604. [DOI] [PubMed] [Google Scholar]
- 53.Boskey AL, Villarreal-Ramirez E. Intrinsically disordered proteins and biomineralization. Matrix Biol. 2016;52–54:43–59. doi: 10.1016/j.matbio.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinha S, Eisenhaber B, Lynn AM. Predicting protein function using homology-based methods. In: Shanker A, editor. Bioinformatics: Sequences, Structures, Phylogeny. Springer; 2018. pp. 289–305. [Google Scholar]
- 55.Akiva A, et al. Minerals in the pre-settled coral Stylophora pistillata crystallize via protein and ion changes. Nat. Commun. 2018;9:1880. doi: 10.1038/s41467-018-04285-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Golbuu Y, Richmond RH. Substratum preferences in planula larvae of two species of scleractinian corals, Goniastrea retiformis and Stylaraea punctata. Mar. Biol. 2007;152:639–644. doi: 10.1007/s00227-007-0717-x. [DOI] [Google Scholar]
- 57.Morse DE, Hooker N, Morse ANC, Jensen RA. Control of larval metamorphosis and recruitment in sympatric agariciid corals. J. Exp. Mar. Biol. Ecol. 1988;116:193–217. doi: 10.1016/0022-0981(88)90027-5. [DOI] [Google Scholar]
- 58.Tebben J, et al. Chemical mediation of coral larval settlement by crustose coralline algae. Sci. Rep. 2015;5:1–11. doi: 10.1038/srep10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaquin T, et al. Exploring coral calcification by calcium carbonate overgrowth experiments. Cryst. Growth Des. 2022;22:5045–5053. doi: 10.1021/acs.cgd.2c00552. [DOI] [Google Scholar]
- 60.Albeck S, Weiner S, Addadi L. Polysaccharides of intracrystalline glycoproteins modulate calcite crystal growth in vitro. Chem. Eur. J. 1996;2:278–284. doi: 10.1002/chem.19960020308. [DOI] [Google Scholar]
- 61.Sun C-Y, et al. Spherulitic growth of coral skeletons and synthetic aragonite: Nature’s three-dimensional printing. ACS Nano. 2017;11:6612–6622. doi: 10.1021/acsnano.7b00127. [DOI] [PubMed] [Google Scholar]
- 62.Sun C-Y, et al. Crystal nucleation and growth of spherulites demonstrated by coral skeletons and phase-field simulations. Acta Biomater. 2021;120:277–292. doi: 10.1016/j.actbio.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Addadi L, Moradian J, Shay E, Maroudas NG, Weiner S. A chemical model for the cooperation of sulfates and carboxylates in calcite crystal nucleation: relevance to biomineralization. Proc. Natl. Acad. Sci. 1987;84:2732–2736. doi: 10.1073/pnas.84.9.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jimenez-Lopez C, Rodriguez-Navarro A, Dominguez-Vera JM, Garcia-Ruiz JM. Influence of lysozyme on the precipitation of calcium carbonate: A kinetic and morphologic study. Geochim. Cosmochim. Acta. 2003;67:1667–1676. doi: 10.1016/S0016-7037(02)01275-9. [DOI] [Google Scholar]
- 65.Gómez-Morales, J., Falini, G. & García-Ruiz, J. M. Biological crystallization. in Handbook of Crystal Growth 873–913 (Elsevier, 2015).
- 66.Ihli J, et al. Visualization of the effect of additives on the nanostructures of individual bio-inspired calcite crystals. Chem. Sci. 2019;10:1176–1185. doi: 10.1039/C8SC03733G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mass T, Drake JL, Peters EC, Jiang W, Falkowski PG. Immunolocalization of skeletal matrix proteins in tissue and mineral of the coral Stylophora pistillata. Proc. Natl. Acad. Sci. USA. 2014;111:12728–12733. doi: 10.1073/pnas.1408621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruidiaz SF, Dreier JE, Hartmann-Petersen R, Kragelund BB. The disordered PCI-binding human proteins CSNAP and DSS1 have diverged in structure and function. Protein Sci. 2021;30:2069–2082. doi: 10.1002/pro.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mass T, et al. Cloning and characterization of four novel coral acid-rich proteins that precipitate carbonates in vitro. Curr. Biol. 2013;23:1126–1131. doi: 10.1016/j.cub.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 70.Laipnik R, et al. Coral acid rich protein selects vaterite polymorph in vitro. J. Struct. Biol. 2019;209:107431. doi: 10.1016/j.jsb.2019.107431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reyes-Bermudez A, Lin Z, Hayward DC, Miller DJ, Ball EE. Differential expression of three galaxin-related genes during settlement and metamorphosis in the scleractinian coral Acropora millepora. BMC Evol. Biol. 2009;9:178. doi: 10.1186/1471-2148-9-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moya A, et al. Carbonic anhydrase in the scleractinian coral Stylophora pistillata characterization, localization, and role in biomineralization. J. Biol. Chem. 2008;283:25475–25484. doi: 10.1074/jbc.M804726200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All alignments, trees, and protein sequences used for orthology inference are available on GitHub (https://github.com/Mass-Lab/Zaquin_Op_Sp_SOM_comparison.git) and are publicly available. The datasets generated during the current study are publicly available in the ProteomeXchange repository under file number PXD034601 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD034601).