Abstract
Fifteen kinds of new insertion sequences (ISs), IS641 to IS643, IS650 to IS658, IS660, IS662, and IS663, and a group II intron (Bh.Int) were identified in the 4,202,352-bp genome of alkaliphilic Bacillus halodurans C-125. Out of 120 ISs identified in the C-125 genome, 29 were truncated, indicating the occurrence of internal rearrangements of the genome. The ISs other than IS650, IS653, IS660, and IS663 generated a 2- to 9-bp duplication of the target site sequence, and the ISs other than IS650, IS653, and IS657 carry 14- to 64-bp inverted repeats. Sequence analysis revealed that six kinds of ISs (IS642, IS643, IS654, IS655, IS657, and IS658) belong to a separate IS family (IS630, IS21, IS256, IS3, IS200/IS605, and IS30, respectively) as a new member. Also, IS651 and IS652 were characterized as new members of the ISL3 family. Significant similarity was found between the transposase (Tpase) sequences between IS650 and IS653 (78.2%), IS651 and IS652 (56.3%), IS656 and IS662 (71.0%), and IS660 and IS663 (44.5%), but the others showed no similarity to one another. Tpases in 28 members of IS651 in the C-125 genome were found to have become diversified. Most of the IS elements widely distributed throughout the genome were inserted in noncoding regions, although some genes, such as those coding for an ATP-binding cassette transporter/permease, a response regulator, and l-indole 2-dehydrogenase, have been mutated through the insertion of IS elements. It is evident, however, that not all IS elements have transposed and caused rearrangements of the genome in the past 17 years during which strain C-125 was subcultured under neutral and alkaline conditions.
Alkaliphilic Bacillus halodurans strain C-125 (JCM9153) was isolated in 1970 and characterized as a β-galactosidase producer (21) and xylanase producer (17, 18). It is the most thoroughly characterized strain, physiologically, biochemically, and genetically, among those in our collection of alkaliphilic Bacillus isolates (19). Generally, alkaliphilic Bacillus strains cannot grow below pH 6.5 but grow well above pH 9.5. The facultative alkaliphilic B. halodurans can grow at pH 7 to 10.5 if sodium ions are supplied at a sufficiently high concentration (1 to 2%) in the medium. Over the past 2 decades, our studies have focused on the enzymology, physiology, and molecular genetics of alkaliphilic microorganisms to elucidate their mechanisms of adaptation to alkaline environments. Industrial applications of these microbes have been investigated, and some enzymes, such as proteases, amylases, cellulases, and xylanases, have been commercialized (19, 46).
Recently, analysis of the entire genome of alkaliphilic B. halodurans strain C-125 was completed and comparison of the genomic sequence with that of Bacillus subtilis has been done in an effort to clarify the mechanisms of adaptation to a highly alkaline environment and thereby further industrial use of alkaliphilic Bacillus strains as a first step (27, 45). Through a series of genome analysis studies, it became clear that the B. halodurans genome contains 112 putative transposase (Tpase) genes. This is one of the notable features of this genome. Insertion sequences (ISs) are small mobile units of DNA consisting of, in general, a unique Tpase gene and terminal inverted repeats (IRs), which serve as the sites for recognition and cleavage by Tpases in transposition reactions (13, 14, 29, 35). To date, a large number of ISs have been classified into 17 families principally based on the amino acid sequence similarities of their Tpases (30). It has been reported that Synechocystis sp. strain PC6803 (23), Escherichia coli MG1655 (6), Mycobacterium tuberculosis (11), Deinococcus radiodurans (51), and Lactococcus lactis (7), whose whole genome sequences have been determined, possess multiple ISs of different families in their genomes. Some ISs form composite transposable elements, i.e., transposons, by flanking a DNA region containing antibiotic resistance genes or catabolic or pathogenic genes (4, 29, 41, 48). It is well recognized that transposition of such mobile elements sometimes results in an insertional mutation or activation of a downstream gene (9, 12, 16, 22, 25, 26, 28, 47). Since ISs and transposons are often associated with transmissible plasmids and bacteriophages, they have become distributed in a wide range of bacteria by horizontal transmission. Thus, ISs have played an important role in evolution by facilitating horizontal gene transfer and also in internal genetic rearrangements in the genome. It is of substantial interest and importance to determine how genetic events occurred by examining the behavior of ISs in the bacterial genome to understand the mechanisms of adaptation to dramatic changes in the environment, especially in the case of adaptation to extreme environments, such as those with high or low pH, high or low temperature, high pressure, or high salinity. In addition, we have a specific interest in determining how the behavior of ISs influences the improvement of enzyme productivity or the stability of enzyme production, because this may contribute to the development of some new theory on the basis of which systematic breeding of industrial strains can be pursued for further industrial application of alkaliphilic Bacillus strains possessing great potential for useful enzyme production.
In this work, we identified and characterized 15 kinds of new ISs and a group II intron in the 4,202,352-bp genome of B. halodurans C-125. Here, we report the distribution and orientation of the members of each element in the genome of strain C-125, the structure and target site sequence of each, and the phylogenetic relationships among them. In addition, we investigated the behavior of IS elements in the C-125 genome over a period of 2 decades by PCR using site-specific primer sets and by examining the digestion patterns of the chromosome obtained using various restriction endonucleases.
MATERIALS AND METHODS
Bacterial strains and media.
B. halodurans C-125, formerly Bacillus sp. strain C-125 (44), lyophilized on 10 November 1983, was regenerated using Horikoshi II medium (pH 9.5) (42). Strain C-125 has been subcultured for 17 years to date, and it was used as a standard strain representative of the current generation. The cells were grown aerobically at 37°C in Horikoshi II medium (pH 7.5 or 9.5).
PFGE.
B. halodurans chromosomal DNA for pulsed-field gel electrophoresis (PFGE) was prepared in agarose plugs by the method previously described (43). Agarose blocks containing the chromosomal DNA were washed in 50 ml of 0.1× Tris-EDTA buffer twice and then equilibrated with the corresponding restriction buffer at 4°C for 1 h. DNA was digested with 100 to 200 U of AscI or Sse8387I (Takara Shuzo, Kyoto, Japan) or I-Ceu I (New England Biolabs) at 37°C overnight in 500 μl of the restriction buffer recommended by the manufacturer. In the case of other restriction endonucleases (SmaI, PacI, PmeI, BssHII, and SwaI), DNA was digested with 60 U of enzyme in 300 μl of reaction mixture overnight. PFGE in 1% pulsed-field-certified agarose was performed by the method previously described (43).
Amplification of ISs from chromosome of B. halodurans
Chromosomal DNA was isolated from B. halodurans C-125 as described previously (38). Each IS region containing a Tpase gene from the chromosome of strain C-125 was amplified by PCR using the various primer sets. PCR was performed using a thermal cycler 9700 (Perkin-Elmer, Norwalk, Conn.) under the following conditions: 25 cycles of 30 s each at 94°C, 30 s at 58°C, and 1 min at 72°C.
Identification of ISs in C-125 genome.
The regions 300 bp up- and downstream of each of the Tpase genes identified in our previous study (45) were searched for IR sequences by using the GENETYX-Mac program (version 10.0) from Software Development Co., Ltd. (Tokyo, Japan). In the cases in which two Tpase genes overlapped or were located close to each other, the region 300 bp upstream of the first Tpase gene and the region 300 bp downstream of the second Tpase gene were searched for IR sequences in a similar manner. When an IR was found in the region flanking a Tpase gene, the regions adjacent to both IRs (IRR and IRL) were searched for direct repeat (DR) sequences to identify target site duplication. When an IR was not found, the genome sequence of strain C-125 was searched for sequences showing nucleotide sequence similarity to the flanking regions 300 bp upstream and 300 bp downstream of the Tpase gene, using the BLAST2N program (2) to confirm the IS region (28). The copy number of each IS was determined through a homology search of the genome of B. halodurans C-125 using the BLAST2N program in the GenomeGambler system (40).
Nucleotide sequence accession numbers.
The B. halodurans C-125 sequence has been deposited in the DDBJ, EMBL, and GenBank databases with accession numbers AP001507 to AP001520.
RESULTS AND DISCUSSION
Identification and characterization of new IS elements and group II intron in C-125 genome.
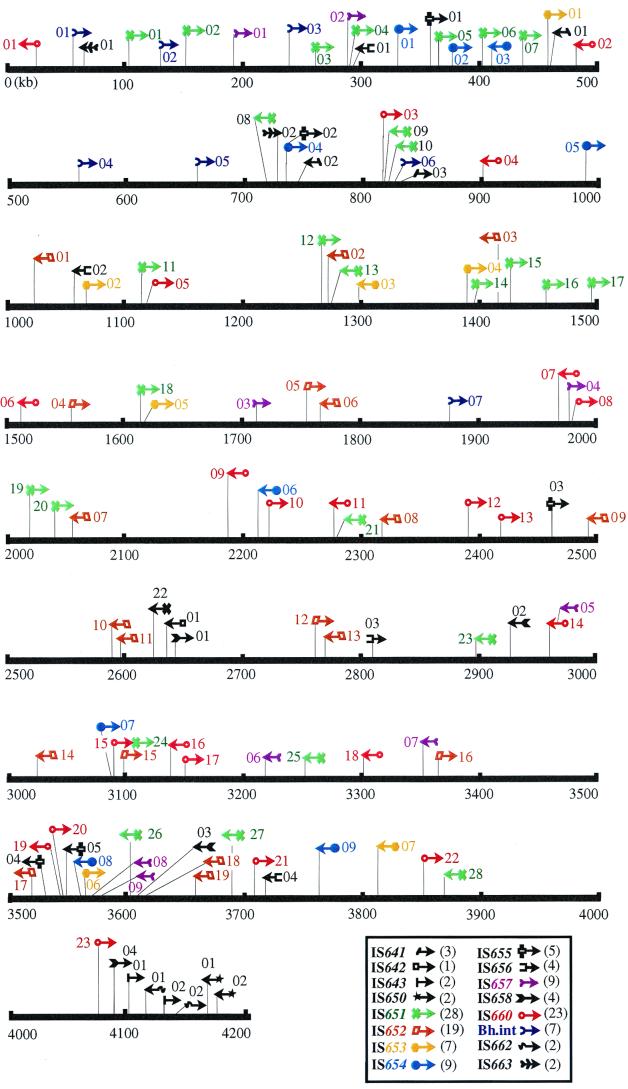
In the previous study (45), we found many kinds of repeated sequences, most of which showed homology with Tpase genes carried by various IS elements. In the present study, we identified and characterized 16 kinds of new elements with or without terminal IRs and with or without target site sequence duplication (TSD), which is a DR. Many of them appear to belong to the known IS families, but a few appeared to belong to new IS families and a group II intron. Members of each of the elements are listed in Table 1. Their locations are shown in Table 1 and Fig. 1.
TABLE 1.
IS elements and a group II intron in the B. halodurans genomea
| IS | Length of TSD (bp) | Direction | IS position in genome | IS formation (bp) |
|---|---|---|---|---|
| IS660-01 | − | 35964–36036 | 1891–end (1963) | |
| − | 36037–36120 | 260–357 | ||
| 36169–36223 | 23–77 | |||
| Bh.Int-01 | None | + | 56387–58269 | 1–end (1883) |
| IS663-01 | − | 58367–58553 | 1–187 | |
| IS651-01 | 8 | + | 108015–109398 | 1–end (1384) |
| Bh.Int-02 | None | + | 130149–132031 | 1–end (1883) |
| IS651-02 | 8 | + | 153091–154475 | 1–end (1384) |
| IS657-01 | 2 | + | 194139–194872 | 1–end (734) |
| Bh.Int-03 | None | + | 242165–244047 | 1–end (1883) |
| IS651-03 | 8 | + | 267837–269220 | 1–end(1384) |
| IS657-02 | 2 | + | 290906–291639 | 1–end (734) |
| IS651-04 | None | + | 291645–293028 | 1–end (1384) |
| IS656-01 | 4 | − | 296015–297572 | 1–end (1558) |
| IS654-01 | 8 | + | 330156–331539 | 1–end(1384) |
| IS655-01 | 3 | + | 356320–357540 | 1–end (1221) |
| IS651-05 | + | 369783–369939 | 1–163 | |
| IS654-02 | 8 | + | 381025–382408 | 1–end (1384) |
| IS651-06 | 8 | + | 408634–410017 | 1–end (1384) |
| IS654-03 | None | − | 415294–416677 | 1–end (1384) |
| IS651-07 | None | + | 443639–445022 | 1–end (1384) |
| IS653-01 | None | + | 461220–463024 | 1–end (1805) |
| IS641-01 | − | 463467–464042 | 569–1159 | |
| IS660-02 | − | 486165–486647 | 1474–end (1963) | |
| 486648–487037 | 1–390 | |||
| Bh.Int-04 | None | + | 565021–566903 | 1–end (1883) |
| Bh.Int-05 | None | + | 662481–664363 | 1–end (1883) |
| IS651-08 | 8 | − | 721388–722771 | 1–end (1384) |
| IS663-02 | None | + | 736789–738768 | 1–end (1980) |
| IS654-04 | + | 743779–743953 | 1–178 | |
| 745176–746383 | 177–end (1384) | |||
| IS655-02 | 3 | + | 743955–745175 | 1–end (1221) |
| IS641-02 | None | − | 746586–747990 | 1–end (1405) |
| IS660-03 | + | 828859–828986 | 32–159 | |
| 828987–829685 | 362–1051 | |||
| 829686–829851 | 1422–587 | |||
| IS651-09 | − | 829916–830032 | 61–178 | |
| IS651-10 | None | − | 830857–832242 | 1–end (1384) |
| Bh.Int-06 | + | 833458–833908 | 1–449 | |
| IS641-03 | 9 | + | 834052–835077 | 1–1026 |
| 835078–835181 | 1302–end (1405) | |||
| IS660-04 | − | 901286–901556 | 1689–end (1963) | |
| 901557–901738 | 1480–1663 | |||
| 901755–902133 | 1–382 | |||
| IS654-05 | 8 | + | 988291–989674 | 1–end (1384) |
| IS652-01 | None | − | 1025486–1026946 | 1–end (1461) |
| IS656-02 | None | − | 1057727–1059284 | 1–end (1558) |
| IS653-02 | None | + | 1069354–1071158 | 1–end (1805) |
| IS651-11 | None | + | 1126321–1127704 | 1–end (1384) |
| IS660-05 | None | + | 1130785–1132747 | 1–end (1963) |
| IS651-12 | 8 | + | 1267250–1268633 | 1–end (1384) |
| IS652-02 | None | − | 1277703–1279163 | 1–end (1461) |
| IS651-13 | − | 1279164–1279214 | 1–51 | |
| IS653-03 | None | − | 1299745–1301549 | 1–end (1805) |
| IS653-04 | None | + | 1391849–1393653 | 1–end (1805) |
| IS651-14 | 8 | + | 1397716–1399099 | 1–end (1384) |
| IS652-03 | 8 | − | 1422289–1423749 | 1–end (1461) |
| IS651-15 | 8 | + | 1433045–1434428 | 1–end (1384) |
| IS651-16 | + | 1465665–1466150 | 1–520 | |
| IS651-17 | 8 | + | 1497488–1498871 | 1–end (1384) |
| IS660-06 | None | − | 1514619–1516581 | 1–end (1963) |
| 1516616–1516781 | 1–167 | |||
| IS652-04 | 8 | + | 1552703–1554163 | 1–end (1461) |
| IS651-18 | 8 | + | 1621997–1623380 | 1–end (1384) |
| IS653-05 | None | + | 1625586–1627390 | 1–end (1805) |
| IS657-03 | 2 | + | 1718440–1719173 | 1–end (734) |
| IS652-05 | 8 | + | 1757990–1759450 | 1–end (1461) |
| IS652-06 | None | − | 1776445–1777905 | 1–end (1461) |
| Bh.Int-07 | + | 1882598–1882735 | 1–139 | |
| IS660-07 | − | 1974095–1974189 | 1867–end (1963) | |
| IS657-04 | 2 | + | 1981014–1981747 | 1–end (734) |
| IS660-08 | + | 1988144–1989106 | 1–962 | |
| IS651-19 | 8 | + | 2019703–2021087 | 1–end (1384) |
| IS651-20 | 8 | + | 2042186–2043569 | 1–end (1384) |
| IS652-07 | 8 | − | 2057902–2059362 | 1–end (1461) |
| IS660-09 | None | − | 2190636–2192598 | 1–end (1963) |
| IS654-06 | 8 | − | 2215773–2217156 | 1–end (1384) |
| IS660-10 | + | 2227035–2227158 | 1840–end (1963) | |
| IS660-11 | − | 2279303–2279682 | 1582–end (1963) | |
| 2279683–2279779 | 1098–1194 | |||
| IS651-21 | − | 2280664–2281124 | 889–1372 | |
| 2281125–2281463 | 1–339 | |||
| IS652-08 | 8 | − | 2324549–2326009 | 1–end (1461) |
| IS660-12 | + | 2393633–2393728 | 1871–end (1963) | |
| IS660-13 | + | 2428054–2428119 | 1897–end (1963) | |
| IS655-03 | 3 | + | 2462716–2463936 | 1–end (1221) |
| IS652-09 | 8 | − | 2496479–2497939 | 1–end (1461) |
| IS652-10 | 8 | − | 2590856–2592317 | 1–end (1461) |
| IS652-11 | 8 | − | 2596787–2598247 | 1–end (1461) |
| IS651-22 | − | 2631772–2631818 | 1–47 | |
| IS642-01 | 2 (TA) | − | 2641522–2642665 | 1–end (1142) |
| IS658-01 | 4 | + | 2645543–2646600 | 1–end (1058) |
| IS652-12 | 8 | + | 2761229–2762689 | 1–end (1461) |
| IS652-13 | None | − | 2771803–2773263 | 1–end (1461) |
| IS656-03 | 4 | + | 2815430–2816987 | 1–end (1558) |
| IS651-23 | None | − | 2899732–2901114 | 1–end (1384) |
| IS658-02 | − | 2930642–2930698 | 649–705 | |
| 2930699–2930798 | 510–609 | |||
| 2930799–2930915 | 1–17 | |||
| IS660-14 | − | 2960602–2962519 | 46–end (1963) | |
| 2963254–2963300 | 1–47 | |||
| IS657-05 | 2 | − | 2962520–2963253 | 1–end (734) |
| IS652-14 | 8 | − | 3035775–3037235 | 1–end (1461) |
| IS654-07 | 8 | + | 3085372–3086755 | 1–end (1384) |
| IS660-15 | + | 3087119–3087363 | 81–328 | |
| 3087364–3087663 | 1605–1942 | |||
| IS651-24 | 8 | + | 3087664–3089047 | 1–end (1384) |
| IS652-15 | 8 | + | 3098551–3100011 | 1–end (1461) |
| IS660-16 | − | 3140937–3140972 | 1–36 | |
| IS660-17 | + | 3152206–3152272 | 10–76 | |
| 3152328–3152403 | 84–159 | |||
| 3152404–3152507 | 1860–end (1963) | |||
| IS657-06 | − | 3227493–3227597 | 232–337 | |
| 3227598–3227654 | 27–83 | |||
| IS651-25 | 8 | − | 3251592–3252975 | 1–end (1384) |
| IS660-18 | − | 3301435–3301917 | 1474–end (1963) | |
| 3305854–3306200 | 75–421 | |||
| IS657-07 | 2 | − | 3352746–3353479 | 1–end (734) |
| IS652-16 | None | + | 3367580–3369040 | 1–end (1461) |
| IS652-17 | 8 | − | 3522324–3523784 | 1–end (1461) |
| IS655-04 | 3 | − | 3529840–3531060 | 1–end (1221) |
| IS660-19 | − | 3548265–3548305 | 1–41 | |
| IS660-20 | + | 3548411–3549000 | 78–670 | |
| 3549013–3549092 | 864–943 | |||
| IS655-05 | 3 | − | 3556078–3557298 | 1–end (1221) |
| IS654-08 | 8 | − | 3569943–3571326 | 1–end (1384) |
| IS653-06 | None | + | 3573033–3574837 | 1–end (1805) |
| IS657-08 | 2 | − | 3576491–3577224 | 1–end (734) |
| IS657-09 | 2 | − | 3587180–3587913 | 1–end (734) |
| IS651-26 | 8 | − | 3606811–3608194 | 1–end (1384) |
| IS658-03 | 4 | − | 3611396–3612453 | 1–end (1058) |
| IS652-18 | None | − | 3616184–3617644 | 1–end (1461) |
| IS652-19 | 8 | − | 3666751–3668211 | 1–end (1461) |
| IS651-27 | 8 | − | 3686646–3688030 | 1–end (1384) |
| IS660-21 | + | 3713536–3713606 | 7–77 | |
| 3713672–3714137 | 78–554 | |||
| 3714138–3714240 | 1861–end (1963) | |||
| IS656-04 | 4 | − | 3735592–3737149 | 1–end (1558) |
| IS654-09 | 8 | − | 3776003–3777386 | 1–end (1384) |
| IS653-07 | None | − | 3824136–3825940 | 1–end (1805) |
| IS660-22 | + | 3856205–3856236 | 4–35 | |
| 3856346–3856432 | 81–166 | |||
| IS651-28 | 8 | − | 3873176–3874558 | 1–end (1384) |
| IS660-23 | + | 4083577–4083650 | 1–77 | |
| 4083718–4083818 | 429–540 | |||
| IS658-04 | 4 | + | 4089048–4090105 | 1–end (1058) |
| IS643-01 | 5 | + | 4105038–4107522 | 1–end (2485) |
| IS662-01 | 4 | − | 4123374–4124939 | 1–end (1566) |
| IS643-02 | + | 4137868–4138646 | 1707–end (2485) | |
| IS662-02 | + | 4144219–4144818 | 1–600 | |
| 4144819–4145300 | 1085–end (1566) | |||
| IS650-01 | − | 4176981–4177118 | 1792–end (1929) | |
| IS650-02 | 2 (TA) | − | 4179133–4181061 | 1–end (1929) |
The DR sequences flanking each intact element member are defined as TSD, in which “none” means that the DR sequences are different in length from those seen in most members of an IS element or are not generated by the insertion event. The partial IS element without a terminal sequence shorter than 100 bp is not basically defined as an IS element in this table.
FIG. 1.
Distribution of IS elements and group II intron in the B. halodurans C-125 genome. Arrows indicate the direction of the ISs, and the number in parentheses is the copy number of each element.
IS elements with IRs that generate TSD.
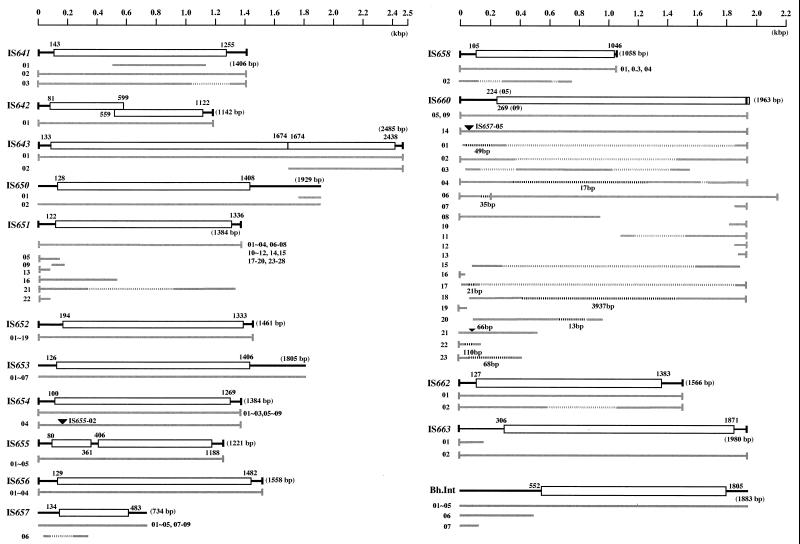
Ten kinds of IS elements were found to have IRs and were flanked by TSD. One of them, at position bp 746586 to 747990 in the genome (Fig. 1; Table 1), has imperfect IRs of 18 bp long, of which the distal 14-bp sequences with an 8-bp palindromic structure match perfectly (Fig. 2). The IS element designated IS641 was found to be flanked by a 9-bp TSD (Fig. 2; Table 2). IS641, 1,405 bp in length, shows 68% identity with the nucleotide sequence of IS4Bsu1 (33) belonging to the IS4 family (24), and the Tpase of IS641 shows 70.3% similarity to that of IS4Bsu1. The DDE motif, which is conserved in most Tpases and other enzymes capable of catalyzing cleavage of DNA strands (14, 29, 35), was found in the Tpase of IS641, i.e., D (124th amino acid [aa]), D (193rd aa), E (293rd aa), and K (300th aa). These findings support the view that IS641 should be categorized as a new member of the IS4 family (Table 2). The genome of strain C-125 has two other copies of IS641 (IS641-01 and IS641-03), with a truncation or deletion in an IS641 segment, respectively (Fig. 3; Table 2).
FIG. 2.
Terminal IRs and TSD of each element identified in the B. halodurans genome. IRs are shown in blue capitals and TSDs are boxed. Red letters indicate the sequence of the B. halodurans genome.
TABLE 2.
New IS elements and group II intron identified in the B. halodurans genome
| IS | Size (bp) | Size of TSDa (bp) | Size of IR (bp) | No. of IS elements
|
Familyc | ||
|---|---|---|---|---|---|---|---|
| Total IS | IS with endsb | Truncated IS | |||||
| IS641 | 1,405 | 9 | 18 | 3 | 2 (0) | 1 | IS4 |
| IS642 | 1,142 | 2 [TA] | 26 | 1 | 1 (0) | 0 | IS630 |
| IS643 | 2,485 | 5 | 64 | 2 | 1 (0) | 1 | IS21 |
| IS650 | 1,929 | 0 | 2 | 1 | 1 | IS650/IS653∗ | |
| IS651 | 1,384 | 8 | 23 | 28 | 22 (5) | 6 | ISL3 |
| IS652 | 1,461 | 8 | 27 | 19 | 19 (6) | 0 | ISL3 |
| IS653 | 1,805 | 0 | 7 | 7 | 0 | IS650/IS653∗ | |
| IS654 | 1,384 | 8 | 31 | 9 | 9 (1) | 0 | IS256 |
| IS655 | 1,221 | 3 | 38 | 5 | 5 (0) | 0 | IS3 |
| IS656 | 1,558 | 4 | 15 | 4 | 4 (1) | 0 | IS656/IS662∗ |
| IS657 | 734 | 2 [TT] | 9 | 8 (0) | 1 | IS200/IS605 | |
| IS658 | 1,058 | 4 | 27 | 4 | 3 (0) | 1 | IS30 |
| IS660 | 1,963 | 0 | 16 | 23 | 6 | 17 | IS1272∗ |
| IS662 | 1,566 | 4 | 15 | 2 | 2 (0) | 0 | IS656/IS662∗ |
| IS663 | 1,980 | 0 | 14 | 2 | 1 | 1 | IS1272∗ |
| Bh.Int | 1,883 | 0 | 7 | 5 | 2 | Group II intron | |
The target site sequence is shown in brackets.
IS elements with two intact ends. Numbers in parentheses show the number of IS elements without a target site duplication.
∗, Proposed new IS families.
FIG. 3.
Structure of each IS element and group II intron identified in the B. halodurans genome. The box shows the Tpase of each element, and the numbers beside each box indicate the position of the Tpase in the element. The gray bars indicate the elements identified in the genome. The gray and black dashed lines indicate deleted and inserted parts, respectively, in the element. The small vertical bar at the end of the element denotes IRs. The black upside-down triangle denotes insertion of another element. The partial IS element without a terminal sequence shorter than 100 bp is not shown.
The IS element at position bp 2641522 to 2642665 (Fig. 1; Table 1) has imperfect IRs that are 26 bp long (Fig. 2). This IS element (1,142 bp in length), designated IS642, was found to be flanked by DRs of a TA sequence (Fig. 2; Table 2). IS642 shows 43.5% identity with the nucleotide sequence of IS630, which duplicates the TA sequence at the target site (30). There are two open reading frames overlapping at position bp 559 to 599 in IS642 (Fig. 3). It is evident that this occurred due to a frameshift mutation, because the first and second open reading frames are both similar to the Tpase of IS630, showing 23.5 and 27.2% similarity, respectively. The DDE motif was found in the Tpase segment encoded by the region straddling the frameshift mutation (data not shown), as in the case of IS630. These results support the view that IS642 is a new member of the IS630 family (Table 2).
In addition, there are eight other new IS elements which carry terminal IRs and generate a TSD: IS643 (2,485 bp; IS21 family [36]), IS651 (1,384 bp; ISL3 family), IS652 (1,461 bp; 43.3% identity to IS651), IS654 (1,384 bp; IS256 family [8]), IS655 (1,221 bp; IS3 family [50]), IS658 (1,058 bp; IS30 family [15]), and IS656 (1,558 bp; 67.7% identity to IS662), and IS662 (1,566 bp). Two ISs (IS656 and IS662) do not show significant similarity to any other IS elements reported to date. These results suggest that IS656 and IS662 can be categorized as members of a new IS family (designated the IS656/IS662 family; Table 2). Note that some intact members of the IS elements described above were found not to be flanked by DRs of a target site sequence (Table 2). This indicates that rearrangements of the genome have occurred through transpositional recombination mediated by IS elements. There exist truncated members of each of the IS elements described above and below (Fig. 3; Table 2), indicating the occurrence of internal rearrangements of the genome, probably through illegitimate recombination.
IS elements with IRs that do not generate TSD.
The IS element designated IS663, with IRs 14 bp long, is present in the C-125 genome (Fig. 2; Table 2). An intact element (1,980 bp) shows 42.2% identity to the nucleotide sequence of IS660 (1,963 bp), with IRs 16 bp long (Fig. 2; Table 2). The putative Tpase of IS663 shows 45.5% similarity to that of IS660, suggesting that these two IS elements are related to each other. IS660 and IS663 show 52.7 and 59.5% identity, respectively, with the nucleotide sequence of an unclassified IS element, IS1272, from Staphylococcus haemolyticus (3). Also, these two ISs (IS660 and IS663) show 42 and 49% identity, respectively, with an unclassified IS element, IS1182 from Staphylococcus aureus (accession no. L43098). These results suggest that IS660 and IS663, as well as IS1272 and IS1182, can be grouped into a new IS family (designated the IS1272 family; Table 2). It is notable that there are many copies of IS660, including various truncated forms, widely distributed throughout the C-125 genome (Fig. 1 and 3), suggesting that IS660 may be the oldest IS element present in the genome and that its wide distribution may have occurred through complicated internal rearrangements of the C-125 genome.
IS elements with no IRs.
Three IS elements, designated IS657, IS650, and IS653, with no IRs were found to be present in the C-125 genome (Fig. 2; Table 2). The first IS element, IS657, was found to be flanked by a 2-bp TSD. IS657 (734 bp) shows 42% identity to the nucleotide sequence of IS605 (10), and the Tpase identified in IS657 is similar to that of IS605, showing 62.5% similarity, although IS605 (1,880 bp) is much longer than IS657. These results support the view that IS657 is a new member of the IS200/IS605 family (5, 10). There exist seven other copies of intact IS657 (IS657-02 to -05 and IS653-07 to -09) and one truncated copy of IS657 (IS657-06) (Table 1; Fig. 2 and 3).
The second IS element, IS650 (1,929 bp), shows 43.9% identity to the nucleotide sequence of the third IS element, IS653 (1,805 bp). A putative Tpase in IS650 shows 78.2% similarity to that of IS653, indicating that these two ISs are closely related to each other. However, these two did not show significant similarity to any other IS elements reported to date, suggesting that they should be categorized as members of a new IS family (designated the IS650/IS653 family; Table 2). There exist six other copies of IS653 (IS653-02 to -07) (Table 1; Fig. 2 and 3).
A group II intron.
Group II introns are catalytic RNAs that function as mobile genetic elements by inserting themselves directly into target sites in double-stranded DNA (1, 31). The element, designated Bh.Int, has no IRs nor TSD (Fig. 2; Table 2). This element (1,883 bp) shows 47.6% identity to the nucleotide sequence of the group II intron of Clostridium difficile (32). The protein coding sequence (CDS) of Bh.Int is similar to the putative reverse transcriptase-maturase-transposase of the group II intron of C. difficile, showing 47.5% similarity. The CDS of Bh.Int also showed significant similarity to group II introns from Sphingomonas aromaticivorans (38.4%) (37) and Pseudomonas putida (25.7%) (accession no. Y18999). Among these putative reverse transcriptase-maturase-transposases, the amino acid sequence GTPQGG is well conserved as a consensus sequence. Thus, IS653 should be categorized as a new member of the group II introns. The C-125 genome contains four other copies of the element (Bh.Int-02 to -05) and two truncated copies of Bh.Int (Bh.Int-06 and -07) (Table 1; Fig. 2 and 3).
The presence of IS family members not identified in genomes of other Bacillus species.
The genome of B. subtilis 168, the entire sequence of which has been determined, has no IS element (27), although a new IS4 family insertion sequence, IS4Bsu1, has just been reported in the case of B. subtilis (natto), which is used as a starter strain for the production of natto (fermented soybeans) (33). IS641 belonging to the IS4 family was also identified in the genome of B. halodurans, which is not taxonomically distant from B. subtilis except for the alkaliphilic phenotype. The IS elements belonging to the families of IS3, IS4, IS6, IS21, IS630, and IS982 and to the group II intron have been reported from other Bacillus strains (Table 3). Interestingly, however, any IS elements belonging to the ISL3, IS256, IS30, IS200, and IS605 families have never been reported from other Bacillus strains to date (Table 3).
TABLE 3.
IS family and group II intron members identified in the genomes of Bacillus species
| IS family | No. of members identified in genomes of:
|
|
|---|---|---|
| B. halodurans strain C-125b | Other Bacillus strainsc | |
| IS3 | 5 (IS655) | 2 |
| IS4 | 3 (IS641) | 12 |
| IS6 | 0 | 4 |
| IS21 | 2 (IS643) | 4 |
| IS30 | 4 (IS658) | 0 |
| IS200/IS605 | 9 (IS657) | 0 |
| IS630 | 1 (IS642) | 1 |
| ISL3 | 47 (IS651; IS652) | 0 |
| IS256 | 9 (IS654) | 0 |
| IS650/IS653a | 9 (IS650; IS653) | 0 |
| IS656/IS662a | 6 (IS656; IS662) | 0 |
| IS1272a | 25 (IS660; IS663) | 0 |
| IS982 | 0 | 2 |
| Group II intron | 7 (Bh.Int) | 2 |
Comparison and phylogenetic analysis of Tpases.
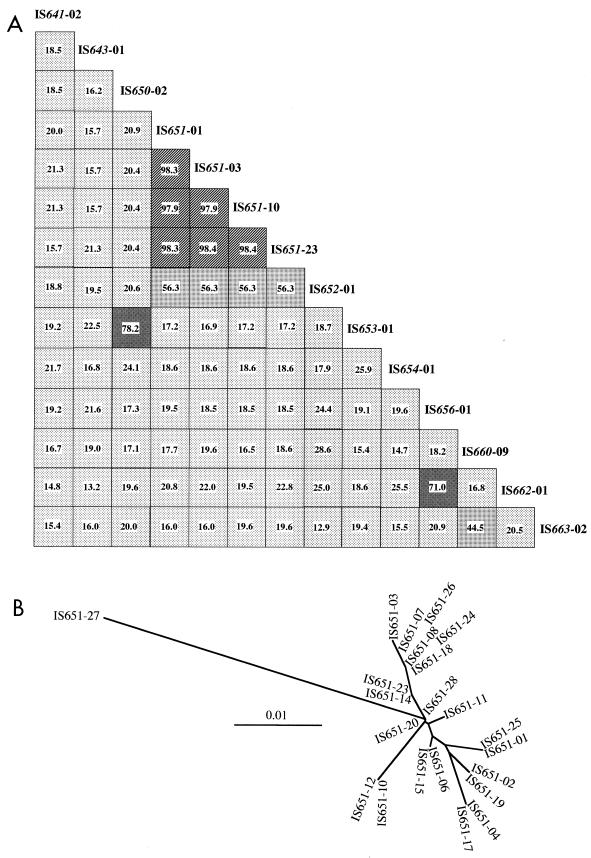
The amino acid sequences of the putative Tpases of the IS elements identified in the C-125 genome were compared. As expected from the nucleotide sequence identity, the Tpase of IS650 showed 78.2% similarity to that of IS653 and the Tpase of IS656 showed 71% similarity to that of IS662 (Fig. 4A). Also, the putative Tpase of IS652 showed relatively high similarity (56.3%) to the Tpases of a series of IS651-related elements and the Tpase of IS660 showed 44.5% similarity to that of IS663 (Fig. 4A). However, the Tpases of the other IS elements (IS641, IS642, IS643, IS654, IS655, IS657, IS658, IS660, and IS663) showed only low similarity to one another, much lower than 28.6%, supporting that these 10 IS elements are derived from a different origin (Fig. 4A). IS651 and IS652 are the most common IS elements in the C-125 genome, as there are 22 copies of the intact form of IS651 and 19 copies of the intact form of IS652 (Tables 1 and 2). The amino acid sequences of the Tpases encoded in the 19 copies of IS652 were identical, except for that of a member (IS652-06) in which a leucine residue was substituted for an isoleucine residue. The amino acid sequences of putative Tpases encoded in the 22 copies of intact IS651, varied, however, with the similarity values ranging from 94.8 to 100% (Fig. 4). The amino acid sequences of all putative Tpases of the 22 copies of intact IS651 were aligned, and a phylogenetic tree was constructed using the NJ algorithm (39). The tree clearly shows relationships among the IS651 members with variety (Fig. 4B).
FIG. 4.
Evolutionary relationships among Tpases in the IS elements identified in the B. halodurans genome. (A) Similarity matrix based on percent similarity among 15 Tpases of IS elements identified in the C-125 genome. The Tpases of the IS elements (IS642, IS655, IS657, and IS658) showed very low similarity to one another and to those of the other IS elements, and therefore they are not shown here. (B) Unrooted phylogenetic tree of the Tpases of the copies of IS651 identified in the C-125 genome. Amino acid sequences of the IS651 Tpases were aligned using the Clustal multiple-alignment program (Clustal X) (49). Sites involving gaps were excluded from all analyses. Consensus sequence segment alignment of the whole region was used to construct a phylogenetic tree for the various IS651 Tpases. A phylogenetic tree was constructed by the neighbor-joining method (39) using the Clustal X program, version 1.64b, and drawn by means of Tree view. Bar = 0.01 Knuc unit.
Alteration of protein-coding regions mediated by IS.
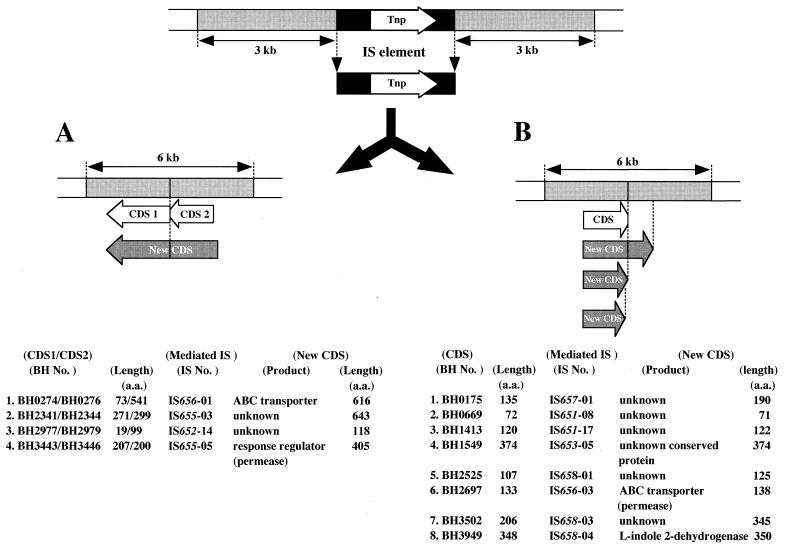
To investigate how protein-coding regions are affected by ISs in the genome, the CDSs in the regions adjacent to each IS were analyzed. The nucleotide sequence of the 3-kb region upstream and that of the 3-kb region downstream of each of all intact IS elements identified in this study were extracted from the entire genome sequence through the ExtremoBase web site (http://www.jamstec.go.jp/jamstec-e/bio/DEEPSTAR/FResearch.html). The 6-kb sequence, from which the IS region was excised (Fig. 5), was searched for CDS by using the BLAST2X program. Although most of the IS elements widely distributed throughout the genome were inserted in noncoding regions, at least 12 CDSs were likely affected by the insertion of seven kinds of IS elements (IS651, IS652, IS653, IS655, IS656, IS657, and IS658). In four examples shown in Fig. 5A, two CDSs (CDS 1 and CDS 2) on both sides of a Tpase gene were identified as originating from one CDS, suggesting that it had been divided into two parts by IS insertion. The gene encoding an ATP-binding cassette transporter (permease) consisting of 616 aa seemed to be divided into two genes encoding BH0274 (73 aa) and BH0276 (541 aa) by IS insertion. Similarly, the gene for a response regulator (a member of the AraC/XylS family) seemed to be divided into two genes encoding BH3443 (207 aa) and BH3446 (200 aa). In addition, two other genes of unknown function were also each presumably divided into two genes, BH2341 (271 aa) and BH2344 (299 aa) in the first case and BH2977 (19 aa) and BH2979 (99 aa) in the second case (Fig. 5A). On the other hand, eight CDSs located upstream of Tpase also seemed to be affected by IS, as shown in Fig. 5B. Six CDSs (BH0175, BH1413, BH2525, BH2697, BH3502, and BH3949) likely occurred by truncation of the original CDS by IS insertion. Two CDSs, BH2697 and BH3949, have been annotated as an ATP-binding cassette transporter (permease) and l-indole 2-dehydrogenase, respectively (45), but the functions of the other four CDSs are still unknown. The gene encoding BH0669, of unknown function, seems to have become 3 bases longer than the original one through the insertion of IS651-08, and in the case of BH1549, the size of the CDS (374 aa) was accidentally the same as the original one in spite of the insertion of IS653-05. Thus, among 89 intact ISs of 16 kinds identified in this study, 12 intact IS elements of 7 kinds consequently seem to have affected a CDS by their insertion. However, it is still unknown how these ISs respond to external signals and how IS insertions are associated with changes in the phenotype of B. halodurans.
FIG. 5.
Pattern of insertion of IS elements into protein-coding regions of the genome. (A) The case in which CDS 1 and CDS 2 identified on both sides of the Tpase coding region in the IS element merge as one CDS upon elimination of the IS element. (B) The case in which an alternation occurred in the C-terminal region of the CDS upon insertion of the IS element.
Changes in the C-125 genome that occurred during the course of subculture for 17 years.
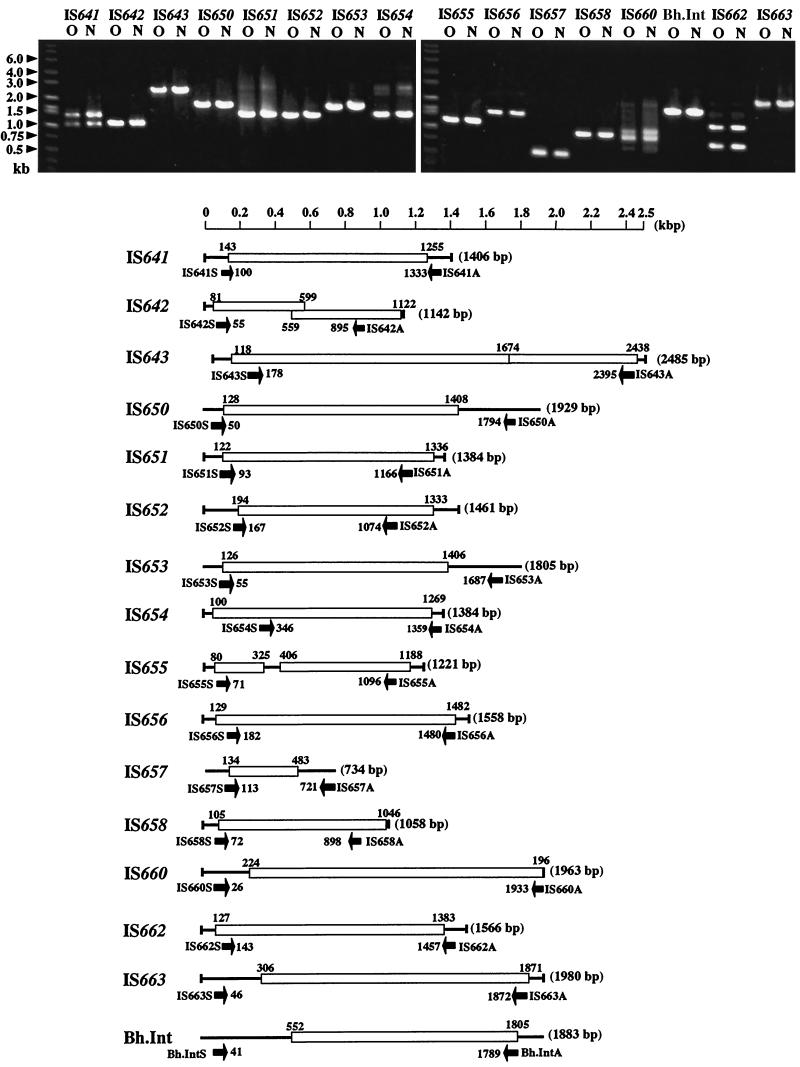
Strain C-125 was isolated from a soil sample by using an alkaline culture medium in 1970 (42), and it was kept in the form of spores on a plate until 1977. This strain was characterized as a β-galactosidase producer and identified as a member of the genus Bacillus in 1977 (20) after being subcultured several times for experiments (21). Thereafter, the strain was kept in the form of spores on a plate again until 1983. In the autumn of 1983, the strain was used in a study involving screening for xylanase production, and then it was lyophilized in an ampule and stored for the purposes of obtaining a patent for alkaline xylanase on 13 November 1983 (17, 18). Thereafter, during the next 2 decades, it was quite often used in various experiments as an enzyme producer and as a standard strain for studies on the mechanisms of adaptation to alkaline environments. Now, we are very intrigued by the question of what kind of changes mediated by IS elements occurred in the genome, comparing the current strain (C-125-00) and the strain lyophilized in 1983 (C-125-83) because strain C-125 has been subcultured alternately under alkaline and neutral conditions during the past 2 decades. Therefore, we examined the pattern of amplification of IS elements from the genome, comparing the two strains, C-125-83 and C-125-00, by PCR using the primer sets shown in Fig. 6. All IS elements identified were amplified from the C-125-83 genome and the C-125-00 genome (Fig. 6), showing exactly the same amplification pattern in both cases, suggesting that no IS element except for indigenous ones in the C-125-83 genome had transposed in the genome during the past 17 years.
FIG. 6.
Comparison of the IS elements amplified by PCR from the chromosomes of the strains C-125-83 and C-125-00. The primer sets used are shown by short solid arrows. Lanes O, strain C-125-83; lanes N, strain C-125-00. The positions of molecular size (in kilobases) markers are on the left.
We designed appropriate site-specific primer sets for IS elements localized in each position in the genome (Table 1) and compared each PCR fragment from the genomes of the two strains (C-125-83 and C-125-00) to investigate what kind of internal rearrangement in the genome had occurred through the action of IS elements. In addition, the patterns of digestion of chromosomal DNA from these two strains with various restriction endonucleases were also compared to check whether any changes had occurred in the genome during the 17-year period of subculture. All IS elements located in noncoding regions of the genome were amplified by PCR with exactly the same pattern between strains C-125-83 and C-125-00 for comparison of the DNA fragments amplified from the two strains using primers specific for each of 11 IS element members (IS641-03, IS643-01, IS651-10, IS652-02, IS653-01, IS654-03, IS655-04, IS656-02, IS657-05, IS660-05, and Bh.Int-03) (data not shown). Also, the amplification patterns of 12 IS elements inserted in CDSs (IS652-14, IS655-03, IS655-05, IS656-01, IS656-03, IS651-08, IS651-17, IS653-05, IS657-01, IS658-01, IS658-03, and IS658-04) were the same between strains C-125-83 and C-125-00. Furthermore, there was no difference between the two strains in terms of the pattern of digestion of chromosomal DNA comparing fragments in the large molecular size range, from 48.5 to 533.5 kb (AscI and I-CeuI), or comparing fragments in the small molecular size range, from 9.42 to 97 kb (PacI, SmaI, BssHII, and SwaI) (data not shown). These results demonstrate that the insertion of IS elements into CDSs and noncoding regions in the genome occurred at least before 1983 and presumably before 1970, when B. halodurans C-125 was isolated because this strain had been kept in the form of spores on a plate, as mentioned above.
The B. halodurans genome contains 120 IS elements, and 91 of them still in the intact form seem to have the potential to transpose themselves into the genome of their host or the genome of another strain. However, there is no sign of transposition of IS in the genome of strain C-125 during the past 17-year period of subculture in the laboratory with the cells grown in Horikoshi II medium under neutral or alkaline conditions. This indicates that the genome of B. halodurans C-125 is quite stable. Therefore, it is of interest to determine when the IS elements jump in the genome and what triggers their transposition. As mentioned above, we have a specific interest in how the behavior of ISs and internal rearrangement in the genome affects enzyme productivity and the stability of enzyme production, especially when systematic breeding of the strain is attempted for industrial applications. As the first step to answer the above questions, we are now looking for the trigger of the transposition of IS elements.
ACKNOWLEDGMENTS
We are grateful to K. Horikoshi for supplying an ampule of the lyophilized strain, B. halodurans C-125. We thank R. Sasaki, H. Oida, M. Tsudome, and H. Uchiyama for their technical assistance.
REFERENCES
- 1.Abarca F M, Toro N. Group II introns in the bacterial world. Mol Microbiol. 2000;38:917–926. doi: 10.1046/j.1365-2958.2000.02197.x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer G L, Thanassi J A, Niemeyer D M, Pucci M J. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1996;40:924–929. doi: 10.1128/aac.40.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. [Google Scholar]
- 5.Beuzon C R, Casadesus J. Conserved structure of IS200 elements in Salmonella. Nucleic Acids Res. 1997;25:1355–1361. doi: 10.1093/nar/25.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Bolotin A, Mauger S, Malarme K, Ehrlich S D, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. [PubMed] [Google Scholar]
- 8.Byrne M E, Rouch D A, Skurray R A. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene. 1989;81:361–367. doi: 10.1016/0378-1119(89)90197-2. [DOI] [PubMed] [Google Scholar]
- 9.Camarena L, Poggio S, Campos A, Bastarrachea F, Osorio A. An IS4 insertion at the glnA control region of Escherichia coli creates a new promoter by providing the −35 region of its 3′-end. Plasmid. 1998;39:41–47. doi: 10.1006/plas.1997.1318. [DOI] [PubMed] [Google Scholar]
- 10.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole S T, Brosch R, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 12.Coucheron D H. An Acetobacter xylinum insertion sequence element associated with inactivation of cellulose production. J Bacteriol. 1991;173:5723–5731. doi: 10.1128/jb.173.18.5723-5731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig N L. Unity in transposition reactions. Science. 1995;270:253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- 14.Craig N L. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 15.Dalrymple B, Caspers P, Arber W. Nucleotide sequence of the prokaryotic mobile genetic element IS30. EMBO J. 1984;3:2145–2149. doi: 10.1002/j.1460-2075.1984.tb02104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall B G. Activation of the bgl operon by adaptive mutation. Mol Biol Evol. 1998;15:1–5. doi: 10.1093/oxfordjournals.molbev.a025842. [DOI] [PubMed] [Google Scholar]
- 17.Honda H, Kudo T, Horikoshi K. Molecular cloning and expression of the xylanase gene of alkaliphilic Bacillus sp. strain C-125 in E. coli. J Bacteriol. 1985;161:784–785. doi: 10.1128/jb.161.2.784-785.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honda H, Kudo T, Ikura Y, Horikoshi K. Two types of xylanases of alkalophilic Bacillus sp. no. C-125. Can J Microbiol. 1985;31:538–542. [Google Scholar]
- 19.Horikoshi K. Alkaliphiles: some applications of their products for biotechnology. Microbiol Mol Biol Rev. 1999;63:735–750. doi: 10.1128/mmbr.63.4.735-750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikura Y, Horikoshi K. Cell free protein synthesizing system of alkalophilic Bacillus no. A-59. Agric Biol Chem. 1978;42:753–756. [Google Scholar]
- 21.Ikura Y, Horikoshi K. Isolation and some properties of β-galactosidase producing bacteria. Agric Biol Chem. 1979;43:85–88. [Google Scholar]
- 22.Kallastu A, Horak R, Kivisaar M. Identification and characterization of IS1411, a new insertion sequence which causes transcriptional activation of the phenol degradation genes in Pseudomonas putida. J Bacteriol. 1998;180:5306–5312. doi: 10.1128/jb.180.20.5306-5312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 24.Klaer R, Kuhn S, Tillmann E, Fritz H J, Starlinger P. The sequence of IS4. Mol Gen Genet. 1981;181:169–175. doi: 10.1007/BF00268423. [DOI] [PubMed] [Google Scholar]
- 25.Kondo K, Horinouchi S. Characterization of an insertion sequence, IS12528, from Gluconobacter suboxydans. Appl Environ Microbiol. 1997;63:1139–1142. doi: 10.1128/aem.63.3.1139-1142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondo K, Horinouchi S. A new insertion sequence IS1452 from Acetobacter pasteurianus. Microbiology. 1997;143:539–546. doi: 10.1099/00221287-143-2-539. [DOI] [PubMed] [Google Scholar]
- 27.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 28.Lewis L A, Lewis D, Persaud V, Gopaul S, Turner B. Transposition of IS2 into the hemB gene of Escherichia coli K-12. J Bacteriol. 1994;176:2114–2120. doi: 10.1128/jb.176.7.2114-2120.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsunami S, Otsubo H, Maeda Y, Otsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987;196:445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- 31.Michel F, Ferat J L. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 32.Mullant P, Pallen M, Wilkins M, Stephen J R, Tabaqchali S. A group II intron in a conjugative transposon from the gram positive bacterium Clostridium difficile. Gene. 1996;174:145–150. doi: 10.1016/0378-1119(96)00511-2. [DOI] [PubMed] [Google Scholar]
- 33.Nagai T, Tran L S P, Inatsu Y, Itho Y. A new IS4 family insertion sequence, IS4Bsu1, responsible for genetic instability of poly-γ-glutamic acid production in Bacillus subtilis. J Bacteriol. 2000;182:2387–2392. doi: 10.1128/jb.182.9.2387-2392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakasone K, Masui N, Takaki Y, Sasaki R, Maeno G, Sakiyama T, Hirama C, Fuji F, Takami H. Characterization and comparative study of the rrn operons of alkaliphilic Bacillus halodurans C-125. Extremophiles. 2000;4:209–214. doi: 10.1007/pl00010713. [DOI] [PubMed] [Google Scholar]
- 35.Plasterk R H. Molecular mechanisms of transposition and its control. Cell. 1993;74:781–786. doi: 10.1016/0092-8674(93)90458-3. [DOI] [PubMed] [Google Scholar]
- 36.Reimmann C, Moore R, Little S, Savioz A, Willetts N S, Haas D. Genetic structure, function and regulation of the transposable element IS21. Mol Gen Genet. 1989;215:416–424. doi: 10.1007/BF00427038. [DOI] [PubMed] [Google Scholar]
- 37.Romine M F, Stillwell L C, Wong K-K, Thurston S J, Sisk E C, Sensen C, Gaasterland T, Fredrikson J K, Saffer J D. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito H, Miura K. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 39.Saitou N, Nei M. A neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;44:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Sakiyama T, Takami H, Ogasawara N, Kuhara S, Doga K, Ohyama A, Horikoshi K. An automated system for genome analysis to support microbial whole-genome shotgun sequencing. Biosci Biotechnol Biochem. 2000;64:670–673. doi: 10.1271/bbb.64.670. [DOI] [PubMed] [Google Scholar]
- 41.Salyers A A, Shoemaker N B, Stevens A M, Li L Y. Conjugate transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takami H, Kobayashi T, Aono R, Horikoshi K. Molecular cloning, nucleotide sequence and expression of the structural gene for a thermostable alkaline protease from Bacillus sp. no. AH-101. Appl Microbiol Biotechnol. 1992;38:101–108. doi: 10.1007/BF00169427. [DOI] [PubMed] [Google Scholar]
- 43.Takami H, Nakasone K, Hirama C, Takaki Y, Masui N, Fuji F, Nakamura Y, Inoue A. An improved physical and genetic map of the genome of alkaliphilic Bacillus sp. C-125. Extremophiles. 1999;3:21–28. doi: 10.1007/s007920050095. [DOI] [PubMed] [Google Scholar]
- 44.Takami H, Horikoshi K. Reidentification of facultatively alkaliphilic Bacillus sp. C-125 to Bacillus halodurans. Biosci Biotechnol Biochem. 1999;63:943–945. doi: 10.1271/bbb.63.943. [DOI] [PubMed] [Google Scholar]
- 45.Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, Kuhara S, Horikoshi K. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takami H, Horikoshi K. Analysis of the genome of an alkaliphilic Bacillus strain from an industrial point of view. Extremophiles. 2000;4:99–108. doi: 10.1007/s007920050143. [DOI] [PubMed] [Google Scholar]
- 47.Takemura H, Horinouchi S, Beppu T. Novel insertion sequence IS1380 from Acetobacter pasteurianus is involved in loss of ethanol-oxidizing ability. J Bacteriol. 1991;173:7070–7076. doi: 10.1128/jb.173.22.7070-7076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan H M. Bacterial catabolic transposon. Appl Microbiol Biotechnol. 1999;51:1–12. doi: 10.1007/s002530051356. [DOI] [PubMed] [Google Scholar]
- 49.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4674–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Timmerman K P, Tu C P. Complete sequence of IS3. Nucleic Acids Res. 1985;13:2127–2139. doi: 10.1093/nar/13.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White O, Eisen J A, Heodelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]