Abstract
Dichelobacter nodosus is the essential causative agent of footrot in sheep. The major D. nodosus-encoded virulence factors that have been implicated in the disease are type IV fimbriae and extracellular proteases. To examine the role of the fimbriae in virulence, allelic exchange was used to insertionally inactivate the fimA gene, which encodes the fimbrial subunit protein, from the virulent type G D. nodosus strain VCS1703A. Detailed analysis of two independently derived fimA mutants revealed that they no longer produced the fimbrial subunit protein or intact fimbriae and did not exhibit twitching motility. In addition, these mutants were no longer capable of undergoing natural transformation and did not secrete wild-type levels of extracellular proteases. These effects were not due to polar effects on the downstream fimB gene because insertionally inactivated fimB mutants were not defective in any of these phenotypic tests. Virulence testing of the mutants in a sheep pen trial conducted under controlled environmental conditions showed that the fimA mutants were avirulent, providing evidence that the fimA gene is an essential D. nodosus virulence gene. These studies represent the first time that molecular genetics has been used to determine the role of virulence genes in this slow growing anaerobic bacterium.
Footrot is a highly contagious disease of the feet of sheep and is characterized by the separation of the keratinous hoof from the underlying epidermal tissue, resulting in severe lameness and loss of body condition (11, 39). The consequences of the disease are very significant for the wool and sheep meat industries, and footrot is among the most significant ovine bacterial diseases, causing economic losses in most producer countries. The disease is dependent on a mixed bacterial infection, but the essential causative agent is Dichelobacter nodosus, a slow-growing, anaerobic, gram-negative rod (2, 37). D. nodosus exhibits a spectrum of virulence ranging from virulent strains, which lead to severe underrunning of the horn of the hoof, to benign strains, which cause a self-limiting interdigital dermatitis (37).
Little is known about the pathogenesis of ovine footrot, although the polar type IV fimbriae (12) and extracellular proteases (23) of D. nodosus have been traditionally considered virulence factors (2). In addition, the vap and vrl genomic islands have been shown to be preferentially associated with virulent isolates (2). Analysis of the role that these proposed virulence factors play in the disease process has been hampered by the lack of a genetic system in D. nodosus. However, we recently reported the successful transformation of several D. nodosus strains (22). In these experiments, a tetracycline resistance gene, tet(M), which was present on a suicide plasmid, was inserted into the chromosome by double-reciprocal crossover events. These studies have provided the essential tools required to enable the use of reverse genetics to examine the pathogenic role of the putative virulence factors of D. nodosus.
The fimbriae of D. nodosus are classified as type IV because of their highly conserved amino-terminal region, polar location, association with twitching motility, and the presence of an N-methylphenylalanine residue as the N-terminal amino acid (41). D. nodosus fimbriae are highly immunogenic, with agglutination reactions involving the fimbrial antigens providing the basis of the classification of D. nodosus into nine major serogroups designated A to I (4). Vaccination of sheep with whole cells of D. nodosus or with purified fimbriae protects against the disease, although this protection is serogroup specific (8, 12, 13). Multivalent recombinant fimbrial vaccines have been prepared by overexpression of each of the nine fimbrial subunit genes in Pseudomonas aeruginosa (9). However, antigenic competition has limited the application of these vaccines (20, 21, 33) so that the most effective strategies are based on univalent and divalent formulations.
The fimA genes encoding the major fimbrial subunit from each of the nine major serogroups have been sequenced, which has allowed the division of D. nodosus isolates into two major classes based on the genetic organisation of the fimbrial gene region (18, 28). In class I strains (serogroups A, B, C, E, F, G, and I), the fimA gene is followed by fimB, which encodes a potential 29.5-kDa membrane protein of unknown function but which is postulated to play a role in the export of fimbrial subunits to the cell surface (18). Strains of serogroups D and H belong to class II and have three additional genes downstream of fimA, namely, fimC, fimD, and fimZ (18). The fimD gene is postulated to be a functional homologue of fimB (18), fimC has sequence similarity to traX from the F plasmid (14), and fimZ may represent a redundant fimbrial subunit (18).
In these studies we decided to determine the role of fimbriae in the disease process by using allelic exchange to disrupt the fimA and fimB genes of a class I strain and by examining the virulence of the resultant mutants in a sheep virulence trial. The results showed that the fimA gene was essential for the production of type IV fimbriae and twitching motility and was also required for natural transformation and protease secretion. When tested in sheep, the fimA mutants were avirulent.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
All D. nodosus strains were derivatives of the type G strain VCS1703A and were routinely grown in a Coy anaerobic chamber (Coy Laboratory Products Inc.) in an atmosphere of 10% H2, 10% CO2, and 80% N2 on Eugon (BBL) yeast extract (EYE) agar with 5% horse blood, with the addition of 1 μg of tetracycline or erythromycin per ml for the selection of transformants, or in EYE broth as described previously (22). D. nodosus strains for use in the sheep virulence trial were grown on hoof agar (42). Escherichia coli strains were derivatives of DH5α (Bethesda Research Laboratories) and were grown in 2YT medium (35).
Transformation of D. nodosus cells.
Overnight 150-ml broth cultures of the required D. nodosus strain were harvested by centrifugation at 6,000 × g for 10 min, and the cell pellet was resuspended in 1 ml of fresh EYE broth. Aliquots (100 μl) of this cell suspension were then mixed with 5 μg of plasmid DNA in Tris-EDTA buffer (35) and left at room temperature in an anaerobic environment for 4 h. Two milliliters of EYE broth was then added to the mixture, and the culture was incubated anaerobically overnight at 37°C. The cultures were then plated onto EYE blood agar containing the appropriate antibiotic to select for transformants or onto EYE blood agar without antibiotics to check viability. The plates were incubated at 37°C in an anaerobic environment for 7 days.
Molecular methods.
Unless otherwise stated, molecular techniques were performed using standard procedures (35). Capillary PCR analysis (22) was used to confirm that the putative transformants were D. nodosus and that they carried the tet(M) gene. DNA sequencing was performed using an ABI 373A automated sequencing apparatus (Applied Biosystems).
Southern hybridization.
Southern hybridization was performed as described previously (22). In addition to the 16S rRNA-and tet(M)-specific probes, probes specific for fimA and fimB were used. These probes were the PCR products of primers 4142 and 5013 and primers 4944 and 4143, respectively (Table 1), using chromosomal DNA from strain VCS1703A as the template.
TABLE 1.
Synthetic oligonucleotides used in this study
| Primer | Nucleotide sequence | Location | Use |
|---|---|---|---|
| 4142 | 5′-CGCGAATCGTCACTATTTCTTT-3′ | aroA | PCR |
| 4143 | 5′-GGCAGTGATGGAGATTGTGATG-3′ | clpB | PCR |
| 4944 | 5′-TCACTTTTTGCTTTCGCTTGTC-3′ | fimB | PCR |
| 5013 | 5′-AAAGGGATACCAAACGAAAAAG-3′ | fimB | PCR |
| 5772 | 5′-GTCATCATCTCCTTGCTGTTC-3′ | fimB | PCR |
| 7379 | 5′-GCTGGTCAAGTTTGCGCGGC-3′ | bprV | RT-PCR |
| 7414 | 5′-AATGGCTTTAGTGGTTTGTGC-3′ | aprV2 | RT-PCR |
| 13240 | 5′-AGCAGTTCCCTTACCATATTC-3′ | fimA | RT-PCR |
| 13241 | 5′-AAAAATCCGTATCGCTGACAAC-3′ | fimA | RT-PCR |
| 13242 | 5′-AGCCACGAACAGCAAGGAGATG-3′ | fimB | RT-PCR |
| 14760 | 5′-TTGTTGTTGCCGTTGTTGAT-3′ | aprV2 | RT-PCR |
| 15202 | 5′-AATAAAGGCGGAAAGTTGAATG-3′ | fimB | RT-PCR |
| 15236 | 5′-GTGATGAGCTCGGAGGATTTA-3′ | aprV5 | RT-PCR |
| 15237 | 5′-AATTTAATGCTGTTATTAGCG-3′ | aprV2 | RT-PCR |
| 15238 | 5′-CCCTGCGGTCGCTTCTATAAACT-3′ | bprV | RT-PCR |
RT-PCR.
D. nodosus cells were grown in EYE broth, and RNA was extracted using TRIzol (Life Technologies) according to the manufacturer's instructions. Reverse transcriptase (RT) reactions were performed at 42°C for 1 h on 5 to 10 μg of total RNA in 20-μl incubation mixtures that contained 5 mM MgCl2, 1× RT buffer, 1 mM each deoxynucleoside triphosphate, 1 U of RNasin (Promega), 15 U of avian myeloblastosis virus RT (Promega), and 3 μM appropriate oligonucleotide primer (Table 1). cDNA was amplified in 25-μl PCRs using a standard protocol and 5 μl of the RT reaction as the template. PCR products were analyzed on 1% agarose gels and sequenced to confirm that they were derived from the correct genes.
Immunoblot analysis.
Whole-cell extracts of D. nodosus strains were prepared by removing an EYE blood agar culture of each strain into 1 ml of phosphate-buffered saline (PBS). The cell suspensions were then centrifuged at 12,000 × g for 5 min, and the cell pellet was resuspended in 200 μl of gel loading buffer and boiled for 5 min. Aliquots (5 μl) were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% acrylamide), and the gels were electroblotted to nitrocellulose using a Mini Protean II apparatus (Bio-Rad). The blots were developed using serogroup G-specific antifimbrial antiserum raised in rabbits, sheep anti-rabbit horseradish peroxidase conjugate, and Renaissance Western blot chemiluminesence reagent (NEN Life Sciences) according to the manufacturer's instructions.
Electron microscopy.
Three-day EYE blood agar cultures of each D. nodosus strain were resuspended in 1 ml of PBS. Cell samples were attached to carbon-coated copper-rhodium grids, negatively stained with phosphotungstic acid, and examined using a JEOL JEM 100S transmission electron microscope.
Twitching motility assays.
Twitching motility assays were performed on EYE containing 1% agar as previously described (5). The cells were stab inoculated to the bottom of the petri dish using a straight wire, and the plates were incubated anaerobically at 37°C for 3 to 5 days. The medium was then compressed by placing paper towels on top of the agar, under pressure, for 30 min. The paper was then removed, and the plates were stained with 0.2% (wt/vol) Coomassie brilliant blue and then destained in 7% (vol/vol) acetic acid–33% (vol/vol) methanol. Cultures that exhibited twitching motility showed large stained zones of growth emanating from the point of inoculation.
Protease assays.
All D. nodosus strains were initially tested by standard diagnostic methods involving growth and elastase production on elastin agar plates (38) and the determination of protease stability using the gelatin-gel assay (30). Total protease activity was subsequently determined using azocasein (Sigma) as the substrate. Briefly, 200 μl of a 44-h culture supernatant or periplasmic extract was mixed with 200 μl of assay buffer (0.02 M Tris-HCl, 5 mM CaCl2, 0.2 M NaCl [pH 8.0]) and 400 μl of 6% (wt/vol) azocasein. A 200-μl sample was withdrawn immediately and mixed with 1 ml of 5% trichloroacetic acid (TCA), this sample served as the reaction blank. The remainder of the sample was incubated at 37°C for 8 h, at which time a further 200-μl sample was removed and mixed with 1 ml of TCA. The samples were centrifuged at 12,000 × g for 5 min, and the absorbance of the supernatant at 334 nm was determined in a Varian DMS 100S UV-visible spectrometer. Standard curves were prepared by incubating elastase (Sigma) with azocasein for several days to allow complete digestion. The resultant mixture was precipitated with TCA in the same proportions as above and treated as a 100% hydrolysate. Dilutions of this mixture in TCA enabled a standard curve to be constructed. One unit of protease activity is defined as the amount of enzyme required to digest 1 μg of azocasein in 1 min.
Periplasmic extracts were prepared by washing the cells from a 150-ml broth culture (44 h) in 10 mM Tris-HCl (pH 8.0) after centrifugation at 6,000 × g for 10 min at room temperature. The cells were resuspended in 30 ml of 20% sucrose–30 mM Tris- HCl (pH 8.0)–1 mM sodium EDTA, incubated at room temperature for 10 min, and then centrifuged as before. The cell pellet was resuspended in 30 ml of ice-cold deionized water, incubated on ice for 10 min, and centrifuged at 6,000 × g for 10 min at 4°C, and the supernatant containing the periplasmic proteins was then removed.
Virulence testing in sheep.
Six-month-old Merino sheep that were free of footrot were obtained from the University of Sydney farm at Marulan in the southern highlands of New South Wales, Australia. Each sheep was identified by a numbered ear tag. The sheep were randomly allocated into five groups of 10 and were housed in an animal house on concrete floors, each group in a separate room maintained at 22°C. These experiments were carried out in a PC2 containment facility in accordance with the guidelines of the Australian Genetic Manipulation Advisory Committee and the Elizabeth Macarthur Agricultural Institute Animal Ethics Committee. The sheep were fed lucerne hay and oats; water was provided ad libitum. The groups of 10 animals were challenged blind with wild-type strain VCS1703A, fimA mutants JIR3727 and JIR3728, and plain agar (negative control) as previously described (11). Briefly, the feet of the animals were predisposed to infection by keeping them on wet foam mats for 4 days before the challenge, to facilitate maceration of the skin. All animals were sampled for D. nodosus with a swab stick applied to the interdigital skin prior to challenge. The sheep were then challenged by applying 4-day-old cultures of the each strain on plugs of 2% hoof agar to the interdigital skin and holding them in place with bandages for 4 days. Each plug contained 8.4 × 105 to 9.5 × 105 CFU of D. nodosus or, for the negative control, uninoculated agar. The mats were removed from the floor 1 week after the start of the challenge, and the animals were again sampled for D. nodosus.
All animals were examined, and their feet were scored for footrot lesions at the start of the trial and then at weekly intervals. A standard lesion scoring method was used (46); this method was a modification of an earlier protocol (10). The total weighted foot score (TWFS) was used to provide an unambiguous overall score for the animal, a score that included information from each of the four feet. Blood samples were collected from the jugular vein of sheep at the start of the trial and each time the feet were scored. The serum was separated from the blood and stored at −20°C until required for enzyme-linked immunosorbent assay (ELISA).
ELISA for Omp and FimA.
Serum samples taken from sheep during the course of the trial were tested for D. nodosus-specific antibodies. The Omp antigen (44) and fimbrial antigen (47) ELISAs were performed essentially as previously described.
RESULTS
The fimA gene is essential for the production of type IV fimbriae, twitching motility, natural transformation, and protease secretion.
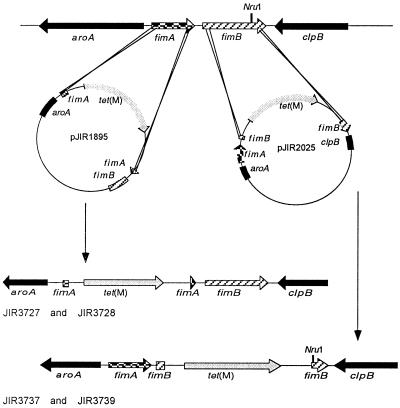
Previous studies showed that the tet(M) gene conferred tetracycline resistance in D. nodosus and could be used to select the progeny of double-crossover events (22). Insertional inactivation with tet(M) was therefore the method of choice for the isolation of fimA mutants. To construct the fimA suicide plasmid pJIR1895 (Fig. 1), an 800-bp fragment containing the first 120 nucleotides of fimA and part of aroA was cloned into pBluescript SKII. A 3.1-kb EcoRI fragment from pVB101 that contained the tet(M) cassette (22) was then added, and a 750-bp fragment containing the last 150 bp of fimA and the first 600 bp of fimB was cloned downstream of tet(M), resulting in a 230-bp deletion within fimA, at the site where tet(M) was inserted. The suicide vector therefore had 800 bp of D. nodosus DNA located upstream of tet(M) and 750 bp downstream of tet(M), sufficient for homologous recombination to occur.
FIG. 1.
Diagrammatic representation of the construction of the fimA and fimB mutants of D. nodosus strain VCS1703A by homologous recombination.
The virulent type G strain VCS1703A was chosen for use in these studies because it was naturally transformable. In fact, in experiments designed to optimize the transformation of D. nodosus, it was determined that all of the D. nodosus strains that were previously thought to be transformable by electroporation were naturally transformable (R. Kennan and J. Rood, unpublished results). VCS1703A, isolated from an outbreak of virulent ovine footrot, was fimbriated and had all of the properties normally associated with virulent isolates of D. nodosus (34) in that it was elastase positive after 7 to 10 days of incubation on elastin agar and was positive in a gelatin-gel protease stability test. In addition, in preliminary pen virulence trials it was shown to produce virulent footrot in sheep (data not shown).
Transformation of strain VCS1703A with the fimA suicide plasmid pJIR1895 produced several tetracycline-resistant colonies. These derivatives were confirmed as D. nodosus by use of a species-specific 16S rRNA gene PCR test (24) and shown by PCR to contain the tet(M) gene. Two independently derived mutants JIR3727 and JIR3728 were selected for further characterization.
Genomic DNA was isolated from VCS1703A, JIR3727, and JIR3728, digested with NruI, and examined in Southern hybridization experiments using fimA- and tet(M)-specific probes. The results showed that in the fimA mutants the band that hybridized with the fimA probe was approximately 3 kb larger than that which hybridized from VCS1703A. As expected, this band also hybridized with the tet(M) probe, which did not hybridize to the wild-type strain (data not shown). Based on these results, it was concluded that JIR3727 and JIR3728 were chromosomal fimAΩtet(M) mutants derived from allelic exchange with the insertionally inactivated suicide vector pJIR1895.
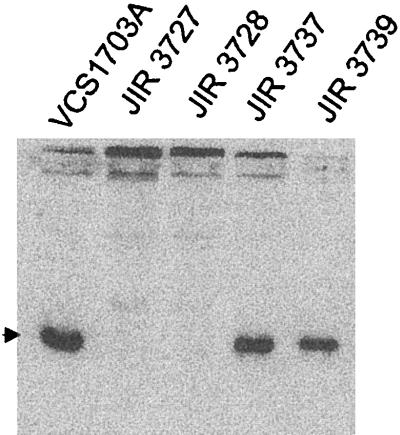
To determine if the mutant strains still produced the fimbrial subunit protein, immunoblot analyses were performed with serogroup G-specific antiserum, using whole-cell extracts of the wild-type and mutant strains. The results (Fig. 2) showed that although a strong type G-reactive 17-kDa FimA band was observed in the wild-type strain, no such band was observed in extracts of the mutants. These results indicated that the type G fimbrial subunits were no longer being produced by the fimA mutants.
FIG. 2.
Western immunoblot of whole-cell extracts of wild-type strains and fimA and fimB mutants. Whole-cell extracts were separated on 12% polyacrylamide gels, electroblotted to nitrocellulose membranes, and developed with type G-specific fimbrial antiserum. The arrowhead marks the position of the fimbrial subunit.
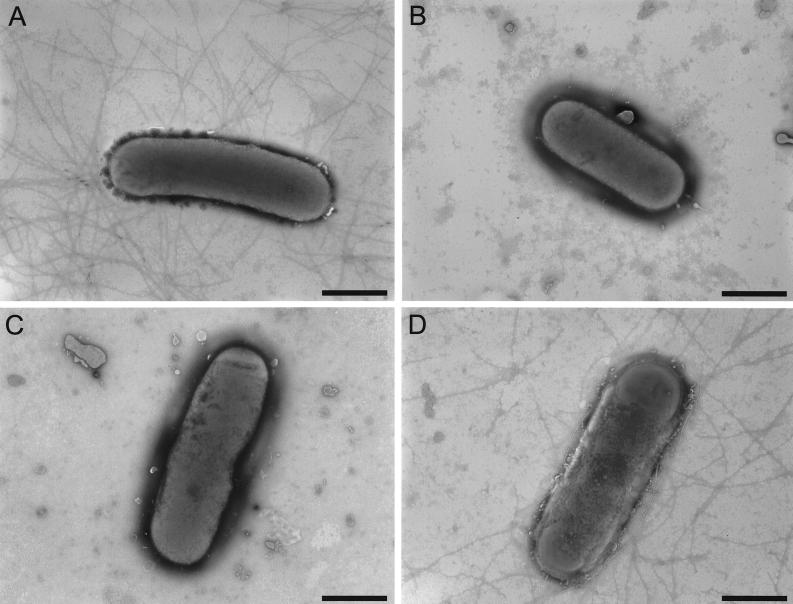
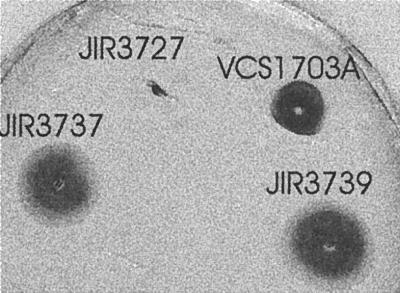
Transmission electron microscopy was used to confirm that type IV fimbriae were no longer produced by the fimA mutants. The results (Fig. 3) showed that although type IV fimbriae were present on the wild-type strain, there were no fimbriae on either of the mutants. In other type IV fimbriate bacteria, the fimbriae impart an unusual type of motility known as twitching motility (17). It has been known for many years that D. nodosus cells also exhibit twitching motility (6). An agar stab twitching motility assay (5) was used to examine the effect of the fimA mutation on the twitching motility process. Unlike the wild-type strain, neither of the fimA mutants exhibited the spreading zones typical of twitching motility (Fig. 4). These experiments clearly showed that the fimA-encoded fimbrial subunit was essential for the production of biologically functional type IV fimbriae.
FIG. 3.
Transmission electron micrographs of D. nodosus cells. Wild-type strain VCS1703A (A), fimA mutant JIR3727 (B), fimA mutant JIR3728 (C), and fimB mutant JIR3737 (D) were grown on EYE blood agar for 3 days, removed with PBS, and negatively stained. Bars represent 1 μm.
FIG. 4.
Twitching motility assays. Cultures were stab inoculated to the bottom of 1% agar plates and incubated at 37°C for 3 to 5 days. The agar was then compressed and stained with Coomassie brilliant blue to reveal the zones of twitching motility. The size of the dark zone around the point of inoculation is indicative of the extent of twitching motility. Representative assays of wild-type strain VCS1703A, fimA mutant JIR3727, and fimB mutants JIR3737 and JIR3739 are shown. The profile obtained with fimA mutant JIR3728 was identical to that of JIR3727.
Type IV fimbriae have been shown to be essential for natural transformation in several bacterial species (15, 16, 40). To see if mutation of the fimA gene affected natural transformation in D. nodosus we performed a series of transformation experiments with strains VCS1703A, JIR3727, and JIR3728. These studies utilized the suicide vector pJIR1836, which contains the erythromycin resistance gene erm(B) flanked by the D. nodosus rrnA promoter and terminator. Natural transformation with strain VCS1703A consistently produced erythromycin-resistant transformants that resulted from allelic exchange between pJIR1836 and one of the three chromosomal rRNA operons. However, despite repeated attempts, no transformants were obtained with either of the fimA mutants. It was concluded that the FimA fimbrial subunit was essential for natural transformation in D. nodosus.
These results had implications for the subsequent sheep virulence studies. Normally, the genetic analysis of putative virulence genes involves not only the virulence testing of mutant strains but the complementation of the chromosomal mutants by a wild-type virulence gene and subsequent virulence testing. Accordingly, since recombinant plasmids could not be introduced into the fimA mutants by natural transformation, attempts were made to electroporate these strains. Unfortunately, despite repeated attempts with different experimental conditions, it was not possible to successfully transform these strains, even by using electroporation. Note that we have also been unable to introduce DNA into D. nodosus by conjugation. Therefore, for technical reasons it was not possible to complement the original fimA mutants. Accordingly, all of the subsequent virulence studies were carried out on both of the independently derived fimA mutants.
Previous studies showed that expression of the fimB gene, which is located immediately downstream of fimA, results from readthrough transcription from the fimA promoter, despite the fact that there is a terminator located between these genes (18). To confirm that the effects we were observing with the fimA mutants were due only to the inactivation of fimA and not to a polar effect on fimB, RT-PCR reactions were performed. The primers used in the RT reactions were located within the deleted region of fimA (primer 13240) and within fimB (primer 13242). A fimA-specific transcript was detected only in VCS1703A as expected (data not shown). By contrast, a fimB-specific transcript was detected in both of the fimA mutants as well as the wild-type (data not shown), indicating that fimB-specific mRNA was being produced and that the fimB gene was still expressed in the fimA mutants. Sequence analysis confirmed that the RT-PCR products observed in the fimB reactions were derived from the fimB gene.
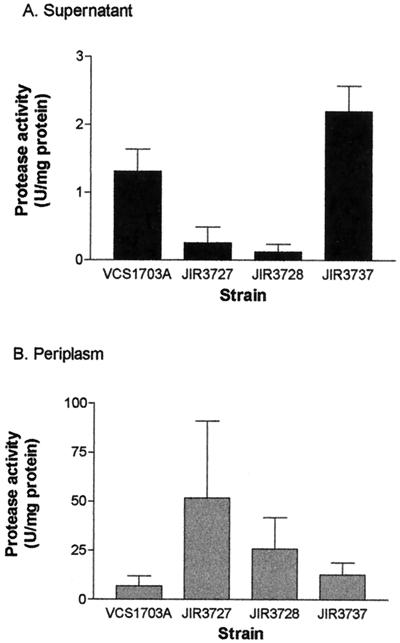
In P. aeruginosa, mutation of the fimbrial subunit gene pilA results in reduced ability to secrete extracellular proteins such as elastase (27). Therefore, routine diagnostic footrot protease tests such as the elastase test and the gelatin-gel protease stability test were performed on the wild-type VCS1703A and the fimA mutants JIR3727 and JIR3728. The results showed that VCS1703A was elastase positive after 7 days of incubation and gelatin-gel positive, whereas the two fimA mutants were negative in both of these tests, suggesting that they no longer produced wild-type levels of extracellular protease. To confirm that the protease genes were still being transcribed in the fimA mutants, RT-PCR experiments were performed on each of the protease genes aprV2, aprV5, and bprV, using separate RT primers that were specific for each protease gene. RT-PCR products specific for each protease gene were obtained in these experiments (data not shown), providing evidence that all three protease genes were transcribed in the fimA mutants. Quantitative protease assays subsequently were carried out on culture supernatants (44 h) and periplasmic extracts of all three strains, using azocasein as the substrate. The results confirmed the initial observations; the fimA mutants had lower levels of protease activity in the supernatant than the wild type (Fig. 5), and these levels were significantly different from the wild-type level (P < 0.05). Periplasmic extracts of the fimA mutants had higher levels of activity than the corresponding wild-type extracts, but these differences were not statistically significant. It was concluded that extracellular proteases are still synthesized in the fimA mutants but are not secreted efficiently.
FIG. 5.
Protease activity of wild-type strain VCS1703A, fimA mutants JIR3727 and JIR3728, and fimB mutant JIR3737. Total protease activity was measured on culture supernatants (A) and periplasmic extracts (B) of each of the strains, using azocasein as the substrate.
The fimB gene is not essential for fimbrial biogenesis.
To examine the role of the fimB gene in fimbrial biogenesis, we decided to construct fimB mutants by allelic exchange. To construct the fimB suicide plasmid pJIR2025 (Fig. 1), a 1-kb PCR fragment, which was derived from VCS1703A chromosomal DNA with primers 4142 and 5013, was blunt-end cloned into pT7Blue. This fragment contained part of aroA, all of fimA, and the first 114 bp of fimB. The tet(M) cassette from pVB101 was cloned into the EcoRI site of pBluescript SKII, and a second PCR fragment, which was derived from the primers 5772 and 4143 and contained the last 350 bp of fimB and the first 360 bp of clpB, was cloned into the SmaI site downstream of tet(M) so that fimB was closest to tet(M). The complete tet(M)-fimB-clpB fragment was then directionally cloned into the pT7Blue derivative with XbaI and XhoI so that the tet(M) gene was located next to the first 114 bp of fimB, thereby creating a 310-bp fimB deletion where tet(M) was inserted. In this construct, pJIR2025, there is 1 kb of D. nodosus DNA upstream of tet(M) and 750 bp downstream.
Transformation of VCS1703A with pJIR2025 produced several tetracycline-resistant colonies that as before were confirmed as D. nodosus and shown to contain the tet(M) gene. Two independently derived transformants, JIR3737 and JIR3739, were selected for further characterization using the same approach as used for the fimA mutants.
Southern hybridization analysis of NruI-digested genomic DNA produced two hybridizing bands with the fimB probe, due to the presence of a NruI site within fimB. However, the smaller, approximately 2-kb band present in the wild-type strain VCS1703A was no longer present in JIR3737 or JIR3739 but was replaced by an approximately 5-kb band, which also hybridized to the tet(M) probe (data not shown), as predicted for an insertionally inactivated fimB gene. To confirm that we had disrupted the fimB gene, RT-PCR was performed with primer 15202, which is located within the deleted region of fimB, and primer 4944. A product was obtained only with the wild-type strain (data not shown), confirming that the fimB gene was not transcribed in the mutants.
Immunoblotting of whole-cell extracts with serogroup G-specific antiserum (Fig. 2) showed that type G-specific fimbrial subunits were still produced by the fimB mutants. Transmission electron microscopy (Fig. 3D) and twitching motility assays (Fig. 4) showed that the fimB mutants produced normal type IV fimbriae that were still capable of imparting twitching motility. In addition, both of the fimB mutants could be transformed with pJIR1836 to erythromycin resistance, indicating that these mutants were still naturally competent. Finally, the fimB mutants were positive at 7 days in the elastase test and were positive in the gelatin-gel test, which indicates that they were secreting functional extracellular proteases, as confirmed by the protease assays (Fig. 5). Note that in these experiments the wild-type strain VCS1703A did not grow quite as well as the mutants, which may account for the fimB mutant having somewhat higher levels of protease activity in the supernatant. Nonetheless, by all of the available phenotypic parameters, the fimB mutants were effectively the same as the wild-type strain.
The fimA gene is an essential virulence gene in ovine footrot.
Wild-type strain VCS1703A and fimA mutants JIR3727 and JIR3728 were virulence tested in sheep in a controlled environment using a standardized method. In summary, groups of 10 sheep were challenged by applying agar cultures of D. nodosus to the interdigital skin of all four feet. Over a 2- to 3-week period, each foot was quantitatively scored on a scale of 0 to 4 for the presence of footrot lesions, and the values were converted to a TWFS, which provides an objective summary of the footrot lesions within a sheep (10, 46). Culturing of foot swabs from the sheep prior to challenge showed them to be free of D. nodosus, while culturing of foot swabs taken 2 weeks after challenge produced D. nodosus isolates only from those sheep challenged with VCS1703A. Characterization of these isolates by serogrouping, elastase, gelatin-gel, and Omp PCR-restriction fragment length polymorphism testing showed them to be identical to VCS1703A.
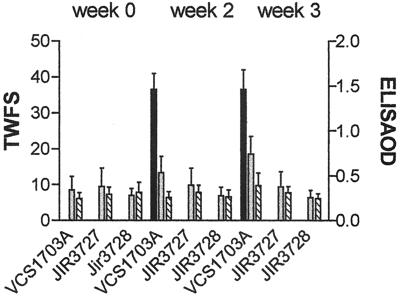
The results of the virulence trials (Fig. 6) clearly showed that only the sheep challenged with the wild-type strain contracted virulent footrot. There was no sign of footrot in either of the groups challenged with the fimA mutants or in the negative control group, which received no D. nodosus cells. These data provide unequivocal evidence that mutation of the fimA gene eliminates the ability to cause ovine footrot. Note that because of the severity of the lesions in the sheep infected with the wild-type strain, for animal ethics reasons these animals had to be treated with antibiotics to eliminate the infection.
FIG. 6.
TWFS and ELISA responses. Sheep were challenged with wild-type strain VCS1703A and fimA mutants JIR3727 and JIR3728 in a blind pen trial. Feet were scored for footrot lesions, (black bars), and blood samples were taken before challenge (week 0) and 2 and 3 weeks after challenge for Omp ELISA (grey bars) and fimbrial ELISA (hatched bars).
Serum samples collected in the course of the sheep trial were analyzed by ELISA for responses to the D. nodosus Omp and fimbrial antigens. The sheep challenged with the fimA mutants produced no response to either antigen, indicating that they had remained uninfected. The sheep challenged with the wild-type strain had a limited immune response to both antigens (Fig. 6). These responses were typical of those seen in sheep 3 weeks postinfection, and it is anticipated that they would have increased further if the sheep had not been treated with antibiotics, as the humoral responses are lesion dependent (45).
In a separate and smaller virulence trial, the virulence of the fimB mutant JIR3737 was tested in a similar manner. The results showed that this strain still had the ability to cause virulent footrot, since more than 10% of the sheep had a TWFS of greater than or equal to 9. However, the average TWFS of 9.4 was lower than that observed with the wild-type strain (TWFS of 36.5), suggesting that the fimB mutant may be slightly attenuated. Further virulence trials would be required to verify this hypothesis. However, since this strain clearly causes virulent footrot, these experiments could not be justified on animal ethics grounds.
DISCUSSION
The type IV fimbriae produced by D. nodosus are regarded as a major virulence factor, as they are highly immunogenic and vaccination with whole cells, purified native fimbriae, or recombinant fimbriae protects against disease (9, 12). We therefore chose to disrupt the fimA fimbrial subunit gene and to examine the effect of this disruption on fimbrial biogenesis and the disease process in sheep. This study represents the first time that genetically defined mutations in putative virulence genes have been constructed and analyzed in this important sheep pathogen.
The results presented here provide evidence that the fimbrial subunit gene is essential for the virulence of D. nodosus in sheep. Virulence testing of two fimA mutants showed they were avirulent, whereas the wild-type strain from which they were derived produced virulent footrot in the same trial, which was conducted under blind conditions. There was no serological response in the sheep infected with the mutants, and no D. nodosus cells were isolated from their feet at the end of the trial, indicating that the mutants did not colonize the ovine hoof. The simplest and most likely explanation for these results is that colonization of the interdigital skin and subsequent penetration of the stratum corneum requires the adhesive activity of type IV fimbriae. However, since these mutants were also altered in their ability to secrete extracellular proteases and other, as yet unknown extracellular proteins, we cannot rule out the possibility that these factors are the major determinants involved in colonization or penetration.
The type II secretion machinery that is responsible for the secretion of many extracellular enzymes and toxins in gram-negative bacteria has several genes that are homologous to type IV fimbrial biogenesis genes (31). Examples include pulGHIJ of Klebsiella oxytoca (32), xcpTUVW of P. aeruginosa (1), and outGHIJ of Erwinia chrysanthemi (26). P. aeruginosa mutants in pilA, which encodes the major pilin subunit, are also defective in protein secretion, leading to the conclusion that the biogenesis of type IV pili (or fimbriae) and protein secretion pathways are shared in this organism (27). The results presented in this paper provide evidence that these pathways are also shared in D. nodosus. Both of the fimA mutants produced little or no extracellular protease or elastase activity, yet in these mutants all three protease genes were still being transcribed and protease activity was detected in periplasmic extracts. It has recently been postulated that type IV fimbriae function in secretion by acting as a piston that pushes secreted proteins through the secretion pore in the outer membrane during extension and retraction of the fimbriae (36). Extension and retraction of the type IV pilus of Neisseria gonorrhoeae has been demonstrated as the process that imparts twitching motility to the bacterial cells (29). The pilT gene of N. gonorrhoeae has been shown to be essential for twitching motility, but not fimbrial biogenesis (48), with pilT mutants still producing fimbriae but not exhibiting twitching motility. Therefore, if in D. nodosus the fimbriae were to function as a piston to push out secreted proteins, this would explain the reduced protease secretion observed in the fimA mutants but still account for the small level of protease activity observed in culture supernatants and the apparent accumulation of proteases in the periplasm. A pilT homologue is yet to be identified in D. nodosus, but the identification and mutation of such a homologue may provide a better understanding of the relationship between fimbriae, twitching motility, protease secretion, and virulence.
The only method by which we are able to introduce recombinant DNA molecules into D. nodosus cells is by a natural transformation process that leads to recombination onto the chromosome. However, since the fimA mutants were no longer naturally transformable, we were unable to complement these mutants to ensure that the phenotypic changes we observed were attributable solely to the disruption of the fimA gene. Two distinct experiments were undertaken to reduce the probability that these changes were not due to the inability to produce the FimA subunit. First, all of the biological tests, including the sheep virulence experiments, were carried out on two independently derived fimA mutants. Identical results were obtained with these mutants, suggesting that it was unlikely that the phenotypic effects resulted from secondary mutations elsewhere on the chromosome. Second, the only gene that could have been affected by a polar fimA mutation was fimB. The RT-PCR results showed that the fimB gene was expressed in the fimA mutants. In addition, the isolation and analysis of independent fimB mutants showed that mutation of fimB had no effect on fimbrial biogenesis, twitching motility, natural transformation, or protease secretion. The virulence tests confirmed that a fimB mutant could still cause virulent footrot in sheep. Accordingly, it was concluded that the phenotypic effects observed in the fimA mutants were the direct result of the inability of the mutants to produce the FimA protein rather than coincident mutations or polar effects on fimB.
Functional type IV fimbrial subunit genes are essential for the natural transformation of N. gonorrhoeae (15), Legionella pneumophila (40), and Pseudomonas stutzeri (16). In addition, other bacteria that are naturally competent possess type IV pilus-related import systems but do not produce fimbriae (3, 25, 43). Our finding that the fimbrial subunits of D. nodosus are essential for natural transformation is in agreement with the observation that the D. nodosus fimA gene can complement a pilA mutant of P. stutzeri and restore pilus production and natural competence (16). Although it has been known for some time that there are common components in the type IV fimbriae assembly apparatus, DNA uptake systems, and type II secretion systems (7, 19), it appears that this is the first report of a bacterium using the same system for all three processes. Identification of further homologues of genes involved in these three processes in D. nodosus will lead to a better understanding of the extent to which these three systems are shared.
ACKNOWLEDGMENTS
We sincerely thank the technical staff of EMAI for assistance with the very complex sheep virulence trials and Khim Hoe for help with electron microscopy.
The project was supported by a grant to J.I.R. from the Australian Research Council.
REFERENCES
- 1.Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol Microbiol. 1992;6:1121–1131. doi: 10.1111/j.1365-2958.1992.tb01550.x. [DOI] [PubMed] [Google Scholar]
- 2.Billington S J, Johnston J L, Rood J I. Virulence regions and virulence factors of the ovine footrot pathogen, Dichelobacter nodosus. FEMS Microbiol Lett. 1996;145:147–156. doi: 10.1111/j.1574-6968.1996.tb08570.x. [DOI] [PubMed] [Google Scholar]
- 3.Breitling R, Dubnau D. A membrane protein with similarity to N-methylphenylalanine pilins is essential for DNA binding by competent Bacillus subtilis. J Bacteriol. 1990;172:1499–1508. doi: 10.1128/jb.172.3.1499-1508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claxton P D. Antigenic classification of Bacteroides nodosus. In: Egerton J R, Yong W K, Riffkin G G, editors. Footrot and foot abscess of ruminants. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 155–166. [Google Scholar]
- 5.Darzins A. The pilG gene product, required for Pseudomonas aeruginosa pilus production and twitching motility, is homologous to the enteric, single-domain response regulator CheY. J Bacteriol. 1993;175:5934–5944. doi: 10.1128/jb.175.18.5934-5944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Depiazzi L J, Richards R B. Motility in relation to virulence of Bacteroides nodosus. Vet Microbiol. 1985;10:107–116. doi: 10.1016/0378-1135(85)90012-4. [DOI] [PubMed] [Google Scholar]
- 7.Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 8.Egerton J R. Significance of Fusiformis nodosus serotypes in resistance of vaccinated sheep to experimental foot-rot. Aust Vet J. 1974;50:59–62. doi: 10.1111/j.1751-0813.1974.tb05252.x. [DOI] [PubMed] [Google Scholar]
- 9.Egerton J R, Cox P T, Anderson B J, Kristo C, Norman M, Mattick J S. Protection of sheep against footrot with a recombinant DNA-based fimbrial vaccine. Vet Microbiol. 1987;14:393–409. doi: 10.1016/0378-1135(87)90030-7. [DOI] [PubMed] [Google Scholar]
- 10.Egerton J R, Roberts D S. Vaccination against ovine foot-rot. J Comp Pathol. 1971;81:179–185. doi: 10.1016/0021-9975(71)90091-0. [DOI] [PubMed] [Google Scholar]
- 11.Egerton J R, Roberts D S, Parsonson I M. The aetiology and pathogenesis of ovine footrot. I. A histological study of the bacterial invasion. J Comp Pathol. 1969;79:207–216. doi: 10.1016/0021-9975(69)90007-3. [DOI] [PubMed] [Google Scholar]
- 12.Elleman T C. Pilins of Bacteroides nodosus: molecular basis of serotypic variation and relationships to other bacterial pilins. Microbiol Rev. 1988;52:233–247. doi: 10.1128/mr.52.2.233-247.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Every D, Skerman T M. Protection of sheep against experimental footrot by vaccination with pili from Bacteroides nodosus. NZ Vet J. 1982;30:156–158. doi: 10.1080/00480169.1982.34921. [DOI] [PubMed] [Google Scholar]
- 14.Firth N, Skurray R A. A protein family associated with filament biogenesis in bacteria. Mol Microbiol. 1995;17:1218–1219. doi: 10.1111/j.1365-2958.1995.mmi_17061218_2.x. [DOI] [PubMed] [Google Scholar]
- 15.Fussenegger M, Rudel T, Barten R, Ryll R, Meyer T F. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene. 1997;192:125–134. doi: 10.1016/s0378-1119(97)00038-3. [DOI] [PubMed] [Google Scholar]
- 16.Graupner S, Frey V, Hashemi R, Lorenz M G, Brandes G, Wackernagel W. Type IV pilus genes pilA and pilC of Pseudomonas stutzeri are required for natural genetic transformation, and pilA can be replaced by corresponding genes from nontransformable species. J Bacteriol. 2000;182:2184–2190. doi: 10.1128/jb.182.8.2184-2190.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrichsen J. Twitching motility. Annu Rev Microbiol. 1983;37:81–93. doi: 10.1146/annurev.mi.37.100183.000501. [DOI] [PubMed] [Google Scholar]
- 18.Hobbs M, Dalrymple B P, Cox P T, Livingstone S P, Delaney S F, Mattick J S. Organization of the fimbrial gene region of Bacteroides nodosus: class I and class II strains. Mol Microbiol. 1991;5:543–560. doi: 10.1111/j.1365-2958.1991.tb00726.x. [DOI] [PubMed] [Google Scholar]
- 19.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 20.Hunt J D, Jackson D C, Brown L E, Wood P R, Stewart D J. Antigenic competition in a multivalent foot rot vaccine. Vaccine. 1994;12:457–264. doi: 10.1016/0264-410x(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 21.Hunt J D, Jackson D C, Wood P R, Stewart D J, Brown L E. Immunological parameters associated with antigenic competition in a multivalent footrot vaccine. Vaccine. 1995;13:1649–1657. doi: 10.1016/0264-410x(95)00145-q. [DOI] [PubMed] [Google Scholar]
- 22.Kennan R M, Billington S J, Rood J I. Electroporation-mediated transformation of the ovine footrot pathogen Dichelobacter nodosus. FEMS Microbiol Lett. 1998;169:383–389. doi: 10.1111/j.1574-6968.1998.tb13344.x. [DOI] [PubMed] [Google Scholar]
- 23.Kortt A A, Riffkin M C, Focareta A, Stewart D J. Amino acid sequence of extracellular acidic protease V5 of Dichelobacter nodosus, the causative organism of ovine footrot. Biochem Mol Biol Int. 1993;29:989–998. [PubMed] [Google Scholar]
- 24.La Fontaine S, Egerton J R, Rood J I. Detection of Dichelobacter nodosus using species-specific oligonucleotides as PCR primers. Vet Microbiol. 1993;35:101–117. doi: 10.1016/0378-1135(93)90119-r. [DOI] [PubMed] [Google Scholar]
- 25.Larson T G, Goodgal S H. Donor DNA processing is blocked by a mutation in the com101A locus of Haemophilus influenzae. J Bacteriol. 1992;174:3392–3394. doi: 10.1128/jb.174.10.3392-3394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindeberg M, Collmer A. Analysis of eight out genes in a cluster required for pectic enzyme secretion by Erwinia chrysanthemi: sequence comparison with secretion genes from other gram-negative bacteria. J Bacteriol. 1992;174:7385–7397. doi: 10.1128/jb.174.22.7385-7397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu H M, Motley S T, Lory S. Interactions of the components of the general secretion pathway: role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and extracellular protein secretion. Mol Microbiol. 1997;25:247–259. doi: 10.1046/j.1365-2958.1997.4561818.x. [DOI] [PubMed] [Google Scholar]
- 28.Mattick J S, Anderson B J, Cox P T, Dalrymple B P, Bills M M, Hobbs M, Egerton J R. Gene sequences and comparison of the fimbrial subunits representative of Bacteroides nodosus serotypes A to I: class I and class II strains. Mol Microbiol. 1991;5:561–573. doi: 10.1111/j.1365-2958.1991.tb00727.x. [DOI] [PubMed] [Google Scholar]
- 29.Merz A J, So M, Sheetz M P. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 30.Palmer M A. A gelatin test to detect activity and stability of proteases produced by Dichelobacter nodosus. Vet Microbiol. 1993;36:113–122. doi: 10.1016/0378-1135(93)90133-r. [DOI] [PubMed] [Google Scholar]
- 31.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pugsley A P, Dupuy B. An enzyme with type IV prepilin peptidase activity is required to process components of the general extracellular protein secretion pathway of Klebsiella oxytoca. Mol Microbiol. 1992;6:751–760. doi: 10.1111/j.1365-2958.1992.tb01525.x. [DOI] [PubMed] [Google Scholar]
- 33.Raadsma H, O'Meara T, Egerton J, Lehrbach P, Schwartzkoff C. Protective antibody titres and antigenic competition in multivalent Dichelobacter nodosus fimbrial vaccines using characterised rRNA antigens. Vet Immunol Immunopathol. 1994;40:253–274. doi: 10.1016/0165-2427(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 34.Rood J I, Howarth P A, Haring V, Billington S J, Yong W K, Liu D, Palmer M A, Pitman D A, Links I, Stewart D A, Vaughan J A. Comparison of gene probe and conventional methods for the differentiation of ovine footrot isolates of Dichelobacter nodosus. Vet Microbiol. 1996;52:127–141. doi: 10.1016/0378-1135(96)00054-5. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sandkvist M. Biology of type II secretion. Mol Microbiol. 2001;40:271–283. doi: 10.1046/j.1365-2958.2001.02403.x. [DOI] [PubMed] [Google Scholar]
- 37.Stewart D J. Footrot of sheep. In: Egerton J R, Yong W K, Riffkin G G, editors. Footrot and foot abscess of ruminants. Boca Raton, Fla: CRC Press; 1989. pp. 5–45. [Google Scholar]
- 38.Stewart D J. The role of elastase in the differentiation of Bacteroides nodosus infections in sheep and cattle. Res Vet Sci. 1979;27:99–105. [PubMed] [Google Scholar]
- 39.Stewart D J, Clark B L, Jarret R G. Differences between strains of Bacteroides nodosus in their effects on the severity of footrot, bodyweight and wool growth on Merino sheep. Aust Vet J. 1984;61:349–352. doi: 10.1111/j.1751-0813.1984.tb07153.x. [DOI] [PubMed] [Google Scholar]
- 40.Stone B J, Kwaik Y A. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol. 1999;181:1395–1402. doi: 10.1128/jb.181.5.1395-1402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 42.Thomas J H. A simple medium for the isolation and cultivation of Fusiformis nodosus. Aust Vet J. 1958;34:411. [Google Scholar]
- 43.Tomb J F, el-Hajj H, Smith H O. Nucleotide sequence of a cluster of genes involved in the transformation of Haemophilus influenzae Rd. Gene. 1991;104:1–10. doi: 10.1016/0378-1119(91)90457-m. [DOI] [PubMed] [Google Scholar]
- 44.Whittington R J, Egerton J R. Application of ELISA to the serological diagnosis of virulent ovine footrot. Vet Microbiol. 1994;41:147–161. doi: 10.1016/0378-1135(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 45.Whittington R J, Nicholls P J. Effects of the severity and duration of lesions on the primary and anamnestic humoral responses of sheep to Dichelobacter nodosus and observations of natural resistance to footrot. Res Vet Sci. 1995;59:128–135. doi: 10.1016/0034-5288(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 46.Whittington R J, Nicholls P J. Grading the lesions of ovine footrot. Res Vet Sci. 1995;58:26–34. doi: 10.1016/0034-5288(95)90084-5. [DOI] [PubMed] [Google Scholar]
- 47.Whittington R J, Saunders V F, Moses E K. Antigens for serological diagnosis of ovine footrot. Vet Microbiol. 1997;54:255–274. doi: 10.1016/s0378-1135(96)01284-9. [DOI] [PubMed] [Google Scholar]
- 48.Wolfgang M, Lauer P, Park H S, Brossay L, Hebert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]