Abstract
The repair of DNA damage is expected to be particularly important to intracellular pathogens such as Mycobacterium tuberculosis, and so it is of interest to examine the response of M. tuberculosis to DNA damage. The expression of recA, a key component in DNA repair and recombination, is induced by DNA damage in M. tuberculosis. In this study, we have analyzed the expression following DNA damage in M. tuberculosis of a number of other genes which are DNA damage inducible in Escherichia coli. While many of these genes were also induced by DNA damage in M. tuberculosis, some were not. In addition, one gene (ruvC) which is not induced by DNA damage in E. coli was induced in M. tuberculosis, a result likely linked to its different transcriptional arrangement in M. tuberculosis. We also searched the sequences upstream of the genes being studied for the mycobacterial SOS box (the binding site for LexA) and assessed LexA binding to potential sites identified. LexA is the repressor protein responsible for regulating expression of these SOS genes in E. coli. However, two of the genes which were DNA damage inducible in M. tuberculosis did not have identifiable sites to which LexA bound. The absence of binding sites for LexA upstream of these genes was confirmed by analysis of LexA binding to overlapping DNA fragments covering a region from 500 bp upstream of the coding sequence to 100 bp within it. Therefore, it appears most likely that an alternative mechanism of gene regulation in response to DNA damage exists in M. tuberculosis.
The repair of DNA damage is likely to be important to pathogens such as Mycobacterium tuberculosis which reside within the very host cells intended to be a defense against infection. Although M. tuberculosis is able to modify the maturation of the normal phagocytic pathway (5, 12, 32), initially, on entering the macrophage, the bacterium is exposed to a variety of reactive oxygen and reactive nitrogen intermediates which are able to damage DNA (1, 2, 25). In addition, DNA repair systems are probably important for successful emergence from the dormant state (21), in which M. tuberculosis persists in the host but does not cause active disease. This possibility is supported by the induction of DNA repair genes in other bacteria during stationary phase (19, 33).
The genome sequence of M. tuberculosis has been analyzed for the presence of homologs of genes known to be involved in DNA repair in Escherichia coli (21). Genes required for nucleotide excision repair, base excision repair, recombination, and SOS repair were all identified, but no homologs of mismatch repair genes could be detected. In addition, no homologs of some individual genes, such as the SOS genes polB and umuD, were found.
The primary response of many bacteria to DNA damage is the induction of a number of genes which are important for DNA repair and the control of cell division. In E. coli, the majority of these genes are part of the so-called SOS response which is regulated by the repressor protein LexA in conjunction with RecA, which acts as an activator (11, 17). Under normal conditions, LexA binds to a specific sequence (the SOS box) upstream of the genes it regulates to repress expression (4, 18). When DNA damage occurs, RecA binds to regions of single-stranded DNA arising from processing of the damage or blockage of replication (27), and in this state it stimulates the autocatalytic cleavage of LexA (16). The cleaved fragments of LexA no longer bind to the SOS boxes (3), resulting in increased transcription of the SOS genes. The degree of induction of a gene depends on the affinity of its SOS box for LexA, the location of the SOS box relative to the promoter, the promoter strength, and the presence of any additional constitutive promoters (11, 28).
The key regulatory elements of the SOS system have been identified in M. tuberculosis (9, 22–24). It has been demonstrated that the recA gene is DNA damage inducible, and the LexA protein has been shown to bind to a specific sequence upstream of each of the recA and lexA genes. We now wished to examine homologs of other members of the E. coli SOS regulon to see if their expression is DNA damage inducible in M. tuberculosis and whether they are regulated by LexA.
MATERIALS AND METHODS
Computer searches.
Searches of the whole M. tuberculosis H37Rv genome were performed using the facilities provided at the TubercuList web site (http://genolist.pasteur.fr/TubercuList/). Searches of 500-bp sequences preceding individual genes were done using the program Findpatterns of the Genetics Computer Group package (8).
Bacterial growth conditions and DNA damage induction.
M. tuberculosis H37Rv was grown in modified Dubos medium (Difco) in tissue culture flasks laid flat in a 37°C incubator. Under these conditions of growth, the doubling time was 25 h. To induce DNA damage, mitomycin C (0.2 μg ml−1) was added to growing cultures (at A600 of 0.4 to 0.6) and incubated for the time indicated.
RNA extraction and cDNA synthesis.
Commercially available kits were used for the isolation of total RNA (Hybaid Ribolyser Blue kit) from bacterial cultures (100 ml), to digest contaminating DNA from the RNA preparations using RNase-free DNase (Roche), and subsequent cleanup procedures (RNeasy Mini Kit; Qiagen). First-strand cDNA synthesis was carried out using Superscript II (Life Technologies) according to the published protocol (24).
Real-time quantitative Taqman PCR assay.
Real-time quantitative PCR was carried out on the ABI Prism 7700 sequence detection system using the Taqman Universal PCR Master Mix (PE Applied Biosystems). The primers and the Taqman probes (carrying both a fluorophore and a quencher) were designed using the Primer Express software and obtained from PE Applied Biosystems. The sequences of the primers and the probes are listed in Table 1. In each case the test gene and the normalizing gene (gnd) were assayed simultaneously along with a set of standard samples for each gene.
TABLE 1.
Sequences of the primers and probes used in the real-time quantitative Taqman PCR assay
| Gene | Sequence (5′→3′)
|
||
|---|---|---|---|
| Forward primer | Reverse primer | Taqman probe | |
| recA | ACCGGCGCGCTGAATA | CGCGGAGCTGGTTGATG | TTCGGGCACCACGGCGATC |
| lexA | ATCCTTGCCGAGGAAGCC | TCACCGATCACCTTGAGCAG | TCCCGCTGCCGCGTGAGC |
| ruvA | CTGATCACCGCGATGATTGT | CGACAGCAGCGTCAGGAATA | CTCGATGACGCTGTACGGGTTTCCC |
| ruvC | GCTCAGGTCACCGCGATG | CGGCCGGTGTCGGTT | CACCAAAATCCTTGCGCTGCAAGCT |
| uvrA | GTCGTATTACGCCGATTTCGA | CGGACTCGGTTTGGGACAT | TGCTGGCGTTCCTGCAACGC |
| ssb | CAACGCCCCGGATCTATG | AGATATTGCACCGGAGGAACA | CCGTCTTTCCATTCGCCGGTCTG |
| dnaB | TGCGTACCGGAGGCTGTATT | CACAGATGCCGCAGAAACAA | CGCGCCCCCAATGACCAGG |
| recN | ACTCGACGTCTCCGAAGAAGG | AGATCAACTGCGGCTTGGC | ACCGGTGAGCTCGCCCACGAATTA |
| dinP | GGGATCAACACCGTTTACCAA | GGACCGAACGTGGACATCA | TTGCACACACCGATTCCGGGC |
| dinG | GGCTTCGGCTACCTTCAGTTC | GATAGGTCGCCATCTCGTCATC | CCGGGCCGCATTGATTGCCT |
| recC | ACCTGCTCGACTTCTTCAAGGAT | TGGACCGGTATCGAGTCCTC | CCGGGCGCTGGACTACACGCT |
| sigA | CCGATCTCGTTGGACCAGA | CCTCGCTGTCTTCGATGAAAT | AGCTGGCTGTCGCCCTCGTCG |
| gnd | GTCCACAACGGCATCGAGTA | GCTGTCCAGATCGCCATTG | TCCGACATGCAGCTCATCGGTGA |
Gel shift assay of LexA binding to individual SOS boxes.
M. tuberculosis LexA was purified from E. coli containing the expression clone pFM18 as described previously (22) and stored at −80°C in 100 mM Tris-Cl (pH 7.5)–500 mM NaCl–1 mM EDTA–1 mM dithiothreitol. For each putative SOS box, an oligonucleotide 24 bases long (containing the particular motif and 6 bases of native sequence on either side [Table 2]) plus its complement were annealed. These double-stranded oligonucleotides were end labeled with [γ-32P] dATP at their 5′ termini, using T4 polynucleotide kinase (New England Biolabs) according to the manufacturer's instructions. Approximately 0.2 pmol of the labeled oligonucleotide was incubated with 2 μl of M. tuberculosis LexA (diluted to 80 nM in 100 mM Tris-Cl [pH 7.5]–100 mM NaCl) and 1 μg of poly(dI-dC) nonspecific competitor DNA in a 20-μl binding reaction [1× binding buffer contained 20 mM HEPES (pH 7.6), 30 mM KCl, 10 mM (NH4)2SO4, 1 mM EDTA, 1 mM DTT, and 0.2% (wt/vol) Tween 20] for 15 min at room temperature. Protein-DNA complexes were resolved from free DNA on an 8% nondenaturing polyacrylamide gel by electrophoresis in 0.5× Tris-borate-EDTA buffer (26) at 180 V for 5 h at 4°C. Gels were dried, and the radioactive bands were visualized by autoradiography.
TABLE 2.
Oligonucleotides used for gel shift assays
| Gene | SOS box label | Sequence of oligonucleotidea |
|---|---|---|
| recA | recA0 | tgaatcGAACaggtGTTCggctac |
| lexA | lexA1a | cctgtcGAACacatGTTtgattct |
| lexA1b | atatccGAACatttGaTCgaagcg | |
| lexA2a | cggttgGAgCcggacTTCcggcgc | |
| lexA2b | gtaatcGctCgcgtGTTCgacact | |
| ruvA | ruvA0 | gctatcGAACgggtGTTCtctcag |
| ruvC | ruvC0 | cgtatcGAACgattGTTCggaaat |
| ruvC2a | atttaccAtCgcacGTTCcatagg | |
| ruvC2b | gtgtgcGAtCgagcGTTtcccgaa | |
| ssb | ssb2a | gaactcGAcCgccaGcTCagcctc |
| ssb2b | ccatacGAAatcatGgTCatcctc | |
| dnaB | dnaB1 | gttgtcGAAtatgcGTTCgggtgc |
| recN | recN2a | cggctgGtgCgcaaGTTCcggttg |
| recN2b | catcgtcAACcgggGTTgggcgct | |
| dinP | dinP2a | ccgctgGAcCgcctGaTCgcattc |
| dinP2b | gcaaagGcACcttgtTTCgccgct | |
| recC | recC1 | gaagccGcACgagaGTTCgccggt |
| recC2a | cgacaaGAAggcccGTaCcgactg | |
| recC2b | gggataGcAgccgaGTTCgggctg |
In each case, the complementary oligonucleotide was annealed with the sequence given and the resulting double-stranded probe was used in the gel shift; the bases matching the consensus SOS box sequence are shown in uppercase.
Gel shift assay of LexA binding to overlapping fragments of upstream DNA.
PCRs were used to generate overlapping DNA fragments spanning at least 500 bp upstream of the coding region and extending at least 100 bp into it for genes ssb and uvrA. Three and two PCR products were generated for ssb and uvrA, respectively, along with a control PCR product containing the SOS box from upstream of the recA gene. The primers used and sizes of the PCR products are given in Table 3. The PCR mixtures contained 0.5 μM each relevant forward and reverse primer, 0.2 mM deoxynucleoside triphosphates, 5% dimethyl sulfoxide (DMSO), 20 ng of total M. tuberculosis DNA (except for recA, for which the template was 2 ng pEJ135 [7]), and 5 U of Pfu Turbo in 1× Pfu buffer (Stratagene). The program used was as follows: 1 cycle of 94°C for 2 min; 25 cycles of 94°C for 30 s, 58 or 55°C (see Table 3) for 30 s, 72°C for 45 s; and 1 cycle of 72°C for 7 min. These PCR products were gel purified; then 0.4 pmol was end labeled with [γ-32P]dATP, and 0.2 pmol was used in binding reactions as described above except that the samples were run on a 6% gel.
TABLE 3.
PCR primers
| PCR product | Sequence
|
|||
|---|---|---|---|---|
| Forward primer | Reverse primer | Annealing temp (°C) | Length of product (bp) | |
| recA-up | GATCTAGACCAGGCTAGCGGTGTTGAG | GACTAGTAACCTTTGCCGTAACTCTTC | 58 | 355 |
| ssb-up1 | ACGGCGGAAAAGTCGAAAAGGTG | GCCGCCTCCCGCCAGATA | 58 | 488 |
| ssb-up2 | GGCCGCACACGACCACAG | GAGCCTACGTAACCGCACCGACAG | 58 | 376 |
| ssb-up3 | GCCGTTCGCGACACTGACATT | TCTCCAAGGACGGGGCTAG | 55 | 272 |
| uvrA-up1 | ATCGTGGCGCCGGGCAGGAAGC | GGATAACCCGGTGAAGACGAT | 55 | 347 |
| uvrA-up2 | CTGTCGCCGTGGGTGAGCA | AGCAGTGGCGCATATGACAACAGT | 55 | 381 |
Reverse transcription (RT)-PCR to test cotranscription of ruvCAB.
The PCR mixture contained 0.25 μg each of forward (AGCGAGGTCAAGGCGGCGGTCACT) and reverse (GCTCGGCGGGCTCGTAGAAATCCA) oligonucleotides, 1 μl of cDNA (or DNA for control), 5 U of Taq polymerase, 1 mM deoxynucleoside triphosphates, and 10% dimethyl sulfoxide in buffer P (Invitrogen PCR Optimizer kit) in a total volume of 50 μl. The program used was as follows: 1 cycle of 94°C for 2 min; 10 cycles of 94°C for 1 min, 63°C for 1 min, and 72°C for 2 min; 20 cycles of 94°C for 1 min, 63°C for 1 min, and 72°C for 2 min plus 20 s per cycle; and 1 cycle of 72°C for 7 min.
RESULTS
Having established that expression of the recA gene is induced by DNA damage in M. tuberculosis (23), we wished to identify other M. tuberculosis DNA damage-inducible genes. In other bacteria the majority of genes which are induced by DNA damage are regulated by the repressor protein LexA (11), and a binding site for mycobacterial LexA has been identified upstream of the M. tuberculosis recA gene and overlapping a putative promoter element (23). Therefore, one approach to identifying other DNA-damage inducible genes would be to search the M. tuberculosis genome sequence (6) for the presence of a mycobacterial SOS box upstream of a coding region. When we performed this search using the sequence GAACN4GTTC and limiting hits to being within 500 bp upstream of a start codon, we identified 35 exact matches which included recA and in addition ruvA and ruvC, genes important for recombination, but also many genes with no known role in DNA repair or recombination. The presence of an SOS box in this region does not necessarily mean that the corresponding gene is regulated by LexA, as the binding site must be in a suitable position relative to the promoter. In addition, it had also been shown that mycobacterial LexA bound to an SOS box found upstream of the lexA gene (22) which had a single mismatch from the consensus. Thus, it may be that our search should allow for hits to have one mismatch from the search sequence; this relaxation of the search constraints resulted in the identification of a further 652 potential sites. Clearly, for this approach to be useful we need to have a better idea of which bases within the SOS box can be altered while maintaining the ability to bind LexA and which cannot; studies are in progress to determine this. Meanwhile, we decided to use the E. coli SOS regulon as a guide to which genes may also be DNA damage inducible in M. tuberculosis and to concentrate on homologs of some of the E. coli SOS genes.
We chose to examine 10 genes, 7 of which are homologs of E. coli SOS genes, in addition to recA as a positive control. These were selected to represent different functions, i.e., regulation (lexA), resolution (ruvA), recombination (recN), excision repair (uvrA), and single-stranded DNA binding (ssb), and two genes originally identified on the basis of their DNA damage inducibility but for which functions have subsequently been identified (dinG [helicase] and dinP/dinB [polymerase IV]). We included ruvC, although it is not an SOS gene in E. coli, because of the SOS box identified upstream of the M. tuberculosis gene. In addition, we decided to study dnaB, which is DNA damage inducible but not part of the SOS response in E. coli (14), and recC, which is a recombination gene that is not DNA damage inducible in E. coli as an expected negative control.
Induction by DNA damage.
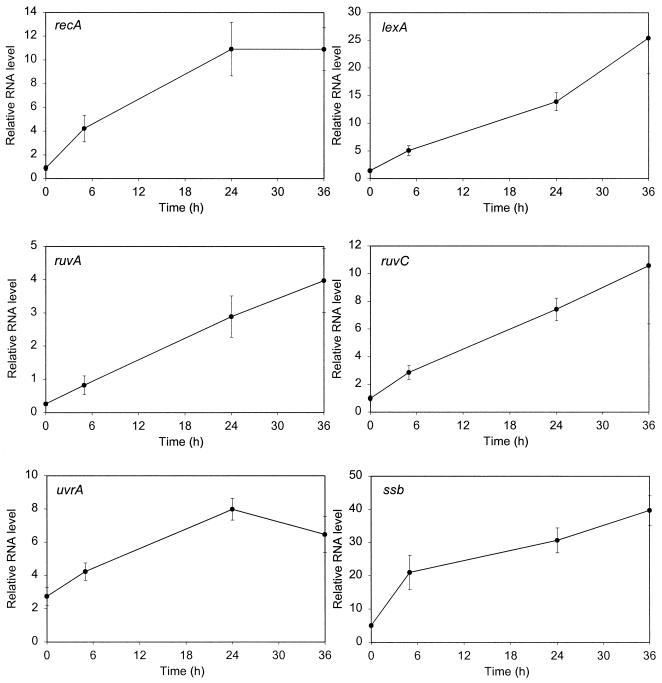
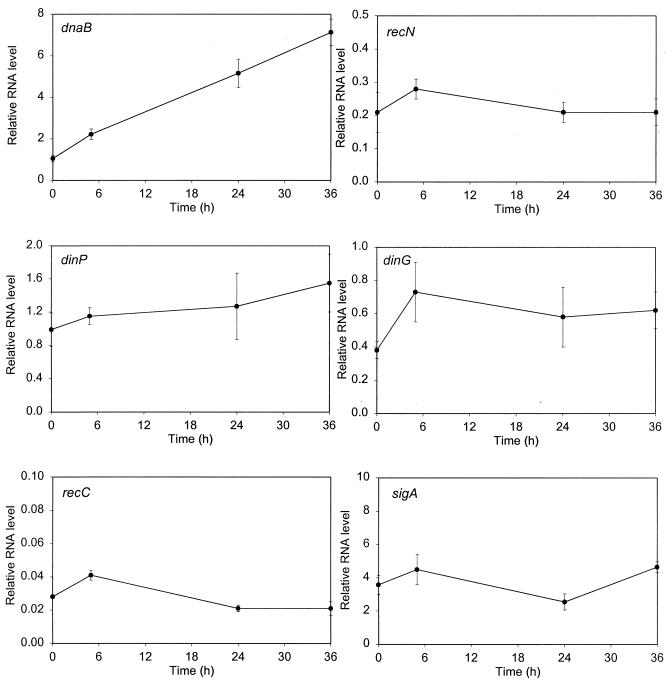
To directly determine whether the selected genes are induced by DNA damage, we examined their expression levels before and after exposure to mitomycin C (0.2 μg/ml), an agent previously shown to be effective at low concentrations in inducing the expression of recA in mycobacteria (23). We also analyzed the expression of sigA, encoding the major sigma factor of M. tuberculosis which had been shown previously to be constitutively expressed (20). We measured the amount of each specific mRNA relative to that of a gene whose expression would not be expected to change under these conditions, gnd (encoding 6-phosphogluconate dehydrogenase), by real-time RT-PCR. This technique is very sensitive and allowed us to examine the expression of multiple genes using the same cultures. In addition to samples from uninduced cultures, we took samples 5, 24, and 36 h after the addition of the DNA-damaging agent, since we had previously discovered that induction of recA in M. tuberculosis requires extended periods of time (23).
The basal expression levels varied among these genes, with those of ssb, uvrA, and sigA being higher than average and those of ruvA, recN, and particularly recC being relatively low. It was clear that a number of these genes were DNA damage inducible (Fig. 1), although the extent of induction at 24 h ranged from approximately 3- to 12-fold (Table 4). However, there were some significant differences in the response of M. tuberculosis to DNA damage compared with that of E. coli. The genes induced by DNA damage in M. tuberculosis included ruvC, which is not induced in E. coli, while three genes (recN, dinP, and dinG) which are SOS genes in E. coli were not, or were barely, induced in M. tuberculosis. As expected, there was little change in the expression of either recC or sigA under these conditions.
FIG. 1.
Gene expression following DNA damage. The amount of mRNA for each gene relative to that of a normalizing gene (gnd) was determined by real-time quantitative RT-PCR using RNA samples from cultures harvested at various time points following the addition of mitomycin C (0.2 μg ml−1). For each gene, at least two assays using three or four samples were performed on each of two independent inductions. The values shown are the means; the error bars indicate the standard deviations. Note that the scales of the y axes vary according to the expression level of the gene being analyzed.
TABLE 4.
Induction by DNA damage of genes being studied in M. tuberculosis compared with E. coli
| Gene | M. tuberculosis LexA bindinga | Induction ratio at 24 h in M. tuberculosis | Inducible in E. coli |
|---|---|---|---|
| recA | + | 12.2 | Yes |
| lexA | + | 9.5 | Yes |
| ruvA | + | 11.1 | Yes |
| ruvC | + | 7.5 | No |
| uvrA | − | 2.9 | Yes |
| ssb | − | 6.1 | Yes |
| dnaB | + | 4.9 | Yes |
| recN | − | 1.0 | Yes |
| dinP | − | 1.3 | Yes |
| dinG | − | 1.5 | Yes |
| recC | − | 0.8 | No |
Identification of potential SOS boxes.
For each of the 10 genes of interest, the 500 bp immediately preceding the start codon was searched for homology to the mycobacterial SOS box GAACN4GTTC, allowing up to three mismatches from the eight defined bases. In every case, numerous sequences with three mismatches were found (data not shown), but only a few sequences with two or fewer mismatches were identified (Table 5). As expected from the genome search, the only genes with sites having no mismatches were recA, ruvA, and ruvC, while single mismatched sites were found upstream of lexA (two sites), dnaB and recC. Several of the genes had two sites each with two mismatches, while there were no sequences with fewer than three mismatches for uvrA and dinG. We decided to label the sites by gene name followed by the number of mismatches and finally, in the case of multiple sites with the same number of mismatches, the letter a or b, depending on its distance from the coding region, e.g. recN2a (Table 5).
TABLE 5.
Potential SOS boxes identified upstream of genes being studied
| Gene | Locationa | Potential SOS box | No. of mismatchesb | SOS box label | LexA bindingc |
|---|---|---|---|---|---|
| recA | −121 | GAACaggtGTTC | 0 | recA0 | + |
| −276 | GcACgccgGaTC | 2 | recA2 | − | |
| lexA | −103 | GAACacatGTTt | 1 | lexA1a | + |
| −236 | GAACatttGaTC | 1 | lexA1b | − | |
| −170 | GAgCcggacTTC | 2 | lexA2a | − | |
| −279 | GctCgcgtGTTC | 2 | lexA2b | + | |
| ruvA | −362 | GAACgggtGTTC | 0 | ruvA0 | + |
| ruvC | −35 | GAACgattGTTC | 0 | ruvC0 | + |
| −94 | cAtCgcacGTTC | 2 | ruvC2a | − | |
| −340 | GAtCgagcGTTt | 2 | ruvC2b | − | |
| uvrA | 3 | − | |||
| ssb | −169 | GAcCgccaGcTC | 2 | ssb2a | − |
| −382 | GAAatcatGgTC | 2 | ssb2b | − | |
| dnaB | −42 | GAAtatgcGTTC | 1 | dnaB1 | + |
| recN | −58 | GtgCgcaaGTTC | 2 | recN2a | − |
| −482 | cAACcgggGTTg | 2 | recN2b | − | |
| dinP | −312 | GAcCgcctGaTC | 2 | dinP2a | − |
| −440 | GcACcttgtTTC | 2 | dinP2b | − | |
| dinG | 3 | − | |||
| recC | −399 | GcACgagaGTTC | 1 | recC1 | − |
| −106 | GAAggcccGTaC | 2 | recC2a | − | |
| −297 | GcAgccgaGTTC | 2 | recC2b | − |
Distance upstream from initiation codon.
From consensus GAACnnnnGTTC.
As determined by gel shift assay using M. tuberculosis LexA (Fig. 3).
LexA binding.
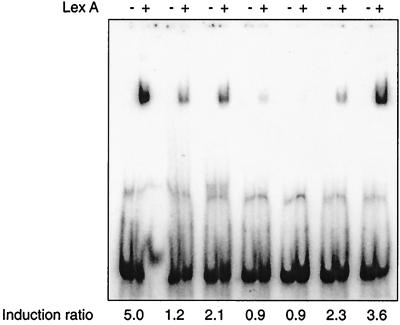
We wished to assess each of these potential SOS boxes for its ability to bind M. tuberculosis LexA. However, we first used a selection of mutated recA SOS boxes with various effects on induction (details to be published elsewhere) to determine that the concentration of LexA used for this in vitro analysis gave binding results which paralleled the deduced effect on binding in vivo (Fig. 2). The in vivo analysis used a transcriptional fusion of lacZ to the LexA-regulated recA promoter containing the various mutations in the SOS box. An induction ratio of 1 or less in this system indicated that LexA did not bind in vivo. Significantly, if concentrations of LexA higher than that shown were used in the gel shift analysis, binding became evident to those sequences which clearly did not bind in vivo, suggesting that such weak interactions were not physiologically relevant. We next assayed each of the identified potential SOS boxes for its ability to bind LexA by gel shift assays using labeled double-stranded oligonucleotides spanning the identified motif and the concentration of LexA determined from the analysis just described. We could then be confident that the LexA binding, or lack of it, seen with our test sequences was significant.
FIG. 2.
Comparison of LexA binding to mutated SOS boxes with their effects on induction in vivo. Double-stranded oligonucleotides containing each mutated SOS box were end labeled with [γ-32P]dATP; following incubation with 8 nM (final concentration) purified M. tuberculosis LexA (lanes marked +) they were assessed for LexA binding by gel shift compared with no-protein controls (lanes marked −). The induction ratio obtained for each mutated SOS box when analyzed using a transcriptional fusion of lacZ to the LexA-regulated recA promoter is indicated below the gel. The wild-type recA0 SOS box, which had been shown previously to bind LexA and to regulate gene expression, is shown in the leftmost pair of tracks. The figure was compiled using Adobe Photoshop.
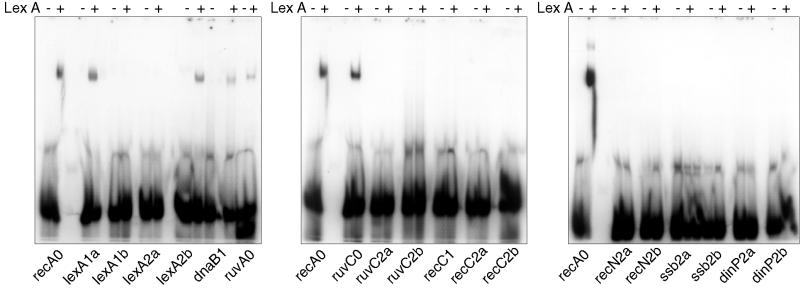
Not surprisingly, LexA bound to all of the sites having no mismatches, although the binding seen with the ruvA0 site seemed weaker than that to the recA0 and ruvC0 sites (Fig. 3). However, while some sites with a single mismatch bound LexA (lexA1a and dnaB1), others did not (lexA1b and recC1). Only one of the sites having two mismatches was bound by LexA (lexA2b). Intriguingly, one of the two changes in this site is the same as the single mismatch found in the recC1 site, which failed to bind. This indicates that bases outside the currently defined motif are important in determining LexA binding. It had been shown previously that the recA2 site did not bind LexA even at a concentration of LexA higher than used here (23), and so this site was not analyzed again. The sites to which LexA did bind were also bound by LexA at the lower concentration of 6 nM rather than 8 nM (data not shown). Thus, the mycobacterial SOS box is not defined well enough for us to accurately predict whether or not LexA will bind. Studies are in progress to define the consensus LexA binding site more precisely.
FIG. 3.
Analysis of LexA binding to potential SOS boxes. Double-stranded oligonucleotides containing each identified motif (indicated below the gel) were end labeled with [γ-32P]dATP; following incubation with 8 nM (final concentration) purified M. tuberculosis LexA (lanes marked +) they were assessed for LexA binding by gel shift compared with no-protein controls (lanes marked −). The recA0 SOS box, which had been shown previously to bind LexA, was included on each gel as a positive control. The figure was compiled using Adobe Photoshop.
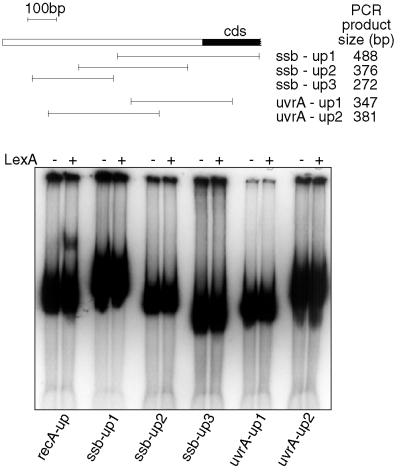
It is noteworthy that expression of two of the genes (ssb and uvrA) which did not appear from the above analysis to have a LexA binding site was nevertheless induced by DNA damage. To assess whether there might be a binding site for LexA upstream of these genes which had not been identified by the computer analysis, we decided to analyze a region spanning at least 500 bp upstream of the coding region and extending at least 100 bp into it for LexA binding. This was done by generating PCR products spanning this region which overlapped by at least 100 bp to ensure that any binding site disrupted at the end of one fragment would be intact on another. These PCR products were then analyzed by gel shift assays using the same molar concentration of fragment and LexA as used before with the oligonucleotides. A similar-size PCR product containing the recA0 site was used as a positive control. This analysis (Fig. 4) confirmed that there was no LexA binding site anywhere within 500 bp upstream of the translation initiation codon for either ssb or uvrA.
FIG. 4.
Analysis of LexA binding to PCR products spanning at least 500 bp upstream of the coding region and extending at least 100 bp into it for ssb and uvrA. The top part shows schematically the locations of the PCR products in relation to the coding sequence and gives their sizes. Shown below are results of the assay for LexA binding to these fragments, done as described for Fig. 2. The recA-up fragment was included as a positive control. The figure was compiled using Adobe Photoshop and Macromedia Freehand.
The uvrA gene is preceded by an open reading frame which is transcribed in the opposite direction, and so it clearly is not part of an operon with expression regulated from a common promoter further upstream. The ssb gene is separated from the gene (rpsF) immediately upstream of it by 107 bp, and rpsF is 222 bp away from the gene preceding it. These distances suggest that it is unlikely that ssb is a downstream gene of an operon, although it could possibly be cotranscribed with rpsF. However, even if ssb was transcribed from a promoter upstream of rpsF, owing to the small size of the rpsF gene (287 bp), almost all of the intergenic region upstream of rpsF (199 of 222 bp) was in any case included in the analysis of LexA binding just described. It is to be noted that to confer regulation, LexA must bind in the region of the promoter and the rpsF promoter is unlikely to be further than 200 bp upstream of the coding region. Thus, the induction of these genes cannot be explained by transcription from a LexA-regulated promoter preceding an upstream open reading frame; therefore, induction in these cases must be controlled by a different, non-LexA dependent mechanism.
The ruvCAB genes form an operon in M. tuberculosis.
As stated above, in M. tuberculosis the ruvC gene as well as ruvA was DNA damage inducible, in contrast to the situation in E. coli. These genes are transcribed independently in E. coli, where the ruvC gene is separated from the ruvAB operon by a separate open reading frame lying in the opposite direction (29, 30). When we examined the sequences encoding the ruv genes in M. tuberculosis, we noticed that not only were the three ruv genes contiguous but also at each junction between pairs of genes the termination codon of one gene overlapped the initiation codon of the next (ATGA). This arrangement suggested that the ruv genes in M. tuberculosis form an operon and are probably translationally coupled. The flanking genes on either side of the ruv genes were transcribed in the opposite orientation.
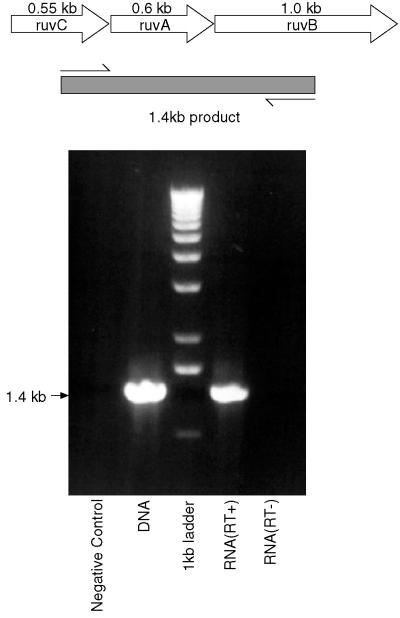
We examined this supposition experimentally by performing RT-PCR using one primer located within ruvC and one from within ruvB. When RNA isolated from M. tuberculosis was reverse transcribed, this primer pair did indeed yield a product of the expected size (Fig. 5), which was the same size as that formed with chromosomal DNA as template. In contrast, if the RT step was not included when RNA was the template, no product was formed, confirming that the PCR product did not arise from contaminating DNA in the RNA preparation. Thus, in M. tuberculosis the genes ruvCAB are cotranscribed and form an operon.
FIG. 5.
The ruvCAB genes are cotranscribed. The top part shows schematically the arrangement of the ruv genes in M. tuberculosis, with the positions of the primers used for RT-PCR and the size of the expected product indicated. The lower part shows that the product obtained using RNA which has been reverse transcribed is the same size as that obtained by PCR using chromosomal DNA, while no product is formed using RNA if the RT step is omitted. The figure was compiled using Adobe Photoshop and Macromedia Freehand.
DISCUSSION
In selecting M. tuberculosis genes for this study, we focused on homologs of well-established members of the SOS regulon in E. coli. These genes perform a range of functions related to DNA repair and recombination. Thus, the genes that we examined included the regulatory genes recA and lexA, a component of the Holliday junction resolvase (ruvA), and a component of the excision repair complex (uvrA). We also investigated the gene for single-stranded DNA binding protein (ssb), a recombination gene (recN), and two genes originally identified solely on the basis of their regulation by LexA (dinG and dinP). The products of these latter two genes have subsequently been ascribed functions, with DinG being identified as a helicase (15) and DinP (also termed DinB) recently being recognized as a mutagenic DNA polymerase (34). These eight genes are all part of the SOS regulon in E. coli. In addition, we chose to analyze another component of the resolvase (ruvC), although it is not an SOS gene in E. coli, because we had identified a perfect match to the M. tuberculosis LexA binding site upstream of this gene. Finally, we included the replicative helicase gene (dnaB), which is DNA damage inducible in E. coli but not LexA regulated (14), and a control recombination gene (recC).
Curiously, there are no sequence homologs in M. tuberculosis for a number of other E. coli SOS genes with known functions. These include the UV mutagenesis gene umuD (although there are homologs of umuC), the gene encoding DNA polymerase II (polB, formerly dinA), the integration host factor subunit gene himA, and the filamentation gene sulA. Nevertheless, while staining cultures of M. tuberculosis treated with DNA-damaging agents for various periods of time to confirm the purity of the culture, we noticed that the cells appeared filamentous, particularly following longer periods of exposure. This observation suggests that there may be an as yet unidentified functional homolog of sulA which is DNA damage inducible. There may also be genes with little or no sequence similarity performing the equivalent functions for some of the other SOS genes. Recently, a number of other LexA-regulated genes have been identified in E. coli, but their functions remain to be determined (10).
Of the nine genes which are DNA damage inducible in E. coli, only six were induced by mitomycin C in M. tuberculosis. In addition, ruvC was induced by DNA damage in M. tuberculosis although it is not DNA damage inducible in E. coli. This latter observation is linked to the discovery that the ruvCAB genes are transcribed as an operon in M. tuberculosis, unlike the gene arrangement in E. coli. The degree of induction seen in M. tuberculosis varied from approximately 3- to 12-fold. Some of these values can be compared with induction ratios quoted for the equivalent E. coli genes although these were determined in a different way: fusions to β-galactosidase were used in a strain lacking functional LexA repressor and related to activity in a strain with wild-type LexA (11). recA was induced 12-fold in M. tuberculosis and 11-fold in E. coli, lexA was induced 9.5- and 6.7-fold, respectively, and uvrA was induced 2.9- and 3.4-fold, respectively. Thus, for the genes which were induced in M. tuberculosis, the induction ratios were similar to those found in E. coli. In contrast, dinP was essentially not induced, with a ratio of 1.3 in M. tuberculosis, compared with 7.3 in E. coli.
The induction ratio for M. tuberculosis recA in this study is higher than the ratio that we previously reported (23). This is almost certainly due to the fact that in the previous work the transcript level was normalized to the amount of rRNA but the induction conditions caused a significant degree of cell death. It has been shown that mycobacterial rRNA is very stable and remains present at relatively high levels even when there is a large reduction in the number of viable cells (13), and so normalizing to rRNA under conditions where cell death is occurring will lead to an underestimation of the specific transcript level. In the present study the transcript level of the gene of interest was instead normalized to an mRNA for a gene whose expression level would not be expected to change under the conditions being investigated.
It is perhaps noteworthy that M. tuberculosis appears to lack a homolog of umuD and that the expression of dinP is not induced by DNA damage. These two genes encode proteins involved in mutagenesis in E. coli, with UmuD′ (the processed form of UmuD) interacting with UmuC to form the error-prone DNA polymerase V (31) and dinP encoding another error-prone polymerase, polymerase IV (34). This relative lack of mutagenic polymerases suggests that M. tuberculosis must rely exclusively on more accurate repair mechanisms for survival following DNA damage.
A finding of particular interest is the apparent lack of correlation of LexA binding to the DNA upstream of a gene with induction of its expression by DNA damage. Excluding ruvA, which is a downstream gene in an operon, six of the genes examined here were induced by mitomycin C in M. tuberculosis. Of these six, four possessed LexA binding sites within 500 bp upstream of the coding region and two did not. These observations suggest that an alternative, non-LexA-dependent mechanism of gene regulation in response to DNA damage must exist in M. tuberculosis. While it is possible that LexA could be controlling the expression of an activator which then acts on genes such as ssb and uvrA, if that is the case, induction would depend on LexA and RecA. Although the ssb and uvrA genes have yet to be tested, it has recently been found using a recA promoter-lacZ transcriptional fusion that the recA promoter remains inducible in a recA deletion mutant of M. tuberculosis (E. O. Davis and K. G. Papavinasasundaram, unpublished data). Thus, even in the case of a gene (recA) which possesses a LexA binding site, it appears that induction in response to mitomycin C can occur independently of LexA and RecA. The data presented here suggest that this alternative mechanism also operates for other DNA damage-inducible genes. To confirm that this is indeed the case, it will be necessary to examine the expression of these genes in the recA deletion mutant of M. tuberculosis following mitomycin C treatment. However, the methodology presented here for the wild type has the disadvantage that it permits the analysis of only a limited number of genes in a rather labor-intensive way. Hence, we plan to use microarrays to examine global gene expression in response to DNA damage in both the wild-type and recA deletion strains of M. tuberculosis. This more universal approach will allow us to determine what proportion of DNA damage-inducible genes are regulated by RecA and/or LexA and how generally the alternative mechanism applies.
ACKNOWLEDGMENTS
We thank M. J. Colston for critical reading of the manuscript and K. G. Papavinasasundaram for helpful discussions.
REFERENCES
- 1.Adams L B, Dinauer M C, Morgenstern D E, Krahenbuhl J L. Comparison of the roles of reactive oxygen and nitrogen intermediates in the host response to Mycobacterium tuberculosis using transgenic mice. Tubercle Lung Dis. 1997;78:237–246. doi: 10.1016/s0962-8479(97)90004-6. [DOI] [PubMed] [Google Scholar]
- 2.Akaki T, Tomioka H, Shimizu T, Dekio S, Sato K. Comparative roles of free fatty acids with reactive nitrogen intermediates and reactive oxygen intermediates in expression of the anti-microbial activity of macrophages against Mycobacterium tuberculosis. Clin Exp Immunol. 2000;121:302–310. doi: 10.1046/j.1365-2249.2000.01298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertrand-Burggraf E, Hurstel S, Daune M, Schnarr M. Promoter properties and negative regulation of the uvrA gene by the LexA repressor and its amino-terminal DNA binding domain. J Mol Biol. 1987;193:293–302. doi: 10.1016/0022-2836(87)90220-8. [DOI] [PubMed] [Google Scholar]
- 4.Brent R, Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci USA. 1981;78:4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemens D L, Horwitz M A. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J Exp Med. 1995;181:257–270. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 7.Davis E O, Sedgwick S G, Colston M J. Novel structure of the recA locus of Mycobacterium tuberculosis implies processing of the gene product. J Bacteriol. 1991;173:5653–5662. doi: 10.1128/jb.173.18.5653-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durbach S I, Andersen S J, Mizrahi V. SOS induction in mycobacteria: analysis of the DNA-binding activity of a LexA-like repressor and its role in DNA damage induction of the recA gene from Mycobacterium smegmatis. Mol Microbiol. 1997;26:643–653. doi: 10.1046/j.1365-2958.1997.5731934.x. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez de Henestrosa A R, Ogi T, Aoyagi S, Chafin D, Hayes J J, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg E, Walker G, Siede W. DNA repair and mutagenesis. Washington, D.C.: American Society for Microbiology Press; 1995. [Google Scholar]
- 12.Hasan Z, Schlax C, Kuhn L, Lefkovits I, Young D, Thole J, Pieters J. Isolation and characterization of the mycobacterial phagosome: segregation from the endosomal/lysosomal pathway. Mol Microbiol. 1997;24:545–553. doi: 10.1046/j.1365-2958.1997.3591731.x. [DOI] [PubMed] [Google Scholar]
- 13.Hellyer T J, DesJardin L E, Hehman G L, Cave M D, Eisenach K D. Quantitative analysis of mRNA as a marker for viability of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:290–295. doi: 10.1128/jcm.37.2.290-295.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinsteuber S, Quinones A. Expression of the dnaB gene of Escherichia coli is inducible by replication-blocking DNA damage in a recA-independent manner. Mol Gen Genet. 1995;248:695–702. doi: 10.1007/BF02191709. [DOI] [PubMed] [Google Scholar]
- 15.Koonin E V. Escherichia coli dinG gene encodes a putative DNA helicase related to a group of eukaryotic helicases including Rad3 protein. Nucleic Acids Res. 1993;21:1497. doi: 10.1093/nar/21.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little J W. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- 17.Little J W, Mount D W. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 18.Little J W, Mount D W, Yanisch-Perron C R. Purified lexA protein is a repressor of the recA and lexA genes. Proc Natl Acad Sci USA. 1981;78:4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loewen P C, Hengge-Aronis R. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 20.Manganelli R, Dubnau E, Tyagi S, Kramer F R, Smith I. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 21.Mizrahi V, Andersen S J. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol Microbiol. 1998;29:1331–1339. doi: 10.1046/j.1365-2958.1998.01038.x. [DOI] [PubMed] [Google Scholar]
- 22.Movahedzadeh F, Colston M J, Davis E O. Characterization of Mycobacterium tuberculosis LexA: recognition of a Cheo (Bacillus-type SOS) box. Microbiology. 1997;143:929–936. doi: 10.1099/00221287-143-3-929. [DOI] [PubMed] [Google Scholar]
- 23.Movahedzadeh F, Colston M J, Davis E O. Determination of DNA sequences required for regulated Mycobacterium tuberculosis RecA expression in response to DNA-damaging agents suggests that two modes of regulation exist. J Bacteriol. 1997;179:3509–3518. doi: 10.1128/jb.179.11.3509-3518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papavinasasundaram K G, Movahedzadeh F, Keer J T, Stoker N G, Colston M J, Davis E O. Mycobacterial recA is cotranscribed with a potential regulatory gene called recX. Mol Microbiol. 1997;24:141–153. doi: 10.1046/j.1365-2958.1997.3441697.x. [DOI] [PubMed] [Google Scholar]
- 25.Rich E A, Torres M, Sada E, Finegan C K, Hamilton B D, Toossi Z. Mycobacterium tuberculosis (MTB)-stimulated production of nitric oxide by human alveolar macrophages and relationship of nitric oxide production to growth inhibition of MTB. Tubercle Lung Dis. 1997;78:247–255. doi: 10.1016/s0962-8479(97)90005-8. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sassanfar M, Roberts J W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 28.Schnarr M, Oertel-Buchheit P, Kazmaier M, Granger-Schnarr M. DNA binding properties of the LexA repressor. Biochimie. 1991;73:423–431. doi: 10.1016/0300-9084(91)90109-e. [DOI] [PubMed] [Google Scholar]
- 29.Sharples G J, Lloyd R G. Resolution of Holliday junctions in Escherichia coli: identification of the ruvC gene product as a 19-kilodalton protein. J Bacteriol. 1991;173:7711–7715. doi: 10.1128/jb.173.23.7711-7715.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahagi M, Iwasaki H, Nakata A, Shinagawa H. Molecular analysis of the Escherichia coli ruvC gene, which encodes a Holliday junction-specific endonuclease. J Bacteriol. 1991;173:5747–5753. doi: 10.1128/jb.173.18.5747-5753.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang M, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Via L E, Deretic D, Ulmer R J, Hibler N S, Huber L A, Deretic V. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J Biol Chem. 1997;272:13326–13331. doi: 10.1074/jbc.272.20.13326. [DOI] [PubMed] [Google Scholar]
- 33.Villarroya M, Perez-Roger I, Macian F, Armengod M E. Stationary phase induction of dnaN and recF, two genes of Escherichia coli involved in DNA replication and repair. EMBO J. 1998;17:1829–1837. doi: 10.1093/emboj/17.6.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner J, Gruz P, Kim S R, Yamada M, Matsui K, Fuchs R P, Nohmi T. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]