SUMMARY
Ovarian cancer (OC) is the most lethal gynecological malignancy, with aggressive metastatic disease responsible for the majority of OC-related deaths. In particular, OC tumors preferentially metastasize to and proliferate rapidly in the omentum. Here, we show that metastatic OC cells experience increased oxidative stress in the omental microenvironment. Metabolic reprogramming, including upregulation of the pentose phosphate pathway (PPP), a key cellular redox homeostasis mechanism, allows OC cells to compensate for this challenge. Inhibition of glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme of the PPP, reduces tumor burden in pre-clinical models of OC, suggesting that this adaptive metabolic dependency is important for OC omental metastasis.
Graphical Abstract
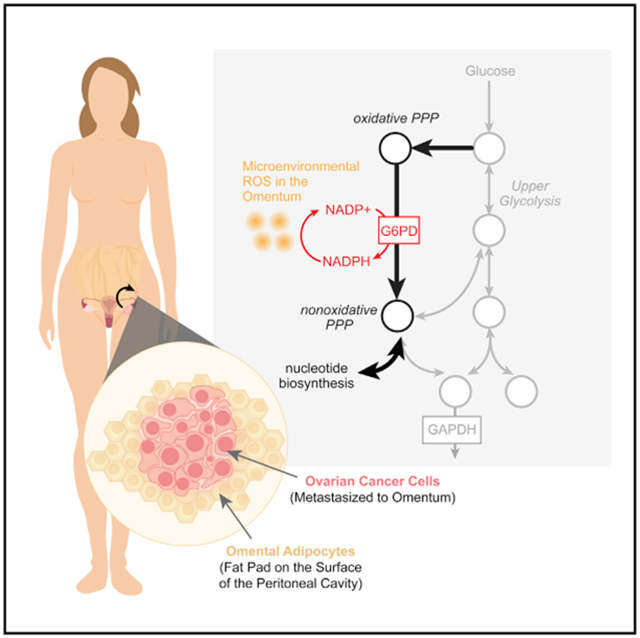
In brief
Bose et al. characterize the importance of G6PD, the rate-limiting enzyme of the oxidative pentose phosphate pathway, in mitigating oxidative stress experienced by ovarian cancer cells metastasizing to the omentum.
INTRODUCTION
Ovarian cancer (OC) remains the most lethal gynecological malignancy, with over 80% of patients diagnosed at stage III or higher, when 5-year survival rates are ~30% (Matulonis et al., 2016). An overwhelming majority of OC deaths are attributed to metastatic disease, with exfoliation from the primary tumor into the peritoneal fluid leading to peritoneal dissemination of cancerous cells throughout the abdominal cavity (Lengyel, 2010). The most clinically relevant and common site of metastatic growth is the omentum, an apron-like layer of adipose tissue that lays over the peritoneal cavity. Omental tumors often grow quite aggressively, leading to significant consequences for patient morbidity and mortality. The precise molecular mechanisms that drive this aggressive phenotype of OC cells in the omental microenvironment are not yet understood but are thus of particular interest.
Alterations in metabolism have recently been proposed as a potential avenue of cellular reprogramming to promote ovarian tumor metastasis; however, the precise nature of these changes is not yet well understood. Evidence suggests that OC cells harness available FA resources in the omental microenvironmental niche (Fidler, 2003; Lengyel, 2010; Nieman et al., 2011). Observed upregulation of proteins critical to cellular fatty acid (FA) metabolism, like FA-binding protein 4 (FABP4), a carrier protein to import free FAs, has provided critical support for this model (Gharpure et al., 2018; Ladanyi et al., 2018; Mukherjee et al., 2020). In addition, adipocytes in the metastatic niche have also been seen to undergo cellular remodeling to support tumor growth and invasion—secreting adipokines to stimulate the adhesion, migration, and invasion of tumor cells, releasing local paracrine factors within the tumor microenvironment, and adapting lipid metabolism to provide free FAs for fuel (Lengyel et al., 2018; Nieman et al., 2013).
The shift toward OC FA metabolism is accompanied by alterations in the omental stroma to create the metastatic niche. To support the increased FA uptake of metastatic OC cells, activated omental adipocytes release reactive oxygen species (ROS) like H2O2 and NO, increasing local oxidative stress (Herroon et al., 2018). Canonically, the cellular antioxidant response is largely reliant on the critical reducing equivalent, nicotinamide adenine dinucleotide phosphate (NADPH). This cofactor is recycled between NADPH and its oxidized form NADP+ to serve as an electron donor for antioxidant defense and reductive biosynthesis. A key enzyme responsible for NADPH generation is glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme of the oxidative pentose phosphate pathway (PPP). This parallel pathway to the glycolytic machinery has been uniquely implicated in as a requirement for NADPH/NADP+ redox homeostasis, mammalian dihydrofolate reductase (DHFR) activity, and folate metabolism in cancer cells (Chen et al., 2019). While G6PD is a potential mediator of chemotherapeutic response (Feng et al., 2020; Yamawaki et al., 2021), the potential role of this redox enzyme in OC omental metastases has not been elucidated.
Here, we investigated alterations in G6PD as an OC adaptation in the omental microenvironment. Both human and mouse omental metastases and in vitro models of omental metastases of OC exhibited higher oxidative stress and upregulated G6PD. Inhibition of G6PD led to increased cell death in the omental microenvironment in vitro and reduced metastases in vivo. Thus, G6PD may play a role in mitigating oxidative stress and promoting growth in OC omental metastases.
RESULTS
Omental metastases exhibit upregulated G6PD expression and activity in the context of higher oxidative stress
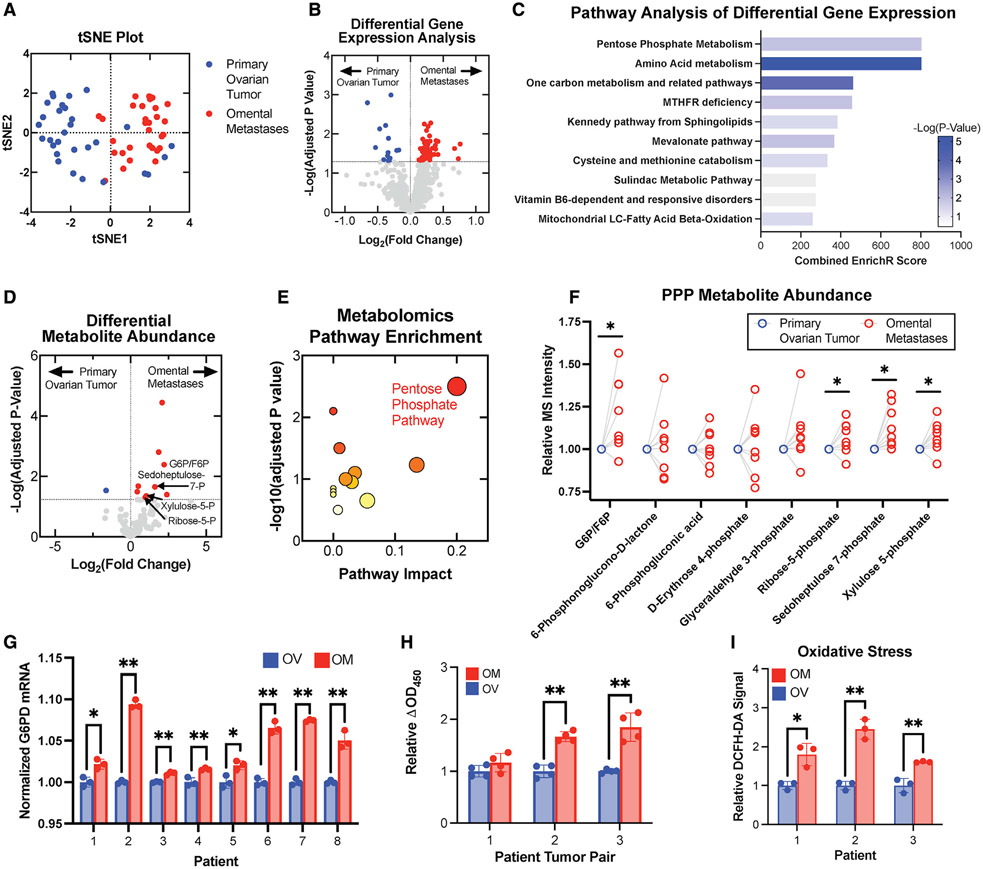
OC cancer cells exhibit a specific tropism and rapid growth rate in the metastatic niche of the omentum. We first studied the transcriptomic profiles of 30 primary ovarian tumors and 30 matched omental metastases from patients with stage III high-grade serous ovarian adenocarcinomas using available NCBI Gene Expression Omnibus datasets (GEO: GSE118828 and GSE30587). Hierarchical clustering and t-distributed stochastic neighbor embedding (t-SNE) plotting (Figures 1A and S1A) suggested distinct transcriptomic profiles for primary tumors versus omental metastases. Paired differential (Figure 1B) and pathway analysis (Figure 1C) of a panel of metabolic genes obtained from the KEGG Database (http://www.kegg.jp) was conducted, and the PPP emerged as an altered pathway in metastatic samples.
Figure 1. The PPP is upregulated in omental metastases compared with primary ovarian cancers.
Gene-expression data revealed metabolic differences of primary ovarian tumors and omental tumors from patients with stage III OC (n = 30).
(A) Distinct clustering in t-SNE plot of metabolic gene data from matched primary ovarian tumors (blue) and omental metastases (red).
(B) Volcano plot of gene-expression changes for paired ovarian and omental tumors highlighted genes significantly upregulated in omental tumors (red) and ovarian tumors (blue).
(C) Pathway analysis of the significantly altered genes.
(D) Volcano plot of metabolite abundances determined by liquid chromatography-mass spectrometry (LC-MS) metabolomic analysis of matched ovarian and omental pairs (n = 8).
(E) Pathway enrichment analysis of metabolomics data; symbol size indicates pathway impact and color indicates p value.
(F) Changes in PPP metabolite abundances are plotted as outlined circles. Blue circles indicate the normalized primary tumor measurements for all tumor pairs (n = 8).
(G) G6PD expression in ovarian and omental metastases (n = 8) was quantified using qPCR.
(H) G6PD activity of tumor pairs (n = 3) was measured via enzymatic assay.
(I) Tumor lysates (n = 3 pairs) were incubated with 25 μM DCFH-DA, and fluorescence was measured at 30 minutes.
All summary bar graphs represent mean ± SD. Statistical significances are noted as *p < 0.05, **p < 0.01 by two-tailed Student’s t tests.
To further characterize the phenotype, we performed metabolomic profiling of matched primary ovarian tumors and omental metastases from eight patients with stage III/IV high-grade serous OC (HGSOC). A targeted analysis reported relative quantities of 124 polar metabolites, and differential analysis of metabolite abundances was performed (Figure 1D). Pathway and enrichment analysis of the statistically significant hits identified alterations in PPP metabolism (Figures 1E and S1B). PPP metabolites emerged as highly differential between primary ovarian tumors and the corresponding omental metastasis (Figure 1F).
In matched patient primary OC-omental metastasis pairs, expression of G6PD, the rate-limiting enzyme of the PPP responsible for generating the key reducing equivalent NADPH, was increased in omental metastases, measured using qPCR (Figure 1G) and immunoblotting (Figures S1C and S1D). G6PD activity measured using an enzymatic assay was similarly increased in omental metastases (Figure 1H). We then used dichlorodihydrofluorescein diacetate (DCFH-DA), a cell-permeable fluorometric probe, to evaluate levels of intracellular ROS in the matched pairs. DCFH-DA assay measurements indicated higher intracellular ROS in extracts from omental metastases compared with primary tumors in these samples (Figure 1I).
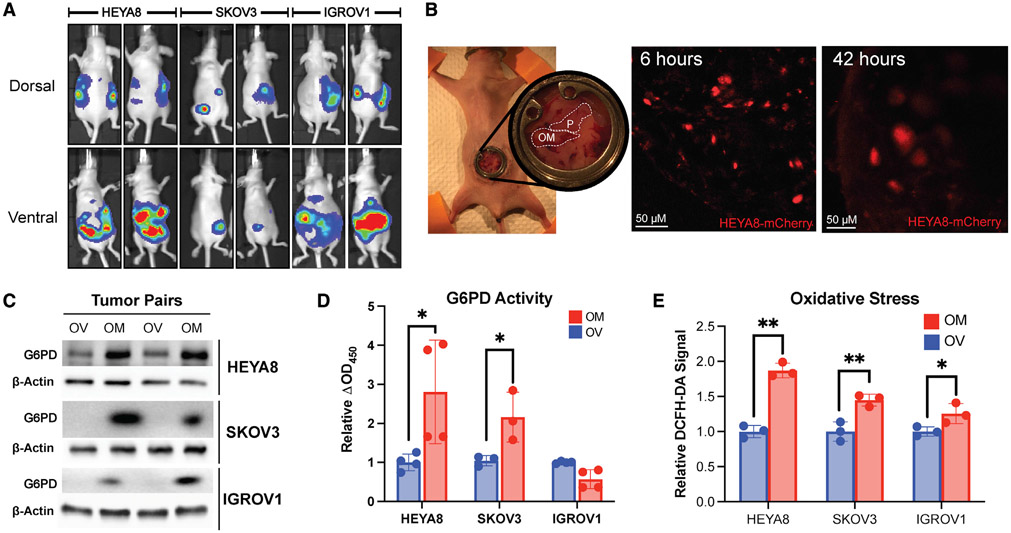
Using an established model of murine OC metastases to replicate the features of primary ovarian tumors and omental metastases, we performed intraperitoneal injections of each of three common HGSOC cell lines—HEYA8, SKOV3, and IGROV1—in immunodeficient mice (Shaw et al., 2004). These cell lines were transduced using lentivirus to stably express mCherry and luciferase. In vitro imaging system (IVIS) imaging of the xenografted nude mice at humane endpoints prior to sacrifice revealed omental metastases present in all cell line models (Figure 2A). To confirm, we used intravital imaging to follow metastatic seeding in the omentum after intraperitoneal injection of HEYA8-mCherry-labeled cells. At 6 h post intraperitoneal injection, disseminated micrometastases were visible in the mouse omentum; by 42 h, growth into larger nodules was seen (Figure 2B).
Figure 2. Murine models of ovarian cancer exhibit omental metastasis.
(A) HEYA8, SKOV3, and IGROV1 tumors were imaged by IVIS (n = 2 per cell line).
(B) Intravital imaging showed metastatic seeding and growth in the omentum at 6 and 42 h.
(C) Immunoblotting for G6PD expression in murine omental tumor samples.
(D) Enzymatic measurement of G6PD activity in primary tumors and omental metastases (n = 4).
(E) Tumor lysates were incubated with 25 μM DCFH-DA, and fluorescence was quantified (n = 3).
The sample size (n) represents the number of technical replicates. All summary bar graphs represent mean ± SD. Statistical significances are noted as *p < 0.05, **p < 0.01 by two-tailed Student’s t tests.
Western blots (WBs) revealed increased G6PD expression in omental metastases (Figure 2C), consistent with our previous observation from human samples (Figure 1G). Enzymatic assays showed increased G6PD activity in omental metastases of HEYA8 and SKOV3 but not in IGROV1, which did not exhibit preferential metastasis to the omentum (Figure 2D). DCFH-DA assays showed higher oxidative stress in omental metastases, congruent with the human samples as well (Figure 2E).
Tumor organoids were also derived from the ovarian primary tumor and from the omental metastases of the three different murine models (Figure S2A). CellROX oxidative stress assays did not show significant differences in oxidative stress between the ovarian and omental tumor organoids cultured in the same organoid media composition (Figure S2B), suggesting that oxidative stress is likely driven by omental microenvironmental factors.
Omental conditioned media increased oxidative stress and G6PD upregulation
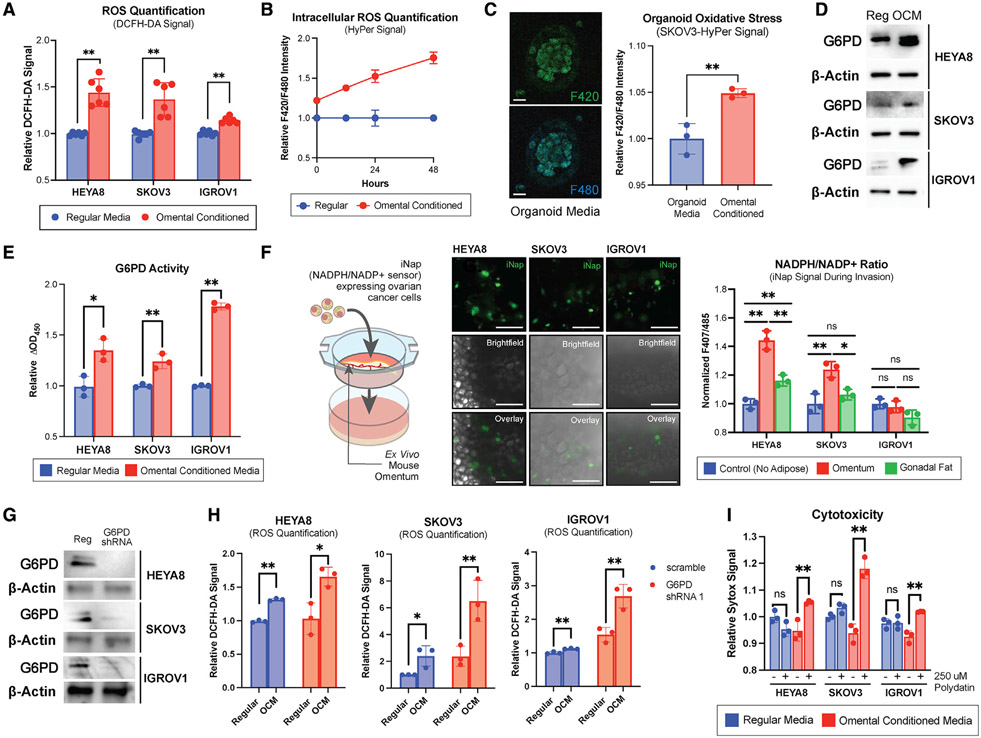
The use of omental conditioned media (OCM) has been extensively validated and characterized in the literature as a model of omental metastases (Clark et al., 2013). We cultured OC cells in media conditioned with omental adipose tissue. Using DCFH-DA as a readout of intracellular ROS, oxidative stress in the OC cells grown in DMEM + 10% FBS was compared with those grown in OCM (Figure 3A). Both HEYA8 and SKOV3 showed higher oxidative stress in OCM-grown conditions, while IGROV1 remained relatively unchanged.
Figure 3. G6PD inhibition sensitizes ovarian cancer cells to the increased oxidative stress of the omental microenvironment.
(A) OC cell lines grown in OCM were incubated with DCFH-DA, and fluorescence was quantified (n = 6).
(B) SKOV3 OC cells were transfected with constructs for the intracellular H2O2 sensor, HyPer (n = 3). Ratiometric fluorescence was measured at 0, 12, 24, and 48 h.
(C) HyPer signal quantification of OC organoids cultured in omental conditioned organoid media (n = 3). Scale bars, 100 μM.
(D and E) G6PD expression via immunoblotting (D) and G6PD activity measured via enzymatic assay (E) in OCM-cultured cells (n = 3).
(F) NADPH/NADP+ ratios of OC cells invading an ex vivo omental culture measured by iNAP, a NADPH/NADP+ ratiometric biosensor (n = 3). Scale bars, 100 μM.
(G) Immunoblotting for G6PD in OC cell lines after lentiviral shRNA knockdown.
(H) Quantification of oxidative stress measured via DCFH-DA signal in WT and G6PD shRNA-expressing OC cells cultured in OCM (n = 3).
(I) Quantification of cytotoxicity measured via Sytox staining of OCM and polydatin-treated OC cells (n = 3).
The sample size (n) represents the number of technical replicates. All summary bar graphs represent mean ± SD. Statistical significances are noted as *p < 0.05, **p < 0.01 by two-tailed Student’s t tests.
Given the rapid dynamics of the redox landscape and multiple layers of cellular regulation, we next sought to characterize oxidative stress using genetically encoded fluorescent biosensors, which could more precisely quantify and track endogenous ROS dynamics. These include rapid interchanges of superoxide anions [O2·−] and hydrogen peroxide (H2O2) and reducing equivalents like NADPH. We first utilized HyPer, a genetically encoded fluorescent biosensor of intracellular H2O2 to study OC cells cultured in OCM. After first establishing that HyPer signal was indeed responsive to oxidative stress induced by exogenous H2O2 (Figure S2C), we used a lentiviral vector to produce a SKOV3 cell line that stably expresses the HyPer biosensor. When grown in OCM, the HyPer signal increased over time, indicating higher oxidative stress in the cells (Figure 3B). Furthermore, to account for potential difference between two-dimensional (2D) and 3D culture, the biosensor-expressing cells were grown in 3D Matrigel to form OC organoids for dynamic metabolic analysis. Using ratiometric excitation at 420 and 480 nm with emission readings at 500 nm, fluorimetry was used to determine dynamic changes in oxidative stress. SKOV3-HyPer organoids exhibited higher oxidative stress when exposed to omental conditioned organoid culture media (Figure 3C).
HEYA8, SKOV3, and IGROV1 OC cells grown in OCM showed higher expression and activity levels of G6PD (Figures 3D and 3E). Furthermore, 10 OC cell lines reflected differing degrees of OCM-induced oxidative stress increases and G6PD activity increases (Figures S2D and S2E), suggesting that G6PD may be employed as a part of the cellular response to oxidative stress.
A key aspect of G6PD’s role in redox homeostasis involves the generation of NADPH, an essential electron donor in all organisms and a critical reducing power for anabolic reactions and redox balance. In order to determine if the process of metastasis into the omentum affected the cellular balance between NADPH and its oxidized counterpart, NADP+, a modified ex vivo 3D coculture model was developed (Krishnan et al., 2015). OC cells expressing iNAP, a ratiometric NADPH/NADP+ biosensor described in the literature (Tao et al., 2017), were cocultured with an omental explant, and the NADPH/NADP+ ratio was measured during the dynamic process of omental invasion. The NADPH/NADP+ ratio was markedly higher in HEYA8 and SKOV3 cells invading the omentum compared with simple migration (Figure 3F). Gonadal fat was also isolated and used as a control explant for invasion. OC cells invading omenta exhibited higher NADPH/NADP+ ratios than those invading gonadal fat, suggesting a greater need for NADPH production in omental-invading cancer cells.
G6PD inhibition sensitized OC cells to oxidative stress
We next investigated if OCM-grown cells experienced higher oxidative stress when compensatory G6PD mechanisms were inhibited. We first established successful lentiviral short hairpin RNA (shRNA) knockdown of G6PD using WB (Figure 3G) and then evaluated intracellular ROS using DCFH-DA. In HEYA8, SKOV3, and IGROV1 cells, shRNA knockdown of G6PD increased oxidative stress, with a combination of growth in OCM and shRNA knockdown responsible for higher oxidative stress than baseline Figure 3H).
Polydatin, a pharmacological inhibitor of G6PD (Mele et al., 2018), was then used to treat OC cells grown in regular media and OCM. Cytotoxicity was significantly increased in OCM-cultured cells treated with polydatin compared with normal culture conditions (Figure 3I). Measures of cellular viability including crystal violet and the formazan-based WST-8 assay reflected similar trends, with less viable cells evident in OCM-polydatin-treated samples (Figures S2F and S2G). In cells treated with polydatin, an increased NADPH/NADP+ ratio was also noted (Figure S2H). Treatment with a known ROS scavenger, N-acetylcysteine (NAC), reversed these cytotoxic effects in vitro, validating that increased oxidative stress caused by OCM growth was indeed responsible for the cell death seen in polydatin-treated cells (Figure S2I).
G6PD inhibition reduces omental metastases in vivo
Having established that G6PD inhibition could sensitize cells to OCM-induced oxidative stress, we next sought to explore its effect on omental metastases in vivo. HEYA8-mCherry-luciferase cells stably expressing G6PD knockdown shRNA were injected into NSG mice. Evaluation of tumor burden using IVIS imaging revealed reduced tumor growth in the G6PD-knockdown cohort (Figures S3A and S3B). At sacrifice, isolation of peritoneal fat deposits and subsequent fluorescence imaging of the mCherry-expressing tumors revealed greater omental tumor burden in the mice injected with G6PD-expressing HEYA8 cells than G6PD-knockdown HEYA8 cells (Figure S3C). Metastatic tumor growth in the gonadal and mesenteric fat was not significantly different between the two cohorts (Figures S3D and S3E).
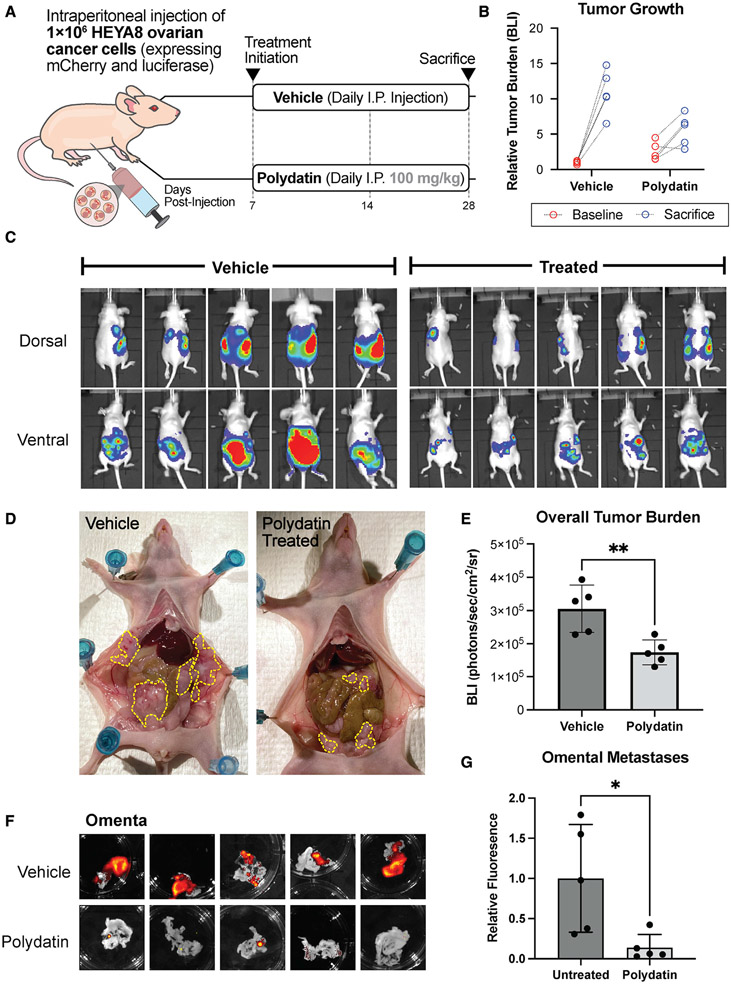
To study the effects of pharmacological G6PD inhibition via polydatin on OC tumor growth and metastasis in vivo, we then injected athymic nude mice with HEYA8 OC cells labeled with mCherry and luciferase tags. Initial tumor engraftment was not significantly different between the treatment and vehicle cohorts (Figures S4A and S4B). Daily doses of 100 mg/kg polydatin were injected intraperitoneally, and a control group of mice were injected with vehicle, as shown in Figure 4A. Tumor growth was followed longitudinally throughout the study using IVIS imaging, during which total bioluminescence (BLI) was quantified (Figure 4B). No adverse effects were noted with polydatin, and weights remained consistent between the vehicle and treatment cohorts (Figure S4E). To evaluate if polydatin treatment affected growth of metastatic tumors in the omentum, mice were imaged and sacrificed 14 days after treatment initiation. IVIS imaging revealed reduced overall tumor burden in the polydatin-treated cohort (Figures 4C and 4D) at the time of sacrifice. Upon gross dissection, primary tumors and associated peritoneal metastases were significantly larger in the vehicle-treated mice than those treated with polydatin (Figure 4E). Isolation of the omentum and subsequent fluorescence imaging for the mCherry-expressing tumors revealed large tumors in the vehicle-treated omenta, with only small puncta visible in the polydatin treated omenta (Figure 4F). Quantification of this difference revealed significantly reduced tumor growth in the omenta of polydatin-treated mice (Figure 4G). Staining of omental tumors was also performed and revealed similar tumor morphology between vehicle and polydatin-treated tumors (Figure S4F), and a reduction in metastases to the mesenteric fat but not gonadal fat (Figures S4C and S4D). Treatment with 30 mg/kg polydatin did not appear to significantly decrease tumor burden at the time of sacrifice nor metastases to the omentum, mesenteric, or gonadal fats (Figure S5).
Figure 4. Polydatin treatment reduces omental metastases in vivo.
(A) Schema for treatment.
(B) BLI was measured at baseline and prior to sacrifice using IVIS (n = 5 per cohort).
(C and D) Reduction in peritoneal metastases by polydatin treatment as measured by (C) IVIS imaging and (D) gross inspection of peritoneal tumor burden at necropsy.
(E) Overall tumor burden as measured as BLI on IVIS in polydatin and vehicle cohorts (**p < 0.01).
(F and G) Fluorescence imaging (F) and quantification (G) of omental metastases (*p < 0.05). All summary bar graphs represent mean ± SD.
DISCUSSION
The Warburg effect is a well-known adaptation of cancer that cancer cells preferentially perform aerobic glycolysis in lieu of the more energy-efficient oxidative phosphorylation. Further studies have revealed other pathways of metabolic reprogramming in cancer, including amino acid and lipid metabolism (Beloribi-Djefaflia et al., 2016; Faubert et al., 2020). As the scope and complexity of our understanding of tumor metabolism expands, important transformations between primary and metastatic tumor metabolism emerge as critical differentiators and barriers to metabolic drug development that would effectively target both. Thus, a deeper understanding of the microenvironmental responses that both drive and enable survival of metastatic cells is particularly valuable. With the advent of new biochemical tools, deeper explorations of the role of oxidative stress in promoting or challenging neoplastic growth may reveal novel avenues to study and potentially target cancer (Hayes et al., 2020).
Here, we report an important role for G6PD in OC metastases to the omentum. While G6PD expression and activity in primary tumors has recently emerged as an area of interest in other cancer types, the metabolic reprogramming of the omental microenvironment—with increased local oxidative stress from immune infiltrates and adipose tissue FA synthesis—requires OC cells to compensate effectively. We found that OC cells in this environment exhibit a dependency on G6PD, perhaps as a mechanism to provide this compensation, with shRNA knockdowns inducing significantly higher stress in the OC environment, and pharmacological inhibition using polydatin increasing cytotoxicity. In vivo, treatment with polydatin significantly reduced metastatic tumor burden in peritoneal fat deposits, including the omentum.
As G6PD is canonically considered an NADPH-producing enzyme, oxidative stress compensation may play a more important role in the metastatic process than previously appreciated in OC. The precise causality of this transformation—whether microenvironmental oxidative stress creates a demand for NADPH, simultaneously triggering FA metabolism, or vice versa—is not well understood and is likely dependent on multiple cellular processes. Indeed, OCM-induced oxidative stress and G6PD activity are heterogeneous in human, murine, and in vitro OC models, implicating multiple cellular mechanisms. In this study, we have focused on understanding the role of the PPP, a redox homeostasis mechanism potentially employed by metastatic, omental OC cells as the demand to neutralize ROS outpaces other NADPH-producing mechanisms.
The importance of the omental microenvironment in driving an aggressive transformation of OC metastases is of clinical relevance, given the burden of OC metastatic disease on patient outcomes and recurrent disease. The reasons underlying the observed increases in oxidative stress responses seen in metastatic cells are likely manifold and will require further studies to fully characterize. G6PD, a key cellular redox homeostasis enzyme, may play an important role in this response, as inhibition of this protein affected OC growth in the omental microenvironment both in vitro and in vivo. Thus, our studies suggest this metabolic state caused by the omental microenvironment poses a context-dependent vulnerability that warrants further investigation, particularly as a potential therapeutic target.
Limitations of the study
The precise mechanism of polydatin has not been well established, and whether inhibitory effects on G6PD mediates observed cardioprotective activity, anti-inflammatory, immunomodulatory, and antioxidative effects remains unclear (Du et al., 2013). In addition, the high polydatin dose used in the animal experiments (100 mg/kg daily) may pose barriers for clinical application in humans. Future investigations using other inhibitors, including the estrogen precursor dehydroepiandrosterone (DHEA), the monocarboxylic acid amide 6-aminonicotinamide (6-AN), or the more specific nonsteroidal inhibitor G6PDi-1 would be of interest (Ghergurovich et al., 2020).
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xiling Shen (xiling.shen@terasaki.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. Raw metabolomics data have been deposited at Mendeley and are publicly available as of the date of publication. DOIs are listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-G6PD antibody | Abcam | RRID:AB_296714 Cat #ab993 |
| Mouse anti-rabbit HRP secondary antibody | Cell Signal | RRID: AB_10892860 Cat #5127 |
| Biological samples | ||
| Human Ovarian Tumor Samples | Duke University School of Medicine Ovarian Cancer Research Biobank | ID: Pro00013710 (Banking Normal and Malignant Gynecologic Tissues Removed at Surgery) |
| Chemicals, peptides, and recombinant proteins | ||
| Polydatin | Sigma-Aldrich | Cat #15721 |
| LentiX Concentrator (100X) | Takara Bio | Cat # 631232 |
| Transdux Virus Transduction Reagent | System Biosciences | Cat # LV850A-1 |
| DCFH-DA (2′,7′-Dichlorofluorescein diacetate) | Millipore Sigma | Cat #35845 |
| Critical commercial assays | ||
| Glucose-6-Phosphate Dehydrogenase Activity Assay Kit | Millipore Sigma | Cat #MAK015 |
| SYTOX™ Blue Dead Cell Stain | Thermofisher Scientific | Cat #S34857 |
| RNeasy RNA Isolation Kit | Qiagen | Cat #74106 |
| RevertAid First Strand cDNA Synthesis Kit | Thermofisher Scientific | Cat #K1621 |
| Deposited data | ||
| High throughput sequencing of primary and metastatic tumor samples from patients with differing stage of serous epithelial ovarian cancer | Shih et al., 2018 | GEO: GSE118828 |
| Expression profiling by array of ovarian tumors and metastases from the omentum | Brodsky et al., 2014 | GEO: GSE30587 |
| Human HGSOC Ovarian Cancer Metabolomics (Primary vs. Omental Metastases) | This Paper | Mendeley Data: https://doi.org/10.17632/ztmzp4dc3y.1 |
| Experimental models: Cell lines | ||
| Human: HEYA8 cells | ATCC | RRID:CVCL_8878 |
| Human: SKOV3 cells | ATCC | RRID:CVCL_0532 |
| Human: IGROV1 cells | ATCC | RRID:CVCL_1304 |
| Human: TykNu cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: Ovaca424 cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: Ovaca432 cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: DOV13 cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: CaOV2 cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: Ovcar8 cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: OV90 cells | Duke Gynecological Oncology Core Resource | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: NOD.Cg-Prkdcscid/J | The Jackson Lab | RRID:IMSR_JAX:001303 |
| Strain #:001303 | ||
| Mouse: Outbred, Athymic Nude | The Jackson Lab | RRID:IMSR_JAX:002019 |
| (homozygous for Foxn1nu) | Strain #:002019 | |
| Oligonucleotides | ||
| pLKO-puro-G6PD shRNA (CAACAGATACAAGAACGTGAA) | RNAi Consortium shRNA Library (MISSION shRNA library) | TRC Clone ID: TRCN0000025817 |
| Actin (ACTB) TaqMan Assay (Hs01060665_g1) | Thermofisher Scientific | Cat #:4453320 |
| G6PD (G6PD) TaqMan Assay (Hs00166169_m1) | Thermofisher Scientific | Cat #:4453320 |
| Recombinant DNA | ||
| pAAV-HyPer-DAAO-NES | Steinhorn et al., 2018 | Addgene Plasmid #119164 |
| pLVX-IRES-Puro-iNap1 plasmid | Tao et al., (2017) (MTA from East China University) | N/A |
| Software and algorithms | ||
| Prism | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| R studio | RStudio | https://www.rstudio.com/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
De-identified human tumor tissues were obtained from Duke University School of Medicine Ovarian Cancer Research Biobank under protocol ID Pro00013710 (Banking Normal and Malignant Gynecologic Tissues Removed at Surgery), following donor’s provision of written informed consent. Eight patients with Stage III/IV HGSOC were identified and corresponding primary ovarian tumors and omental metastases were obtained. Further details can be found in Table S2. Tumor samples were isolated at the time of tumor resection and debulking surgery, and snap-frozen in liquid nitrogen at time of collection.
Animal studies
Athymic, nude (Foxn1nu) or NOD.Cg-Prkdcscid/J (NOD.scid) 6–8-week-old, female mice were obtained from the Jackson Laboratory and maintained in compliance with Institutional Animal Care and Use Committee (IACUC) guidelines. All animal care and protocols were in accordance with the Duke Animal Care and Use Program. Mice were group housed (3-5 mice per cage) in temperature controlled facility with a 12:12 h light:dark cycle. Food and water were provided ad libitum.
Cell culture studies
Ovarian cancer cell lines were obtained as described in the key resources table and were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS) (Corning Life Sciences) and 1% penicillin/streptomycin/L-glutamine (Corning Life Sciences). Cultures were maintained at 37°C in 5% CO2 incubator and were periodically screened for Mycoplasma contamination. HEYA8, SKOV3, and IGROV1 ovarian cancer cells were obtained from ATCC, and no further authentication was performed. The remaining cell lines were obtained and authenticated by the Duke Gynecological Oncology Core Resource, which performed polymorphic short tandem repeat (STR) profiling through the Duke University DNA Analysis Facility.
METHOD DETAILS
Human studies
For human sample assays, tissue was ground using mortar and pestle while maintained in liquid nitrogen, and ~20 mg of tissue was used for DCFH-DA assays, G6PD enzyme activity assays, and WBs. For metabolomics analysis, metabolite extraction from ground tissue normalized by weight was performed in cold extraction buffer (40% methanol: 40% acetonitrile: 20% water) solution. Extraction was performed by vortexing and centrifugation at 16,000 x g for 10 min, after which the supernatant was isolated and 15% NH4HCO3 was added to neutralize acids and precipitate proteins. Following centrifugation, the supernatant was applied to a speed vac and a concentrated metabolite pellet was obtained. Following reconstitution, the sample was used for LC-MS performed at the Rutgers Metabolomics Core Service.
Animal studies
Mice were injected with 1x106 ovarian cells intraperitoneally (n = 5 mice/group) for drug treatment studies or in the flank for organoid development. Where indicated, polydatin treatment (100 mg/kg) or 90% PBS +10% DMSO (as vehicle) was administered daily for 14 days. Bioimaging was performed at different time points/once per week starting one week post tumor engraftment using an in vivo IVIS (IVIS Kinetic). Peritoneal metastases were evaluated based on mCherry signals by an OV100 microscope (Olympus) after sacrifice and dissection. All studies were performed in compliance with institutional guidelines under an IACUC-approved protocol. Tumor specimens were isolated and either snap-frozen for further analysis or processed for immunostaining or organoid development.
Cell culture experiments
Constructs for HyPer-DAAO (Thomas Michel, Addgene) and iNAP (Yiping Yi, gift), two genetically encoded metabolic biosensors were transfected using Lipofectamine 2000 and were maintained in DMEM +10% FBS until fluorescent expression was seen under a fluorescence microscope (Olympus OV100).
Polydatin (Sigma-Aldrich) was dissolved in DMSO and aliquoted to stock solutions of 250 mM kept at −20, made fresh prior to cell culture experiments. Omental conditioned media was generated by isolating fresh omentum from sacrificed C57BL/6J mice. After 3 washes in PBS to remove residual clots from dissection, omenta were cultured in DMEM without FBS or penicillin/streptomycin for 48 h. The adipose tissue was then removed, and the media was passed through a 45 μM filter prior to use in cell culture applications.
Lentiviral mCherry/HyPer-DAAO/luciferase expression and shRNA knockdown
To generate lentivirus, HEK293T cells were transfected with psPAX2 and pMDG.2 packaging plasmids and the lentiviral expression construct. Viral supernatant was collected after 48 and 72 h and concentrated using LentiX concentrator (Takara Bio). This virus was used to transduce cells with Transdux (System Bio). To generate mCherry-luciferase expressing cell lines, cells were stably infected with lentiviral constructs and FACS was used to select based on mCherry-expression. To generate HyPer-DAAO expressing cell lines, HyPer-DAAO was cloned into a lentiviral vector backbone prior to lentiviral generation using HEK293T cells. Bacterial glycerol stocks of G6PD shRNAs in a pLKO-puromycin backbone were obtained from Mission (Millipore Sigma) and were used to generate lentivirus. Following infection of OC cells, selection was performed using 5 ug/mL puromycin.
Organoid cultures
In brief, organoids were established according to previously described methods (de Witte et al., 2020). Following isolation, tumor tissue was stored in AdDF+++ (Advanced DMEM/F12 containing 1x Glutamax, 10 mM HEPES and antibiotics), prior to being washed 3x in ice-cold PBS. Tissue was then minced finely in a small amount of HBSS and centrifuged to form a cell pellet. Digestion was carried out using AdDF+++ supplemented with 5 μM RHO/ROCK pathway inhibitor (Abmole Bioscience, Y-27632) containing 0.5–1.0 mg/mL collagenase (Sigma, C9407) at 37°C for 1 h, after which cells were spun down. The remaining cell pellet was resuspended in Matrigel (Corning Sciences, Inc.) and plated in 25 μL domes in 24 well plate. Organoid culture media composition is shown in Table S1.
Ex vivo omental invasion As’say
Using omenta isolated from sacrificed C57BL/6J mice, we performed an ex vivo assay as previously described (Krishnan et al., 2015). In short, after washing the omentum in HBSS, a small amount of Matrigel was applied to the bottom of a transwell plate and the omentum was gently placed upon it. Similarly, a thin strip of gonadal fat was used as a control and placed on the bottom of a transwell insert in a similar fashion. After 10 min at 37 degrees, DMEM +10% FBS +1% penicillin/streptomycin was applied to the bottom chamber, and serum-free DMEM was applied to the top chamber. OC cells expressing a fluorescence biosensor were applied to the top chamber and the fluorescence was quantified using a plate reader (Varioskan and Incucyte) imaging during the invasion process.
DCFH-DA assay
Within cells, the acetate groups of DCFH-DA are first cleaved by cellular esterizes to produce the intermediate DCFH, which then undergoes two electron oxidation reactions by intracellular ROS to generate the fluorogenic molecule DCF (Aranda et al., 2013). Cells were incubated in serum-free DMEM containing 10 μM DCFH-DA solution for 30 min prior to fluorescence readings at an excitation wavelength of 485 nm and an emission wavelength of 530 nm.
G6PD activity assay
The G6PD enzyme activity (Sigma Aldrich) was performed according to the supplier’s instructions and as described previously (Jiang et al., 2011; Tian et al., 1998). Briefly, G6PD activity in tumor lysate was measured by the rate of conversion of NADP + to NADPH in the presence of a fluorogenic NADPH sensor. The resulting fluorescent signal (excitation/emission = 540/590 nm) was monitored using a fluorescence microplate reader (Varioskan) in kinetic mode for 1 h. Enzyme activities were normalized to protein concentration, measured using BCA Assay Kit (Pierce).
Cytotoxicity/viability assays
Cells were plated at appropriate confluence in a 96-well plate (Corning Life Sciences), and allowed to seed for 24 h, after which media was aspirated and replaced with appropriate treatment media. For viability assays, cells were allowed to grow for 48 h, after which media was again aspirated and cells were incubated with Crystal Violet stain for 30 min. Stain was removed and wells were washed with PBS until only adherent cells appeared stained under light microscopy. Plate was allowed to air dry and appropriate solubilization agent was added, after which absorbance at 570 nm was read (Varioskan). For cytotoxicity assays, cells were allowed to grow for 72 h, after which media was aspirated and cells were incubated with 2.5 μM Sytox Blue Reagant dissolved in PBS for 30 minutes at RT. Fluorescence was measured at excitation/emission of 405 nm/488 nm (Varioskan).
Immunostaining
Immediately following dissection, tumors were isolated and washed in PBS, prior to immersion in 30% sucrose overnight. After fixation in 4% neutral-buffered formalin, tumors were then embedded in Tissue-Tek® O.C.T. Compound (Sakura Finetek) and sectioned with a cryostat at 20 μm. Tumor sections were blocked and probed first with G6PD antibody (Abcam) followed by incubation with an HRP-conjugated secondary antibody (R&D Systems). The sections were then stained with DAB and counterstained with hematoxylin. For Ki67 immunohistochemistry, sections were incubated with anti-Ki67 antibody (Abcam), followed by immunostaining with a fluorescent-labeled secondary antibody (Invitrogen). Slides were counterstained using DAPI (Sigma-Aldrich). Finally, H&E staining was also performed as previously described (Fischer et al., 2008). Briefly, slides were immersed in a hematoxylin solution, followed by a series of rinses and counterstaining with eosin prior to application of a clear coverslip. These slides were imaged using color microscopy (Zeiss Axio Z2) or confocal microscopy (Leica SP8 Confocal).
Quantitative PCR
qPCR experiments were performed as previously described. Briefly, human tissue samples were ground using mortar and pestle in liquid nitrogen, and RNA was isolated using RNeasy kit (Qiagen) and converted to cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher). We performed qPCR with a Taqman Gene Expression Assay using a QuantStudio 12K Flex machine (Applied Biosystems). Primers were obtained from ThermoFisher for Actin (Catalog #4331182) and G6PD (Catalog #4331182).
Western blotting
Immunoblotting experiments were performed as previously described. Briefly, whole-cell lysates were generated by lysing cells in 50 mM Tris pH 7.5, 150 mM NaCl, 0.5% NP40, 1 mM EDTA, 10% glycerol, protease inhibitor mix (Roche) and phosphatase inhibitor cocktail (Sigma). Total protein amount was measured using a BCA assay, after which protein loading was normalized across samples. A Varioskan plate reader was used (Molecular Devices, Sunnyvale, CA, USA).
QUANTIFICATION AND STATISTICAL ANALYSIS
Computational analysis
Two NCBI Gene Expression Omnibus (GEO) databases (GEO: GSE118828 and GSE30587) were identified that contain transcriptomic profiles of 30 primary ovarian tumors and 30 matched omental metastases from patients with Stage III high grade serous ovarian adenocarcinomas. High throughput RNA sequencing data was normalized to the primary tumor means and log2 transformed. Microarray expression data was analyzed using the Bioconductor package in R Studio, and normalized data was pooled for metanalysis. Hierarchical clustering was performed. The 20 patients with the greatest variation between transcriptomic profiles were identified by calculations of Euclidean distance between vectors of gene expression in omental metastasis and primary tumors. This cohort was used to perform pathway analysis in EnrichR (R package).
Statistical analysis
Statistics were performed using GraphPad Prism7 (GraphPad Software) or R and are specified in each figure legend. All data were tested for normality. Paired and unpaired two-tailed Student’s t-tests were used as appropriate for each experiment. A p value of <0.05 was considered significant, the level of significance is indicated as follows: * = p < 0.05, ** = p < 0.01; n.s. indicates no significance. All data are presented as mean ± SD.
Supplementary Material
Highlights.
The omental microenvironment is a high-oxidative-stress metastatic niche for OC cells
G6PD, a key redox-maintaining PPP enzyme, is upregulated in omental metastases
G6PD inhibition increases OC oxidative stress in the omental microenvironment
Pharmacological G6PD inhibition reduces omental metastases in vivo
ACKNOWLEDGMENTS
This work was supported by National Cancer Institute grants NIH-U01CA217514 and U01CA214300 as well as National Institutes of Health F30 fellowship 1F30CA257365-01. We would like to thank Dr. Yiping Yi (East China University) for providing the NADPH/NADP+ sensor iNAP for use in this study and Dr. Thomas Michel (Harvard Medical School) for providing the intracellular hydrogen peroxide HyPer-DAAO sensor. We would like to thank Regina Whitaker and the Duke OB/GYN clinical team for their assistance obtaining clinical samples. Services, results, and/or products in support of the research project were generated by the Rutgers Cancer Institute of New Jersey Metabolomics Shared Resource, supported in part with funding from NCICCSG P30CA072720-5923.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111012.
DECLARATION OF INTERESTS
X.S. is a co-founder of Xilis, Inc. This manuscript does not have any overlap with Xilis’s commercial interests.
REFERENCES
- Aranda A, Sequedo L, Tolosa L, Quintas G, Burello E, Castell J, and Gombau L (2013). Dichloro-dihydro-fluorescein diacetate (DCFH-DA) assay: a quantitative method for oxidative stress assessment of nanoparticle-treated cells. Toxicol. Vitro 27, 954–963. [DOI] [PubMed] [Google Scholar]
- Beloribi-Djefaflia S, Vasseur S, and Guillaumond F (2016). Lipid metabolic reprogramming in cancer cells. Oncogenesis 5, e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky AS, Fischer A, Miller DH, Vang S, MacLaughlan S, Wu HT, Yu J, Steinhoff M, Collins C, Smith PJ, and Raphael BJ (2014). Expression profiling of primary and metastatic ovarian tumors reveals differences indicative of aggressive disease. PloS one 9, e94476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang Z, Hoshino A, Zheng HD, Morley M, Arany Z, and Rabinowitz JD (2019). NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nature Metab. 1, 404–415. [PMC free article] [PubMed] [Google Scholar]
- Clark R, Krishnan V, Schoof M, Rodriguez I, Theriault B, Chekmareva M, and Rinker-Schaeffer C (2013). Milky spots promote ovarian cancer metastatic colonization of peritoneal adipose in experimental models. Am. J. Pathol 183, 576–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Witte CJ, Valle-Inclan JE, Hami N, Lohmussaar K, Kopper O, Vreuls CPH, Jonges GN, van Diest P, Nguyen L, and Clevers H (2020). Patient-derived ovarian cancer organoids mimic clinical response and exhibit heterogeneous inter-and intrapatient drug responses. Cell Rep. 31, 107762. [DOI] [PubMed] [Google Scholar]
- Du QH, Peng C, and Zhang H (2013). Polydatin: a review of pharmacology and pharmacokinetics. Pharmaceutical biology 51 (11), 1347–1354. [DOI] [PubMed] [Google Scholar]
- Faubert B, Solmonson A, and DeBerardinis RJ (2020). Metabolic reprogramming and cancer progression. Science 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Li X, Sun W, Sun M,Li Z, Sheng H, Xie F, Zhang S, and Shan C (2020). Targeting G6PD reverses paclitaxel resistance in ovarian cancer by suppressing GSTP1. Biochem. Pharmacol 178, 114092. [DOI] [PubMed] [Google Scholar]
- Fidler IJ (2003). The pathogenesis of cancer metastasis: the ’seed and soil’ hypothesis revisited. Nat. Rev. Cancer 3, 453–458. 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Fischer AH, Jacobson KA, Rose J, and Zeller R (2008). Hematoxylin and eosin staining of tissue and cell sections. Cold Spring Harb. Protoc 2008. pdb. prot4986. [DOI] [PubMed] [Google Scholar]
- Gharpure KM, Pradeep S, Sans M, Rupaimoole R, Ivan C, Wu SY, Bayraktar E, Nagaraja AS, Mangala LS, Zhang X, and Haemmerle M (2018). FABP4 as a key determinant of metastatic potential of ovarian cancer. Nat. Commun 9, 2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghergurovich JM, García-Carñaveras JC, Wang J, Schmidt E, Zhang Z, TeSlaa T, Patel H, Chen L, Britt EC, Piqueras-Nebot M, and Gomez-Cabrera MC (2020).A small molecule G6PD inhibitor reveals immune dependence on pentose phosphate pathway. Nature chemical biology 16, 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Dinkova-Kostova AT, and Tew KD (2020). Oxidative stress in cancer. Cancer Cell 38, 167–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herroon MK, Rajagurubandara E, Diedrich JD, Heath EI, and Podgorski I (2018). Adipocyte-activated oxidative and ER stress pathways promote tumor survival in bone via upregulation of Heme Oxygenase 1 and Survivin. Sci. Rep 8, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, and Yang X (2011). p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol 13, 310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Clark R, Chekmareva M, Johnson A, George S, Shaw P, Seewaldt V, and Rinker-Schaeffer C (2015). In vivo and ex vivo approaches to study ovarian cancer metastatic colonization of milky spot structures in peritoneal adipose. JoVE. 10.3791/52721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladanyi A, Mukherjee A, Kenny HA, Johnson A, Mitra AK, Sundaresan S, Nieman KM, Pascual G, Benitah SA, Montag A, et al. (2018). Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 37, 2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel E (2010). Ovarian cancer development and metastasis. Am. J. Pathol 177, 1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel E, Makowski L, DiGiovanni J, and Kolonin MG (2018). Cancer as a matter of fat: the crosstalk between adipose tissue and tumors. Trends Cancer 4, 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, and Karlan BY (2016). Ovarian cancer. Nat. Rev. Dis. Prim 2, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele L, Paino F, Papaccio F, Regad T, Boocock D, Stiuso P, Lombardi A, Liccardo D, Aquino G, and Barbieri A (2018).A new inhibitor of glucose-6-phosphate dehydrogenase blocks pentose phosphate pathway and suppresses malignant proliferation and metastasis in vivo. Cell Death Dis. 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Chiang C-Y, Daifotis HA, Nieman KM, Fahrmann JF, Lastra RR, Romero IL, Fiehn O, and Lengyel E (2020). Adipocyte-induced FABP4 expression in ovarian cancer cells promotes metastasis and mediates carboplatin resistance. Cancer Res. 80, 1748–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, et al. (2011). Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nature Med. 17, 1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman KM, Romero IL, Van Houten B, and Lengyel E (2013). Adipose tissue and adipocytes support tumorigenesis and metastasis. Biochim. Biophys. Acta 1831, 1533–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw TJ, Senterman MK, Dawson K, Crane CA, and Vanderhyden BC (2004). Characterization of intraperitoneal, orthotopic, and metastatic xenograft models of human ovarian cancer. Mol. Ther 10, 1032–1042. [DOI] [PubMed] [Google Scholar]
- Shih AJ, Menzin A, Whyte J, Lovecchio J, Liew A, Khalili H, Bhuiya T, Gregersen PK, and Lee AT (2018). Identification of grade and origin specific cell populations in serous epithelial ovarian cancer by single cell RNA-seq. PLoS one 3, e0206785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhorn B, Sorrentino A, Badole S, Bogdanova Y, Belousov V, and Michel T (2018). Chemogenetic generation of hydrogen peroxide in the heart induces severe cardiac dysfunction. Nature communications 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Zhao Y, Chu H, Wang A, Zhu J, Chen X, Zou Y, Shi M, Liu R, and Su N (2017). Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat. Methods 14, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W-N, Braunstein LD, Pang J, Stuhlmeier KM, Xi Q-C, Tian X, and Stanton RC (1998). Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J. Biol. Chem 273, 10609–10617. [DOI] [PubMed] [Google Scholar]
- Yamawaki K, Mori Y, Sakai H, Kanda Y, Shiokawa D, Ueda H, Ishiguro T, Yoshihara K, Nagasaka K, and Onda T (2021). Integrative analyses of gene expression and chemosensitivity of patient-derived ovarian cancer spheroids link G6PD-driven redox metabolism to cisplatin chemoresistance. Cancer Lett. 521, 29–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. Raw metabolomics data have been deposited at Mendeley and are publicly available as of the date of publication. DOIs are listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-G6PD antibody | Abcam | RRID:AB_296714 Cat #ab993 |
| Mouse anti-rabbit HRP secondary antibody | Cell Signal | RRID: AB_10892860 Cat #5127 |
| Biological samples | ||
| Human Ovarian Tumor Samples | Duke University School of Medicine Ovarian Cancer Research Biobank | ID: Pro00013710 (Banking Normal and Malignant Gynecologic Tissues Removed at Surgery) |
| Chemicals, peptides, and recombinant proteins | ||
| Polydatin | Sigma-Aldrich | Cat #15721 |
| LentiX Concentrator (100X) | Takara Bio | Cat # 631232 |
| Transdux Virus Transduction Reagent | System Biosciences | Cat # LV850A-1 |
| DCFH-DA (2′,7′-Dichlorofluorescein diacetate) | Millipore Sigma | Cat #35845 |
| Critical commercial assays | ||
| Glucose-6-Phosphate Dehydrogenase Activity Assay Kit | Millipore Sigma | Cat #MAK015 |
| SYTOX™ Blue Dead Cell Stain | Thermofisher Scientific | Cat #S34857 |
| RNeasy RNA Isolation Kit | Qiagen | Cat #74106 |
| RevertAid First Strand cDNA Synthesis Kit | Thermofisher Scientific | Cat #K1621 |
| Deposited data | ||
| High throughput sequencing of primary and metastatic tumor samples from patients with differing stage of serous epithelial ovarian cancer | Shih et al., 2018 | GEO: GSE118828 |
| Expression profiling by array of ovarian tumors and metastases from the omentum | Brodsky et al., 2014 | GEO: GSE30587 |
| Human HGSOC Ovarian Cancer Metabolomics (Primary vs. Omental Metastases) | This Paper | Mendeley Data: https://doi.org/10.17632/ztmzp4dc3y.1 |
| Experimental models: Cell lines | ||
| Human: HEYA8 cells | ATCC | RRID:CVCL_8878 |
| Human: SKOV3 cells | ATCC | RRID:CVCL_0532 |
| Human: IGROV1 cells | ATCC | RRID:CVCL_1304 |
| Human: TykNu cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: Ovaca424 cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: Ovaca432 cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: DOV13 cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: CaOV2 cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: Ovcar8 cells | Duke Gynecological Oncology Core Resource | N/A |
| Human: OV90 cells | Duke Gynecological Oncology Core Resource | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: NOD.Cg-Prkdcscid/J | The Jackson Lab | RRID:IMSR_JAX:001303 |
| Strain #:001303 | ||
| Mouse: Outbred, Athymic Nude | The Jackson Lab | RRID:IMSR_JAX:002019 |
| (homozygous for Foxn1nu) | Strain #:002019 | |
| Oligonucleotides | ||
| pLKO-puro-G6PD shRNA (CAACAGATACAAGAACGTGAA) | RNAi Consortium shRNA Library (MISSION shRNA library) | TRC Clone ID: TRCN0000025817 |
| Actin (ACTB) TaqMan Assay (Hs01060665_g1) | Thermofisher Scientific | Cat #:4453320 |
| G6PD (G6PD) TaqMan Assay (Hs00166169_m1) | Thermofisher Scientific | Cat #:4453320 |
| Recombinant DNA | ||
| pAAV-HyPer-DAAO-NES | Steinhorn et al., 2018 | Addgene Plasmid #119164 |
| pLVX-IRES-Puro-iNap1 plasmid | Tao et al., (2017) (MTA from East China University) | N/A |
| Software and algorithms | ||
| Prism | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| R studio | RStudio | https://www.rstudio.com/ |