Abstract
Many species of gram-positive bacteria produce branched peptidoglycan precursors resulting from the transfer of various l-amino acids or glycine from amino acyl-tRNA to the ɛ-amino group of l-lysine. The UDP-MurNAc-pentapeptide:l-alanine ligase and alanyl-tRNA synthetase genes from Enterococcus faecalis were identified, cloned, and overexpressed in Escherichia coli. The purified enzymes were necessary and sufficient for tRNA-dependent addition of l-alanine to UDP-MurNAc-pentapeptide in vitro. The ligase belonged to the Fem family of proteins, which were initially identified genetically as factors essential for methicillin resistance in Staphylococcus aureus.
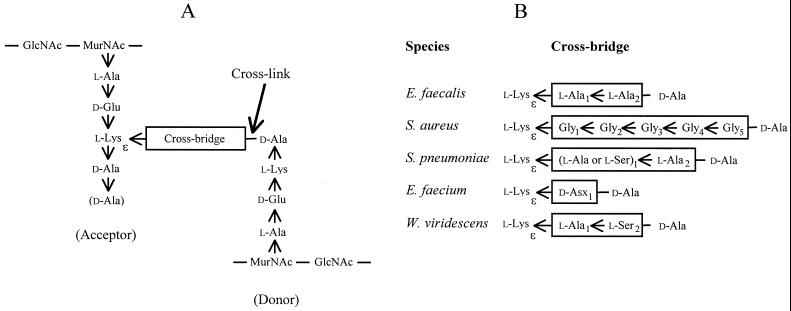
In gram-positive bacteria, the ɛ-amino group of l-lysine in the pentapeptide stem of peptidoglycan precursors is often replaced by amino acid chains of various lengths and compositions (19) (Fig. 1). These amino acids form cross bridges between l-Lys3 and d-Ala4 following cross-linking of the peptide stems from nascent chains of peptidoglycan by the d,d-transpeptidases. The ligases for the addition of glycine and l-amino acids to the ɛ-amino group of l-lysine use aminoacyl-tRNAs as substrates, whereas d-amino acids are added in a tRNA-independent reaction (15, 20).
FIG. 1.
Structure of cross-linked peptidoglycan. (A) Fragment of the primary structure of peptidoglycan. The position of the peptide bond formed by the d,d-transpeptidases (penicillin-binding proteins) between the acceptor and donor peptide stems is indicated (cross-link). The C-terminal d-Ala residue of the pentapeptide acceptor stem (shown in parentheses) is generally cleaved by d,d-carboxypeptidases. (B) Sequence of the cross bridges in various bacterial species. The amino acid residues are added to the ɛ-amino group of l-Lys of peptidoglycan precursors by various ligases discussed in the text. d-Asp is incorporated into the peptidoglycan precursors of E. faecium and is secondarily partially amidated (d-Asx may be d-Asp or d-Asn), according to Schleifer and Kandler (19). Arrows indicate CO→NH orientation of peptide bonds.
In Staphylococcus aureus, the femA, femB, and fmhB genes were shown to be essential for incorporation of glycine into the side chain of peptidoglycan precursors (18, 21). The femAB locus was initially identified as a factor essential for methicillin resistance (fem) based on random insertional inactivation of chromosomal genes and a screen for reduced expression of resistance mediated by the penicillin binding protein 2A (PBP2A) (2, 5). Inactivation of femA or femB was subsequently reported to prevent incorporation of glycine residues at positions 2 to 5 or positions 4 to 5 of the penta-glycine cross bridge since muropeptides cross-linked by one or three glycine residues were detected in the corresponding mutants (4, 21). Inactivation of fmhB, formerly femX, is lethal, but the construction of a mutant conditionally expressing fmhB under the control of a xylose-inducible promoter showed that the gene was essential for synthesis of branched peptidoglycan precursors (18). These results indicated that the fem gene products were required for incorporation of glycine at positions 1 (FmhB), 2 and 3 (FemA), and 4 and 5 (FemB) of the cross bridge, although the catalytic activity of the proteins was not directly assessed (18). Similarly, inactivation of two fmhB homologues in Streptococcus pneumoniae, designated murM (fibA) and murN (fibB), reduced addition of l-Ala or l-Ser to the ɛ-amino group of l-Lys and subsequent addition of a second l-Ala residue, respectively (6, 7, 23). Overall, disruption of the murMN operon reduced the proportion of branched peptide stems in the peptidoglycan from 89 to 33% (6). In contrast to what occurs in S. aureus (18), direct cross-linking of l-Lys to d-Ala occurs in S. pneumoniae, and the murMN operon was accordingly reported to be unessential (6). However, production of branched peptidoglycan precursors was necessary for expression of penicillin resistance (6) and appeared to affect mainly the activity of mosaic PBP2x (23).
In enterococci, streptococci, and staphylococci, the ligation reactions are catalyzed by membrane-associated enzymes acting exclusively or preferentially on late peptidoglycan precursors linked to the undecaprenyl-phosphate lipid carrier, namely undecaprenyl-PP-MurNAc-pentapeptide (lipid I) and undecaprenyl-PP-MurNAc(pentapeptide)-GlcNAc (lipid II) (14). This conclusion is supported by the detection of only small amounts (<4%) of UDP-MurNAc-hexapeptide in Staphylococcus haemolyticus and S. aureus in spite of high-level accumulation (ca. 12-fold) of UDP-MurNAc-pentapeptide in bacteria treated with ramoplanin, which blocks lipid II synthesis (3). Similarly, moderate accumulation of l-Ala-containing UDP-MurNAc-hexapeptide was detected in penicillin-treated Enterococcus faecalis (12). Finally, UDP-MurNAc-pentapeptide was not a substrate of the ligases of S. aureus and Streptococcus pyogenes, suggesting that the enzymes act at a later step of peptidoglycan synthesis (8). In contrast, Weissella viridescens, formerly Lactobacillus viridescens, produces a soluble UDP-MurNAc-pentapeptide:l-alanine ligase (15). The enzyme was purified and characterized in 1970 (16), but the corresponding gene was identified only recently (8). In this study, the alanyl-tRNA synthetase, UDP-MurNAc-pentapeptide:l-alanine ligase, and tRNA from W. viridescens were partially purified to develop an assay for ligation of l-alanine to UDP-MurNAc-pentapeptide. This assay was used to identify the alanyl-tRNA synthetase and ligase genes for incorporation of the first l-alanine of the E. faecalis cross bridge.
MATERIALS AND METHODS
In vitro UDP-MurNAc-hexapeptide synthesis.
The assay was performed in a total volume of 15 μl containing Tris-HCl (50 mM, pH 7.2), MgCl2 (12.5 mM), ATP (2.5 mM), l-[14C]alanine (67 μM, 93 mCi/mmol; ICN, Costa Mesa, Calif.), UDP-MurNAc-pentapeptide (67 μM) purified from S. aureus as previously described (1), and fractions containing tRNA (4 μg), alanyl-tRNA synthetase, and UDP-MurNAc-pentapeptide:l-alanine ligase. The reaction mixture was incubated 30 min at 30°C and the reaction was stopped by heating at 96°C for 2 min. l-[14C]alanine was separated from [14C]UDP-MurNAc-hexapeptide by descending paper chromatography (Whatman no. 4 filter paper) with a mobile phase composed of isobutyric acid and 1 M ammonia (5:3, vol/vol). The radioactive spots were located and quantified with a radioactivity scanner (model Multi-Tracermaster LB285; EG and G Wallac/Berthold, Courtaboeuf, France). l-Alanine and UDP-MurNAc-hexapeptide were also separated by high-pressure liquid chromatography (HPLC) at 30°C on a Hypersil C18 column (3 μm, 4.6 by 250 mm; Interchrom, Montluçon, France) at a flow rate of 0.5 ml/min with a 0 to 4% acetonitrile gradient in 10 mM ammonium acetate, pH 5.0 (3). Products were detected by the absorbance at 262 nm and, when appropriate, by liquid scintillation with a Radioflow Detector (LB508; Perkin-Elmer, Courtaboeuf, France) coupled to the HPLC apparatus (L-62000A; Merck, Nogent-sur-Marne, France).
Partial purification of W. viridescens UDP-MurNAc-pentapeptide:l-alanine ligase, alanyl-tRNA synthetase, and tRNA.
W. viridescens CIP 102810 T (Institut Pasteur collection), formerly designated L. viridescens ATCC 12706 (15), was grown to late exponential phase in lactobacillus MRS broth (Difco, Elancourt, France). Bacteria were ground with alumina in a chilled mortar at 4°C. Soluble proteins were loaded onto a DEAE Sepharose Fast Flow column (XK26/70; Pharmacia, Saclay, France) and eluted with a linear (0 to 300 mM) KCl gradient as previously described (16). Fraction I contained UDP-MurNAc-pentapeptide:l-alanine ligase and alanyl-tRNA synthetase activity (elution between 170 and 200 mM KCl). A 10-ml fraction eluting at ca. 250 mM KCl was treated with phenol, sodium acetate (pH 5.2) was added to a final concentration of 0.3 M, and nucleic acids were precipitated with 3 volumes of ethanol at −20°C for 30 min. The preparation was centrifuged in aliquots in a microcentrifuge at 4°C for 20 min. The pellets were washed with ethanol, dried under vacuum, and resuspended in a total volume of 10 ml of water. The resulting preparation, designated fraction II, contained tRNA (2 μg/μl), as judged by agarose gel electrophoresis and absorbance measurements.
Solid ammonium sulfate was added to the above-mentioned DEAE fraction I to a final concentration of 1.4 M, and soluble proteins were recovered by centrifugation. The preparation was loaded onto a phenyl sepharose 6 Fast Flow (high sub) column (XK50/30; Pharmacia) equilibrated with 50 mM potassium phosphate (pH 7.0) containing 1.4 M ammonium sulfate. Elution was performed with a linear 1.4 to 0 M ammonium sulfate gradient. UDP-MurNAc-pentapeptide:l-alanine ligase and alanyl-tRNA synthetase activities eluted at 900 to 800 mM (fraction III, 9 mg of protein) and 300 to 260 mM (fraction IV, 40 mg of protein) ammonium sulfate, respectively.
Plasmid construction.
Three E. faecalis genes encoding proteins related to the Bacillus subtilis alanyl-tRNA synthetase (alaSEfa) and to the S. aureus FmhB protein (orf1 and orf2) were identified based on sequence comparison at The Institute for Genomic Research (TIGR) website (http://www.tigr.org). Genomic DNA from E. faecalis JH2-2 (9) was prepared (11), and the alaSEfa, orf1, and orf2 genes were amplified by PCR using the pwo DNA polymerase (Roche, Meylan, France). The PCR products were purified by agarose gel electrophoresis and cloned into vector pCRblunt (Invitrogen, Groningen, The Netherlands) generating plasmids pDA18(alaSEfa), pDA4(orf1), and pDA15(orf2).
The alanyl-tRNA synthetase gene was subcloned into vector pTrcHis60 (17) to generate a translational fusion with 6 codons specifying His at the 3′ end of the gene. The resulting plasmid construct, designated pDA26, was obtained by cloning the BspHI-BamHI fragment of pDA18 between the NcoI and BglII sites of pTrcHis60.
Plasmid pDA28(orf1) was generated by ligating the SacI-SmaI fragment of pDA4 with pTrc99A (Pharmacia) digested with the same enzymes placing orf1 under the control of the ptrc promoter of the vector. Similarly, pDA29(orf2) was obtained by cloning orf2 of pDA15 into pTrc99A using SacI and SmaI.
Derivatives of pCRblunt were propagated in Escherichia coli Top10 (Invitrogen). E. coli JM83 (22) was the host for other plasmids. All cultures of E. coli strains were performed at 37°C in brain heart infusion (BHI) broth or agar (Difco) containing ampicillin (100 μg/ml) or kanamycin (50 μg/ml) for plasmid selection.
Purification of the E. faecalis alanyl-tRNA synthetase.
E. coli harboring recombinant plasmid pDA26(alaSEfa) was grown to an optical density at 600 nm of ca. 0.3 in 1 liter of BHI broth containing 100 μg of ampicillin per ml, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and incubation was continued for 2 h. Bacteria were harvested by centrifugation (8,000 × g for 20 min at 4°C), washed in 100 ml of 50 mM Tris-HCl, pH 7.5, and resuspended in 5 ml of the same buffer containing 2 mM 2-mercaptoethanol and 300 mM KCl (buffer A). Bacteria were disrupted by sonication with a Branson Sonifier 450 for 2 min with cooling, the extract was centrifuged at 27,000 × g for 30 min at 4°C, and the supernatant (crude extract) was mixed with 4 ml of Ni2+-nitrilotriacetate-agarose resin (Pharmacia) previously equilibrated with buffer A. After 2 h of incubation at 4°C, the resin was recovered by centrifugation and washed with buffer A containing increasing concentrations of imidazole (0, 10, 15, 20, 25, 30, 40, 100, and 200 mM). Proteins eluting at 200 mM imidazole were loaded onto a Superdex 75 HR10/30 gel filtration column (Pharmacia) equilibrated with 50 mM Tris-HCl, pH 7.5, containing 2 mM 2-mercaptoethanol.
Preparation of E. faecalis crude extracts containing ORF1 and ORF2.
E. coli JM83/pDA28(orf1) and JM83/pDA29(orf2) were grown to an optical density at 600 nm of 0.7 in 1 liter of BHI broth containing 100 μg of ampicillin per ml, and induction was performed with 1 mM IPTG for 2 h. Bacteria were collected by centrifugation (8,000 × g for 20 min at 4°C), washed in 100 ml of 50 mM Tris-HCl, pH 7.2, and resuspended in 5 ml of the same buffer containing 2 mM 2-mercaptoethanol (buffer B). Bacteria were disrupted by sonication and centrifuged (27,000 × g for 30 min at 4°C), and the supernatants were collected (crude extracts).
Purification of E. faecalis ORF2.
The crude extract from E. coli JM83/pDA29(orf2) was loaded onto a 10-ml Bio-Scale Q anion exchange column (Bio-Rad, Ivry sur Seine, France) equilibrated in buffer B at a flow rate of 1 ml/min, and elution was performed with a 0 to 2 M NaCl gradient in buffer B. A 5-ml fraction eluting at ca. 320 mM NaCl (fraction 1) was saved for further purification steps.
Solid ammonium sulfate was added to fraction 1 to a final concentration of 1.8 M, precipitated proteins were removed by centrifugation (27,000 × g for 30 min at 4°C), and the supernatant (fraction 2) was loaded at a flow rate of 1 ml/min onto an alkyl Superose HR5/5 column (Pharmacia) equilibrated with buffer C (50 mM potassium phosphate, pH 7.0, 2 mM 2-mercaptoethanol) containing 1.8 M ammonium sulfate. A 1.8 to 0 M ammonium sulfate gradient was applied in buffer C, and proteins eluting between 1.17 and 0.99 M were pooled (fraction 3).
Eight milliliters of buffer C were added to fraction 3 (2 ml) and the proteins were loaded onto a HiTrap heparin affinity column (1 ml; Pharmacia) equilibrated in buffer C at a flow rate of 0.17 ml/min. Elution was performed with a 0 to 2 M NaCl gradient in buffer C, providing a 0.25-ml fraction eluting at ca. 900 mM NaCl (fraction 4).
Gel filtration was performed on fraction 4 with a Superdex 75 HR10/30 column (Pharmacia) equilibrated with buffer C containing 200 mM NaCl at a flow rate 0.5 ml/min. Fraction 5 eluted between 8.5 and 9.0 ml.
Mass spectrometry.
Samples of UDP-MurNAc-hexapeptide produced by the W. viridescens and E. faecalis ligases were isolated by HPLC, lyophilized, and dissolved in H2O:CH3CN (50:50, vol/vol). A drop of 10% ammonium hydroxide was added to improve ionization, and the sample was injected at a flow rate of 20 μl/min into an electrospray Micromass LCT mass spectrometer in the negative mode.
RESULTS
Synthesis of UDP-MurNAc-hexapeptide by W. viridescens extracts.
Three fractions containing partially purified tRNA, alanyl-tRNA synthetase, and UDP-MurNAc-pentapeptide:l-alanine ligase were obtained by ion exchange and hydrophobic-interaction chromatography (Table 1; Materials and Methods). Addition of l-[14C]Ala to UDP-MurNAc-pentapeptide required these three fractions, Mg2+, and ATP, as previously described (16). Formation of the radioactive UDP-MurNAc-hexapeptide was abolished by RNase. The assay developed with partially purified enzymes and tRNA from W. viridescens was subsequently used to identify the alanyl-tRNA synthetase and UDP-MurNAc-pentapeptide:l-alanine ligase genes of E. faecalis based on cloning and expression in E. coli (Table 1). Such heterologous systems for branched peptidoglycan synthesis were expected to be functional since aminoacyl-tRNA synthetases and ligases are not strictly specific for the tRNA issued from the same species (8, 13).
TABLE 1.
Synthesis of UDP-MurNAc-hexapeptide by W. viridescens and E. coli extractsa
| Source of:
|
Hexapeptide production | |
|---|---|---|
| UDP-MurNAc-pentapeptide:l-alanine ligase | Alanyl-tRNA synthetase | |
| W. viridescens fraction I | W. viridescens fraction I | + |
| W. viridescens fraction III | W. viridescens fraction IV | + |
| W. viridescens fraction III | None | − |
| None | W. viridescens fraction IV | − |
| W. viridescens fraction III | E. coli JM83/pDA26(alaSEfa) crude extract | + |
| E. coli JM83/pDA28(orf1) crude extract | W. viridescens fraction IV | − |
| E. coli JM83/pDA29(orf2) crude extract | W. viridescens fraction IV | + |
Addition of l-[14C]alanine to UDP-MurNAc-pentapeptide required tRNA (W. viridescens fraction II) and was suppressed if RNase was added to the reaction mixture. l-[14C]alanine and [14C]UDP-MurNAc-hexapeptide were separated by paper chromatography.
Overproduction and purification of the E. faecalis alanyl-tRNA synthetase.
The partial E. faecalis genome sequence available at the TIGR website contained a single open reading frame encoding a protein displaying high-level amino acid identity with the alanyl-tRNA synthetases from B. subtilis (61%) and E. coli (45%). The E. faecalis gene identified by sequence comparison (alaSEfa) was amplified by PCR and cloned into vector pTrcHis60 to generate a translational fusion with six histidine codons at the 3′ end of the gene. The resulting plasmid construct, pDA26(alaSEfa), specified an active alanyl-tRNA synthetase since the crude E. coli extract harboring the plasmid could replace W. viridescens fraction IV in the assay for UDP-MurNAc-hexapeptide synthesis (Table 1). The E. faecalis alanyl-tRNA synthetase containing the C-terminal six-His tag was purified in two steps based on affinity chromatography on a nickel column and exclusion chromatography. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed the expected ca. 100-kDa protein band estimated to be >95% pure (data not shown).
Identification of the E. faecalis UDP-MurNAc-pentapeptide:l-alanine ligase gene.
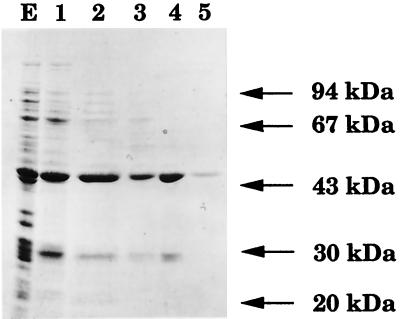
The partial E. faecalis genome sequence contained two open reading frames (orf1 and orf2) encoding polypeptides related to the S. aureus FmhB protein. These open reading frames were amplified and cloned into the expression vector pTrc99A. Both recombinant plasmids, pDA28(orf1) and pDA29(orf2), directed the synthesis of soluble proteins of ca. 46 kDa (Fig. 2 and data not shown) in agreement with the calculated molecular mass of the deduced products of orf1 (48,321 Da) and orf2 (46,023 Da). The crude extract from E. coli pDA29(orf2) contained UDP-MurNAc-pentapeptide:l-alanine ligase activity (Table 1). Addition of l-[14C]Ala to UDP-MurNAc-pentapeptide was not observed with the crude extract from E. coli JM83/pDA28(orf1).
FIG. 2.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of ORF2 purification steps. The E. faecalis UDP-MurNAc-pentapeptide:l-alanine ligase was purified from E. coli JM83/pDA29(orf2) crude extract (lane E) by anion exchange chromatography (lane 1) and by ammonium sulfate precipitation (lane 2), followed by hydrophobic-interaction (lane 3), affinity (lane 4), and exclusion (lane 5) chromatographies. Lanes 1 to 5 refer to fractions 1 to 5, respectively, as described in Materials and Methods.
Purification of the E. faecalis UDP-MurNAc-pentapeptide:l-alanine ligase.
The ligase was purified by anion exchange, hydrophobic-interaction, affinity, and exclusion chromatographies (Fig. 2; Materials and Methods). Precipitation of the protein was the major difficulty in the elaboration of the purification procedures. This difficulty was overcome by avoiding dialysis and maintaining the ionic strength above 250 mM.
Characterization of the product of the E. faecalis and W. viridescens ligases.
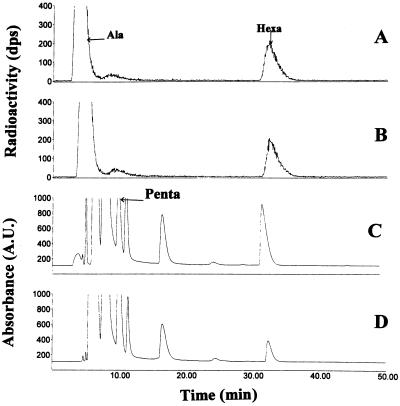
The [14C]UDP-MurNAc-hexapeptides produced by the W. viridescens and E. faecalis enzymes were analyzed by HPLC coupled to a radioactive flow detector (Fig. 3A and B). The HPLC profiles revealed a single radioactive peak in addition to l-[14C]alanine. The products of the W. viridescens and E. faecalis ligases had the same retention time and eluted after UDP-MurNAc-pentapeptide as expected (Fig. 3A and B).
FIG. 3.
HPLC chromatograms showing production of [14C]UDP-MurNAc-hexapeptide by W. viridescens fraction III (A) and purified E. faecalis ORF2 (B). The standard assay (Materials and Methods) contained purified AlaSEfa (1.8 μg) and W. viridescens fraction III (0.17 μg of protein) or ORF2 (1 μg). The standard assay was scaled up for purification of UDP-MurNAc-hexapeptides produced by the W. viridescens (C) or E. faecalis (D) ligases. The reaction mixture (60 μl) contained Tris-HCl (50 mM, pH 7.2), MgCl2 (12.5 mM), ATP (2.5 mM), l-alanine (0.75 mM), UDP-MurNAc-pentapeptide (0.6 mM), tRNA (8 μg, W. viridescens fraction II), purified AlaSEfa (7.2 μg), and W. viridescens fraction III (30 μg of protein) or ORF2 (12 μg). Ala, l-alanine; Hexa, UDP-MurNAc-hexapeptide; Penta, UDP-MurNAc-pentapeptide.
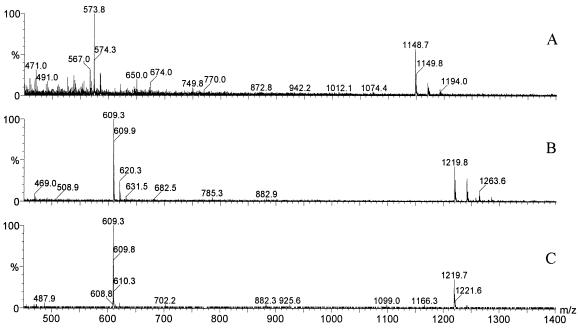
For mass spectrometry analysis, the standard assay for UDP-MurNAc-hexapeptide synthesis was scaled up, l-[14C]alanine was replaced by l-alanine, and the products were isolated by HPLC (Fig. 3C and D). The molecular mass of UDP-MurNAc-pentapeptide was determined to be 1,149 Da from the peaks at m/z 1,148.7 and 573.8 that were assigned to be [M-H]− and [M-2H]2− ions, respectively (Fig. 4A). The molecular mass of the product of the reaction catalyzed by the W. viridescens ligase was found to be 1,220 Da from peaks at m/z 1,219.8 and 609.3, which were assigned to be [M-H]− and [M-2H]2− ions, respectively (Fig. 4B). The same peaks (1,219.7 and 609.3) were detected in the mass spectrum for the product obtained with the E. faecalis ORF2 (Fig. 4C). These molecular mass assignments were confirmed by the presence of [M+Na-2H]− and [M+Na-3H]2− adduct ions. These molecular masses, 1,149 and 1,220 Da, obtained by electrospray mass spectrometry, match the predicted values of 1,149 and 1,220 Da for UDP-MurNAc-pentapeptide and UDP-MurNAc-hexapeptide, respectively. Together, these results indicate that the W. viridescens and E. faecalis ligases catalyze the same reaction.
FIG. 4.
Negative-ion mass spectra obtained for UDP-MurNAc-pentapeptide (A) and UDP-MurNAc-hexapeptide produced by the W. viridescens (B) or E. faecalis (C) ligases.
Comparison of the UDP-MurNAc-pentapeptide:l-alanine ligases of W. viridescens and E. faecalis
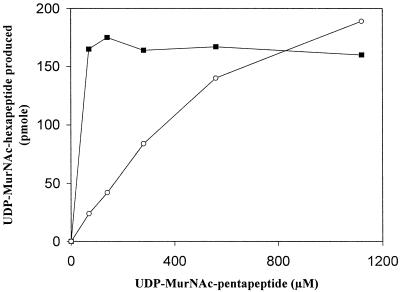
Synthesis of UDP-MurNAc-hexapeptide by the W. viridescens enzyme was independent of the concentration of UDP-MurNAc-pentapeptide in the 70 to 1,120 μM range (Fig. 5) in agreement with the reported Km value of 0.2 μM (16). In contrast, the amount of UDP-MurNAc-hexapeptide produced by the E. faecalis ligase increased with the concentration of UDP-MurNAc-pentapeptide up to the highest concentration tested (1,120 μM) (Fig. 5), implying a Km greater than 200 μM. Overall, the E. faecalis enzyme appeared catalytically less active than the W. viridescens enzyme since similar amounts of proteins were used to generate the data in Fig. 5, although the W. viridescens enzyme was not extensively purified. The difference in the activities of the two enzymes could be at least partially due to the different Km for UDP-MurNAc-pentapeptide. Impurities present in the W. viridescens enzyme preparation may also be involved since an unknown high-molecular-mass factor (>200 kDa) was reported to accelerate the reaction catalyzed by the W. viridescens enzyme four- to eightfold (8).
FIG. 5.
The assay for UDP-MurNAc-hexapeptide synthesis contained various concentrations of UDP-MurNAc-pentapeptide, purified AlaSEfa (1.8 μg), and purified E. faecalis ORF2 ligase (0.16 μg) (circle) or W. viridescens fraction III (0.17 μg of protein) (square). The reaction mixture (15 μl) also contained Tris-HCl (50 mM, pH 7.2), MgCl2 (12.5 mM), ATP (2.5 mM), l-[14C]alanine (67 μM, 93 mCi/mmol), and tRNA (4 μg). Incubation was for 30 min at 30°C.
Assay for UDP-MurNAc-heptapeptide synthesis.
HPLC chromatograms revealed only two radioactive peaks corresponding to UDP-MurNAc-hexapeptide and l-alanine (Fig. 3A and B). This observation suggests that the W. viridescens and E. faecalis ligases added a single l-alanine to UDP-MurNAc-pentapeptide. To confirm this result, unlabeled UDP-MurNAc-hexapeptide was prepared as described above (Fig. 3C and D) and incubated with various extracts containing ligase activity under the standard assay conditions described in Materials and Methods. [14C]UDP-MurNAc-heptapeptide was not detected with the purified E. faecalis ligase or with the partially purified W. viridescens ligase (fraction III), confirming that these enzymes do not add more than one l-alanine to UDP-MurNAc-pentapeptide.
DISCUSSION
The E. faecalis gene encoding the ligase for addition of the first amino acid in the side chain of peptidoglycan precursors (Fig. 1) was identified on the basis of sequence homology with the S. aureus fmhB gene, purification of the encoded protein (Fig. 2), and demonstration of UDP-MurNAc-pentapeptide:l-alanine ligase activity (Fig. 3 and 4). Synthesis of UDP-MurNAc-hexapeptide was obtained in vitro by using extensively purified alanyl-tRNA synthetase and ligase, both produced in E. coli, which does not possess the synthesis pathway for branched peptidoglycan precursors. Thus, incorporation of the first l-alanine into the side chain of peptidoglycan precursors does not require any additional enzyme in E. faecalis. In contrast, previous identification of the proteins involved in similar pathways in S. aureus (18) and S. pneumoniae (7) were based only on gene inactivation and determination of the structure of the cross bridge in mature peptidoglycan. In agreement with this, synthesis of the UDP-MurNAc-hexapeptide has been recently obtained by using purified ligase from W. viridescens (8).
The assay for UDP-MurNAc-hexapeptide synthesis proved useful in identifying the E. faecalis ligase (Table 1), although this enzyme is likely to preferentially act in vivo on the lipid intermediates, as reported for several membrane-associated ligases of gram-positive cocci (enterococci, staphylococci, and streptococci) (3, 8, 14). W. viridescens is an exception since it produces a soluble ligase for addition of l-alanine to cytoplasmic UDP-MurNAc-pentapeptide (8, 15). The difference in the apparent Km values of the E. faecalis and W. viridescens ligases for UDP-MurNAc-pentapeptide is consistent with this notion since it implies that the two enzymes may preferentially use different substrates in vivo (Fig. 5; see also Results). Localization of the ligase from gram-positive cocci at the inner surface of the membrane may optimize interaction with the lipid intermediates. The nature of the association with the membrane remains unknown. Binding to other peptidoglycan biosynthesis enzymes may be involved since the E. faecalis ligase does not appear to contain a membrane anchor composed of clustered hydrophobic amino acids. In addition, the ligase, as overproduced in E. coli, was soluble, indicating that the enzyme was not strongly associated with the lipid bilayer in this host (Fig. 2 and data not shown).
Comparison of pairs of sequences and phylogenetic analysis (data not shown) indicated that the E. faecalis ligase is closely related (39% identity) to MurM, which adds l-Ala or l-Ser at the first position of the cross bridge in S. pneumoniae (7). FmhB belonged to the same lineage, suggesting that the ligases adding the first amino acid in the cross bridges form a cluster of related sequences and may therefore be orthologs. ORF1 from E. faecalis and MurN from S. pneumoniae may also be orthologs (37% identity) and belong to a distinct lineage also including FemA and FemB. These observations are clearly limited and should be critically evaluated since the structure and mode of synthesis of the cross bridge can be extremely variable in related bacteria (Fig. 1) (19). For example, ligases from species belonging to the same genus, such as E. faecalis and E. faecium, may incorporate amino acids of the l or d configuration (Fig. 1) by distinct pathways involving mechanistically unrelated ligases (15, 20). This observation is surprising since it implies that the essential d,d-transpeptidases from related species use different acceptors bearing l- or d-amino acids in the cross-linking reaction (Fig. 1).
The ligases for branched peptidoglycan synthesis are attractive targets for the design of new antibiotics (10). The spectra of such antibiotics would be expected to be narrow since the pathway is not present in many bacteria, including Pseudomonas aeruginosa and members of the family Enterobacteriaceae, but this may be advantageous by limiting the overall spread of resistance. Inhibitors of the ligases for branched peptidoglycan synthesis may also be useful to restore β-lactam activity against resistant strains since the integrity of the side chain of the acceptor appears to be essential for the activities of PBP2x in S. pneumoniae (7) and of PBP2A in S. aureus (18), although murMN and femAB are not essential operons for these two organisms, respectively.
ACKNOWLEDGMENTS
This work was supported by Wyeth-Ayerst Research and by the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires (MENRT).
E. faecalis genome sequence data were kindly provided by The Institute for Genomic Research (TIGR) as publicly released at http: //www.tigr.org. We thank K. Tabei and M. M. Siegel for MS analysis.
REFERENCES
- 1.Arthur M, Depardieu F, Cabanie L, Reynolds P, Courvalin P. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol Microbiol. 1998;30:819–830. doi: 10.1046/j.1365-2958.1998.01114.x. [DOI] [PubMed] [Google Scholar]
- 2.Berger-Bachi B, Barberis-Maino L, Strassle A, Kayser F H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989;219:263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- 3.Billot-Klein D, Shlaes D, Bryant D, Bell D, Legrand R, Gutmann L, Van Heijenoort J. Presence of UDP-N-acetylmuramyl-hexapeptides and -heptapeptides in enterococci and staphylococci after treatment with ramoplanin, tunicamycin, or vancomycin. J Bacteriol. 1997;179:4684–4688. doi: 10.1128/jb.179.15.4684-4688.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Jonge B L, Sidow T, Chang Y S, Labischinski H, Berger-Bachi B, Gage D A, Tomasz A. Altered muropeptide composition in Staphylococcus aureus strains with an inactivated femA locus. J Bacteriol. 1993;175:2779–2782. doi: 10.1128/jb.175.9.2779-2782.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lencastre H, Tomasz A. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2590–2598. doi: 10.1128/aac.38.11.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filipe S R, Pinho M G, Tomasz A. Characterization of the murMN operon involved in the synthesis of branched peptidoglycan peptides in Streptococcus pneumoniae. J Biol Chem. 2000;275:27768–27774. doi: 10.1074/jbc.M004675200. [DOI] [PubMed] [Google Scholar]
- 7.Filipe S R, Tomasz A. Inhibition of the expression of penicillin resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc Natl Acad Sci USA. 2000;97:4891–4896. doi: 10.1073/pnas.080067697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hegde S S, Shrader T E. FemABX family members are novel nonribosomal peptidyltransferases and important pathogen-specific drug targets. J Biol Chem. 2001;16:6998–7003. doi: 10.1074/jbc.M008591200. [DOI] [PubMed] [Google Scholar]
- 9.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopp U, Roos M, Wecke J, Labischinski H. Staphylococcal peptidoglycan interpeptide bridge biosynthesis: a novel antistaphylococcal target? Microb Drug Resist. 1996;2:29–41. doi: 10.1089/mdr.1996.2.29. [DOI] [PubMed] [Google Scholar]
- 11.Le Bouguénec C, de Cespédès G, Horaud T. Presence of chromosomal elements resembling the composite structure Tn3701 in streptococci. J Bacteriol. 1990;172:727–734. doi: 10.1128/jb.172.2.727-734.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandelstam P, Loercher R, Strominger J. A uridine diphosphoacetylmuramyl hexapeptide from penicillin-treated Streptococcus faecalis. J Biol Chem. 1962;237:2683–2688. [PubMed] [Google Scholar]
- 13.Matsuhashi M, Dietrich C P, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell wall. III. The role of soluble ribonucleic acid and of lipid intermediates in glycine incorporation in Staphylococcus aureus. J Biol Chem. 1967;242:3191–3206. [Google Scholar]
- 14.Matsuhashi M, Dietrich C P, Strominger J L. Incorporation of glycine into the cell wall glycopeptide in Staphylococcus aureus: role of sRNA and lipid intermediates. Proc Natl Acad Sci USA. 1965;54:587–594. doi: 10.1073/pnas.54.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plapp R, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. XVII. Biosynthesis of peptidoglycan and of interpeptide bridges in Lactobacillus viridescens. J Biol Chem. 1970;245:3667–3674. [PubMed] [Google Scholar]
- 16.Plapp R, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. XVIII. Purification and properties of l-alanyl transfer ribonucleic acid-uridine diphosphate-N-acetylmuramyl-pentapeptide transferase from Lactobacillus viridescens. J Biol Chem. 1970;245:3675–3682. [PubMed] [Google Scholar]
- 17.Pompeo F, van Heijenoort J, Mengin-Lecreulx D. Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the bifunctional GlmU protein from Escherichia coli: site-directed mutagenesis and characterization of the mutant enzymes. J Bacteriol. 1998;180:4799–4803. doi: 10.1128/jb.180.18.4799-4803.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohrer S, Ehlert K, Tschierske M, Labischinski H, Berger-Bachi B. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc Natl Acad Sci USA. 1999;96:9351–9356. doi: 10.1073/pnas.96.16.9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staudenbauer W, Strominger J L. Activation of d-aspartic acid for incorporation into peptidoglycan. J Biol Chem. 1972;247:5095–5102. [PubMed] [Google Scholar]
- 21.Stranden A M, Ehlert K, Labischinski H, Berger-Bachi B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J Bacteriol. 1997;179:9–16. doi: 10.1128/jb.179.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vieira J, Messing J. The pUC plasmids, and M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 23.Weber B, Ehlert K, Diehl A, Reichmann P, Labischinski H, Hakenbeck R. The fib locus in Streptococcus pneumoniae is required for peptidoglycan crosslinking and PBP-mediated beta-lactam resistance. FEMS Microbiol Lett. 2000;188:81–85. doi: 10.1111/j.1574-6968.2000.tb09172.x. [DOI] [PubMed] [Google Scholar]