Summary
Background
Recent studies have associated several microRNAs (miRNAs) with childhood obesity and energy homeostasis, suggesting that an individual miRNA profile could be used as an early predictor to estimate the response to weight loss interventions in the design of precision nutrition.
Objective
To investigate associations between the expression of circulating adiposity‐related miRNAs and the response to a weight loss intervention.
Methods
A total of 51 Spanish girls (age 7–16 years) with abdominal obesity underwent 8 weeks of a multidisciplinary intervention for weight loss. Participants were stratified into two groups in accordance with changes in body mass index (BMI) standard deviation score: low‐responders (LR) and high‐responders (HR). The expression of 39 circulating miRNAs (c‐miRNAs) was evaluated in plasma of all subjects before the intervention.
Results
Six miRNAs were differentially expressed between LR and HR. However, after adjustment for Tanner stage, the association was maintained only for miR‐126‐3p and miR‐221‐3p with a higher expression in HR group compared to LR group. After the intervention, miR‐221‐3p expression decreased in all subjects with a significant difference in the change within groups. However, changes in miR‐126‐3p levels were not significant. The expression of miR‐221‐3p was positively correlated with body weight, BMI and waist circumference, and negatively correlated with quantitative insulin sensitivity check index.
Conclusions
Bioinformatic analysis evidenced that miR‐221‐3p participates in several obesity‐related pathways, and more interestingly, this miRNA targets several candidate genes to childhood obesity according to DisGeNet database. Thus, miR‐221‐3p could be used for predicting the response to a multidisciplinary intervention for weight loss in young girls.
Keywords: microRNA, miR‐126‐3p, paediatric obesity, precision nutrition
Abbreviations
- miRNAs
MicroRNAs
- c‐miRNAs
circulating microRNAs
- HR
high‐responders
- LR
low‐responders
- WC
waist circumference
- BMI‐SDS
standard deviation score for body mass index
- HOMA‐IR
homeostasis model assessment of insulin resistance
- QUICKI
quantitative insulin sensitivity check index
- ANCOVA
analysis of covariance
- SAT
subcutaneous adipose tissue
- PLA2G7
phospholipase A2 G7
- ADIPOR1
ADIPOnectin receptor‐1
- PPARGC1A
peroxisome proliferator‐activated receptor gamma coactivator 1‐alpha
1. INTRODUCTION
Obesity in children usually develops from early age and predicts negative health consequences in adult life, including a variety of chronic diseases or syndromes such as hypertension, coronary artery diseases, diabetes mellitus and cerebral stroke. 1 In this context, a meta‐analysis, including 23 studies showed that children with high body mass index (BMI) are 5 times more likely to have obesity in adulthood compared to normal‐weight children. 2
Because the presence of risk factors in this age is associated with the subsequent development of metabolic diseases in adulthood, 3 it has been postulated that the prevention of obesity in children must reduce obesity‐related diseases in adults. However, the actual mechanisms involved in the development of childhood obesity and its consequences are not completely understood, which impedes the timely management of these complications. 3
It is well established that gene expression can be modulated by epigenetic factors, including microRNAs (miRNAs), a class of short non‐coding RNAs. MiRNAs are important regulators of gene expression and promising candidates as disease biomarkers. 4 , 5 MiRNAs bind to a complementary sequence of mRNA, and are capable to influence multiple pathways, including insulin signalling, inflammation pathways, adipokine expression, adipogenesis, lipid metabolism and regulation of satiety, therefore, regulating up to 60% of all human genes. 6 The combined effect of targeting several genes within a common pathway may exert powerful effects on cellular processes such as growth, differentiation, metabolism and apoptosis. 7 , 8 Thus, the role of miRNAs as effective biomarkers for diagnosing and evaluating the risk of obesity and its related comorbidities has been established in the recent years. The majority of research, however, has been conducted in adults and there is a lack of evidence in childhood obesity. 9
In this regard, a systematic review identified 65 miRNAs associated with childhood obesity; however, only few of them were reported by at least two studies. 10 Moreover, a genetic model, including PLIN1 genes and four PLIN1‐targeting miRNAs (hsa‐miR‐4777‐3p, hsa‐miR‐642b‐3p, hsa‐miR‐3671‐1 and hsa‐miR‐551b‐2) explained 12.7% of the variance in childhood obesity risk. In addition, only the four miRNAs together explained 10.1% of the risk of developing obesity. 11 Previously, our group has characterized differences in DNA methylation levels of PTPRS and PER3 and miRNA coding sequences associated in children with obesity. 12 , 13 Moreover, we found differential DNA methylation patterns in response to a lifestyle intervention. 14 A recent study has suggested that the individual miRNAs profile could be used as a predicting factor to estimate the response to weight loss intervention. 15
On the other hand, recent studies suggest that the individual miRNA profile could be used as a predicting factor to estimate the response to weight loss interventions. 16 , 17 In this regard, the aim of the present study was to characterize differences in the circulating levels of obesity‐related miRNAs in girls with obesity (children and adolescents) that followed a lifestyle intervention for obesity management. We have compared the baseline miRNA profile between high‐responders (HR) and low‐responders (LR) to the intervention, analysed the miRNA expression changes induced by the intervention, and performed target gene prediction and pathway enrichment analyses for the differential miRNAs.
2. METHODS
2.1. Subjects
The Intervention of Grupo de Estudio Navarro de Obesidad Infantil (IGENOI) is a randomized controlled trial (ClinicalTrials.gov, Identifier: NCT03147261) carried out in Pamplona, Spain. Moreover, the study followed the ethical standards recognized in the Declaration of Helsinki (Fortaleza, Brazil, October 2013), and was approved and supervised by the Human Research Ethics Committee of the University of Navarra (reference number 044/2014). All participants and their parents or legal guardians provided written informed consent.
A total of 126 participants were recruited, and 121 of them had met the inclusion criteria. This group of children and adolescents (7–16 years old) with abdominal obesity defined as a waist circumference (WC) above the 90th percentile according to national reference data 18 were enrolled in the 2‐year family‐based lifestyle intervention study (an 8‐week programme and a follow‐up of 22 months). Subjects were recruited from the Paediatric Endocrinology Departments at ‘Clínica Universidad de Navarra’ and ‘Complejo Hospitalario de Navarra’ and from primary healthcare centres in Pamplona. Subjects with pre‐diabetes or food intolerance, following special diets, regular alcohol consumption, major psychiatric illness, eating disorders, or medical therapy were excluded.
Of the 121 participants, 114 (70 girls and 44 boys) concluded the 8‐week programme. Of these 114 patients, plasma samples were available for 51 girls and 25 boys. Thus, due to the reduced sample of boys and aiming to reduce sex‐bias, only girls' samples were analysed in the present study. Subjects were distributed according to the response based on change in ‘Standard Deviation Score for Body Mass Index’ (BMI‐SDS, median equal to 0.32). Thus, girls with a reduction in adiposity equal or lower to 0.32 in BMI‐SDS were considered LR (n = 26) and those with a decrease higher than 0.32 in BMI‐SDS, as HR (n = 25) (Figure S1).
2.2. Lifestyle intervention
A multidisciplinary team conformed by dietitians, pediatricians, nurses, physical activity experts and laboratory technicians were involved in the development of the study protocol. The 8‐week programme consists of nutritional education based on mediterranean diet style and an increment in physical activity to at least 200 minutes per week. Further aspects of the design of the IGENOI study have previously been detailed elsewhere. 19
2.3. Anthropometric, clinical and biochemical measurements
All variables were carried out at baseline and after the 8‐week programme. Anthropometric measurements were performed by trained personnel according to standard procedures and calibrated tools, while the participants were barefoot and wore light clothing.
Height was obtained with a Harpenden's stadiometer of 1 mm precision (Seca 220, Vogel & Halke, Hamburg, Germany). Body weight and body fat were evaluated using a digital scale BC‐418 segmental body composition analyser (Tanita, Tokyo, Japan). BMI was calculated as weight–height squared (kg/m2). The SD of BMI was calculated according to specific values for sex and age derived from Spanish reference data. 18 A non‐stretchable measuring tape (Type SECA 200) was used for measuring WC by standard procedures (Type SECA 200). Blood pressure was measured using an electronic sphygmomanometer (OMRON M6, Hoofddorp, The Netherlands) on the right arm after the subjects had rested quietly for 15 min. Pubertal status was determined using Tanner stage 20 and was evaluated by a pediatrician at baseline.
Venous blood samples were obtained by specialized trained nurses at the hospital after an overnight fast. Glucose, insulin and lipid profile were determined by standard autoanalyser techniques. Insulin resistance was calculated from the homeostasis model assessment of insulin resistance (HOMA‐IR) and insulin sensitivity from the quantitative insulin sensitivity check index (QUICKI). Leptin levels were measured by enzyme‐linked immunosorbent assay (ELISA) (R&D Systems, Minneapolis, MN).
Information regarding energy intake was collected by trained dieticians at baseline and after the 8‐week programme using a semi‐quantitative 136‐item Food‐Frequency Questionnaire previously validated in Spain. 21
3. miRNA EXPRESSION ANALYSIS
Total RNA was extracted from 200 μl ethylenediaminetetraacetic acid (EDTA)‐plasma using the miRNeasy Serum/Plasma kit (Qiagen. Hilden, Germany) according to the manufacturer's recommendations. To analyse the robustness of RNA extraction, cel‐miR‐39 was added to the thawed RNA sample. Total RNA (4 μl) was reverse transcribed in 10 μl reaction using the miRCURY LNA Universal RT miRNA PCR, polyadenylation and cDNA synthesis kit II (Qiagen). UniSp6, a cDNA synthesis control, was added in the reverse transcription reaction to determine the effectiveness of this process.
3.1. Search for miRNAs‐related to pediatric obesity
The election of the target miRNAs‐related to pediatric obesity and weight loss was based on available literature 1 , 2 , 3 and also on the search for miRNAs potentially associated with obesity in humans using the miRWalk 2.0 database. 4
3.2. Relative expression of the 48 miRNAs
Relative expression of the 48 miRNAs was analysed in plasma from all children using the Custom Pick‐&‐Mix miRNA PCR Panel v5 (Qiagen). In addition to the target miRNAs, cel‐miR‐39, haemolysis controls and a blank were also included in each plate, as shown in Table S1.
RT‐qPCR experiments were performed using a CFX384 Real‐time system (Bio‐Rad, Hercules, CA). The following cycling conditions were used: 95°C for 2 min, followed by 40 cycles at 95°C for 10 s and 56°C for 1 min. Relative expressions were calculated using the 2ΔΔCq method. 22 All individual RNA samples were analysed in a custom assay panel of 48 miRNAs. The miRNAs with complete data were used for the global mean method for normalization of the data, since this was found to be the most stable normalizer. 16
Haemolysis was measured by the ratio between hsa‐miR‐451a and hsa‐miR‐23a‐3p. 23 The difference in expression values between these 2 miRNAs provides a good measure of the haemolysis degree, with values >5 suggesting erythrocyte miRNA contamination. 23 Only those miRNAs whose assay cut‐off was <35 cycles and were expressed in at least 20% of the total sample, were taken into account. 24
3.3. Validation of miR‐126‐3p and miR‐221‐3p expression
miRNAs differentially expressed between LR and HR in the first analysis were selected to have their expression levels analysed in baseline and post‐intervention plasma samples. The following cycling conditions were used: 95°C for 2 min, followed by 40 cycles at 95°C for 10 s and 56°C for 1 min. Relative expressions were calculated using the 2ΔΔCq method.
qPCR experiments were performed in a CFX384 real‐time system (Bio‐Rad), using the following cycling conditions: 95°C for 2 min, followed by 40 cycles at 95°C for 10 s and 56°C for 1 min. Each sample was assayed in triplicate, and a negative control was included in each experiment. Relative quantifications of the miRNAs of interest were performed by using the 2ΔΔCq method. The reference gene selection software NormFinder was used to assess the best reference gene using intra‐ and intergroup (response to diet) variation. Results indicate miR‐425‐5p as the best combination of reference genes for normalization. NormFinder allows estimating not only the overall expression variation of the candidate genes but also take into account both intra‐ and intergroup variations. Genes with the lowest variability value are the most stable. 25
3.4. miR‐221‐3p target prediction and functional enrichment analysis
Potential targets for hsa‐miR‐221‐3p were searched using miRWalk 3.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/, accessed 29th October 2020). To better understand the biological relevance of the miR‐221‐3p target genes, a network analysis was performed using PathDIP (accessed 29th October 2020). 26 A hypergeometric test was used to calculate the statistical significance of the enriched pathways, and p‐values were corrected for multiple testing using the Benjamini–Hochberg procedure, which provides a false discovery rate (FDR) adjusted‐p‐value (q‐value). KEGG pathways associated with a q‐value <0.05 were considered significantly enriched.
To prioritize and identify disease candidate genes, we compared the hsa‐miR‐221‐3p target genes to experimentally validate and computationally predict genes associated with pediatric obesity (C2362324) found at DisGeNET database. The DisGeNET database is a comprehensive platform that integrates information concerning human disease‐associated genes. 27 This database integrates data from expert‐curated repositories, text mining data extracted from scientific literature, experimentally validated data and referred data. 27 The result data were imported into Cytoscape 3.8.1 for analysis and visualization of the network.
3.5. Statistical analysis
Normalized data (RQ expression levels) were initially analysed with an estimation and comparison of expression levels between groups. Normal distribution of data was assessed using the Kolmogorov–Smirnov and Shapiro–Wilk tests. Variables with normal distribution are presented as mean ± standard deviation (SD). Variables with skewed distribution were log‐transformed prior to analysis and presented as median (25th–75th percentiles). Categorical data are shown as percentages. Student's unpaired or paired t‐test was used for comparisons between groups (‡) or within (†) group, respectively. In addition, data were adjusted for Tanner stage using analysis of covariance (ANCOVA).
Changes in variables (0–8 weeks) were calculated as ‘variable at week 8’ minus ‘variable at baseline’. Pearson's correlation coefficients were used to describe the associations between miR‐221‐3p with all parameters. Associations were assessed using multivariable linear regression analysis adjustment for Tanner stage (data not shown), however, similar results were obtained.
STATA 12.0 for Windows (version 12.0, College Station, TX: StataCorp LP, USA) was used for statistical analysis. The statistical significance level was p < 0.05. The network visualization of miRNA‐target genes was generated using Cytoscape v.3.7.1. 28 One heatmap plot of the correlation values was produced using MORPHEUS web tool (Morpheus, https://software.broadinstitute.org/morpheus). Graphics were performed using Ploty chart studio (https://chart-studio.plotly.com).
4. RESULTS
4.1. Anthropometric, clinical and biochemical characteristics of subjects included in the study
The characteristics of the girls with abdominal obesity distributed according to the response (change in BMI‐SDS ≤0.32, LR = 26 and> 0.32, HR = 25) are summarized in Table 1. No differences in the characteristics were found between the two groups at baseline.
TABLE 1.
Baseline characteristics of low (LR) and high (HR) responders to the 8‐week lifestyle intervention
| LR (n = 26) | HR (n = 25) | p‐value | |
|---|---|---|---|
| Tanner % (I/II/III/IV/V) | 27/15/12/8/38 | 32/16/20/12/20 | 0.687 |
| Age (years) | 11 ± 2 | 11 ± 2 | 0.697 |
| Weight (kg) | 68 ± 19 | 65 ± 17 | 0.633 |
| Height (cm) | 151 ± 13 | 151 ± 12 | 0.828 |
| BMI (kg/m2) | 29 ± 5 | 28 ± 4 | 0.547 |
| BMI‐SDS | 3 ± 1 | 3 ± 1 | 0.652 |
| Fat mass (%) | 40 ± 5 | 38 ± 6 | 0.344 |
| Waist circumference (cm) | 86 ± 12 | 85 ± 9 | 0.672 |
| Glucose (mg/dL | 87 ± 6 | 89 ± 7 | 0.199 |
| Insulin (μU/mL) | 20 ± 21 | 19 ± 11 | 0.816 |
| HOMA‐IR | 4 ± 5 | 4 ± 3 | 0.919 |
| QUICKI | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.607 |
| Total cholesterol (mg/dL) | 164 ± 28 | 155 ± 27 | 0.311 |
| HDL‐cholesterol (mg/dL) | 45 ± 7 | 46 ± 11 | 0.778 |
| LDL‐cholesterol (mg/dL) | 101 ± 23 | 91 ± 24 | 0.157 |
| Triglycerides (mg/dL) | 98 ± 39 | 93 ± 55 | 0.720 |
| Leptin (ng/mL) | 49 ± 21 | 47 ± 18 | 0.738 |
| Total energy intake (kcal/day) | 2545 ± 549 | 2800 ± 779 | 0.199 |
Note: Data are mean ± SD.
Abbreviations: BMI, body mass index; BMI‐SDS, standard deviation score for BMI; BP, blood pressure; HOMA‐IR, homeostasis model assessment for insulin resistance; QUICKI, quantitative insulin sensitivity check index; HDL, high‐density lipoprotein; LDL, low‐density lipoprotein.
Table 2 shows the changes in anthropometric, clinical and biochemical variables after the lifestyle intervention. Participants of both groups showed a significant decrease in adiposity. Specifically, HR subjects significantly reduced body weight, BMI, BMI‐SDS, fat mass and WC in comparison with LR subjects. In the HR group, LDL‐cholesterol and triglycerides were reduced and a greater improvement in total cholesterol (p = 0.039) and leptin (p = 0.014) were found in this group. After the intervention, both groups reduced HDL‐cholesterol and energy intake (LR subjects −673 kcal/day and HR subjects −866 kcal/day).
TABLE 2.
Change in anthropometric, clinical and biochemical variables of low (LR) and high (HR) responders to the 8‐week lifestyle intervention
| Δ | LR (n = 26) | p‐value ‡ | HR (n = 25) | p‐value ‡ | p‐value † |
|---|---|---|---|---|---|
| Weight (kg) | −0.65 ± 1.26 | 0.014 | −3.63 ± 1.57 | <0.001 | <0.001 |
| Height (cm) | 0.81 ± 0.58 | <0.001 | 1.00 ± 0.64 | <0.001 | 0.261 |
| BMI (kg/m2) | −0.55 ± 0.53 | <0.001 | −1.98 ± 0.76 | <0.001 | <0.001 |
| BMI‐SDS | −0.10 ± 0.26 | 0.058 | −0.68 ± 0.29 | <0.001 | <0.001 |
| Fat mass (%) | −0.78 ± 1.39 | 0.008 | −2.36 ± 2.40 | <0.001 | 0.006 |
| Waist circumference (cm) | −2.38 ± 4.06 | 0.006 | −5.11 ± 3.08 | <0.001 | 0.010 |
| Glucose (mg/dL) | −1.05 ± 6.84 | 0.491 | −2.53 ± 8.03 | 0.187 | 0.533 |
| Insulin (μU/mL) | −0.02 ± 10.15 | 0.993 | −1.61 ± 5.29 | 0.295 | 0.610 |
| HOMA‐IR | −0.12 ± 2.70 | 0.852 | −0.45 ± 1.35 | 0.248 | 0.681 |
| QUICKI | −0.01 ± 0.02 | 0.251 | 0.01 ± 0.02 | 0.083 | 0.029 |
| Total cholesterol (mg/dL) | −3.20 ± 19.78 | 0.478 | −18.00 ± 23.32 | 0.004 | 0.039 |
| HDL‐cholesterol (mg/dL) | −3.89 ± 6.53 | 0.018 | −4.44 ± 7.59 | 0.024 | 0.814 |
| LDL‐cholesterol (mg/dL) | 0.47 ± 14.23 | 0.886 | −10.02 ± 19.43 | 0.049 | 0.071 |
| Triglycerides (mg/Ll) | 0.26 ± 36.78 | 0.976 | −17.67 ± 26.29 | 0.011 | 0.099 |
| Leptin (ng/mL) | −3.86 ± 18.01 | 0.515 | −20.37 ± 8.76 | <0.001 | 0.014 |
| Total energy (kcal/day) | −673 ± 5618 | <0.001 | −866 ± 862 | <0.001 | 0.199 |
Note: Data are mean ± SD.
Abbreviations: BMI, body mass index; BMI‐SDS, standard deviation score for BMI; BP, blood pressure; HOMA‐IR, homeostasis model assessment for insulin resistance; LAP, lipid accumulation product; QUICKI, quantitative insulin sensitivity check index; TyG, triglyceride‐glucose index.
Paired t‐tests to compare changes within groups.
Unpaired t‐test to compare changes between groups.
Bold values are represents p value lower than 0.05.
4.2. Circulating miRNA (c‐miRNA) expression analysis at baseline
The circulating levels of 39 c‐miRNAs were compared between LR and HR subjects at baseline. Of these 39 miRNAs, 36 (88.9%) had detectable expression in plasma in more than 20% of the samples with Cq <35, allowing reliable statistical analyses (Figure 1). Six of these miRNAs, miR126‐3p, miR‐138‐5p, miR‐33b‐5p, miR‐210‐3p, miR‐29b‐3p and miR‐221‐3p, were significantly upregulated in HR compared to LR at baseline (Figure 2A). However, after adjustment for Tanner stage, the association remained significant only for miR‐126‐3p and miR‐221‐3p (Figure 2B).
FIGURE 1.
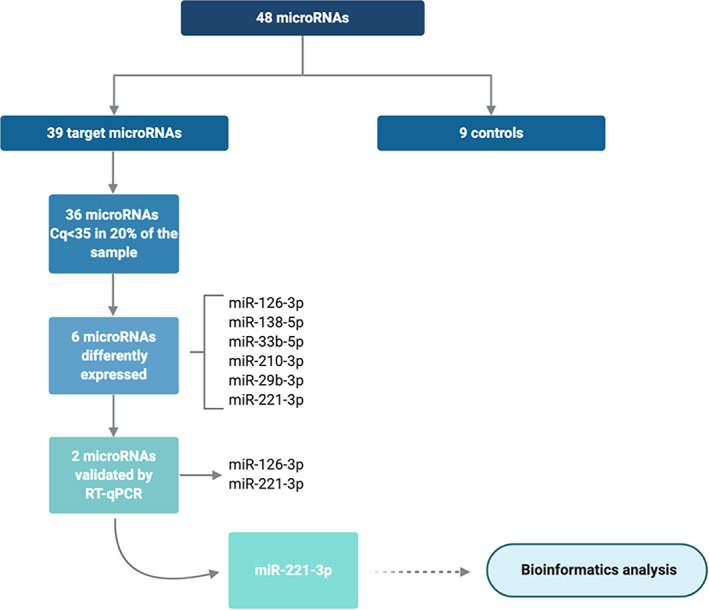
Flowchart of the analysis and main results
FIGURE 2.
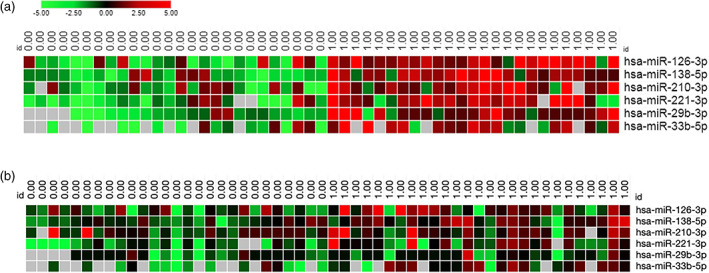
Distinct miRNA expression profile in low and high‐responders. (A) Heatmap of the six miRNAs differentially expressed in low and high‐responders before Tanner score adjustment. (B) Heatmap of the six miRNAs differentially expressed in low and high‐responders after Tanner score adjustment. Each column represents individual samples in each group (0 = low‐responders and 1 = high‐responders) and each row represents an individual miRNA. Expression levels of miRNAs are shown in red (upregulated) and green (downregulated), with brighter shades indicating higher fold differences in relation to the calibrator sample. Lack of difference in expression levels is represented in black
4.3. Validation of miR‐126‐3p and miR‐221‐3p expression
Since miR‐126‐3p and miR‐221‐3p remained significantly different after adjustment for Tanner stage, they were selected to investigate their expression at baseline and after the 8‐week lifestyle intervention.
At baseline, the circulating levels of miR‐221‐3p were significantly higher in the HR group compared to LR subjects (1.26 ± 0.45 vs. 0.97 ± 0.40; p = 0.015). Interestingly, after the 8‐week lifestyle intervention, miR‐221‐3p levels decreased in both groups (p < 0.001), and a significant difference in the change was observed within groups (HR: −0.75 ± 0.54 vs. LR −0.41 ± 0.54; p = 0.014) (Figure 3). For miR‐126‐3p, there were differences in baseline expression between groups (HR: 2.72 ± 3.05 vs. LR: 1.06 ± 1.13; p = 0.021). However, miR‐126‐3p levels were not different after 8‐week lifestyle intervention in both group (p > 0.050) without a significant difference within groups (p = 0.196) (data not shown).
FIGURE 3.
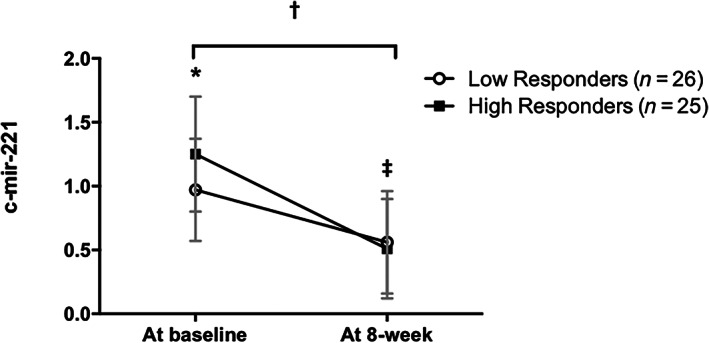
Plasma c‐mir‐221 abundance at baseline and after an 8‐week lifestyle intervention in low and high‐responders (change in 0.32 SDS‐BMI). Data are mean ± SD; p < 0.05; *difference at baseline (p = 0.015); ‡difference pre and post within group (in both groups p < 0.001); †difference pre and post between groups (p = 0.014). Data adjusted for Tanner stage using analysis of covariance (ANCOVA)
4.4. Correlation analysis between anthropometric, clinical and biochemical characteristics and the expression of miR‐221‐3p
The associations between changes in miR‐221‐3p levels and changes in anthropometric, clinical and biochemical parameters are shown in Table 3. We observed a positive association between miR‐221‐3p and changes in body weight (p = 0.018), BMI (p = 0.041) and WC (p = 0.010) and an inverse correlation with QUICKI (p = 0.028). Data remain statistically significant in multiple linear regression analysis after adjustment for Tanner Status.
TABLE 3.
Association between changes in expressed‐miR‐221‐3p and changes in anthropometric, clinical and biochemical variables
| Δ | Δ c‐miR‐221 | |||||
|---|---|---|---|---|---|---|
| Crude model | Adjusted model | |||||
| β | R 2 | p‐value | β | R 2 | p‐value | |
| Weight (kg) | 0.090 | 0.110 | 0.018 | 0.084 | 0.106 | 0.031 |
| Height (cm) | −0.163 | 0.031 | 0.215 | −0.105 | 0.021 | 0.454 |
| BMI (kg/m2) | 0.165 | 0.083 | 0.041 | 0.184 | 0.113 | 0.026 |
| BMI‐SDS | 0.352 | 0.064 | 0.074 | 0.369 | 0.083 | 0.062 |
| Fat mass (%) | 0.045 | 0.029 | 0.236 | 0.056 | 0.056 | 0.142 |
| Waist circumference (cm) | 0.052 | 0.129 | 0.010 | 0.050 | 0.133 | 0.014 |
| Glucose (mg/dL) | 0.009 | 0.012 | 0.499 | 0.004 | 0.017 | 0.743 |
| Insulin (μU/mL) | 0.019 | 0.079 | 0.118 | 0.016 | 0.003 | 0.226 |
| HOMA‐IR | 0.073 | 0.051 | 0.113 | 0.060 | 0.002 | 0.229 |
| QUICKI | −11.292 | 0.150 | 0.028 | −11.162 | 0.095 | 0.047 |
| Total cholesterol (mg/dL) | 0.005 | 0.041 | 0.219 | 0.006 | 0.102 | 0.164 |
| HDL‐cholesterol (mg/dL) | 0.016 | 0.036 | 0.259 | 0.012 | 0.045 | 0.430 |
| LDL‐cholesterol (mg/dL) | 0.006 | 0.030 | 0.316 | 0.007 | 0.073 | 0.241 |
| Triglycerides (mg/dL) | 0.000 | 0.000 | 0.998 | 0.003 | 0.045 | 0.421 |
| Leptin (ng/mL) | 0.003 | 0.007 | 0.719 | 0.000 | 0.120 | 0.952 |
| Total energy (kcal/day) | 0.000 | 0.047 | 0.137 | 0.000 | 0.087 | 0.061 |
Abbreviations: BMI, body mass index; BMI‐SDS, standard deviation score for BMI; BP, blood pressure; HOMA‐IR, homeostasis model assessment for insulin resistance; QUICKI, quantitative insulin sensitivity check index.
Bold values are represents p value lower than 0.05.
4.5. Target gene prediction and pathway enrichment analysis of miR‐221‐3p
A bioinformatic target gene prediction of miR‐221‐3p was accomplished using different databases of miRNA‐gene interactions in the miRWalk environment. Following this methodology, 14 233 genes were found as putative targets of miR‐221‐3p. Of these genes, 315 were retrieved from experimentally validated interactions. In order to identify key genes, we referred to DisGeNET, a comprehensive discovery platform database containing known human gene‐disease association, to validate our results. The search on the DisGeNET database retrieved 191 childhood obesity‐related genes. Of these 191 genes, 136 were miR‐221‐3p target genes. Moreover, six of these genes were experimentally validated target genes of miR‐221‐3p (ELAVL2, MKKS, MMP2, PEX1, SIRT1 and SOCS3). These findings are shown in Figure 4.
FIGURE 4.
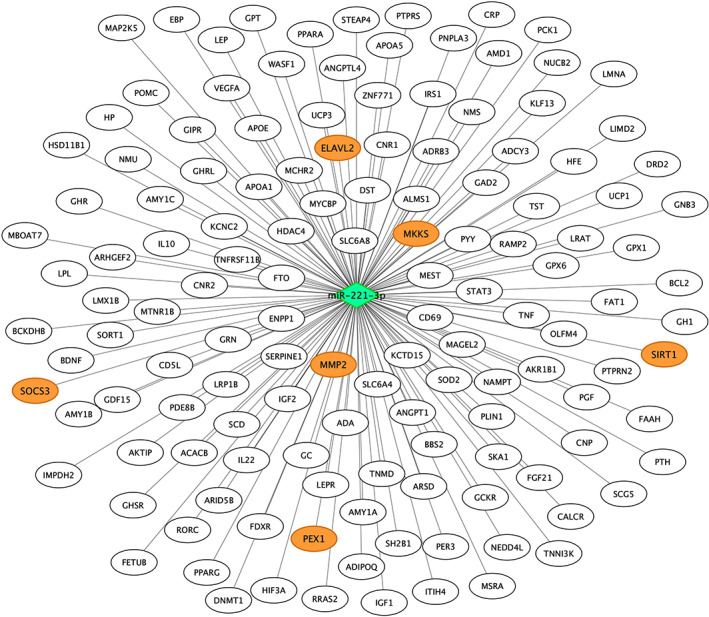
Experimentally validated target genes of miR‐221‐3p. Genes coloured in orange are those previously associated with pediatric obesity, according to DisGeNet database. Validated target genes were retrieved from miRWalk database. The diamond represents the miRNA and the circles represent the target genes
Functional enrichment analyses were performed using KEGG Pathways in the PathDIP environment. A total of 304 pathways were significantly over‐represented (q‐values <0.05) in the analysed lists of 14 233 target genes. The results from the pathway enrichment analysis based on predicted/validated miR‐221‐3p targets showed several key metabolic pathways, including PI3K‐Akt, MAPK, sphingolipid, mTOR, AMPK, phospholipase D, Wnt and insulin signalling pathways (Table 4). In addition, the six validated genes also participate in pathways related to obesity; however, they seem to have complementary functions (Figure 5).
TABLE 4.
Top pathways regulated by miR‐221‐3p target genes
| Rank | Pathway name | p‐value | q‐value (FDR: BH‐method) |
|---|---|---|---|
| 1 | Pathways in cancer | 1.84E‐60 | 5.91E‐58 |
| 2 | PI3K‐Akt signalling | 2.46E‐41 | 2.63E‐39 |
| 3 | Endocytosis | 3.41E‐38 | 2.74E‐36 |
| 4 | MAPK signalling | 5.49E‐38 | 3.52E‐36 |
| 5 | Rap1 signalling | 8.76E‐33 | 4.69E‐31 |
| 6 | cAMP signalling | 2.02E‐31 | 9.29E‐30 |
| 7 | Regulation of Actin cytoskeleton | 6.87E‐31 | 2.76E‐29 |
| 8 | Focal adhesion | 5.64E‐30 | 2.01E‐28 |
| 9 | MiRNAs in cancer | 1.05E‐29 | 3.37E‐28 |
| 10 | Neuroactive ligand‐receptor interaction | 4.96E‐28 | 1.45E‐26 |
| 18 | Sphingolipid signalling | 1.35E‐21 | 2.28E‐20 |
| 19 | mTOR signalling | 2.16E‐21 | 3.47E‐20 |
| 20 | AMPK signalling | 5.93E‐21 | 9.07E‐20 |
| 24 | Phospholipase D signalling | 1.59E‐19 | 2.04E‐18 |
| 26 | Wnt signalling | 5.38E‐19 | 6.40E‐18 |
| 27 | Insulin signalling | 5.77E‐19 | 6.61E‐18 |
Note: q‐values: p‐values corrected for multiple testing using the Benjamini–Hochberg method.
FIGURE 5.
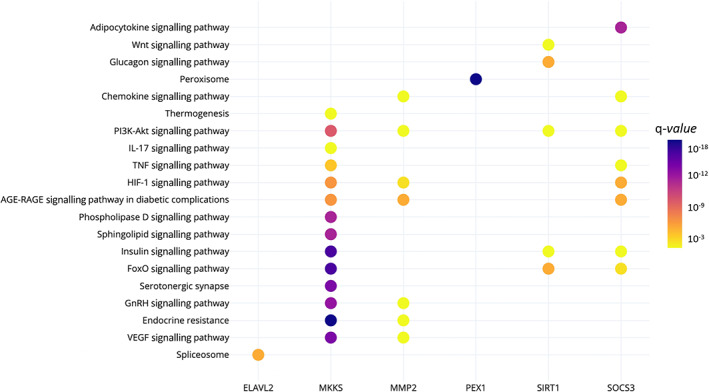
KEGG pathways in which the miR‐221‐3p target genes previously associated with paediatric obesity participates. The colour of the dots represents the pathway's q‐value. The y‐axis represents the KEGG pathways and the x‐axis shows the six target genes that participate in each selected pathway
5. DISCUSSION
The results from the current study have shown marked dysregulation in the expression level of miR‐221‐3p depending on the response to the lifestyle intervention in girls with obesity. In addition, the present research also provides novel understandings into the complex molecular mechanisms involved in obesity and weight loss, revealing pathways that may be regulated by miR‐221‐3p. This miRNA potentially modulates the expression of genes from several pathways related to adiposity, metabolism and inflammation, such as lipolysis regulation in adipocytes. More interestingly, we found that six candidate genes to pediatric obesity were experimentally validated target genes of miR‐221‐3p, suggesting a role for this miRNA in pediatric obesity pathogenesis. In this context, a previous study performed by our group including adults with obesity had evidenced that miR‐221‐3p was differentially expressed in plasma of non‐responders compared to responders to a low‐fat diet intervention. 16
There is accumulating evidence for differential miRNA expression between women and men across a variety of tissues, and the sex‐biased expression of miRNAs could have functional implication. 29 For example, miR‐221 and let‐7g are expressed more prominently in the plasma of women compared to men, and could be sex‐specific biomarkers of metabolic syndrome. 30 In this context, the expression of five miRNAs‐related to cardiometabolic health (miR‐24‐3p, miR‐361‐3p. miR‐3605‐5p, miR‐486‐5p and miR‐199b‐3p) was different between males and females in a sample of Hispanic adolescents (60% males). 31
The high expression levels of miR‐122 and miR222 conferred a 3.11‐fold increase in the risk for obesity in children with Mayan ethnicity. Moreover, the increased expression levels of these miRNAs were also associated with higher BMI, waist to height ratio (WHR), fat percentage, triglycerides and metabolic index; and low levels of serum high‐density lipid levels. 32
Some studies have also assessed the relationship between obesity‐related miRNAs and physical exercise. In this context, miR‐126, miR‐21, miR‐146a, miR‐221 and miR‐223 participates in molecular signalling pathways related to cellular adaptation to exercise in people with obesity. 33
In our study, six miRNAs, including miR‐221‐3p were significantly upregulated in HR compared to LR, although only miR126‐3p and miR221‐3p remain significant after adjustment for the Tanner stage. In the Parr and cols' study, it evaluated the abundance of miR‐221‐3p, and the authors did not find differences in expression at baseline in HR compared to LR, however, the abundance of miR‐221‐3p were modulated with a 16‐week intervention. 17 MiR‐221‐3p has been reported to affect adipocyte differentiation, metabolism and insulin signalling. 34 , 35 , 36 , 37 MiR‐221 expression is usually upregulated in obesity and is induced upon adipose tissue inflammation. 35 , 36 , 37 Previous studies have associated miR‐221‐3p with metabolic disease and it is induced upon inflammatory stimulation of adipocytes. 36 , 38 MiR221‐3p has been shown to target ANGPTL8 and to reduce adipocyte ANGPTL8 protein expression in subcutaneous adipose tissue (SAT). A significant negative correlation between ANGPTL8 and miR‐221‐3p has been identified in pre‐surgery SAT samples of subjects with morbid obesity undergoing bariatric surgery. Moreover, miR‐221‐3p showed a significant positive correlation with the mRNA of the inflammatory gene phospholipase A2 G7 (PLA2G7) in human SAT. 36
Another study has demonstrated that miR‐221 suppresses adiponectin receptor‐1 (ADIPOR1) expression and interferes with adiponectin signalling and insulin sensitivity of adipocytes. 38 In the same way, an inverse relationship between the expression levels of miR‐221‐3p and its target gene peroxisome proliferator‐activated receptor gamma coactivator 1‐alpha (PPARGC1A) has been found both in vivo and in vitro. 39 Previous studies have shown that PGC‐1α functions as a transcriptional regulator that can modulate mitochondrial biogenesis and function by targeting mitochondrial genes during adaptive thermogenesis.
miR‐221‐3p also has an important role in the terminal differentiation of human white adipocytes and their lipid composition. Increased expression of this miRNA inhibited terminal differentiation of adipocytes and reduced triglyceride storage. Notably, miR‐221‐3p overexpression inhibited de novo lipogenesis, reduced diacylglycerols and increased the concentrations of ceramides and sphingomyelins. This was accompanied by a suppression of ATP citrate lyase, sphingomyelin phosphodiesterase and acid ceramidase. On the contrary, a miR‐221‐3p inhibitor increased triglyceride storage. 40 Besides indicating that miR‐221‐3p is dysregulated in response to the lifestyle intervention in girls with obesity, the present study also provides new insights into the complex molecular mechanisms involved in weight loss and obesity by revealing pathways that may be regulated by miR‐221‐3p (Table 4). This miRNA potentially regulates genes from several significant KEGG pathways, including cancer, PI3K‐Akt, endocytosis, MAPK, cAMP, neuroactive ligand‐receptor interaction, sphingolipid, phospholipase D, Wnt and insulin signalling, which have been previously associated with metabolic disorders.
‘Some limitations of the present study should be considered. First, the reduced sample size which could lead to lack of power to detect small differences in miRNA expression between groups. Second, the exclusion of boys in the study in order to reduce sex bias. Third, the lack of an independent cohort to validate our findings. Fourth, even though a hypothesis driving approach was performed, the possibility of type I or type II errors due to multiple comparisons cannot be excluded. Therefore, these limitations should be considered when interpreting the results’.
In conclusion, this research provides novel information showing that circulating levels of miR‐221‐3p differ between LR and HR to a weight loss intervention in a group of girls with obesity. In addition, this miRNA regulates genes implicated in important pathways related to metabolism, adiposity, inflammation and obesity development. Moreover, our findings demonstrated the potential use of c‐miRNAs in the design of precision nutrition solutions for tackling pediatric obesity. However, further research with larger cohorts of participants and longer duration of intervention will be important for determining whether miRNAs regulate the cellular mechanisms that control weight loss, and therefore, become accurate biomarkers for obesity development.
CONFLICT OF INTEREST
None of the authors of this manuscript have a conflict of interest.
AUTHOR CONTRIBUTIONS
Ana Ojeda‐Rodríguez and Taís Silveira Assmann contributed to the data collection, performed the statistical analyses and wrote the manuscript; Ana Ojeda‐Rodríguez, Taís Silveira Assmann and Lucia Alonso‐Pedrero performed experiments. Maria Cristina Azcona‐Sanjulian, Fermín I. Milagro and Amelia Marti were responsible of the follow‐up, design and financial management and editing of the manuscript. All the authors actively participated in the manuscript preparation, as well as revise and approved the final manuscript.
Supporting information
Figure S1. Flowchart of intervention.
Table S1. Sequence of 48 included microRNAs.
ACKNOWLEDGEMENTS
We thank Dra. Lydia Morell‐Azanza and Dra. Mara Chueca for the recruitment and nutritional assistance, and Dra. Mirian Samblas for laboratory support. We acknowledge the financial support from MERCK Foundation, ORDESA Laboratories‐FEI‐AEPgrant (Sant Boi de Llobregat; Barcelona, España) and CIBEROBN (grant number: CB12/03/30002).
Ojeda‐Rodríguez A, Assmann TS, Alonso‐Pedrero L, Azcona‐Sanjulian MC, Milagro FI, Marti A. Circulating miRNAs in girls with abdominal obesity: miR‐221‐3p as a biomarker of response to weight loss interventions. Pediatric Obesity. 2022;17(8):e12910. doi: 10.1111/ijpo.12910
Ana Ojeda‐Rodríguez and Taís Silveira Assmann contributed equally to this study.
Funding information CIBEROBN, Grant/Award Number: CB12/03/30002 (https://www.ciberobn.es/); ORDESA Laboratories (https://www.ordesalab.com/); MERCK Foundation (https://www.fundacionmercksalud.com/)
REFERENCES
- 1. Ng M, Fleming T, Robinson M, et al. Global, regional and national prevalence of overweight and obesity in children and adults 1980‐2013: a systematic analysis. Lancet. 2014;384(9945):766‐781. doi: 10.1016/S0140-6736(14)60460-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simmonds M, Burch J, Llewellyn A, et al. The use of measures of obesity in childhood for predicting obesity and the development of obesity‐related diseases in adulthood: a systematic review and meta‐analysis. Health Technol Assess. 2015;19(43):1‐336. doi: 10.3310/hta19430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cui X, You L, Zhu L, et al. Change in circulating microRNA profile of obese children indicates future risk of adult diabetes. Metabolism. 2018;78:95‐105. doi: 10.1016/j.metabol.2017.09.006 [DOI] [PubMed] [Google Scholar]
- 4. Lu T, Rothenberg M. MicroRNA. J Allergy Clin Immunol. 2018;14(4):1202‐1207. doi: 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marques‐Rocha JL, Samblas M, Milagro FI, Bressan J, Martínez JA, Marti A. Noncoding RNAs, cytokines, and inflammation‐related diseases. FASEB J. 2015;29(9):3595‐3611. doi: 10.1096/fj.14-260323 [DOI] [PubMed] [Google Scholar]
- 6. Bartel D. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281‐297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 7. Da Costa Martins PA, De Windt LJ. MicroRNAs in control of cardiac hypertrophy. Cardiovasc Res. 2012;93(4):563‐572. doi: 10.1093/cvr/cvs013 [DOI] [PubMed] [Google Scholar]
- 8. Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469(7330):336‐342. doi: 10.1038/nature09783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oses M, Sanchez JM, Portillo MP, Aguilera CM, Labayen I. Circulating miRNAs as biomarkers of obesity and obesity‐associated comorbidities in children and adolescents: a systematic review. Nutrients. 2019;11(12):1‐13. doi: 10.3390/nu11122890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flórez CAR, García‐Perdomo HA, Escudero MM. MicroRNAs associated with overweight and obesity in childhood: a systematic review. MicroRNA. 2020;9(4):255‐265. doi: 10.2174/2211536609666191209152721 [DOI] [PubMed] [Google Scholar]
- 11. Yllmaz SG, Bozkurt H, Ndadza A, et al. Childhood obesity risk in relationship to Perilipin 1 (PLIN1) gene regulation by circulating microRNAs. OMICS. 2020;24(1):43‐50. doi: 10.1089/omi.2019.0150 [DOI] [PubMed] [Google Scholar]
- 12. Samblas M, Milagro FI, Mansego ML, Marti A, Martinez JA. PTPRS and PER3 methylation levels are associated with childhood obesity: results from a genome‐wide methylation analysis. Pediatr Obes. 2018;13(3):149‐158. doi: 10.1111/ijpo.12224 [DOI] [PubMed] [Google Scholar]
- 13. Mansego ML, Garcia‐Lacarte M, Milagro FI, Marti A, Martinez JA. DNA methylation of miRNA coding sequences putatively associated with childhood obesity. Pediatr Obes. 2017;12(1):19‐27. doi: 10.1111/ijpo.12101 [DOI] [PubMed] [Google Scholar]
- 14. Moleres A, Campión J, Milagro FI, et al. Differential DNA methylation patterns between high and low responders to a weight loss intervention in overweight or obese adolescents: the EVASYON study. FASEB J. 2013;27(6):2504‐2512. doi: 10.1096/fj.12-215566 [DOI] [PubMed] [Google Scholar]
- 15. Catanzaro G, Filardi T, Sabato C, et al. Tissue and circulating microRNAs as biomarkers of response to obesity treatment strategies. J Endocrinol Invest. 2020;44:1159‐1174. doi: 10.1007/s40618-020-01453-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Assmann TS, Riezu‐Boj JI, Milagro FI, Martínez JA. Circulating adiposity‐related microRNAs as predictors of the response to a low‐fat diet in subjects with obesity. J Cell Mol Med. 2020;24(5):2956‐2967. doi: 10.1111/jcmm.14920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parr EB, Camera DM, Burke LM, Phillips SM, Coffey VG, Hawley JA. Circulating microrna responses between “high” and “low” responders to a 16‐Wk diet and exercise weight loss intervention. PLoS One. 2016;11(4):e0152545. doi: 10.1371/journal.pone.0152545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Serra‐Majem L, Aranceta‐Bartrina J, Ribas‐Barba L, Pérez‐Rodrigo C, Gracía‐Closas C. Obesidad infantil y juvenil en España. Resultados del Estudio enKid (1998‐2000). Med clin. 2003;121(19):725‐732. [DOI] [PubMed] [Google Scholar]
- 19. Ojeda‐Rodríguez A, Zazpe I, Morell‐azanza L, Chueca J. Improved diet quality and nutrient adequacy in children and adolescents with abdominal obesity after a lifestyle intervention. Nutrients. 2018;10(10):1500. doi: 10.3390/nu10101500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tanner JM, Whitehouse RH. Clinical longitudinal standards for height, weight, height velocity, weight velocity, and stages of puberty. Arch Dis Child. 1976;51(3):170‐179. doi: 10.1136/adc.51.3.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De La Fuente‐Arrillaga C, Vázquez Ruiz Z, Bes‐Rastrollo M, Sampson L, Martínez‐González MA. Reproducibility of an FFQ validated in Spain. Public Health Nutr. 2010;13(9):1364‐1372. doi: 10.1017/S1368980009993065 [DOI] [PubMed] [Google Scholar]
- 22. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real‐time PCR experiments. Clin Chem. 2009;55(4):611‐622. doi: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 23. Blondal T, Jensby Nielsen S, Baker A, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59(1):S1‐S6. doi: 10.1016/j.ymeth.2012.09.015 [DOI] [PubMed] [Google Scholar]
- 24. Gevaert AB, Witvrouwen I, Vrints CJ, et al. MicroRNA profiling in plasma samples using qPCR arrays: recommendations for correct analysis and interpretation. PLoS One. 2018;13(2):1‐13. doi: 10.1371/journal.pone.0193173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andersen CL, Jensen JL, Ørntoft TF. Normalization of real‐time quantitative reverse transcription‐PCR data: a model‐based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245‐5250. doi: 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- 26. Rahmati S, Abovsky M, Pastrello C, Jurisica I. PathDIP: an annotated resource for known and predicted human gene‐pathway associations and pathway enrichment analysis. Nucleic Acids Res. 2017;45(D1):D419‐D426. doi: 10.1093/nar/gkw1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piñero J, Bravo Á, Queralt‐Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease‐associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833‐D839. doi: 10.1093/nar/gkw943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models. Genome Res. 2003;13(11):2498‐2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dai R, Ansar AS. Sexual dimorphism of miRNA expression: a new perspective in understanding the sex bias of autoimmune diseases. Ther Clin Risk Manag. 2014;10:151‐163. doi: 10.2147/TCRM.S33517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang YT, Tsai PC, Liao YC, Hsu CY, Juo SHH. Circulating microRNAs have a sex‐specific association with metabolic syndrome. J Biomed Sci. 2013;20(1):1. doi: 10.1186/1423-0127-20-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karere GM, Cox LA, Bishop AC, et al. Sex differences in MicroRNA expression and cardiometabolic risk factors in Hispanic adolescents with obesity. J Pediatr. 2021;235:138‐143.e5. doi: 10.1016/j.jpeds.2021.03.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. González‐Arce LM, Lara‐Riegos JC, Pérez‐Mendoza GJ, et al. High expression levels of circulating microRNA‐122 and microRNA‐222 are associated with obesity in children with Mayan ethnicity. Am J Hum Biol. 2020;33:e23540. doi: 10.1002/ajhb.23540 1–12. [DOI] [PubMed] [Google Scholar]
- 33. Ehtesham N, Shahrbanian S, Valadiathar M, Mowla SJ. Modulations of obesity‐related microRNAs after exercise intervention: a systematic review and bioinformatics analysis. Mol Biol Rep. 2021;48(3):2817‐2831. doi: 10.1007/s11033-021-06275-3 [DOI] [PubMed] [Google Scholar]
- 34. Skårn M, Namløs HM, Noordhuis P, Wang MY, Meza‐Zepeda LA, Myklebost O. Adipocyte differentiation of human bone marrow‐derived stromal cells is modulated by microRNA‐155, microRNA‐221, and microRNA‐222. Stem Cells Dev. 2012;21(6):873‐883. doi: 10.1089/scd.2010.0503 [DOI] [PubMed] [Google Scholar]
- 35. Peng J, Zhou Y, Deng Z, et al. miR‐221 negatively regulates inflammation and insulin sensitivity in white adipose tissue by repression of sirtuin‐1 (SIRT1). J Cell Biochem. 2018;119(8):6418‐6428. doi: 10.1002/jcb.26589 [DOI] [PubMed] [Google Scholar]
- 36. Mysore R, Ortega FJ, Latorre J, et al. MicroRNA‐221‐3p regulates angiopoietin‐like 8 (ANGPTL8) expression in adipocytes. J Clin Endocrinol Metab. 2017;102(11):4001‐4012. doi: 10.1210/jc.2017-00453 [DOI] [PubMed] [Google Scholar]
- 37. Meerson A, Traurig M, Ossowski V, Fleming JM, Mullins M, Baier LJ. Human adipose microRNA‐221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNF‐α. Diabetologia. 2013;56(9):1971‐1979. doi: 10.1007/s00125-013-2950-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lustig Y, Barhod E, Ashwal‐Fluss R, et al. RNA‐binding protein PTB and MicroRNA‐221 coregulate AdipoR1 translation and adiponectin signaling. Diabetes. 2014;63(2):433‐445. doi: 10.2337/db13-1032 [DOI] [PubMed] [Google Scholar]
- 39. Li X, Ballantyne LL, Yu Y, Funk CD. Perivascular adipose tissue–derived extracellular vesicle miR‐221‐3p mediates vascular remodeling. FASEB J. 2019;33(11):12704‐12722. doi: 10.1096/fj.201901548R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ahonen MA, Asghar MY, Parviainen SJ, et al. Human adipocyte differentiation and composition of disease‐relevant lipids are regulated by miR‐221‐3p. Biochim Biophys Acta ‐ Mol Cell Biol Lipids. 2021;1866(1):158841. doi: 10.1016/j.bbalip.2020.158841 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Flowchart of intervention.
Table S1. Sequence of 48 included microRNAs.


