Abstract
Objective
To explore whether total cholesterol (TC), high-density lipoprotein (HDL), low-density lipoprotein (LDL), and triglyceride (TG) are mediators in the pathway of body mass index (BMI) on serum urate and determine the proportion of the mediation effect.
Methods
This study used observational and two-sample Mendelian randomization (MR) analyses to explore the mediation effects of TC, HDL, LDL, and TG in the pathway of BMI on serum urate. We determined the size and the extent to which these lipids mediate any effect of BMI on serum urate.
Results
Observational analysis results showed that HDL and TG can partially explain the association of BMI on serum urate, and the proportion of mediation effect was 10.2% and 8.9%, respectively. MR results demonstrated that TG has a causal effect on serum urate (β = 0.22, 95% CI: 0.15, 0.29; p = 2.28×10–10.) and its proportion of mediation effect was 14.1%. TC, HDL, and LDL are not the mediators in the pathway of BMI on serum urate in MR estimates.
Conclusion
To a certain extent, TG mediates the effect of BMI on serum urate, and the risk of gout may be reduced by controlling both BMI and TG.
Keywords: body mass index (BMI), serum urate, lipid traits, mediation analysis, Mendelian randomization
Introduction
Gout is a chronic disease caused by the deposition of monosodium urate (MSU) crystals (1). The global prevalence of gout is on the rise: the result of the Global Burden of Disease Study in 2017 showed that the age-standardized prevalence rates of gout in men and women in 1990 were 747.48/100,000 and 233.52/100,000, but the number had risen to 790.90/100,000 and 253.49/100,000 in 2017, respectively (2). In 2010, the global disability-adjusted life years for gout were 11,400, an increase of 38,000 compared to those in 1990 (3). Now, gout has become the most common inflammatory arthritis in developed countries (4). Elevated serum urate is the direct cause of MSU crystal deposition and the development of gout (5). However, the proportion of patients who initiate and continue urate-lowering therapy is very low, ranging from 10% to 46%, worldwide (2, 6). Studies have suggested that mature adipocytes and adipose tissue produce and secrete urate. Moreover, obesity increases the expression of xanthine oxidoreductase mRNA and promotes serum urate secretion in adipose tissue (7, 8). Researchers have shown that obesity is an important risk factor for hyperuricemia (5, 9). Dalbeth and colleagues found that people with a mean body mass index (BMI) of 30.8 kg/m2 had a higher level of serum urate than those with a mean BMI of 21.8 kg/m2 (10). Furthermore, studies suggested that greater BMI leads to higher levels of total cholesterol (TC), low-density lipoprotein (LDL), and triglyceride (TG) but lower high-density lipoprotein (HDL) levels (11). Moreover, hyperuricemia is usually related to lipid metabolism disorders (12). Thus, the association of BMI with serum urate may be mediated by these lipids. However, to our knowledge, there is no published literature about the mediation analysis of the above lipids in the pathway of BMI on serum urate.
In mediation analysis, three parameters are typically estimated: total effect (the effect of exposure on outcome through all potential pathways), direct effect (the effect of exposure on outcome when controlling for mediators), and indirect effect (the effect of exposure on outcome that acts through mediators).
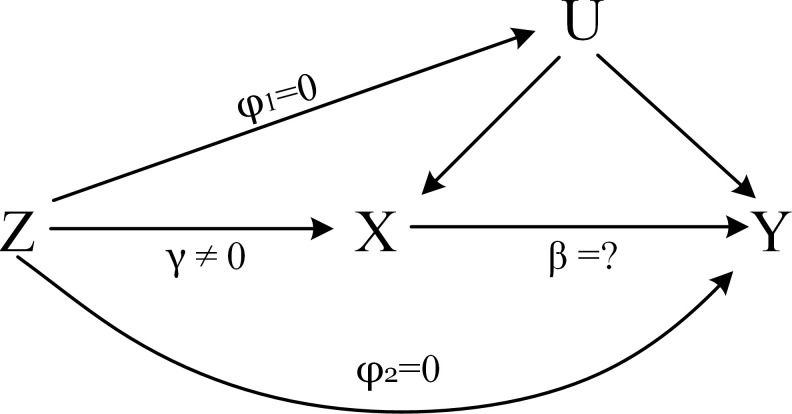
Mendelian randomization (MR) such as the two-step Mendelian randomization (13) and multivariable Mendelian randomization (14) has been used to investigate mediation effects in recent years. MR uses genetic variants, normally single nucleotide polymorphisms (SNPs), as instrumental variables to estimate the causal effect of an exposure on an outcome free from bias due to unknown confounders and reverse causality (15). Figure 1 describes the core assumptions of MR (16).
Figure 1.
The core assumptions of MR. Z represents the genetic instrument (SNPs), X the exposure (BMI, TC, HDL, LDL, and TG), Y the outcome (serum urate), and U denotes the confounders of the relationship of X–Y. Assumptions: 1. Genetic variation is closely related to the exposure of interest, γ ≠ 0; 2. Genetic variation has nothing to do with confounding, φ 1 = 0; 3. Genetic variation only affects the outcome through X, φ 2 = 0.
MR has previously been used to investigate the causal effects of BMI on serum urate, TC, HDL, LDL, and TG and also the effects of these four lipids on serum urate. Studies have shown the association of BMI, lipids, and serum urate (17–19), but their mediation effects are still to be determined. In this study, we investigated the role of TC, HDL, LDL, and TG in mediating the association of BMI with serum urate.
Due to the low rates of urate-lowering therapy in gout patient, understanding the changes of these four lipids in the risk of BMI on gout and preventing these risk factors are veryimportant public health aims and are primary prevention measures for higher-risk populations.
Materials and methods
Overall study design
We first established the association of BMI with serum urate and then explored the mediation effect of the lipid traits. The data analysis of this study was organized into two parts: part 1 involves observational analysis using the product of coefficients (20) of the data from the National Health and Nutrition Examination Survey (NHANES), and part 2 involves two-sample MR analysis with summary-level genetic data. This involved the two-step two-sample MR (13) and multivariable Mendelian randomization (14). We first used the two-step MR to identify which of the lipids are possible mediators. Then, multivariable MR was used to calculate the direct effect of BMI and mediators on serum urate in order to calculate the size and proportion of the mediation effect.
Data sources
Observational analysis
NHANES is a population-based survey designed to collect information on the health and nutrition of the US household population. Since data collection for 2019–2020 has not been completed, the data from the NHANES 2017–2018 survey were selected for this study. In this study, we obtained 3,036 participants, of which 819 individuals were excluded due to missing data, and 2,217 were eventually included. We collected age, gender, education level (the highest level or level of education completed by adults 20 years and older), marital status, family annual income, place of birth, race, BMI, TC, HDL, LDL, TG, and serum urate. Among the 2,217 participants included, men accounted for 47.9%, and women accounted for 52.1%. The mean age (standard deviation) of the participants was 51.32 (17.65), 41.3% were under 60 years old, and 52.7% were over 60 years old. The average levels of BMI, TC, HDL, LDL, TG, and urate were 29.82 kg/m2 (7.49), 185.75 mg/dl (41.09), 53.87 mg/dl (15.53), 110.38 mg/dl (36.19), 107.38 mg/dl (63.66), and 5.53 mg/dl (1.50), respectively.
Two-sample MR analysis
Summary-level data were made publicly available and ethical approval was obtained in the original studies ( Table 1 ). Summary-level data for BMI were derived from a recently published meta-analysis of GWAS, including 681,275 European descent individuals from the Genetic Investigation of Anthropometric Traits consortium (GIANT) (21). Genetic association with HDL (N = 403,943), LDL (N = 440,546), and TG (N = 441,016) were obtained from the UK Biobank (22). The average age of these participants was 56.9 (range: 39–73) years old and 54.2% were women. The mean values of the lipid concentrations (standard deviation) were as follows: HDL = 1.45 (0.38) mmol/L, LDL = 3.57 (0.87) mmol/L, and TG (median) = 1.50 (1.11) mmol/L. Summary-level data for TC were obtained from the Global Lipids Genetics Consortium (GLGC) involving 187,365 participants, and these participants were from multiple ethnic populations including European, East Asian, South Asian, and African (23). For serum urate, summary-level data were from the Global Urate Genetics Consortium genome-wide association study of serum urate involving 110,347 participants of European ancestry (24). The serum urate concentration ranged from 3.9 to 6.1 mg/dl (median = 5.2 mg/dl). All SNPs used in each two-sample MR analysis are provided in the Supplementary File .
Table 1.
GWAS cohorts used in this study.
| Phenotype | First author (year) | Sample size | Consortium |
|---|---|---|---|
| BMI | Yengo (2018) | 681,275 | GIANT |
| TC | Willer (2013) | 187,365 | GLGC |
| HDL | Richardson (2020) | 403,943 | UK Biobank |
| LDL | Richardson (2020) | 440,546 | UK Biobank |
| TG | Richardson (2020) | 441,016 | UK Biobank |
| Serum urate | Kottgen (2013) | 110,347 | GUGC |
Statistical analysis
Association of BMI on serum urate
In the observational analysis, we used multivariable linear regression to estimate the association of BMI with serum urate. We adjusted confounders including age, gender, annual family income, education, marital status, race, and place of birth (25–27).
In the two-sample MR analysis, the causal effects were investigated using the ratio method with standard errors derived using the delta method (28). We used the random-effects inverse variance weighted (IVW) method to pool MR estimates across individual SNPs (29).
Mediation by TC, HDL, LDL, and TG
In the observational mediation analysis, when investigating the degree to which the effects of BMI on serum urate are mediated through each lipid trait (TC, HDL, LDL, and TG) individually, we used the product of coefficients method to estimate the indirect effect (20). This involved first estimating the association of BMI on each lipid trait individually and then multiplying this with the direct effect of those lipid traits on serum urate after adjusting for BMI. We used multivariable linear regression to estimate the association of BMI with each lipid trait after adjusting for confounders (age, gender, annual family income, education, marital status, race, and place of birth). We then estimated the direct effect of each lipid trait on serum urate after adjusting for BMI, age, gender, annual family income, education, marital status, race, and place of birth. The two estimates were multiplied together to estimate the indirect effect with confidence intervals derived using the non-parametric bootstrap method (simulation times = 1,000). Finally, we estimated the proportion of the effect of BMI on serum urate that was mediated by each lipid trait by dividing the indirect effect by the total effect.
In the two-sample MR analysis, we used the two-step two-sample MR (13) to identify the potential mediator. The first step involved estimating the causal effects of BMI on TC, HDL, LDL, and TG, and the second step involved estimating the causal effects of each lipid trait on serum urate. Then, BMI and the mediators (the lipid trait was not only influenced by BMI but also affected by serum urate) determined by the above steps were simultaneously included in the multivariable Mendelian randomization model (14) to estimate their direct effects on serum urate. We estimated the indirect effect by multiplying the effect of BMI on mediators with the direct effect of mediators on serum urate. The proportion of mediation was calculated by dividing the indirect effect by the total effect.
In the selection of SNPs to address the potential weak instrument bias, we included SNPs that were genome-wide significantly ( p< 5×10–8). All F statistics used in MR were above 30, suggesting that a weak instrument bias was unlikely (more details seen in the Supplementary File ). To further ensure the independence of the instruments, SNPs that were in linkage disequilibrium (LD) were excluded from the instrument variable set using the clumping algorithm (r 2 threshold > 0.01 and window size = 5,000 kb).
Sensitivity analysis
In the observational analysis, 819 individuals were excluded due to data missing, and the proportion of missing values was about 26%. To avoid possible selection bias, we imputed values for those subjects with missing data using the MissForest method (30). This method can successfully handle missing values in the range of 10%–30%, especially in datasets including different types of variables (30). We then performed the observational analysis again for 3,036 subjects with complete data.
In the two-sample MR, a range of sensitivity analyses was carried out, including MR-Egger, weighted median estimator (WME), and MR pleiotropy residual sum and outlier (MR-PRESSO) (31–33). MR-Egger provides a valid test of pleiotropy, and the estimated value of the intercept in MR-Egger can be interpreted as an estimate of the average pleiotropic effect across the SNPs. Moreover, the estimator of WME is consistent even when up to 50% of the information comes from invalid SNPs. MR-PRESSO was used to identify horizontal pleiotropic SNPs and then estimated the causal effect excluding these SNPs. Furthermore, mediation will be estimated with bias if serum urate has an effect on BMI or the four lipid traits (means reverse causality). We tested the reverse causal effect of BMI, TC, HDL, LDL, TG, and serum urate with each other using the IVW and MR-Egger methods.
In this study, the estimates were betas. To account for multiple testing, we employed a Bonferroni-corrected threshold of p<0.0056 (0.05/9 to correct for one exposure and four lipids in relation to serum urate). All statistical analysis was performed using R (version 4.1.0.) software. We used “TwoSampleMR (0.5.5)” package and “mediation” package to implement MR and observational mediation analyses.
Results
Observational association of BMI with mediators and outcome
After adjusting for age, gender, annual family income, education, marital status, race, and place of birth, the total effect of BMI on serum urate was 0.058 (95% CI: 0.051, 0.065; p< 2×10–16), on TC −0.22 (95% CI: −0.46, 0.015; p=0.66), on HDL −0.62 (95% CI: −0.69, −0.54; p< 2×10–16), on LDL 0.091 (95% CI: −0.11, 0.30; p =0.38), and on TG 1.55 (95% CI: 1.21, 1.89; p< 2×10–16).
Observational mediation analysis
After adjusting for age, gender, annual family income, education, marital status, race, place of birth, and BMI, the average mediation effects of HDL and TG on serum urate were 0.0059 (95% CI: 0.0029, 0.010; p< 2×10–16) and 0.0052 (95% CI: 0.0035, 0.010; p< 2×10–16), respectively. The mediated effects of HDL and TG accounted for the total effect of BMI on UA was 10.2% and 8.9%, respectively. There was no evidence suggesting that TC and LDL have mediated effects ( Table 2 ).
Table 2.
The results of observational mediation analysis.
| Estimation | 95% CI lower | 95% CI upper | p-value | |
|---|---|---|---|---|
| TC ACME ADE Total effect Prop.Mediated HDL |
−0.00031 0.058 0.058 −0.0046 |
−0.0010 0.050 0.049 −0.017 |
0.00 0.070 0.070 0.00 |
0.13 p<2 × 10−16 2 × 10−16 0.13 |
| ACME | 0.0059 | 0.0029 | 0.010 | <2 × 10−16 |
| ADE | 0.052 | 0.045 | 0.060 | <2 × 10−16 |
| Total effect | 0.058 | 0.051 | 0.060 | <2 × 10−16 |
| Prop.Mediated | 0.102 | 0.049 | 0.15 | <2 × 10−16 |
| LDL | ||||
| ACME ADE Total effect Prop.Mediated |
0.00013 0.058 0.058 0.0020 |
−0.00020 0.051 0.051 −0.0034 |
0.000 0.060 0.070 0.010 |
0.44 <2 × 10−16 <2 × 10−16 0.44 |
| TG ACME ADE Total effect Prop.Mediated |
0.0052 0.053 0.058 0.089 |
0.0035 0.046 0.051 0.058 |
0.010 0.060 0.060 0.12 |
<2 × 10−16 <2 × 10−16 <2 × 10−16 <2 × 10−16 |
ACME, average causal mediation effect; ADE, average direct effect.
In the observational sensitivity analysis with 3,036 individuals, the results were consistent with the above, where the proportion of the mediated effects for HDL and TG was 10.7% and 5.1%. Moreover, there was also no evidence suggesting that LDL has a mediated effect ( Table 3 ).
Table 3.
The sensitivity of observational mediation analysis.
| Estimation | 95% CI lower | 95% CI upper | p-value | |
|---|---|---|---|---|
| TC ACME ADE Total effect Prop.Mediated HDL |
0.0002 0.058 0.058 0.0024 |
−0.00009 0.052 0.052 −0.0015 |
0.00 0.06 0.06 0.01 |
0.22 <2 × 10−16 <2 × 10−16 0.22 |
| ACME | 0.0065 | 0.0041 | 0.010 | <2 × 10−16 |
| ADE | 0.052 | 0.046 | 0.060 | <2 × 10−16 |
| Total effect | 0.059 | 0.054 | 0.060 | <2 × 10−16 |
| Prop.Mediated | 0.107 | 0.068 | 0.14 | <2 × 10−16 |
| LDL | ||||
| ACME ADE Total effect Prop.Mediated |
0.00036 0.058 0.058 0.0066 |
−0.00025 0.052 0.052 −0.0043 |
0.000 0.060 0.060 0.020 |
0.26 <2 × 10−16 <2 × 10−16 0.26 |
| TG ACME ADE Total effect Prop.Mediated |
0.0030 0.055 0.058 0.051 |
0.0015 0.050 0.052 0.025 |
0.00 0.060 0.060 0.080 |
<2 × 10−16 <2 × 10−16 <2 × 10−16 <2 × 10−16 |
ACME, average causal mediation effect; ADE, average direct effect.
MR association of BMI with mediators and outcome
The two-sample MR showed that greater BMI was a risk factor for a higher level of serum urate. For each standard deviation of BMI increased, serum urate increased by 0.30 mg/dl (95% CI: 0.25, 0.34; p = 2.37×10–35). There was a causal relationship between BMI and the lipids. A greater BMI was associated with lower levels of HDL (β = −0.30, 95% CI: −0.32, −0.27; p = 1.39×10–104) and LDL (β = −0.20, 95% CI: −0.30, −0.098; p = 1.04×10–4) and a higher level of TG (β = 0.21, 95% CI: 0.18, 0.25; p = 1.15×10–39.). There was no evidence for a causal link between BMI and TC. The results of the sensitivity analysis are consistent ( Table 4 ). Table 5 shows the estimates of TC, HDL, LDL, and TG on serum urate. Among the four lipids, only TG has a causal relationship with serum urate (β = 0.22, 95% CI: 0.15, 0.29; p = 2.28×10–10.). For the relationship between HDL and serum urate, the intercept of MR-Egger was statistically significant. Thus, we adopted the MR-Egger’s result. Since TC, HDL, and LDL had no significant causal effect on serum urate, it indicated that only TG may be the mediator of BMI on serum urate.
Table 4.
The effect of BMI on serum urate and lipid traits.
| Outcome | Method | nSNPs | Betas (95% CI) | p-value |
|---|---|---|---|---|
| Urate | IVW | 851 | 0.30 (0.25, 0.34) | 2.37 × 10−35 |
| MR-Egger | 851 | 0.34 (0.21, 0.48) | 6.06 × 10−7 | |
| Egger-intercept | −0.00076 | 0.45 | ||
| WME | 851 | 0.31 (0.25, 0.36) | 1.37 × 10−32 | |
| MR-PRESSO | 838 | 0.32 (0.29, 0.35) | 6.71 × 10−65 | |
| TC | IVW | 856 | −0.030 (−0.075, 0.015) | 0.18 |
| MR-Egger | 856 | −0.16 (−0.28, −0.042) | 0.0071 | |
| Egger-intercept | 0.0022 | 0.018 | ||
| WME | 856 | −0.039 (−0.078, 0.00020) | 0.054 | |
| MR-PRESSO | 820 | −0.029 (−0.070, 0.012) | 0.17 | |
| HDL | IVW | 964 | −0.30 (−0.32, −0.27) | 1.39 × 10−104 |
| MR-Egger | 964 | −0.31 (−0.38, −0.23) | 3.93 × 10−14 | |
| Egger-intercept | 0.00014 | 0.81 | ||
| WME | 964 | −0.29 (−0.31, −0.28) | 5.18 × 10−197 | |
| MR-PRESSO | 863 | −0.32 (−0.32, −0.30) | 1.69 × 10−203 | |
| LDL | IVW | 964 | −0.068 (−0.10, −0.034) | 9.45 × 10−5 |
| MR-Egger | 964 | −0.20 (−0.30, −0.098) | 1.04 × 10−4 | |
| Egger-intercept | 0.0020 | 0.0066 | ||
| WME | 964 | −0.060 (−0.079, −0.042) | 5.24 × 10−10 | |
| MR-PRESSO | 908 | −0.049 (−0.054, −0.035) | 4.40 × 10−12 | |
| TG | IVW | 964 | 0.21 (0.18, 0.25) | 1.15 × 10−39 |
| MR-Egger | 964 | 0.18 (0.091, 0.28) | 1.11 × 10−4 | |
| Egger-intercept | 0.00047 | 0.50 | ||
| WME | 964 | 0.23 (0.21, 0.245) | 5.62 × 10−105 | |
| MR-PRESSO | 853 | 0.25 (0.23, 0.27) | 4.36 × 10−142 |
Table 5.
Effect of lipid traits on serum urate.
| Exposure | Method | nSNPs | Betas (95% CI) | p-value |
|---|---|---|---|---|
| TC | IVW | 109 | −0.035 (−0.096, 0.026) | 0.26 |
| MR-Egger | 109 | 0.019 (−0.10, 0.14) | 0.75 | |
| Egger-intercept | −0.0029 | 0.31 | ||
| WME | 109 | −0.048 (−0.097, 0.001) | 0.055 | |
| MR-PRESSO | 101 | −0.020 (−0.077, 0.037) | 0.49 | |
| HDL | IVW | 208 | −0.090 (−0.14, −0.039) | 0.00047 |
| MR-Egger | 208 | −0.00056 (−0.074, 0.072) | 0.99 | |
| Egger-intercept | −0.0032 | 0.0013 | ||
| WME | 208 | −0.059 (−0.12, 0.00069) | 0.054 | |
| MR-PRESSO | 203 | −0.080 (−0.13, −0.035) | 0.00057 | |
| LDL | IVW | 79 | −0.0027 (−0.13, 0.12) | 0.97 |
| MR-Egger | 79 | 0.071 (−0.16, 0.30) | 0.55 | |
| Egger-intercept | −0.024 | 0.46 | ||
| WME | 79 | −0.043 (−0.14, 0.057) | 0.39 | |
| MR-PRESSO | 74 | −0.065 (−0.14, 0.0075) | 0.084 | |
| TG | IVW | 174 | 0.22 (0.15, 0.29) | 2.28 × 10−10 |
| MR-Egger | 174 | 0.11 (0.0024, 0.22) | 4.66 × 10−2 | |
| Egger-intercept | 0.0033 | 0.013 | ||
| WME | 174 | 0.15 (0.077, 0.23) | 1.73 × 10−4 | |
| MR-PRESSO | 164 | 0.19 (0.14, 0.24) | 1.84 × 10−10 |
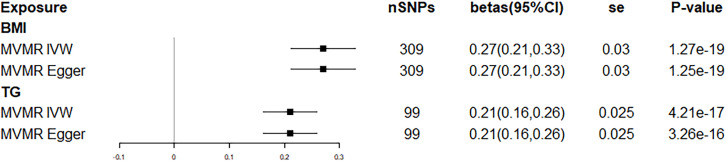
We used BMI and TG as independent variables in the multivariable Mendelian randomization model to estimate their direct effect on serum urate. The direct effects of BMI and TG on serum urate obtained by the IVW method were 0.27 (95% CI: 0.21, 0.33; p = 1.27×10–19) and 0.21 (95% CI: 0.16, 0.26, p=4.21×10−17), respectively; the direct effect estimated by MR-Egger was consistent with the IVW method ( Figure 2 ). The mediated effect of TG was 0.045, the total effect of BMI on serum urate was 0.32, and the proportion of mediated effect of TG was 14.1%. Reverse MR analysis suggested that there was no reverse association between BMI, TC, HDL, LDL, TG, and serum urate ( Table 6 ).
Figure 2.
Forest plot of multivariable Mendelian randomization.
Table 6.
The results of reverse MR.
| Methods | nSNPs | Betas (95% CI) | p-value | |
|---|---|---|---|---|
| Urate–BMI | IVW | 25 | −0.022 (−0.054, 0.0092) | 0.16 |
| MR-Egger | 25 | |||
| Slope | −0.031 (−0.085, 0.022) | 0.26 | ||
| Intercept | 0.00097 | 0.68 | ||
| Urate–TC | IVW | 33 | 0.031 (−0.038, 0.10) | 0.38 |
| MR-Egger | 33 | |||
| Slope | 0.070 (−0.040, 0.18) | 0.22 | ||
| Intercept | −0.0045 | 0.38 | ||
| Urate–TG | IVW | 28 | 0.071 (−0.039, 0.18) | 0.21 |
| MR-Egger | 28 | |||
| Slope | −0.0069 (−0.18, 0.16) | 0.94 | ||
| Intercept | 0.0010 | 0.26 | ||
| Urate–HDL | IVW | 28 | −0.026 (−0.062, 0.010) | 0.16 |
| MR-Egger | 28 | |||
| Slope | 0.014 (−0.041, 0.068) | 0.63 | ||
| Intercept | −0.0051 | 0.074 | ||
| Urate–LDL | IVW | 28 | −0.0040 (−0.047, 0.039) | 0.86 |
| MR-Egger | 28 | |||
| Slope | 0.030 (−0.037, 0.097) | 0.38 | ||
| Intercept | −0.0044 | 0.20 |
The Bonferroni method was used to correct the significance level of the causal association between exposures and serum urate, with p<0.0056 (0.05/9) being statistically significant.
Discussion
In this study, observational and MR analyses supported that TG can explain the effect of BMI on serum urate to a certain extent. A potential mechanism is that TG synthesis accelerates the de-novo synthesis of ribose-5-phosphate to phosphoribosyl pyrophosphate through the common metabolic pathway of NADP–NADPH, resulting in increased serum urate production (34). In the observational analysis, we suggested that the mediating effect of TG accounted for 8.9% of the total effect (BMI-serum urate). For increasing the reliability of the results, we further took the two-step two-sample MR and multivariable Mendelian randomization to prove the mediation effect. Moreover, the two-sample MR showed that the proportion of mediation effect of TG was about 14.1%. Although the point estimates were different, both methods indicated the same conclusions. Moreover, existing studies have shown that greater BMI was associated with a higher level of TG and serum urate and the level of TG was also positively correlated with serum urate (12, 17, 19), which were consistent with our findings.
The point estimates of MR were slightly larger than the estimates of the observational study likely due to MR using genetic variants as instrumental variables to estimate the causal effect of exposure mediators and outcome. The influence of a gene on its phenotype has a lifetime effect. However, an observational study focuses on the effect of exposure on the outcome at a certain point in its investigation. These may explain the relatively large point estimates in MR. Additionally, this could be due to unknown confounding in the observational analysis. The studies by Alice R. Carter and others also suggested that the estimation of MR is greater than in observational studies (13, 35).
In the estimation of HDL-mediated effect, the results of the observational study showed that the proportion of HDL-mediated effect was 10.2%, while in MR, only the IVW approach suggested that serum urate decreased with the increase of HDL. The results of MR-Egger and WME showed that there was no causal effect between HDL and serum urate. Since the intercept term in the MR-Egger model was statistically significant, it indicated that the SNPs included in the MR model existed in horizontal pleiotropy, but the IVW method could not correct for the impact of the horizontal pleiotropy; for this, we adopted the result of the MR-Egger method here. The different results between observational analysis and MR may be due to the absence of unmeasured confounding in the multivariable linear regression model. In the analysis of LDL, observational and MR results suggested there was no causal relationship between LDL and serum urate. It suggested that LDL may not be the mediator in the pathway of BMI on serum urate.
In the two-sample MR analysis, there was some overlap between the samples of exposure and mediators, which could bias the estimates from the two-sample MR toward the confounded observational association, for example, weak instrument bias. Fortunately, bias from the sample overlap can be minimized by using strong instruments (F statistic greater than 10 for the instrument–exposure association) (36, 37). Moreover, the robust strength of our instruments for BMI (mean F statistic of 59) likely minimized any potential impact of sample overlap bias in our results.
In recent years, the incidence and prevalence of gout are increasing worldwide in close relation to the epidemic of obesity and metabolic syndrome. Therefore, it is very important to control BMI and its mediators on serum urate at the same time to reduce the risk of obesity-related gout. Our results suggested that reducing the level of TG may alleviate the increased prevalence of gout which is caused by the increasing BMI to a certain extent. More importantly, there is evidence indicating that statins can reduce the level of TG in our body (38); studies have also confirmed that there is a rare variant in APOC3 having a marked effect on TG levels, which provides a potential target for the development of drugs to reduce the level of TG (39, 40). This means that controlling BMI and TG simultaneously to reduce the occurrence of gout is a practical preventive measure.
Strengths and limitations
This study used observational and two-sample MR analyses to determine the mediators and the size of the mediation effect. We have used these two methods, each with different potential sources of biases, to improve the reliability of our results through triangulation (41). For unmeasured confounding and reverse causality in observational analysis, we used MR to correct the biases. In MR analysis, we identified the mediators by using the two-step two-sample MR, and then multivariable Mendelian randomization was used to estimate the direct effect of exposure and mediator on outcome.
Both methods used in this study assumed that the relationship of BMI, TC, HDL, LDL, TG, and serum urate is linear, and whether there is a non-linear relationship needs to be further explored. Another important limitation of our study is that we assumed that there is no interaction between BMI and the four lipids. In view of the mediated effects, TG can only explain 14.1% of the effect of BMI on serum urate, indicating that there are still other factors that can explain the effect of BMI on serum urate, and these factors may be the future direction of follow-up research.
Conclusions
In this study, observational and MR analyses were used to analyze the mediated effect of TC, HDL, LDL, and TG in the pathway of BMI on serum urate. Our results indicated that TG mediated the effect of BMI on serum urate to a certain extent, and the risk of gout may be reduced by controlling both BMI and TG.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.cdc.gov/nchs/nhanes/index.htm https://gwas.mrcieu.ac.uk/.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
TW had full access to all the data in the study and took responsibility for the integrity of the data and the accuracy of the data analysis. TW and LP designed the study. LP drafted the manuscript. LP and JJ performed the statistical analysis. TW, SH, JW, and XG critically revised the manuscript for important intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (grant numbers 81872715, 82073674, 82103949); the Basic Research Project of Shanxi Province, China (grant number 20210302124186); and the Major Science and Technology Project of Shanxi Province (grant numbers: 202102130501003, 202005D121008).
Acknowledgments
This work was made possible by the generous sharing of GWAS summary statistics and the observational data from NHANES. The authors acknowledge the use of summary-level data from the GIANT consortium, GUGC consortium, and UK Biobank consortium. We also thank the NHANES for offering individual data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.938891/full#supplementary-material
References
- 1. Dalbeth N, Choi HK, Joosten LAB, Khanna PP, Matsuo H, Perez-Ruiz F, et al. Gout. Nat Rev Dis Primers (2019) 5(1):69. doi: 10.1038/s41572-019-0115-y [DOI] [PubMed] [Google Scholar]
- 2. Xia Y, Wu Q, Wang H, Zhang S, Jiang Y, Gong T, et al. Global, regional and national burden of gout, 1990-2017: A systematic analysis of the global burden of disease study. Rheumatol (Oxford) (2020) 59(7):1529–38. doi: 10.1093/rheumatology/kez476 [DOI] [PubMed] [Google Scholar]
- 3. Smith E, Hoy D, Cross M, Merriman TR, Vos T, Buchbinder R, et al. The global burden of gout: Estimates from the global burden of disease 2010 study. Ann Rheum Dis (2014) 73(8):1470–6. doi: 10.1136/annrheumdis-2013-204647 [DOI] [PubMed] [Google Scholar]
- 4. Dehlin M, Jacobsson L, Roddy E. Global epidemiology of gout: Prevalence, incidence, treatment patterns and risk factors. Nat Rev Rheumatol (2020) 16(7):380–90. doi: 10.1038/s41584-020-0441-1 [DOI] [PubMed] [Google Scholar]
- 5. So A, Thorens B. Uric acid transport and disease. J Clin Invest (2010) 120(6):1791–9. doi: 10.1172/JCI42344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuo C-F, Grainge MJ, Zhang W, Doherty M. Global epidemiology of gout: Prevalence, incidence and risk factors. Nat Rev Rheumatol (2015) 11(11):649–62. doi: 10.1038/nrrheum.2015.91 [DOI] [PubMed] [Google Scholar]
- 7. Tsushima Y, Nishizawa H, Tochino Y, Nakatsuji H, Sekimoto R, Nagao H, et al. Uric acid secretion from adipose tissue and its increase in obesity. J Biol Chem (2013) 288(38):27138–49. doi: 10.1074/jbc.M113.485094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheung KJ, Tzameli I, Pissios P, Rovira I, Gavrilova O, Ohtsubo T, et al. Xanthine oxidoreductase is a regulator of adipogenesis and PPARγ activity. Cell Metab (2007) 5(2):115–28. doi: 10.1016/j.cmet.2007.01.005 [DOI] [PubMed] [Google Scholar]
- 9. Dalbeth N, Merriman TR, Stamp LK. Gout. Lancet (2016) 388(10055):2039–52. doi: 10.1016/s0140-6736(16)00346-9 [DOI] [PubMed] [Google Scholar]
- 10. Dalbeth N, Allan J, Gamble GD, Horne A, Woodward OM, Stamp LK, et al. Effect of body mass index on serum urate and renal uric acid handling responses to an oral inosine load: Experimental intervention study in healthy volunteers. Arthritis Res Ther (2020) 22(1):259. doi: 10.1186/s13075-020-02357-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klop B, Elte JW, Cabezas MC. Dyslipidemia in obesity: Mechanisms and potential targets. Nutrients (2013) 5(4):1218–40. doi: 10.3390/nu5041218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Y, Zhang M, Yu X, Wei F, Chen C, Zhang K, et al. Association of hypertension and hypertriglyceridemia on incident hyperuricemia: An 8-year prospective cohort study. J Transl Med (2020) 18(1):409. doi: 10.1186/s12967-020-02590-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Relton CL, Davey Smith G. Two-step epigenetic mendelian randomization: A strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol (2012) 41(1):161–76. doi: 10.1093/ije/dyr233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burgess S, Thompson SG. Multivariable mendelian randomization: The use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol (2015) 181(4):251–60. doi: 10.1093/aje/kwu283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith GD, Ebrahim S. 'Mendelian randomization': Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol (2003) 32(1):1–22. doi: 10.1093/ije/dyg070 [DOI] [PubMed] [Google Scholar]
- 16. Gao X, Wang H, Wang T. Review on correction methods related to the pleiotropic effect in mendelian randomization. Chin J Epidemiol (2019) 40 (3):360–5 doi: 10.3760/cma.j.issn.0254-6450. [DOI] [PubMed] [Google Scholar]
- 17. Larsson SC, Burgess S, Michaelsson K. Genetic association between adiposity and gout: A mendelian randomization study. Rheumatol (Oxford) (2018) 57(12):2145–8. doi: 10.1093/rheumatology/key229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu L, Borges MC, Hemani G, Lawlor DA. The role of glycaemic and lipid risk factors in mediating the effect of BMI on coronary heart disease: A two-step, two-sample mendelian randomisation study. Diabetologia (2017) 60(11):2210–20. doi: 10.1007/s00125-017-4396-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu X, Wang T, Huang S, Zeng P. Evaluation of the causal effects of blood lipid levels on gout with summary level GWAS data: two-sample mendelian randomization and mediation analysis. J Hum Genet (2020) 66(5):465–73. doi: 10.1038/s10038-020-00863-0 [DOI] [PubMed] [Google Scholar]
- 20. MacKinnon DP, Lockwood CM, Hoffman JM, West SG, Sheets V. A comparison of methods to test mediation and other intervening variable effects. psychol Methods (2002) 7(1):83–104. doi: 10.1037//1082-989x.7.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet (2018) 27(20):3641–9. doi: 10.1093/hmg/ddy271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richardson TG, Sanderson E, Palmer TM, Ala-Korpela M, Ference BA, Davey Smith G, et al. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable mendelian randomisation analysis. PloS Med (2020) 17(3):e1003062. doi: 10.1371/journal.pmed.1003062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet (2013) 45(11):1274–83. doi: 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kottgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet (2013) 45(2):145–54. doi: 10.1038/ng.2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Assari S, Nikahd A, Malekahmadi MR, Lankarani MM, Zamanian H. Race by gender group differences in the protective effects of socioeconomic factors against sustained health problems across five domains. J Racial Ethn Health Disparities (2016) 4:884–94. doi: 10.1007/s40615-016-0291-3 [DOI] [PubMed] [Google Scholar]
- 26. Masood M, Aggarwal A, Reidpath DD. Effect of national culture on BMI: A multilevel analysis of 53 countries. BMC Public Health (2019) 19(1):1212. doi: 10.1186/s12889-019-7536-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vishnu A, Belbin GM, Wojcik GL, Bottinger EP, Gignoux CR, Kenny EE, et al. The role of country of birth, and genetic and self-identified ancestry, in obesity susceptibility among African and Hispanic americans. Am J Clin Nutr (2019) 110(1):16–23. doi: 10.1093/ajcn/nqz098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thompson JR, Minelli C, Del Greco MF. Mendelian randomization using public data from genetic consortia. Int J Biostat (2016) 12(2):1–11. doi: 10.1515/ijb-2015-0074 [DOI] [PubMed] [Google Scholar]
- 29. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol (2013) 37(7):658–65. doi: 10.1002/gepi.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stekhoven DJ, Buhlmann P. MissForest–non-parametric missing value imputation for mixed-type data. Bioinformatics (2012) 28(1):112–8. doi: 10.1093/bioinformatics/btr597 [DOI] [PubMed] [Google Scholar]
- 31. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol (2015) 44(2):512–25. doi: 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol (2016) 40(4):304–14. doi: 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet (2018) 50(5):693–8. doi: 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nejatinamini S, Ataie-Jafari A, Qorbani M, Nikoohemat S, Kelishadi R, Asayesh H, et al. Association between serum uric acid level and metabolic syndrome components. J Diabetes Metab Disord (2015) 14:70. doi: 10.1186/s40200-015-0200-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carter AR, Gill D, Davies NM, Taylor AE, Tillmann T, Vaucher J, et al. Understanding the consequences of education inequality on cardiovascular disease: Mendelian randomisation study. BMJ (2019) 365:l1855. doi: 10.1136/bmj.l1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pierce BL, Burgess S. Efficient design for mendelian randomization studies: Subsample and 2-sample instrumental variable estimators. Am J Epidemiol (2013) 178(7):1177–84. doi: 10.1093/aje/kwt084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample mendelian randomization. Genet Epidemiol (2016) 40(7):597–608. doi: 10.1002/gepi.21998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wurtz P, Wang Q, Soininen P, Kangas AJ, Fatemifar G, Tynkkynen T, et al. Metabolomic profiling of statin use and genetic inhibition of HMG-CoA reductase. J Am Coll Cardiol (2016) 67(10):1200–10. doi: 10.1016/j.jacc.2015.12.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Timpson NJ, Walter K, Min JL, Tachmazidou I, Malerba G, Shin SY, et al. A rare variant in APOC3 is associated with plasma triglyceride and VLDL levels in europeans. Nat Commun (2014) 5:4871. doi: 10.1038/ncomms5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Drenos F, Smith GD, Ala-Korpela M. Metabolic characterization of a rare genetic variation within APOC3 and its lipoprotein lipase-independent effects. (2016) 9(3):231–9. doi: 10.1161/CIRCGENETICS.115.001302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lawlor DA, Tilling K, Davey Smith G. Triangulation in aetiological epidemiology. Int J Epidemiol (2016) 45(6):1866–86. doi: 10.1093/ije/dyw314 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.cdc.gov/nchs/nhanes/index.htm https://gwas.mrcieu.ac.uk/.