Abstract
Bacterial cell-to-cell communication facilitates coordinated expression of specific genes in a growth rate-II and cell density-dependent manner, a process known as quorum sensing. While the discovery of a diffusible Escherichia coli signaling pheromone, termed autoinducer 2 (AI-2), has been made along with several quorum sensing genes, the overall number and coordination of genes controlled by quorum sensing through the AI-2 signal has not been studied systematically. We investigated global changes in mRNA abundance elicited by the AI-2 signaling molecule through the use of a luxS mutant that was unable to synthesize AI-2. Remarkably, 242 genes, comprising ca. 5.6% of the E. coli genome, exhibited significant transcriptional changes (either induction or repression) in response to a 300-fold AI-2 signaling differential, with many of the identified genes displaying high induction levels (more than fivefold). Significant induction of ygeV, a putative ς54-dependent transcriptional activator, and yhbH, a ς54 modulating protein, suggests ς54 may be involved in E. coli quorum sensing.
Many bacteria have evolved the ability to condition culture medium by secreting low-molecular-weight signaling pheromones in association with growth phase to control expression of specific genes, a process termed quorum sensing (19). Physiological processes controlled by quorum sensing occur in diverse species of bacteria and include bioluminescence (17), antibiotic biosynthesis (4), pathogenicity (34), and plasmid conjugal transfer (18). While acyl-homoserine lactones (HSL) appear to be the predominant quorum signal (or autoinducer [AI]) used by host-associated gram-negative bacteria, discovery of a second signaling pathway in the marine bacterium Vibrio harveyi (6, 8, 41) revealed an alternate AI, termed AI-2, which regulates bioluminescence in conjunction with AI-1 (N-(3-hydroxybutanoyl)-l-homoserine lactone) (7).
Importantly, AI-2 (or AI-2-like) activity has been observed in virtually all strains of pathogenic and nonpathogenic Escherichia coli and Salmonella enterica serovar Typhimurium (16, 40–42), requiring the luxS gene for synthesis (43). The physiological role of AI-2 in E. coli has not been clearly elucidated, but initial findings indicate that inhibition of chromosomal replication was subject to a quorum sensing mechanism (52). More recently, quorum sensing in E. coli has been implicated in regulating the expression and activity of SdiA, a LuxR-type transcriptional activator of the cell division genes ftsQAZ, through AI-2 (15, 39). In addition, extracellular factors which accumulate in enterohemorrhagic E. coli O157:H7 culture supernatants bind to the N-terminal region of SdiA for controlling the expression of virulence factors in a quorum-dependent fashion (25). Besides possible roles in cell division and pathogenesis, quorum sensing in E. coli was postulated to play a role in stationary phase gene expression (23, 27, 39), perhaps in a bimodal fashion with the stationary phase sigma factor rpoS or with other yet-to-be-determined quorum signals.
Recently, the application of global identification methodologies (e.g., DNA microarrays) has resulted in identification of quorum-regulated processes as well as the characterization of quorum circuit architecture in Streptococcus pneumoniae and Pseudomonas aeruginosa (16, 51). Therefore, a systematic investigation of native, quorum-mediated genes in E. coli was performed here to quantitatively analyze the global transcriptional pattern in response to the extracellular AI-2 signal molecule. To this end, DNA microarray analysis was utilized to quantify changes in transcription for every open reading frame (ORF) of E. coli strain W3110 in response to AI-2 signaling molecule.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
E. coli strains used in this study were W3110 (F− λ− IN(rrnD-rrnE) rph-1) (22), E. coli Genetic Stock Center, New Haven, Conn., and MDAI2, a luxS::Tcr derivative of W3110 (16). V. harveyi strains BB152 (luxL::Tn5 AI-1−, AI-2+) and BB170 (luxN::Tn5 sensor 1−, sensor 2+) for determination of AI-2 activity (41) were kindly provided by B. L. Bassler. Plasmid pGFPuv-ftsQ2p for quantifying quorum-regulated ftsQp2 expression is described elsewhere (15). Luria-Bertani (LB) medium contained 5 g of yeast extract (Sigma Chemical Co.) per liter, 10 g of Bacto tryptone (Difco) per liter, and 10 g of NaCl per liter and was supplemented with 50 mM glucose. Autoinducer bioassay medium and LM medium (l-marine) are given in detail elsewhere (8, 37).
Growth conditions.
Overnight E. coli MDAI2 cultures grown aerobically in LB broth plus supplemental glucose (50 mM) were subcultured into 200 ml of LB plus 50 mM glucose (1% [vol/vol] inoculum). Cultures were grown aerobically at 30°C to an optical density at 600 nm (OD600) of 1.0 followed by centrifugation (2,500 × g at 4°C) and gentle resuspension in ∼1 ml of fresh LB. Resuspended cells were split equally into two parallel flasks each containing 100 ml of conditioned medium (positive or negative for AI-2) plus 50 mM glucose (prepared as outlined below) such that the culture OD600 was maintained at ∼1.0. Aerobic growth ensued for 20 min, at which time 5-ml samples were collected for total RNA extraction.
Preparation of cell-free culture fluids and conditioned medium.
W3110 (luxS+) and MDAI2 (luxS) overnight cultures, grown aerobically at 30°C in LB plus 50 mM glucose, were used to inoculate 500 ml (1% inoculum) of fresh LB plus 50 mM glucose. Cultures were grown to an OD600 of 3.0 (∼8 h), and glucose analysis (YSI glucose analyzer model 2700) was used to confirm identical growth patterns in addition to growth rate calculations based on culture OD600. Cell-free culture fluids or conditioned medium (CM) was prepared by centrifugation of 500-ml E. coli cultures for 10 min (10,000 × g at 4°C). Cleared supernatants were passed through 0.22-μm vacuum-driven Millipore filters and were stored at −20°C. Prior to use in AI-2 signaling experiments, CM was supplemented with 50 mM glucose and was assayed for AI-2 activity to confirm signaling conditions (positive or negative for AI-2). V. harveyi BB152 cell-free culture fluids were prepared analogously to obtain positive (+AI-2) control samples as reported previously (41).
AI activity assay.
E. coli cell-free culture fluids were tested for the presence of AI-2 using the V. harveyi reporter strain BB170, which responds only to AI-2 (41). Luminescence assays were performed as outlined elsewhere (41), and luminescence was measured as a function of V. harveyi cell density by quantitating light production with a luminometer (EG & G Berthold). Data reported as fold activation were obtained by dividing the light produced by the reporter after addition of E. coli culture fluid by the light output of the reporter when growth medium alone was added (15).
Growth stimulation assays.
Overnight cultures of W3110 and MDAI2 grown in LB were used to inoculate (1%, vol/vol) one of the following: LB plus 10% CM (+AI-2); LB plus 10% CM (+AI-2) plus 0.8% glucose; LB plus 10% CM (−AI-2); or LB plus 10% CM (−AI-2) plus 0.8% glucose. These experiments were performed in triplicate. OD600 measurements were taken every 60 min over a 9-h period and used to calculate the specific growth rate for exponentially growing batch cultures. The specific growth rates were determined, with most accuracy for the first four data points.
RNA isolation and labeling.
Harvested cells were centrifuged (5,000 × g at 4°C) and resuspended in lysozyme-containing buffer (5 μg μl−1 of lysozyme (Sigma) in 1× Tris-EDTA Buffer [Sigma], pH 8.0) at room temperature for 5 min. Total RNA was purified from 3 × 109 to 5 × 109 cells using a Qiagen RNeasy mini kit. RNA was eluted with diethyl pyrocarbonate (DEPC) water (Sigma) and quantified by measuring absorbance (A260) with a spectrophotometer (Beckman DU 640). RNA samples (∼75 to 80 μg) were concentrated to ∼15 μl using a Microcon YM-30 filter (Millipore). Total purified RNA was labeled with either Cy3-dUTP or Cy5-dUTP as outline previously (49). Briefly, total RNA (∼15 μl) was mixed with random hexamer primers (Amersham Pharmacia) and combined with 1X labeling mixture (first-strand buffer [Gibco-BRL], deoxynucleoside triphosphates (low TTP; Pharmacia), RNAsin (Promega), and DEPC water) and incubated at room temperature for 10 min. Labeling with Cy-3-dUTP (or Cy-5-dUTP) during a reverse transcriptase reaction using Superscript II (Gibco-BRL) was at 42°C for 1 h in the dark. After labeling, NaOH was added to the sample to hydrolyze RNA template and was incubated (65°C for 15 min) followed by neutralization with HC1 and Tris (pH 7.6). Following Microcon MY-30 filter purification, control and experimental samples were combined, pulse centrifuged (<1 min), washed, and concentrated by filter centrifugation prior to direct hybridization to glass microarrays.
Microarray hybridization procedures.
Glass DNA microarrays (University of Wisconsin Gene Expression Center), consisting of full-length PCR products (spotted one time) from all E. coli ORFs according to Blattner et al. (9), were used to quantify relative mRNA levels by parallel two-color hybridization according to protocols described elsewhere (49). The number of features on the slide, therefore, consisted of the 4,290 annotated ORFs plus ∼200 control spots, including fragmented E. coli genomic DNA, nonspecific salmon sperm DNA, 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate), and six different yeast ORF PCR products. Slides were 36 spots by 36 spots by 4 panels, with each spot averaging 100 μ in diameter. Briefly, labeled probes were mixed with salmon sperm DNA, yeast tRNA, and PerfectHyb buffer (Sigma) prior to overnight hybridization to a DNA microarray (∼12 h at 50°C). Arrays were washed with 0.2X SSC and 0.1% Sodium dodecyl sulfate (Sigma) for 2 min. Subsequent 2-min washes (three times) were with 0.2X SSC followed by dipping (C9. 10 times) in 0.05X SSC. Arrays were centrifuged to dry the surface and then scanned immediately using a GMS418 scanner (Genetic Microsystems) at 10-μm resolution and Arrayscan software (Genetic Microsystems). The resulting 16-bit TIFF images were analyzed using SCANALYZE software, publicly available at http://rana.stanford.edu/software/. RNA samples from experimental flasks (+AI-2 cultures) were first labeled with Cy5, and control RNA (−AI-2 culture) was labeled with Cy3. Reverse labeling of the RNA (experiment, Cy3; control, Cy5) was then performed on a second array and used to verify all transcriptional induction ratios. Analogously to procedures of LaRossa and colleagues (50), further validation of microarray data was made via independent measures of RNA transcript level by RNA dot blots as outlined previously (15).
Data selection and analysis.
Microarray data whose intensities were reproducibly higher than that of the background level were selected for analysis to eliminate expression ratios that were extremely high or low due to undetectable signal in control or experimental samples (26). Induction ratios (Cy5 for experimental RNA relative to Cy3 for control RNA) as well as background intensities for each dye were independently distributed in approximately normal fashion. Induction ratios were obtained by dividing background-corrected signal intensities of experimental samples by background-corrected intensities of control samples. Intensities used to calculate induction ratios were normalized as a percentage of the total of intensities of all the spots (Cy5 or Cy3) on the array, thereby accounting for the specific activity of the probes. That is, all Cy5 spots were normalized to the average Cy5 signal and vice versa for Cy3. Signals that were higher under control conditions were inverted to permit direct comparison between induction and repression ratios. Correlation among reverse-labeled repeat hybridizations with forward-labeled samples ranged from 0.756 to 0.998, and the standard deviation of duplicate (but reverse-labeled) induction ratios ranged from 0.08 to 0.45, providing a measure of reproducibility.
RESULTS
Generation of AI-2 signaling differential.
Our overall objective was to identify all of the E. coli ORFs that exhibit a significant increase or decrease (more than twice the standard deviation [SD] of the mean induction ratio) in mRNA abundance caused by a differential in AI-2 signaling activity. MDAI2 (luxS) cells, which were unable to produce AI-2 as confirmed using a V. harveyi AI-2 activity assay (41), were grown to an OD600 of 1.0 (ca. 3 to 4 h), split evenly, and resuspended in either CM exhibiting high AI-2 activity (+AI-2) or identically generated CM deficient in AI-2 signaling activity (−AI-2) (see Materials and Methods). This procedure resulted in a greater than 300-fold difference in AI-2 signaling activity experienced by experimental cultures compared to that for negative controls (Table 1). Comparative measurements of transcript abundance were made by extracting RNA from cells immediately following 20 min of exposure to CM (+AI-2) or identically generated CM (−AI-2). The final cell densities (at 20 min) of experimental and control cultures were identical (OD600 of ca. 1.2; 17% change), suggesting that transcriptional changes were not substantially influenced by growth rate or cell density differences of control and experimental samples. Lastly, we performed growth stimulatory assays (see Materials and Methods) to determine whether the culture growth rates were affected by the presence of AI-2. The results (not shown) confirmed that there was no significant difference in growth rate between cultures exposed to CM (+AI-2) or CM (−AI-2). Thus, it was concluded that under the conditions tested here, AI-2 does not stimulate or inhibit cell growth.
TABLE 1.
AI-2 activity of CM experienced by MDAI2 luxS cells
| CM (+AI-2 or −AI-2) | Fold AI-2 activitya |
|---|---|
| CM stock solutions | |
| CM generated from W3110 luxS+ | 1,520 |
| CM generated from MDAI2 luxS | 5 |
| Experimental samples | |
| MDAI2 cells at an OD600 of 1.0 | 7 |
| MDAI2 cells (control, t = 0 min) | 6 |
| MDAI2 cells (exp, t = 0 min) | 1,498 |
| MDAI2 cells (control, t = 20 min)b | 5 |
| MDAI2 cells (exp, t = 20 min)b | 1,514 |
Fold AI-2 activity represents the induction of luminescence (RLU) in V. harveyi reporter strain BB170 (sensor 1−, sensor 2+) from E. coli cell-free culture samples in relation to luminescence (RLU) with fresh medium alone. Values are reported as the averages of three independent activity measurements.
Samples used for RNA isolation and Cy3 or Cy5 labeling and subsequent hybridization to microarray. OD600S of these cultures were almost identical (OD600 = 1.2). Fold induction results from three replicate experiments agreed within 10%. exp, experimental.
Identification of quorum-regulated genes in E. coli.
Using DNA microarrays, we identified 242 genes representing approximately 5.6% of the entire genome that were upregulated (154 total genes) or repressed (88 total genes) more than 2.3-fold (corresponding to 2 SD above the mean induction ratio over the entire array) in the presence of AI-2. A total of 139 genes changed more than 2.9-fold (3 SD), and 23 genes changed more than 5-fold, including frwC (33.0-fold), yeiK (25.4-fold), and yidS (21.3-fold). On the contrary, 25 genes were more than 5-fold repressed, notably b2650 (27.8-fold), thiH (19.2-fold), and b2247 (15.2-fold). The entire dataset of induction ratios for all 4,290 annotated E. coli ORFs can be accessed at http://www.umbi.umd.edu/∼cab/bentley/AI-2_array.html.
Involvement of AI-2 in multiple physiological processes.
Importantly, we observed that a large number of the responding genes comprised three broad functional categories, according to Riley and Labedan (35). To be specific, we identified 22 genes involved in cell division, DNA processing, and morphological (cell shape) processes (Table 2), which was consistent with previous findings that a quorum sensing mechanism regulates DNA replication and cell division (48, 52). Twenty-three genes were involved in processes known to be quorum-regulated in other gram-negative bacteria, such as virulence, biofilm and exopolysaccharide formation, cell motility, and other surface-associated phenomena (Table 3) (14, 21, 31, 32, 45). Lastly, a cluster of 28 genes involved in small-molecule metabolism included genes which, to date, have not been implicated in cell-cell communication but may provide a link between central metabolism and quorum signaling, perhaps for the production and degradation of AI-2 itself (Table 4). Interestingly, addition of AI-2 did not significantly affect any of the currently known quorum sensing genes in E. coli under the timing and conditions studied here (Table 5) but did significantly alter expression of 10 putative signal transduction-associated genes (Table 6). Also, a large percentage (ca. 60%) of the genes exhibiting 2.9-fold changes (3 SD) were putative ORFs with no currently known function (Table 7).
TABLE 2.
Cell division, DNA processing, and morphological genes responding to AI-2
| Gene | Gene product and/or functiona | Induction ratio (fold) |
|---|---|---|
| bolA | Possible regulator of murein genes | +10.2 |
| tra8_3 | IS30 transposase | +4.4 |
| himA | Integration host factor, alpha subunit; site-specific recombination | +3.9 |
| mreD | Rod-shape-determining protein | +3.6 |
| ompA | Outer membrane protein 3a | +3.2 |
| pin | Inversion of adjacent DNA; at locus of e14 element | +3.2 |
| holE | DNA polymerase III, theta subunit | +3.1 |
| nohB | Bacteriophage DNA packaging protein | +3.0 |
| dnaQ | DNA polymerase III, epsilon subunit | +2.8 |
| csrA | Carbon storage regulator; controls glycogen synthesis, gluconeogenesis, cell size and surface properties | +2.8 |
| mrdB | Rod-shape-determining membrane protein; sensitivity to radiation and drugs | +2.6 |
| b2505 | Putative outer membrane lipoprotein | −2.7 |
| cspC | Cold shock protein, transcription antiterminator | −2.7 |
| lpp | Murein lipoprotein | −2.7 |
| yfjN | Putative cell division protein | −2.7 |
| cspI | Cold shock-like protein | −2.8 |
| osmB | Osmotically inducible lipoprotein | −2.9 |
| ftsE | ATP-binding component of a membrane-associated complex involved in cell division | −3.1 |
| cspE | Cold shock protein, chromatin partitioning, and nucleoid structure | −3.1 |
| dicB | Inhibition of cell division | −3.2 |
| murD | UDP-N-acetylmuramoylalanine-d-glutamate ligase | −3.4 |
| gef | Gef protein interferes with membrane function when in excess, cell killing | −6.6 |
Genome information is from Blattner et al. (9).
TABLE 3.
Genes comprising quorum-regulated processes (i.e., virulence, biofilm formation, motility, surface, and outer membrane-associated functions)
| Gene or B number | Gene product and/or functiona | Induction ratio (fold) |
|---|---|---|
| hha | Hemolysin expression-modulating protein | +11.1 |
| wzb | Probable protein-tyrosine-phosphatase | +6.2 |
| ompG | Outer membrane porin protein | +5.1 |
| smpA | Small membrane protein A | +4.5 |
| yadK | Putative fimbrial protein | +3.8 |
| rcsB | Positive response regulator for colanic capsule biosynthesis (sensor, RcsC) | +3.5 |
| yadN | Putative fimbrial-like protein | +3.5 |
| crl | Transcriptional regulator of cryptic csgA gene for curli surface fibers | +3.5 |
| rfaJ | UDP-d-glucose:(galactosyl)lipopolysaccharide glucosyltransferase | +3.4 |
| rnk | Regulator of nucleoside diphosphate kinase | +3.1 |
| motB | Enables flagellar motor rotation, linking torque machinery to cell wall | +3.1 |
| b1502 | Putative adhesin; similar to FimH protein | +3.0 |
| tap | Methyl-accepting chemotaxis protein IV, peptide sensor receptor | +2.9 |
| pstC | High-affinity phosphate-specific transport system, cytoplasmic membrane component | +2.8 |
| yehA | Putative type 1 fimbrial protein | +2.7 |
| rfaY | Lipopolysaccharide core biosynthesis | +2.5 |
| rfaD | ADP-l-glycero-d-mannoheptose-6-epimerase | −2.7 |
| uhpT | Hexose phosphate transport protein | −2.7 |
| cheW | Positive regulator of CheA protein activity | −2.7 |
| b1629 | Putative membrane protein | −2.8 |
| evgS | Putative sensor for regulator EvgA and homologous to B. pertussis bvgS | −2.8 |
| fliP | Flagellar biosynthesis | −2.9 |
| nlpC | Lipoprotein | −2.9 |
| flgN | Protein of flagellar biosynthesis | −3.7 |
Genome information is from Blattner et al. (9). Bold-faced genes exist within known operons
TABLE 4.
Small molecule metabolism induced by AI-2 signaling
| Gene | Gene product and/or functiona | Induction ratio (fold) |
|---|---|---|
| frwC | PTS system, fructose-like enzyme II component | +33.0 |
| fpr | Ferredoxin-NADP reductase | +10.9 |
| ntpA | dATP pyrophosphohydrolase | +4.5 |
| bioH | Biotin biosynthesis; reaction prior to pimeloyl Coenzyme A | +3.7 |
| lysP | Lysine-specific permease | +3.6 |
| edd | 6-Phosphogluconate dehydratase | +3.3 |
| trpR | Regulator for trp operon and aroH; trp aporepressor | +3.2 |
| gpmB | Phosphoglyceromutase 2 | +3.2 |
| marB | Multiple antibiotic resistance protein | +3.1 |
| psiF | Induced by phosphate starvation | +2.9 |
| dgt | Deoxyguanosine triphosphate triphosphohydrolase | +2.8 |
| glcB | Malate synthase G | +2.8 |
| caiF | Transcriptional regulator of cai operon | +2.8 |
| tdcA | Transcriptional activator of tdc operon | +2.7 |
| tsx | Nucleoside channel; receptor of phage T6 and colicin K | +2.6 |
| rfbC | dTDP-6-deoxy-d-glucose-3,5 epimerase | −2.7 |
| potB | Spermidine/putrescine transport system permease | −3.1 |
| wcaH | GDP-mannose mannosyl hydrolase | −3.1 |
| usg | Putative PTS system enzyme II A component | −3.3 |
| pheL | Leader peptide of chorismate mutase-P-prephenate dehydratase | −3.5 |
| carB | Carbamoyl-phosphate synthase large subunit | −3.7 |
| potA | ATP-binding component of spermidine/putrescine transport | −5.1 |
| mtlR | Repressor for mtl | −5.1 |
| ilvL | ilvGEDA operon leader peptide | −5.4 |
| hyfA | Hydrogenase 4 Fe-S subunit | −5.6 |
| ugpC | ATP-binding component of sn-glycerol 3-phosphate transport system | −6.2 |
| thiH | Thiamin biosynthesis, thiazole moiety | −19.2 |
Genome information is from Blattner et al. (9). Bold-faced genes exist within a known operon.
TABLE 5.
Response of known E. coli quorum sensing genes to AI-2
| Quorum-regulated gene | Function or description | Fold increase | Reference |
|---|---|---|---|
| ftsQ | Cell division | 1.1 | 48 |
| ftsA | Cell division | 1.0 | 48 |
| ftsZ | Cell division | 1.4 | 48 |
| sdiA | Positive regulator of cell division | 2.0 | 39 |
| rpoS | Stationary phase sigma factor | 1.2 | 39 |
| cysK | O-acetylserine lyase A | 1.5 | 3 |
| astD | Putative succinylglutamate-semialdehyde dehyrogenase | 1.3 | 3 |
| tnaB | Glutamate:succinate semialdehyde aminotransferase | 1.1 | 3 |
| gabT | Low-affinity tryptophan permease | 1.4 | 3 |
TABLE 6.
Putative signal transduction-associated genes that significantly respond to the AI-2 quorum signal
| Gene or B number | Probable gene producta | Induction ratio (fold) |
|---|---|---|
| ylcA | Putative two-component transcriptional regulator | +4.4 |
| b2380 | Putative sensor protein | +3.9 |
| ygeV | Putative ς54-dependent transcriptional regulator | +3.6 |
| b2248 | Putative regulator | +3.6 |
| ybiF | Putative transmembrane subunit | +3.4 |
| yqhC | Putative AraC-type regulatory protein | +2.7 |
| yhhM | Putative receptor | +2.6 |
| yhbH | Probable ς54 modulation protein | +2.5 |
| ycgE | Putative transcriptional regulator | −2.6 |
| ycjZ | Putative transcriptional regulator, LysR type | −3.3 |
Genome information is from Blattner et al. (9).
TABLE 7.
Genes of unknown class or function significantly induced or repressed by AI-2 quorum signaling
| Gene or B number | Induction ratio (fold) |
|---|---|
| yeiK | +25.4 |
| yidS | +21.3 |
| pqqL | +12.7 |
| yghB | +11.3 |
| ycbR | +9.8 |
| yaeT | +9.8 |
| b1506 | +9.7 |
| ydfE | +9.7 |
| yggE | +9.6 |
| b0100 | +7.8 |
| b2145 | +7.0 |
| ybcJ | +6.5 |
| b3254 | +6.2 |
| yccE | +5.4 |
| ydaC | +4.9 |
| yaiV | +4.8 |
| ybiM | +4.7 |
| yddM | +4.5 |
| b1543 | +4.3 |
| yagH | +4.3 |
| b2432 | +4.2 |
| b2875 | +4.2 |
| yhbP | +4.1 |
| ydfA | +4.1 |
| b2326 | +4.0 |
| b2792 | +3.9 |
| yebG | +3.9 |
| yqgB | +3.8 |
| ybhH | +3.7 |
| yhcO | +3.6 |
| b2868 | +3.5 |
| yhiE | +3.5 |
| ydiB | +3.4 |
| yibA | +3.3 |
| ygeF | +3.3 |
| b1152 | +3.3 |
| yecN | +3.3 |
| yjhC | +3.2 |
| ycbG | +3.2 |
| b0245 | +3.1 |
| yfeH | +3.1 |
| b1364 | +3.1 |
| b1155 | +3.1 |
| b1582 | +3.0 |
| b1445 | +3.0 |
| yebA | +3.0 |
| yabI | +3.0 |
| b1567 | +3.0 |
| b1438 | −3.0 |
| b2859 | −3.0 |
| ydcD | −3.1 |
| yfiK | −3.1 |
| ybeB | −3.1 |
| ybiK | −3.1 |
| ydjA | −3.2 |
| yqjC | −3.3 |
| b4250 | −3.3 |
| yjgX | −3.5 |
| yqjH | −3.6 |
| b1963 | −3.7 |
| ycjC | −3.9 |
| yifM | −3.9 |
| b2390 | −3.9 |
| b2511 | −4.2 |
| b2748 | −4.4 |
| b1553 | −4.7 |
| ybgF | −5.7 |
| yaiC | −5.9 |
| ygaF | −6.1 |
| yifN | −6.8 |
| b2254 | −6.8 |
| b2363 | −7.7 |
| yecE | −7.7 |
| ymgC | −8.4 |
| b2247 | −15.2 |
| b2650 | −27.8 |
Genome information is from Blattner et al., (9).
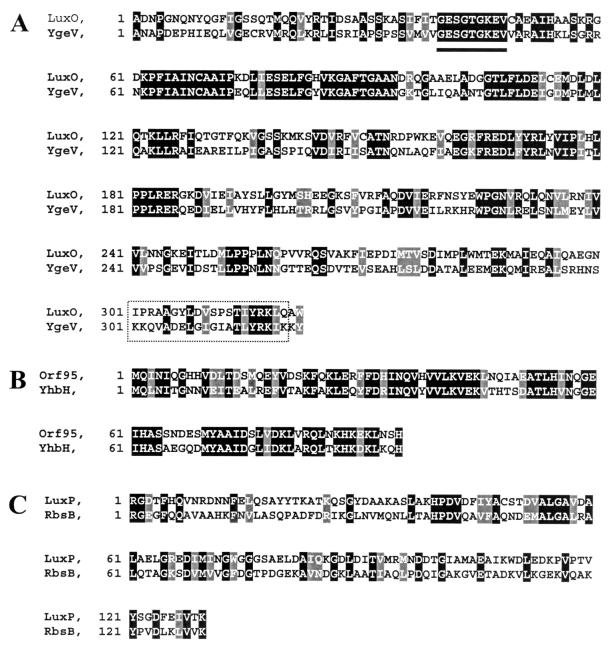
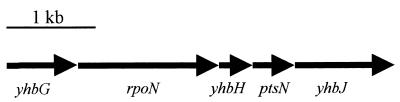
Interestingly, a putative ς54-dependent transcriptional activator, ygeV, whose gene product contained a putative ς54 interaction domain as well as a DNA-binding helix-turn-helix (HTH) motif, exhibited significant homology (61% identity) to the LuxO quorum response regulator of V. harveyi (28) and was upregulated 3.6-fold. Alignment of the central portions of LuxO and YgeV as well as several other AI-2 responding proteins is depicted in Fig. 1. In addition, yhbH, encoding a putative ς54 modulating protein and having 85% identity to ORF95 of V. harveyi (Fig. 1), was observed to increase 2.5-fold (Table 6). Interestingly, rpoN ς54 levels were relatively unchanged (decreased 1.08-fold), suggesting that corresponding ς54 levels are either unchanged in response to AI-2 or regulated at the translational level, perhaps by YbhH-mediated modulation. Of note, the genetic organization of the rpoN chromosomal region of E. coli (Fig. 2) (24) is almost identical to that of the V. harveyi rpoN region, as will be discussed later. Finally, we found that expression of rbsB, encoding a ribose binding protein homologous to LuxP (Fig. 1) and thought to bind AI-2 directly (28), was relatively unchanged (1.4-fold). To determine whether ribose had a direct effect on AI-2 quorum sensing in E. coli, W3110 cells harboring a ftsQA p2 promoter probe plasmid that is positively regulated by SdiA (39) and responds to AI-2 (15) were exposed to varying concentrations of ribose. Plasmid-bearing cells were grown in LB medium plus 50 mM glucose, and experimental cultures were supplemented with 2 g of either l- or d-ribose per liter. Results demonstrated that l-ribose moderately stimulated expression of the quorum-regulated ftsQA genes through the p2 promoter relative to controls containing no l-ribose (Fig. 3), while exposure to d-ribose resulted in nearly identical ftsQA induction levels as negative controls (data not shown). Interestingly, we found that addition of l-ribose at similar concentrations (ca. 1 to 2 g/liter) resulted in moderate induction of the lux genes in the V. harveyi AI-2 reporter assay (ca. 300- to 400-fold activation; data not shown), suggesting l-ribose might act as an analogue or precursor that can trigger the AI-2-stimulated quorum response.
FIG. 1.
Alignment of LuxO of V. harveyi with a putative ς54-dependent transcriptional activator of E. coli (A), YgeV ORF95 of V. harveyi with YhbH of E. coli (B), both putative ς54-modulating proteins, and LuxP of V. harveyi with the ribose periplasmic binding protein of E. coli, RbsB (C). Amino acids that match the consensus generated for the two sequences are boxed in black. The glycine-rich region encoding the nucleotide binding domain common of ς54-interacting proteins is underlined, while the putative HTH DNA binding domains for LuxO and YgeV are boxed by a dashed line.
FIG. 2.
Genetic organization of the E. coli rpoN chromosomal region. The genetic organization of this region is similar to that described for the rpoN region of Vibrio cholerae and V. harveyi. yhbG encodes a probable ATP-binding cassette (ABC) transporter protein, yhbH encodes a putative ς54 regulatory protein much like orf95 of V. harveyi, ptsN encodes a PTS system nitrogen regulator, and yhbJ has no known function (24).
FIG. 3.
Induction of ftsQA expression through the p2 promoter by W3110/pGFPuv-ftsQ2p reporter cells in LB medium plus 50 mM glucose or LB medium plus 50 mM glucose and supplemented with l-ribose (2 g/liter). AU, arbitrary units.
Independent measure of RNA levels confirms AI-2 stimulatory effect on gene expression.
Previously, quantitative reverse transcription-PCR (50) and lacZ transcriptional fusions (45) were used to independently validate microarray expression data. In a similar manner, total RNA dot blotting was performed as previously outlined (15) and used to quantitatively verify the expression level changes of representative genes identified by microarray analysis. For these confirmatory experiments, RNA samples were harvested from a replicate experiment (see Materials and Methods), and induction ratios were compared between the two methodologies (Table 8). Reassuringly, the expression changes of 8 genes (out of 9 tested) were confirmed using total RNA dot blotting, which represented agreement (ca. 89%) similar to that previously reported (50). The lone discrepancy, thiH, was significantly repressed (19.2-fold) according to comprehensive transcript profiling but was upregulated (2.8-fold) according to the dot blotting procedure. The source of this difference, while not definitively resolved, was possibly due to high background signal in the film-based development associated with RNA blotting.
TABLE 8.
Fold induction of transcripts in response to AI-2 quorum signal as determined by microarray probing and RNA dot blotting
| Gene | Fold change witha:
|
|
|---|---|---|
| RNA dot blotting | Microarray probing | |
| ftsQ | 1.0 | 1.1 |
| sdiA | 1.0 | 2.0 |
| ompA | 4.7 | 3.2 |
| rpoS | 1.3 | 1.2 |
| rcsB | 4.3 | 3.5 |
| thrS | 4.5 | 3.1 |
| ygeV | 3.9 | 3.6 |
| thrS | 27.2 | 33.0 |
| thiH | 2.75 | 19.2 |
RNA dot blotting (and microarray) results were obtained as outlined in Materials and Methods. Reported values are intensity obtained from experimental RNA samples divided by intensity obtained from control RNA and are the averages of three replicate samples. Microarray and blotting RNA samples were from replicate experiments.
DISCUSSION
To determine the transcriptional response of E. coli to the AI-2 quorum signal, cells deficient in AI-2 production (W3110 luxS::Tcr) (15) were exposed for 20 min to medium conditioned by either AI-2-producing (W3110 luxS+) or non-AI-2-producing (W3110 luxS) cells. Generation of a 300-fold differential in AI-2, confirmed by an AI-2 activity assay (41), led to the discovery that almost 6% of the E. coli genome (242 genes) was modulated more than 2.3-fold (2 SD) in response to AI-2-regulated quorum sensing. Consistent with these results, it has been conservatively estimated that 3 to 5% of all genes in P. aeruginosa are regulated by acyl-HSL quorum signaling, as was partially demonstrated by screening a library of lacZ promoter probes which uncovered 270 genes showing more than twofold stimulation (70 showed more than fivefold stimulation) (51).
It is now well known that V. harveyi uses a species nonspecific signaling pathway mediated by AI-2 for regulating lux gene expression (7, 8, 41). Recognition of AI-2 by LuxP, homologous to the ribose binding protein of E. coli, has been proposed to transmit the signal to the LuxU phosphorelay protein via interaction with a hybrid sensor kinase, LuxQ (7, 8, 28). In turn, the signal is relayed to a central regulator, LuxO, which, upon interaction with ς54, indirectly represses the lux operon (28). In this study it was observed that AI-2 induced a gene encoding a ς54-dependent transcriptional activator (ygeV) as well as a ς54 modulator (yhbH), leading to our postulation that E. coli may employ ς54 during quorum sensing in a fashion analogous to that of V. harveyi. The striking similarity of the rpoN chromosomal regions of V. harveyi and E. coli along with the observed increase in yhbH expression (2.5-fold) suggests similar regulatory controls exist. Finally, in addition to regulating light production, LuxO and ς54 regulate siderophore production and colony morphology, demonstrating that multiple processes are regulated by quorum sensing in V. harveyi (28). In the present study, quorum-regulated genes existed in several functional classes, and while systematic determination of quorum-controlled processes still remains, it is clear that AI-2 also affects multiple processes in E. coli and perhaps enables population-wide coordination of these events.
Determination of quorum-regulated processes in E. coli has been elusive, as discovery of a quorum signal was only recently made. However, the finding that an extracellular factor exhibited inhibitory activity during initiation of DNA replication (52) provided preliminary evidence that E. coli employed a quorum sensing mechanism. Consistent with this observation, a gene encoding the integration host factor alpha subunit (himA) involved in the replication of the E. coli chromosome, and holE, a gene encoding the θ subunit of DNA polymerase III (necessary for elongation), were upregulated 3.9- and 3.1-fold, respectively, in response to AI-2. A role for quorum sensing in cell division was first demonstrated by SdiA-mediated changes in ftsQAZ expression from the p2 upstream promoter induced by CM (20, 39, 48) and later attributed to AI-2 (15). In this study, expression of sdiA, a luxR-type transcriptional regulator, was observed to increase only slightly (2.0-fold) in response to AI-2, indicating either that AI-2 does not significantly affect sdiA expression or that the effect occurs on a different time scale than that tested here. Accordingly, expression of ftsQ, ftsA, and ftsZ were relatively unchanged (1.1-fold, 1.0-fold, and 1.4-fold, respectively), although ftsE, encoding an ATP-binding component of a membrane-associated complex involved in cell division, decreased 3.1-fold. While the results presented here appear to contradict the earlier findings of increased AI-2-mediated ftsQAZ transcription through p2, we conclude that the p2 construct alone behaves differently than the p1 and p2 promoters acting in concert. This observation is supported by experiments using CM that showed that an extracellular factor stimulated ftsQA expression fivefold from P2ftsQ but only two- to threefold from both P1ftsQ and P2ftsQ together (39). Bassler and colleagues also reported that fusions of both p1 and p2 promoters to lacZ were not significantly altered by the presence of AI-2. Therefore, that we earlier observed ftsQAZ expression changes stimulated by AI-2 through promoter p2 alone suggests that under physiological conditions ftsQAZ expression is influenced by overlapping regulation from the neighboring rpoS-dependent P1ftsQ promoter. In fact, it has been documented that the two ftsQA promoters are regulated differentially, with expression from P2ftsQ occurring throughout growth and dependent on sdiA, while that from P1ftsQ (a gearbox promoter) increases as the growth rate declines and is dependent on rpoS (1, 39, 48).
Additional evidence that quorum sensing positively regulates cell division was the observed 3.2-fold decrease in expression of dicB, an inhibitor of the synthesis and activity of FtsZ. While sdiA is known to positively regulate cell division, inhibition of division can occur via derepression of dicB, whose gene product cooperates with MinC to inhibit FtsZ assembly, blocking septation at all potential division sites (13). Therefore, repression of dicB by the AI-2 quorum signal might exert additional positive control over cell division. Further, rcsB, another luxR-type transcriptional regulator protein known to affect colanic acid capsular polysaccaride synthesis, demonstrated a 3.5-fold increase in transcription. The role of rcsB in activating the ftsA and ftsZ genes (10), perhaps through P1ftsQ or P2ftsQ, coupled with its increased transcription induced by AI-2, suggests that a quorum regulatory mechanism governs these distinct processes. In further support of a role for AI-2 in exopolysaccharide biosynthesis was the increased transcription of wzb (6.2-fold increase), a gene found within the colanic acid gene cluster and, in conjunction with wzc, that is known to participate in the export of the extracellular polysaccharide colanic acid from the cell to the medium (46).
Using RegulonDB software (36) available at http://www.cifn.unam.mx/regulondb/, we obtained a predicted 81-bp promoter of rcsB, which was subsequently input to GRASP-DNA software (38) available at http://www-bioeng.ucsd.edu/∼grasp/home.html and was used to identify homologous putative DNA-protein binding sites. This analysis revealed significant homologous regulatory regions upstream of rcsB and ompG, which was interesting, as ompG expression was similarly upregulated (5.1-fold). Additionally, the threonyl-tRNA synthetase, thrS, having ca. 46% identity to a short segment of luxU (data not shown), was observed to increase 3.1-fold. A putative 81-bp promoter of thrS had upstream promoter homology with rfaJ, a lipopolysaccharide biosynthesis gene which was similarly upregulated (3.7-fold). Of note, thrS mRNA has been shown to accumulate with increasing growth rate (11), which might be a consequence of its response to AI-2, which also accumulates in a growth-rate-dependent fashion (15). Similarly, operon structure could be probed using RegulonDB. All of the AI-2-responding genes were examined using the RegulonDB graphical interface, and while many of the genes occurred within predicted (as opposed to known) operons, only the cheAW-motAB, potABCD, and rfaQGPSBIJYZK operons contained multiple genes responding to AI-2. In the first two cases, two genes responded similarly (e.g., rfaY and rfaJ, and potA and potB), while in the last case the responses of cheW and motB were in different directions. In all cases putative promoters are located between the identified genes, so a differential response might be expected. Interestingly, it has been shown that mutation of the att operon of Agrobacterium tumefaciens, a 10-kb region of 9 ORFs bearing strong homology to the pot operon of gram-negative bacteria, resulted in avirulence and inability to attach to plant cells (31). However, the ability of att mutants to bind to host cells was restored by the addition of conditioned medium during incubation of the bacteria with the host, suggesting that either efflux or uptake of an extracellular factor necessary for attachment through the spermidine pathway was blocked in mutant strains. This is very interesting in light of the 3.1- and 5.1-fold decrease in potAB expression in response to E. coli CM containing AI-2. This approach demonstrates that combination of global expression data with powerful bioinformatic algorithms, such as GRASP-DNA and RegulonDB, can elucidate potential regulatory overlap from transcriptional data.
Transcription of several other exopolysaccharide (rcsB, rfaD, rfaJ, rfaY, and rnk)- and virulence (hha and evgS)-related genes responded to AI-2. Outer surface polysaccharides are important components in the virulence of many pathogens, as they mediate direct interaction between bacteria and their immediate environment. While the E. coli strain studied here was not virulent, this was not entirely surprising, as many pathogenic gram-negative bacteria regulate virulence via quorum sensing (14). For example, hha, encoding the regulator of the hemolysin operon and reported to mediate the environmental regulation of virulence factors in P. aeruginosa (51), increased 11.1-fold in response to AI-2. Interestingly, ompA expression increased 3.2-fold, consistent with existing evidence that OmpA, in addition to maintaining outer membrane integrity, might play an important role in virulence of Pasteurella haemolytica (30). Of note, ompA expression was reported to decrease by 59% in an E. coli hha mutant (5). Also, the putative E. coli virulence gene, evgS, which constitutes a two-component system with the luxR-type regulator evgA that is structurally and functionally similar to the bvgAS two-component regulator of virulence factors in Bordatella pertussis (44), was repressed 2.8-fold. Finally, csrA, a global repressor of glycogen biosynthesis that alters stability of specific mRNA targets (29), increased 2.8-fold. While csrA has not been directly associated with quorum sensing in E. coli, structural and functional homologues regulate invasion genes in S. enterica serovar Typhimurium (2) and extracellular enzymes and N-(3-oxohexanoyl)-l-homoserine lactone quorum signals and pathogenicity in Erwinia carotovora (rsmA) (12). Additionally, csrA has been documented to affect cell size and surface properties, which is in agreement with the transcriptional changes of several murein sacculus-associated morphological genes. These genes include bolA (10.2-fold), an ftsZ-dependent regulator of the murein genes (1), and mreD (3.6-fold), encoding a rod-shape-determining protein as well as several exoskeletal (fimbriae, flagella, and curli surface fibers) genes, such as yadK (3.8-fold), yadN (3.5-fold), crl (3.5-fold), b1502 (3.0-fold), yehA (2.7-fold), fliP (−2.7-fold), and flgN (−3.7-fold). The coupling of morphological gene expression to AI-2 quorum signaling might ensure that the cytoskeletal framework be temporally regulated in association with growth phase and cell cycle progression.
Overall, our results yield significant insight into possible AI-2-coordinated changes in gene regulation that might temporally and spatially unify processes such as cell division, morphogenesis, and cell surface architecture. Interestingly, as many as 10 known sensors and/or transcriptional regulators as well as 10 putative signal transduction genes responded to increased AI-2 signaling, which, along with several other candidate genes and processes, warrant further study in the context of AI-2-stimulated quorum regulation. It is clear that quorum sensing is a complex signaling circuit that is built upon transducing elements that allow integration and channeling of multiple environmental cues, and elucidation of AI-2-controlled genes is a critical first step in mapping the metabolic pathways that define the E. coli quorum circuit.
ACKNOWLEDGMENTS
We thank B. Bassler for supplying strains used in this study.
This research was supported by the U.S. Army Engineering, Research, and Development Center, Edgewood, Md. (grant no. DAAM01-96-0037).
REFERENCES
- 1.Aldea M, Garrido T, Pla J, Vicente M. Division genes in Escherichia coli are expressed coordinately to cell septum requirements by gearbox promoters. EMBO J. 1990;9:3787–3794. doi: 10.1002/j.1460-2075.1990.tb07592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altier C, Suyemoto M, Lawhon S D. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect Immun. 2000;68:6790–6797. doi: 10.1128/iai.68.12.6790-6797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baca-DeLancey R R, South M M, Ding X, Rather P N. Escherichia coli genes regulated by cell-to-cell signaling. Proc Natl Acad Sci USA. 1999;96:4610–4614. doi: 10.1073/pnas.96.8.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bainton N J, Stead P, Chhabra S R, Bycroft B W, Salmond G P, Stewart G S, Williams P. N-(3-oxohexanoyl)-L-homoserine lactone regulates carbapenem antibiotic production in Erwinia carotovora. Biochem J. 1992;288:997–1004. doi: 10.1042/bj2880997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balsalobre C, Johansson J, Uhlin B E, Juarez A, Munoa F J. Alterations in protein expression caused by the hha mutation in Escherichia coli: influence of growth medium osmolarity. J Bacteriol. 1999;181:3018–3024. doi: 10.1128/jb.181.10.3018-3024.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassler B L, Greenberg E P, Stevens A M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassler B L, Wright M, Showalter R E, Silverman M R. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 8.Bassler B L, Wright M, Silverman M R. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 9.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 10.Carballes F, Bertrand C, Bouche J P, Cam K. Regulation of Escherichia coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB. Mol Microbiol. 1999;34:442–450. doi: 10.1046/j.1365-2958.1999.01605.x. [DOI] [PubMed] [Google Scholar]
- 11.Comer M M, Dondon J, Graffe M, Yarchuk O, Springer M. Growth rate-dependent control, feedback regulation and steady-state mRNA levels of the threonyl-tRNA synthetase gene of Escherichia coli. J Mol Biol. 1996;261:108–124. doi: 10.1006/jmbi.1996.0445. [DOI] [PubMed] [Google Scholar]
- 12.Cui Y, Chatterjee A, Liu Y, Dumenyo C K, Chatterjee A K. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J Bacteriol. 1995;177:5108–5115. doi: 10.1128/jb.177.17.5108-5115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Boer P A, Crossley R E, Rothfield L I. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci USA. 1990;87:1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Kievit T R, Iglewski B H. Bacterial quorum sensing in pathogenic relationships. Infect Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLisa M P, Valdes J J, Bentley W E. Mapping stress-induced changes in autoinducer AI-2 production in chemostat-cultivated Escherichia coli K-12. J Bacteriol. 2001;183:2918–2928. doi: 10.1128/JB.183.9.2918-2928.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Saizieu A, Gardes C, Flint N, Wagner C, Kamber M, Mitchell T J, Keck W, Amrein K E, Lange R. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol. 2000;182:4696–4703. doi: 10.1128/jb.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 18.Fuqua W C, Winans S C. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Lara J, Shang L H, Rothfield L I. An extracellular factor regulates expression of sdiA, a transcriptional activator of cell division genes in Escherichia coli. J Bacteriol. 1996;178:2742–2748. doi: 10.1128/jb.178.10.2742-2748.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glessner A, Smith R S, Iglewski B H, Robinson J B. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J Bacteriol. 1999;181:1623–1629. doi: 10.1128/jb.181.5.1623-1629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill C W, Harnish B W. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huisman G W, Kolter R. Sensing starvation: a homoserine lactone-dependent signaling pathway in Escherichia coli. Science. 1994;265:537–539. doi: 10.1126/science.7545940. [DOI] [PubMed] [Google Scholar]
- 24.Jones D H, Franklin F C, Thomas C M. Molecular analysis of the operon which encodes the RNA polymerase sigma factor sigma 54 of Escherichia coli. Microbiology. 1994;140:1035–1043. doi: 10.1099/13500872-140-5-1035. [DOI] [PubMed] [Google Scholar]
- 25.Kanamaru K, Tatsuno I, Tobe T, Sasakawa C. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 2000;38:805–816. doi: 10.1046/j.1365-2958.2000.02171.x. [DOI] [PubMed] [Google Scholar]
- 26.Khodursky A B, Peter B J, Cozzarelli N R, Botstein D, Brown P O, Yanofsky C. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:12170–12175. doi: 10.1073/pnas.220414297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazazzera B A. Quorum sensing and starvation: signals for entry into stationary phase. Curr Opin Microbiol. 2000;3:177–182. doi: 10.1016/s1369-5274(00)00072-2. [DOI] [PubMed] [Google Scholar]
- 28.Lilley B N, Bassler B L. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol. 2000;36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 29.Liu M Y, Yang H, Romeo T. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J Bacteriol. 1995;177:2663–2672. doi: 10.1128/jb.177.10.2663-2672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahasreshti P J, Murphy G L, Wyckoff III J H, Farmer S, Hancock R E, Confer A W. Purification and partial characterization of the OmpA family of proteins of Pasteurella haemolytica. Infect Immun. 1997;65:211–218. doi: 10.1128/iai.65.1.211-218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthysse A G, Yarnall H A, Young N. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J Bacteriol. 1996;178:5302–5308. doi: 10.1128/jb.178.17.5302-5308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLean R J, Whiteley M, Stickler D J, Fuqua W C. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol Lett. 1997;154:259–263. doi: 10.1111/j.1574-6968.1997.tb12653.x. [DOI] [PubMed] [Google Scholar]
- 33.Mukherjee A, Cui Y, Liu Y, Dumenyo C K, Chatterjee A K. Global regulation in Erwinia species by Erwinia carotovora rsmA, a homologue of Escherichia coli csrA: repression of secondary metabolites, pathogenicity and hypersensitive reaction. Microbiology. 1996;142:427–434. doi: 10.1099/13500872-142-2-427. [DOI] [PubMed] [Google Scholar]
- 34.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 35.Riley M, Labedan B. Escherichia coli gene products: physiological functions and common ancestries. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2277–2294. [Google Scholar]
- 36.Salgado H, Santos-Zavaleta A, Gama-Castro S, Millan-Zarate D, Blattner F R, Collado-Vides J. RegulonDB (version 3.0): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 2000;28:65–67. doi: 10.1093/nar/28.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salmond G P, Bycroft B W, Stewart G S, Williams P. The bacterial ‘enigma’: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 38.Schilling C H, Held L, Torre M, Saier M H., Jr GRASP-DNA: a web application to screen prokaryotic genomes for specific DNA-binding sites and repeat motifs. J Mol Microbiol Biotechnol. 2000;2:495–500. [PubMed] [Google Scholar]
- 39.Sitnikov D M, Schineller J B, Baldwin T O. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc Natl Acad Sci USA. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sperandio V, Mellies J L, Nguyen W, Shin S, Kaper J B. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surette M G, Bassler B L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Surette M G, Bassler B L. Regulation of autoinducer production in Salmonella typhimurium. Mol Microbiol. 1999;31:585–595. doi: 10.1046/j.1365-2958.1999.01199.x. [DOI] [PubMed] [Google Scholar]
- 43.Surette M G, Miller M B, Bassler B L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Utsumi R, Katayama S, Taniguchi M, Horie T, Ikeda M, Igaki S, Nakagawa H, Miwa A, Tanabe H, Noda M. Newly identified genes involved in the signal transduction of Escherichia coli K-12. Gene. 1994;140:73–77. doi: 10.1016/0378-1119(94)90733-1. [DOI] [PubMed] [Google Scholar]
- 45.Van Dyk T K, Wei Y, Hanafey M K, Dolan M, Reeve M J, Rafalski J A, Rothman-Denes L B, LaRossa R A. A genomic approach to gene fusion technology. Proc Natl Acad Sci USA. 2001;98:2555–2560. doi: 10.1073/pnas.041620498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vincent C, Doublet P, Grangeasse C, Vaganay E, Cozzone A J, Duclos B. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J Bacteriol. 1999;181:3472–3477. doi: 10.1128/jb.181.11.3472-3477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Bodman S B, Majerczak D R, Coplin D L. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc Natl Acad Sci USA. 1998;95:7687–7692. doi: 10.1073/pnas.95.13.7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X D, de Boer P A, Rothfield L I. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991;10:3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei Y, Lee J M, Richmond C, Blattner F R, Rafalski J A, LaRossa R A. High-density microarray-mediated gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:545–5456. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei Y, Lee J M, Smulski D R, LaRossa R A. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:2265–2272. doi: 10.1128/JB.183.7.2265-2272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whiteley M, Lee K M, Greenberg E P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Withers H L, Nordstrom K. Quorum-sensing acts at initiation of chromosomal replication in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:15694–15699. doi: 10.1073/pnas.95.26.15694. [DOI] [PMC free article] [PubMed] [Google Scholar]