Abstract
Klebsiella pneumoniae is able to grow anaerobically with citrate as a sole carbon and energy source by a fermentative pathway involving the Na+-dependent citrate carrier CitS, citrate lyase, and oxaloacetate decarboxylase. The corresponding genes are organized in the divergent citC and citS operons, whose expression is strictly dependent on the citrate-sensing CitA-CitB two-component system. Evidence is provided here that the citrate fermentation genes are subject to catabolite repression, since anaerobic cultivation with a mixture of citrate and glucose or citrate and gluconate resulted in diauxic growth. Glucose, gluconate, and also glycerol decreased the expression of a chromosomal citS-lacZ fusion by 60 to 75%, whereas a direct inhibition of the citrate fermentation enzymes was not observed. The purified cyclic AMP (cAMP) receptor protein (CRP) of K. pneumoniae bound to two sites in the citC-citS intergenic region, which were centered at position −41.5 upstream of the citC and citS transcriptional start sites. Binding was apparently stimulated by the response regulator CitB. These data indicate that catabolite repression of the citrate fermentation genes is exerted by CRP and that in the absence of repressing carbon sources the cAMP-CRP complex serves to enhance the basal, CitB-dependent transcription level.
Klebsiella pneumoniae, a member of the Enterobacteriaceae, is able to grow with citrate as a sole carbon and energy source under anoxic conditions (for a review, see reference 4). Citrate fermentation involves uptake by a Na+-dependent citrate carrier (32, 44), cleavage into acetate and oxaloacetate by citrate lyase, and decarboxylation of oxaloacetate to pyruvate by the Na+ ion pump oxaloacetate decarboxylase (12, 13). The conversion of pyruvate to acetate, formate, CO2, and H2 (7, 38) is catalyzed by enzymes generally involved in anaerobic pyruvate catabolism. The citrate-specific fermentation genes form a cluster of two divergent operons (5, 6). The citCDEFG operon encodes citrate lyase ligase (CitC), the citrate lyase subunits (CitD, CitE, and CitF), and triphosphoribosyl-dephospho-coenzyme A synthase (CitG), which catalyzes the formation of a precursor of the citrate lyase prosthetic group (36, 37). The citS-oadGAB-citAB operon encodes the citrate carrier CitS, the oxaloacetate decarboxylase subunits (OadG, OadA, and OadB), and a two-component system composed of the sensor kinase CitA and the response regulator CitB (Fig. 1A). The CitA-CitB system was shown to be essential for the expression of both operons (6). CitA presumably functions as a sensor of extracellular citrate, since the periplasmic domain of this membrane-bound protein binds citrate with high affinity (Kd ≈ 6 μM) and specificity (23). CitB acts as a transcriptional activator of the citS and the citC operons and binds to two sites in the 193-bp citC-citS intergenic region (Fig. 1) in a phosphorylation-dependent fashion (24).
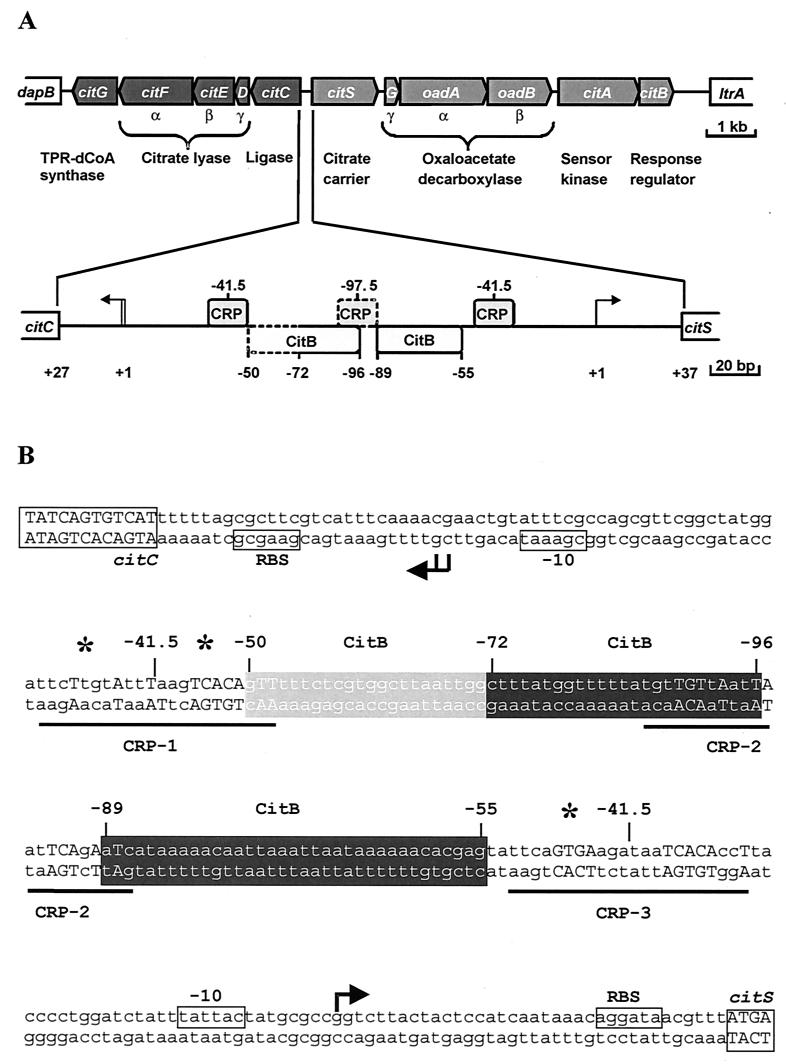
FIG. 1.
Organization of the citrate-specific fermentation genes in K. pneumoniae. The 13-kb DNA cluster encompassing 11 genes involved in citrate fermentation is shown in panel A. The lower part shows an enlargement of the citC-citS intergenic region. The arrows indicate transcription start sites. The positions of the CitB binding sites as deduced from DNase I footprints and of putative CRP binding sites are indicated. The sequence of the citC-citS intergenic region is shown in panel B. Indicated are the coding regions, putative ribosome binding sites (RBS), the transcriptional start sites as determined by primer extension (25), the −10 regions, the CitB binding sites, the putative CRP binding sites, and the hypersensitive sites observed in DNase I footprints (asterisks).
Maximal expression of a chromosomally integrated translational citS′-′lacZ fusion requires citrate, anoxic conditions, and Na+ ions (6). Whereas the inducing effect of citrate can be attributed to the CitA-CitB system, the proteins responsible for the regulatory effects of Na+ and O2 are not yet known. In citrate minimal media containing glucose or glycerol in addition, expression of citS′-′lacZ was strongly reduced (6), supporting previous evidence for catabolite repression of the citrate fermentation genes (7, 10).
In enterobacteria, the cyclic AMP (cAMP) receptor protein (CRP) is a key player in catabolite repression (3). A comparison of the citC-citS intergenic DNA sequence with the palindromic CRP consensus binding site (2) revealed three potential binding sites for CRP (Fig. 1), one centered at −41.5 bp in front of the citS transcription start site, another at −41.5 bp in front of the citC transcription start site, and the third in between. The −41.5 sites fulfil the exact spacing requirement of class II CRP-dependent promoters (9, 15), where CRP alone is sufficient to activate transcription.
In this work we analyzed the effects of glucose, gluconate, and glycerol on citrate fermentation. Under anaerobic conditions, glucose is taken up by the phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) and fermented to the products characteristic of butanediol fermentation (1). The occurrence of this fermentation type in the K. pneumoniae strain used in this work was confirmed by the detection of the characteristic product acetoin. Gluconate uptake and fermentation have not been studied in detail but most likely involve H+ symport as in Escherichia coli (20, 31) and the Entner-Doudoroff pathway. Glycerol, which presumably is taken up a by a glycerol facilitator (46), is fermented by K. pneumoniae to 1,3-propanediol and acetate as major products (14, 21, 22). Several of the glycerol fermentation genes, which form the dha regulon, have been cloned and sequenced (11, 41–43).
Here we show that glucose, gluconate, and glycerol, despite the differences in uptake and catabolism, inhibit the expression of the citrate fermentation genes, thus representing an example of catabolite repression under fermentative conditions. In addition, evidence for the involvement of the CRP-cAMP system in this process is provided.
MATERIALS AND METHODS
Bacterial strains and culture media
K. pneumoniae ATCC 13882, which is designated the wild type, was grown as described previously (6). E. coli DH5α (Bethesda Research Laboratories) was used as a host for all cloning procedures. E. coli BL21(DE3), which contains the T7 RNA polymerase gene under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lacUV5 promoter (39), served as host for overproduction of the CRP protein from the pET expression plasmid. The E. coli strains were routinely grown at 37°C either in Luria-Bertani (LB) medium or in 2× YT medium (34). K. pneumoniae strain MB1 (6), which contains a chromosomally integrated translational citS′-′lacZ fusion, was grown in minimal media containing citrate, glycerol, or both as described previously (6). For growth experiments, a minimal medium containing 100 mM potassium phosphate buffer (pH 7.0), 50 mM NaCl, 10 mM (NH4)2SO4, 1 mM Mg2SO4, and 0.4% trace element solution (180 mM CaCl2, 0.77 mM CoCl2, 0.505 mM MnCl2, 0.413 mM Na2MoO4, and 7.2 mM FeSO4) was used. Unless indicated otherwise, potassium citrate, glucose, potassium gluconate, or glycerol was added as a carbon source (20 mM). Anaerobic cultures were grown in flasks with septa at 37°C under nitrogen (150 kPa) without shaking. Precultures were washed in 100 mM potassium phosphate buffer (pH 7.0) prior to inoculation. The initial optical density at 600 nm (OD600) after inoculation was approximately 0.05.
β-Galactosidase assays and determination of citrate.
The β-galactosidase assays of strain MB1 grown under various conditions were performed with permeabilized cells essentially as described by Miller (25). The citrate content of culture supernatants was determined spectrophotometrically at 365 nm using a coupled assay containing 2 U of malate dehydrogenase (pig heart; Boehringer Mannheim), 1 U of lactate dehydrogenase (hog muscle; Boehringer Mannheim), and 0.2 to 0.4 U of citrate lyase (Aerobacter aerogenes; Boehringer Mannheim) in a 50 mM glycylglycine buffer at pH 7.9 in the presence of 2 mM ZnCl2 and 0.5 mM NADH (5).
Construction of pET-CRP.
Chromosomal DNA of K. pneumoniae ATCC 13882 was used as template for the PCR with Vent DNA polymerase (New England Biolabs). The primers pCRP-fo (5′-AGATATACATATGGTGCTTGGCAAACCGCAAAC-3′) and pCRP-re (5′-GTGGTGCTCGAGTTAACGGGTGCCGTAGACGACG-3′) introduced an NdeI restriction site at the initiation codon and an XhoI restriction site immediately downstream of the stop codon (restriction sites are underlined), respectively. The coding sequence of the primers was designed according to the nucleotide sequence of the K. aerogenes crp gene (GenBank accession number M68973). The resulting 655-bp product was cleaved with NdeI and XhoI and subsequently cloned into pET24b (Novagen) cut with the same enzymes. The resulting plasmid pET-CRP was sequenced with T7 promoter and T7 terminator primers. Two positions were found to differ from that of the published K. aerogenes sequence: C instead of A at position 12 and A in the place of C at position 507. These differences, however, do not result in an altered amino acid sequence.
Purification of CRP and CitBHis.
E. coli BL21(DE3) containing the expression plasmid pET-CRP was grown in LB medium including kanamycin (50 μg/ml). The cultures were incubated at 37°C and 180 rpm until the OD600 reached a value between 0.6 and 0.8. Then, IPTG was added to a final concentration of 1 mM, and incubation was continued for another 3 h. Cells were harvested by centrifugation, washed once in 20 ml of buffer A plus 0.2 M NaCl (buffer A consists of 20 mM sodium phosphate [pH 7.2], 2 mM EDTA, and 5 mM β-mercaptoethanol), and resuspended in the same buffer (5 g [wet weight]/ml). After addition of the protease inhibitor diisopropyl fluorophosphate (1 mM) and 0.2 mg of DNase I/ml the cells were disrupted by three passages through a French pressure cell at 108 MPa. Intact cells and cell debris were removed by centrifugation (30 min at 27,000 × g). The cell extract was ultracentrifuged (60 min at 150,000 × g), and the resulting supernatant was used for purification of CRP by affinity chromatography with cAMP agarose (Sigma A0144) as described previously (47). Purification was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and protein concentrations were determined with the Bradford assay kit (Bio-Rad Laboratories). Buffer exchange was performed by gel filtration (PD-10 columns; Pharmacia). CitBHis was purified by Ni2+ chelate affinity chromatography as described previously (24).
DNA-protein interaction studies.
For gel retardation assays of CRP, a 348-bp MluI-SphI DNA fragment covering the entire intergenic region between citC and citS was labeled with the Klenow fragment of E. coli DNA polymerase I and suitable α-32P-radiolabeled nucleotides according to the protocol of the manufacturer. Unincorporated nucleotides were removed using the QIAquick-spin PCR purification kit (Qiagen). An equal volume of a serial dilution of CRP in TKMD buffer (50 mM Tris-HCl [pH 7.5], 200 mM KCl, 5 mM MgCl2, 5 mM dithiothreitol) containing 200 μM cAMP was mixed with the DNA premixture, which contained the radiolabeled fragment (86 fmol; ∼280,000 cpm), competitor DNA (0.2 μg of sonicated salmon sperm DNA/μl), and tracking dye (0.02% xylenecyanol FF) in 2× footprint buffer (80 mM Tris-HCl [pH 7.2], 12 mM MgCl2, 2 mM dithiothreitol, 20% glycerol). The mixture was incubated for 20 min at room temperature and subsequently loaded onto a 10% native polyacrylamide gel (ratio of acrylamide to bisacrylamide, 75:1 [wt/wt]) with 0.5× TBE buffer (TBE buffer is 89 mM Tris base, 89 mM boric acid, 2.5 mM EDTA; final pH 8.3) containing 20 μM cAMP as gel and running buffer. Electrophoresis was carried out at a constant voltage of 180 V and cooled with tap water. The gel was subsequently dried and exposed to a phosphorimager screen (Molecular Dynamics).
The DNase I footprint assays with CRP and CitBHis were carried out essentially as described for CitBHis (24). Constant amounts of the labeled 348-bp MluI-SphI DNA fragment (∼300,000 cpm) were incubated with CRP (final concentration, 3.7 or 2.0 μM) and phosphorylated CitBHis (final concentration, 0.4 or 1.6 μM). CitBHis was phosphorylated with ATP, and the kinase domain of CitA was fused to the maltose binding protein (MalE-CitAC) as previously described (23, 24). The reaction mixtures contained CRP and/or CitBHis, competitor DNA (20 ng of sonicated salmon sperm DNA/μl), 1 mM cAMP, and the labeled DNA fragment in 1× footprint buffer supplemented with 10 mM CaCl2 and 100 mM KCl. A total of 7.5 U of DNase I was added to start the reaction. A control reaction was carried out with buffer instead of protein solution. If no CRP was present in the reaction mixture, cAMP was omitted. The samples were separated on a 6% denaturing polyacrylamide gel containing 7 M urea.
RESULTS
Influence of preculture conditions on anaerobic growth with citrate.
In a first set of experiments, we studied the effect of the preculture conditions on the growth characteristics of K. pneumoniae during citrate fermentation. Precultures were grown either anaerobically on citrate minimal medium or aerobically on citrate or glucose minimal medium. Following inoculation with these precultures, fermentative growth on citrate minimal medium started after a lag phase of 0.5, 5, and 10 h, respectively (data not shown). This result confirmed that the citrate fermentation enzymes are not constitutive but are induced under anoxic conditions in the presence of citrate. The reduced lag phase of cells precultured under oxic conditions with citrate might be due to a slightly increased concentration of the CitA-CitB two-component system, allowing for a faster induction upon a shift to anoxic conditions. Comparable induction patterns were previously reported for citrate fermentation by Salmonella enterica serovar Typhimurium (45).
Influence of glucose, gluconate, and glycerol on anaerobic citrate utilization.
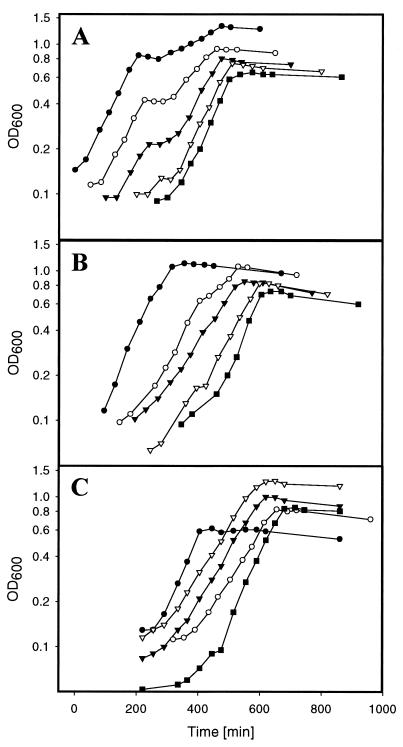
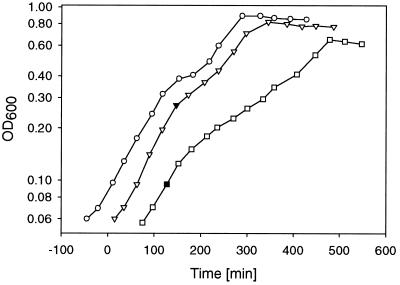
To test the influence of glucose, gluconate, and glycerol on citrate fermentation, different concentrations of these carbon sources were added to citrate minimal medium before inoculation with a preculture grown anaerobically with citrate as the sole carbon and energy source. As shown in Fig. 2A, the presence of glucose led to typical diauxic growth curves. The start of the intermediate lag phase was strictly dependent on the glucose concentration, showing that glucose is used preferentially to citrate. If gluconate was added to citrate minimal medium, growth was also diauxic and showed preferential use of gluconate. However, the diauxic effect was less pronounced than with glucose (Fig. 2B). When glycerol was present in addition to citrate, no diauxic growth was observed, but the growth rate was slower than in the presence of either substrate alone (Fig. 2C).
FIG. 2.
Anaerobic growth of the K. pneumoniae wild type in citrate minimal medium containing different concentrations of glucose (A), gluconate (B), and glycerol (C). The inoculating culture had been grown anaerobically on citrate minimal medium. For the sake of better readability, the individual growth curves are displayed with time differences. Symbols indicate the presence of the following concentrations of glucose, gluconate, or glycerol as noted above: (A) solid circles, 8 mM; open circles, 4 mM; solid triangles, 2 mM; open triangles, 1 mM; solid squares, 0 mM; (B) solid circles, 20 mM; open circles, 8 mM; solid triangles, 4 mM; open triangles, 2 mM; filled squares, 1 mM; (C) open triangles, 20 mM; solid triangles, 10 mM; open circles, 5 mM; solid circles, 0 mM. The solid squares in panel C indicate the presence of glycerol minimal medium without citrate.
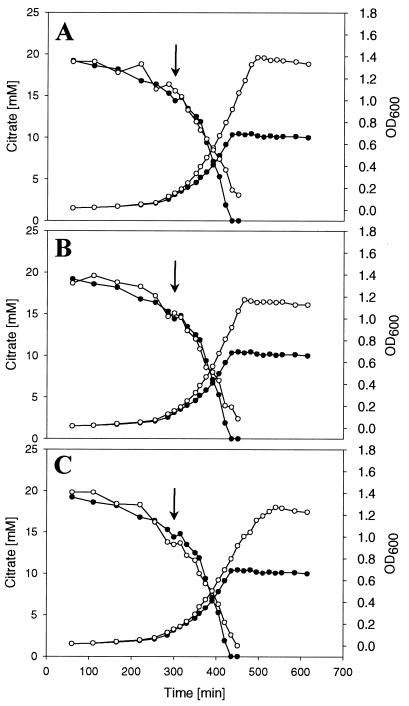
The data reported above clearly showed that citrate fermentation is subject to catabolite repression. In several cases it was shown that catabolite repression is due to direct inhibition of catabolic enzymes by unphosphorylated enzyme IIAGlc, e.g., in the case of lactose permease (26, 28) or glycerol kinase (27). We therefore tested whether the addition of glucose, gluconate, or glycerol to citrate-fermenting cells inhibited the further catabolism of citrate. As shown in Fig. 3, the addition of the three different carbon sources led to an only slightly reduced citrate consumption, indicating that none of the proteins specifically involved in citrate fermentation, i.e., the citrate carrier CitS, citrate lyase, and oxaloacetate decarboxylase, is directly inhibited. The small reduction of citrate consumption is probably due to reduced concentrations of these enzymes as a consequence of reduced synthesis (see below).
FIG. 3.
Anaerobic growth and citrate consumption of the K. pneumoniae wild type in citrate minimal medium (solid circles) or in citrate minimal medium with either 4 mM glucose (A), 4 mM gluconate (B), or 10 mM glycerol (C) (indicated in panels A through C by open circles). The additional carbon sources were added at an OD600 of about 0.15 (arrows).
Influence of glucose, gluconate, and glycerol on citS′-′lacZ expression.
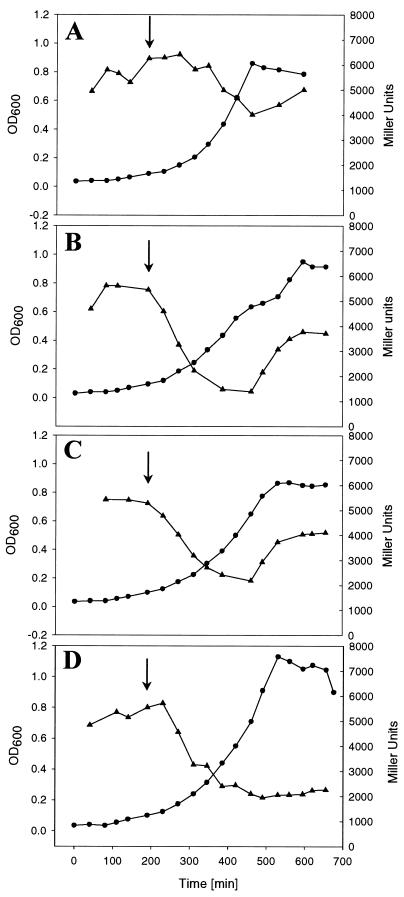
The above results indicated that catabolite repression of citrate fermentation by glucose, gluconate, and glycerol was at the level of enzyme synthesis. Therefore, the influence of the alternative carbon sources on the expression of the citrate fermentation genes was tested by adding them to a culture of K. pneumoniae strain MB1 already growing exponentially on citrate minimal medium. This strain carries a chromosomally integrated citS′-′lacZ fusion (6), which served as reporter for the determination of the expression of the citS operon. As shown in Fig. 4B, addition of 4 mM glucose led to a rapid decrease in β-galactosidase activity. Within 3 h, activity dropped from 5,500 to 1,500 Miller units and increased again to ∼4,000 Miller units when glucose had been consumed. Although less pronounced, the growth curve was diauxic as in the experiment shown in Fig. 2A. If glucose was added at a higher optical density (OD600 > 0.15), the diauxic effect disappeared, possibly because all citrate had already been consumed when catabolite repression was relieved. In a control culture, where 4 mM citrate was added instead of glucose, β-galactosidase activity constantly remained between 4,000 and 6,000 Miller units up to the end of growth (Fig. 4A). When 4 mM gluconate was added as an additional carbon source, β-galactosidase activity was reduced from 5,500 to 2,200 Miller units within 3 to 4 h and then increased again to 4,000 Miller units (Fig. 4C). Although there was no intermediate lag phase visible in the growth curve, the increase in β-galactosidase activity showed that gluconate had been consumed and catabolite repression had been relieved. Similar to gluconate, addition of 10 mM glycerol led to a decrease of β-galactosidase activity from 5,500 to about 2,000 Miller units within 3 h and then remained constant at this level up to the end of growth (Fig. 4D). The lack of an increase in β-galactosidase activity in this experiment is due to the high concentration of glycerol added, resulting in a complete consumption of citrate before glycerol has been consumed. In summary, all carbon sources tested reduced expression of the citS operon to about 25 to 40% of the level measured in their absence.
FIG. 4.
β-Galactosidase activity in Miller units (triangles) and OD600 (circles) of K. pneumoniae strain MB1 during anaerobic growth in citrate minimal medium with either 4 mM glucose (B), 4 mM gluconate (C), or 10 mM glycerol (D) added at an OD600 of 0.1 (arrows). In a control culture (A), 4 mM citrate was added at an OD600 of 0.1 (arrow).
Independent evidence for the inhibition of expression of the citrate fermentation genes by glycerol was obtained previously by Northern blot experiments. The level of the transcripts of the citS operon and of the citC operon in RNA isolated from cells grown anaerobically with citrate and glycerol was significantly lower than in RNA isolated from cells grown anaerobically with citrate as the sole carbon and energy source (6, 24).
Influence of cAMP on diauxic growth.
The results described above showed that the citrate fermentation genes are subject to catabolite repression. Since in enterobacteria the CRP-cAMP system plays a central role in catabolite repression and since three potential CRP binding sites are present in the citC-citS intergenic region, studies were performed to determine whether this system also regulates expression of the K. pneumoniae citrate fermentation genes. First, the effect of exogenously added cAMP on growth of the K. pneumoniae wild type in citrate minimal medium containing 2 mM glucose was tested. To this end, three cultures were inoculated with a preculture grown anaerobically in citrate minimal medium. To two of these cultures, cAMP (20 mM) was added at an OD600 of 0.095 and 0.271, respectively, while the third one served as control and received no cAMP (Fig. 5). Since 2 mM glucose supported growth to an OD600 of approximately 0.4, cAMP addition preceded glucose depletion. The addition of cAMP at an OD600 of 0.271 clearly diminished the diauxic effect observed in the control culture, indicating an enhanced expression of the citrate fermentation genes prior to the complete consumption of glucose. If cAMP was added at an OD600 of 0.095, the diauxic effect was also abolished, but in addition, the growth rate and the final cell density were significantly reduced. The reason for this inhibition is unknown; it may be caused by an interference of citrate fermentation and glucose fermentation or by other components of the cAMP-CRP regulon.
FIG. 5.
Influence of cAMP (20 mM) on growth of the K. pneumoniae wild type in citrate minimal medium containing 2 mM glucose. The addition of cAMP is indicated by solid symbols. For the sake of better readability, the individual growth curves are displayed with a time difference of 1 h.
The cAMP effect and the presence of three putative CRP binding sequences in the citC-citS intergenic region (Fig. 1) indicated an involvement of CRP in catabolite repression of the citrate fermentation genes. To demonstrate this, several attempts were made to study the effect of a crp deletion on the expression of the citrate fermentation genes. Unfortunately, our efforts to construct a crp deletion mutant of the K. pneumoniae strain used in this study failed (data not shown). Since we were not able to analyze the role of CRP in the expression of the citrate fermentation genes in vivo, the ability of purified CRP to bind to the potential binding sites in the citC-citS promoter region was tested.
Purification of CRP from K. pneumoniae.
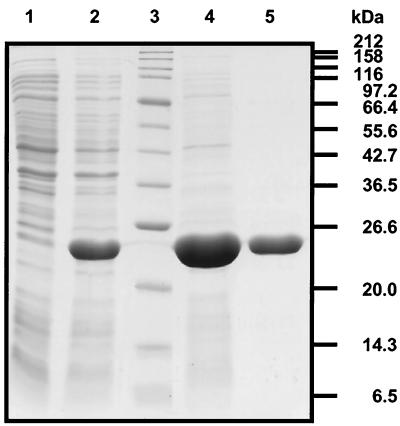
The crp gene of K. pneumoniae ATCC 13882 was amplified by PCR from chromosomal DNA using Vent DNA polymerase. The resulting 655-bp product was cloned into the expression vector pET24b with the restriction sites that had been introduced with the primers. Sequence analysis of two of the resulting pET-CRP plasmids revealed two positions that differed from the published sequence of K. aerogenes crp (29). These differences did not cause a change at the protein level, however. For CRP overproduction, pET-CRP was transferred into E. coli BL21(DE3), which carries the gene for the T7 RNA polymerase under the control of an IPTG-inducible lacUV5 promoter. After induction with IPTG, a dominant protein band was observed after SDS-PAGE of whole-cell extracts (Fig. 6). Its apparent size of about 25 kDa corresponded well with the predicted molecular mass of CRP (23.7 kDa). Fractionation of the cells revealed that most of the protein was present in the supernatant obtained after ultracentrifugation of cell extract, indicating that CRP had been obtained in a soluble form. Purification of CRP was achieved by affinity chromatography with cAMP agarose as a matrix. The eluted protein was essentially pure as estimated by Coomassie-stained polyacrylamide gels (Fig. 6). It should be mentioned that a very small proportion of the purified protein was probably E. coli CRP, which differs, however, from K. pneumoniae CRP in only one position (29). The corresponding proteins were shown to be completely interchangeable with respect to activation of the E. coli lacZ and the K. pneumoniae hutU promoters (30).
FIG. 6.
Coomassie-stained SDS-PAGE gel containing the following samples: whole-cell lysate of E. coli BL21(DE3)/pET-CRP prior to (lane 1) and 3 h after (lane 2) IPTG induction, protein standards (lane 3), and purified CRP protein (lanes 4 and 5) after affinity chromatography on cAMP agarose.
Binding of CRP to the citC-citS intergenic region.
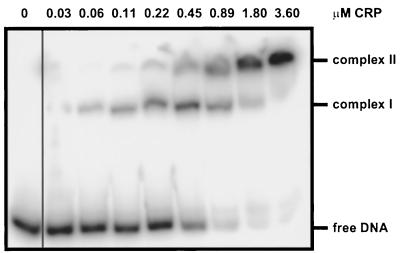
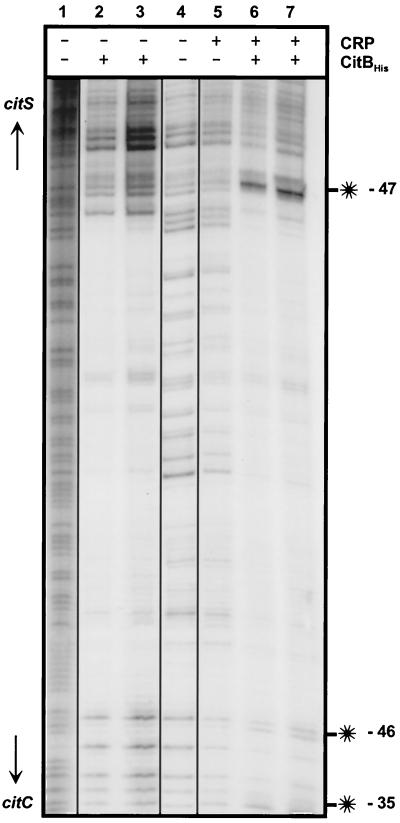
Purified CRP was used for gel retardation assays with radioactively labeled 348-bp MluI-SphI DNA fragments encompassing the citC-citS intergenic region. As shown in Fig. 7, two specific protein-DNA complexes were observed, indicating that CRP was bound to two of three putative binding sites present on this fragment. Half-maximal binding required about 0.5 μM CRP. To define the CRP binding sites more exactly, DNase I footprint experiments were performed with the same DNA fragment used in the gel retardation assay. With CRP concentrations up to 3.7 μM, hardly visible hypersensitive sites were present at positions −35 and −46 upstream of the citC transcription start site (Fig. 8, lane 5). When the DNase I footprint experiment was performed with 2.0 μM CRP in the presence of either 0.4 μM CitBHis or 1.6 μM CitBHis (Fig. 8, lanes 6 and 7), the hypersensitive sites at positions −35 and −46 upstream of the citC transcription start site were easily visible; in addition, a strong hypersensitive site at position −47 upstream of the citS transcription start site appeared. Since none of the hypersensitive sites was observed in DNase I footprints obtained with CitBHis alone (Fig. 8, lanes 2 and 3), they must be caused by the presence of CRP.
FIG. 7.
Gel retardation assay with CRP and the 348-bp MluI-SphI DNA fragment covering the intergenic region between citC and citS. The concentration of CRP is indicated on top of the figure.
FIG. 8.
DNase I footprint assay with CRP and CitBHis. The 348 bp MluI-SphI fragment covering the citC-citS intergenic region was labeled by fill-in of the 3′-recessed ends created by MluI with Klenow polymerase. Lane 1, G+A sequencing ladder; lane 2, 0.4 μM CitBHis; lane 3, 1.6 μM CitBHis; lane 4, no protein; lane 5, 3.7 μM CRP; lane 6, 2.0 μM CRP plus 0.4 μM CitBHis; lane 7, 2.0 μM CRP plus 1.6 μM CitBHis. The hypersensitive sites (indicated with asterisks) are located at positions −47 relative to the citS transcription start site (top) and at −35 and −46 relative to the citC transcription start site (bottom).
The location of the hypersensitive sites strongly indicated that CRP binds to the two postulated sites centered at −41.5 upstream of the citC and citS transcription start sites, but not to the one in between. This is in accord with the result of the gel retardation assay, which indicated two CRP binding sites to be present in the citC-citS intergenic region, and with the fact that the putative CRP binding site, which is not occupied, strongly overlaps with the CitB binding site (Fig. 1). The influence of CitBHis on the appearance of the hypersensitive sites indicated that CRP binding is stimulated by CitBHis, in particular to the site in front of the citS promoter.
DISCUSSION
Catabolite repression has been studied extensively in enterobacteria, in particular in E. coli, during the last decades. Nearly all studies have focused exclusively on catabolite repression during aerobic growth. The results reported in this study provide an example of catabolite repression under anaerobic, fermentative conditions. Three different carbon sources, i.e., glucose, gluconate, and glycerol, taken up by fundamentally different transport systems and metabolized by different pathways, were shown to inhibit the expression of the citrate fermentation genes. The strongest repression was exerted by glucose and resulted in typical diauxic growth curves (Fig. 2A). The repression by gluconate was weaker, but still led to diauxie (Fig. 2B). The repressing effect of glycerol was in the same range as that of gluconate, but in this case, no diauxie was observed (Fig. 2C). In enterobacteria, catabolite repression is prevalently exerted by the cAMP-CRP complex and we provide evidence that this holds true also for the citrate fermentation genes in K. pneumoniae. First, the diauxic effect caused by glucose could be impaired by addition of cAMP; second, CRP was shown to bind to two sites in the citC-citS intergenic region, which are centered at −41.5 upstream of the citC and the citS transcription start sites. This position was previously demonstrated to be one of the optimal sites for transcriptional activation by the cAMP-CRP complex (15). Interestingly, the binding of the cAMP-CRP complex was easily observed in a gel retardation assay, whereas the emergence of hypersensitive sites in DNase I footprints was extremely weak with the cAMP-CRP complex alone but strongly supported by the simultaneous presence of the response regulator CitB. The latter observation indicated that binding of the cAMP-CRP complex to the citC-citS intergenic region is stimulated by the previous binding of phosphorylated CitB (24). Since the expression of the citrate fermentation genes was only partially inhibited by the alternative carbon sources (Fig. 4), it seems possible that the cAMP-CRP complex serves to enhance a basal, CitB-dependent transcription level in the absence of repressing carbon sources. Since the citAB genes are positively autoregulated by cotranscription with citS and oadGAB (6), a reduced transcription of the citS operon due to a low cAMP-CRP concentration would result in lowered levels not only of the dissimilatory proteins (CitS and oxaloacetate decarboxylase) but also of the regulatory proteins CitA and CitB. As a consequence, transcription of the citrate fermentation genes will decrease even further.
The mechanism described above implies that the repressing carbon sources reduce the cAMP-CRP concentration. It was recently shown that not only glucose and other PTS sugars, but also non-PTS substrates such as lactose or gluconate, in fact lead to lower levels of both cAMP and CRP (17). In the case of cAMP, it is believed that enzyme IIAGlc, a protein of the PTS and a key player in catabolite repression of enterobacteria (33), activates adenylate cyclase, the cAMP biosynthetic enzyme, when it is present in the phosphorylated state. PTS substrates cause dephosphorylation of enzyme IIAGlc mainly by phosphoryl transfer to the sugars, whereas non-PTS sugars can elicit dephosphorylation of IIAGlc by a reduction of the PEP/pyruvate ratio (16). For gluconate, however, it was shown that despite a lowered PEP/pyruvate ratio, enzyme IIAGlc was predominantly in the phosphorylated state (16). Therefore, another mechanism seems to be effective for the gluconate-induced reduction of the cAMP level. The lowering of the CRP concentration in the presence of repressing carbon sources seems to be due to complex autoregulation of the crp gene (18, 19, 40).
Based on the findings described above, a reduced concentration of the cAMP-CRP complex is also postulated to be the cause of catabolite repression by glucose, gluconate, and glycerol during citrate fermentation in K. pneumoniae. However, further experiments are required to prove this hypothesis, such as measurements of the concentrations of cAMP, CRP, PEP, and pyruvate and of the phosphorylation state of enzyme IIAGlc under the fermentative conditions used in our studies. As the fermentation of glucose, gluconate, and glycerol involves the formation of pyruvate from PEP, it seems likely that these carbon sources lead to a different PEP/pyruvate ratio than fermentation of citrate, where pyruvate is formed from oxaloacetate and PEP required for biosynthetic purposes has to be generated from pyruvate by PEP synthetase. Irrespective of the outcome of such studies, the data provided here and by others (8, 35) show that the sophisticated mechanism of CRP-mediated catabolite repression in enterobacteria works not only under aerobic but also under anaerobic, fermentative conditions.
ACKNOWLEDGMENT
This work was supported by a grant from the Swiss National Foundation for Scientific Research to M.B.
REFERENCES
- 1.Bender R A. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. Variations on a theme by Escherichia; pp. 4–9. [Google Scholar]
- 2.Berg O G, von Hippel P H. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988;200:709–723. doi: 10.1016/0022-2836(88)90482-2. [DOI] [PubMed] [Google Scholar]
- 3.Botsford J L, Harman J G. Cyclic AMP in prokaryotes. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bott M. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch Microbiol. 1997;167:78–88. [PubMed] [Google Scholar]
- 5.Bott M, Dimroth P. Klebsiella pneumoniae genes for citrate lyase and citrate lyase ligase: localization, sequencing, and expression. Mol Microbiol. 1994;14:347–356. doi: 10.1111/j.1365-2958.1994.tb01295.x. [DOI] [PubMed] [Google Scholar]
- 6.Bott M, Meyer M, Dimroth P. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol Microbiol. 1995;18:533–546. doi: 10.1111/j.1365-2958.1995.mmi_18030533.x. [DOI] [PubMed] [Google Scholar]
- 7.Brewer C R, Werkman C H. The anaerobic dissimilation of citric acid by Aerobacter aerogenes. Enzymologia. 1939;6:273–281. [Google Scholar]
- 8.Buchet A, Nasser W, Eichler K, Mandrand-Berthelot M. Positive co-regulation of the Escherichia coli carnitine pathway cai and fix operons by CRP and the CaiF activator. Mol Microbiol. 1999;34:562–575. doi: 10.1046/j.1365-2958.1999.01622.x. [DOI] [PubMed] [Google Scholar]
- 9.Busby S, Ebright R H. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 10.Dagley S, Dawes E A. Citric acid metabolism of Aerobacter aerogenes. J Bacteriol. 1953;66:259–265. doi: 10.1128/jb.66.3.259-265.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel R, Bobik T A, Gottschalk G. Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes. FEMS Microbiol Rev. 1998;22:553–566. doi: 10.1111/j.1574-6976.1998.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 12.Dimroth P. Primary sodium ion translocating enzymes. Biochim Biophys Acta. 1997;1318:11–51. doi: 10.1016/s0005-2728(96)00127-2. [DOI] [PubMed] [Google Scholar]
- 13.Dimroth P. Sodium ion transport decarboxylases and other aspects of sodium ion cycling in bacteria. Microbiol Rev. 1987;51:320–340. doi: 10.1128/mr.51.3.320-340.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forage R G, Foster M A. Glycerol fermentation in Klebsiella pneumoniae: functions of the coenzyme B12-dependent glycerol and diol dehydratases. J Bacteriol. 1982;149:413–419. doi: 10.1128/jb.149.2.413-419.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaston K, Bell A, Kolb A, Buc H, Busby S. Stringent spacing requirements for transcription activation by CRP. Cell. 1990;62:733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- 16.Hogema B M, Arents J C, Bader R, Eijkemans K, Yoshida H, Takahashi H, Aiba H, Postma P W. Inducer exclusion in Escherichia coli by non-PTS substrates: the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol Microbiol. 1998;30:487–498. doi: 10.1046/j.1365-2958.1998.01053.x. [DOI] [PubMed] [Google Scholar]
- 17.Hogema B M, Arents J C, Inada T, Aiba H, van Dam K, Postma P W. Catabolite repression by glucose 6-phosphate, gluconate and lactose in Escherichia coli. Mol Microbiol. 1997;24:857–867. doi: 10.1046/j.1365-2958.1997.3991761.x. [DOI] [PubMed] [Google Scholar]
- 18.Ishizuka H, Hanamura A, Inada T, Aiba H. Mechanism of the down-regulation of cAMP receptor protein by glucose in Escherichia coli: role of autoregulation of the crp gene. EMBO J. 1994;13:3077–3082. doi: 10.1002/j.1460-2075.1994.tb06606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishizuka H, Hanamura A, Kunimura T, Aiba H. A lowered concentration of cAMP receptor protein caused by glucose is an important determinant for catabolite repression in Escherichia coli. Mol Microbiol. 1993;10:341–350. doi: 10.1111/j.1365-2958.1993.tb01960.x. [DOI] [PubMed] [Google Scholar]
- 20.Izu H, Kawai T, Yamada Y, Aoshima H, Adachi O, Yamada M. Characterization of the gntT gene encoding a high-affinity gluconate permease in Escherichia coli. Gene. 1997;199:203–210. doi: 10.1016/s0378-1119(97)00368-5. [DOI] [PubMed] [Google Scholar]
- 21.Johnson E A, Burke S K, Forage R G, Lin E C C. Purification and properties of dihydroxyacetone kinase from Klebsiella pneumoniae. J Bacteriol. 1984;160:55–60. doi: 10.1128/jb.160.1.55-60.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson E A, Levine R L, Lin E C C. Inactivation of glycerol dehydrogenase of Klebsiella pneumoniae and the role of divalent cations. J Bacteriol. 1985;164:479–483. doi: 10.1128/jb.164.1.479-483.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaspar S, Perozzo R, Reinelt S, Meyer M, Pfister K, Scapozza L, Bott M. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol Microbiol. 1999;33:858–872. doi: 10.1046/j.1365-2958.1999.01536.x. [DOI] [PubMed] [Google Scholar]
- 24.Meyer M, Dimroth P, Bott M. In vitro binding of the response regulator CitB and of its carboxy-terminal domain to A + T-rich DNA target sequences in the control region of the divergent citC and citS operons of Klebsiella pneumoniae. J Mol Biol. 1997;269:719–731. doi: 10.1006/jmbi.1997.1076. [DOI] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 26.Nelson S O, Wright J K, Postma P W. The mechanism of inducer exclusion: direct interaction between purified IIIGlc of the phosphoenolpyruvate:sugar phosphotransferase system and the lactose carrier of Escherichia coli. EMBO J. 1983;2:715–720. doi: 10.1002/j.1460-2075.1983.tb01490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novotny M J, Frederickson W L, Waygood E B, Saier M H., Jr Allosteric deregulation of glycerol kinase by enzyme IIIglc of the phosphotransferase system in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1985;162:810–816. doi: 10.1128/jb.162.2.810-816.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osumi T, Saier M H. Regulation of lactose permease activity by the phosphoenolpyruvate:sugar phosphotransferase system: evidence for direct binding of the glucose-specific enzyme III to the lactose permease. Proc Natl Acad Sci USA. 1982;79:1457–1461. doi: 10.1073/pnas.79.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osuna R, Bender R A. Klebsiella aerogenes catabolite gene activator protein and the gene encoding it. J Bacteriol. 1991;173:6626–6631. doi: 10.1128/jb.173.20.6626-6631.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osuna R, Boylan S A, Bender R A. In vitro transcription of the histidine utilization (hutUH) operon from Klebsiella aerogenes. J Bacteriol. 1991;173:116–123. doi: 10.1128/jb.173.1.116-123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porco A, Peekhaus N, Bausch C, Tong S, Isturiz T, Conway T. Molecular genetic characterization of the Escherichia coli gntT gene of GntI, the main system for gluconate metabolism. J Bacteriol. 1997;179:1584–1590. doi: 10.1128/jb.179.5.1584-1590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pos K M, Dimroth P. Functional properties of the purified Na+-dependent citrate carrier of Klebsiella pneumoniae: evidence for asymmetric orientation of the carrier protein in proteoliposomes. Biochemistry. 1996;35:1018–1026. doi: 10.1021/bi951609t. [DOI] [PubMed] [Google Scholar]
- 33.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sawers G. A novel mechanism controls anaerobic and catabolite regulation of the Escherichia coli tdc operon. Mol Microbiol. 2001;39:1285–1298. doi: 10.1111/j.1365-2958.2001.02316.x. [DOI] [PubMed] [Google Scholar]
- 36.Schneider K, Dimroth P, Bott M. Biosynthesis of the prosthetic group of citrate lyase. Biochemistry. 2000;39:9438–9450. doi: 10.1021/bi000401r. [DOI] [PubMed] [Google Scholar]
- 37.Schneider K, Dimroth P, Bott M. Identification of triphosphoribosyl-dephospho-CoA as precursor of the citrate lyase prosthetic group. FEBS Lett. 2000;483:165–168. doi: 10.1016/s0014-5793(00)02105-0. [DOI] [PubMed] [Google Scholar]
- 38.Steuber J, Krebs W, Bott M, Dimroth P. A membrane-bound NAD(P)+-reducing hydrogenase provides reduced pyridine nucleotides during citrate fermentation by Klebsiella pneumoniae. J Bacteriol. 1999;181:241–245. doi: 10.1128/jb.181.1.241-245.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Studier F W, Moffatt B A. Use of T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 40.Tagami H, Inada T, Kunimura T, Aiba H. Glucose lowers CRP* levels resulting in repression of the lac operon in cells lacking cAMP. Mol Microbiol. 1995;17:251–258. doi: 10.1111/j.1365-2958.1995.mmi_17020251.x. [DOI] [PubMed] [Google Scholar]
- 41.Tobimatsu T, Azuma M, Hayashi S, Nishimoto K, Toraya T. Molecular cloning, sequencing and characterization of the genes for adenosylcobalamin-dependent diol dehydratase of Klebsiella pneumoniae. Biosci Biotechnol Biochem. 1998;62:1774–1777. doi: 10.1271/bbb.62.1774. [DOI] [PubMed] [Google Scholar]
- 42.Tobimatsu T, Azuma M, Matsubara H, Takatori H, Niida T, Nishimoto K, Satoh H, Hayashi R, Toraya T. Cloning, sequencing, and high level expression of the genes encoding adenosylcobalamin-dependent glycerol dehydrase of Klebsiella pneumoniae. J Biol Chem. 1996;271:22352–22357. doi: 10.1074/jbc.271.37.22352. [DOI] [PubMed] [Google Scholar]
- 43.Tong I T, Liao H H, Cameron D C. 1,3-Propanediol production by Escherichia coli expressing genes from the Klebsiella pneumoniae dha regulon. Appl Environ Microbiol. 1991;57:3541–3546. doi: 10.1128/aem.57.12.3541-3546.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van der Rest M E, Siewe R M, Abee T, Schwarz E, Oesterhelt D, Konings W N. Nucleotide sequence and functional properties of a sodium-dependent citrate transport system from Klebsiella pneumoniae. J Biol Chem. 1992;267:8971–8976. [PubMed] [Google Scholar]
- 45.Wifling K, Dimroth P. Isolation and characterization of oxaloacetate decarboxylase of Salmonella typhimurium, a sodium ion pump. Arch Microbiol. 1989;152:584–588. doi: 10.1007/BF00425491. [DOI] [PubMed] [Google Scholar]
- 46.Willard B L. Ph.D. thesis. Madison: University of Wisconsin—Madison; 1994. [Google Scholar]
- 47.Zhang X, Gunasekera A, Ebright Y W, Ebright R H. Derivatives of CAP having no solvent-accessible cysteine residues, or having a unique solvent-accessible cysteine residue at amino acid 2 of the helix-turn-helix motif. J Biomol Struct Dyn. 1991;9:463–473. doi: 10.1080/07391102.1991.10507929. [DOI] [PubMed] [Google Scholar]