Abstract
In the β-proteobacterium Azoarcus evansii, the aerobic metabolism of 2-aminobenzoate (anthranilate), phenylacetate, and benzoate proceeds via three unprecedented pathways. The pathways have in common that all three substrates are initially activated to coenzyme A (CoA) thioesters and further processed in this form. The two initial steps of 2-aminobenzoate metabolism are catalyzed by a 2-aminobenzoate-CoA ligase forming 2-aminobenzoyl-CoA and by a 2-aminobenzoyl-CoA monooxygenase/reductase (ACMR) forming 2-amino-5-oxo-cyclohex-1-ene-1-carbonyl-CoA. Eight genes possibly involved in this pathway, including the genes encoding 2-aminobenzoate-CoA ligase and ACMR, were detected, cloned, and sequenced. The sequence of the ACMR gene showed that this enzyme is an 87-kDa fusion protein of two flavoproteins, a monooxygenase (similar to salicylate monooxygenase) and a reductase (similar to old yellow enzyme). Besides the genes for the initial two enzymes, genes for three enzymes of a β-oxidation pathway were found. A substrate binding protein of an ABC transport system, a MarR-like regulator, and a putative translation inhibitor protein were also encoded by the gene cluster. The data suggest that, after monooxygenation/reduction of 2-aminobenzoyl-CoA, the nonaromatic CoA thioester intermediate is metabolized further by β-oxidation. This implies that all subsequent intermediates are CoA thioesters and that the alicyclic carbon ring is not cleaved oxygenolytically. Surprisingly, the cluster of eight genes, which form an operon, is duplicated. The two copies differ only marginally within the coding regions but differ substantially in the respective intergenic regions. Both copies of the genes are coordinately expressed in cells grown aerobically on 2-aminobenzoate.
2-Aminobenzoic acid (anthranilic acid) plays an important role in the synthesis and degradation of many N-heterocyclic compounds such as tryptophan, indole, indoleacetic acid, and derived compounds. As a consequence of its wide occurrence, 2-aminobenzoate is a common substrate for many microorganisms that are able to cleave aromatic rings.
There are three established aerobic pathways by which 2-aminobenzoate is converted to either catechol or gentisate (reviewed in reference 26). These compounds are central intermediates of aerobic aromatic metabolism which are further oxidized and cleaved, via either ortho- or meta-cleavage pathways. Notably, the aromatic ring is cleaved oxygenolytically, and coenzyme A (CoA) thioesters of organic acids come into play only after ring cleavage, e.g., as 3-oxo-adipyl-CoA and succinyl-CoA. Such rules also apply to the aerobic metabolism of other aromatic substrates. These pathways have been discussed and reviewed elsewhere (14).
It has been found that 2-aminobenzoate, benzoate, and phenylacetate are metabolized aerobically via nonconventional routes in the bacterium Azoarcus evansii (4, 13, 28). This organism is a member of the β-Proteobacteria and grows with any of these substrates either aerobically or anoxically, with nitrate as the terminal electron acceptor of respiration. Evidently, the metabolism of aromatic substrates under denitrifying conditions cannot involve oxygen-dependent reactions, which are characteristic for the aerobic metabolism of aromatic compounds. Anaerobic metabolism of these aromatic acids is initiated by the formation of CoA thioesters by specifically induced CoA ligases. The pathways involved in anaerobic aromatic metabolism have been reviewed recently (18, 19).
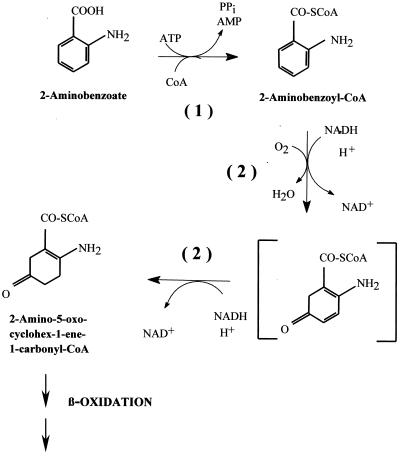
Surprisingly, A. evansii induces a similar set of three CoA ligase isoenzymes acting on 2-aminobenzoate, benzoate, and phenylacetate when it is shifted to aerobic growth on these substrates (3, 5, 28). There is increasing evidence that these compounds are aerobically metabolized throughout via CoA thioesters by unique pathways that are poorly understood at present. The two initial steps in 2-aminobenzoate metabolism are shown in Fig. 1.
FIG. 1.
Initial steps in the aerobic degradation of 2-aminobenzoate in the β-proteobacterium A. evansii. Reaction 1, 2-aminobenzoate-CoA ligase (EC 6.2.1.32); reaction 2, ACMR (EC 1.14.13.40), with two partial reactions, monooxygenation and reduction. The chemical structure of an assumed enzyme-bound intermediate is shown in brackets. Reaction 1 is catalyzed by AbmG, reaction 2 by AbmA, and β-oxidation presumably by AbmB, AbmC, and AbmD.
More specifically, 2-aminobenzoate is converted to 2-aminobenzoyl-CoA by 2-aminobenzoate-CoA ligase (AMP forming) (EC 6.2.1.32). This monomeric 65-kDa enzyme has been purified and characterized from A. evansii, and the N-terminal amino acid sequence has been determined (3). 2-Aminobenzoyl-CoA is subsequently hydroxylated at C-5, and the aromatic ring is reduced by two electrons to yield a nonaromatic product, 2-amino-5-oxo-cyclohex-1-ene-1-carbonyl-CoA. This reaction is catalyzed by the flavoenzyme 2-aminobenzoyl-CoA monooxygenase/reductase (ACMR) (EC 1.14.13.40), a homodimeric protein of 170 kDa. The reaction requires 2 NADH molecules and 1 O2 molecule. ACMR and the reaction it catalyzes have been studied experimentally and theoretically in some detail (12, 13, 17, 22, 23, 24, 30). In addition, its N-terminal amino acid sequence has been determined (2).
The subsequent reactions converting the first nonaromatic intermediate to intermediates of the central carbon metabolism have not yet been studied. The main reason is that synthesis of sufficient amounts of the intermediate CoA thioesters, the substrates required for further enzyme assays, requires great effort and expense. To obtain insight into this novel pathway, we cloned and sequenced the genes involved.
MATERIALS AND METHODS
Materials.
Chemicals and biochemicals were obtained from Roche Diagnostics (Mannheim, Germany), Fluka (Neu-Ulm, Germany), Merck (Darmstadt, Germany), Roth (Karlsruhe, Germany), Sigma-Aldrich (Deisenhofen, Germany), and Bio-Rad (Munich, Germany). Enzymes used were obtained from MBI Fermentas (St. Leon-Rot, Germany) and Amersham Pharmacia Biotech (Freiburg, Germany). Nitrocellulose and positively charged nylon membranes used for Southern blotting and phage hybridization were obtained from Schleicher and Schuell (Dassel, Germany), and Amersham Pharmacia Biotech.
Strains and culture conditions.
A. evansii KB740 (DSM 6898) was grown at 30°C in chemically defined medium aerobically or anaerobically (with 5 mM nitrate) with 2 mM 2-aminobenzoate (11, 24, 33), or anaerobically with 5 mM benzoate and 10 mM nitrate (31). Cells were harvested at 4°C at an optical density at 578 nm (OD578) of 0.1 when used for RNA isolation. Large-scale aerobic growth with 2-aminobenzoate was carried out using a mineral salt medium (11) containing 5 mM 2-aminobenzoate as the carbon and energy source. Cultivation was carried out in a 200-liter fermentor with stirring at 200 rpm, at an air gassing rate of 40 liters/min. A 5% inoculum was used, and 2-aminobenzoate (5 mM) was refed once.
Escherichia coli strain XL1 blue, MRF′[Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)], and E. coli strain XLOLR, Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 thi-1 recA1 gyrA96 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)] Su− λr, were grown at 37°C in Luria-Bertani (LB) medium (29). Antibiotics were added to E. coli cultures as follows (final concentrations): ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; tetracycline, 20 μg/ml.
Preparation of cell extracts and protein determination.
Cell extracts were prepared as described previously (12). Protein was generally quantified by the method of Bradford (10), and bovine serum albumin was used as the standard.
Purification of ACMR, enzyme assay, and determination of the N-terminal amino acid sequence.
ACMR was purified as described previously (24). Its activity was measured by using N-ethylmaleimide as described by Langkau et al. (24). The N-terminal amino acid sequence of the purified enzyme was determined as described by Davies (15).
Cloning, transformation, amplification, and purification of nucleic acids.
Standard protocols were used for DNA cloning, transformation, amplification, and purification (7, 29). Total RNA used in primer extension experiments was isolated using the hot-phenol method (1). Total RNA used for reverse transcription (RT)-PCR was isolated using an RNeasy Total RNA Kit (Qiagen, Hilden, Germany). Plasmid DNA was prepared by the method of Birnboim and Doly (8). Large plasmids (up to 30 kb) were prepared as described in the Plasmid Mini Kit instruction manual (Qiagen). A λ-ZAP Express gene library of Sau3A-digested genomic DNA from A. evansii was prepared according to the ZAP Express Cloning Kit instruction manual (Stratagene, Amsterdam, The Netherlands). PCR products used as probes in screening or Southern blot analysis were labeled with digoxigenin-11-dUTP (Roche Diagnostics) by PCR. Probes were detected with alkaline phosphatase (AP)-conjugated anti-digoxigenin Fab fragments and CSPD (Roche Diagnostics) in Southern blot analysis, followed by a 30-min exposure to X-ray film (Hyperfilm MP; Amersham). For screening of hybridizing λ clones, labeled probe was detected with anti-digoxigenin-AP, nitroblue tetrazolium chloride (NBT), and X-Phosphate (5-chloro-4-bromo-3-indoyl-phosphate toluidine salt) (Biomol, Hamburg, Germany).
Cloning of the DNA containing the genes for aerobic 2-aminobenzoate metabolism.
Two degenerate oligonucleotides, F1 (ATGCG CATCG TSTGC ATCGG) and R1 (CCGAA GGTRT CGTAS GGGCG GTT), were designed on the basis of the determined N-terminal amino acid sequence of ACMR and the codon usage of A. evansii. The primers were used in PCR to amplify a 123-bp digoxigenin-labeled probe (P1). The probe was used for screening the λ-ZAP Express gene library. Primers F1a (GCACT TGCGC ACCCA TTGCG) and R1a (GGTCC CATTC GCGCA TGTT) were derived from the 5′ ends of the cloned sequences and used to amplify a second probe (P1a) by PCR. This probe was used to screen for 5′ extension clones. New primers were also derived from the 3′ ends of the sequences obtained and were used to amplify four additional probes, P2 (F2, CGGCG GCGAC GTGTT CGAGA; R2, CGGCG GCATC GCGGC CGCCC), P3 (F3, CGGCG AGATG GCGAC CGACA T; R3, CCCAT CACCT CGCGG TAAAC C), P4 (F4, CGGTG ATCCC GGACT GACCC; R4, CGCCG GATCG GTGTC GGCGC), and P5 (F5, CGGCT TCAAG GAGGC CTTGG; R5, CCTTA CATGC GGCAG CCGCG C), which were used to complete the gene cluster by chromosome walking. Recombinant plasmids were maintained in E. coli XL1 blue. Missing parts of the sequence were amplified by PCR using primers derived from intergenic regions and were sequenced directly.
Investigation of the observed sequence heterogeneity via colony PCR.
Colony PCR was carried out using A. evansii cells from single colonies. Specific forward primers were derived from the 5′ noncoding regions of the type I and II sequences (pI [GCACT TGCGC ACCCA TTGCG] and pII [GAAGA GCAGC CCCGC GAGCG C], respectively). A common reverse primer for both sequences (pR [GGTCC CATTCG CGCAT GTTGT]) was derived from the 5′ end of open reading frame 1 (ORF 1).
Expression of ACMR in E. coli.
Two primers were derived upstream and downstream of the gene coding for type I ACMR (ORF 1I); they carried restriction sites for EcoRI and SalI, respectively (EcoRI-forw, CGGTG CCGGG GAATT CGGAC GGAGG AGGC; SalI rev, GCGGC GCGTC GACTT CTCTG TTGCG GCCG). The ACMR gene was amplified by PCR and cloned into the pGex vector (Amersham Pharmacia Biotech). The recombinant plasmid was transferred to E. coli XL1 blue, and expression of ACMR was induced at an OD578 of 0.5 by addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (for 3 h at 30°C) to the LB-ampicillin medium. Cells were harvested at 4°C, and activity was measured in the cell extract.
RT-PCR.
Contaminating DNA in total-RNA preparations was removed with 1 U of fast protein liquid chromatography (FPLC)-pure DNase I (Amersham Pharmacia Biotech) per μg of RNA (for 10 min at 37°C). Complete removal of DNA was tested by PCR (with pI and pR). A cDNA library was provided by an RT reaction using a hexanucleotide random priming mix (Roche Diagnostics). The cDNA was used in subsequent PCRs with specific primers. To corroborate the simultaneous expression of both gene clusters, specific primers were derived from the varying intergenic regions [5′orf1(II)for, GCGTC CAGAT GGAGG AGACC] or from differing areas of the coding regions [orf1(I)for, CCGCG TGGTC GAACG CAA; orf1(I)rev, CAGCC CGTGC GCCTT CCAGA; orf1(II)rev, GAGGC CGTGC GCCTT CCACA; orf7(I)for, CAGCC CGATG CTGGT TGCAT; orf7(II)for, GCCTG CTGGT TCGCG ATCAT; orf7(I/II)rev, GCGGC GGGCT GCCGA TGAA] of ORF 1 and ORF 7. To confirm the expression of the gene clusters as an operon, primers were derived from the coding or intergenic regions of each ORF and used in PCR to amplify fragments including the intergenic sequence between two ORFs [orf1(I)for, GGCGG CGGCG TCGAG AA; orf2(I)rev, CGCGG CGAAA TCGGT GATA; orf1(II)for, CCGAC CACGC GAACA GCATC A; orf2(II)rev, TCGCC TCGAG CGCTG CACCG; orf2for, GGTTG CCGGC GGCGA AGTGA; orf3rev, GCCGT CGATC GCGGC GATGA; interorf2/3for, TTCCG CGCGT CTCGA CCGTC; orf3/4rev, GCGGT CCATG GCGTG GGGCC; orf4for, CGCGA CCGAG GTGCA GAAGC; interorf5/6rev, AACAA GGGGA AGGGG CCAGA TC; inter orf5/6for, GACTCC GAGCC CGCAT GTCCA; interorf6/7rev, CGCAG CCGGG CCGTC GCAAG; orf6for, GCACG AGCAA TGGGT CGTCG G; orf7rev, CGGCC CTTGT CGAGG ATCT; orf7for, CGCAC GATCG CGCCG TACAA A; orf8rev, GCGAC CTTGA AACTG CCGTA]. For a control, a cDNA library was amplified with RNA isolated from cells grown anaerobically on 2-aminobenzoate. PCR was performed with primer sets orf1(I)for–orf1(I)rev and 5′orf1(II)for–orf1(II)rev. For a positive control, two primers were used to amplify the gene expressing the BcrD subunit of benzoyl-CoA reductase, which is expressed during anaerobic growth on 2-aminobenzoate (bcr-for, GGCAT CGACG TCGGC TCGGT; bcr-rev, CCGCT CCAGC AGGCT CAC).
DNA sequencing and computer analysis.
Plasmid DNA was purified as described in the E.Z.N.A. Plasmid Miniprep Kit I manual (PeqLab, Erlangen, Germany). DNA sequences were determined either in our laboratories by use of an ALFexpress automated sequencer (Amersham Pharmacia Biotech) or by J. Alt-Mörbe (Labor für DNA-Analytik, Freiburg, Germany). DNA and amino acid sequences were analyzed by use of the BLAST network service (National Center for Biotechnology Information, Bethesda, Md.)
Mapping of the 5′ end of mRNA.
Start sites of mRNAs encoded by the 2-aminobenzoate metabolism gene clusters were mapped by primer extension (9, 25) using a Cy5-labeled primer derived from the 5′ end of ORF 1 (PE1, GGATAGCGAAATACAGCCC CGC). Extension products were analyzed by use of an ALFexpress sequencer.
Nucleotide sequence accession numbers.
The sequence data reported here were submitted to GenBank (accession numbers AF320253 and AF320254).
RESULTS
Reinvestigation of ACMR.
Previous unexplained findings prompted us to reinvestigate ACMR. The enzyme was purified from A. evansii cells grown aerobically with 2-aminobenzoate as the sole carbon and energy source. Forty-one amino acids of the N terminus of ACMR were determined by Edman degradation. From previous work (2), the first 24 amino acids (aa) were known, with only one uncertain amino acid residue at position 5, possibly a cysteine. In addition, the N-terminal amino acid sequence of 2-aminobenzoate-CoA ligase had been determined previously (3) (Table 1). The N-terminal amino acid sequence of ACMR was reconfirmed; further down, however, the sequence exhibited mixtures of 2 aa each at positions 25 (V and A) and 36 (A and P) (Table 1). This finding suggested a heterogeneous protein sample.
TABLE 1.
N-terminal amino acid sequences of 2-aminobenzoate-CoA ligase and ACMR, as determined by Edman degradationa
| Enzyme (method) | Amino acid sequenceb |
|---|---|
| ACMR (Edman degradation) | MRIVc IGGGP AGLYF AILMK KLNP(V/A) HEIrV IerNr (A/P)YDTF G |
| ACMR (deduced from ORF 1, type I) | MRIVC IGGGP AGLYF AILMK KLNPV HEIR V VERNR AYDTF G |
| ACMR (deduced from ORF 1, type II) | MRIVC IGGGP AGLYF AILMK KLNPA HEIRV IERNR PYDTF G |
| 2-AB-CoA ligasec (Edman degradation) | TSHVD TFARD XLPPX eQQ |
| 2-AB-CoA ligase (deduced from ORF 7, type I) | TSHVD TFARD RLPPP ELQ |
| 2-AB-CoA ligase (deduced from ORF 7, type II) | TSHVD TFARD RLPPP EQQ |
Deduced amino acid sequences of ORFs 1 and 7 (ACMR and 2-aminobenzoate-CoA ligase, respectively), which exist as duplicate copies I and II with minor differences (boldfaced amino acids), are included.
Lowercase letters, residues of uncertain identity; X, unknown amino acid; V/A and A/P, mixtures of V and A and of A and P, respectively.
2-AB-CoA ligase, 2-aminobenzoate-CoA ligase.
Screening for genes involved in aerobic 2-aminobenzoate metabolism.
A 123-bp labeled DNA probe (P1) was obtained in PCR assays with chromosomal DNA using two degenerate primers derived from the N-terminal amino acid sequence of ACMR. This probe was checked by sequencing and used to screen a λ-ZAP library for the ACMR gene and additional genes involved in the aerobic 2-aminobenzoate pathway. The first screen resulted in a 4.5-kb DNA fragment (clone I) containing the complete ACMR gene (ORF 1), 400 bp of upstream DNA, and two further ORFs downstream, ORF 2 and ORF 3.
Cloning and sequencing of two possible catabolic operons.
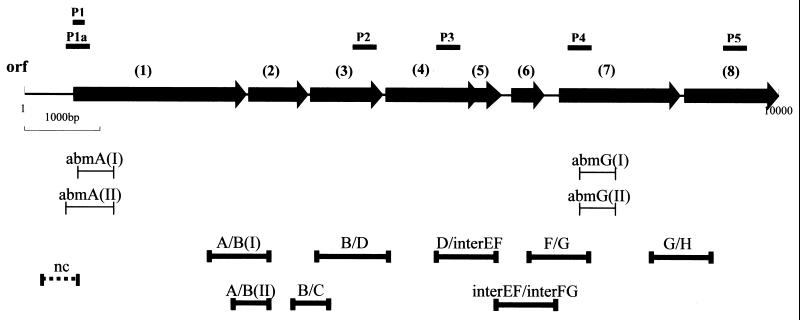
The first clone obtained was extended by chromosome walking using labeled PCR products derived from both flanks of the known sequence. A DNA fragment of 10.5 kb was cloned and sequenced, and altogether 8 ORFs were identified. These ORFs were grouped in the same orientation with only short intergenic regions (Fig. 2). Surprisingly, a second type of clone was obtained during chromosome walking, carrying a second gene cluster of 8 ORFs with very similar, but not completely identical, sequences. The ORFs in the two gene clusters were organized identically. While the DNA sequences within equivalent ORFs were highly similar, the sequences of intergenic regions differed markedly between the two clusters. The noncoding regions upstream of the two variants of ORF 1 also differed markedly. These sequence variations within the noncoding regions were utilized to derive PCR primers to fill in the remaining gaps in the sequences of the two gene clusters. The intergenic regions were also useful in identifying the two variant gene clusters, if necessary. In total, two DNA fragments of approximately 10.5 kb each were sequenced. The order of genes and other properties of the two putative operons are shown in Fig. 2. The operons and genes are referred to as types I and II, respectively.
FIG. 2.
Common organization of the two gene clusters, types I and II, coding for proteins involved in aerobic 2-aminobenzoate metabolism (abm). ORFs: 1, abmA (2.32 kb), encoding ACMR; 2, abmB (0.81 kb), encoding β-hydroxyacyl-CoA dehydrogenase; 3, abmC (0.85 kb), encoding enoyl-CoA hydratase; 4, abmD (1.2 kb), encoding acyl-CoA dehydrogenase; 5, abmE (0.4 kb), encoding a putative translation regulator protein; 6, abmF (0.48 kb), encoding a MarR-like regulator protein; 7, abmG (1.62 kb), encoding 2-aminobenzoate-CoA-ligase; 8, abmH (1.14 kb), encoding a putative substrate-binding protein of the ABC-transporter system. The positions of probes P1 to P5, which were used in library screening, are indicated above the arrows representing the ORFs. Bars immediately below the gene cluster diagram indicate locations of the amplified RT-PCR products of type I and type II sequences that are shown in Fig. 3. Boldfaced bars at the bottom indicate locations of the amplified RT-PCR products shown in Fig. 9.
Demonstration of simultaneous expression of both gene clusters and regulation by oxygen.
The DNA sequences in the ORF 1 coding regions (coding for ACMR) of the two gene clusters were nearly identical. Some significant differences were still observed; mostly they occurred in the wobble positions of codons, but sometimes they caused conservative amino acid exchanges.
The first 41 aa residues of ACMR determined by Edman degradation showed 2 aa residues each at positions 25 (V and A) and 36 (A and P), with V25 and A36 in clearly lesser amounts (one-third). The mixture of 2 aa observed at these positions is consistent with the N-terminal amino acid sequences deduced from the ACMR genes of type I (coding for the variants V25 and A36) and type II (coding for the variants A25 and P36), respectively (Table 1). A third amino acid mixture of valine and isoleucine expected at position 31 (valine in type I and isoleucine in type II) was not detected in Edman degradation; only isoleucine was observed at this position. In this case, the valine signal at position 31 from variant I was probably overlaid by the preceding V at position 30. These results clearly indicated that two similar ACMR genes are present in the genome, that both variants are functional, and that they are expressed simultaneously.
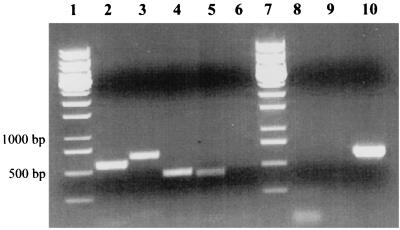
To support this conclusion, RT-PCR was carried out. DNA-free total RNA was prepared from cells grown aerobically on 2-aminobenzoate. Complete removal of DNA was tested by PCR. Using a hexanucleotide mixture for random priming, a cDNA library was built by RT. Specific primers were derived from varying regions of both gene clusters and used in a PCR assay with the cDNA library. As a control, all assays were performed using total RNA preparations from cells grown anaerobically on 2-aminobenzoate. PCR fragments of ACMR types I and II, as well as fragments of ORF 7 (2-aminobenzoate-CoA ligase) of both gene clusters, were obtained with the cDNA of aerobically grown cells (Fig. 3; compare with Fig. 2).
FIG. 3.
Demonstration of the simultaneous expression of both abm operons during aerobic growth on 2-aminobenzoate. RT-PCR was performed using primers specific for abmA (the ACMR gene) and abmG (the CoA ligase gene) of type I and type II gene clusters, respectively (see Fig. 2). As a control, RNA from cells grown anoxically under denitrifying conditions with 2-aminobenzoate was used. abmA expression could not be detected under anoxic growth conditions. Lanes 1 and 7, molecular standards; lanes 2 through 5, PCR products of abmA(I) (lane 2), abmA(II) (lane 3), abmG(I) (lane 4), and abmG(II) (lane 5); lane 6, negative control using a forward primer binding upstream of ORF 1 in the nontranscribed region and a reverse primer binding in ORF 1 (Fig. 2, nc); lane 8, abmA(I) RNA prepared from anoxically grown cells; lane 9, abmA(II) RNA prepared from anoxically grown cells; lane 10, positive control, showing amplification of bcrD from cells grown anoxically on 2-aminobenzoate. For conditions and primers, see Materials and Methods.
To show that expression of the genes was controlled by oxygen, mRNA from cells grown anaerobically with 2-aminobenzoate under denitrifying conditions was tested for the presence of ACMR transcript. Neither of the mRNA products of the two ACMR genes could be detected in anaerobically grown cells (Fig. 3). As a positive control, the gene coding for the delta subunit of benzoyl-CoA reductase, bcrD, was amplified (Fig. 3). Benzoyl-CoA reductase is required for anoxic 2-aminobenzoate metabolism. This showed that the ACMR genes are expressed only under aerobic conditions.
Search for plasmids, colony PCR, and Southern blot experiments.
Experiments were performed to exclude some trivial causes for the two similar copies of DNA sequences observed, such as impure cultures containing two similar Azoarcus strains or the existence of a plasmid and a chromosomal copy of variants I and II.
The existence of plasmids in A. evansii was tested by two methods. DNA preparations were analyzed by agarose gel electrophoresis for the existence of plasmids, without any positive result. In order to exclude the possibility that megaplasmids were lost, the preparation protocol was modified to include large plasmids, but no indication of the existence of megaplasmids was obtained. Also, CsCl density gradient centrifugation of total DNA preparations resulted in only one chromosomal DNA band. The existence of both ACMR genes in the genome of A. evansii was confirmed by PCR. Specific primers were derived from the varying intergenic regions of both types of sequences and used in PCR. Both types of sequences could be amplified in the chromosomal DNA preparation, as expected, and their heterogeneity was confirmed by sequencing.
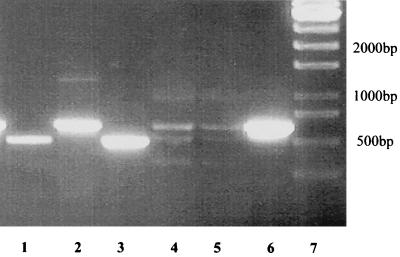
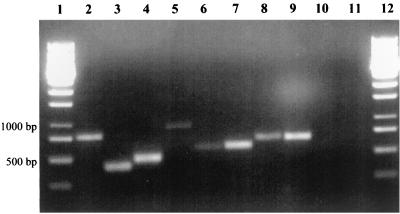
To exclude the possibility of contamination of the A. evansii strain, KB740, by a closely related strain, resulting in a mixed gene library, single colonies were tested for the presence of the two variants of ORF 1. Both single-colony PCR and PCR with DNA preparations of cultures derived from single colonies gave the same result. Both types of gene clusters were always present (Fig. 4).
FIG. 4.
Colony PCR showing the presence of two copies of similar genes on the A. evansii genome. Specific primers were derived from the differing intergenic regions of gene cluster types I and II and were used in PCR experiments. Lane 1, colony PCR with primers specific for type I; lane 2, colony PCR with primers specific for type II; lane 3, PCR with a type I subclone and primers specific for type I; lane 4, PCR with a type I subclone and primers specific for type II (negative control); lane 5, PCR with a type II subclone and primers specific for type I (negative control); lane 6, PCR with a type II subclone and primers specific for type II; lane 7, DNA standard. Small amounts of several products were found in negative controls (lanes 4 and 5). These products were of no reproducible size and therefore were considered nonspecific.
Southern blot analysis was performed to corroborate the existence of two similar but distinct sequences of ORF 1. By PCR, a labeled 1.3-kb probe of the 5′ end of the ACMR type I gene (ORF 1I) was obtained. Total DNA was cut with two endonucleases, PstI and SacI, and analyzed by Southern blot hybridization. In both cases, three fragments were expected if only one sequence of ORF 1 (type I) existed. Instead, four fragments were obtained after restriction with SacI, one of which deviated from the expected size (data not shown). After restriction with PstI, five fragments were obtained, including the three expected and two additional fragments (data not shown). This indicated that, in addition to the type I sequence, the second (type II), which differed mainly in the noncoding intergenic regions, was present. These regions probably contain different restriction sites giving rise to further fragments.
Organization and characterization of the gene clusters.
As indicated above, the organization of the 8 ORFs was identical in the two gene clusters (Fig. 2). In the noncoding regions, the duplicated gene clusters differed by 35% in nucleotide sequence and by 15% in length (average values). In the coding regions, the nucleotide sequence differed by only 9.6%. As in the ACMR gene, the differences in the other coding DNA sequences mostly affected the wobble positions of codons but sometimes caused conservative amino acid exchanges. The deduced amino acid sequences were 93.8% identical and 99.2% similar (average values). The two clusters exhibited similar G+C contents of approximately 67%, similar to the overall G+C content of the organism (67.5%) (6). The shortest intergenic region in the two fragments was 4 bp, and the longest was 185 bp. ORF 4 and ORF 5 overlapped. Within a range of about 700 bp upstream of ORF 1 (I and II), no further ORFs were found.
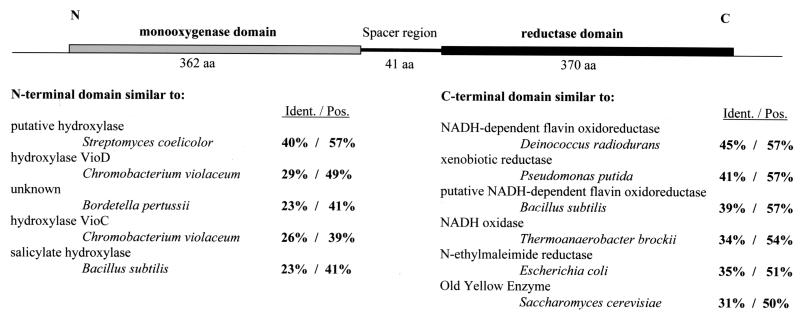
Similarities of the ORF 1 sequences with sequences in DNA and protein databases are shown in Fig. 5, and those of ORFs 2 to 8 are shown in Table 2. The ORFs are designated abm, for aminobenzoate metabolism.
FIG. 5.
Primary structure of ACMR (ORF 1 product; AbmA), consisting of two domains and a short spacer region, and percent identities (Ident.) and similarities (Pos.) with enzymes in the databases. Databases and accession numbers are as follows: Streptomyces coelicolor putative hydroxylase, EMBL CAB42932.1; Chromobacterium violaceum hydroxylase VioD, DBJ BAA84785.1; Bordetella pertussis unknown protein, GB ACC46266; C. violaceum hydroxylase VioC, DBJ BAA84784.1; Bacillus subtilis salicylate hydroxylase, GB AAA64522; Deinococcus radiodurans NADH-dependent flavin oxidoreductase, GB AAF11740.1; Pseudomonas putida xenobiotic reductase, GB AFF02538.1; B. subtilis putative NADH-dependent flavin oxidoreductase, SP P54550; Thermoanaerobacter brockii NADH oxidase, PIR S35706; E. coli N-ethylmaleimide reductase, SP P77258; Saccharomyces cerevisiae old yellow enzyme, DBJ BAA20121.
TABLE 2.
Similarities of the deduced amino acid sequences of ORFs 2 through 8 with sequences in DNA and protein databases
| ORF (protein) | Similar protein | % Identity | % Similarity | ACR no. |
|---|---|---|---|---|
| 2 (AbmB)a | Domain of daunorubicin-doxorubicin polyketide synthase (Streptomyces peucetius) | 37 | 49 | L35560, GB AAA65204 |
| d-β-hydroxybutyrate dehydrogenase (Rhodobacter sp. DSMZ 12077) | 36 | 47 | AF037323, GB AAD42688 | |
| Ketoacyl reductase (Streptomyces galilaeus) | 37 | 47 | D14040, DBJ BAA03128 | |
| 3-Hydroxybutyrate dehydrogenase (Pseudomonas aeruginosa) | 36 | 50 | AE004626, GB AAG05391 | |
| 3 (AbmC) | Putative acyl-CoA isomerase [Streptomyces coelicolor A3(2)] | 33 | 45 | AL390975, EMBL CAC01345 |
| PhaB (Pseudomonas putida) | 29 | 45 | AF029714, GB AAC24330 | |
| Enoyl-CoA isomerase (E. coli) | 27 | 45 | X97452, EMBL CAA66096 | |
| 2-Cyclohexenylcarbonyl-CoA isomerase (Streptomyces collinus) | 31 | 44 | AF268489, GB AAF73478 | |
| 4 (AbmD) | Butyryl-CoA dehydrogenase (Thermoanaerobacterium thermosaccharolyticum) | 41 | 52 | Z92974, EMBL CAB07496 |
| Probable acyl-CoA dehydrogenase (P. aeruginosa) | 36 | 51 | AE004683, GB AAG05940 | |
| Acyl-CoA dehydrogenase (Mycobacterium leprae) | 34 | 53 | U00012, GB AAA85936 | |
| Acyl-CoA dehydrogenase (Bacillus halodurans) | 31 | 49 | AP001520, DBJ BAB07518 | |
| 5 (AbmE) | Translational inhibitor protein p14.5 (Homo sapiens) | 30 | 51 | X95384, EMBL CAA64670 |
| Hypothetical protein (Synechocystis sp.) | 34 | 47 | D64005, DBJ BAA10755 | |
| Conserved hypothetical protein (P. aeruginosa) | 30 | 49 | AE004951, GB AAG08777 | |
| 14.5-kDa translational inhibitor protein (Rattus sp.) | 28 | 45 | D49363, DBJ BAA08359 | |
| 6 (AbmF) | Transcriptional regulator, MarR family (Deinococcus radiodurans) | 36 | 57 | AE001965, GB AAF10732 |
| Similar to transcriptional regulator (MarR family) (Bacillus subtilis) | 31 | 51 | Z99111, EMBL CAB13207 | |
| Transcriptional regulator, putative, MarR family (Thermotoga maritima) | 44 | 58 | AE001749, GB AAD35898 | |
| Transcriptional regulator (MarR family) (B. halodurans) | 27 | 49 | AP001514, DBJ BAB05895 | |
| 7 (AbmG) | Acetate-CoA ligase (ACS-5) (Archaeoglobus fulgidus) | 37 | 51 | AE001037, GB AAB90267 |
| Putative aryl-CoA ligase EncN (Streptomyces maritimus) | 33 | 51 | AF254925, GB AAF81733 | |
| Aryl-CoA ligase (Alcaligenes sp. strain BR60) | 40 | 53 | AF041042, GB AAC38458 | |
| Benzoate-CoA ligase (Rhodopseudomonas palustris) | 33 | 48 | L42322, GB AAA92151 | |
| 4-Hydroxybenzoate-CoA ligase (R. palustris) | 32 | 46 | U02033, GB AAA62604 | |
| 8 (AbmH) | Putative ABC transporter substrate binding protein (R. palustris) | 50 | 63 | U75364, GB AAC13366 |
| Urea/short-chain amide ABC transporter, periplasmic binding protein (D. radiodurans) | 25 | 36 | AE001863, GB AAF12459 | |
| Branched-chain amino acid ABC transporter, periplasmic binding protein (T. maritima) | 26 | 40 | AE001771, GB AAD36211 |
Note that the enzymes similar to the ORF 2 product probably act on 3-hydroxyacyl-CoA compounds rather than on the corresponding free acids, as suggested by the database entries.
ORF 1 (abmA).
ORF 1 (abmA) codes for ACMR. Obviously, both variants are expressed during growth on 2-aminobenzoate, since both types of protein were found in the purified enzyme. The type I gene may be expressed to a lesser extent, since the amino acids derived from that gene (V25 and A36) were present in lower concentrations than those derived from the type II gene (A25 and P36) (see above). The N-terminal half of ORF 1 showed high similarities to known monooxygenases such as salicylate 1-monooxygenases (27); the C-terminal half showed similarities to various NADH-dependent, flavin-containing oxidoreductases of the old-yellow-enzyme type (20) (Fig. 5). The two halves are joined by a short linker region which showed no similarity to entries in the current databases. The structure of these domains and sequence similarities are shown in Fig. 5. The N-terminal region of the first protein domain (aa 3 to 16) contains a potential flavin adenine dinucleotide (FAD) binding site. Other potential FAD or NADH binding sites may be predicted, for instance, aa 80 to 96 (Fig. 6A). Another potential flavin nucleotide binding site was identified at aa 692 to 728 (Fig. 6B). The calculated molecular mass of the gene products, 87 kDa, was close to the experimentally determined mass of ACMR, 85 kDa. The calculated isoelectric points (IEP) of the two variants were 6.0 and 6.2 (types I and II, respectively); the experimental IEP of the ACMR subunits was 5.3 ± 0.2 (13). The ACMR type I gene was cloned into the pGex vector and expressed in E. coli XL1 blue. ACMR activity could be measured in the cell extract (∼50 nmol of NADH oxidized min−1 mg of protein−1), but >95% of the protein was found inactive in inclusion bodies (data not shown).
FIG. 6.
Alignment of the sequences of the putative nucleotide binding domains of ACMR with those of several NADH-dependent flavin oxidoreductases. DR2190, D. radiodurans (AAF11740); CAA47660, T. brockii; FadH1, Pseudomonas aeruginosa (AAG06480); baiCD, Clostridium sp. (AAF22846); baiH, Eubacterium sp. (AAC45417); xenA, P. putida (AAF02538); YqjM, Halobacterium sp. (AAG19363); AF0248, Archaeoglobus fulgidus (AAB90979).
ORFs 2 to 4 (abmBCD).
ORFs 2 to 4 of the gene clusters code for enzymes of a β-oxidation process. ORF 2 (abmB) codes for a 27-kDa putative 3-hydroxyacyl-CoA dehydrogenase. A characteristic domain for short-chain alcohol dehydrogenases was found in the ORF 2 gene products between aa 151 and 179 (Table 3). ORF 3 (abmC) codes for a 30-kDa putative enoyl-CoA hydratase/isomerase. A sequence motif typical for such enzymes was identified between aa 121 and 141 of the derived gene product (Table 3). ORF 4 (abmD) codes for a 43-kDa putative acyl-CoA dehydrogenase. A characteristic sequence motif for acyl-CoA dehydrogenases is located between aa 128 and 140 in the derived gene product (Table 3). The 3′ ends of the ORF 4 genes in the type I and II gene clusters differed markedly.
TABLE 3.
Characteristic domains identified for AbmB, AbmC, AbmD, and AbmG
| Protein for which domain is characteristic | Motifa | PROSITE no. |
|---|---|---|
| Short-chain alcohol dehydrogenase (AbmB) | [LIVSPADNK]-X12-Y-[PSTAGNCV]-[STAGNQCIVM]-[STAGC]-K-[PC]-[SGH]-[LIVMSTAGD]-X2-[LIVMFYW]-X4-[GSACQRHM] | PS00061 |
| Enoyl-CoA hydratase/isomerase (AbmC) | [LIVM]-[STA]-X-[LIVM]-[DENQRHSTA]-G-X3-[AG]-X6-[LIVMST]-X-[CSTA]-[DQHP]-[LIVMFY] | PDOC00150 |
| Acyl-CoA dehydrogenase (AbmD) | [GAC]-[LIVM]-[ST]-E-X2-[GSAN]-G-[ST]-D-X2-[GSA] | PDOC00070 |
| CoA ligase, nucleotide binding region (AbmG) | [LIVMFY]-X2-[STG]-[STAG]-G-[ST]-[STEI]-[SG]-X-[PASLIVM]-[KR] | PDOC00427 |
Possible amino acids are surrounded by brackets. X, no amino acid consensus identified for motif.
ORF 5 (abmE) and ORF 6 (abmF).
ORF 5 codes for a putative translation inhibitor protein of 14.6 kDa. No intergenic region was present between ORF 4 and ORF 5 in either gene cluster; rather, the coding sequences of the two ORFs overlapped by 10 bp. ORF 6 codes for a MarR-like transcription regulator protein with a molecular mass of 18.4 kDa.
ORF 7 (abmG).
ORF 7 codes for the first enzyme of the pathway, 2-aminobenzoate-CoA ligase. The calculated molecular mass of the protein was approximately 59 kDa; the experimentally determined molecular mass was 65 kDa. A second 2-aminobenzoate-CoA ligase, which seemed to be predominantly expressed under anoxic conditions, has been found in A. evansii (3). This anaerobically expressed 2-aminobenzoate-CoA ligase showed a molecular mass of 60 kDa and exhibited an almost identical N-terminal amino acid sequence in Edman degradation (3). ORF 7 showed high similarity to various organic acid-CoA ligases. The deduced N-terminal amino acid sequence was identical to that determined by Edman degradation, with one exception. At position 17, a mixture of L (ORF 7I) and Q (ORF 7II) was to be expected. Since in position 18 another Q was present and the type I operon seemed to be less abundantly expressed than the type II operon, the Q signal may have overlain the L signal at position 17. A potential AMP-binding region typical for CoA ligases was found in the derived gene products between aa 197 and 208 (Table 3).
ORF 8 (abmH).
ORF 8 codes for a putative 41 kDa-substrate binding protein of an ABC transporter system. Curiously, the genes for the other components of such a transporter system were not found downstream of ORF 8. Rather, in the downstream regions of the type I and II gene clusters (about 1.5 kb sequenced), a possible ORF coding for another substrate binding protein of an ABC transporter system, and oriented in the opposite direction, was detected.
Possible promoter regions and primer extension experiments.
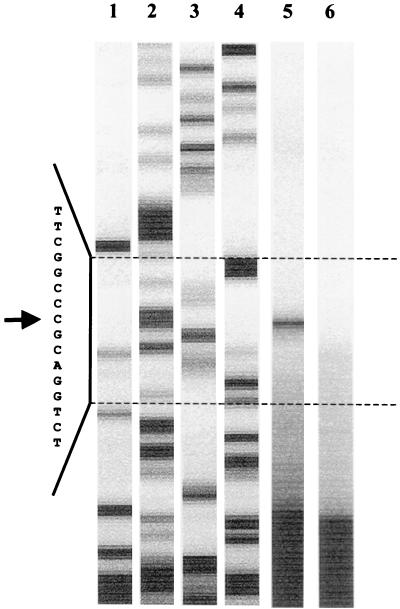
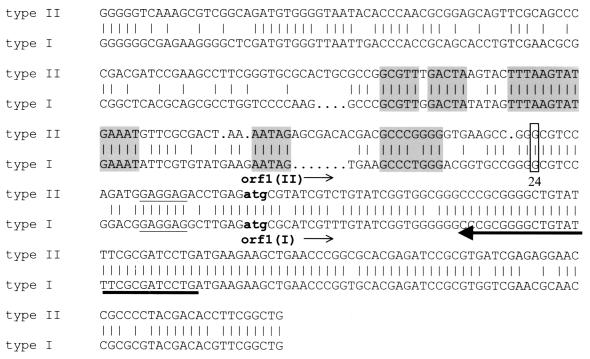
A probable transcription start upstream of ORF 1 was identified via primer extension using a primer derived from the common 5′ end of ORF 1 of types I and II. Total RNA was isolated from cells grown aerobically on 2-aminobenzoate and used for primer extension assays. As a control, RNA from cells grown anaerobically with benzoate and nitrate was used. The cDNA products were analyzed on a sequencing gel. Because of the significant differences between the upstream regions of type I and II gene clusters, two distinct signals were expected. Surprisingly, only one signal was detected 24 bp upstream of the translation start of ORF 1, indicating a conserved transcription start in the two gene clusters. Figure 7 shows the experiment with the type II operon. The same transcription start was observed in sequencing reactions with the type I operon (data not shown). The regions of the observed mRNA start sites are indeed highly similar in the two gene clusters. Furthermore, several conserved regions were also found for both gene clusters in the assumed −10 and −35 regions and in regions further upstream, which may represent promoter and operator box consensus sequences (Fig. 8).
FIG. 7.
Mapping of the mRNA starts of the 2-aminobenzoate catabolic operons by primer extension. By using a common primer in the experiment, only one common transcription start was observed for the type I and II operons. Lanes 1 to 4, sequencing reaction with the type II operon (A, C, G, and T, respectively); lane 5, primer extension reaction with RNA from cells grown aerobically with 2-aminobenzoate; lane 6, negative control with RNA from cells grown anaerobically with benzoate and nitrate. Shown is the gel-like representation produced by the ALFexpress sequencer. Letters and arrow (indicating the transcription start site) represent the reverse sequence complementary to the coding sequence (see Fig. 8).
FIG. 8.
Alignment of the 5′ regions of ORF 1II (upper lines) and type I operons (lower lines). Conserved regions, which may indicate promoter or regulator sites, are shaded. Lowercase letters (atg), translation start codons. Underlined letters, possible ribosome binding sequence. The mapped transcription starts are boxed. Arrow points to the location of the primer used for the primer extension experiment.
Expression of the gene clusters as operons.
RT-PCR was carried out to demonstrate that the 8 genes of each gene cluster are transcribed into one mRNA product and are expressed coordinately. A cDNA library of total RNA from cells grown aerobically on 2-aminobenzoate was prepared and used in PCRs. Primers were derived from the coding and/or intergenic regions of each gene, and PCR-fragments covering the intergenic regions between the respective ORFs were amplified. It could be confirmed that all 8 ORFs were expressed as an operon, referred to as the abm operon (Fig. 9).
FIG. 9.
Demonstration of the expression of abm gene clusters as operons. RT-PCR was performed using primers derived from the 3′ part of one gene (forward primer) and the 5′ part of the following gene (reverse primer), or from intergenic regions between two ORFs. Most of the primers were derived from sequences that were identical in the two gene clusters. The amplified PCR products included the intergenic regions between two ORFs and therefore demonstrate their coordinated transcription into one mRNA product. Positions of the fragments are shown in Fig. 2. Lanes 1 and 12, molecular standards; lane 2, abmA/B(I); lane 3, abmA/B(II); lane 4, abmB/C; lane 5, abmB/D(I); lane 6, abmD/interEF(II); lane 7, interEF/interFG(II); lane 8, abmF/G; lane 9, abmG/H; lane 10, negative control, as indicated in Fig. 2 (nc); lane 11, negative control, without cDNA.
DISCUSSION
β-Oxidation involved in 2-aminobenzoate metabolism.
The aerobic metabolism of 2-aminobenzoate in A. evansii proceeds via CoA thioester intermediates. The first two enzymes and intermediates of this pathway have been characterized, whereas the subsequent steps are unknown. Here we show that two very similar clusters of eight genes code for proteins that are most likely involved in this metabolism (Fig. 2; Table 2). Besides the genes coding for the first two enzymes of the pathway, three genes encoding enzymes involved in a β-oxidation-like pathway were found. This suggests that the product of ACMR is further metabolized by β-oxidation involving three additional pathway-specific enzymes (Fig. 1). One of these enzymes, encoded by abmC (a putative enoyl-CoA hydratase/isomerase), may be involved in hydrolytic ring opening using a putative alicyclic 3-oxoacyl-CoA intermediate. Some members of the enoyl-CoA hydratase (crotonase) family also function as hydrolases acting on 3-oxoacyl-CoA compounds. These enzymes cleave the carbon-carbon bond between C-2 and C-3 hydrolytically, releasing C-3 as –COOH (16). The other gene products encoded by the gene clusters include two proteins that are likely to be involved in regulation, and one potential substrate binding protein of an ABC transport system was found (Table 2).
Discrepancies with previous cloning and sequencing results.
In a previous attempt to clone and sequence the genes involved in aerobic 2-aminobenzoate metabolism, oligonucleotides derived from N-terminal amino acid sequences of the first two enzymes of the pathway were used for screening a gene library. The experiments indicated that the genes for ACMR and 2-aminobenzoate-CoA-ligase were carried on a plasmid (2). The putative genes for the CoA ligase and ACMR were sequenced, together with other ORFs of unknown function which showed low similarity to viral proteins. It is now obvious that the previously published sequences were incorrect; due to N-terminal similarity with other proteins a false DNA fragment may have been cloned and sequenced. The apparent localization of those sequences on a plasmid, as well as the similarity of the derived amino acid sequences to those of various phage proteins, suggests that a nonfunctional prophage was unintentionally sequenced.
Duplicated gene cluster and indication for operon structure.
Surprisingly, the gene cluster containing the genes for 2-aminobenzoate metabolism exists in duplicate, with minor differences in the coding regions and major differences in the intergenic regions. Contamination of the cultures or gene libraries is unlikely, although these possibilities were seriously considered. All our experiments suggest that both clusters are located on a chromosome, although large plasmids cannot be ruled out, and that they are expressed simultaneously during aerobic growth with 2-aminobenzoate. The 8 ORFs of each gene cluster were shown to form an operon (Fig. 9). The exact locations of the two operons and the distance between them remain to be determined.
Duplicated operons are rare in bacteria. Known examples are the tfd gene cluster for chlorophenol and chlorocatechol metabolism (21) and the cbd gene clusters coding for autotrophic CO2 fixation in Ralstonia eutropha (formerly Alcaligenes eutrophus) (32). However, unlike the gene clusters responsible for aerobic 2-aminobenzoate metabolism, the tfd gene clusters did not show a conserved organization of the ORFs, and the percentage of similar amino acids was significantly lower (15 to 62%). Furthermore, the tfd gene clusters are located on a common catabolic plasmid and vary in their G+C content. Therefore, these clusters are considered to be of independent evolutionary origin rather than to be due to a recent duplication incident. In contrast, the cbd genes, which code for several Calvin cycle enzymes, are duplicated and clustered within two very similar operons. One copy is localized on the chromosome, the other on a megaplasmid. The two gene clusters show the same organization of ORFs, and sequencing has revealed approximately 93% base identity. The situation of the 2-aminobenzoate metabolic gene clusters reported here is similar to that of the cbd genes, with the exception that both copies are probably located on the chromosome.
Several experimental results corroborate the existence of duplicated and slightly varying gene clusters. (i) Southern hybridization experiments showed more bands than would be expected if only one copy were present (data not shown). (ii) When varying intergenic regions were used for deducing specific primers, two copies of the genes were revealed by PCR experiments. (iii) PCR with DNA prepared from several cultures derived from single colonies, as well as colony PCR, always resulted in two variants of amplified sequences (Fig. 4). (iv) N-terminal amino acid sequencing of ACMR revealed 2 aa each at positions 25 and 36, which could be attributed to the two different gene products expected from the amino acid sequences encoded by the two operons (Table 1). (v) RT-PCR supported the existence and simultaneous expression of the two gene clusters (Fig. 3) and also corroborated the expression of both clusters as operons (Fig. 9).
Possible explanation for observed protein heterogeneity.
The observed duplication of the genes provides possible explanations for previous odd biochemical results (12). The supposedly homodimeric ACMR (170 kDa) showed only one N-terminal amino acid sequence (24 aa tested [Table 1]) but migrated as a double band on native polyacrylamide gel electrophoresis (PAGE) gels, and the protein eluted in three similarly active fractions from a MonoQ column. When analyzed by native PAGE, one MonoQ fraction contained only the slower-migrating protein, the second fraction contained only the faster-migrating protein, and the third fraction contained a mixture of both proteins. Repeat chromatography of the mixed fraction again resulted in splitting into three active fractions. This suggested the presence of three equally active isoforms of the enzyme, αα, αα′, and α′α′. The subunits of the αα′ isoform can apparently recombine to give the αα and α′α′ forms. These results may now be explained by assuming that the α subunit corresponds to one of the two variants of ORF 1 and the α′ subunit corresponds to the other variant. The calculated amino acid compositions, sizes, and IEP of the two variants of ORF 1 (ACMR types I and II) differ slightly (masses of 86.9 and 87.1 kDa, and IEP of 6.0 and 6.2, respectively). These slight differences may cause the observed differences in electrophoretic mobility on native PAGE gels, as well as the different behaviors of the isoforms during chromatography on MonoQ. This interpretation implies that both types of ORF 1 are expressed, as was confirmed by the detection of both variants by N-terminal amino acid sequencing and RT-PCR experiments.
Structure of ACMR.
All previous studies of the structure and function of ACMR have indicated that it is a bifunctional enzyme with two catalytic centers, a monooxygenase (ACM) and a reductase (ACR) center (24). With limiting concentrations of NADH, the enzyme catalyzed only the hydroxylation of the substrate. NADH-dependent reduction of N-ethylmaleimide to N-ethylsuccinimide was a convenient side reaction used for the enzyme assay. For both reactions, monooxygenation and reduction, enzyme-bound FAD is required, albeit with different roles. For hydroxylation, a 4a-FAD-hydroperoxide is the key intermediate; for reduction, FADH2 transfers the hydride equivalent from NADH (17).
This model is fully supported by sequence information indicating that the protein indeed is a fusion protein consisting of an N-terminal monooxygenase (ACM) half and a C-terminal reductase (ACR) half (Fig. 5). These halves are linked by a spacer region showing no homologies to known sequences in databases. We assume that the genes coding for the monooxygenase and reductase functions were initially separated and that a mutation caused the loss of the stop codon of the original monooxygenase gene, leading to the observed gene fusion. The original intergenic region between the monooxygenase and reductase genes would now form the spacer region between the domains. This fusion event apparently occurred prior to the duplication of the operons, since it is conserved in both variants. A similar event may have caused the loss of the intergenic region between ORF 4 and ORF 5 (Fig. 2). The intergenic region was apparently added to the C terminus of ORF 4, resulting in quite different C termini for ORF 4 in the type I and II operons.
Several possible nucleotide binding sites were identified in both domains of ACMR (Fig. 6). The FAD-content of the functional dimer was determined as 1.5 to 3.2. Therefore, the enzyme contains at least 2 mol of FAD/mol of native enzyme (12), but a content of 4 mol of FAD/mol of native enzyme cannot be excluded. Two moles of NADH is needed to metabolize 1 mol of 2-aminobenzoate (Fig. 1). The functions of those monooxygenases in the databases that are most similar to the N-terminal ACM-domain of ACMR are not known; the closest known oxygenase is salicylate 1-monooxygenase (27). Interestingly, the reductase domain of ACMR shows high similarity to the E. coli “N-ethylmaleimide reductase,” which catalyzes the same side reaction as ACR (reduction of N-ethylmaleimide) but is of unknown physiological function. All these reductases show similarity to old yellow enzyme (EC 1.6.99.1), the first flavoenzyme studied, whose function in yeast is still at issue (Fig. 5). Therefore, ACR might be regarded as a member of the old-yellow-enzyme protein family (20).
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (Bonn, Germany) and the Fonds der Chemischen Industrie (Frankfurt, Germany).
Thanks are due to Hermann Schägger (Frankfurt, Germany) for N-terminal amino acid sequencing of ACMR and to Juliane Alt-Mörbe (Freiburg, Germany) for part of the DNA sequencing. Thanks are also due to Christian Völzing for his contribution during the earlier stages of this work.
REFERENCES
- 1.Aiba H, Adhya S, deCombrugge B. Evidence for two functional gal promoters in intact Escherichia coli. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Altenschmidt U, Eckerskorn C, Fuchs G. Evidence that enzymes of a novel aerobic 2-aminobenzoate metabolism in denitrifying Pseudomonas are coded on a small plasmid. Eur J Biochem. 1990;194:647–653. doi: 10.1111/j.1432-1033.1990.tb15664.x. [DOI] [PubMed] [Google Scholar]
- 3.Altenschmidt U, Oswald B, Fuchs G. Purification and characterization of benzoyl-CoA ligase and 2-aminobenzoyl-CoA ligase from a denitrifying Pseudomonassp. J Bacteriol. 1991;173:5494–5501. doi: 10.1128/jb.173.17.5494-5501.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altenschmidt U, Oswald B, Steiner E, Herrmann H, Fuchs G. New aerobic benzoate oxidation pathway via benzoyl-coenzyme A and 3-hydroxybenzoyl-coenzyme A in a denitrifying Pseudomonassp. J Bacteriol. 1993;175:4851–4858. doi: 10.1128/jb.175.15.4851-4858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altenschmidt U, Fuchs G. Novel aerobic 2-aminobenzoate metabolism. Purification and characterization of 2-aminobenzoate-CoA ligase, localization of the gene on a 8-kbp plasmid, and cloning and sequencing of the gene from a denitrifying Pseudomonassp. Eur J Biochem. 1992;205:721–727. doi: 10.1111/j.1432-1033.1992.tb16835.x. [DOI] [PubMed] [Google Scholar]
- 6.Anders H-J, Kaetzke A, Kämpfer P, Ludwig W, Fuchs G. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K172 and KB740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the β subclass of the Proteobacteria. Int J Syst Bacteriol. 1995;45:327–333. doi: 10.1099/00207713-45-2-327. [DOI] [PubMed] [Google Scholar]
- 7.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987. [Google Scholar]
- 8.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boorstein W R, Craig E A. Primer extension analysis from RNA. Methods Enzymol. 1989;180:347–369. doi: 10.1016/0076-6879(89)80111-9. [DOI] [PubMed] [Google Scholar]
- 10.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Braun K, Gibson D. Anaerobic degradation of 2-aminobenzoate (anthranilic acid) by denitrifying bacteria. Appl Environ Microbiol. 1984;48:102–107. doi: 10.1128/aem.48.1.102-107.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buder R, Fuchs G. 2-Aminobenzoyl-CoA monooxygenase/reductase, a novel type of flavoenzyme: purification and some properties of the enzyme. Eur J Biochem. 1989;185:629–635. doi: 10.1111/j.1432-1033.1989.tb15159.x. [DOI] [PubMed] [Google Scholar]
- 13.Buder R, Ziegler K, Fuchs G, Langkau B, Ghisla S. 2-Aminobenzoyl-CoA monooxygenase/reductase, a novel type of flavoenzyme. Studies on the stoichiometry and the course of the reaction. Eur J Biochem. 1989;185:637–643. doi: 10.1111/j.1432-1033.1989.tb15160.x. [DOI] [PubMed] [Google Scholar]
- 14.Dagley S. Microbial metabolism of aromatic compounds, p 483–505. In: Moo-Young M, editor. Comprehensive bio/technology. Oxford, United Kingdom: Pergamon Press; 1985. [Google Scholar]
- 15.Davies L. Basic methods in molecular biology. New York, N.Y: Elsevier; 1986. [Google Scholar]
- 16.Haller T, Buckel T, Rétey J, Gertl J A. Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli. Biochemistry. 2000;39:4622–4629. doi: 10.1021/bi992888d. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann S, Hultschig C, Eisenreich W, Fuchs G, Bacher A, Ghisla S. NIH shift in flavin-dependent monooxygenation: mechanistic studies with 2-aminobenzoyl-CoA monooxygenase/reductase. Proc Natl Acad Sci USA. 1999;96:7831–7836. doi: 10.1073/pnas.96.14.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harwood C S, Burchhardt G, Herrmann H, Fuchs G. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev. 1999;22:439–458. [Google Scholar]
- 19.Heider J, Fuchs G. Anaerobic metabolism of aromatic compounds. Eur J Biochem. 1997;243:577–596. doi: 10.1111/j.1432-1033.1997.00577.x. [DOI] [PubMed] [Google Scholar]
- 20.Karplus P A, Fox K M, Massey V. Flavoprotein structure and mechanism. 8. Structure-function relations for old yellow enzyme. FASEB J. 1995;9:1518–1526. doi: 10.1096/fasebj.9.15.8529830. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli C M, Leveau J H, Zehnder A, van der Meer J R. Characterization of a second tfd gene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in Ralstonia eutrophaJM134(pJP4) J Bacteriol. 2000;182:4165–4172. doi: 10.1128/jb.182.15.4165-4172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langkau B, Ghisla S. Kinetic and mechanistic studies on the reactions of 2-aminobenzoyl-CoA monooxygenase/reductase. Eur J Biochem. 1995;230:686–697. doi: 10.1111/j.1432-1033.1995.0686h.x. [DOI] [PubMed] [Google Scholar]
- 23.Langkau B, Ghisla S, Buder R, Ziegler K, Fuchs G. 2-Aminobenzoyl-CoA monooxygenase/reductase, a novel type of flavoenzyme: identification of the reaction products. Eur J Biochem. 1990;191:365–371. doi: 10.1111/j.1432-1033.1990.tb19131.x. [DOI] [PubMed] [Google Scholar]
- 24.Langkau B, Vock P, Massey V, Fuchs G, Ghisla S. 2-Aminobenzoyl-CoA monooxygenase/reductase: evidence for two distinct loci catalyzing substrate monooxygenation and hydrogenation. Eur J Biochem. 1995;230:676–685. [PubMed] [Google Scholar]
- 25.Leuthner B, Heider J. Anaerobic toluene catabolism of Thauera aromatica: the bbsoperon codes for enzymes of β-oxidation of the intermediate benzylsuccinate. J Bacteriol. 2000;182:272–277. doi: 10.1128/jb.182.2.272-277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lochmeyer C, Koch J, Fuchs G. Anaerobic degradation of 2-aminobenzoate via benzoyl-CoA and cyclohex-1-ene-carboxyl-CoA in a denitrifying bacterium. J Bacteriol. 1992;174:3621–3628. doi: 10.1128/jb.174.11.3621-3628.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massey V, Hemmerich P. Flavin and pteridine monooxygenases. In: Boyer P D, editor. The enzymes. 3rd ed. XII. New York, N.Y: Academic Press; 1975. pp. 191–252. [Google Scholar]
- 28.Mohamed M. Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii. J Bacteriol. 2000;182:286–294. doi: 10.1128/jb.182.2.286-294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Torres R, Bruice T. Theoretical investigation of the [1,2]-sigmatropic hydrogen migration in the mechanism of oxidation of 2-aminobenzoyl-CoA by 2-aminobenzoyl-CoA monooxygenase/reductase. Proc Natl Acad Sci USA. 1999;96:14748–14752. doi: 10.1073/pnas.96.26.14748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tschech A, Fuchs G. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch Microbiol. 1987;148:213–217. doi: 10.1007/BF00414814. [DOI] [PubMed] [Google Scholar]
- 32.Windhövel U, Bowien B. On the operon structure of the cfx gene clusters in Alcaligenes eutrophus. Arch Microbiol. 1990;154:85–91. doi: 10.1007/BF00249183. [DOI] [PubMed] [Google Scholar]
- 33.Ziegler K, Buder R, Winter J, Fuchs G. Activation of aromatic acids and aerobic 2-aminobenzoate metabolism in a denitrifying Pseudomonasstrain. Arch Microbiol. 1989;151:171–176. [Google Scholar]