Abstract
Portal tracts are key intrahepatic structures where leukocytes accumulate during immune responses. They contain the blood inflow, which includes portal blood from the gut, and lymphatic and biliary outflow of the liver, and as such represent a key interface for potential pathogen entry to the liver. Myeloid cells residing in the interstitium of the portal tract might play an important role in the surveillance or prevention of pathogen dissemination; however, the exact composition and localization of this population has not been explored fully. Our in‐depth characterization of portal tract myeloid cells revealed that in addition to T lymphocytes, portal tracts contain a heterogeneous population of MHCIIhigh myeloid cells with potential antigen presenting cell (APC) function. These include a previously unreported subset of CSF1R‐dependent CX3CR1+ macrophages that phenotypically and morphologically resemble liver capsular macrophages, as well as the two main dendritic cell subsets (cDC1 and cDC2). These cells are not randomly distributed, but each subset forms interconnected networks intertwined with specific components of the portal tract. The CX3CR1+ cells were preferentially detected along the outer border of the portal tracts, and also in the portal interstitium adjacent to the portal vein, bile duct, lymphatic vessels and hepatic artery. cDC1s abounded along the lymphatic vessels, while cDC2s mostly surrounded the biliary tree. The specific distributions of these discrete subsets predict that they may serve distinct functions in this compartment. Overall, our findings suggest that portal tracts and their embedded cellular networks of myeloid cells form a distinctive lymphoid compartment in the liver that has the potential to orchestrate immune responses in this organ.
Keywords: cDC, Clec4f, intrahepatic dendritic cell, intrahepatic immunity, monocyte‐derived macrophage, portal triad, portal vein
This study provides an in‐depth characterisation of liver myeloid cells and reveals that most MHCIIhigh myeloid cells are localised around portal tracts. Our analysis reveals that these cells include a population that we identified as a previously unreported subset of CSF1R‐dependent CX3CR1+ macrophages that phenotypically and morphologically resemble liver capsular macrophages, type 1 conventional dendritic cells (cDC1) and type 2 dendritic cells (cDC2). These cells are not randomly distributed, but each subset forms interconnected networks intertwined with specific components of the portal tract suggesting that they may serve distinct functions in this compartment that contribute to orchestrate immune responses in this organ.
INTRODUCTION
The liver possesses unique immune properties that set it apart from other organs. In addition to its tolerance‐inducing properties observed in experimental models and various clinical settings, effective immune defense against pathogens occurs in this organ. While T cell‐mediated immunity is critical for eliminating hepatotropic pathogens and for long‐term antigen‐specific protection, recent studies have highlighted the key role of macrophages in preventing pathogen dissemination. The liver harbors two known distinct resident macrophage populations strategically positioned at key sites of pathogen entry, allowing them to deliver rapid optimal responses that limits dissemination. 1 , 2 , 3 Kupffer cells (KC), the most populous and well‐studied liver‐resident macrophage, are located in the sinusoidal lumen, where they are exposed to blood‐borne pathogens. Their uneven distribution and higher density closer to portal tracts has been reported to be optimized for maximal pathogen clearance. 1 We have recently characterized a second liver‐resident macrophage subset that occupies the hepatic capsule. 2 These liver capsular macrophages (LCMs) are phenotypically and ontogenetically distinct from KCs as they expressed CX3CR1 and have a monocytic origin. One of their key roles is to prevent the dissemination of bacteria from the peritoneal cavity through the liver capsule by attracting neutrophils that mediate pathogen clearance. 2 The liver is also potentially exposed to gut‐derived microbiota and pathogens through the biliary tree that carries bile secreted by hepatocytes into the gut lumen. 4 The biliary tree radiates through the liver within portal tracts, a key hepatic structure comprising juxtaposed branches derived from the portal vein, hepatic artery and biliary tree. As an interface between the liver parenchyma and both the portal circulation and the biliary tree, the portal tracts therefore also represent an important potential pathogen entry point into the liver.
Surprisingly, while several studies have shown that portal tracts of the steady state liver contain a significant immune cell population of diverse subtypes, its exact spatial composition has not been characterized. We therefore sought to characterize this compartment in depth, revealing that it carries a rich network of specialized macrophages and antigen presenting cells (APCs) throughout the liver, with the potential to orchestrate immune responses within this organ.
RESULTS
Portal tracts channel embedded liver lymphatics and harbor a heterogeneous population of MHCIIhigh cells
To better visualize cells in portal tracts, we optimized an immunofluorescence confocal microscopy (ICM) approach and imaged the liver of reporter Alb‐Cre x ROSA26 mT/mG transgenic mice, in which hepatocytes express EGFP while all other cells express tdTomato. Thick vibratome slices (150 μm) of in situ fixed liver were stained for cytokeratin 19 (CK19) expression to identify bile duct epithelial cells (BECs), which are located exclusively within portal tracts. CK19 staining revealed large portal tracts throughout the mouse liver branching out into smaller structures (Figure 1a). The 3D microscopic structure of a typical portal tract was reconstituted by stacking a series of high magnification cross‐section views, illustrating its three characteristic components derived from the portal vein (PV), bile duct (BD) and hepatic artery (HA) (Figure 1b, left image). Small vessels exhibiting higher levels of tdTomato fluorescence observed surrounding the CK19+ bile duct represent peri‐bile ductular capillaries supplying the bile duct and portal interstitium (Figure 1b ROI, arrowheads). To highlight lymphatic vessels, LYVE‐1 staining was performed to identify lymphatic endothelial cells (LECs) (Figure 1c, d). The juxtaposition of lymphatics with the blood vasculature in portal tracts was visualized in greater 3‐dimensional detail by imaging longitudinal portal tract cross sections (Figure 1d). Large LYVE‐1high lymphatic vessels followed the portal tract as it traverses the hepatic parenchyma, collecting lymph fluid that drains from the hepatic sinusoids via smaller diameter vessels (Figure 1d). Confirming the view that portal tracts serve as a conduit for intrahepatic lymphocytes trafficking through the liver, TCRβ+ T lymphocytes were readily observed within the portal interstitium, making intimate contacts with other portal interstitial cells, as well as the various vascular components of the portal tract (Figure 1e arrow; Figure 1f).
Figure 1.
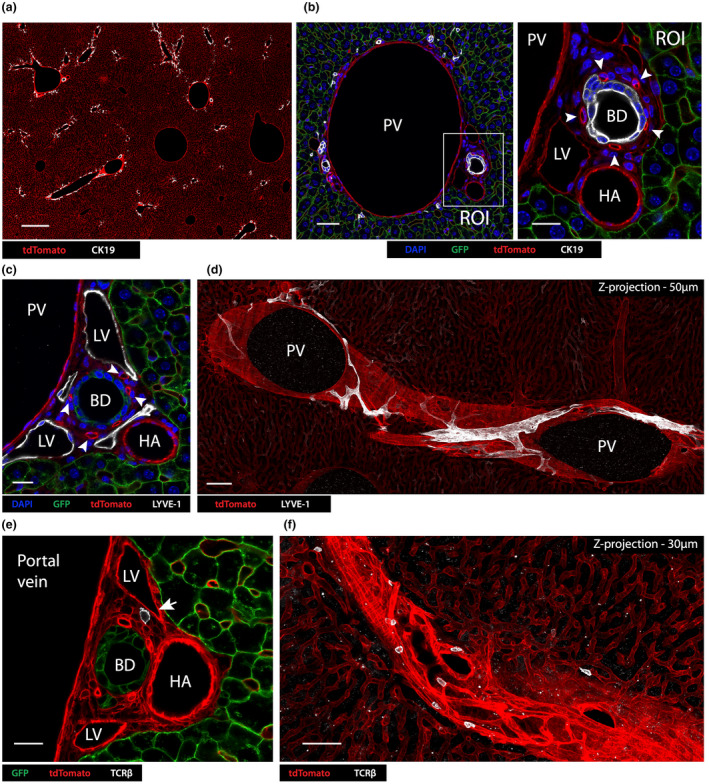
Portal tracts contain an embedded lymphatic vasculature and T lymphocytes. (a) Confocal immunofluorescence image at low magnification (scale bar 400 μm) of an Alb‐Cre x ROSA mT/mG mouse liver expressing tdTomato (red) in all non‐hepatocyte cells, stained with anti‐CK19 (white) to identify BECs in portal tracts. GFP expressed on hepatocytes has been excluded from this image. (b) High magnification view of a portal tract with a large portal vein (scale bar 50 μm); A region of interest (ROI; right panel, scale bar 20 μm) shows a higher magnification view of the bile duct (BD) and hepatic artery (HA). This area also includes a lymphatic vessel (LV). Arrows indicate the bile ductular capillary network. (c) Confocal immunofluorescence image of Alb‐Cre x ROSA mT/mG mouse portal tract stained for LYVE‐1 to detect LVs (tdTomato – red; GFP – green; DAPI – blue; LYVE‐1 – white; scale bar 20 μm). (d) Confocal 3D image by 2‐dimensional z‐projection (50 μm) of an Alb‐Cre x ROSA mT/mG mouse liver at low power (scale bar 100 μm) stained for LYVE‐1 (white). GFP+ hepatocytes were excluded to reveal the vasculature. (e) Confocal image showing a high‐power view of an Alb‐Cre x ROSA mT/mG mouse liver portal tract stained with TCRβ to identify T cells (scale bar = 20 μm; hepatocytes – green; TCRβ – white; other cells – red). (f) 2‐Dimensional z‐projection (30 μm sections, scale bar = 50 μm) showing a longitudinal section of an Alb‐Cre x ROSA mT/mG portal tract. White – TCRβ+ T cells. Images are representative of two independent experiments with n = 4 mice.
As the liver capsule and hepatic sinusoids are surveyed by specific liver‐resident macrophage subsets, we sought to determine whether portal tracts contain myeloid cell subsets potentially surveying this compartment. To assess the steady state distribution of professional APCs throughout the whole liver, Alb‐Cre x R26mT/mG mouse livers were stained for MHCII and imaged using immunofluorescence confocal microscopy. Interestingly, while a few isolated MHCIIhigh cells were detected throughout the liver parenchyma/sinusoidal compartment, most were associated with portal tracts (Figure 2a), where they were detected in the interstitial perivascular area surrounding and in‐between the portal vein, hepatic artery, bile duct, peri‐bile ductular capillaries and lymphatic vessels (Figure 2b). MHCIIhigh cells surrounded the portal vasculature and their dendrites extended in close apposition to the vessels (Figure 2c). Many distinct cells were observed interacting with other MHCIIhigh cells (Figure 2c, ROI). Portal tract MHCIIhigh cells displayed variable morphology. While some cells were elongated and had long dendrites, resembling CX3CR1+ liver capsular macrophages (LCMs), others had a less extended and more compact body (Figure 2c, ROI). These two subsets could be distinguished through their different expression of F4/80 (Figure 2d). “LCM‐like” cells expressed F4/80 (ROI1), while smaller cells were F4/80− (ROI2). These findings suggest that portal tracts represent a unique compartment harboring a rich and heterogeneous population of MHCIIhigh myeloid cells made of distinct subsets and in close proximity to lymph and blood circulations and bile ducts.
Figure 2.
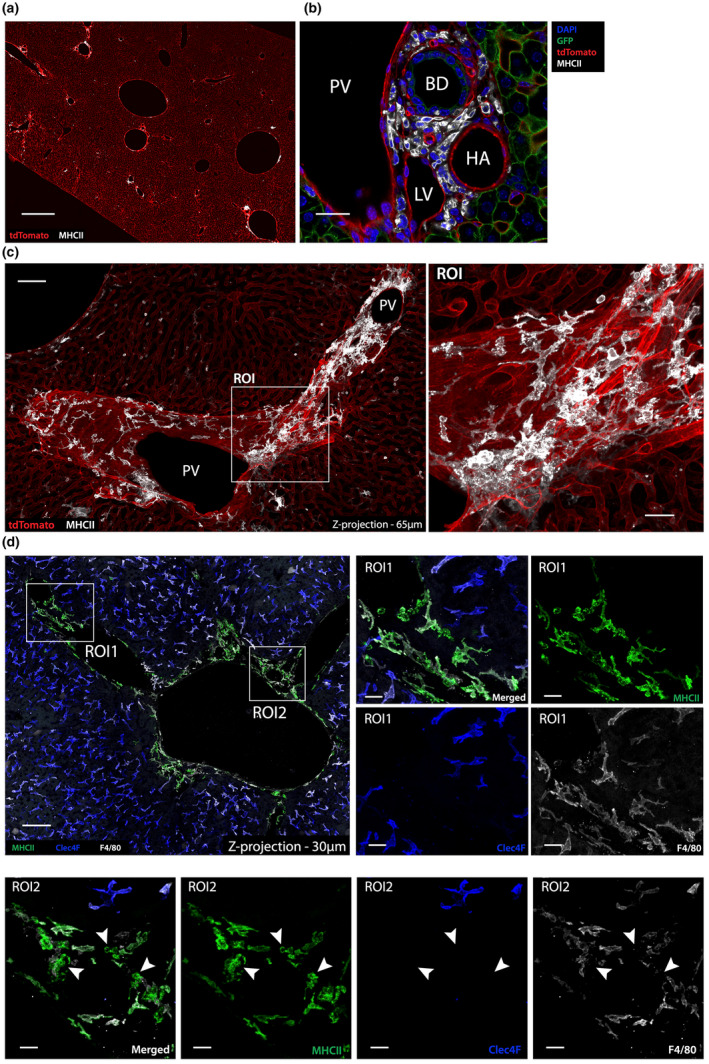
MHCIIhigh myeloid cells are enriched within portal tracts. (a) Confocal image showing a low power view (scale bar = 500 μm) of an Alb‐Cre x ROSA mT/mG mouse liver stained for MHCII (white). GFP expression by hepatocytes was excluded. (b) High‐power view of a portal tract in an Alb‐Cre x ROSA mT/mG mouse liver showing that MHCIIhigh cells are located in the portal tract interstitium between the bile duct (BD), hepatic artery (HA), lymphatic vessels (LV) and portal vein (PV) (DAPI – blue; GFP – green; tdTomato – red; MHCII – white; scale bar 30 μm). (c) 2‐Dimensional z‐projection (65 μm) of a series of confocal images showing a longitudinal portal tract section of an Alb‐Cre x ROSA mT/mG mouse liver (GFP+ hepatocytes excluded), revealing the large network formed by MHCIIhigh cells (white) closely associated with the portal tract vasculature (scale bar 75 μm). (d) Left panel: 2‐dimensional z‐projection (30 μm) of a series of confocal images of a C57BL/6 mouse liver portal tract stained with MHCII and various macrophage markers. Middle and right panels: ROI1 from left panel, showing a portal tract region with MHCIIhigh cells (green) that co‐express F4/80 (white) but lacked CLEC4F (blue), suggesting that they are macrophages distinct from KCs. ROI2 (scale bar 10 μm) shows a portal tract region with MHCIIhigh cells (green) that do not express F4/80 or CLEC4F (arrowheads), suggesting that non‐macrophage MHCIIhigh cells, potentially DCs, are also present within steady state portal tracts. Images are representative of n = 4 mice from two independent experiments.
Steady state portal tracts contain a population of CSF1R signaling‐dependent MHCIIhighCX3CR1 + cells displaying monocyte‐derived macrophage phenotype
We first sought to characterize the F4/80+ cell subset associated with portal tracts. These cells lacked the KC marker CLEC4F and expressed higher levels of MHCII (Figure 2d), indicating they were not KCs. Their LCM‐like morphology and F4/80 expression suggested that these cells were macrophages. Portal tract F4/80+ cells co‐expressed CD64 and GFP (CX3CR1) in the livers of Cx3cr1 +/gfp reporter mice (Figure 3a), providing further evidence in support of their macrophage lineage. 2 Expression of the endogenous CX3CR1 protein in these cells was confirmed by in vivo staining of C57BL/6 livers with a conjugated anti‐CX3CR1 antibody (data not shown). As LCMs are highly dependent on CSF1R signaling for survival, 2 , 5 we next assessed the CSF1R dependence of CX3CR1+ macrophages in portal tracts. Cx3cr1 +/gfp mice were treated with a non‐depleting anti‐CSF1R blocking mAb or isotype control mAb, and 4 days later the livers were stained and imaged by ICM (Figure 3b, c). Treatment depleted most LCMs as well as CX3CR1+ macrophages in portal tracts (Figure 3c, d), demonstrating that portal tract macrophages were, like LCMs, highly CSF1R dependent. In‐depth analysis of images revealed that most CX3CR1+ portal tract macrophages formed an interconnected cellular network that was largely associated with portal veins and the outer boundaries of the portal tract (Supplementary figure 1a–c). However, some of these macrophages were also detected in the portal tract interstitium, where they could be observed establishing intimate contacts with the lymphatic vasculature (Supplementary figure 1a, ROI), hepatic artery (Supplementary figure 1c, ROI), the bile duct (Supplementary figure 1b, ROI) and bile duct‐associated capillaries (Supplementary figure 1c, ROI).
Figure 3.
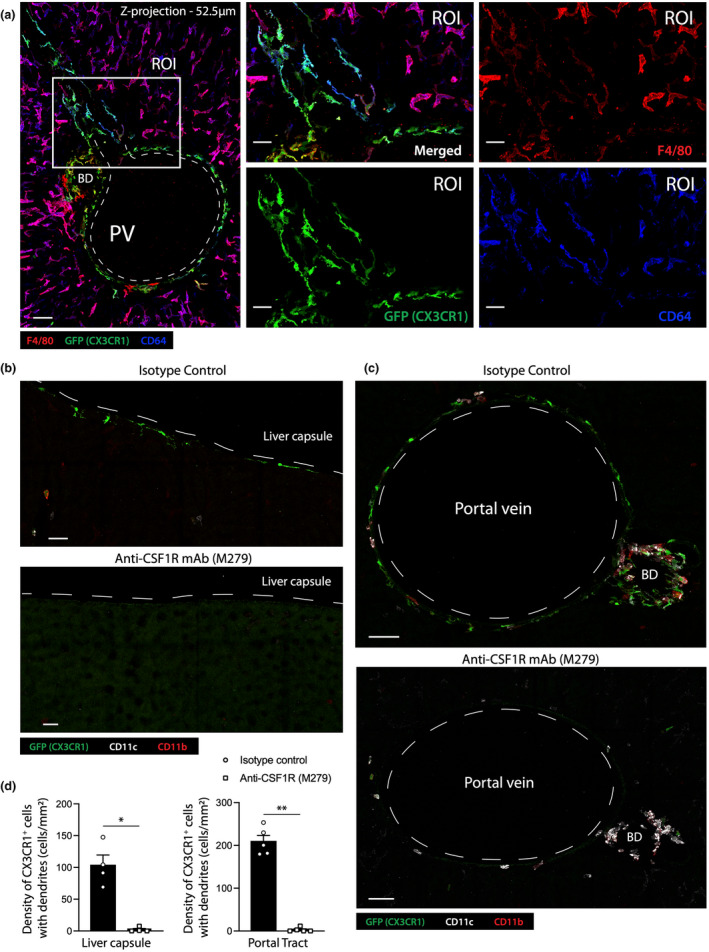
Portal tract CX3CR1+ macrophages are CSF‐1R signaling dependent. (a) Left panel: 2‐dimensional z‐projection (52.5 μm) of a series of confocal images showing a longitudinal cross section of a Cx3cr1 +/gfp mouse portal tract (F4/80 – red; CX3CR1 – green; CD64 – blue; scale bar 50 μm). An ROI from the left image was magnified to generate the four images on the right to highlight the phenotype and morphology of portal tract macrophages (scale bar 25 μm). (b, c) Confocal images of livers of C57BL/6 mice treated with anti‐isotype control, (b) or anti‐CSF1R blocking antibody (c) (top images, scale bar 20 μm; bottom images, scale bar 50 μm). (d) Quantification of the density of CX3CR1+ cells with dendrites in the liver capsule and portal tracts following treatment with anti‐CSF1R blocking antibody or isotype control. Data represent the mean ± SEM of at least n = 4 mice from two independent experiments. P‐values were calculated by the Student's t‐test. *P < 0.05, **P < 0.01. All images are representative of n = 4 mice from two independent experiments.
Steady state portal tracts are enriched in XCR1 + cDC1s and CD11b + cDC2s
Although CSF1R‐blocking treatment effectively depleted F4/80+ subsets of CX3CR1+ macrophages, some MHCIIhigh cells remained in the portal tracts (Figure 3c), likely corresponding to the MHCIIhighF4/80− cell subset described above (Figure 2d). The phenotype of these cells, combined with their CSF1R‐independence, suggested they were likely DCs. To confirm their DC lineage, we investigated the expression of CD11c, a common cDC marker generally expressed by all DCs, 6 by assessing the expression of GFP in the livers of steady state Itgax‐Cre.GFP reporter mice stained to identify macrophages. In addition to CX3CR1+CD64+ macrophages, the portal tracts contained CX3CR1− cells that lacked CD64, but expressed CD11c (Supplementary figure 2), indicating that they were most likely derived from the DC lineage. DCs are typically subdivided into two distinct subsets with different functions: type 1 conventional DCs (cDC1s) that specialize in cross‐presentation of cell‐associated antigens to CD8+ T cells, and type 2 conventional DCs (cDC2s) that cannot cross‐present antigen but are more specialized in the presentation of exogenous antigens to CD4+ T cells. 7 These two subsets can be distinguished by their expression of CD11b: while cDC1s lack CD11b, most cDC2s express CD11b. To assess the cDC types contained in the portal tracts, the livers were co‐stained for CD11c, CD11b and MHCII. Portal tracts contained both CD11c+CD11b−, likely to be cDC1s (Figure 4a, arrowheads), and CD11c+CD11b+, most likely cDC2s (Figure 4a, arrows). To confirm that MHCIIhiCD11c+CD11b− were cDC1s, we imaged the portal tracts of two reporter lines, Xcr1 +/venus and Cd207‐EGFP, after staining for CD11b and CD11c. XCR1 is a chemokine receptor expressed exclusively by cross‐presenting cDC1s but not by cDC2s. 8 Likewise, langerin (CD207) is a lectin expressed by some cDC1s but not expressed by cDC2s. All CD11c+CD11b− cells detected in the portal tracts of Xcr1 +/venus mouse livers expressed Venus (Figure 4b), while a significant proportion of CD11c+CD11b− cells expressed GFP in Cd207‐EGFP mice (Figure 4c). In contrast, MHCIIhiCD11c+CD11b+ cells did not express Venus or EGFP in Xcr1 +/venus or Cd207‐EGFP mice, respectively (Figure 4b, c). These findings provide additional evidence that MHCIIhighCD11c+CD11b− were cDC1s, while MHCIIhighCD11c+CD11b+ were cDC2s. Most portal tract XCR1+ cDC1 co‐expressed CD103 (integrin αE), a phenotype described for migratory cDC1s detected in most peripheral tissues (Figure 4d). Interestingly, CD8α, a marker generally detected in non‐migratory cDC1s residing in secondary lymphoid organs, 9 , 10 , 11 was also expressed by some XCR1+ cDC1s in portal tracts (Figure 4e). Conversely, further phenotypic analysis of portal tract cDC2s revealed that they expressed CD301a (CLEC10A) (Figure 4f), a marker identifying a subpopulation of pro‐inflammatory cDC2s, termed cDC2B, specialized in promoting Th1, Th2 and Th17 responses. 7 , 12 Collectively, these results indicate that MHCIIhigh cells in portal tracts are heterogenous and include CX3CR1+ monocytic macrophages as well as XCR1+ cDC1s and CD11b+ cDC2s.
Figure 4.
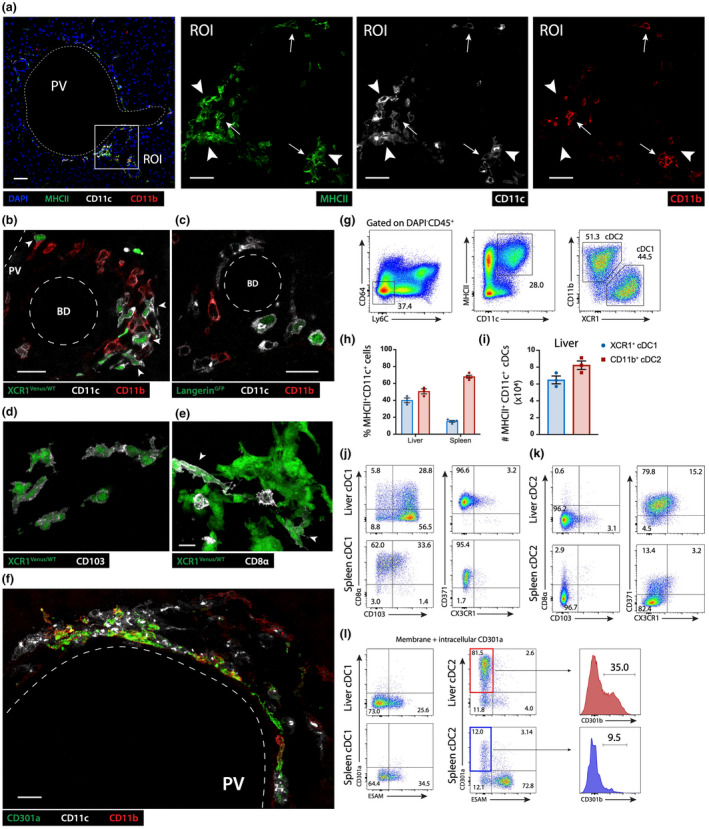
Portal tract MHCIIhigh APCs are a heterogenous population containing CX3CR1+ macrophages, XCR1+ cDC1 and CD301a+ cDC2B. (a) Left image: Confocal image of a C57BL/6 mouse portal tract (DAPI – blue; MHCII – green; CD11b – red; CD11c – white, scale bar 50 μm). A ROI was magnified to generate the three images on the right (arrowhead – CD11c+CD11b− cells; arrow – CD11c+CD11b+ cells; scale bars 20 μm). (b, c) Confocal images showing a high‐power view of a Xcr1 +/venus (b) or Cd207 gfp reporter mouse, (c) portal tract, Venus and GFP expression (green), respectively driven by the XCR1 or langerin promoters (BD – Bile duct; PV – portal vein; CD11b – red; CD11c – white; scale bars 20 μm). (d, e) High magnification confocal image of a Xcr1 +/venus reporter mouse portal tract, (d) CD103+ − white; (e) CD8α – white (arrowhead – Venus expressing, CD8α positive cell); (f) Confocal image of a C57BL/6 mouse portal tract showing CD301a+ CD11c+ cells identified as cDC2s (CD11b – red; CD11c – white; CD301a – green; scale bar 20 μm). (a–f) All images are representative of n = 4 mice from two independent experiments. (g–k) Flow cytometric analysis of the total number and phenotype of conventional DCs isolated from livers and spleens of steady state C57BL/6 mice. (g) Gating strategy used to identify cDC1 and cDC2 subsets. (h, i) Proportion and total number of cDC1s and cDC2s isolated from the liver or spleen. (j–l) Phenotypic analysis of cDC1s (j, l) and cDC2s (k, l) isolated from the liver and spleen. Data in h and i represent the mean ± SEM of n = 3 mice. All data and FACS plots are representative of at least n = 3 mice from two independent experiments. ESAM, endothelial cell‐selective adhesion molecule.
Phenotypic differences exist between hepatic cDCs and their splenic counterparts
XCR1+ cDC1 and CD11b+ cDC2 subsets from liver were compared with those from spleen via flow cytometric analysis. XCR1+ cDC1s represented approximately 40% of MHCIIhighCD11chigh cells in the liver and 20% in the spleen, while cDC2s made up approximately 50% of MHCIIhighCD11chigh cells in the liver and 70% in the spleen (Figure 4g, h). C57BL/6 mouse livers contained an average total number of ~7 × 104 XCR1+ cDC1s and ~8 × 104 CD11b+ cDC2s in the steady state (Figure 4i). Consistent with our imaging findings described above, most liver XCR1+ cDC1s expressed CD103, while 30% co‐expressed CD8α (Figure 4j). In contrast, the vast majority of XCR1+ cDC1s in the spleen were CD103− CD8α+ (Figure 4j). Consistent with the phenotype of splenic cDC1 and cDC2 described previously, 7 , 9 , 12 intrahepatic and splenic XCR1+ cDC1s expressed CD205 (DEC‐205) and CD371 (Clec12A) but lacked CD301b, CD301a or CX3CR1 (Figure 4j), while both intrahepatic and splenic CD11b+ cDC2s lacked CD103, CD8α, CD205 and CX3CR1 expression (Figure 4k). Approximately 75% of CD11b+ cDC2s in the liver expressed CD301a, and approximately 30% of CD301a+ cDC2s co‐expressed CD301b, identifying this population as cDC2B (Figure 4k; Supplementary figure 3). In contrast, only 5% of splenic cDC2s expressed CD301a (Figure 4j). Finally, the vast majority of intrahepatic cDC2s expressed CD371, while almost none of the splenic cDC2 expressed this marker. Conversely, while most splenic cDC2s expressed high levels of endothelial cell‐selective adhesion molecule (ESAM), most intrahepatic cDC2s did not express this marker (Figure 4j).
CX3CR1 + MHCIIhigh macrophages, XCR1 + cDC1s and CD301a + cDC2s form a continuous cellular network within portal tracts
To assess whether CX3CR1+ macrophages, XCR1+ cDC1s and CD301a+ cDC2s occupy a specific niche within the portal tract or were randomly positioned, we assessed their distribution in large volumes of liver tissue from Cx3cr1 +/gfp , Xcr1 +/Venus, and C57BL/6 mice stained with CD301a, following Ce3D clearing. 13 The vast majority of CX3CR1+CD64+ macrophages that were not associated with the liver capsule were closely associated with portal tracts (Figure 5a, e). A higher magnification view of a portal tract cross‐section confirmed our previous observations that CX3CR1+CD64+ macrophages appear to line the portal vein and bile duct, as well as showing interactions with the portal lymphatic vasculature, hepatic artery and peri‐bile ductular capillaries (Figures 2 and 5a). Except for a few XCR1+ cells scattered throughout the liver, the majority of XCR1+ cells clustered within portal tracts, where they formed an interconnected network throughout the liver (Figure 5b, f). Higher‐power views showed that XCR1+ cells were located within the interstitium of portal tracts and were excluded from the hepatic lobules supplied by the sinusoids (Figure 5c). Further detailed analysis revealed that this cDC1 network was intertwined with the LYVE‐1high lymphatic vasculature (Figure 5c). While many XCR1+ cells directly contacted the lymphatic vasculature, some were detected inside the lymphatic vessel lumen (Figure 5c, arrows). Like hepatic cDC1s, hepatic CD301a+ cDC2Bs also formed an interconnected cellular network within the portal tracts (Figure 5d, g). In contrast to cDC1s, most hepatic CD301a+ cDC2Bs were not associated with the lymphatic vasculature but were preferentially localized around the bile ducts and hepatic arteries (Figure 5d). As the biliary tree is supplied by a myriad of arterioles and capillaries and is directly adjacent to the hepatic arterial vasculature (Figure 1a–d), it was difficult to distinguish whether cDC2s were associated with the arterial vasculature, the biliary tree, or both. Interestingly, CX3CR1+ macrophages were observed physically interacting with cDC1s and cDC2s within the portal tracts (Figure 6), suggesting that these APC populations possess the potential to communicate with each other to direct immune responses in this compartment.
Figure 5.
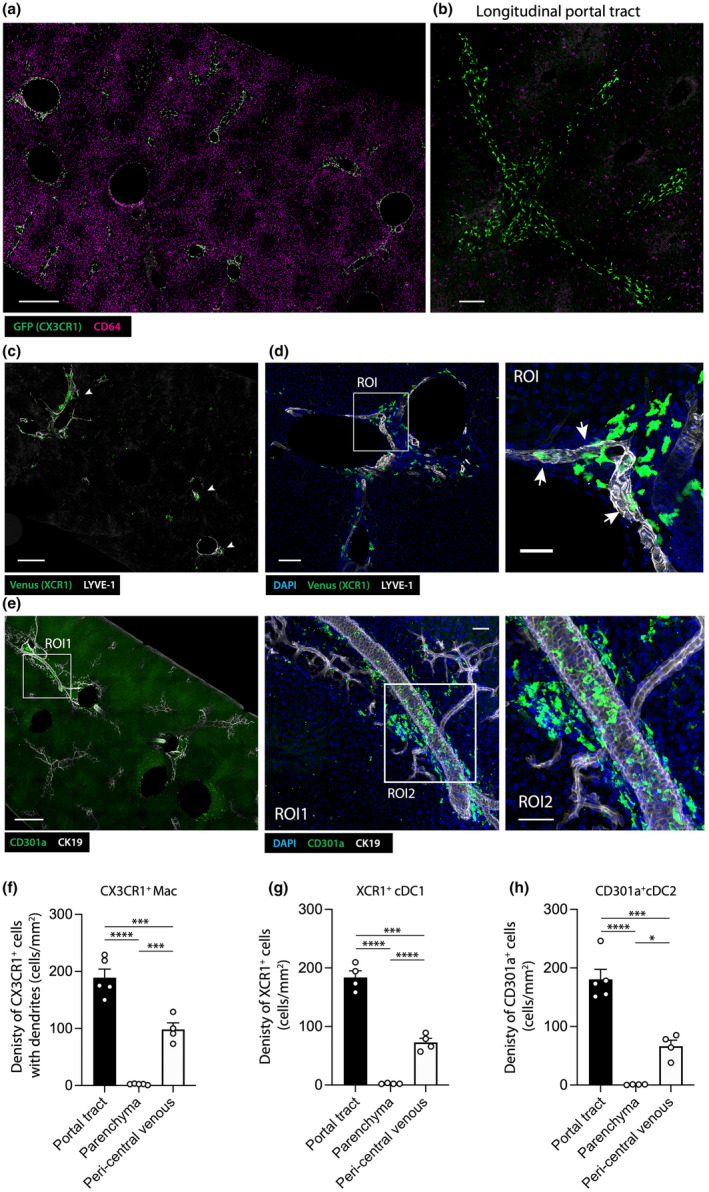
CX3CR1+ macrophages, XCR1+ cDC1s and CD301a+ cDC2s are not randomly distributed in hepatic portal tracts but form interconnected networks. (a) Left panel: confocal image showing a large view of a Ce3D cleared liver derived from a Cx3cr1 +/gfp mouse (GFP (CX3CR1) – green; CD64 – magenta; scale bar 500 μm). (b) Right panel: confocal image showing a longitudinal portal tract cross section, highlighting the networks formed by CX3CR1+ portal tract macrophages (scale bar 100 μm). (c, d) Confocal images showing a large view of a Ce3D cleared Xcr1 +/venus mouse liver (LYVE‐1 – white; DAPI – blue; Venus – green); (c) scale bar 500 μm, arrowheads indicate XCR1+ cell clusters associated with portal tracts; (d) left panel: scale bar 100 μm; right panel showing magnified ROI from left panel, scale bar 40 μm, arrowheads indicate XCR1+ cells in direct contact with LYVE‐1+ lymphatic vessels. (e) Left panel: confocal image showing low power view of a Ce3D cleared C57BL/6 mouse liver (CD301a – green; CK19 – white; scale bar 500 μm). Middle panel: magnified image of ROI1 from left panel (scale bar 100 μm); Right panel magnified image of ROI2 from middle panel (scale bar 100 μm). (a–e) All images are representative of n = 4 mice from two independent experiments. (f–h) The density of (f) CX3CR1+ cells with dendrites, (g) XCR1+ cells and (h) CD301+ cells in portal tract, parenchymal and peri central venous regions of the steady state liver. P‐values were calculated by one‐way ANOVA with multiple comparisons, *P < 0.05, ***P < 0.001, ****P < 0.0001. Data show the mean ± SEM of n = 4 mice and is representative of two independent experiments.
Figure 6.
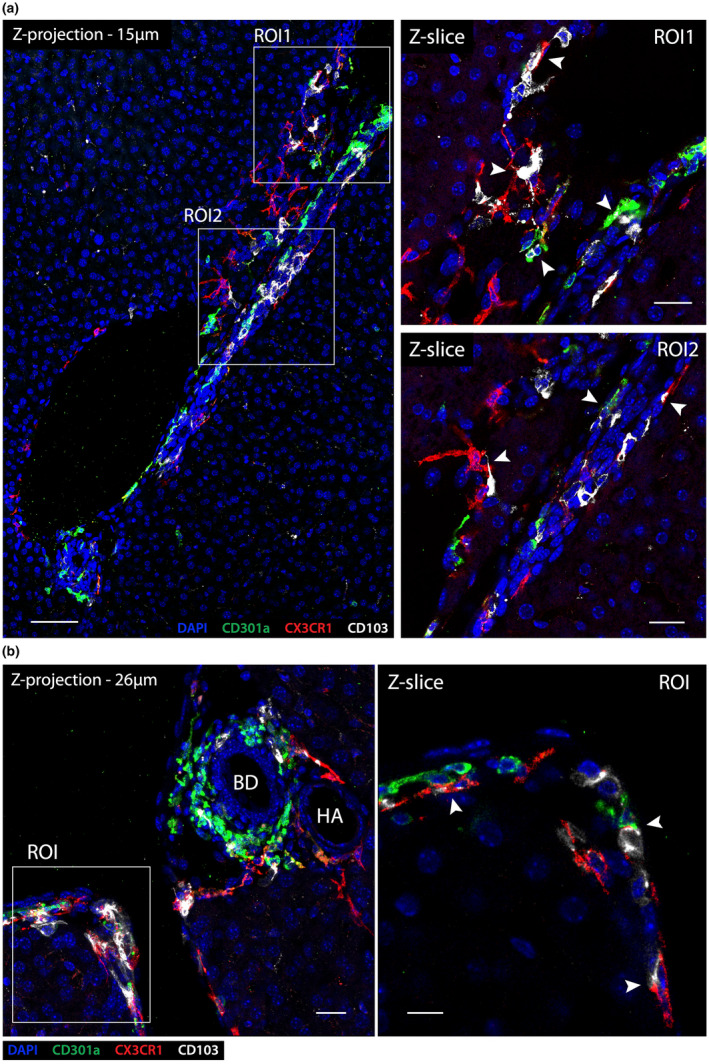
CX3CR1+ macrophages, CD103+ cDC1s and CD301a+ cDC2s interact in portal tracts. (a) Left panel: z‐projection (15 μm) of a longitudinal cross section of a portal tract from a C57BL/6 mouse liver in the steady state (DAPI – blue; CD301a – green; CX3CR1 – red; CD103 – white; scale bar 70 μm). Right panels: higher magnification image of ROI1 (upper) and ROI2 (lower) from left panel, shown in single Z‐slices. Arrowheads denote direct cell–cell interactions between CX3CR1+ macrophages, CD103+ cDC1s and CD301a+ cDC2s within the portal interstitium; scale bars 25 μm. (b) Left panel: z‐projection (26 μm) of a C57BL/6 portal tract cross section at high magnification (bile duct – BD; hepatic artery – HA, CX3CR1+ macrophages – red; CD103+ cDC1s – white; CD301a+ cDC2s – green; scale bar 20 μm). Right panels: higher magnification image of ROI1 (right upper) and ROI2 (right lower) from left panel, shown in single Z‐slices. Arrowheads indicate direct cell–cell interactions between CX3CR1+ macrophages, CD103+ cDC1s and CD301a+ cDC2s; scale bar 10 μm. Images are representation of n = 4 mice from two independent experiments.
Finally, analysis of Alb‐Cre/R26mT/mG mouse livers revealed that in addition to portal tracts, some MHCIIhigh cells were also associated with large central veins throughout the liver (Figure 5f–h; Supplementary figure 3). MHCIIhigh cells associated with central veins did not form a continuous network as they did along the portal tracts, but instead formed isolated clusters. These clusters were positioned between the final layer of hepatocytes and the central vein endothelium (Supplementary figure 3a), and contained CX3CR1+ macrophages, as well as XCR1+ cDC1s and CD301a+ cDC2s (Figure 5f–h; Supplementary figure 3b–d).
DISCUSSION
Our study highlights that the portal tract interstitium forms a significant continuous compartment embedded within the liver parenchyma. With its myriad of branches and subbranches throughout the entire organ, this compartment comprises a substantial volume. These findings show that this compartment contains a dense and heterogeneous population of MHCIIhigh cells that include macrophages expressing the monocytic marker CX3CR1, XCR1+ cDC1s with potential cross‐presentation properties, and CD301a+ cDC2s, specialized in driving helper T cell responses (Supplementary figure 4). These cells are not randomly distributed within the portal tracts, but instead occupy very specific niches in this compartment. Each population appears preferentially associated with distinct and specific portal tract structures: CX3CR1+ macrophages with portal veins, bile ducts and regions delineating the portal tract interstitium from the hepatic parenchyma; XCR1+ cDC1 are crowded along the lymphatic vasculature; and CD301a+ cDC2s tend to be associated with the biliary tree/arterial vasculature. Both macrophages and cDC subsets appear to interact and establish frequent contacts with each other, forming a large interconnected myeloid cell network that is intertwined with the lymphatic and blood vasculature, as well as with the biliary tree.
While the role of the specific positioning of these cellular networks embedded within the portal tracts was not investigated in the current study, it is likely optimized to serve important immune functions. Portal tracts are one of the main sites where leukocytes traffic and accumulate during both acute and chronic immune responses in the liver. 14 , 15 , 16 , 17 , 18 The size and type of periportal infiltrates are commonly used to score the severity of liver inflammation and disease activity in liver diseases; 17 , 18 however, the mechanistic immunological significance of these observations remains unclear. Portal tracts have generally been considered as neutral conduits, the interstitium of which contain fibroblasts and connective tissue, along which leukocytes traffic with the flow of lymph fluid through the liver. The regular influx of transiting immune cells explains why they are often associated with portal tracts. Furthermore, it would also explain the periportal leukocyte infiltrates observed in liver disease, as the increased influx of immune cells during intrahepatic immune responses would lead to “traffic congestions” in portal tracts and massive numbers of leukocytes would accumulate in these areas. However, this current view is hypothetical and not based on mechanistic evidence. This existing model fails to explain why portal tracts have a rich MHCIIhigh immune cell content and channel an embedded lymphatic vasculature. The presence of these populations, with organized relationships to key portal tract structures, suggests that rather than being inert conduits, portal tracts represent key areas in which active interactions can occur between lymphocytes and APCs.
This study allowed us to revisit the current understanding of portal tracts in mice, including the characterization of its micro‐anatomical structure and detailed analyses of its immune cell content. Our findings led us to identify and characterize a previously unreported subset of CX3CR1+ macrophages residing in this area, which we term “portal tract macrophages”. Portal tract macrophages phenotypically and morphologically resemble the recently identified liver capsular macrophage (LCMs). 2 They expressed common macrophage markers, F4/80, CD64, CD11b and CX3CR1, and were highly dependent on CSF1R signaling for survival, suggesting that they could be derived from blood monocytes, although future research using fate mapping approaches will be required to confirm the precise origin of CX3CR1+ portal tract macrophages. LCMs form a network within the liver capsule; 2 similarly, portal tract macrophages form a network in the portal interstitium, where they specifically localize close to the portal vein, lymphatic vessels and bile ducts. This distribution suggests that portal tract macrophages sample portal blood, liver lymph fluid and the biliary system, and could play important roles in preventing infection of the liver from pathogens that might gain access via portal blood that enters the liver from the GI tract or the biliary system. 4 In addition to their phenotypic resemblance to LCMs, CX3CR1+ portal tract macrophages display a phenotype also ascribed to MHCIIhighCX3CR1highLYVE‐1low interstitial macrophages detected in several murine peripheral tissues including the lung, heart, fat and dermis 19 and reported to be closely associated with sympathetic neurons and nerve fibers. 19 As sympathetic and parasympathetic nerve fibers that innervate the liver are contained in portal tracts, 20 it is likely that some MHCIIhighCX3CR1+ portal tract macrophages are positioned along the liver nerve fibers and regulate inflammation in these areas. As portal tract macrophages are distributed throughout the entire interstitium and along the outer border of portal tracts, it is unlikely that nerve fibers dictate the position of all portal tract macrophages within the portal tract. It is, however, possible that portal tract macrophages include distinct subsets and that some of these resemble perineural macrophages detected in other tissues. Our findings regarding portal tract macrophages are consistent with the conclusions of a recent study that was published during the preparation of this manuscript. 21 That study employed single cell transcriptomics, spatial transcriptomics and spatial proteomics to characterize the myeloid cell subsets residing in the murine liver at steady state and diseased state and investigated their respective niches. Although the two studies use different approaches, they identify similar populations of myeloid cells in portal tracts and our main findings are remarkably similar, further consolidating their conclusions. Despite sharing many key findings, each study brings its own additional complementary information. Notably, the published study showed that portal tract macrophages included at least two subsets occupying distinct niches: one associated with bile ducts, termed lipid associated macrophages (LAMs), and the other with the portal vein. Our study reveals macrophages occupying the same niches described in the other study, and we therefore conclude that the populations described in the two studies are the same cells. Further, we demonstrate that they express CX3CR1 and CSF1R‐dependent, aspects not addressed in the other study. The complex interactions of portal tract myeloid cells with each other and their specific arrangement along different components of the portal tract is an important additional finding of the current study that hints to the function of these populations.
Several reports have established that the liver contains a large population of dendritic cells, specialized in antigen presentation and the activation of naïve T cells. 22 , 23 Early studies suggested that intrahepatic DCs were immature, and the immaturity and/or distinct subtype of intrahepatic DCs led some investigators to propose that donor‐derived liver DCs might induce an incomplete T cell activation resulting in tolerance and that this mechanism might explain the spontaneous acceptance of liver allografts. 24 , 25 However, most of these studies were performed before the classification of cDCs and their subdivision into cDC1 and cDC2 subsets. In addition, these studies primarily utilized flow cytometry to analyze intrahepatic leukocytes isolated from dissociated liver tissue, and therefore lacked spatial information about cDC positioning within the liver. Before this study, the most detailed characterization of hepatic cDC subsets in mice combined flow cytometric analysis of cells isolated from collagenase‐digested liver tissues with immunofluorescence histology. That study showed that hepatic cDCs included CD103+ cDC1s, some of which expressed CD8α, and CD11b+ cDC2s. 26 Some cells with a cDC1 and cDC2 phenotype were observed close to portal tracts; however, detailed characterization of hepatic myeloid cells and their distribution with regard to portal tracts was not performed. Our findings confirm that hepatic XCR1+ cDC1s expressed CD103, DEC‐205, and a significant proportion co‐express CD8α and langerin. XCR1+ cDC1s are antigen presenting cells specialized in the cross presentation of cell‐associated exogenous antigens for priming CD8+ T cell responses to intracellular pathogens and cancer. 7 Recent evidence suggests that XCR1+ cDC1s play important roles in priming anti‐tumor CD4+ T cell responses, as well as the transfer of CD4+ T cell help to anti‐viral and anti‐tumor CD8+ T cells. 27 , 28 , 29 Hepatic XCR1+ cDC1s were recently demonstrated to contribute toward liver pathology in a mouse model of non‐alcoholic steatohepatitis (NASH), and while the precise mechanism was not determined, it likely involves influencing the intrahepatic T cell response. While currently not completely understood, it is likely that hepatic XCR1+ cDC1s play important roles in priming and driving intrahepatic CD4+ and CD8+ T cell responses.
cDC2s are specialized in capturing and presenting exogenous antigens on MHCII molecules for priming and directing the differentiation of CD4+ T helper cell responses. CD11b+ cDC2s have previously been identified within the liver12, 26, 30 however, their distribution within the liver and the cellular network they form in portal tracts was, until now, not understood. cDC2s localized to barrier tissues in other peripheral tissues of the body, including the lung, skin and GI tract, play important roles in regulating type II immune responses to parasites, helminths and fungi through promoting CD4+ Th2 responses and type 2 innate lymphoid cell activation. 7 cDC2s have also been demonstrated to play important roles in regulating type III immune and antibody responses to extracellular bacteria 7 and type I immune responses to viruses. 31 The biliary tree is in direct contact with the GI tract, and it is therefore tempting to speculate that this network of bile duct associated cDC2s plays important roles in protecting the liver from infection by pathogens that could gain access via the GI tract. It was recently identified that the expression of ESAM and CD301a/CD301b can be used to distinguish two subtypes of cDC2 in mice, known as cDC2A and cDC2B, respectively. 12 Brown and colleagues went on to show that cDC2B have a higher pro‐inflammatory potential compared with cDC2A in steady state conditions. It has been suggested that DCs in the liver are more tolerogenic compared with their counterparts in lymphoid and other peripheral tissues; 24 , 25 , 32 it is thus interesting to note that our analysis revealed that the liver primarily contains cDC2s with a CD301a+ESAM− cDC2B phenotype, and potentially represent a more immunogenic subset with the potential to promote inflammation within the liver. Such cells could contribute to the pathogenesis of several diseases that predominantly target the portal tract bile ducts, such as primary biliary cholangitis, primary sclerosing cholangitis, some drug hepatoxicities and liver allograft rejection.
In summary, our findings suggest that hepatic portal tracts contain a heterogeneous population of MHCIIhigh cells that include a specialized subset of CSF1R‐dependent CX3CR1+ macrophages, as well as the two main subsets of cDCs: cDC1s and cDC2s. These three types of cells are not randomly distributed but form distinct, though interconnected, cellular networks that are associated with portal tracts. The positioning of these networks in areas of high lymphocyte traffic suggest they might possess important immune regulatory functions.
METHODS
Mice
C57BL6 mice were purchased from Australian BioResources (ABR, Moss Vale, NSW, Australia) and housed under specific pathogen free (SPF) conditions in the Centenary Institute animal facility. Alb‐Cre x ROSA mT/mG mice were generated by crossing Albumin‐Cre mice with mT/mG mice. 33 , 34 Cx3cr1 gfp/gfp CD45.1+/+ knock‐in reporter mice 35 were purchased from The Jackson Laboratories (Maine, USA). The Xcr1 venus/venus knock‐in reporter mouse line 8 were maintained at the Centenary Institute under SPF conditions. Cd207‐EGFP transgenic mice were kindly provided by Bernard Malissen (Marseille Luminy, France). Itgax‐Cre.GFP transgenic reporter mice were kindly obtained from Anselm Enders (ANU, Canberra, Australia). The transgenic/reporter mouse lines were maintained as homozygous at the Centenary Institute. All knock‐in reporter mice (Cx3cr1 gfp and Xcr1 venus) used in all our experiments were heterozygous mice resulting from crossing homozygous mice with C57BL/6 mice. All procedures were performed in strict accordance with the recommendations of the Australian code of practice for the care and use of animals for scientific purposes, under protocols approved by the Sydney Local Health District Animal Ethics Committee (Protocols 2020–005, 2021–013, 2016–006, 2016–041).
In vivo antibody mediated CSF1R blockade
Cx3cr1 +/gfp mice were injected i.v. with 400 μg M279 anti‐mouse CSF1R blocking mAb (Amgen, Macquarie Park, Australia) or rat IgG isotype control (ICN Biomedicals Inc, Aurora, OH, USA) in sterile Dulbecco's PBS (dPBS) (Thermo Fisher Scientific, North Ryde, Australia) 4 days before animal killing as described by MacDonald et al. 2010. 5
Isolation of myeloid cells from the liver and spleen
Livers were retrogradely perfused in situ with 5 mL of 37°C HBSS containing Ca and Mg (Thermo Fisher Scientific, North Ryde, Australia) and 1 mg mL−1 collagenase type IV (Sigma, Australia) via the IVC, following transection of the portal vein to allow outflow of perfusate. Gallbladders were excised, and the livers were then removed, chopped, and incubated at 37°C for 30 min in HBSS with Ca and Mg, containing 1 mg mL−1 collagenase type IV (Sigma, Australia) and 1 μg mL−1 DNase type I (Roche, Basel, Switzerland). The livers were gently pressed through an 80‐gauge stainless‐steel mesh sieve in RPMI (Thermo Fisher Scientific, North Ryde, Australia) containing 2% FCS (Hyclone, Australia), then centrifuged at 440 × g for 5 min at 4°C. The pellet was resuspended in dPBS containing 33% isotonic Percoll (GE Life Sciences, Australia) and centrifuged at 700 × g for 12 min at room temperature with no brake. The cell pellet was washed with RPMI +2% FCS. Red blood cells were lysed following 1 min incubation with red cell removal buffer (150 mM ammonium chloride, 0.1 mM EDTA disodium, 12 mM sodium hydrogen carbonate in triple distilled water) (Sigma, Australia), before a final wash with RPMI +2% FCS. Spleens were collected in HBSS with Ca and Mg, finely chopped and digested with collagenase type IV (1 mg mL−1) and DNase type I (1 μg mL−1) for 30 min at 37°C. Spleen tissue remnants were gently pressed through an 80‐gauge stainless steel mesh sieve, washed and resuspended in RPMI +2% FCS.
Immunofluorescence confocal microscopy
Livers were retrogradely perfused in situ with cold PBS, then fixed with cold PBS 2% PFA after transecting the portal vein. After excising the gallbladder, the livers were removed and incubated in PBS 2% PFA overnight or for 8 h at 4°C in the dark, then washed with PBS and embedded in 2% agarose. 150 μm sections were cut using a vibratome (Leica VT1200, Germany) and placed in blocking buffer containing 4% BSA, 5% goat or donkey serum, 5% mouse serum and 0.3% Triton‐X 100 in PBS overnight at 4°C in the dark with gentle agitation. The sections were stained with fluorophore‐conjugated primary antibodies in blocking buffer overnight at 4°C in the dark with gentle agitation, then washed 3 times in dPBS with 0.1% Triton‐X 100 for at least 6 h. When unconjugated purified antibodies were used, tissue sections were additionally stained with the appropriate anti‐Ig species Alexafluor‐conjugated antibody overnight at 4°C in the dark with gentle agitation, then washed again 3 times in dPBS with 0.1% Triton‐X 100 for 6 h. To reveal nuclear DNA, some sections were further incubated with 1 μg mL−1 DAPI for 1 h at 4°C in the dark, then washed 3 times for 30 min. All tissue samples were mounted with Dabco antifade mounting medium for imaging. 36 In some experiments, thick liver sections (> 300 μm) were optically cleared before image acquisition using the Ce3D clearing protocol described previously by Germain and colleagues. 13 Images were acquired using an inverted confocal microscope with a motorized stage for tiled imaging (Leica DMi8, SP8 scanning head unit). Images were processed and analyzed using Imaris 9 (Bitplane) software. For quantification of cell densities, the cells were manually counted, and the area was calculated using the surface function in Imaris 9 software.
Flow cytometry
Cells isolated from dissociated tissues were incubated with anti‐FcγII receptor 2.4G2 monoclonal antibody (supernatant generated in house) for 20 min and stained with conjugated antibodies as described previously. 2 For intracellular staining, the cells were fixed in 1% PFA for 20 min and stained with conjugated antibodies for 2 h in FACSwash containing 0.25% saponin. Stained cells were passed through a filter and DAPI was added to a final concentration of 0.1 μg mL−1 immediately before acquisition. Cell counting was performed using AccuCount Blank beads (Sphero, Chicago, USA). Flow cytometric acquisition was performed using an LSR‐Fortessa flow cytometer (Becton Dickinson, North Ryde, NSW, Australia), with data acquisition using FACSdiva (Becton Dickinson, North Ryde, NSW, Australia). Analysis was performed using Flowjo 10 software (Becton Dickinson, North Ryde, NSW, Australia) on a Macintosh computer (Apple, Cupertino, CA, USA).
Antibodies
The following primary conjugated antibodies were used for flow cytometry and unless otherwise stated were purchased from Biolegend (San Diego, USA): CD3‐FITC (145‐2C11), CD19‐PerCP‐Cy5.5 (1D3), CD11b‐BV650 (M1/70), CD11c‐APC‐Cy7 (N418), CD45‐BUV395 (30‐F11), CD64‐PE‐Cy7 (X54‐5/7.1), CD103‐PE (2E7), CD205‐AF647 (NLDC‐145), CD301a‐PE (LOM‐8.7), CD301b‐AF647 (URA‐1), CX3CR1‐BV711 (SA011F13), MHCII‐PB (M5/114.15.2), Ly6C‐BV605 (HK1.4), XCR1‐BV785 (ZET). The following primary antibodies were used for IF histology and unless otherwise stated were purchased from Biolegend: CK19 unconjugated (EPNCIR127B) (Abcam), LYVE‐1 unconjugated (223322) (R&D systems), MHCII‐AF647 (M5/114.15.2), CD11b unconjugated (M1/70), CD11c‐AF647 (N418) (BD), TCRβ‐APC (H57‐597), CLEC4F‐AF647 (3E3F9), F4/80 unconjugated (hybridoma supernatant, in‐house), GFP unconjugated (Cat. #A‐11122) (Invitrogen), CD64 unconjugated (AT152‐9) (Bio‐Rad), CD103 unconjugated (Cat. #AF1990), CD8α‐AF647 (53–6.7), CD301a unconjugated (LOM‐8.7), CD31 unconjugated (Cat. #550274) (BD). The following secondary antibodies were used for IF histology and unless otherwise stated were purchased from Invitrogen: anti‐rat IgG‐AF555, anti‐rat IgG‐AF647, anti‐rabbit IgG‐AF488, anti‐rabbit IgG‐AF647, anti‐goat IgG‐AF405 (Abcam), anti‐goat IgG‐AF647.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 9.3.1 (GraphPad Software, San Diego, California, USA). Data are presented as mean ± SEM. Significance was determined by the Student's t‐test for comparison between two groups or one‐way ANOVA with Tukey's multiple comparisons between different groups. Statistical significance was set at P < 0.05.
AUTHOR CONTRIBUTIONS
Kieran English: Conceptualization; data curation; formal analysis; investigation; methodology; validation; visualization; writing – original draft; writing – review and editing. Sioh‐Yang Tan: Conceptualization; data curation; formal analysis; methodology; visualization. Rain Kwan: Methodology; validation. Lauren Holz: Resources; validation. Frederic Sierro: Conceptualization; methodology; validation; writing – review and editing. Claire M McGuffog: Methodology. Tsuneyasu Kaisho: Resources. William Ross Heath: Conceptualization; resources; writing – review and editing. Kelli MacDonald: Conceptualization; resources; writing – review and editing. Geoffrey W McCaughan: Conceptualization; resources; writing – review and editing. David Bowen: Conceptualization; funding acquisition; supervision; writing – original draft; writing – review and editing. Patrick Bertolino: Conceptualization; formal analysis; funding aquisition; supervision; writing ‐ original draft; writing ‐ review and editing.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
Supporting information
ACKNOWLEDGMENTS
We thank the Centenary Institute Animal Facility and “The Advanced Cytometry Facility of the Centenary Institute and the University of Sydney” for their excellent technical support. We particularly thank Angela Fontaine for sharing with us her imaging expertise and for giving us advice and assistance for confocal microscopy. This work was supported by an NHMRC Australia Project Grant (APP APP1146677). KE was the recipient of a PhD Scholarship from the University of Sydney. Open access publishing facilitated by The University of Sydney, as part of the Wiley ‐ The University of Sydney agreement via the Council of Australian University Librarians.
Contributor Information
David G Bowen, Email: d.bowen@centenary.org.au.
Patrick Bertolino, Email: p.bertolino@centenary.org.au.
DATA AVAILABILITY STATEMENT
Data are available on request from the authors.
REFERENCES
- 1. Gola A, Dorrington MG, Speranza E, et al. Commensal‐driven immune zonation of the liver promotes host defence. Nature 2021; 589: 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sierro F, Evrard M, Rizzetto S, et al. A liver capsular network of monocyte‐derived macrophages restricts hepatic dissemination of intraperitoneal bacteria by neutrophil recruitment. Immunity 2017; 47: e376. [DOI] [PubMed] [Google Scholar]
- 3. English K, Bowen DG, Bertolino P. Zone defence – the gut microbiota position macrophages for optimal liver protection. Immunol Cell Biol 2021; 99: 565–569. [DOI] [PubMed] [Google Scholar]
- 4. Wu T, Zhang Z, Liu B, et al. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large‐scale study. BMC Genomics 2013; 14: 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. MacDonald KP, Palmer JS, Cronau S, et al. An antibody against the colony‐stimulating factor 1 receptor depletes the resident subset of monocytes and tissue‐ and tumor‐associated macrophages but does not inhibit inflammation. Blood 2010; 116: 3955–3963. [DOI] [PubMed] [Google Scholar]
- 6. Steinman RM, Kaplan G, Witmer MD, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. V. Purification of spleen dendritic cells, new surface markers, and maintenance in vitro . J Exp Med 1979; 149: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anderson DA, Dutertre CA, Ginhoux F, Murphy KM. Genetic models of human and mouse dendritic cell development and function. Nat Rev Immunol 2021; 21: 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamazaki C, Sugiyama M, Ohta T, et al. Critical roles of a dendritic cell subset expressing a chemokine receptor, XCR1. J Immunol 2013; 190: 6071–6082. [DOI] [PubMed] [Google Scholar]
- 9. Vremec D, Pooley J, Hochrein H, Wu L, Shortman K. CD4 and CD8 expression by dendritic cell subtypes in mouse thymus and spleen. J Immunol 2000; 164: 2978–2986. [DOI] [PubMed] [Google Scholar]
- 10. Sung SS, Fu SM, Rose CE Jr, Gaskin F, Ju ST, Beaty SR. A major lung CD103 (αE)‐β7 integrin‐positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J Immunol 2006; 176: 2161–2172. [DOI] [PubMed] [Google Scholar]
- 11. Eisenbarth SC. Dendritic cell subsets in T cell programming: location dictates function. Nat Rev Immunol 2019; 19: 89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown CC, Gudjonson H, Pritykin Y, et al. Transcriptional basis of mouse and human dendritic cell heterogeneity. Cell 2019; 179: e824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li W, Germain RN, Gerner MY. Multiplex, quantitative cellular analysis in large tissue volumes with clearing‐enhanced 3D microscopy (Ce3D). Proc Natl Acad Sci USA 2017; 114: E7321–E7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benechet AP, De Simone G, Di Lucia P, et al. Dynamics and genomic landscape of CD8+ T cells undergoing hepatic priming. Nature 2019; 574: 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Billerbeck E, Wolfisberg R, Fahnoe U, et al. Mouse models of acute and chronic hepacivirus infection. Science 2017; 357: 204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trivedi S, Murthy S, Sharma H, et al. Viral persistence, liver disease, and host response in a hepatitis C‐like virus rat model. Hepatology 2018; 68: 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bach N, Thung SN, Schaffner F. The histological features of chronic hepatitis C and autoimmune chronic hepatitis: a comparative analysis. Hepatology 1992; 15: 572–577. [DOI] [PubMed] [Google Scholar]
- 18. Freni MA, Artuso D, Gerken G, et al. Focal lymphocytic aggregates in chronic hepatitis C: occurrence, immunohistochemical characterization, and relation to markers of autoimmunity. Hepatology 1995; 22: 389–394. [PubMed] [Google Scholar]
- 19. Chakarov S, Lim HY, Tan L, et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 2019; 363: eaau0964. [DOI] [PubMed] [Google Scholar]
- 20. Jensen KJ, Alpini G, Glaser S. Hepatic nervous system and neurobiology of the liver. Compr Physiol 2013; 3: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guilliams M, Bonnardel J, Haest B, et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 2022; 185: e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Steiniger B, Klempnauer J, Wonigeit K. Phenotype and histological distribution of interstitial dendritic cells in the rat pancreas, liver, heart, and kidney. Transplantation 1984; 38: 169–174. [DOI] [PubMed] [Google Scholar]
- 23. Prickett TC, McKenzie JL, Hart DN. Characterization of interstitial dendritic cells in human liver. Transplantation 1988; 46: 754–761. [DOI] [PubMed] [Google Scholar]
- 24. Xia S, Guo Z, Xu X, Yi H, Wang Q, Cao X. Hepatic microenvironment programs hematopoietic progenitor differentiation into regulatory dendritic cells, maintaining liver tolerance. Blood 2008; 112: 3175–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pillarisetty VG, Shah AB, Miller G, Bleier JI, DeMatteo RP. Liver dendritic cells are less immunogenic than spleen dendritic cells because of differences in subtype composition. J Immunol 2004; 172: 1009–1017. [DOI] [PubMed] [Google Scholar]
- 26. Krueger PD, Kim TS, Sung SS, Braciale TJ, Hahn YS. Liver‐resident CD103+ dendritic cells prime antiviral CD8+ T cells in situ . J Immunol 2015; 194: 3213–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferris ST, Durai V, Wu R, et al. cDC1 prime and are licensed by CD4+ T cells to induce anti‐tumour immunity. Nature 2020; 584: 624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eickhoff S, Brewitz A, Gerner MY, et al. Robust anti‐viral immunity requires multiple distinct T cell‐dendritic cell interactions. Cell 2015; 162: 1322–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hor JL, Whitney PG, Zaid A, Brooks AG, Heath WR, Mueller SN. Spatiotemporally distinct interactions with dendritic cell subsets facilitates CD4+ and CD8+ T cell activation to localized viral infection. Immunity 2015; 43: 554–565. [DOI] [PubMed] [Google Scholar]
- 30. Deczkowska A, David E, Ramadori P, et al. XCR1+ type 1 conventional dendritic cells drive liver pathology in non‐alcoholic steatohepatitis. Nat Med 2021; 27: 1043–1054. [DOI] [PubMed] [Google Scholar]
- 31. Bosteels C, Neyt K, Vanheerswynghels M, et al. Inflammatory type 2 cDCs acquire features of cDC1s and macrophages to orchestrate immunity to respiratory virus infection. Immunity 2020; 52: e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goddard S, Youster J, Morgan E, Adams DH. Interleukin‐10 secretion differentiates dendritic cells from human liver and skin. Am J Pathol 2004; 164: 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double‐fluorescent Cre reporter mouse. Genesis 2007; 45: 593–605. [DOI] [PubMed] [Google Scholar]
- 34. Yakar S, Liu JL, Stannard B, et al. Normal growth and development in the absence of hepatic insulin‐like growth factor I. Proc Natl Acad Sci USA 1999; 96: 7324–7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jung S, Aliberti J, Graemmel P, et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 2000; 20: 4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson GD, Davidson RS, McNamee KC, Russell G, Goodwin D, Holborow EJ. Fading of immunofluorescence during microscopy: a study of the phenomenon and its remedy. J Immunol Methods 1982; 55: 231–242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on request from the authors.


