Abstract
Bacterial mechanisms for the uptake of peptides and their hydrolysis to amino acids are known in great detail, whereas much less is known about the fates of the peptide-derived amino acids. We show that the addition of l-threonine-containing di- or tripeptides results in reduction of the growth of Corynebacterium glutamicum, with concomitant high intracellular accumulation of l-threonine to up to 130 mM. Using transposon mutagenesis and isolation of mutants with increased Thr peptide sensitivity, nine open reading frames (ORFs) were identified, almost all encoding hypothetical proteins of unknown function. Three ORFs encode membrane proteins. Their individual functional characterizations in the wild-type background led to the identification of thrE. Upon thrE overexpression, growth is no longer sensitive to the presence of the Thr peptide, and l-threonine is exported at a rate of 3.8 nmol min−1 mg of dry weight−1, whereas the rate of export of a thrE inactivation mutant is reduced to 1.1 nmol min−1 mg of dry weight−1. In addition to l-threonine, l-serine is also a substrate for the exporter. The exporter exhibits nine predicted transmembrane-spanning helices with long charged C and N termini and with an amphipathic helix present within the N terminus. All these data suggest that the carrier encoded by thrE serves to export small molecules such as l-threonine and that the carrier is a prototype of a new translocator family. Homologues of ThrE are present in Mycobacterium tuberculosis and Streptomyces coelicolor.
As is evident from current genome analyses, a substantial number of bacterial genes encode membrane transport proteins. These proteins enable the controlled solute exchange between the cell and its environment. For instance, in Mycobacterium tuberculosis, about 120 gene products might encode transporters (9), and in Escherichia coli, about 300 candidates are present (5). However, at least half of these putative transport proteins are functionally undefined. Of course, many of the transport proteins are necessary to import nutrients such as carbohydrates, ions, amino acids, or peptides. However, in addition, some carriers are known to act as exporters. In most situations these export carriers catalyze the extrusion of noxious substances. Examples are the very well known multidrug resistance carriers (40), the metal resistance carriers (27), and the substrate-product exchange carriers (37). In addition, recent studies have shown that there are also export carriers with rather unexpected substrates, such as sugars (6, 24) and amino acids (1, 11, 49). Although in many situations the primary function of these latter carriers is still unknown, there is at least good evidence that one of the amino acid exporters naturally serves for the export of basic amino acids. This new exporter is LysE of Corynebacterium glutamicum (49), which is necessary during growth on complex medium or in the presence of peptides rich in l-lysine or l-arginine (4). Under such special growth conditions, l-lysine or l-arginine might accumulate to toxic levels, a situation which is prevented by their export. Homologues of the protein are widespread and occur in bacteria and archaea (50).
Studies on the peptide uptake systems of Staphylococcus aureus (29), Streptococcus faecalis, and E. coli (35) indicate that in these bacteria, peptide use can be accompanied by the efflux of their constituent amino acids. It thus appears that more exporters exist which accept amino acids as substrates. The efflux of selected amino acids also occurs with Lactococcus lactis grown on milk (18) or in the presence of milk-derived peptides (21). With C. glutamicum there is evidence that the efflux of l-glutamate (16), l-isoleucine (52), and l-threonine (32) is at least in part actively driven.
Studies with C. glutamicum are significant because of its enormous economic impact, since it is used worldwide for the production of l-glutamate and l-lysine. Together with Mycobacterium and Nocardia spp., they comprise the Corynebacterium-Mycobacterium-Nocardia (CMN) bacteria. These bacteria possess a mycolic acid layer which is thought to contribute significantly to the flux properties of the cell envelope and which is a major barrier to antibiotic access to Mycobacterium (28). We here describe the identification of a carrier exporting l-threonine from C. glutamicum; this exporter represents a previously unknown family of membrane transport proteins. The approach that we used to identify this exporter is based on the well-known peptide utilization of bacteria. It might therefore be suited to the isolation of further exporters with amino acids or amino-acid-related compounds as substrates, thus reducing the gap between putative and identified membrane transport proteins.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. As the standard medium for E. coli, Luria broth was used. C. glutamicum was precultivated on brain heart infusion (BHI) medium (Difco). The minimal medium used for C. glutamicum was CGXII (19). To induce amino acid export, cells were cultivated in CGXII containing 1 mM tripeptide Thr-Thr-Thr, 1 mM tripeptide Ser-Ser-Ser, or 2 mM dipeptide Lys-Ala (13). When appropriate, ampicillin (50 μg ml−1) or kanamycin (15, 25, or 50 μg ml−1) was added to the medium. A C. glutamicum ΔilvA strain received 300 mg of l-isoleucine liter−1. E. coli was grown at 37°C, and C. glutamicum was grown at 30°C.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant characteristicsa | Reference(s) or sourceb |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αMCR | endA1 supE44 recA1 gyrA96 relA1 U169Δ(mrr-hsdRMS-mcrBC) | 14 |
| GM2929 | dam-13::Tn9 dcm-6 hsdR2 recF143 mcrA mcrB | 31 |
| C. glutamicum | ||
| R127 | Restriction deficient | 23 |
| ATCC 14752 | Wild type | ATCC |
| 13032::thrE | Wild type with thrE (ORF53) disrupted by pK18mob | This work |
| 13032::ORF22 | Wild type with ORF22 disrupted by pK18mob | This work |
| 13032::ORF81 | Wild type with ORF81 disrupted by pK18mob | This work |
| 14752 ΔilvA | Wild type with a deletion of a 241-nt BglII fragment of ilvA | This work |
| R127 ΔilvA | R127 with a deletion of a 241-nt BglII fragment of ilvA | This work |
| 13032 ΔthrE | Wild type with a deletion of a 968-nt SacII-EcoRV fragment of thrE | This work |
| Plasmids | ||
| pCGL0040 | Donor of Tn5531 (IS1207 Kmr); AproriVE.c. | U53587 |
| pET2 | Promoter-probe vector | 46, 48 |
| pZ1 | Shuttle vector; KmroriVE.c. oriVC.g. | 25 |
| pK18mob | Integration vector; KmroriVE.c. oriT | 44 |
| pK19mobsacB | Integration vector; KmroriVE.c. oriT sacB | 44 |
| pK19mobsacBΔilvA | pK19mobsacB with ilvA with a deletion of a 242-bp internal fragment | 42 |
| pK19mobsacBΔthrE | pK19mobsacB with thrE with a deletion of a 968-bp internal fragment | This work |
| pK18mobthrEint | pK18mob with a 411-bp internal fragment of thrE | This work |
| pK18mobORF22int | pK18mob with a 274-bp internal fragment of ORF22 | This work |
| pK18mobORF81int | pK18mob with a 368-bp internal fragment of ORF81 | This work |
| pUC18thrE | pUC18 with a 1.9-kb PCR fragment containing thrE | This work |
| pZ1thrE | pZ1 with a 1.9-kb SacI-XbaI insert from pUC18thrE | This work |
| pZ1ORF22 | pZ1 with a 1.0-kb PCR fragment containing ORF22 | This work |
| pZ1ORF81 | pZ1 with a 1.5-kb PCR fragment containing ORF81 | This work |
| pET2thrE | pET2 with a 267-bp BamHI-KpnI PCR fragment containing the promoter region of thrE | This work |
Kmr, kanamycin resistant; Apr, ampicillin resistant. E.c., E. coli; C.g., C. glutamicum.
ATCC, American Type Culture Collection.
Transposon mutagenesis, screening for threonine-sensitive mutants, and localization of transposon insertion sites.
The Tn5531-containing plasmid pCGL0040 was isolated from E. coli GM2929, and C. glutamicum ATCC 14752 ΔilvA was transformed with the plasmid by electroporation. Transposon insertion mutants were selected by plating on LBHIS (Luria broth with brain heart infusion) containing 15 μg of kanamycin ml−1 (23). The resulting colonies were transferred to CGXII agar plates containing 300 mg of l-isoleucine liter−1, 25 μg of kanamycin ml−1, and either 2 mM Thr-Thr-Thr or no peptide. Mutants that were able to grow normally on CGXII minimal medium without any addition of Thr-Thr-Thr but that exhibited retarded growth in the presence of peptide were retrieved from the master plate and retested in liquid CGXII medium. For that purpose, mutant strains were precultivated on BHI medium containing 25 μg of kanamycin ml−1. CGXII minimal medium (containing 300 mg of l-isoleucine liter−1 and 25 μg of kanamycin ml−1) was inoculated to an initial optical density at 600 nm (OD600) of 0.1. Growth was monitored in parallel in liquid CGXII medium without any addition of peptide or with 2 mM Thr-Thr-Thr. Mutants that grew more slowly in the presence of Thr-Thr-Thr were stored in glycerol at −70°C for further studies.
For the localization of the transposon insertion locus in the C. glutamicum ATCC 14752 ΔilvA chromosome, genomic DNA from the transposon mutants was isolated as described previously (12). The insertion loci of Tn5531 were identified by cloning of transposon-chromosome junctions into pUC18 and subsequent DNA sequencing with oligonucleotides Tn5531-Eco (5′-CGGGTCTACACCGCTAGCCCAGG-3′) and Tn5531-Xba (5′-CGGTGCCTTATCCATTCAGG-3′) as primers as described by Ankri et al. (3).
Construction of plasmids.
All plasmid constructions were made in E. coli DH5αMCR. Open reading frames (ORFs) ORF22, ORF81, and ORF53 (thrE) were cloned from strain ATCC 13032 by PCR. Plasmids pZ1ORF22 and pZ1ORF81 were obtained by ligating the corresponding PCR fragments into the ScaI site of pZ1. To construct pZ1thrE, the PCR fragment was first cloned into the SmaI site of pUC18. The resulting plasmid, pUC18thrE, was digested with SacI and XbaI, and the thrE-containing insert obtained was blunted and ligated into the ScaI site of pZ1. Plasmids pK18mobORF22int and pK18mobORF81int were obtained by ligating the corresponding internal fragments made by PCR into the SmaI site of pK18mob. To construct pK18mobthrEint, pUC18thrE was digested with ClaI and EcoRV (see Fig. 3) to yield a 411-bp internal fragment of thrE. This fragment was blunted and cloned into the SmaI site of pK18mob. The promoter region of thrE was cloned into vector pET2 via its BamHI and KpnI sites.
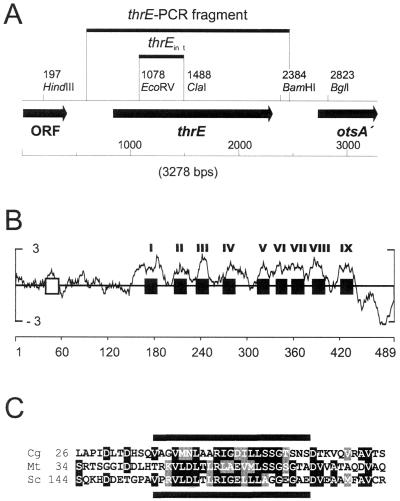
FIG. 3.
Overview of the thrE locus of C. glutamicum and structural properties of ThrE. (A) DNA fragments used for thrE overexpression and inactivation as well as adjacent genes. (B) Average local hydrophobicity at each residue according to the algorithm of Kyte and Doolittle (22) using a window of 13 amino acids, as plotted on the vertical axis, versus the residue number on the horizontal axis. The transmembrane-spanning helices predicted by use of the neuronal network program PHD.htm (41) are highlighted as black squares and numbered I to IX. The amphipathic helix at the beginning of the protein is highlighted as an open box. (C) Part of a sequence alignment of ThrE of C. glutamicum (Cg) with putative proteins of M. tuberculosis (Mt) and S. coelicolor (Sc) in the region of the amphipathic helix, which is indicated by the thick lines. The numbers specify the amino acid positions at the start of the peptide stretches shown. Identical amino acid residues (black background) and conserved amino acid residues (gray shading) are indicated.
Construction of strains.
C. glutamicum ATCC 13032 was transformed by electroporation (47). To obtain thrE, ORF22, and ORF81 insertion mutants of C. glutamicum ATCC 13032, nonreplicating plasmids pK18mobthrEint, pK18mobORF22int, and pK18mobORF81int, respectively, were transferred to C. glutamicum ATCC 13032. The correct integration of the vector into the chromosome of the obtained insertion mutants, 13032::thrE, 13032::ORF22, and 13032::ORF81, was verified by PCR analysis. Deletion mutant C. glutamicum ATCC 13032 ΔthrE was constructed as follows. Vector pUC18thrE was restricted with EcoRV and KspI, blunted, and religated. From this vector, a fragment with a deletion of 968 nucleotides (nt) of the thrE coding region was excised as a SacI-XbaI fragment; the latter was subsequently blunted and ligated with SmaI-digested pK19mobsacB. C. glutamicum ATCC 13032 was transformed with the resulting vector, pK19mobsacBΔthrE, and chromosomal deletion was carried out using the method described by Schäfer et al. (44) and verified by PCR analysis. Construction of ilvA deletion mutants of C. glutamicum ATCC 14752 and R127 was performed as described previously for strain ATCC 13032 (42) and verified by PCR analysis.
Primer extension.
Total RNA was isolated from C. glutamicum using extraction with hot acidic phenol (12). The transcription start site of thrE was determined by primer extension using SuperScript II reverse transcriptase (Gibco BRL) and primers labeled with [32P]ATP. In parallel, the respective DNA (pET2pthrE) was sequenced using 32P-labeled primers and a Thermosequenase (Amersham Pharmacia Biotech, Uppsala, Sweden) kit. The sequencing reaction mixtures and primer extension products were heated at 95°C for 4 min, and 2-μl samples were loaded onto a polyacrylamide gel.
Assay of amino acid export.
For the determination of amino acid export rates, pregrown cells (BHI medium) were washed once with 0.9% NaCl, transferred into prewarmed CGXII minimal medium containing 1 mM Thr-Thr-Thr or 1 mM Ser-Ser-Ser at an initial OD600 of 2.0, and cultivated for 2 h at 30°C. The cells were harvested by centrifugation (5,000 × g, 10 min) and washed once with ice-cold CGXII minimal medium. Amino acid excretion was initiated by resuspending the cells in prewarmed CGXII minimal medium (30°C) containing the appropriate peptide at the concentration given above. The resulting cell density (OD600) was 8 to 10, corresponding to 2.4 to 3.0 mg of dry weight ml−1. The cells were incubated at 30°C and stirred rapidly with a magnetic stirrer. Samples for silicone oil centrifugation (20) were taken every 15 min over a period of 2 h. The separation of cellular and extracellular fractions as well as the quantification of the amino acids as their o-phthaldialdehyde derivatives via high-pressure liquid chromatography were carried out as described previously (49). The intracellular volume used for calculations was 2 μl mg of dry weight−1.
In the presence of carbonyl cyanide m-chlorophenylhydrazone (CCCP), the determination of export rates was modified. The cells were precultivated, loaded with the appropriate peptide, and harvested as described above. After the cells were washed in ice-cold export buffer (100 mM morpholineethanesulfonic acid [MES], Tris [pH 7.5], 100 mM NaCl, 100 mM KCl, 500 mM mannitol, 20 mM glucose), amino acid excretion was initiated by resuspension of the cells in the same but prewarmed (30°C) buffer to a cell density (OD600) of 8 to 10. The CCCP concentration used was 20 μM. For extracellular amino acid determinations, samples of 200 μl were taken every 3 min over a period of 20 min and centrifuged (10,000 × g, 1 min), and amino acids in the supernatants were quantified. The diffusion constant, Kd, was calculated according to the equation v = Kd[Thr]in, where v is the rate of l-threonine efflux in nanomoles per minute per milligram of dry weight and [Thr]in is the intracellular l-threonine concentration in nanomoles per microliter.
Sequence analysis.
DNA sequencing was performed by using standard automated cycle sequencing protocols.
Nucleotide sequence accession numbers.
The sequence data have been submitted to the GenBank database under accession numbers AF326510 (thrE), AF326511 (ORF22), and AF326512 (ORF81).
RESULTS
Increase in intracellular l-threonine levels results in growth delay.
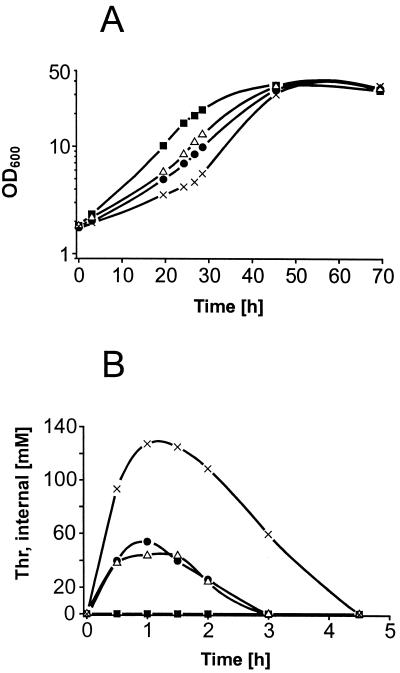
The deletion of the basic amino acid exporter of C. glutamicum results in growth arrest in the presence of lysine-containing peptides (49). This is due to the accumulation of peptide-derived l-lysine up to an extremely high intracellular concentration, more than 1 M. To address whether such an effect of impaired growth could serve as a basis for the isolation of an l-threonine export-deficient mutant, we assayed the response of the wild-type derivative C. glutamicum R127 to the addition of threonine-containing peptides (Fig. 1A). The addition of 1 mM Thr-Ala or Ala-Thr dipeptide resulted in a significant growth delay. The strongest growth reduction was obtained in the presence of 1 mM Thr-Thr-Thr. In a separate experiment, the intracellular l-threonine concentrations were quantified by silicone oil centrifugation (Fig. 1B). Whereas without the addition of peptide the l-threonine concentration was below 1 mM, the presence of the dipeptides resulted in about 50 mM intracellular l-threonine, and with the tripeptide a concentration of up to 130 mM was obtained.
FIG. 1.
Consequences of l-threonine peptide addition to C. glutamicum for growth and the intracellular l-threonine concentration. (A) Growth without peptide addition (▪) and in response to the addition of 1 mM Ala-Thr (●), Thr-Ala (▵), or Thr-Thr-Thr (×). (B) Time course for the intracellular l-threonine concentration within the first 5 h. Symbols are as described for panel A.
As is already known from the overexpression of threonine biosynthesis genes (39), we found an extracellular accumulation of l-isoleucine for the high intracellular l-threonine concentrations. To achieve an even more elevated level of l-threonine accumulation, ilvA, encoding threonine dehydratase (26), the key enzyme of isoleucine synthesis, was deleted. This led to an approximately twofold increase in the intracellular l-threonine level (data not shown). Interestingly, the increased intracellular l-threonine concentration was at best only transiently present for 4.5 h (Fig. 1B). Nevertheless, the growth delay lasted for more than 20 h. These experiments show that peptide use can dramatically alter the growth behavior of the cell. Furthermore, the correlation of high intracellular l-threonine concentration with retarded growth suggests the suitability of the observed effect for isolating mutants deficient in l-threonine export as clones characterized by a strong sensitivity to the tripeptide Thr-Thr-Thr.
Isolation of threonine peptide-sensitive mutants and analysis of transposon insertion loci.
Using transposon Tn5531 (3) and strain C. glutamicum ATCC 14752 ΔilvA, a transposon mutant bank was constructed. This strain had to be used because no transposon mutants were obtained with C. glutamicum R127 ΔilvA. The strain used was confirmed to exhibit the Thr-Thr-Thr-dependent growth delay (data not shown). A total of 2,000 Kmr clones were tested individually for increased peptide sensitivity on agar plates. After retesting of 150 potential candidates, 21 clones remained which grew more slowly than the parent strain in the presence of peptide but normally in its absence. These clones were finally cultivated in liquid medium (CGXII with or without 2 mM Thr-Thr-Thr). Nine mutants repeatedly displayed retarded growth in the presence of Thr-Thr-Thr.
The upstream and downstream sequences of the transposon insertion loci of these mutants were cloned, sequenced, and finally analyzed using the BLASTX program (2). In one mutant, the fourth ORF within the dapAB operon of C. glutamicum was interrupted. This ORF is presumed to be involved in gene expression (34) and has now been identified in a large variety of bacteria, but its explicit function is still unknown. In a second mutant, the product of the interrupted ORF exhibited a high similarity (46%) to OtsB (trehalose-6-phosphate phosphatase) of M. tuberculosis. In all other mutants, the transposon was located within or close to an ORF whose derived gene product did not show any similarities to proteins of known function. Since we were interested in the actively driven component of l-threonine efflux, we focused on mutants 22, 53, and 81, since in these mutants the transposon was inserted into ORFs which could encode membrane proteins.
Analysis of ORF22 and ORF81.
The deduced amino acid sequence of ORF22 revealed, among others, similarity to a putative amino acid transporter of Bacillus halodurans. We therefore first studied ORF22 in detail. For this purpose, a defined mutant of wild-type C. glutamicum ATCC 13032 was constructed using intergeneric gene transfer (44). Growth of the strain with ORF22 disrupted in the presence of 2 mM Thr-Thr-Thr in a liquid culture confirmed the growth delay in the type strain (Fig. 2A). However, in assays where pregrown cells were loaded with l-threonine and export rates were determined (see Materials and Methods), the inactivation mutant exhibited no decrease in l-threonine export compared to the control. This result suggests a function of the ORF different from catalysis of l-threonine export. The deduced gene product shares a similarity of 48% over a stretch of 61 aminoacyl residues with aquaporin of Rattus rattus. There is evidence that these water channel proteins and related proteins of the MIP family of channel proteins are involved in adaptation to osmotic stress conditions (8). We investigated the growth of the strain with ORF22 disrupted and of a strain with ORF22 overexpressed in the presence of up to 0.75 M NaCl and in media containing different osmolytes (glycine betaine and proline). As no growth alteration could be detected, an involvement of ORF22 in osmoregulation is unlikely.
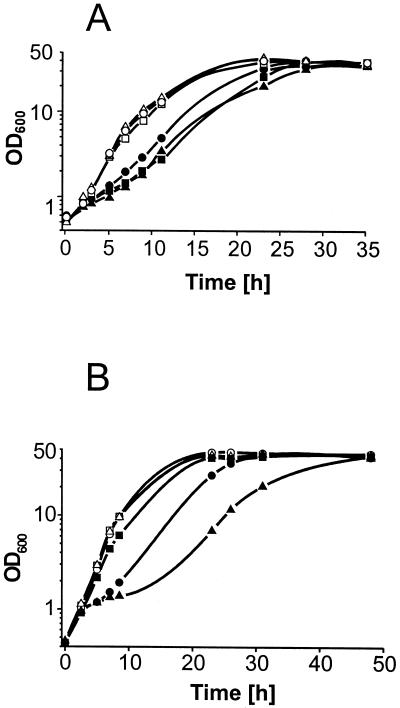
FIG. 2.
(A) Growth of the wild type (circles), of strain 13032::ORF22 (triangles), and of strain 13032::ORF81 (squares) without (open symbols) and with (solid symbols) 2 mM Thr-Thr-Thr. (B) Growth of C. glutamicum 13032(pZ1thrE) (squares) and 13032::thrE (triangles) compared to that of the control strain 13032(pZ1) (circles) without (open symbols) and with (solid symbols) the addition of 2 mM threonine tripeptide.
Similar studies carried out with a set of C. glutamicum ATCC 13032-derived strains with ORF81 interrupted or overexpressed again confirmed growth retardation in the presence of Thr-Thr-Thr (Fig. 2A), but the l-threonine export rate was not altered.
Characterization of ORF53 (thrE) and its gene product.
We therefore focused on ORF53 which, according to the subsequent functional analyses, was termed thrE (threonine exporter). The growth of strain 13032::thrE was indistinguishable from that of the wild type in the absence of Thr-Thr-Thr but was reduced in its presence (Fig. 2B). Overexpression of thrE in strain 13032(pZ1thrE) counteracted the negative Thr-Thr-Thr effect. Growth was even better than that of the wild type in the presence of the peptide, indicating a dose-effect relationship for thrE expression.
An overview of the thrE locus with adjacent ORFs and selected fragments used for strain constructions is given in Fig. 3A. thrE is 1,467 nt long. The thrE gene product is predicted to be a hydrophobic protein of 489 amino acids with a molecular weight of 51,697. The only homologues in databases are Rv3737 of M. tuberculosis (9), with 29.6% identical amino acids, and a putative membrane protein of Streptomyces coelicolor (38), with 22.0% identical amino acids. A hydrophobicity analysis revealed pronounced local hydrophobicities within the stretch of the protein between aminoacyl residues 160 and 430 (Fig. 3B). Accordingly, application of the transmembrane prediction procedure PHD.htm (41) distinguished nine transmembrane-spanning helices within this stretch, placing the extremely long amino-terminal end in the periplasm and the carboxy-terminal end in the cytoplasm. Extensive alignments and database searches showed that ThrE does not belong to any characterized transporter family (43).
Upstream of thrE is a small ORF (399 nt) which might encode a regulator. It has a helix-turn-helix motif and exhibits an identity of 19.5% with the hypothetical transcriptional regulator MTH1328 of Methanobacterium thermoautotrophicum (45). The deduced amino acid sequence of the truncated ORF downstream of thrE exhibits a high identity, up to 63%, with trehalose-6-phosphate synthases (otsA) of several microorganisms.
Determination of the transcriptional start site of thrE.
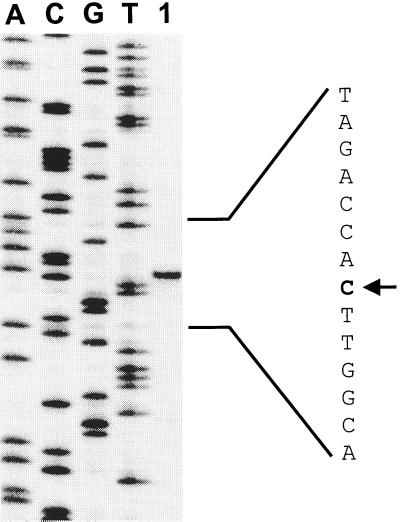
To define the thrE gene, its transcription initiation site was determined. For this purpose, a 267-bp BamHI-KpnI fragment was cloned into the promoter-probe vector pET2. The resulting plasmid made C. glutamicum resistant to chloramphenicol at an MIC of up to 40 μg/ml, indicating that the thrE promoter is of low strength (33). The result of the primer extension experiment with the sequencing reaction carried out in parallel with the same primer is shown in Fig. 4. The same initiation site was determined with a different primer. In front of thrE, an appropriate −10 hexamer is present, whereas a distinct −35 motif is not apparent. This is a typical feature of C. glutamicum promoters (34).
FIG. 4.
Mapping of the transcriptional start site of thrE by primer extension analysis. The primer extension product was run in lane 1. The sequencing ladder (ACGT) of the coding strand was generated using the same primer as that used for primer extension. The transcriptional start site is indicated by the arrow.
Expression of thrE correlates with export and is energy dependent.
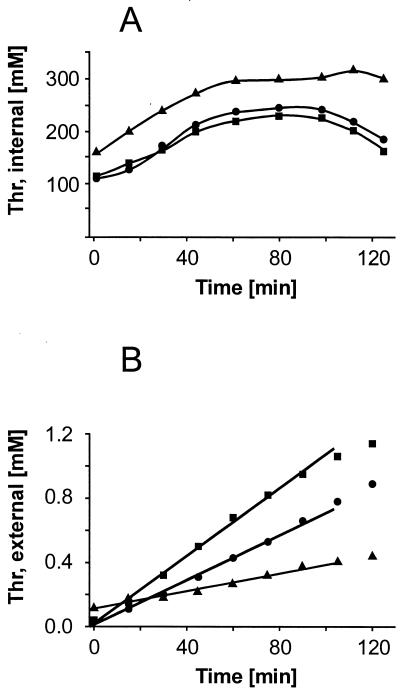
In order to functionally characterize thrE and to quantify its contribution to total cellular l-threonine efflux, export rates were determined with recombinant strains. A precondition for this test is the presence of a high internal l-threonine concentration, since the concentration is normally on the order of 1 mM (Fig. 1B). We therefore tested various conditions in order to achieve a greatly increased internal concentration remaining as constant as possible over an extended period of time. This is the case when cells are incubated for 2 h at 30°C with 1 mM Thr-Thr-Thr in CGXII minimal medium and, after being rinsed with cold medium, are transferred to identical fresh medium. In this way, a high internal l-threonine concentration is obtained at the start of the efflux experiment (Fig. 5A). Under these conditions, the efflux rate for the thrE-overexpressing strain is 3.8 nmol min−1 mg of dry weight−1, and that of the wild type is 2.7 nmol min−1 mg of dry weight−1 (Fig. 5B). This clear difference with almost identical internal concentrations is evidence that the thrE gene product catalyzes the export of l-threonine from the cell. The linear increase could indicate saturation of the exporter at 100 mM. The export rate is reduced to 1.1 nmol min−1 mg of dry weight−1 for the thrE inactivation mutant, even though in this strain the internal threonine concentration may increase to about 300 mM, probably due to the absence of thrE.
FIG. 5.
Intracellular l-threonine concentration and export in recombinant C. glutamicum strains. The internal and external l-threonine concentrations are shown. The strains are C. glutamicum 13032(pZ1thrE) (▪), 13032::thrE (▴), and 13032(pZ1) (control) (●).
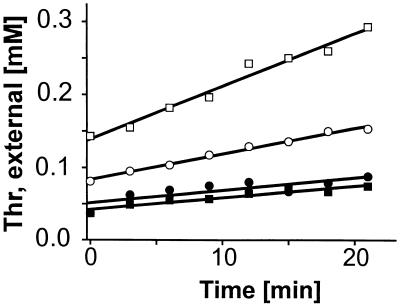
Interestingly, the thrE inactivation mutant still excreted l-threonine at a substantial rate. Since the remaining efflux might be due in part to diffusion (32) or other, still-unknown active carriers, we used the proton ionophore CCCP to distinguish between these possibilities. Threonine export rates were determined as before, but in the presence of 20 μM CCCP. In this experiment, we used shorter measurement times to ensure a high internal l-threonine concentration, since deenergization might also influence peptide uptake. The measurements, made under slightly different conditions (see Materials and Methods), confirmed the export rates for the wild type and the inactivation mutant (Fig. 6). For both strains, however, the efflux rate was reduced to 0.6 nmol min−1 mg of dry weight−1 with CCCP addition. Since active export is abolished under these conditions, the residual efflux is likely to represent passive diffusion. The calculated diffusion constant, Kd, is 0.004 μl min−1 mg of dry weight−1. This experiment shows that the ThrE-mediated export of l-threonine is dependent on the proton motive force. It furthermore indicates that an additional active carrier catalyzing l-threonine export is present in C. glutamicum and verifies that passive diffusion, as a third component (32), contributes to total l-threonine efflux.
FIG. 6.
Effect of the proton ionophore CCCP on l-threonine export in C. glutamicum. Extracellular l-threonine accumulation by 13032::thrE (circles) is compared to that of control strain 13032(pZ1) (squares) without (open symbols) and with (solid symbols) the addition of 20 μM CCCP.
Substrate specificity of ThrE.
It is well-known that the l-threonine uptake systems of E. coli, encoded by tdcC and sstT, also catalyze l-serine uptake (30, 46). We were therefore interested in analyzing whether this is also true of the new carrier which translocates l-threonine from the interior of the cell to the environment of the cell. We once again added peptides (Ser-Ala and Ser-Ser-Ser) to make these measurements possible. With the tripeptide it was possible to achieve an internal concentration, comparable to that of l-threonine, of about 180 mM l-serine (data not shown). The calculated export rates obtained from the linear increase in extracellular l-serine accumulation were 1.9, 1.4, and 0.6 nmol min−1 mg of dry weight−1 for the overexpressing strain, the wild type, and the deletion mutant, respectively.
We also quantified glycine efflux by the three strains with different thrE expression levels. Efflux was determined in the same experiment as that shown in Fig. 5, since intracellular l-threonine is partly degraded to glycine in C. glutamicum (39). However, the level of glycine export in the isogenic strains was almost identical (0.9 nmol min−1 mg of dry weight−1) to that of l-lysine export (about 1.1 nmol min−1 mg of dry weight−1), which was quantified as a control.
DISCUSSION
This study offers an approach to identifying amino acid exporter genes. Its successful application resulted in the identification of thrE. The use of peptide-sensitive mutants is indirect compared to a recent approach in which the cysteine metabolite exporter of E. coli was identified by screening of a gene bank for increased extracellular cysteine accumulation (11). Therefore, some of the target genes inactivated might be more indirectly related to peptide sensitivity. This situation is already discernible from the fact that Thr peptide addition resulted in a short transient increase in intracellular l-threonine levels but a long-lasting growth lag (Fig. 1). These results indicate that in addition to the direct consequences of elevated l-threonine levels, an intracellular pulse of free amino acid initiates a chain of cellular events comparable, for instance, to the stringent response of enterobacteria. ORF22 and ORF70, exhibiting weak identities with aquaporin and trehalose-6-phosphate phosphatase, might be related to such secondary effects as osmotic processes, although we have no experimental evidence to support this possibility.
The polypeptide sequence of the thrE gene product does not exhibit significant identities with known translocators. However, there are two putative proteins in M. tuberculosis and S. coelicolor which share more than 36% similar aminoacyl residues with ThrE. Obviously, ThrE is the prototype of a new translocator family of hitherto-unknown structure. In addition to the predicted nine transmembrane-spanning helices, the proteins are characterized by exceptional N- and C-terminal extensions. With 166 amino acids, the N terminus of ThrE is unusually long. It might be localized toward the periplasmic side, and it is rich in charged amino acids. With 13 positively and 15 negatively charged aminoacyl residues, it carries almost half of all the charged residues in ThrE. Interestingly, in all three homologues, a conserved amphipathic helix is present in this part (Fig. 3C), reminiscent of a similar structure in the long N terminus of ProW of E. coli (15). The C terminus of ThrE displays an even greater charge density. Of the 51 aminoacyl residues, 16 are positively charged and 4 are negatively charged. Such a strong preponderance of charged residues in the C-terminal region is known for the proline betaine transporter ProP of E. coli (10) and the glycine betaine uptake carrier BetP of C. glutamicum (36), where the extension is thought to play a role in regulation of the carrier activity.
ThrE actively exports l-threonine to the extracellular environment. However, there is also a diffusion component of efflux. In a mutant strain of C. glutamicum with deregulated biosynthesis (39), l-threonine excretion was attributed to active export and, to a minor extent, to diffusion (32). The identification of thrE enables the different efflux routes to be quantified in the wild type in detail. At an intracellular concentration in the range of 170 mM l-threonine, at least three separate components contribute to total l-threonine efflux. The major component, amounting to 59%, is the export driven by ThrE. This is evident from the analysis of the inactivation mutant. However, part of the remaining translocation is still dependent on the proton motive force. After CCCP addition (Fig. 6), the efflux due to passive diffusion contributes 22%. Therefore, a still-unknown carrier is expected to catalyze the remaining 19% of export. Since together with LysE we have now already found two novel export carriers (49; this work), it would not be surprising if there were other export carriers as well. Thus, for example, an assumed l-isoleucine transporter in C. glutamicum (52) could also export l-threonine, since it is known that the branched-chain amino acid import system LIV-I of Pseudomonas aeruginosa also accepts l-threonine with a low affinity (17).
A pertinent question is, of course, what the natural function of the discovered exporter might be. With respect to the basic amino acid exporter LysE of C. glutamicum, the absence of degrading activities for l-lysine and l-arginine and the control of lysE expression by these amino acids (4) are in agreement with the idea that LysE naturally serves to export these two amino acids. A special threonine-degrading activity, like that of the threonine dehydrogenase (tdh) in E. coli (7), is not present in C. glutamicum (unpublished results). The fact that the thrE deletion strain displays any phenotype at all at high internal l-threonine concentrations excludes a basic function of ThrE. However, the exporter could be required under special conditions. In this regard, it is interesting that C. glutamicum can be isolated only from soil samples contaminated with bird feces (51). In a special environment, such as the bird intestine, the export of selected low-molecular-weight compounds might be advantageous. This scenario, together with the fact that ThrE also accepts l-serine as a substrate, a feature typical of l-threonine uptake carriers like TdcC and SstT of E. coli (30, 46), leads us to assume that ThrE of C. glutamicum is structurally designed for the export of small solutes that have a structure similar to that of l-threonine.
ACKNOWLEDGMENTS
We thank M. Pátek for carrying out the primer extension experiment.
We thank Degussa AG for financial support.
REFERENCES
- 1.Aleshin V V, Zakataeva N P, Livshits V A. A new family of amino-acid-efflux proteins. Trends Biochem Sci. 1999;24:133–135. doi: 10.1016/s0968-0004(99)01367-5. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ankri S, Serebrijski I, Reyes O, Leblon G. Mutations in the Corynebacterium glutamicum proline biosynthetic pathway: a natural bypass of the proA step. J Bacteriol. 1996;178:4412–4419. doi: 10.1128/jb.178.15.4412-4419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellmann A, Vrljic M, Pátek M, Sahm H, Krämer R, Eggeling L. Regulation and specificity of the LysE-mediated export of amino acids by Corynebacterium glutamicum. Microbiology. 2001;147:1765–1774. doi: 10.1099/00221287-147-7-1765. [DOI] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Bost S, Silva F, Belin D. Transcriptional activation of ydeA, which encodes a member of the major facilitator superfamily, interferes with arabinose accumulation and induction of the Escherichia coli arabinose PBAD promoter. J Bacteriol. 1999;181:2185–2191. doi: 10.1128/jb.181.7.2185-2191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boylan S A, Dekker E E. l-Threonine dehydrogenase. Purification and properties of the homogenous enzyme from Escherichia coli K-12. J Biol Chem. 1981;256:1809–1815. [PubMed] [Google Scholar]
- 8.Calamita G, Kempf B, Bonhivers M, Bishai W R, Bremer E, Agre P. Regulation of the Escherichia coli water channel aqpZ. Proc Natl Acad Sci USA. 1998;95:3627–3631. doi: 10.1073/pnas.95.7.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 10.Culham E D, Lansby B, Marangoni A G, Milner J L, Steer B A, van Nues R W, Wood J M. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J Mol Biol. 1993;229:268–276. doi: 10.1006/jmbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- 11.Daßler T, Maier T, Winterhalter C, Böck A. Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway. Mol Microbiol. 2000;36:1101–1112. doi: 10.1046/j.1365-2958.2000.01924.x. [DOI] [PubMed] [Google Scholar]
- 12.Eikmanns B J, Thum-Schmitz N, Eggeling L, Lüdtke K, Sahm H. Nucleotide sequence, expression and transcriptional analysis of gltA gene encoding citrate synthase. J Gen Microbiol. 1994;140:1817–1828. doi: 10.1099/13500872-140-8-1817. [DOI] [PubMed] [Google Scholar]
- 13.Erdmann A, Weil B, Krämer R. Lysine secretion by wild-type Corynebacterium glutamicum triggered by dipeptide uptake. J Gen Microbiol. 1993;139:3115–3122. [Google Scholar]
- 14.Grant S G N, Jessee J, Bloom F R, Hanahan S. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haardt M, Bremer E. Use of phoA and lacZ fusions to study the membrane topology of ProW, a component of the osmoregulated ProU transport system of Escherichia coli. J Bacteriol. 1996;178:5370–5381. doi: 10.1128/jb.178.18.5370-5381.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoischen C, Krämer R. Evidence for an efflux carrier system involved in the secretion of glutamate by Corynebacterium glutamicum. Arch Microbiol. 1989;151:342–347. [Google Scholar]
- 17.Hoshino T, Kose-Terai K, Sato K. Solubilization and reconstitution of the Pseudomonas aeruginosa high affinity branched-chain amino acid transport system. J Biol Chem. 1992;267:21313–21318. [PubMed] [Google Scholar]
- 18.Juillard V, Le Bars D, Kunji E R S, Konings W N, Gripon J, Richard J. Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth on milk. Appl Environ Microbiol. 1995;61:3024–3030. doi: 10.1128/aem.61.8.3024-3030.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keilhauer C, Eggeling L, Sahm H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klingenberg M, Pfaff E. Means of terminating reactions. Methods Enzymol. 1977;10:680–684. [Google Scholar]
- 21.Kunji E R S, Mierau I, Poolman B, Konings W N, Venema G, Kok J. Fate of peptides in peptidase mutants of Lactococcus lactis. Mol Microbiol. 1996;21:123–131. doi: 10.1046/j.1365-2958.1996.6231339.x. [DOI] [PubMed] [Google Scholar]
- 22.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 23.Liebl W, Bayerl A, Stillner U, Schleifer K H. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Micobiol Lett. 1989;65:299–304. doi: 10.1016/0378-1097(89)90234-6. [DOI] [PubMed] [Google Scholar]
- 24.Liu J Y, Miller P F, Willard J, Olson E R. Functional and biochemical characterization of Escherichia coli sugar efflux transporters. J Biol Chem. 1999;274:22977–22984. doi: 10.1074/jbc.274.33.22977. [DOI] [PubMed] [Google Scholar]
- 25.Menkel E, Thierbach G, Eggeling L, Sahm H. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl Environ Microbiol. 1989;55:684–688. doi: 10.1128/aem.55.3.684-688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Möckel B, Eggeling L, Sahm H. Threonine dehydratases of Corynebacterium glutamicum with altered allosteric control: their generation and biochemical and structural analysis. Mol Microbiol. 1994;13:833–842. doi: 10.1111/j.1365-2958.1994.tb00475.x. [DOI] [PubMed] [Google Scholar]
- 27.Nies D H. Microbial heavy-metal resistance. Appl Microbiol Biotechnol. 1999;51:730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 28.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 29.Nisbet T M, Payne J W. The characteristics of peptide uptake in Streptococcus faecalis: studies on the transport of natural peptides and antibacterial phosphonopeptides. J Gen Microbiol. 1982;128:1357–1364. doi: 10.1099/00221287-128-6-1357. [DOI] [PubMed] [Google Scholar]
- 30.Ogawa W, Kim Y-M, Mizushima T, Tsuchiya T. Cloning and expression of the gene for the Na+-coupled serine transporter from Escherichia coli and characteristics of the transporter. J Bacteriol. 1998;180:6749–6752. doi: 10.1128/jb.180.24.6749-6752.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer B R, Marinus M G. The dam and dcm strains of Escherichia coli—a review. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- 32.Palmieri L, Berns D, Krämer R, Eikmanns M. Threonine diffusion and threonine transport in Corynebacterium glutamicum and their role in threonine production. Arch Microbiol. 1996;165:48–54. [Google Scholar]
- 33.Pátek M, Bilic M, Krumbach K, Eikmanns B, Sahm H, Eggeling L. Identification and transcriptional analysis of the dapB-ORF2-dapA-ORF4 operon of Corynebacterium glutamicum, encoding two enzymes involved in l-lysine synthesis. Biotechnol Lett. 1997;19:1113–1117. [Google Scholar]
- 34.Pátek M, Eikmanns B J, Pátek J, Sahm H. Promoters from Corynebacterium glutamicum: cloning, molecular analysis and search for a consensus motif. Microbiology. 1996;142:1297–1309. doi: 10.1099/13500872-142-5-1297. [DOI] [PubMed] [Google Scholar]
- 35.Payne J W, Bell G. Direct determination of the properties of peptide transport systems in Escherichia coli, using a fluorescent-labeling procedure. J Bacteriol. 1979;137:447–455. doi: 10.1128/jb.137.1.447-455.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peter H, Burkovski A, Krämer R. Osmo-sensing by N- and C-terminal extensions of the glycine betaine uptake system BetP of Corynebacterium glutamicum. J Biol Chem. 1998;273:2567–2574. doi: 10.1074/jbc.273.5.2567. [DOI] [PubMed] [Google Scholar]
- 37.Poolman B, Molenaar D, Smid E J, Ubbink T, Abee T, Renauld P P, Konings W N. Malolactic fermentation: electrogenic uptake and malate/lactate antiport generate metabolic energy. J Bacteriol. 1991;173:6030–6037. doi: 10.1128/jb.173.19.6030-6037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 39.Reinscheid D J, Kronemeyer W, Eggeling L, Eikmanns B J, Sahm H. Stable expression of hom1-thrB in Corynebacterium glutamicum and its effect on the carbon flux to threonine and related amino acids. Appl Environ Microbiol. 1994;60:126–132. doi: 10.1128/aem.60.1.126-132.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg M F, Callaghan R, Ford R C, Higgins C F. Structure of the multidrug resistance P-glycoprotein to 2.5 nm resolution determined by electron microscopy and image analysis. J Biol Chem. 1997;272:10685–10694. doi: 10.1074/jbc.272.16.10685. [DOI] [PubMed] [Google Scholar]
- 41.Rost B, Casadio R, Fariselli P, Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahm H, Eggeling L. d-Pantothenate synthesis in Corynebacterium glutamicum and use of panBC and genes encoding l-valine synthesis for d-pantothenate overproduction. Appl Environ Microbiol. 1999;65:1973–1979. doi: 10.1128/aem.65.5.1973-1979.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saier M H. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 45.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H-M, Dubois J, Aldredge T, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sumantran V N, Schweizer H P, Datta P A. A membrane-associated threonine permease encoded by the tdcC gene of Escherichia coli. J Bacteriol. 1990;172:4288–4294. doi: 10.1128/jb.172.8.4288-4294.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Rest M E, Lange C, Molenaar D. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl Microbiol Biotechnol. 1999;52:541–545. doi: 10.1007/s002530051557. [DOI] [PubMed] [Google Scholar]
- 48.Vašicová P, Abrhámová Z, Nešvera J, Pátek M, Sahm H, Eikmanns B. Integrative and autonomously replicating vectors for analysis of promoters in Corynebacterium glutamicum. Biotechnol Techniques. 1998;12:743–746. [Google Scholar]
- 49.Vrljic M, Garg J, Bellmann A, Wachi S, Freudl R, Malecki M J, et al. The LysE superfamily: topology of the lysine exporter LysE of Corynebacterium glutamicum, a paradigm for a novel superfamily of transmembrane solute translocators. J Mol Microbiol Biotechnol. 1999;1:327–336. [PubMed] [Google Scholar]
- 50.Vrljic M, Sahm H, Eggeling L. A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol Microbiol. 1996;22:815–826. doi: 10.1046/j.1365-2958.1996.01527.x. [DOI] [PubMed] [Google Scholar]
- 51.Woodruff H B. A soil microbiologist's odyssey. Annu Rev Microbiol. 1981;35:1–28. doi: 10.1146/annurev.mi.35.100181.000245. [DOI] [PubMed] [Google Scholar]
- 52.Zittrich S, Krämer R. Quantitative discrimination of carrier-mediated excretion of isoleucine from uptake and diffusion in Corynebacterium glutamicum. J Bacteriol. 1994;176:6892–6899. doi: 10.1128/jb.176.22.6892-6899.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]