Abstract
Klebsiella pneumoniae is an opportunistic pathogen, which concerns public health systems worldwide, as multiple antibiotic‐resistant strains are frequent. One of its pathogenicity factors is the Type VI Secretion System (T6SS), a macromolecular complex assembled through the bacterial membranes. T6SS injects effector proteins inside target cells. Such effectors confer competitive advantages or modulate the target cell signaling and metabolism to favor bacterial infection. The VgrG protein is a T6SS core component. It may present a variable C‐terminal domain carrying an additional effector function. Kp52.145 genome encodes three VgrG proteins, one of them with a C‐terminal extension (VgrG4‐CTD). VgrG4‐CTD is 138 amino acids long, does not contain domains of known function, but is conserved in some Klebsiella, and non‐Klebsiella species. To get insights into its function, recombinant VgrG4‐CTD was used in pulldown experiments to capture ligands from macrophages and lung epithelial cells. A total of 254 proteins were identified: most of them are ribosomal proteins. Cytoskeleton‐associated and proteins involved in the phagosome maturation pathway were also identified. We further showed that VgrG4‐CTD binds actin and induces actin remodeling in macrophages. This study presents novel clues on the role of K. pneumoniae T6SS in pathogenesis.
Keywords: bacterium–host interactions, cytoskeleton, protein–protein interactions, proteomics, T6SS
1. INTRODUCTION
Klebsiella pneumoniae is a gram‐negative encapsulated member of the Enterobacteriaceae family. This species is ubiquitous in nature, and frequently associated with animals and humans. 1 On one hand, there are the classical pathogenic strains mainly affecting immune‐compromised or hospitalized individuals, causing infections in the urinary and respiratory tracts, as well as bacteremia. Frequently, those strains are multiple antibiotic‐resistant. 2 In another hand, there are hypervirulent strains that may cause acute community‐acquired infections. Moreover, K. pneumoniae virulence and resistance determinants are often encoded in mobile genetic elements, and there are reports of strains that are simultaneously hypervirulent and multiple antibiotic‐resistant. 3 , 4 , 5 Considering the limited therapeutical alternatives for the treatment of such infections, there is an urgent need for research and development of new strategies to control the K. pneumoniae infections. In this context, broadening the knowledge of pathogen biology and host–pathogen molecular interactions may provide new insights.
K. pneumoniae virulence factors include the polysaccharide capsule, lipopolysaccharide, siderophores, fimbriae, a phospholipase D family protein, type II and type VI secretions systems, among others. 6 , 7 , 8 , 9
Gram‐negative bacteria encode and utilize secretion systems to translocate proteins from their cytoplasm to the extracellular medium. 10 , 11 Secretion systems are macromolecular protein complexes that enable the transport of large and relatively hydrophilic molecules which do not cross the membrane by other mechanisms. Some of these systems are formed by two different complexes inserted in the inner and outer membranes, while other systems form continuous tubes that cross both membranes. Some systems may even traverse a third membrane, thus translocating molecules from the bacterial cytoplasm directly into the target cell. 10 , 11
The type VI secretion system (T6SS) is composed of proteins from at least 13 protein families that assemble to form a central tube, a contractile outer sheath, inner and outer membranes anchoring complexes, chaperones, and a cytoplasmic ATPase. 12 , 13 The central tube is formed by hexameric rings of Hcp (hemolysin‐coregulated protein/TssD), which are stacked from the bacterial cytoplasm and are eventually propelled to the extracellular compartment as a consequence of a conformational change in VipA/VipB dodecamers (TssB and TssC, respectively). 14 , 15 VipA/VipB dodecamers form a cytoplasmatic sheath around the Hcp tube. 16 , 17 , 18 , 19
At the tip of the central Hcp tube is the spike‐like trimer of VgrG (TssI) protein, often associated with PAAR repeat‐containing proteins. 20 , 21 , 22 , 23 , 24 , 25 So far, it has been shown that Hcp, VgrG, and PAAR proteins may recruit effectors, either by covalent interactions or by noncovalent associations. Upon sheath contraction, the Hcp tube and VgrG/PAAR tip are propelled through the membranes. These components and associated effectors are then released. 23 , 26 , 27 , 28
The VgrG family proteins typically have sequences ranging from ~750 to 1200 amino acids. The ~750 amino acids enclose conserved domains. On the other hand, there is a variable C‐terminal region that often confers an effector role to the protein. 29 , 30 , 31 , 32 , 33 For example, the C‐terminal regions of Vibrio cholerae and Aeromonas hydrophyla VgrG proteins present a domain capable of inducing reorganization of the actin cytoskeleton of the host cells. 24 , 33 , 34 In other species, VgrGs contain domains conferring lipase activity or peptidoglycan binding, for instance. 32 Thus, among the T6SS proteins, VgrG is one of the components directly related to the functional activity of the complex.
There are thousands of K. pneumoniae genomes sequenced so far, and most of them contain genes encoding T6SS proteins. However, there is extensive diversity in gene content and organization in those loci among strains. 7 , 35 , 36 , 37 It has been shown that K. pneumoniae T6SS might be used to outcompete bacteria and fungi. 37 , 38 , 39 It is expressed during bacterial infection of macrophages, 40 and affects fimbriae expression, cell adhesion, invasion and intestinal colonization. 39 Moreover, it has been shown that K. pneumoniae T6SS is regulated by temperature, oxygen tension, pH, osmolarity, iron levels, polymyxins, and human defensin 3, 37 , 41 in a mechanism dependent on the two‐component system PhoPQ. 37 In the Kp52.145 strain, it has been pointed out that VgrG4 is an important antibacterial effector. 37
In this strain, there are three genes coding for VgrG family proteins (BN49_RS06025, BN49_RS14055, and BN49_RS18800). All of them contain the following conserved domains: Phage GPD domain, Rhs element, and DUF2345. It has been shown that the DUF2345 domain of VgrG4 (BN49_RS18800) is sufficient to intoxicate bacteria and yeast. 37 VgrG4 also codes for 138 additional amino acids at its C‐terminal (VgrG4‐CTD). VgrG4‐CTD does not present significant similarity to other proteins of known function, except other VgrG family proteins. We believe that understanding the function of the VgrG4‐CTD is a key for the elucidation of T6SS role in the pathogenesis of this species.
Herein, we employed mass spectrometry‐based proteomics to identify K. pneumoniae VgrG4‐CTD ligands and generate hypotheses regarding the role of this domain in the virulence of K. pneumoniae. We found that VgrG4‐CTD binds ribosome and cytoskeleton‐associated proteins from macrophages and epithelial cells, as well as phagosome maturation proteins in macrophages. Recombinant VgrG4‐CTD binds actin, and induces alterations in actin microfilaments network. These results suggest that K. pneumoniae VgrG4 might be involved in pathogenesis, in addition to the interspecies competition.
2. EXPERIMENTAL PROCEDURES
2.1. Protein sequence analysis
VgrG4 protein sequence is under accession WP_046043552 in RefSeq database. Conserved domains were identified with Pfam 34.0. Similarity searches were performed using the online NCBI Blastp tool.
2.2. VgrG4‐CTD recombinant expression and purification
vgrG4‐CTD was cloned into pET28a+ using NdeI and BamHI restriction sites, by Genscript. This construct allows for the expression of VgrG4‐CTD fused at its N‐terminal to a 6×‐Histidine tag and a thrombin cleavage site (Figure S1). The plasmid was transformed into chemically competent Escherchia coli BL21 (DE3) cells by standard thermal‐shock procedures. Transformants were selected by plating into LB‐agar culture medium supplemented with 50 μg/ml kanamycin.
Bacteria were grown up to OD600nm ~ 0.6 to 0.8, then protein expression was induced by the addition of 0.5 mM IPTG (isopropyl β‐d‐1‐thiogalactopyranoside) at 37°C, 120 rpm, for 4 h (Figure S2A). Bacterial cells were collected by centrifugation at 5000 × g for 30 min at 4°C, then resuspended in phosphate buffer 20 mM pH 7.4, NaCl 500 mM, imidazole 20 mM, and 1 mM phenylmethanesulfonyl fluoride. Cells were lysed by sonication in an ice‐cold bath, and soluble proteins were collected as the supernatant after a 14 000 × g centrifugation for 30 min at 4°C.
The soluble protein extract was injected into a nickel‐affinity column HisTrap FF 5 ml (GE Healthcare) at the fast protein liquid chromatographer AKTA (GE Healthcare). Phosphate buffer 20 mM pH 7.4, NaCl 500 mM, and imidazole 20 mM were used as the chromatographic buffer. His‐trapped protein was eluted along an imidazole gradient from 20 mM to 0.5 M, under 1.5 ml min‐1 mobile phase flux (Figure S2B). Protein identity was confirmed by mass spectrometry. Protein purity was estimated by gel electrophoresis at >95%, thus not requiring additional purification steps. Cleavage of histidine‐tag using thrombin resulted in protein degradation (data not shown). Therefore, we used the 6×‐Histidine tagged protein on further experiments.
2.3. Eukaryotic cell culture
THP‐1 (ATCC Cat# TIB‐202, RRID:CVCL_0006), a cell line of monocytes derived from peripheral blood, were routinely grown in RPMI‐Roswell Park Memorial Institute Medium 1640 (LGC Biotech) supplemented with 10% SFB‐fetal bovine serum (Cripion) in 5% CO₂ at 37°C. Monocytes were treated with 80 mM PMA (Phorbol 12‐Myristate 13‐Acetate, SIGMA‐ALDRICH) for 24 h to differentiate to macrophages. All experiments were performed with THP‐1‐derived macrophages. A549 lung epithelial cells (ATCC Cat# CRM‐CCL‐185, RRID:CVCL_0023) were maintained in Dulbecco's Modified Eagle Medium F12 (LGC Biotechnology) supplemented with 10% SFB (Cripion) in a 5% CO₂ incubator at 37°C. RAW 264.7 (ATCC Cat# SC‐6003, RRID:CVCL_UL71) macrophages from an adherent blood cell line from Mus musculus BALB/c mice were maintained in Dulbecco's Modified Eagle Medium High‐Glucose (LGC Biotechnology) supplemented with 10% SFB (Cripion) in a 5% CO₂ incubator at 37°C.
2.4. Protein extraction
The 107 cells were treated following the protocol of the ProteoExtract Transmembrane Protein Extraction Kit (Calbiochem) to obtain both a soluble and a membrane‐enriched extract. Extraction is performed in two steps, using differential solubility property of membrane proteins—as described subsequently. Cells were washed in PBS twice and then released from the bottle with 10 mM EDTA pH 8. Cells were centrifuged for 10 min at 300 × g at 4°C and the supernatant discarded. The cell pellet was resuspended in 1 ml of Extraction Buffer 2A, and 5 μl of protease inhibitors were added. The sample was transferred to a microtube, incubated for 10 min at 4°C, and centrifuged for 5 min at 10 000 × g at 4°C. The supernatant was transferred to a new microtube (soluble fraction of the protein extract). Then, the pellet was resuspended in 200 μl of Extraction Buffer 2B. Such buffer is indicated as a highly efficient extraction agent, facilitating the recovery of transmembrane proteins of difficult extraction. It consists of 1:1 Buffer II:Tm‐PEK reagent B. Further, we added 5 μl of protease inhibitors, and the sample was incubated for 2 h at room temperature on FANEM Orbital 255‐B Stirrer. The sample was then centrifuged for 15 min at 16 000 × g at 4°C. The supernatant was transferred to a microtube (membrane‐enriched protein fraction). We performed biological triplicates from three independent cultures of both A549 cells and THP‐1 macrophages.
Protein concentration was measured by the Qubit fluorimetric method. The 10 μg of each protein extract was subjected to a 12.5% polyacrylamide gel electrophoresis to separate the proteins according to their molecular weight. Proteins were stained with Coomassie Blue R‐250.
2.5. Pull‐down assays
Latex beads of 3 μm (Sigma) were used as a support for VgrG4‐CTD protein binding. The 1.4×106 beads were incubated with 250 μg of the VgrG4‐CTD protein in 50 mM MES buffer pH 6.1. Samples were kept at 4°C overnight for protein adsorption. Then, samples were centrifuged for 15 min at 5000 × g at 4°C, and the supernatant was discarded. The pellet containing coated beads was resuspended in 200 mg/ml glycine solution in 50 mM MES pH 6.1 to fulfill putative empty binding sites on the beads surface. Beads were then collected by centrifugation for 15 min at 5000 × g at room temperature. As a control, “empty beads” were prepared ‐ which underwent the entire procedure described above, but the VgrG4‐CTD protein solution was replaced with 50 mM MES buffer pH 6.1. VgrG4‐CTD coated beads, as well as control beads, were incubated with the following protein extracts: A549 soluble fraction, A549 membrane‐enriched extract, THP‐1 soluble, THP‐1 membrane‐enriched extract—in biological triplicates, summing 12 samples. For each binding assay, ~1.4×106 beads were incubated with 125 μg of each protein extract in 50 mM MES buffer pH 6.1, for 3 h, at room temperature, with orbital shaking, and then kept overnight at 4°C. Samples were centrifuged for 15 min at 5000 × g at room temperature. Beads were then washed twice with PBS to remove proteins that did not bind to VgrG4‐CTD and were collected by centrifugation for 15 min at 5000 × g at room temperature. Samples were immediately processed as described subsequently.
2.6. Proteomic analysis
A bottom‐up proteomics approach was performed for the identification of VgrG4‐CTD ligands. Pellets containing the beads and bound proteins were resuspended in 50 μl of 50 mM NH₄HCO3, and heated for 10 min at 80°C to promote protein denaturation. Then, proteins were treated with 100 mM DTT (Di‐Thio‐Threitol) for 30 min at 60°C to reduce the disulfide bonds, and then with 300 mM IAA (IodoAcetoAmide) for 30 min in the dark, at room temperature for alkylation of cysteines. Proteins were trypsin digested with Trypsin Gold MS‐grade (Promega) 0.2 mg/ml at 37°C overnight. The 5% formic acid was added to interrupt the reaction. Samples were centrifuged for 15 min at 5000 × g at room temperature and the supernatant containing the soluble peptides was collected and dried on a Speed Vac System (THERMO SAVANT's ISS110) for 50 min. Peptides were resuspended in 20 μl of 0.1% trifluoroacetic acid (TFA), desalted on C18 ziptips (Millipore) according to the manufacturer's protocol. Peptides were speed‐vac dried again, for 20 min, and finally resuspended in 20 μl of 3% acetonitrile (ACN) and 0.1% TFA.
2.7. Mass spectrometry
Peptides were analyzed on LC–MS/MS to assign unique peptide sequences and perform protein identification—at the Proteomics and Mass Spectrometry Unit (UEMP) of the Federal University of Rio de Janeiro, Brazil. Samples were automatically injected into the Waters Nano Acquity System (Waters, Milford, MA) and peptides were desalted and separated throughout a 0%–50% ACN gradient on a C18 column. The ESI‐Q‐TOF mass spectrometry apparatus (Q‐Tof Micro‐Waters Corporation) was used for data acquisition. Instrument control was conducted in the MassLynx program (Version 4.1, Waters). All data were processed using the ProteinLynx Global server (version 2.5, Waters) where the mass/charge value of each precursor (MS) and fragment (MS/MS) were determined from the mass spectra from these chromatograms using the Q‐Tof LockSpray system (Waters, Milford, MA).
2.8. Protein identification
The quality of the mass spectra was evaluated manually in the MassLynx (Waters) program, and the mass lists were generated by the ProteinLynx Global Server (Waters) program. Proteins were identified using MASCOT Server (www.matrixscience.com), considering the following parameters: Database: SwissProt version September 2017, Taxonomy: Homo sapiens, Enzyme: Trypsin, One missed cleavage, Fixed modification: Carbamidomethyl (C), Variable modifications: Oxidation (M), Peptide tolerance ±: 0.2, MS/MS tolerance ±: 0.2, Peptide charge: +2, +3, +4, Data format: Micromass (.PKL), Instrument: ESI‐QUAD‐TOF. Protein categorization and enrichment analysis were performed according to Gene Ontology (http://geneontology.org/) and Panther (http://pantherdb.org), which provide information on molecular function, biological process, and cellular component. Protein–protein network analysis was performed with String (https://string-db.org/) and Cytoscape 3.8.2.
2.9. Bacterial culture
The wild‐type K. pneumoniae strain Kp52.145 was obtained from the collection of the Pasteur Institute. K. pneumoniae strain was grown in LB Miller broth medium and incubated at 150 rpm, at 37°C for 16 h. Then, they were spiked 1:1000 in LB medium and incubated at 37°C until O.D.600nm ~ 0.7.
2.10. Intracellular delivery of VgrG4‐CTD using PULSin reagent
A total of 1 × 105 RAW 264.7 cells were plated in coverslips‐containing 24‐well plates (KASVI) in DMEM/High‐Glucose +10% FBS medium. Plates were incubated at 37°C with 5% CO₂ atmosphere for 16 h for cell adhesion. PULSin‐VgrG4‐CTD complex was prepared according to standard procedures recommended by the manufacturer (Polyplus). Cells were stimulated with the PULSin‐VgrG4‐CTD complex or with pure PULSin reagent for 4 h at 37°C with 5% CO₂ atmosphere. Nonstimulated cells were analyzed as a control.
2.11. Evaluation of actin filaments by fluorescence microscopy
We analyzed RAW 264.7 cells stimulated with VgrG4‐CTD versus nonstimulated. For infection conditions, K. pneumoniae at the multiplicity of infection (MOI) 50:1 was used. Plates were centrifuged to synchronize the infection, and samples were incubated for 1 h at 37°C and 5% CO₂ atmosphere. Then, cells were fixed in 4% paraformaldehyde for 20 min, at 4°C. Cells were washed in PBS and permeabilized with PBS/Triton X‐100 0.5% for 15 min. This procedure was repeated 3 times. Then, actin filaments were stained with Atto 488 Phalloidin (Sigma‐Aldrich) 12.5 pmol, for 30 min, in the dark. Cells were 3× washed in PBS. Nuclei were evidenced by 4,6‐diamidino‐2‐phenylindole staining (Sigma‐Aldrich); slides were mounted with Permafluor (Thermo Scientific) and analyzed via the AxiObserver Z1 Colibri microscope.
2.12. Protein binding assays in microplates
To corroborate the capacity of K. pneumoniae VgrG4‐CTD to bind to actin, tubulin, vimentin, and cytokeratin, 70 μg/ml of recombinant VgrG4‐CTD in 100 μl 0.05 M sodium carbonate–bicarbonate buffer (pH 9.6) was used to coat the wells of a polystyrene microplate (Costar 3590, Corning). The microtiter plate was incubated overnight at 4°C. The wells were then washed with PBS/0.05% Tween 20 (PBS/T) and blocked for 1 h with 100 μl 5% BSA in PBS at 4°C. After three washing steps with PBS/T, 100 μl of increasing concentrations of A549 or THP‐1 protein extracts were added to the wells and incubated at 4°C for 3 h. The wells were then rinsed 3 times with PBS/T and incubated with the antiatom (RRID:AB_2893453), antivimentin (RRID:AB_2893452), anticytokeratin (Santa Cruz Biotechnology Cat# sc‐56 371, RRID:AB_2133461), or antitubulin (Santa Cruz Biotechnology Cat# sc‐5274, RRID:AB_2288090) antibodies, diluted 1:300, for 1 h at 4°C. After washing with PBS/T, rabbit anti‐mouse IgG peroxidase conjugate (Sigma; 1:1000) was added and incubated for an additional 1 h at 4°C. Peroxidase activity was revealed with 3,3′,5,5’‐Tetramethylbenzidine (TMB) chromogenic substrate at 0.18 μg/ml in sodium acetate/citric acid pH 4 buffer for 1 h. The reaction was stopped with 100 μl of 2.5 N sulfuric acid solution and read at 450 nm with an EON microplate reader (Biotek). Control wells coated with BSA were included in all binding assays.
2.13. G‐actin purification
G‐actin from chicken skeletal muscles was previously extracted with acetone. The acetone powder extract was solubilized in low temperature by stirring with 20 ml of buffer A per gram of powder. Buffer A consists of 2.5 mM Tris–HCl buffer, 0.2 mM CaCl2 pH 8.0. The supernatant was passed through several layers of gauze and centrifuged for 10 min at 14 500 rpm at 4°C.
2.14. Protein–protein interaction assessed by Ni2+‐affinity chromatography
Binding interactions between actin and His6‐tagged VgrG4‐CTD proteins were assayed by copurification on Nickel Sepharose Fast Flow resin. The resin was washed with 5 volumes of water, then charged with 2 volumes of 500 mM NiCl2, washed with 5 volumes of water, and equilibrated with 5 volumes of the reaction buffer (20 mM Tris pH 7.5 and 500 mM NaCl). Five μg of each protein were mixed in the reaction buffer. Then, the protein mixture was added to the Ni2+ resin and incubated at room temperature for 1 h under rotational agitation. The reactions were centrifuged to collect the resin, and the supernatant was removed and analyzed as the “unbound” fraction. The resin was then washed twice with 2 volumes of reaction buffer supplemented with 20 mM imidazole. The suspension was centrifuged to collect the resin, and the supernatants were removed and analyzed as the “wash 1” and “wash 2” fractions. Resin‐bound proteins were eluted with reaction buffer supplemented with 250 mM imidazole. The suspension was centrifuged to collect the resin, and the supernatant was removed and analyzed as the “bound” fraction. All fractions were analyzed by SDS‐PAGE and stained with Coomassie brilliant blue.
2.15. Fluorescence spectroscopy
The fluorescence spectra were acquired in a RF‐5301PC Shimadzu fluorescence spectrophotometer and processed using “Fluorescence Spectroscopy for RF‐531PC Version 2.04” software. The samples were excited at 295 nm and the observed emission range was 300 to 450 nm. The processed spectra were analyzed in SigmaPlot 10.0 software from Systat Software Inc. VgrG4‐CTD was in 20 mM phosphate buffer, 200 mM NaCl pH 7.4. Its concentration was fixed in 2 μM and titrated with increasing actin concentrations ranging from 0.5 to 12 μM.
3. RESULTS AND DISCUSSION
3.1. The C‐terminal region of VgrG4 is conserved in some Klebsiella isolates and other species
Kp52.145 VgrG4 is 899 amino acids long. Three conserved domains have been identified (Figure 1A): Phage‐GPD (position 35–355), T6SS_VgrG (position 490–592), and DUF2345 (position 612–759). This domain architecture is usual in some VgrG family proteins, according to Pfam database, as well as literature. 42 , 43 We did not find any conserved domain, nor structure, related to the sequence after DUF2345. Herein, the sequence comprised between amino acids 761–899 is designated VgrG4‐CTD.
FIGURE 1.
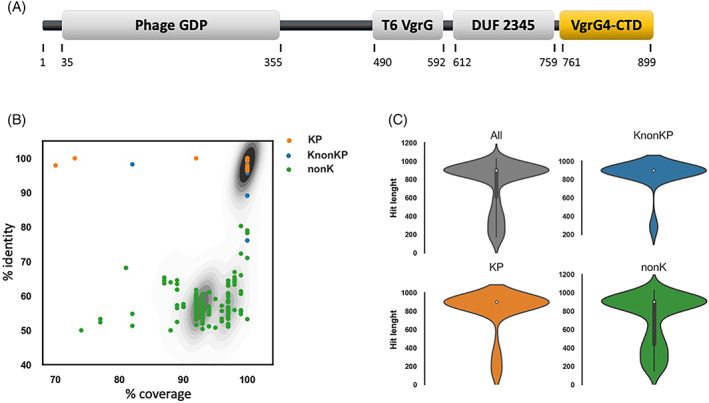
VgrG4‐CTD sequence does not contain known domains but is conserved in several K. pneumoniae isolates (KP), Klebsiella non‐K. pneumoniae (KnonKP), and also non‐Klebsiella bacteria (nonK). (A) Schematic representation of the VgrG4 amino acid sequence. Functional domains according to NCBI Conserved Domain Database are represented as gray boxes: Phage GPD, T6SS VgrG, and DUF 2345. The C‐terminal region (amino acids 761–899) does not contain any known domain, and is highlighted in a yellow box (VgrG4‐CTD). (B) Scatter plot showing the % identity, and % coverage of Blastp hits of VgrG4‐CTD. Contour plot in the background represents the density of hits falling in the same position of the plot. (C) Violin plots showing the density of VgrG4‐CTD hits according to hit length (amino acids).
On one hand, there is no sequence of known function related to VgrG4‐CTD. On the other hand, we found hundreds of protein sequences, mostly annotated as VgrG family proteins, containing sequences similar to VgrG4‐CTD. There are proteins containing VgrG4‐CTD similar sequences in at least 102 K. pneumoniae (>96% identity and > 70% coverage), 38 Klebsiellas non‐K. pneumoniae (>76% identity and > 82% coverage), and 229 non‐Klebsiella (> 50% identity and > 74% coverage) (Figure 1B). Approximately 73% of those proteins containing VgrG4‐CTD‐similar sequences comprise >750 amino acids, thus indicating they may correspond to evolved VgrGs (Figure 1C). 24 , 29 The lack of structural and functional information about VgrG4‐CTD prompted us to investigate more.
3.2. VgrG4‐CTD interactome network was identified
Recombinant VgrG4‐CTD was expressed in the soluble fraction and purified (Figure S2). Cleavage of histidine‐tag using thrombin resulted in protein degradation (data not shown). Therefore, we used the 6×‐Histidine tagged protein on further experiments.
A previous study showed that DUF2345 from VgrG4 is enough for antibacterial competition. 37 Thus, we hypothesized whether the CTD region could be important for interaction with host cells. Literature reports that K. pneumoniae surface components modulate immune and inflammatory responses in macrophages and lung epithelial cells. 44 , 45 , 46 , 47 These cells function as barriers and defense mechanisms of the host upon pathogen infection. Therefore, we decided to investigate if VgrG4‐CTD targets those cells. Protein extracts were obtained in biological triplicates from A549 lung epithelial cells and macrophage‐derived from the human monocyte cell line THP‐1, and used as a source of ligands for VgrG4‐CTD.
Soluble and membrane‐enriched fractions were quantitatively, and qualitatively evaluated. All samples presented a variety of protein bands, ranging from ~10 to ~120 kDa. Protein expression profiles of THP‐1 × A549 cells were dissimilar, as expected. For both cell types, membrane‐enriched and soluble fractions presented a distinct pattern, thus justifying the need for processing both fractions of each sample (Figure S3).
Beads were covered with an excess of VgrG4‐CTD, and empty sites were blocked. Then, treated beads were incubated with the different protein extracts. As control of unspecific binding, noncovered beads were incubated with glycine and then with the protein extracts. After washing steps, bound proteins were recovered, and processed for mass spectrometry identification (Figure 2A). Proteins containing at least one unique peptide were considered identified. It is crucial to note that unique peptides are exclusively found in one protein among all protein sequences available in the database. Therefore, the reliable identification of a unique peptide is enough proof of its presence in a sample. 48
FIGURE 2.
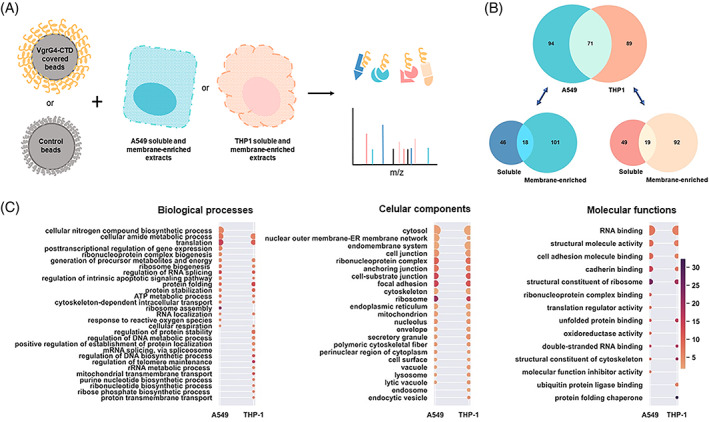
VgrG4‐CTD ligands. (A) Methodological approach used to characterize the VgrG4‐CTD interactome network. Briefly, latex beads covered with VgrG4‐CTD were incubated with A549 or THP‐1 proteins extracts (soluble and membrane‐enriched fractions). Bound proteins were eluted, and mass spectrometry identified. (B) Venn diagrams showing the number of proteins identified in each sample. (C) Gene ontology terms enriched in A549 or THP‐1 putative ligands of VgrG4‐CTD. Terms associated to biological processes, cellular components and molecular functions are shown. The size of the circles are proportional to the number of proteins associated to the term (ranging from 7 to 113), and the color scale is related to the enrichment factor (from 1.7 to 32).
Finally, proteins identified in pulldown assays using VgrG4‐CTD‐coated beads, but not identified from control beads are the VgrG4‐CTD putative ligands. Proteomic analysis revealed 254 proteins associated with VgrG4‐CTD. From those, 165 were identified from lung epithelial cells and 160 from macrophage extracts. Seventy‐one proteins were identified from both cell types (Figure 2B).
In all, 64 proteins were identified in the A549 soluble fraction and 119 in the membrane‐enriched fraction. In the replicates of THP‐1, 68 proteins were identified in the soluble fraction and 111 in the membrane‐enriched fraction (Figure 2B). Not surprisingly, some proteins were identified in both soluble and membrane‐enriched fractions. Such dual identification may be due to their high abundance, formation of macromolecular complexes, partial solubilization in the first step of the extraction procedure, or even a mixture of different protein isoforms that varies in subcellular localization. For example, the cytoskeleton is anchored in the plasma membrane. Thus, it is reasonable that its components are found in both fractions.
The experimental approach performed allows the identification of proteins interacting directly or indirectly with VgrG4‐CTD. It means that VgrG4‐CTD does not necessarily bind directly to each protein identified, but it may interact with at least one protein of the complexes trapped in the assay. Thus, the diversity of proteins identified is expected.
3.3. VgrG4‐CTD interactome in A549 lung epithelial cells
The proteins in the soluble fraction were identified with 1–32 peptides per protein, sequence coverage up to 55%, and False Discovery Rate (FDR) <1% (Tables S1 and S2). In membrane‐enriched protein extracts, VgrG4‐CTD ligands were identified with up to 35 peptides, protein sequence coverage up to 74%, and FDR <0.48% (Table S2).
Gene Ontology (GO) terms associated with A549 proteins were identified (Figure 2C). Regarding terms related to biological processes, the highest number of hits (56) was assigned to “nitrogen compound biosynthesis” (56), followed by “amide metabolism” (44) and “translation” (41). The most enriched terms were “ribosome assembly” (~21×), “translation” (~15×), and “regulation of RNA splicing” (~12×). Accordingly, the most enriched term for cellular component analysis is “ribosome” (~21×), and for a molecular function is “structural component of ribosome” (~24×).
Indeed, numerous ribosomal proteins were identified. Diverse elongation factors and nuclear proteins were also found. The identification of these proteins suggests that VgrG4‐CTD might be a bacterial nucleomodulin and induce global changes in protein expression profile. Nucleomodulins comprise a family of effector proteins targeting processes as chromatin dynamics, histone modification, DNA methylation, RNA splicing, DNA replication, cell cycle, and cell signaling pathways (reviewed by Reference 49). We identified nucleolin as part of the VgrG4‐CTD interactome network. Interestingly, the E. coli T3SS nucleomodulin effector EspF induces a significant redistribution of nucleolin, suppressing host ribosome biogenesis, and promoting increased bacterial access to nutritional resources, as well as pathogen persistence. 50 There are also reports of intracellular pathogens such as Legionella pneumophila that manipulate the host cell translation machinery, 51 and some human pathogens secrete bacterial effectors that act as modifiers of the host chromatin landscape to subvert host metabolism. 51 , 52 Moreover, several plant pathogens hijack the host nuclear processes targeting their nuclear proteins. 53 , 54 Thus, K. pneumoniae may employ similar strategies using its T6SS effectors.
It is also noteworthy the identification of cytoskeleton‐associated proteins. GO terms such as “cytoskeleton” (40 hits, ~2.2× enriched), “structural constituent of cytoskeleton” (8 hits, ~10× enriched), “polymeric cytoskeletal fiber” (8 hits, ~10× enriched), and “cytoskeleton‐dependent intracellular transport” (10 hits, ~7× enrichment) are highlighted (Figure 2C). Proteins identified include actin, alpha and beta‐tubulin, myosin, and vimentin (Table 1). These proteins belong to the microtubules, microfilaments, and intermediate filaments, and are somehow closely interconnected inside the cells. Their identification suggests that VgrG4‐CTD can modulate cytoskeletal rearrangements to favor bacterial infection.
TABLE 1.
Cytoskeleton structural and associated proteins identified as putative VgrG4‐CTD interacting partners.
| Accession | Protein description | A549‐S | A549‐M | THP‐1‐S | THP‐1‐M |
|---|---|---|---|---|---|
| ACTB_HUMAN | Actin, cytoplasmic 1 | x | x | x | x |
| ACTN1_HUMAN | Alpha‐actinin‐1 | x | |||
| ACTN4_HUMAN | Alpha‐actinin‐4 | x | x | ||
| ARP3B_HUMAN | Actin‐related protein 3B | x | x | ||
| ARPC4_HUMAN | Actin‐related protein 2/3 complex subunit 4 | x | x | ||
| CNPY2_HUMAN | Protein canopy homolog 2 | x | |||
| COF1_HUMAN | Cofilin‐1 | x | x | ||
| FLNA_HUMAN | Filamin‐A | x | x | ||
| GELS_HUMAN | Gelsolin | x | |||
| ITB2_HUMAN | Integrin beta‐2 | x | x | ||
| K1C10_HUMAN | Keratin, type I cytoskeletal 10 | x | x | ||
| K1C18_HUMAN | Keratin, type I cytoskeletal 18 | x | |||
| K1C9_HUMAN | Keratin, type I cytoskeletal 9 | x | x | ||
| K2C1_HUMAN | Keratin, type II cytoskeletal 1 | x | x | x | |
| K2C8_HUMAN | Keratin, type II cytoskeletal 8 | x | x | ||
| KRT81_HUMAN | Keratin, type UU cuticular Hb1 | x | |||
| KTN1_HUMAN | Kinectin | x | |||
| LMNA_HUMAN | Prelamin‐A/C | x | x | ||
| LMNB1_HUMAN | Lamin‐B1 | x | |||
| ML12A_HUMAN | Myosin regulatory light chain 12A | x | x | ||
| MYH9_HUMAN | Myosin‐9 | x | x | ||
| MYL6_HUMAN | Myosin light polypeptide 6 | x | x | ||
| SPTN1_HUMAN | Spectrin alpha chain, nonerytrocytic 1 | x | |||
| TAGL2_HUMAN | Transgelin‐2 | x | |||
| TBA1B_HUMAN | Tubulin alpha‐1B chain | x | x | x | |
| TBA1C_HUMAN | Tubulin alpha‐1C chain | x | |||
| TBB2A_HUMAN | Tubulin beta‐2A chain | x | |||
| TBB5_HUMAN | Tubulin beta chain | x | x | x | |
| TCPA_HUMAN | T‐complex protein 1 subunit alpha | x | |||
| TCPB_HUMAN | T‐complex protein 1 subunit beta | x | x | ||
| TCPD_HUMAN | T‐complex protein 1 subunit delta | x | |||
| TCPE_HUMAN | T‐complex protein 1 subunit epsilon | x | x | ||
| TCPG_HUMAN | T‐complex protein 1 subunit gamma | x | x | ||
| TCPZ_HUMAN | T‐complex protein 1 subunit zeta | x | |||
| TLN1‐HUMAN | Talin‐1 | x | |||
| TPM1_HUMAN | Tropomyosin alpha‐1 chain | x | |||
| VIME_HUMAN | Vimentin | x |
Abbreviations: A549‐S, the soluble protein extract from epithelial A549 cells; A549‐M, the membrane‐enriched protein extract from epithelial A549 cells; THP‐1‐S, the soluble protein extract from THP‐1 macrophages; THP‐1‐M, the membrane‐enriched protein extract from THP1 macrophages.
Several pathogens modulate the host cell cytoskeleton for their benefit. 55 , 56 , 57 K. pneumoniae interfere with host cell processes avoiding phagosome–lysosome fusion, evading phagocytosis, and transcellular cross epithelial barriers. 58 The molecular mechanisms of such modulation are not yet depicted, but our findings suggest that VgrG4 is involved. We found that VgrG4‐CTD associate with Talin. Curiously, it has been shown that a talin‐mediated integrin activation induces F‐actin polymerization, and promotes K. pneumoniae internalization in A549 cells. 47 In another hand, it has been reported that E. coli and Aeromonas hydrophila VgrG1 interact with actin. 33 , 59 , 60 , 61 Thus, even without sequence conservation to previously known actin‐binding domains, VgrG4‐CTD probably have an analogous role to previously characterized T6SS effectors.
Another possibility is that VgrG4‐CTD modulates cell–cell and cell–extracellular matrix adhesions, as “cadherin binding,” “focal adhesion,” “cell–substrate junction,” and “cell adhesion molecule binding” are also terms highly enriched. Interestingly, the serine–threonine phosphatase 1A identified negatively regulates the formation of functional tight junctions, modulating the formation of Par3 scaffold through association with 14–3‐3 protein (also identified as part of VgrG4‐CTD interactome network). 62
It is noteworthy that the proteins identified are highly interconnected. Analysis of protein–protein interactions among the putative VgrG4‐CTD ligands showed there are 155 nodes, and 703 edges in the network (Figure 3A). It means an average number of neighbors ~15 per protein—quite more than expected. Thus, it indicates that the approach used captured large protein complexes that VgrG4‐CTD interacts with.
FIGURE 3.
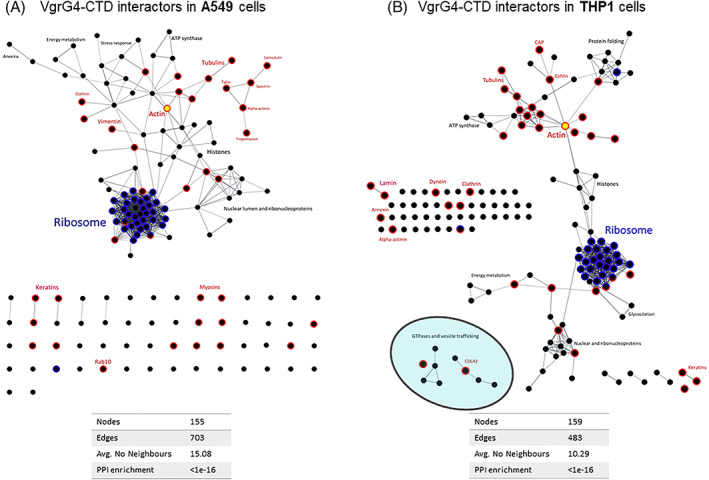
Protein–protein interactions (PPI) among putative ligands of VgrG4‐CTD in A549 (A) or THP‐1 (B). PPI networks were depicted from String database and visualized in Cytoscape. Nodes (dots) represent identified proteins. Edges represent physical interactions between proteins, considering only high confidence interactions determined by experiments. Ribosome‐related proteins are represented in blue, cytoskeleton‐associated proteins in red. Actin is highlighted in yellow.
3.4. VgrG4‐CTD putative ligands in THP‐1 macrophages
In THP‐1 extracts, proteins were identified with up to 66 peptides, and sequence coverage up to 90% (Tables S3 and S4). To have a global view of those proteins, GO terms have been assigned and enrichment analysis performed. Overall, we observed several terms also identified when analyzing epithelial cells (Figure 2C). Although proteins identified in THP‐1 and A549 were not the same ones, terms associated with cellular components were similarly represented. The majority of the proteins are ribosomal and translation‐related, involved in cell adhesion, or cytoskeleton‐associated. Despite the previously mentioned proteins, the identification of six subunits of T‐complex chaperonin is intriguing. T‐complex is a molecular chaperone known to play a role in the folding of actin and tubulin 63 —reinforcing that VgrG4‐CTD somehow targets those filaments.
Another protein identified in both cell types is annexin A2 (AnxA2). AnxA2 has a role in the inflammatory response to K. pneumoniae infection, facilitating TLR4 internalization, activation of TRAM‐dependent signaling, and release of anti‐inflammatory cytokines. 64 With regards to intracellular membrane trafficking and vesicles transport, small GTPases from Rab and Rac families were also identified: Rab1A, Rab7A, Rab31, Rap1A, and Rac2. Depending on their activation state (GDP or GTP bound states), these proteins recruit a different set of effectors involved in vesicle formation and trafficking, autophagosome assembly, as well as cellular processes such as migration and adhesion. Several bacterial effectors target those proteins. For instance, it has been shown that two Mycobacterium tuberculosis effectors bind Rab1A to inhibit the autophagy of infected cells. 65 A Salmonella typhimurium effector induces a posttranslational modification in Rab7, affecting the host endocytic vesicular transport pathway and bacterial survival. 66 Shigella flexineri escapes from lysosomal degradation by delivering an effector that inactivates Rab31. 67 Thus, understanding how K. pneumoniae and VgrG4‐CTD modulate those proteins will certainly contribute to improved knowledge of its pathogenesis. Apart from similarities between A549 and THP‐1 ligands, terms such as “endosome” (17 hits, ~2.2× enriched), and “endocytic vesicle” (13 hits, 5× enriched) were primarily found in THP‐1.
Concerning molecular functions, “protein folding chaperone” was the most enriched term (~32×) among THP‐1 ligands, although not found in A549. “Ubiquitin protein ligase binding” (15 hits, ~6.5× enriched) was also exclusively found in THP‐1 samples. Regarding biological processes, “regulation of telomere maintenance” (~16×), “mitochondrial transmembrane transport” (~12×), “regulation of protein stability” (~7.6×), “proton transmembrane transport” (~6.7×), “purine nucleotide biosynthetic process” (~6.7×), “ribose phosphate biosynthetic process” (~6.2×), and “posttranscriptional regulation of gene expression” (~6×) were represented only among THP‐1 proteins.
Protein–protein interaction analysis revealed 159 nodes, 483 edges, and a mean of ~10 interacting partners per protein (Figure 3). There are proteins working as hubs connecting subnetworks. Consistently with the several known roles of cytoskeleton, actin is one of those hubs.
3.5. VgrG4‐CTD interacts with cytoskeleton filaments
The proteomic approach employed suggests that VgrG4‐CTD binds cytoskeleton‐associated proteins (such as actin, vimentin, alpha‐actinin, tubulin, myosin, cytokeratin, among others, Table 1). To validate mass spectrometry data, we performed protein–protein binding assays on microplates. For those experiments, the recombinant VgrG4‐CTD protein was used to coat the wells that were subsequently incubated with A549 or THP‐1 protein extracts. Bound actin, tubulin, vimentin, and cytokeratin were immunodetected. This experiment is somehow similar to the pull‐down experiments using beads, however different blocking agents, controls, and detection methods were used. The results obtained confirm that VgrG4‐CTD interacts with actin, vimentin, and cytokeratin (Figure 4A). Using this method we did not detect the interaction with tubulin.
FIGURE 4.
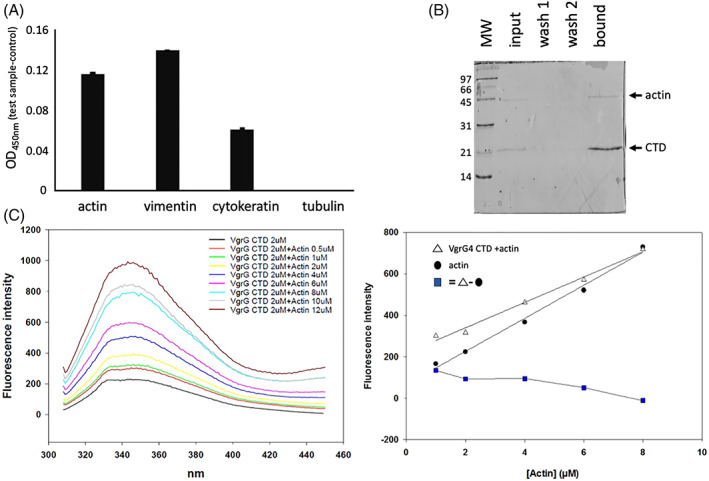
VgrG4‐CTD interacts with actin and other cytoskeleton structural proteins. (A) VgrG4‐CTD was adsorbed in microplate wells. Empty binding sites were blocked with BSA. THP‐1 protein extracts were incubated. Nonbound proteins were washed. Actin, tubulin, cytokeratin and vimentin primary antibodies were used to detect them. Secondary antibody conjugated with peroxidase and TMB substrate were added to reveal the interaction. Increased values at 450 nm indicate higher amounts of protein bound. (B) Purified actin was mixed with the recombinant VgrG4‐CTD‐His6 and then incubated with Ni2+‐affinity resin. The input, washes, and elution samples were analyzed by SDS‐PAGE. Low‐range molecular weight (Biorad) is shown (kDa). (C) Fluorescence spectrum of VgrG4‐CTD incubated with increasing concentration of actin. The correlation between actin concentration and the peak of fluorescence emission is shown. White triangles curve represent the fluorescence intensity of the different Actin concentrations with VgrG4‐CTD. Black circles curve denotes pure actin. Blue squares indicate the subtraction of free actin intensity values from the titration curve of actin with VgrG4‐CTD.
Then, we asked whether the VgrG4‐CTD directly binds actin. Thus, we performed a copurification experiment. We incubated the purified actin with the recombinant VgrG4‐CTD‐His6 and then the mixture with Ni2+‐affinity resin. SDS‐PAGE analysis of the bound and free fractions revealed that untagged actin was retained with VgrG4‐CTD‐His6 on the Ni2+ resin, suggesting a physical interaction between the two proteins (Figure 4B). As a control experiment, we have incubated pure actin with Ni2+‐resin in the absence of VgrG4‐CTD. The unbound fraction (flow‐through), wash, and eluate (bound fraction) have been analyzed by SDS‐PAGE. Actin band was detected exclusively in the unbound fraction (data not shown). Thus, it confirmed that pure actin does not bind directly to Ni2+‐resin.
As the proportion of VgrG4‐CTD recovered was higher than actin, we decided to further investigate the interaction between VgrG4‐CTD and microfilaments. We measured the intrinsic fluorescence of VgrG4‐CTD in the presence of increasing concentrations of purified actin. Molecules absorb light and emit fluorescence according to their structure. In proteins, tryptophan, tyrosine, and phenylalanine amino acids are responsible for fluorescence emission. Changes in the chemical environment affect this spectroscopic property. As protein–protein interactions usually induce conformational changes and affect the solvent exposure of aromatic residues, it implies alterations of fluorescence emission. We observed that VgrG4‐CTD suppresses actin fluorescence intensity (Figure 4C), thus suggesting that VgrG4‐CTD directly binds actin.
3.6. VgrG4‐CTD induces actin remodeling in macrophages
It is well known that several bacteria manipulate the host cytoskeleton for their benefit. 68 , 69 It has been shown that microfilaments and microtubules are required to K. pneumoniae strain Ca0438 invade and translocate across intestinal epithelium 58 and to promote K. pneumoniae strain 3091 internalization into bladder epithelial cells. 70 Moreover, it has been previously shown that both actin and tubulin filaments are required for Kp43816 phagocytosis by MH‐S cells. 58 Kp43816 strain harbors a VgrG4‐CTD homolog. Our proteomic results identified several proteins from microfilaments (actin, alpha‐actinin, filamin, etc.) as putative ligands of VgrG4‐CTD, and actin as a probable direct ligand. Thus, we checked whether VgrG4‐CTD could interfere with actin filaments organization in eukaryotic cells. RAW 264.7 macrophages were stimulated with VgrG4‐CTD using the PULSin protein delivery reagent. PULSin contains a cationic amphiphile molecule that forms noncovalent complexes with proteins of interest. Complexes are then internalized via anionic cell‐adhesion receptors, then released and disassembled into the cytoplasm. This process is not toxic and delivers functional proteins. Noninfected cells stimulated with VgrG4‐CTD presented a distinct pattern of actin filaments dispersion, with more cellular protrusions than nonstimulated cells (Figure 5). Moreover, we observed that cells infected with K. pneumoniae (which expresses VgrG4) also present such protuberances. These data showed that the pulldown assay was able to identify functionally relevant proteins in the VgrG4‐CTD interactome network. Additionally, it shows that there are phenotypic alterations in host cells due to VgrG4‐CTD interaction with actin.
FIGURE 5.
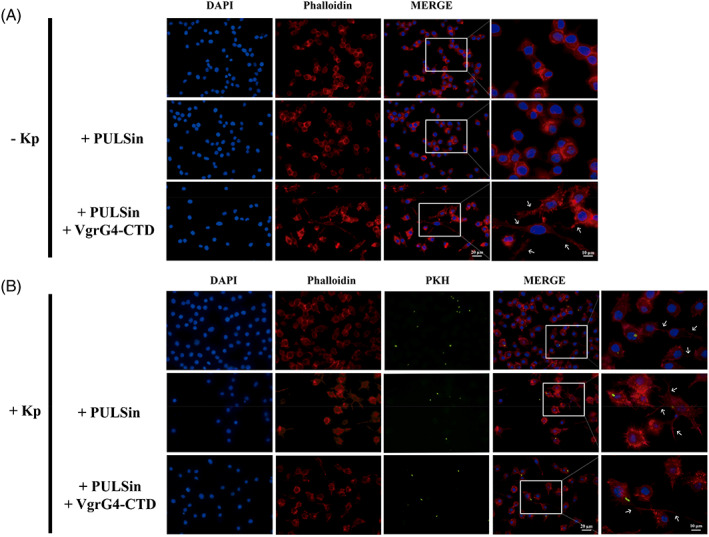
VgrG4‐CTD induces actin remodeling in RAW 264.7 macrophages. Intracellular delivery of VgrG4‐CTD disturbs actin filaments in RAW 264.7 macrophages. Cells were treated with PULSin reagent carrying VgrG4‐CTD for 4 h, as indicated. As a control, cells were stimulated with the pure PULSin reagent. Cell cultures were then infected with KPH26‐stained K. pneumoniae Kp52.145 (green) in MOI 50:1 for 1 h. Phalloidin‐stained Actin filaments (red) and nuclei (blue) are evidenced. Arrows point to the cellular protrusions observed in VgrG4‐CTD stimulated or infected cells. MOI, multiplicity of infection.
4. CONCLUSIONS
Proteomic and interactome approaches have been successfully used as discovery‐based nonbiased methodologies to give clues on molecular and cellular processes. The results suggest that VgrG4‐CTD have a role in the interaction of K. pneumoniae with host cells. VgrG4‐CTD interacts with ribosomal proteins, and cytoskeleton. It does bind actin and induces actin remodeling in macrophages. These results provide a ground basis for future studies.
AUTHOR CONTRIBUTIONS
Talyta do Nascimento Soares and Letícia Miranda Santos Lery performed pull‐down experiments, mass spectrometry data acquisition and analysis, protein–protein binding experiments, and wrote the manuscript. Verônica Silva Valadares and Gisele Cardoso Amorim worked on recombinant protein expression and purification, as well as fluorescence spectroscopy measurements. Mayara de Mattos Lacerda de Carvalho, Fábio Ceneviva Lacerda Almeida, Marcia Berrêdo‐Pinho, and Paulo Mascarello Bisch participated in study design. Letícia Miranda Santos Lery and Paulo Ricardo Batista performed sequence analysis, as well as conceived, designed, and coordinated the study. All authors read and approved the final manuscript.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/prot.26344.
Supporting information
Figure S1 Recombinant VgrG4‐CTD. A) Schematic representation of the plasmid used for the recombinant protein expression. B) Recombinant VgrG4‐CTD primary sequence. The histidine tag and the thrombin recognition site are shadowed in gray
Figure S2 VgrG4‐CTD purification on a nickel affinity column. A) Polyacrylamide electrophoresis showing (1) protein profile of E. coli expressing VgrG4‐CTD (2) nonbound fraction (3 and 4) column washings (5 and 6) VgrG4‐CTD eluted in 500 mM imidazole buffer. B) Chromatographic profile of VgrG4‐CTD in a nickel affinity column. The red arrows point to VgrG4‐CTD band and peak
Figure S3 Protein profile of the cell lysates used in pulldown experiments. M: membrane‐enriched fractions, C: soluble fractions. The molecular weights of the Kaleidoscope prestained protein standard (Biorad) are shown in kDa.
ACKNOWLEDGMENTS
This work was supported by Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Inova Fiocruz/Fundação Oswaldo Cruz. The authors thank André Alves Dias, Camila Oliveira da Silva, and Fabrício da Mota Ramalho Costa for their help with cell culture; and the Unidade de Espectrometria de Massas e Proteômica (UEMP) for the use of the mass spectrometry facility.
do Nascimento Soares T, Silva Valadares V, Cardoso Amorim G, et al. The C‐terminal extension of VgrG4 from Klebsiella pneumoniae remodels host cell microfilaments. Proteins. 2022;90(9):1655‐1668. doi: 10.1002/prot.26344
Funding information Conselho Nacional de Desenvolvimento Científico e Tecnológico; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior; Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro; Programa Inova Fiocruz/Fundação Oswaldo Cruz
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Brisse S, Grimont F, Grimont PAD. The Genus Klebsiella . In: Dworkin M et al., eds. The Prokaryotes. Springer; 2006. [Google Scholar]
- 2. Lee CR, Lee JH, Park KS, et al. Antimicrobial resistance of Hypervirulent Klebsiella pneumoniae: epidemiology, Hypervirulence‐associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112(27):E3574‐E3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344‐359. [DOI] [PubMed] [Google Scholar]
- 5. Lan P, Jiang Y, Zhou J, Yu Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;25:26‐34. [DOI] [PubMed] [Google Scholar]
- 6. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629‐661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lery LM, Frangeul L, Tomas A, et al. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 2014;12:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tomás A, Lery L, Regueiro V, et al. Functional genomic screen identifies Klebsiella pneumoniae factors implicated in blocking nuclear factor κB (NF‐κB) signaling. J Biol Chem. 2015;290(27):16678‐16697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang G, Zhao G, Chao X, Xie L, Wang H. The characteristic of virulence, biofilm and antibiotic resistance of Klebsiella pneumoniae. Int J Environ Res Public Health. 2020;17(17):6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coulthurst SJ. The type VI secretion system ‐ a widespread and versatile cell targeting system. Res Microbiol. 2013;164(6):640‐654. [DOI] [PubMed] [Google Scholar]
- 11. Coulthurst S. The type VI secretion system: a versatile bacterial weapon. Microbiology. 2019;165:503‐515. [DOI] [PubMed] [Google Scholar]
- 12. Bönemann G, Pietrosiuk A, Mogk A. Tubules and donuts: a type VI secretion story. Mol Microbiol. 2010;76(4):815‐821. [DOI] [PubMed] [Google Scholar]
- 13. Pukatzki S, Ma AT, Sturtevant D, et al. Identification of a conserved bacterial protein secretion system in vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103(5):1528‐1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV‐mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28(4):315‐325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pell LG, Kanelis V, Donaldson LW, Lynne Howell P, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long‐tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci USA. 2009;106(11):4160‐4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brackmann M, Wang J, Basler M. Type VI secretion system sheath inter‐subunit interactions modulate its contraction. EMBO Rep. 2018;19(2):225‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kudryashev M, Wang RYR, Brackmann M, et al. Structure of the type VI secretion system contractile sheath. Cell. 2015;160(5):952‐962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang J, Brackmann M, Castaño‐Díez D, et al. Cryo‐EM structure of the extended type VI secretion system sheath‐tube complex. Nat Microbiol. 2017;2(11):1507‐1512. [DOI] [PubMed] [Google Scholar]
- 19. Salih O, He S, Planamente S, et al. Atomic structure of type VI contractile sheath from Pseudomonas aeruginosa . Structure. 2018;26(2):329‐336.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hachani A, Allsopp LP, Oduko Y, Filloux A. The VgrG proteins are “à la carte” delivery systems for bacterial type VI effectors. J Biol Chem. 2014;289(25):17872‐17884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hachani A, Wood TE, Filloux A. Type VI secretion and anti‐host effectors. Curr Opin Microbiol. 2016;29:81‐93. [DOI] [PubMed] [Google Scholar]
- 22. Leiman PG, Basler M, Ramagopal UA, et al. Type VI secretion apparatus and phage tail‐associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106(11):4154‐4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR‐repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500(7462):350‐353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike‐like protein into target cells where it cross‐links Actin. Proc Natl Acad Sci USA. 2007;104(39):15508‐15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cianfanelli FR, Alcoforado Diniz J, Guo M, de Cesare V, Trost M, Coulthurst SJ. VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 2016;12(6):e1005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma J, Pan Z, Huang J, Sun M, Lu C, Yao H. The hcp proteins fused with diverse extended‐toxin domains represent a novel pattern of antibacterial effectors in type VI secretion systems. Virulence. 2017;8(7):1189‐1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Silverman JM, Agnello DM, Zheng H, et al. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell. 2013;51(5):584‐593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Whitney JC, Beck CM, Goo YA, et al. Genetically distinct pathways guide effector export through the type VI secretion system. Mol Microbiol. 2014;92(3):529‐542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pukatzki S, McAuley SB, Miyata ST. The type VI secretion system: translocation of effectors and effector‐domains. Curr Opin Microbiol. 2009;12(1):11‐17. [DOI] [PubMed] [Google Scholar]
- 30. Schwarz S, Singh P, Robertson JD, et al. VgrG‐5 is a Burkholderia type VI secretion system‐exported protein required for multinucleated giant cell formation and virulence. Infect Immun. 2014;82(4):1445‐1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Durand E, Derrez E, Audoly G, et al. Crystal structure of the VgrG1 Actin cross‐linking domain of the vibrio cholerae type VI secretion system. J Biol Chem. 2012;287(45):38190‐38199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Durand E, Cambillau C, Cascales E, Journet L. VgrG, Tae, Tle, and beyond: the versatile arsenal of type VI secretion effectors. Trends Microbiol. 2014;22(9):498‐507. [DOI] [PubMed] [Google Scholar]
- 33. Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of Actin. J Bacteriol. 2010;192(1):155‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma AT, Mekalanos JJ. In vivo Actin cross‐linking induced by vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci USA. 2010;107(9):4365‐4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sarris PF, Zoumadakis C, Panopoulos NJ, Scoulica EV. Distribution of the putative type VI secretion system core genes in Klebsiella spp. Infect Genet Evol. 2011;11(1):157‐166. [DOI] [PubMed] [Google Scholar]
- 36. Shrivastava S, Mande SS. Identification and functional characterization of gene components of type VI secretion system in bacterial genomes. PLoS One. 2008;3(8):e2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Storey D, McNally A, Åstrand M, et al. Klebsiella pneumoniae type VI secretion system‐mediated microbial competition is PhoPQ controlled and reactive oxygen species dependent. PLoS Pathog. 2020;16(3):e1007969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu L, Ye M, Li X, et al. Identification and characterization of an antibacterial type VI secretion system in the Carbapenem‐resistant strain. Front Cell Infect Microbiol. 2017;7:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hsieh PF, Lu YR, Lin TL, Lai LY, Wang JT. Klebsiella pneumoniae type VI secretion system contributes to bacterial competition, cell invasion, type‐1 fimbriae expression, and in vivo colonization. J Infect Dis. 2019;219(4):637‐647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bent ZW, Poorey K, LaBauve AE, Hamblin R, Williams KP, Meagher RJ. A rapid spin column‐based method to enrich pathogen transcripts from eukaryotic host cells prior to sequencing. PLoS One. 2016;11(12):e0168788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barbosa VAA, Lery LMS. Insights into Klebsiella pneumoniae type VI secretion system transcriptional regulation. BMC Genomics. 2019;20(1):506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Repizo GD, Espariz M, Seravalle JL, Salcedo SP. Bioinformatic analysis of the type VI secretion system and its potential toxins in the. Front Microbiol. 2019;10:2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shyntum DY, Venter SN, Moleleki LN, Toth I, Coutinho TA. Comparative genomics of type VI secretion systems in strains of Pantoea ananatis from different environments. BMC Genomics. 2014;15:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. March C, Moranta D, Regueiro V, et al. Klebsiella pneumoniae outer membrane protein A is required to prevent the activation of airway epithelial cells. J Biol Chem. 2011;286(12):9956‐9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moranta D, Regueiro V́, March C, et al. Klebsiella pneumoniae capsule polysaccharide impedes the expression of beta‐defensins by airway epithelial cells. Infect Immun. 2010;78(3):1135‐1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Teng Y, Miao J, Shen X, et al. The modulation of MiR‐155 and MiR‐23a manipulates Klebsiella pneumoniae adhesion on human pulmonary epithelial cells via integrin α5β1 signaling. Sci Rep. 2016;6:31918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang X, Li J, Chen S, et al. Knockdown of HMGN2 increases the internalization of Klebsiella pneumoniae by respiratory epithelial cells through the regulation of α5β1 integrin expression. Int J Mol Med. 2016;38(3):737‐746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao Y, Lin YH. Whole‐cell protein identification using the concept of unique peptides. Genom Proteom Bioinform. 2010;8(1):33‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hanford HE, Von Dwingelo J, Abu Kwaik Y. Bacterial nucleomodulins: a coevolutionary adaptation to the eukaryotic command center. PLoS Pathog. 2021;17(1):e1009184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dean P, Scott JA, Knox AA, Quitard S, Watkins NJ, Kenny B. The enteropathogenic E. coli effector EspF targets and disrupts the nucleolus by a process regulated by mitochondrial dysfunction. PLoS Pathog. 2010;6(6):e1000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rolando M, Gomez‐Valero L, Buchrieser C. Bacterial remodelling of the host epigenome: functional role and evolution of effectors methylating host histones. Cell Microbiol. 2015;17(8):1098‐1107. [DOI] [PubMed] [Google Scholar]
- 52. Cornejo E, Schlaermann P, Mukherjee S. How to rewire the host cell: a home improvement guide for intracellular bacteria. J Cell Biol. 2017;216(12):3931‐3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bierne H, Cossart P. When bacteria target the nucleus: the emerging family of nucleomodulins. Cell Microbiol. 2012;14(5):622‐633. [DOI] [PubMed] [Google Scholar]
- 54. Canonne J, Rivas S. Bacterial effectors target the plant cell nucleus to subvert host transcription. Plant Signal Behav. 2012;7(2):217‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barbieri JT, Riese MJ, Aktories K. Bacterial toxins that modify the actin cytoskeleton. Annu Rev Cell Dev Biol. 2002;18:315‐344. [DOI] [PubMed] [Google Scholar]
- 56. Radhakrishnan GK, Splitter GA. Modulation of host microtubule dynamics by pathogenic bacteria. Biomol Concepts. 2012;3(6):571‐580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stebbins CE. Structural insights into bacterial modulation of the host cytoskeleton. Curr Opin Struct Biol. 2004;14(6):731‐740. [DOI] [PubMed] [Google Scholar]
- 58. Cano V, March C, Insua JL, et al. Klebsiella pneumoniae survives within macrophages by avoiding delivery to lysosomes. Cell Microbiol. 2015;17(11):1537‐1560. [DOI] [PubMed] [Google Scholar]
- 59. Aubert DF, Hu S, Valvano MA. Quantification of type VI secretion system activity in macrophages infected with Burkholderia cenocepacia. Microbiology (Reading). 2015;161(11):2161‐2173. [DOI] [PubMed] [Google Scholar]
- 60. Flaugnatti N, le TTH, Canaan S, et al. A phospholipase A1 antibacterial type VI secretion effector interacts directly with the C‐terminal domain of the VgrG spike protein for delivery. Mol Microbiol. 2016;99(6):1099‐1118. [DOI] [PubMed] [Google Scholar]
- 61. Zheng J, Shin OS, Cameron DE, Mekalanos JJ. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 2010;107(49):21128‐21133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Traweger A, Wiggin G, Taylor L, Tate SA, Metalnikov P, Pawson T. Protein phosphatase 1 regulates the phosphorylation state of the polarity scaffold Par‐3. Proc Natl Acad Sci USA. 2008;105(30):10402‐10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Llorca O, Martín‐Benito J, Ritco‐Vonsovici M, et al. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi‐native conformations. EMBO J. 2000;19(22):5971‐5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang S, Yu M, Guo Q, et al. Annexin A2 binds to endosomes and negatively regulates TLR4‐triggered inflammatory responses via the TRAM‐TRIF pathway. Sci Rep. 2015;5:15859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Strong EJ, Ng TW, Porcelli SA, Lee S. Mycobacterium tuberculosis PE_PGRS20 and PE_PGRS47 proteins inhibit autophagy by interaction with Rab1A. mSphere. 2021;6(4):e0054921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mohapatra G, Gaur P, Prabhakar M, et al. A SUMOylation‐dependent switch of RAB7 governs intracellular life and pathogenesis of Salmonella Typhimurium . J Cell Sci. 2019;132(1):jcs222612. [DOI] [PubMed] [Google Scholar]
- 67. Sun J, Wang X, Lin H, et al. Shigella escapes lysosomal degradation through inactivation of Rab31 by IpaH4.5. J Med Microbiol. 2021;70(7):1382. [DOI] [PubMed] [Google Scholar]
- 68. Gruenheid S, Finlay BB. Microbial pathogenesis and cytoskeletal function. Nature. 2003;422(6933):775‐781. [DOI] [PubMed] [Google Scholar]
- 69. Kudryashova E, Heisler DB, Kudryashov DS. Pathogenic mechanisms of Actin cross‐linking toxins: peeling away the layers. Curr Top Microbiol Immunol. 2017;399:87‐112. [DOI] [PubMed] [Google Scholar]
- 70. Oelschlaeger TA, Tall BD. Invasion of cultured human epithelial cells by Klebsiella pneumoniae isolated from the urinary tract. Infect Immun. 1997;65(7):2950‐2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Recombinant VgrG4‐CTD. A) Schematic representation of the plasmid used for the recombinant protein expression. B) Recombinant VgrG4‐CTD primary sequence. The histidine tag and the thrombin recognition site are shadowed in gray
Figure S2 VgrG4‐CTD purification on a nickel affinity column. A) Polyacrylamide electrophoresis showing (1) protein profile of E. coli expressing VgrG4‐CTD (2) nonbound fraction (3 and 4) column washings (5 and 6) VgrG4‐CTD eluted in 500 mM imidazole buffer. B) Chromatographic profile of VgrG4‐CTD in a nickel affinity column. The red arrows point to VgrG4‐CTD band and peak
Figure S3 Protein profile of the cell lysates used in pulldown experiments. M: membrane‐enriched fractions, C: soluble fractions. The molecular weights of the Kaleidoscope prestained protein standard (Biorad) are shown in kDa.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


