Abstract
Genetic manipulation of whole‐plant transpiration rate (TR) response to increasing atmospheric vapor pressure deficit (VPD) is a promising approach for crop adaptation to various drought regimes under current and future climates. Genotypes with a non‐linear TR response to VPD are expected to achieve yield gains under terminal drought, thanks to a water conservation strategy, while those with a linear response exhibit a consumptive strategy that is more adequate for well‐watered or transient‐drought environments. In wheat, previous efforts indicated that TR has a genetic basis under naturally fluctuating conditions, but because TR is responsive to variation in temperature, photosynthetically active radiation, and evaporative demand, the genetic basis of its response VPD per se has never been isolated. To address this, we developed a controlled‐environment gravimetric phenotyping approach where we imposed VPD regimes independent from other confounding environmental variables. We screened three nested association mapping populations totaling 150 lines, three times over a 3‐year period. The resulting dataset, based on phenotyping nearly 1400 plants, enabled constructing 63‐point response curves for each genotype, which were subjected to a genome‐wide association study. The analysis revealed a hotspot for TR response to VPD on chromosome 5A, with SNPs explaining up to 17% of the phenotypic variance. The key SNPs were found in haploblocks that are enriched in membrane‐associated genes, consistent with the hypothesized physiological determinants of the trait. These results indicate a promising potential for identifying new alleles and designing next‐gen wheat cultivars that are better adapted to current and future drought regimes.
1. INTRODUCTION
Anthropogenic climate change is driving an increase in atmospheric vapor pressure deficit (VPD, or evaporative demand) in major agricultural hotspots across the globe, leading to major losses of productivity (Ficklin & Novick, 2017; Yuan et al., 2019; Lopez et al., 2021). Because VPD increases drive higher transpiration rates (TR) from crop canopies, productivity penalties typically occur as a result of these losses not being matched by adequate water supply either through irrigation or precipitation (Kimm et al., 2020; Lobell et al., 2013). For grain crops such as wheat (Triticum aestivum L.) grown on stored soil moisture under terminal drought, such losses most often take place due to exacerbated exposure of the highly sensitive flowering and seed‐fill phases to water deficit (Zheng et al., 2013; Zheng et al., 2016).
Recently, two independent modeling approaches have shown that cultivars exhibiting a limitation on TR as VPD increased (i.e., sensitivity to VPD), could lead to substantive yield increases in Mediterranean‐type environments under current and future climate change scenarios (Collins et al., 2021; Sadok et al., 2019). In both cases, the yield gains arose from a water‐conservation phenotype resulting from a reduction in excessive TR losses during times of the day where evaporative demand is highest (VPD‐sensitivity), which leads to larger amounts of soil moisture available during critical reproductive phases in comparison to more ‘consumptive’ phenotypes (VPD‐insensitivity).
Since this VPD response was hypothesized to be a potential ‘trait’ that was quantitatively related to yield performance in the systems analysis of Sinclair et al. (2005), efforts have been made to identify such a water‐saving phenotype in various crops (for review, see Sinclair et al., 2017 and Monnens & Sadok, 2020). In wheat, Rebetzke et al. (2003) identified substantial phenotypic variability in leaf‐level conductance among wheat breeding lines, with evidence suggesting direct involvement of gene action, but direct relationships with VPD variation were not investigated due to multiple confounding variables in the field. More recently, substantive phenotypic variability for whole‐plant TR response curves to increasing VPD was established under controlled environments, with findings suggesting that water‐saving responses are typically associated with drought‐adapted cultivars (Schoppach et al., 2017; Tamang et al., 2019). The mechanistic basis of such a strategy was found to be associated with a reduction in root hydraulic conductance likely to be mediated by smaller metaxylem vessel size and a lack of root mercury‐sensitive aquaporins (Schoppach et al., 2014; Steinemann et al., 2015) that are possibly modulated by root auxin levels (Sadok & Schoppach, 2019; Schoppach et al., 2016).
For these results to be translatable for breeding purposes, a major bottleneck is the limited ability to phenotype for this trait in such a fashion to enable genetic mapping. In a first step to test the feasibility of the approach, Schoppach et al. (2016) screened a mapping population of 145 double‐haploid lines for their TR dynamics over the course of a single day under naturally fluctuating conditions, based on an hourly manual weighting of pots over a 7‐hour period. This led to identifying a major QTL controlling whole‐plant TR located on chromosome 5A, which explained more than 25% of the genotypic variance (Schoppach et al., 2016). However, this preliminary effort did not enable identifying the genetic basis of TR sensitivity to VPD per se, since evaporative demand was co‐varying with temperature (20.9–38.5°C) and the number of datapoints based on which TR response curves were built was too limited to enable detection of VPD‐sensitivity (i.e., change in slope of TR response to VPD).
Given that temperature itself can have VPD‐independent effects on TR through temperature‐dependent mechanisms that influence stomatal K+ channels, aquaporin regulation, changes in cuticle permeance or/and water viscosity over such ranges (Sadok et al., 2021 and references therein), the genetic mapping of Schoppach et al. (2016) could not be directly ascribed to VPD effects. This is a major limitation to any breeding efforts based on this trait, because if no genetic basis is found underlying TR responses to VPD per se, then this would indicate that this eco‐physiological trait is genetically intractable. Additionally, temperature effects have been shown to cause a shift from a water‐saving to a consumptive TR response to VPD in several species (Riar et al., 2015; Shekoofa et al., 2015, 2016), which makes it necessary to phenotype TR response independent from co‐variation in temperature. Otherwise, this would substantially complicate any effort at a genetic dissection of TR response to VPD, since it would lead to uncovering markers that are not directly relevant to this response.
To circumvent these limitations, we developed a controlled‐environment gravimetric phenotyping platform deployed across three adjacent walk‐in growth chambers where TR response curves could be built independent from co‐variation in temperature, air mixing, photosynthetically active radiation (PAR) and soil moisture availability (Tamang & Sadok, 2018). The imposition of target VPD regimes in this system is enabled by ad hoc humidification/ de‐humidification set‐ups that are programmed in coordination with the growth chamber's settings such that each genotype is characterized by 21 TR versus VPD datapoints per experimental run. This system was successfully tested on maize (27 lines, Tamang & Sadok, 2018), barley (26 lines, Sadok & Tamang, 2019) and wheat (58 lines, Tamang et al., 2019), revealing substantial phenotypic variability in each one of these crops.
Leveraging this resource, the first goal of this investigation was to develop a phenotyping protocol for screening TR response curves to increasing VPD for larger populations, in order to enable for the first time, a genetic mapping effort for this trait. To this end, we used three nested association mapping (NAM) families chosen in order to maximize diversity, using in total 150 lines. The second goal was to conduct a genetic mapping effort and suggest potential candidate genes to gain insights into the underlying genetic basis of this trait. To achieve this, a total of three independent experiments were conducted over the course of 3 years, leading to genotypes being typically characterized by very high‐resolution (63‐point) TR response curves to increasing VPD based on the phenotyping of nearly 1400 plants. We discuss the relevance of the findings and implications for breeding.
2. MATERIALS AND METHODS
2.1. Genetic material
A total of 150 spring wheat lines (Triticum aestivum L.), representing three families (FAM14, FAM22 and FAM24) were selected from the recently developed Spring Wheat Multiparent Introgression Population (SWMIP, Sallam et al., 2020). This population consists of 25 families that were developed out of crosses between 25 landraces assembled from across the globe and the common parent RB07 (Anderson et al., 2009), a commercial cultivar with wide adaptation released by the University of Minnesota wheat breeding program. Out of the total lines that were available in each family (68, 80, and 87 for FAM14, FAM22 and FAM24 respectively), 50 lines per family were randomly sampled for this study. These three families were selected because each one of them had a parent, namely PI 220455 (FAM14), PI 430750 (FAM22) and PI 519465 (FAM24) that contrasted sharply in its TR response curves to increasing VPD relative to the common parent RB07 (Tamang et al., 2019; Figure S1).
2.2. Phenotyping strategy and growth conditions
A total of three independent phenotyping experiments were undertaken across three years, each conducted over a two‐month period (July–August 2017, August–September 2018, and April–May 2019), resulting in a total of nine replicates for each genotype (three replicates per experiment). In each experiment, three staggered plantings took place one week apart, where 50 lines from one of the three NAM families, replicated three times, were planted along with the recurrent parent RB07. This sequencing was necessary due to the space limitations of the gravimetric platform developed for phenotyping TR response curves to increasing VPD, which translated into a maximal throughput of 54 plants per day (Tamang et al., 2019, see section 2.3 for details). However, this sequencing did not significantly impact the response curves of the check cultivar across all experiments. The logistics of the phenotyping process required that each group of 50 genotypes be screened over a period of three consecutive days in a given week. In each experiment, all plants were phenotyped at approximately the same age (32–34, 38–40, and 34–36 days in Exps. 1, 2, and 3, respectively). In a given week, phenotyping was initiated when 50% or more of the plants reached the jointing stage (Zadok's scale of 31).
In all experiments, plants were grown in a glasshouse at the University of Minnesota, Saint Paul, MN, USA (44° 59′18.0′’N 93° 10′50.5′’W). For each genotype, three replicate plants were seeded at a depth of 2.5 cm in 3.8 cylindrical pots filled with premium garden mix (Plaisted Companies Inc., Elkriver) to which a slow‐release fertilizer (Osmocot Plus, Everris, 15‐9‐12 NPK) was added. Plants were watered every 3 days during the first 2 weeks, every second day during the 3–4 weeks and daily during the fifth week of their growing period. Pots were rotated every 2–3 days inside the greenhouse to minimize differences in growth rates due to spatial heterogeneities in environmental conditions.
Inside the glasshouse, air temperature, relative humidity and VPD conditions were recorded every 15 min by means of three pocket sensors (EL‐USB‐2‐LCD, Lascar Electronics) placed at three locations at the canopy height. A 14‐h photoperiod (0600–2000 h) was maintained during the growth of the plants in all experiments. Supplemental lighting was provided by eight 1000‐W HPS‐Metal Halide lightbulbs that were programmed to automatically activate when the incident irradiance fell below 300 W m−2 s−1 for 30 min. Details of the growth conditions experienced by the plants are provided in Table 1.
TABLE 1.
Average temperature and vapor pressure deficit (VPD) conditions (± SE) experienced by the plants during their growth inside the glasshouse
| Daytime conditions | Nighttime conditions | ||||||
|---|---|---|---|---|---|---|---|
| Exp a . | Planting b | T (± SE) | RH (± SE) | VPD (±SE) | T (±SE) | RH (±SE) | VPD (± SE) |
| Exp 1 | Week 1 | 34.6 ± 2.5 | 39.2 ± 5.5 | 3.6 ± 0.7 | 21.6 ± 0.9 | 71.2 ± 5.2 | 0.8 ± 0.2 |
| Week 2 | 33.7 ± 2.8 | 41.1 ± 6.4 | 3.4 ± 1.8 | 21.1 ± 0.9 | 73.4 ± 5.4 | 0.7 ± 0.2 | |
| Week 3 | 32.7 ± 2.7 | 42.0 ± 6.9 | 3.2 ± 0.7 | 20.9 ± 0.9 | 74.1 ± 5.4 | 0.7 ± 0.2 | |
| Exp 2 | Week 1 | 31.6 ± 1.8 | 47.6 ± 8.7 | 2.6 ± 0.6 | 21.6 ± 1.0 | 75.8 ± 4.3 | 0.6 ± 0.2 |
| Week 2 | 31.9 ± 2.1 | 48.3 ± 8.4 | 2.6 ± 0.6 | 21.9 ± 1.1 | 75.0 ± 4.5 | 0.7 ± 0.2 | |
| Week 3 | 31.4 ± 2.2 | 48.8 ± 9.0 | 2.6 ± 0.6 | 21.4 ± 1.3 | 75.7 ± 4.4 | 0.6 ± 0.2 | |
| Exp 3 | Week 1 | 26.9 ± 2.1 | 31.7 ± 7.6 | 2.6 ± 0.5 | 16.9 ± 0.7 | 61.5 ± 5.5 | 0.8 ± 0.2 |
| Week 2 | 26.9 ± 1.9 | 33.6 ± 8.1 | 2.5 ± 0.5 | 17.0 ± 0.7 | 65.4 ± 5.5 | 0.7 ± 0.2 | |
| Week 3 | 26.7 ± 2.1 | 37.0 ± 8.5 | 2.4 ± 0.5 | 17.2 ± 0.8 | 68.0 ± 5.5 | 0.6 ± 0.2 | |
Experiment number.
Planting group.
2.3. Phenotyping transpiration rate response curves to increasing vapor pressure deficit
The phenotyping effort was conducted using the Gravimetric Phenotyping (GraPh) system (Tamang & Sadok, 2018). The platform consists of (i) 54 balances with a resolution of 0.01 g connected to data loggers, (ii) nine temperature and RH sensors, and (iii) humidity control equipment (12 industrial humidifiers and three industrial de‐humidifiers) deployed inside three walk‐in, adjacent growth chambers as fully described elsewhere (Sadok & Tamang, 2019; Tamang et al., 2019; Tamang & Sadok, 2018). Briefly, during the phenotyping sequence, plants are exposed to a stepwise increase in VPD levels, each 60 min‐long, from ~1.5 to 3 kPa under a constant temperature of 30°C and photosynthetically active radiation (PAR = 500 μmol m−2 s−1) at canopy height. The first step, consisting of the lowest VPD level, was achieved by turning the humidifiers on for the first hour after which they were turned off. Over the next 6 h, the dehumidifier automatically decreased RH 80%–30% in a stepwise manner, that is, each hour, resulting in the environmental regime detailed in Table 2.
TABLE 2.
Key environmental conditions imposed on the mapping population during the phenotyping of transpiration rate (TR) response curves to increasing vapor pressure deficit (VPD) in the GraPh platform
| Exp a . | Week | Day | T (± SE) | VPD1 (± SE) | VPD2 (± SE) | VPD3 (± SE) | VPD4 (± SE) | VPD5 (± SE) | VPD6 (± SE) | VPD7 (± SE) |
|---|---|---|---|---|---|---|---|---|---|---|
| Exp 1 | Week 1 | 1 | 30.2 ± 0.0 | 1.7 ± 0.1 | 2.0 ± 0.1 | 2.1 ± 0.2 | 2.3 ± 0.1 | 2.5 ± 0.1 | 2.8 ± 0.1 | 2.9 ± 0.1 |
| 2 | 30.2 ± 0.1 | 1.7 ± 0.1 | 2.0 ± 0.1 | 2.2 ± 0.1 | 2.3 ± 0.1 | 2.5 ± 0.1 | 2.8 ± 0.1 | 2.9 ± 0.1 | ||
| 3 | 30.0 ± 0.1 | 1.6 ± 0.1 | 2.0 ± 0.1 | 2.1 ± 0.2 | 2.3 ± 0.2 | 2.5 ± 0.1 | 2.7 ± 0.1 | 2.8 ± 0.1 | ||
| Week 2 | 1 | 30.2 ± 0.1 | 1.6 ± 0.1 | 2.0 ± 0.1 | 2.2 ± 0.1 | 2.4 ± 0.0 | 2.6 ± 0.1 | 2.8 ± 0.0 | 3.0 ± 0.0 | |
| 2 | 30.4 ± 0.1 | 1.6 ± 0.2 | 2.0 ± 0.1 | 2.2 ± 0.1 | 2.5 ± 0.1 | 2.7 ± 0.0 | 2.9 ± 0.1 | 3.0 ± 0.1 | ||
| 3 | 30.2 ± 0.3 | 1.7 ± 0.2 | 1.9 ± 0.2 | 2.1 ± 0.2 | 2.3 ± 0.2 | 2.5 ± 0.1 | 2.8 ± 0.1 | 2.9 ± 0.1 | ||
| Week 3 | 1 | 30.0 ± 0.3 | 1.5 ± 0.2 | 2.0 ± 0.1 | 2.1 ± 0.2 | 2.3 ± 0.2 | 2.5 ± 0.2 | 2.9 ± 0.2 | 2.9 ± 0.2 | |
| 2 | 30.0 ± 0.1 | 1.5 ± 0.2 | 2.0 ± 0.2 | 2.2 ± 0.1 | 2.3 ± 0.0 | 2.5 ± 0.0 | 2.8 ± 0.1 | 3.0 ± 0.1 | ||
| 3 | 30.0 ± 0.2 | 1.5 ± 0.2 | 1.9 ± 0.2 | 2.1 ± 0.2 | 2.3 ± 0.1 | 2.5 ± 0.1 | 2.7 ± 0.1 | 2.8 ± 0.1 | ||
| Exp 2 | Week 1 | 1 | 30.3 ± 0.4 | 1.7 ± 0.2 | 2.1 ± 0.1 | 2.3 ± 0.2 | 2.5 ± 0.1 | 2.7 ± 0.1 | 2.9 ± 0.2 | 2.9 ± 0.1 |
| 2 | 30.5 ± 0.4 | 1.5 ± 0.2 | 2.0 ± 0.1 | 2.3 ± 0.1 | 2.6 ± 0.1 | 2.8 ± 0.1 | 2.9 ± 0.1 | 2.8 ± 0.1 | ||
| 3 | 30.6 ± 0.3 | 1.7 ± 0.1 | 2.1 ± 0.0 | 2.3 ± 0.1 | 2.5 ± 0.6 | 2.8 ± 0.1 | 3.0 ± 0.1 | 2.9 ± 0.1 | ||
| Week 2 | 1 | 30.4 ± 0.6 | 1.6 ± 0.2 | 2.1 ± 0.1 | 2.3 ± 0.2 | 2.5 ± 0.2 | 2.7 ± 0.2 | 2.9 ± 0.1 | 2.9 ± 0.1 | |
| 2 | 30.3 ± 0.4 | 1.5 ± 0.1 | 2.0 ± 0.1 | 2.2 ± 0.1 | 2.5 ± 0.1 | 2.7 ± 0.1 | 2.9 ± 0.1 | 3.0 ± 0.1 | ||
| 3 | 30.3 ± 0.5 | 1.6 ± 0.1 | 2.0 ± 0.1 | 2.2 ± 0.1 | 2.4 ± 0.2 | 2.7 ± 0.1 | 2.9 ± 0.1 | 3.0 ± 0.1 | ||
| Week 3 | 1 | 30.2 ± 0.8 | 1.7 ± 0.2 | 2.0 ± 0.2 | 2.3 ± 0.2 | 2.5 ± 0.2 | 2.7 ± 0.2 | 2.9 ± 0.9 | 3.1 ± 0.3 | |
| 2 | 30.1 ± 0.4 | 1.6 ± 0.1 | 2.1 ± 0.2 | 2.3 ± 0.2 | 2.5 ± 0.1 | 2.7 ± 0.1 | 2.9 ± 0.1 | 3.0 ± 0.1 | ||
| 3 | 30.3 ± 0.7 | 1.7 ± 0.2 | 2.1 ± 0.2 | 2.3 ± 0.2 | 2.5 ± 0.2 | 2.7 ± 0.2 | 3.0 ± 0.2 | 3.1 ± 0.4 | ||
| Exp 3 | Week 1 | 1 | 30.1 ± 0.6 | 1.6 ± 0.1 | 2.1 ± 0.1 | 2.2 ± 0.1 | 2.4 ± 0.2 | 2.7 ± 0.2 | 2.9 ± 0.2 | 3.0 ± 0.2 |
| 2 | 30.4 ± 0.4 | 1.6 ± 0.1 | 1.9 ± 0.2 | 2.1 ± 0.2 | 2.2 ± 0.2 | 2.5 ± 0.2 | 2.8 ± 0.2 | 2.9 ± 0.2 | ||
| 3 | 28.7 ± 0.4 | 1.6 ± 0.1 | 2.1 ± 0.1 | 2.2 ± 0.1 | 2.4 ± 0.1 | 2.7 ± 0.1 | 2.9 ± 0.1 | 3.0 ± 0.2 | ||
| Week 2 | 1 | 29.9 ± 0.3 | 1.6 ± 0.2 | 1.9 ± 0.2 | 2.2 ± 0.2 | 2.5 ± 0.1 | 2.6 ± 0.1 | 2.8 ± 0.1 | 2.9 ± 0.2 | |
| 2 | 30.0 ± 0.5 | 1.7 ± 0.1 | 2.0 ± 0.2 | 2.1 ± 0.2 | 2.3 ± 0.2 | 2.6 ± 0.1 | 2.8 ± 0.1 | 2.9 ± 0.2 | ||
| 3 | 30.0 ± 0.4 | 1.7 ± 0.1 | 1.9 ± 0.1 | 2.1 ± 0.1 | 2.4 ± 0.1 | 2.6 ± 0.1 | 2.9 ± 0.1 | 2.9 ± 0.2 | ||
| Week 3 | 1 | 30.4 ± 0.5 | 1.6 ± 0.1 | 1.9 ± 0.1 | 2.1 ± 0.1 | 2.3 ± 0.1 | 2.6 ± 0.1 | 2.8 ± 0.1 | 2.9 ± 0.2 | |
| 2 | 30.4 ± 0.4 | 1.6 ± 0.1 | 2.0 ± 0.1 | 2.1 ± 0.1 | 2.3 ± 0.1 | 2.6 ± 0.1 | 2.8 ± 0.1 | 3.0 ± 0.2 | ||
| 3 | 29.8 ± 0.3 | 1.5 ± 0.1 | 1.8 ± 0.1 | 2.0 ± 0.1 | 2.3 ± 0.1 | 2.5 ± 0.1 | 2.7 ± 0.1 | 2.8 ± 0.1 |
Experiment number.
Approximately 6 h prior to transferring the plants to the phenotyping platform, pots were slowly watered until dripping and then were left (for 6 h) to drain any excess water. Immediately after watering, aluminum foil was tightly wrapped around the base of the plant tillers and sealed around the pot edge to nullify direct water evaporation from the soil. A total of 54 plants were then transferred at approximately 18:00 h to the GraPh system, where each replicate plant was randomly placed in one of three chambers (18 plants per chamber) and exposed to temperature, VPD and PAR levels of 30°C, 1.7 kPa and 500 μmol m−2 s−1 respectively for 2 h before lights were turned off at 20:00 h. During the nighttime, plants were subjected to a temperature and VPD values of 20.3°C and 0.7 kPa, respectively until the phenotyping experiment began at 6:30 h the following morning, following the sequence outlined in Table 2.
At the end of the last VPD step, plants were removed from the growth chambers and transferred to the lab, where leaf areas (LA) were measured destructively using a LA meter (model LI3100C, LI‐COR) so that transpiration rates could be calculated on a LA basis (mg H2O m−2 s−1, Fletcher et al., 2007). Leaf samples were then oven‐dried at 65°C for 10 days and weighted afterwards to compute specific LA (SLA, cm2 g−1). Overall, combining the three independent experiments, a total of 1386 plants were phenotyped, typically resulting in 63 transpiration versus VPD datapoints per genotype in the 1.5–3.1 kPa range.
3. DATA ANALYSIS
3.1. Extraction of phenotypes from TR response curves to VPD
Phenotypic and environmental data obtained via the GraPh platform were processed as in Tamang and Sadok (2018) and Tamang et al. (2019), using scripts written in R (R Development Core Team, 2017). Briefly, TR time courses with a temporal resolution of 1 min were constructed for each individual plant in each experiment. Average TR values and their corresponding VPDs were then computed for each VPD step, while excluding the first 15 min during which TR was still acclimating to the new VPD regime (Schoppach & Sadok, 2012). We tested whether TR was at a steady‐state regime during the remaining 45 min by regressing TR against time in each VPD step (Tamang et al., 2019). Less than 3% of the analyzed 8802 TR time courses had significantly non‐zero slopes, indicating that TR of the vast majority of plants were at steady‐state during phenotyping.
In a second step, regression analyses were conducted to characterize TR versus VPD response curves for each genotype in each experiment. Two regression models, one linear and one segmented were compared in each case and the best fitting formalism for a given genotype was determined based on an extra sum‐of‐squares F test (p < 0.05). The linear model resulted in two parameters, the slope and y‐intercept:
| (1) |
while the segmented one produced four parameters:
| (2) |
where slope1, y‐intercept1, VPDBP and slope2 are respectively the first slope, its y‐intercept, the VPD breakpoint (BP)—that is, the VPD at which the slope of TR response to VPD changes—, are the second slope of regression. This method relied on an iterative procedure (maximum number of iterations = 1000) by starting with initial conditions provided by the user. The iteration tested primarily on whether the slopes of the two linear segments derived from linear regression were significantly different (P < 0.05; Motulsky, 1999).
Depending on the experiment, the relative proportions of linear: segmented response curves were 128:20, 113:35 and 102:45 for experiments 1, 2 and 3, respectively. Because the two fits presented different sets of parameters with different potential biological interpretations, it was impossible to conduct a genetic analysis on the two groups of genotypes based on the same set of traits. To circumvent this limitation and enable a joint analysis of data from the two groups, the formalisms presented in Equation 1 and Equation 2 were used to calculate, for each genotype within each experiment, four TR values at four regularly spaced VPD levels spanning the entire range imposed (1.5, 2.0, 2.5 and 3.0 kPa), yielding the corresponding traits: TR1.5, TR2.0, TR2.5, and TR3.0.
3.2. Best linear unbiased estimates of trait values and repeatabilities
Best linear unbiased estimates (BLUEs) for each of the traits were calculated using the lme4 R package (Bates et al., 2015). The experimental effect in the fitted mixed model was treated as a random effect and the replicates within each experiment were nested while lines were treated as fixed effect. Broad‐sense repeatability (r) of each trait was calculated based on Bajgain et al. (2021) and Sallam et al. (2020) as follows:
| (3) |
where σ2 L∶F represent the variance of lines nested within families, σ2 F represent the variance between families, σ2e is the pooled residual variance and E is the number of experiments. Within‐family broad sense heritabilities (H2) were calculated using the formula and reported in Table S1.
3.3. Linkage disequilibrium, genetic relatedness, association mapping and candidate gene analysis
The procedure for genotyping the SWMIP population and generation of consensus linkage map is fully described in Sallam et al. (2020). For the present study, genotypic data was subset to the three families and filtered to remove monomorphic markers, markers with >20% missing data and markers with minor allele frequency of <5% (Anderson et al., 2010). This procedure retained 13,590 Single Nucleotide Polymorphisms (SNP) for downstream analyses of association mapping. Linkage disequilibrium (LD) values among these 13,590 SNPs were estimated as pairwise squared correlation coefficients (r2) across all SWMIP, where any two SNPs with an r2 value of >0.2 were declared to be in LD (Lin et al., 2012; Sallam et al., 2020). Principal Component Analyses (PCA) were conducted in R using the prcomp function to characterize genetic relatedness among the three families.
Data from two lines out of the 150 was excluded from the genetic mapping procedure due to data losses, so that the association mapping was conducted on data from 148 lines. The analysis conducted to identify SNPs significantly associated with the traits was conducted on data obtained by calculating BLUE values as described above. For the four TR traits, since models were fitted at the experimental level combining replicates, the total number of values used to calculate BLUE values were three. For rest of the traits, this number was nine. SNPs were identified by implementing two different GWAS models as discussed in Sallam et al. (2020) and Bajgain et al. (2021). The first model was implemented in the GWAS function of the rrBLUP R package (Endelman, 2011) which takes genetic kinship (K model) into account. The second model was implemented using gwas2 function of R package NAM (Xavier et al., 2015) which accounts for both family stratification and genetic kinship (N model). In both models, significant SNPs were identified using the R package qvalue (Storey & Tibshirani, 2003) with a false discovery rate (FDR) of 0.05. This package calculates q‐values and local FDR values from a list of p values resulting from the simultaneous testing for significance for all SNPs. If multiple SNPs were identified within the same haplotype block, only the SNP with the lowest p value was reported. The percentage of phenotypic variance explained by each SNP (R2) was computed as implemented in the R/qtl package (Broman et al., 2003).
For the candidate gene analysis, all the genes within the haplotype block where the SNPs localized were scanned for protein coding genes along with their annotations using IWGSC RefSeq v1.0 available in Ensembl Plants Biomart (https://plants.ensembl.org/biomart). For SNPs not within any haplotype blocks, a 100 kb region around the SNP was scanned.
3.4. Other statistical analyses
Stability analyses of TR plateaus at each VPD step and regression fits of TR response curves to VPD (linear and segmented) were conducted using the statistical software Prism (version 9.2.0., GraphPad software Inc.). Analysis of variance on TR traits were carried out in R using the aov function. The best‐fit model was selected using the Akaike information criterion (AIC) model selection method implemented in R using the AICcmodavg package.
4. RESULTS
4.1. Diversity in transpiration response curves to increasing VPD across the three families
As exemplified on Figure 1 and fully detailed in Table S1, there was a substantial diversity in TR responses to VPD among the genotypes of the study.
FIGURE 1.
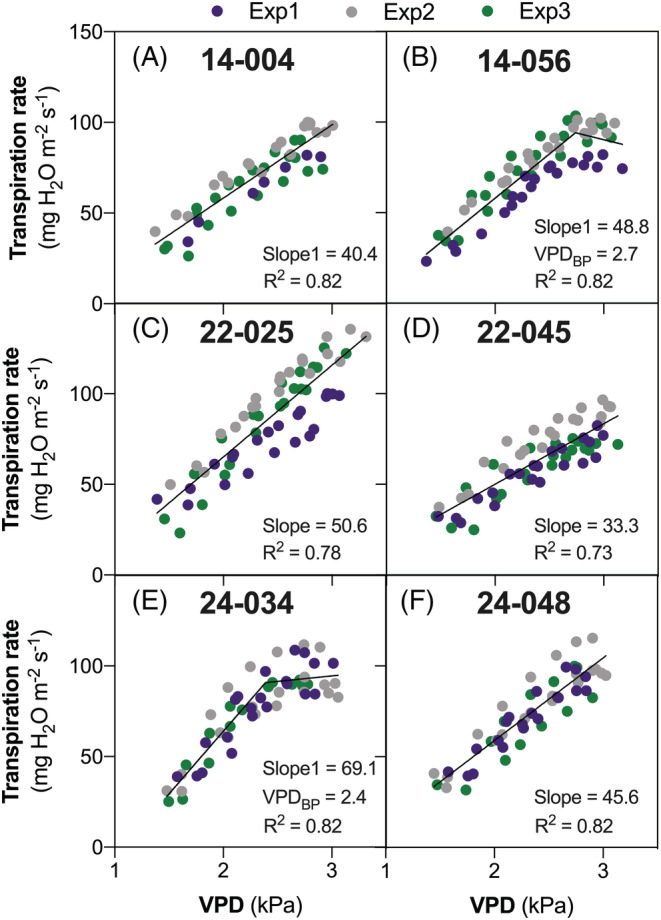
Examples of whole‐plant transpiration rate (TR) response curves to temperature‐independent increases in atmospheric vapor pressure deficit (VPD) for six lines selected from the three families (two lines per family). Panels A–B, C–D, and E–F represent responses of two genotypes from FAM14 (14–004 and 14–056), FAM22 (22–025 and 22–045) and FAM24 (24–034 and 24–048), respectively, including data from all three independent experiments. Linear (slope) and segmented (Slope1, Slope2, VPDBP) regression parameters, along with their coefficients of determination (R2) are indicated.
For each experiment, regression fits were of a good quality, with coefficients of determination (R2) averaging 0.80 (median = 0.83). Due to larger scatter associated with increased sampling size, these averaged 0.64 (median = 0.65) when combining all experiments as exemplified in Figure 1. The percentage of linear versus segmented responses was comparable across the three families, and a similar observation applies to the ranges of the regression parameters (Figure 2).
FIGURE 2.
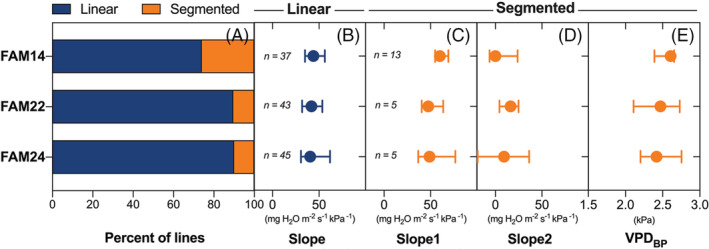
Variation in best fits (linear vs. segmented, panel A) and their parameters (panels B–E) for transpiration rate (TR) response curves to increasing vapor pressure deficit (VPD) across the three nested association mapping (NAM) families. In panels B–E, horizontal segments (blue and orange for linear and segmented fits, respectively) reflect the range, while the circle's position indicates the median value for the parameter of interest. The letter n reflects the number of genotypes best described by a linear or segmented response in each family. Parameters slope, Slope1, Slope2 and VPDBP are fully described in the materials and methods.
Independent from the family, and combining data from all experiments, the majority (85%) of the 148 genotypes presented a linear TR response to VPD with slopes that ranged from 30.5 to 61.7 mg H2O m−2 s−1 kPa−1, a 2‐fold variation. The responses of the remaining 15% (22 genotypes) were best described by a segmented fit, where Slope1 and Slope2 ranged from 36.7 to 76.5 and 19.0 to 36.2 mg H2O m−2 s−1 kPa−1 respectively, while VPDBP values ranged from 2.1 to 2.8 kPa (Figure 2).
4.2. Frequency distributions and repeatability estimates
Combining data from all three families, a significant genotypic variability was found for TR1.5 (p < 0.0001), TR2.0 (p < 0.0001), TR2.5 (p < 0.004) and TR3.0 (p < 0.006) as illustrated in Figure 3. Phenotypic values varied nearly two‐fold for all TR traits, whereby TR1.5, TR2.0, TR2.5 and TR3.0. respectively ranged from 25.5 to 55.7, 43.9 to 78.2, 58.7 to 101.5, and 73.4 to 124.8 mg H2O m−2 s−1. Broad‐sense repeatabilities ranged from 0.35 to 0.45 depending on the TR trait (Figure 3), and were lower than those found for leaf dry mass (LDM), LA, and SLA, which exhibited repeatabilities of 0.88 (LDM and LA) and 0.57 (SLA). TR traits correlated significantly with biomass traits such as LDM, LA, and SLA (Figure 4).
FIGURE 3.
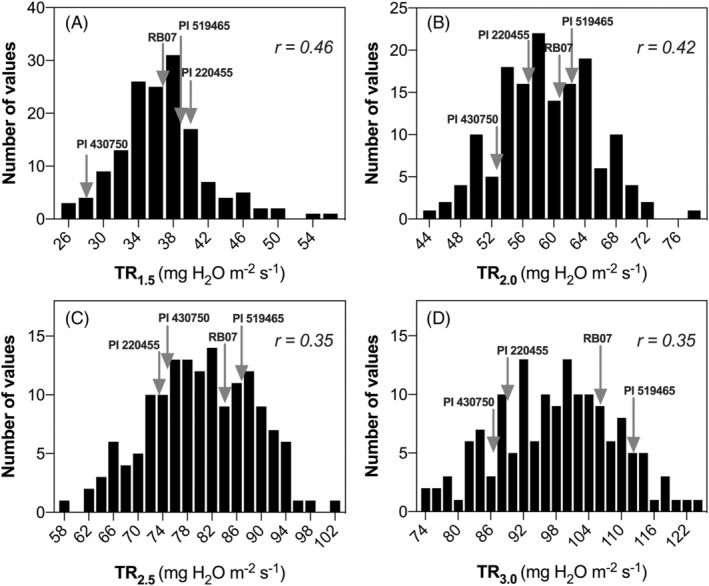
Frequency distribution of the four TR traits. Panels A, B, C, and D represent data for TR1.5, TR2.0. TR2.5 and TR3.0 respectively. In each panel, the values of four parents are highlighted by the gray arrows. Broad sense repeatability (r) values are reported in each panel.
FIGURE 4.
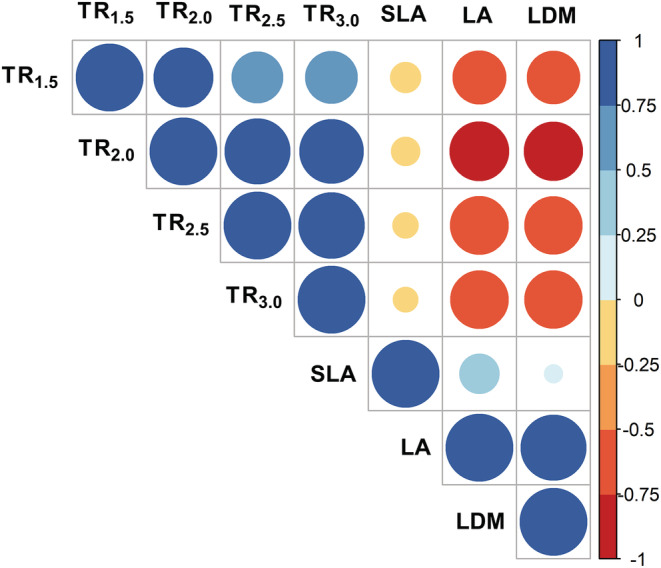
Correlation matrix between the traits considered in the analysis. Positive and negative correlations are indicated as shades of blue and red. The color, shade, and size of the circles are respectively proportional to the direction and value of the correlation coefficients, as indicated by the scale on the right‐hand side of the figure.
4.3. Genome‐wide association mapping of markers associated with transpiration rate response to increasing VPD
The genetic kinships between the families, visualized as PCA plots (Figure 5) revealed that lines derived from the same parent cluster together. This separation between groups indicates successful development of the NAM families and as a result makes it possible to deploy GWAS methods, which use family structure as a covariate (N model).
FIGURE 5.
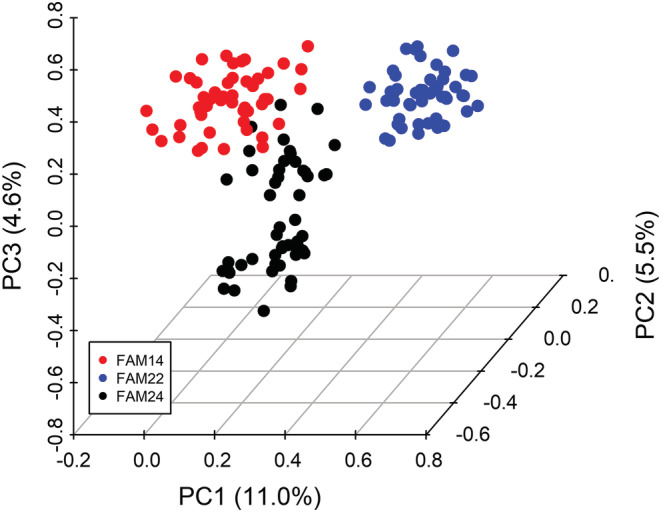
Scatter plot of the first three principal components generated from the genetic data of the 150 lines used in this study. The three NAM families are color‐coded as outlined in the insert.
The two GWAS methods (N and K models) yielded a total of 27 unique and significant markers, which were identified for the four transpiration and three biomass‐related traits. Among these markers, three were exclusively identified by the K model, 13 by the N model and 11 by both N + K models (Table 3). Out of the 27 markers, seven were not found to be contained in any haplotype blocks, while 20 were in 15 different haplotype blocks (Table S2), spanning 10 chromosomes. Among the 15 haplotype blocks, two had more than one marker, with HaploBlock_5A_573.05_589.3 containing the highest number of six markers (Table S2). The SNPs with the highest percent of variation explained (PVE) were identified for LA, DW, and SLA, consistently with the higher repeatabilities of these traits (Table 3).
TABLE 3.
List of significant markers associated with the studied traits along with their allele type, minor allele frequency (MAF), percent of variation explained (PVE), −log10 p value, additive effect of the parental alleles and the type of GWAS method (N, K, or both) that identified the markers
| Trait | Markers | Alleles | MAF | PVE | −log10 p value | Additive effect | Method | |||
|---|---|---|---|---|---|---|---|---|---|---|
| RB07 | PI 220455 | PI 430750 | PI 519465 | |||||||
| TR1.5 | S5A_588416899a | C/A | 0.06 | 13.42 | 5.99 | 1.38 | 0.00 | −2.13 | 0.75 | N + K |
| S5A_591993956 | A/G | 0.09 | 11.32 | 5.17 | 1.89 | 0.00 | −1.89 | 0.00 | N + K | |
| S5A_595425852b | G/A | 0.12 | 15.17 | 6.69 | 2.13 | 0.00 | −1.78 | −0.34 | N + K | |
| S5D_194957929 | T/C | 0.22 | 9.85 | 4.60 | −0.91 | 2.99 | −1.79 | −0.28 | N | |
| TR2.0 | S5A_584821710a | A/G | 0.11 | 14.82 | 6.55 | 1.88 | 0.00 | −2.72 | 0.84 | N + K |
| S5A_591993956 | A/G | 0.09 | 12.71 | 5.71 | 2.43 | 0.00 | −2.43 | 0.00 | N + K | |
| S5A_595425852b | G/A | 0.12 | 17.14 | 7.49 | 2.63 | 0.00 | −2.44 | −0.19 | N + K | |
| S7D_500209644 | C/T | 0.09 | 10.37 | 4.80 | 2.16 | −0.24 | −1.92 | 0.00 | N + K | |
| TR2.5 | S5A_579969562a | A/G | 0.14 | 12.34 | 5.57 | 2.68 | 2.68 | −3.27 | 1.91 | N |
| S5A_586600382a | G/T | 0.09 | 10.97 | 5.03 | 2.76 | 0.00 | −2.76 | 0.00 | K | |
| S5A_591993956 | A/G | 0.09 | 10.61 | 4.89 | 3.09 | 0.00 | −3.09 | 0.00 | N + K | |
| S5A_595425852b | G/A | 0.12 | 14.09 | 6.26 | 3.12 | 0.00 | −3.28 | 0.16 | N + K | |
| S7D_500209644 | C/T | 0.09 | 11.97 | 5.42 | 3.24 | −0.53 | −2.71 | 0.00 | N + K | |
| TR3.0 | S5A_588416899a | C/A | 0.06 | 9.20 | 4.35 | 2.81 | 0.00 | −3.49 | 0.68 | K |
| S5A_591993956 | A/G | 0.09 | 8.67 | 4.14 | 3.58 | 0.00 | −3.58 | 0.00 | K | |
| S5A_595425852b | G/A | 0.12 | 11.89 | 5.39 | 3.62 | 0.00 | −3.93 | 0.32 | K | |
| S7D_500209644 | C/T | 0.09 | 10.19 | 4.73 | 4.02 | −1.29 | −2.73 | 0.00 | K | |
| LA | S2B_72586852 | A/G | 0.10 | 6.97 | 3.48 | −16.34 | −3.33 | 19.67 | 0.00 | K |
| S5A_570788577c | G/T | 0.14 | 8.85 | 4.21 | −19.84 | 0.00 | 20.77 | −0.93 | N + K | |
| S5A_573046932a | G/T | 0.08 | 7.52 | 3.69 | −20.08 | 0.00 | 20.08 | 0.00 | K | |
| S5A_579969562a | A/G | 0.14 | 25.27 | 10.94 | −31.71 | 5.43 | 36.52 | −10.24 | N + K | |
| S5A_591993956 | A/G | 0.09 | 21.12 | 9.14 | −36.10 | 0.00 | 36.10 | 0.00 | N + K | |
| S5A_595425852b | G/A | 0.12 | 23.54 | 10.18 | −26.47 | 0.00 | 43.66 | −17.19 | N + K | |
| S5A_608360107b | A/G | 0.22 | 7.98 | 3.87 | −13.85 | −10.36 | 25.64 | −1.43 | N | |
| S7D_500209644 | C/T | 0.09 | 8.45 | 4.05 | −19.94 | −5.87 | 25.81 | 0.00 | N + K | |
| LDM | S5A_570788577c | G/T | 0.14 | 9.71 | 4.54 | −0.08 | 0.00 | 0.09 | −0.01 | N + K |
| S5A_579969562a | A/G | 0.14 | 21.03 | 9.11 | −0.11 | 0.02 | 0.13 | −0.04 | N + K | |
| S5A_591993956 | A/G | 0.09 | 19.35 | 8.40 | −0.13 | 0.00 | 0.13 | 0.00 | K | |
| S5A_595425852b | G/A | 0.12 | 21.44 | 9.28 | −0.10 | 0.00 | 0.15 | −0.05 | N + K | |
| S7D_500209644 | C/T | 0.09 | 7.90 | 3.84 | −0.07 | −0.03 | 0.10 | 0.00 | K | |
| SLA | S1B_15589918 | T/C | 0.41 | 10.02 | 4.66 | −1.43 | −1.06 | −5.91 | 8.40 | N |
| S1B_16432170 | G/T | 0.37 | 7.78 | 3.79 | −0.98 | −1.91 | −4.93 | 7.81 | N | |
| S1B_17145879 | T/C | 0.12 | 11.50 | 5.24 | −3.48 | 0.97 | −7.54 | 10.04 | N + K | |
| S1B_645514214 | T/C | 0.24 | 9.93 | 4.63 | −3.28 | 0.24 | −4.47 | 7.50 | N | |
| S1B_6474600 | T/C | 0.15 | 12.68 | 5.70 | −5.28 | −3.99 | 0.00 | 9.27 | N + K | |
| S1B_8175738 | T/C | 0.09 | 9.69 | 4.53 | −2.98 | −0.14 | −7.78 | 10.91 | N | |
| S1B_9571874 | C/T | 0.16 | 20.89 | 9.05 | −7.11 | −2.35 | 0.00 | 9.46 | N + K | |
| S4B_4128125 | G/C | 0.18 | 8.40 | 4.03 | −6.37 | −2.20 | −5.02 | 13.59 | N | |
| S5A_584821710a | A/G | 0.11 | 7.57 | 3.71 | −5.12 | 0.00 | 1.29 | 3.83 | N + K | |
| S6A_568743993 | T/C | 0.17 | 7.39 | 3.64 | −12.89 | −6.87 | −16.60 | 36.35 | N | |
| S6A_602923768 | C/T | 0.07 | 7.65 | 3.75 | −6.23 | 0.00 | 0.00 | 6.23 | N + K | |
| S6A_603354559 | A/G | 0.12 | 8.57 | 4.10 | −7.80 | 0.00 | 0.00 | 7.80 | N | |
| S6D_459750797 | C/G | 0.15 | 9.32 | 4.39 | −4.58 | −5.77 | −3.60 | 13.95 | N | |
| S7A_484464079 | T/C | 0.15 | 9.41 | 4.43 | −3.22 | −2.54 | −7.51 | 13.27 | N | |
| S7A_64614962 | A/G | 0.12 | 7.53 | 3.70 | −3.07 | −2.24 | −3.42 | 8.73 | N | |
| S7B_66021438 | G/C | 0.11 | 8.05 | 3.90 | −6.00 | −8.04 | 0.00 | 14.04 | N | |
Note: Bolded markers indicate SNPs that are detected for several traits and marker identifiers followed by the same lower‐case letters (a, b, and c) are within the same haplotype block. The absence of these letters indicate that the corresponding SNP is either the only SNP in the given haplotype block or not associated with any haplotype block.
Overall, a total of eight significant SNPs were found to be associated with the four transpiration traits (TR1.5, TR2.0, TR2.5, and TR3.0.), individually explaining 8.7%–17.1% of the observed phenotypic variation (average PVE = 12%). Among these eight SNPs, four were identified in two or more TR traits and five were detected in both N and K GWAS models (Table 3). With the exception of two markers, located on chromosomes 5D (for TR1.5) and 7D (for TR2.0, TR2.5 and TR3.0), all the remaining ones were found to be located in a major region of chromosome 5A, also shared with SNPs associated with LA and SLA (Table 3, Figure 6).
FIGURE 6.
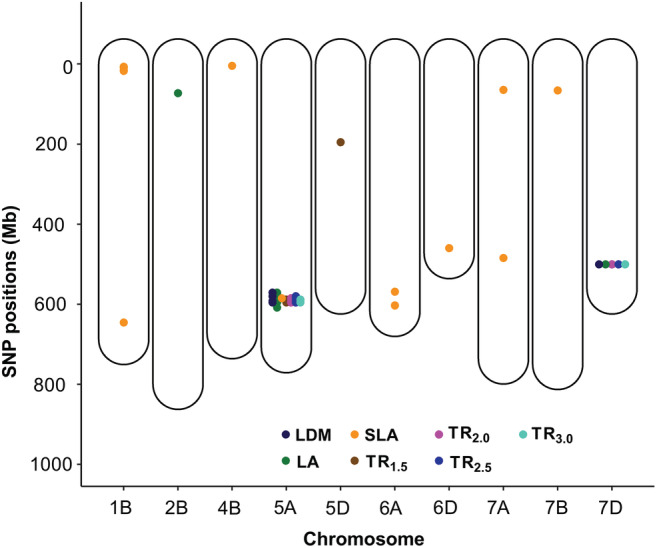
Genetic map of statistically significant markers associated with the studied traits. Each point in the map represents the marker's physical position in mega base pairs, with colors corresponding to different traits. Only those chromosomes where significant markers are found are shown (10 out of 21 wheat chromosomes).
In this region of chromosome 5A, the TR‐associated marker S5A_595425852 consistently had the highest PVE (11.9 to 17.1) and for this SNP, the allelic effect of parent RB07 was consistently positive (+2.13–+3.62), while that of the parent PI 430750 was consistently negative (−1.78–−3.93). The allelic effects of parents PI 220455 and PI 519465 were either null or weak, respectively (Table 3).
As indicated on Figure 7, some of the transpiration SNPs were found to be associated with specific VPD conditions. Specifically, high‐VPD transpiration traits TR2.0, TR2.5, and TR3.0 were found to be associated with an SNP on chromosome 7D (S7D_500209644) that was not detected for the lower‐VPD transpiration trait (TR1.5). Similarly, an SNP detected for TR1.5 on chromosome 5D (S5D_194957929) was found to be specific to this trait only, that is, it was associated only with TR under moderate (1.5 kPa) VPD conditions (Table 3, Figure 7).
FIGURE 7.
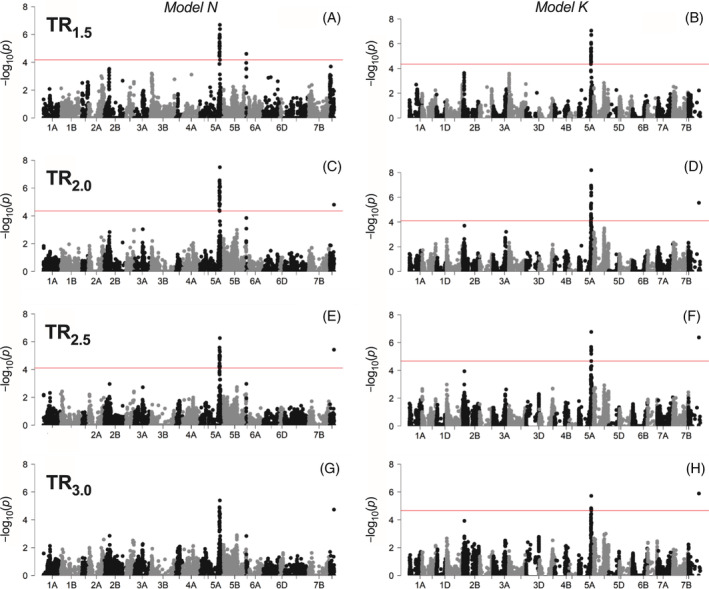
Manhattan plots showing significant SNPs associated with traits controlling transpiration rate (TR) response to increasing vapor pressure deficit (VPD). Results are shown based on two genome‐wide association models, N (left) and K (right) for TR1.5 (A, B), TR2.0 (C, D), TR2.5 (E, F) and TR3.0 (G, H). In each panel (except G), the horizontal line indicates the threshold level above which SNPs are declared as significantly associated with the trait with an FDR of 0.05.
Specific leaf area‐associated SNPs were found to be highly unique to this trait, with 15 out of 16 SNPs being specifically associated with this trait. Seven of these SNPs concentrate in a region located on chromosome 1B, including one (S1B_9571874) explaining 20.9% of the phenotypic variation (Table 3). The rest of the unique SNPs cover chromosomes 4B, 6A, 6D, 7A, and 7B, but have much lower explanatory power (Table 3, Figure 6).
4.4. Candidate gene identification
For the four major SNPs identified (S5A_579969562, S5A_595425852, S5D_194957929 and S7D_500209644), the corresponding haploblocks were examined for candidate gene identification (Table S3). HaploBlock_5D_66.76_290.6, where the SNP S5D_194957929 localizes, has the highest number of genes (665) while HaploBlock_5A_573.05_589.3 and HaploBlock_5A_595.43_608.36, where SNPs S5A_579969562 and S5A_595425852 reside, have a total of 112 and 87 genes, respectively. The SNP S7D_500209644 turned out not to be contained in any haplotype block and only has one gene within 100 kb from it. As reported in Figure 8, the largest class of genes associated with all three SNPs represents the gene products and protein complexes embedded in the cell membrane.
FIGURE 8.
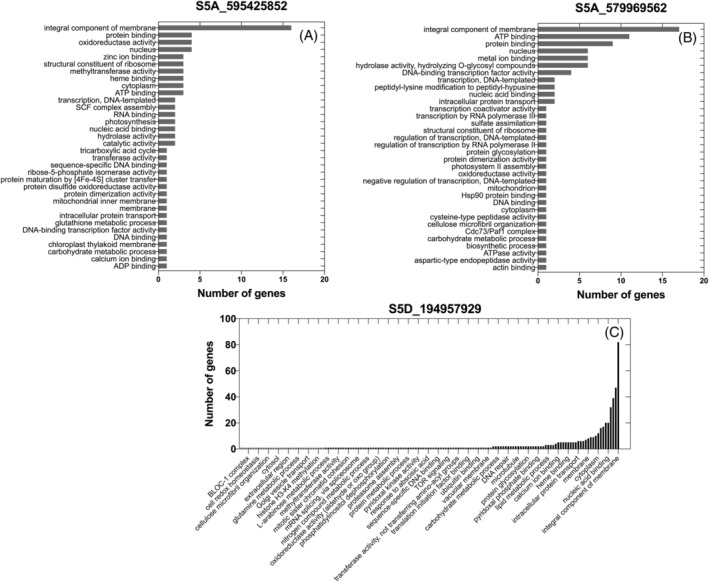
Gene ontology term distribution in haploblocks where three major SNPs associated with TR response to VPD are located on chromosomes 5A (A, B) and 5C
5. DISCUSSION
5.1. Relevance of the VPD‐associated SNPs identified
To the best of our knowledge, this is the first study that identified the genetic basis of TR response to VPD per se in plants, that is, independent from confounding environmental variables such as temperature, PAR, air mixing, and soil drying. In this regard, the study indicates that a biophysical trait, namely whole‐plant TR sensitivity to VPD has a genetic basis in wheat. In the literature, while there is a very limited set of studies that undertook an effort of identifying the genetic basis of whole‐plant TR, these took place under naturally fluctuating environments that did not separate out interacting effects on TR arising from co‐variation in temperature, PAR and air mixing along with VPD (Kholova et al., 2012; Schoppach et al., 2016). Because water use strategies are driven by TR response to VPD, it is critical to confirm that such behaviors have a genetic basis if they are to be used as a basis for a breeding effort.
In this respect, the present study revealed a major region on chromosome 5A, which appears to be a ‘hotspot’ for traits controlling whole‐plant TR and evaporative surface (Figure 6, Table 3). More specifically, this region harbors an SNP of a particular interest (S5A_595425852) that explained up to 17.1% of the observed phenotypic variance, with the RB07 and PI 430750 alleles consistently exhibiting positive and negative effects. These effects are in alignment with the rationale for selecting PI 430750 as the parent that exhibited the lowest slope for TR response to VPD, in comparison to RB07 (Figure S1, Tamang et al., 2019). The haploblock where this SNP resides harbors various gene families, the majority of which are integral parts of a membrane (Figure 8A). While further studies are needed to determine the exact functions of these genes, such a finding is consistent with the need for recruitment of intercellular, trans‐membrane aquaporins for water movement during TR response to VPD (Maurel et al., 2016; Sadok & Sinclair, 2010; Schoppach et al., 2014; Sinclair et al., 2008). Additionally, the haploblock is associated with three photosynthesis‐related genes, (GO:0009535, GO:0019684, GO:0015979), which functions could be related to TR response to VPD given the coupling between transpiration and CO2 fixation through the stomata.
Another SNP of interest in this region is S5A_579969562 (Figure 8B). The profile of the haploblock harboring this gene is similar to that of S5A_595425852. It contains genes which the majority of are membrane‐associated, but additionally, it presents two auxin‐responsive genes (GO:0006351). Such findings are in alignment with Schoppach et al. (2016) who identified a major QTL controlling TR under naturally fluctuating condition on chromosome 5A. This peak region was mapped to genes responsive to auxin, and the subsequent finding was that auxin differentially accumulated in the root tissues of genotypes from that population, expressing contrasting levels of TR (Sadok & Schoppach, 2019). Interestingly, the haploblock of this SNP also harbors the Vrn‐A1 gene, which is known to affect plant development (Dixon et al., 2019). If confirmed, our findings would indicate that Vrn‐A1 could also influence transpiration, presumably through pleiotropic effects.
The investigation also revealed that some SNPs are associated with either low VPD (TR1.5, S5D_194957929) or higher VPDs only (S7D_500209644 for TR2.0, TR2.5, and TR3.0), which are respectively located on chromosomes 5D and 7D (Figures 6, 7, Table 3). While S7D_500209644 was not found to be in any haploblock, the one containing S5D_194957929 presented a very rich gene ontology profile (Figure 8C), which highlighted membrane‐associated genes as the biggest class, similar to the two previously discussed SNPs. This SNP was also found to be associated with at least one ABA‐responsive gene (GO:0009737), an observation that could be interpreted as consistent with this SNP being associated with a specific VPD level, since differential VPD levels are associated with different ABA accumulation levels in the plant (Kholová et al., 2010; Kudoyarova et al., 2011). If confirmed, these SNPs offer a potential avenue for further fine‐tuning the design of cultivars adapted to specific VPD regimes.
5.2. Advantages and limits of the phenotyping approach
Due to unavoidable space and logistical limitations, the three‐year phenotyping approach deployed in this study relied each time on a sequential planting in order to phenotype the three mapping populations. This resulted in the challenge of exposing each one of the three cohorts of a given experiment to sensibly different growth conditions, as these were grown under naturally fluctuating conditions in a greenhouse (Table 1). Despite this, reasonable repeatabilities were found for the traits of interest (Figure 3), indicating that future efforts of phenotyping the entire mapping population simultaneously is likely to yield higher repeatabilities, and perhaps more SNPs. Relieving this logistical bottleneck is probably critical for the future, to perform more routine and risk‐averse studies given the relatively complex nature of the phenotypes being investigated (response curves), which tend to be sensitive to changes in growth conditions (Sadok et al., 2021). With such larger phenotyping set‐ups, the highest throughput for screening TR versus VPD response curves, achieved in this investigation (150 lines), could be perhaps increased 2–3 folds. Such effort might be particularly needed in future efforts, as our results indicate that VPD sensitivity, meaning the expression of a significant change in slope in TR response to increasing VPD, is a relatively rare trait, which was expressed by only 15% of the genotypes of the study (Figure 2). This likely requires the screening of a large number of genotypes in order to be able to maximize phenotypic variation.
One potential limit of the proposed approach is its reliance on controlled, rather than fluctuating environmental conditions for phenotyping TR responses to VPD. We argue that this approach is critical for a number of reasons. First, ever since the hypothesis of TR sensitivity to VPD was proposed as a promising avenue for improving crop productivity in water‐limited environments in the systems analysis of Sinclair et al. (2005), the vast majority of the efforts looking for this trait in crops failed to impose VPD as the only source of variation (see review by Sinclair et al., 2017). As recent research efforts highlighted the benefits of breeding intentionally for this trait (Collins et al., 2021; Gaffney et al., 2015; Sadok et al., 2019; Ye et al., 2020), the inability to identify a genetic basis for TR sensitivity to VPD per se would mean that this trait is not a viable breeding target.
A second argument in favor of the controlled environment approach lies in the importance of untangling the effects on TR arising from VPD from those resulting from co‐variation in PAR or, even more importantly, temperature. Variation in temperature conditions within and between days could strongly mask differences between genotypes, and in fact, may trigger a switch from a VPD‐sensitive (i.e., a water‐saving) phenotype to a VPD‐insensitive (i.e., consumptive) one, and to complicate things, in a genotype‐dependent fashion (Sadok et al., 2021). In this context, our results provide a parsimonious basis for interpreting the physiological relevance of candidate genes/markers as outlined in the above section. The relatively low number of SNPs identified underlying an a priori complex trait might be the direct result of the phenotyping approach, which has the benefit of pointing directly to those that directly underlie TR response to VPD, rather than other confounding variables.
AUTHOR CONTRIBUTIONS
Conceptualization: B.G.T. and W.S.; Formal analysis; B.G.T. and W.S.; Writing‐original draft: B.G.T. and W.S.; Funding acquisition: W.S., B.J.S, J.A.A.; Writing‐review & editing: B.G.T., J.A.A., B.J.S., W.S.
Supporting information
Figure S1 Diversity in whole‐plant transpiration rate (TR) response curves to increasing vapor pressure deficit (VPD) across the selected parents of the three families phenotyped in the study. Linear and segmented regression fit parameters are indicated in each panel, where Slope is the slope of the linear regression, while Slope1, Slope2, and VPDBP represent the first slope, the second slope and the VPD breakpoint (BP), that is, the VPD at which the slope of TR response to VPD changes. Data is from Tamang et al. (2019).
Table S1: Regression parameters (linear vs. segmental) of TR response to VPD response curves for all the genotypes.
Table S2: Haplotype blocks with significant SNPs associated with the studied traits in three wheat NAM families.
Table S3: List of genes within haplotype block of SNPs.
ACKNOWLEDGMENTS
W.S. was supported by USDA NIFA through the Minnesota Agricultural Experiment Station (Project No. MIN‐13‐124). This research was funded by the Minnesota Wheat Research and Promotion Council (Projects: #00070003, 00076909, 00084893). We thank Reina Nielsen for her help during the phenotyping experiment and Ahmad Sallam for earlier discussions on the genetic analysis.
Tamang, B.G. , Monnens, D. , Anderson, J.A. , Steffenson, B.J. & Sadok, W. (2022) The genetic basis of transpiration sensitivity to vapor pressure deficit in wheat. Physiologia Plantarum, 174(5), e13752. Available from: 10.1111/ppl.13752
Edited by: K. Noguchi
Funding information Minnesota Wheat Research and Promotion Council, Grant/Award Number: #00070003 00076909 00084893; USDA NIFA ‐ Minnesota Agricultural Experiment Station, Grant/Award Number: MIN‐13‐124
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.
REFERENCES
- Anderson, C.A. , Pettersson, F.H. , Clarke, G.M. , Cardon, L.R. , Morris, A.P. & Zondervan, K.T. (2010) Data quality control in genetic case‐control association studies. Nature Protocols, 5(9), 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.A. , Linkert, G.L. , Busch, R.H. , Wiersma, J.J. , Kolmer, J.A. , Jin, Y. et al. (2009) Registration of “RB07” wheat. Journal of Plant Registrations, 3(2), 175–180. [Google Scholar]
- Bajgain, P. , Sallam, A.H. , Annor, G. , Conley, E. , Steffenson, B.J. , Muehlbauer, G.J. et al. (2021) Genetic characterization of flour quality and bread‐making traits in a spring wheat nested association mapping population. Crop Science, 61(2), 1168–1183. [Google Scholar]
- Bates, D. , Mächler, M. , Bolker, B. & Walker, S. (2015) Fitting linear mixed‐effects models using lme4. Journal of Statistical Software, 67, 1–48. [Google Scholar]
- Broman, K.W. , Wu, H. , Sen, S. & Churchill, G.A. (2003) R/QTL: QTL mapping in experimental crosses. Bioinformatics, 19(7), 889–890. [DOI] [PubMed] [Google Scholar]
- Collins, B. , Chapman, S. , Hammer, G. & Chenu, K. (2021) Limiting transpiration rate in high evaporative demand conditions to improve Australian wheat productivity. In Silico Plants, 3(1), 1–16. [Google Scholar]
- Dixon, L.E. , Karsai, I. , Kiss, T. , Adamski, N.M. , Liu, Z. , Ding, Y. et al. (2019) Vernalization1 controls developmental responses of winter wheat under high ambient temperatures. Development, 146(3), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman, J.B. (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. The Plant Genome, 4(3), 250–255. [Google Scholar]
- Ficklin, D.L. & Novick, K.A. (2017) Historic and projected changes in vapor pressure deficit suggest a continental‐scale drying of the United States atmosphere. Journal of Geophysical Research: Atmospheres, 122, 2061–2079. [Google Scholar]
- Fletcher, A.L. , Sinclair, T.R. & Allen, L.H. (2007) Transpiration responses to vapor pressure deficit in well watered “slow‐wilting” and commercial soybean. Environmental and Experimental Botany, 61(2), 145–151. [Google Scholar]
- Gaffney, J. , Schussler, J. , Löffler, C. , Cai, W. , Paszkiewicz, S. , Messina, C. et al. (2015) Industry‐scale evaluation of maize hybrids selected for increased yield in drought‐stress conditions of the US Corn Belt. Crop Science, 55(4), 1608–1618. [Google Scholar]
- Kholová, J. , Hash, C.T. , Kakkera, A. , Kocová, M. & Vadez, V. (2010) Constitutive water‐conserving mechanisms are correlated with the terminal drought tolerance of pearl millet [Pennisetum glaucum (L.) R. Br.]. Journal of Experimental Botany, 61(2), 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholová, J. , Nepolean, T. , Tom Hash, C. , Supriya, A. , Rajaram, V. , Senthilvel, S. et al. (2012) Water saving traits co‐map with a major terminal drought tolerance quantitative trait locus in pearl millet [Pennisetum glaucum (L.) R. Br.]. Molecular Breeding, 30(3), 1337–1353. [Google Scholar]
- Kimm, H. , Guan, K. , Gentine, P. , Wu, J. , Bernacchi, C.J. , Sulman, B.N. et al. (2020) Redefining droughts for the U.S. Corn Belt: the dominant role of atmospheric vapor pressure deficit over soil moisture in regulating stomatal behavior of maize and soybean. Agricultural and Forest Meteorology, 287, 107930. [Google Scholar]
- Kudoyarova, G. , Veselova, S. , Hartung, W. , Farhutdinov, R. , Veselov, D. & Sharipova, G. (2011) Involvement of root ABA and hydraulic conductivity in the control of water relations in wheat plants exposed to increased evaporative demand. Planta, 233(1), 87–94. [DOI] [PubMed] [Google Scholar]
- Lin, C.‐Y. , Xing, G. & Xing, C. (2012) Measuring linkage disequilibrium by the partial correlation coefficient. Heredity, 109(6), 401–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell, D.B. , Hammer, G.L. , McLean, G. , Messina, C. , Roberts, M.J. & Schlenker, W. (2013) The critical role of extreme heat for maize production in the United States. Nature Climate Change, 3(5), 497–501. [Google Scholar]
- López, J. , Way, D.A. & Sadok, W. (2021) Systemic effects of rising atmospheric vapor pressure deficit on plant physiology and productivity. Global Change Biology, 27(9), 1704–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel, C. , Verdoucq, L. & Rodrigues, O. (2016) Aquaporins and plant transpiration. Plant, Cell and Environment, 39(11), 2580–2587. [DOI] [PubMed] [Google Scholar]
- Monnens, D. & Sadok, W. (2020) Whole‐plant hydraulics, water saving, and drought tolerance: a triptych for crop resilience in a drier world. Annual Plant Reviews Online, 3(4), 661–698. [Google Scholar]
- Motulsky, H. (1999) Analyzing data with GraphPad prism. San Diego CA: GraphPad Software Inc. www.graphpad.com [Google Scholar]
- R Core Team . (2017) R: a language and environment for statistical computing. Vienna: Austria. https://www.R-project.org/ [Google Scholar]
- Rebetzke, G.J. , Condon, A.G. , Richards, R.A. & Farquhar, G.D. (2003) Gene action for leaf conductance in three wheat crosses. Australian Journal of Agricultural Research, 54(4), 381. [Google Scholar]
- Riar, M.K. , Sinclair, T.R. & Prasad, P.V.V. (2015) Persistence of limited‐transpiration‐rate trait in sorghum at high temperature. Environmental and Experimental Botany, 115, 58–62. [Google Scholar]
- Sadok, W. , Lopez, J.R. & Smith, K.P. (2021) Transpiration increases under high‐temperature stress: potential mechanisms, trade‐offs and prospects for crop resilience in a warming world. Plant, Cell & Environment, 44(7), 2102–2116. [DOI] [PubMed] [Google Scholar]
- Sadok, W. & Schoppach, R. (2019) Potential involvement of root auxins in drought tolerance by modulating nocturnal and daytime water use in wheat. Annals of Botany, 124(6), 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadok, W. , Schoppach, R. , Ghanem, M.E. , Zucca, C. & Sinclair, T.R. (2019) Wheat drought‐tolerance to enhance food security in Tunisia, birthplace of the Arab spring. European Journal of Agronomy, 107, 1–9. [Google Scholar]
- Sadok, W. & Sinclair, T.R. (2010) Genetic variability of transpiration response of soybean [Glycine max (L.) Merr.] shoots to leaf hydraulic conductance inhibitor AgNO3. Crop Science, 50(4), 1423–1430. [Google Scholar]
- Sadok, W. & Tamang, B.G. (2019) Diversity in daytime and night‐time transpiration dynamics in barley indicates adaptation to drought regimes across the middle‐east. Journal of Agronomy and Crop Science, 205(4), 372–384. [Google Scholar]
- Sallam, A.H. , Manan, F. , Bajgain, P. , Martin, M. , Szinyei, T. , Conley, E. et al. (2020) Genetic architecture of agronomic and quality traits in a nested association mapping population of spring wheat. The Plant Genome, 13(3), e20051. [DOI] [PubMed] [Google Scholar]
- Schoppach, R. , Fleury, D. , Sinclair, T.R. & Sadok, W. (2017) Transpiration sensitivity to evaporative demand across 120 years of breeding of Australian wheat cultivars. Journal of Agronomy and Crop Science, 203(3), 219–226. [Google Scholar]
- Schoppach, R. & Sadok, W. (2012) Differential sensitivities of transpiration to evaporative demand and soil water deficit among wheat elite cultivars indicate different strategies for drought tolerance. Environmental and Experimental Botany, 84, 1–10. [Google Scholar]
- Schoppach, R. , Taylor, J.D. , Majerus, E. , Claverie, E. , Baumann, U. , Suchecki, R. et al. (2016) High resolution mapping of traits related to whole‐plant transpiration under increasing evaporative demand in wheat. Journal of Experimental Botany, 67(9), 2847–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppach, R. , Wauthelet, D. , Jeanguenin, L. & Sadok, W. (2014) Conservative water use under high evaporative demand associated with smaller root metaxylem and limited trans‐membrane water transport in wheat. Functional Plant Biology, 41(3), 257–269. [DOI] [PubMed] [Google Scholar]
- Shekoofa, A. , Rosas‐Anderson, P. , Carley, D.S. , Sinclair, T.R. & Rufty, T.W. (2016) Limited transpiration under high vapor pressure deficits of creeping bent grass by application of Daconil‐action. Planta, 243(2), 421–427. [DOI] [PubMed] [Google Scholar]
- Shekoofa, A. , Rosas‐Anderson, P. , Sinclair, T.R. , Balota, M. & Isleib, T.G. (2015) Measurement of limited‐transpiration trait under high vapor pressure deficit for peanut in chambers and in field. Agronomy Journal, 107(3), 1019–1024. [Google Scholar]
- Sinclair, T.R. , Devi, J. , Shekoofa, A. , Choudhary, S. , Sadok, W. , Vadez, V. et al. (2017) Limited‐transpiration response to high vapor pressure deficit in crop species. Plant Science, 260, 109–118. [DOI] [PubMed] [Google Scholar]
- Sinclair, T.R. , Hammer, G.L. & van Oosterom, E.J. (2005) Potential yield and water‐use efficiency benefits in sorghum from limited maximum transpiration rate. Functional Plant Biology, 32(10), 945–952. [DOI] [PubMed] [Google Scholar]
- Sinclair, T.R. , Zwieniecki, M.A. & Holbrook, N.M. (2008) Changes in plant–soil hydraulic pressure gradients of soybean in response to soil drying. The Annals of Applied Biology, 152(1), 49–57. [Google Scholar]
- Steinemann, S. , Zeng, Z. , McKay, A. , Heuer, S. , Langridge, P. & Huang, C.Y. (2015) Dynamic root responses to drought and rewatering in two wheat (Triticum aestivum) genotypes. Plant and Soil, 391(1–2), 139–152. [Google Scholar]
- Storey, J.D. & Tibshirani, R. (2003) Statistical significance for genome wide studies. Proceedings of the National Academy of Sciences of the United States of America, 100(16), 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang, B.G. & Sadok, W. (2018) Nightly business: links between daytime canopy conductance, nocturnal transpiration and its circadian control illuminate physiological trade‐offs in maize. Environmental and Experimental Botany, 148, 192–202. [Google Scholar]
- Tamang, B.G. , Schoppach, R. , Monnens, D. , Steffenson, B.J. , Anderson, J.A. & Sadok, W. (2019) Variability in temperature‐independent transpiration responses to evaporative demand correlate with nighttime water use and its circadian control across diverse wheat populations. Planta, 250(1), 115–127. [DOI] [PubMed] [Google Scholar]
- Xavier, A. , Xu, S. , Muir, W.M. & Rainey, K.M. (2015) NAM: association studies in multiple populations. Bioinformatics, 31(23), 3862–3864. [DOI] [PubMed] [Google Scholar]
- Ye, H. , Song, L. , Schapaugh, W.T. , Ali, M.L. , Sinclair, T.R. , Riar, M.K. et al. (2020) The importance of slow canopy wilting in drought tolerance in soybean. Journal of Experimental Botany, 71(2), 642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, W. , Zheng, Y. , Piao, S. , Ciais, P. , Lombardozzi, D. , Wang, Y. et al. (2019) Increased atmospheric vapor pressure deficit reduces global vegetation growth. Science. Advances, 5(8), eaax1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, B. , Biddulph, B. , Li, D. , Kuchel, H. , & Chapman, S. (2013) Quantification of the effects of VRN1 and Ppd‐D1 to predict spring wheat (Triticum aestivum) heading time across diverse environments. Journal of Experimental Botany, 64(12), 3747–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, B. , Chenu, K. & Chapman, S.C. (2016) Velocity of temperature and flowering time in wheat ‐ assisting breeders to keep pace with climate change. Global Change Biology, 22(2), 921–933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Diversity in whole‐plant transpiration rate (TR) response curves to increasing vapor pressure deficit (VPD) across the selected parents of the three families phenotyped in the study. Linear and segmented regression fit parameters are indicated in each panel, where Slope is the slope of the linear regression, while Slope1, Slope2, and VPDBP represent the first slope, the second slope and the VPD breakpoint (BP), that is, the VPD at which the slope of TR response to VPD changes. Data is from Tamang et al. (2019).
Table S1: Regression parameters (linear vs. segmental) of TR response to VPD response curves for all the genotypes.
Table S2: Haplotype blocks with significant SNPs associated with the studied traits in three wheat NAM families.
Table S3: List of genes within haplotype block of SNPs.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.


