Summary
Eukaryotic genomes contain a vast diversity of transposable elements (TEs). Formerly often described as selfish and parasitic DNA sequences, TEs are now recognised as a source of genetic diversity and powerful drivers of evolution. However, because their mobility is tightly controlled by the host, studies experimentally assessing how fast TEs may mediate the emergence of adaptive traits are scarce.
We exposed Arabidopsis thaliana high‐copy TE lines (hcLines) with up to c. eight‐fold increased copy numbers of the heat‐responsive ONSEN TE to drought as a straightforward and ecologically highly relevant selection pressure.
We provide evidence for increased drought tolerance in five out of the 23 tested hcLines and further pinpoint one of the causative mutations to an exonic insertion of ONSEN in the ribose‐5‐phosphate‐isomerase 2 gene. The resulting loss‐of‐function mutation caused a decreased rate of photosynthesis, plant size and water consumption.
Overall, we show that the heat‐induced transposition of a low‐copy TE increases phenotypic diversity and leads to the emergence of drought‐tolerant individuals in A. thaliana. This is one of the rare empirical examples substantiating the adaptive potential of mobilised stress‐responsive TEs in eukaryotes. Our work demonstrates the potential of TE‐mediated loss‐of‐function mutations in stress adaptation.
Keywords: adaptation, Arabidopsis thaliana, drought tolerance, experimental evolution, loss‐of‐function mutation, transposable elements
Introduction
Plants are constantly exposed to fluctuating environments. To successfully reproduce, they rely on mechanisms that allow them to react and adapt to suboptimal growth conditions. Genetic variation is a prerequisite for adaptation and the evolution of new traits. There is evidence that severe stresses can not only trigger the formation of small‐scale mutations (Belfield et al., 2021; Lu et al., 2021) but also increase genetic diversity through the stress‐induced activation of transposable elements (TEs) (McClintock, 1950; Lisch, 2013; Negi et al., 2016). In eukaryotic genomes, TEs are highly abundant (Wells & Feschotte, 2020) and the proportion of TE‐derived sequences in plants can reach up to > 80% in cases of some crops such as maize (Stitzer et al., 2021) or wheat (Wicker et al., 2018). In contrast with single nucleotide polymorphisms (SNPs), TEs can not only efficiently knock‐out genes (Van Houwelingen et al., 1998; Ram et al., 2019) but also bring their flanking regions under epigenetic control or render them stress responsive (Butelli et al., 2012; Grandbastien, 2015; Makarevitch et al., 2015; Roquis et al., 2021). By moving in response to the environment, TEs are therefore believed to be of particular importance for generating the phenotypic diversity needed for rapid adaptation to challenging environments (Torkamanzehi et al., 1992; Walser et al., 2006; Naito et al., 2009; González et al., 2010; Hof et al., 2016; Rey et al., 2016; Li et al., 2018; Esnault et al., 2019; Baduel et al., 2021).
In plants, the majority of TEs are class I elements belonging to the subclass of long terminal repeat (LTR)‐retrotransposons that move through the reverse transcription of an RNA‐intermediate in a copy‐and‐paste mechanism (Wicker et al., 2007; Schulman, 2013). To ensure a limited mutation rate and to safeguard genome stability, TE mobility is usually restricted by epigenetic silencing mechanisms, which in plants involves the RNA‐directed DNA methylation (RdDM) pathway (Matzke & Mosher, 2014). As a consequence, only few TE families have been observed transposing in planta (McClintock, 1953; Grandbastien et al., 2005; Picault et al., 2009; Lanciano et al., 2017; Masuta et al., 2018; Benoit et al., 2019) and this even in plants free of DNA methylation (He et al., 2022). The understanding to what extent TEs may play a role in adaptation in this kingdom therefore largely relies on population genomics data and correlative studies (Quadrana et al., 2016; Li et al., 2018; Stritt et al., 2018; Baduel et al., 2021) rather than on experimental validation.
In this context, we study here the functional impact of the 5 kb Arabidopsis thaliana retrotransposon ONSEN (AtCOPIA78), one of the best characterised TE families in plants. Equipped with heat‐responsive elements in its LTRs, ONSEN can sense the heat stress‐response of its host and utilise it to initiate its own copy‐and‐paste lifecycle (Tittel‐Elmer et al., 2010; Ito et al., 2011; Cavrak et al., 2014). While transgenerational transposition events of ONSEN in wild‐type (wt) plants are too rare to be observed in real‐time (Hayashi et al., 2020), transcripts and reverse transcribed extrachromosomal DNA copies of ONSEN in the Col‐0 wild‐type (wt) can already be detected following a heat shock at 37°C for 12 h (Ito et al., 2011; Cavrak et al., 2014). Moreover, genomic copy numbers of ONSEN are known to vary between natural accessions of A. thaliana (Masuda et al., 2016; Quadrana et al., 2016) suggesting an ongoing heat‐dependent mobility of ONSEN in wild populations.
To overcome the limitation of the low transposition frequency of active TEs in natural accessions, we previously developed a method to amplify ONSEN in wt plants. By transiently reducing TE silencing by the inhibition of DNA‐methyltransferases (Baubec et al., 2009) and RNA polymerase II in combination with a heat shock for 24 h at 37°C, we were able to increase ONSEN activity, which resulted in the integration and inheritance of novel genomic ONSEN copies in Arabidopsis Col‐0 wt plants (please refer to Materials and Methods section and Thieme et al., 2017). The copy numbers of the selected ONSEN high‐copy TE lines (hcLines) were stable over three generations of selfing suggesting genetic stability (Thieme et al., 2017). Notably, the distribution of novel ONSEN copies in these hcLines is not random but reflects a distinct insertion bias towards exons and H2A.Z enriched regions (Roquis et al., 2021) confirming previous findings in TE accumulation lines (Quadrana et al., 2019).
The transposition of ONSEN is known to result in transcriptomic changes (Ito et al., 2011; Roquis et al., 2021) and new phenotypes such as altered seed colour (Thieme et al., 2017) or abscisic acid insensitivity (Ito et al., 2016). However, while the heat‐dependent mobility of ONSEN could therefore create the raw material for evolution and was proposed to confer a unique adaptive potential to global warming in A. thaliana (Quadrana et al., 2016), its immediate adaptive potential and fitness effects have not been tested experimentally. Due to climate change and temperature increase, drought is predicted to constitute one of the most severe environmental constraints to which plants will have to adapt in the near future (Exposito‐Alonso et al., 2019; Brás et al., 2021). We used here our collection of hcLines to experimentally test whether and how the heat‐induced transposition of ONSEN may help individuals to survive in warmer and therefore water‐limited environments.
Materials and Methods
Plant material
ONSEN hcLines were generated by treating A. thaliana Col‐0 plants with a combination of a heat shock and drugs that inhibit TE silencing, as described previously (Thieme et al., 2017). Briefly, Col‐0 seeds were germinated and grown under long day conditions (16 h light) at 24°C (day) 22°C (night) on half‐strength Murashige and Skoog (½MS) medium with 1% sucrose and 0.5% Phytagel, pH 5.8. To reduce TE silencing and increase the rate of ONSEN transposition, seedlings were grown analogously on ½MS medium supplied with a combination sterile filtered zebularine (Z, 40 μM) and α‐amanitin (A, 5 mg ml−1). After 7 d of growth on control ½MS or medium supplied with A and Z, seedlings were ether exposed to control stress (CS; 24 h at 6°C followed by 24 h at normal conditions) or heat stress (HS; 24 h at 6°C followed by 24 h at 37°C), then transferred to soil and selfed to obtain the S1 generation. Individual S1 plants originating from plants that were either only exposed to CS or HS or additionally treated with A and Z (AZ) were separated and repeatedly self‐fertilised until we obtained the S4 generation. In this study we used 23 ONSEN hc‐lines originating from 13 plants that were treated with HS + AZ, five independent control lines that were either only exposed to CS (two lines), HS (two lines) or CS + AZ (one line) and the Col‐0 wild‐type that was propagated on soil. The rpi2‐1 mutant (SALK_022117) (Xiong et al., 2009) was obtained from the Nottingham Arabidopsis Stock Centre (Alonso et al., 2003).
qPCR for ONSEN copy numbers
To determine the average ONSEN copy numbers of the hcLines and controls used in this study, we extracted DNA of the aboveground parts of at least 24 pooled individuals per line of the S4 generation grown for 8 d under sterile conditions on ½MS medium (1% sucrose, 0.5% Phytagel, pH 5.8) under long day conditions (16 h light) at 24°C (day) 22°C (night) using the DNeasy Plant Kit (Qiagen). ONSEN copy numbers were determined by qPCR using 12 ng total DNA using the KAPA SYBR FAST master mix universal on a C1000 Touch (Bio‐Rad) machine. ACTIN2 (At3g18780) was used to normalise DNA levels and DNA of Col‐0 served as a control. Three technical replicates were used and data were analysed using the Bio‐Rad CFX Manager 3.1 software. Sequences of oligos are listed in Supporting Information Table S1.
Identification, expression analysis and visualisation of ONSEN and T‐DNA insertions
Novel ONSEN insertions of hcLine31 were identified and characterised recently (Roquis et al., 2021) by whole‐genome sequencing. Briefly, reads were trimmed using trimmomatic (Bolger et al., 2014) with the parameters ILLUMINACLIP: TruSeq3:2:30:10 LEADING:20 TRAILING:20 SLIDINGWINDOW:4:20 and MINLEN:36. ONSEN insertions were detected and analysed previously using Transposable Insertion Finder v.1.6 (Nakagome et al., 2014) with the TAIR10 version of the A. thaliana Col‐0 reference genome (Berardini et al., 2015) as described in detail (Roquis et al., 2021). Processed genomic reads were mapped to the TAIR10 version of the A. thaliana genome using bwa mem (v.0.7.17‐r1188) (Li & Durbin, 2009) with the ‐M parameter set. To validate the presence and zygosity of the ONSEN and T‐DNA insertions in RPI2 in the segregating F2 populations, we designed primers (mto_007 and mto_067) spanning the predicted insertion sites of ONSEN and the T‐DNA (based on SIGnAL) and combined them with primers specific to ONSEN (mto_196) or the T‐DNA (LBb1.3 mto_063) (Fig. 3a; Table S1). For the PCRs we used a standard Taq DNA polymerase (Sigma Aldrich) and limited the elongation time to 90 s so that an homozygous insertion of the 5‐kb ONSEN TE or the T‐DNA would prevent the formation of a PCR product. To genotype the F2 populations, we used DNA from the homozygous parental plants (wt, hcLine31 and rpi2‐1 (SALK_022117)).
Fig. 3.
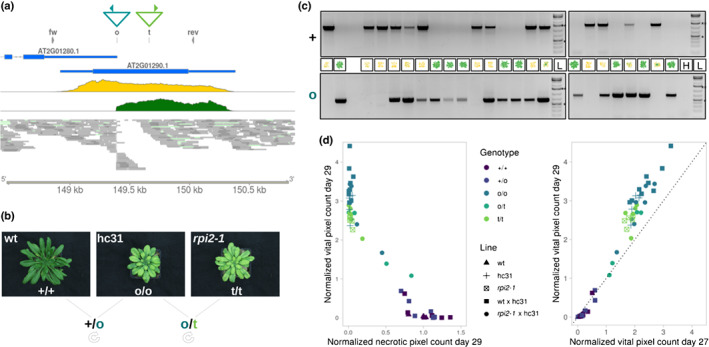
An ONSEN insertion in RPI2 leads to an increased drought tolerance of Arabidopsis thaliana hcLine31 (hc31). (a) Triangles indicate the insertion site of ONSEN (turquoise) and the location of the T‐DNA in the rpi2‐1 mutant (green) on chromosome 2. Primer locations used for the genotyping in (c) and (d) are depicted as filled triangles. Annotation track (blue), RNA‐seq coverage of the wt (yellow) and hcLine31 (green) and aligned genomic reads from hcLine31 are shown. (b) Images and crossing scheme of the wt, hcLine31 and rpi2‐1. Genotypes are depicted with + (wt), o (ONSEN), and t (T‐DNA). (c) Genotypes and phenotypes (day 29) of the segregating F2 population of hcLine31 × wt. A wt and a hcLine31 (first two lanes) are shown as references. Primers specific for the wt (upper gel, +) and the ONSEN insertion (lower gel, o) (please refer to (a)) were used. H, water control; L, GeneRuler 1 kb plus ladder. 0.5 kb (*) and 1.5 kb (**) bands are marked. (d) Pixel counts of living tissue after 2 d of recovery (day 29) in relation to necrotic tissues on the same day (left panel) and compared with vital pixel counts before recovery (day 27) (right panel). Shapes indicate the plant line (parental or segregating F2 individuals of the crosses from panel (b) and colours indicate the genotype of RPI2.
RNA‐seq data of one representative biological replicate of the Col‐0 wt and hcLine31 exposed to CS and whole‐genome sequencing data of hcLine31 were obtained from and analysed according to a previous report (Roquis et al., 2021). Briefly, reads were trimmed using trimmomatic (Bolger et al., 2014) as described above. Processed RNA‐seq reads were aligned to the TAIR10 version of the A. thaliana genome using Star (v.2.7.9a) (Dobin et al., 2013) using the same parameters such as Roquis et al. (2021) but with ‐alignIntronMax set to 10 000. The insertion site of ONSEN was visualised using the generated bam files and the packages gviz (v.1.28.3) (Hahne & Ivanek, 2016), rtracklayer (v.1.44.4) (Lawrence et al., 2009) and the annotation package txdb.athaliana.biomart.plantsmart28 (v.3.2.2) (Carlson & Maintainer, 2015) in R (v.3.6.3) (R Core Team, 2020) in Rstudio (v.1.1.456) (RStudio Team, 2016).
Drought assay
To obtain comparable and robust results, we ran one comprehensive drought experiment in which we tested the S4 generation of hcLines, the control lines and the segregating F2 generations of crosses between hcLine31 and Col‐0, and hcLine31 and rpi2‐1 (SALK_022117) in parallel. We included five replicates for each high‐copy and control line and for the parents of the cross between rpi2‐1 and hcLine31, and tested 22 F2 individuals of the cross of hcLine31 with the wt and 16 F2 individuals of the cross of hcLine31 and rpi2‐1 (SALK_022117). Seeds were sown in pots filled with Einheitserde that was incubated with a solution (75 mg l−1) of the insecticide Kohinor (Leu + Gygax AG) and kept at 4°C for 3 d. After stratification, pots were moved into a Hiros climate chamber (Clitec GmbH, Küssnacht am Rigi, Switzerland) set to short day conditions with 10 h light (LED Valoya Ns12 C75/65, c. 120 μmol m−2 s−1) at 22°C : 19°C, day : night, with 60% humidity. After 10 d of growth, seedlings were picked into pots filled with equal amounts of Kohinor‐treated soil and grown under well watered conditions for 36 d. The position of pots was frequently shifted to ensure similar growth conditions. Before watering was suspended, pots were again saturated with water and weighted to obtain the maximal water content. At 1 d later (day 1 of the experiment), top‐view pictures were taken with a Canon EOS 70D camera on a tripod at the following settings: 5.6 s shutter opening, 1/60 shutter speed, ISO 200. This procedure was repeated three times at an interval of 7 d until day 22 of the experiment. Due to technical issues, nine out of 193 images (affecting one to two biological replicates of seven different lines and one biological replicate of the F2 of the cross of hcLine31 and rpi2‐1) from day 15 are missing in the analysis. On day 27, pots were again weighted, top‐view pictures were taken, and drought stress was stopped by filling trays with water and allowing the pots to absorb water over night. After 2 d of regeneration under well watered conditions, final pictures of the plants were taken (day 29). To account for different zoom levels during the course of the experiment, we took photographs of a white label that later served as a calibrator to normalise predicted vital and necrotic leaf areas. At 1 d after the last photographs were taken, one leaf of each plant from the segregating F2 populations was sampled for DNA extractions and genotyping. Pots were removed from the climate chamber and dried for 8 wk at room temperature to obtain the dry weight to calculated the water content of each pot. Pictures and weight were determined on 2 d successively (except for day 27); therefore, we extrapolated the weight measurements to determine the water content on the exact day pictures were taken.
Machine‐learning‐based prediction of necrotic and vital leaf areas
We used the pixel classification tool of Ilastik (v.1.3.3post3) (Berg et al., 2019) with all 13 features for colour/intensity, edge, texture of sigma 0.3, 0.7 and 10.00 selected. We defined three different pixel classes: ‘background’, ‘necrotic’ and ‘vital’. We performed an iterative manual training to gradually improve the accuracy of the prediction and finally used 24 images of plants at different stages of the experiment to train the model. For the disc assay, we combined one to three leaf discs that were punched from vital and necrotic leaves onto soil of a single pot that did not contain a plant. We took three photographs of each combination. To cover a broad spectrum of possibilities, leaf discs were shuffled and/or moved on the pot between pictures. Therefore some discs were photographed multiple times but in different combinations and/or positions. Similar to the model training with living and necrotic tissues, we used five images to train ilastik to detect background and the white labels that served as a scale to normalise pixel counts between different days of the experiment. We then processed and exported all image files in ilastik with the following settings: source, ‘simple segmentation’; convert to datatype, ‘floating 32 bit’; format, ‘tif’, and used the ‘getHistogram’ function of ImageJ (v.1.53g, Java 1.8) (Schneider et al., 2012) to extract the pixel counts of the areas predicted by ilastik. Pixel counts of vital and necrotic leaves were then normalised using the predicted areas of the size references for each time point of the drought experiment using R.
SPAD value and C : N ratio
We grew S4 generation plants under the same conditions and watering regime as for the drought experiment and used a chlorophyll meter SPAD‐502 chlorophyll meter (Ling et al., 2011) to determine the Soil Plant Analysis Development (SPAD) values of three or six leaves of six wt and hcLine31 plants that were grown as triplicates in two independent experiments. SPAD values were measured before the occurrence of necrotic leaves 15 d after watering was suspended.
To determine the carbon/nitrogen ratios of the wt and hcLine31 we grew S3 generation plants under well watered conditions on Einheitserde under short day conditions in a Sanyo MLR‐350 growth chamber with an 8 h : 16 h, 20°C : 18°C, day : night cycle for 14 wk. We sampled one leaf from each of four plants of the wt and hcLine31, inactivated them for 30 s in a microwave and dried them for 8 d at 60°C. Plant material was then ground for 3 min at 30 Hz with an oscillating mill (MM400; Retsch GmbH, Haan, Germany). Then, 2 mg of plant material were put in tin capsule and C : N ratios were analysed with a thermal conductivity detector by the Basel Stable Isotope Laboratory.
RPI2 analysis in natural accessions
We used the vcf file produced by The 1001 Genomes Consortium (Alonso‐Blanco et al., 2016) to extract SNPs for 1135 sequenced accessions of A. thaliana. To limit the effect of the phylogenetic relationship in further analyses, we used the function ‘–relatedness2’ from vcftools (Danecek et al., 2011) to keep only ecotypes with a kinship coefficient k < 0.5. For the remaining ecotypes, bioclimatic variables (https://www.worldclim.org/data/worldclim21.html) and Global Aridity Index (https://cgiarcsi.community/data/global‐aridity‐and‐pet‐database/) were extracted using the R packages raster (v.3.5‐2 (Hijmans & van Etten, 2012)) and rgdal (v.1.5‐27 (Keitt et al., 2010)). We fitted a linear mixed model with the R package lme4 (v.1.1‐27 (Bates et al., 2015)) to test the association between aridity levels averaged over May to August and SNPs in RPI2 harbouring a minor allele frequency (maf) > 0.3. We added the admixture groups defined by The 1001 Genomes Consortium (Alonso‐Blanco et al., 2016) as a random effect to account for population structure. We used Qgis (v.3.16; https://www.qgis.org/en/site/) to display RPI2 alleles in Eurasia. For the rest of the manuscript, all statistical analyses were performed in R and parametric or nonparametric tests were used according to the sample size and data distribution.
Survival probability of ONSEN in RPI2
TIPs in RPI2 were screened in the data published by Baduel et al. (2021). We used forward‐in‐time models to examine the short‐term fate of a TE inserting in RIP2, simulating a population of 100 diploid individuals undergoing drought events. We used the SliM3 simulator (Haller & Messer, 2019) to model a population with a selfing rate of 99% (Richard et al., 1989), and ran the simulations for 50 generations. Assuming a generation time of c. 2 months and 6 months of suitable growth conditions per year, this simulation should cover c. 16 yr of a natural population. The TE insertion only had a fitness effect at the homozygous state (dominance coefficient h = 0) and was initially sampled during the first generation from a single genome (over 200). The selective advantage (s) of individuals carrying the TE insertion at the homozygous state during the drought events was set at +50%, +100%, +200% or +500% relative to the wild‐type. During normal conditions, this coefficient was set at −10%, −20%, −50% or −90% relative to the wild‐type. We applied a model of hard selection, with a population size being a function of the average fitness, with a maximal population size of 100 (carrying capacity). This was included as a way to consider the varying effects of drift in small populations, which may affect the odds for a deleterious allele to rise in frequency. We simulated one to four drought events at regular intervals. Each of these events could last two, three or four generations. We ran 1000 simulations for each combination of parameters. Scripts are freely available at https://github.com/YannBourgeois/Slim‐simu‐TE.
Results
ONSEN hcLines differ in growth
We grew the S4 generation of 23 of the aforementioned hcLines originating from 13 independent heat stressed and transiently de‐methylated Col‐0 plants (please refer to Materials and Methods section and Thieme et al., 2017) and validated by quantitative PCR (qPCR) an up to c. 8 fold copy number increase (therefore up to c. 64 stably inserted ONSEN copies; Fig. 1a) compared with the wt. However, in some hcLines (e.g. hcLine9), no additional ONSEN copies were detectable by qPCR even though some were previously validated by sequencing (Roquis et al., 2021) indicating a higher sensitivity of the TE‐detection pipeline compared with the qPCR.
Fig. 1.
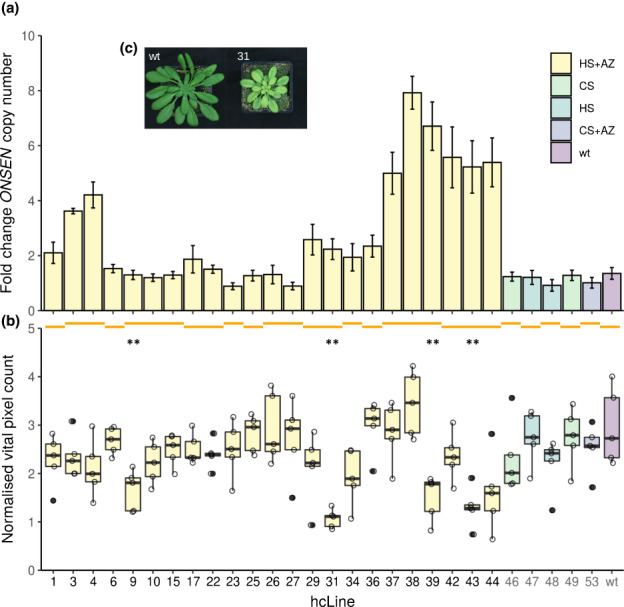
ONSEN copy numbers and size of Arabidopsis thaliana hcLines. Box colours indicate the history of the lines: control stress (CS), heat stress (HS), chemical de‐methylation (AZ) and wild‐type propagated on soil (wt). Orange lines spanning multiple hcLines indicate their origin from a common parent. Control lines and the wt are indicated by grey tick labels. (a) Fold change of ONSEN copy numbers measured by qPCR compared with the wt (n = 3 technical replicates ±SD). (b) Vital pixel count of hcLines and controls before the occurrence of necrotic leaves (day 8). Significant differences to the wt are indicated. Horizontal line defines median, hinges represent 25th and 75th percentiles, whiskers extend to 1.5× IQR and outliers are shown as filled dots. n = 5 biological replicates. Wilcoxon test: **, P < 0.01. (c) Representative pictures of the wt and hcLine31 showing significant differences in size (please refer to (b)) on day 8 of the experiment.
As the hcLines were originally exposed to a combination of heat and the drugs zebularine (Z) and alpha‐amanitin (A) (HS + AZ) (please refer to the Materials and Methods section and Thieme et al., 2017), we harnessed a set of control lines to account for a potential ONSEN‐independent phenotypic variation caused by epi/genetic changes induced by the heat stress or the chemical demethylation. Namely, we included a Col‐0 wt plant that was propagated on soil and five independent controls, that is lines that originated from plants that were also grown in vitro but only exposed to CS (two lines), HS (two lines) or CS plus chemical demethylation (CS + AZ; one line) (Thieme et al., 2017). In accordance with previous observations (Thieme et al., 2017; Roquis et al., 2021), and the strict heat dependence of ONSEN (Ito et al., 2011; Cavrak et al., 2014) we did not detect an increase in genomic ONSEN copy numbers in these control lines when compared with the wt (Fig. 1a).
We suspended watering after 36 d of growth and recorded plant development and water loss of the pots every week by taking top‐view pictures and by weighting the pots. To quantify the growth and the degree of drought‐induced leaf senescence, we trained the image‐based interactive learning and segmentation toolkit (ilastik) (Berg et al., 2019) to specifically detect living (from this point forwards vital) and necrotic leaf segments. We first tested the accuracy of the prediction by placing one to three punched leaf discs of necrotic or vital segments onto a pot that did not contain a plant. After processing the images with ilastik, we obtained a linear increase of vital and necrotic pixel counts according to the number of segments placed onto the pots (Fig. S1), confirming the reliability of the method. For the rest of the study, we therefore used vital pixel counts as a proxy for plant size. To assess size variations between the hcLines, we analysed the pixel counts of predicted vital areas before the appearance of necrotic leaves 8 d after watering was suspended (Figs 1b, 2a). Although heat stress and the AZ drug treatment have been shown to induce epi/genetic mutations (Liu et al., 2015; Belfield et al., 2021; Roquis et al., 2021), we did not observe differences in growth among our five control lines. However, in accordance with the fact that transposition is predominantly associated with fitness loss of the host (Wilke & Adams, 1992; Boissinot et al., 2006; Chuong et al., 2017; Roquis et al., 2021), we found a significantly reduced number of vital pixels compared with the wt for four hcLines (Wilcoxon test P < 0.01; Fig. 1b,c).
Fig. 2.
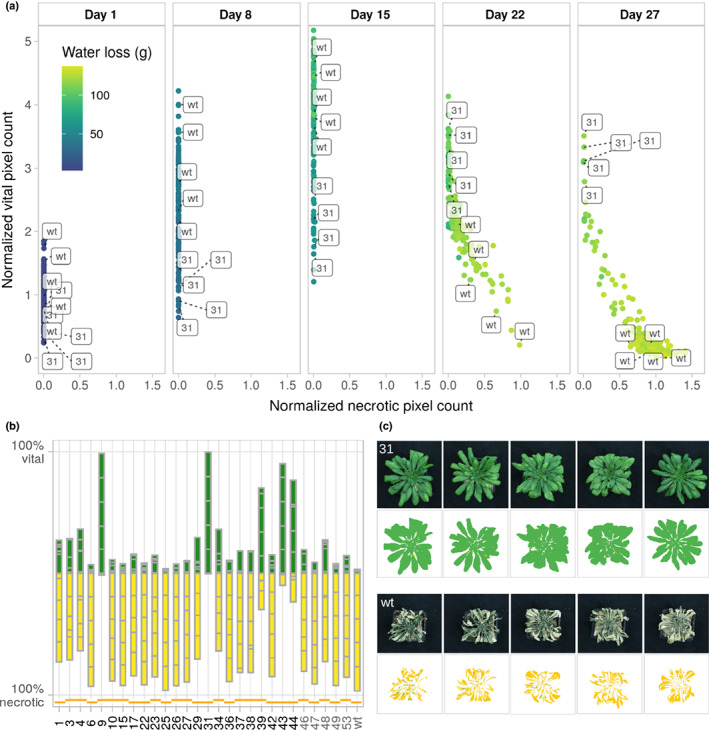
Drought tolerance of Arabidopsis thaliana ONSEN hcLines. (a) Pixel counts of living and necrotic tissues of the six control and 23 ONSEN high‐copy lines a during the drought stress. Each dot represents one individual plant and the colour code indicates the cumulative water loss over time. n = 3–5 biological replicates per line are shown. (b) Percentage of vital (green) and necrotic (yellow) tissues of all five replicates per line 2 d after recovery (day 29). The contribution of each replicate to the total amount of vital and necrotic pixels is indicated with grey bars. Orange lines spanning multiple hcLines indicate their origin from a common parent. Control lines and the wt are indicated by grey tick labels. (c) Original (upper) and processed (ilastik, lower) images of the five replicates of hcLine31 (31) (upper) and wt (lower) on day 29. Predicted vital leaf segments are depicted in green, necrotic segments in yellow.
Notably, in some cases we also observed a strong size variation between hcLines originating from the same parent (e.g. hcLine37 and hcLine38 vs hcLine39) indicating an expected (Matsunaga et al., 2015) and previously reported (Roquis et al., 2021) genetic segregation of the lines (Fig. 1b). We did not find any significant global correlation between ONSEN copy numbers and plant size (P > 0.05, Fig. S2). Taken together, these observations suggest that single TE insertions rather than the overall ONSEN load were responsible for the observed phenotypic variation.
An ONSEN ‐mediated loss‐of‐function mutation leads to an increased drought tolerance of hcLine31
The machine learning approach allowed us to further track the dynamics of growth and drought‐induced necrosis of the individual plants. None of the plants had necrotic leaves until 15 d after watering was suspended, (Fig. 2a). After 27 d, we resumed watering and recorded leaf areas after 2 d of recovery (day 29). In contrast with the high drought‐induced mortality of the five control lines and the wt, five out of the 23 hcLines (hcLine9, 31, 39, 43 and 44) originating from four independent parental lines were more stress tolerant (mean vital leaf area > 50%) (Fig. 2b). Although the water limitation was lethal for the wt (0.03% mean vital area), hcLine31 did not show any signs of necrosis (97.9% mean vital area, Fig. 2a–c). Because hcLine31 displayed the most drought‐tolerant phenotype (Fig. 2b,c), we selected it to characterise the functional link between the heat‐induced insertion of novel ONSEN copies and the observed increase in drought tolerance. Note that because wt plants and many hcLines died from drought stress before the end of the experiment, no other fitness‐related traits (e.g. seed production) were assessed.
We then harnessed whole‐genome re‐sequencing data that had been previously used to locate all transposon‐insertion polymorphisms (TIPs) of hcLine31 (Roquis et al., 2021). In accordance with its insertion bias towards actively transcribed regions in the A. thaliana genome (Quadrana et al., 2019; Roquis et al., 2021), six out of the 10 ONSEN TIPs detected in hcLine31 were located in exons (Table 1). The consistent vitality of all five replicates of hcLine31 indicated that a homozygous mutation was underlying the observed drought tolerance of hcLine31. We also speculated that the causal insertion was located in the exon of a gene. Therefore, we considered only three (in At1G58602, At2G01290 and At5G03435) out of the 10 novel ONSEN copies as candidate insertions. Based on our phenotypic observation, we selected the insertion in the ribose‐5‐phosphate‐isomerase 2 (RPI2, At2G01290) as our top candidate, as RPI2 is involved in chloroplast photosynthetic capacity (Xiong et al., 2009). The link between drought stress on plant photosynthesis is indeed well documented (e.g. Reddy et al., 2004) and we therefore tested experimentally whether the mutation of RPI2 was underlying the high survival rate of hcLine31.
Table 1.
Novel ONSEN insertions in Arabidopsis thaliana hcLine31.
| chr | Coordinates | Context | ID | Description | Zygosity |
|---|---|---|---|---|---|
| 1 | 21761950–55 | Exon | AT1G58602 | LRR and NB‐ARC domains‐containing disease resistance protein | (−/−) |
| 2 | 149387–91 | Exon | AT2G01290 | Cytosolic ribose‐5‐phosphate isomerase | (−/−) |
| 3 | 19300993–97 | Exon | AT3G52020 | Serine carboxypeptidase‐like 39 | (+/−) |
| 3 | 22631755–59 | Exon | AT3G61150 | Homeodomain GLABROUS 1; HD‐ZIP IV family | (+/−) |
| 5 | 853776–80 | Exon | AT5G03435 | Ca2+‐dependent plant phosphoribosyltransferase family protein | (−/−) |
| 5 | 10632816–20 | TE | AT5G28626 | AT5TE38720; SADHU; Sadhu noncoding retroTE family | (−/−) |
| 5 | 18850327–31 | Promoter | AT5G46490 | Disease resistance protein (TIR‐NBS‐LRR class) family | (−/−) |
| 5 | 21050239–43 | Intron | AT5G51800 | Protein kinase superfamily protein | (−/−) |
| 5 | 21602030–34 | Exon | AT5G53240 | Hypothetical protein (DUF295) | (+/−) |
| 5 | 22846432–36 | Intron | AT5G56400 | FBD, F‐box, Skp2‐like and Leucine Rich Repeat domains‐containing protein | (+/−) |
Location, description (Araport11) and zygosity (−/− homozygous ONSEN, +/− heterozygous) of predicted ONSEN insertion sites extracted from Roquis et al. (2021).
First, we verified the impact of the ONSEN insertion on the expression of RPI2 by analysing previously published RNA‐seq data of hcLine31 (Roquis et al., 2021) and found a premature transcriptional stop coinciding with the detected exonic ONSEN insertion in RPI2 (Fig. 3a). Knowing about the loss‐of‐function mutation of RPI2 in hcLine31, we then crossed hcLine31 to a Col‐0 wt plant (Fig. 3b) and assessed the drought tolerance and genotypes of RPI2 of individual plants in the segregating F2 generation obtained from self‐fertilisation. In accordance with our hypothesis that the loss‐of‐function of RPI2 would lead to an increased drought tolerance of hcLine31, we found that solely F2 individuals carrying a homozygous ONSEN insertion survived the drought stress (Fig. 3c).
To conclusively verify these findings by combining the ONSEN insertion with another independent recessive allele of rpi2, we harnessed the mutant rpi2‐1 (SALK_022117) that carries a homozygous T‐DNA insertion in the exon of RPI2 that also leads to a knock‐out of the gene (Xiong et al., 2009). Accordingly, a cross between hcLine31 and the rpi2‐1 mutant should only lead to drought‐tolerant offspring. Indeed, all F2 individuals of rpi2‐1 x hcLine31 that were either homozygous for the ONSEN or the T‐DNA insertion or that carried both the ONSEN and the T‐DNA‐insertions in RPI2 survived the drought stress and showed continued growth 2 d after recovery (Fig. 3d). These results unequivocally demonstrated that the ONSEN insertion in RPI2, that results in a recessive loss‐of‐function mutation, was responsible for the increased drought tolerance of hcLine31.
Reduced size and water use explains desiccation tolerance of hcLine31
As mentioned above a previous study indicated that the loss of RPI2 leads to chloroplast dysfunction and reduced chlorophyll content (Xiong et al., 2009). To test whether the photosynthesis was indeed affected in hcLine31, we assessed carbon/nitrogen (C : N) ratios and SPAD values, which are directly linked to photosynthetic activity (Ling et al., 2011; Otori et al., 2017), following growth under well watered conditions and before the emergence of necrotic leaves. SPAD and C : N ratios were significantly reduced (Wilcoxon test, P < 0.05) in hcLine31 compared with the wt (Fig. 4a; Table S2). In accordance with (Xiong et al., 2009), this indicated a reduced photosynthetic capacity in hcLine31 before the onset of the drought stress. This raised a question about the mechanistic link between the ONSEN insertion in RPi2 and the high survival rate under water limitation. As noted earlier, compared with the wt, we noticed a reduced size of hcLine31 under well watered conditions (Fig. 1b,c) and that drought‐induced symptoms in hcLine31 occurred significantly later (Fig. 2a). We therefore hypothesised that the hampered photosynthetic capacity resulting in a slower growth and therefore reduced water consumption of hcLine31 underlies the high survival rate of this line. Therefore we looked for a general link between plant size and drought tolerance. We first fitted a linear model in which water loss (before the occurrence of necrotic leaves at day 8 of the experiment) was entered as the response variable and the plant lines, vital pixel count and their interaction as explanatory variables. Although the overall model was significant and explained a large part of the variance (R 2 = 0.72, P < 2.2e−16), we did not detect a significant contribution of the plant line nor of the interaction between the line and vital pixel count, indicating that all hcLines, control lines and the wt had a similar efficiency regarding water use. However, we found that vital pixel count had a significant effect on water loss (P = 5.19e−11). This was further confirmed by a reduced model in which the line and interaction effects were dropped (Fig. 4b; R 2 0.71, P < 0.001). Therefore, these analyses indeed suggested that the ONSEN insertion in RPI2 resulted in reduced photosynthetic capacity leading to slower growth and a reduced water consumption, allowing hcLine31 to escape severe drought stress.
Fig. 4.
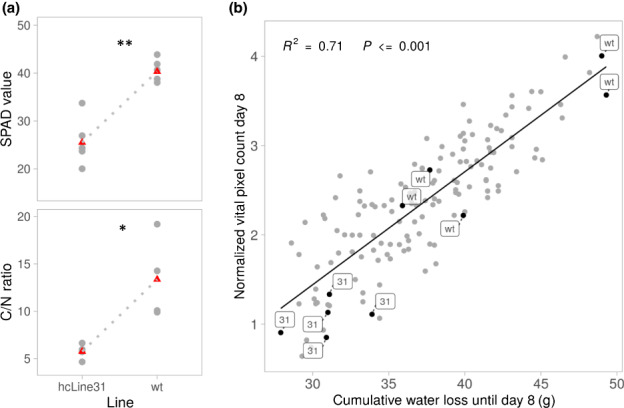
Physiological differences between Arabidopsis thaliana hcLine31 and wt plants. (a) SPAD meter values (upper) and C : N ratio (lower) of the two lines. n = 4–6 biological replicates. Means are marked with red triangles, Wilcoxon test: *, P < 0.05; **P < 0.01. (b) Relationship between pixel counts of living tissues and cumulative water loss until day 8 before the occurrence of necrotic leaves. Linear regression model, adjusted R‐squared: 0.71. P < 0.001.
Based on these observations we also tested whether RPI2 could play a more general quantitative role in the adaptation to arid climate in A. thaliana natural populations. We therefore further tested the link between RPI2 and tolerance to aridity by extracting the Global Aridity Index in summer (GAIsummer; Global Aridity Index averaged over April to August) for each locality of 596 genetically distinct A. thaliana ecotypes (kinship coefficient < 0.5 and no missing data for subsequent analyses) from the 1001 genome project (Alonso‐Blanco et al., 2016; Table S3).
We identified 11 SNPs with a maf > 0.3 in RPI2. Although some of these SNPs were found to be in strong linkage disequilibrium (Fig. 5a), one of them located in the regulatory region of RPI2 (chr2 at nt150495) showed a stronger association with GAIsummer (Fig. 5a). Our linear mixed model analysis revealed that this SNP was significantly associated with higher GAIsummer even when accounting for population structure (4.3% of the variance explained; Fig. 5b). Accordingly, we detected a high frequency of this allele in areas with high aridity levels in July (Fig. 5c). In summary, these findings suggest that our candidate ONSEN insertion in hcLine31 boosts the native function of RPI2 with respect to adaptation to drought.
Fig. 5.
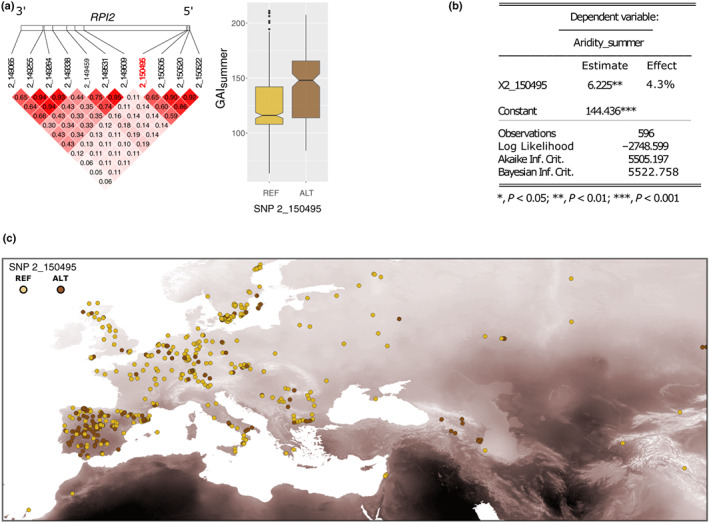
The association between aridity and RPI2 variations in natural populations of Arabidopsis thaliana. (a) Linkage disequilibrium among the 11 SNPs located in RPI2 (left panel) and the association between SNP 2_150495 and Global Aridity Index in summer (GAIsummer; right panel). (b) Linear mixed model results based on 596 accessions. (c) Occurrence of the reference and alternative alleles of SNP 2_150495 in Eurasia. The intensity of the background map displays aridity levels in July from low (light) to high (dark).
Survival probability of ONSEN in RPI2
This type of insertion for ONSEN or other heat‐induced TEs could provide a selective advantage in the face of global warming (Ito et al., 2016; Quadrana et al., 2019; Baduel et al., 2021). However, because the knock‐out of RPI2 leads to a reduced photosynthetic activity, one may expect large‐scale mutations providing such a strong effect on growth under well watered conditions to be purged by natural selection. In fact, we did not find any natural TE insertions in RPI2 (Baduel et al., 2021). To further determine if and under which conditions large‐scale mutations with strong phenotypic effects could reach a high frequency in A. thaliana populations, we simulated different scenarios of temporary drought events (Fig. S3). Four parameters were applied to control the simulations: the number of drought events (one to four), the length of each drought event (two to four generations) and the selective advantage of the insertion during drought (s+) and normal conditions (s−). We used a model of ‘hard’ selection, in which the population size varied as a function of the average fitness. When drought events were rare and short, no TIP was found in the population after 50 generations. However, the frequency of the TIP increased with higher values of s+ and lower values of s−, but required frequent and long episodes of drought events. This confirmed that a mutation with high deleterious effects under optimal growth conditions is hardly maintained over time (Fig. S3). However, it also shows that this mutation reaches a much higher frequency when the population encounters repeated drought events.
Discussion
In the context of climate change, assessing how fast plants will adapt to longer periods of severe heat and drought stress is crucial both for conservation biology and food security (Loarie et al., 2009; Exposito‐Alonso et al., 2019; Brás et al., 2021). Selection from standing variation, as opposed to the emergence of new mutations (Hermisson & Pennings, 2017), is classically expected to lead to faster evolution (Barrett & Schluter, 2008). TEs however, may challenge this prediction due to the combined effect of their stress‐inducible activity and large mutagenic properties (Baduel et al., 2021). Here, we experimentally confirmed with a real‐time setup that a single novel insertion of ONSEN resulting in a loss‐of‐function of RPI2, can indeed rapidly lead to a selective advantage upon drought. More specifically, we document a case in which a decrease of the photosynthetic capacity under well watered conditions, by reducing plant growth and water consumption, leads to an increased survival rate under water limitation. Reducing evapotranspiration and growth are known evolutionary strategies of plants to survive drought (Kusaka et al., 2005; Borrell et al., 2014) and believed to be adaptive in ever‐drier environments (Rauschkolb et al., 2022). Accordingly, variations in the growth scaling of natural accessions of A. thaliana have been shown to be linked to abiotic parameters such as temperature and precipitation (Vasseur et al., 2018). Here, we also show that RPI2, the gene whose loss‐of‐function is causative for the distinct drought tolerance of hcLine31, is quantitatively associated with the aridity level in wild A. thaliana population.
In A. thaliana, several studies have established a functional link between loss‐of‐function alleles and adaptive traits (e.g. Johanson et al., 2000; Kroymann et al., 2003; Gujas et al., 2012) also in the context of adaptation to drought (Monroe et al., 2018; Xu et al., 2019). Together with this large body of evidence, our study substantiates the ‘less is more’ hypothesis (Olson, 1999) that is, that in contrast with intuitive expectations gene loss may fuel evolution. Indeed, in contrast with the wt, all five individuals of hcLine31 survived the severe drought stress applied in our experiment, indicating a drastic gain of fitness under these conditions. Conversely, the reduced growth of hcLine31 underlying the increased drought tolerance may be linked to a high fitness cost under more humid conditions. As our terminal drought experiment resulted in the premature death of most plants, we were not able to assess fitness‐related traits such as biomass or seed production and instead simulated under which conditions an insertion causing such a strong phenotype could be maintained in a population. We found that a severe and repeated water limitation would be needed and that such distinct, TE‐mediated adaptive effects may therefore be transient and thus difficult to capture with population genomics data. This may explain why no natural insertions of ONSEN were observed in RPI2. Conversely, positively selected loss‐of‐function mutations leading to a reduced plant size have been reported previously in A. thaliana (Barboza et al., 2013) and the absence of natural TE insertions in RPI2 could also indicate that this transposition event did not take place in the wild or only occurred in marginal populations not yet sampled. Indeed, TE‐mediated mechanisms leading to large‐effect mutations might be especially important in less‐adapted, frequently stressed populations in which drastically altered phenotypes could be advantageous. In agreement with this hypothesis, Baduel et al. (2021) recently pointed towards a link between positive selection of weak alleles of the largest subunit of RNA polymerase V, a key component of RdDM, and a globally relaxed silencing of TEs in A. thaliana ecotypes growing under extreme conditions.
We only performed the in‐depth and labour‐intensive molecular characterisation for one of our hcLines. Therefore, whether the observed high frequency of five independent drought‐tolerant hcLines could be explained by the insertion preference of ONSEN towards exons and H2A.Z enriched regions that are shown to be associated with responsive genes (Coleman‐Derr & Zilberman, 2012; Quadrana et al., 2019; Roquis et al., 2021), remains speculative. Because the insertion preference of ONSEN has also been demonstrated in a population of epiRILs (Johannes et al., 2009; Quadrana et al., 2019), therefore by a different approach than the transient drug treatment (Thieme et al., 2017), we conclude that our findings using hcLines in the Col‐0 background are solid and highly relevant for the understanding the role of ONSEN in natural accessions. Although at a lower frequency, ONSEN indeed transposes in the wild (Masuda et al., 2016; Quadrana et al., 2016). Although we did not observe major phenotypic changes among the six included control lines that were only heat stressed or demethylated we can not rule out that stochastic small‐scale mutations (Liu et al., 2015) or reported epialleles (Roquis et al., 2021) caused by the drug treatment have contributed to the observed high frequency of drought‐tolerant hcLines. In this regard, other exogenous TEs such as the tobacco Tnt1 retrotransposon (Grandbastien et al., 1989) that is used as a mutagen in plants including Arabidopsis (Lucas et al., 1995) and known to efficiently knock‐out genes could serve as a control in experiments systemically addressing the specific capacity of ONSEN to generate drought‐tolerant individuals. Whereas the comparison of the evolutionary potential of individual TE families requires a larger number of observations, our current study also provides the foundation for selection experiments with populations of hcLines whose TE‐composition has not however been shaped by natural selection. These types of experiments will allow us to extrapolate the overall gain or loss of plant fitness following a heat‐induced transposition of an endogenous TE.
In conclusion our study substantiates that stress‐induced TE mobility can rapidly lead to an increased stress tolerance of the host as formerly suggested by population genomics data (Quadrana et al., 2016). Because TE activity is family dependent and can be triggered by various abiotic and biotic stresses (for review Negi et al., 2016), these findings also support the hypothesis that TEs are more likely to rapidly modulate gene expression and traits than classical point mutations (Uzunović et al., 2019). In line with models of rapid adaptation through large mutation effects (Orr, 2005) and more recent works in A. thaliana (e.g. Fulgione et al., 2022) our study demonstrates that TEs can provide a powerful means by which plants can keep pace with the rapidly changing environmental conditions.
Competing interests
None declared.
Author contributions
Conceptualisation, MT; data acquisition, MT, BK, AB; data analysis, MT, ACR, YB; funding acquisition, MT, EB, ACR; writing – original draft, MT; writing – review and editing, MT, ACR, EB, AB, BK, YB.
Supporting information
Fig. S1 Leaf disc assay to validate the accuracy of the pixel counts for the machine learning‐based prediction using ilastik.
Fig. S2 Pixel count of vital tissues in all tested lines before the occurrence of necrotic leaves and the fold change of ONSEN copy numbers measured using qPCR. Linear regression model.
Fig. S3 Simulated frequency of a TE insertion in a population of Arabidopsis thaliana exposed to drought stress.
Table S1 Names and sequences of oligos used for the qPCR to determine ONSEN copy numbers and for genotyping of RPI2.
Table S2 Raw data of pixel counts and weight, SPAD values and C : N‐measurements.
Table S3 SNPs in RIP2 and bioclimatic variables for the subset of 596 ecotypes.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We would like to thank Anja Schmutz for her assistance with R, Mahendra Mariadassou for his kind support with statistics. We further want to thank Ansgar Kahmen and Thomas Boller for their involvement at the beginning of the project. Finally, we want to thank the editor for handling our manuscript and Christoph Stritt and the three reviewers for their valuable comments on our work. This work was supported by University of Zürich Research Priority Programs (URPP) Evolution in Action (to MT, BK, ACR), European Commission PITN‐GA‐2013‐608422‐IDP BRIDGES (to MT), Freiwillige Akademische Gesellschaft Basel (to MT), SNSF 31003A_182785 (to ACR, BK) and European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program 725701, BUNGEE (to EB). Open access funding provided by University of Zurich. Open access funding provided by Universitat Zurich.
Data availability
Raw and segmented images and ilastik classifications were uploaded to Figshare (doi: https://doi.org/10.6084/m9.figshare.17159231). Seeds are available upon request to michael.thieme@botinst.uzh.ch.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R et al. 2003. Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Alonso‐Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KMM, Cao J, Chae E, Dezwaan TMM, Ding W et al. 2016. 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana . Cell 166: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baduel P, Leduque B, Ignace A, Gy I, Gil J, Loudet O, Colot V, Quadrana L. 2021. Genetic and environmental modulation of transposition shapes the evolutionary potential of Arabidopsis thaliana . Genome Biology 22: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza L, Effgen S, Alonso‐Blanco C, Kooke R, Keurentjes JJB, Koornneef M, Alcázar R. 2013. Arabidopsis semidwarfs evolved from independent mutations in GA20ox1, ortholog to green revolution dwarf alleles in rice and barley. Proceedings of the National Academy of Sciences, USA 110: 15818–15823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RDH, Schluter D. 2008. Adaptation from standing genetic variation. Trends in Ecology & Evolution 23: 38–44. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker BM, Walker SC. 2015. Fitting linear mixed‐effects models using lme4. Journal of Statistical Software 67: 1–48. [Google Scholar]
- Baubec T, Pecinka A, Rozhon W, Mittelsten SO. 2009. Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. The Plant Journal 57: 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfield EJ, Brown C, Ding ZJ, Chapman L, Luo M, Hinde E, van Es SW, Johnson S, Ning Y, Zheng SJ et al. 2021. Thermal stress accelerates Arabidopsis thaliana mutation rate. Genome Research 31: 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit M, Drost HG, Catoni M, Gouil Q, Lopez‐Gomollon S, Baulcombe D, Paszkowski J. 2019. Environmental and epigenetic regulation of Rider retrotransposons in tomato. PLoS Genetics 15: e1008370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E. 2015. The arabidopsis information resource: making and mining the ‘gold standard’ annotated reference plant genome. Genesis 53: 474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M et al. 2019. ilastik: interactive machine learning for (bio)image analysis. Nature Methods 16: 1226–1232. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. 2006. Fitness cost of LINE‐1 (L1) activity in humans. Proceedings of the National Academy of Sciences, USA 103: 9590–9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell AK, Mullet JE, George‐Jaeggli B, van Oosterom EJ, Hammer GL, Klein PE, Jordan DR. 2014. Drought adaptation of stay‐green sorghum is associated with canopy development, leaf anatomy, root growth, and water uptake. Journal of Experimental Botany 65: 6251–6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brás TA, Seixas J, Carvalhais N, Jagermeyr J. 2021. Severity of drought and heatwave crop losses tripled over the last five decades in Europe. Environmental Research Letters 16: 65012. [Google Scholar]
- Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato‐Recupero G, Martin C. 2012. Retrotransposons control fruit‐specific, cold‐dependent accumulation of anthocyanins in blood oranges. Plant Cell 24: 1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Maintainer BP. 2015. txdb.athaliana.biomart.plantsmart28: annotation package for TxDb object(s) . R package v.3.2.2. [WWW document] URL https://bioconductor.org/packages/release/data/annotation/html/TxDb.Athaliana.BioMart.plantsmart28.html [accessed 24 March 2021].
- Cavrak VV, Lettner N, Jamge S, Kosarewicz A, Bayer LM, Mittelsten SO. 2014. How a retrotransposon exploits the plant's heat stress response for its activation. PLoS Genetics 10: e1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nature Reviews. Genetics 18: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman‐Derr D, Zilberman D. 2012. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genetics 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST et al. 2011. The variant call format and VCFtools . Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. Star: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Lee M, Ham C, Levin HL. 2019. Transposable element insertions in fission yeast drive adaptation to environmental stress. Genome Research 29: 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito‐Alonso M, Gómez Rodríguez R, Barragán C, Capovilla G, Chae E, Devos J, Dogan ES, Friedemann C, Gross C, Lang P et al. 2019. Natural selection on the Arabidopsis thaliana genome in present and future climates. Nature 573: 126–129. [DOI] [PubMed] [Google Scholar]
- Fulgione A, Neto C, Elfarargi AF, Tergemina E, Ansari S, Göktay M, Dinis H, Döring N, Flood PJ, Rodriguez‐Pacheco S et al. 2022. Parallel reduction in flowering time from de novo mutations enable evolutionary rescue in colonizing lineages. Nature Communications 13: 1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J, Karasov TL, Messer PW, Petrov DA. 2010. Genome‐wide patterns of adaptation to temperate environments associated with transposable elements in Drosophila . PLoS Genetics 6: e1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien MA. 2015. LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochimica et Biophysica Acta 1849: 403–416. [DOI] [PubMed] [Google Scholar]
- Grandbastien MA, Audeon C, Bonnivard E, Casacuberta JM, Chalhoub B, Costa APP, Le QH, Melayah D, Petit M, Poncet C et al. 2005. Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenetic and Genome Research 110: 229–241. [DOI] [PubMed] [Google Scholar]
- Grandbastien M‐A, Spielmann A, Caboche M. 1989. Tnt1, a mobile retroviral‐like transposable element of tobacco isolated by plant cell genetics. Nature 337: 376–380. [DOI] [PubMed] [Google Scholar]
- Gujas B, Alonso‐Blanco C, Hardtke CS. 2012. Natural Arabidopsis brx loss‐of‐function alleles confer root adaptation to acidic soil. Current Biology 22: 1962–1968. [DOI] [PubMed] [Google Scholar]
- Hahne F, Ivanek R. 2016. Visualizing genomic data using Gviz and Bioconductor . In: Mathé E, Davis S, eds. Statistical genomics. Methods in molecular biology, vol. 1418. New York, NY, USA: Humana Press, 335–351. [DOI] [PubMed] [Google Scholar]
- Haller BC, Messer PW. 2019. SLiM 3: forward genetic simulations beyond the Wright–Fisher model. Molecular Biology and Evolution 36: 632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Takehira K, Nozawa K, Suzuki T, Masuta Y, Kato A, Ito H. 2020. ONSEN shows different transposition activities in RdDM pathway Mutants. Genes & Genetic Systems 95: 183–190. [DOI] [PubMed] [Google Scholar]
- He L, Huang H, Bradai M, Zhao C, You Y, Ma J, Zhao L, Lozano‐Durán R, Zhu J‐K. 2022. DNA methylation‐free Arabidopsis reveals crucial roles of DNA methylation in regulating gene expression and development. Nature Communications 13: 1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS. 2017. Soft sweeps and beyond: understanding the patterns and probabilities of selection footprints under rapid adaptation. Methods in Ecology and Evolution 8: 700–716. [Google Scholar]
- Hijmans RJ, van Etten J. 2012. raster: geographic analysis and modeling with raster data . R package v.3.5‐2. [WWW document] URL https://cran.r‐project.org/web/packages/raster/index.html [accessed 15 October 2021].
- Hof AEV, Campagne P, Rigden DJ, Yung CJ, Lingley J, Quail MA, Hall N, Darby AC, Saccheri IJ. 2016. The industrial melanism mutation in British peppered moths is a transposable element. Nature 534: 102–105. [DOI] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J. 2011. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472: 115–119. [DOI] [PubMed] [Google Scholar]
- Ito H, Kim J‐M, Matsunaga W, Saze H, Matsui A, Endo TA, Harukawa Y, Takagi H, Yaegashi H, Masuta Y et al. 2016. A stress‐activated transposon in Arabidopsis induces transgenerational abscisic acid insensitivity. Scientific Reports 6: 23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes F, Porcher E, Teixeira FK, Saliba‐Colombani V, Simon M, Agier N, Bulski A, Albuisson J, Heredia F, Audigier P et al. 2009. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genetics 5: e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347. [DOI] [PubMed] [Google Scholar]
- Keitt TH, Bivand R, Pebesma E, Rowlingson B. 2010. rgdal: bindings for the ‘Geospatial’ data abstraction library . R package v.1.5‐27. [WWW document] URL https://cran.r‐project.org/web/packages/rgdal/index.html [accessed 17 September 2021].
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell‐Olds T. 2003. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proceedings of the National Academy of Sciences, USA 100: 14587–14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaka M, Lalusin AG, Fujimura T. 2005. The maintenance of growth and turgor in pearl millet (Pennisetum glaucum [L.] Leeke) cultivars with different root structures and osmo‐regulation under drought stress. Plant Science 168: 1–14. [Google Scholar]
- Lanciano S, Carpentier MC, Llauro C, Jobet E, Robakowska‐Hyzorek D, Lasserre E, Ghesquiere A, Panaud O, Mirouze M. 2017. Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLoS Genetics 13: e1006630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Gentleman R, Carey V. 2009. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25: 1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z‐W, Hou X‐H, Chen J‐F, Xu Y‐C, Wu Q, González J, Guo Y‐L. 2018. Transposable elements contribute to the adaptation of Arabidopsis thaliana . Genome Biology and Evolution 10: 2140–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Q, Huang W, Jarvis P. 2011. Use of a SPAD‐502 meter to measure leaf chlorophyll concentration in Arabidopsis thaliana . Photosynthesis Research 107: 209–214. [DOI] [PubMed] [Google Scholar]
- Lisch D. 2013. How important are transposons for plant evolution? Nature Reviews. Genetics 14: 49–61. [DOI] [PubMed] [Google Scholar]
- Liu CH, Finke A, Diaz M, Rozhon W, Poppenberger B, Baubec T, Pecinka A. 2015. Repair of DNA damage induced by the cytidine analog zebularine requires ATR and ATM in Arabidopsis . Plant Cell 27: 1788–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loarie SR, Duffy PB, Hamilton H, Asner GP, Field CB, Ackerly DD. 2009. The velocity of climate change. Nature 462: 1052–1055. [DOI] [PubMed] [Google Scholar]
- Lu Z, Cui J, Wang L, Teng N, Zhang S, Lam HM, Zhu Y, Xiao S, Ke W, Lin J et al. 2021. Genome‐wide DNA mutations in Arabidopsis plants after multigenerational exposure to high temperatures. Genome Biology 22: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas H, Feuerbach F, Kunert K, Grandbastien MA, Caboche M. 1995. RNA‐mediated transposition of the tobacco retrotransposon Tnt1 in Arabidopsis thaliana . EMBO Journal 14: 2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross‐Ibarra J, Springer NM. 2015. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genetics 11: e1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Nozawa K, Matsunaga W, Masuta Y, Kawabe A, Kato A, Ito H. 2016. Characterization of a heat‐activated retrotransposon in natural accessions of Arabidopsis thaliana . Genes & Genetic Systems 91: 293–299. [DOI] [PubMed] [Google Scholar]
- Masuta Y, Kawabe A, Nozawa K, Naito K, Kato A, Ito H. 2018. Characterization of a heat‐activated retrotransposon in Vigna angularis . Breeding Science 68: 168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga W, Ohama N, Tanabe N, Masuta Y, Masuda S, Mitani N, Yamaguchi‐Shinozaki K, Ma JF, Kato A, Ito H. 2015. A small RNA mediated regulation of a stress‐activated retrotransposon and the tissue specific transposition during the reproductive period in Arabidopsis . Frontiers in Plant Science 6: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. 2014. RNA‐directed DNA methylation: an epigenetic pathway of increasing complexity. Nature Reviews. Genetics 15: 394–408. [DOI] [PubMed] [Google Scholar]
- McClintock B. 1950. The origin and behavior of mutable loci in maize. Proceedings of the National Academy of Sciences, USA 36: 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1953. Induction of instability at selected loci in maize. Genetics 38: 579–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JG, Powell T, Price N, Mullen JL, Howard A, Evans K, Lovell JT, McKay JK. 2018. Drought adaptation in Arabidopsis thaliana by extensive genetic loss‐of‐function. eLife 7: e41038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR. 2009. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134. [DOI] [PubMed] [Google Scholar]
- Nakagome M, Solovieva E, Takahashi A, Yasue H, Hirochika H, Miyao A. 2014. Transposon Insertion Finder (TIF): a novel program for detection of de novo transpositions of transposable elements. BMC Bioinformatics 15: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi P, Rai AN, Suprasanna P. 2016. Moving through the stressed genome: emerging regulatory roles for transposons in plant stress response. Frontiers in Plant Science 7: 1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MV. 1999. Molecular evolution '99 when less is more: gene loss as an engine of evolutionary change. American Journal of Human Genetics 64: 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. 2005. The genetic theory of adaptation: a brief history. Nature Reviews. Genetics 6: 119–127. [DOI] [PubMed] [Google Scholar]
- Otori K, Tanabe N, Maruyama T, Sato S, Yanagisawa S, Tamoi M, Shigeoka S. 2017. Enhanced photosynthetic capacity increases nitrogen metabolism through the coordinated regulation of carbon and nitrogen assimilation in Arabidopsis thaliana . Journal of Plant Research 130: 909–927. [DOI] [PubMed] [Google Scholar]
- Picault N, Chaparro C, Piegu B, Stenger W, Formey D, Llauro C, Descombin J, Sabot F, Lasserre E, Meynard D et al. 2009. Identification of an active LTR retrotransposon in rice. The Plant Journal 58: 754–765. [DOI] [PubMed] [Google Scholar]
- Quadrana L, Etcheverry M, Gilly A, Caillieux E, Madoui MA, Guy J, Bortolini Silveira A, Engelen S, Baillet V, Wincker P et al. 2019. Transposition favors the generation of large effect mutations that may facilitate rapid adaption. Nature Communications 10: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Silveira AB, Mayhew GF, Leblanc C, Martienssen RA, Jeddeloh JA, Colot V. 2016. The Arabidopsis thaliana mobilome and its impact at the species level. eLife 5: e15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2020. R: a language and environment for statistical computing . [WWW document] URL https://www.r‐project.org [accessed 4 July 2022].
- Ram H, Soni P, Salvi P, Gandass N, Sharma A, Kaur A, Sharma TR. 2019. Insertional mutagenesis approaches and their use in rice for functional genomics. Plants 8: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauschkolb R, Henres L, Lou C, Godefroid S, Dixon L, Durka W, Bossdorf O, Ensslin A, Scheepens JF. 2022. Historical comparisons show evolutionary changes in drought responses in European plant species after two decades of climate change. Basic and Applied Ecology 58: 26–38. [Google Scholar]
- Reddy AR, Chaitanya KV, Vivekanandan M. 2004. Drought‐induced responses of photosynthesis and antioxidant metabolism in higher plants. Journal of Plant Physiology 161: 1189–1202. [DOI] [PubMed] [Google Scholar]
- Rey O, Danchin E, Mirouze M, Loot C, Blanchet S. 2016. Adaptation to global change: a transposable element‐epigenetics perspective. Trends in Ecology & Evolution 31: 514–526. [DOI] [PubMed] [Google Scholar]
- Richard H, Abbott J, Gomes MF. 1989. Population genetic structure and outcrossing rate of Arabidopsis thaliana . Heredity 62: 411–418. [Google Scholar]
- Roquis D, Robertson M, Yu L, Thieme M, Julkowska M, Bucher E. 2021. Genomic impact of stress‐induced transposable element mobility in Arabidopsis . Nucleic Acids Research 49: 10431–10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team . 2016. RStudio: integrated development environment for R . [WWW document] URL https://www.rstudio.com [accessed 4 July 2022].
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman AH. 2013. Retrotransposon replication in plants. Current Opinion in Virology 3: 604–614. [DOI] [PubMed] [Google Scholar]
- Stitzer MC, Anderson SN, Springer NM, Ross‐Ibarra J. 2021. The genomic ecosystem of transposable elements in maize. PLoS Genetics 17: e1009768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritt C, Gordon SP, Wicker T, Vogel JP, Roulin AC. 2018. Recent activity in expanding populations and purifying selection have shaped transposable element landscapes across natural accessions of the mediterranean grass Brachypodium distachyon . Genome Biology and Evolution 10: 304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme M, Lanciano S, Balzergue S, Daccord N, Mirouze M, Bucher E. 2017. Inhibition of RNA polymerase II allows controlled mobilisation of retrotransposons for plant breeding. Genome Biology 18: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel‐Elmer M, Bucher E, Broger L, Mathieu O, Paszkowski J, Vaillant I. 2010. Stress‐induced activation of heterochromatic transcription. PLoS Genetics 6: e1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamanzehi A, Moran C, Nicholas FW. 1992. P element transposition contributes substantial new variation for a quantitative trait in Drosophila melanogaster . Genetics 131: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunović J, Josephs EB, Stinchcombe JR, Wright SI, Parsch J. 2019. Transposable elements are important contributors to standing variation in gene expression in Capsella Grandiflora . Molecular Biology and Evolution 36: 1734–1745. [DOI] [PubMed] [Google Scholar]
- Van Houwelingen A, Souer E, Spelt K, Kloos D, Mol J, Koes R. 1998. Analysis of flower pigmentation mutants generated by random transposon mutagenesis in Petunia hybrida . The Plant Journal 13: 39–50. [DOI] [PubMed] [Google Scholar]
- Vasseur F, Exposito‐Alonso M, Ayala‐Garay OJ, Wang G, Enquist BJ, Vile D, Violle C, Weigel D. 2018. Adaptive diversification of growth allometry in the plant Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 115: 3416–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser JC, Chen B, Feder ME. 2006. Heat‐shock promoters: targets for evolution by P transposable elements in Drosophila . PLoS Genetics 2: 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JN, Feschotte C. 2020. A field guide to eukaryotic transposable elements. Annual Review of Genetics 54: 539–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Gundlach H, Spannagl M, Uauy C, Borrill P, Ramírez‐González RH, De Oliveira R, Mayer KFX, Paux E, Choulet F. 2018. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biology 19: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua‐Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O et al. 2007. A unified classification system for eukaryotic transposable elements. Nature Reviews. Genetics 8: 973–982. [DOI] [PubMed] [Google Scholar]
- Wilke CM, Adams J. 1992. Fitness effects of Ty transposition in Saccharomyces cerevisiae . Genetics 131: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Defraia C, Williams D, Zhang X, Mou Z. 2009. Deficiency in a cytosolic ribose‐5‐phosphate isomerase causes chloroplast dysfunction, late flowering and premature cell death in Arabidopsis . Physiologia Plantarum 137: 249–263. [DOI] [PubMed] [Google Scholar]
- Xu Y‐C, Niu X‐M, Li X‐X, He W, Chen J‐F, Zou Y‐P, Wu Q, Zhang YE, Busch W, Guo Y‐L. 2019. Adaptation and phenotypic diversification in Arabidopsis through loss‐of‐function mutations in protein‐coding genes. Plant Cell 31: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Leaf disc assay to validate the accuracy of the pixel counts for the machine learning‐based prediction using ilastik.
Fig. S2 Pixel count of vital tissues in all tested lines before the occurrence of necrotic leaves and the fold change of ONSEN copy numbers measured using qPCR. Linear regression model.
Fig. S3 Simulated frequency of a TE insertion in a population of Arabidopsis thaliana exposed to drought stress.
Table S1 Names and sequences of oligos used for the qPCR to determine ONSEN copy numbers and for genotyping of RPI2.
Table S2 Raw data of pixel counts and weight, SPAD values and C : N‐measurements.
Table S3 SNPs in RIP2 and bioclimatic variables for the subset of 596 ecotypes.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
Raw and segmented images and ilastik classifications were uploaded to Figshare (doi: https://doi.org/10.6084/m9.figshare.17159231). Seeds are available upon request to michael.thieme@botinst.uzh.ch.


