Significance
Ketone bodies, intermediates in energy metabolism and signaling, have attracted significant attention due to their role in health and disease. We performed an around-the-clock study on ketone bodies and ketogenesis with mice on different diets. We found that caloric restriction, a dietary intervention that improves metabolism and longevity, induced high-amplitude daily rhythms in blood β-hydroxybutyrate (βOHB). The blood βOHB rhythms resulted from rhythmic ketogenesis in the liver controlled by the interaction between the circadian clock and peroxisome-proliferator-activated-receptor α transcriptional networks. This interaction resulted in transcriptional reprogramming of beta-oxidation and ketogenesis enzymes. We found that the reprogramming is impaired in circadian clock–mutant mice. The circadian clock–gated ketogenesis contributes to the diet impact on health and longevity.
Keywords: aging, metabolism, caloric restriction, circadian rhythms, fatty acid metabolism
Abstract
Ketone bodies are energy-rich metabolites and signaling molecules whose production is mainly regulated by diet. Caloric restriction (CR) is a dietary intervention that improves metabolism and extends longevity across the taxa. We found that CR induced high-amplitude daily rhythms in blood ketone bodies (beta-hydroxybutyrate [βOHB]) that correlated with liver βOHB level. Time-restricted feeding, another periodic fasting–based diet, also led to rhythmic βOHB but with reduced amplitude. CR induced strong circadian rhythms in the expression of fatty acid oxidation and ketogenesis genes in the liver. The transcriptional factor peroxisome-proliferator-activated-receptor α (PPARα) and its transcriptional target hepatokine fibroblast growth factor 21 (FGF21) are primary regulators of ketogenesis. Fgf21 expression and the PPARα transcriptional network became highly rhythmic in the CR liver, which implicated the involvement of the circadian clock. Mechanistically, the circadian clock proteins CLOCK, BMAL1, and cryptochromes (CRYs) interfered with PPARα transcriptional activity. Daily rhythms in the blood βOHB level and in the expression of PPARα target genes were significantly impaired in circadian clock–deficient Cry1,2−/− mice. These data suggest that blood βOHB level is tightly controlled and that the circadian clock is a regulator of diet-induced ketogenesis.
Ketone bodies—3-β-hydroxybutyrate (βOHB), acetoacetate, and acetone—are metabolic intermediates derived from acetyl-coenzyme A (acetyl-CoA) (1, 2). During fasting, ketone bodies produced by the liver circulate in the bloodstream and serve as an energy source for other tissues such as the brain, heart, and skeletal muscles (3, 4). βOHB, the main ketone body in the bloodstream, serves not only as a source for energy production but also as a signaling molecule (5). Ketone bodies in the bloodstream are elevated in response to prolonged fasting and are reduced back upon refeeding (2, 6, 7). Previous studies proposed that ketone bodies may have some positive impact on metabolism and physiology (8, 9). Several diets known to increase blood ketone bodies, such as intermittent fasting or high-fat ketogenic diets, are used as experimental therapy for neurologic disorders (10), diabetes (11), cardiometabolic diseases, and other conditions (12).
Acetyl-CoA is converted to ketone bodies in a chain of reactions known as ketogenesis. Acetyl-CoA for ketone production is generated predominantly in the liver through beta-oxidation of fatty acids in the mitochondria (13, 14). Ketogenesis is regulated by several metabolic signaling pathways such as fibroblast growth factor 21 (FGF21) (15, 16), mechanistic target of rapamycin (mTOR) (17), and the peroxisome-proliferator-activated-receptor α (PPARα) transcriptional network (18, 19). PPARα is a transcriptional factor and a master regulator of fasting metabolism (20). It controls the expression of enzymes involved in fatty acid transport and oxidation as well as ketone biosynthesis. The PPARα transcriptional network is considered a central controller of ketogenesis. Indeed, fasting-induced ketone production is impaired in PPARα-deficient animals (21).
Calorie restriction (CR) is the most studied and reproducible dietary intervention that has multiple metabolic benefits and extends lifespan in many organisms (22–24). Little is known regarding the control of ketogenesis in CR, and the existing literature is not consistent. Increase, decrease, and no change in blood ketone bodies in CR has been reported by different groups (25–27); unfortunately, the time of day when the analysis was performed has not been reported, but it can be one of the contributing factors. CR is a periodic fasting diet, and it is tightly connected with circadian clocks and rhythms (28–32). The circadian clock is an evolutionary conserved biological time-keeping system that synchronizes organisms with the 24-h light/dark cycle (32). The mammalian circadian clock is a hierarchy of oscillators operating in most of the tissues. The peripheral oscillators are synchronized by signals from the central oscillator in the Suprachiasmatic Nucleus SCN of the hypothalamus. A molecular circadian oscillator is organized as several interconnected transcriptional–translational feedback loops formed by the following transcriptional regulators: CLOCK, BMAL1, PERs, CRYs, REV-ERBs, and RORs (33–36). Circadian rhythms play an important role in human health, and clock disruption is associated with multiple diseases, including cardiometabolic diseases and diabetes (37–39). Here we investigated whether CR impacts ketogenesis in mammals and whether the circadian clock contributes to the regulation of ketogenesis. We found that CR induced strong daily rhythms in liver ketogenesis and blood βOHB level. The rhythms were associated with daily rhythms in the PPARα transcriptional network, and they were significantly disrupted in the circadian clock–deficient mice.
Results
CR induced daily rhythms in blood ketone bodies independently from mTORC1 activity rhythms.
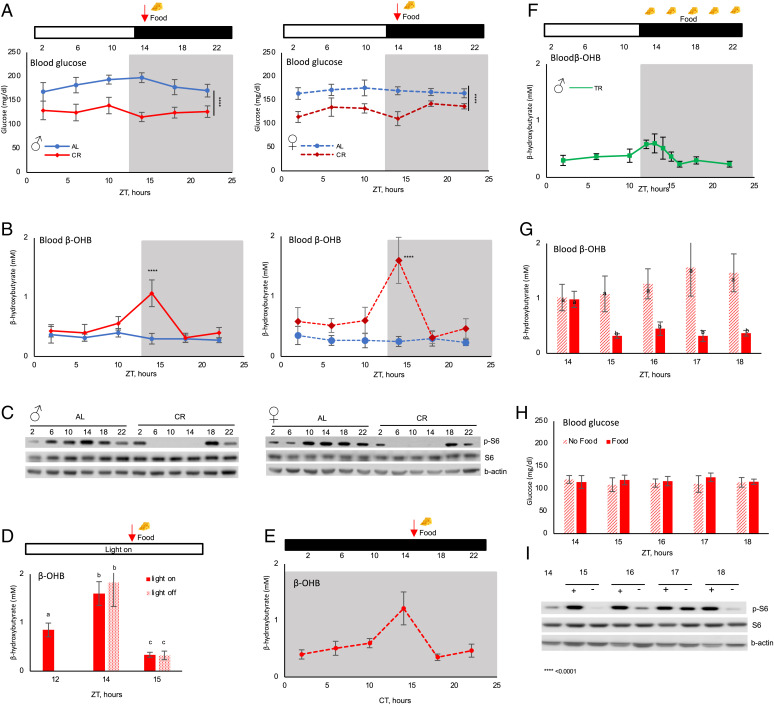
Male and female mice were randomly assigned to ad libitum (AL) and 30% CR groups. CR mice received 70% of their AL food intake as a single meal once per day, 2 h after the light was turned off. Blood glucose and ketone bodies were analyzed around the clock with a 4-h resolution. As expected (40), both male and female CR mice had significantly reduced blood glucose across the day (Fig. 1A). βOHB is a major circulating blood ketone body (1, 2). βOHB was low across the day in AL mice (Fig. 1B). No significant changes were observed even during the light/resting phase, when food consumption is reduced (41). Mice are nocturnal, but with AL access to food, they eat small amounts during the light/rest phase (40), which may be sufficient to prevent ketogenesis. There was no significant difference in blood βOHB between AL males and females (SI Appendix, Fig. 1). CR induced high-amplitude rhythms of blood βOHB (Fig. 1B) in both males and females, with the peak observed at Zeitgeber Time14 (ZT14), just before the feeding time. At the peak, females had significantly higher blood βOHB compared to males (SI Appendix, Fig. 1); therefore, males and females were analyzed separately throughout the study. It was hypothesized that blood ketone bodies and glucose are in reverse correlation (7, 42). However, according to our data, blood glucose and ketone levels can be uncoupled. Indeed, blood glucose was near stable across the day and blood βOHB was highly rhythmic.
Fig. 1.
CR-induced circadian rhythms in blood βOHB. Around-the-clock blood glucose (A) and βOHB (B) in male (Right) and female (Left) mice. Mice were on AL (solid blue line) or 30% CR (dashed red line) diets. The food was provided to CR mice at ZT14 (indicated by arrow). n = 6 per time point per diet. (C) Western blotting for indicated proteins in the liver of AL and CR mice around the clock, pooled samples; n = 3 per diet per time point. (D) Blood βOHB when the light was not turned off at ZT12; n = 8 per time point. (E) Blood βOHB in CR mice that were placed in constant darkness for 48 h; n = 6–8 per time point. (F) Blood βOHB in TRF mice around the clock. The food was provided at ZT14 and removed at ZT2; n = 6 per time point. βOHB (G) and glucose (H) in the blood of CR mice that were provided (solid bars) or not provided (dashed bars) with the food at ZT14. Bars with different letters are statistically different from each other; n = 6. (I) Western blotting for indicated proteins in the liver of CR mice that were provided (+) or not provided (−) with the food; n = 3 per diet per time point. The light was on at ZT0 and off at ZT12. White and black bars represent light and dark. Gray panels on the graphs indicate the dark phase of the day. ZT is Zeitgeber Time, CT is Circadian Time. Significant difference between the diet, ****P < 0.0001, at indicated time point.
The mechanistic target of rapamycin complex 1 (mTORC1), a nutrient sensor and master regulator of metabolism, is implicated in the control of ketogenesis. It has been proposed that fasting-induced suppression of mTORC1 contributes to the increased production of ketone bodies (17). We compared mTORC1 activity in the liver of AL and CR mice across the day (Fig. 1C). mTORC1 activity was rhythmic in AL mice and rhythms were enhanced in CR mice, in agreement with our previous data (43). There was only a partial correlation between mTORC1 activity and blood βOHB rhythms. In AL mice, mTORC1 activity was low during the light phase and high during the dark phase, while the βOHB level was stable across the day. In CR mice, mTORC1 activity was low and βOHB level was high at ZT14 in agreement with the proposed ketogenesis-inhibiting effect of mTORC1 (17). At the same time, mTORC1 activity was low between ZT2 and ZT10, but there was no significant increase in blood βOHB level (Fig. 1 B and C). Thus, the down-regulation of mTORC1 during the light (rest/fasting) phase of the day may be necessary, but it is not sufficient to induce ketogenesis. We concluded that the rhythms in mTORC1 signaling cannot alone explain the blood βOHB rhythms in CR mice.
CR-induced daily rhythms in blood βOHB.
The rapid changes in blood βOHB around ZT14 in CR mice coincided with a time when the light was turned off (ZT12) and food was provided (ZT14). We investigated whether light and/or food play a direct role in blood βOHB rhythms. First, the light was not turned off at ZT12 and the food was still provided at ZT14. Blood ketone bodies were assayed at ZT12, ZT14, and ZT15. Blood βOHB followed the same kinetics independently whether the light was on or off (Fig. 1D): fast increase before the feeding and fast reduction after the feeding. Thus, the kinetics of blood βOHB around feeding time was not driven by the light-to-dark transition in a Pavlovian reflex–like manner. To further investigate βOHB rhythms and light cycle in CR, we placed the mice in constant darkness. The daily meal was still provided at the same time of day. Blood βOHB level was assayed after 48 h in constant darkness. The rhythms in blood βOHB were preserved in constant darkness (Fig. 1E). CR is a combination of a reduction in caloric intake and a periodic feeding/fasting cycle. To address the relative contribution of these factors, the blood βOHB level was assayed in mice that were subjected to time-restricted feeding (TRF), when unlimited food was provided for 12 h. The food was provided at ZT14, at the same time when the food was provided to CR mice, and it was removed at ZT2. AL mice eat the food around the clock, but the majority of the food is consumed during the dark phase of the day and restricting the food availability to the dark phase does not cause misalignment between feeding and light/dark cycles as judged by the clock and metabolic gene expression (40, 44). Previously we reported that TRF mice consumed approximately the same amount of food as AL mice (40, 44); therefore, the mice were not calorie-restricted but periodically fasted for approximately 12 h. TRF resulted in blood βOHB rhythms with the peak at around ZT14 (Fig. 1F). Interestingly, the kinetics were similar for both TRF and CR diets, but the amplitude of the rhythms was significantly smaller in TRF mice compared with CR mice. Thus, both the reduced caloric intake and periodic fasting contributed to the rhythms in blood βOHB.
To test whether the meal was necessary for reduction in blood βOHB in CR mice at ZT18, one group of CR mice was provided with the food and another did not receive the meal at the expected feeding time (Fig. 1G and SI Appendix, Fig. 1). Within 1 h, blood βOHB was rapidly reduced to AL level in fed CR mice, while it was not reduced in unfed CR mice. Thus, food was necessary for the reduction of βOHB in CR mice. Notably, blood βOHB was not increased significantly for approximately 20 h after the meal (Fig. 1B), and it rapidly increased in 4 h just before the meal, between ZT10 and ZT14 (Fig. 1 B and D). It was not further increased in the next few hours even if the mice were not provided with the food (Fig. 1G). Thus, the time-dependent induction of blood βOHB in CR was tightly regulated. Interestingly, there was no difference in blood glucose between fed and unfed CR mice (Fig. 1H), further confirming that blood glucose and βOHB levels are uncoupled. mTORC1 activity in the liver of fed and unfed CR mice is shown in Fig. 1I. As expected, ribosomal protein S6 phosphorylation was induced in 1 h after the meal and it was high in the next few hours. Surprisingly, S6 phosphorylation was also induced in the liver of unfed CR mice, with the maximum in 3 h after the expected feeding time. At the peak, S6 phosphorylation was comparable between fed and unfed states. Thus, the induction of mTORC1 was not sufficient to suppress blood βOHB, which is in agreement with the results from reference (45) and suggests that additional mechanisms must be involved in the regulation of βOHB rhythms.
CR reprograms rhythms in fatty acid oxidation and ketogenesis in the liver.
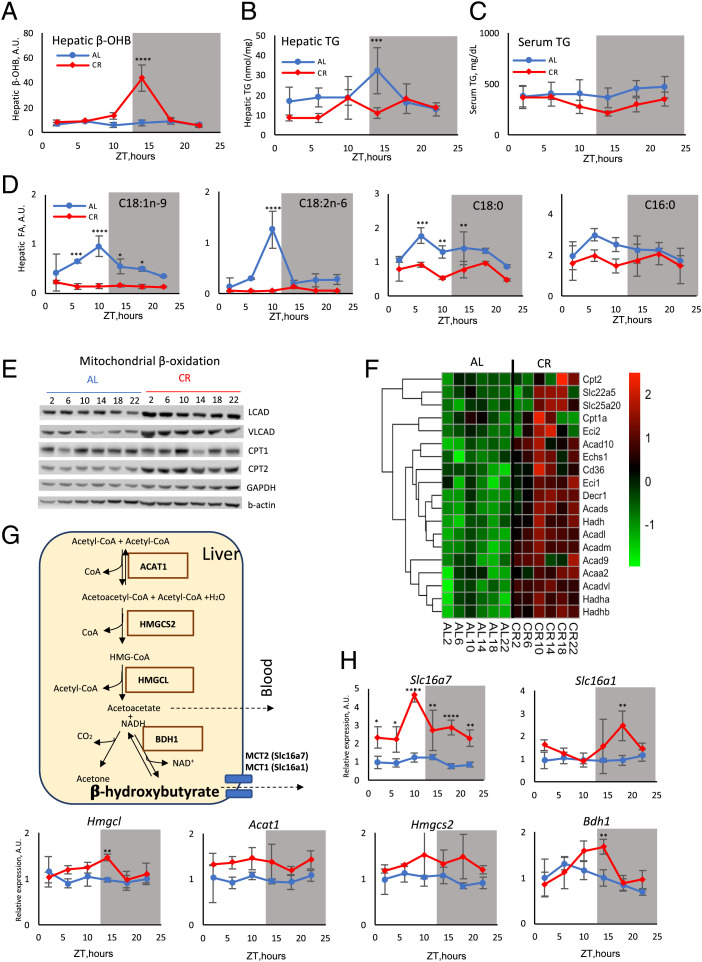
βOHB is produced predominantly by the liver with some contribution from other tissues such as the kidney and intestine (1). Therefore, we hypothesized that CR regulates blood βOHB through the control of its production in the liver. Liver βOHB was assayed in AL and CR mice around the clock (Fig. 2A). βOHB was low across the day in AL mice, and it was increased in CR mice at ZT10 and ZT14 (Fig. 2A) in agreement with our expectation. During fasting the adipose tissue releases fatty acids (46, 47), which are the main source for βOHB production. After the uptake by the liver, these fatty acids can be oxidized in mitochondria or converted back to triglycerides (TG). We assayed serum and hepatic TG around the clock (Fig. 2 B and C) and did not find a significant difference between AL and CR. Next, we assayed the level of individual free fatty acids (FFAs) in the liver. CR caused the reduction of several major FFAs such as C16:0, C18:0, C18:1, and C18:2 at some time points (Fig. 2D). Thus, increased βOHB in the liver did not correlate directly with the levels of TG or FFAs.
Fig. 2.
CR-induced circadian rhythms in liver ketogenesis. βOHB (A), triglycerides in the liver (B), triglycerides in the serum (C), and selected indicated FFAs (D) in the liver of AL (solid blue line) or CR (dashed red line) mice; n = 3–6 per diet per time point. (E) Western blotting for indicated beta-oxidation proteins in the liver of AL and CR mice around the clock; pooled samples, n = 3 per diet per time point. (F) mRNA expression (RNAseq) for indicated beta-oxidation genes in the liver of AL and CR mice around the clock; n = 3 per diet per time point. (G) Scheme of ketogenesis in the liver mitochondria; enzymes are shown in solid boxes. (H) The expression (RNAseq) of ketogenesis genes in the liver of AL (blue line) or CR (red line) mice; n = 3 per diet per time point. The light was on at ZT0 and off at ZT12. Gray panels on the graphs indicate the dark phase of the day. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Significant difference between the diets.
Beta-oxidation is a chain of cyclic reactions when fatty acids are broken down to acetyl-CoA. The expression of enzymes responsible for fatty acids activation, transport, and beta-oxidation was significantly induced in CR mice dependent on the time of the day (Fig. 2E). Messenger RNA (mRNA) expression of fatty acid oxidation genes was analyzed with RNA sequencing (RNAseq), and the data are presented as a heat map in Fig. 2F. Seven of these genes became highly rhythmic in the CR liver with the peak at ZT10 to ZT14, while only two genes were rhythmic in AL mice (SI Appendix, Table 1). CR-induced changes in the expression of fatty acid catabolism enzymes could contribute to the increased ketogenesis between ZT10 and ZT14.
CR-induced beta-oxidation will result in an increased production of acetyl-CoA (48), but elevated hepatic acetyl-CoA alone cannot determine the rate of ketogenesis (1). Four enzymes direct the condensation of acetyl-CoA to βOHB (Fig. 2G). The expression of Acat1 was not significantly affected by CR. The expression of Hmgcl, Hmgcs2, and Bdh1 was significantly up-regulated at ZT10 to ZT14 (Fig. 2H and SI Appendix, Table 3). Hmgcs2 and Bdh1 encode two rate-limiting enzymes in ketogenesis, and the up-regulation of their expression was in good agreement with the increased βOHB production. Several transporters are implicated in the βOHB transport from mitochondria to the cytoplasm and from the cell (1, 49, 50). The expression of the Slc16A7 gene, which encodes the MCT2 protein, was significantly up-regulated in the CR liver across the day with the peak at ZT10. The expression of Slc16A1 (MCT1) and Slc16A6 (MCT7) was only modestly increased at ZT18 or was not affected by CR, respectively (Fig. 2H and SI Appendix, Fig. 2). Thus, CR induced transcriptional reprogramming to synchronize in time the expression of multiple enzymes involved in fatty acid beta-oxidation, ketogenesis, and transport, which may contribute to the rhythms of βOHB production.
Circadian clock proteins interfere with PPARα transcriptional activity.
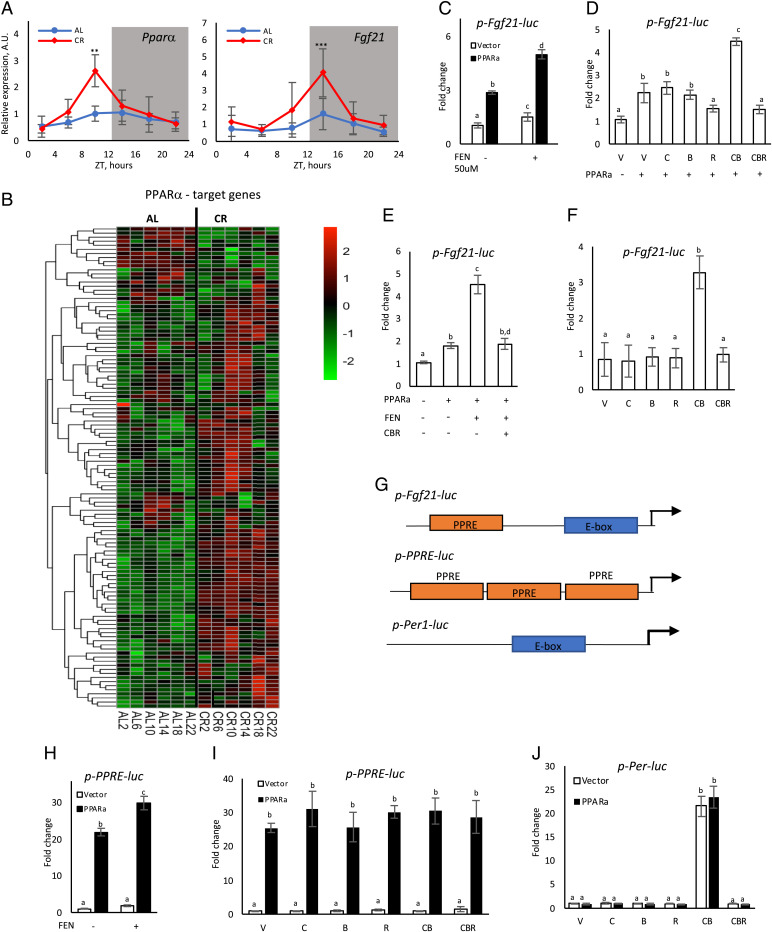
Many of the beta-oxidation and ketogenesis enzymes are direct targets of the PPARα transcriptional factor (19, 51, 52). PPARα plays an important role in the control of ketogenesis during fasting (20, 53–55), and we hypothesized that CR rhythms in ketogenesis were regulated by PPARα. The expression of Pparα mRNA was assayed in the liver of AL and CR mice by RT-PCR. The expression of Pparα was arrhythmic in the AL liver and it became rhythmic in the CR liver with the peak at ZT12, just before the induction of ketogenesis (Fig. 3A, Left, and SI Appendix, Table 2). To investigate whether PPARα transcriptional activity in vivo was impacted by CR we compared the expression of 117 known PPARα target genes (56) using RNAseq (Fig. 3B and SI Appendix, Fig. 3). The expression of 75% of the PPARα targets was significantly affected in the CR liver compared with the AL liver (SI Appendix, Table 3). Rhythmic analysis (SI Appendix, Table 2) revealed that 16 genes were rhythmic in AL mice. Six of these genes lost the rhythms in CR mice, 10 were still rhythmic in CR mice, and 38 genes became rhythmic in CR mice. Thus, 41% of PPARα target genes were rhythmic in CR mice compared to only 14% in AL mice, and approximately 50% of the targets that were rhythmic in CR had peaks at approximately ZT10 to ZT14. One of the PPARα transcriptional targets is FGF21, which is a liver-produced hormone (57). FGF21 is produced during fasting and has been implicated in the control of ketogenesis (16, 58). The expression of Fgf21 mRNA was low and arrhythmic in the liver of AL mice and became rhythmic with the peak at ZT16 in CR mice (Fig. 3A, Right, and SI Appendix, Table 2). Thus, the increased Fgf21 expression correlated with the high level of liver and blood βOHB. These data suggest that CR impacted the daily rhythms of PPARα transcriptional activity.
Fig. 3.
Circadian clock proteins interfere with PPARα transcriptional network. (A) mRNA expression of Pparα (Left) and Fgf21 (Right) in the liver of AL (blue) and CR (red) mice around the clock; n = 3–6 per diet per time point. (B) Heat map expression (RNAseq) of PPARα known target genes on AL and CR diets; n = 3 per diet per time point. (C–F, H–J) HEK293 cells were transiently transfected with combinations of luciferase reporter and transcriptional factors expressing plasmids. The experiments were done in triplicate. Bars with different letters are significantly different from each other. (C) PPARα induced the activity of the Fgf21-luc promoter reporter. (D) The Fgf21-luc reporter was coexpressed with PPARα and plasmids expressing the following proteins: V – vector, C – CLOCK, B – BMAL1, and R – CRY1. (E) Cells transfected with indicated plasmids were treated with fenofibrate (FEN). (F) Fgf21-luc reporter cotransfected with plasmids expressing indicated circadian clock proteins. (G) Scheme of the promoter regions in the indicated reporter plasmids. PPRE, PPARα responsive element; E-box, circadian E-box element. (H) PPRE-luc was cotransfected with PPARα and treated with FEN. (I) The PPRE-luc reporter was cotransfected with indicated plasmids and without (open bars) or with (black bars) PPARα. (J) Cells were transfected with the Per-luc reporter and indicated plasmids without (open bars) or with (black bars) of PPARα.
Robust 24-h rhythms in the expression of PPARα target genes induced by CR led us to the hypothesis that the circadian clock, which is involved in CR mechanisms (59, 60), might contribute to the rhythms in ketogenesis by interacting with the PPARα transcriptional network in the liver. To test this hypothesis, we investigated whether clock proteins interfere with PPARα transcriptional activity. The Fgf21pro1-luc reporter expresses the luciferase gene under control of the Fgf21 promoter (61). Human Embryonic Kiney (HEK293) cells were cotransfected with Fgf21pro1-luc and the PPARα-expressing plasmid. As expected, the coexpression of PPARα strongly induced the Fgf21 promoter (Fig. 3C). Treatment of the cells with fenofibrate, a known agonist of PPARα, further increased the activity of the Fgf21 promoter (Fig. 3C). The cotransfection of plasmids expressing circadian clock proteins impacted PPARα-dependent regulation of the Fgf21 promoter (Fig. 3D). The transfection of CLOCK or BMAL1 expressing plasmids had no significant effect on the PPARα-dependent induction of the Fgf21 promoter. CLOCK and BMAL1 formed a transcriptional hetero-complex and, when BMAL1 and CLOCK were coexpressed together with PPARα, the promoter was additionally induced. The coexpression of CRY1 suppressed the promoter induction. Finally, the coexpression of all three circadian proteins together inhibited the PPARα-dependent induction of the Fgf21 promoter. Similar results were obtained when the cells were treated with the PPARα agonist (Fig. 3E). Thus, circadian clock proteins interfered with both basal and agonist-dependent transcription of the Fgf21 promoter. We analyzed the Fgf21 promoter for circadian E-box elements and identified one circadian E-box element located at approximately 50 bps from the predicted start site for transcription (Fig. 3G). We tested whether CLOCK, BMAL1, and CRY1 can directly regulate transcription from the Fgf21 promoter. The coexpression of CLOCK and BMAL1 resulted in a strong induction of the Fgf21 promoter, and the coexpression of CRY1 inhibited it (Fig. 3F). Thus, the Fgf21 promoter was regulated by circadian clock transcriptional factors even if exogenous PPARα was not expressed, which argues for possible promoter interference mechanisms.
Circadian clock proteins may interfere with PPARα-dependent transcription on the Fgf21 promoter through several different mechanisms. They can directly or indirectly bind to PPARα and modulate its activity. Indeed, it was reported that CRYs can directly interact with PPARδ in skeletal muscles (62). Alternatively, circadian clock proteins can regulate PPARα-dependent transcription through the promoter interference mechanism when transcriptional factors do not physically interact but instead can bind to the same promoter (63), and indeed PPARα and CRYs co-occupy many genome loci (64). If CRY1 inhibits the Fgf21 promoter through direct/indirect binding to PPARα, then one must expect that the suppression of transcription will not depend on the promoter. Contrary to that, the promoter interference mechanism implicates that the regulation will be promoter specific. PPAR Responsive Element (PPRE) plasmid has three copies of PPARα-responsive elements in front of a minimal promoter (Fig. 3G). The promoter was strongly induced by PPARα and by the treatment with the PPARα agonist (Fig. 3H). Notably, this artificial PPRE promoter does not have an E-box element (65), and the cotransfection of plasmids expressing circadian proteins did not significantly affect the PPRE promoter (Fig. 3I, white bars). The cotransfection of any combination of circadian clock proteins also did not interfere with the PPARα-dependent induction of the PPRE promoter (Fig. 3I, black bars). These results support the hypothesis that the regulation of the Fgf21 promoter by circadian clock proteins occurred through the promoter interference mechanism. We also investigated whether clock proteins and PPARα interact on the promoters of circadian clock genes. Per1-Luc plasmid expresses the luciferase reporter under the control of the Period1 promoter (66) (Fig. 3G), and this promoter is strongly regulated by circadian clock transcriptional factors. PPARα did not impact the Per1 promoter on its own or in a combination with circadian clock proteins (Fig. 3J), further confirming that circadian clock proteins and PPARα interfered with each other’s transcriptional activity in a promoter-dependent manner.
CR-induced ketogenesis is disrupted in circadian clock mutant mice.
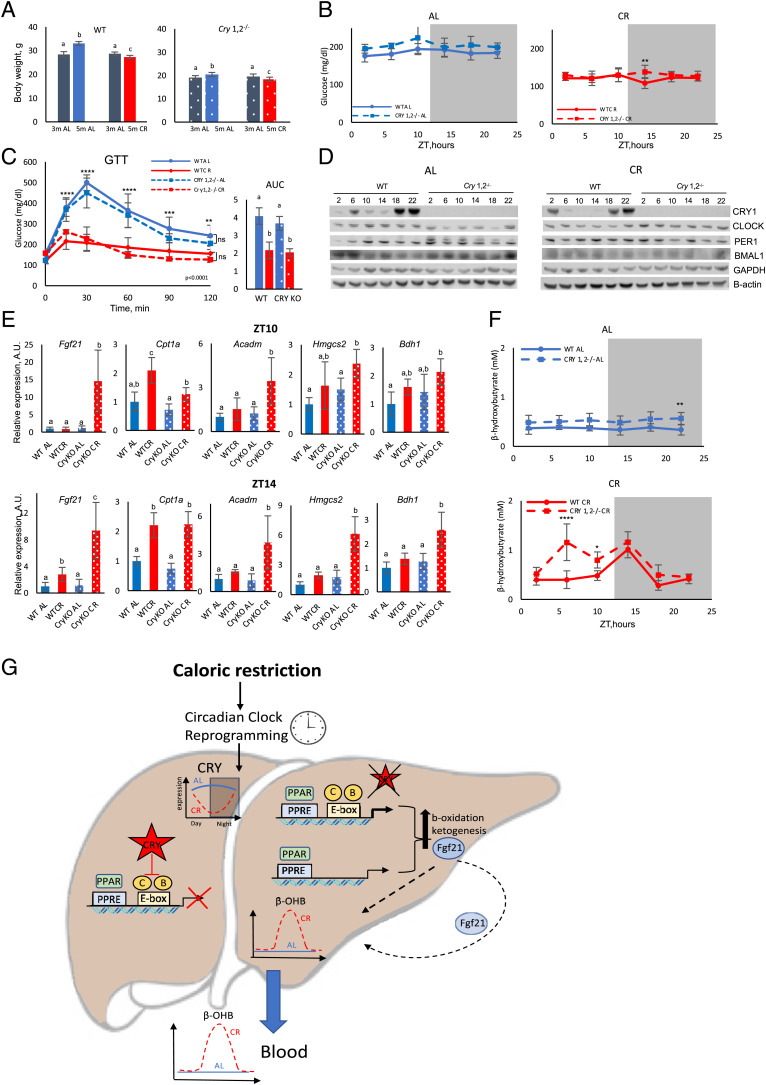
If the circadian clock controls the diet-induced rhythms of ketogenesis, then one might expect that the rhythms will be disrupted in circadian clock–mutant mice. Mice with total body knockout of both the Cry1 and Cry2 genes (Cry1,2−/−) have disrupted circadian rhythms in behavior and gene expression (67, 68). Wild-type and Cry1,2−/− mice were placed on 30% CR. Both wild-type and Cry1,2−/− mice lost approximately 10% of body weight (Fig. 4A); note that Cry1,2−/− mice are smaller compared with wild-type mice (69). One of the well-documented beneficial effects of CR is improved glucose homeostasis. Both wild-type and clock mutants on the CR diet had a significantly reduced blood glucose level across the day (Fig. 4B). Glucose tolerance was also significantly improved in both genotypes (Fig. 4C). Thus, Cry1,2−/− mice responded to 30% CR similarly to wild-type mice. CRY1 expression was significantly down-regulated in the liver of CR mice at ZT6 and ZT10 (Fig. 4D and SI Appendix, Fig. 4). Interestingly, the low expression of CRY1 at around ZT6 to ZT14 coincided with the increased expression of Fgf21 and other PPARα transcriptional targets in the liver of CR wild-type mice (Figs. 3B and 4D and SI Appendix, Figs. 3 and 4). We compared the expression of several PPARα transcriptional targets such as Fgf21, fatty acid beta-oxidation enzymes: Cpt1a and Acadm; ketogenesis enzymes: Hmgsc2 and Bdh1; and ketone body transporter Slc16a7 in the liver of wild type and Cry1,2−/− mice at ZT10 (Fig. 4E, Top, and SI Appendix, Fig. 5) and ZT14 (Fig. 4E, Bottom, and SI Appendix, Fig. 5). All tested genes were induced by CR in both genotypes, but the induction was significantly higher in the Cry1,2−/− mice (except for Cpt1a and Mct2). Interestingly, at ZT18, 4 h after the feeding, there was no difference between wild-type and Cry1,2−/− mice in the expression of these genes (SI Appendix, Fig. 6). Thus, CRYs contributed to the control of ketogenesis-associated genes in a manner dependent on the time of the day. Analysis of available Chromatin Immunoprecipitation Sequensing (ChIP-Seq) databases confirmed that the circadian clock proteins CLOCK, BMAL1, and CRYs were presented on the promoter regions of tested genes (64, 70), where they might interfere with the ability of PPARα to regulate transcription, which further supports the cross talk of circadian and PPARα transcriptional networks in the regulation of ketogenesis.
Fig. 4.
CR-induced circadian rhythms in ketogenesis are impaired in clock-deficient Cry1,2−/− mice. (A) The effect of CR on body weight of wild-type (WT) and Cry1,2−/− mice. Bars with different letters are significantly different from each other; n = 10 per diet point per age per genotype. (B) Blood glucose around the clock in AL (Left) and CR (Right) wild-type (solid line) and Cry1,2−/− (dashed line) mice; n = 6 per diet per time point per genotype. (C) Glucose tolerance test (GTT) in wild-type (solid) and Cry1,2−/− (dashed) mice on AL (blue) or CR (red) diets; n = 6 per diet per genotype. AUC, area under the curve. (D) Western blotting for indicated circadian clock proteins in the liver of wild-type and Cry1,2−/− mice on AL (Left) and CR (Right) diets. Numbes on the top are ZT hours. Pooled samples, n = 3 per diet per time point per genotype. (E) The mRNA expression of PPARα target genes in the liver of wild-type (solid bars) and Cry1,2−/− (dotted bars) mice on AL (blue) or CR (red) diets at ZT10 (Top) and ZT14 (Bottom); n = 6 per diet per time point per genotype. Bars that do not have common letters are significantly different (P < 0.05) from each other. (F) βOHB in the blood of wild-type (solid) and Cry1,2−/− (dashed) mice on AL (Top) and CR (Bottom) diets; n = 6 per time point per genotype. (G) Model of the cross talk between the circadian clock and PPARα in control of ketogenesis rhythms. In CR mice, PPARα is activated and drives the expression of its target genes, including genes involved in ketogenesis. Some of these genes also have circadian E-box elements in their promoters. The circadian clock transcriptional factors CLOCK, BMAL1, and CRYs interfere with the ability of PPARα to regulate the transcription of these genes. The expression of CRY1 is highly rhythmic in the liver of CR mice; therefore, it periodically releases the inhibition of the PPARα-controlled promoters, which results in the rhythms in ketogenesis. See more details in the text. *Significant difference between the genotypes at the indicated time point.
To test the physiological effect of CRY deficiency on ketogenesis, we assayed blood βOHB in wild-type and Cry1,2−/− mice (Fig. 4F). There was a tendency for an increased βOHB level in Cry1,2−/− mice compared with wild-type mice on the AL diet, but it did not reach statistical significance (Fig. 4F, Top). On the CR diet, blood βOHB in wild-type and Cry1,2−/− mice followed different kinetics. βOHB was induced significantly earlier in Cry1,2−/− mice compared with wild-type mice. A high level of blood βOHB was detected already at ZT6, and it was high at ZT10 and ZT14 (Fig. 4F, Bottom, and SI Appendix, Fig. 7). Upon refeeding, blood βOHB was reduced to AL levels with comparable kinetics in both genotypes. It is known that circadian clock deficiency is associated with disrupted daily feeding rhythms on the AL diet (41). To exclude the possibility that the difference in the blood βOHB kinetics in CR mice was due to some difference in feeding patterns, we compared the daily food intake between wild-type and Cry1,2−/− mice on the CR diet. Cry1,2−/− mice consumed all the provided food in 2 h after feeding, which agreed with the wild-type mice. Thus, the different kinetics in blood βOHB between wild-type and clock mutants was not due to a difference in feeding rhythms and rather correlated with different patterns of induction of fat metabolism and ketogenesis-associated genes in the liver of wild-type and circadian clock–mutant mice.
Discussion
Ketone bodies play an important role in fasting metabolism as a source for energy production and as signaling molecules (1, 2, 5, 7, 71). High levels of ketone bodies were found in the plasma of mice subjected to calorie restriction in several studies (26). Contrary to that finding, no increase or even reduction in blood ketone bodies in CR mice was also reported (25, 27). We found that CR induced high-amplitude daily rhythms in blood βOHB with a peak between ZT12 and ZT14; thus, the difference in the above-cited studies might be due to the time of sample collection. CR-induced rhythms in ketogenesis, which support the role of the circadian clock as a regulator of ketogenesis, were significantly disturbed in circadian clock–mutant Cry1,2−/− mice. Notably, both wild-type and circadian-mutant mice on the CR diet consumed all the provided food in 2 h. Thus, mice of both genotypes fasted for the same duration of time. However, βOHB was induced much earlier in Cry1,2−/− mice compared with wild-type mice, suggesting that the circadian clock inhibits ketone bodies production. Interestingly, mice with liver-specific deletion of the circadian clock gene Per2 (LPer2−/−) also have disrupted ketogenesis in a restricted feeding paradigm (72). In this study, blood βOHB was elevated in the food-restricted wild-type mice before the feeding time (72), which was similar to our data for the wild-type mice under CR (Fig. 1B). Blood βOHB was not elevated in LPer2−/− mice under restricted feeding (72). In contrast, βOHB was strongly increased in the blood of Cry1,2−/− mice under CR (Fig. 4F). The expression of the Cpt1 and Hmgcs2 genes was induced in the liver of food-restricted wild-type mice (72), which is similar to the induction of the expression of these genes by CR (Fig. 4E). The induction was attenuated in LPer2−/− mice (72). We propose that PER2 regulates ketogenesis through a direct control of the Cpt1 gene and indirect control of the Hmgcs2 gene. Interestingly, Cpt1 expression was not affected in Cry1,2−/− mice (Fig. 4E), and CPT1 expression was not induced by CR on the protein level; therefore, our data argue against the leading role of CPT1 in CR ketogenesis rhythms. Instead, CRYs deficiency impacts the regulation of several other PPARα transcriptional targets known to be involved in ketogenesis such as Fgf21, AcadM, Hmgsc2, and Bdh1. Most likely, the rhythms were a consequence of changes in the expression of multiple genes rather than one target. Interestingly, PERs and CRYs form a complex and act as a negative arm of the circadian clock feedback loop by suppressing CLOCK/BMAL1-dependent transcription. At the same time, PER2 cooperates with PPARα on the Cpt1a promoter and CRY1 negatively interferes with PPARα-induced transcription on the Fgf21 promoter. Thus, the role of PER2 and CRYs in the transcriptional control of ketogenesis is not identical and warrants further study.
How can the circadian clock control CR-induced rhythms in ketogenesis? Mechanisms of ketogenesis are well studied in fasting (2, 20). The reduction of blood glucose and insulin, an increase of blood and liver FFAs, an activation of PPARα and FGF21, and a down-regulation of mTORC1 have all been reported as contributors to ketogenesis (16, 17, 54, 55, 58). We did not find any correlation between βOHB rhythms with rhythms in blood glucose, TG, FFAs, and mTORC1 signaling. βOHB rhythms correlated with CR-induced rhythms in the expression of the genes involved in fatty acid oxidation, ketogenesis, and βOHB transport. Many of these genes are PPARα targets, and we proposed the model of the cross talk between the circadian clock and PPARα in control of ketogenesis rhythms (Fig. 4G). Circadian clock transcriptional factors can bind to the promoter regions of the genes involved in ketogenesis, where they interfere with PPARα. CRYs block the ability of PPARα to activate the transcription of these genes. Under AL, almost constant eating, PPARα is not activated; therefore, clock-dependent inhibition does not result in strong expression rhythms of PPARα target genes. Under CR, fatty acids are released from the adipose tissues and transported to the liver, where they serve as substrates for ketogenesis, and some fatty acids serve as cofactors for PPARα. As a result, PPARα is activated but CRYs binding to the promoters prevent PPARα-dependent transcription. The expression of CRY1 is rhythmic, it is reduced during the light phase, and the inhibition of the PPARα-controlled promoters is released. Thus, the rhythmic expression of the circadian clock genes leads to strong rhythms in the expression of PPARα-controlled ketogenesis genes and, ultimately, to the circadian rhythms in ketogenesis. In agreement with the model, the lowest expression of CRY1 between ZT6 and ZT14 correlated with the increased PPARα transcriptional activity (Figs. 3B and 4D and SI Appendix, Fig. 3 and 4), which was followed by the peak of βOHB. Notably, only a fraction of PPARα-controlled genes will be regulated through such mechanisms; indeed, only 41% of known PPARα transcriptional targets became rhythmic in CR mice (SI Appendix, Table 2). Interestingly, based on bioinformatical analysis of ChIP-Seq data, only a fraction of PPARα binding sites in the genome are co-occupied by CRYs (64). According to the proposed model, the genes that are regulated by the circadian clock and PPARα must have both a circadian E-box and a PPARα-responsive element in their promoters. The Fgf21 promoter has both elements and was regulated by both circadian transcriptional factors and by PPARα in the reporter experiment (Fig. 3). The expression of Fgf21 in the liver also followed the model-predicted pattern (Figs. 3 A and 4E). It was proposed that Fgf21 might also be under circadian control in fasting (73–75), but it was never experimentally tested. Our data support the hypothesis on the circadian regulation of Fgf21 expression. We performed a transcriptional factor enrichment analysis for PPARα transcriptional targets using ChEA3 (76). As expected, PPARα was highly enriched in these genes (SI Appendix, Table 4). The circadian transcriptional factors CLOCK and BMAL1 were not enriched in the targets rhythmic under AL feeding, but they were enriched in arrhythmic genes. NR1D1 was enriched in rhythmic genes but not in arrhythmic genes, and NR1D2 was not enriched. Under the CR diet, CLOCK, BMAL1, and NR1D1 were significantly enriched in rhythmic targets but not for arrhythmic genes. CLOCK and NR1D1 were also significantly enriched (BMAL1 was close to enrichment; P = 0.05484) for PPARα transcriptional targets, whose expression was affected by CR. Only NR1D1 was enriched in the targets not affected by CR. Thus, the enrichment of CLOCK and BMAL1 strongly correlates with rhythmicity in response to CR, in agreement with the model.
Notably, in addition to the proposed promoter interference model, other mechanisms may be involved in the regulation of PPARα and its transcriptional targets by the circadian clock. The cross talk between the circadian clock and the PPARα pathway was previously reported in other tissues. It was shown that Ppara expression is regulated by BMAL1 and CLOCK (77, 78). In turn, PPARα regulates the transcription of the Bmal1 gene (78–80). When it is overexpressed in cell culture, PPARα can be precipitated in the complex with PER2, but whether this interaction is direct or indirect and exists in vivo is not known (81). In skeletal muscles CRYs directly bind to PPARδ and inhibit its activity, which regulates the exercise capacity (62). This material will be the subject for an independent study regarding whether these mechanisms will contribute to the control of ketogenesis in addition to the proposed promoter interference mechanism.
Thus, during CR the circadian clock, through CRYs, gates ketogenesis to a narrow time window. Various ketogenic diets are popular now and have a proven positive impact on physiology and metabolism (8, 9). What might be a physiological importance of this tight regulation of ketogenesis by the circadian clock? First, to prevent ketoacidosis, a modest increase in blood βOHB is beneficial, while a high level of blood ketone bodies may be harmful. Indeed, ketoacidosis is one of the serious complications of type 1 diabetes (82). Second, chronic up-regulation of ketone bodies may have a negative effect on glucose homeostasis, cardiovascular physiology, and the nervous system (83, 84). Third, βOHB rhythms may be important in ketone body–related signaling. Indeed, chronic up-regulation of βOHB could impair the ability of cells to respond. In mouse models, ketogenic diets have a positive impact on health and longevity, but the effects are not as strong as the effects of CR. It is tempting to speculate that the stronger longevity benefits of CR compared with KD may be due to rhythms in ketone bodies versus chronic up-regulation.
In summary, we found that the circadian clock controls CR-induced rhythms in ketogenesis by reprogramming the PPARα transcriptional network. The study extends current knowledge on mechanisms of ketogenesis, which was studied predominantly in the context of acute fasting. We propose that the rhythms in blood ketone bodies may contribute to the health and longevity benefits of CR.
Limitations of the study
The study was focused on the liver as the main site of ketone bodies production. βOHB can be produced by other tissues such as the kidney and intestine (1). There is also a possibility that CR and the clock regulate βOHB tissue uptake and/or oxidation. Indeed, the circadian clock transcriptional factor BMAL1 regulates the expression of Bdh1 in mouse heart (85). The circadian clock also controls the rhythms of Nicotine Amide Dinucleotide (NAD+) (86), the metabolite that serves as a cofactor for sirtuins, deacetylases that regulate the activity of ketogenic enzymes (87). The study did not address the potential role of blood glucocorticoid rhythms. Glucocorticoids are important players in fasting metabolism. The cross talk between the circadian clock and glucocorticoid production and signaling is known (81, 88). Whether glucocorticoids contribute to daily rhythms in βOHB needs to be addressed in future studies. All these factors can contribute to the circadian regulation of ketogenesis and need to be addressed in future research.
Materials and Methods
Animals.
All experiments involving animals were conducted in accordance with federal and university guidelines, and all procedures were approved by Institutional Animal Care nd Use Committee (IACUC) at Cleveland State University. C57BL/6J male and female mice were aged 12–13 wk at the start of the experiments. Cry1,2−/− mice were previously described (68). Mice were maintained on 12 h light: 12 h dark cycle, light on at 7:00 AM (ZT0), and light off at 7:00 PM (ZT12). Animals were fed a 5008 LabDiet (proteins 26.5%, fat 16.9%, carbohydrates 56.5%). CR mice received 70% of food once per day at ZT14 (2 h after the light was turned off). TR mice received food at ZT14 (2 h after the light was turned off), and food was withdrawn at ZT2 (2 h after the light was turned on) every day; in other words, TR mice were fed AL food for 12 h per day. Mice had unlimited access to water and were maintained on the diets 2 mo before tissue collection. For the constant darkness experiment, CR mice were placed in constant darkness for 48 h and food was provided once a day at ZT14, 2 h after the light was supposed to be off. Blood glucose and ketones were measured on a second day of the complete darkness and the measurement started at ZT18, 4 h after the feeding, and continued until ZT14, before the next feeding. The feeding and blood collection were done under red light. Liver tissues were collected across the day and immediately frozen and stored at −80 °C. At ZT14, tissues were collected before feeding.
Blood glucose and β-hydroxybutyrate detection.
Blood for the analysis was collected through the tail vein nick. Glucose was measured using the CVS Advanced Health Blood Glucose Meter (CVS Pharmacy, Woonsocket, RI) with CVS Health Advanced Glucose Meter Test Strips (CVS Pharmacy, Woonsocket, RI). Blood ketones were measured as the βOHB level using the Precision Xtra Blood Glucose and Ketone meter (Abbott Laboratories).
Glucose tolerance test.
AL mice were fasted for 22 h (from ZT16 until ZT14). CR mice received their last meal at ZT14 the previous day and were fasted for the same amount of time (22 h). At ZT14, mice were subjected to intraperitoneal injection of glucose (1.2 g/kg). Blood glucose was measured via tail vein at 0, 15, 30, 60, 90, and 120 min post injection. The glucose tolerance test was conducted after 6 to 8 wk on the diets.
Cell culture and transfection.
HEK293 cells were cultured at 37 °C in a humidified incubator with 5% CO2 and regularly tested for mycoplasma infection. Cells were maintained in DMEM with 4.5 g/L glucose, supplemented with 10% FBS, 1% L-glutamine, and 1% of penicillin and streptomycin.
Cells were transfected in 6-well plates with the indicated amounts of the plasmids (the final DNA amount was adjusted to 2,500 ng with a pcDNA3 vector) using Xreme X-tremeGENE 360 Transfection Reagent (Sigma-Aldrich, St. Louis, MO) according to the manufacturer’s protocol. A total of 1 ng pcDNA3-β Galactosidase was added to normalize transfection efficiency. Cells were collected for analysis 24 h after transfection. PPARα ligand treatment was done with 50 μM fenofibrate (Sigma-Aldrich, St. Louis, MO). pcHA-Bmal1, pcHA-Clock, and Cry1 were previously described (89, 90). pGL3-mFgf21pro1-luc was a gift from Seiichi Oyadomari (Addgene plasmid number 101797). pSG5 PPARα was a gift from Bruce Spiegelman (Addgene plasmid number 22751). PPRE ×3-TK-luc was a gift from Bruce Spiegelman (Addgene plasmid number 1015).
Luciferase activity detection was performed with the Luciferase Assay System (Promega) according to the manufacturer’s protocol using Victor3 Wallac multichannel reader (Perkin-Elmer). For β-Galactosidase detection, cell lysates were incubated with 1 mg/mL ONPG solution for 1 h and Optical density (OD) were measured at 420 nm.
RNA isolation and qPCR analysis.
Total RNA was extracted from frozen liver tissue using TRIzol according to the manufacturer’s instructions. qRT-PCR was performed using the iTaq Universal SYBR Green Supermix (Bio-Rad). Gene expression was normalized on 18S rRNA and b-actin. Fold change was determined by the ΔΔCt method. The sequence of primers is shown in the SI Appendix.
Western blot analysis.
Total liver lysates were prepared with lysis buffer (1 M Tris base pH 7.5, 5 M NaCl, 0.5 M EGTA, 0.5 M EDTA, Triton-X, 0.1 M Na4P2O7, 1 M β-glycerophosphate, 1 M Na3VO4) containing protease and phosphatase inhibitor mixtures (Sigma-Aldrich, St. Louis, MO). Next, 30 µg protein was loaded equally to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Thermo Fisher Scientific Inc., Waltham, MA). Quantification of images were done using Image Studio Lite software (LI-COR Biosciences, Lincoln, NE). The source of primary antibodies is shown in SI Appendix.
RNAseq.
Total RNA was extracted from 20 mg liver tissue using the RNeasy Mini Kit (Qiagen, 74104) according to the manufacturer’s protocol. Concentration was determined by NanoDrop 2000 (Thermo Fisher Scientific, Inc., Waltham, MA). RNA with an RNA integrity number > 7 was used for further analysis. Three biologic samples per each time point for the AL and CR groups were sent for mRNA sequencing to Novogene, which performed bioinformatic analysis from library preparation to gene expression quantification. The average read depth was 50 M per sample. The mapping rates for all the samples were greater than 90%. Based on calculated Fragments Per Kilobase of transcript per Million mapped reads (FPKM) (gene expression values) distributions, we did not observe that any samples were systemically different from others. Differential expression analysis was completed using Deseq2 and heat maps were generated using R studio. The false discovery rate–adjusted P value cutoff for differential expression analysis was less than 0.05 with a log 2-fold change value greater than 0.7 or less than −0.7. The transcriptomics data are available through the Gene Expression Omnibus repository (GSE211975).
Triglycerides and NEFA measurement.
Serum and liver triglycerides were measured using the Triglyceride Quantification Kit (Sigma-Aldrich, St. Louis, MO, number MAK266).
Liver βOHB and fatty acid measurement.
Samples were analyzed using Agilent GC system 7890B coupled with the Agilent MSD 5977A Mass Spectrometer (Agilent Technologies Inc., Santa Clara, CA). Chromatographic separation was achieved with Agilent J&W HP-5ms in a (5%-phenyl)-methylpolysiloxane column (30 m × 250 µm × 0.25 µm; Agilent Technologies Inc., Santa Clara, CA). Metabolite identification was performed based on retention times of appropriate standards and the National Institute of Standards and Technology library. Details of the sample preparation and gas chromatography-mass spectrometry analysis are included in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the NIH/National Institute on Aging grant R01AG039547 and funds from the Center for Gene Regulation in Health and Disease (Cleveland State University) to R.K. We thank Jillian Kodger and Igor Radzikh for technical assistance with mass spectrometry.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205755119/-/DCSupplemental.
Data, Materials, and Software Availability.
Row experimental data have been deposited to Mendeley (10.17632/8jrxyxkcvw.2). The transcriptomics data are available through the Gene Expression Omnibus repository [GSE211975 (91)].
References
- 1.Puchalska P., Crawford P. A., Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab. 25, 262–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laffel L., Ketone bodies: A review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes Metab. Res. Rev. 15, 412–426 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Yurista S. R., et al. , Therapeutic potential of ketone bodies for patients with cardiovascular disease: JACC state-of-the-art review. J. Am. Coll. Cardiol. 77, 1660–1669 (2021). [DOI] [PubMed] [Google Scholar]
- 4.García-Rodríguez D., Giménez-Cassina A., Ketone bodies in the brain beyond fuel metabolism: From excitability to gene expression and cell signaling. Front. Mol. Neurosci. 14, 732120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Z., et al. , Metabolic regulation of gene expression by histone lysine β-hydroxybutyrylation. Mol. Cell 62, 194–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson A. M., Williamson D. H., Physiological roles of ketone bodies as substrates and signals in mammalian tissues. Physiol. Rev. 60, 143–187 (1980). [DOI] [PubMed] [Google Scholar]
- 7.Balasse E. O., Féry F., Ketone body production and disposal: Effects of fasting, diabetes, and exercise. Diabetes Metab. Rev. 5, 247–270 (1989). [DOI] [PubMed] [Google Scholar]
- 8.Newman J. C., et al. , Ketogenic diet reduces midlife mortality and improves memory in aging mice. Cell Metab. 26, 547–557.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Cabo R., Mattson M. P., Effects of intermittent fasting on health, aging, and disease. N. Engl. J. Med. 381, 2541–2551 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Levy R. G., Cooper P. N., Giri P., Weston J., Ketogenic diet and other dietary treatments for epilepsy. Cochrane Database Syst. Rev. 3, CD001903 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Accurso A., et al. , Dietary carbohydrate restriction in type 2 diabetes mellitus and metabolic syndrome: Time for a critical appraisal. Nutr. Metab. (Lond.) 5, 9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter D. G., Schugar R. C., Crawford P. A., Ketone body metabolism and cardiovascular disease. Am. J. Physiol. Heart Circ. Physiol. 304, H1060–H1076 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGarry J. D., Foster D. W., Regulation of hepatic fatty acid oxidation and ketone body production. Annu. Rev. Biochem. 49, 395–420 (1980). [DOI] [PubMed] [Google Scholar]
- 14.Cahill G. F. Jr., Fuel metabolism in starvation. Annu. Rev. Nutr. 26, 1–22 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Potthoff M. J., et al. , FGF21 induces PGC-1α and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc. Natl. Acad. Sci. U.S.A. 106, 10853–10858 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badman M. K., et al. , Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 5, 426–437 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Sengupta S., Peterson T. R., Laplante M., Oh S., Sabatini D. M., mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez J. C., Gil-Gómez G., Hegardt F. G., Haro D., Peroxisome proliferator-activated receptor mediates induction of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene by fatty acids. J. Biol. Chem. 269, 18767–18772 (1994). [PubMed] [Google Scholar]
- 19.Pawlak M., Lefebvre P., Staels B., Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 62, 720–733 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Kersten S., et al. , Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. J. Clin. Invest. 103, 1489–1498 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montagner A., et al. , Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut 65, 1202–1214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colman R. J., et al. , Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat. Commun. 5, 3557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ungvari Z., Parrado-Fernandez C., Csiszar A., de Cabo R., Mechanisms underlying caloric restriction and lifespan regulation: Implications for vascular aging. Circ. Res. 102, 519–528 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson K. A., et al. , Evaluating the beneficial effects of dietary restrictions: A framework for precision nutrigeroscience. Cell Metab. 33, 2142–2173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anson R. M., et al. , Intermittent fasting dissociates beneficial effects of dietary restriction on glucose metabolism and neuronal resistance to injury from calorie intake. Proc. Natl. Acad. Sci. U.S.A. 100, 6216–6220 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin A. L., Zhang W., Gao X., Watts L., Caloric restriction increases ketone bodies metabolism and preserves blood flow in aging brain. Neurobiol. Aging 36, 2296–2303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo J., Bakshi V., Lin A. L., Early shifts of brain metabolism by caloric restriction preserve white matter integrity and long-term memory in aging mice. Front. Aging Neurosci. 7, 213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato S., et al. , Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell 170, 664–677.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acosta-Rodríguez V. A., de Groot M. H. M., Rijo-Ferreira F., Green C. B., Takahashi J. S., Mice under caloric restriction self-impose a temporal restriction of food intake as revealed by an automated feeder system. Cell Metab. 26, 267–277.e2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckel-Mahan K. L., et al. , Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel S. A., Velingkaar N., Makwana K., Chaudhari A., Kondratov R., Calorie restriction regulates circadian clock gene expression through BMAL1 dependent and independent mechanisms. Sci. Rep. 6, 25970 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bass J., Takahashi J. S., Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ko C. H., Takahashi J. S., Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 15, R271–R277 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Dibner C., Schibler U., Albrecht U., The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koronowski K. B., Sassone-Corsi P., Communicating clocks shape circadian homeostasis. Science 371, eabd0951 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asher G., Sassone-Corsi P., Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Green C. B., Takahashi J. S., Bass J., The meter of metabolism. Cell 134, 728–742 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panda S., Circadian physiology of metabolism. Science 354, 1008–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Velingkaar N., et al. , Reduced caloric intake and periodic fasting independently contribute to metabolic effects of caloric restriction. Aging Cell 19, e13138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turek F. W., et al. , Obesity and metabolic syndrome in circadian clock mutant mice. Science 308, 1043–1045 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Courchesne-Loyer A., et al. , Inverse relationship between brain glucose and ketone metabolism in adults during short-term moderate dietary ketosis: A dual tracer quantitative positron emission tomography study. J. Cereb. Blood Flow Metab. 37, 2485–2493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tulsian R., Velingkaar N., Kondratov R., Caloric restriction effects on liver mTOR signaling are time-of-day dependent. Aging (Albany NY) 10, 1640–1648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatori M., et al. , Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selen E. S., Wolfgang M. J., mTORC1 activation is not sufficient to suppress hepatic PPARα signaling or ketogenesis. J. Biol. Chem. 297, 100884 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longo V. D., Mattson M. P., Fasting: Molecular mechanisms and clinical applications. Cell Metab. 19, 181–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stern J. H., Rutkowski J. M., Scherer P. E., Adiponectin, leptin, and fatty acids in the maintenance of metabolic homeostasis through adipose tissue crosstalk. Cell Metab. 23, 770–784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mezhnina V., et al. , CR reprograms acetyl-CoA metabolism and induces long-chain acyl-CoA dehydrogenase and CrAT expression. Aging Cell 19, e13266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hugo S. E., et al. , A monocarboxylate transporter required for hepatocyte secretion of ketone bodies during fasting. Genes Dev. 26, 282–293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sass J. O., Fukao T., Mitchell G. A., Inborn errors of ketone body metabolism and transport: An update for the clinic and for clinical laboratories. J. Inborn Errors Metab. Screen. 6, 1–7 (2018). [Google Scholar]
- 51.Gulick T., Cresci S., Caira T., Moore D. D., Kelly D. P., The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc. Natl. Acad. Sci. U.S.A. 91, 11012–11016 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aoyama T., et al. , Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor α (PPARalpha). J. Biol. Chem. 273, 5678–5684 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Djouadi F., et al. , A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator-activated receptor α-deficient mice. J. Clin. Invest. 102, 1083–1091 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leone T. C., Weinheimer C. J., Kelly D. P., A critical role for the peroxisome proliferator-activated receptor α (PPARalpha) in the cellular fasting response: The PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. U.S.A. 96, 7473–7478 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hashimoto T., et al. , Defect in peroxisome proliferator-activated receptor α-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J. Biol. Chem. 275, 28918–28928 (2000). [DOI] [PubMed] [Google Scholar]
- 56.de la Calle Arregui C., et al. , Limited survival and impaired hepatic fasting metabolism in mice with constitutive Rag GTPase signaling. Nat. Commun. 12, 3660 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kharitonenkov A., et al. , FGF-21 as a novel metabolic regulator. J. Clin. Invest. 115, 1627–1635 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inagaki T., et al. , Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 5, 415–425 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Acosta-Rodríguez V. A., Rijo-Ferreira F., Green C. B., Takahashi J. S., Importance of circadian timing for aging and longevity. Nat. Commun. 12, 2862 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duregon E., Pomatto-Watson L. C. D. D., Bernier M., Price N. L., de Cabo R., Intermittent fasting: From calories to time restriction. Geroscience 43, 1083–1092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyake M., et al. , Skeletal muscle-specific eukaryotic translation initiation factor 2α phosphorylation controls amino acid metabolism and fibroblast growth factor 21-mediated non-cell-autonomous energy metabolism. FASEB J. 30, 798–812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jordan S. D., et al. , CRY1/2 selectively repress PPARδ and limit exercise capacity. Cell Metab. 26, 243–255.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kondratov R. V., Kondratova A. A., Gorbacheva V. Y., Vykhovanets O. V., Antoch M. P., Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 20, 1868–1873 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kriebs A., et al. , Circadian repressors CRY1 and CRY2 broadly interact with nuclear receptors and modulate transcriptional activity. Proc. Natl. Acad. Sci. U.S.A. 114, 8776–8781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim J. B., Wright H. M., Wright M., Spiegelman B. M., ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc. Natl. Acad. Sci. U.S.A. 95, 4333–4337 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zylka M. J., Shearman L. P., Weaver D. R., Reppert S. M., Three period homologs in mammals: Differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20, 1103–1110 (1998). [DOI] [PubMed] [Google Scholar]
- 67.van der Horst G. T., et al. , Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Vitaterna M. H., et al. , Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. U.S.A. 96, 12114–12119 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaudhari A., Gupta R., Patel S., Velingkaar N., Kondratov R., Cryptochromes regulate IGF-1 production and signaling through control of JAK2-dependent STAT5B phosphorylation. Mol. Biol. Cell 28, 834–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koike N., et al. , Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Newman J. C., Verdin E., Ketone bodies as signaling metabolites. Trends Endocrinol. Metab. 25, 42–52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chavan R., et al. , Liver-derived ketone bodies are necessary for food anticipation. Nat. Commun. 7, 10580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Staiger H., Keuper M., Berti L., Hrabe de Angelis M., Häring H. U., Fibroblast growth factor 21-metabolic role in mice and men. Endocr. Rev. 38, 468–488 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Oishi K., Uchida D., Ishida N., Circadian expression of FGF21 is induced by PPARalpha activation in the mouse liver. FEBS Lett. 582, 3639–3642 (2008). [DOI] [PubMed] [Google Scholar]
- 75.Andersen B., Beck-Nielsen H., Højlund K., Plasma FGF21 displays a circadian rhythm during a 72-h fast in healthy female volunteers. Clin. Endocrinol. (Oxf.) 75, 514–519 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Keenan A. B., et al. , ChEA3: Transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 47, W212–W224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oishi K., Shirai H., Ishida N., CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor α (PPARalpha) in mice. Biochem. J. 386, 575–581 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Canaple L., et al. , Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol. Endocrinol. 20, 1715–1727 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Schmutz I., Ripperger J. A., Baeriswyl-Aebischer S., Albrecht U., The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 24, 345–357 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Inoue I., et al. , CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J. Atheroscler. Thromb. 12, 169–174 (2005). [DOI] [PubMed] [Google Scholar]
- 81.Lamia K. A., et al. , Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ehrmann D., et al. , Risk factors and prevention strategies for diabetic ketoacidosis in people with established type 1 diabetes. Lancet Diabetes Endocrinol. 8, 436–446 (2020). [DOI] [PubMed] [Google Scholar]
- 83.Grandl G., et al. , Short-term feeding of a ketogenic diet induces more severe hepatic insulin resistance than an obesogenic high-fat diet. J. Physiol. 596, 4597–4609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kinzig K. P., Honors M. A., Hargrave S. L., Insulin sensitivity and glucose tolerance are altered by maintenance on a ketogenic diet. Endocrinology 151, 3105–3114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Young M. E., et al. , Cardiomyocyte-specific BMAL1 plays critical roles in metabolism, signaling, and maintenance of contractile function of the heart. J. Biol. Rhythms 29, 257–276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone-Corsi P., Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shimazu T., et al. , SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 12, 654–661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oster H., et al. , The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 4, 163–173 (2006). [DOI] [PubMed] [Google Scholar]
- 89.Kondratov R. V., et al. , BMAL1-dependent circadian oscillation of nuclear CLOCK: Posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 17, 1921–1932 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kondratov R. V., Shamanna R. K., Kondratova A. A., Gorbacheva V. Y., Antoch M. P., Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J. 20, 530–532 (2006). [DOI] [PubMed] [Google Scholar]
- 91.V. Mezhnina et al., Circadian clock controls rhythms in ketogenesis by interfering with PPARa transcriptional network. NCBI: GEO http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE211975. Deposited 31 August 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Row experimental data have been deposited to Mendeley (10.17632/8jrxyxkcvw.2). The transcriptomics data are available through the Gene Expression Omnibus repository [GSE211975 (91)].