Abstract
The COVID-19 pandemic has significantly increased the development of the development of point-of-care (POC) diagnostic tools because they can serve as useful tools for detecting and controlling spread of the disease. Most current methods require sophisticated laboratory instruments and specialists to provide reliable, cost-effective, specific, and sensitive POC testing for COVID-19 diagnosis. Here, a smartphone-assisted Sensit Smart potentiostat (PalmSens) was integrated with a paper-based electrochemical sensor to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A disposable paper-based device was fabricated, and the working electrode directly modified with a pyrrolidinyl peptide nucleic acid (acpcPNA) as the biological recognition element to capture the target complementary DNA (cDNA). In the presence of the target cDNA, hybridization with acpcPNA probe blocks the redox conversion of a redox reporter, leading to a decrease in electrochemical response correlating to SARS-CoV-2 concentration. Under optimal conditions, a linear range from 0.1 to 200 nM and a detection limit of 1.0 pM were obtained. The PNA-based electrochemical paper-based analytical device (PNA-based ePAD) offers high specificity toward SARS-CoV-2 N gene because of the highly selective PNA-DNA binding. The developed sensor was used for amplification-free SARS-CoV-2 detection in 10 nasopharyngeal swab samples (7 SARS-CoV-2 positive and 3 SARS-CoV-2 negative), giving a 100% agreement result with RT-PCR.
Keywords: COVID-19, SARS-CoV-2, Portable potentiostat, Paper-based device, PNA, Point-of-care testing
Graphical abstract
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the latest threat to global health and economies. Due to rapid transmission through direct contact, aerosol spread and new variants [1], cases now exceed 346 million with approximately 5.5 million deaths worldwide as of January 2022 [2]. Although vaccines have been approved, it will take time to keep the COVID-19 pandemic under control and there will continue to be a need for diagnostic testing to monitor breakthrough cases [[3], [4], [5]]. Thus, diagnostics continue to play a crucial role in limiting the infection spread and identifying individuals who are candidates for therapies. Current COVID-19 diagnostics rely primarily on molecular tests and immunological assays [6]. Molecular tests use either reverse transcriptase real-time polymerase chain reaction (RT-PCR), which is a highly sensitive and specific diagnostic tool, or isothermal techniques like loop mediated isothermal amplification (LAMP) [7]. However, these approaches can only be performed in a laboratory setting due to the requirement of specialized handling and sophisticated equipment. Thus, the lack of resources has become the major obstacle for using RT-PCR testing in developing countries [8]. Immunological assays use antibody-antigen reactions for detecting the target viral protein (antigen) or immune response (antibody) against the SARS-CoV-2 [9]. Immunoassays have been miniaturized in several formats such as lateral flow device and electrochemical sensor at point-of-care (POC) diagnostics [10]. For example, Grant et al. developed a half-strip lateral flow assay for SARS-CoV-2 antigen detection [11]. Yakoh et al. developed the electrochemical paper-based analytical device for SARS-CoV-2 detection that relied on a label-free immunoassay strategy in clinical serum samples [12]. Fabiani et al. developed a magnetic bead (MB)-based electrochemical biosensor using labeled immunoassay strategy for SARS-CoV-2 detection in saliva samples [13]. Also, Samper et al. developed electrochemical capillary-flow device for determination of IgG antibody against SARS-CoV-2 in human blood samples [14]. However, these devices either require long analysis times (>1 h) and/or are prone to false-positives [15]. False-positives are frequently caused by non-specific reactions between the target and the immobilized proteins and is a leading source of inaccuracy [16]. Consequently, a rapid, sensitive, selective, accurate approach for COVID-19 diagnosis is still needed for POC testing to control the pandemic.
As mentioned above, sensor selectivity and specificity are major challenges for immunological assays, while the nucleic acid-based assays via RT-PCR are too costly for routine use. Thus, the use of nucleic acids (DNA, RNA) as well as analogues such as peptide nucleic acid (PNA) as a biorecognition element are proposed as an alternative to enhance biosensor selectivity and specificity [17]. PNA offers particular advantages due to its unique electrostatically neutral structure that contributes to its stronger binding affinity towards DNA/RNA targets without compromising specificity [18,19]. Among several available PNA systems, our pyrrolidinyl peptide nucleic acid (acpcPNA) [19,20] offers outstanding characteristics that make them a powerful sensing probe. The rigid D-prolyl-2-aminocyclopentanecarboxylic acid (ACPC) backbone [21] of acpcPNA leads to the stronger binding affinity and sequence specificity toward DNA compared to many systems including the original PNA [22]. Given the benefits of PNA probe over conventional oligonucleotide probes, it has been used as a probe for RT-LAMP [23] and RT-PCR [24] with colorimetric assay for highly specific determination of SARS-CoV-2.
Electrochemical detection can be performed in a reagent-free format, providing user-friendly operation for a variety of protein and nucleic acid analytes [25]. The reagent-free-based biosensor relies on the target-induced conformational change that triggers a change in the electrochemical response either of the DNA bases or of a redox active indicator such as methylene blue (MB) [26,27], [Ru (NH3)6]3+ [28,29] and [Fe(CN)6]3−/4− [30,31]. Among redox active indicators, the common negatively complex of [Fe (CN)6]3−/4− has been extensively used due to its property of high chemical stability, inexpensive, and fast electrochemical kinetics [32]. Moreover, the repulsive force from the same negatively charged motif between the redox indicator and the DNA hybridization generates the change in electrochemical response. It is exceedingly useful to monitor and detect DNA hybridization using an electrochemical sensor [30,33]. Among electrochemical sensing substrates, paper is readily available, inexpensive, and possesses desirable characteristics of natural capillary-driven fluid flow and disposability, making it ideal for the proposed diagnostic tool [34].
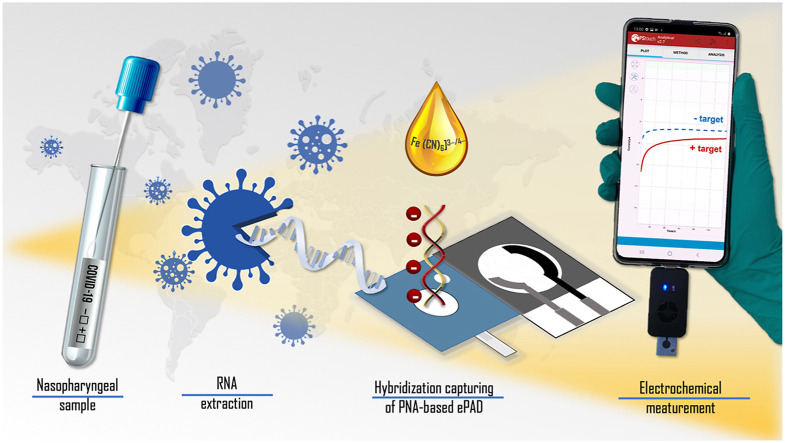
Here, a paper-based electrochemical paper-based analytical device (ePAD) featuring the high affinity and specific acpcPNA probe coupled with a smartphone potentiostat was developed to detect SARS-CoV-2. The platform offers rapid, sensitive, specific, and portable sensing of the SARS-CoV-2 nucleocapsid N gene (Scheme 1 ). For the diagnostic step, the electrochemical signal is measured using amperometry to monitor the signal of [Fe(CN)6]3−/4− with a smartphone application. The amperometric response decreases upon the hybridization of target cDNA with the acpcPNA probe designed to recognize the SARS-CoV-2 N gene. We further demonstrate the high selectivity of electrochemical paper-based analytical device (ePAD) originates from the acpcPNA probe affinity towards its complementary N gene target due to the unique rigid and electrostatically neutral backbone of the acpcPNA probe [35]. Finally, the biosensor was evaluated for SARS-CoV-2 detection in clinical nasopharyngeal swab samples without amplification, showing potential in COVID-19 diagnosis with amplification-free system.
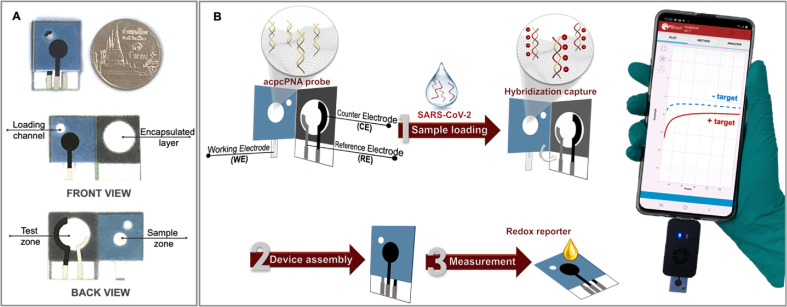
Scheme 1.
(A) Schematic illustration of the proposed PNA-based ePAD with front and back views; (B) the detection principle and procedure for SARS-CoV-2 detection using the PNA-based ePAD sensor connected with a smartphone-based potentiostat.
2. Materials and methods
This section mainly describes the methodology for SARS-CoV-2 detection via PNA-based ePAD sensing, including devices fabrication and electrochemical measurement. The details of acpcPNA synthesis, the ePAD fabrication, and the sample preparation were presented in the supporting information (S2–S5).
2.1. Immobilization of acpcPNA probe on the device
Taking the advantage of the presence of abundant polyol functional groups on the cellulose paper, periodate oxidation was used to create aldehyde groups for the covalent attachment to the amino (-NH2) group of the acpcPNA probe. The covalent immobilization of the acpcPNA probe on the backside of the working ePAD was performed via a facile two-step reaction sequence: (i) periodate oxidation of the hydroxyl group of cellulose, generating an aldehyde group and (ii) imine formation between the aldehyde group and the amino group of the acpcPNA probe followed by the reduction with sodium cyanoborohydride to form a stable covalent bond [30,36]. Briefly, 3 μL of 2.10 M LiCl dissolved in 0.04 M NaIO4 was directly dropcast on the sample zone and the oxidation reaction was allowed to perform in the dark for 15 min. The residual inorganic salt was removed by washing with Milli-Q water and the modified paper was allowed to dry at room temperature (25 °C). For the covalent immobilization of the acpcPNA probe, 3 μL of a solution consisting of 1 μM acpcPNA and 1 mg/mL NaBH3CN in DMF was added to the aldehyde-modified paper and the reaction proceeded in the dark under humid conditions for 12 h at 25 °C. Once the reaction was completed, the unbound acpcPNA probe was removed by washing with a mixture of Milli-Q water: acetonitrile (1:1 v/v) and allowed to dry at room temperature for 30 min. The device was stored at 4 °C until use. No additional materials or reagents (e.g. graphene oxide and glutaraldehyde) were required.
2.2. DNA hybridization
To conduct SARS-CoV-2 (N gene) detection using the COVID-19 PNA-based ePAD for SARS-CoV-2 (N gene) detection, 3 μL of the SARS-CoV-2 N gene DNA was directly added to the sample zone containing the immobilized complementary acpcPNA probe and allowed to hybridize for 30 min at room temperature in a humidity chamber to prevent evaporation. Next, the unhybridized DNA was removed by washing with 0.1 M PBS (pH 7.4).
2.3. Electrochemical measurement
All electrochemical measurements were performed using 5 mM of hexacyanoferrate (III)/(II) ([Fe (CN)6]3−/4−) in 0.1 M KNO3 as a redox indicator. The smartphone equipped with Sensit Smart electrochemical workstation from Palmsens (the Netherlands) and a Pstouch app were employed using amperometry by applying +0.05 V for 2 min.
For electrochemical characterizations, the Nyquist plot for electrochemical impedance spectroscopy (EIS) was recorded in the frequency range of 0.01 Hz–100 kHz with the alternating current (AC) potential of +0.1 V (vs Ag/AgCl) and logarithmic scale of 10 points per decade. Cyclic voltammetry (CV) measurements were conducted between −0.4 and + 0.6 V (vs Ag/AgCl) with potential step of 0.01 V (vs Ag/AgCl) and scan rate of 0.1 V/s.
3. Results and discussion
3.1. Characterization of the acpcPNA-modified electrode surface
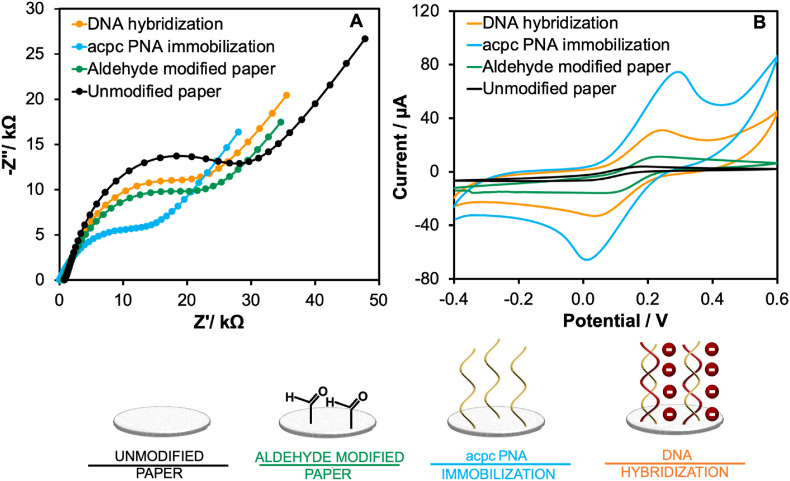
The acpcPNA probe immobilization is a crucial step for specificity toward the target SARS-CoV-2 DNA capturing. The polyol functional groups of unmodified cellulose paper were modified toward the active aldehyde group (called as aldehyde modified paper) through an oxidation reaction. After that, the acpcPNA probe was subsequently immobilized onto the aldehyde-modified paper via imine formation, generating acpcPNA immobilized paper for detecting the target DNA. Therefore, the probe immobilization was confirmed by investigating the electrochemical behavior of the device at each stage of the fabrication using EIS and CV. EIS enables discrimination of a small change on the electrode surface by monitoring the electron transfer efficiency of a redox couple (generally [Fe (CN)6]3−/4− solution) at the electrode/electrolyte interface (the charge transfer resistance, Rct) [37,38]. Here, the Randles equivalent circuit model was fitted to the EIS Nyquist plot as shown in Fig. 1A. The unmodified paper-based electrode showed the largest Rct (20.45 ± 0.80 kΩ) due to its insulating nature that hinders electron transfer between the electrode surface and the redox reporter. In contrast, the Rct (17.38 ± 0.65 kΩ) of the aldehyde modified paper-based electrode decreased due to electroactive functional group [30,39], showing the successful paper modification. Following acpcPNA probe immobilization, the Rct value decreased further to 11.56 ± 0.92 kΩ. This result might be attributed to the electrocatalytic property of the nitrogenous bases (guanine (G) and adenine (A)) in the acpcPNA probe, leading to the improved electron transfer [[40], [41], [42], [43]]. The result demonstrates successful acpcPNA immobilization. Once the SARS-CoV-2 (N gene) DNA was hybridized with its complementary acpcPNA probe, the Rct value (20.29 ± 0.55 kΩ) increased since the anionic PNA-DNA duplex impeded the electron transfer at the interface, indicating the successful DNA hybridization.
Fig. 1.
Electrochemical characterization of each modification step including unmodified paper substrate (black line), aldehyde-modified paper substrate (green line), acpcPNA immobilization (blue line) and DNA hybridization (orange line) via (A) EIS and (B) CV techniques. (All EIS Nyquist plots were fitted with the Randles equivalent circuit). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Additionally, these results are consistent with CV results, as shown in Fig. 1B. In comparison to unmodified paper-based electrodes, the peak current of aldehyde-modified electrodes increased to 11.13 ± 0.90 μA, and the peak potential shifted positive by +0.20 V vs Ag/AgCl against the unmodified paper-based electrode (peak current of 5.69 ± 0.43 μA and peak potential of 0.17 V vs Ag/AgCl). After the acpcPNA probe immobilization, the peak current increased to 71.38 ± 1.67 μA, and the peak potential continually shifted positive to +0.30 V vs Ag/AgCl due to the presence of the electroactive G and A bases that increased electron transfer kinetics [44]. A decrease in peak current (30.80 ± 0.34 μA) was observed after hybridization with the target cDNA, giving a negative shift of the peak potential (+0.24 V vs Ag/AgCl) because of the repulsion between the anionic phosphate DNA backbone and the anionic Fe(CN)6 3−/4-. According to the EIS and CV results, immobilization of the acpcPNA probe on the cellulose paper was accomplished, and the hybridization with the DNA target occurred on the paper surface. Moreover, laser scanning confocal microscopy (LSCM) and the attenuated total reflection fourier transform infrared spectroscopy (ATR-FTIR) were employed to investigate the surface roughness and the change in the functional group of each probe immobilization step, respectively, as discussed in the supporting information (S6).
3.2. SARS-CoV-2 DNA detection using smartphone-based amperometric measurements
After hybridization was complete, the sample zone was folded onto the test zone to create a closed format device for electrochemical measurement. The device was designed to protect the user against direct contact with the biohazardous fluid. Next the device was inserted into the connector of the Sensit Smart potentiostat and the potentiostat was connected to the smartphone. Finally, [Fe(CN)6] 3−/4− was directly added to the sample zone. The solution initially flowed toward the bottom zone and covered the three electrodes to complete the electrical circuit. Subsequently, the amperometric signal was measured.
3.3. Assay optimization
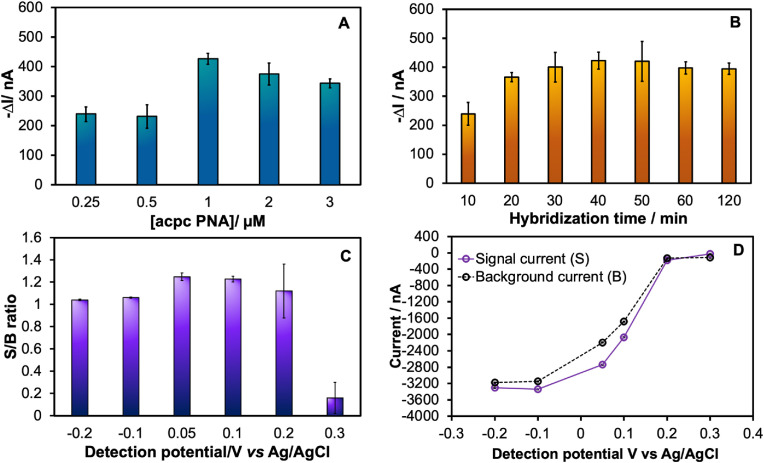
To achieve the best performance for SARS-CoV-2 testing, parameters affecting the PNA-based device potential toward the SARS-CoV-2 diagnostic, including the acpcPNA probe concentration, hybridization time, and detection potential, were systematically investigated using a complementary synthetic DNA oligonucleotide as a model for SARS-CoV-2 N gene.
3.3.1. acpcPNA probe concentration
Since the concentration that determines the amounts of the immobilized acpcPNA probe may affect the DNA hybridization capacity, this parameter was investigated at first. The acpcPNA probe concentration was studied in the range of 0.25–3 μM at a detection potential of +0.1 V vs Ag/AgCl for 120 s, while the target DNA concentration was fixed at 50 nM with 60 min hybridization time. The signal obtained after the targeted DNA hybridization at different probe concentrations (Fig. 2A) increased and reached a maximum at 1 μM acpcPNA probe, and then decreased at higher probe concentrations likely due to steric hindrance that prevents efficient hybridization. Therefore, 1 μM acpcPNA probe was chosen as the optimal concentration.
Fig. 2.
Assay optimization of the important parameters for SARS-CoV-2 DNA based detection including: (A) acpcPNA concentrations; (B) hybridization times; (C) detection potentials for measuring 50 nM SARS-CoV-2 DNA (N gene) via amperometric technique at 120 s sampling times; (D) the amperometric current response for 5 mM [Fe(CN)6]3-/4- in presence (S) and in absence (B) of 50 nM SARS-CoV-2 target. The error bar of all experiment represented the standard deviation of three repetitive measurements using three independent devices (n = 3).
3.3.2. Hybridization time
The hybridization time was also investigated. Different hybridization times ranging from 10 to 120 min were evaluated using 50 nM target DNA. As shown in Fig. 2B, the ΔI value gradually increased with the increasing hybridization time up to 30 min and then became constant. Thus, the hybridization of acpcPNA probe with target DNA in this study was completed in 30 min and was selected as an optimal hybridization time.
3.3.3. Detection potential
The detection potential was investigated in the range of −0.2 to +0.3 V vs Ag/AgCl for 120 s sampling time. The current in the presence (S) or absence of SARS-CoV-2 target (B) for 5 mM of [Fe(CN)6]3-/4−was used to obtain signal to background (S/B) values. The S/B ratio (Fig. 2C) increased significantly with the increasing detection potential up to +0.05 V vs Ag/AgCl and then decreased with detection potential over +0.05 V vs Ag/AgCl. This result can be attributed to the increase in both the signal and background current response with increasing the detection potential as shown in Fig. 2D. The increase might occur from the faster reduction kinetic rate of Fe (III) toward Fe (II) at high detection potential. A larger current differential between signal and background current was chosen to acquire a high sensitivity for the target DNA detection. Therefore, a detection potential of +0.05 V vs Ag/AgCl was used for further experiments.
3.4. Analytical performance
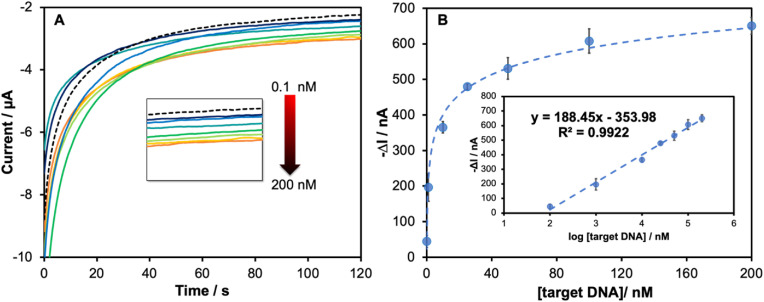
After optimizing each parameter, the analytical performance of the system was determined. The amperograms (Fig. 3A) displayed a gradual decrease in the current as a function of the target DNA concentration. A linear correlation (Fig. 3B) between ΔI and the logarithmic SARS-CoV-2 N gene concentration was observed in the range of 0.1 nM–200 nM with good correlation coefficient (R2 = 0.9922). The limit of detection (LOD) as calculated by 3SDblank/slope was found to be 1.0 pM. The analytical performance of the developed PNA-based ePAD for SARS-CoV-2 detection is compared to the previous reports for SARS-CoV-2 detection in Table S2. Although the previous electrochemical sensors for detecting the specific antibody (IgG, IgM) [12,14] and antigen (nucleocapsid (N) and spike (S) protein) [13] of SARS-CoV-2 protein provide lower LOD than the proposed sensor, their assays rely on biological recognition elements which reduce the sensor stability and increase the device cost. Moreover, the nucleic acid-based sensors for detecting specific genomes of SARS-CoV-2 (N gene, S gene, and RdRp gene) [[45], [46], [47], [48], [49]] also show high performance with lower LOD. There is some previous report using EIS biosensor showing high LOD (10 nM) for the RdRP gene [48]. However, the assays still require the multistep label-based assay and sample amplification step, resulting in long analysis time. Therefore, the PNA-based ePAD sensor is compensated by being a label-free system using small volume of sample (3 μL) without the need of complicated pretreatment steps. In addition, the current limit of detection is close to existing rapid antigen assays, making it useful for clinical applications. Additionally, the overall analysis time of the proposed PNA-based ePAD sensing was accomplished within 42 min (including 10 min of sample extraction, 30 min of hybridization time and 2 min of measurement time). This was faster than other DNA-based methods (e.g. RT-PCR or other DNA sensing), which require at least 3 h of total analysis time. Therefore, the proposed DNA-based sensing offers simplicity, rapid analysis, affordability, and can be used for on-site measurement via a portable smartphone-based potentiostat.
Fig. 3.
(A) Amperograms and (B) calibration plot of the current response at 120 s in a function of SARS-CoV-2 DNA concentrations in the range from 0.1 nM to 200 nM showing the enlarged amperogram in each different DNA concentration in the inset (A); linear calibration plot as a function of logarithmic concentration of SARS-CoV-2 DNA in the inset (B) (n = 3).
The stability of the PNA-based ePAD was also evaluated by comparing storage at 4 °C versus a desiccator at room temperature. The sensor after storage was tested by measuring the current response after hybridization with the target DNA (100 nM) as shown in Fig. S4. The current remained at 96% (stored at RT) and 102% (stored at 4 °C) of the initial response after 12 and 16 days of storage, respectively. The results demonstrate excellent storage stability for the proposed PNA-based ePAD, showing the applicability of the device distribution worldwide during pandemics [50].
3.5. Specificity
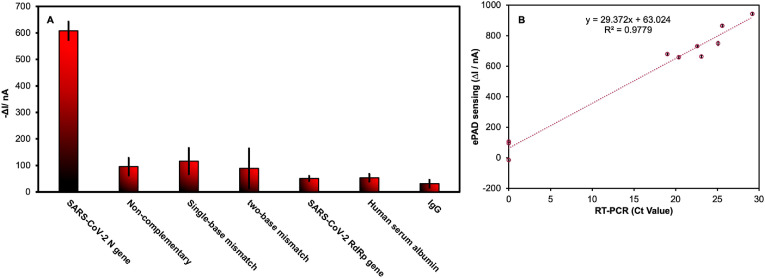
The specificity of the PNA-based ePAD was investigated using non-target DNA with sequences similar to the target SARS-CoV-2 N gene. Four control mismatched DNA sequences, including non-complementary DNA, single-base mismatch DNA, two-base mismatch DNA, and SARS-CoV-2 RdRp genes were tested. As shown in Fig. 4A, the high signal response was only observed in the presence of the target SARS-CoV-2 DNA (N gene), whereas all other the control DNA sequences produced responses that were barely different from the background, indicating high specificity for the target DNA sequence. Besides the mismatch DNA test, other potential proteins including human serum albumin (HSA) and IgG expressed in respiratory secretion were investigated at the maximum concentration of 15.6 mg/mL [51]. As a result, HSA and IgG provided insignificant ΔI changes for the target SARS-CoV-2 DNA, demonstrating that the washing step after sample incubation on PNA-based ePAD can be effectively avoided nonspecific adsorption of these HSA and IgG proteins.
Fig. 4.
(A) ΔI value in the presence of SARS-CoV-2 (N gene), non-complementary, single-base mismatch, two-base mismatch, SARS-CoV-2 (RdRp gene) at 100 nM and 15.6 mg/mL of HSA and IgG (n = 3); (B) correlation of SARS-CoV-2 (N gene) detection in nasopharyngeal swab samples using ePAD sensing (y-axis) vs RT-PCR assay (x-axis).
3.6. Sample analysis
To verify the applicability of the ePAD for detecting SARS-CoV-2, artificial saliva samples were spiked with various concentrations of the standard DNA to concentrations of 1 nM, 10 nM and 100 nM. As shown in Table S3, the satisfactory recovery and error values were found to be in the range of 96.5%–104.5% and 1.9%–6.9%, respectively. The recovery percentage of over 100% might be caused by the different media and viscosity of saliva [52,53]. 10 clinical nasopharyngeal swab samples were collected, including seven samples from COVID-19 patients and three from healthy volunteers. After RNA extraction, the samples were tested with our proposed ePAD sensing platform and compared to RT-PCR (Table 1 ). Additionally, the scatterplot between the ΔI value obtained from the ePAD sensor and Ct value obtained from the RT-PCR assay from 10 nasopharyngeal swab samples was also in good agreement with a good regression coefficient (R2 = 0.977) as shown in Fig. 4B. The result shows the cluster of low values is clearly separated from the cluster of higher values. It is useful to discriminate between each the COVID-19-negative test and the COVID-19-positive test in real sample analysis [54]. Regarding this result, the ΔI value 600 nA was exhaustively considered as the COVID-19-positive test using the proposed PNA-based ePADs. The amperometric response of all clinical sample measurements using the ePAD (Fig. S5) was clear between positive (COVID-19 patients) and negative (healthy volunteers). The percentage of sensitivity and specificity was found to be 100% concerning the real sample.
Table 1.
SARS-CoV-2 (N gene) detection in nasopharyngeal swab samples using the proposed PNA-based ePAD comparing with the standard RT-PCR assay.
| Sample | Proposed systema | RT-PCRb | ||
|---|---|---|---|---|
| Patient #1 | (+) | (ΔI = 943 ± 4.3 nA) | (+) | (Ct = 29.19) |
| Patient #2 | (+) | (ΔI = 730 ± 4.4 nA) | (+) | (Ct = 22.60) |
| Patient #3 | (+) | (ΔI = 865 ± 7.2 nA) | (+) | (Ct = 25.59) |
| Patient #4 | (+) | (ΔI = 658 ± 5.0 nA) | (+) | (Ct = 20.36) |
| Patient #5 | (+) | (ΔI = 679 ± 4.6 nA) | (+) | (Ct = 19.00) |
| Patient #6 | (+) | (ΔI = 663 ± 7.6 nA) | (+) | (Ct = 23.08) |
| Patient #7 | (+) | (ΔI = 749 ± 3.3 nA) | (+) | (Ct = 25.08) |
| Patient #8 | (−) | (ΔI = 106 ± 2.6 nA) | (−) | (N/A) |
| Patient #9 | (−) | (ΔI = 95 ± 7.2 nA) | (−) | (N/A) |
| Patient #10 | (−) | (ΔI = −15 ± 0.96 nA) | (−) | (N/A) |
(+) and (−) signs represent the positive and negative tested patients, respectively,
RT-PCR test was performed at virology laboratory, Faculty of Medicine Ramathibodi Hospital, Mahidol University.
4. Conclusions
Here, a paper-based electrochemical DNA sensor based on a specific binding of immobilized acpcPNA probe against target SARS-CoV-2 (N gene) sequence was developed for COVID-19 diagnosis. This DNA sensor was integrated with a portable Sensitsmart electrochemical potentiostat and smartphone app. The whole assay was performed in 42 min. Under optimal conditions, the proposed sensing platform showed a wide linear range from 0.1 nM to 200 nM with an LOD of 1 pM, demonstrating the high sensitivity for SARS-CoV-2 DNA sensing. Moreover, the proposed sensing offers highly specific SARS-CoV-2 N gene detection against other relevant DNA sequences, emphasizing the beneficial use of acpcPNA probe. Finally, the ePAD was applied for the detection of the target DNA in human saliva samples with satisfactory recovery in the range of 96.5%–104.5%. In addition, we successfully demonstrated the applicability of the proposed PNA-based ePAD for amplification-free detection of SARS-CoV-2 (N gene) in RNA samples extracted from nasopharyngeal swab samples, with the satisfying sensitivity and specificity values of 100%. Therefore, the developed biosensor will be promising as an alternative tool for screening COVID-19 infections.
Ethical statement
All results were proven for conceptualization, and all further experiments related with humans was applied by the Institutional Research Committee (IRB. number MURA2021/967) of the Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand.
Credit authors statement
Atchara Lomae: Conceptualization, Methodology, Investigation, Validation, Data curation, Formal analysis, Writing-original draft. Pattarachaya Preechakasedkit: Investigation, Writing-original draft. Orakan Hanpanich: Investigation, Writing-original draft, Tugba Ozer: Conceptualization, Investigation, Writing-review & editing. Charles S. Henry: Investigation, Writing-review & editing. Atsushi Maruyama: Investigation, Writing-review & editing. Ekawat Pasomsub: Investigation, Resource. Angsana Phuphuakrat: Investigation, Resource. Sirirat Rengpipat: Supervision. Tirayut Vilaivan: Investigation, Resource, Writing-review & editing. Orawon Chailapakul: Funding acquisition, Writing-review & editing, Supervision. Nipapan Ruecha: Project administration, Conceptualization, Writing-original draft, Writing-review & editing, Supervision. Nattaya Ngamrojanavanich: Conceptualization, Funding acquisition, Writing-review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research project is supported by the Second Century Fund (C2F), Chulalongkorn University and A-STEP (JPMJTR21U2 to A.M.) from Japan Science and Technology Agency (JST). The author acknowledges bilateral grant funded by the National Science and Technology Development Agency (NSTDA) through NSTDA-TÜBİTAK Joint Research Program (RES6434823032) for Thailand and The Scientific and Technological Research Council of Turkey (No.120N615) for Turkey. Additional support was provided by National Research Council of Thailand (NRCT) (N41A640073). CSH was supported by a grant from the National Institutes of Health (R01EB031510).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2022.123992.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Li X., Zai J., Wang X., Li Y. Potential of large “first generation” human‐to‐human transmission of 2019‐nCoV. J. Med. Virol. 2020;92 doi: 10.1002/jmv.25693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) 2022. Weekly Epidemiological Update on COVID-19 - 25 January 2022.https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---25-january-2022 (accessed January 28, 2022) [Google Scholar]
- 3.Sun X., Andoh E.A., Yu H. A simulation-based analysis for effective distribution of COVID-19 vaccines: a case study in Norway. Transp. Res. Interdiscip. Perspect. 2021;11 doi: 10.1016/j.trip.2021.100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binagwaho A., Mathewos K., Davis S. Equitable and effective distribution of the COVID-19 vaccines – a scientific and moral obligation. Int. J. Health Pol. Manag. 2021 doi: 10.34172/ijhpm.2021.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgos R.M., Badowski M.E., Drwiega E., Ghassemi S., Griffith N., Herald F., Johnson M., Smith R.O., Michienzi S.M. The race to a COVID-19 vaccine: opportunities and challenges in development and distribution. Drugs Context. 2021;10:1–10. doi: 10.7573/dic.2020-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh B., Datta B., Ashish A., Dutta G. A comprehensive review on current COVID-19 detection methods: from lab care to point of care diagnosis. Sensors Int. 2021;2 doi: 10.1016/j.sintl.2021.100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 8.Freeman W.M., Walker S.J., Vrana K.E. Quantitative RT-PCR: pitfalls and potential. Biotechniques. 1999;26 doi: 10.2144/99261rv01. [DOI] [PubMed] [Google Scholar]
- 9.Liu X., Wang J., Xu X., Liao G., Chen Y., Hu C.-H. Patterns of IgG and IgM antibody response in COVID-19 patients. Emerg. Microb. Infect. 2020;9 doi: 10.1080/22221751.2020.1773324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan R., Li G., Liu S., Zhang X., Liu J., Su Z., Wu Y. Emerging nanolabels-based immunoassays: principle and applications in food safety. TrAC, Trends Anal. Chem. 2021;145 doi: 10.1016/j.trac.2021.116462. [DOI] [Google Scholar]
- 11.Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S., Weigl B.H., Nichols K.P. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal. Chem. 2020;92 doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- 12.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabiani L., Saroglia M., Galatà G., de Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samper I.C., Sánchez-Cano A., Khamcharoen W., Jang I., Siangproh W., Baldrich E., Geiss B.J., Dandy D.S., Henry C.S. Electrochemical capillary-flow immunoassay for detecting anti-SARS-CoV-2 nucleocapsid protein antibodies at the point of care. ACS Sens. 2021;6:4067–4075. doi: 10.1021/acssensors.1c01527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh K., Kawamitsu T., Osaka Y., Sato K., Suzuki Y., Kiriba C., Saito Y., Hirose R., Ichihara H., Saito M., Mitsuke Y., Kuzumi K., Miyashita H., Tsutani H. False positive results in severe acute respiratory coronavirus 2 (SARS-CoV-2) rapid antigen tests for inpatients. J. Infect. Chemother. 2021;27 doi: 10.1016/j.jiac.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaniha K., Kinjo T., Akamine M., Setoguchi M., Tateyama M., Fujita J. False-positive for SARS-CoV-2 antigen test in a man with acute HIV infection. J. Infect. Chemother. 2021;27 doi: 10.1016/j.jiac.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morales M.A., Halpern J.M. Guide to selecting a biorecognition element for biosensors. Bioconjugate Chem. 2018;29:3231–3239. doi: 10.1021/acs.bioconjchem.8b00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun H., Kong J., Zhang X. Application of peptide nucleic acid in electrochemical nucleic acid biosensors. Biopolymers. 2021 doi: 10.1002/bip.23464. [DOI] [PubMed] [Google Scholar]
- 19.Ananthanawat C., Vilaivan T., Hoven V.P., Su X. Comparison of DNA, aminoethylglycyl PNA and pyrrolidinyl PNA as probes for detection of DNA hybridization using surface plasmon resonance technique. Biosens. Bioelectron. 2010;25 doi: 10.1016/j.bios.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 20.Vilaivan T. Pyrrolidinyl PNA with α/β-Dipeptide backbone: from development to applications. Acc. Chem. Res. 2015;48:1645–1656. doi: 10.1021/acs.accounts.5b00080. [DOI] [PubMed] [Google Scholar]
- 21.Suparpprom C., Srisuwannaket C., Sangvanich P., Vilaivan T. Synthesis and oligodeoxynucleotide binding properties of pyrrolidinyl peptide nucleic acids bearing prolyl-2-aminocyclopentanecarboxylic acid (ACPC) backbones. Tetrahedron Lett. 2005;46 doi: 10.1016/j.tetlet.2005.02.126. [DOI] [Google Scholar]
- 22.Poomsuk N., Vilaivan T., Siriwong K. Insights into the structural features and stability of peptide nucleic acid with a D-prolyl-2-aminocyclopentane carboxylic acid backbone that binds to DNA and RNA. J. Mol. Graph. Model. 2018;84 doi: 10.1016/j.jmgm.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Bat-Ochir C., Kim Y.-S., Kim H.G., Lee S.S., Lee H.W., Park H.K. Development, evaluation of the PNA RT-LAMP assay for rapid molecular detection of SARS-CoV-2. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-00041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi W.-S., Jeong J.H., Nicolas H.D.G., Oh S., Antigua K.J.C., Park J.-H., Kim B., Yoon S.-W., Shin K.S., Choi Y.K., Baek Y.H., Song M.-S. Peptide nucleic acid (PNA)-Enhanced specificity of a dual-target real-time quantitative polymerase chain reaction (RT-qPCR) assay for the detection and differentiation of SARS-CoV-2 from related viruses. Diagnostics. 2020;10:775. doi: 10.3390/diagnostics10100775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jesadabundit W., Jampasa S., Patarakul K., Siangproh W., Chailapakul O. Enzyme-free impedimetric biosensor-based molecularly imprinted polymer for selective determination of L-hydroxyproline. Biosens. Bioelectron. 2021;191 doi: 10.1016/j.bios.2021.113387. [DOI] [PubMed] [Google Scholar]
- 26.Kelley S.O., Barton J.K., Jackson N.M., Hill M.G. Electrochemistry of methylene blue bound to a DNA-modified electrode. Bioconjugate Chem. 1997;8:31–37. doi: 10.1021/bc960070o. [DOI] [PubMed] [Google Scholar]
- 27.Han S., Liu W., Yang S., Wang R. Facile and label-free electrochemical biosensors for MicroRNA detection based on DNA origami nanostructures. ACS Omega. 2019;4:11025–11031. doi: 10.1021/acsomega.9b01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das J., Lee J.-A., Yang H. Ultrasensitive detection of DNA in diluted serum using NaBH 4 electrooxidation mediated by [Ru(NH 3) 6] 3+ at Indium−Tin oxide electrodes. Langmuir. 2010;26:6804–6808. doi: 10.1021/la904089e. [DOI] [PubMed] [Google Scholar]
- 29.Steichen M., Decrem Y., Godfroid E., Buess-Herman C. Electrochemical DNA hybridization detection using peptide nucleic acids and [Ru(NH3)6]3+ on gold electrodes. Biosens. Bioelectron. 2007;22:2237–2243. doi: 10.1016/j.bios.2006.10.041. [DOI] [PubMed] [Google Scholar]
- 30.Srisomwat C., Teengam P., Chuaypen N., Tangkijvanich P., Vilaivan T., Chailapakul O. Pop-up paper electrochemical device for label-free hepatitis B virus DNA detection. Sensor. Actuator. B Chem. 2020;316 doi: 10.1016/j.snb.2020.128077. [DOI] [Google Scholar]
- 31.Raziq A., Kidakova A., Boroznjak R., Reut J., Öpik A., Syritski V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai Y., Zheng Y., Swain G.M., Proshlyakov D.A. Equilibrium and kinetic behavior of Fe(CN) 6 3−/4− and cytochrome c in direct electrochemistry using a film electrode thin-layer transmission cell. Anal. Chem. 2011;83:542–548. doi: 10.1021/ac102113v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faria H.A.M., Zucolotto V. Label-free electrochemical DNA biosensor for zika virus identification. Biosens. Bioelectron. 2019;131:149–155. doi: 10.1016/j.bios.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 34.Boonkaew S., Jang I., Noviana E., Siangproh W., Chailapakul O., Henry C.S. Electrochemical paper-based analytical device for multiplexed, point-of-care detection of cardiovascular disease biomarkers. Sensor. Actuator. B Chem. 2021;330 doi: 10.1016/j.snb.2020.129336. [DOI] [Google Scholar]
- 35.Vilaivan T. Pyrrolidinyl PNA with α/β-Dipeptide backbone: from development to applications. Acc. Chem. Res. 2015;48 doi: 10.1021/acs.accounts.5b00080. [DOI] [PubMed] [Google Scholar]
- 36.Pelton R. Bioactive paper provides a low-cost platform for diagnostics. TrAC, Trends Anal. Chem. 2009;28:925–942. doi: 10.1016/j.trac.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayorga-Martinez C.C., Chamorro-García A., Serrano L., Rivas L., Quesada-Gonzalez D., Altet L., Francino O., Sánchez A., Merkoçi A. An iridium oxide nanoparticle and polythionine thin film based platform for sensitive Leishmania DNA detection. J. Mater. Chem. B. 2015;3:5166–5171. doi: 10.1039/c5tb00545k. [DOI] [PubMed] [Google Scholar]
- 38.Lisdat F., Schäfer D. The use of electrochemical impedance spectroscopy for biosensing. Anal. Bioanal. Chem. 2008;391:1555–1567. doi: 10.1007/s00216-008-1970-7. [DOI] [PubMed] [Google Scholar]
- 39.Jaušovec D., Vogrinčič R., Kokol V. Introduction of aldehyde vs. carboxylic groups to cellulose nanofibers using laccase/TEMPO mediated oxidation. Carbohydr. Polym. 2015;116:74–85. doi: 10.1016/j.carbpol.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Gao Y.S., Xu J.K., Lu L.M., Wu L.P., Zhang K.X., Nie T., Zhu X.F., Wu Y. Overoxidized polypyrrole/graphene nanocomposite with good electrochemical performance as novel electrode material for the detection of adenine and guanine. Biosens. Bioelectron. 2014;62:261–267. doi: 10.1016/j.bios.2014.06.044. [DOI] [PubMed] [Google Scholar]
- 41.Sarıoğulları H., Şenocak A., Basova T., Demirbaş E., Durmuş M. Effect of different SWCNT-BODIPY hybrid materials for selective and sensitive electrochemical detection of guanine and adenine. J. Electroanal. Chem. 2019;840:10–20. doi: 10.1016/j.jelechem.2019.03.045. [DOI] [Google Scholar]
- 42.Chen S.-M., Wang C.-H. Electrocatalytic properties of guanine, adenine, guanosine-5′-monophosphate, and ssDNA by Fe(II) bis(2,2′:6′,2″-terpyridine), Fe(II) tris(1,10-phenanthroline), and poly-Fe(II) tris(5-amino-1,10-phenanthroline) Bioelectrochemistry. 2007;70:452–461. doi: 10.1016/j.bioelechem.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Zhang M., Gan F., Cheng F. Electrochemical behaviors and simultaneous determination of guanine and adenine based on highly ordered Pd-nanowire arrays-modified glassy carbon electrode. Anal. Methods. 2015;7:4988–4994. doi: 10.1039/C5AY00608B. [DOI] [Google Scholar]
- 44.Abbaspour A., Mehrgardi M.A. Electrocatalytic oxidation of guanine and DNA on a carbon paste electrode modified by cobalt hexacyanoferrate films. Anal. Chem. 2004;76:5690–5696. doi: 10.1021/ac049421f. [DOI] [PubMed] [Google Scholar]
- 45.Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar M.S., Nandeshwar R., Lad S.B., Megha K., Mangat M., Butterworth A., Knapp C.W., Knapp M., Hoskisson P.A., Corrigan D.K., Ward A.C., Kondabagil K., Tallur S. Electrochemical sensing of SARS-CoV-2 amplicons with PCB electrodes. Sensor. Actuator. B Chem. 2021;343 doi: 10.1016/j.snb.2021.130169. [DOI] [Google Scholar]
- 47.Ang W.L., Lim R.R.X., Ambrosi A., Bonanni A. Rapid electrochemical detection of COVID-19 genomic sequence with dual-function graphene nanocolloids based biosensor. FlatChem. 2022;32 doi: 10.1016/j.flatc.2022.100336. [DOI] [Google Scholar]
- 48.Hwang C., Park N., Kim E.S., Kim M., Kim S.D., Park S., Kim N.Y., Kim J.H. Ultra-fast and recyclable DNA biosensor for point-of-care detection of SARS-CoV-2 (COVID-19) Biosens. Bioelectron. 2021;185 doi: 10.1016/j.bios.2021.113177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao H., Liu F., Xie W., Zhou T.C., OuYang J., Jin L., Li H., Zhao C.Y., Zhang L., Wei J., Zhang Y.P., Li C.P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021;327 doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozer T., Henry C.S. Paper-based analytical devices for virus detection: recent strategies for current and future pandemics. TrAC, Trends Anal. Chem. 2021;144 doi: 10.1016/j.trac.2021.116424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bose M.E., McCaul K.C., Mei H., Sasman A., He J., Kramp W., Shively R., Yan K., Henrickson K.J. Simulated respiratory secretion for use in the development of influenza diagnostic assays. PLoS One. 2016;11 doi: 10.1371/journal.pone.0166800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pungjunun K., Yakoh A., Chaiyo S., Praphairaksit N., Siangproh W., Kalcher K., Chailapakul O. Laser engraved microapillary pump paper-based microfluidic device for colorimetric and electrochemical detection of salivary thiocyanate. Microchim. Acta. 2021;188:140. doi: 10.1007/s00604-021-04793-2. [DOI] [PubMed] [Google Scholar]
- 53.Petruzzi L., Maier T., Ertl P., Hainberger R. Quantitative detection of C-reactive protein in human saliva using an electrochemical lateral flow device. Biosens. Bioelectron. X. 2022;10 doi: 10.1016/j.biosx.2022.100136. [DOI] [Google Scholar]
- 54.Panraksa Y., Amin A.G., Graham B., Henry C.S., Chatterjee D. Immobilization of Proteinase K for urine pretreatment to improve diagnostic accuracy of active tuberculosis. PLoS One. 2021;16 doi: 10.1371/journal.pone.0257615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.