Summary
Background
Neuroblastoma (NB) represents the most frequent form of extra-cranial solid tumour of infants, responsible for 15% of childhood cancer deaths. Nucleolin (NCL) prognostic value in NB was investigated.
Methods
NCL protein expression was retrospectively evaluated in tumour samples of NB patients at diagnosis and after chemotherapy. NCL prognostic value at mRNA level was assessed in a cohort of 20 patients with stage 4 NB (qPCR20, n=20, discovery dataset) and in the MultiPlatform786 including 786 patients of all stages (validation dataset). Overall and event-free survival curves were plotted by Kaplan-Meier method and compared by log-rank test.
Findings
NCL protein, down-modulated after chemotherapy in association with features of neuroblastic differentiation,resulted statistically significantly overexpressed in NB tumours and higher in stage 4 compared to stage 1,2,3 patients. In the stage 4 patients cohort qPCR20, patients with high NCLmRNA expression revealed a statisticallysignificant lower survival probability than those with low NCL expression (OS: HR 4.1 95%CI 1.2–13.8;p=0.0215[Log-rank test], EFS: HR 4.1 95%CI 1.2–14.0, p=0.0197[Log-rank test]). In the MultiPlatform786 (n=786), multivariate analysis suggested thatNCL expression has a statistically significant prognostic value even in the model adjusted for established prognostic markers. NCL expression significantly stratified also patients with >18 months and stage 4 tumour(OS: HR 1.8 95%CI 1.2–2.7, p=0.0009[Log-rank test]; EFS: HR 1.7 95%CI 1.1–2.5, p=0.002[Log-rank test]), patients with>18 months stage 4 with MYCN non amplified tumour[EFS: HR 2.3 95%CI 1.2–4.7, p=0.01[Log-rank test]), and patients with MYCN non amplified and MYC high [OS: HR 11.9 95%CI 2.3–62.4, p=0.003[Log-rank test]; EFS: HR 7.2 95%CI 1.6–33.4, p=0.01[Log-rank test]) . A statistically significant correlation between NCL and MYCN, MYC, and TERT was found in independent datasets (MultiPlatform786 (n=786) and Agilent394 (n=394). Gene set enrichment analysis revealed a statisticallysignificant positive enrichment of MYC target genes and genes involved in telomerase maintenance.
Interpretation
NCL is a novel and independent (adjusting for age, INSS stage, and MYCN status) prognostic marker for NB.
Funding
IMH-EuroNanoMed II-2015 and AIRC-IG.
Keywords: Neuroblastoma, Nucleolin, Biomarker, Prognostic value
Research in context.
Evidence before this study
Neuroblastoma (NB) represents the most frequent form of extra-cranial solid tumour of infants, responsible for 15% of childhood cancer deaths. Despite intensive multimodal treatments, refractory and/or recurrent high-risk NB patients still have very poor prognosis, making mandatory the identification of novel prognostic biomarkers and more active and less toxic therapies. To assess the existing evidence of NCL for cancers and neuroblastoma, we performed a Pubmed search using the following keywords: “NCL and cancer” or “NCL and neuroblastoma”. Nucleolin(NCL) is a physiological protein expressed in all cells of the body, but its up-regulation is observed in several adult solid and blood cancers. On the contrary, little is reported about NCL in pediatric tumours, including NB.
Added value of this study
In this manuscript, our findings indicate that NCL is an unfavourable and independent(adjusting for age, INSS stage, and MYCN status) biomarker for NB.
Implications of all the available evidence
The results obtained demonstrate the potential clinical utility of NCL in most of the cases of High-Risk (HR)-NB patients (such as patients older than 18 months with stage 4 tumour, patients older than 18 months with stage 4 and MYCN non amplified tumour, and patients with MYCN non amplified and MYC high), whose 5-year survival rate is still poor and stratification with established clinical and molecular factors still remains difficult from the clinical perspective. NCL expression is able tosignificantly stratify this subset of patients, making NCL a suitable candidate factor for inclusion in future risk stratification schemas of HR-NB patients.Interestingly, NCL expression is also able to stratify NB patients characterized by favourable prognosis, indicating the possibility that NCL levels may improve actual NB pre-treatment risk stratification.
Alt-text: Unlabelled box
Introduction
Nucleolin (NCL) is one of the most abundant proteins of the nucleolus, involved in the controlof DNA and RNA metabolism, ribosome biogenesis, rRNA synthesis and processing, chromatin organization and stability, cytokinesis, cell proliferation, angiogenesis, apoptosis regulation, stress response and microRNA processing.1, 2, 3 Recently, it was demonstrated that NCL is also implicated in pathological conditions, especially in tumorigenesis and viral infection, rendering NCL a potential target for the development of anti-viral and anti-tumour strategies.2,3Up-regulated NCL, observed in several solid and blood cancers, might contribute to tumorigenesis by increasing rRNA synthesis and the assembly of functional ribosomes. Interestingly, NCL is not located only in the nucleolus, but also in the cytoplasm and on the cell membrane andthis subcellular localization is correlated with its functions.2,3 Anti-tumour strategies based on NCL targeting are reported.2,4,5 For instance,the 26-mer DNA aptamer with G-quadruplex structure AS1411 was demonstrated to target,at the molecular level, various pathway of NCL located in different cellular counterparts.2 After being renamed to ACT-GRO-777, its therapeutic efficacy was also demonstrated in the clinical setting(ClinicalTrials.gov NCT00740441). Furthermore, cell surface NCL is a functional receptor for endostatin, of which it mediates the anti-angiogenic and anti-tumour activities.6
Recently, we have demonstrated that NCL islocalized on the cell surface of neuroblastoma (NB) cells derived from patients suffering with metastatic bone marrow infiltration, both at recurrence and at onset, and on NB primary tumour masses, supporting NCL as a new marker and a useful cellular target molecule in the clinic.7 On the contrary,little is known about the potential prognostic value of NCL inpaediatric tumours, only partially reported in NB.8
NB is a heterogeneous paediatricsolid tumour of the sympathetic nervous system, accounting for 7–8% of childhood cancers and for 15% of deaths from paediatric cancer.9 Despite extensive treatment modalities, the 5-year survival rate for high-risk (HR) NB patients is less than 50%10,11 and the identification of novel prognostic biomarkers and more active and less toxic therapies are urgently needed.
Like many paediatric malignancies, NB usually harbours fewer somatic genomic aberrations and mutations than adult tumours.12 Genome wide studies have confirmed that no single genetic aberration is responsible for the development of all NBs. Instead, sporadic disease is driven by the combination of multiple low frequency mutations and deleterious chromosomal events.13 To date, although a number of other commonly over-expressed proteins and low frequency somatic mutations have been implicated in tumour progression and drug resistance,14 ALK mutations and MYCN amplification are the only validated drivers of NB.14,15 Moreover, attempts to stratify those HR-NB patients with particularly poor outcome, and defined as “ultra-high risk” NB patients,16 are currently underway. The identification of new biomarkers allowing this subgroup stratification would be fundamental to determine the appropriate treatment. In this retrospective study, we suggest the potential prognostic value of NCL in NB, and we propose NCL as a novel molecular target in NB.
Methods
Ethics
This retrospective study was approved by the Ethics Committee of IRCCS Istituto G. Gaslini(Ethics Committee number 104, approved June 14th, 2011).
Patient samples and immunohistochemistry analysis
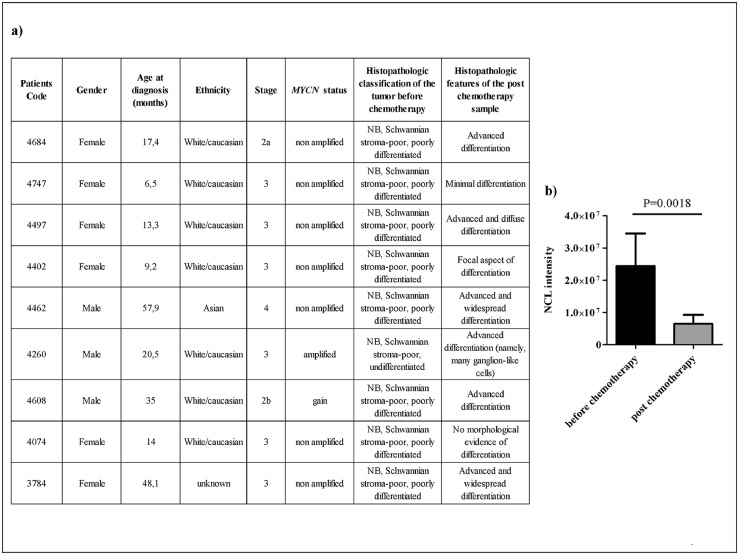
On the basis of availability, samples from primary peripheral neuroblastic tumours – Schwannianstroma-poor, poorly differentiated – collected at diagnosis and after induction chemotherapy from 16 patients (International Neuroblastoma Staging System (INSS)stages 1, 2, 3 and 4) (Table 1S and Figure 7a) were provided by the BIT (Integrated Tissue-Genomic Biobank), Tissue Section of the IRCCS Istituto G.Gaslini, Genoa, Italy.
Figure 7.
NCL expression after induction chemotherapy. NCL protein expression was evaluated by immunohistochemistry staining on formalin-fixed tumour sections derived from NB patients (n = 9)before and after induction chemotherapy. a) Induction of maturation and differentiation. b) NCL intensity in tumours before and after chemotherapy. Column: mean ± standard deviation.P value is shown on top of the plot.
Collection and manipulation of human samples were approved by the competent Ethic Committee and informed consent was obtained from each patient in accordance with the Declaration of Helsinki. For immunohistochemistry (IHC) staining, fully automated ImmunostainerBenchMark® ULTRA Roche from Ventana Medical Systems was used. Paraffin-embedded tissues sections (3 μm) were de-paraffinized and incubated with anti-NCL moAb([EPR7952] ab129200, Abcam) diluted 1:750 in Dako Real™ Antibody Diluent (Dako). The ultraView Universal DAB detection kit from Ventana was used to detect the binding of primary antibody. Sections were counterstained with Hematoxylin II (Ventana). For NCL quantification, sections from each experimental group were scanned using the whole-slide morphometric analysis scanning platform AperioScanscope CS (Leica Biosystems, Nussloch, Germany). All the slides were scanned at the maximum available magnification (40×) and stored as digital high-resolution images on the workstation associated with the instrument. Digital slides were inspected with Aperio Image Scope v.11 software (Leica Biosystems) at 20× magnification and 10 fields with an equal area were selected for the analysis at 40× magnification. The protein expression was assessed with the Positive Pixel Count algorithm embedded in the Aperio Image Scope software and reported as intensity of the positively stained pixels out of the total pixels in the image.
Study design and patients
In this retrospective study, the gene expression dataset referred to as qPCR20, relative to 20 stage 4 patients alive or dead at 10 years follow-up and measured by qPCR method, was used as an independent validation set17 to assess the NCL expression prognostic valueat mRNA level. The gene expression profiles of 786 distinct primary tumour NB samples analysed by Illumina, Agilent, and Affymetrix platforms were used for the evaluation of NCL expression and survival analyses.18 The dataset, referred to as MultiPlatform786, included patient's molecular and clinical data such as age at diagnosis, stage, MYCN status, overall survival (OS) and event-free survival (EFS). Gene expression profiles from additional 394 tumour samples, referred to as Agilent394, analysed by the Agilent customized 4 × 44 k oligonucleotide microarray were used to validate the correlation between NCL and MYCN, MYC, and TERT mRNA expression.19 Gene expression and associated clinical data were collected in the original manuscript19 or from the gene expression omnibus (GEO) at the accession GSE120572. The primary endpoint was the OS defined as the time (in years) from disease diagnosis to patient death or the last follow-up if the patient survived.
Statistics
The Graph Pad Prism version 8.0 for Windows http://www.graphpad.com/ (Graph Pad Software, San Diego, CA, USA) was used for analyses and the limit for statistical significance was set at p<0.05. OS and EFS curves were plotted by the Kaplan-Meier method and compared with the log-rank test by GraphPad Prism. Hazard ratio and 95% confidence interval (CI) of each Kaplan-Meier plot was calculated by Cox proportional hazard regression model. Log-rank p-value lower than 0.05 were considered statistically significant. Hazard ratios with 95%CI not including 1 were considered significant. Multivariate analysis with a Cox proportional hazards regression model was used to assess the prognostic effect of NCL expression in the context of concomitant effects of other known prognostic factors (i.e., age at diagnosis, INSS stage, and MYCN status). Patients with missing data were discarded from the survival analysis. The Survival R package (R 3.1.2) was used for the computation of the Cox regression models.20,21 Expression data were z-scored to standardize data across samples.22 Subsequently, cut-off values distinguishing between low or high NCL expression levels were identified by elbow method.23 Elbow is an empirical method that, given a numerical variable, selects one or more cut-off values whose slope change is evident. For cut-off values, the stratification power of NCL expression was evaluated within 6 populations of NB patients defined based on age at diagnosis, stage and MYCN status, and additional clinically relevant subgroups of patients defined by a combination of established prognostic markers. The significance of the differences of expression between couples of patients’ subsets was assessed by unpaired t test. Correlation between NCL and MYCN, MYC, and TERT expression was assessed by Pearson method.
Gene set enrichment analysis (GSEA), a well-known computational method used to assess whether curated sets of functionally-related genes show a statistically significant and concordant modulation between two biological states,24 was used for interpreting gene expression data.25,26 Hallmark (H), collection 2 chemical and genetic perturbations (C2.CGP), and collection 2 reactome (C2.REATCOME) retrieved from the Molecular Signature Database (MSigDB) v7.2 database,27 were used in the analysis. An enrichment score (ES) and normalized enrichment score (NES) were automatically calculated by GSEA for each gene set. An empirical phenotype permutation test using 1000 permutations was carried out to estimate the statistical significance of a NES and to obtain the nominal p-value (NOM p-val). False Discovery Rate q-value (FDR q-value) of a gene set was automatically assessed by GSEA for accounting the increased probability of false positive findings in the context of multiple hypothesis testing. Gene ranking was assessed by Diff_of_Classes metric. Gene sets containing lower than 10 or higher than 250 genes were filtered out from subsequent analyses. Gene sets with NOM p-val lower than 0.001 and FDR q-value lower than 0.05 were considered statistically significantly enriched.24
Role of funders
Funders did not have any role in the study design, data collection, data analyses, interpretation, or writing the manuscript.
Results
NCL protein is expressed on patient-derived neuroblastoma tumours
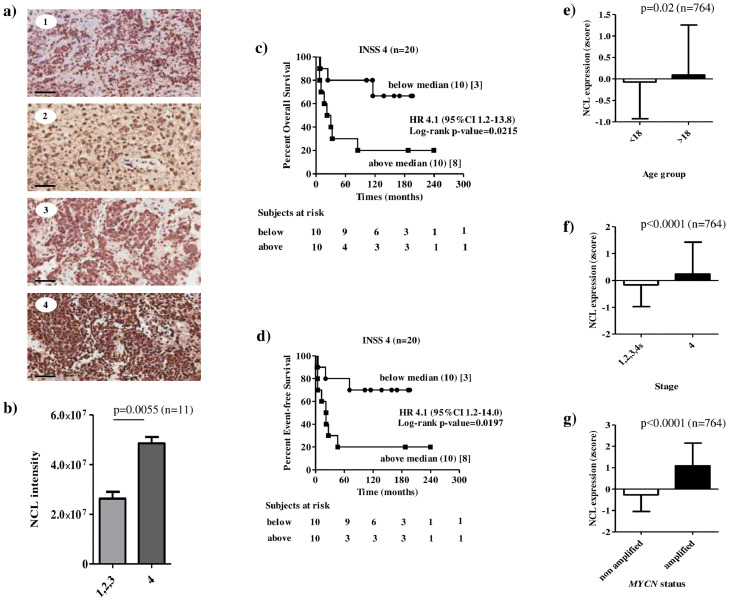
The expression of constitutive NCL was firstly evaluated on formalin-fixed samples derived from Schwannian-stroma poor, poorly differentiated, INSS stage 1, 2, 3 and 4 NB tumours of 11 patients (Table 1S). The schematic representation of the analyses carried out in the present study is shown in Figure 1. IHC evaluations displayed a marked NCL expression in all NB examined (Figure 2). Interestingly, NCL positivity, found not only in the nucleus of the tumour cells but also at the cytoplasm and cell membrane levels (images 1–4) (Figure 2a), was statistically significantly more expressed in stage 4 tumours(image 4) than stage 1, 2 and 3 (images 1, 2 and 3) tumours (Figure 2a and b).
Figure 1.
Workflow of the experiments and analyses performed in the study. The expression of constitutive NCL protein was evaluated on formalin-fixed samples derived from 16 primary peripheral neuroblastictumours –Schwannianstroma-poor, poorly differentiated samples. Comparison of NCL intensity was carried out between tumour and non-neoplastic control material or between tumour samples before and after chemotherapy. The gene expression profile of acohort of INSS stage 4 NB tumours measured by RT-qPCR (qPCR20) was used to assess the prognostic value of NCL in human NB. Kaplan-Meier plots were generated using the median expression value of NCL(0.4429) as cut-off. In the MultiPlatform786,multivariate analysis was carried out to assess the added prognostic value of NCL after adjusting the prognostic model with impact of established prognostic markers for NB. Stratification of two cut-offs identified by elbow method was assessed for overall or event-free survival in selected clinically relevant subsets of patients. To determine whether known sets of functionally related genes were statistically significantly enriched in the gene expression profile of tumours with high or low NCL expression GSEA was used. To explore the possible co-regulating mechanisms between NCL and well-known prognostic genes in NB, a correlation study between NCL and MYCN, MYC or TERT in the MultiPlatform786 dataset was performed and the findings confirmed in the independent Agilent394 dataset including the gene expression profile of 394 NB tumours.
Figure 2.
Constitutive expression of NCL in Schwannian-stroma poor NB patients at diagnosis and prognostic value of NCL expression. a) NCL protein expression was evaluated by immunohistochemistry staining on formalin-fixed tumour sections (n = 11). Representative images ofINSS stage 1 (image 1), INSS stage 2 (image 2), INSS stage 3 (image 3) and INSS stage 4 (image 4) tumours, and of healthy liver (image 5).Brown: NCL expression. Bar: 60 μm. b) NCL intensity in INSS stages 1, 2, 3 and 4tumours. Column: mean ± standard deviation.c-d) Prognostic value of NCL expression in a cohort of 20 NB patients with INSS stage 4 tumour. Kaplan–Meier estimates and significance of the difference analysis based on NCL gene expression. Overall (c) and Event-free (d) survivalsof patients with high (above median) and low (below median)NCLexpression are shown. The median value of NCL expression was 0.4429. Curves were compared by log-rank test, and p values are reported. Each Kaplan–Meier plot reports the hazard ratio (HR) and 95% confidence interval (CI). The number of subjects at risk is displayed under the Kaplan–Meier plots. (e-g) Mean and standard deviation plots of NCL in the populations defined by (e) age at diagnosis, (f) INSS stage, and (g) MYCN status of the MultiPlatform786 dataset (n = 786). Expression values were z-scored prior to any visualization. Significance of the difference between the means has been assessed by unpaired t test. A probability smaller than 0.05 was considered a statistically significant difference. P value and number of patients are shown on top of the plot.
High NCL expression is associated with poor patients’ prognosis in neuroblastoma
To assess the prognostic value of NCL in human NB, an independent cohort of 20 stage 4 NB patients (qPCR20) was used. RT-qPCR analyses revealed that patients with high NCL expression showed a statisticallysignificant lower overall and event-free survival than those with low NCL expression (Figure 2c and d).The prognostic value of NCL expression at mRNA was then assessed in the MultiPlatform786 including gene expression profiles of 786 primary tumours relative to NB patients of all stages. In this dataset, 22 patients were excluded from subsequent analyses as corresponding data were missing. Multivariate analysis with a Cox proportional hazards regression model showed that high levels of NCL is associated with poor patient overall and event-free survival, independently from age at time of diagnosis, tumour stage, and MYCN status (OS: HR 1.16 95%CI 1.002–1.3, p = 0.046[Cox proportional hazard regression]; EFS: HR 1.17 95%CI 1.03–1.33, p = 0.012[Cox proportional hazard regression]; Table 1).
Table 1.
Multivariate analysis of overall and event-free survival in the Merged786 dataset.
| Covariate | HR | 95% CI | P value |
|---|---|---|---|
| Overall survival | |||
| NCL | 1.16 | (1.002 - 1.3) | 0.046 |
| Age at diagnosis (≥ 18 months vs < 18 months) | 3.34 | (2.3 - 4.8) | ˂0.0001 |
| INSS stage (4 vs 1, 2, 3, 4s) | 3.38 | (2.3 - 4.8) | ˂0.0001 |
| MYCN status (Amplified vs non amplified) | 2.45 | (1.7 - 3.5) | ˂0.0001 |
| Event-free survival | |||
| NCL | 1.17 | (1.03 - 1.33) | 0.012 |
| Age at diagnosis (≥18 months vs < 18 months) | 2 | (1.5 - 2.6) | ˂0.0001 |
| INSS stage (4 vs 1, 2, 3, 4s) | 1.99 | (1.5 - 2.6) | ˂0.0001 |
| MYCN status (Amplified vs non amplified) | 1.54 | (1.1 - 2.1) | 0.007 |
Multivariate analysis of overall and event-free surival was assessed by Cox proportional hazards regression.
Patients with missing data were excluded from the analysis. NCL expression was z-scored.
HR: Hazard ration. CI: Confidence interval. INSS: international neuroblastoma staging system.
To assess the extent to which NCL is associated with unfavourable clinical characteristics, we evaluated the distribution of NCL expression in the subsets defined by age at diagnosis, INSS stage and MYCN status. Analysis revealed that NCL mRNA expression was statistically significantly higher in age at diagnosis >18 months (p=0.02[Unpaired Student's t test], Figure 2e), in stage 4tumours (p<0.0001[Unpaired Student's t test], Figure 2f), reflecting the IHC results (Figure 2b), and in MYCN amplified tumours (p<0.0001[Unpaired Student's t test], Figure 2g), respect to age at diagnosis ˂18 months, stage 1, 2, 3 and 4s patients, and MYCN non-amplified tumours, respectively.
We can thus conclude that higher NCL expression in NB tumours is an independent and unfavourable(adjusting for age, INSS stage, and MYCN status) prognostic factor.
NCL expression can stratify clinically relevant subsets of patients
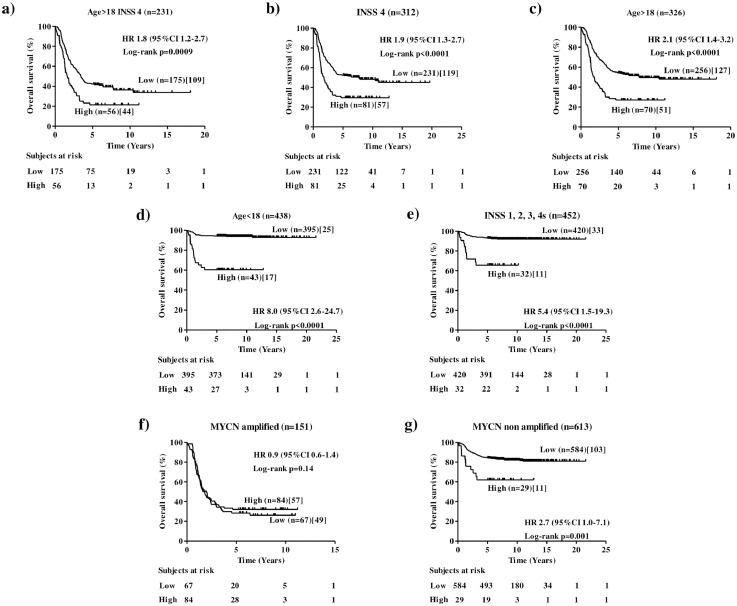
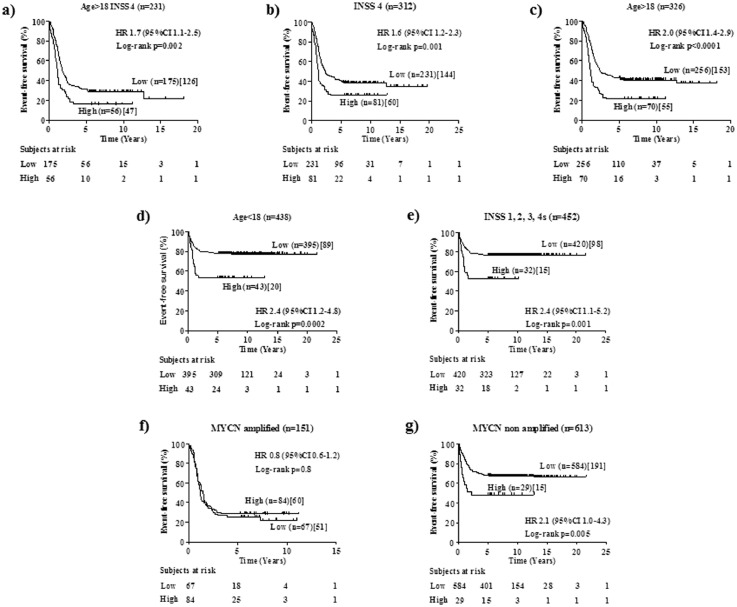
The clinical utility of a new marker is related to its ability to statistically significantly stratify relevant subgroups of patients.18 To assess the clinical relevance of NCL expression in human NB, cut-off values of 0.838 and 1.474 separating MultiPlatform786 profiles of patients into two distinct groups with high or low NCL expression were identified by the elbow method to simplify subsequent analyses. For both cut-off values, Kaplan-Meier analysis showed that NCL expression was able to statistically significantly stratifythe subset of high-risk patients older than 18 months with stage 4tumours (Age >18, Stage 4) in terms of both overall survival (OS: HR 1.8 95%CI 1.2–2.7, p=0.0009[Log-rank test], Figure 3a; HR 1.8 95%CI 1.0–3.0, p=0.007[Log-rank test], Figure 1Sa) and event-free survival(EFS: HR 1.7 95%CI 1.1–2.5, p=0.002[Log-rank test], Figure 4a; HR 1.6 95%CI 1.0–2.6,p=0.001[Log-rank test], Figure 2Sa) that is a group of high-risk patients for whom stratification is traditionally difficult.28
Figure 3.
Overall survival analysis of clinically relevant sub-groups of patients stratified by the NCL expression (cut-off 0.838) in the MultiPlatform786 dataset. Kaplan-Meier curves show overall survival of NB patients with high or low NCL expression. NCL expression cut-off of 0.838 was determined with the elbow method. Expression values were z-scored. Plots are relative to (a) high-risk patients older than 18 months with INSS stage 4tumour, (b) INSS stage 4 patients, (c) age at diagnosis greater than 18 months, (d) age at diagnosis lower than 18 months, (e) stage 1, 2, 3 and 4s patients, (f) MYCNamplified tumours, (g) MYCN non amplified tumours. Plots are entitled with the characteristics of the patients in the sub-population. Survival curves were compared by log-rank test. Number of patients with low or high NCL expressions (brackets) and those who succumbed to disease (square brackets) are reported. Each plot reports the hazard ratio (HR) and 95% confidence interval (CI). The number of subjects at risk is displayed under the Kaplan–Meier plots.
Figure 4.
Event-free survival analysis of clinically relevant sub-groups of patients stratified by the NCL expression (cut-off 0.838) in the MultiPlatform786 dataset. Kaplan-Meier curves show event-free survival of NB patients with high or low NCL expression. NCL expression cut-off of 0.838 was determined with the elbow method. Expression values were z-scored. Plots are relative to (a) high-risk patients older than 18 months with INSS stage 4 tumour, (b) INSS stage 4 patients, (c) age at diagnosis greater than 18 months, (d) age at diagnosis lower than 18 months, (e) stage 1, 2, 3 and 4s patients, (f) MYCNamplified tumours, (g) MYCN non amplified tumours. Plots are entitled with the characteristics of the patients in the sub-population. Survival curves were compared by log-rank test. Number of patients with low or high NCL expressions (brackets) and those who succumbed to disease (square brackets) are reported. Each plot reports the hazard ratio (HR) and 95% confidence interval (CI).
For both cut-off values, analyses also showed that NCL expression statistically significantly stratifies patients with stage 4tumoursfor both overall survival (OS: HR 1.995%CI 1.3–2.7, p<0.0001[Log-rank test], Figure 3b; HR 2.195%CI 1.3–3.4, p<0.0001[Log-rank test], Figure 1Sb) and event-free survival (EFS: HR 1.6 95%CI 1.2–2.3, p=0.001[Log-rank test], Figure 4b; HR 1.8 95%CI 1.2–2.8, p=0.006[Log-rank test], Figure 2Sb) and those with age at diagnosis >18 months for both overall survival (OS: HR 2.1 95%CI 1.4–3.2, p<0.0001[Log-rank test], Figure 3c; HR 1.8 95%CI 1.0–3.0, p=0.002[Log-rank test], Figure 1Sc) and event-free survival (EFS: HR 2.0 95%CI 1.4–2.9, p<0.0001[Log-rank test], Figure 4c; HR 1.7 95%CI 1.1–2.7, p=0.005[Log-rank test], Figure 2Sc). Interestingly, NCL expression was also able to stratify patients with age at diagnosis <18 months for both overall survival (OS: HR 8.0 95%CI 2.6–24.7, p<0.0001[Log-rank test], Figure 3d; HR 14.9 95%CI 2.9–76.2, p<0.0001[Log-rank test], Figure 1Sd) and event-free survival (EFS: HR 2.4 95%CI 1.2–4.8, p=0.0002[Log-rank test], Figure 4d; HR 4.2 95%CI 1.6–11.0, p<0.0001[Log-rank test], Figure 2SD), and stages 1, 2, 3 and 4stumoursfor both overall survival (OS: HR 5.4 95%CI 1.5–19.3, p<0.0001[Log-rank test], Figure 3e; HR 6.5 95%CI 0.9–45.4, p<0.0001[Log-rank test], Figure 1Se) and event-free survival (EFS: HR 2.4 95%CI 1.1–5.2, p=0.001[Log-rank test], Figure 4e; HR 3.0 95%CI 0.9–10.0, p=0.001[Log-rank test], Figure 2Se), independently from MYCN status (Figure 3S), suggesting a potential prognostic role of NCL expression also for the patients with better prognosis. Finally, NCL expression was not able to stratify patients with MYCN amplified population (OS: HR 0.9 95%CI 0.6–1.4, p=0.14[Log-rank test], Figure 3f; HR 1.1 95%CI 0.7–1.6, p=0.86[Log-rank test], Figure 1Sf) and event-free survival (EFS: HR 0.8 95%CI 0.6–1.2, p=0.8[Log-rank test], Figure 4f; HR 1.1 95%CI 0.7–1.6, p=0.6[Log-rank test], Figure 2Sf), but it was able to statistically significantly stratify patients with MYCN nonamplified tumour for both overall survival (OS: HR 2.7 95%CI 1.0–7.1, p=0.001[Log-rank test], Figure 3g; HR 4.5 95%CI 0.7–28.2, p=0.003[Log-rank test], Figure 1Sg) and event-free survival (EFS: HR 2.1 95%CI 1.0–4.3, p=0.005[Log-rank test], Figure 4g; HR 3.8 95%CI 0.9–16.2, p=0.0002[Log-rank test], Figure 2Sg).
Patients with MYCN non amplified tumours accounts for 50% of all high-risk NB cases.29 Since confidence interval for hazard ratio was higher than 1 in all subsets, we considered 0.838 the most promising cut-off and, hereinafter, we used this cut-off for subsequent analyses. Multivariate analysis in the subset of patients with MYCN non amplified tumour showed that high levels of NCLis still associated with poor patient overall and event-free survival, independently from age at time of diagnosis and tumour stage in this subset of patients (OS: HR 2 95%CI 1.1–3.7, p = 0.02[Cox proportional hazard regression]; EFS: HR 1.8 95%CI 1.1–3.1, p = 0.02[Cox proportional hazard regression]; Table2S).
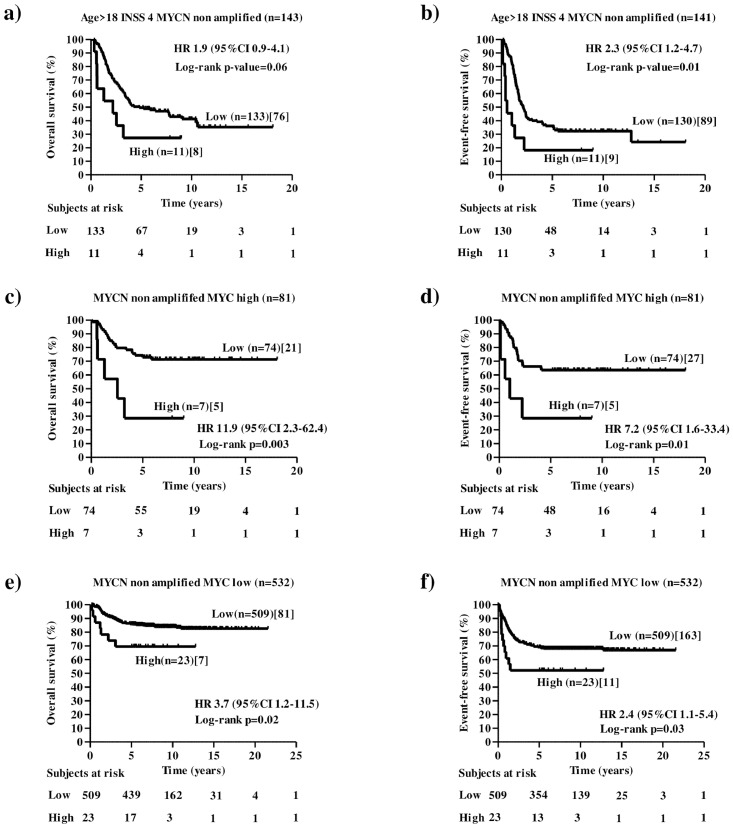
The ability of NCL of stratifying other relevant subsets of high-risk NB patients with MYCN non amplified tumours was then assessed. Patients older than 18 months with stage 4 and MYCN non amplified tumours identify a separate subset of high-risk NB cases. Survival analysis on this subset of patients showed that NCL marginally statistically significantly stratifies patients for overall survival (OS: HR 1.9 95%CI 0.9–4.1, p=0.06[Log-rank test], Figure 5a) and statistically significantly stratifies them for event-free survival (EFS: HR 2.3 95%CI 1.2–4.7, p=0.01[Log-rank test], Figure 5b).
Figure 5.
Overall and event-free survival analysis of other clinically relevant sub-groups of patients stratified by the NCL expression (cut-off 0.838) in the MultiPlatform786 dataset. Kaplan-Meier curves show overall and event-free survival of NB patients with high or low NCL expression. NCL expression cut-off of 0.838 was determined with the elbow method. Expression values were z-scored. Plots are relative to:patients older than 18 months with stage 4 and MYCN non amplified tumour (a and b), patients with MYCNnon amplified tumours and high levels of MYC expression (c and d), and low-risk patients with MYCN non amplified tumours and low levels of MYC expression (e and f). High or low MYC expression was determined by cut-off 1.107997 using elbow method. Plots are entitled with the characteristics of the patients in the sub-population. Survival curves were compared by log-rank test. Number of patients with low or high NCL expressions (brackets) and those who succumbed to disease (square brackets) are reported. Each plot reports the hazard ratio (HR) and 95% confidence interval (CI). The number of subjects at risk is displayed under the Kaplan–Meier plots.
Furthermore, high levels of MYC expression without a concomitant MYCN amplification identifies a separate subset of high-risk NB cases.30,31 Elbow method identified 1.10079 as candidate cut-off for differentiating patients with high or low levels of MYC expression. Survival analysison the subset of patients with MYCN non amplified tumours and high levels of MYC based on 1.10079 cut-off and NCL cut-off 0.838 showed that NCL statistically significantly stratifies these patients for both overall survival (OS: HR 11.9 95%CI 2.3–62.4, p=0.003[Log-rank test], Figure 5c) and event-free survival (EFS: HR 7.2 95%CI 1.6–33.4, p=0.01[Log-rank test], Figure 5d). Furthermore, NCL was able to statistically significantly stratify also the subset of low-risk patients with MYCN non amplified tumours and low levels of MYC for both overall survival (OS: HR 3.7 95%CI 1.2–11.5, p=0.02[Log-rank test], Figure 5e) and event-free survival (EFS: HR 2.4 95%CI 1.1–5.4, p=0.03[Log-rank test], Figure 5f).
Altogether, these findings suggest the potential clinical utility of NCL levels as a prognostic marker.
NCL significantly correlates with MYCN, MYC or TERT expression
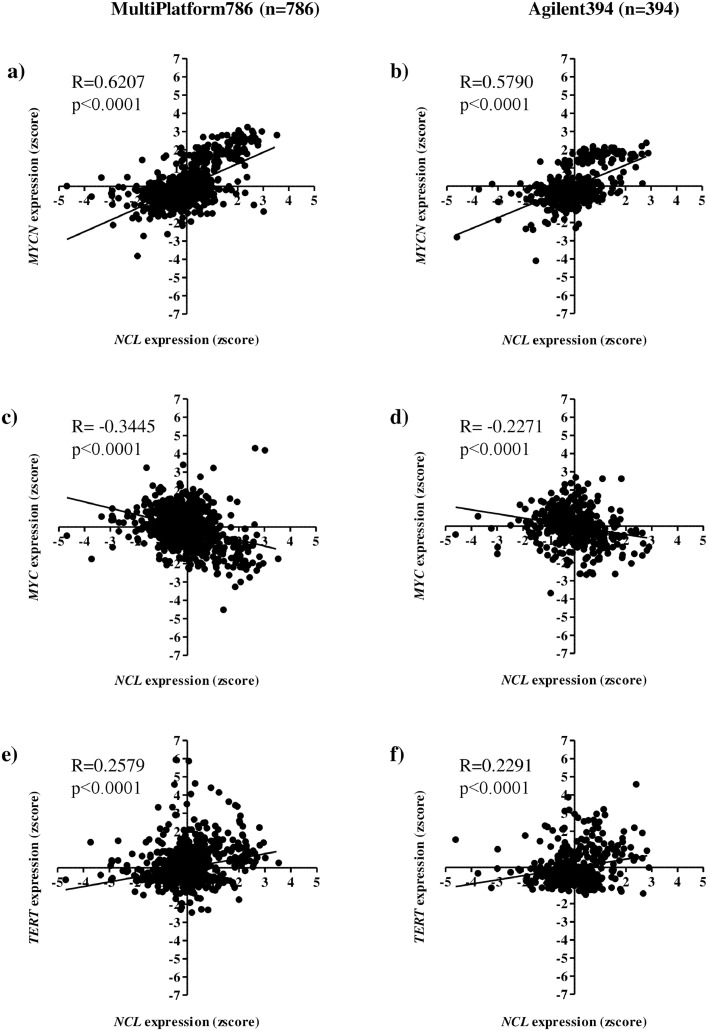
Similar or opposite gene expressions may indicate that genes are co-regulated by common biological mechanisms.32 To assess the possible co-regulating mechanisms between NCL and well-known prognostic genes in NB, we defined the correlation between NCL and MYCN, MYC or TERT mRNA expression in the MultiPlatform786 dataset. Statistically significant correlations for all genes were found (p<0.0001[Pearson correlation test], Figure 6a, 6c and 6e): strongly positive between NCL and MYCN (0.6< R <0.79, Figure 6a), weak negative between NCL and MYC (-0.39<R<-0.2, Figure 6c), and weak positive between NCL and TERT (0.2<R<0.39, Figure 6e). Interestingly, the statistically significant correlations obtained in MultiPlatform786 were validated in Agilent394 dataset (p<0.0001[Pearson correlation test], Figure 6b, 6d and 6f): moderate positive between NCL and MYCN (0.4< R <0.59, Figure 6b), weak negative between NCL and MYC (-0.39<R<-0.2, Figure 6d), and weak positive between NCL and TERT (0.2<R<0.39, Figure 6f), thereby corroborating the involvement of NCL in molecular mechanisms of other well-known unfavourable prognostic genes in NB. The summary of the correlation with relative 95%CI between NCL and MYCN, MYC, or TERT gene in the two independent datasets is shown in Table 3S.
Figure 6.
Correlation analysis between expression levels of NCL and of well-known prognostic genes in two independent datasets. Scatter plots are relative to the correlation between NCL and MYCN (a-b), MYC (c-d) and TERT (e-f) expression in the MultiPlatform786 and Agilent394 data sets, respectively. The number of patients in each dataset is reported between brackets. Each dot represents a patient. Black line indicates the regression line. Pearson correlation and its significance is shown. R>0 or R<0 indicate a positive or negative correlation between two genes, respectively.
Positive enrichment of MYC target genes and genes involved in telomerase maintenance by high NCL expression
We used GSEA24 to determine whether known sets of functionally related genes were statistically significantly enriched in the gene expression profile of tumours with high or low NCL expression. To perform an unbiased and robust analysis, GSEA was run using the H collection, representing 50 well-defined biological states or processes and the C2.CGP and C2.CP.REACTOME collections, consisting of 2613 and 1078 sets of genes curated by domain experts starting from publications in PubMed or from the REACTOME database, respectively. Sixty gene sets were positively enriched in the tumour samples with high NCL expression (Nom p-value<0.001[empirical phenotype-based permutation test] and FDR q-value < 0.05, Table 4S). Statistically significant gene sets were involved in the transcription regulation, RNA processing, MYC targets, and telomerase maintenance. Enrichment plots (Figure 4S) evidenced a clear up-regulation of MYC targets and telomere maintenance, confirming a robust association between high NCL expression and genes that are well-established markers of unfavourable NB patients’ prognosis.
NCL is down-regulated in patient-derived neuroblastoma tumours after chemotherapy
The expression of NCL was also evaluated on formalin-fixed samples derived from Schwannian-stroma poor, undifferentiated or poorly differentiated NB tumours, before and after induction chemotherapy. Independently from the therapeutic protocol used (i.e., HR-1.033 or European Low and Intermediate Risk Neuroblastoma (LINES),34 chemotherapy induced maturation and differentiation in the majority of the tumours (Figure 7a). In the analysed series, an association between neuroblastic differentiation and a statistically significant down-regulation of NCL protein was found (Figure 7b), suggesting that high NCL protein expression might be a potential marker of undifferentiated/poorly differentiated NB cells.
Discussion
In this retrospective study,nucleolin (NCL) is presented as an independent(adjusting for age, INSS stage, and MYCN status) prognostic marker for neuroblastoma (NB).
Patients’ stratification is conventionally used at diagnosis to assign a pre-treatment risk group to patients with NB.35 Risk group is used by clinicians to provide the most appropriate treatment protocol.35 INSS stage, age at diagnosis, histologic category, grade of tumour differentiation, MYCN status, 11q aberration, and ploidy are the clinical and molecular factors currently used in NB pre-treatment risk assignment.35,36
Although the overall survival of patients with NB has improved in the last years, more than 50% of high-risk patients still undergo a relapse and die,16,36 making mandatory the exploration of new prognostic factors able to improve patients’ stratification. To this end, NB tumour biology has been dissected from the perspective of multiple omics including genome,12 transcriptome,18,37 metabolome,38 and mirnome.39 Promising prognostic markers have been described from these extensive investigations of the tumour biology and patient stratification is a fundamental step for assessing the clinical utility of new biomarkers in NB.18 However, despite several biomarkers have been proposed only few of them have been included in NB pre-treatment risk stratification schema.36,40
In the present work, we show that patients with high NCL expression had statistically significantly inferior overall and event-free survival than those with low NCL expression in two datasets of 20 and 786 NB patients. The 20 NB patients’ dataset was initially used to evaluate the prognostic value of NCL in the subset of patients with INSS stage 4, following the findings obtained by IHC staining, in which we demonstrated that NCL was statistically significantly more expressed in stage 4 tumours than stage 1, 2 and 3 tumours. The 786 NB patients’ dataset was used to validate the prognostic value of NCL in an independent set of data and to assess (i) the stratification ability of NCL in clinically relevant subset of patients, (ii)the correlation of NCL expression with well-known prognostic genes, and (iii)the gene set enrichment analysis.
Furthermore, using data coming from distinct omics, gene expression platforms, and timings of disease development, independent evidence of the prognostic value of NCL expression in NB is provided.Indeed, multivariate analysis showed that the prognostic value of NCL expression is preserved independently from MultiPlatform786 available established prognostic features, such as age at diagnosis, MYCN status, and INSS stage, or fromage at diagnosis and INSS stage in the subset of patients with MYCN non amplified.
Biomarker-driven stratification is central to define the most appropriate patient risk assignment and treatment. To this aim, patients were divided into two groups based on the two NCL expression cut-offs of 0.838 or 1.474 using the elbow method. We found statistically significant differences in clinical and biologic characteristics between NB patients with high or low NCL mRNA expression, also showing that high expression of NCL is associated with the worse clinical and molecular characteristics of patients, thereby corroborating the unfavourable prognostic value of NCL.
The clinical utility of evaluating NCL expression cut-offs of 0.838 or 1.474 in NB patient has been assessed by investigating the stratification ability in patient groups, such as age > 18 months, stage 4, MYCN amplificationor MYC high, characterized by severe prognosis. Our analyses report the statisticallysignificant stratification of three out of four groups. MYCN amplified remains a subset of patients for which a statistically significant stratification has not been reported to date. However, the results obtained demonstrate the potential clinical utility of NCL expression in most of the cases of high-risk NB patients. For instance, patients older than 18 months with stage 4tumours, patients older than 18 months with stage 4 and MYCN non amplified tumours, as well as patients with MYCN non amplified and MYC high representingtwo subsets of high-risk patients with the highest percentage of deceases and relapses and whose stratification with established clinical and molecular factors still remains difficult from the clinical perspective.16,30 Interestingly, herein we demonstrate that NCL expression is able to statistically significantly stratify these subsets of patients, making NCL a suitable candidate factor for inclusion in future risk stratification schemas of high-risk NB patients.
Furthermore, NB patients characterized by favourable prognosis, such as age < 18 months, stage 1, 2, 3 or 4s and with MYCN not amplified tumours, have been assessed. As a note, NCL expression was able to stratify also these groups, indicating the possibility that the evaluation of NCL levels may improve actual NB pre-treatment risk stratification.Although further validation of the NCL cut-off will be needed, taken together, our findings indicate that NCL is an unfavourable and independent (adjusting for age, INSS stage, and MYCN status) biomarker with potential clinical utility in NB.
Along with MYCN, also TERT and MYC are well-known genes whose deregulation has been associated with unfavourable NB patient prognosis.41,42 The expression of NCL, which is a target gene of both MYCN and MYC,43,44 positively correlates with MYCN expression and negatively correlates with MYC expression. These results seem to be in agreement with the inverse correlation of MYCN and MYC expression in NB subtypes.41 Interestingly, herein we demonstrate that NCL expression positively correlates also with TERT expression, suggesting a potential co-regulation of these genes, which was further supported by GSEA analyses on available sets of genes in public data bases. Because these results provided unbiased and robust evidence of a statistically significant co-regulation of NCL and genes involved in telomerase maintenance or MYC target genes, our findings indicate that NCL might have a role in NB development and progression.
As stated before, biomarkers have many potential applications in oncology, including risk assessment, screening, differential diagnosis, prediction of response to treatment, and monitoring disease progression. Moreover, they are crucial for developing personalized therapeutic approaches. Despite the critical role that biomarkers play at all stages of disease, there is a substantial attrition in the number of biomarkers that reach the patient, suggesting that a quite worrisome roadblock is preventing successful clinical translation of biomarkers. For this reason, it is important that biomarkers undergo rigorous evaluation, including analytical and clinical validations, besides assessment of clinical utility, prior to incorporation into routine clinical care. In this context, the evaluation of NCL expression could help to identify a subset of NB with very bad prognosis in the group of high-risk patients both at diagnosis and after chemotherapy. Indeed, NCL protein, statistically significantly more expressed in INSS stage 4 tumours, was down-modulated after chemotherapy, in association with morphological features of neuroblastic differentiation. Prognostic and therapeutic indications could be thus derived by the evaluation of possible differential expression of NCL in the primary tumours before and after chemotherapy. Of particular relevance could be to follow the NCL modifications on bone marrow tumour cells, evaluating the evolution of the disease by bone marrow biopsies, aspirates or bone trephine biopsies. In addition, by utilizing a simple, not expensive IHC test, NCL could be evaluated as prognostic marker in the patients with localized disease with favourable genomic and histopathological profiles that in a few cases relapse and die.45,46
Finally, NCLprotein has been demonstrated to be highly expressed on the cell surface of NB cells derived from patients suffering with metastatic bone marrow tumour infiltration, both at recurrence and at onset, and on NB primary tumour masses.7 NCL is also expressed on the cell surface of endothelial cells and pericytes of angiogenic tumour blood vessels.4,47 These features support NCL as a new marker and a useful cellular target molecule in the clinic and increase the possibility to develop NCL-based therapeutic strategy also in NB.5,7
In conclusion, NCL can be considered as is an unfavourable and independent (adjusting for age, INSS stage, and MYCN status) biomarker for NB. The findings hereinpresented indicate the potential clinical utility of NCL in most of the cases of High-Risk (HR)-NB patients. NCL is, indeed, able to stratify these subsets of patients, becoming a suitable candidate factor for inclusion in future risk stratification schemas of HR-NB patients. As a note of caution, alimitation in this study is represented by the lack of functional studies, which will provide the means to ascertain whether NCL is reactive or causative, and validate its prognostic valueprospectively. Moreover, additional validation by comparing NCL status with other pre-treatment risk assignment classification factors (i.e., ploidy histologic category,11q loss of heterozygosityand tumour grade) and with other known NB prognostic biomarkers (i.e.,1q loss of heterozygosity, serum LDH, and serum ferritin) still has to be performed. This could ultimately assist the design of future stratifications or intervention strategies, and clinical decision support systems.
Contributors
DC, MP and FP contributed to the conception and design of the study. DC, CB, VB, RT, FM, BC, PP, DR, JNM, AE, LA, MC, AG, ARS, MVC, MP and FP contributed to data acquisition, data analysis and data interpretation. DC, CB, EG, EC, NAF, verified the data in the study. All authors contributed to the writing of the manuscript drafts and approved the final version of the manuscript. FP had responsibility for the decision to submit for publication.
Data sharing statement
Gene expression profile of neuroblastoma primary tumours and associated patients' characteristics are immediately available in the supplementary material of the Cangelosi et al. manuscript18 or in the qPCR20 dataset accessible upon request from the date this work is published by contacting FP (fabiopastorino@gaslini.org).
Declaration of interests
JNM is share-holder of TREAT U, SA. MC is Advisory Board Eusapharma member.The remaining authors declare no competing interests.
Acknowledgements
We thank the BIT-Gaslini Bio-Bank (Integrated Tumour Bio-Bank of Gaslini Institute, Tissue Section), IRCCS Istituto G. Gaslini, Genoa, Italy for providing the human tumour samples;Comanducci F., Verroca M., Erminio G. and Fragola M. for technical assistance.
This study was supported by Italian ministry of Health under the frame of EuroNanoMed II-2015 (ER-2015–2360441-Eranet to F.P.) and AssociazioneItaliana per la RicercasulCancro (AIRC), Investigator Grant, IG 2020 – ID. 24397 – P.I. Fabio Pastorino.E.C. is recipients of AssociazioneOncologiaPediatrica E Neuroblastoma (OPEN) ONLUS fellowship.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104300.
Contributor Information
Mirco Ponzoni, Email: mircoponzoni@gaslini.org.
Fabio Pastorino, Email: fabiopastorino@gaslini.org.
Appendix. Supplementary materials
References
- 1.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112(Pt 6):761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 2.Berger CM, Gaume X, Bouvet P. The roles of nucleolin subcellular localization in cancer. Biochimie. 2015;113:78–85. doi: 10.1016/j.biochi.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 3.Jia W, Yao Z, Zhao J, Guan Q, Gao L. New perspectives of physiological and pathological functions of nucleolin (NCL) Life Sci. 2017;186:1–10. doi: 10.1016/j.lfs.2017.07.025. [DOI] [PubMed] [Google Scholar]
- 4.Gilles ME, Maione F, Cossutta M, et al. Nucleolin targeting impairs the progression of pancreatic cancer and promotes the normalization of tumor vasculature. Cancer Res. 2016;76(24):7181–7193. doi: 10.1158/0008-5472.CAN-16-0300. [DOI] [PubMed] [Google Scholar]
- 5.Romano S, Fonseca N, Simoes S, Goncalves J, Moreira JN. Nucleolin-based targeting strategies for cancer therapy: from targeted drug delivery to cytotoxic ligands. Drug Discov Today. 2019;24(10):1985–2001. doi: 10.1016/j.drudis.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Shi H, Huang Y, Zhou H, et al. Nucleolin is a receptor that mediates antiangiogenic and antitumor activity of endostatin. Blood. 2007;110(8):2899–2906. doi: 10.1182/blood-2007-01-064428. [DOI] [PubMed] [Google Scholar]
- 7.Brignole C, Bensa V, Fonseca NA, et al. Cell surface Nucleolin represents a novel cellular target for neuroblastoma therapy. J Exp Clin Cancer Res. 2021;40(1):180. doi: 10.1186/s13046-021-01993-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F, Zhou S, Qi D, et al. Nucleolin is a functional binding protein for salinomycin in neuroblastoma stem cells. J Am Chem Soc. 2019;141(8):3613–3622. doi: 10.1021/jacs.8b12872. [DOI] [PubMed] [Google Scholar]
- 9.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nature reviews Cancer. 2003;3(3):203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 10.Matthay KK, Maris JM, Schleiermacher G, et al. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078. doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 11.Wienke J, Dierselhuis MP, Tytgat GAM, Kunkele A, Nierkens S, Molenaar JJ. The immune landscape of neuroblastoma: challenges and opportunities for novel therapeutic strategies in pediatric oncology. Eur J Cancer. 2021;144:123–150. doi: 10.1016/j.ejca.2020.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high-risk neuroblastoma. Nature genetics. 2013;45(3):279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tolbert VP, Coggins GE, Maris JM. Genetic susceptibility to neuroblastoma. Curr Opin Genet Dev. 2017;42:81–90. doi: 10.1016/j.gde.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perri P, Ponzoni M, Corrias MV, Ceccherini I, Candiani S, Bachetti T. A focus on regulatory networks linking MicroRNAs, transcription factors and target genes in neuroblastoma. Cancers (Basel) 2021;13(21) doi: 10.3390/cancers13215528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barr EK, Applebaum MA. Genetic predisposition to neuroblastoma. Children (Basel) 2018;5(9) doi: 10.3390/children5090119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgenstern DA, Bagatell R, Cohn SL, et al. The challenge of defining “ultra-high-risk” neuroblastoma. Pediatr Blood Cancer. 2019;66(4):e27556. doi: 10.1002/pbc.27556. [DOI] [PubMed] [Google Scholar]
- 17.Morandi F, Scaruffi P, Gallo F, et al. Bone marrow-infiltrating human neuroblastoma cells express high levels of calprotectin and HLA-G proteins. PLoS One. 2012;7(1):e29922. doi: 10.1371/journal.pone.0029922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cangelosi D, Morini M, Zanardi N, et al. Hypoxia predicts poor prognosis in neuroblastoma patients and associates with biological mechanisms involved in telomerase activation and tumor microenvironment reprogramming. Cancers (Basel) 2020;12(9) doi: 10.3390/cancers12092343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ackermann S, Cartolano M, Hero B, et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science. 2018;362(6419):1165–1170. doi: 10.1126/science.aat6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dondero A, Morini M, Cangelosi D, et al. Multiparametric flow cytometry highlights B7-H3 as a novel diagnostic/therapeutic target in GD2neg/low neuroblastoma variants. J Immunother Cancer. 2021;9(4) doi: 10.1136/jitc-2020-002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molenaar JJ, Koster J, Zwijnenburg DA, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483(7391):589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 22.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5(2):73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ognibene M, Pagnan G, Marimpietri D, et al. CHL1 gene acts as a tumor suppressor in human neuroblastoma. Oncotarget. 2018;9(40):25903–25921. doi: 10.18632/oncotarget.25403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cangelosi D, Muselli M, Parodi S, et al. Use of attribute driven incremental discretization and logic learning machine to build a prognostic classifier for neuroblastoma patients. BMC Bioinformat. 2014;15(suppl 5):S4. doi: 10.1186/1471-2105-15-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cangelosi D, Resaz R, Petretto A, et al. A proteomic analysis of GSD-1a in mouse livers: evidence for metabolic reprogramming, inflammation, and macrophage polarization. J Proteome Res. 2019;18(7):2965–2978. doi: 10.1021/acs.jproteome.9b00309. [DOI] [PubMed] [Google Scholar]
- 27.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosse KR, Maris JM. Advances in the translational genomics of neuroblastoma: from improving risk stratification and revealing novel biology to identifying actionable genomic alterations. Cancer. 2016;122(1):20–33. doi: 10.1002/cncr.29706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otte J, Dyberg C, Pepich A, Johnsen JI. MYCN function in neuroblastoma development. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.624079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang LL, Teshiba R, Ikegaki N, et al. Augmented expression of MYC and/or MYCN protein defines highly aggressive MYC-driven neuroblastoma: a children's oncology group study. Br J Cancer. 2015;113(1):57–63. doi: 10.1038/bjc.2015.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmerman MW, Liu Y, He S, et al. MYC drives a subset of high-risk pediatric neuroblastomas and is activated through mechanisms including enhancer hijacking and focal enhancer amplification. Cancer Discov. 2018;8(3):320–335. doi: 10.1158/2159-8290.CD-17-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics. 2008;91(3):243–248. doi: 10.1016/j.ygeno.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Garaventa A, Poetschger U, Valteau-Couanet D, et al. Randomized trial of two induction therapy regimens for high-risk neuroblastoma: HR-NBL1.5 international society of pediatric oncology european neuroblastoma group study. J Clin Oncol. 2021;39(23):2552–2563. doi: 10.1200/JCO.20.03144. [DOI] [PubMed] [Google Scholar]
- 34.Bagatell R, Beck-Popovic M, London WB, et al. Significance of MYCN amplification in international neuroblastoma staging system stage 1 and 2 neuroblastoma: a report from the International Neuroblastoma Risk Group database. J Clin Oncol. 2009;27(3):365–370. doi: 10.1200/JCO.2008.17.9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tolbert VP, Matthay KK. Neuroblastoma: clinical and biological approach to risk stratification and treatment. Cell Tissue Res. 2018;372(2):195–209. doi: 10.1007/s00441-018-2821-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohn SL, Pearson AD, London WB, et al. The international neuroblastoma risk group (INRG) classification system: an INRG Task Force report. J Clin Oncol. 2009;27(2):289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viprey VF, Gregory WM, Corrias MV, et al. Neuroblastoma mRNAs predict outcome in children with stage 4 neuroblastoma: an European HR-NBL1/SIOPEN study. J Clin Oncol. 2014;32(10):1074–1083. doi: 10.1200/JCO.2013.53.3604. [DOI] [PubMed] [Google Scholar]
- 38.Verly IRN, van Kuilenburg ABP, Abeling N, et al. 3-Methoxytyramine: an independent prognostic biomarker that associates with high-risk disease and poor clinical outcome in neuroblastoma patients. Eur J Cancer. 2018;90:102–110. doi: 10.1016/j.ejca.2017.11.025. [DOI] [PubMed] [Google Scholar]
- 39.Morini M, Cangelosi D, Segalerba D, et al. Exosomal microRNAs from longitudinal liquid biopsies for the prediction of response to induction chemotherapy in high-risk neuroblastoma patients: a proof of concept SIOPEN study. Cancers (Basel) 2019;11(10) doi: 10.3390/cancers11101476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokol E, Desai AV. The evolution of risk classification for neuroblastoma. Children (Basel) 2019;6(2) doi: 10.3390/children6020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westermann F, Muth D, Benner A, et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008;9(10):R150. doi: 10.1186/gb-2008-9-10-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ackermann S, Fischer M. Telomere maintenance in pediatric cancer. Int J Mol Sci. 2019;20(23) doi: 10.3390/ijms20235836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulte JH, Horn S, Otto T, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int J Cancer. 2008;122(3):699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- 44.Greasley PJ, Bonnard C, Amati B. Myc induces the nucleolin and BN51 genes: possible implications in ribosome biogenesis. Nucleic Acids Res. 2000;28(2):446–453. doi: 10.1093/nar/28.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lerone M, Ognibene M, Pezzolo A, et al. Molecular genetics in neuroblastoma prognosis. Children (Basel) 2021;8(6) doi: 10.3390/children8060456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irwin MS, Naranjo A, Zhang FF, et al. Revised neuroblastoma risk classification system: a report from the children's oncology group. J Clin Oncol. 2021;39(29):3229–3241. doi: 10.1200/JCO.21.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christian S, Pilch J, Akerman ME, Porkka K, Laakkonen P, Ruoslahti E. Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J Cell Biol. 2003;163(4):871–878. doi: 10.1083/jcb.200304132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.