Abstract
Helicobacter pylori, a gram-negative bacterium associated with gastritis, peptic ulceration, and gastric adenocarcinoma in humans, secretes a protein toxin, VacA, that causes vacuolar degeneration of epithelial cells. Several different families of H. pylori vacA alleles can be distinguished based on sequence diversity in the “middle” region (i.e., m1 and m2) and in the 5′ end of the gene (i.e., s1 and s2). Type s2 VacA toxins contain a 12-amino-acid amino-terminal hydrophilic segment, which is absent from type s1 toxins. To examine the functional properties of VacA toxins containing this 12-amino-acid segment, we analyzed a wild-type s1/m1 VacA and a chimeric s2/m1 VacA protein. Purified s1/m1 VacA from H. pylori strain 60190 induced vacuolation in HeLa and Vero cells, whereas the chimeric s2/m1 toxin (in which the s1 sequence of VacA from strain 60190 was replaced with the s2 sequence from strain Tx30a) lacked detectable cytotoxic activity. Type s1/m1 VacA from strain 60190 formed membrane channels in a planar lipid bilayer assay at a significantly higher rate than did s2/m1 VacA. However, membrane channels formed by type s1 VacA and type s2 VacA proteins exhibited similar anion selectivities (permeability ratio, PCl/PNa = 5). When an equimolar mixture of the chimeric s2/m1 toxin and the wild-type s1/m1 toxin was added to HeLa cells, the chimeric toxin completely inhibited the activity of the s1/m1 toxin. Thus, the s2/m1 toxin exhibited a dominant-negative phenotype similar to that of a previously described mutant toxin, VacA-(Δ6–27). Immunoprecipitation experiments indicated that both s2/m1 VacA and VacA-(Δ6–27) could physically interact with a c-myc epitope-tagged s1/m1 VacA, which suggests that the dominant-negative phenotype results from the formation of heterooligomeric VacA complexes with defective functional activity. Despite detectable differences in the channel-forming activities and cytotoxic properties of type s1 and type s2 VacA proteins, the conservation of type s2 sequences in many H. pylori isolates suggests that type s2 VacA proteins retain an important biological activity.
Helicobacter pylori is a gram-negative bacterium that colonizes the gastric mucosa of humans. Colonization with these organisms consistently induces gastric mucosal inflammation and is associated with an increased risk for peptic ulcer disease, gastric adenocarcinoma, and gastric lymphoma (6, 16).
The only cytotoxin known to be secreted into the extracellular space by H. pylori is the vacuolating cytotoxin, VacA (5, 37). The hallmark of VacA activity is the formation of prominent intracellular vacuoles when the toxin is added to cultured cells (30). These vacuoles represent hybrid compartments derived from late endosomes and lysosomes (35). The mechanism of VacA-induced vacuole formation is not yet completely understood but is thought to involve alterations in membrane trafficking along the endosomal-lysosomal pathway (37) and seems to be dependent on the formation of anion-selective channels in cellular membranes (11, 24, 53, 56, 57). One current model suggests that vacuolation is somehow related to an influx of anions through VacA channels formed in the membranes of endosomes (11, 24, 53, 56, 57). In addition to causing formation of intracellular vacuoles, VacA interferes with the process of antigen presentation (36), increases the permeability of polarized epithelial monolayers (42), induces apoptosis (18, 43), and interacts with a cellular protein associated with intermediate filaments (13). The results of these studies suggest that VacA is a multifunctional toxin.
The vacA gene encodes a 140-kDa precursor protein which is cleaved at both its N and C termini to yield the mature 88-kDa secreted VacA cytotoxin monomer (7, 10, 38, 48, 55). These 88-kDa monomers assemble into complex flower-shaped oligomeric structures (8, 31). Upon exposure to acidic or alkaline pH, VacA oligomers dissociate into the component monomers, which are capable of reassembling into oligomeric structures under neutral-pH conditions (8, 34, 62). Exposure of the purified oligomeric toxin to acidic or alkaline pH (activation) results in enhanced internalization of the toxin by cells and markedly increases its cytotoxic activity (14, 33).
There is a high level of sequence diversity among vacA genes from different H. pylori strains, and several families of vacA alleles are recognized (1). Two families (s1 and s2) can be differentiated based on analysis of sequences at the 5′ end of the vacA gene, including the portion that encodes the VacA amino-terminal signal sequence, and two additional families (m1 and m2) can be differentiated based on analysis of vacA “midregions” (1). Various s1, m1, and m2 subfamilies of vacA alleles have also been described (1, 22, 51). Analysis of H. pylori isolates from multiple unrelated persons indicates that recombination among vacA alleles has occurred commonly (52), but the main families of vacA sequences (s1, s2, m1, and m2) have nevertheless remained relatively intact. This suggests that various in vivo selective forces favor preservation of these structures.
The classification of vacA alleles according to families, particularly according to s1 or s2 types, seems to correlate with the risk for clinical disease. Numerous studies have concluded that peptic ulceration occurs more commonly among patients infected with H. pylori strains containing a type s1 vacA allele than among patients infected with strains containing a type s2 vacA allele (1, 15, 19, 22, 27, 45, 51, 59). This association is less apparent in many Asian countries than in Europe and the Americas (41). To account for the association of certain vacA genotypes with peptic ulcer disease in Western countries, at least three possible explanations have been suggested. First, strains that contain type s1 vacA alleles more frequently contain the cag pathogenicity island and more frequently express the BabA2 adhesin (a Lewis-b binding factor) than do strains that contain type s2 vacA alleles, which suggests that multiple bacterial factors could contribute to ulcerogenesis (1, 19, 39, 45). In particular, products of the cag pathogenicity island contribute to induction of a gastric mucosal inflammatory response, which seems to play an important role in the pathogenesis of peptic ulcer disease (39, 44, 45). Second, strains that contain type s1 vacA alleles transcribe and express higher levels of VacA than do strains with type s2 vacA alleles (17), and type s1 VacA signal sequences may be more efficient than type s2 signal sequences in transporting the VacA protoxin across the cytoplasmic membrane. Finally, it is possible that mature VacA proteins containing a type s1 amino terminus cause gastric epithelial damage to a greater extent than do VacA proteins containing a type s2 amino terminus (1, 17, 28). Gastric epithelial damage induced by type s1 VacA may contribute to the pathogenesis of peptic ulceration (20, 55).
Several studies have suggested that type s2 VacA proteins are relatively noncytotoxic in vitro compared to type s1 VacA proteins (1, 17, 28). In one recent study, Letley and Atherton constructed a recombinant H. pylori strain that secreted a chimeric s2/m1 VacA protein and reported that broth culture supernatant from this strain lacked cytotoxic activity (28). The functional properties of type s2 VacA toxins have not yet been examined in any detail, in part because these proteins are typically secreted at low levels by wild-type H. pylori strains and are thus difficult to purify in reasonable quantities. In this study, we tested the hypothesis that type s1 and type s2 VacA proteins differ in the capacity to form anion-selective membrane channels. We report that a 12-amino-acid hydrophilic amino-terminal segment, present in type s2 but absent from type s1 VacA proteins, diminishes the capacity of VacA to form membrane channels and induce cytotoxic effects. In addition, we report that an s2/m1 chimeric toxin can inhibit the vacuolating activity of wild-type s1/m1 VacA and thus exhibits a dominant-negative phenotype.
MATERIALS AND METHODS
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. H. pylori strains 60190 (ATCC 49503) and Tx30a (ATCC 51932) contain prototypes for two highly divergent families of vacA alleles (designated type s1/m1 and type s2/m2, respectively) (1, 10). H. pylori cells were grown on Trypticase soy agar plates containing 5% sheep blood at 37°C in ambient air containing 5% CO2. Liquid cultures were grown in sulfite-free brucella broth containing either 5% fetal bovine serum or 0.5% activated charcoal.
TABLE 1.
Bacterial strains and plasmids used in this study
| H. pylori strain or plasmid | Genotype or description | Reference |
|---|---|---|
| 60190 (ATCC 49503) | vacA s1/m1 | 10 |
| Tx30a (ATCC 51932) | vacA s2/m2 | 1 |
| VM022 | 60190 with sacB-kan replacing BsmFI-to-EcoNI region of vacA | 61 |
| VM083 | vacA s2/m1; VM022 with vacA gene restored, including the s2 region from pA177 | This study |
| VM084 | vacA s1/m1; VM083 transformed with pAV202 | This study |
| VT330 | vacA s1/m1-c-myc; VM022 with vacA gene restored, including the c-myc epitope from plasmid pVT330 | This study |
| AV452 | Expresses VacA-(Δ6-27) | 61 |
| Plasmids | ||
| pA148 | 6.3-kb EcoRV-XhoI fragment containing the entire vacA gene from H. pylori strain 60190 cloned into pBluescript | 3 |
| pA167 | pA148 digested with Kpnl and BglII; ends blunted and religated | This study |
| pA176 | StuI site introduced into pA167 at vacA codons 12 and 13 | This study |
| pA177 | StuI-to-BclI fragment of pA176 replaced with StuI-to-BclI-digested vacA PCR product from H. pylori Tx30a | This study |
| pAV202 | Derivative of H. pylori strain 60190 VXC-1 (17) containing nucleotides 49 through 1679 of GenBank accession no. U05676 (including 883 nucleotides from the 5′ end of vacA) and containing a chloramphenicol resistance determinant in the cysS-vacA intergenic region | This study |
| pA178 | Site-specific mutagenesis of pA167, resulting in introduction of a StuI restriction site at vacA codons 349 to 351 | This study |
| pVT330 | Oligonucleotides encoding the c-myc epitope inserted into the StuI site of pA178 | This study |
VacA purification.
VacA was purified from H. pylori broth culture supernatants as described previously (8), except that the buffer was phosphate-buffered saline (PBS) (pH 7.5) containing 1 mM EDTA and 0.02% sodium azide. Briefly, broth culture supernatant proteins were concentrated by precipitation with a 50% saturated solution of ammonium sulfate, and the oligomeric form of VacA was isolated by fractionation using a Superose 6 HR 16/50 gel filtration column.
Cell culture.
HeLa and Vero cells were grown in minimal essential medium (modified Eagle's medium containing Earle's salts; MEM) containing 10% fetal bovine serum. Serial dilutions of concentrated H. pylori broth culture supernatants or purified VacA were incubated with cultured cells in a microtiter format, as described previously (9). Purified VacA preparations were routinely acid activated by adjusting them to pH 3 by the addition of 250 mM hydrochloric acid before they were added to cell culture wells (8, 14). After incubation for 24 h, the cells were examined by inverted light microscopy. Samples that induced vacuolation in >50% of the cells were scored as positive for the vacuolating cytotoxin phenotype (1). In some experiments, vacuolation was also quantified by neutral red uptake assay (9).
Introduction of a type s2 vacA sequence into H. pylori 60190.
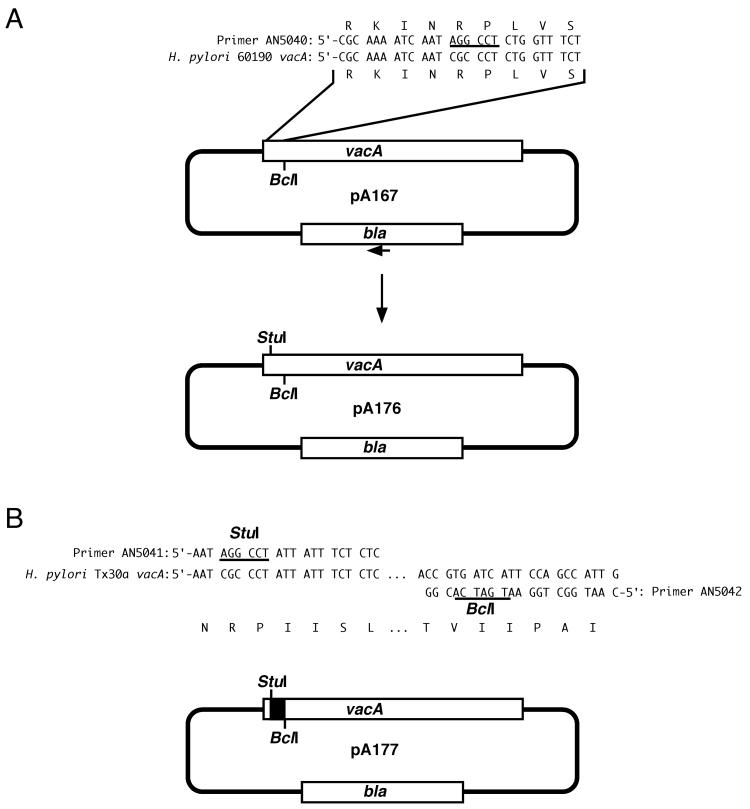
We first constructed a plasmid (pA167) that contains an ≈3-kb fragment including the 5′ end of vacA from H. pylori strain 60190 (Table 1). A StuI site was introduced into pA167 at a site encoding amino acids 12 and 13 of the VacA signal sequence, thereby generating plasmid pA176 (Fig. 1). This was accomplished using the Gene Editor site-directed mutagenesis kit (Promega) with oligonucleotide AN5040 (5′-CGCAAAATCAATAGGCCTCTGGTTTCT) and a selection oligonucleotide as described in the kit. A 130-nucleotide vacA fragment containing an s2 sequence was then PCR amplified from H. pylori Tx30a using primers AN5041 (5′-AATAGGCCTATTATTTCTCTC) and AN5042 (5′-CAATGGCTGGAATGATCACGG). Both the PCR product and pA176 were digested with StuI and BclI, and thereafter, the s2-containing segment from H. pylori Tx30a was cloned into the digested pA176 to yield pA177 (Fig. 1). H. pylori VM022, which contains a sacB-kan cassette within vacA (4, 61), was then transformed with pA177, and a sucrose-resistant, kanamycin-sensitive colony was selected. PCR and DNA sequence analysis confirmed that a double-crossover event had occurred in the chromosome of this strain (VM083) and that the strain now contained a chimeric s2/m1 vacA allele. To restore the original type s1 vacA sequence, strain VM083 was transformed with pAV202 (containing a fragment of vacA from H. pylori 60190, described in Table 1), and chloramphenicol-resistant colonies were selected. PCR and DNA sequence analysis of one such colony (VM084) confirmed that a type s1 vacA sequence, identical to that in wild-type strain 60190, was present.
FIG. 1.
Construction of plasmids used to generate H. pylori strain VM083. (A) A StuI restriction site (underlined) was introduced into plasmid pA167 using oligonucleotide primer AN5040 and the selection oligonucleotide (small arrow) from the Gene Editor kit (Promega) to generate plasmid pA176, as described in Materials and Methods. (B) A 130-nucleotide vacA fragment containing an s2 vacA sequence was PCR amplified from H. pylori strain Tx30a using primers AN5041 and AN5042. The 111-nucleotide StuI-to-BclI restriction fragment from this PCR product (solid box) was introduced into pA176 to generate pA177. Natural transformation of H. pylori VM022 with pA177 yielded H. pylori strain VM083. H. pylori VM083 expresses a chimeric s2/m1 VacA toxin.
Amino-terminal sequence analysis.
VacA purified from culture supernatant of H. pylori strain VM083 was electrophoresed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride paper (Bio-Rad) by electroblotting it for 30 min at 50 V in 10 mM 3-cyclohexylamino-1-propanesulfonic acid buffer (pH 11). After Coomassie blue staining, the VacA band was excised, and amino-terminal sequence analysis was performed with a PE Biosystems Procise 492 protein sequencing apparatus in the Peptide Sequencing and Amino Acid Analysis Shared Resource of the Vanderbilt University School of Medicine.
Introduction of DNA encoding a c-myc epitope tag into the vacA allele of H. pylori 60190.
To facilitate the expression of an epitope-tagged form of VacA, a unique StuI restriction site was first introduced using the Gene Editor site-directed mutagenesis kit (Promega) into plasmid pA167 to generate plasmid pA178 (Table 1). (Introduction of the StuI restriction site was accompanied by missense mutations at codons 312, 314, 315, and 317, replacing each of the wild-type codons with alanine-encoding codons.) Complementary primers (AN5341 [5′-CCGAACAGAAACTGATATCTGAAGAAGATCTAG] and AN5342 [5′-CTAGATCTTCTTCAGATATCAGTTTCTGTTCGG]) encoding the c-myc epitope (EQKLISEEDL) were annealed and ligated into the unique StuI site of plasmid pA178. Sequence analysis of the resulting plasmid, pVT330 (Table 1), revealed a single copy of the c-myc epitope-encoding sequence in the proper orientation. Plasmid pVT330 was then transformed into H. pylori strain VM022. Transformants in which the sacB-kan cassette was replaced by a vacA–c-myc sequence were selected by growth on plates containing 7.5% sucrose. Sequence analysis of PCR-amplified DNA from one of the transformants (H. pylori strain VT330) (Table 1) verified that the vacA allele now encoded a c-myc epitope. The predicted amino acid sequence of VacA residues 310 to 320 produced by H. pylori strain VT330 is as follows (the c-myc epitope is underlined): GAANAAQAEQKLISEEDLASSQ.
Biotinylation of VacA.
Purified VacA was concentrated, and the buffer was exchanged with 25 mM HEPES (pH 7.2) using Centriprep 30 centrifugal concentrators (Amicon). The protein concentration was determined using the Micro BCA protein assay (Pierce). Aliquots of the concentrated toxin were placed in microcentrifuge tubes containing a 1/10 volume of sodium bicarbonate (7.5% [wt/vol]). N-Hydroxysuccinimidobiotin (NHS-biotin; Pierce) dissolved in dimethyl sulfoxide was added at molar ratios of NHS-biotin to VacA ranging from 1:1 to 5:1, and the reaction mixtures were incubated at 25°C for 1 h. The reactions were stopped by addition of a 1/10 volume of hydroxylamine (10 mg per ml). The biotinylated toxin was then separated from unincorporated biotin by gel filtration chromatography using a Micro Bio-Spin chromatography column (Bio-Rad) containing Bio-Gel P-6 equilibrated in 25 mM HEPES containing 150 mM sodium chloride and bovine serum albumin (100 μg per ml). Under these conditions, biotinylation of VacA from H. pylori strain 60190 resulted in minimal loss of vacuolating activity. Biotinylated VacA was detected in immunoblotting studies by use of streptavidin-conjugated horseradish peroxidase (Life Technologies) and enhanced chemiluminescence (Amersham Pharmacia Biotech).
Immunoprecipitations of c-myc-tagged VacA.
Equimolar mixtures of biotinylated and c-myc-tagged VacA were diluted in 1 ml of PBS (pH 7) containing 0.05% Tween 20 and 2% ammonium sulfate (PBS-T-AS) to yield a final concentration of 2 μg of each toxin species per ml. When necessary, the toxins were adjusted to pH 3 (acid activated) by dropwise addition of 250 mM HCl and then neutralized upon dilution in PBS-T-AS. Anti-c-myc monoclonal antibody (9E10; Roche) (5 μg) was added to the VacA preparation, and the mixture was incubated at 4°C for 1 h. Protein G-Sepharose beads (Zymed) (25 μl), washed twice with PBS-T-AS, were added to the toxin-antibody mixture and incubated for an additional hour at 4°C. The beads were then washed three times in PBS-T-AS. The immunoprecipitated proteins were separated from the beads by boiling the beads in SDS-PAGE sample buffer and were analyzed by immunoblot analysis.
Electrophysiologic analysis of VacA channel-forming activity.
Planar lipid bilayers, composed of egg phosphatidylcholine-dioleoylphosphatidylserine-cholesterol (55:15:30 mol%) dissolved in n-decane, were prepared as described previously (11, 24, 61). Purified acid-activated VacA toxins were added to the lipid bilayers in a buffer consisting of 5 mM citric acid (pH 4.0) and 2 mM EDTA, with the salt composition as described in the figure legends and tables. For experiments using mixtures of different VacA proteins, the two VacA species (each 30 nM) were mixed together at neutral pH, and the mixture was then acidified to pH 3 and maintained at this pH for 1 h before being added to planar lipid bilayers. Membrane currents were measured as described previously (61). The potential is indicated relative to the cis side, defined as the chamber to which the protein was added. Permeability ratios were determined from the Goldman-Hodgkin-Katz equation (21), after the membrane voltage for zero current (reversal potential) in asymmetric salt concentrations was measured. Statistical significance was analyzed using Student's t test.
RESULTS
Construction and cell culture analysis of a chimeric s2/m1 VacA.
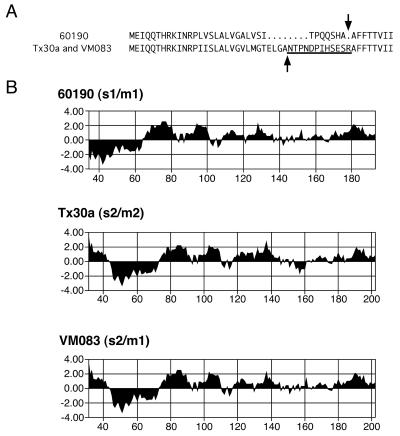
Culture supernatants from wild-type H. pylori strains expressing type s1 VacA toxins induce vacuolation of HeLa and Vero cells, whereas supernatants from wild-type strains containing s2 VacA proteins lack vacuolating activity for these cell types (1, 17). To test experimentally whether the presence of a type s2 VacA sequence diminshes toxin activity, we introduced a 111-nucleotide type s2 sequence (derived from H. pylori strain Tx30a) into the vacA allele of H. pylori 60190, as described in Materials and Methods, thereby replacing the type s1 sequence with a type s2 sequence. An H. pylori strain (VM083) containing this chimeric s2/m1 vacA sequence expressed and secreted VacA at a level similar to that of wild-type H. pylori 60190, and the chimeric toxin could be purified as large oligomeric structures with a molecular mass greater than 900 kDa. Amino-terminal sequence analysis of the mature secreted s2/m1 chimeric protein from strain VM083 revealed that the protein underwent cleavage of a 30-amino-acid amino-terminal signal sequence (Fig. 2A). The site of signal sequence cleavage was different from that used in the wild-type strain 60190 but identical to that used in wild-type strain Tx30a (Fig. 2A) (1, 10). Notably, the mature secreted forms of both the chimeric s2/m1 VacA protein and the s2/m2 VacA protein produced by wild-type strain Tx30a contain a 12-amino-acid extension (NTPNDPIHSESR) at the amino terminus compared with the amino-terminal sequence of wild-type s1/m1 VacA from H. pylori 60190 (Fig. 2A). This results in a marked change in the predicted hydrophobicity of this region, such that mature secreted type s2 VacA proteins contain a hydrophilic amino terminus whereas type s1 VacA proteins contain a hydrophobic amino terminus (Fig. 2B).
FIG. 2.
Sequence diversity in the amino termini of s1- and s2-type toxins. (A) Amino-terminal amino acid sequences of type s1 (from H. pylori strain 60190) and s2 (from H. pylori strains Tx30a and VM083) VacA toxins. The arrows indicate the sites at which the amino-terminal signal sequences are cleaved. The 12-amino-acid amino-terminal extension found in s2-type toxins is underlined. (B) Predicted hydrophilicity of VacA toxins produced by H. pylori strains 60190, Tx30a, and VM083. The analysis is limited to the amino-terminal portion of each mature toxin. The amino acid numbering is based on the initiating methionine of the protoxins as amino acid 1. The 12-amino-acid amino-terminal extension found in s2 toxins is predicted to increase the hydrophilicity of the VacA amino terminus.
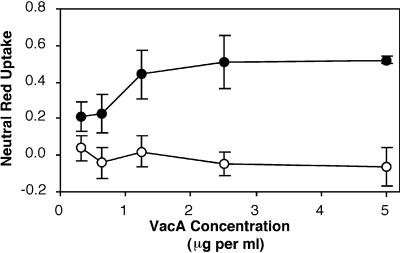
We next compared the cytotoxic activities of wild-type 60190 VacA and the chimeric s2/m1 VacA protein. Purified oligomeric toxins of each species were acid activated (a procedure which enhances cytotoxic activity) (14, 33) and then added to the neutral-pH medium overlying HeLa cells. In contrast to the wild-type VacA from strain 60190, the purified s2/m1 protein lacked detectable vacuolating activity as determined by both microscopic examination (data not shown) and analysis by a neutral-red uptake assay (Fig. 3). To verify that the lack of toxin activity was indeed a result of the presence of the s2 sequence rather than due to some other unrecognized mutation in a different region of vacA, we transformed H. pylori strain VM083 with plasmid pAV202 (containing the original s1 vacA sequence from wild-type H. pylori strain 60190), as described in Materials and Methods. PCR and sequence analysis confirmed that a double-crossover event had occurred in the bacterial chromosome, resulting in restoration of the original s1 vacA sequence. This strain, VM084, exhibited intact vacuolating activity (data not shown). Furthermore, introduction of an s1 sequence into the wild-type s2/m2 vacA allele of strain Tx30a resulted in expression of a chimeric s1/m2 VacA protein that exhibited vacuolating activity on Vero cells (data not shown), in contrast to the wild-type s2/m2 VacA from strain Tx30a, which lacked activity. These data confirm that the presence of a 12-amino-acid amino-terminal extension, consistently found in type s2 VacA proteins, confers a vacuolation-negative phenotype (1, 28).
FIG. 3.
Vacuolating activities of VacA toxins from strains 60190 (s1/m1) and VM083 (s2/m1). Purified, acid-activated VacA preparations from H. pylori strains 60190 (●) and VM083 (○) were incubated with HeLa cells in MEM containing 10 mM ammonium chloride for 16 h at 37°C. Vacuolating activity was quantified using a neutral red uptake assay (9). The results represent the mean (± standard deviation) net absorbance at 540 nm from triplicate samples. VacA from strain 60190 induced cell vacuolation, whereas the chimeric s2/m1 VacA from strain VM083 did not.
Electrophysiologic properties of channels formed by wild-type and chimeric VacA toxins.
Several recent studies have suggested that the vacuolating-cytotoxic effects of type s1 VacA are dependent on the capacity of the toxin to form anion-selective membrane channels (11, 53, 56, 57) and that a unique 32-amino-acid hydrophobic segment located at the amino terminus of type s1 VacA proteins plays an important role in membrane channel formation (32, 61). Consequently, we hypothesized that the hydrophilic s2 amino-terminal extension might interfere with the capacity of VacA to form membrane channels. To test this hypothesis, purified VacA preparations from H. pylori strains 60190 and VM083 were each incubated with planar lipid bilayers at pH 4. Type s1/m1 VacA from H. pylori strain 60190 induced a macroscopic current of 100 pA after incubation with bilayers for 51.5 ± 30.7 min, a result consistent with previous findings (24, 61). Addition to lipid bilayers of the s2/m1 toxin from strain VM083 resulted in a macroscopic current that was detectable only after a much longer delay than with the s1/m1 toxin under identical conditions (Table 2) (P < 0.005). Both of the VacA toxins examined formed channels with similar anion selectivities (Table 2). The currents formed by both toxins could be inhibited by 4,4′-diisothiocyanatostilbene-2, 2′-disulfonic acid, an inhibitor of anion transporters known to inhibit channels formed by VacA (2, 57) (data not shown). These data indicate that the presence of a type s2 12-amino-acid amino-terminal hydrophilic extension alters the capacity of VacA to form membrane channels.
TABLE 2.
VacA channel-forming activity
| VacA samplea | Time to 100 pAb | PCl/PNac |
|---|---|---|
| s1/m1 | 51.5 ± 30.7 (n = 12) | 5.17 ± 0.89 (n = 11) |
| s2/m1 | 275 ± 120 (n = 12) | 4.80 ± 0.93 (n = 10) |
| s1/m1 + s2/m1 | 30.1 ± 7.4 (n = 10) | 5.35 ± 1.01 (n = 10) |
VacA toxins were purified from H. pylori strains 60190 (s1/m1) and VM083 (s2/m1). The toxins were acid activated and added to lipid bilayers at a concentration of 30 nM. In experiments using mixtures of different toxins (s1/m1 plus s2/m1), each toxin was present at a concentration of 30 nM.
Time (in minutes) required to achieve a current of 100 pA. Acid-activated VacA was added to lipid bilayers in 100 mM sodium chloride, 5 mM citric acid, and 2 mM EDTA at pH 4.0, with an applied voltage of −50 mV. The results represent the means and standard deviations. The rate of macroscopic current formation was significantly higher for the s1/m1 VacA from wild-type strain 60190 than for the s2/m1 chimeric toxin (Student's t test; P < 0.005). Type s1/m1 VacA induced a detectable current at 30 min in 8 of the 12 experiments, whereas type s2/m1 VacA failed to induce current at this time point in any experiment. In contrast to s1/m1 VacA, the s2/m1 chimeric VacA toxin failed to induce a macroscopic current in five experiments (after periods of 210, 300, 360, 360, and 570 min) before the membrane ruptured. These results were excluded from our analysis.
Permeability ratios were determined from the Goldman-Hodgkin-Katz equation (21) after measuring the membrane voltage for zero current (reversal potential) in asymmetric salt concentrations. Acid-activated VacA was added at a concentration of 30 nM. The cis-side buffer consisted of 200 mM sodium chloride, 5 mM citric acid, and 2 mM EDTA at pH 4.0; the trans-side buffer was identical except that it contained 100 mM sodium chloride. The results represent the means and standard deviations.
We recently reported that a mutant s1/m1 toxin containing a deletion within the amino-terminal hydrophobic region, VacA-(Δ6–27), lacks detectable vacuolating activity and is capable of altering the channel-forming activity of wild-type s1/m1 toxin by a mechanism believed to involve the formation of heteromeric structures (61). Specifically, mixtures of wild-type s1/m1 VacA and VacA-(Δ6–27) formed channels in lipid bilayers significantly more slowly than did wild-type s1/m1 VacA alone, and channels formed by the mixture of toxins exhibited an altered anion selectivity (61). Therefore, we examined the electrophysiologic properties of channels formed by mixtures of s1/m1 VacA and the chimeric s2/m1 toxin. In contrast to the results we reported previously with mixtures of wild-type s1/m1 VacA and VacA-(Δ6–27), mixtures of s2/m1 VacA and s1/m1 toxin produced a macroscopic current at least as fast as the s1/m1 toxin alone, and the channels exhibited an anion selectivity that was indistinguishable from that of channels formed by s1/m1 VacA (Table 2). These results suggested that the s2/m1 and VacA-(Δ6–27) toxins interact differently with wild-type s1/m1 VacA or that perhaps s2/m1 VacA, in contrast to VacA-(Δ6–27), is unable to interact with wild-type s1/m1 VacA.
Dominant-negative phenotype of the type s2/m1 VacA toxin.
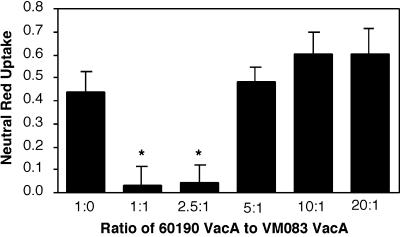
It is widely believed that the cytotoxic activity of VacA is dependent upon the capacity of VacA to assemble into oligomeric structures (8, 11, 31, 33, 53, 61, 64). Therefore, we hypothesized that the nontoxic s2/m1 VacA protein might alter the cytotoxic activity of wild-type s1 VacA. Acid-activated wild-type s1/m1 VacA from strain 60190 was added to the neutral-pH culture medium overlying HeLa cells in the presence of various concentrations of acid-activated chimeric s2/m1 toxin from strain VM083. When the two proteins were present in equimolar concentrations, s2/m1 VacA completely inhibited the activity of the s1/m1 VacA from strain 60190 (Fig. 4). Significant inhibition was also detected when the ratio of s1/m1 to s2/m1 toxins was 2.5 to 1. An acidified buffer control lacked any inhibitory activity, and nonacidified s2/m1 VacA failed to inhibit the activity of the acid-activated s1/m1 toxin (data not shown). Thus, the acid-activated s2/m1 chimeric protein exhibited a dominant-negative phenotype. This dominant-interfering activity of s2/m1 VacA was similar to that described previously for VacA-(Δ6–27) (61).
FIG. 4.
Inhibition of type s1/m1 VacA activity by the type s2/m1 toxin from H. pylori strain VM083. Acid-activated s1/m1 VacA from H. pylori strain 60190 (5 μg/ml) was mixed with various concentrations of acid-activated s2/m1 VacA from H. pylori strain VM083 in the neutral-pH medium (MEM containing 10 mM ammonium chloride) overlying HeLa cells and incubated for 16 h at 37°C. Vacuolating activity was quantified using a neutral red uptake assay (9). The results represent the mean (± standard deviation) net absorbance at 540 nm from triplicate samples. The asterisks denote significant differences (P < 0.05) compared with wild-type s1/m1 VacA alone.
Protein-protein interactions between different toxin species.
The most likely mechanism by which VacA-(Δ6–27) and the chimeric s2/m1 VacA inhibit the activity of s1/m1 VacA is believed to involve the formation of heteromeric complexes with reduced activity (58, 61). However, the formation of heterooligomeric VacA complexes has not yet been demonstrated experimentally. To determine whether either VacA-(Δ6–27) or the chimeric s2/m1 VacA could physically interact with s1/m1 VacA, we needed to develop a means for distinguishing between different forms of VacA. To do this, we altered the type s1/m1 vacA allele in H. pylori strain 60190 as described in Materials and Methods so that the resulting strain, VT330, would express a modified toxin displaying a c-myc epitope in a location predicted to be surface exposed in the VacA oligomer (8, 55). This c-myc epitope-tagged form of VacA was secreted, formed oligomeric structures, and was indistinguishable from wild-type s1/m1 VacA with respect to HeLa cell-vacuolating activity [including being inhibited by both VacA-(Δ6–27) and s2/m1 VacA (data not shown)]. Moreover, the c-myc-tagged VacA, but not the wild-type s1/m1 VacA, was recognized in both enzyme-linked immunosorbent assays and immunoblot assays by a monoclonal anti-c-myc antibody and could be immunoprecipitated by the anti-c-myc antibody (data not shown). Thus, the c-myc-tagged VacA protein was functionally indistinguishable from wild-type s1/m1 VacA from H. pylori 60190, but the epitope-tagged VacA could be specifically recognized by the anti-c-myc antibody.
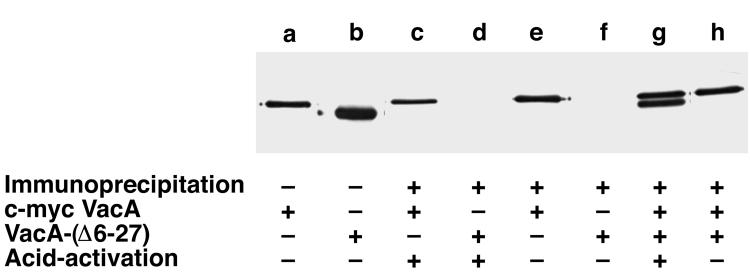
For our initial study of VacA protein-protein interactions, we selected c-myc-tagged VacA and VacA-(Δ6–27) for analysis. Because of the considerable difference in mass between the c-myc-tagged VacA and VacA-(Δ6–27) (Fig. 5, lanes a and b), the two toxins could be easily distinguished by immunoblotting using anti-VacA antisera. As expected, when incubated with either toxin alone, the anti-c-myc antibody immunoprecipitated c-myc-tagged VacA (Fig. 5, lanes c and e) but failed to immunoprecipitate VacA-(Δ6–27) (Fig. 5, lanes d and f). Similarly, when the anti-c-myc antibody was incubated with a non-acid-activated mixture of c-myc-tagged VacA and VacA-(Δ6–27), only the c-myc-tagged VacA was immunoprecipitated (Fig. 5, lane h). However, when mixtures of c-myc-tagged VacA and VacA-(Δ6–27) were acid activated (i.e., converted to monomers) and then neutralized to allow reannealing of the monomers (8, 34, 62), the anti-c-myc antibody immunoprecipitated both c-myc-tagged VacA and VacA-(Δ6–27) (Fig. 5, lane g). Thus, c-myc-tagged s1/m1 VacA and and VacA-(Δ6–27) could form heteromeric structures.
FIG. 5.
Interactions of VacA-(Δ6–27) with c-myc-tagged s1/m1 VacA. c-myc-tagged VacA and nonbiotinylated VacA-(Δ6–27) could be readily distinguished by size differences in immunoblots using anti-VacA antiserum and an alkaline-phosphatase-conjugated secondary antibody (lanes a and b). As expected, the anti-c-myc antibody 9E10 immunoprecipitated c-myc-tagged VacA but not VacA-(Δ6–27) (lanes c to f). VacA-(Δ6–27) was successfully immunoprecipitated by the anti-c-myc antibody 9E10 if mixtures of c-myc-tagged VacA and VacA-(Δ6–27) were acid activated prior to the immunoprecipitation (lane g) but not in the absence of acid activation (lane h). +, present; −, absent.
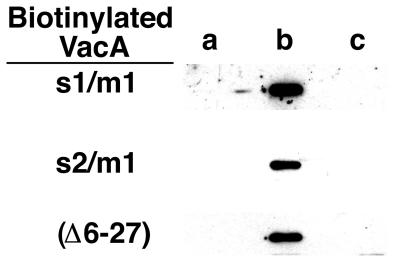
In order to detect interactions between the c-myc-tagged VacA and either the chimeric s2/m1 VacA or wild-type s1/m1 (which do not differ considerably in mass), various forms of purified VacA [wild-type s1/m1, s2/m1 VacA, and VacA-(Δ6–27)] were biotinylated as described in Materials and Methods. As expected, the anti-c-myc antibody failed to immunoprecipitate any of the acid-activated biotinylated toxins alone (Fig. 6, lane a). Similarly, the anti-c-myc antibody failed to immunoprecipitate any of the biotinylated VacA species from non-acid-activated mixtures of c-myc-tagged VacA and biotinylated VacA (Fig. 6, lane c). The biotinylated toxins were only immunoprecipitated by the anti-c-myc antibody from acid-activated mixtures of c-myc-tagged VacA and biotinylated VacA (Fig. 6, lane b). Based on these experiments, we conclude that s1/m1 VacA, s2/m1 VacA, and VacA-(Δ6–27) can each interact with c-myc-tagged s1/m1 VacA. The formation of heteromeric complexes only following acid activation of VacA species is consistent with the known capacity of s1/m1 VacA oligomers to disassemble at acidic pH and subsequently reassemble into oligomeric structures upon neutralization (8, 34, 62).
FIG. 6.
Interactions of VacA-(Δ6–27), s1/m1 VacA, and chimeric s2/m1 VacA with c-myc-tagged s1/m1 VacA. Three species of purified VacA were biotinylated as described in Materials and Methods. Samples used for immunoprecipitations included acid-activated biotinylated VacA (lane a), acid-activated mixtures of biotinylated VacA and c-myc-tagged VacA (lane b), and untreated mixtures of biotinylated VacA and c-myc-tagged VacA (lane c). Immunoprecipitations using anti-c-myc antibody 9E10 were performed as described in Materials and Methods. Immunoprecipitated proteins were separated by SDS-PAGE and transferred to nitrocellulose, and biotinylated VacA was detected with streptavidin-conjugated horseradish peroxidase and enhanced chemiluminescence. Biotinylated VacA species were successfully immunoprecipitated only if mixtures of biotinylated VacA and c-myc VacA were acid activated prior to the immunoprecipitation (lane b).
DISCUSSION
There is considerable variation among H. pylori strains in production of vacuolating-toxin activity (1, 17, 30). This variation can be attributed in part to variation in VacA amino acid sequences (1, 17, 22, 51). Previous studies have provided evidence that sequence variation in the midregion of VacA (e.g., m1 versus m2), as well as sequence variation at the amino terminus of VacA (e.g., s1 versus s2), help to determine the level of toxic activity (1, 25, 28, 40). In the present study, we compared the actions of a wild-type s1/m1 toxin and a chimeric s2/m1 toxin. The amino acid sequences of these two toxins were identical, except that the latter toxin contained a type s2 segment in place of the original type s1 segment. One striking difference between the s2/m1 and s1/m1 toxins was that they underwent amino-terminal signal sequence cleavage at two different sites. Consequently, the mature secreted s2/m1 toxin contained a 12-amino-acid hydrophilic amino-terminal extension which was absent from the s1/m1 toxin. Our results confirm that the presence of this 12-amino-acid hydrophilic extension, characteristic of type s2 toxins, abolishes the capacity of VacA to induce cytoplasmic vacuolation in cultured cells (1, 28).
We hypothesized that the 12-amino-acid amino-terminal extension present in the type s2/m1 toxin might alter functions specified by the amino-terminal portion of the s1/m1 VacA protein. In particular, we reasoned that the s2 extension (being hydrophilic) might alter the conformation or mobility of a hydrophobic segment located at the amino terminus of type s1 VacA proteins. The amino-terminal type s1 VacA hydrophobic region is capable of inserting itself into membranes and dimerizing when expressed in Escherichia coli as part of a TOXCAT fusion protein (32, 46). Deletion of a portion of this region from s1/m1 VacA, as in VacA-(Δ6–27), results in a toxin that is defective in both vacuolating cytotoxic activity and the capacity to form membrane channels (61). Furthermore, various truncations or substitution mutations within this hydrophobic region ablate vacuolating-cytotoxin activity for HeLa cells when analyzed in a transient transfection system (12, 61, 63). When taken together, these previous results all indicate that the amino-terminal hydrophobic portion of type s1 VacA plays an important role in both the toxin's cytotoxic activity and its capacity to form membrane channels. Therefore, we hypothesized that the s1/m1 and s2/m1 VacA proteins might differ in the capacity to form membrane channels.
To test this possibility, we compared the capacities of the s1/m1 and s2/m1 VacA toxins to induce channels in planar lipid bilayers. Our results indicate that the chimeric s2/m1 toxin forms membrane channels at a rate significantly lower than that of the s1/m1 toxin. Once formed, the channels produced by wild-type s1/m1 VacA and the chimeric s2/m1 VacA exhibit similar anion selectivities. Thus, the presence of the type s2 amino-terminal segment inhibits efficient formation of membrane channels but does not alter the anion selectivity of the channels that do form. It seems likely that differences in the capacities of s1 and s2 toxins to form channels in planar lipid bilayers are relevant to the different capacities of these proteins to exert cytotoxic effects on eukaryotic cells. Interestingly, the defects in channel formation exhibited by s2/m1 VacA in the present study are not identical to the defects detected previously with VacA-(Δ6–27) (61). VacA-(Δ6–27) formed channels that were less anion selective than the channels formed by either wild-type s1/m1 VacA or s2/m1 VacA (61), and some preparations of VacA-(Δ6–27) lacked any detectable channel-forming activity (unpublished data). Presumably, a more drastic alteration in VacA-lipid interactions occurs due to the deletion of the amino-terminal hydrophobic region in VacA-(Δ6–27) than occurs due to the presence of the type s2 hydrophilic extension in s2/m1 VacA.
We reported previously that the mutant toxin VacA-(Δ6–27), when mixed in an equimolar ratio with wild-type s1/m1 VacA, inhibited the cytotoxic activity of the wild-type toxin (61). In the present study, we have identified a second form of VacA (s2/m1 VacA) that exhibits a similar dominant-negative phenotype. Thus, the addition of a hydrophilic amino-terminal extension in s2/m1 VacA and deletion of a large segment of an amino-terminal hydrophobic region in VacA-(Δ6–27) each altered VacA structure in ways that permitted the emergence of a dominant-negative phenotype. Presumably, an important characteristic of both of these dominant-negative mutant toxins is their retained capacity to assemble into oligomeric structures. In future studies, it will be important to carefully test other inactive mutant forms of VacA for the presence of a similar dominant-negative phenotype. Previous work using VacA inactivated by treatment with diethyl pyrocarbonate (61), as well as preliminary studies with additional inactive forms of VacA (containing deletions elsewhere in VacA, containing certain single-amino-acid substitutions, or belonging to the s2/m2 family), reveal that some inactive forms of VacA either fail to exhibit such a phenotype or possess an inhibitory effect considerably less prominent than that exhibited by VacA-(Δ6–27) or s2/m1 VacA (M. S. McClain and T. L. Cover, unpublished data). Further analysis will be required to decipher why there is variability among inactive forms of VacA in the potencies of dominant-negative effects.
A dominant-negative phenotype is most commonly observed when a nonfunctional mutant protein interferes with the proper assembly, folding, or function of oligomeric protein complexes (58). Therefore, we hypothesized that both s2/m1 VacA and VacA-(Δ6–27) are capable of interfering with the assembly or function of oligomeric structures containing wild-type s1/m1 VacA. In support of this hypothesis, experiments in the present study demonstrate for the first time that VacA-(Δ6–27) and s2/m1 VacA are indeed capable of interacting with s1/m1 VacA to form heterooligomeric complexes. Such heteromeric complexes formed only if the different VacA species were each acid activated and then shifted to neutral pH, i.e., conditions that promote disassembly of VacA oligomers followed by oligomer reassembly (8, 34, 62). Similarly, s2/m1 VacA and VacA-(Δ6–27) exhibited a dominant-negative phenotype in cell culture assays only if these proteins were first acid activated. This correlation supports a model in which the dominant-negative phenotype results from formation of heteromeric structures with defective activity and is consistent with the hypothesis that VacA oligomerization is required for cytotoxic activity.
Why heteromeric VacA complexes (containing both wild-type s1/m1 VacA and dominant-negative mutant toxins) might be inactive remains incompletely understood. We noted in a previous study that mixtures of VacA-(Δ6–27) and wild-type s1/m1 VacA formed channels less efficiently than did the wild-type s1/m1 VacA alone and that channels formed by such mixtures had an altered anion selectivity compared to wild-type s1/m1 VacA channels (61). Therefore, we speculated that alterations in VacA channel function might account for the dominant-negative phenotype of VacA-(Δ6–27) (61). In contrast, we have tested the capacities of mixtures of s2/m1 and s1/m1 VacA to form channels and have discovered that such mixtures form channels at least as efficiently as wild-type s1/m1 VacA alone. These results suggest that, although the process of VacA-induced cell vacuolation seems to be dependent on the formation of membrane channels (11, 24, 53, 56, 57), channel formation alone is not sufficient for VacA to induce cell vacuolation. Further work will be required to elucidate the precise mechanism by which s2/m1 VacA exerts its inhibitory action. Although it is possible that s2/m1 VacA somehow interferes with channel formation in a subtle manner that cannot be detected with the present lipid bilayer assays, it seems more likely that s2/m1 VacA blocks a step in the cellular intoxication process that is distinct from membrane channel formation. We speculate that VacA heterooligomers containing s2/m1 VacA might be defective in their intracellular trafficking and localization and thus might differ from s1/m1 homooligomers in their ability to induce vacuolation. Alternatively, heterooligomers containing s2/m1 VacA might be defective in interacting with an important but not-yet-identified intracellular target.
The presence of the 12-amino-acid “type s2” amino-terminal extension markedly alters the functional properties of type s2 toxins in two different in vitro assay systems (with endpoints of vacuolating cytotoxicity or membrane channel formation) compared with those of a type s1 toxin. Nevertheless, H. pylori strains encoding type s2 toxins are found commonly in human stomachs, and strains lacking vacA alleles or containing nonsense mutations in vacA seem to be quite rare. This suggests that, despite exhibiting apparent defects in in vitro assays, type s2 VacA toxins likely serve important functions in vivo which confer a selective advantage over vacA-null strains. In accordance with this hypothesis, Salama et al. (47) recently reported that an H. pylori strain (SS1) encoding a type s2/m2 VacA toxin (60) colonized a mouse model significantly more efficiently than did the isogenic vacA-null mutant strain.
We speculate that the ability of type s2 VacA proteins to form anion-selective membrane channels, even in the absence of cell-vacuolating activity, is an important function of VacA in vivo. For example, it has been suggested that such channels might promote the release of small metabolites, such as bicarbonate and pyruvate, from gastric epithelial cells in vivo, which might be favorable for growth of H. pylori in the gastric mucus layer (53). Analysis of the channel-forming properties of type s2/m2 forms of VacA has not been conducted in any detail due to difficulties in purifying sufficient quantities of such toxins, but our preliminary investigations indicate that s2/m2 toxins can form anion-selective membrane channels with properties similar to those formed by the s2/m1 VacA protein analyzed in this study (data not shown). In addition to channel-forming activity, type s2 VacA proteins may also have other important biological functions in vivo, similar to those reported for type s1/m1 VacA toxins (13, 18, 36, 42, 43).
The dominant-negative phenotype of the s2/m1 VacA protein could also have important implications for growth of H. pylori in vivo. For example, since expression of certain forms of VacA seems to enhance the capacity of H. pylori to colonize the gastric mucosa in a mouse model (47), it seems possible that the capacity of H. pylori (expressing fully active forms of VacA) to colonize the stomach could be attenuated in the presence of dominant-negative forms of VacA. H. pylori strains expressing type s2/m1 forms of VacA occur quite rarely in human stomachs compared to strains expressing other forms of VacA (s1/m1, s1/m2, and s2/m2). Nevertheless, strains expressing s2/m1 forms of VacA have occasionally been isolated from human stomachs (29), and infection with multiple H. pylori strains is not uncommon (23, 26, 54). Based on the results of the present study, we predict that s2/m1 toxins will exhibit a dominant-negative phenotype in a mixed infection and might be capable of inhibiting colonization by H. pylori strains expressing type s1/m1 forms of VacA. Thus, the capacity of secreted proteins from one bacterium to alter the actions of proteins from other bacteria may represent an important form of bacterial competition. In support of the hypothesis that dominant-negative forms of bacterial proteins may have important actions in vivo, two recent reports indicated that administration of dominant-negative mutant forms of anthrax protective antigen were effective in blocking the activity of anthrax toxin in vivo (49, 50). The mechanism of these dominant-negative mutants of protective antigen is presumed to be similar to that proposed here for VacA, i.e., involving the formation of dysfunctional heterooligomeric structures. Thus, dominant-negative forms of secreted bacterial proteins not only may contribute to bacterial competition but also may eventually be used as a novel therapeutic approach for modifying the courses of various infectious diseases.
ACKNOWLEDGMENTS
We thank Beverly Hosse and Donna Choate for technical assistance and Wayne Schraw, James Graham, and Mark Forsyth for helpful discussions. DNA oligonucleotides were synthesized by the Vanderbilt University DNA Chemistry Core Facility, DNA sequencing was performed by the Vanderbilt University DNA Sequencing Laboratory, and amino-terminal amino acid sequencing was performed by the Vanderbilt University Peptide Sequencing and Amino Acid Analysis Shared Resource.
This work was supported by NIH grants AI39657, RR07720, HL48807, and DK53623 and by the Medical Research Department of the Department of Veterans Affairs.
REFERENCES
- 1.Atherton J C, Cao P, Peek R M, Jr, Tummuru M K, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 2.Cabantchik Z I, Greger R. Chemical probes for anion transporters of mammalian cell membranes. Am J Physiol. 1992;262:C803–C827. doi: 10.1152/ajpcell.1992.262.4.C803. [DOI] [PubMed] [Google Scholar]
- 3.Cao P, Cover T L. High-level genetic diversity in the vapD chromosomal region of Helicobacter pylori. J Bacteriol. 1997;179:2852–2856. doi: 10.1128/jb.179.9.2852-2856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Copass M, Grandi G, Rappuoli R. Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect Immun. 1997;65:1949–1952. doi: 10.1128/iai.65.5.1949-1952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cover T L. The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 6.Cover T L, Berg D E, Blaser M J, Mobley H L T. H. pylori pathogenesis. In: Groisman E A, editor. Principles of bacterial pathogenesis. San Diego, Calif: Academic Press; 2001. pp. 510–558. [Google Scholar]
- 7.Cover T L, Blaser M J. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- 8.Cover T L, Hanson P I, Heuser J E. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. J Cell Biol. 1997;138:759–769. doi: 10.1083/jcb.138.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cover T L, Puryear W, Pérez-Pérez G I, Blaser M J. Effect of urease on HeLa cell vacuolation induced by Helicobacter pylori cytotoxin. Infect Immun. 1991;59:1264–1270. doi: 10.1128/iai.59.4.1264-1270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cover T L, Tummuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 11.Czajkowsky D M, Iwamoto H, Cover T L, Shao Z. The vacuolating toxin from Helicobacter pylori forms hexameric pores in lipid bilayers at low pH. Proc Natl Acad Sci USA. 1999;96:2001–2006. doi: 10.1073/pnas.96.5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Bernard M, Burroni D, Papini E, Rappuoli R, Telford J, Montecucco C. Identification of the Helicobacter pylori VacA toxin domain active in the cell cytosol. Infect Immun. 1998;66:6014–6016. doi: 10.1128/iai.66.12.6014-6016.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bernard M, Moschioni M, Napolitani G, Rappuoli R, Montecucco C. The VacA toxin of Helicobacter pylori identifies a new intermediate filament-interacting protein. EMBO J. 2000;19:48–56. doi: 10.1093/emboj/19.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Bernard M, Papini E, de Filippis V, Gottardi E, Telford J, Manetti R, Fontana A, Rappuoli R, Montecucco C. Low pH activates the vacuolating toxin of Helicobacter pylori, which becomes acid and pepsin resistant. J Biol Chem. 1995;270:23937–23940. doi: 10.1074/jbc.270.41.23937. [DOI] [PubMed] [Google Scholar]
- 15.De Gusmao V R, Nogueira Mendes E, De Magalhaes Queiroz D M, Aguiar Rocha G, Camargos Rocha A M, Ramadan Ashour A A, Teles Carvalho A S. vacA genotypes in Helicobacter pylori strains isolated from children with and without duodenal ulcer in Brazil. J Clin Microbiol. 2000;38:2853–2857. doi: 10.1128/jcm.38.8.2853-2857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsyth M H, Atherton J C, Blaser M J, Cover T L. Heterogeneity in levels of vacuolating cytotoxin gene (vacA) transcription among Helicobacter pylori strains. Infect Immun. 1998;66:3088–3094. doi: 10.1128/iai.66.7.3088-3094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galmiche A, Rassow J, Doye A, Cagnol S, Chambard J C, Contamin S, de Thillot V, Just I, Ricci V, Solcia E, Van Obberghen E, Boquet P. The N-terminal 34 kDa fragment of Helicobacter pylori vacuolating cytotoxin targets mitochondria and induces cytochrome c release. EMBO J. 2000;19:6361–6370. doi: 10.1093/emboj/19.23.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerhard M, Lehn N, Neumayer N, Boren T, Rad R, Schepp W, Miehlke S, Classen M, Prinz C. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci USA. 1999;96:12778–12783. doi: 10.1073/pnas.96.22.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghiara P, Marchetti M, Blaser M J, Tummuru M K, Cover T L, Segal E D, Tompkins L S, Rappuoli R. Role of the Helicobacter pylori virulence factors vacuolating cytotoxin, CagA, and urease in a mouse model of disease. Infect Immun. 1995;63:4154–4160. doi: 10.1128/iai.63.10.4154-4160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman D E. Potential, impedance, and rectification in membranes. J Gen Physiol. 1943;27:37–60. doi: 10.1085/jgp.27.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han S R, Schreiber H J, Bhakdi S, Loos M, Maeurer M J. vacA genotypes and genetic diversity in clinical isolates of Helicobacter pylori. Clin Diagn Lab Immunol. 1998;5:139–145. doi: 10.1128/cdli.5.2.139-145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirschl A M, Richter M, Makristathis A, Pruckl P M, Willinger B, Schutze K, Rotter M L. Single and multiple strain colonization in patients with Helicobacter pylori-associated gastritis: detection by macrorestriction DNA analysis. J Infect Dis. 1994;170:473–475. doi: 10.1093/infdis/170.2.473. [DOI] [PubMed] [Google Scholar]
- 24.Iwamoto H, Czajkowsky D M, Cover T L, Szabo G, Shao Z. VacA from Helicobacter pylori: a hexameric chloride channel. FEBS Lett. 1999;450:101–104. doi: 10.1016/s0014-5793(99)00474-3. [DOI] [PubMed] [Google Scholar]
- 25.Ji X, Fernandez T, Burroni D, Pagliaccia C, Atherton J C, Reyrat J M, Rappuoli R, Telford J L. Cell specificity of Helicobacter pylori cytotoxin is determined by a short region in the polymorphic midregion. Infect Immun. 2000;68:3754–3757. doi: 10.1128/iai.68.6.3754-3757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jorgensen M, Daskalopoulos G, Warburton V, Mitchell H M, Hazell S L. Multiple strain colonization and metronidazole resistance in Helicobacter pylori-infected patients: identification from sequential and multiple biopsy specimens. J Infect Dis. 1996;174:631–635. doi: 10.1093/infdis/174.3.631. [DOI] [PubMed] [Google Scholar]
- 27.Kidd M, Lastovica A J, Atherton J C, Louw J A. Heterogeneity in the Helicobacter pylori vacA and cagA genes: association with gastroduodenal disease in South Africa? Gut. 1999;45:499–502. doi: 10.1136/gut.45.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letley D P, Atherton J C. Natural diversity in the N terminus of the mature vacuolating cytotoxin of Helicobacter pylori determines cytotoxin activity. J Bacteriol. 2000;182:3278–3280. doi: 10.1128/jb.182.11.3278-3280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letley D P, Lastovica A, Louw J A, Hawkey C J, Atherton J C. Allelic diversity of the Helicobacter pylori vacuolating cytotoxin gene in South Africa: rarity of the vacA s1a genotype and natural occurrence of an s2/m1 allele. J Clin Microbiol. 1999;37:1203–1205. doi: 10.1128/jcm.37.4.1203-1205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leunk R D, Johnson P T, David B C, Kraft W G, Morgan D R. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol. 1988;26:93–99. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 31.Lupetti P, Heuser J E, Manetti R, Massari P, Lanzavecchia S, Bellon P L, Dallai R, Rappuoli R, Telford J L. Oligomeric and subunit structure of the Helicobacter pylori vacuolating cytotoxin. J Cell Biol. 1996;133:801–807. doi: 10.1083/jcb.133.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClain M S, Cao P, Cover T L. Amino-terminal hydrophobic region of Helicobacter pylori vacuolating cytotoxin (VacA) mediates transmembrane protein dimerization. Infect Immun. 2001;69:1181–1184. doi: 10.1128/IAI.69.2.1181-1184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClain M S, Schraw W, Ricci V, Boquet P, Cover T L. Acid-activation of Helicobacter pylori vacuolating cytotoxin (VacA) results in toxin internalization by eukaryotic cells. Mol Microbiol. 2000;37:433–442. doi: 10.1046/j.1365-2958.2000.02013.x. [DOI] [PubMed] [Google Scholar]
- 34.Molinari M, Galli C, de Bernard M, Norais N, Ruysschaert J M, Rappuoli R, Montecucco C. The acid activation of Helicobacter pylori toxin VacA: structural and membrane binding studies. Biochem Biophys Res Commun. 1998;248:334–340. doi: 10.1006/bbrc.1998.8808. [DOI] [PubMed] [Google Scholar]
- 35.Molinari M, Galli C, Norais N, Telford J L, Rappuoli R, Luzio J P, Montecucco C. Vacuoles induced by Helicobacter pylori toxin contain both late endosomal and lysosomal markers. J Biol Chem. 1997;272:25339–25344. doi: 10.1074/jbc.272.40.25339. [DOI] [PubMed] [Google Scholar]
- 36.Molinari M, Salio M, Galli C, Norais N, Rappuoli R, Lanzavecchia A, Montecucco C. Selective inhibition of Ii-dependent antigen presentation by Helicobacter pylori toxin VacA. J Exp Med. 1998;187:135–140. doi: 10.1084/jem.187.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montecucco C, Papini E, de Bernard M, Telford J L, Rappuoli R. Helicobacter pylori vacuolating cytotoxin and associated pathogenic factors. In: Alouf J E, Freer J H, editors. The comprehensive sourcebook of bacterial protein toxins. San Diego, Calif: Academic Press; 1999. pp. 264–286. [Google Scholar]
- 38.Nguyen V Q, Caprioli R M, Cover T L. Carboxy-terminal proteolytic processing of Helicobacter pylori vacuolating toxin. Infect Immun. 2001;69:543–546. doi: 10.1128/IAI.69.1.543-546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogura K, Maeda S, Nakao M, Watanabe T, Tada M, Kyutoku T, Yoshida H, Shiratori Y, Omata M. Virulence factors of Helicobacter pylori responsible for gastric diseases in Mongolian gerbil. J Exp Med. 2000;192:1601–1610. doi: 10.1084/jem.192.11.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pagliaccia C, de Bernard M, Lupetti P, Ji X, Burroni D, Cover T L, Papini E, Rappuoli R, Telford J L, Reyrat J M. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc Natl Acad Sci USA. 1998;95:10212–10217. doi: 10.1073/pnas.95.17.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan Z J, Berg D E, van der Hulst W, Su W W, Raudonikiene A, Xiao S D, Dankert J, Tytgat G N, van der Ende A. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J Infect Dis. 1998;178:220–226. doi: 10.1086/515601. [DOI] [PubMed] [Google Scholar]
- 42.Papini E, Satin B, Norais N, de Bernard M, Telford J L, Rappuoli R, Montecucco C. Selective increase of the permeability of polarized epithelial cell monolayers by Helicobacter pylori vacuolating toxin. J Clin Investig. 1998;102:813–820. doi: 10.1172/JCI2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peek R M, Jr, Blaser M J, Mays D J, Forsyth M H, Cover T L, Song S Y, Krishna U, Pietenpol J A. Helicobacter pylori strain-specific genotypes and modulation of the gastric epithelial cell cycle. Cancer Res. 1999;59:6124–6131. [PubMed] [Google Scholar]
- 44.Peek R M, Jr, Miller G G, Tham K T, Perez-Perez G I, Zhao X, Atherton J C, Blaser M J. Heightened inflammatory response and cytokine expression in vivo to cagA+ Helicobacter pylori strains. Lab Investig. 1995;73:760–770. [PubMed] [Google Scholar]
- 45.Rudi J, Kolb C, Maiwald M, Kuck D, Sieg A, Galle P R, Stremmel W. Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J Clin Microbiol. 1998;36:944–948. doi: 10.1128/jcm.36.4.944-948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russ W P, Engelman D M. TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc Natl Acad Sci USA. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salama N R, Otto G, Tompkins L, Falkow S. Vacuolating cytotoxin of Helicobacter pylori plays a role during colonization in a mouse model of infection. Infect Immun. 2001;69:730–736. doi: 10.1128/IAI.69.2.730-736.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 49.Sellman B R, Mourez M, Collier R J. Dominant-negative mutants of a toxin subunit: an approach to therapy of anthrax. Science. 2001;292:695–697. doi: 10.1126/science.109563. [DOI] [PubMed] [Google Scholar]
- 50.Singh Y, Khanna H, Chopra A P, Mehra V. A dominant negative mutant of Bacillus anthracis protective antigen inhibits anthrax toxin action in vivo. J Biol Chem. 2001;276:22090–22094. doi: 10.1074/jbc.M010222200. [DOI] [PubMed] [Google Scholar]
- 51.Strobel S, Bereswill S, Balig P, Allgaier P, Sonntag H G, Kist M. Identification and analysis of a new vacA genotype variant of Helicobacter pylori in different patient groups in Germany. J Clin Microbiol. 1998;36:1285–1289. doi: 10.1128/jcm.36.5.1285-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suerbaum S, Smith J M, Bapumia K, Morelli G, Smith N H, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szabo I, Brutsche S, Tombola F, Moschioni M, Satin B, Telford J L, Rappuoli R, Montecucco C, Papini E, Zoratti M. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J. 1999;18:5517–5527. doi: 10.1093/emboj/18.20.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor N S, Fox J G, Akopyants N S, Berg D E, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter F M, et al. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol. 1995;33:918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Telford J L, Ghiara P, Dell'Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce M F, Censini S, Covacci A, Xiang Z, et al. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994;179:1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tombola F, Carlesso C, Szabo I, de Bernard M, Reyrat J M, Telford J L, Rappuoli R, Montecucco C, Papini E, Zoratti M. Helicobacter pylori vacuolating toxin forms anion-selective channels in planar lipid bilayers: possible implications for the mechanism of cellular vacuolation. Biophys J. 1999;76:1401–1409. doi: 10.1016/S0006-3495(99)77301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tombola F, Oregna F, Brutsche S, Szabo I, Del Giudice G, Rappuoli R, Montecucco C, Papini E, Zoratti M. Inhibition of the vacuolating and anion channel activities of the VacA toxin of Helicobacter pylori. FEBS Lett. 1999;460:221–225. doi: 10.1016/s0014-5793(99)01348-4. [DOI] [PubMed] [Google Scholar]
- 58.Unger T, Mietz J A, Scheffner M, Yee C L, Howley P M. Functional domains of wild-type and mutant p53 proteins involved in transcriptional regulation, transdominant inhibition, and transformation suppression. Mol Cell Biol. 1993;13:5186–5194. doi: 10.1128/mcb.13.9.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Doorn L J, Figueiredo C, Megraud F, Pena S, Midolo P, Queiroz D M, Carneiro F, Vanderborght B, Pegado M D, Sanna R, De Boer W, Schneeberger P M, Correa P, Ng E K, Atherton J, Blaser M J, Quint W G. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/s0016-5085(99)70065-x. [DOI] [PubMed] [Google Scholar]
- 60.van Doorn N E, Namavar F, Sparrius M, Stoof J, van Rees E P, van Doorn L J, Vandenbroucke-Grauls C M. Helicobacter pylori-associated gastritis in mice is host and strain specific. Infect Immun. 1999;67:3040–3046. doi: 10.1128/iai.67.6.3040-3046.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vinion-Dubiel A D, McClain M S, Czajkowsky D M, Iwamoto H, Ye D, Cao P, Schraw W, Szabo G, Blanke S R, Shao Z, Cover T L. A dominant negative mutant of Helicobacter pylori vacuolating toxin (VacA) inhibits VacA-induced cell vacuolation. J Biol Chem. 1999;274:37736–37742. doi: 10.1074/jbc.274.53.37736. [DOI] [PubMed] [Google Scholar]
- 62.Yahiro K, Niidome T, Kimura M, Hatakeyama T, Aoyagi H, Kurazono H, Imagawa K, Wada A, Moss J, Hirayama T. Activation of Helicobacter pylori VacA toxin by alkaline or acid conditions increases its binding to a 250-kDa receptor protein-tyrosine phosphatase beta. J Biol Chem. 1999;274:36693–36699. doi: 10.1074/jbc.274.51.36693. [DOI] [PubMed] [Google Scholar]
- 63.Ye D, Blanke S R. Mutational analysis of the Helicobacter pylori vacuolating toxin amino terminus: identification of amino acids essential for cellular vacuolation. Infect Immun. 2000;68:4354–4357. doi: 10.1128/iai.68.7.4354-4357.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye D, Willhite D C, Blanke S R. Identification of the minimal intracellular vacuolating domain of the Helicobacter pylori vacuolating toxin. J Biol Chem. 1999;274:9277–9282. doi: 10.1074/jbc.274.14.9277. [DOI] [PubMed] [Google Scholar]